Abstract
Endophytic and epiphytic bacteria and fungi that live in association with macroalgae produce compounds that favor the growth of the host, being in some cases more efficient than those produced by the terrestrial microbiome. This review collects information from articles published in Scopus, ScienceDirect, PubMed, and Wiley Online Library. Articles were organized according to their antimicrobial properties, synthesis of hydrolytic enzymes, production of other bioactive compounds by bacteria and fungi, and their application. The information collected showed that bacteria and fungi associated with macroalgae have the ability to inhibit bacteria, fungi, yeasts, and protozoa that affect aquaculture, public health, and the food industry, reporting that the pyrenocines A, B, E, and S isolated from Phaeosphaeria sp. Inhibited pathogenic protozoa. Additionally, other compounds identified as alkaloids, steroids, triterpenoids, and flavonoids could act by altering the morphology and physiology of pathogenic microorganisms, which can be applied in the food, pharmaceutical, paper, chemical, textile, and cosmetic industries. In addition, these microorganisms can synthesize enzymes such as xylanase, amylase, cellulase, pectinase, agarase, lignocellulose, chitinase, gelatinase, asparaginase, glutaminase, and lipase, which can be used to reduce oxidation and enzymatic browning, improve digestibility and functionality of feed, synthesis of chitin oligomers with antimicrobial properties, bioremediation of agricultural residues and industrial effluents, and production of hydrolysates.
1 Introduction
Hosts from the marine biome are the most abundant, representing 70% of what exists on the planet (Silva et al., 2020). In addition, there are terrestrial flora, polar vegetation, and tropical forests, which live in association with endophytic microbes (Sarasan et al., 2020), or epiphytic or free-living (Darienko et al., 2021), including bacteria (Deutsch et al., 2021) and fungus (Kjer et al., 2010; Zainee et al., 2021) within the endophytes group (Cha et al., 2021). The most representative within the group of fungus are filamentous Ascomycota (Eurotiales, Capnodiales, Hypocreales, and Pleosporales) as the most numerous (Lucini et al., 2022), basimetric yeasts (Sporidiobolales), and, within the bacteria, Arcobacter, Bacteroidetes, Flavobacteriaceae, Mycoplasma, Labrenzia, Phyllobacteriaceae and Xanthomonadaceae (Hollants et al., 2011).
The presence of bacterial and fungal species as endophytes and epiphytes (Darienko et al., 2021) depends on host conditions such as the growth of the frond, the erect part of variable size and shape with an attachment organ (Pereira, 2021), types of thallus tissues, different heights of the tide, different stages of the life cycle (Bonthond et al., 2022), or the high rate of growth and sporulation of the host (Kamat et al., 2020), as well as geographical differences (Chen et al., 2022), geographic location (Ismail et al., 2016), multiple abiotic parameters (temperature, light, nutrient pollution, salinity, and pH) (Pereira et al., 2017; Deutsch et al., 2021; Wong Chin et al., 2021; Lucini et al., 2022), and anthropomorphic influences (Flewelling et al., 2015) (Figure 1). The latter generates a condition of fragility, favoring the presence of microorganisms in the cell wall, although the endophyte penetration mechanism is unknown (Pereira et al., 2017).
