Abstract
Okadaic acid (OA), produced by dinoflagellates during harmful algal blooms, is a principal diarrhetic shellfish poisoning toxin. This toxin poses a potential threat to bivalves with economic values. To better understand the toxicity mechanism of OA to bivalves, in this study, oxidative stress biomarkers (superoxide dismutase, SOD; catalase, CAT; glutathione S-transferase, GST; malondialdehyde, MDA) and the expression of detoxification genes (heat shock protein 70, HSP70; heat shock protein 90, HSP90; cytochrome P450, CYP450) were assessed in the gills of scallops Chlamys farreri after 24 h, 48 h and 96 h exposure to OA. In addition, the digestive glands of scallops exposed to OA for 96 h were dissected for an iTRAQ based quantitative proteomic analysis. The results of OA exposure experiments showed that OA induces oxidative stress and significant enhancement of the expression of detoxification genes in scallops. The proteomics analysis revealed that 159 proteins altered remarkably in OA-treated scallops, and these proteins were involved in phagosomes, regulation of actin cytoskeleton, adherens junction, tight junction, and focal adhesion. Amino acid biosynthesis, carbon metabolism, pentose phosphate pathway, fructose and mannose metabolism in the digestive glands were also significantly impacted. Our data shed new insights on the molecular responses and toxicity mechanisms of C. farreri to OA.
Introduction
In recent years, harmful algal blooms (HABs) have occurred widespread in the ocean, with higher frequency and longer periods than that in the past (Dolah, 2000; Anderson et al., 2012; Gobler et al., 2017). The emergence of HABs poses a major threat to human health, fisheries resources, and marine ecosystems worldwide (Van Dolah and Ramsdell, 2001; Hoagland and Scatasta, 2006; Hallegraeff et al., 2021). One main hazard of HABs is the release of a variety of shellfish toxins, including diarrhetic shellfish toxins (DST), neurotoxic shellfish toxins (NST), paralytic shellfish toxins (PST), and forgetful shellfish toxins (AST) (Christian and Luckas, 2008).
Diarrhetic shellfish toxins (DST) is one of the most threatening shellfish toxins in the world. The lipophilicity allows DST to accumulate in animal tissues and to be biomagnified in the food web (Gerssen et al., 2010; Lloyd et al., 2013; Valdiglesias et al., 2013; Liu et al., 2019). Numerous reports have referred to public safety and health incidents caused by diarrheal shellfish poisoning (DSP) around the world (Yasumoto et al., 1978; de Carvalho et al., 2019; Young et al., 2019). Okadaic acid (OA) and its analogs, a typical DST, are responsible for DSP in humans (Garcia et al., 2005). Studies on mammalian or in vitro mammalian cell lines have shown that OA inhibits the serine and threonine phosphatases PP1 and PP2A (Bialojan and Takai, 1988; Honkanan et al., 1994; Maynes et al., 2001), leads to oxidative damage (Kamat et al., 2013; Vieira et al., 2013) and cytoskeleton destruction (Kreienbuhl et al., 1992; Fiorentini et al., 1996; Opsahl et al., 2013), affects the cell cycle (Messner et al., 2001; Valdiglesias et al., 2010a,b), neurotoxicity (Kamat et al., 2013) and apoptosis (Jayaraj et al., 2009; Ferron et al., 2014; Fu et al., 2019). In bivalves, laboratory exposure experiments have also shown that OA cause DNA damage in clams Ruditapes decussatus (Braga et al., 2021), lead to oxidative stress in oysters Crassostrea gigas and mussels Mytilus galloprovincesis (Romero-Geraldo and Hernandez-Saavedra, 2014; Prego-Faraldo et al., 2017), and decrease the immunity of oysters C. gigas, mussels Perna perna and clams Anomalocardia brasiliana (Mello et al., 2010; Prado-Alvarez et al., 2013). Overall, although the mode and mechanism of action of OA on mammals have been well studied, current studies on the impacts of OA on bivalves are limited and mostly confined to the physiological level.
The scallops Chlamys farreri is an important fishery and aquaculture species widely distributed in the subtropical western Pacific, playing an important role in the coastal ecosystem (Smaal et al., 2001). Moreover, C. farreri can accumulate shellfish toxins and therefore is widely used in algal toxin research (Li et al., 2017). As little is known about the toxic effects and the molecular mechanisms underlying the effects of OA on bivalves, it is necessary to carry out studies using omics for more global information. In this study, we combined traditional physiology and iTRAQ proteomics to (1) determine the physiological responses of C. farreri to OA and (2) elucidate the potential mechanisms of OA toxicity on C. farreri from the molecular perspective.
Materials and Methods
Reagent
Okadaic acid (OA) (NRC CRM-OA-d) was obtained from the Institute for Marine Biosciences (National Research Centre, Halifax Regional Municipality, NS, Canada), and stored in the dark at 4°C until the experiment. Stock solutions (20 μg/L) were prepared in dimethyl sulfoxide (DMSO, Sinopharm Chemical Reagent, China).
Experimental Conditions
Experimental scallops, C. farreri (shell length: 4–6 cm) were collected from a local farm (Yantai, Shandong, China). After cleaning shells, the scallops were transported to the Muping Coastal Environmental Research Station and acclimated in filter-sterilized seawater (temperature 15.3 ± 0.2°C, pH 8.12 ± 0.07, salinity 31.2 ± 0.5‰, mean ± SD) for 14 days. The scallops were fed daily with commercial algal blends during the acclimation period.
After acclimation, three treatment groups, including blank control [filter-sterilized seawater (FSSW)], solvent control (dimethyl sulfoxide, DMSO, 0.01%, v/v), and exposure groups (OA at nominal concentrations of approximately 10–20 μg OA eq. 100 g–1 shellfish meat) were conducted. The exposure concentration of OA used in this study is close to the European Food Safety Authority (EFSA) regulation limit for human consumption of shellfish (16 μg OA eq.100 g–1 shellfish meat) (Authority, 2008). The experiment was carried out in an 80L aquarium with 40 organisms in each aquarium. Three duplicate aquariums were used in each treatment (a total of 120 individuals in each treatment). In the OA treatment, the scallops were directly injected with 100 μL of 20 μg/L OA. The scallops in the blank control and solvent control were injected with 100 μL FSSW and DMSO, respectively. After injection, scallops were put back into the corresponding aquarium and sampled at 24 h, 48 h, and 96 h. No scallop mortality was observed during the experiment. Sampling time points were set according to the standardized method proposed by USEPA (2002). it can be proved that Chlamys farreri is a potential indicator organism that can respond quickly in environmental monitoring events.
At each sampled time point, the gills and digestive glands of the scallops were carefully cut off on the ice and immediately frozen in liquid nitrogen and stored at –80°C for subsequent analysis. The gills of the bivalves are more closely exposed to the environment, more susceptible to exogenous substances (Regoli and Principato, 1995; Magara et al., 2018). In addition, the digestive gland is one of the most important tissues for assessing the health status of bivalves and can be associated with functional damage to cells (Teng et al., 2021).
Oxidative Stress Biomarkers
To measure oxidative stress biomarkers, 10-15 individuals (gills) were dissected at each time point and pooled into 5 independent replicates to minimize biological variation. After sampling, the gills were used to prepare a tissue homogenate as follows: the gills were homogenized in phosphate buffer (50 mM potassium dihydrogen phosphate; 50 mM potassium phosphate dibasic; 1 mM EDTA; pH 7.0) using an IKA homogenizer (Ultra Turrax IKA T10 basic, Staufen, Germany). The homogenates were then centrifuged at 10,000 g for 20 min at 4°C to obtain supernatants. To investigate oxidative stress in scallops, superoxide dismutase (SOD), catalase (CAT), and glutathione S-transferase (GST) activities were determined using the commercial kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Malondialdehyde content was measured also using commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Subsequently, protein concentrations were determined according to the Bradford method using bovine γ-globulin as a standard (Bradford, 1976).
Real-Time Quantitative PCR
A total of 6–9 individual samples (gills) were dissected and pooled into 3 independent samples for real-time quantitative polymerase chain reaction (PCR) analysis (qRT-PCR). In this study, the genes heat shock protein 70 (HSP70), heat shock protein 90 (HSP90), and cytochrome P450 (CYP450) were selected for qRT-PCR. In addition, β-actin was used as an internal control to quantify the expression of selected genes in scallops. The primers designed for qRT-PCR are listed in Supplementary Table 1. Total RNA was extracted from gills of scallops using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s instructions. The cDNA was synthesized from DNase I-treated (Promega, United States) RNA and then combined with 10 μL of 2 × Master Mix (Applied Biosystems, United States), 4.8 μL of DEPC-treated H2O, and 0.4 μL (0.2 μM) of each forward and reverse primer to a final volume of 20 μL. The qRT-PCR was performed on the Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, United States) using standard protocols. The specificity of the qPCR products was analyzed by a dissociation curve analysis of the amplification products. The expression of selected genes was analyzed using the comparative 2–ΔΔCT method (Livak and Schmittgen, 2001).
iTRAQ-Based Quantitative Proteomic Analysis
After 96 h exposure, six samples of digestive glands in solvent control and OA treatment with three biological replicates per treatment were used in the proteomic analysis with an equal mass (300 mg wet weight). Protein extraction, iTRAQ labeling, and LC-MS/MS analysis were conducted based on standard proteomics methods (Han et al., 2013). More detailed information about the quantitative proteomic analysis is described in the Supplementary Material.
