Abstract
The Beibu Gulf is considered as one of China’s four major fishing grounds, although the substantial overexploitation of fisheries has led to the collapse of many fish stocks, and to changes to spawning grounds in recent decades. Classifying fish eggs is an important way to monitor the recruitment process and identify the spawning sites of fish. However, the lack of a basis for morphological identification and difficulties in correctly identifying fish eggs based on morphological characteristics has limited scientific studies. In the present study, we identified fish eggs using molecular detection of cytochrome c oxidase subunit I and cytochrome b fragments. Ichthyoplankton surveys were conducted in the spring and late autumn–winter of 2020 in the eastern Beibu Gulf. Among the DNA extracted from the 873 chosen fish eggs, we successfully obtained 541 high-quality cytochrome c oxidase subunit I sequences and 41 high-quality cytochrome b sequences. We successfully identified 212 fish eggs (36.4%) from 32 species; 291 eggs (50.0%) showed ambiguous species delimitation, and 79 eggs (13.6%) could not be identified. Among the identified species, we found 25 species in spring and 25 species in late autumn–winter, out of which 18 species occurred in both seasons. We also obtained high resolution photographs of fish eggs at the species level for further morphological analysis and identification. The present study confirms the efficacy of using molecular methods to identify fish species from eggs and provides valuable information for protecting the spawning ground of economically valuable fish and for managing fishery resources.
Introduction
Eggs represent the earliest phase in the life cycle of fish. Because eggs cannot move unaided, they can provide valuable information about the reproductive biology, preferred spawning times, spawning sites, and recruitment success rates within fish populations (Baumgartner et al., 2004; Cao et al., 2007; Hou et al., 2021a; Takeuchi et al., 2021). Information on the composition and spatial and temporal distribution of fish eggs is vital for determining closed fishing zones, fishing moratoriums, fishery resource management strategies, marine protection areas, and even environmental evaluation of sea/fresh water-related construction and engineering (Rakocinski et al., 1996; Cao et al., 2007; Oliveira and Ferreira, 2008; Ahern et al., 2018). Although the importance of the fish egg pool has been long recognized, species-level information on the occurrence of fish eggs and their distributional patterns is lacking because of the difficulties in identifying fish egg stages (Baumgartner et al., 2004; Bui et al., 2011; Burrows et al., 2018; Hou et al., 2021a).
The paucity of species-level information on fish eggs occurs mainly because of the complex dynamic changes in the ontogeny of fish eggs (Zhang et al., 1985; Cao et al., 2007; Ikeda et al., 2014). There are nearly 30 stages of embryonic development, from fertilized eggs to newly hatched larvae (Zhang et al., 2006; Cao et al., 2007). In the early egg stages before early gastrula, there are strong morphological similarities among different species, while after the muscle burl stages potential morphological classification characteristics may be detected at the taxa level (Cao et al., 2007; Ikeda et al., 2014; Wan and Zhang, 2016). It is not easy to collect all the morphological ontogenetic stages of fish eggs and larvae within a limited time period through field surveys, especially for oceanic fish which are difficult to hatch and rear to juvenile/adult stages to confirm the species (Zhang et al., 1985; Ikeda et al., 2014; Wan and Zhang, 2016). Thus, the marked ontogenetic changes that occur within and among the fish species and the lack of morphological data make it difficult to precisely identify fish eggs at the species level.
In the last decade, molecular identification methods have been widely applied to identify fish eggs, including use of mitochondrial DNA fragments such as cytochrome c oxidase subunit I (COI) (Valdez-Moreno et al., 2010; Lewis et al., 2016; Leyva-Cruz et al., 2016; Ahern et al., 2018; Kerr et al., 2020; Chen et al., 2021), cytochrome b (cytb) (Liu et al., 2018), mtDNA PCR restriction fragment length polymorphism (RFLP) (Karaiskou et al., 2007; Stéphanie et al., 2010; Bui et al., 2011; Fitzcharles, 2012; Lelievre et al., 2012), and mtDNA control region (D-loop) (Shao et al., 2002). The DNA barcode method (COI) has been widely applied to define fish species, and facilitate the identification of fish eggs and larvae. For instance, Leyva-Cruz et al. (2016) identified fish eggs from 42 taxa collected from the waters surrounding Banco Chinchorro in the Mexican Caribbean. Burrows et al. (2018) applied a DNA barcode to identify fish eggs in the Gulf of Mexico, and allocated 709 eggs to 62 species. Kerr et al. (2020) successfully identified 564 fish eggs in 89 taxa collected from northwestern Cuba and across the Florida Straits using DNA barcodes. These studies demonstrate that molecular techniques can be used effectively to identify fish eggs, and they illustrate the potential for searching for spawning sites and relevant studies.
The Beibu Gulf (17–21.75°N, 105.67–110.17° E), situated in the northwestern South China Sea (SCS), covers an area of 12.88 × 103 km2 with an average depth of 38 m. It has a number of habitats, including mangrove forests, coral reefs, and numerous estuaries (such as the Red River and Nanliu River). This heterogeneity provides suitable habitats for a high diversity of marine organisms. More than 600 fish species have been reported in the Beibu Gulf, with 60 commercially important fish taxa supporting a substantial fishery (Sun et al., 2010; Wang et al., 2011; Sun and Chen, 2013). The Beibu Gulf is one of China’s four major fishing grounds, playing an important role in food security, the economy, and employment (Qiu et al., 2008). However, the marine system in the Beibu Gulf has been heavily disturbed and degraded, especially in the last few decades, owing to global climate change and various human activities, including overexploitation, illegal fishing, coastal mangrove damage, and habitat loss. Consequently, the fish diversity in the Beibu Gulf has been threatened, the fish trophic levels and assemblages have changed significantly, and fishery resources and catch rates have declined substantially (Sun and Lin, 2004; Qiu et al., 2008; Wang et al., 2010; Zhang et al., 2020).
Ranked as one of the most important fishery grounds in China and SCS, the Beibu Gulf is also an important spawning and nursing ground for numerous fish species. Studies on fish eggs and larvae can be traced to 1959–1962 and 1997–1999, although they did not analyze the fish egg and larvae separately (Jia et al., 2004; Zhou et al., 2008, 2011). Jia et al. (2004) and Zhou et al. (2008) described the fish eggs and larvae abundance and/or composition in partial season. Zhou et al. (2011) morphologically identified 153 taxa of fish eggs and larvae in the eastern Beibu Gulf, although only 11 taxa were identified at the species level from fish eggs. Hou et al. (2021b) used molecular identification of 53 species of fish larvae fixed in formalin from the Beibu Gulf. However, owing to the difficulties of identifying fish egg stages, information on most spawning sites of fish in the Beibu Gulf on the basis of ichthyoplankton surveys is still limited.
Given the present conditions of fishery resources in the Beibu Gulf, there is an urgent need to document the current spawning grounds of fish species, from a perspective of both fishery management and fish biodiversity protection perspectives. We undertook ichthyoplankton surveys in the spring and late autumn–winter of 2020 in the eastern Beibu Gulf. In the present study, our aims were to: (i) identify the pelagic eggs collected in these surveys using molecular techniques; (ii) determine the diversity of fish eggs during the study period; and (iii) evaluate the effectiveness of the molecular method in identifying fish eggs in the Beibu Gulf. Our findings will contribute to improved management and conservation of fish species and their spawning grounds in the Beibu Gulf.
Materials and Methods
Study Area and Sample Collection
Two surveys were conducted in the area of the eastern Beibu Gulf (17.75–21.25°N, 107.25–109.75°E) in the spring (April) and late autumn–winter (November–December) of 2020 (Figure 1). Ichthyoplankton samples were collected by zooplankton nets (0.8 m diameter, 2.7 m long, 505 μm mesh, with a cod-end container mesh of 400 μm). Four zooplankton net deployments were made at each survey station: two simultaneous horizontal tows at fixed depth (10 min at 1.5–2.2 knots), and two vertical hauls (∼ sea bed to the surface, hauled at 1.5 m/s). Specimens from one horizontal and one vertical tow were quickly preserved in 75% ethanol–sea water solution, and then stored in cold storage (<0°C). The other specimens were preserved in 4% formalin solution. The horizontal and vertical nets were fitted with General Oceanic flowmeters for estimating filtered water volume.