Figure 1
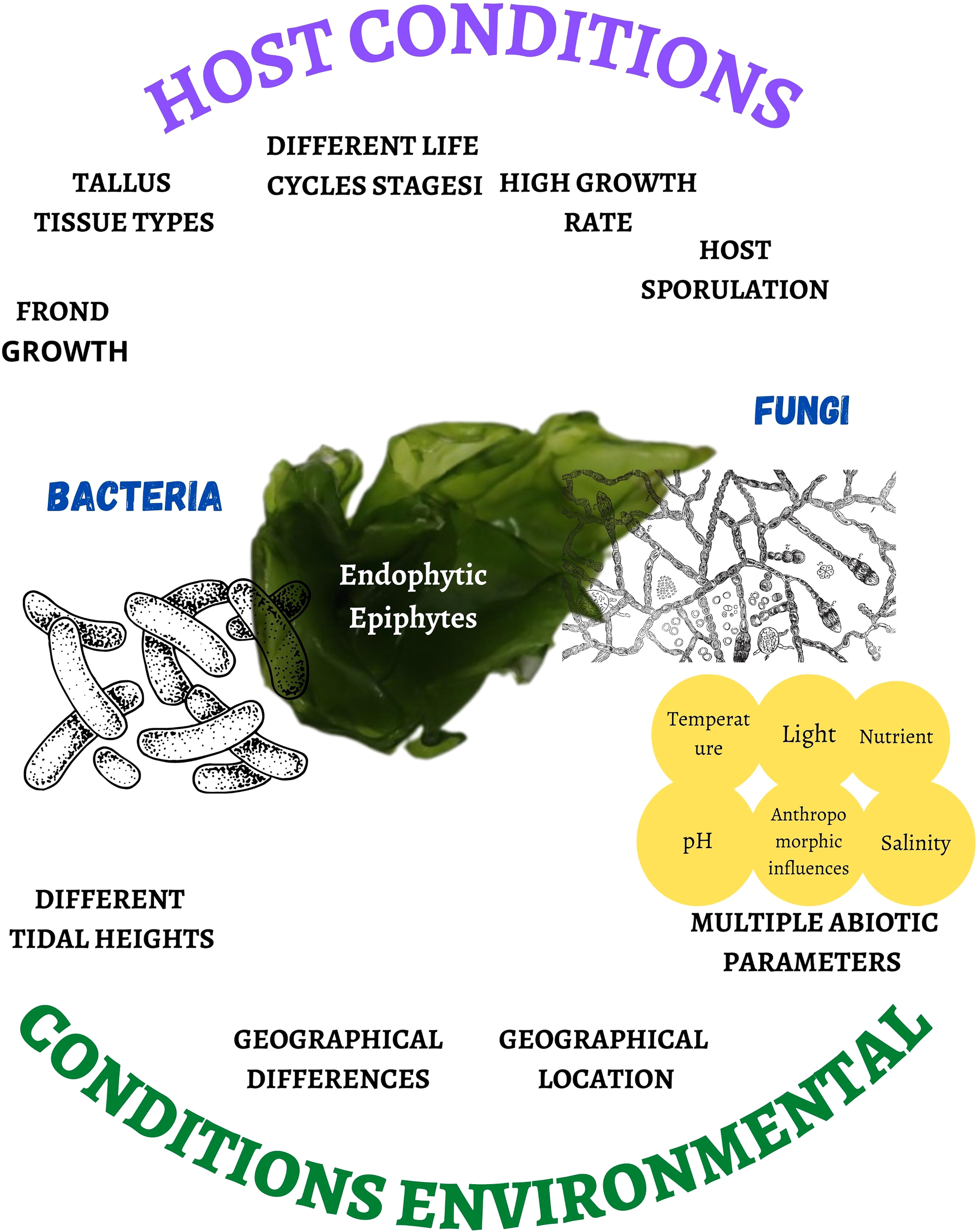
Host and environmental conditions that affect the proliferation rate and biodiversity of bacteria and fungi associated with marine macroalgae.
This association and understanding of intercellular chemical signaling pathways between macroalgae and associated bacteria and fungi (Menaa et al., 2020) allows macroalgae to be protected from marine pathogens during the stages of wound repair and from susceptible protoplasts through the production of highly bioactive metabolites (Deutsch et al., 2021), such as pyrenocines (Vallet et al., 2018), specific vitamins (Villarreal-Gómez et al., 2010), or others, which also aid to withstand high temperatures and high minerals concentrations (Deutsch et al., 2021). These improve the ability of microalgae to tolerate stress (Gonçalves et al., 2020; Wong Chin et al., 2021), allow the exchange of nutrients via photosynthetic pathway (Handayani et al., 2019; Cha et al., 2021), or induce the synthesis of molecules to increase the growth and development of the host macroalgae (Wichard and Beemelmanns, 2018).
