Abstract
Nowadays, fish production through aquaculture and wild capture is escalating due to the higher fish protein demand for human consumption. Thus, the needs for fish products to be improved in scale and quality are becoming increasingly important to meet the nutrition requirements. The negative impacts caused by pathogens infection both in hatchery and grow-out ponds, have resulted in low aquaculture productivity and economic losses for the farmers. Therefore, a systematic study exploring relevant omics datasets through a systems biology approach will enable scientist to describe the complexity and characteristics of interactions in the host-pathogen network, leading to identifying new targets or biomarkers for diseases infection caused by bacteria, viruses, and parasites. The use of different approaches and emerging technologies as a powerful tool in obtaining a new knowledge for sustainable aquaculture production has been increasingly applied by many researchers over the years. In this paper, several omics approaches used to assess the fish immune response were reviewed. A brief description is provided for the omics approaches applicable to aquacultures, such as genomics, transcriptomics, proteomics, and metabolomics. Each tool used are contributes to develop better treatment and prevention strategies to overcome aquaculture disease due to the microbial infections. In genomics and transcriptomics analysis, their parallel approach involving gene expression studies play important role in identifying immune-related genes of the host, response to infectious diseases. While for proteomics and metabolomics approaches, these omics will provide information on the biomarker discovery and understanding the immune-related biosynthetic pathway during pathogen infection. Additionally, examples of the integrative analysis of multi-omics tools required for researchers to better understand the biological systems of aquatic animals especially fish, will also be provided.
Introduction
The dramatic increase in seafood products’ demands in the past few decades has affected the fisheries capture industry and global aquaculture production. In addition, the continuously growing number of humans has resulted in the severe needs for a sustainable resource for high-quality protein in food supplies (FAO, 2020a; Waiho et al., 2020). According to the Food and Agriculture Organization (FAO), the global fish production expected to increase from 179 million tons in 2018 to 204 million tons in 2030. The total aquaculture production in 2018 had reach 114.5 million tons of live weight production, or approximately USD 263.6 billion out of the total sales value (FAO, 2020b). In fulfilling this demand, disease-related problems usually occur in aquaculture systems due to intensive fish practices such as the poor biosecurity measurement by the farmer and culturing high fish stoking density that often leads to low water quality which eventually cause fish mortality (Liu et al., 2018; Opiyo et al., 2018). Disease outbreaks also arise due to uncontrolled transmission from wild stocks, accidental transfers of infected stocks between farms and management failure in identifying unhealthy fish stocks. If care is not taken, in some cases, this might result in the mass mortality of cultured fish, which could potentially lead to the collapse of the aquaculture industry.
Therefore, to overcome these biological constraints and meet the ever-growing demands, novel tools in biotechnological areas, such as the recently emerged omics technologies, i.e., genomics, transcriptomics, proteomics, and metabolomics, have been used to improve our knowledge of aquaculture safety, production, quality, and health. In aquaculture research, the application of various omics approaches includes genomics, the study of DNA variations; transcriptomics, analysis of genome characterization by gene expression (mRNA); proteomics, the study of cell and tissue expression of protein; and metabolomics, the study of chemical processes involving small molecules known as metabolites (Alfaro and Young, 2016; Chandhini and Rejish Kumar, 2018; Ye et al., 2018) are needed. These omics technologies provide detailed information of the organisms that help identify the molecular mechanisms of action and subsequently understand the immune response pathways of interest (Essock et al., 2019; Paul et al., 2019). Moreover, all the data and information obtained from these different biological levels can be integrated to create a general view of what is happening in the whole organism, which is an improvement over the single omics approach. By integrating multiple omics platforms, this interactions between different biological systems components can be utilized to understand the biological interactions’ complexity and how the cells and organism, can adapt, function, growth and develop during disease progression (Pinu et al., 2019). Although some omics approaches are already being applied in the aquaculture sector, their applications seem limited. Therefore, this review aimed to summarize briefly, and highlight the potential application of four different omics approaches which are genomics, transcriptomics, proteomics and metabolomics, used to evaluate the fish immune response by providing examples of omics tools from relevant studies in the published literature. This review also focuses on providing basic and useful explanation of molecular mechanisms during microbial infection which play important roles in discovering and understanding the fish immune responses.
Fish Immune System
The immune system is a system of biological structures and processes that defends the body against foreign substances such as viruses, bacteria, and parasitic agents. The system protects the organism against various diseases by distinguish between the self and non-self-cells (Wakim and Grewal, 2021). The fish immune systems’ structures and functions must be well understood to develop new technologies and enhanced productivity. In all organisms, the immune system possesses layered of defense mechanisms to prevent the entrance of pathogens (Sen et al., 2016). The immune system provides fish with the ability to resist pathogenic agents, by these two main components known as innate and adaptive immunity (Ashfaq et al., 2019). According to Marshall et al. (2018), innate immunity constitutes the first line of host defense after the invasion of pathogens. It comprises physical barriers that initiated the response within minutes after invasion. The physical barriers, includes the skin, gills and gut, preventing the pathogen invasion by producing mucosal structures to interfere and trap the pathogens. This mucosal structure consists not only a variety of antimicrobial factors but also other potent molecules such as lysozyme, complement protein, lectins, immunoglobulins, and others (Cabillon and Lazado, 2019). In addition, innate immunity consists with non-specific humoral components and non-specific cellular components. The humoral component consists of complement system, lysozyme, cytokines, transferrin, antimicrobial peptides, and lectins. While for non-specific cellular components, it consists of macrophages, monocytes, neutrophils, granulocytes, mast cells, c-reactive protein, toll-like receptors, non-specific cytotoxic cells and natural killer-like cells (Firdaus-Nawi and Zamri-Saad, 2016; Abram et al., 2017). In one of the humoral components of innate immunity, the complement system composed of approximately 30 inactive circulating proteins and membrane-bound receptors (Smith et al., 2019). These proteins can be activated by three pathways, known as the classical pathway (activated by antigen-antibody complexes), alternative pathway (activated by molecules on the surface of microorganisms in an antibody-independent manner) and lectin pathway (activated by binding of mannose-binding lection to bacterial surface carbohydrates) (Abram et al., 2017; Matsushita, 2018). In addition to the complement system, a wide variety of soluble defense factors protect against the microbial intruder, such as microbial growth inhibitor (e.g., transferrin and interferon), enzyme inhibitor (e.g., protease inhibitors like α2-macroglobulin), antimicrobial peptide (e.g., lysozyme), agglutinins (e.g., C-reactive protein), chemokines, and as well as cytokines (Roy et al., 2017; Rodrigues et al., 2020). In invertebrates and other primitive vertebrates, the innate immune response is known as the vital and basis of their immune defense against pathogenic organism (Milutinović and Kurtz, 2016). For fish, the innate immune system is the fundamental defense mechanism that essentially equal the role of adaptive immune system in higher vertebrates (Romo et al., 2016). In adaptive immunity, the system will be activated if pathogens successfully evaded the physical barrier and the innate immunity (Firdaus-Nawi and Zamri-Saad, 2016). The adaptive immune system plays a significant role for elimination against recurrent of infection by recognizing and responding to specific pathogens. There are two major components in adaptive immunity which are the humoral response consists of B-cell lymphocytes produced immunoglobulins, while cellular response consists of T-cell lymphocytes (Ashfaq et al., 2019). Although fish possess innate and adaptive immune systems, the former seems stronger in fish than the latter. To summarize the fish immune system components, Figure 1 shows the general components of the fish immune response against pathogen invasion.
FIGURE 1
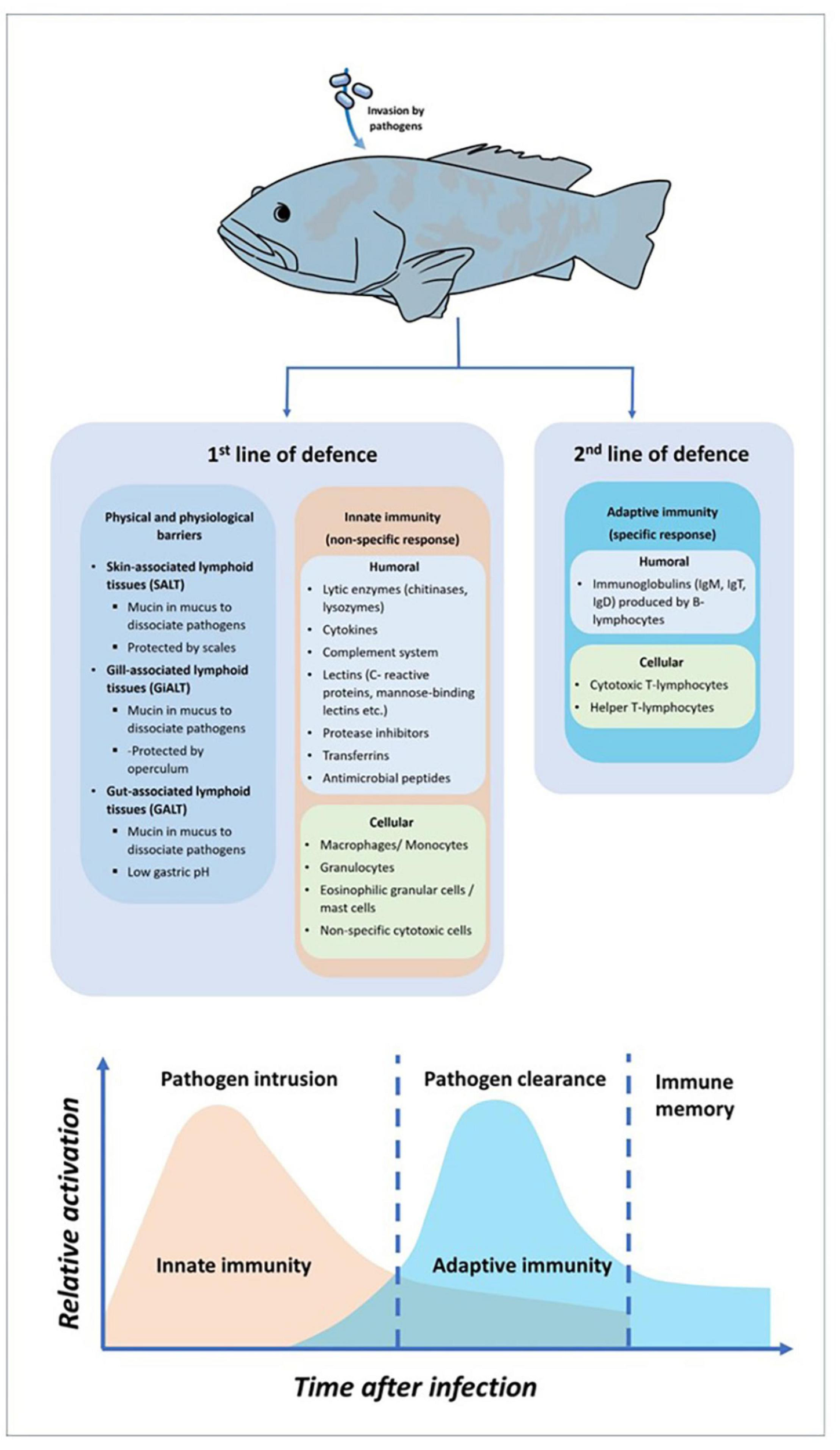
General components of fish immune response against pathogens invasion.
Several specialized organs are designed to detect and react against any foreign microbes that enter the fish body. Apart from thymus, the most well-known immune organ in fish is the head kidney, formed by two Y-shaped arms that spread underneath the gills. Both thymus and head kidney are known as the primary lymphoid organs in fish immune system (Cabillon and Lazado, 2019). Although the entire kidney contained with immune cells, both lymphopoiesis and hematopoiesis were actively occurred in the anterior kidney (head kidney) as it contained with the highest concentration of developing B and T lymphocytes, macrophages granulocytes and antibody-secreting cells (ASC) (Stosik et al., 2018). Furthermore, thymus that located under the upper gill operculum, consists of immature and developed T lymphocytes that involved in macrophage function and kill invader cells (Firdaus-Nawi and Zamri-Saad, 2016). Similar to mammalian immune system, fish also own secondary lymphoid organs such as kidney, spleen, liver and mucosal-associated lymphoid tissues (MALT). The spleen is a dark brown color pulp occupies with reticular cell network and leukocytes such as lymphocytes and macrophages (Sales et al., 2017; Udroiu and Sgura, 2017). The functions of spleen are including for hematopoiesis support, antibody production, erythrocytic destruction, blood filtration and antigen degradation during an immune response (Lewis et al., 2019). As for MALT, this mucosal structure can be sub-categorized into three lymphoid tissues, designated as gut-associated lymphoid tissue (GALT), gill-associated lymphoid tissue (GiALT) and skin-associated lymphoid tissue (SALT). These three tissues play a crucial role in fish immunity response where they will obstruct the pathogens invasion by trapping them before a variety of antimicrobial factors such as lysozyme, lectins, complement protein, immunoglobulins and antibacterial peptides were produced and destruct the pathogens (Salinas and Magadán, 2017). It has been described that the majority of MALT can be present in fish tissues, including the skin, gills and gut (Khansari et al., 2018).