Protein Pilot Software v. 5.0 (AB SCIEX®, United States) was used to analyze the acquired MS/MS raw data, and protein identification was performed using the Paragon algorithm (Shilov et al., 2007). Functional annotations of the differentially expressed proteins (DEPs) were conducted using the Blast2GO program. Differentially expressed proteins were annotated into three categories based on Gene Ontology (GO) terms: biological process (BP), molecular function (MF), and cellular component (CC). The protein pathway was annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Then, the GO enrichment terms and pathways of DEPs in the background of identified proteins were determined via a hypergeometric test (p < 0.05). To gain a deeper understanding of the pathways significantly enriched after OA treatment, protein-protein network analysis was performed using Cytoscape 3.7.1
Statistical Analysis
Experimental data are represented as the means ± standard error of mean and were analyzed using Graphpad prism 7.0. The normality of the data was verified by the Shapiro-Wilk test, and the homogeneity of the variances was analyzed by Levene’s test. One-way ANOVA and Tukey’s post hoc test was performed to analyze whether the differences between each group were significant. P < 0.05 was considered significant difference.
Results
No significant difference was detected in the physiological indexes between the solvent control group and the control group, thus only experimental results derived from the control group were analyzed and discussed.
Oxidative Stress Biomarkers
In the scallops, SOD and CAT activities were significantly stimulated by OA treatment at 96 h, increasing by 1.55-fold and 1.3-fold compared to the control, respectively (Figures 1A,C). Meanwhile, GST activity was enhanced by 1.13-fold after the 48 h exposure to OA (Figure 1B). A significant increase in MDA content was also observed in the OA group in the 48 h and 96 h treatments (Figure 1D).
FIGURE 1
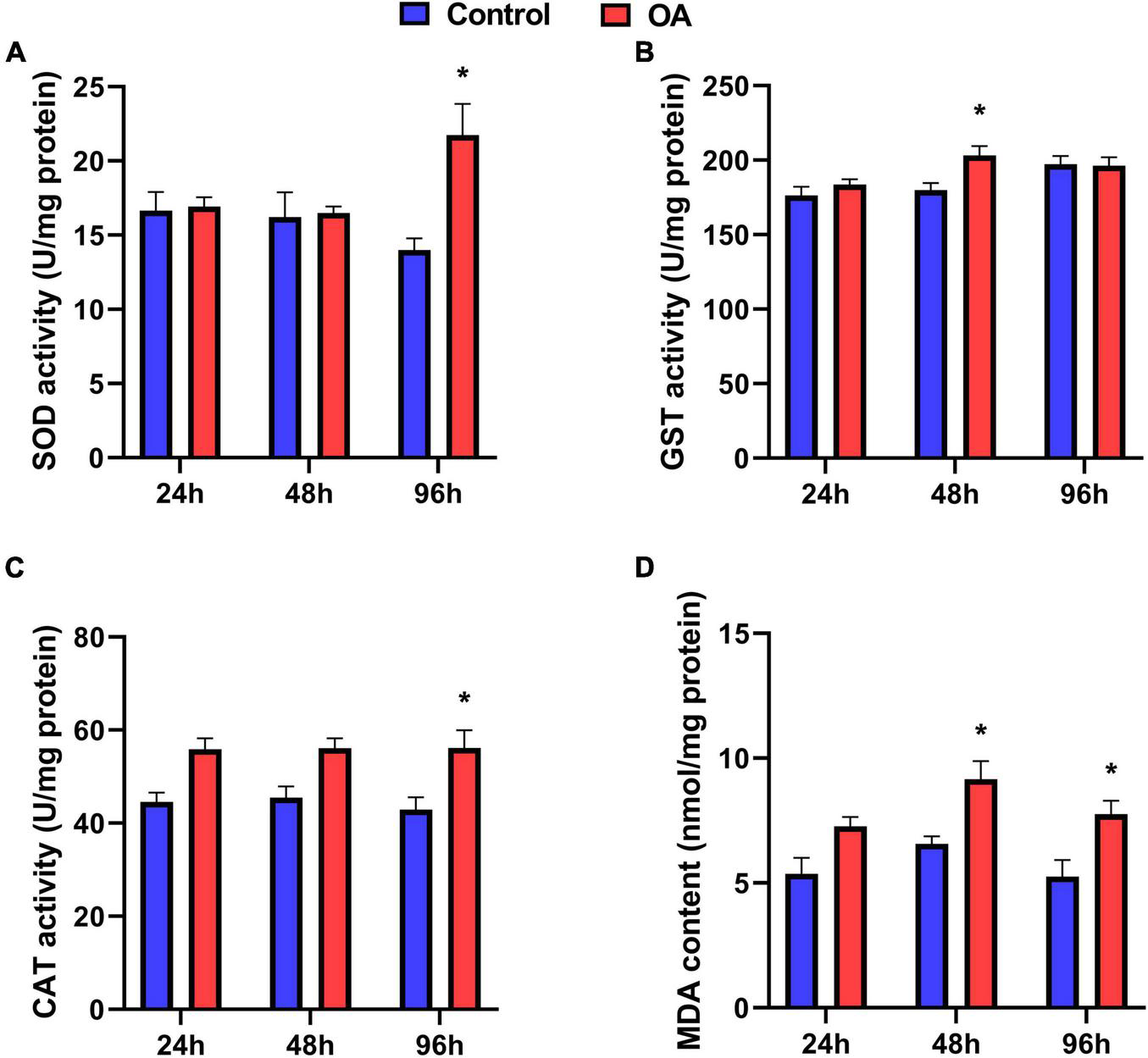
Activities of SOD (A), GST (B), CAT (C), and MDA content (D) of C. farreri exposed to OA (n = 5). Asterisks indicate significant differences between the control and OA-exposed group within each fixed sampling period, and different letters indicate significant differences among different sampling periods within each treatment (p < 0.05).
Real-Time Quantitative PCR Analyses
Among OA treatments, a significant increase in mRNA expression of HSP70 was observed in the scallops at 48 h and 96 h compared to those at 24 h (Figure 2A). Unexpectedly, the mRNA expression of HSP70 is significantly suppressed by OA in the 24 h treatment (Figure 2A). Although with no statistical difference, the expression of HSP90 transcripts showed a similar temporal trend to HSP70 after OA injection (Figure 2B). In addition, the expression of CYP450 increased significantly at 96 h in the OA-treated group, which was 4.61 times higher than that at 24 h (Figure 2C).
FIGURE 2
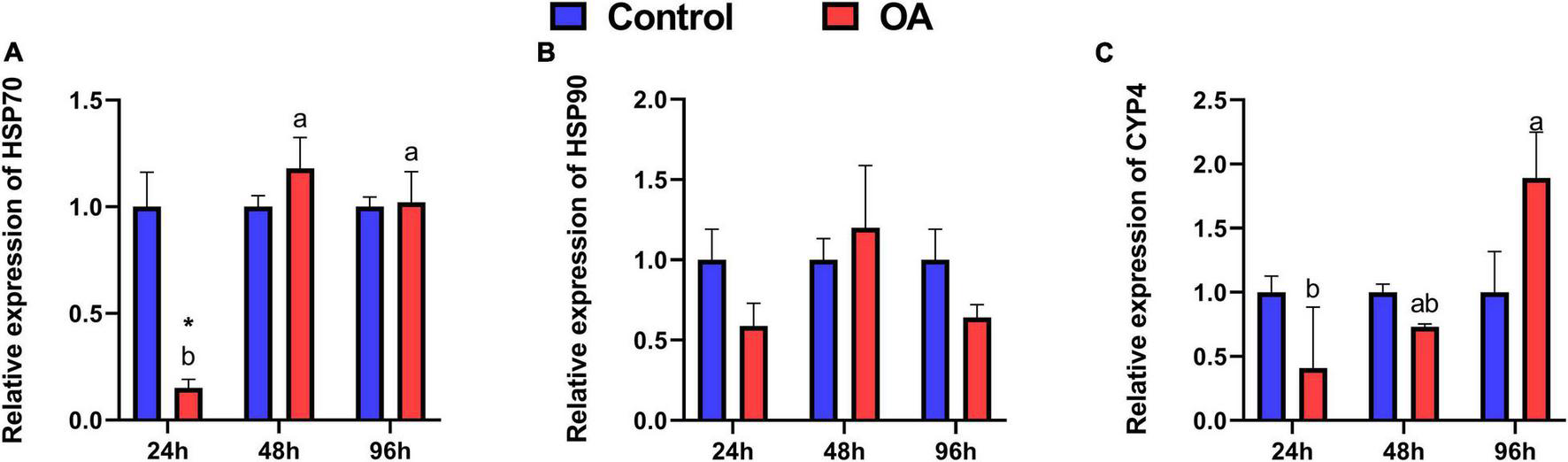
Relative expression levels of HSP70 (A), HSP90 (B), and CYP450 (C) transcripts in the C. farreri after 24 h, 48 h, and 96 h exposure to OA (n = 3). Asterisks indicate significant differences between the control and OA-exposed group within each fixed sampling period, and different letters indicate significant differences among different sampling periods within each treatment (p < 0.05).