Additionally, microalgae are involved in the degradation of cellulose, lignin, and polymeric organic matter within oxic and anoxic sediments, participating in the carbon cycle in marine environments (Perkins et al., 2021) or degrading chitin as an abundant resource, as it forms part of the shells of marine crustaceans (Venkatachalam et al., 2015), or breaking down proteins. Microalgae also use part of their wide variety of enzymes (Suryanarayanan et al., 2012) and metabolites, which they produce even under adverse conditions (Wong Chin et al., 2021), that are novel and structurally unique (Manomi et al., 2015). These functional properties are more efficient than the terrestrial microbiome (Patyshakuliyeva et al., 2020). It is also known that healthy macroalgae release part of the carbon fixed during photosynthesis as a secretion called mucilage, which is rich in substances of interest that act as an attraction factor for colonizing microorganisms, such as fungi (Teixeira et al., 2019b). However, some of the endophytes and epiphytes are harmful (Aumack et al., 2011).
This great variety of metabolic pathways confers adaptation and survival to the habitat (Bogas et al., 2022), performing an important role in the biological processes of its hosts (Liu et al., 2022b), which can be understood by metaomic technologies (Singh and Reddy, 2016), such as transcriptomic, proteomic, and genomic analysis (Balabanova et al., 2018), in order to have an understanding of the functionality of proteins and enzymes for future applications in different fields of study. These functional properties make macroalgal endophytes a promising new source of biologically active metabolites (Kjer et al., 2010; Espinoza et al., 2021; Cooper and Walker, 2022) and unique in hydrolytic enzymes (Sarasan et al., 2020). They are very important in biotechnological use (Espinoza et al., 2021), by meeting the demands of the pharmaceutical industry (Sarasan et al., 2017). They can also be used to combat infections such as SARS, dengue, and Zika virus (Deshmukh et al., 2018), as new natural antibiotic compounds (Zainee et al., 2021), in anticancer treatments using chrysin production (Kamat et al., 2020), as an apoptotic agent (Sahoo et al., 2021), and they have anti-inflammatory and antioxidant capacities (Teixeira et al., 2019a).
This review discloses the antimicrobial properties of bacteria and fungi associated with marine macroalgae against pathogenic bacteria, fungi, and protozoa or food spoilers. In addition, the production of enzymes such as agarolytic, cellulase, ligninase, chitinase, and others is discussed, as well as their applications.
2 Bacteria associated with macroalgae
2.1 Antimicrobial properties
Currently, the demand for food without synthetic preservatives and the demand for use of natural products has led to studies focusing on different sources that allow food to be preserved from deterioration as part of food safety and quality (Arshad and Batool, 2017). Given this situation, plants are the main source of antimicrobials as part of the terrestrial biome; however, the search for beneficial microorganisms that can produce antibiotics, organic acids, or bacteriocins (Cesa-Luna et al., 2020) are considered untapped reserves of promising biomolecules (Danquah et al., 2022), especially bacteria that are associated with macroalgae as part of the marine biome.
Table 1 summarizes the information that has been reported for the last 7 years, which shows the sampling site, the host macroalgae, the isolated strains, the codes access referring to these sequences, the extracts used, and the microorganisms inhibited. The most important information from the results has been reported, and 50% of the studies were carried out in India and the other 50% in Israel, Egypt, and Morocco.