In general, studies of the fish immune response have been limited due to its complexity and by the lack of suitable reagents for conducting more classical immunology assays (Salinas and Magadán, 2017). Nevertheless, by implementing genomics, transcriptomics, proteomics, and metabolomics, we hope that these approaches will benefit fish immune studies. A compilation of various omics tools used in studies related to the fish immune response is shown in Tables 1–4. These interdisciplinary methods can develop high throughput technologies that facilitate data acquisition to contribute to novel findings.
TABLE 1
| Fish species | Microbial challenge | Condition/sampling | Tissue analyzed | Platform | Main findings | References |
| Rainbow trout | Flavobacterium psychrophilum (bacteria) | Infected vs. uninfected (days 1 and 5 post infection) | Whole body | RNA-seq, Illumina HiSeq 2000 | 556 differentially expressed of long non-coding RNAs were identified related to protein-coding gene involved in fish signaling molecules immunity such as complement component, chemokines and cytokines | Paneru et al. (2016) |
| Salmonid species; Oncorhynchus kisutch, Salmo salar and Oncorhynchus mykiss | Piscirickettsia salmonis (bacteria) | Infected fish was maintained until no mortality observed (survival and dead fish) | Caudal fin | Illumina HiSeq 2500 | Identified 21 genes associate with bacterial resistant such as innate immunity, inflammation, kinase activity, GTP hydrolysis, helicase activity, lipid metabolism and cytoskeletal dynamics | Yáñez et al. (2019) |
| Spotted sea bass (Lateolabrax maculatus) | Vibrio harveyi (bacteria) | Infected vs. uninfected (0, 12, 24, 48 and 72 h post-infection) | Head kidney | Qrt-PCR, RNA-seq | Tlr genes (tlr1-1, tlr1-2, tlr2-2, tlr5, and tlr7) play important roles in immune response against Vibrio infection | Fan et al. (2019) |
| Japanese flounder (Paralichthys olivaceus) | Edwardsiella tarda (bacteria) | Infected vs. uninfected (0, 8 and 48 h post-infection) | Blood, gill and kidney | RT-qPCR, Bioanalyzer 2100, Illumina HiSeq 4000 | About 44 hsp40 genes were up or down-regulated after infection proved the differential expression genes help in disease resistance | Yan et al. (2021) |
| Channel catfish | Edwardsiella tarda (bacteria) | Infected vs. control (2 weeks post-infection) | Blood | GeneSeek, Affymetrix Genotyping Console | 37 genes located at 3 quantitative trait loci (QTL) were identify for immunity function including genes for antigen processing and presentation and MHC I | Shi et al. (2018) |
| Yellow catfish (Pelteobagrus fulvidraco) | Escherichia coli Lipopolysaccharide (bacteria) | Infected vs. uninfected (6 h post-infection) | Head kidney | Applied Biosystems 3730 analyzer, qRT-PCR | From a total of 300 putative expressed sequence taq (EST) clones, 27 immune related genes were identified, involved in pattern recognition receptors, antimicrobial peptide, complement and cytokines | Liu P.F. et al. (2016) |
| Grass carp | Aeromonas hydrophila (bacteria) and grass carp reovirus (GCRV) | -Uninfected vs. infected (2, 24, 48, 72, 96, and 120 h after viral infection) -Uninfected vs. infected (4, 8, 12, 24, 48, and 72 h after bacterial infection) | Spleen, gill, hepatopancreas and intestine tissue Spleen | RNA-seq, Illumina HiSeq 2000 RNA-seq, Illumina NextSeq 500 | A total of 64 complement system were identified. Among them, 16 genes in spleen, 19 genes in hepatopancreas, 13 genes in intestine and 8 genes in gill recorded the strong immune response after viral infection, while 17 genes in spleen showed strong immune response after bacterial infection | Liao et al. (2019) |
| Atlantic salmon | Infectious pancreatic necrosis virus (IPNV) | IPNV resistant vs. IPNV susceptible (days 1, 7, and 20 post-infection) | Whole body | RNA-seq, RT-qPCR | Susceptible fish showed ineffective genes related to inflammatory systems and cytokine, while in resistance fish, it demonstrates a more moderate immune response including the expression of genes related to macrophage system in modulating inflammatory response, which contribute to fish survival | Robledo et al. (2016) |
| Atlantic cod (Gadus morhua) | – | Commercially caught fish | Fin, muscle, thymus, spleen, blood | Illumina HiSeq 2000, Celera Assembler for genome sequence, qPCR | Evolution caused the major loss of MHC II genes function in fish, while MHC I genes were expand | Malmstrøm et al. (2016) |
Summary of fish immune response study using genomics approach.
Genomics
Genomics research focuses on whole or near-whole genome studies that aim to sequence, assemble, annotate, and analyze nucleotide sequences to explain the genomic structures and functions of the organism. Using genomics approaches such as gene knockdown, gene overexpression and gene mapping, the molecular pathways responsible for physiological or behavioral phenotypes can provide important information about diseases and genes traits (Suravajhala et al., 2016). In aquaculture, this method has contributed to disease resistance studies (Yáñez et al., 2019), selection of desirable production traits (Tsai et al., 2017), genetically improved stocks and in sustainability of food security (Bernatchez et al., 2017). In genomic studies, technologies such as next-generation sequencing technologies (NGS) have been rapidly developed because of the high demand for tools that can produce a large volume of data (sequences) in the fastest and most inexpensive way per run. Multiple technologies for NGS are already available, such as those from Illumina (Solexa Sequencing Technology), Roche (454 Life Sciences), Helicos Biosciences, Applied Biosystems (Thermo Fisher), Pacific Biosciences, Ion Torrent semiconductor sequencing (Thermo Fisher) and Oxford Nanopore Technologies. Overall, with these technology systems, analyses of rapid DNA sequencing, gene expression profiling, DNA variation discovery and whole genome de novo assembly can be achieved to understand the potential gene functions of specific intracellular mechanisms (Cheifet, 2019).
Genomics mechanism action can be explained in a simple workflow, started with DNA extraction and sequences analysis for genes profiling by genome assembly. The genes sequences were then analyzed for comparative genomics before network analysis by genome-scale modeling was conducted for validation and model prediction. Several studies using NGS technologies have enhanced the assembly and annotation of expressed sequence tags (ESTs) and gene expression barcoding, which are important for gene expression studies’ characterization. The increased number of studies on the potential of immune-related genes in fish have been performed, for example in spotted sea bass, Lateolabrax maculatus infected with Vibrio harveyi. It has been found that immune-related genes or molecular biomarker known as the toll-like receptor (TLR) genes in spotted sea bass can be identified by observing differential gene expression between the head kidney of healthy and infected fish. From 16 TLR genes that were identified based on transcriptomic and genomic database of spotted sea bass, five TLR genes were highly up-regulated including TLR1-1, TLR1-2, TLR2-2, TLR5 and TLR7, while others TLR genes were down-regulated upon V. harveyi infection (Fan et al., 2019). Meanwhile, the comparative analysis between Atlantic cod teleost species genomes, revealed the lacks functional of specific antibody responses in major histocompatibility complex II (MHC II) and CD4+ T cells, as well as the absence of the entire Gadiformes lineage is because the Atlantic cod teleost species had adopt alternative strategies to deal with infectious diseases such as the highly expanded number of specific genes including Toll-like receptors, antimicrobial peptides, and MHC I over the years (Buonocore and Gerdol, 2016; Malmstrøm et al., 2016). In another study, Liao et al. (2019) characterized and identified the complement system genes in grass carp, Ctenopharyngodon idella, before and after infection with grass carp reovirus (GCRV) or Aeromonas hydrophila, which plays essential roles in the fish immune response against infection. The RNA-seq datasets of 64 complement members were profiled using BLASTN profiling and it was identified that genes from spleen (16 genes), hepatopancreas (19 genes) and intestine tissues (13 genes) showed the strong immune responses, compared to some complement immune responses in gills (8 genes) after GCRV infection. Meanwhile for grass crap infected with A. hydrophila, about 17 genes from the spleen were identified to have the strong immune responses toward bacterial infection.
The use of others high-throughput genomics resources, such as ESTs clones from the cDNA library have helped identified essential genes and gene function related to immune response genes in yellow catfish, Pelteobagrus fulvidraco infected with Escherichia coli lipopolysaccharide (LPS) (Liu P.F. et al., 2016). From EST analysis, about 224 ESTs sequences were identified with high sequence quality that generated 95 unigenes. These 95 unigenes were divided into nine groups, including 27 immune-related gene, 24 metabolism related genes, 10 stress response related genes, nine respiratory and energy metabolism related genes, seven cytoskeleton related genes, seven transport related genes, seven transcription and translation regulation genes and three involved in cell cycle and apoptosis genes, while 59 genes were unknown. Further analysis for expression of immune related genes by the qRT-PCR analysis suggested that 16 differentially expressed genes were up-regulated, such as TLR20, Cathepsin L and NK-lysin (antimicrobial protein) in infected yellow catfish as compared to the control. The up-regulated of these genes, indicates the involvement, in the activation of innate immune response against LPS (Liu P.F. et al., 2016). Following was rainbow trout experimental challenged with Flavobacterium psychrophilum, the rainbow trout showed a number of immune-related gene sequences identified for complement component, chemokines, cytokines and several signaling molecules involved immunity after KEGG annotation, provides potential for genetic markers in disease resistance (Paneru et al., 2016). In other problems related to infectious diseases, quantitative traits locus (QTL) analysis, genome-wide identification and characterization of specific immune related genes are regarded as beneficial tools for better understand the host defense system. Several genomic resources that involve the identification of important QTL are those on catfish infected with Edwardsiella ictaluri (Shi et al., 2018), Atlantic salmon infected with infectious pancreatic necrosis virus (IPNV) (Robledo et al., 2016) and salmonid species such as Oncorhynchus kisutch, Salmo salar and Oncorhynchus mykiss infected with Piscirickettsia salmonis (Yáñez et al., 2019). The QTL identification among these fish species is essential for marker-assisted selection issues related to fish resistance against specific pathogens such as QTL on linkage group 21 can regulate the resistance toward IPNV infection in Atlantic salmon, while in catfish the identification of several immune related genes at QTL (MHC I and antigen processing and presentation) could activated the immunity function of the catfish. In another immune related study, Japanese flounder, Paralichthys olivaceus, showed 88% differential expression patterns of immune response gene after bacterial challenged (E. tarda), where the most abundance gene was known as the heat shock protein (hsp40) gene. This finding provides valuable evidence that hsp40 genes are responsible in participating in disease defense system of the fish (Yan et al., 2021). It has been reported from previous study that hsp protein played a role in humoral and cellular responses of innate immunity in aquatic organisms (Huang et al., 2018). A summary of related fish immune studies and tools is presented in Table 1.
Transcriptomics
Transcriptomics studies using NGS platforms have been applied for understanding the immune response of fish and the fundamental molecular mechanisms of fish immunity in response to pathogen infection (Salinas and Magadán, 2017). Usually, transcriptomics studies provide information about what seems to be happening in an organism under specific biological circumstances (Lowe et al., 2017). The relative abundance of RNA transcripts offers a better view of active cell expression than the genomic approach (Ye et al., 2018). The introduction of a new technology which applied a wide transcriptome profiling using a high-throughput sequenced data, known as RNA sequencing (RNA-seq) and based on the next-generation sequencing (NGS) platform, provides advantages in quantifying and mapping transcriptomes without requiring any previous information of specific gene or sequence information (Anamika et al., 2016). Several NGS platforms such as the ABI SOLiD Gene Sequencer, Solexa/Illumina Genome Analyzer, Ion Torrent and Pacific Biosciences provide rapid and high-throughput data, also offer cost-effective technologies in sequencing the transcriptome (RNA-seq).