FIGURE 1
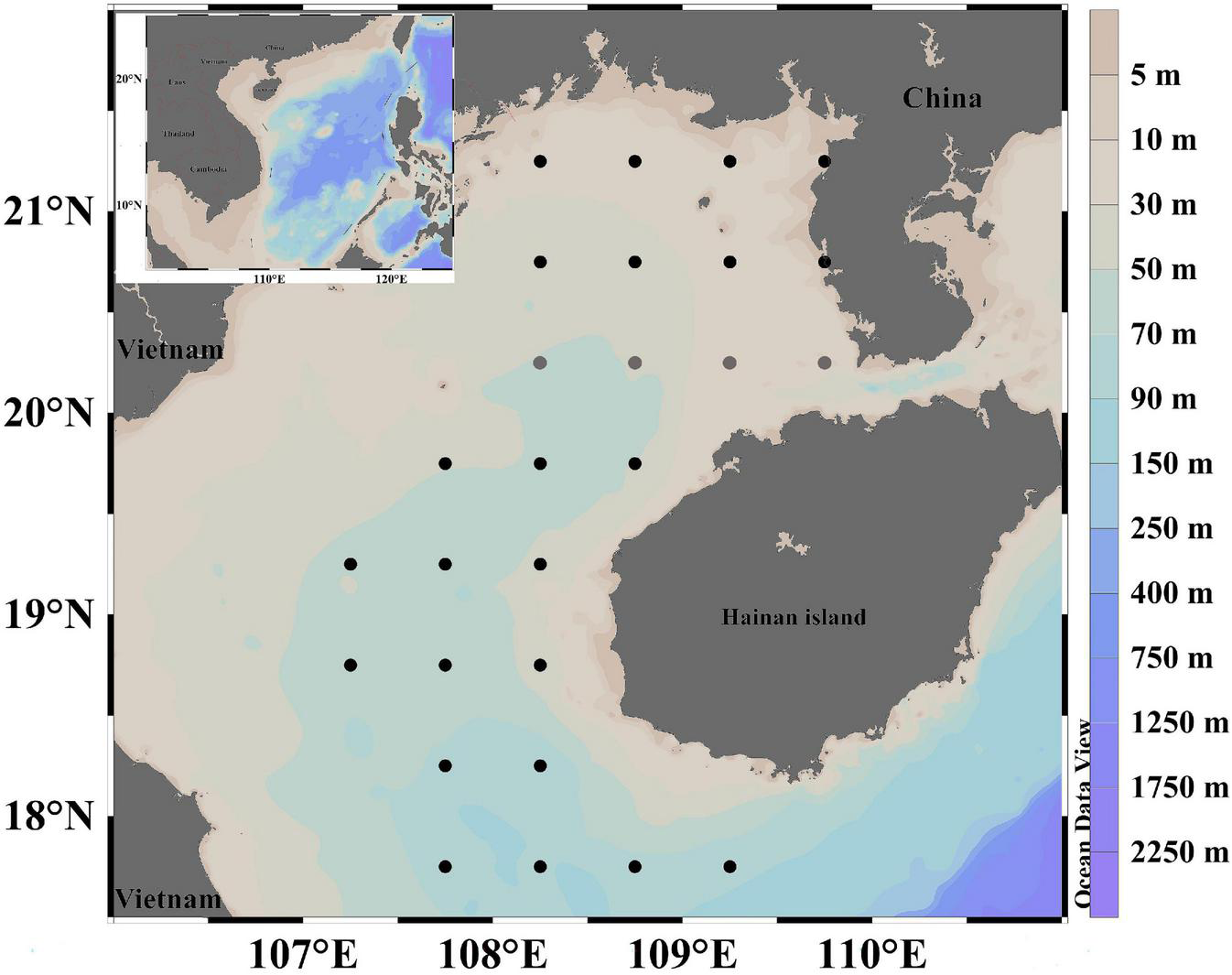
Sampling stations of fish eggs in the eastern Beibu Gulf. The black dots indicate the stations where the spring and late autumn-winter survey was conducted, the gray dots indicate the stations where only the spring survey was conducted.
Photography and Molecular Experiment
In the laboratory, all fish eggs from each station were examined using a stereomicroscope. Because we could not amplify and sequence all the collected eggs, we randomly selected partial samples for molecular identification. Up to 30 eggs were randomly selected at a survey station, if there were more than 30 eggs, and photographed. If there were <30, all the eggs were selected, and 15 eggs were selected each time; this was done twice in each station to give a total of 30 eggs. We also selected eggs with different morphological features to identify more fish species or taxa.
Each egg was first numbered, rehydrated in hydrogen peroxide for 5–8 min for cleaning, and then photographed using a Zeiss microscope (Axioplan 2 imaging E; Figure 2). The primer treating processes and DNA extraction followed Hou et al. (2020). Total genomic DNA was extracted using an Axygen DNA Extraction Kit (Axygen, Shanghai, China). A partial fragment of the 5’-end of COI sequences (∼648 bp) was firstly amplified, using the universal primers FishF1 and FishR1 (Ward et al., 2005). If COI sequences were not obtained, then fragments of cytb were amplified, using the primers L14724 and H15915 (Xiao et al., 2001). The polymerase chain reaction (PCR) conditions of COI were as follows: 94°C for 3 min, 35 cycles at 94°C for 30 s, 51°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 10 min (Hou et al., 2020). The PCR conditions of cytb were as follows: 96°C for 5 min; 35 cycles at 96°C for 30 s, 58°C for 30 s, 72°C for 1.5 min; and a final extension at 72°C for 5 min. The amplification products of PCR reactions were purified using 1% low-melting agarose by electrophoresis, and sequenced bidirectionally on an ABI 3730 XL DNA system, following the manufacturer’s protocols.
FIGURE 2
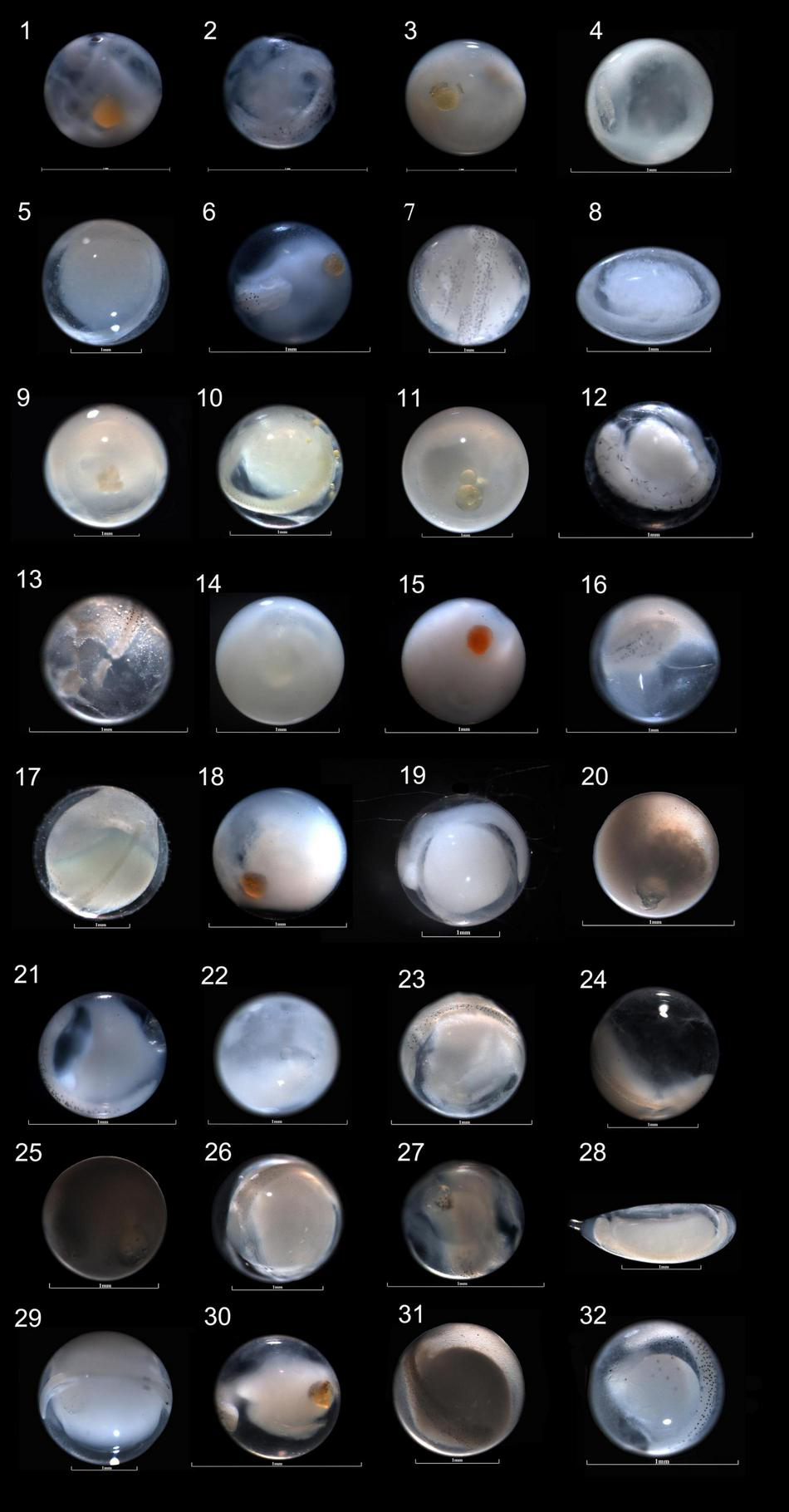
Photographs of the egg specimens that were identified to species level by COI and cytb sequences in the eastern Beibu Gulf. (1) Acropoma japonicum, GDYH16088, 0.923 mm; (2) Atropus Atropos, GDYH16147, 0.719 mm; (3) Branchiostegus albus, GDYH15795, 1.106 mm; (4) Carangoides malabaricus, GDYH13119, 0.790 mm; (5) Chirocentrus dorab, GDYH13103, 1.784 mm; (6) Chrysochir aureus, GDYH16116, 0.766 mm; (7) Coryphaena hippurus, GDYH13118, 1.641 mm; (8) Encrasicholi heteroloba, GDYH12984, 1.178 × 0.792 mm; (9) Fistularia petimba, GDYH14150, 1.912 mm; (10) Hilsa kelee, GDYH13042, 1.234 mm; (11) Ilisha melastoma, GDYH13028, 1.401 mm; (12) Leiognathus ruconius, GDYH15838, 0.707 mm; (13) Megalaspis cordyla, GDH12789, 1.085 mm; (14) Mene maculate, GDYH13073, 1.085 mm; (15) Moolgarda perusii, GDYH16099, 0.833 mm; (16) Nemipterus marginatus, GDYH12936, 0.731 mm; (17) Oxyporhamphus micropterus, GDYH13048, 2.407 mm; (18) Parapercis lutevittata, GDYH15760, 0.976 mm; (19) Parexocoetus mento, GDYH13132, 1.719 mm; (20) Penhia anea, GDYH12757, 0.833 mm; (21) Penhia microcephalus, GDYH16123, 0.867 mm; (22) Penhia pawak, GDYH12932, 0.859 mm; (23) Pomadasys maculatus, GDYH12791, 0.897 mm; (24) Saurida elongate, GDYH15780, 1.430 mm; (25) Scomberomorus commerson, GDYH12756, 1.193 mm; (26) Seriolina nigrofasciata, GDYH12880, 1.393 mm; (27) Sillago sihama, GDYH15562, 0.782 mm; (28) Stolephorus waitei, GDYH12913, 1.950 × 0.736 mm; (29) Tentoriceps cristatus, GDYH13107, 2.006 mm; (30) Terapon jarbua, GDYH15866, 0.726 mm; (31) Trachicephalus uranoscopus, GDYH12765, 1.583 mm; and (32) Scomberoides tol, GDYH13111, 0.899 mm. Among them, egg specimens of serial number 1–31 were identified by COI sequences, and 32 was identified by cytb sequence.