Table 1
| Sampling site | Host macroalgae | Isolated strains | Access code | Extract solvent | Bioactive compounds: Application | Inhibited microorganisms | Reference |
|---|---|---|---|---|---|---|---|
| Inhibition of pathogenic bacteria | |||||||
| Marine and freshwater sources, Israel | Jania Rubens | Bacillus safensis | NR | SE | NR | Photobacterium damselae Streptococcus iniae Aeromonas salmonicida | (Deutsch et al., 2021) |
| Laurencia papillosa | Bacillus safensis | NR | |||||
| Bacillus safensis | MT186284 | ||||||
| NR | |||||||
| Padina pavonica | Bacillus pumilus | MT116792 | |||||
| MT186599 | |||||||
| Gracilaria sp. | Bacillus safensis | MT232982 | |||||
| Mandapam, Gulf of Mannar, India | Ulva reticulata | Pseudomonas stutzeri | NR | Hexane | phenol, 3, 5-bis (1, 1-dimethylethyl): NT | Staphylococcus aureus | (Dhanya et al., 2016) |
| Escherichia coli | |||||||
| Egyptian Red Sea, El Quseir, Egypt | Cystoseira myrica | Catenococcus thiocycli | OK584768 | Methanolics | Heptacosane; 2-methyloctacosane: NT |
Escherichia coli Salmonella typhi Klebsiella pneumoniae Micrococcus luteus Proteus mirabilis | (Hagaggi and Abdul-Raouf, 2022) |
| Indian Peninsula and Arabian Sea, India | Hypnea valentiae |
Shewanella algae
Bacillus amyloliquefacien |
MH157093 MH157092 |
Ethyl acetate | NR | Methicillin resistant Staphylococcus aureus (MRSA) Vancomicin resistant Enterococcus faecalis |
(Kizhakkekalam and Chakraborty, 2020) |
| Northwestern Atlantic coast of Morocco, southwest of El Jadida, Morocco | Carpodesmia tamariscifolia | KC179a,b Streptomyces albidoflavusc |
NR MW555177 |
Butanolica Hexaneb |
Desferrioxamina Bc Desferrioxamina B2c: NT |
Methicilin resistant Staphylococcus aureus (MRSA)b,c Acinetobacter baumanniia,c Pseudomonas aeruginosac |
(Katif et al., 2022) |
| Someshwar beach, located in Mangalore, district of Dakshina Kannada, Karnataka, India | Ulva lactuca | Bacillus subtilisa | NR | Chloroform Ethyl acetate |
Alkaloidsa Esteroidsa,b Triterpenoidsa,b Flavonoidsa,b: NT |
Enterococcus faecalis Klebesella pneumoniae | (Habbu et al., 2016) |
| Enterobacter cloacaeb | NR | ||||||
| Inhibition of pathogenic fungi | |||||||
| Egyptian Red Sea, El Quseir, Egypt | Cystoseira myrica | Catenococcus thiocycli | OK584768 | Ethyl acetate | 2-methyloctacosane | Aspergillus niger Candida albicans | (Hagaggi and Abdul-Raouf, 2022) |
| Someshwar beach, located in Mangalore, district of Dakshina Kannada, Karnataka, India | Ulva lactuca |
Bacillus subtilisa
Enterobacter cloacaeb |
NR NR |
Chloroform Ethyl acetate |
Triterpenoidsa,b Flavonoidsa,b Esteroidsb: NT |
Aspergillus nigera
Aspergillus flavusb Candida albicansa,b |
(Habbu et al., 2016) |
| Inhibition of pathogenic protozoa | |||||||
| Marine and freshwater sources, Israel | Jania Rubens | Bacillus velezensis | MT187620 | SE | NR | Saprolegnia parasítica | (Deutsch et al., 2021) |
| Bacillus subtilis | MT192301 | ||||||
| Bryopsis plumosa | Bacillus subtilis | MT187638 | |||||
| Pithophora spp. | Pseudomonas alcaligenes | MT186697 | |||||
| Ulva sp. | Bacillus velezensis | MT178235 | |||||
| Enteromorpha ralfsii | Bacillus subtilis | MN134034 | |||||
| Bacillus pumilus | MT272978 | ||||||
Bacteria associated with macroalgae and their inhibition capacity against pathogenic microorganisms.
*NR, Not reported; NE, No extract; NT, Not tested; a,b,c: Endophytic species related to the solvent, bioactive compound, and pathogen.