Similar to genomics, transcriptomics mechanism of action can be started with the extraction of RNA and sequencing analysis. The transcriptome was then profile by transcriptome assembly and comparative analysis of differentially expressed genes were conducted using gene cloning. For validation, the genes were analyze by the co-expressed network analysis. To date, transcriptomics analyses in fish have been focused on identifying differential gene expression between control and challenged groups (Ye et al., 2018). Studies incorporating transcriptomics analyses have provided valuable insights into gene expression’s characterization in fish when specific pathogens infect the fish. This was shown in a study by Tso and Lu (2018), where the gene expression profile of the brain tissue of grouper in response to nervous necrosis virus (NNV) provided information associated with transcriptome expression in the immune cell signaling, surface receptor expression and interferon response after gene ontology analysis. In other transcriptional resources, microarray and RNA-seq were used to analyze gene expression in Atlantic salmon, S. salar challenged with infectious salmon anemia virus (ISAV) (Dettleff et al., 2017), Chinese tongue sole, Cynoglossus semilaevis challenged with V. harveyi (Xu et al., 2019), turbot, Scophthalmus maximus challenged with Vibrio anguillarum (Gao et al., 2016) and sockeye salmon challenged with both infectious hematopoietic necrosis virus (IHNV) and salmon louse, Lepeophtheirus salmonis (Long et al., 2019). All the findings are summarized in Table 2. A transcriptome profiling study on rainbow trout infected with Yersinia ruckeri give insight on molecular mechanism in understanding the fish immunological responses. Analysis via RNAseq toward spleen samples identified 2083 up-regulated and 415 down-regulated of differentially expressed genes in infected fish compared to control. An in-depth assessment toward DEGs associated with immune system were conducted using KEGG pathway enrichment analysis and it was revealed that these genes were associated with immunological pathways such as toll-like receptor signaling, cytokine-cytokine receptor interaction, NOD-like receptor signaling, MAPK signaling pathway and RIG-I-like receptor signaling. Overall, these findings offer a better understand of immune responses, leading to the identification of several important immune-related genes against pathogen infection (Wang et al., 2021).
TABLE 2
| Fish species | Microbial challenge | Condition/sampling | Tissue analyzed | Platform | Main findings | References |
| Chinese tongue sole (Cynoglossus semilaevis) | Vibrio harveyi (bacteria) | Infected (24 h) vs. uninfected (0 h) | Liver, spleen and kidney | Bioanalyzer 2100 system, Illumina HiSeq, RT-qPCR | Identified about 713 genes related to immune response against Vibrio infection | Xu et al. (2019) |
| Grouper (Epinephelus fuscoguttatus) | Vibrio vulnificus (bacteria) | Infected vs. uninfected (31 days post-infection) | Gill, whole body | Illumina HiSeqTM 4000 | Data in brief: Provides transcriptome sequences data for better understanding of host immune response | Jazamuddin et al. (2018) |
| Grass crap (Ctenopharyngodon idella) | Aeromonas hydrophila (bacteria) | Infected vs. uninfected (6 h post-infection) | Spleen | HiSeqTM 2500 (Illumina), RT-qPCR | Identified significant gene clusters involved in complement, apoptosis, phagocytosis, antigen processing and presentation and antioxidant in infected fish | Yang et al. (2016) |
| Common carp (Cyprinus carpio) | Aeromonas hydrophila (bacteria) | Infected A. hydrophila vs. infected XX1 vs. control (0, 7, 14, 21, 28 days post-infection) | Spleen and liver | RT-qPCR | Live attenuated vaccine from A. hydrophila strain XX1 proved the effectiveness for inducing innate and specific immune response in fish | Jiang et al. (2016) |
| Atlantic salmon (Salmo salar L.) | Piscirickettsia salmonis (bacteria) | Infected vs. uninfected (5 days and 35 days post-infection) | Head kidney | RNA-seq, Illumina MiSeq sequencer | Differentially expressed genes signaling the NF-κB pathway to regulate innate and inflammatory response of infected fish | Rozas-Serri et al. (2018) |
| Gray mullet (Mugil cephalus) | Lactococcus garvieae (bacteria) | Infected vs. uninfected (24 h post-infection) | Head kidney, spleen | Illumina HiSeqTM 2000, qPCR | Differentially expressed genes identified for complement, TLR, cytokine, hematopoietic cell lineage, antigen processing and presentation and T cell receptor signaling pathway in infected fish | Byadgi et al. (2016) |
| Turbot (Scophthalmus maximus) | Vibrio anguillarum (bacteria) | Infected (1, 4, 12 h post-infection) vs. control (0 h) | Intestine | RNA-seq, Illumina HiSeq 2500, Agilent 2100 Bioanalyzer system | Differentially expressed genes were identified and categorized into pathogen attachment and recognition, mucus barrier modification, immune activation and apoptosis/antioxidant that act to reduce or suppress pathogen entry and host immune response respectively | Gao et al. (2016) |
| Rainbow trout (Oncorhynchus mykiss) | Yersinia ruckeri (bacteria) | Infected vs. uninfected (24 h post-infection) | Spleen | Illumina Hiseq 2500 | Differentially expressed genes associated with 20 different immunological KEGG pathways were identified including toll-like receptor signaling, cytokine receptor interaction, NOD-like receptor signaling and MAPK signaling pathway | Wang et al. (2021) |
| Atlantic salmon (Salmo salar) | Infectious salmon anemia virus (ISAV) | ISAV susceptible vs. ISAV resistance (47 days post-infection) | Spleen | RNA-seq, Illumina Miseq | Differential expression genes related to immune response, stress and protein folding were identified. Different expression of viral segment in resistant fish demonstrated ability to overcome virus replication | Dettleff et al. (2017) |
| Malabar grouper (Epinephelus malabaricus) | Nervous necrosis virus (NNV) | Infected-NNV vs. inactivated-NNV (9 days post-infection) | Brain tissue | RNA-seq, Illumina NextSeq 500 | Differential expressed genes for immune cell activity signaling, surface receptor expression and interferon induced response were up-regulated (T-cell, LAG3, PD-L1) in infected fish demonstrate the potential of fish to persistent infection | Tso and Lu (2018) |
| Sockeye salmon (Oncorhynchus nerka) | Lepeophtheirus salmonis (parasite) and infectious hematopoietic necrosis virus (IHNV) | Infected L. salmonis vs. IHNV vs. L. salmonis + IHNV vs. control (35 days post-infection) | Anterior kidney | RNA-seq, RT-qPCR | The co-infected fish having the lower survival rate due to the osmoregulatory consequences of both infections, where the presence of IHNV partially impaired the fish recovery from L. salmonis | Long et al. (2019) |
| Orange-spotted grouper (Epinephelus coioides/Ec) | Red-spotted grouper nervous necrosis virus (RGNVV) | Infected vs. uninfected (24 h post-infection) | Grouper spleen cell line | RNA-seq, qRT-PCR (Roche 480 Real Time Detection System) | BAG5 homolog gene was identified in red-spotted grouper. Ec-BAG5 response to RGNNV infection and activated the interferon and inflammation responses. | Zou et al. (2020) |
Summary of fish immune response study using transcriptomics approach.
In other studies, the transcriptomics approach was used to expand our fundamental knowledge and understanding of certain metabolic pathways of specific fish species, for example by using Illumina HiSeqTM 2500, which is an RNA-seq technology, to investigate the immune response of grass carp, Ctenopharyngodon idellus infected by A. hydrophila that cause intestinal hemorrhages. It was identified that 105 differently expressed genes (DEGs) are involved in the grass carp immune response, categorized into apoptosis, phagocytosis, cytokines, complement system, antigen processing, cell adhesion molecules, antioxidant enzymes and pattern recognition receptors (Yang et al., 2016). Meanwhile, in our previous study, Illumina HiSeqTM 4000 was used to obtain the raw transcriptome data before it was trimmed to a total of 437,186,232 clean read sequences. With this transcriptome data, the immune-related genes that play roles in the immune response mechanisms of Epinephelus fuscoguttatus against Vibrio vulnificus infection can be determined (Jazamuddin et al., 2018). As in Epinephelus coioides, gene Bcl-2-associated anthanogene 5 (BAG5) which known as a molecular chaperone can bind to the Bcl-2 and modulate the cell survival of the fish. The expression pattern of BAG5 gene was investigated following the infection of red-spotted grouper nervous necrosis virus (RGNNV) toward grouper spleen cells line. The qRT-PCR was applied for gene expression profiling analysis and transcript levels of BAG5 gene showed significantly up-regulated in infected cell line compared with healthy cell line (Zou et al., 2020). In another study, transcriptomics data of Atlantic salmon infected with P. salmonis were processed by library constructing and sequencing using the Illumina MiSeq sequencer. The data was then analyzed thru De novo assembly, clustering and gene annotation. Based on the differential gene expression analysis, several immune related genes involved in the pro-inflammatory cytokine response were regulated such as the DC-SIGN and TLR5 gene. The results also revealed the strategies of the host organism to maintain survival by increase the IFN-inducible (e.g., GBP-1 and IRF-1) response and inflammatory responses, while inhibit the anti-oxidative, decreased lysosomal protease activity and degradation of ER stress associated proteins (Rozas-Serri et al., 2018). For gray mullet, Mugil cephalus infected with Lactococcus garvieae, the transcriptomics data were analyzed using Illumina HiSeqTM 2000. From the transcriptomics profiling of the gray mullet head kidney and spleen, a large number of differentially expressed genes were identified, not only related to inflammatory activation, but also in the complement system, cytokine receptor interactions, hematopoietic cell lineages and toll-like receptors. It was also observed that, during L. garvieae infection, the lack of IL-8 and the up-regulation of MHC I and TLR2 indicate the innate immune response in gray mullet (Byadgi et al., 2016). A list of these immune fish studies and the transcriptomics approach’s tools are provided in Table 2.
Proteomics
Although the applications of proteomics in aquaculture seem to be limited as compared to genomics and transcriptomics, as time has passed, proteomics studies have steadily gained more attention from researchers. As one of the most powerful tools, the proteomics approach has been increasingly used over the last decade for drug discovery research, biomarker identification, understanding pathogenicity mechanisms and altering protein expression patterns in response to different diseases (Boccio et al., 2016; Aslam et al., 2017). Proteomics also contributes to understand intra- and extracellular processes, where differential protein expression such as protein abundance, protein post-translational modifications and protein–protein interactions can be observed at certain times and in different environments (Aslam et al., 2017). In some studies, the advantages of the proteomics approach in terms of understanding biological mechanisms could be used as a novel way to comprehend the characteristics of protein functions in aquaculture organisms. Nowadays, the most popular technologies in protein identification are based on peptide mass fingerprinting using mass spectrometry (MS) analysis, such as matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) and electrospray ionization mass spectrometry ESI-MS (Boccio et al., 2016). These technologies generally involve further separation by labeling approaches using stable isotopes such as isotope-coded affinity tags (ICAT), stable isotope labeling with amino acids in cell culture (SILAC) and isobaric tags for relative and absolute quantification (iTRAQ). While for the three-dimensional structures study of the protein, the x-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy technologies are two widely used techniques that might be helpful to understand its biological mechanism (Aslam et al., 2017).
Proteomic workflow usually started with the total protein extraction before the proteins were separated either using the gel-based or shotgun approaches. In gel-based approach, several methods can be used for identifying and quantifying expressed proteins such as, 2D gel electrophoresis, protein spot selection and protein digestion. Meanwhile, in shotgun approach, method such as peptide clean-up and mass spectrometry were usually used. Many studies on fish have focused on proteome characterizations in the liver, blood or skin while evaluating the response to hormones (Dietrich et al., 2020), stress (Naderi et al., 2018), pollutants (Roland et al., 2016) and pathogens infection (Saleh et al., 2019). Numerous studies have shown other promising applications of proteomics, such as the identification of antigenic proteins (Sheng et al., 2018) and the detection of protein contents in fishery products, for quality parameter (Li et al., 2018). A number of fish species have been investigated regarding proteome changes after infection with pathogens. Saleh et al. (2019) reported that there were proteome changes in skin mucus of common carp, Cyprinus carpio, following infection with Ichthyophthirius multifiliis. Using the nano-LC ESI MS/MS approaches, about 19 immune related protein and signal transduction protein were differentially expressed between infected and healthy common carp. Significantly different expression levels were observed not only in epithelial chloride channel protein, but also in granulin, lysozyme C, galactose-specific lectin nattection and protein-glutamine gamma-glutamyltransferase 2 which likely involved in lectin pathway activation that contributed to common carp innate immune system activation. In yellow catfish, Pelteobagrus fulvidraco, several immune related proteins were identified, involved in the humoral stress response, such as the complement component, lectins, immunoglobins, and antibacterial peptides in response to E. ictaluri infection. Proteomics analysis that conducted on skin mucus of infected and non-infected yellow catfish, by label-free LCMS/MS approach demonstrated the potential of this technique in building protein expression profiles, which contribute to the differentially expressed proteins (DEPs) identification for candidate biomarker of disease resistance in yellow catfish. About 133 DEPs, including 76 up-regulated and 57 down-regulated proteins were identified, which most of its were involved in immune system pathway (Xiong et al., 2020). In another species of catfish known as Pangasianodon hyphophthalmus infected with E. ictaluri, several proteins were identified involve in some pathway related to immune responses, such as natural killer cell mediated cytotoxicity, antigen processing and presentation, leukocyte transendothelial migration, calcium signaling pathway, apoptosis, and nucleotide oligomerization domain, NOD-like receptor signaling pathway. The proteomics data analysis was conducted using nanoUPLC system called LC-IMS (Ion Mobility Separation)-QTOF-MS (Nhu et al., 2020).
