Abstract
Molting is a key biological process in crustaceans, with impacts on their growth, development and reproduction. Extracellular vesicles (EVs) serve as bio-cargo carrying such as nucleic acids, proteins, and lipids which mediate intercellular communication and participate in various cell biological processes. In this study, we obtained hemocyte transcriptome data during the intermolt, premolt, and postmolt stages of the mud crab Scylla paramamosain. We analyzed the differentially expressed genes in the three stages of molt and identified a number of immune-related genes and structural cuticle genes. We then isolated and characterized the EVs from the hemolymph of S. paramamosain and sequenced their miRNA. In total, we characterized 89 EVs microRNAs (miRNAs) targeting 1447 genes that are potentially involved in the molting process of S. paramamosain. The EVs miRNAs mainly regulated the immune-related genes and affected the molting process by mediating ecdysone signaling and insulin signaling pathways during the mud crab molt cycle. This is the first study of EVs miRNAs and their interaction with mRNA to identify important candidate genes associated with, or regulating, the molting process. This study provides a better understanding of, and novel insights into, the molting cycle of crustaceans and offers baseline information for further studies in the mechanisms regulating molt.
Introduction
The mud crab S. paramamosain, inhabits estuaries and coastal waters in Southeast Asia and is one of the most economically important crustaceans in Chinese mariculture (Vay, 2001). Molting is a key biological process in the growth, development and reproduction of crustaceans (Chang and Mykles, 2011). Over the entire life cycle, S. paramamosain molt 18 times, including five developmental larval molts, several growth‐related molts, and one last adult reproductive molt (Liu et al., 2021). Four major molt stages can be distinguished according to morphological features, including intermolt (InM), premolt (PrM), ecdysis (E) and postmolt (PoM) (Liu et al., 2021). The InM stage is the longest, and energy accumulates and muscles regenerate during this stage. Mud crabs generate a new exoskeleton during the PrM stage. The E stage is very short, taking from only a few seconds to a few minutes. During the E and PoM stages, the crab absorbs a large amount of water, expands its body, and hardens the new soft exoskeleton as quickly as possible (Chang and Mykles, 2011).
Molt is a precise procedure and is controlled by a complex multi-hormone system including ecdysone, the prohormone of the steroid molting hormone, and ecdysteroids, a class of steroid molting hormones (Gong et al., 2015). 20-hydroxyecdysone (20E) has been shown to be a physiologically active ecdysteroid (Gong et al., 2015). Other hormones, such as insulin and juvenile hormone, are also involved in the molting process (Song and Zhou, 2020; Yuan et al., 2020). Crustacean eyestalks are considered to constitute the molt control center and contain the X-organ/sinus gland (XO/SG) complex, which releases a number of peptide hormones such as molt-inhibiting hormone and crustacean hyperglycemic hormone into the hemolymph (Mykles, 2021). The hepatopancreas plays a vital role in energy storage and breakdown, nutrient accumulation, and carbohydrate and lipid metabolism during molt (Xu et al., 2020). Other core genes also participate in the molting process, such as those coding for ecdysone receptor (EcR) (Song et al., 2017), cuticle-related enzymes (chitinases, chitin deacetylase, and chitin synthase) (Zhou et al., 2018), and the structural proteins of the cuticle (Strus et al., 2015). The open circulatory system of crustaceans carries the hemolymph, which contains the hemocytes, oxygen, hormones, and nutrients, etc. The concentration of ecdysteroid in the hemolymph fluctuates dramatically during the molt cycle (Chang and Mykles, 2011). Thus, in addition to the XO-SG complex, the roles of the hemolymph and its contents should be borne in mind when considering the mud crab molting process.
Previously, researches on the molt of mud crabs have mainly focused on the effects of environmental factors on the molting process, endocrine regulation, and functional gene expression levels during the molt cycle (Abehsera et al., 2018; González et al., 2020; Minh Nhut et al., 2020; Abehsera et al., 2021). In addition, several reports of the transcriptomes of different crustacean tissues during molting cycle have been published, such as the gills (Li et al., 2019), eyestalks (Lv et al., 2017), hepatopancreas (Xu et al., 2020; Su et al., 2021), and muscles (Tian and Jiao, 2019). Moreover, the proteomes of the hepatopancreas of mud crabs during the molting cycle have also been reported (Liu et al., 2021). microRNAs (miRNAs) are small non-coding RNAs (18–25 nucleotides in length) which can bind to the 3’ untranslated region (3’UTR) of target mRNAs and regulate the target genes. miRNAs are considered to play various important roles in many fundamental biological processes in animals, including cell differentiation, proliferation, development, apoptosis, metabolism, signal transduction, immunity, and evolution (Saliminejad et al., 2019). Some studies have investigated the roles of miRNAs in S. paramamosain, including miRNA modulation of networks underlying osmoregulation (Wang et al., 2018) and WSSV infection (Lai et al., 2020). However, there is a lack of information on the miRNAs modulation during molting cycles of mud crab. Combining information on the mRNAs and protein profiles will better explain the complex biological processes of molting at the miRNA level.
Extracellular vesicles (EVs) are endocytic in origin, produced by various donor cells and released into the extracellular environment (Kalluri and LeBleu, 2020). They serve as mediators in intercellular communications and transport bio-cargoes, including lipids, proteins, mRNAs, and miRNAs. Numerous studies have shown that EVs are involved in multiple biological and pathological processes, such as reproduction and development, cancer progression, cardiovascular diseases, pathogen infection, and immune responses (Kalluri and LeBleu, 2020). miRNAs can be packaged into EVs to modulate the expression of specific target genes in recipient cells (Bartel, 2004). Recent studies on mud crabs have demonstrated the novel role of exosome miRNAs after viral infection and clarified the mechanisms underlying exosome-mediated apoptosis during the innate immune response (Gong et al., 2020; Gong et al., 2021). However, little information was known regarding the function of EVs miRNAs during the molt process.
In recent years, the study of EVs and the development of miRNAs high-throughput sequencing technology have provided new ideas for the study of the molting process of mud crabs. This study isolated and characterized the EVs in the hemolymph of S. paramamosain and characterized the EVs miRNAs and the target genes potentially involved in S. paramamosain molt. This is the first study of EVs miRNAs and their interactions with mRNA to identify important candidate genes associated with, or playing regulatory roles in, the molting process. This work provides novel insights into the molt cycle of S. paramamosain, and contributes to a better understanding of it. It provides a basic resource for further studies into the mechanisms regulating molt.
Materials and method
Sample collection
Healthy mud crabs (weighing 300 ± 11.2 g) were captured at random from a local crab farm (Beihai, Guangxi, China). Prior to the experiment, crabs were cultured for a week in 0.5 t plastic tanks before sampling, to acclimate to laboratory conditions (25‰ salinity, temperature 28°C). Three molt stages were identified based on morphological features: PoM, InM, and PrM. Three individuals from each stage were sampled. Hemolymph samples were collected using 5 mL syringes containing 1 mL EDTA (Solarbio, Beijing, China) and centrifuged at 500 × g for 5 min to collect the supernatant and hemocytes. All the samples were immediately frozen in liquid nitrogen for storage. The mud crabs were processed according to the Guidelines for the Ethical Review of Laboratory Animal Welfare, People’s Republic of China National Standard GB/T 35892-2018.
mRNA sequencing and analysis
Total RNA was extracted from the mud crab hemocytes using a PureLink RNA Mini Kit (Ambion, California, USA) according to the manufacturer’s protocols. RNA integrity was tested using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto USA). Samples with a RIN ≥ 7 were used to construct libraries using a TruSeq Stranded mRNA LTSample Preparation Kit (Illumina, San Diego, USA). Transcriptome sequencing was then performed by OE Biotech Co., Ltd. (Shanghai, China) on an Illumina platform HiSeq 2500.
Raw reads containing ploy-N and low-quality reads were removed to obtain clean reads. Unigenes were annotated using a variety of databases: NCBI nonredundant (NR), SwissProt, Clusters of orthologous groups for eukaryotic complete genomes (KOG), the Gene ontology (GO) classification, and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The fragments per kilobase million (FPKM) of each unigene was calculated using bowtie2 and eXpress. Differentially expressed genes (DEGs) were identified using DESeq (https://bioconductor.org/packages/release/bioc/html/DESeq2.html), in which unigenes with a threshold P value < 0.05 and foldChange > 2, or foldChange < 0.5 were considered to be significantly differentially expressed. Heatmap and principal component analyses (PCA) were conducted using R version 3.5.2 (https://cran.r-project.org/bin/windows/base/old/3.5.2/). Venn diagrams were drawn using the Venny 2.1 program (http://bioinfogp.cnb.csic.es/tools/venny/).
Isolation and analysis of EVs
Mud crab EVs were isolated from 10 mL hemolymph samples taken from three individuals from each molt stage and separated to collect the supernatants. The supernatants were centrifuged at 10,000 × g for 30 min to remove cell debris, filtered (pore size 0.22 μm), and subjected to ultracentrifugation (Beckman, Optima XE, Duarte, USA) at 120,000 × g for another 70 min. The resulting EVs pellets were resuspended in 200 μl DEPC PBS.
A NanoSight NS300 (Malvern Instruments Ltd, Malvern, UK) was used to quantify EVs and to generate EVs size distribution profiles. The EVs were also imaged using a JEOL JEM-1230 transmission electron microscope (JEOL, Tokyo, Japan). The mud crab EVs were characterized using three phylogenetically conserved EVs markers: CD81 (diluted 1:500), HSP70 (diluted 1:2000), and TSG101 (diluted 1:1000). Calnexin (diluted 1:2000) and GAPDH (diluted 1:10,000) were used as negative controls.
EVs miRNA sequencing and analysis
miRNA was extracted from EVs using a mirVana miRNA Isolation Kit and sequenced by OE biotech Co., Ltd. (Shanghai, China). Low-quality reads, reads with 5’ primer contaminants, poly (A) reads, reads without 3’ adapters and insert tags, and reads shorter than 15 nt and longer than 41 nt, were all filtered from the raw data. Known miRNAs were identified using the miRBase v21 database (http://www.mirbase.org/) and novel miRNAs were predicted using mirdeep2 v2.0.0.8 (http://www.mirbase.org/). Differentially expressed miRNAs with P < 0.05 were identified, calculated using the DEG algorithm in the R package. The target genes of differentially expressed miRNAs were predicted using the miRanda software (http://www.microrna.org/). GO enrichment and KEGG pathway enrichment analysis of differentially expressed miRNA target genes were performed using R. The interactions between miRNA and target genes were visualized using Cytoscape_3.3 (https://cytoscape.org/download.html).