2.1.1 Bioactive compounds and mechanism of action.
Dhanya et al. (2016), observed that P. stutzeri presented a high inhibitory capacity on S. aureus and E. coli. This inhibitory action may be related to phenol, 3, 5-bis (1, 1-dimethylethyl), which has been evidenced by Abdullah et al. (2011), where this partially lipophilic phenolic compound could cross the cell membrane by passive diffusion, acidifying the cytoplasm, causing alteration of the bacterial membrane, generating the escape of essential intracellular constituents, denaturing the proteins (Campos et al., 2009; Ecevit et al., 2022), and causing cell death (Ecevit et al., 2022) by bacterial lysis (Ochieng et al., 2023). Furthermore, the presence of phenolic compounds synthesized by these endophytic bacteria exerts an inhibitory effect on the biosynthesis of nucleic acids and on their metabolic processes (Flewelling et al., 2015).
Hagaggi and Abdul-Raouf (2022) mentioned that heptacosane and 2-methyloctacosane were the major components detected in C. thiocycli, being the first related to antibacterial activity (Magina et al., 2009) and the second to antimicrobial (Res, 2018). Bankeu et al. (2018) showed that heptacosane moderately inhibits ureases that confer pathogenicity to Helicobacter pylori (Rego et al., 2018). 2-methyloctacosane, which is a volatile compound (Sharon P. et al., 2021) and a cyclic alkane (Amin et al., 2022) of the octacosane group, could alter membrane permeability, inhibit enzymes, and perturb bacterial morphology, which in some cases can cause irreversible damage by triggering the secretion of proteins and genetic material (Magina et al., 2009).
Kizhakkekalam and Chakraborty (2020) showed that S. algae and B. amyloliquefaciens inhibited methicillin resistant S. aureus (MRSA) and vancomycin resistant Enterococcus faecalis, due to the destabilization of the cell membrane permeability (Le and Desbois, 2017) because of the presence of polyketides and lipopeptides (Aleti et al., 2015) synthesized from pks-1 and nrps genes (Kizhakkekalam and Chakraborty, 2020). In addition, lipopeptides as antibiotic substances increase the amphipath city of calcium ions forming daptomycin micelles upon interaction with membranes, causing solute secretion in Gram positive bacteria (Gray and Wenzel, 2020).
The inhibition A. baumannii and P. aeruginosa by Streptomyces albidoflavus (Katif et al., 2022) is probably due to a local increase in pluronic micelles on the surface of Gram-negative bacteria, causing an alteration in membrane permeability (Purro et al., 2018), due to the presence of desferrioxamine B and B2 (Katif et al., 2022), a partial structure of butyldiamine that acts as an iron chelator (Purro et al., 2018).
Habbu et al. (2016) demonstrated inhibitory action of B. subtilis and E. cloacae on E. faecalis, K. pneumoniae, and C. albicans; regarding molds, B. subtilis inhibited Aspergillus niger and E. cloacae inhibited A. flavus. Phytochemical investigations of the two endophytes, when fractionated, revealed the presence of alkaloids, steroids, triterpenoids, and flavonoids attributed to antimicrobial activity, although their mechanism of action has not been detailed for each compound identified.
Regardless of the compounds mentioned, endophytic bacteria can produce lytic enzymes (chitinases) or volatile compounds that exert antimicrobial action (Figure 2); these endophytic and pathogenic bacteria can alter the pH and compete with each other for space and nutrients causing indirect inhibition of the pathogen (Ortiz-Ojeda et al., 2020).
Figure 2

Mechanisms of action of bioactive compounds synthesized by bacteria associated with macroalgae and their inhibition capacity in pathogenic bacteria, fungi, and protozoa or food spoiler.