Proteome Analysis
A total of 159 DEPs (Supplementary Materials, Supplementary Table 2) were detected in the digestive glands of the scallops exposed to OA, compared to the solvent controls (fold change > 1.2 as up-regulated or < 0.83 as down-regulated, p < 0.05). In these DEPs, 104 of the 159 DEPs were up-regulated and 55 were down-regulated (Figure 3A). At the same time, all the differences in protein GO enrichment (BP, CC, MF) and pathway KEGG enrichment results and significant (p < 0.05) numbers were summarized, and intuitively display through the column figure (Figure 3B). Specifically, 855 proteins were annotated with biological process (BP), and 239 terms were significantly enriched. In contrast, 156 proteins were annotated with cell component (CC), and 58 terms were significantly enriched; 284 proteins were annotated with molecular function, including 94 terms were significantly enriched. Furthermore, KEGG analysis showed that there are 9 pathways were significantly enriched.
FIGURE 3
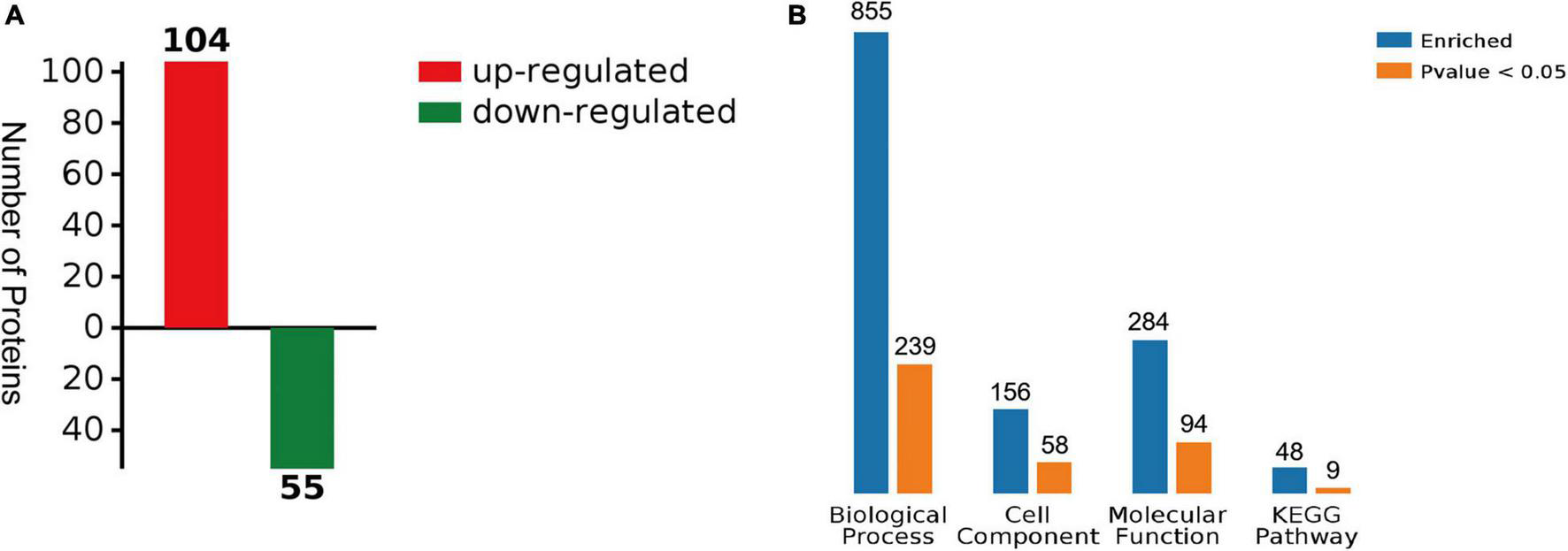
Summary of differentially expressed proteins (DEPs). (A) The number of DEPs between solvent control and OA treatment. (B) Summary of differences and significant numbers of protein GO enrichment (BP, CC, MF) and pathway KEGG enrichment results (p < 0.05).
Differentially expressed proteins (DEPs) were classified into various categories based on gene ontology (GO) terms (Figure 4). According to the category of biological process (Figure 4A), the DEPs were mostly annotated to the “single-organism metabolic process,” “organonitrogen compound metabolic process,” and “small molecule metabolic process.” The DEPs covered a wide range of molecular functions (Figure 4B), mainly including “catalytic activity,” “anion binding,” “nucleotide binding,” and “nucleoside phosphate binding.” Based on the category of cellular components (Figure 4C), the DEPs were mainly located in “intracellular,” “intracellular part,” and “cytoplasm.”
FIGURE 4
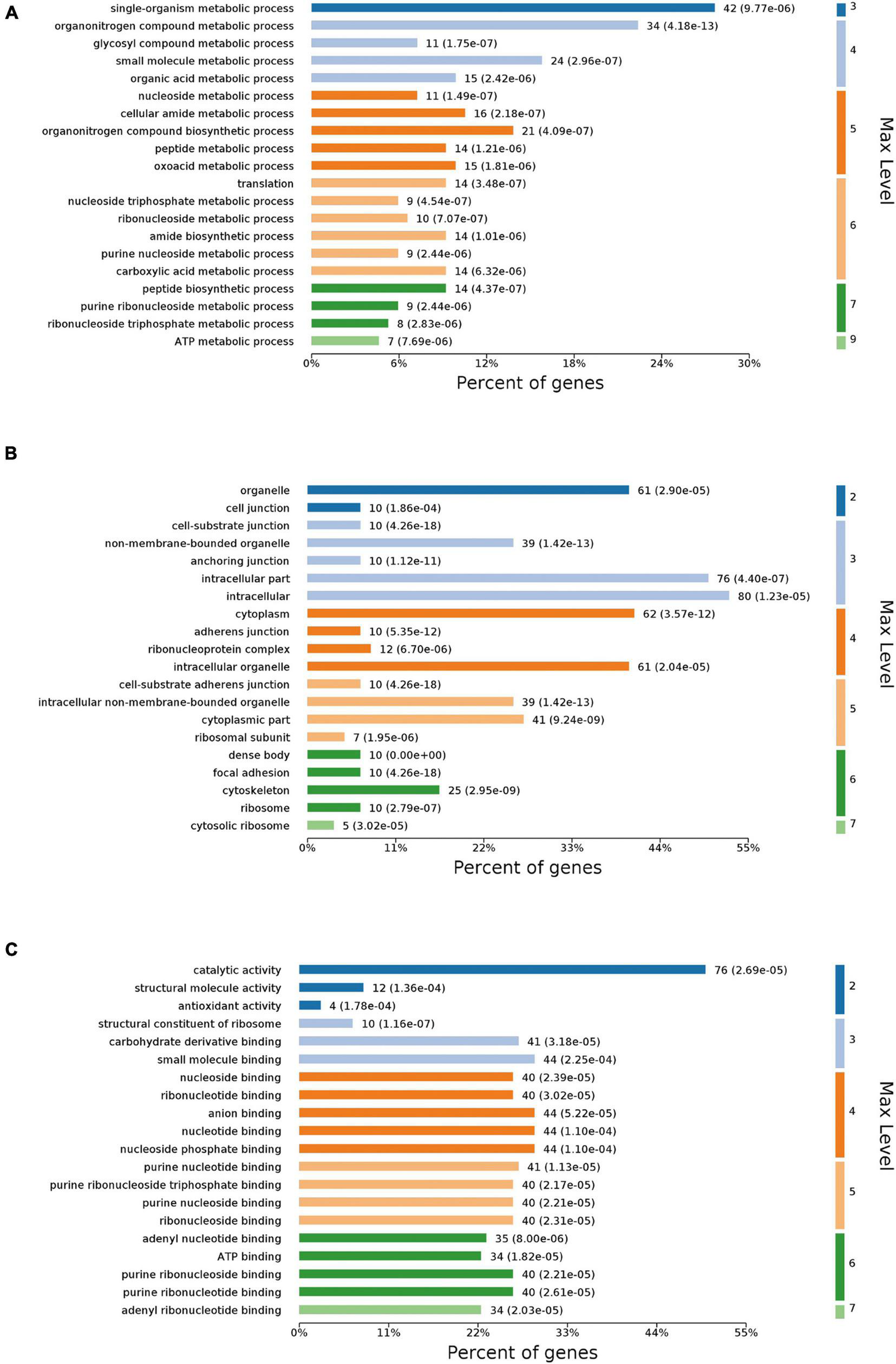
GO annotation of DEPs. DEPs were assigned to second-tier GO categories associated with three terms: biological process (A), cellular component (B), and molecular function (C).
To further investigate the main pathways of the scallop’s response to OA, KEGG enrichment analysis and protein-protein interaction (PPI) networks were also performed on the DEPs. As shown in Figures 5, 6, OA treatment led to DEPs involved in several cellular processes in the scallops, such as phagosomes, regulation of actin cytoskeleton, adherens junction, tight junction, and focal adhesion. In addition, DEPs were also significantly enriched in pathways of amino acid biosynthesis, carbon metabolism, pentose phosphate pathway, fructose and mannose metabolism.