Data Analyses
The tracer files were initially checked and sequences assembled using SEQMAN in Lasergene v 7.0 (DNASTAR Inc., Madison, WI, United States). High-quality sequences were aligned and manually edited using MEGA v6.0 (Tamura et al., 2013). Fish egg identification was performed in the Barcode of Life Data System (BOLD, V4). We retained reference sequences of both the best and second-best interspecific matches in BOLD and documented the percentages of sequence matches. Three criteria similar to those used in Hubert et al. (2015) were adopted in our study: (i) sequences of the COI fragment with >98% similarity to the best sequence match and <98% similarity to the nearest neighbor species, exhibiting >2% genetic divergence between sequences and the nearest neighbor species were unambiguously tagged with that species name. If the top 99 matches were blasted to single species within the above threshold, the sequences were delimited to the matches species (Case I). (ii) If the genetic divergence was more than 98% similarity, or less than the 2% threshold to the best matched species and nearest neighbor species, the two matched species were in the same genus, and the sequences were delimited to genus level; if not in the same genus, and the sequences were identified as species uncertain (Case II). (iii) Sequences with matches <98% were deemed unidentified (Case III).
The sequences of the cytb fragment were blasted in the GenBank nucleotide database using the Basic Local Alignment Search Tool (BLAST).1 Sequences with >99% similarity were tagged with that species name. Genetic distance was calculated in MEGA6 based on the Kimura 2-parameter model (K2P, Kimura, 1980). A neighbor-joining (NJ) tree based on the K2P model was reconstructed to illustrate lineage diversity via the phylogenetic topology. The NJ tree was constructed with 1,000 bootstrap replicates using MEGA v6.0.
To accumulate description morphological characteristics of fish eggs in the waters of the eastern Beibu Gulf, the egg diameter and oil diameter were measured (to the nearest 0.001 mm). To assess whether the sampling effort was sufficient to describe the species richness of fish eggs, species accumulation curves were created for each survey using the “random” method as an accumulator function to determine the means and standard deviations of species accumulation curves from random subsampling of the data without replacement (Gotelli and Colwell, 2001). The species accumulation curves were fitted in the “vegan” package implemented in R version 4.0.5 (Oksanen et al., 2020).
Results
Sequence Information
In all, 13,984 individual alcohol-preserved fish eggs were collected in spring, including 11,239 individual eggs in horizontal trawls, and 2,745 individuals in vertical trawls. A further 527 alcohol-preserved fish eggs were collected and preserved in late autumn–winter, including 392 individuals eggs in horizontal trawls, and 135 in vertical trawls. The mean total abundance in spring was 191.49 ind./100 m3 in horizontal tows and 1067.55 ind./100 m3 in vertical hauls. The mean total abundance in late autumn-winter was 8.75 ind./100 m3 in horizontal tows and 3.08 ind./100 m3 in vertical hauls (Supplementary Figure 1). A total of 873 fish eggs were chosen for DNA extraction and molecular identification. The COI fragment in the fish eggs was first sequenced. If it failed to be amplified after three attempts, the cytb fragment was then sequenced. Owing to the delays and difficulties in the laboratory experiments because of the coronavirus disease outbreak, 291 eggs (33.33%) failed to yield high-quality sequences. In summary, we finally obtained high-quality COI sequences from 541 fish eggs and 41 cytb sequences (66.67%).
Molecular Identification of Fish Eggs
Using the 2% genetic divergence and 98% similarity thresholds to represent species boundaries in the BOLD platform, approximately 89.5% of the COI sequences (484/541) had a match of 88–100%. Among the 484 COI sequences, 193 sequences (∼35.7%) were successfully categorized as Case I, and were assigned to 31 species. A total of 291 (∼53.8%) sequences were categorized as Case II, and 57 sequences (∼10.5%) were categorized as Case III (Figure 3 and Supplementary Table 1). Using an identification threshold of more than 99%, 19 cytb sequences were successfully assigned to five species, i.e., Ilisha melastoma (Bloch and Schneider, 1801), Terapon jarbua (Forsskål, 1775), Fistularia petimba (Lacepède, 1803), Scomberomorus commerson (Lacepède, 1800), and Scomberoides tol (Cuvier, 1832), and 22 sequences could not be identified at the species level (Supplementary Table 2). In total, 212 fish eggs (36.4%) were successfully identified to 32 species (Table 1), 291 eggs (50.0%) had ambiguous species delimitation, and 79 eggs (13.6%) could not be identified using the BOLD and NCBI platforms.
FIGURE 3
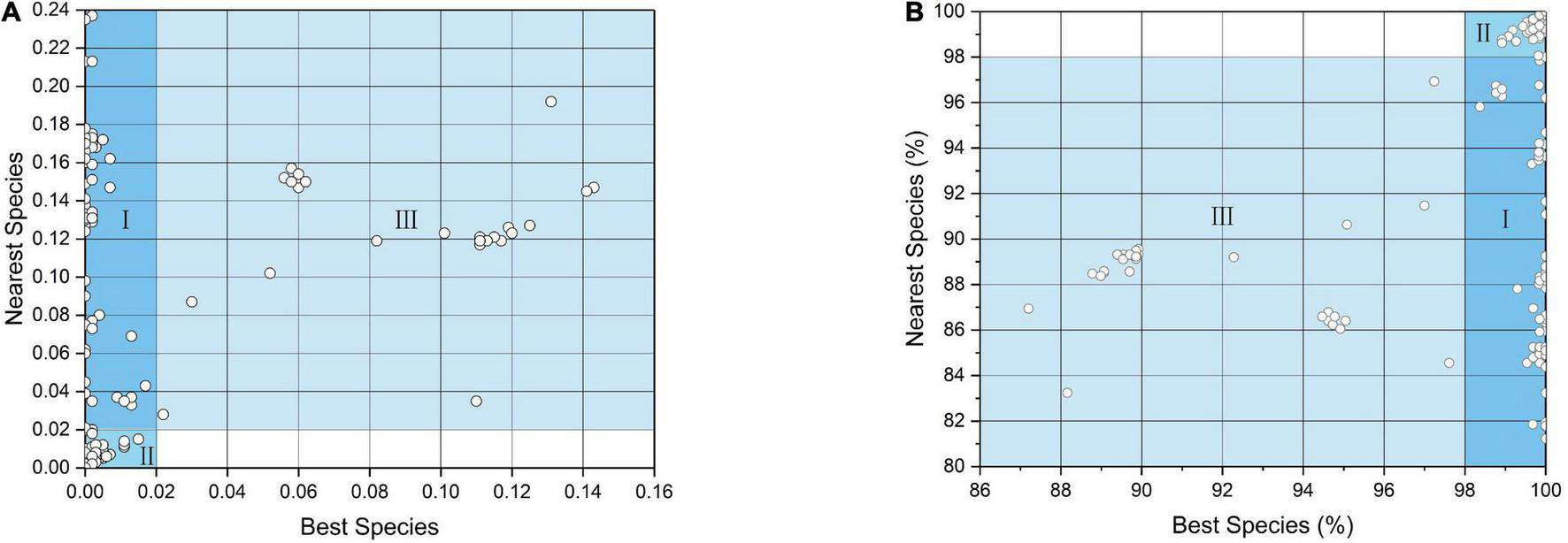
The best match compared with the nearest neighbor for each specimen. (A) Genetic distance, (B) similarity percentage. Case I, delimited to species level; Case II, delimited to genus or uncertain; Case III, unidentified.
TABLE 1
| Family | Species | Common name | Number of specimens identified by COI/Cytb | Egg diameter/mm | Oil diameter/mm | Spring | Late autumn-winter | Gene used | Habitat |
| Acropomatidae | Acropoma japonicum | Glowbelly | 5 | 0.923–0.950 | 0.244–0.251 | + | + | COI | Bathypelagic |
| Carangidae | Megalaspis cordyla | Torpedo scad | 4 | 0.760–1.085 | 0.23 | + | COI | Reef–associated | |
| Carangidae | Atropus atropos | Cleftbelly trevally | 4 | 0.719–0.748 | 0.166–0.209 | + | + | COI | Pelagic–neritic |
| Carangidae | Seriolina nigrofasciata | Blackbanded trevally | 2 | 1.393–1.393 | + | COI | Reef–associated | ||
| Carangidae | Scomberoides tol | Needlescaled queenfish | 2 | 0.853–0.899 | 0.156 | + | Cytb | Reef–associated | |
| Carangidae | Carangoides malabaricus | Malabar trevally | 2 | 0.790–0.869 | + | + | COI | Reef–associated | |
| Chirocentridae | Chirocentrus dorab | Dorab wolf–herring | 11 | 1.656–2.039 | + | + | COI | Reef–associated | |
| Clupeidae | Hilsa kelee | Kelee shad | 5 | 1.101–1.242 | 0.041–0.122* | + | COI | Pelagic–neritic | |
| Coryphaenidae | Coryphaena hippurus | Common dolphinfish | 12 | 1.446–1.641 | 0.260–0.463 | + | + | COI | Pelagic–neritic |
| Engraulidae | Encrasicholina heteroloba | Shorthead anchovy | 22 | 1.073–1.408 ×0.692–0.842 | + | + | COI | Reef–associated | |
| Engraulidae | Stolephorus waitei | Spotty–face anchovy | 35 | 1.727–2.158 ×0.772–1.047 | 0.053–0.177 | + | + | COI | Pelagic–neritic |
| Exocoetidae | Parexocoetus mento | African sailfin flyingfish | 6 | 1.685–1.788 | + | + | COI | pelagic–neritic | |
| Fistulariidae | Fistularia petimba | Red cornetfish | 2/3 | 1.759–1.976 | 0.513 | + | + | COI/Cytb | Reef–associated |
| Haemulidae | Pomadasys maculatus | Saddle grunt | 2 | 0.848–0.897 | + | + | COI | Reef–associated | |
| Hemiramphidae | Oxyporhamphus micropterus | Bigwing halfbeak | 18 | 2.112–2.694 | + | + | COI | Pelagic–oceanic | |
| Latilidae | Branchiostegus albus | Null | 3 | 1.059–1.106 | 0.221–0.257 | + | + | COI | Benthopelagic |
| Leiognathidae | Leiognathus ruconius | Deep pugnose ponyfish | 4 | 0.660–0.732 | + | + | COI | Demersal | |
| Menidae | Mene maculata | Moonfish | 1 | 1.085 | + | COI | Reef–associated | ||
| Mugilidae | Moolgarda perusii | Longfinned mullet | 3 | 0.799–0.919 | 0.198–0.200 | + | COI | Reef–associated | |
| Nemipteridae | Nemipterus marginatus | Red filament threadfin bream | 1 | 0.731 | + | COI | Demersal | ||
| Pinguipedidae | Parapercis lutevittata | Yellow–striped sandperch | 1 | 0.976 | 0.223 | + | COI | Demersal | |
| Pristigasteridae | Ilisha melastoma | Indian ilisha | 10/1 | 1.125–1.443 | 0.227–0.348 | + | + | COI/Cytb | Pelagic–neritic |
| Sciaenidae | Chrysochir aureus | Reeve’s croaker | 10 | 0.766–0.846 | 0.129–0.214 | + | COI | benthopelagic | |
| Sciaenidae | Pennahia anea | Donkey croaker | 1 | 0.833 | 0.229 | + | COI | Demersal | |
| Sciaenidae | Pennahia macrocephalus | Big–head pennah croaker | 3 | 0.848–0.897 | 0.206 | + | COI | Demersal | |
| Sciaenidae | Pennahia pawak | Pawak croaker | 1 | 0.859 | 0.099 | + | COI | Benthopelagic | |
| Scombridae | Scomberomorus commerson | Narrow–barred Spanish mackerel | 1/12 | 1.116–1.257 | 0.238–0.450 | + | COI/Cytb | Pelagic–neritic | |
| Sillaginidae | Sillago sihama | Silver sillago | 4 | 0.709–0.802 | 0.138–0.164 | + | + | COI | Reef–associated |
| Synanceiidae | Trachicephalus uranoscopus | Stargazing stonefish | 4 | 1.499–1.590 | + | COI | Demersal | ||
| Synodontidae | Saurida elongata | Slender lizardfish | 4 | 1.427–1.511 | + | + | COI | Demersal | |
| Terapontidae | Terapon jarbua | Jarbua terapon | 9/1 | 0.683–0.775 | 0.143–0.264 | + | + | COI/Cytb | Demersal |
| Trichiuridae | Tentoriceps cristatus | Crested hairtail | 6 | 1.764–2.006 | + | + | COI | Benthopelagic |
The fish eggs identified in species level using molecular methods.