2.2 Hydrolytic enzymes
Regarding the potential for polymeric hydrolysis of epiphytic bacteria (Alteromonas, Pseudoalteromonas, Psychrobacter, Shewanella, and Vibrio sp.; Brevundimonas, Paracoccus, Roseomonas, and Sphingomonas sp.; Bacillus, Staphylococcus, and Alkalihalobacillus sp.; Cellulomonas, Cellulosimicrobium, Isoptericola, Kocuria, Micrococcus, and Streptomyces sp.) isolated from macroalgae (Ulva sp., Sargassum polycystum, Pterocladia musciformis, Dictyota sp., and Padina antillarum) from the central west coast of India, an enzyme activity of 61.3%, 59.7%, 58.8%, and 52.2% on xylan, starch, cellulose, and pectin, respectively, was shown (Kumar et al., 2022).
Additionally, Sánchez Hinojosa et al. (2018) demonstrated that epiphytic bacteria Cellulophaga sp., Colwellia sp., Lacinutrix sp., Olleya sp., Paraglaciecola sp., Pseudoalteromonas sp., and Winogradskyella sp. isolated from Himantothallus grandifolius, Pantoneura plocamioides, and Plocamium cartilagineum originating from different parts of the Rey Jorge island, Antarctic Peninsula presented a high agarolytic activity at 20°C; however the isolates Olleya sp. and different species of Pseudoalteromonas showed the highest activities at low temperature (4°C), demonstrating their potential as biocatalysts for industrial biotechnological applications.
2.3 Production of other bioactive compounds
In another study, Harikrishnan et al. (2021) identified compounds such as 1H-Indole, 3-ethyl-2-(2-pyridyl)-, 2H-Benzo(d,E)isoquinoline-1,3(1H,3H)-dione, 2-(2-acetoxyethyl)- 6-(4-morpholyl)-, arsenic acid, tris(trimethylsilyl), ester, Thiocarbamic acid, N,N-dimethyl,S-1,3-diphenyl-2-butenyl-ester, Phenol, 2,5-bis(1,1- dimethylethyl)- and Tris(tert-butyldimethylsiloxy)arsane from Pichia kudriavzevii, Cystobasidium minutum, Grammothele fuligo, Candida railenensis, and Rigidoporus vinctus isolated from macroalgae Chaetomorpha, Rhizoclonium, Undaria, Ulva, and Enteromorpha. Even though the bioactivity of these compounds is unknown, it could be related to their antibacterial activity and antioxidant capacity.
3 Fungi associated with macroalgae
3.1 Antimicrobial properties
Resistance to antimicrobial compounds by pathogenic microbes has had a major impact on food production worldwide, boosting a constant need for new discoveries, where marine fungi have been identified as a novel source of natural fungal products (Jakubczyk and Dussart, 2020). However, their synthesis on a commercial scale must be achieved by improving the potentiality of the strain, optimizing growth conditions, and incorporating omic techniques, to be offered as suitable substitutes for ineffective and expensive antimicrobials (Rani et al., 2021).
Table 2 compiles information from the last 10 years including the sampling site, host macroalgae, isolated strains, codes access referring to these sequences, extract used, and microorganisms inhibited from the main reported results. It can be observed that 33.3% of the studies were carried out in India, followed by Indonesia (22%) and other countries such as Malaysia, Brazil, Canada, Scotland and France. Green macroalgae were collected in the highest proportion for isolation, and the presence of associated fungi of the species was identified.