FIGURE 5
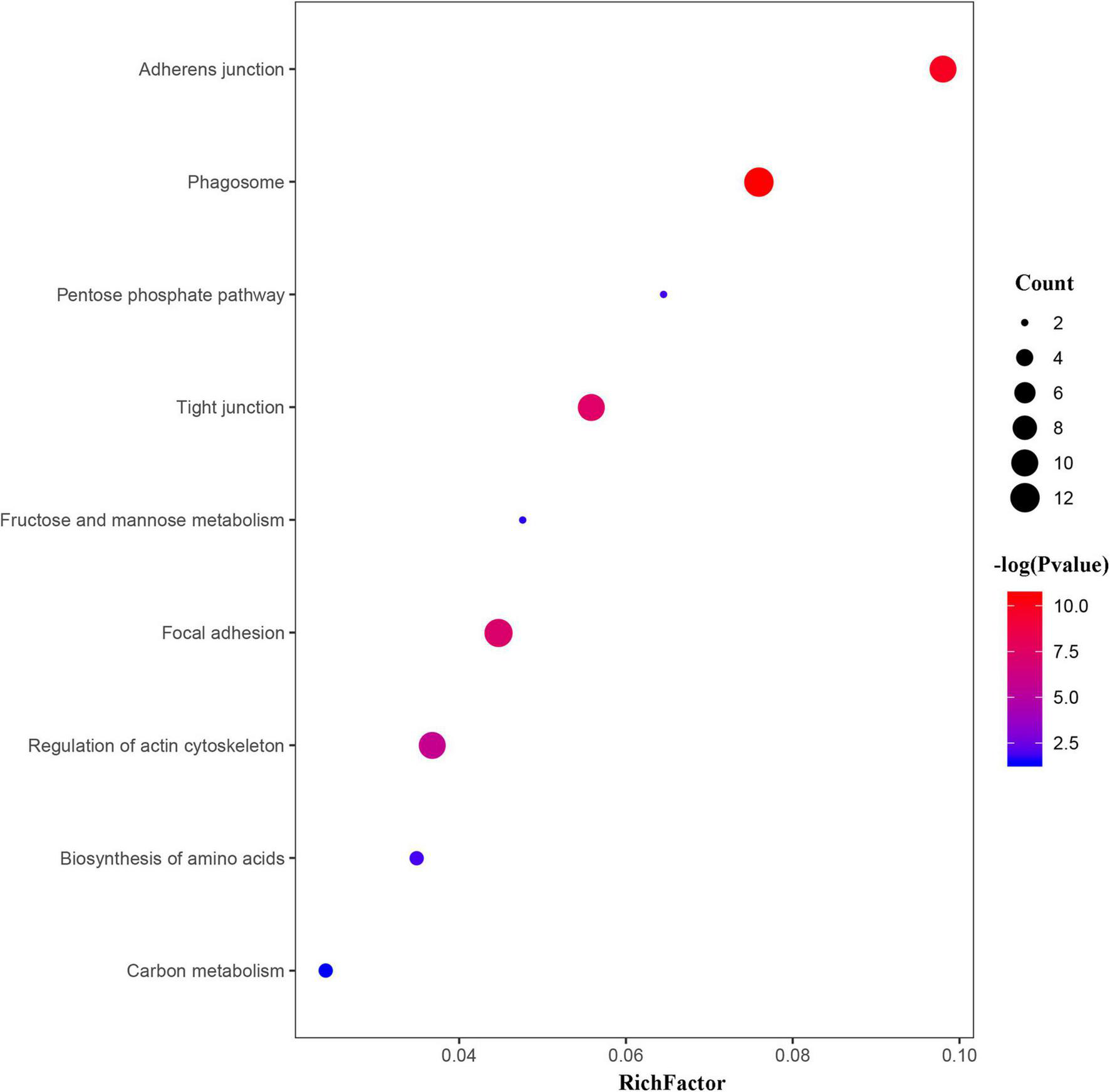
Bubble chart showing the significantly enriched KEGG pathways in C. farreri exposed to OA (p < 0.05). In the bubble chart, the vertical axis is the functional classification or pathway and the horizontal axis is the proportion of the different proteins in this functional type compared to the proportion of the identified protein. The color of the circle indicates the enrichment significance P-value, and the size of the circle indicates the number of differential proteins in functional classes or pathways.
FIGURE 6
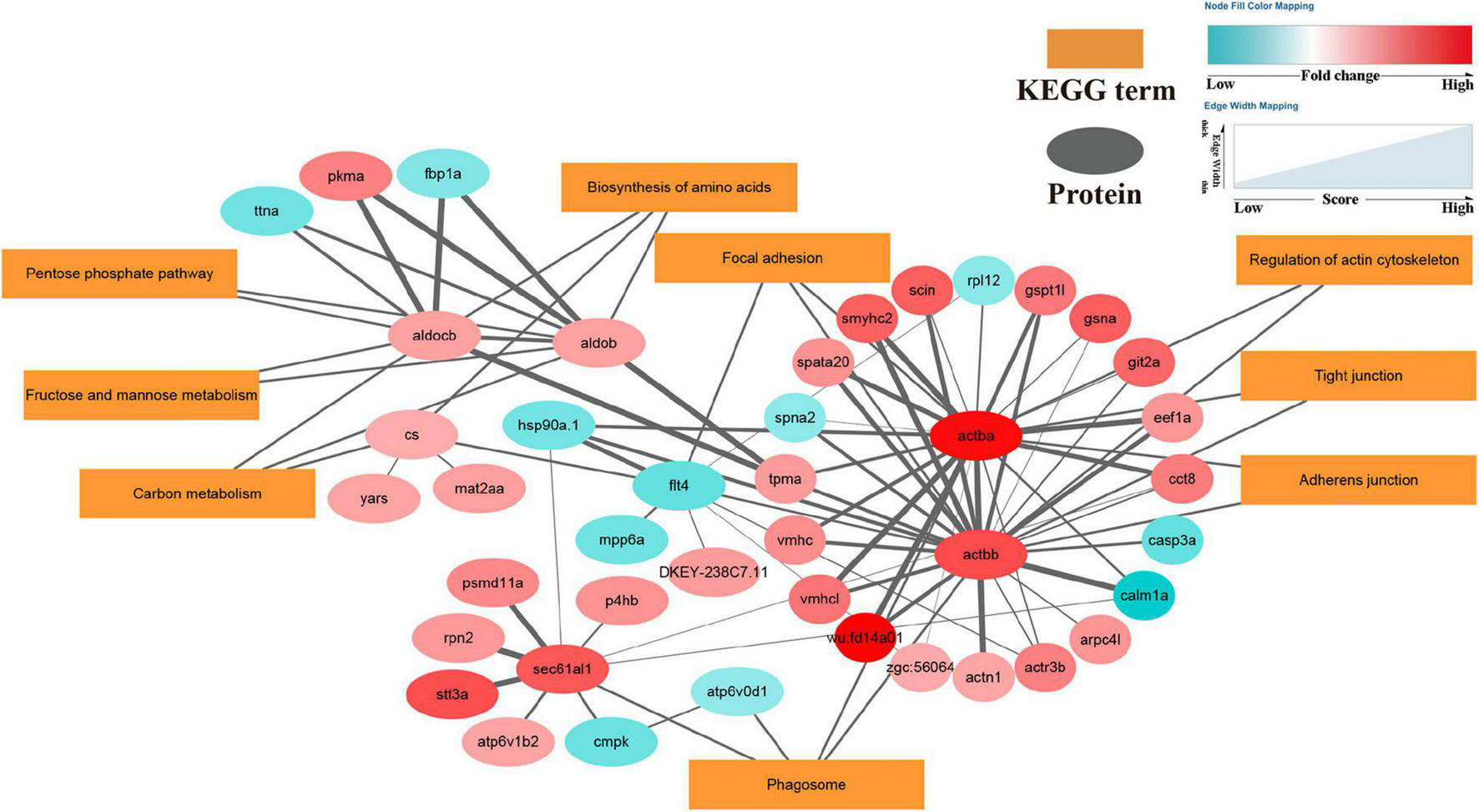
DEPs enriched in the protein-protein interaction network in OA-exposed scallops. Proteins are shown as small elliptical nodes (blue nodes represent down-regulated proteins and red nodes represent up-regulated proteins) and are connected with associated KEGG pathways (rectangular Nodes). The size of the line between nodes represents the scoring value of the interaction between the two nodes, and the greater the score, the more reliable.