Information of egg diameter, oil diameter, the occurrence season and habitat of adult are also shown. * indicate numerous oil globules occur in each fish egg.
The 32 species that were identified belonged to 24 families. The most abundant families were Carangidae (5 species, 14 individuals), Sciaenidae (4 species, 15 individuals), and Engraulidae (2 species, 57 individuals). Among these, the eggs of 15 economically important species were identified, i.e., Sillago sihama (Forsskål, 1775), Pomadasys maculatus (Bloch, 1793), Megalaspis cordyla (Linnaeus, 1758), Chirocentrus dorab (Forsskål, 1775), Chrysochir aureus (Richardson, 1846), Pennahia pawak (Richardson, 1846), Pennahia macrocephalus (Tang, 1937), Pennahia anea (Bloch, 1793), Branchiostegus albus (Dooley, 1978), Mene maculate (Bloch and Schneider, 1801), T. jarbua, Saurida elongata (Temminck and Schlegel, 1846), Coryphaena hippurus (Linnaeus, 1758), S. commerson, and F. petimba.
The NJ tree of COI sequences suggested that the identified species of fish eggs based on the BOLD and GenBank databases generated 80 independent lineages (Figure 4). The NJ tree of cytb sequences generated nine independent lineages (Supplementary Figure 2), suggesting that at least 81 distinct species/taxa were collected and occurred in the study period. In spring, the fish eggs of 25 fish taxa were successfully identified at the species level, and the same number were identified in late autumn–winter. Among them, the eggs of 18 fish species occurred in both the spring and late autumn–winter surveys, and seven species occurred only in spring or in late autumn-winter (Table 1). Using the habitat data for adult fish species cited in Fishbase,2 11, 8, 7, 4, 1, and 1 species were reef-associated, demersal, pelagic-neritic, benthopelagic, bathypelagic, and pelagic-oceanic, respectively, indicating that the spawning activities of multiple ecological groups of fish occurred in the semi-closed gulf. The species accumulation curves of fish eggs were non-asymptotic, indicating that substantially more taxa would have been encountered with increased sampling effort in both seasons (Figure 5).
FIGURE 4
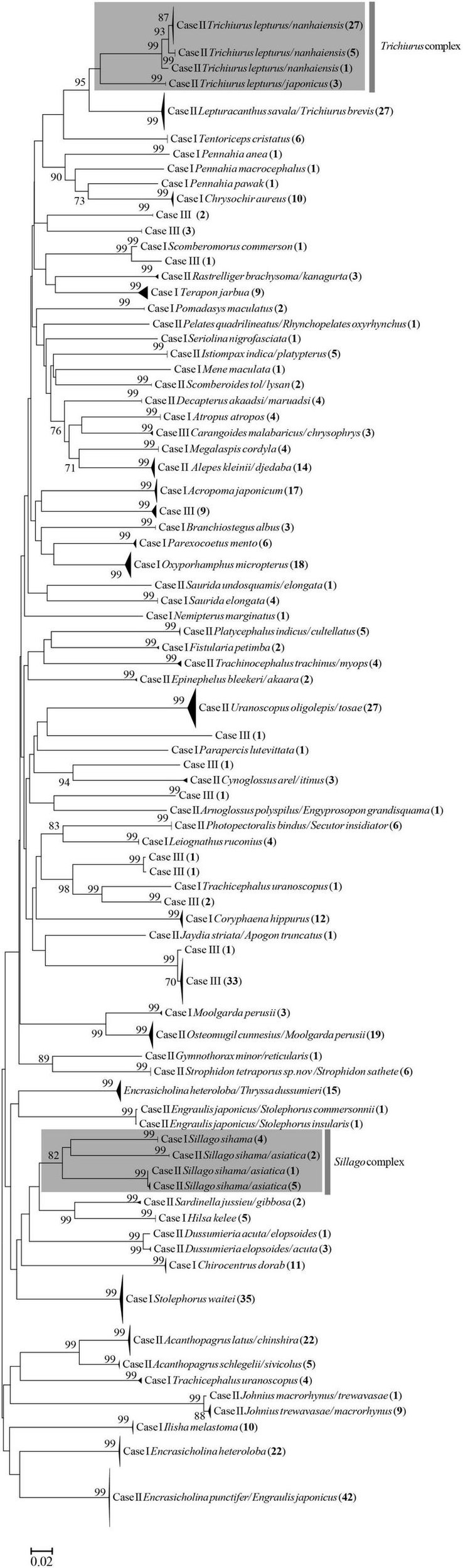
The neighbor-joining tree of COI sequences of fish eggs. The bootstrap values are given at nodes.
FIGURE 5
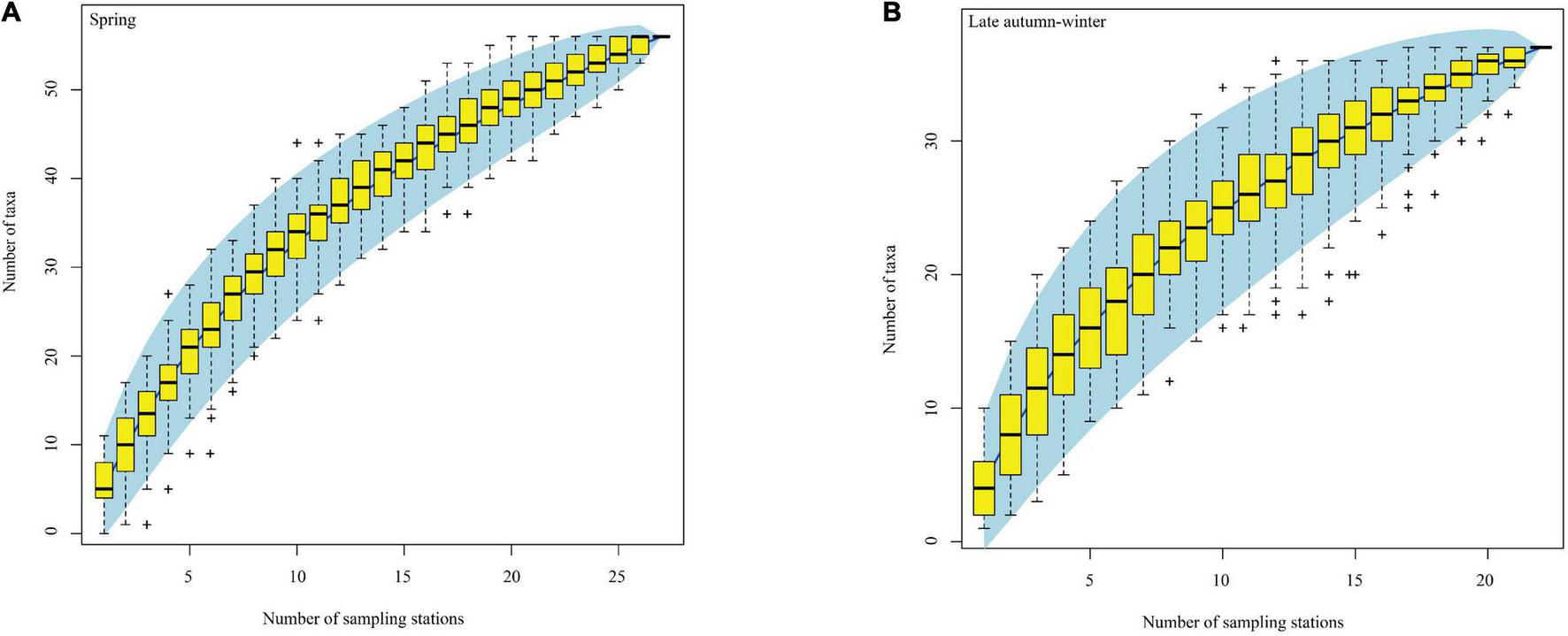
Species accumulation curves of fish eggs taxa identified by molecular methods in the eastern Beibu Gulf. (A) Spring; (B) late autumn-winter. The light blue area indicates 95% confidence intervals. Yellow boxes are the interquartile ranges, the central bold mark on each box indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers, and the outliers are plotted individually using the “+”symbol.