Results
De novo assembly and functional annotation of the mRNA transcriptomes of hemocytes in the three molt stages of S. paramamosain
The transcriptome data were obtained from the mud crab hemocytes in the InM (Figure 1A), PrM (Figure 1B), and PoM stages (Figure 1C). A total of 62.33 G clean reads with an average length of 1362.17 bp were generated (Supplementary Table 1). The original sequencing data were uploaded to the NCBI BioProject database with accession number: PRJNA842705. The assembled unigenes were searched against the NCBI, NR, KEGG, SwissProt, KOG, GO, and Pfam databases, using BLAST+ (E-value, 10-6) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download). Of the 31,713 assembled unigenes: 14,921 (47.05%) were annotated to NR; 9,376 (29.57%) to SwissProt; 4,225 (13.32%) to KEGG; 8,265 (26.06%) to KOG; 8,651 (27.28%) to GO; and 8,833 (27.85%) to Pfam.
Figure 1

Three molt stages of S. paramamosain. (A) intermolt (InM, all parts of the body were hard). (B) premolt (PrM, the old and new carapaces were separated completely). (C) postmolt (PoM, parts of the body were flaccid).
Analysis of the DEGs in the hemocytes of the three molt stages of S. paramamosain
Based on similarities in gene expression patterns, all of the DEGs in the samples could be clustered into three groups, as shown by the PCA plot (Figure 2A). Pairwise comparisons of gene expression were made between consecutive molt stages to gain insight into global gene expression levels in the hemocytes during molt. The analysis of the DEGs showed that a total of 630 unigenes were differentially expressed between the InM and PoM stages, of which 492 were upregulated and 138 were downregulated. A total of 2139 unigenes were differentially expressed between the PoM and PrM stages (745 upregulated and 1394 downregulated). A total of 1157 unigenes, were differentially expressed between the PrM and InM stages (608 upregulated and 549 downregulated) (Figure 2B).
Figure 2
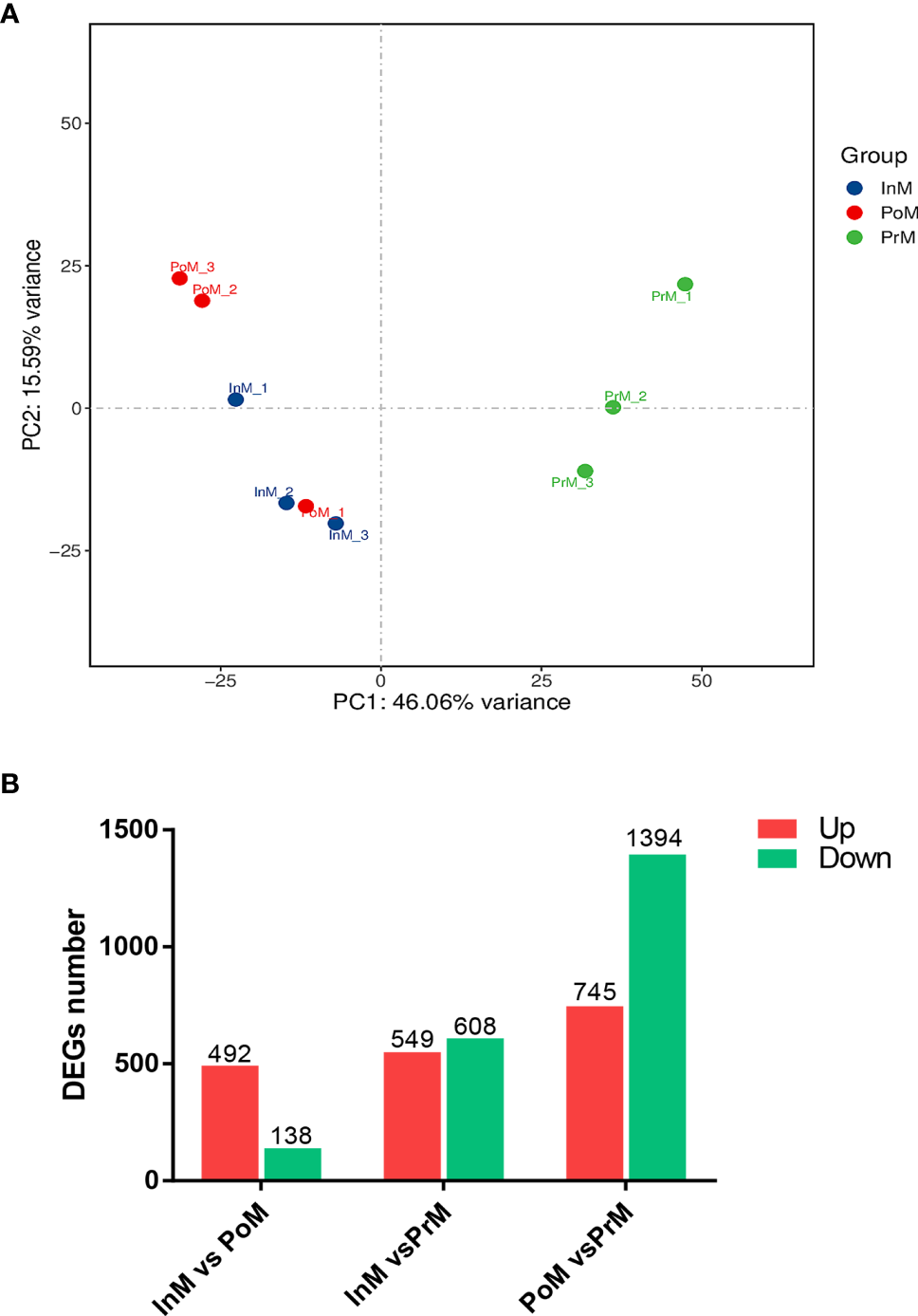
mRNA expression overview of molt stages of S. paramamosain. (A) PCA analysis between adjacent molting stages in S. paramamosain. (B) Differentially expressed genes between adjacent molting stages in S. paramamosain. The values in red and green indicate the numbers of upregulated and downregulated mRNA. PoM, postmolt; InM, intermolt; PrM, premolt.
Of the DEGs, 37 unigenes were differentially expressed in all three molt stages InM vs PoM, PoM vs PrM, and InM vs PrM, including the cuticle proteins and several hormone-related genes (Figure 3). 445 unigenes were only differentially expressed between InM and PoM stages, including insect cuticle protein, broad-complex core protein, prophenoloxidase (proPO), and crustin (Figure 3). 468 genes were only differentially expressed between the PrM and InM stages, including several exoskeleton-related genes such as Di-N-acetylchitobiase, chitinase domain-containing protein, and dynein heavy chain. Between the PoM and PrM stages, 765 shared DEGs were detected, notably those related to immunity and exoskeleton-related genes (Figure 3). In addition, we found 560 DEGs only at the intersections of PoM vs PrM and InM vs PrM, including exoskeleton-related genes (chitin binding-like protein, cuticle protein, early cuticle protein, etc.) and immune-related genes (inhibitor of apoptosis protein, scygonain, etc.) (Figure 3).
Figure 3
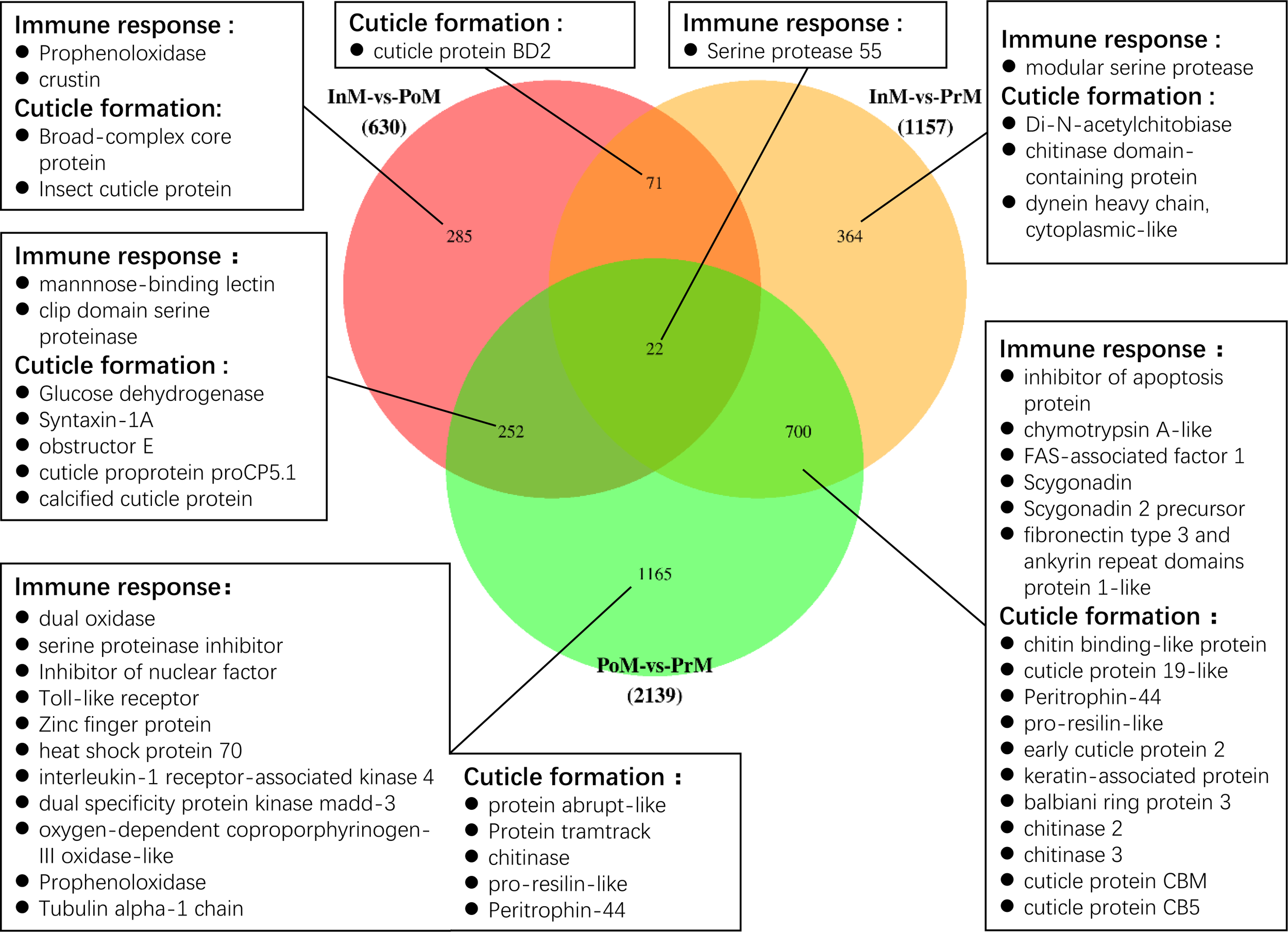
Venn diagram of the DEGs among three molting stages. The numbers of the unique and common DEGs are indicated. The genes related to immune response and cuticle formation have been listed.