Discussion
Oxidative Stress Biomarkers
When exposed to environmental stress, organisms usually produce excessive ROS, which leads to oxidative stress (Lushchak, 2011). Antioxidant enzymes play a central role in the cellular defense mechanism against oxidative stress (Valko et al., 2006; Lushchak, 2011). As the first line of the antioxidant defense system, SOD catalyzes superoxide anion disproportionation to oxygen and hydrogen peroxide, while CAT prevents the formation of excessive H2O2 through decomposing H2O2 once it is formed (Halliwell, 1974). As a phase II detoxification enzyme, GST protects cells and tissues against oxidative stress by catalyzing the conjugation of the reduced form of glutathione to various xenobiotic substrates (Hayes and Strange, 1995). These antioxidant enzymes interact with each other to maintain the balance between ROS production and ROS scavenging in an organism. In this study, all three antioxidant enzymes (SOD, CAT, and GST) were generally stimulated in the gills of scallops, indicating that the antioxidant system was activated in response to the OA exposure, which may have caused oxidative stress to the scallops. Similar to our results, increased antioxidant enzymes activity was also observed in the bivalves Nodipecten subnodosus (Campa-Cordova et al., 2009) and Anomalocardia flexuosa (Leite et al., 2021), when exposed to Prorocentrum lima (a dinoflagellate producing DSP toxins).
Lipid peroxidation (LPO) is a major indicator of cellular oxidative damage level in organisms and is a major contributor to the loss of cell function under OA exposure (Cossu et al., 2000; Alves de Almeida et al., 2007). Lipid peroxidation usually occurs when the production of ROS exceeds the capacity of the antioxidant system (Halliwell, 1996; Lushchak, 2011). The main product of lipid peroxidation is MDA, so generally, the extent of LPO can be assessed indirectly by the amount of MDA (Ohkawa et al., 1979). In this study, the LPO level increased significantly in the scallops after 48 h and 96h exposure of OA, indicating that the increasing antioxidant enzyme activities could not maintain the balance between ROS production and elimination, eventually leading to the LPO.
Alterations in Gene Expression of the Stress-Related Proteins
As molecular chaperones, HSPs help organisms combat environmental stresses and maintain cellular homeostasis by promoting protein folding and preventing aggregation of denatured proteins (Johnston and Kucey, 1988; Liu et al., 2014; Johnston et al., 2018). CYP450 is a monooxygenase that promotes the biotransformation and metabolic detoxification of endogenous and exogenous toxic compounds (Snyder, 2000; Xu et al., 2005). HSPs and CYPs genes can be activated by various forces in aquatic organisms, which are widely used as biomarkers for toxicology research (Chaty et al., 2004; Rewitz et al., 2006; Chae et al., 2009; Liu et al., 2013; Ding et al., 2018). In the present study, the expression of both HSPs and CYP450 genes increased with the OA exposure time, supporting their roles in stress response and detoxification process to OA. The time-dependent changes in genes to OA are also in line with the result of previous studies involving other environmental stresses (Liu et al., 2018; Nguyen et al., 2018; Nie et al., 2020; Zhang et al., 2020). We hypothesize that overexpression of the HSPs gene may counteract the proteins damaged by the ROS after OA exposure, while the enhanced synthesis of CYP450 may contribute to complete the biotransformation of OA. In short, expressional changes of these genes reflect pressures of OA on the scallop tissues from the molecular aspect.
Proteomic Responses of Scallops to Okadaic Acid Exposure
As major organelles in the translation process, ribosomes are essential for protein production in cells (Wimberly et al., 2000; Sonenberg and Hinnebusch, 2009). In the OA-treated scallops of this study, 11 ribosome-associated DEPs exhibited significant over-expression (Supplementary Table 2), while significant enrichment of the amino acid biosynthetic pathway was suggested by the KEGG analysis and PPI network. The up-regulation of these proteins or pathways suggests that protein synthesis is effectively enhanced in the scallops exposed to OA, which possibly compensates for the proteins damaged by oxidative stress (Tomanek, 2010; Dasuri et al., 2013).
In the current study, the KEGG pathway analysis shows that phagosome is one of the important pathways in the scallops’ response to OA (Figure 5). In the invertebrate immune system, phagocytosis is a necessary mechanism for the removal of invading microorganisms, dead cells, and other relatively large particles (Mydlarz et al., 2006). The internalization of phagocytosis of large particles is initiated by the combination of particles with cell surface receptors, followed by the reconstitution and reorganization of the cytoskeleton to form phagosomes, which ultimately complete the internalization of particles (Castellano et al., 2001; Freeman and Grinstein, 2014). As an important component of phagocytosis, the formation of phagosomes is a prerequisite for the degradation of intracellular macromolecules (Mizushima, 2007; Xie and Klionsky, 2007; Lancaster et al., 2021). In addition, we also detected the significantly altered abundance of ATP6V0D1, ATP6V1B2, and other lysosome-associated proteins in the OA treatments, suggesting that OA affects the formation of phagocytic lysosomes. Previous studies have shown that some functions of phagosomes occur only in specific membrane domains. For example, phagosomes can only undergo acidification and fuse with lysosomes to produce immunocompetent phagolysosomes (Vieira et al., 2002; Luzio et al., 2007). Moreover, endoplasmic reticulum (ER)-associated proteins, such as RPN2 and Sec61 subunits known as ER marker proteins, were also identified in the phagosome pathway of the scallops. Previous studies have reported that the ER might contribute to the formation of the phagosome (Muller-Taubenberger et al., 2001; Gagnon et al., 2005; Mao et al., 2020). In summary, our results indicated that altered phagocytosis activity is an important way for scallops to respond to OA exposure, which could remove damaged proteins and apoptotic cells caused by OA, thereby reducing inflammation and tissue damage (Ellis et al., 2011; Song et al., 2015).
Our proteomic results showed that a total of 25 cytoskeleton-related proteins changed significantly, and 23 DEPs were up-regulated significantly. KEGG analysis also showed that the regulation of actin cytoskeleton in the OA-treated scallops was changed. In eukaryotic cells, the cytoskeleton is a complex reticular system of protein components such as microtubules, microfilaments, and intermediate fibers (Schliwa and Vanblerkom, 1981), playing a key role in maintaining cell structure, adhesion, migration, differentiation, division, and organelle transport (Hall, 1998). The result of our proteomic study is in accordance with the previous physical and omics studies showing that DST induced extensive exert cytotoxic effects by modulating the cytoskeleton (Espina and Rubiolo, 2008; Hanana et al., 2012; Huang et al., 2015; He et al., 2019). Changes in cytoskeleton-related proteins may also be related to cell connections, such as tight junctions and focal adhesion, thereby affecting the transport of macromolecules between cells and the signal transmission between cells and the extracellular matrix (Schneeberger and Lynch, 1992; Fanning et al., 1998). In this study, OA treatment led to significant alteration in the expression of proteins from tight junction, adherens junction, and focal adhesion pathways. Tight junctions and focal adhesion are two well-known cell connections associated with the cytoskeleton. The former links the plasma membranes of adjacent cells together and maintains normal cell function by acting as a barrier between cells to prevent the diffusion of soluble substances as well as membrane proteins and plasmids (Hartsock and Nelson, 2008). The latter plays a key role in regulating cell migration, proliferation, and apoptosis by mediating mechanical and biochemical signaling between cells and the extracellular matrix (Takeichi, 1995; Gumbiner, 2005; Mitra et al., 2005; Hartsock and Nelson, 2008; Burridge, 2017). Overall, the alteration of cytoskeletal proteins and related cell connections may compensate for the increased protein turnover and altered phagocytosis activity caused by OA.
The glycolytic pathway is the universal metabolic pathway that provides energy and converting energy metabolism to glycolysis has been shown to inhibit apoptosis and thus promote cell survival (Gohil et al., 2010; Slavov et al., 2014; Gao et al., 2020). In this study, the fructose-bisphosphate aldolase B (ALDOB) and fructose-bisphosphate aldolase C-B (ALDOCB) were significantly up-regulated. The two proteins are fructose diphosphate aldolase that catalyzes the formation of dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP) from fructose-1,6-bisphosphate (FBP) in the glycolytic pathway (Bauer and Levenbook, 1969). In addition, our KEGG analysis showed significant enrichment in the pentose phosphate pathway and fructose and mannose metabolism (Figure 5). The pentose phosphate pathway and fructose and mannose metabolism are essential branches of glycolysis, and both pathways produce intermediate products of glycolysis that converge into the glycolytic pathway (Jiang et al., 2014). Moreover, data obtained in this study demonstrate that OA exposure significantly enhanced the activity of citrate synthase (CS). Citrate synthase is the downstream process after the glycolytic pathway, which catalyzes the condensation of acetyl coenzyme A with oxaloacetate produced by glycolysis or other isomerization reactions to synthesize citrate in the tricarboxylic acid cycle. The up-regulated expression of CS further confirms the activation of the glycolytic pathway (Champe et al., 2005). In general, after exposure to OA, scallops may increase the energy produced by glycolysis to meet the additional energy demand caused by changes in biological processes such as amino acid biosynthesis and regulation of the actin cytoskeleton (Tomanek and Zuzow, 2010; Huang et al., 2015).