Morphological Characteristics of Fish Eggs
The egg diameters of 582 fish eggs identified by COI and cytb sequences were measured. Most of the eggs were <2.500 mm in diameter, with 29.55% belonging to the 0.630–1.000 mm size class and 60.48% belonging to the 1.010–2.000 mm size class. Altogether, 97.42% of the eggs were <2.500 mm, while the remaining 2.58% belonged to 2.510–3.190 mm and 4.350–4.920 mm classes (Figure 6). For the 32 identified species, the shapes of eggs were spherical for 30 species, 1 species was elliptical (Figure 2, 8th), and 1 species was elliptical with a knob (Figure 2, 28th). The egg diameters ranged from 0.660 to 2.694 mm. The smallest eggs at the species level were those of Leiognathus ruconius (Hamilton, 1822), and the largest were those of Oxyporhamphus micropterus (Valenciennes, 1847), while the smallest eggs at the family level were those of Leiognathidae, and the largest were those of Hemiramphidae (Figure 7). The average egg diameters of 16 fish species were <1.000 mm (i.e., 50%), while 10 of 24 families were <1.000 mm (38.5%; Figure 7). This indicated that a large proportion of fish species in the eastern Beibu Gulf produced small eggs. Most of the eggs had a transparent and smooth chorion. However, a chorion surface structure was observed in two species. The eggs of O. micropterus had numerous short spines (ca. 0.030–0.050 mm) uniformly scattered (Figure 2, 17th), while the eggs of Parexocoetus mento (Valenciennes, 1847) had many long filaments all over the surface (ca. 0.410–1.540 mm; Figure 2, 19th).
FIGURE 6
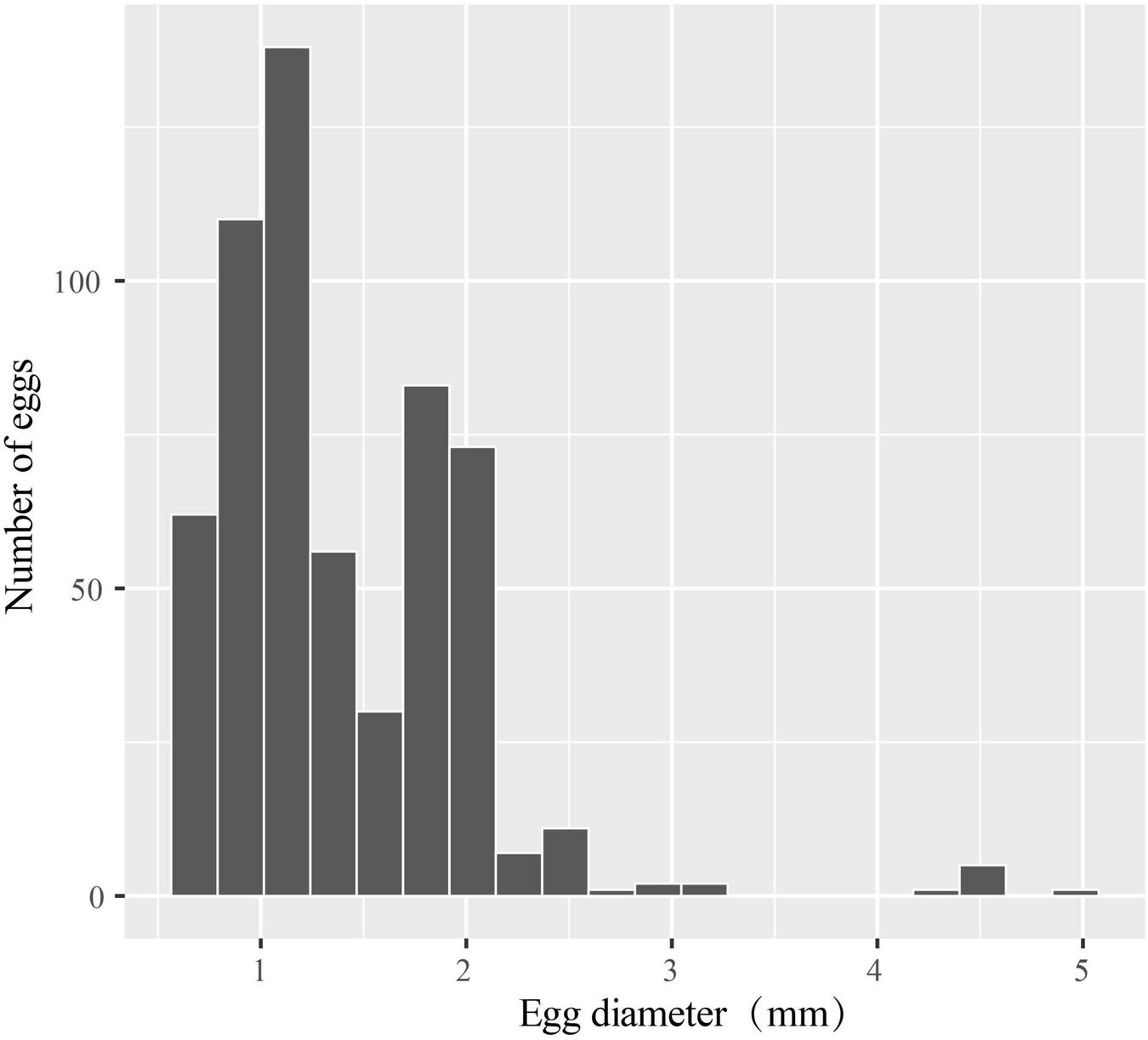
The egg diameter distribution of identified fish eggs in the eastern Beibu Gulf.
FIGURE 7
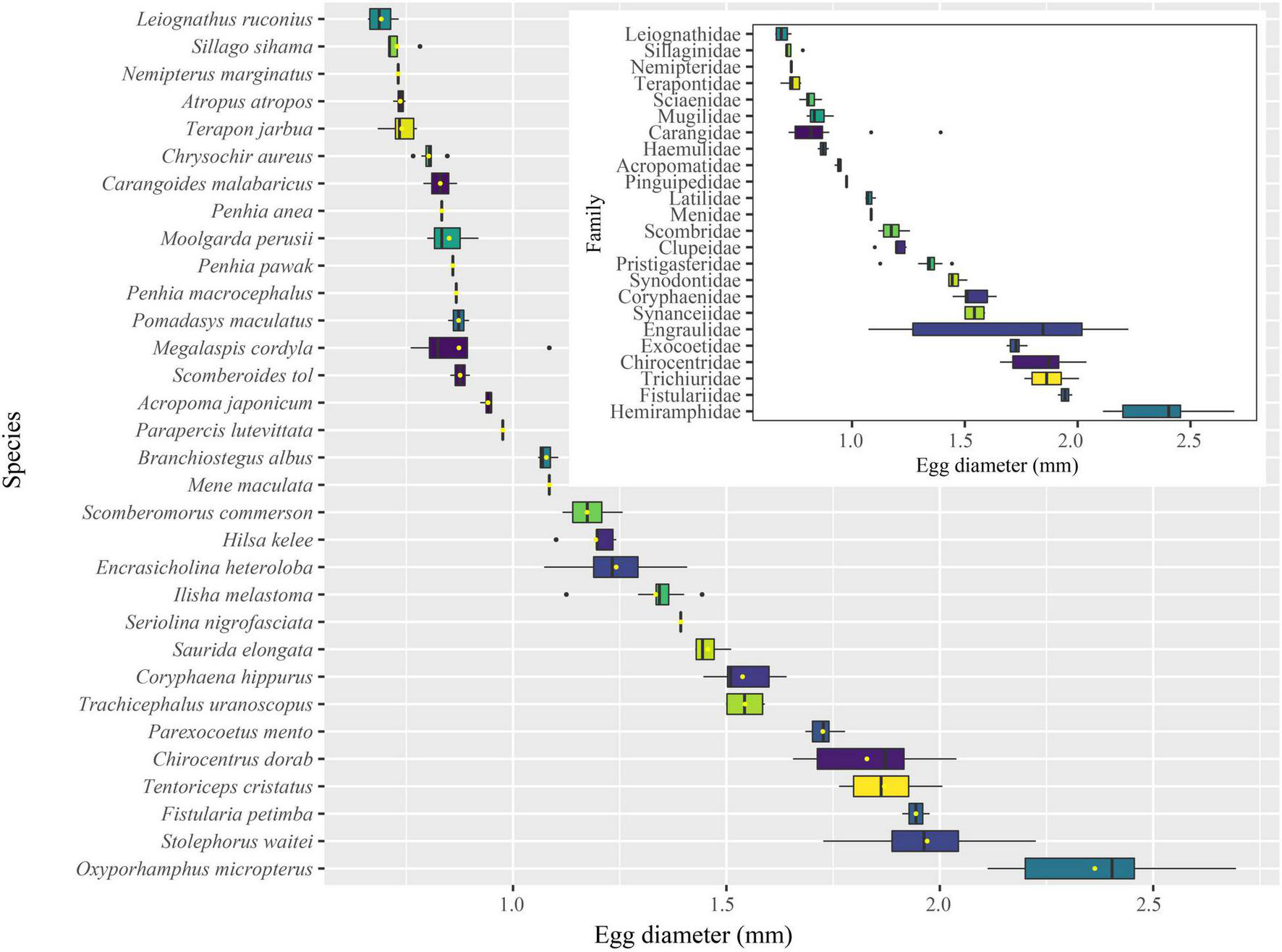
The egg diameter of identified fish eggs at the species level.