Concerning changes in skin mucosal proteins relevant to immune response to infection, greater amberjack, Seriola dumerili infected with Neobenedenia girellae (Fernández-Montero et al., 2021) and common carp infected with Ichthyophthirius multifliis (Saleh et al., 2018), showed differential expression of skin mucosal proteins as the potential components in the response to pathogen infections. In amberjack fish, lower differential expressed proteins (1251 DEPs) were identified in infected fish as compared to healthy fish (1377 DEPs). The functional analysis of these proteins showed that proteins were identified for antimicrobial peptides and ribosomal proteins. Meanwhile in common carp, several novel proteins were identified after infected with I. multifiliis, such as lumican, dermatopontin, olfactomedin 4, I cytoskeletal 18 and papilin. Therefore, the findings in both studies would represents the potential search for biomarkers, in helping better understanding and monitoring the interaction between the host and pathogens. Both studies had used LC-MS/MS platform for proteomics data analysis. A similar study using iTRAQ combined with 2D LC-MS/MS demonstrated differentially expressed proteomes in the kidney of Atlantic salmon infected with Aeromonas salmonicida. Here, about 4009 distinct proteins were identified, before only 140 proteins were chosen for protein–protein interaction analysis. After proteins analysis network, only 39 proteins were considered as potential biomarker including hemoglobin subunit beta and hemoglobin subunit alpha-4 for Atlantic salmon immune responses. This finding has confirmed the presence of proteins biomarkers in kidney of Atlantic salmon in response to bacterial infection (Liu et al., 2017).
In other fish species, proteomic analysis revealed the up-regulation of differentially expressed proteins related to the molecular mechanism of cryptocaryonosis in serum of Takifugu rubripes against Cryptocaryon irritans. From the total of 2088 differentially expressed genes, eight specific proteins including integrin beta-1-like isoform X2, H2A.V, glucokinase-like, H4, histone H1-like, histone H2AX-like, histone H2B 1/2-like and myosin-9 isoform X1 were overexpressed and considered as potential biomarker for fish immune response against infection after STRING-based functional analysis (Liu et al., 2020). In zebrafish, Danio rerio, two majors’ proteins were up-regulated in fish plasma during the spring viremia of crap virus (SVCS) infection. It was revealed that both proteins; known as grass carp reovirus-induced gene (gig) and vitellogenin were involved in the immune response in fish against viral challenge after functional characterization analysis (Medina-Gali et al., 2018). Meanwhile, in the liver of rainbow trout infected with A. salmonicida, proteomics analysis was conducted using quadrupole-Orbitrap mass spectrometer. Protein abundance profiling were listed and revealed several differential expressed proteins involved in innate immune response, metabolic system and growth system. Among 2433 filtered proteins, 109 proteins showed significant differential proteins in response to bacterial infection, which include the protein for complement system, acute phase response proteins and molecules proteins for metabolic re-adjustments. It was also revealed that, from the gene ontology analyses, many proteins significant enrichment to immune system process, homeostatic process, response to stress and vesicle mediated transport (Causey et al., 2018). A summary of proteomic studies in fish immunity and the tools used are presented in Table 3.
TABLE 3
| Fish species | Microbial challenge | Condition/sampling | Tissue analyzed | Platform | Main findings | References |
| Atlantic salmon (Salmo salar) | Aeromonas salmonicida (bacteria) | Infected vs. uninfected (0, 7, and 14 days post-infection) | Kidney | iTRAQ, 2D LC-MS/MS | 39 significantly regulated proteins obtained involved in biological regulation, immune response, cellular process, biological adhesion, metabolic process, multicellular organismal process and developmental process | Liu et al. (2017) |
| Rainbow trout | Aeromonas salmonicida (bacteria) | Infected vs. uninfected (48 h post-infection) | Liver | LC-MS | Up-regulated proteins involved in complement system and acute phase response proteins were identified following bacterial challenge, including eight novel proteins from C3 proteins | Causey et al. (2018) |
| Zebrafish (Danio. Rerio) | Spring viremia of carp virus (SVCV) | Infected vs. uninfected (1, 2, 5 days post-infection) | Blood | LC-MS/MS | The presence of two major proteins; grass carp reovirus-induced gene (gig2) and vitellogenins in infected fish plasma indicated the potential of protein biomarkers for antiviral response in fish | Medina-Gali et al. (2018) |
| Common crap (Cyprinus carpio) | Ichthyophthirius multifiliis (parasite) | Infected vs. uninfected (1 day and 9 days post-infection) | Skin mucus | Nano-LC ESI MS/MS, QTOF mass spectrometry for SWATH | About 19 immune related proteins were identified involved in protease inhibitor activity, lectin pathways activation and proteolysis regulation which plays important role in activation of fish innate immune response on the skin mucus | Saleh et al. (2019) |
| Common carp (Cyprinus carpio) | Ichthyophthirius multifiliis (parasite) | Infected vs. uninfected (1 and 9 days post-infection) | Skin mucus | LC-MS/MS, nano-HPLC Ultimate 3000 RSLC system, qTOF | 25 differentially expressed proteins were identified in infected fish, which some were novel proteins such as lumican, papilin, olfactomedin 4 and I cytoskeletal 18 | Saleh et al. (2018) |
| Takifugu rubripes | Cryptocaryon irritans (parasite) | Infected vs. uninfected (4 days post-infection) | Blood | Orbitrap coupled to UPLC/MS | 8 specific proteins including integrin beta-1-like isoform X2, H2A.V, glucokinase-like, H4, histone H1-like, histone H2AX-like, histone H2B 1/2-like and myosin-9 isoform X1 were overexpressed to induce and considered as potential biomarker for fish immune response against infection | Liu et al. (2020) |
| Greater amberjack (Seriola dumerili) | Neobenedenia girellae (parasite) | Infected vs. uninfected (15 days post-infection) | Skin mucus | LC-MS/MS | A vast of proteins were identified involved in immune response such as antimicrobial peptides, serine-type proteases and metalloproteases in infected fish | Fernández-Montero et al. (2021) |
Summary of fish immune response study using proteomics approach.
Metabolomics
Although metabolomics technology is a relatively new emerging approach, its aquaculture potential might contribute significantly to novel findings. In addition, increased understanding of biochemical pathways involving immunological responses may provide crucial information regarding fish health threats. In aquaculture, metabolomics analyses are sometimes used to describe the effects of fish diet and pollutants on fish (Wei et al., 2016; Ussery et al., 2018). In addition, metabolomics analyses are also extended to resolve issues related to fish production, such as in farmed Mandarin fish, Siniperca chuatsi (Xiao et al., 2020), as well as post-harvest quality control, such as in the gilthead sea bream, Sparus aurata (Melis et al., 2014). Moreover, metabolomics analyses can be used to investigate diseases, such as the metabolic profiling of Atlantic salmon infected with A. salmonicida (Liu Q.N. et al., 2016). Compared to other omics, researchers and scientists rarely explore the applications of metabolomics in aquaculture research. The most common metabolomics tools used are mass spectrometry (MS) and nuclear magnetic resonance (NMR). Furthermore, MS technology is often combined with chromatography such as gas chromatography-mass spectrometry (GC-MS), liquid chromatography-mass spectrometry (LC-MS) or capillary electrophoresis coupled with mass spectrometry (CE-MS) for the analysis of metabolites and compounds (Jorge et al., 2016). Although the NMR-based approach has dominated as the analytical platform of metabolomics studies, the number of metabolomics studies using MS-based techniques has been on the rise because of its high sensitivity and accuracy during the detection of metabolites in a sample (Zhang et al., 2020).
Metabolomics workflow started with the extraction of samples by chemical solvent, solid phase extraction or hydrodistillation method. The sample was then analysis using selected analytical platform such as NMR or MS tools, before data analysis was conducted by multi-variate analysis and statistical analysis. Based on GC-MS metabolomics analysis conducted to study tilapia infected with E. tarda, glucose was the key metabolite in fish survival. By exogenous glucose into the fish, it promotes biosynthesis of stearic acid and palmitic acid which attenuated the TCA cycle before elevated the fish immune response against bacterial infection (Zeng et al., 2017). In another study conducted on tilapia infected with Streptococcus iniae, serine and proline were identified as two metabolites that play important role for fish survival after exogenous of L-leucine into the fish. It was suggested that serine can elevated the interleukin 1-beta (Il-1β) and interleukin-8 (Il-8) in fish and promotes phagocytosis of macrophages, similar with L-leucine that promotes macrophages activity to kill bacterial pathogens (Du et al., 2017). Another study on yellow grouper plasma infected with Streptococcus agalactiae showed that the decreased of malic acid and the increased of adenosine indicated key metabolites in distinguishing fish from bacterial infection and non-infection (Wang et al., 2016). For crucian carp, Carassius carassius, the differential responses toward bacterial infection could be partially due to the different temperatures. Based on the metabolomics analysis using GC-MS, the increased of palmitate, threonine, and taurine at 30°C condition, enhances the crucian carp survival, as well as regulate the innate immune response as compared to crucian carp grown in 20°C. Among the genes involved in crucian carp innate immunity during bacterial infection at 30°C were lysozyme, complement system, Toll like receptor family and cytokines (Jiang et al., 2019). These results highlight that, after exposure to an infectious pathogen, crucial metabolites are up-regulated and act as biomarkers of fish’s immune response. Additionally, our previous study on brown marble grouper (E. fuscoguttatus) infected with V. vulnificus showed higher production of metabolites from omega-9 fatty acids such as palmitoleic acid, oleic acid, 6,9-octadecenoic acid, cis-erucic acid, 8,11-eicosadienoic acid and 5,8,11-eicosatrienoic using a GC-MS approach (Nurdalila et al., 2019, 2020). Based on this result, metabolomics profiling has the potential to investigate fish metabolic responses toward disease infection. A similar study was conducted by Mayalvanan (2019) on grouper infected with V. vulnificus using LC-qTOF-MS-based analysis. The up-regulated metabolites identified in the surviving fish were mostly amino acids such as lysine, aspartic acid, and leucine. Thus, the method can be potentially used for diagnostic and therapeutic studies.
Nowadays, many researchers use metabolomics to monitor the health conditions of various aquatic organisms, and for the evaluation and exploration of mechanisms of resistance and efficacy in fish infected with pathogens (Low et al., 2017). In one study, crucian carp grown in different temperature (18°C and 33°C) were infected with Aeromonas sobrial. By GC-MS analysis, it was found that at 33°C, maltose metabolite was found to be the most suppressed compound in the spleen of infected crucian carp. Further experiment of exogenous maltose into the fish had showed the down-regulated production of cytokines and the increased of complement component and lysozyme involved in fish humoral innate immune responses (Jiang et al., 2020). In yellow drum fish, skin metabolome involves in the immune responses can be identified using metabolomics approach. Here, by using GC-MS based metabolic profiling, the metabolomic mechanisms were identified in yellow drum’s skin in response to Cryptocaryon irritans infection. The results demonstrated that an at 24 h of post-infection, C. irritans caused a strong metabolic stress on the fish, while at 72 h of post-infection, the restoration metabolic state took place indicates their resistance mechanism activation. During this 72 h post-infection about 17 metabolites were identified for potential biomarkers including 15-hydroxy-eicosatetraenoic acid (15-HETE) and adenine for anti-inflammatory responses, while betaine and delta-valerolactam for stimulation of antibodies production (Maha et al., 2019). A study conducted by Liu Q.N. et al. (2016) on the kidneys of Atlantic salmon infected against A. salmonicida, analysis using NMR comprehensively investigated the metabolic changes between infected and non-infected Atlantic salmon. Four metabolic pathways, namely glycolysis/gluconeogenesis, the citrate cycle, the urea cycle, and tryptophan metabolism were constructed based on significant metabolites (alanine, aspartate, betaine, choline, fumarate, glycine, glycerophosphocholine and valine) observed between the two groups (infected and non-infected). After analysis, it was concluded that, although these four metabolic pathways did not directly contribute to the innate immune response in Atlantic salmon, their peculiar activations might link to the disturbed production of intermediate protein synthesis, which benefits viral genome replication. Table 4 shows several fish immune studies and tools using the metabolomics approach.