Conclusion
In this study, even though the concentration used in this study was well below those tolerated by the scallops, OA still resulted in significant changes in the physiology and proteomics of the scallops. OA not only induced oxidative stress and altered stress gene expression in scallops but also significantly changed biological processes like biosynthesis of amino acids, energy metabolism, cytoskeleton functions, and phagosome in scallops. Our findings provide global information for the potential mechanisms of OA toxicity on bivalves.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
XW performed part of the experiment, analyzed the data, and drafted the manuscript. DW performed part of the experiment. TZ assisted with data analysis. QZ contributed to the review and editing. JZ did the conceptualization, funding acquisition, and review. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA23050303), the National Key Research and Development Program of China (2018YFC1406503), and the Youth Innovation Promotion Association, Chinese Academy of Sciences (No. 2019216).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.792050/full#supplementary-material
Footnotes
References
1
Alves de Almeida E. Celso Dias, Bainy A. Paula, de Melo Loureiro A. Regina Martinez G. et al (2007). Oxidative stress in Perna perna and other bivalves as indicators of environmental stress in the Brazilian marine environment: antioxidants, lipid peroxidation and DNA damage.Comp. Biochem. Physiol. A Mol. Integr. Physiol.146588–600. 10.1016/j.cbpa.2006.02.040
2
Anderson D. M. Cembella A. D. Hallegraeff G. M. (2012). Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management.Ann. Rev. Mar. Sci.4143–176. 10.1146/annurev-marine-120308-081121
3
Authority E. F. S. (2008). Marine biotoxins in shellfish - okadaic acid and analogues - scientific opinion of the panel on contaminants in the food chain.EFSA. J.6:589. 10.2903/j.efsa.2008.589
4
Bauer A. C. Levenbook L. (1969). Fructose diphosphate aldolase during growth and development of blowfly phormia regina (meigen).Comp. Biochem. Physiol.28619–624. 10.1016/0010-406x(69)92093-3
5
Bialojan C. Takai A. (1988). Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases - specificity and kinetics.Biochem. J.256283–290. 10.1042/bj2560283
6
Bradford M. M. (1976). Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding.Anal. Biochem.72248–254. 10.1016/0003-2697(76)90527-3
7
Braga A. C. Marcal R. Marques A. Guilherme S. Vilarino O. Martins J. M. L. et al (2021). Invasive clams (Ruditapes philippinarum) are better equipped to deal with harmful algal blooms toxins than native species (R.decussatus): evidence of species-specific toxicokinetics and DNA vulnerability.Sci. Total Environ.767:144887. 10.1016/j.scitotenv.2020.144887
8
Burridge K. (2017). Focal adhesions: a personal perspective on a half century of progress.FEBS J.2843355–3361. 10.1111/febs.14195
9
Campa-Cordova A. I. Nunez-Vazquez E. J. Luna-Gonzalez A. Romero-Geraldo M. J. Ascencio F. (2009). Superoxide dismutase activity in juvenile Litopenaeus vannamei and Nodipecten subnodosus exposed to the toxic dinoflagellate Prorocentrum lima.Comp. Biochem. Physiol. C Toxicol. Pharmacol.149317–322. 10.1016/j.cbpc.2008.08.006
10
Castellano F. Chavrier P. Caron E. (2001). Actin dynamics during phagocytosis.Semin. Immunol.13347–355. 10.1006/smim.2001.0331
11
Chae Y. J. Pham C. H. Lee J. Bae E. Yi J. Gu M. B. (2009). Evaluation of the toxic impact of silver nanoparticles on Japanese medaka (Oryzias latipes).Aquat. Toxicol.94320–327. 10.1016/j.aquatox.2009.07.019
12
Champe P. C. Harvey R. A. Ferrier D. R. (2005). Biochemistry.Philadelphia, PA: Lippincott Williams & Wilkins.
13
Chaty S. Rodius F. Vasseur P. (2004). A comparative study of the expression of CYP1A and CYP4 genes in aquatic invertebrate (freshwater mussel, Unio tumidus) and vertebrate (rainbow trout, Oncorhynchus mykiss).Aquat. Toxicol.6981–93. 10.1016/j.aquatox.2004.04.011
14
Christian B. Luckas B. (2008). Determination of marine biotoxins relevant for regulations: from the mouse bioassay to coupled LC-MS methods.Anal. Bioanal. Chem.391117–134. 10.1007/s00216-007-1778-x
15
Cossu C. Doyotte A. Babut M. Exinger A. Vasseur P. (2000). Antioxidant biomarkers in freshwater bivalves, Unio tumidus, in response to different contamination profiles of aquatic sediments.Ecotoxicol. Environ. Safe45106–121. 10.1006/eesa.1999.1842
16
Dasuri K. Zhang L. Keller J. N. (2013). Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis.Free Radical Biol. Med.62170–185. 10.1016/j.freeradbiomed.2012.09.016
17
de Carvalho I. L. Pelerito A. Ribeiro I. Cordeiro R. Núncio M. S. Vale P. (2019). Paralytic shellfish poisoning due to ingestion of contaminated mussels: a 2018 case report in Caparica (Portugal).Toxicon X.4:100017. 10.1016/j.toxcx.2019.100017
18
Ding J. N. Zhang S. S. Razanajatovo R. M. Zou H. Zhu W. B. (2018). Accumulation, tissue distribution, and biochemical effects of polystyrene microplastics in the freshwater fish red tilapia (Oreochromis niloticus).Environ. Pollut.2381–9. 10.1016/j.envpol.2018.03.001
19
Dolah F. M. V. (2000). Marine algal toxins: origins, health effects, and their increased occurrence.Environ. Health Perspect.108133–141. 10.1289/ehp.00108s1133
20
Ellis R. P. Parry H. Spicer J. I. Hutchinson T. H. Pipe R. K. Widdicombe S. (2011). Immunological function in marine invertebrates: responses to environmental perturbation.Fish Shellfish Immunol.301209–1222. 10.1016/j.fsi.2011.03.017
21
Espina B. Rubiolo J. A. (2008). Marine toxins and the cytoskeleton: pectenotoxins, unusual macrolides that disrupt actin.FEBS J.2756082–6088. 10.1111/j.1742-4658.2008.06714.x
22
Fanning A. S. Jameson B. J. Jesaitis L. A. Anderson J. M. (1998). The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton.J. Biol. Chem.27329745–29753. 10.1074/jbc.273.45.29745
23
Ferron P. J. Hogeveen K. Fessard V. Le Hegarat L. (2014). Comparative analysis of the cytotoxic effects of okadaic acid-group toxins on human intestinal cell lines.Mar. Drugs124616–4634. 10.3390/md12084616
24
Fiorentini C. Matarrese P. Fattorossi A. Donelli G. (1996). Okadaic acid induces changes in the organization of F-actin in intestinal cells.Toxicon34937–945. 10.1016/0041-0101(96)00025-6
25
Freeman S. A. Grinstein S. (2014). Phagocytosis: receptors, signal integration, and the cytoskeleton.Immunol. Rev.262193–215. 10.1111/imr.12212
26
Fu L. L. Zhao X. Y. Ji L. D. Xu J. (2019). Okadaic acid (OA): toxicity, detection and detoxification.Toxicon1601–7. 10.1016/j.toxicon.2018.12.007
27
Gagnon E. Bergeron J. J. Desjardins M. (2005). ER-mediated phagocytosis: myth or reality?J. Leukoc. Biol.77843–845. 10.1189/jlb.0305129
28
Gao Z. Dlamini M. B. Ge H. Jiang L. Geng C. Li Q. et al (2020). ATF4-mediated autophagy-dependent glycolysis plays an important role in attenuating apoptosis induced by Cr (VI) in A549 cells.Toxicol. Lett.331178–187. 10.1016/j.toxlet.2020.06.015
29
Garcia C. Truan D. Lagos M. Santelices J. P. Diaz J. C. Lagos N. (2005). Metabolic transformation of dinophysistoxin-3 into dinophysistoxin-1 causes human intoxication by consumption of O-acyl-derivatives dinophysistoxins contaminated shellfish.J. Toxicol. Sci.30287–296. 10.2131/jts.30.287
30
Gerssen A. Pol-Hofstad I. E. Poelman M. Mulder P. P. J. van den Top H. J. Dde Boer J. (2010). Marine toxins: chemistry, toxicity, occurrence and detection, with special reference to the dutch situation.Toxins2878–904. 10.3390/toxins2040878
31
Gobler C. J. Doherty O. M. Hattenrath-Lehmann T. K. Griffith A. W. Kang Y. Litaker R. W. (2017). Ocean warming since 1982 has expanded the niche of toxic algal blooms in the North Atlantic and North Pacific oceans.Proc. Natl. Acad. Sci. U.S.A.1144975–4980. 10.1073/pnas.1619575114
32
Gohil V. M. Sheth S. A. Nilsson R. Wojtovich A. P. Lee J. H. Perocchi F. et al (2010). Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis.Nat. Biotechnol.28249–255. 10.1038/nbt.1606