Of the DEGs, several immune related genes were discovered, such as antimicrobial peptides (AMPs), genes involved in the Toll pathways and proPO cascades (Supplementary Table 4 and Figure 3). The AMPs of crustin were significantly upregulated at the InM stage, and the scygonadin was upregulated at the PrM stage. The proPO transcripts were upregulated in the InM and PrM stages. The genes involved in the Toll pathway (clip domain serine proteinase, mannnose-binding lectin, and Serine protease 55) were all significantly upregulated in the InM stage. In the PrM stage, most of the Toll pathways genes were upregulated (Pro-resilin-like, inhibitor of apoptosis protein, FAS-associated factor 1-like) with the exception of Serine protease 55. Besides the immune related genes, we also discovered a bunch of genes involved in the cuticle formation and these genes were the most actively expressed in the PrM stage (Supplementary Table 4 and Figure 3).
GO and KEGG analysis of the DEGs of hemocytes in the three molt stages of S. paramamosain
The DEGs were related to the GO and KEGG databases to obtain complete functional information classifying the molting process of mud crabs. GO analysis revealed that the upregulated DEGs of the InM vs PoM stages were mostly enriched in the molecular functions relating to serine-type endopeptidase activity and the biological processes involved in innate immune responses. The downregulated DEGs of the InM vs PoM stages were mostly enriched in molecular functions related to the structural constituents of the cuticle and the biological process of RNA polyadenylation. In the molecular function category between the InM and PrM stages, a large number of upregulated DEGs were associated with polysaccharide binding, and the downregulated DEGs were involved in serine-type endopeptidase activity. Within the biological processes between the InM and PrM stages, upregulated DEGs were mostly enriched in defense responses to fungus attacks, and the downregulated DEGs were associated with the carnitine biosynthetic process. In the analysis of the PoM vs PrM stages, the largest cluster of upregulated DEGs was associated with the biological process category of positive regulation of inflammatory response and the molecular function of polysaccharide binding. Meanwhile, the biological process category innate immune response and the molecular function serine-type endopeptidase activity were the most abundant categories among the downregulated DEGs (Figure 4A).
Figure 4
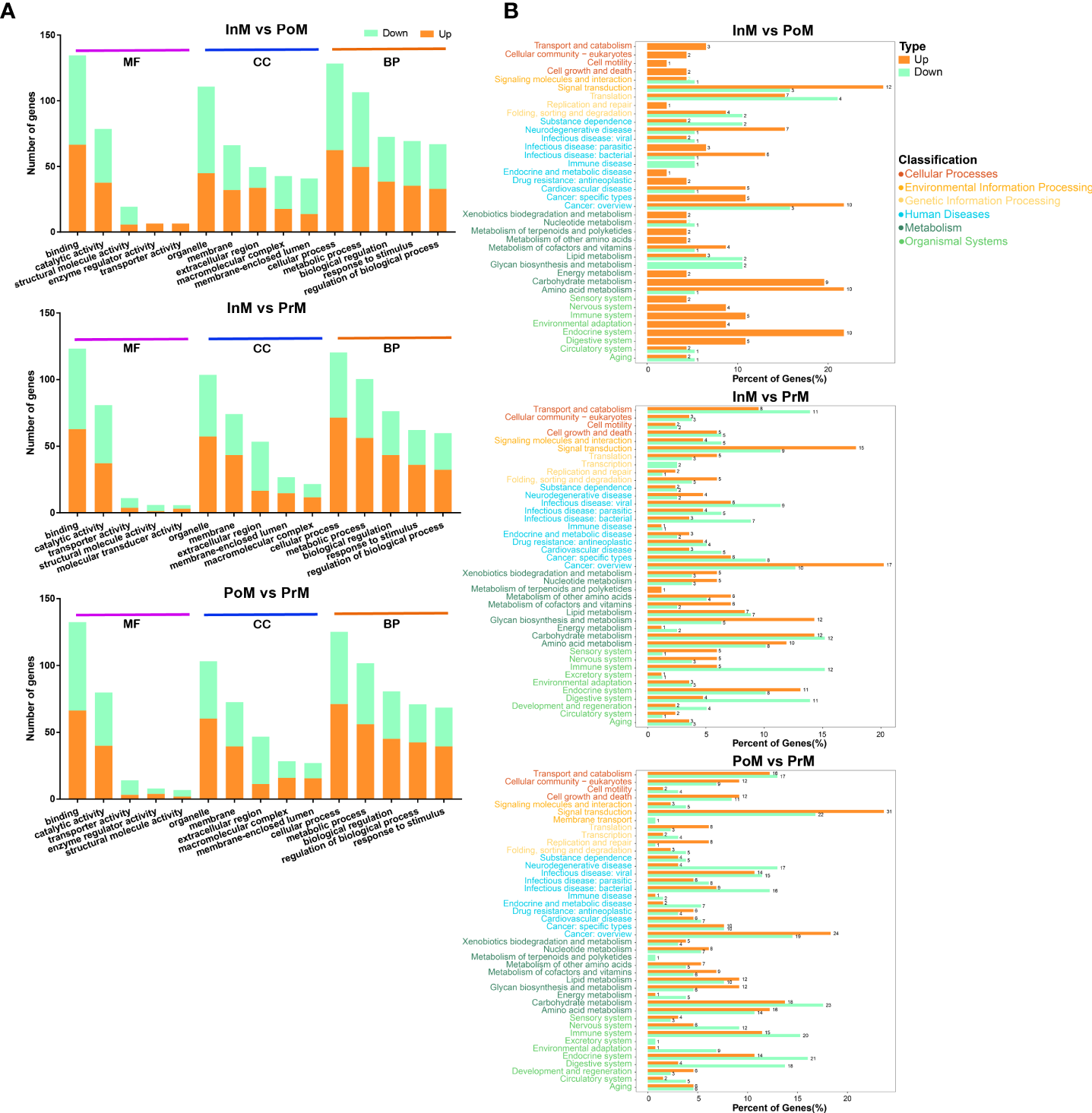
Enrichment of DEGs between the molting stages in GO and KEGG annotation. (A) Enriched GO terms of DEGs between the molting stages. MF, Molecular Function; CC, cellular component; BP, biological process. (B) KEGG enrichment analysis of DEGs between the molting stages. PoM, postmolt; InM, intermolt; PrM, premolt.
In the KEGG pathway analysis of DEGs between the InM and PoM stages, the top five were enriched in Riboflavin metabolism, Prion diseases, Arginine and proline metabolism, Glycolysis/Gluconeogenesis, and Arginine biosynthesis. The top five KEGG pathways for DEGs between the InM and PrM stages were enriched in Neuroactive ligand-receptor interaction, Glycolysis/Gluconeogenesis, Chemical carcinogenesis, Glycosphingolipid biosynthesis-globo and isoglobo series, and Complement and coagulation cascades. The top five KEGG pathways of DEGs between the InM and PoM stages were enriched in Complement and coagulation cascades, Glycolysis/Gluconeogenesis, Other glycan degradation, Glycosphingolipid biosynthesis-globo and isoglobo series, and Lysosome (Figure 4B).
The involvement of EVs in the molting process of mud crabs
To characterize the EVs of mud crabs during the molting process, we isolated EVs from the hemolymph of the InM, PrM, and PoM stages. The cup-shaped structure and size of the isolated EVs were detected using a transmission electron microscope (Figure 5A) and nanoparticle tracking analysis (NTA) (Figure 5B). Three exosome markers (CD81, HSP70, and TSG010) were detected using western blot to further ascertain that the isolated particles were in fact EVs (Figure 5C).