TABLE 4
| Fish species | Microbial challenge | Condition/sampling | Tissue analyzed | Platform | Main findings | References |
| Brown marble grouper (Epinephelus fuscoguttatus) | Vibrio vulnificus (bacteria) | Infected vs. uninfected (31 days post-infection) | Gill, liver, spleen, body tissue | LC-qTOF-MS | Amino acids such as glutamine, alanine, phenylalanine and tyrosine were found highly abundance in survived-infected fish, which highlight the functions of these metabolites in immune related pathways | Mayalvanan (2019) |
| Tilapias | Edwardsiella tarda (bacteria) | D-glucose + infected vs. infected (6 days post-infection) | Liver | GC-MS | Glucose enhances fish survival while promotes stearic acid and palmitic acid biosynthesis which reduced the TCA cycle process to potentiate fish against bacterial infection | Zeng et al. (2017) |
| Tilapias | Streptococcus iniae (bacteria) | L-leucine + infected vs. infected (5 days) | Liver | GC-MS | Based on metabolomics analysis, two metabolites, serine and proline were identified helping exogenous L-leucine in elevates immune response of infected fish | Du et al. (2017) |
| Yellow grouper | Streptococcus agalactiae (bacteria) | Plasma mixed with bacterial vs. plasma (control) (24 h) | Plasma | GC-MS | The increased of adenosine and decreased of malic acid, were the key metabolites in attenuated the TCA cycle while elevated the purine metabolism fish survival | Wang et al. (2016) |
| Brown marble grouper (Epinephelus fuscoguttatus) | Vibrio vulnificus (bacteria) | Infected vs. uninfected (5, 10, 15, 20 and 21 days post-infection) | Whole body | GC-MS | Fatty acid from omega 9 (ω-9) group (oleic acid, palmitoleic acid, cis-erucic acid, 8,11-eicosadienoic acid, 6,9-octadecenoic acid, and 5,8,11-eicosatrienoic acid were highly abundant in challenged group. The changes demonstrate a potential biomarker for fish resistance against bacterial infection | Nurdalila et al. (2019, 2020) |
| Crucian carp (Carassius auratus) | Aeromonas sobrial (bacteria) | Maltose + infected vs. infected (15 days post-infection) | Spleen | GC-MS | Maltose proven to reduce fish mortality by downregulated the cytokines while enhanced the lysozyme and complement component (C3) activity during bacterial infection | Jiang et al. (2020) |
| Atlantic salmon (Salmo salar) | Aeromonas salmonicida (bacteria) | Infected vs. uninfected (7 and 14 days post-infection) | Kidney | NMR | The abundant of metabolites such as alanine, valine, aspartate, betaine, fumarate, glycerophosphocholine and choline that involved in tryptophan metabolism, citrate cycle, glycolysis and urea cycle in survived fish, offer better understanding of infection mechanisms | Liu Q.N. et al. (2016) |
| Crucian carp (Carassius carassius) | Edwardsiella tarda (bacteria) | Infected at 20°C vs. infected at 30°C (15 days) | Spleen | GC-MS | The decreased abundant of taurine, threonine and palmitate represents the metabolic characteristic of infected fish between low and high temperature. At higher temperature, the TCA cycle was elevated and caused the increasing of fish immune response | Jiang et al. (2019) |
| Yellow drum (Nibea albiflora) | Cryptocaryon irritans (parasite) | Infected vs. uninfected (24 and 72 h post-infection) | Skin | LC-MS | 17 metabolites identified as potential biomarkers such as adenine, betaine and L-glutamate, which involved not only in innate immune responses but also in physiological role and another metabolic pathway | Maha et al. (2019) |
Summary of fish immune response study using metabolomics approach.
Introduction of Multi-Omics Integration
As stated above, omics approaches have given us high throughput databases and more holistic methodologies than traditional methods. These omics approaches are known for investigating molecular pathways and the identification and quantification of differentially expressed genes or molecules in animals exposed to infectious diseases. Worthy of note that a single omics study cannot entirely reveal the complexities of a living system (Boccio et al., 2016). Thus, by integrating multi-omics data, a general view of the principles of a biological system and predicting the outcome of certain responses in certain living organisms can be obtained (Bersanelli et al., 2016). The new improved experimental methodologies and computational approaches that being used in omics data analysis can generate new knowledge and understand the complexity of biological processes. The support from the high-throughput technologies, able researchers to produce a large database that cost affordable and easy to outreach within the internet. Although the integration of omics in aquaculture is still in its infancy, the emerging field of integrative data is proliferating due to the growth of omics research studies such as genomics, transcriptomics, proteomics, and metabolomics.
Multi-omics technologies have been widely used in aquaculture research studies. The genome, transcriptome, proteome and metabolome from different layers of omics, often be used to reflect the characterization of an organism at different biomolecule levels. The data generated from this omics technologies can be linked together using bioinformatics approach. Although single omics could produce relevant information data, however by integrating different omics data, it helps researchers to extract more insightful information for better understanding of biological system and molecular processes in an organism. For example, the integration between proteomic and transcriptomic analysis, the protein changes during microbial infection conditions can be provided, after genes is regulated at the translation levels via proteomic analysis (Li et al., 2020). Furthermore, the requirement of genomic or transcriptomic technologies in providing genetic information about the tissues will contribute to better understanding of fish genetic and protein functional at different conditions. In another example, as genomics and transcriptomics technologies continue to grow, fish genomes are expected to be easily sequenced with these technologies (Pathak et al., 2019). The transcriptome data can reflect the genomes that were expressed by showing how the genes were active in different cells. Through omics integration technologies, metabolites and proteins can correlate in the same metabolic pathway. The metabolomic profiling gives data-rich information of metabolic changes that influenced by the proteome expressed in a cellular physiology. Simple diagram showing multi-omics integration interaction can be refer in Figure 2.
FIGURE 2
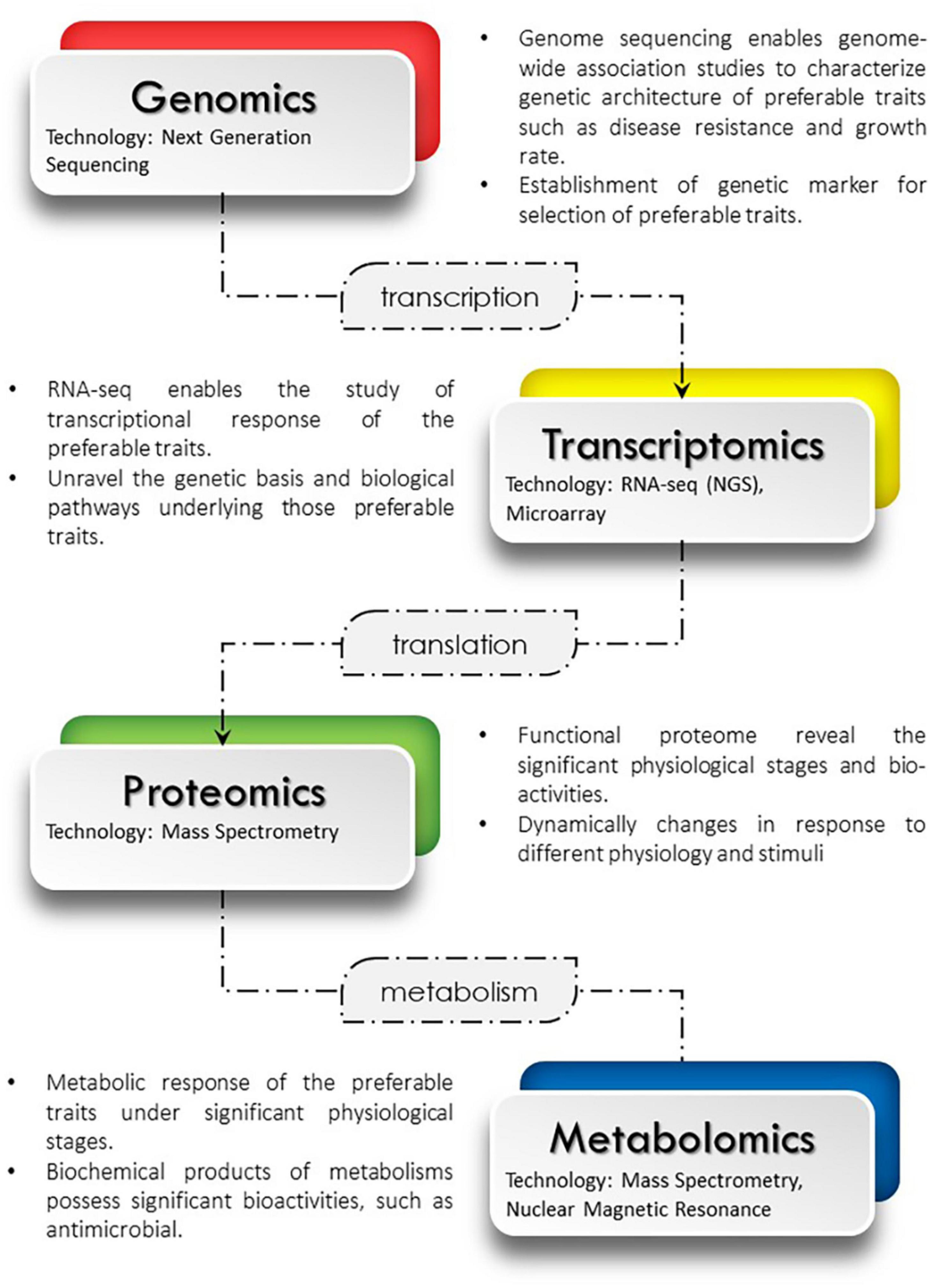
Multi-omics integration of genomics, transcriptions, proteomics and metabolomics.
Case Study of Integration Omics for Aquatic Animals
In fish immune responses, integrating data from different omics studies provide a better understanding in molecular mechanisms of fish immune system, not only regarding gene expression and gene regulation, but also in terms of protein expression and metabolite production. For example, a study by Li et al. (2020) successfully integrated transcriptomics and proteomics data to investigate the response of candidate genes and proteins of grass carp, associated with inducing intestinal mucosal immunity after vaccination with a double-targeted DNA vaccine of Vibrio mimicus. By using RNA-seq and iTRAQ-coupled LCMS/MS, a total of 5,339 DEGs and 1,173 DEPs were identified, before integrated data analysis revealed that about 250 DEPs were matched with DEGs at both transcriptome and proteome levels. Further analysis via Gene Ontology and KEGG pathway analysis found that about 50 DEPs/DEGs were enriched in immune-related pathway such as during (i) antigen processing and presentation, (ii) complement and coagulation cascades, (iii) phagosome (iv) nucleotide oligomerization domain (NOD)-like receptors (NLRs) and (v) mitogen-activate protein kinase (MAPK) signaling pathways. Overall, this integrated transcriptomics and proteomics analysis identified five activated immune related pathways that essential in enhancing the intestinal mucosal immune response of immunized grass carp. The integration of transcriptomic and proteomic data has also been studied in rainbow trout O. mykiss against A. salmonicida. By using iTRAQ and Illumina-Hiseq technologies, differential gene expression and protein expression were identified between infected and non-infected rainbow trout. A total of 1,036 genes and 133 proteins were analyzed, and the correlation between the transcriptomic and proteomic data indicated that 17 proteins showed consistent expression at the RNA and protein levels. From these results, several proteins identified as IL-13 receptor alpha-1 (IL-13Rα1), GTPase IMAP family member 7 (GIMAP7), ferritin, CD209, VDAC2, proteasome subunit alpha type (PSMA1) and annexin ANXA11s could have the potential to act as a biomarker during aminoacyl-tRNA biosynthesis, nucleotide metabolism and proteasome in rainbow trout when infected with A. salmonicida (Long et al., 2015).