33
Gumbiner B. M. (2005). Regulation of cadherin-mediated adhesion in morphogenesis.Nat. Rev. Mol. Cell Biol.6622–634. 10.1038/nrm1699
34
Hall A. (1998). Rho GTPases and the actin cytoskeleton.Science279509–514. 10.1126/science.279.5350.509
35
Hallegraeff G. Enevoldsen H. Zingone A. (2021). Global harmful algal bloom status reporting.Harmful Algae102101992–101992. 10.1016/j.hal.2021.101992
36
Halliwell B. (1974). Superoxide-dismutase, catalase and glutathione peroxidase - solutions to problems of living with oxygen.New Phytol.731075–1086. 10.1111/j.1469-8137.1974.tb02137.x
37
Halliwell B. (1996). Antioxidants in human health and disease.Annu. Rev. Nutr.1633–50. 10.1146/annurev.nutr.16.1.33
38
Han G. D. Zhang S. Marshall D. J. Ke C. H. Dong Y. W. (2013). Metabolic energy sensors (AMPK and SIRT1), protein carbonylation and cardiac failure as biomarkers of thermal stress in an intertidal limpet: linking energetic allocation with environmental temperature during aerial emersion.J. Exp. Biol.2163273–3282. 10.1242/jeb.084269
39
Hanana H. Talarmin H. Pennec J. P. Droguet M. Morel J. Dorange G. (2012). Effect of okadaic acid on cultured clam heart cells: involvement of MAPkinase pathways.Biol. Open1, 1192–1199. 10.1242/bio.20122170
40
Hartsock A. Nelson W. J. (2008). Adherens and tight junctions: structure, function and connections to the actin cytoskeleton.Biochim. Biophys. Acta Biomembr.1778660–669. 10.1016/j.bbamem.2007.07.012
41
Hayes J. D. Strange R. C. (1995). Invited commentary potential contribution of the glutathione-s-transferase supergene family to resistance to oxidative stress.Free Radic. Res.22193–207. 10.3109/10715769509147539
42
He Z.-B. Duan G.-F. Liang C.-Y. Li H.-Y. Liu J.-S. Yang W.-D. (2019). Up-regulation of Nrf2-dependent antioxidant defenses in Perna viridis after exposed to Prorocentrum lima.Fish Shellfish Immunol.90173–179. 10.1016/j.fsi.2019.05.003
43
Hoagland P. Scatasta S. (2006). The economic effects of harmful algal blooms.Ecol. Harmful Algae189391–402. 10.1007/978-3-540-32210-8_30
44
Honkanan R. E. Codispoti B. A. Tse K. Boynton A. L. (1994). Characterization of natural toxins with inhibitory activity against serine/threonine protein phosphatases.Toxicon32339–350. 10.1016/0041-0101(94)90086-8
45
Huang L. Zou Y. Weng H.-W. Li H.-Y. Liu J.-S. Yang W.-D. (2015). Proteomic profile in Perna viridis after exposed to Prorocentrum lima, a dinoflagellate producing DSP toxins.Environ. Pollut.196350–357. 10.1016/j.envpol.2014.10.019
46
Jayaraj R. Gupta N. Rao P. V. L. (2009). Multiple signal transduction pathways in okadaic acid induced apoptosis in HeLa cells.Toxicology256118–127. 10.1016/j.tox.2008.11.013
47
Jiang P. Du W. J. Wu M. A. (2014). Regulation of the pentose phosphate pathway in cancer.Trends Biochem. Sci.5592–602. 10.1007/s13238-014-0082-8
48
Johnston C. L. Marzano N. R. van Oijen A. M. Ecroyd H. (2018). Using single-molecule approaches to understand the molecular mechanisms of heat-shock protein chaperone function.J. Mol. Biol.4304525–4546. 10.1016/j.jmb.2018.05.021
49
Johnston R. Kucey B. (1988). Competitive inhibition of HSP70 gene expression causes thermosensitivity.Science2421551–1554. 10.1126/science.3201244
50
Kamat P. K. Rai S. Nath C. (2013). Okadaic acid induced neurotoxicity: an emerging tool to study Alzheimer’s disease pathology.Neurotoxicology37163–172. 10.1016/j.neuro.2013.05.002
51
Kreienbuhl P. Keller H. Niggli V. (1992). Protein phosphatase inhibitors okadaic acid and calyculin-A alter cell-shape and F-actin distribution and inhibit stimulus-dependent increases in cytoskeletal actin of human neutrophils.Blood802911–2919.
52
Lancaster C. E. Fountain A. Dayam R. M. Somerville E. Sheth J. Jacobelli V. et al (2021). Phagosome resolution regenerates lysosomes and maintains the degradative capacity in phagocytes.J. Cell Biol.220:e202005072. 10.1083/jcb.202005072
53
Leite IdP Sandrini-Neto L. Squella F. L. Alves T. P. Schramm M. A. Calado et al (2021). Toxin accumulation, detoxification and oxidative stress in bivalve (Anomalocardia flexuosa) exposed to the dinoflagellate Prorocentrum lima.Aquat. Toxicol.232:105738. 10.1016/j.aquatox.2020.105738
54
Li Y. Sun X. Hu X. Xun X. Zhang J. Guo X. et al (2017). Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins.Nat. Commun.8:1721. 10.1038/s41467-017-01927-0
55
Liu H.-H. He J.-Y. Chi C.-F. Shao J. (2014). Differential HSP70 expression in Mytilus coruscus under various stressors.Gene543166–173. 10.1016/j.gene.2014.04.008
56
Liu X. Ji K. Jo A. Moon H. B. Choi K. (2013). Effects of TDCPP or TPP on gene transcriptions and hormones of HPG axis, and their consequences on reproduction in adult zebrafish (Danio rerio).Aquat. Toxicol.134104–111. 10.1016/j.aquatox.2013.03.013
57
Liu Y. Yu R.-C. Kong F.-Z. Li C. Dai L. Chen Z.-F. et al (2019). Contamination status of lipophilic marine toxins in shellfish samples from the Bohai Sea.China Environ. Pollut.249171–180. 10.1016/j.envpol.2019.02.050
58
Liu Z. P. Gu W. B. Tu D. D. Zhu Q. H. Zhou Y. L. Wang C. et al (2018). Effects of both cold and heat stress on the liver of the giant spiny frog (Quasipaa spinosa): stress response and histological changes.J. Exp. Biol.221:11. 10.1242/jeb.186379
59
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method.Methods25402–408. 10.1006/meth.2001.1262
60
Lloyd J. K. Duchin J. S. Borchert J. Quintana H. F. Robertson A. (2013). Diarrhetic shellfish poisoning. Washington, USA, 2011.Emerg. Infect. Dis.191314–1316. 10.3201/eid1908.121824
61
Lushchak V. I. (2011). Environmentally induced oxidative stress in aquatic animals.Aquat. Toxicol.10113–30. 10.1016/j.aquatox.2010.10.006
62
Luzio J. P. Pryor P. R. Bright N. A. (2007). Lysosomes: fusion and function.Nat. Rev. Mol. Cell Biol.8622–632. 10.1038/nrm2217
63
Magara G. Elia A. C. Syberg K. Khan F. R. (2018). Single contaminant and combined exposures of polyethylene microplastics and fluoranthene: accumulation and oxidative stress response in the blue mussel, Mytilus edulis.J. Toxicol. Environ. Health Part A81761–773. 10.1080/15287394.2018.1488639
64
Mao F. Mu H. Wong N.-K. Liu K. Song J. Qiu J. et al (2020). Hemocyte phagosomal proteome is dynamically shaped by cytoskeleton remodeling and interorganellar communication with endoplasmic reticulum during phagocytosis in a marine invertebrate, Crassostrea gigas.Sci. Rep.10:6577. 10.1038/s41598-020-63676-3
65
Maynes J. T. Bateman K. S. Cherney M. M. Das A. K. Luu H. A. Holmes C. F. B. et al (2001). Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.J. Biol. Chem.27644078–44082. 10.1074/jbc.M107656200
66
Mello D. F. de OliveiraProença L. A. Barracco M. A. (2010). Comparative study of various immune parameters in three bivalve species during a natural bloom of dinophysis acuminata in santa catarina island, brazil.Toxins21166–1178. 10.3390/toxins2051166
67
Messner D. J. Ao P. Jagdale A. B. Boynton A. L. (2001). Abbreviated cell cycle progression induced by the serine/threonine protein phosphatase inhibitor okadaic acid at concentrations that promote neoplastic transformation.Carcinogenesis221163–1172. 10.1093/carcin/22.8.1163
68
Mitra S. K. Hanson D. A. Schlaepfer D. D. (2005). Focal adhesion kinase: in command and control of cell motility.Nat. Rev. Mol. Cell Biol.656–68. 10.1038/nrm1549
69
Mizushima N. (2007). Autophagy: process and function.Genes Dev.212861–2873. 10.1101/gad.1599207
70
Muller-Taubenberger A. Lupas A. N. Li H. W. Ecke M. Simmeth E. Gerisch G. (2001). Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis.EMBO J.206772–6782. 10.1093/emboj/20.23.6772
71
Mydlarz L. D. Jones L. E. Harvell C. D. (2006). Innate immunity environmental drivers and disease ecology of marine and freshwater invertebrates.Annu. Rev. Ecol. Evol. Syst.37251–288. 10.1146/annurev.ecolsys.37.091305.110103
72
Nguyen M. T. Somogyvari M. Soti C. (2018). HSP90 stabilizes SIRT1 orthologs in mammalian cells and C-elegans.Int. J. Mol. Sci.19:3661. 10.3390/ijms19113661