Figure 5
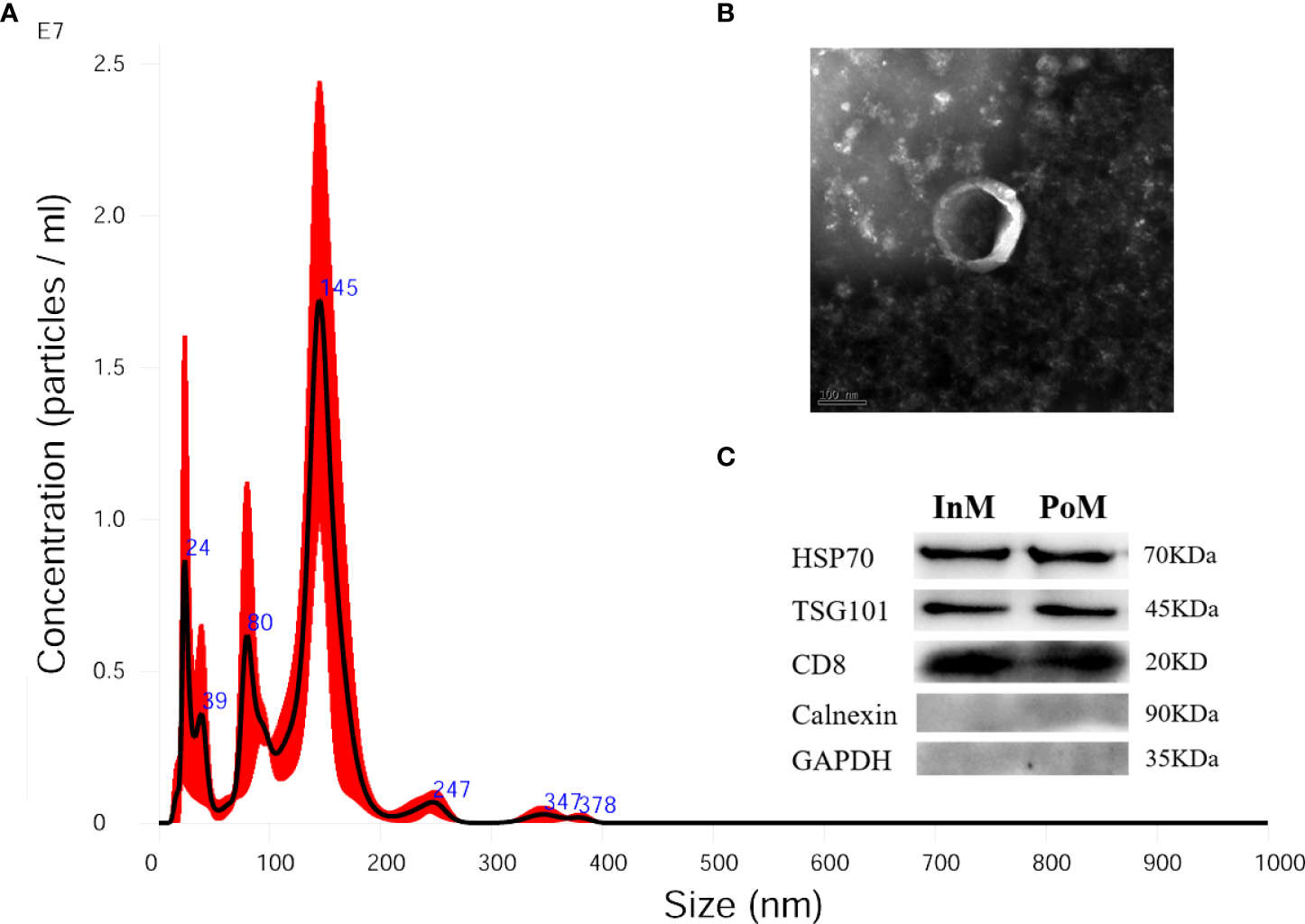
Isolated EVs from hemolymph of S. paramamosain. (A) Nanoparticle tracking analysis (NTA) showing a representative size distribution profile of mud crab EVs. (B) Transmission electron microscopy (TEM) from mud crab EVs; scale bar represents 100 nm. (C) Western blotting analysis (WB) showing mud crab EVs positive for CD81, HSP70 and TSG101; Calnexin and GAPDH served as negative control; the molecular weight for standard is indicated in kilodaltons (kDa).
EVs miRNA expression profiling in the molting process of mud crabs
The miRNA profiles of EVs in the InM, PrM, and PoM stages were sequenced (Supplementary Table 2) and 329.95 M raw reads were obtained. To understand and illustrate the relationship between InM, PoM and PrM, we performed hierarchical clustering of the DE miRNAs from all the samples (Figure 6 A) After removing sequences less than 15 nt or longer than 41 nt (Figure 6B), reads with Q20 less than 80%, and those containing N bases, 171.11 M reads were obtained. To characterize the miRNAs, all of the sequences were compared in the miRbase 22.0 database to discover known miRNAs. A total of 379 miRNAs were identified as previously known. The original sequencing data were uploaded to the NCBI BioProject database with accession number: PRJNA842845.
A total of 61 EVs miRNAs were differentially expressed between the InM and PrM stages (42 upregulated and 19 downregulated), using the criteria of at least a 2-fold difference and a P < 0.05. In addition, 65 differentially expressed miRNAs were expressed comparing PoM and InM. Among them, 44 were upregulated and 21 downregulated. Comparing InM and PoM, 24 EVs miRNAs were differentially expressed, 10 upregulated and 14 downregulated (Figures 6C, D).
Figure 6
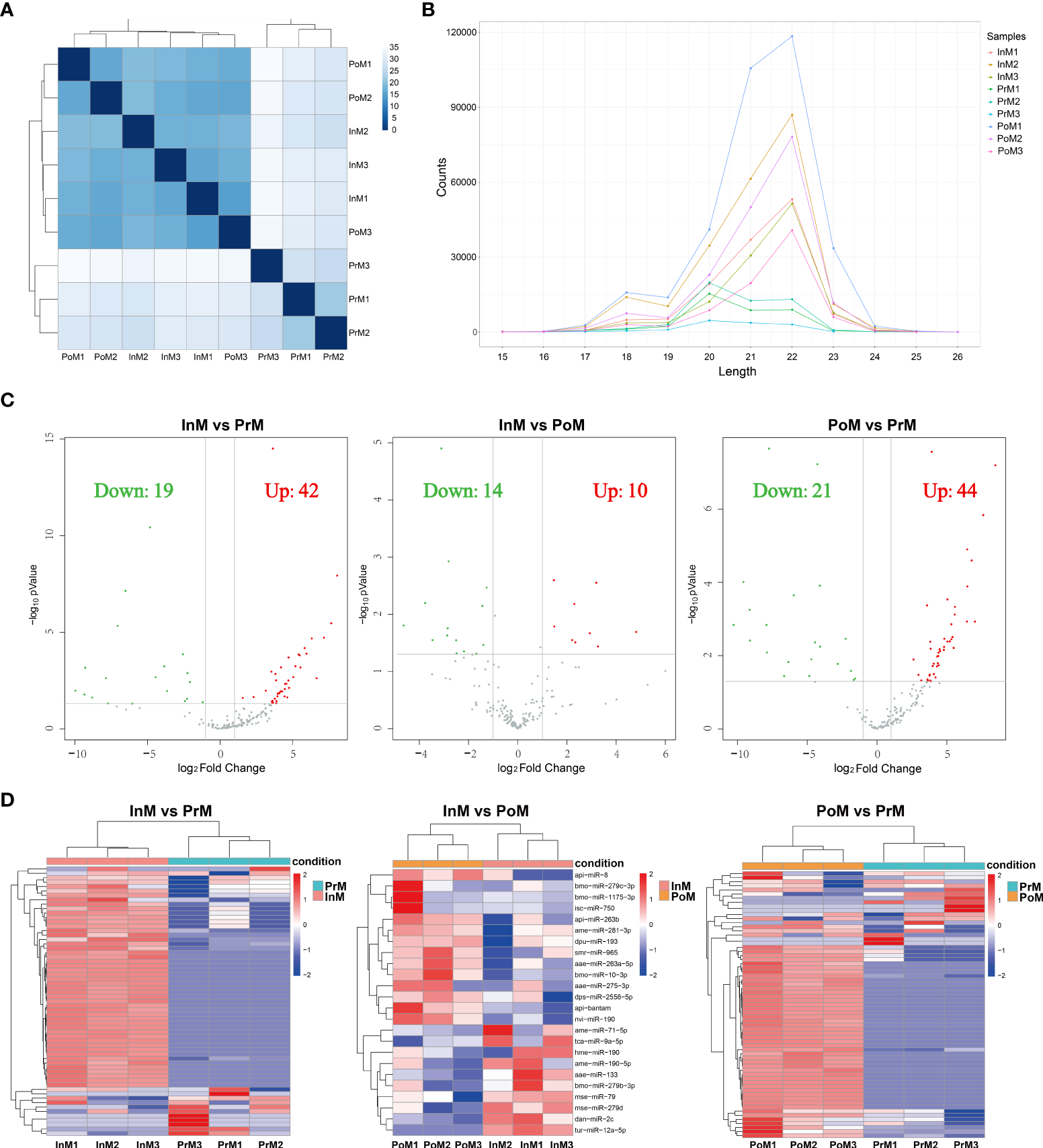
Annotation and analysis of mRNAs. (A) The DE miRNAs correlation heatmap of sample clustering. (B) Length distribution of miRNAs. (C) Volcano plot of the DE miRNAs between adjacent molting stages of S. paramamosain. The horizontal axis shows the log2 Fold change, and the vertical axis represents -log10 pvalue. The grey color represents the unchanged, red color represents upregulated, and the green color represents the downregulated DE miRNAs. PoM, postmolt; InM, intermolt; PrM, premolt. (D) Heatmap diagram of the DE miRNAs between adjacent molting stages of S. paramamosain. Each row represents one miRNA. The horizontal axis represents the sample clusters. Each column represents one biological replicate. The red and blue segments indicate relatively high and low expression level. PoM, postmolt; InM, intermolt; PrM, premolt.
Bioinformatics analysis of EVs miRNA target genes and functional enrichment analysis of miRNA target genes and pathways
RNAhybrid (v2.1.2) (x) + svm_light (v6.01) (x), Miranda (v3.3a) (x), and TargetScan (v7.0) (x) were used to predict target genes. The intersection of the results of these three methods were considered as miRNA-target genes. A total of 89 EVs miRNAs targeting 1447 genes were predicted, covering a wide range of functions (Supplementary Table 3).
GO analysis showed that miRNA-target genes were considerably enriched in the GO terms of biological processes (Figure 7A) (e.g., ubiquitin-dependent protein catabolic process, positive regulation of transcription by RNA polymerase II, and negative regulation of apoptotic process), cell components (e.g., cytoplasm, nucleus, and cytosol), and molecular function (e.g., metal ion binding, ATP binding, and DNA binding).