Discussion
Sequencing Success Rate of Fish Eggs
Preserving fish eggs in formaldehyde allows the visual characteristics of development for morphological classification to be retained, but this method can cause fragmentation and denaturation of DNA (Akimoto et al., 2002; Srinivasan et al., 2002; Karaiskou et al., 2007). Ethanol-preserved eggs may shrink, obscuring the developmental stages, lose pigmentation, and turn opaque (Espeland and Sannæs, 2018). Because there is little morphology literature on fish eggs in the Beibu Gulf, we preferred to preserve the eggs in ethanol with the primary aim of identifying them. In the present study, the sequencing success rate of fish eggs was low (66.78%). The high failure rate was most likely due to poor preservation at sea, the long time needed to pick out eggs preserved in low concentrations in ethanol, and the delayed DNA extraction process affected by the coronavirus epidemic in 2020. In previous studies, the sequencing success rate for fish eggs varied considerably, i.e., Ahern et al. (2018) selected 6,422 eggs out of which 2,354 were sequenced successfully (COI/16S fragments, 36.66%); Liu et al. (2018) selected 8,983 drifting eggs and obtained 7,933 cytb sequences (88.31%); and Chen et al. (2021) selected 641 eggs and obtained 397 high-quality sequences (61.93%). It is considered that a low ethanol concentration and long storage in ethanol (>6 months) both negatively affect DNA stability and lead to DNA degradation (Zimmermann et al., 2008; Michaud and Foran, 2011; Marquina et al., 2021). However, highly concentrated ethanol (95–100%) was found to induce albefaction, brittleness, dissolution of oil globules, and damage to the morphology (Marquina et al., 2021; Zheng et al., 2021). In this study, we preserved the specimens in 75% ethanol–sea water solution to balance a relatively good morphology preservation and sequencing success rate. This ethanol concentration can also reduce potential bacterial pollution in the specimens and reduce the amplification of DNA fragments of non-target species. However, timely DNA extraction should be conducted to increase the success rate in future studies.
Performance of Molecular Techniques for Identifying Fish Eggs
In the present study, we identified 81 distinct taxa using COI and cytb sequences, out of which 32 taxa (39.51%) were identified at the species level (Figures 2, 4). However, nearly 36 taxa (44.44%) were ambiguous, and could only be identified at the genus or family level (Figure 4 and Supplementary Table 1). The high ratio (53.89% of COI sequences) of ambiguous identification potentially limited the performance of species identification in the informatics platforms, BOLD and NCBI (Meiklejohn et al., 2019; Pentinsaari et al., 2020). For the two platforms, BOLD mines barcode sequences from GenBank periodically, and sequences in BOLD are submitted to GenBank when preparing for publication. The percentage of sequences on BOLD originating from GenBank are lower than the barcodes in GenBank, and include unverified sequences mined from GenBank. In addition, the taxonomic classifications or scientific name changes for fish specimens that are submitted in BOLD can be updated and amended freely before being submitted to NCBI. The database managers regularly undertake taxonomic checks in BOLD. Therefore, BOLD provided a more informative platform for the fish eggs in this study than GenBank, given the contemporary COI sequencing of candidate species in the present study. Despite using the delineation criteria for those sequences with GenBank accession numbers, the high ratio of Case II findings introduced caution about fish species identification.
As mitochondrial DNA is maternally inherited, DNA barcoding has limitations in identifying species associated with incomplete lineage sorting, introgression hybridization, or ancestral polymorphism. In addition, the method can be affected by misidentification in the reference libraries or in the query sequences in the platforms (Pentinsaari et al., 2020). The misidentifications that occur during the process of developing the DNA barcode library of fish are normal because of morphological characteristics, i.e., phenotypic plasticity, new species, cryptic diversity, genotypic variation, or different life stages. In the present study, ambiguous delineation mainly appeared in the taxa or genera Alepes, Acanthopagrus, Johnius, Istiompax, Scomberoides, Rastrelliger, Decapterus, Carangoides, Saurida, Platycephalus, Epinephelus, Uranoscopus, Cynoglossus, Sillago, and Sardinella, and families Apogonidae, Bothidae, Clupeidae, Theraponidae, Trichiuridae, Mugilidae, and Engraulidae, among which misidentifications, cryptic diversity, and new species were common (Mat Jaafar et al., 2012; Qin et al., 2013; Gon et al., 2015; Tucker et al., 2016; Fricke, 2018; Hata and Motomura, 2018; Chao et al., 2019; Frable et al., 2019; Huang et al., 2020; Prokofiev, 2021; Xiao et al., 2021). Developing a reliable DNA barcode library for these taxa, with correct species identifications via morphology, or using multiple DNA fragments is recommended in future studies (Chakraborty et al., 2006; Hsu et al., 2009).
Species Composition, Spawning Periods, and Occurrence in Spring and Late Autumn–Winter
On the basis of the COI and cytb sequences obtained, we successfully identified 32 species in 81 taxa, belonging to 30 genera and 24 families (Table 1). Five and four identified fish species from their eggs (29 sequences, 13.48%) were Carangidae and Sciaenidae, respectively, indicating that these two families were the dominant fish groups in the eastern Beibu Gulf. The other 23 species belonged to 22 families (29 sequences, 86.52%), suggesting a high species diversity distribution pattern in the study area. In previous studies, the dominant species were Acropoma japonicum (Günther, 1859), Photopectoralis bindus (Valenciennes, 1835) (also known as Leiognathus bindus), Trachurus japonicus (Temminck and Schlegel, 1844), Evynnis cardinalis (Lacepède, 1802) (also known as Parargyrops edita), and P. macrocephalus (also known as Argyrosomus microcephalus) based on the quarterly otter trawl surveys (Wang et al., 2011), and A. japonicum, E. cardinalis, T. japonicus, Decatperus maruadsi, and L. ruconius using acoustic measurement with ordinary kriging in the Beibu Gulf (Wang et al., 2012; Sun et al., 2019). In the present study, the eggs of the dominant fish species (A. japonicum, L. ruconius, and P. microcephalus) were also detected using the DNA barcode method (Table 1, Figure 4, and Supplementary Table 1). For D. maruadsi and P. bindus, 4 and 6 COI sequences of Decatperus sp. and Leiognathidae sp. were categorized as Case II (i.e., D. akaadsi/maruadsi and P. bindus/Secutor insidiator, respectively) following the criteria. These eggs may need to be analyzed further in combination with the local DNA barcode library (Hou et al., 2018, 2020). For E. cardinalis, although the fish’s spawning period occurred in the winter season, we did not collect eggs from this species in either of the two surveys, indicated that more surveys should be conducted. In addition, given that we only used zooplankton nets to collect drifting and suspended eggs in surveys, we chose a partial set of specimens for molecular identification, and we had a high failure rate of amplification, it is likely that the species diversity of the egg pool in the eastern Beibu Gulf was underestimated, and a random sampling error may have occurred. Accordingly, the composition of the fish community was biased, and did not include the entire fish community known from the waters. In addition, a strong La Niña phenomenon in 2020 affected fish reproduction in the autumn and winter seasons, which correspondingly reduced the eggs of potential fish species that were collected. However, we did find several unexpected and important taxa, i.e., the first identification of Stolephorus waitei (Jordan and Seale, 1926) eggs with morphological characteristics of an elliptical shape, with a knob on the egg membrane (Gao et al., 2016); the potentially new species Strophidon tetraporus sp. nov (Case II, Supplementary Table 1; Huang et al., 2020); and the economically important species B. albus, C. aureus, M. cordyla, M. maculate, and S. sihama, and genus Pennahia. The spawning ground of these species are not well-known in the Beibu Gulf.
Among the 32 species identified, 25 species were found in spring and 25 species in late autumn–winter, out of which 18 species (56.25%) were found in both surveys. This finding indicated that these species spawned both in spring and late autumn–winter, which are the main spawning seasons in the Beibu Gulf. The Beibu Gulf is located in a subtropical/tropical sea area, with a sea surface temperature ranging between 18 and 32°C through the year (Huang et al., 2008; Gao et al., 2017; Shen et al., 2018). The typical hatching time of pelagic fish eggs in this sea area is between 18 and 70 h (Zhang et al., 1985). Although these planktonic fish eggs can disperse some distance on the ocean current, the main current in the Beibu Gulf is circular, because of the dynamics of coastal water, mixing with oceanic water, and a cold upwelling water mass (Su, 2005; Gao et al., 2017). Thus the complicated hydrological conditions do not transport the fish eggs a long distance prior to hatching. In addition, with a background trend of ocean warming, the hatching time of fertilized fish eggs is becoming shorter (Pauly and Pullin, 1988). Therefore, most eggs collected in the current study probably participated in local spawning events, and the sampling sites can be determined as the spawning grounds of these species.
The average egg diameters of 50% of the fish species (38.5% families) were <1 mm (Figure 7). Small eggs hatch into small fish larvae, which tend to have a high mortality rate in conditions of marine environmental change (Ware, 1975; Fuiman, 2003). This indicates that the population recruitment and assemblage structure in this marine area may fluctuate substantially, which corresponds to the current fishery status in the Beibu Gulf (Sun and Lin, 2004; Wang et al., 2010). In the present study, the smallest eggs were from L. ruconius, which have become the dominant species following the decline of large economically valuable fish species. The increase in fish eggs from this family cannot be regarded as a manifestation of improved fish resources (Jia et al., 2004).
Conclusion
Although our surveys and collection period were limited, we were able to use molecular identification to discern 32 species in 81 taxa, belonging to 24 families in the present study, which facilitates the delimitation of fish eggs based on morphological features. The number of taxa identified corresponded to nearly 27% of teleost fish referred to in the recent fish assemblage status study by Wang et al. (2010, 2011)) in the Beibu Gulf. The use of informatics platforms for species identification generated many ambiguous and unidentified taxa. To overcome this limitation, substantial efforts shall be made to improve the completeness of DNA barcode reference libraries for fish in the Beibu Gulf. However, we identified, for the first time, the spawning sites of economically important fish species proved by molecular methods. Our study provides new insights into the ichthyoplankton in the eastern Beibu Gulf, and will benefit the protection of spawning grounds and fishery management.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The animal study was reviewed and approved by the Guangdong Ocean University.
Author contributions
GH, JW, and HZ analyzed the data and completed the first draft. BF, CP, and JW performed the field survey. GH, YC, CP, and JW conducted the laboratory experiments. GH and HZ provided guidance on the data analysis and structure. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key R&D Program of China (No. 2018YFD0900906), the National Natural Science Foundation of China (Nos. 31702347 and 42090044), the Youth Innovation Promotion Association CAS (No. 2020211), the Undergraduate Innovation and Entrepreneurship Training Program of Guangdong province (S202010566005), and the Start-up Project of Guangdong Ocean University (R19006).