In another omics integrated study, metabolomics and proteomics methods were used to elucidate and understand the effects of ammonia toxicity in marine medaka, Oryzias melastigma. From the metabolic analysis, multiple changes in metabolites such as amino acids, carbohydrates and energy metabolism products were identified. During the proteomic analysis, about 23 altered proteins were identified in related to immune stress, apoptosis, oxidative stress, nervous system, cytoskeleton assembly and locomotor system. From these comparative metabolomic and proteomic analysis, it was found that ammonia toxicity can affected the carbohydrates composition, amino acids composition and energy metabolism in marine medaka, caused immune stress, oxidation stress and apoptosis in the fish (Zhu et al., 2018). A similar study by Zhu et al. (2019) showed the integration of transcriptomics and metabolomics tools in detecting the differential responses in the liver of Nile tilapia, Oreochromis niloticus due to acute ammonia exposure. Overall finding using integrated analysis, at 6 h exposure, about 2258 of differential expressed genes (DEGs) and 31 metabolites were identified in Nile tilapia liver, while at 24 h of exposure, only 315 DEG and 36 metabolites were detected. The significant differences of DEGs can be observed in a different hour due to many processes related to urea biosynthesis, mitochondrial function, and energy conservation during ammonia toxicity in tilapia. For metabolomic analysis, 31 and 36 metabolites were detected at 6 h and 24 h of ammonia exposure. It was found that the down-regulated metabolites such as L-malic acid, fumaric acid and D-(glycerol 1-phosphate) at both hour after ammonia exposure also involved in the mitochondria function and energy production. Moreover, L-malic acid and fumaric acid also involved in urea synthesis during the TCA cycle to reduce the ammonia-nitrogen stress.
In Nile tilapia, RNA-seq transcriptomic and iTRAQ proteomic approaches have been used, demonstrating differential expressed genes and differential expressed proteins in different organs. The transcriptome data from gut and liver was analyses using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) for functional annotation and classification of genes. Based on the results, it was found that immune transcripts were higher in gut than liver. While, for proteomic analysis, proteome data from bile and intestinal mucus were also classified and functional annotated using GO and KEGG. Here, more immune proteins were presence in bile than intestinal mucus. Both data was then overlap and it was determined that 62 common immune genes between bile and liver were involved in complement system, acute phase reaction and immune cell response. The finding conclude that homeostasis and inflammation were activated in all the organs analyzed (Wu et al., 2016). Additionally, Wen et al. (2019) used metabolite, transcriptomic and proteomic studies to understand the cold-tolerance mechanisms pathways utilized by pufferfish, Takifugu fasciatus, as the fish is known as warm water fish. Under cold stress conditions, 40 differential metabolites were identified, include 31 up-regulated and 9-regulated metabolites. The combination of transcriptomics, metabolomics and proteomics data analysis had identified 14 differentially co-expressed genes and proteins (DEGs/DEPs) and 17 differentially expressed metabolites (DEMs). Overall, the interaction network involved are such as membrane transport, signal transduction, fatty acids metabolism, and DNA damage and defense of pufferfish. These results offer a foundation for other researchers to understand the molecular mechanisms capabilities of pufferfish under cold stress.
While in Litopenaeus vannamei or known as Pacific white shrimp, the metabolome and transcriptome data we used to analyses the molecular mechanism of choline dietary that played important roles in glycerophospholipid metabolism and decreased the oxidant damage of L. vannamei. Malondialdehyde (MDA) is a key metabolite for lipid peroxidation, and as MDA contents in shrimp were decreased by choline dietary, it also decreases oxidative damage in the hepatopancreas of L. vannamei. However, the excessive of choline supplement might inhibit the digestion of protein and reduce the protein deposition which cause a loss in whole-body crude protein in shrimp that affect the shrimp growth. In transcriptomic analysis, gene expression of carboxypeptidase A1-like and glutathione S-transferase 1-like were up-regulated, while trypsin 1-like gene was down-regulated in shrimp given with choline supplement. It was known that carboxypeptidase serves as degradation proteins in digestive tract, while glutathione S-transferase act as a protein that protect aquatic organism from oxidative damage. For trypsin, the protein is important in hydrolysis and food digestion. After metabolome analysis, L-proline metabolite identified up-regulated in the sample proven that this metabolite was involved in protein or food digestion likes trypsin in transcriptome analysis. In comparison of gene expressions, genes such as apolipoprotein D-like, bile salt-activated lipase-like and glutathione S-transferase 1-like were up-regulated in non-given choline supplement. Here apolipoprotein D was identified as a potential biomarker in shrimp as the gene involved in oxidative stress during cell body protection (Huang et al., 2020). Table 5 shows several studies related to the integration of omics approaches used for aquatic organisms.
TABLE 5
| Studies | Aquatic organisms | Platforms | Main finding | References |
| Integration of transcriptomic and proteomic in response to double-targeted DNA vaccine of Vibrio mimicus | Grass carp | RNA-seq and iTRAQ-coupled LCMS/MS | From 250 associated differentially expressed proteins (DEPs) and differentially expressed genes (DEGs) identified in fish intestines, 50 were involved in immune-related associated DEPs and DEGs such as phagosome, complement coagulation cascades, NLRs, MAPK pathways and antigen processing and presentation | Li et al. (2020) |
| Integration of transcriptomic and proteomic data in response to Aeromonas salmonicida | Rainbow trout (Oncorhynchus mykiss) | iTRAQ and Illumina-Hiseq technologies | Upregulated proteins, namely CD209, ferritin, PSMA1, VDAC2, IL13Rα1, GIMAP7 and two ANXA11s were identified as potential biomarkers for rainbow trout immune response as DEGs and DEPs were involved in immune system process and signaling molecules and interaction pathway. | Long et al. (2015) |
| Metabolomic and proteomic analysis in response to ammonia toxicity exposure | Marine medaka (Oryzias melastigma) | 2-DE gel analysis, H-NMR spectroscopy | Comparative metabolomic and proteomic analysis, found that ammonia toxicity can affected the carbohydrates, amino acids composition and energy metabolism in marine medaka, caused not only immune stress, oxidation stress, apoptosis but also impaired the fish nervous system | Zhu et al. (2018) |
| Transcriptomic and proteomic analysis of fish gut-liver immunity during homeostasis and inflammation | Nile tilapia (Oreochromis niloticus) | RNA-seq, Illumina HiSeq 2000, iTRAQ, HPLC, LC-MS/MS | More immune transcripts were found in gut than liver, while more immune proteins were found in bile than intestinal mucus. | Wu et al. (2016) |
| Metabolomic, transcriptomic and proteomics analysis in response to cold stress responses | Pufferfish (Takifugu fasciatus) | UHPLC-Q-TOF MS, Bioanalyzer 2100, Illumina HiSeq 2500, RT-qPCR, iTRAQ | Interaction analysis of DEGs/DEPs/DEMs mainly regulated the antioxidant capacity, immune response and growth. The DEGs/DEPs interaction provide potential indicators for cold tolerant traits, while DEMs can be used as potential feed additives to enhance the fish immunity | Wen et al. (2019) |
| Transcriptomic and metabolomic response in fish liver due to exposure of acute ammonia | Nile Tilapia (Oreochromis niloticus) | Illumina HiSeq 2500, GC-TOF-MS | DEGs were identified involved for urea biosynthesis, mitochondrial function, and energy conservation after ammonia toxicity in tilapia. The down-regulated metabolites such as L-malic acid, fumaric acid and D-(glycerol 1-phosphate) correlated in mitochondria function and energy production. Moreover, L-malic acid and fumaric acid also involved in urea synthesis during the TCA cycle to reduce the ammonia-nitrogen stress. | Zhu et al. (2019) |
| Transcriptomic and metabolomic analysis in response to dietary choline chloride | Pacific White Shrimp (Litopenaeus vannamei) | Agilent 2100 Bioanalyzer, Illumina Novaseq 6000, LC-MS | The choline dietary decreased the shrimp’s oxidant damage, while excessive dietary inhibits the protein digestion, protein deposition and growth in shrimp. Both transcriptomic and metabolomic analysis showed the apolipoprotein D served the potential in biomarker in shrimp to identified oxidative stress activity | Huang et al. (2020) |
The Integration of omics approach on aquatic organisms.
Since a large amount of data is generated by the integration of omics studies, significant statistical and computational efforts are required on researchers. The data produced from different layers of omics approaches must be integrated and analyzed. The complexity of the biological system itself and the limitation of high throughput technologies make integrative analysis even more challenging. Thus, the development of integrative analysis methods from multilayer datasets has become one of the most relevant problems that need to be tackled by computational scientists and bioinformaticians (Bersanelli et al., 2016). The large numbers of biological variables have also resulted in the developing different types of software tools, which are invaluable for a better understanding of genomics, transcriptomics, proteomics, and metabolomics (Boccio et al., 2016). Moreover, these provide opportunities and make it relatively easier for researchers to obtain and discover new vaccination, anti-bacterial, anti-viral and anti-parasite products in the aquaculture field (Ye et al., 2018).
Conclusion and Future Perspectives
In summary, omics technologies have become powerful tools in the development of aquaculture, not only for fish health, but also for fish larval production, nutrition, therapeutics, and food safety. Economic losses caused by fish mortality due to pathogen infections must be resolved quickly so that the aquaculture industry will not be badly affected. The use of difference omics approaches is expected to better understand the fish immune mechanisms by investigating gene expression, genome annotation, protein networks, metabolic changes and more. In addition, with continuous studies on the analytical capability of bioinformatics being carried out, these methods enable a large number of omics data to be acquired more effectively and efficiently. Moreover, integrative approaches used in omics technologies have the potential to characterize phenotypes fully, thus leading to novel discoveries in the biological context of the intensive aquaculture sector for enhanced production, disease prevention and improved efficiency demanded aquaculture food sources for human consumption. These approaches may also lead to future discoveries on effective vaccinations or drugs for aquatic organisms.
Statements
Author contributions
MN, C-FL, C-MC, HB, and SB wrote and edited the manuscript. All authors reviewed and approved the final version for submission.
Funding
This work was supported by Universiti Kebangsaan Malaysia Research University Grants, GUP-2017-073 and DIP 2015-024 awarded to SB.
Acknowledgments
The authors would like to thank the staff and students of Metabolomics laboratory for kindly supporting and giving suggestions to the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Abram Q. H. Dixon B. Katzenback B. A. (2017). Impacts of low temperature on the teleost immune system.Biology6:39. 10.3390/biology6040039
2
Alfaro A. C. Young T. (2016). Showcasing metabolomic applications in aquaculture: a review.Rev. Aquac.10135–152. 10.1111/raq.12152
3
Anamika K. Verma S. Jere A. Desai A. (2016). Transcriptomic Profiling Using Next Generation Sequencing – Advances, Advantages, and Challenges.London: IntechOpen.