Figure 7
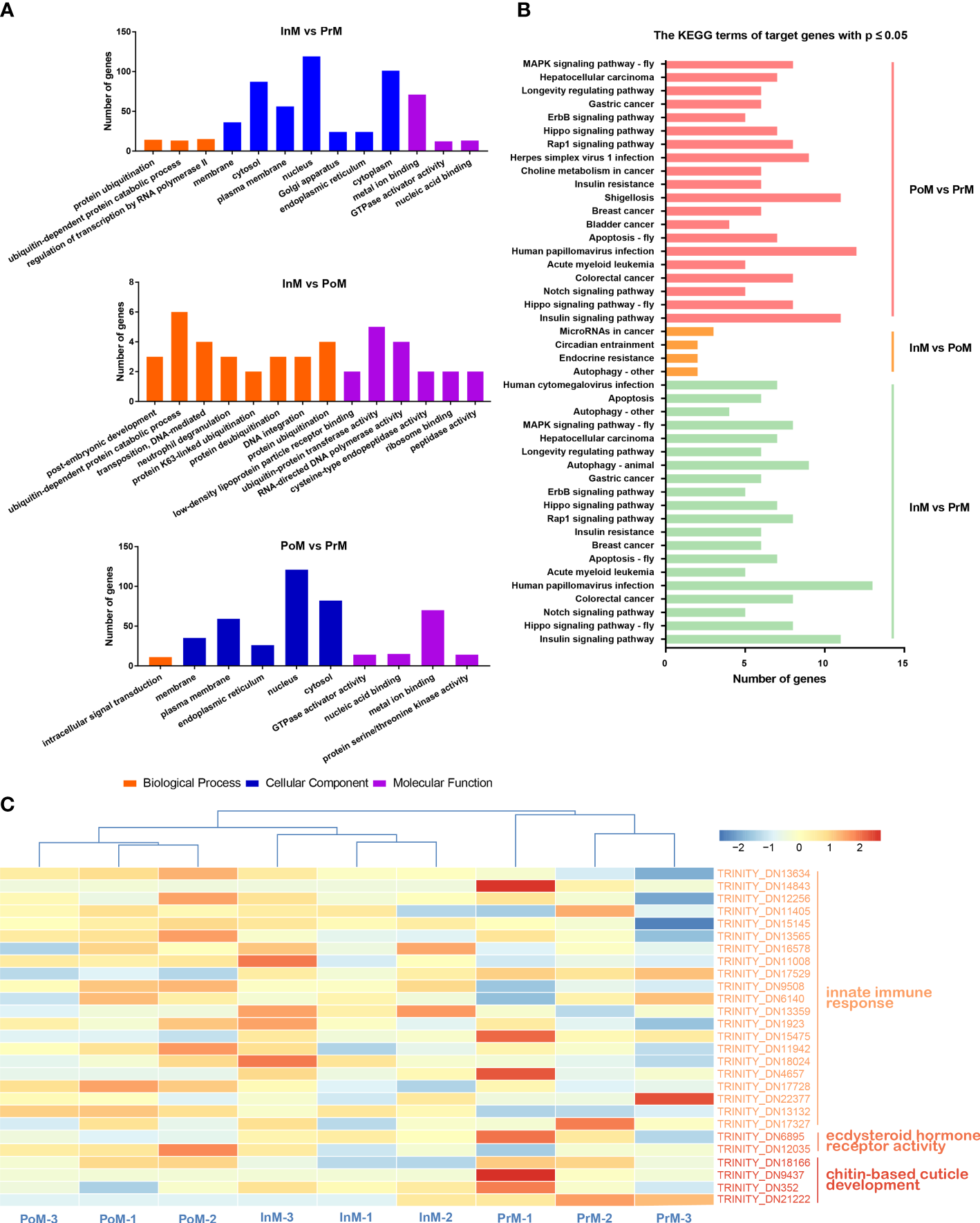
Functional enrichment analysis of miRNA target genes and pathways. (A) GO enrichment analysis of miRNA target genes. (B) KEGG enrichment analysis of the miRNA target genes between adjacent molting stages of S. paramamosain. PoM, postmolt; InM, intermolt; PrM, premolt. (C) Heatmap of miRNA target genes involved in innate immune response, ecdysteroid hormone receptor activity and chitin-base cuticle development in molting stages in S. paramamosain. Colored keys represent the log10(FPKM+1) of target genes between adjacent molt stages. The red color represents upregulation and the blue color represents downregulation. Each column represents one biological replicate and each row represents a miRNA target gene.
KEGG enrichment analysis showed that the 1447 predicted putative target genes were grouped into 238 KEGG terms (Figure 7B). Among these pathways, the Insulin signaling pathway, Hippo signaling pathway, Notch signaling pathway, Colorectal cancer, and Human papillomavirus infection were the five main pathways in the InM vs PrM comparison. Insulin signaling pathway, Hippo signaling pathway, Notch signaling pathway, Colorectal cancer, and Acute myeloid leukemia were the top five pathways in PoM vs PrM. Between InM vs PoM, the most enriched pathways were Autophagy-other, Endocrine resistance, Circadian entrainment, and MicroRNAs in cancer.
In addition, we identified several target genes related to innate immune response, ecdysteroid hormone receptor activity, and chitin-based cuticle development (Figure 7C and Table 1), including tumor necrosis factor, anti-lipopolysaccharide factor (ALF), dual oxidase 1, EcR, and ecdysone-induced protein 74EF (Eip74EF).
Table 1
| miRNA ID | Target genes ID | NR ID | NR annotation |
|---|---|---|---|
| miR-34 | TRINITY_DN13634 | MPC18128.1 | Zinc finger E-box-binding homeobox protein zag-1 |
| miR-133 | TRINITY_DN14843 | AWK27045.1 | X-linked inhibitor of apoptosis protein |
| miR-2556 | TRINITY_DN12256 | XP_027239479.1 | uncharacterized protein LOC113830469 |
| miR-6056 | TRINITY_DN11405 | AYD41594.1 | tumor necrosis factor, partial |
| miR-307 | TRINITY_DN15145 | SLM84439.1 | Toll-like receptor 2 |
| miR-282 | TRINITY_DN13565 | AVN99055.1 | SRSF protein kinase 3-like, partial |
| miR-34 | TRINITY_DN16578 | MPC54724.1 | Serine proteinase stubble |
| miR-184 | TRINITY_DN11008 | AJR22372.1 | serine proteinase inhibitor-2 |
| miR-14 | TRINITY_DN17529 | MPC29207.1 | Serine protease 44 |
| miR-34 | TRINITY_DN9508 | MPC36616.1 | Peptidyl-prolyl cis-trans isomerase |
| miR-6056 | TRINITY_DN6140 | QCU71416.1 | Nuclear factor interleukin 3-regulated transcription factor |
| miR-14 | TRINITY_DN13359 | AQV03739.1 | i-type lysozyme |
| miR-34 | TRINITY_DN1923 | ROT68746.1 | hypothetical protein C7M84_013112 |
| miR-34 | TRINITY_DN15475 | AVN99053.1 | glycogen phosphorylase, partial |
| miR-34 | TRINITY_DN11942 | MPC20397.1 | Germinal-center associated nuclear protein |
| miR-995 | TRINITY_DN18024 | XP_027228265.1 | fibrocystin-L-like |
| miR-92 | TRINITY_DN4657 | AXN93677.1 | dual oxidase 1 |
| miR-375 | TRINITY_DN17728 | XP_027229017.1 | DNA mismatch repair protein Msh2-like |
| miR-6016 | TRINITY_DN22377 | AHK59786.1 | C-type lectin |
| miR-34 | TRINITY_DN13132 | AHB62419.1 | anti-lipopolysaccharide factor |
| miR-34 | TRINITY_DN17327 | AYV97198.1 | alpha-2 macroglobulin |
| miR-34 | TRINITY_DN6895 | AGB34182.1 | ecdysone receptor isoform 2a |
| miR-14 | TRINITY_DN12035 | XP_027206747.1 | ecdysone-induced protein 74EF-like |
| miR-34 | TRINITY_DN18166 | XP_027209046.1 | dynein heavy chain, cytoplasmic-like |
| miR-34 | TRINITY_DN9437 | XP_027228021.1 | uncharacterized protein LOC113819973 |
| miR-2 | TRINITY_DN352 | XP_027217714.1 | uncharacterized protein LOC113810207 |
| miR-6056 | TRINITY_DN21222 | MPC32089.1 | Chondroitin proteoglycan 2 |
EVs miRNAs with putative target genes in relation to innate immune response, ecdysteroid hormone receptor activity, and chitin-based cuticle development.
Interaction analysis of EVs miRNAs and hemocyte mRNAs in the molting process of mud crabs
To further explore the potential network of miRNAs and mRNAs, the interaction networks of miRNAs-mRNAs were constructed based on the DEGs of hemocytes and differentially expressed EVs miRNAs (Figure 8 and Table 2). In total, 50 pairs of differentially expressed miRNAs and mRNAs displaying negative correlations were found. In most cases, the miRNAs were found to target only one mRNA, such as miR-1175 targeting nitric oxide synthase, and miR-278 targeting Retrovirus-related Pol polyprotein from type-2 retrotransposable element R2DM. It was also found that a single differentially expressed miRNA could participate in the regulation of multiple DEGs. For example, miR-34 had 15 target mRNAs, and miR-6056 had seven target genes.