Acknowledgments
We would like to thank the editor and reviewers for their constructive comments on our manuscript. We thank numerous survey ship members for their help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.806208/full#supplementary-material
References
1
Ahern A. L. M. Gómez-Gutiérrez J. Aburto-Oropeza O. Saldierna-Martínez R. J. Johnson A. F. Harada A. E. et al (2018). DNA sequencing of fish eggs and larvae reveals high species diversity and seasonal changes in spawning activity in the southeastern Gulf of California.Mar. Ecol. Prog. Ser.592159–179. 10.3354/meps12446
2
Akimoto S. Kinoshita S. Sezaki K. Mitami I. (2002). Identification of alfonsino and related fish species belonging to the genus Beryx with mitochondrial 16S rRNA gene and its application on their pelagic eggs.Fish. Sci.681242–1249. 10.1046/j.1444-2906.2002.00561.x
3
Baumgartner G. Nakatani K. Gomes L. C. Bialetzki A. Sanches P. V. Makrakis M. C. (2004). Identification of spawning sites and natural nurseries of fishes in the upper Paraná River, Brazil.Environ. Biol. Fish.71115–125. 10.1007/s10641-004-0098-z
4
Bui A. O. V. Castonguay M. Ouellet P. Sévigny J. M. (2011). Searching for Atlantic cod (Gadus morhua) spawning sites in the northwest Gulf of St Lawrence (Canada) using molecular techniques.ICES J. Mar. Sci.68911–918. 10.1093/icesjms/fsr016
5
Burrows M. Browing J. S. Breitbart M. Murawski S. A. (2018). DNA barcoding reveals clear delineation between spawning sites for neritic versus oceanic fishes in the Gulf of Mexico.Fish. Oceanogr.28228–239. 10.1111/fog.12404
6
Cao W. Chang J. Qiao Y. Duan Z. (2007). Fish Resources of Early Life History Stages in Yangtze River.Beijing: China waterpower press.
7
Chakraborty A. Aranishi F. Iwatsuki Y. (2006). Genetic differentiation of Trichiurus japonicus and T. lepturus (Perciformes: trichiuridae) based on mitochondrial DNA analysis.Zool. Stud.45419–427.
8
Chao N. L. Chang C. W. Chen M. H. Guo C. C. Lin B. A. Liou Y. Y. et al (2019). Johnius taiwanensis, a new species of Sciaenidae from the Taiwan Strait, with a key to Johnius species from Chinese waters.Zootaxa4651259–270. 10.11646/zootaxa.4651.2.3
9
Chen W. T. Zhu S. L. Yang J. P. Li X. H. Li Y. F. Li J. (2021). DNA barcoding reveals the temporal community composition of drifting fish eggs in the lower Hongshui River, China.Ecol. Evol.1111507–11514. 10.1002/ece3.7943
10
Espeland S. H. Sannæs H. (2018). Estimating cod egg developmental stage based on DNA concentration.ICES J. Mar. Sci.75825–830. 10.1093/icesjms/fsx172
11
Fitzcharles E. M. (2012). Rapid discrimination between four Antarctic fish species, genus Macrourus, using HRM analysis.Fish. Res.12166–170. 10.1016/j.fishres.2012.02.002
12
Frable B. W. Tucker S. J. Walker H. (2019). A new species of grouper, Epinephelus craigi (Perciformes: epinephelidae), from the South China Sea.Ichthyol. Res.66215–224. 10.1007/s10228-018-0669-9
13
Fricke R. (2018). Two new species of stargazers of the genus Uranoscopus (Teleostei: uranoscopidae) from the western Pacific Ocean.Zootaxa4476157–167. 10.11646/zootaxa.4476.1.15
14
Fuiman L. A. (2003). “Special considerrations of fish eggs and larvae,” in Fishery Science: The Unique Contributions of Early Life Stages, edsFuimanL. A.WernerR. G. (Hoboken, NJ: Blackwell Science), 1–32.
15
Gao D. Wan R. Ma Q. Zhang X. Bian X. (2016). Development of eggs and larvae of Stolephorus commersonnii and taxonomic key to fish eggs of the Clupeidae and Engraulidae off China.Mar. Biol. Res.12255–267. 10.1080/17451000.2015.1125004
16
Gao J. Wu G. Ya H. (2017). Review of the circulation in the Beibu gulf, South China Sea.Cont. Shelf Res.138106–119. 10.1080/17451000.2015.1125004
17
Gon O. Liao Y. C. Shao K. T. (2015). A new species of the cardinalfish genus Jaydia (Teleostei: apogonidae) from the Philippines.Zootaxa3980286–292. 10.11646/zootaxa.3980.2.9
18
Gotelli N. J. Colwell R. K. (2001). Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness.Ecol. Lett.4379–391. 10.1046/j.1461-0248.2001.00230.x
19
Hata H. Motomura H. (2018). Stolephorus continentalis, a new anchovy from the northwestern South China Sea, and redescription of Stolephorus chinensis (Günther 1880)(Clupeiformes: engraulidae).Ichthyol. Res.65374–382. 10.1007/s10228-018-0621-z
20
Hou G. Chen W. T. Lu H. S. Cheng F. Xie S. G. (2018). Developing a DNA barcode library for perciform fishes in the South China Sea: species identification, accuracy and cryptic diversity.Mol. Ecol. Resour.18137–146. 10.1111/1755-0998.12718
21
Hou G. Xu Y. Chen Z. Zhang K. Huang W. Wang J. et al (2021a). Identification of eggs and spawning zones of Hairtail fishes Trichiurus (Pisces: trichiuridae) in northern South China Sea, using DNA Barcoding.Front. Env. Sci.9:703029. 10.3389/fenvs.2021.703029
22
Hou G. Chen Y. Wang S. Wang J. Zhang H. (2021b). Formalin-fixed fish larvae could be effectively identified by DNA Barcodes: a case study on thousands of specimens in South China Sea.Front. Mar. Sci.8:634575. 10.3389/fmars.2021.634575
23
Hou G. Wang J. Chen Z. Zhou J. Huang W. Zhang H. (2020). Molecular and morphological identification and seasonal distribution of eggs of four Decapterus fish species in the northern South China Sea: a key to conservation of spawning ground.Front. Mar. Sci.7:970. 10.3389/fmars.2020.590564
24
Hsu K. C. Shih N. T. Ni I. H. Shao K. T. (2009). Speciation and population structure of three Trichiurus species based on mitochondrial DNA.Zool. Stud.48851–865.
25
Huang W. C. Mohapatra A. Thu P. T. Chen H. M. Liao T. Y. (2020). A review of the genus Strophidon (Anguilliformes: muraenidae), with description of a new species.J. Fish. Biol.971462–1480. 10.1111/jfb.14514
26
Huang Y. Li Y. Shan H. Li Y. (2008). Seasonal variations of sea surface temperature, chlorophyll a and turbidity in Beibu Gulf, MODIS imagery study.J. Xiamen Univ.47856–863.
27
Hubert N. Espiau B. Meyer C. Planes S. (2015). Identifying the ichthyoplankton of a coral reef using DNA barcodes. Mol. Ecol. Resour.15, 57–67. 10.1111/1755-0998.12293
28
Ikeda T. Hirai A. Tabata S. Onishi Y. Mito S. (2014). “Key to fish eggs in Japan,” in An Atlas of the Early Stage Fishes In Japan, 2nd Edn, ed.OkiyamaM. (Kanagawa: Tokai University Press), 1–108.
29
Jia X. P. Li Y. Z. Li C. H. Qiu Y. S. Gan J. L. (2004). Fishery Ecology Environment and Fishery Resources in the South China Sea Exclusive Economic Zone Adn Continental Shelf.Beijing: Science Press.
30
Karaiskou N. Triantafyllidis A. Alvarez P. Lopes P. Garcia-Vazquez E. Triantaphyllidis C. (2007). Horse mackerel egg identification using DNA methodology.Mar. Ecol.28429–434. 10.1111/j.1439-0485.2007.00190.x
31
Kerr M. Browning J. Bønnelycke E. M. Zhang Y. Hu C. Armenteros M. et al (2020). DNA barcoding of fish eggs collected off northwestern Cuba and across the Florida Straits demonstrates egg transport by mesoscale eddies.Fish. Oceanogr.29340–348. 10.1111/fog.12475
32
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol.16, 111–120. 10.1007/BF01731581
33
Lelievre S. Jerome M. Maes G. E. Vaz S. Calaivany S. Verrez-Bagnis V. (2012). Integrating molecular identification of pelagic eggs with geostatistical mapping to improve the delineation of North Sea fish spawning grounds.Mar. Ecol. Prog. Ser.445161–172. 10.3354/meps09443
34
Lewis L. A. Richardson D. E. Zakharov E. V. Hanner R. (2016). Integrating DNA barcoding of fish eggs into ichthyoplankton monitoring programs.Fish. Bull. Natl Oceanic Atmos. Adm.114153–168. 10.7755/FB.114.2.3
35
Leyva-Cruz E. Vásquez-Yeomans L. Carrillo L. Valdez-Moreno M. (2016). Identifying pelagic fish eggs in the southeast Yucatan Peninsula using DNA barcodes.Genome591117–1129. 10.1139/gen-2015-0151
36
Liu M. D. Wang D. W. Gao L. Tian H. W. Liu S. P. Chen D. P. et al (2018). Species diversity of drifting fish eggs in the Yangtze River using molecular identification.PeerJ6:e5807. 10.7717/peerj.5807
37
Marquina D. Buczek M. Ronquist F. Łukasik P. (2021). The effect of ethanol concentration on the morphological and molecular preservation of insects for biodiversity studies.PeerJ9:e10799. 10.7717/peerj.10799
38
Mat Jaafar T. N. A. Taylor M. I. Mohd Nor S. A. De Bruyn M. Carvalho G. R. (2012). DNA barcoding reveals cryptic diversity within commercially exploited Indo-Malay Carangidae (Teleosteii: perciformes).PLoS One7:e49623. 10.1371/journal.pone.0049623
39
Meiklejohn K. A. Damaso N. Robertson J. M. (2019). Assessment of BOLD and GenBank- their accuracy and reliability for the identification of biological materials.PLoS One14:e0217084. 10.1371/journal.pone.0217084
40
Michaud C. L. Foran D. R. (2011). Simplified field preservation of tissues for subsequent DNA analyses.J. Forensic Sci.56846–852. 10.1111/j.1556-4029.2011.01771.x
41
Oksanen J. Blanchet F. G. Friendly M. Kindt R. Wagner H. H. (2020). Package ‘vegan’: Community Ecology Package. R package Version 2.5-6.