4
Ashfaq H. Soliman H. Saleh M. El-Matbouli M. (2019). CD4: a vital player in the teleost fish immune system.Vet. Res.50:1. 10.1186/s13567-018-0620-0
5
Aslam B. Basit M. Nisar M. A. Khurshid M. Rasool M. H. (2017). Proteomics: technologies and their applications.J. Chromatogr. Sci.55182–196. 10.1093/chromsci/bmw167
6
Bernatchez L. Wellenreuther M. Araneda C. Ashton D. T. Barth J. M. I. Beacham T. D. et al (2017). Harnessing the power of genomics to secure the future of seafood.Trends Ecol. Evol.32665–680. 10.1016/j.tree.2017.06.010
7
Bersanelli M. Mosca E. Remondini D. Giampieri E. Sala C. Castellani G. et al (2016). Methods for the integration of multi-omics data: mathematical aspects.BMC Bioinform.17:15. 10.1186/s12859-015-0857-9
8
Boccio P. D. Rossi C. M. Cicalini I. Sacchetta P. Pieragostino D. (2016). Integration of metabolomics and proteomics in multiple sclerosis: from biomarkers discovery to personalized medicine.Proteomics Clin. Appl.10470–484. 10.1002/prca.201500083
9
Buonocore F. Gerdol M. (2016). Alternative adaptive immunity strategies: coelacanth, cod and shark immunity.Mol. Immunol.69157–169. 10.1016/j.molimm.2015.09.003
10
Byadgi O. Chen Y. C. Barnes A. C. Tsai M. A. Wang P. C. Chen S. C. (2016). Transcriptome analysis of grey mullet (Mugil cephalus) after challenge with Lactococcus garvieae.Fish Shellfish Immunol.58593–603. 10.1016/j.fsi.2016.10.006
11
Cabillon N. A. R. Lazado C. C. (2019). Mucosal barrier functions of fish under changing environmental conditions.Fishes4:2. 10.3390/fishes4010002
12
Causey D. R. Pohl M. A. N. Stead D. A. Martin S. A. M. Secombes C. J. Macqueen D. J. (2018). High-throughput proteomic profiling of the fish liver following bacterial infection.BMC Genomics19:719. 10.1186/s12864-018-5092-0
13
Chandhini S. Rejish Kumar V. J. (2018). Transcriptomics in aquaculture: current status and applications.Rev. Aquac.111379–1397. 10.1111/raq.12298
14
Cheifet B. (2019). Where is genomics going next?Genome Biol.20:17. 10.1186/s13059-019-1626-2
15
Dettleff P. Moen T. Santi N. Martinez V. (2017). Transcriptomic analysis of spleen infected with infectious salmon anemia virus reveals distinct pattern of viral replication on resistant and susceptible Atlantic salmon (Salmo salar).Fish Shellfish Immunol.61187–193. 10.1016/j.fsi.2017.01.005
16
Dietrich M. A. Irnazarow I. Inglot M. Słowiñska M. Judycka S. Ciereszko A. (2020). Hormonal stimulation of carp (Cyprinus carpio L.) males triggers changes in the sperm proteome.Aquaculture530:735791. 10.1016/j.aquaculture.2020.735791
17
Du C. Yang M. Li M. Y. Yang J. Peng B. Li H. et al (2017). Metabolic mechanism for l-Leucine-induced metabolome to eliminateStreptococcus iniae. J. Proteome Res.16, 1880–1889. 10.1021/acs.jproteome.6b00944
18
Essock B. T. Soderblom E. J. Orihuela B. Moseley M. A. Rittschof D. (2019). Hypothesis testing with proteomics: a case study using wound healing mechanisms in fluids associated with barnacle glue.Front. Mar. Sci.6:343. 10.3389/fmars.2019.00343
19
Fan H. Wang L. Wen H. Wang K. Qi X. Li J. et al (2019). Genome-wide identification and characterization of toll-like receptor genes in spotted sea bass (Lateolabrax maculatus) and their involvement in the host immune response to Vibrio harveyi infection.Fish Shellfish Immunol.92782–791. 10.1016/j.fsi.2019.07.010
20
FAO (2020a). Fisheries Statistical Collections: Global Aquaculture Production.Rome: Food and Agriculture Organization of the United Nations.
21
FAO (2020b). The State of World Fisheries and Aquaculture 2020. In brief. Sustainability in Action.Rome: FAO.
22
Fernández-Montero Á Torrecillas S. Montero D. Acosta F. Prieto-Álamo M. J. Abril N. et al (2021). Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae - an immunological approach.Fish Shellfish Immunol.110100–115. 10.1016/j.fsi.2021.01.001
23
Firdaus-Nawi M. Zamri-Saad M. (2016). Major components of fish immunity: a review.Pertanika J. Trop. Agric. Sci.39393–420.
24
Gao C. Fu Q. Su B. Zhou S. Liu F. Song L. et al (2016). Transcriptomic profiling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot (Scophthalmus maximus) following Vibrio anguillarum challenge.Dev. Comp. Immunol.65159–168. 10.1016/j.dci.2016.07.007
25
Huang M. Lin H. Xu C. Yu Q. Wang X. Qin J. G. et al (2020). Growth, metabolite, antioxidative capacity, transcriptome, and the metabolome response to dietary choline chloride in Pacific white shrimp Litopenaeus vannamei.Animals10:2246. 10.3390/ani10122246
26
Huang X. Li S. Gao Y. Zhan A. (2018). Genome-wide identification, characterization and expression analyses of heat shock protein-related genes in a highly invasive ascidian Ciona savignyi.Front. Physiol.9:1043. 10.3389/fphys.2018.01043
27
Jazamuddin F. M. Aizat W. M. Goh H. H. Low C. F. Baharum S. N. (2018). Transcriptome data of Epinephelus fuscoguttatus infected by Vibrio vulnificus.Data Brief.16466–469. 10.1016/j.dib.2017.11.024
28
Jiang M. Chen Z. Zheng J. Peng B. (2019). Metabolites-enabled survival of crucian carps infected by Edwardsiella tarda in high water temperature.Front. Immunol.10:1991. 10.3389/fimmu.2019.01991
29
Jiang M. Yang L. F. Zheng J. Chen Z. G. Peng B. (2020). Maltose promotes crucian carp survival against Aeromonas sobrial infection at high temperature.Virulence11877–888. 10.1080/21505594.2020.1787604
30
Jiang X. Zhang C. Zhao Y. Kong X. Pei C. Li L. et al (2016). Immune effects of the vaccine of live attenuated Aeromonas hydrophila screened by rifampicin on common carp (Cyprinus carpio L).Vaccine343087–3092. 10.1016/j.vaccine.2016.04.075
31
Jorge T. F. Mata A. T. António C. (2016). Mass spectrometry as a quantitative tool in plant metabolomics.Phil. Trans. R. Soc. A Math. Phys. Eng. Sci.374:20150370. 10.1098/rsta.2015.0370
32
Khansari A. R. Balasch J. C. Vallejos-Vidal E. Parra D. Reyes-López F. E. Tort L. (2018). Comparative immune and stress-related transcript response induced by air exposure and Vibrio anguillarum bacterin in rainbow trout (Oncorhynchus mykiss) and gilthead seabream (Sparus aurata) mucosal surfaces.Front. Immunol.9:856. 10.3389/fimmu.2018.00856
33
Lewis S. M. Williams A. Eisenbarth S. C. (2019). Structure-function of the immune system in the spleen.Sci. Immunol.4:eaau6085. 10.1126/sciimmunol.aau6085
34
Li J. N. Zhao Y. T. Cao S. L. Wang H. Zhang J. J. (2020). Integrated transcriptomic and proteomic analyses of grass carp intestines after vaccination with a double-targeted DNA vaccine of Vibrio mimicus.Fish Shellfish Immunol.98641–652. 10.1016/j.fsi.2019.10.045
35
Li Y. Zhang Y. Li H. Zhao W. Guo W. Wang S. (2018). Simultaneous determination of heat stable peptides for eight animal and plant species in meat products using UPLC- MS/MS method.Food Chem.245125–131. 10.1016/j.foodchem.2017.09.066
36
Liao Z. Wan Q. Yuan G. Su J. (2019). The systematic identification and mRNA expression profiles post viral or bacterial challenge of complement system in grass carp Ctenopharyngodon della.Fish Shellfish Immunol.86107–115. 10.1016/j.fsi.2018.11.032
37
Liu G. Ye Z. Liu D. Zhao J. Sivaramasamy E. Deng Y. et al (2018). Influence of stocking density on growth, digestive enzyme activities, immune responses, antioxidant of Oreochromis niloticus fingerlings in biofloc systems.Fish Shellfish Immunol.81416–422. 10.1016/j.fsi.2018.07.047
38
Liu P. Du Y. Meng L. Li X. Liu Y. (2017). Proteomic analysis in kidneys of Atlantic salmon infected with Aeromonas salmonicida by iTRAQ.Dev. Comp. Immunol.72140–153. 10.1016/j.dci.2017.02.013
39
Liu P. Xia Y. Hua X. Fan K. Li X. Zhang Z. et al (2020). Quantitative proteomic analysis in serum of Takifugu rubripes infected with Cryptocaryon irritans.Fish Shellfish Immunol.104213–221. 10.1016/j.fsi.2020.06.008
40
Liu P. F. Du Y. Meng L. Li X. Liu Y. (2016). Metabolic profiling in kidneys of Atlantic salmon infected with Aeromonas salmonicida based on 1-H NMR.Fish Shellfish Immunol.58292–301. 10.1016/j.fsi.2016.08.055
41
Liu Q. N. Xin Z. Z. Chai X. Y. Jiang S. H. Li C. F. Zhang H. B. et al (2016). Characterization of immune-related genes in the yellow catfish Pelteobagrus fulvidraco in response to LPS challenge.Fish Shellfish Immunol.56248–254. 10.1016/j.fsi.2016.05.019
42
Long A. Garver K. A. Jones S. R. M. (2019). Synergistic osmoregulatory dysfunction during salmon lice (Lepeophtheirus salmonis) and infectious hematopoietic necrosis virus co-infection in sockeye salmon (Oncorhynchus nerka) smolts.J. Fish Dis.42869–882. 10.1111/jfd.12989
43
Long M. Zhao J. Li T. Tafalla C. Zhang Q. Wang X. et al (2015). Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. Salmonicida.J. Proteomics12241–54. 10.1016/j.jprot.2015.03.031
44
Low C. F. Rozaini M. Z. H. Musa N. Baharum S. N. (2017). Current knowledge of metabolomic approach in infectious fish disease studies.J. Fish Dis.401267–1277. 10.1111/jfd.12610
45
Lowe R. Shirley N. Bleackley M. Dolan S. Shafee T. (2017). Transcriptomics technologies.PLoS Comput. Biol.13:e1005457. 10.1371/journal.pcbi.1005457
46
Maha I. F. Xie X. Zhou S. Yu Y. Liu X. Zahid A. et al (2019). Skin metabolome reveals immune responses in yellow drum Nibea albiflora to Cryptocaryon irritans infection.Fish Shellfish Immunol.94661–674. 10.1016/j.fsi.2019.09.027
47
Malmstrøm M. Matschiner M. Tørresen O. K. Star B. Snipen L. G. Hansen T. F. et al (2016). Evolution of the immune system influences speciation rates in teleost fishes.Nat. Genet.481204–1212. 10.1038/ng.3645
48
Marshall J. S. Warrington R. Watson W. Lim H. L. (2018). An introduction to immunology and immunopathology.Allergy Asthma Clin. Immunol.14:49. 10.1186/s13223-018-0278-1
49
Matsushita M. (2018). The complement system of agnathans.Front. Immunol.9:1405. 10.3389/fimmu.2018.01405
50
Mayalvanan Y. (2019). Metabolomics study of the response of Epinephelus fuscoguttatus towards vibriosis. Master’s thesis. Bangi: Universiti Kebangsaan Malaysia.