Figure 8
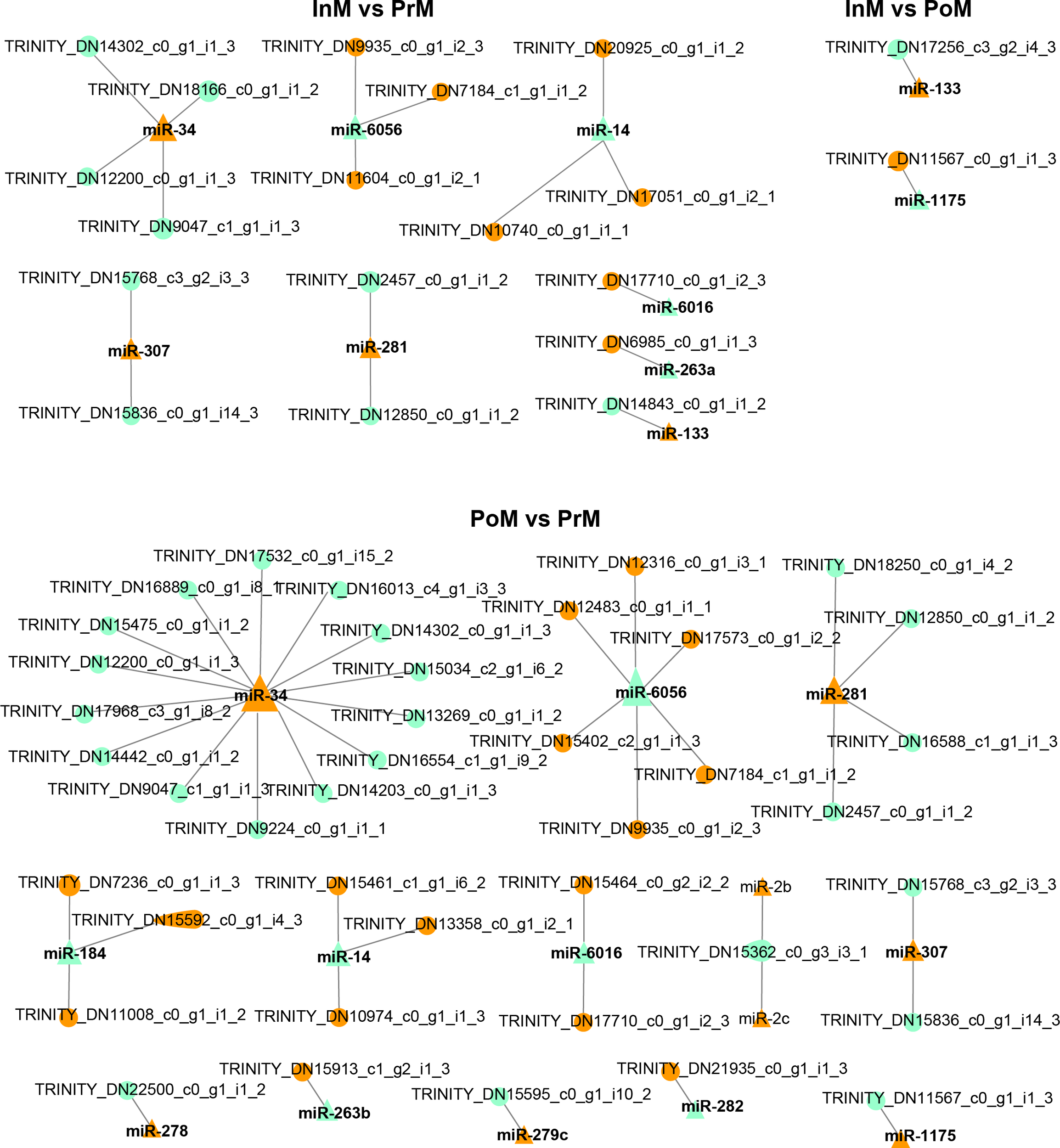
Interaction analysis of mRNA and EVs miRNAs. The triangles are miRNA, the circles are mRNA, orange color represents upregulated, and light green color represents downregulated, and the size represents connectivity.
Table 2
| miRNA id | Regulation | Gene id | log2FC | p-value | Regulation | NR annotation | |
|---|---|---|---|---|---|---|---|
| InM vs PoM | |||||||
| miR-1175 | Down | TRINITY_DN11567_c0_g1_i1_3 | 1.17 | 7.01E-06 | Up | nitric oxide synthase | |
| miR-133 | Up | TRINITY_DN17256_c3_g2_i4_3 | -2.40 | 0.000339 | Down | Sacsin | |
| InM vs PrM | |||||||
| miR-6056 | Down | TRINITY_DN11604_c0_g1_i2_1 | 2.73 | 0.000601 | Up | Protein prune 2 | |
| miR-133 | Up | TRINITY_DN14843_c0_g1_i1_2 | -9.34 | 0.000529 | Down | X-linked inhibitor of apoptosis protein | |
| miR-6016 | Down | TRINITY_DN17710_c0_g1_i2_3 | 3.26 | 1.31E-05 | Up | WSC domain-containing protein 1 | |
| miR-14 | Down | TRINITY_DN17051_c0_g1_i2_1 | 1.90 | 0.00011 | Up | PREDICTED: prostaglandin E2 receptor EP4 subtype-like | |
| miR-281 | Up | TRINITY_DN12850_c0_g1_i1_2 | -1.73 | 8.05E-05 | Down | Mitochondrial carnitine/acylcarnitine carrier protein | |
| miR-34 | Up | TRINITY_DN9047_c1_g1_i1_3 | -3.78 | 6.06E-05 | Down | hypothetical protein C7M84_012357 | |
| miR-6056 | Down | TRINITY_DN9935_c0_g1_i2_3 | 4.01 | 2.81E-08 | Up | hypothetical protein | |
| miR-281 | Up | TRINITY_DN2457_c0_g1_i1_2 | -1.60 | 0.000911 | Down | hypothetical protein | |
| miR-263 | Down | TRINITY_DN6985_c0_g1_i1_3 | 2.33 | 0.000165 | Up | hypothetical protein | |
| miR-14 | Down | TRINITY_DN20925_c0_g1_i1_2 | 6.25 | 0.000965 | Up | hypothetical protein | |
| miR-34 | Up | TRINITY_DN14302_c0_g1_i1_3 | -11.35 | 1.56E-13 | Down | Four-domain proteases inhibitor | |
| miR-34 | Up | TRINITY_DN18166_c0_g1_i1_2 | -1.05 | 0.000255 | Down | dynein heavy chain, cytoplasmic-like | |
| miR-307 | Up | TRINITY_DN15836_c0_g1_i14_3 | -2.36 | 3.28E-06 | Down | beta-actin | |
| miR-6056 | Down | TRINITY_DN7184_c1_g1_i1_2 | 2.73 | 6.04E-05 | Up | – | |
| miR-34 | Up | TRINITY_DN12200_c0_g1_i1_3 | -1.76 | 3.87E-10 | Down | – | |
| miR-307 | Up | TRINITY_DN15768_c3_g2_i3_3 | -3.95 | 4.44E-09 | Down | – | |
| miR-14 | Down | TRINITY_DN10740_c0_g1_i1_1 | 3.68 | 0.00051 | Up | – | |
| PoM vs PrM | |||||||
| miR-6016 | Down | TRINITY_DN17710_c0_g1_i2_3 | 3.06 | 0.002804 | Up | WSC domain-containing protein 1 | |
| miR-6016 | Down | TRINITY_DN15464_c0_g2_i2_2 | 1.96 | 0.002464 | Up | uncharacterized protein LOC113804112 | |
| miR-184 | Down | TRINITY_DN15592_c0_g1_i4_3 | 1.61 | 0.000508 | Up | transmembrane and coiled-coil domains protein 1-like isoform X2 | |
| miR-14 | Down | TRINITY_DN13358_c0_g1_i2_1 | 1.11 | 0.00184 | Up | transcription factor CSL | |
| miR-34 | Up | TRINITY_DN17532_c0_g1_i15_2 | -1.68 | 0.001763 | Down | serine/threonine-protein phosphatase 6 regulatory subunit 3-like isoform X2 | |
| miR-184 | Down | TRINITY_DN11008_c0_g1_i1_2 | 2.02 | 0.000523 | Up | serine proteinase inhibitor-2 | |
| miR-6056 | Down | TRINITY_DN17573_c0_g1_i2_2 | 1.01 | 0.002449 | Up | rho GTPase-activating protein 29-like | |
| miR-278 | Up | TRINITY_DN22500_c0_g1_i1_2 | -5.65 | 0.001643 | Down | Retrovirus-related Pol polyprotein from type-2 retrotransposable element R2DM | |
| miR-34 | Up | TRINITY_DN16889_c0_g1_i8_1 | -2.38 | 3.48E-09 | Down | putative RB13-6 antigen | |
| miR-34 | Up | TRINITY_DN15034_c2_g1_i6_2 | -1.25 | 0.000187 | Down | putative phosphatidylinositol 3-kinase catalytic subunit type 3 isoform X1 | |
| miR-14 | Down | TRINITY_DN10974_c0_g1_i1_3 | 1.23 | 1.16E-06 | Up | putative CCR4-NOT transcription complex subunit 6-like isoform X1 | |
| miR-34 | Up | TRINITY_DN16554_c1_g1_i9_2 | -1.24 | 0.000996 | Down | Protein timeless | |
| miR-6056 | Down | TRINITY_DN12483_c0_g1_i1_1 | 1.20 | 0.00261 | Up | Protein Shroom4 | |
| miR-34 | Up | TRINITY_DN13269_c0_g1_i1_2 | -2.70 | 9.22E-08 | Down | Phosphatidylinositol transfer protein alpha isoform | |
| miR-1175 | Up | TRINITY_DN11567_c0_g1_i1_3 | -2.10 | 7.90E-08 | Down | nitric oxide synthase | |
| miR-34 | Up | TRINITY_DN14203_c0_g1_i1_3 | -1.63 | 3.99E-07 | Down | M-phase inducer phosphatase-like isoform X1 | |
| miR-281 | Up | TRINITY_DN12850_c0_g1_i1_2 | -1.49 | 3.42E-05 | Down | Mitochondrial carnitine/acylcarnitine carrier protein | |
| miR-34 | Up | TRINITY_DN16013_c4_g1_i3_3 | -2.84 | 9.58E-09 | Down | membrane metallo-endopeptidase-like 1 isoform X5 | |
| miR-34 | Up | TRINITY_DN9047_c1_g1_i1_3 | -7.15 | 7.15E-14 | Down | hypothetical protein C7M84_012357 | |
| miR-6056 | Down | TRINITY_DN9935_c0_g1_i2_3 | 3.73 | 2.76E-06 | Up | hypothetical protein | |
| miR-34 | Up | TRINITY_DN9224_c0_g1_i1_1 | -2.26 | 5.91E-05 | Down | hypothetical protein | |
| miR-282 | Down | TRINITY_DN21935_c0_g1_i1_3 | 1.65 | 0.000719 | Up | hypothetical protein | |
| miR-281 | Up | TRINITY_DN2457_c0_g1_i1_2 | -2.11 | 1.20E-06 | Down | hypothetical protein | |
| miR-281 | Up | TRINITY_DN16588_c1_g1_i1_3 | -2.56 | 0.000327 | Down | hypothetical protein | |
| miR-184 | Down | TRINITY_DN7236_c0_g1_i1_3 | 1.56 | 0.000374 | Up | hypothetical protein | |
| miR-14 | Down | TRINITY_DN15461_c1_g1_i6_2 | 1.96 | 0.000244 | Up | hypothetical protein | |
| miR-6056 | Down | TRINITY_DN12316_c0_g1_i3_1 | 1.28 | 0.00014 | Up | hormone receptor 96 | |
| miR-34 | Up | TRINITY_DN15475_c0_g1_i1_2 | -1.07 | 0.001777 | Down | glycogen phosphorylase, partial | |
| miR-34 | Up | TRINITY_DN14302_c0_g1_i1_3 | -9.61 | 7.69E-12 | Down | Four-domain proteases inhibitor | |
| miR-34 | Up | TRINITY_DN17968_c3_g1_i8_2 | -2.15 | 0.000903 | Down | Endothelin-converting enzyme-like 1 | |
| miR-2c | Up | TRINITY_DN15362_c0_g3_i3_1 | -4.03 | 0.000622 | Down | Cell wall-associated hydrolase | |
| miR-2b | Up | TRINITY_DN15362_c0_g3_i3_1 | -4.03 | 0.000622 | Down | Cell wall-associated hydrolase | |
| miR-307 | Up | TRINITY_DN15836_c0_g1_i14_3 | -3.45 | 5.50E-13 | Down | beta-actin | |
| miR-263b | Down | TRINITY_DN15913_c1_g2_i1_3 | 6.19 | 0.0023 | Up | Ankyrin repeat and SAM domain-containing protein 3 | |
| miR-6056 | Down | TRINITY_DN15402_c2_g1_i1_3 | 1.16 | 0.000715 | Up | probable Ras GTPase-activating protein | |
| miR-281 | Up | TRINITY_DN18250_c0_g1_i4_2 | -1.78 | 0.003144 | Down | kinase D-interacting substrate of 220 kDa B-like | |
| miR-6056 | Down | TRINITY_DN7184_c1_g1_i1_2 | 3.08 | 2.62E-06 | Up | – | |
| miR-34 | Up | TRINITY_DN12200_c0_g1_i1_3 | -1.67 | 4.90E-11 | Down | – | |
| miR-34 | Up | TRINITY_DN14442_c0_g1_i1_2 | -3.48 | 4.19E-07 | Down | – | |
| miR-307 | Up | TRINITY_DN15768_c3_g2_i3_3 | -5.22 | 1.60E-15 | Down | – | |
| miR-279c | Up | TRINITY_DN15595_c0_g1_i10_2 | -1.72 | 0.001414 | Down | – | |
The interaction networks of EVs miRNAs and mRNAs in S. paramamosain molt stages.