42
Oliveira E. C. D. Ferreira E. J. (2008). Spawning areas, dispersion and microhabitats of fish larvae in the Anavilhanas Ecological Station, rio Negro, Amazonas State, Brazil.Neotrop. Ichthyol.6559–566. 10.1590/s1679-62252008000400003
43
Pauly D. Pullin R. S. (1988). Hatching time in spherical, pelagic, marine fish eggs in response to temperature and egg size.Environ. Biol. Fish.22261–271. 10.1007/BF00004892
44
Pentinsaari M. Ratnasingham S. Miller S. E. Hebert P. D. (2020). BOLD and GenBank revisited-Do identification errors arise in the lab or in the sequence libraries?PLoS One15:e0231814. 10.1371/journal.pone.0231814
45
Prokofiev A. (2021). To the taxonomy of the stargazers of the genus Uranoscopus of the Indo-Pacific waters with a description of three new species (Uranoscopidae).J. Ichthyol.61655–679. 10.1134/S0032945221050131
46
Qin Y. Song N. Zou J. Zhang Z. Cheng G. Gao T. et al (2013). A new record of a flathead fish (Teleostei: platycephalidae) from China based on morphological characters and DNA barcoding.Chin. J. Oceanol. Limnol.31617–624. 10.1007/s00343-013-2186-z
47
Qiu Y. Zeng X. Chen T. Yuan W. Wang Y. (2008). Fishery Resources and Management in South China Sea.Beijing: The Ocean Press.
48
Rakocinski C. F. Lyczkowski-Shultz J. Richardson S. L. (1996). Ichthyoplankton assemblage structure in Mississippi Sound as vevealed by canonical correspondence analysis.Estuar. Coast. Shelf Sci.43237–257. 10.1006/ecss.1996.0067
49
Shao K. T. Chen K. C. Wu J. H. (2002). Identification of marine fish eggs in Taiwan using light microscopy, scanning electric microscopy and mtDNA sequencing.Mar. Freshw. Res.53355–365. 10.1071/MF01141
50
Shen C. Y. Yan Y. R. Zhao H. Pan J. Y. Devlin A. T. (2018). Influence of monsoonal winds on chlorophyll-α distribution in the Beibu Gulf.PLoS One13:e0191051. 10.1371/journal.pone.0191051
51
Srinivasan M. Sedmak D. Jewell S. (2002). Effect of fixatives and tissue processing on the content and integrity of nucleic acids-sciencedirect.Am. J. Pathol.1611961–1971. 10.1016/S0002-9440(10)64472-0
52
Stéphanie L. Véronique V. B. Marc J. Sandrine V. (2010). PCR-RFLP analyses of formalin-fixed fish eggs for the mapping of spawning areas in the Eastern Channel and Southern North Sea.J. Plankton Res.321527–1539. 10.1093/plankt/fbq067
53
Su J. (2005). The Offshore Hydrology in China.Beijing: China Ocean Press, 285–296.
54
Sun D. F. Zhu W. C. Ai H. Li N. N. Dong L. N. Shi Y. R. et al (2010). Taxonomic diversity of fish species in Beibu Gulf.Guangdong Agric. Sci.374–7. 10.16768/j.issn.1004-874x.2010.06.099
55
Sun D. R. Chen Z. (2013). Fish Categories Books in the South China Sea, Vol. 1. Beijing: Maritime Press.
56
Sun D. R. Lin Z. J. (2004). Variations of major commercial fish stocks and strategies for fishery management in Beibu Gulf.J. Trop. Oceanogr.2362–68. 10.3724/sp.j.1118.2012.00062
57
Sun M. S. Chen Z. J. Zhang J. Zhang K. (2019). Analysis of fish abundance and distribution pattern in the Beibu Gulf using fishery acoustic measurement combined with ordinary Kriging method.Fresen. Environ. Bull.289058– 9069.
58
Takeuchi A. Higuchi T. Watanabe S. Miller M. J. Yama R. Fukuba T. et al (2021). Several possible spawning sites of the Japanese eel determined from collections of their eggs and Preleptocephali.Fish. Sci.87339–352. 10.1007/s12562-021-01519-4
59
Tamura K. Stecher G. Peterson D. Filipski A. Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0.Mol. Biol. Evol.302725–2729. 10.1093/molbev/mst197
60
Tucker S. J. Kurniasih E. M. Craig M. T. (2016). A new species of grouper (Epinephelus; Epinephelidae) from the Indo-Pacific.Copeia104658–662. 10.1643/CI-16-398
61
Valdez-Moreno M. Vásquez-Yeomans L. Elías-Gutiérrez M. Ivanova N. V. Hebert P. D. N. (2010). Using DNA barcodes to connect adults and early life stages of marine fishes from the Yucatan Peninsula, Mexico: potential in fisheries management.Mar. Freshw. Res.61665–671. 10.1071/MF09222
62
Wan R. J. Zhang R. J. (2016). Fish Eggs, Larvae and Juveniles in the Offshore Waters of China and Their Adjacent Waters.Shanghai: Shanghai Scientific and Technological Press.
63
Wang X. Qiu Y. Du F. Lin Z. Sun D. Huang S. (2010). Fish community pattern and its relation to environmental factors in the Beibu Gulf.J. Fish. China341579–1586. 10.3724/SP.J.1231.2010.06827
64
Wang X. Qiu Y. Du F. Lin Z. Sun D. Huang S. (2011). Spatio-temporal variability of fish diversity and dominant species in the Beibu Gulf.J. Fish. Sci. China18427–436. 10.3724/SP.J.1011.2011.00415
65
Wang X. Qiu Y. Du F. Lin Z. Sun D. Huang S. (2012). Dynamics of demersal fish species diversity and biomass of dominant species in autumn in the Beibu Gulf, northwestern South China Sea.Acta Ecol. Sin.32333–342. 10.5846/stxb201011291700
66
Ward R. D. Zemlak T. S. Innes B. H. Last P. R. Hebert P. D. (2005). DNA barcoding Australia’s fish species.Philos. Trans. R. Soc. B3601847–1857. 10.1098/rstb.2005.1716
67
Ware D. (1975). Relation between egg size, growth, and natural mortality of larval fish.J. Fish. Board Can.322503–2512. 10.1139/f75-288
68
Xiao J. G. Yu Z. S. Song N. Gao T. X. (2021). Description of a new species, Sillago nigrofasciata sp. nov. (Perciformes, Sillaginidae) from the southern coast of China.ZooKeys101185–100. 10.3897/zookeys.1011.57302
69
Xiao W. Zhang Y. Liu H. (2001). Molecular systematics of Xenocyprinae (Teleostei : cyprinidae): taxonomy, biogeography, and coevolution of a special group restricted in east Asia.Mol. Phylogenet. Evol.18163–173. 10.1006/mpev.2000.0879
70
Zhang H. F. Liu X. C. Zhong L. Y. Xing W. Y. Li L. Huang G. G. et al (2006). Embryonic development, morphological development of larva, juvenile and young fish of Epinephelus coioides.J. Fish. Sci. China13689–699.
71
Zhang K. Cai Y. Liao B. Jiang Y. E. Sun M. Su L. et al (2020). Population dynamics of threadfin porgy Evynnis cardinalis, an endangered species on the IUCN red list in the Beibu Gulf, South China Sea.J. Fish Biol.971–11. 10.1111/jfb.14398
72
Zhang R. Lu S. Zhao C. Chen L. Zang Z. Zhang X. (1985). Fish Eggs and Larvae in the Offshore Waters of China.Shanghai: Shanghai Scientific and Technological Press.
73
Zheng Y. J. Li X. Q. Yang Z. X. Cai W. X. Lou Q. S. Tao W. (2021). The identification of fish eggs of two species, the ovate sole Solea ovata and black porgy Acanthopagrus schlegelii.J. Fish. Biol.991746–1751. 10.1111/jfb.14854
74
Zhou M. Y. Lin Y. S. Hou T. T. (2008). “The abundance and distribution of ichthyoplankton in the Gulf of Tonkin in summer, 2006,” in Proceedings of the Symposium of Marine Science Research in Beibu Gulf, edsHuJ.YangS.et al (Beijing: China Ocean Press), 264–271.
75
Zhou M. Lin Y. Yang S. Cao W. Zheng L. (2011). Composition and ecological distribution of ichthyoplankton in eastern Beibu Gulf.Acta Ecol. Sin.3094–105. 10.1007/s13131-011-0095-6
76
Zimmermann J. Hajibabaei M. Blackburn D. C. Hanken J. Cantin E. Posfai J. et al (2008). DNA damage in preserved specimens and tissue samples: a molecular assessment.Front. Zool.5:18. 10.1186/1742-9994-5-18
Summary
Keywords
fish eggs, molecular species identification, spawning site, fish conservation, Beibu Gulf
Citation
Hou G, Chen Y, Wang J, Pan C, Lin J, Feng B and Zhang H (2022) Molecular Identification of Species Diversity Using Pelagic Fish Eggs in Spring and Late Autumn-Winter in the Eastern Beibu Gulf, China. Front. Mar. Sci. 8:806208. doi: 10.3389/fmars.2021.806208
Received
31 October 2021
Accepted
08 December 2021
Published
28 January 2022
Volume
8 - 2021
Edited by
Huang Wei, Second Institute of Oceanography, Ministry of Natural Resources, China
Reviewed by
Jiaguang Xiao, Third Institute of Oceanography, Ministry of Natural Resources, China; Liandong Yang, Institute of Hydrobiology, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2022 Hou, Chen, Wang, Pan, Lin, Feng and Zhang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Feng, fengb@gdou.edu.cnHui Zhang, zhanghui@qdio.ac.cn
†Lead author
This article was submitted to Marine Fisheries, Aquaculture and Living Resources, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.