73
Nie X. P. Wang Y. Zhao H. J. Guo M. H. Liu Y. C. Xing M. W. (2020). As3+ or/and Cu2+ exposure triggers oxidative stress imbalance, induces inflammatory response and apoptosis in chicken brain.Ecotoxicol. Environ. Safe.203:8. 10.1016/j.ecoenv.2020.110993
74
Ohkawa H. Ohishi N. Yagi K. (1979). Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction.Anal. Biochem.95351–358. 10.1016/0003-2697(79)90738-3
75
Opsahl J. A. Ljostveit S. Solstad T. Risa K. Roepstorff P. Fladmark K. E. (2013). Identification of dynamic changes in proteins associated with the cellular cytoskeleton after exposure to okadaic acid.Mar. Drugs111763–1782. 10.3390/md11061763
76
Prado-Alvarez M. Florez-Barros F. Mendez J. Fernandez-Tajes J. (2013). Effect of okadaic acid on carpet shell clam (Ruditapes decussatus) haemocytes by in vitro exposure and harmful algal bloom simulation assays.Cell Biol. Toxicol.29189–197. 10.1007/s10565-013-9246-1
77
Prego-Faraldo M. V. Vieira L. R. Eirin-Lopez J. M. Méndez J. Guilhermino L. (2017). Transcriptional and biochemical analysis of antioxidant enzymes in the mussel Mytilus galloprovincialis during experimental exposures to the toxic dinoflagellate Prorocentrum lima.Mar. Environ. Res.129304–315. 10.1016/j.marenvres.2017.06.009
78
Regoli F. Principato G. (1995). Glutathione, glutathione-dependent and antioxidant enzymes in mussel, Mytilus galloprovincialis, exposed to metals under field and laboratory conditions: implications for the use of biochemical biomarkers.Aquat. Toxicol.31143–164. 10.1016/0166-445X(94)00064-W
79
Rewitz K. F. Styrishave B. Lobner-Olesen A. Andersen O. (2006). Marine invertebrate cytochrome P450: emerging insights from vertebrate and insect analogies.Comp. Biochem. Physiol. C Toxicol. Pharmacol.143363–381. 10.1016/j.cbpc.2006.04.001
80
Romero-Geraldo R. D. Hernandez-Saavedra N. Y. (2014). Stress gene expression in Crassostrea gigas (Thunberg, 1793) in response to experimental exposure to the toxic dinoflagellate Prorocentrum lima (Ehrenberg) Dodge, 1975.Aquacult. Res.451512–1522. 10.1111/are.12100
81
Schliwa M. Vanblerkom J. (1981). Structural interaction of cytoskeletal components.J. Cell Biol.90222–235. 10.1083/jcb.90.1.222
82
Schneeberger E. E. Lynch R. D. (1992). Structure, function, and regulation of cellular tight junctions.Am. J. Physiol.262L647–L661. 10.1152/ajplung.1992.262.6.L647
83
Shilov I. V. Seymourt S. L. Patel A. A. Loboda A. Tang W. H. Keating S. P. et al (2007). The paragon algorithm, a next generation search engine that uses sequence temperature values sequence temperature values and feature probabilities to identify peptides from tandem mass spectra.Mol. Cell Proteomics61638–1655. 10.1074/mcp.T600050-MCP200
84
Slavov N. Budnik B. A. Schwab D. Airoldi E. M. van Oudenaarden A. (2014). Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis.Cell Rep.7705–714. 10.1016/j.celrep.2014.03.057
85
Smaal A. Van Stralen M. Schuiling E. (2001). The interaction between shellfish culture and ecosystem processes.Can. J. Fish. Aquat. Sci.58991–1002. 10.1139/f01-026
86
Snyder M. J. (2000). Cytochrome P450 enzymes in aquatic invertebrates: recent advances and future directions.Aquat. Toxicol.48529–547. 10.1016/s0166-445x(00)00085-0
87
Sonenberg N. Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets.Cell136731–745. 10.1016/j.cell.2009.01.042
88
Song L. S. Wang L. L. Zhang H. Wang M. Q. (2015). The immune system and its modulation mechanism in scallop.Fish Shellfish Immunol.4665–78. 10.1016/j.fsi.2015.03.013
89
Takeichi M. (1995). Morphogenetic roles of classic cadherins.Curr. Opin. Cell Biol.7619–627. 10.1016/0955-0674(95)80102-2
90
Teng J. Zhao J. Zhu X. Shan E. Wang Q. (2021). Oxidative stress biomarkers, physiological responses and proteomic profiling in oyster (Crassostrea gigas) exposed to microplastics with irregular-shaped PE and PET microplastic.Sci. Total Environ.786:147425. 10.1016/j.scitotenv.2021.147425
91
Tomanek L. (2010). Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs.J. Exp. Biol.213971–979. 10.1242/jeb.038034
92
Tomanek L. Zuzow M. J. (2010). The proteomic response of the mussel congeners Mytilus galloprovincialis and M. trossulus to acute heat stress: implications for thermal tolerance limits and metabolic costs of thermal stress.J. Exp. Biol.2133559–3574. 10.1242/jeb.041228
93
USEPA (2002). Methods for Measuring the Acute Toxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms.EPA-821-R-02-012, 5th Edn. Washington, DC: US Environmental Protection Agency.
94
Valdiglesias V. Garcia-Leston J. Pasaro E. Mendez J. Laffon B. (2010a). Cell cycle alterations and apoptosis assessment in SHSY5Y human neuroblastoma cells exposed to okadaic acid.Toxicol. Lett.196S345–S345. 10.1016/j.toxlet.2010.03.1090
95
Valdiglesias V. Mendez J. Pasaro E. Cemeli E. Anderson D. Laffon B. (2010b). Assessment of okadaic acid effects on cytotoxicity, DNA damage and DNA repair in human cells.Mutat. Res.68974–79. 10.1016/j.mrfmmm.2010.05.004
96
Valdiglesias V. Veronica Prego-Faraldo M. Pasaro E. Mendez J. Laffon B. (2013). Okadaic acid: more than a diarrheic toxin.Mar. Drugs114328–4349. 10.3390/md11114328
97
Valko M. Rhodes C. J. Moncol J. Izakovic M. Mazur M. (2006). Free radicals, metals and antioxidants in oxidative stress-induced cancer.Chem. Biol. Interact.1601–40. 10.1016/j.cbi.2005.12.009
98
Van Dolah F. M. Ramsdell J. S. (2001). Review and assessment of in vitro detection methods for algal toxins.J. AOAC Int.841617–1625. 10.1093/jaoac/84.5.1617
99
Vieira A. C. Rubiolo J. A. Lopez-Alonso H. Cifuentes J. M. Alfonso A. Bermudez R. et al (2013). Oral toxicity of okadaic acid in mice: study of lethality, organ damage, distribution and effects on detoxifying gene expression.Toxins52093–2108. 10.3390/toxins5112093
100
Vieira O. V. Botelho R. J. Grinstein S. (2002). Phagosome maturation: aging gracefully.Biochem. J.366689–704. 10.1042/bj20020691
101
Wimberly B. T. Brodersen D. E. Clemons W. M. Morgan-Warren R. J. Carter A. P. Vonrhein C. et al (2000). Structure of the 30S ribosomal subunit.Nature407327–339. 10.1038/35030006
102
Xie Z. P. Klionsky D. J. (2007). Autophagosome formation: core machinery and adaptations.Nat. Cell Biol.91102–1109. 10.1038/ncb1007-1102
103
Xu C. J. Li C. Y. T. Kong A. N. T. (2005). Induction of phase I, II and III drug metabolism/transport by xenobiotics.Arch. Pharm. Res.28249–268. 10.1007/bf02977789
104
Yasumoto T. Oshima Y. Yamaguchi M. (1978). Ocurrence of a new type of shellfish poisoning in tohoku district.Bull. Jpn. Soc. Sci. Fish.441249–1255. 10.2331/suisan.44.1249
105
Young N. Robin C. Kwiatkowska R. Beck C. Mellon D. Edwards P. et al (2019). Outbreak of diarrhetic shellfish poisoning associated with consumption of mussels, United Kingdom, May to June 2019.Eurosurveillance242–6. 10.2807/1560-7917.ES.2019.24.35.1900513
106
Zhang C. N. Lu K. L. Wang J. H. Qian Q. Yuan X. Y. Pu C. C. (2020). Molecular cloning, expression HSP70 and its response to bacterial challenge and heat stress in Microptenus salmoides.Fish Physiol. Biochem.462389–2402. 10.1007/s10695-020-00883-9
Summary
Keywords
okadaic acid, Chlamys farreri , oxidative stress, detoxification genes, iTRAQ
Citation
Wang X, Wang D, Zhang T, Zhang Q and Zhao J (2021) iTRAQ-Based Quantitative Proteomic Analysis Reveals Toxicity Mechanisms in Chlamys farreri Exposed to Okadaic Acid. Front. Mar. Sci. 8:792050. doi: 10.3389/fmars.2021.792050
Received
09 October 2021
Accepted
17 November 2021
Published
08 December 2021
Volume
8 - 2021
Edited by
Youji Wang, Shanghai Ocean University, China
Reviewed by
Wenguang Liu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), China; Jingjing Miao, Ocean University of China, China
Updates
Copyright
© 2021 Wang, Wang, Zhang, Zhang and Zhao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianqian Zhang, qqzhang@yic.ac.cnJianmin Zhao, jmzhao@yic.ac.cn
This article was submitted to Aquatic Physiology, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.