Discussion
Molting is one of the most essential events in the growth and development of S. paramamosain. The hemolymph of mud crabs circulates in an open circulatory system and plays an important role in facilitating the molt process. EVs carry nucleic acids around the body (Kalluri and LeBleu, 2020) and participate in various aspects of cell biology (Kalluri and LeBleu, 2020). To better understand the physiological regulation of molt, it is vital to identify the transcriptional regulatory genes and EVs miRNAs associated with the molting process.
In this study, we found that most of the immune-related genes were upregulated at the InM and PrM stages in the hemocyte transcriptome data. The AMP of crustin was significantly upregulated at the InM stage. Crustins are cysteine-rich antibacterial peptides which are widely discovered in decapoda crustaceans with a broad spectrum of antibacterial activity (Rekha et al., 2018). This result was similar to that of three AMPs (crustins, SpHyastatin, and lysozyme), which were upregulated in the InM stage in the hepatopancreas cDNA library of S. paramamosain (Xu et al., 2020). However, the AMP of scygonadin was upregulated at the PrM stage. Scygonadin was originally considered as a male-specific anionic AMP in mud crabs (Wang et al., 2007). It may also play a role in the different developmental stages of S. paramamosain from embryogenesis to maturation (Xu et al., 2011). The induced expression of AMPs acted as a mechanism of innate prophylactic immunity in Drosophila melanogaster during molting (An et al., 2012). Furthermore, we discovered that the EVs miRNA miR-34 could target ALF. The first ALF isoform was identified in the amoebocytes of the horseshoe crabs Tachypleus tridentatus and Limulus polyphemus (Tanaka et al., 1982). It could bind to lipopolysaccharide (LPS) to inhibit the coagulation cascade (Sruthy and Philip, 2021). The proPO activation system is also an vital part of the immune system in crustaceans (Cerenius and Söderhäll, 2004). We discovered several proPO transcripts significantly upregulated in the InM and PrM stages, which suggested the involvement of the proPO system in the molting process of mud crabs. Toll pathway have been well studied for its importance in innate immunity (Lemaitre and Hoffmann, 2007). Plenty of work involving crucial components of Toll pathway have been conducted in S. paramamosain (Chen et al., 2018). In our study, we discovered that the genes involved in the Toll pathway were upregulated in the InM and PrM. The actively expressed genes of immune related genes suggested a stronger immune reaction in crabs requiring protection at InM stage and during active cuticle formation at the PrM stage. However, fewer immune related genes were identified at the PoM stage in present study.
In this study, the KEGG analysis of the target genes showed that the insulin signaling pathway was the most enriched pathway in the different stages of molt. Insulin signaling regulates growth rate and the steroid hormone 20E regulates the growth period (Yuan et al., 2020), and these are considered to be the two principle factors determining final body size in insects (Buhler et al., 2018; Yuan et al., 2020). 20E is the principal steroid hormone regulating molt and metamorphosis (Yoo et al., 2021). EcR binds with 20E and directs crustacean metamorphosis by triggering a transcriptional cascade (Yoo et al., 2021). Eip74EF acts at the top of these hierarchies to coordinate the induction of target genes (Sun et al., 2004). In our study, we explored the involvement of EVs during the molting process of mud crabs. We discovered that the miR-34 and miR-14 in the EVs might modulate the Eip74EF and EcR respectively during the various molt stages. Previous studies have shown that miR-14 directly targets the mRNA of EcR (Chen and Fu, 2021). miR-34 is highly conserved in insects and has been shown to interact with the 3’ UTR of the Eip74EF mRNA through the predicted binding sites. miR-34 has been considered a key miRNA in the integration of age-associated physiology (Liu et al., 2012). Moreover, the results of our analysis of the miRNA-mRNA interactions, besides showing the presence of miR-14 and miR-34, showed that miR-281 was also related to EcR. It has been shown in Bombyx mori that the expression of miR-281 is suppressed by treatment with 20E and that miR-281 participates in developmental regulation through the suppression of EcR expression (Jiang et al., 2013). Our results showed that the miRNAs carried in the hemolymph EVs of S. paramamosain function in regulating the molting process not only through the insulin signaling pathway but also through ecdysone signaling during the mud crab molt cycle.
Conclusions
In conclusion, a stronger immune reaction at the susceptible PoM and InM stages of molt was found in order to protect mud crabs S. paramamosain from infection, and that cuticle formation genes were more active at the PrM stage. This is the first study of EVs miRNAs and their interactions with mRNAs to identify important candidate genes associated with, or which play, regulatory roles in the molting process. EVs miRNAs from the hemolymph of S. paramamosain mainly regulated the immune-related genes and were involved in the molting process through the mediation of the ecdysone signaling and insulin signaling pathways during the molt cycle.
Funding
This work was funded by the Youth Foundation of Guangxi Zhuang Autonomous Region (2021JJB150073); Innovation Driven Development Foundation of Guangxi (AD19245135 & AD19245161); the General Program of Guangxi Zhuang Autonomous Region (Grant No. 2021GXNSFAA220030); China Postdoctoral Scientific Foundation (2021M701798); the Fund of Hainan Provincial Key Laboratory of Tropical Maricultural Technologies (Grant No. TMTOF202104); the Guangxi Key Laboratory of Beibu Gulf Marine Biodiversity Conservation, Beibu Gulf University (Grant No. 2022KA04); Scientific Research and Technology Development Program of Guangxi (Grant No. AB16380105), the National Modern Agricultural Industry Technology System Guangxi Innovation Team (Grant No. nycytxgxcxtd-14-06).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA842705; https://www.ncbi.nlm.nih.gov/genbank/, PRJNA842845.
Ethics statement
The animal study was reviewed and approved by ethics committee of Fourth Institute of Oceanography, Ministry of Natural Resources.