51
Medina-Gali R. Belló-Pérez M. Ciordia S. Mena M. C. Coll J. Novoa B. et al (2018). Plasma proteomic analysis of zebrafish following spring viremia of carp virus infection.Fish Shellfish Immunol.86892–899. 10.1016/j.fsi.2018.12.035
52
Melis R. Cappuccinelli R. Roggio T. Anedda R. (2014). Addressing marketplace gilthead sea bream (Sparus aurata L.) differentiation by 1H NMR-based lipid fingerprinting.Food Res. Int.63258–264. 10.1016/j.foodres.2014.05.041
53
Milutinović B. Kurtz J. (2016). Immune memory in invertebrates.Semin. Immunol.28328–342. 10.1016/j.smim.2016.05.00
54
Naderi M. Keyvanshokooh S. Ghaedi A. Salati A. P. (2018). Effect of acute crowding stress on rainbow trout (Oncorhynchus mykiss): a proteomics study.Aquaculture495106–114. 10.1016/j.aquaculture.2018.05.038
55
Nhu T. Q. Hang B. B. T. Cornet V. Oger M. Bach L. T. Dao A. N. L. et al (2020). Single or combined dietary supply of Psidium guajava and Phyllanthus amarus extracts differentially modulate immune responses and liver proteome in striped catfish (Pangasianodon hyphophthalmus).Front. Immunol.11:797. 10.3389/fimmu.2020.00797
56
Nurdalila A. A. Mayalvanan Y. Baharum S. N. (2019). Metabolite profiling of Epinephelus fuscoguttatus infected with vibriosis reveal omega 9 as potential metabolite biomarker.Fish Physiol. Biochem.451203–1215. 10.1007/s10695-019-00633-6
57
Nurdalila A. A. Natnan M. E. Baharum S. N. (2020). The effects of amino acids and fatty acids on the disease resistance of Epinephelus fuscoguttatus in response to Vibrio vulnificus infection.3 Biotech10:544. 10.1007/s13205-020-02543-4
58
Opiyo M. A. Marijani E. Muendo P. Odede R. Leschen W. Charo-Karisa H. (2018). A review of aquaculture production and health management practices of farmed fish in Kenya.Int. J. Vet. Sci. Med.6141–148. 10.1016/j.ijvsm.2018.07.001
59
Paneru B. Al-Tobasei R. Palti Y. Wiens G. D. Salem M. (2016). Differential expression of long non-coding RNAs in three genetic lines of rainbow trout in response to infection with Flavobacterium psychrophilum.Sci. Rep.6:36032. 10.1038/srep36032
60
Pathak A. K. Rashid I. Nagpure N. S. Kumar R. Pati R. Singh M. et al (2019). FisOmics: a portal of fish genomic resources.Genomics1111923–1928. 10.1016/j.ygeno.2019.01.003
61
Paul V. J. Freeman C. J. Agarwal V. (2019). Chemical ecology of marine sponges: new opportunities through “-Omics”.Integr. Comp. Biol.59765–776. 10.1093/icb/icz014
62
Pinu F. R. Beale D. J. Paten A. M. Kouremenos K. Swarup S. Schirra H. J. et al (2019). Systems biology and multi-omics integration: viewpoints from the metabolomics research community.Metabolites9:76. 10.3390/metabo9040076
63
Robledo D. Taggart J. B. Ireland J. H. McAndrew B. J. Starkey W. G. Haley C. S. et al (2016). Gene expression comparison of resistant and susceptible Atlantic salmon fry challenged with Infectious Pancreatic Necrosis virus reveals a marked contrast in immune response.BMC Genomics17:279. 10.1186/s12864-016-2600-y
64
Rodrigues M. V. Zanuzzo F. S. Koch J. F. A. Oliveira C. A. F. Sima P. Vetvicka V. (2020). Development of fish immunity and the role of β-glucan in immune responses.Molecules25:5378. 10.3390/molecules25225378
65
Roland K. Kestemont P. Dieu M. Raes M. Silvestre F. (2016). Using a novel “Integrated Biomarker Proteomic” index to assess the effects of freshwater pollutants in European eel peripheral blood mononuclear cells.J. Proteomics137:83e96. 10.1016/j.jprot.2016.01.007
66
Romo R. M. Pérez-Martínez D. Ferrer C. C. (2016). Innate immunity in vertebrates: an overview.Immunology148125–139. 10.1111/imm.12597
67
Roy S. Kumar V. Kumar V. Behera B. K. (2017). Acute phase proteins and their potential role as an indicator for fish health and in diagnosis of fish diseases.Protein Pept. Lett.2478–89. 10.2174/0929866524666161121142221
68
Rozas-Serri M. Peña A. Maldonado L. (2018). Transcriptomic profiles of postsmolt Atlantic salmon challenged with Piscirickettsia salmonis reveal a strategy to evade the adaptive immune response and modify cell-autonomous immunity.Dev. Comp. Immunol.81348–362. 10.1016/j.dci.2017.12.023
69
Saleh M. Kumar G. Abdel-Baki A. A. Dkhil M. A. El-Matbouli M. Al-Quraishy S. (2018). Quantitative shotgun proteomics distinguishes wound-healing biomarker signatures in common carp skin mucus in response to Ichthyophthirius multifiliis.Vet. Res.49:37. 10.1186/s13567-018-0535-9
70
Saleh M. Kumar G. Abdel-Baki A. A. S. Dkhil M. A. El-Matbouli M. Al-Quraishy S. (2019). Quantitative proteomic profiling of immune responses to Ichthyophthirius multifiliis in common carp skin mucus.Fish Shellfish Immunol.84834–842. 10.1016/j.fsi.2018.10.078
71
Sales C. F. Silva R. F. Amaral M. G. C. Domingos F. F. T. Ribeiro R. I. M. A. Thomé R. G. et al (2017). Comparative histology in the liver and spleen of three species of freshwater teleost.Neotrop. Ichthyol.15:e160041. 10.1590/1982-0224-20160041
72
Salinas I. Magadán S. (2017). Omics in fish mucosal immunity.Dev. Comp. Immunol.7599–108. 10.1016/j.dci.2017.02.010
73
Sen R. Nayak L. De R. K. (2016). A review on host–pathogen interactions: classification and prediction.Eur. J. Clin. Microbiol. Infect. Dis.351581–1599. 10.1007/s10096-016-2716-7
74
Sheng X. Liu M. Liu H. Tang X. Xing J. Zhan W. (2018). Identification of immunogenic proteins and evaluation of recombinant PDHA1 and GAPDH as potential vaccine candidates against Streptococcus iniae infection in flounder (Paralichthys olivaceus).PLoS One13:e0195450. 10.1371/journal.pone.0195450
75
Shi H. Zhou T. Wang X. Yang Y. Wu C. Liu S. et al (2018). Genome-wide association analysis of intra-specific QTL associated with the resistance for enteric septicemia of catfish.Mol. Genet. Genomics2931365–1378. 10.1007/s00438-018-1463-0
76
Smith N. C. Rise Matthew L. Christian Sherri L. (2019). A comparison of the innate and adaptive immune systems in cartilaginous fish, ray-finned fish, and lobe-finned fish.Front. Immunol.10:2292. 10.3389/fimmu.2019.02292
77
Stosik M. P. Tokarz-Deptuła B. Deptuła W. (2018). Specific humoral immunity in Osteichthyes.Cent. Eur. J. Immunol.43335–340. 10.5114/ceji.2018.80054
78
Suravajhala P. Kogelman L. J. A. Kadarmideen H. N. (2016). Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare.Genet. Sel. Evol.48:38. 10.1186/s12711-016-0217-x
79
Tsai H. Y. Matika O. Edwards S. M. Roberto A. S. Hamilton A. Guy D. R. et al (2017). Genotype imputation to improve the cost-efficiency of genomic selection in farmed Atlantic salmon.G371377–1383. 10.1534/g3.117.040717
80
Tso C. H. Lu M. W. (2018). Transcriptome profiling analysis of grouper during nervous necrosis virus persistent infection.Fish Shellfish Immunol.76224–232. 10.1016/j.fsi.2018.03.009
81
Udroiu I. Sgura A. (2017). The phylogeny of the spleen.Q. Rev. Biol.92411–443. 10.1086/695327
82
Ussery E. Bridges K. N. Pandelides Z. Kirkwood A. E. Bonetta D. Venables B. J. et al (2018). Effects of environmentally relevant metformin exposure on Japanese medaka (Oryzias latipes).Aquat. Toxicol.20558–65. 10.1016/j.aquatox.2018.10.003
83
Waiho K. Fazhan H. Ishak S. D. Kasan N. A. Liew H. J. Norainy M. H. et al (2020). Potential impacts of COVID-19 on the aquaculture sector of Malaysia and its coping strategies.Aquac. Rep.18:100450. 10.1016/j.aqrep.2020.100450
84
Wakim S. Grewal M. (2021). Introduction to the Immune System. Available online at: https://bio.libretexts.org/@go/page/16845(accessed on 28 April 2021).
85
Wang D. Sun S. Li S. Lu T. Shi D. (2021). Transcriptome profiling of immune response to Yersinia ruckeri in spleen of rainbow trout (Oncorhynchus mykiss).BMC Genomics22:292. 10.1186/s12864-021-07611-4
86
Wang Z. Li M. Peng B. Cheng Z. Li H. Peng X. (2016). GC-MS-based metabolome and metabolite regulation in serum-resistant Streptococcus agalactiae. J. Proteome Res.15, 2246–2253. 10.1021/acs.jproteome.6b00215
87
Wei Y. Liang M. Mai K. Zheng K. Xu H. (2016). The effect of ultrafiltered fish protein hydrolysate levels on the liver and muscle metabolic profile of juvenile turbot (Scophthalmus maximus L.) by 1 H NMR-based metabolomics studies.Aquac. Res.483515–3527. 10.1111/are.13178
88
Wen X. Hu Y. Zhang X. Wei X. Wang T. Yin S. (2019). Integrated application of multi-omics provides insights into cold stress responses in pufferfish Takifugu fasciatus.BMC Genomics20:563.
89
Wu N. Song Y. L. Wang B. Zhang X. Y. Zhang X. J. Wang Y. L. et al (2016). Fish gut-liver immunity during homeostasis or inflammation revealed by integrative transcriptome and proteome studies.Sci. Rep.6:36048. 10.1038/srep36048
90
Xiao M. Qian K. Wang Y. Bao F. (2020). GC-MS metabolomics reveals metabolic differences of the farmed Mandarin fish Siniperca chuatsi in recirculating ponds aquaculture system and pond. Sci. Rep.10:6090. 10.1038/s41598-020-63252-9
91
Xiong Y. Dan C. Ren F. Su Z. Zhang Y. Mei J. (2020). Proteomic profiling of yellow catfish (Pelteobagrus fulvidraco) skin mucus identifies differentially-expressed proteins in response to Edwardsiella ictaluri infection.Fish Shellfish Immunol.10098–108. 10.1016/j.fsi.2020.02.059
92
Xu H. Xu X. Li X. Wang L. Cheng J. Zhou Q. et al (2019). Comparative transcriptome profiling of immune response against Vibrio harveyi infection in Chinese tongue sole. Sci. Data.6, 224. 10.1038/s41597-019-0231-2
93
Yan W. Qiao Y. Qu J. Liu X. Zhang Q. Wang X. (2021). The hsp40 gene family in Japanese flounder: identification, phylogenetic relationships, molecular evolution analysis, and expression patterns.Front. Mar. Sci.7:596534. 10.3389/fmars.2020.596534
94
Yáñez J. M. Yoshida G. M. Parra Á. Correa K. Barría A. Bassini L. N. et al (2019). Comparative genomic analysis of three salmonid species identifies functional candidate genes involved in resistance to the intracellular bacterium Piscirickettsia salmonis. Front. Genet.10:665. 10.3389/fgene.2019.00665
95
Yang Y. Yu H. Li H. Wang A. (2016). Transcriptome profiling of grass carp (Ctenopharyngodon idellus) infected with Aeromonas hydrophila.Fish Shellfish Immunol.51329–336. 10.1016/j.fsi.2016.02.035
96
Ye H. Lin Q. Luo H. (2018). Applications of transcriptomics and proteomics in understanding fish immunity.Fish Shellfish Immunol.77319–327. 10.1016/j.fsi.2018.03.046
97
Zeng Z. H. Du C. C. Liu S. R. Li H. Peng X. X. Peng B. (2017). Glucose enhances tilapia against Edwardsiella tarda infection through metabolome reprogramming. Fish Shellfish Immunol.61, 34–43. 10.1016/j.fsi.2016.12.010
98
Zhang X. Li Q. Xu Z. Dou J. (2020). Mass spectrometry-based metabolomics in health and medical science: a systematic review.RSC Adv.103092–3104. 10.1039/c9ra08985c
99
Zhu L. Gao N. Wang R. Zhang L. (2018). Proteomic and metabolomic analysis of marine medaka (Oryzias melastigma) after acute ammonia exposure.Ecotoxicology27267–277. 10.1007/s10646-017-1892-2
100
Zhu Z. X. Jiang D. L. Li B. J. Qin H. Meng Z. N. Lin H. R. et al (2019). Differential transcriptomic and metabolomic responses in the liver of Nile tilapia (Oreochromis niloticus) exposed to acute ammonia.Mar. Biotechnol.21488–502. 10.1007/s10126-019-09897-8
101
Zou Z. Zheng Q. Cai J. Tang J. Xia L. Li P. F. et al (2020). Identification of BAG5 from orange-spotted grouper (Epinephelus coioides) involved in viral infection.Dev. Comp. Immunol.1:103916. 10.1016/j.dci.2020.103916
Summary
Keywords
fish immune response, omics, genomics, transcriptomics, proteomics, metabolomics
Citation
Natnan ME, Low C-F, Chong C-M, Bunawan H and Baharum SN (2021) Integration of Omics Tools for Understanding the Fish Immune Response Due to Microbial Challenge. Front. Mar. Sci. 8:668771. doi: 10.3389/fmars.2021.668771
Received
18 February 2021
Accepted
27 May 2021
Published
21 June 2021
Volume
8 - 2021
Edited by
Seyed Hossein Hoseinifar, Gorgan University of Agricultural Sciences and Natural Resources, Iran
Reviewed by
Luis Mercado, Pontifical Catholic University of Valparaíso, Chile; Tawheed Hasan, Sylhet Agricultural University, Bangladesh
Updates
Copyright
© 2021 Natnan, Low, Chong, Bunawan and Baharum.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Syarul N. Baharum, nataqain@ukm.edu.my
This article was submitted to Marine Fisheries, Aquaculture and Living Resources, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.