Author contributions
YQ and SZ contributed to conception and design of the study. XM, YX, and XC organized the database. LH performed the statistical analysis. XM and LH wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.971648/full#supplementary-material
References
1
Abehsera S. Bentov S. Li X. Weil S. Manor R. Sagi S. et al . (2021). Genes encoding putative bicarbonate transporters as a missing molecular link between molt and mineralization in crustaceans. Sci. Rep.11 (1), 11722. doi: 10.1038/s41598-021-91155-w
2
Abehsera S. Zaccai S. Mittelman B. Glazer L. Weil S. Khalaila I. et al . (2018). CPAP3 proteins in the mineralized cuticle of a decapod crustacean. Sci. Rep.8 (1), 2430. doi: 10.1038/s41598-018-20835-x
3
An S. Dong S. Wang Q. Li S. Gilbert L. I. Stanley D. et al . (2012). Insect neuropeptide bursicon homodimers induce innate immune and stress genes during molting by activating the NF-κB transcription factor relish. PloS One7 (3), e34510. doi: 10.1371/journal.pone.0034510
4
Bartel D. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
5
Buhler K. Clements J. Winant M. Vulsteke V. Callaerts P. (2018). Growth control through regulation of insulin signalling by nutrition-activated steroid hormone in Drosophila. Development145(21), dev165654. doi: 10.1242/dev.165654
6
Cerenius L. Söderhäll K. (2004). The prophenoloxidase-activating system in invertebrates. Immunol. Rev.198, 116–126. doi: 10.1111/j.0105-2896.2004.00116.x
7
Chang E. S. Mykles D. L. (2011). Regulation of crustacean molting: A review and our perspectives. Gen. Comp. Endocrinol.172 (3), 323–330. doi: 10.1016/j.ygcen.2011.04.003
8
Chen Y. Aweya J. J. Sun W. Wei X. Gong Y. Ma H. et al . (2018). SpToll1 and SpToll2 modulate the expression of antimicrobial peptides in Scylla paramamosain. Dev. Comp. Immunol.87, 124–136. doi: 10.1016/j.dci.2018.06.008
9
Chen X. Fu J. (2021). The microRNA miR-14 regulates egg-laying by targeting EcR in honeybees (Apis mellifera). Insects12 (4), 351. doi: 10.3390/insects12040351
10
Gong Y. Kong T. Ren X. Chen J. Lin S. Zhang Y. et al . (2020). Exosome-mediated apoptosis pathway during WSSV infection in crustacean mud crab. PloS Pathog.16 (5), e1008366. doi: 10.1371/journal.ppat.1008366
11
Gong Y. Wei X. Sun W. Ren X. Chen J. Aweya J. J. et al . (2021). Exosomal miR-224 contributes to hemolymph microbiota homeostasis during bacterial infection in crustacean. PloS Pathog.17 (8), e1009837. doi: 10.1371/journal.ppat.1009837
12
Gong J. Ye H. Xie Y. Yang Y. Huang H. Li S. et al . (2015). Ecdysone receptor in the mud crab Scylla paramamosain: A possible role in promoting ovarian development. J. Endocrinol.224 (3), 273–287. doi: 10.1530/joe-14-0526
13
González M. Betancourt J. L. Rodríguez-Ramos T. Estrada M. P. Carpio Y. Ramos L. (2020). Induction of spawning in pacific white shrimp Litopenaeus vannamei (Boone 1931) by injection of its molt inhibiting hormone isoform II produced in E.coli. Aquaculture Res.51 (8), 31003108. doi: 10.1111/are.14644
14
Jiang J. Ge X. Li Z. Wang Y. Song Q. Stanley D. W. et al . (2013). MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform b in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol.43 (8), 692–700. doi: 10.1016/j.ibmb.2013.05.002
15
Kalluri R. LeBleu V. S. (2020). The biology, function, and biomedical applications of exosomes. Science367 (6478), aau6478. doi: 10.1126/science.aau6977
16
Lai Y. Jin Q. Zhu F. (2020). Differential expression of microRNAs in mud crab Scylla paramamosain in response to white spot syndrome virus (WSSV) infection. Fish shellfish Immunol.105, 1–7. doi: 10.1016/j.fsi.2020.06.055
17
Lemaitre B. Hoffmann J. (2007). The host defense of drosophila melanogaster. Annu. Rev. Immunol.25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
18
Li J. Sun J. Dong X. Geng X. Qiu G. (2019). Transcriptomic analysis of gills provides insights into the molecular basis of molting in Chinese mitten crab (Eriocheir sinensis). PeerJ7, e7182. doi: 10.7717/peerj.7182
19
Liu L. Fu Y. Xiao L. Liu X. Fang W. Wang C. (2021). iTRAQ-based quantitative proteomic analysis of the hepatopancreas in Scylla paramamosain during the molting cycle. Comp. Biochem. Physiol. Part D Genomics Proteomics40, 100870. doi: 10.1016/j.cbd.2021.100870
20
Liu N. Landreh M. Cao K. Abe M. Hendriks G. J. Kennerdell J. R. et al . (2012). The microRNA miR-34 modulates ageing and neurodegeneration in drosophila. Nature482 (7386), 519–523. doi: 10.1038/nature10810
21
Lv J. Zhang L. Liu P. Li J. (2017). Transcriptomic variation of eyestalk reveals the genes and biological processes associated with molting in Portunus trituberculatus. PloS One12 (4), e0175315. doi: 10.1371/journal.pone.0175315
22
Minh Nhut T. Mykles D. L. Elizur A. Ventura T. (2020). Ecdysis triggering hormone modulates molt behaviour in the redclaw crayfish Cherax quadricarinatus, providing a mechanistic evidence for conserved function in molt regulation across pancrustacea. Gen. Comp. Endocrinol.298, 113556. doi: 10.1016/j.ygcen.2020.113556
23
Mykles D. L. (2021). Signaling pathways that regulate the crustacean molting gland. Front. Endocrinol. (Lausanne)12. doi: 10.3389/fendo.2021.674711
24
Rekha R. Vaseeharan B. Ishwarya R. Anjugam M. Alharbi N. S. Kadaikunnan S. et al . (2018). Searching for crab-borne antimicrobial peptides: Crustin from portunus pelagicus triggers biofilm inhibition and immune responses of artemia salina against GFP tagged vibrio parahaemolyticus Dahv2. Mol. Immunol.101, 396–408. doi: 10.1016/j.molimm.2018.07.024
25
Saliminejad K. Khorram Khorshid H. R. Soleymani Fard S. Ghaffari S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol.234 (5), 5451–5465. doi: 10.1002/jcp.27486
26
Song Y. Villeneuve D. L. Toyota K. Iguchi T. Tollefsen K. E. (2017). Ecdysone receptor agonism leading to lethal molting disruption in arthropods: Review and adverse outcome pathway development. Environ. Sci. Technol.51 (8), 4142–4157. doi: 10.1021/acs.est.7b00480
27
Song J. Zhou S. (2020). Post-transcriptional regulation of insect metamorphosis and oogenesis. Cell Mol. Life Sci.77 (10), 1893–1909. doi: 10.1007/s00018-019-03361-5
28
Sruthy K. S. Philip R. (2021). Anti-lipopolysaccharide factor from crucifix crab Charybdis feriatus, cf-ALF2: Molecular cloning and functional characterization of the recombinant peptide. Probiotics Antimicrob. Proteins13 (3), 885–898. doi: 10.1007/s12602-020-09716-w
29
Strus J. Znidaršič N. Blejec A. Tušek-Žnidarič M. (2015). Crustacean cuticle: Synthesis and remodeling of a dynamic extracellular matrix during molt cycle. Microscopy Microanalysis21 (S3), 895–896. doi: 10.1017/S143192715005279
30
Su S. Munganga B. P. Tian C. Li J. Yu F. Li H. et al . (2021). Comparative analysis of the intermolt and postmolt hepatopancreas transcriptomes provides insight into the mechanisms of Procambarus clarkii molting process. Life (Basel)11 (6), 480. doi: 10.3390/life11060480
31
Sun G. Zhu J. Raikhel A. S. (2004). The early gene E74B isoform is a transcriptional activator of the ecdysteroid regulatory hierarchy in mosquito vitellogenesis. Mol. Cell Endocrinol.218 (1-2), 95–105. doi: 10.1016/j.mce.2003.12.014
32
Tanaka S. Nakamura T. Morita T. Iwanaga S. (1982). Limulus anti-LPS factor: an anticoagulant which inhibits the endotoxin mediated activation of limulus coagulation system. Biochem. Biophys. Res. Commun.105 (2), 717–723. doi: 10.1016/0006-291x(82)91493-0
33
Tian Z. Jiao C. (2019). Molt-dependent transcriptome analysis of claw muscles in Chinese mitten crab Eriocheir sinensis. Genes Genomics41 (5), 515–528. doi: 10.1007/s13258-019-00787-w
34
Vay L. L. (2001). Ecology and management of mud crab Scylla spp. Asian Fish Sci.14 (2), 101111. doi: 10.33997/j.afs.2001.14.2.001
35
Wang K. J. Huang W. S. Yang M. Chen H. Y. Bo J. Li S. J. et al . (2007). A male-specific expression gene, encodes a novel anionic antimicrobial peptide, scygonadin, in Scylla serrata. Mol. Immunol.44 (8), 1961–1968. doi: 10.1016/j.molimm.2006.09.036
36
Wang H. Wei H. Tang L. Lu J. Mu C. Wang C. (2018). Identification and characterization of miRNAs in the gills of the mud crab (Scylla paramamosain) in response to a sudden drop in salinity. BMC Genomics19, 609. doi: 10.1186/s12864-018-4981-6
37
Xu Z. Liu A. Li S. Wang G. Ye H. (2020). Hepatopancreas immune response during molt cycle in the mud crab, Scylla paramamosain. Sci. Rep.10 (1), 13102. doi: 10.1038/s41598-020-70139-2
38
Xu W. F. Qiao K. Huang S. P. Peng H. Huang W. S. Chen F. Y. et al . (2011). The expression pattern of scygonadin during the ontogenesis of Scylla paramamosain predicting its potential role in reproductive immunity. Dev. Comp. Immunol.35 (10), 1078–1090. doi: 10.1016/j.dci.2011.03.028
39
Yoo B. Kim H. Y. Chen X. Shen W. Jang J. S. Stein S. N. et al . (2021). 20-hydroxyecdysone (20E) signaling regulates amnioserosa morphogenesis during drosophila dorsal closure: EcR modulates gene expression in a complex with the AP-1 subunit, jun. Biol. Open10 (8). doi: 10.1242/bio.058605
40
Yuan D. Zhou S. Liu S. Li K. Zhao H. Long S. et al . (2020). The AMPK-PP2A axis in insect fat body is activated by 20-hydroxyecdysone to antagonize insulin/IGF signaling and restrict growth rate. Proc. Natl. Acad. Sci.117 (17), 9292–9301. doi: 10.1073/pnas.2000963117
41
Zhou Z. K. Gu W. B. Wang C. Zhou Y. L. Tu D. D. Liu Z. P. et al . (2018). Seven transcripts from the chitinase gene family of the mud crab Scylla paramamosain: Their expression profiles during development and moulting and under environmental stresses. Aquaculture Res.49 (8), 3296–3308. doi: 10.1111/are.13793
Summary
Keywords
extracellular vesicles (EVs), interaction analysis, molting, Scylla paramamosain , miRNA
Citation
Qiao Y, Ma X, Huang L, Zhong S, Xing Y and Chen X (2022) Interaction analysis of hemolymph extracellular vesicles miRNA and hemocytes mRNA reveals genes and pathways associated with molting in Scylla paramamosain. Front. Mar. Sci. 9:971648. doi: 10.3389/fmars.2022.971648
Received
06 July 2022
Accepted
10 August 2022
Published
05 September 2022
Volume
9 - 2022
Edited by
Hongyu Ma, Shantou University, China
Reviewed by
Huan Wang, Ningbo University, China; Yin Zhang, Shantou University, China
Updates
Copyright
© 2022 Qiao, Ma, Huang, Zhong, Xing and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowan Ma, bowl_88@hotmail.com
†These authors have contributed equally to this work
This article was submitted to Aquatic Physiology, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.