Table 2
| Sampling site | Host macroalgae | Isolated strains | Access code | Extract solvent | Bioactive compounds: Application | Inhibited microorganisms | Reference |
|---|---|---|---|---|---|---|---|
| Inhibition of pathogenic bacteria | |||||||
| Rocky coastline of Konkan and Goa, India | Chaetomorpha sp. | Chaetomium globosuma | MH645800.1 | Ethyl acetate | Azelaic acid, azetidine and furo-pyranse: NT |
Escherichia colia,b,c,d,e
Staphylococcus aureusb,c,d Pseudomona aeruginosaa,b,c,d,e Xantomonas campestrisa,b,c,d,e Bacillus subtilisb,c,d Bacillus megateriumb,c,d |
(Kamat et al., 2020) |
| Chaetomorpha sp. | Aspergillus unguis | MH654996b | |||||
| MH654997c | |||||||
| MH655006d | |||||||
| MH655000e | |||||||
| Nirvana beach, Padang, West Sumatra, Indonesia |
Padina sp. | Trichoderma harzianuma | NR | Ethyl acetate | NR |
Escherichia coli
a
Staphylococcus aureus a |
(Handayani et al., 2019) |
| Kovalam coast, Trivandrum, Kerala, India |
Sargassum wightii
Caulerpa peltata |
Aspergillus flavus a | MH469530 | Raw fungal extract | NR |
Escherichia coli
a,b,c,d,e
Edwardsiella tarda a,b,c,d,e Pseudomonas aeruginosa a,b,c,d,e Aeromonas hydrophila a,b,c,d,e Vibrio cholera a,b,c,d,e Vibrio parahaemolyticus a,b,c,d,e Vibrio alginolyticus a,b,c,d,e Vibrio harveyi a,b,c,d,e Vibrio proteolyticcus a,b,c,d,e Bacillus cereus c,d,e Staphylococcus aureus c,d,e |
(Sarasan et al., 2020) |
|
Sargassum wightii
Dictyota dichotoma Halymenia venusta Grateloupia lithophila |
Penicillium chrysogenumb | MH469504 | |||||
| Hypnea musiformis | Aspergillus terreusc | MH469513 | |||||
| Chaetomorpha antennina | Aspergillus versicolor d | MH469523 MH469532 |
|||||
|
Sargassum wightii
Hypnea musiformis |
Hortea werneckii e | MH469501 | |||||
| Munambam beach, Cochin and Varkala beach, Trivandrum, Kerala, India | Rhizoclonium | Cystobasidium minutuma | MT372471 | Ethyl acetate | Phenol, 2,4-bis(1,1-dimethylethyl)-; Arsenous acid, tris(trimethylsilyl) ester; silicic acid, diethyl bis(trimethylsil) ester and butylated hydroxytoluene a,c,c,d,e: NT | Pseudomona aeruginosa a,b,c,d | (Harikrishnan et al., 2021) |
| Ulva | Candida railenensisb | MK920215 | |||||
| Enteromorpha | Rigidoporus vinctusc | MK920214 | |||||
| Undaria | Grammothele fuligod | MT372472 | |||||
| Chaetomorpha | Pichia kudriavzeviie | MT372473 | |||||
| Johor coast (Pulau Merambong, Nusajaya, Tanjung Dato, and Tanjung Lompa), Malaysia | Sargassum polycystum | Trichoderma viride a | NR | NE | NR |
Staphylococcus aureus
a,b
Shigella sp.a,b Escherichia colia,b Serratia marcescensa,b Salmonella typhia,b Klebsiella pneumoniaea |
(Zainee et al., 2021) |
| Ulva Lactuca | Pythium sp.b | ||||||
| Rocky coastlines of Praia Dura, Ubatuba, São Paulo, Brazil | Bostrychia tenella | Penicillium waksmanii a | NR | n-hexane fraction | Cytochalasin Dc: NT |
Staphylococcus aureus
a,b,c
Klebsiella pneumoniae a,c |
(De Felício et al., 2015) |
|
Acremonium implicatum
b
Xylaria sp.c |
|||||||
| L’Etete, New Brunswick, Canada | Devaleraea ramentacea | Aureobasidium pullulans a | NR | Methanol | NR |
Staphylococcus aureus
a,b,c,d,e,f
Pseudomonas aeruginosa a |
(Flewelling et al., 2013) |
| Botrytis sp.b | NR | ||||||
| Palmaria palmata | Penicillium crustosum c | KF572147 | |||||
| Botrytis sp.d | NR | ||||||
| Polysiphonia lanosa | Botryotinia fuckeliana e | KF572148 | |||||
| Ascophyllum nodosum | Aspergillus sp.f | NR | |||||
| Indonesia | Euchema sp. | Aspergillus elegans | NR | Ethyl acetate | (+)-Epi-Epoformin: NT |
Escherichia coli
Staphylococcus aureus Bacillus subtilis |
(Rajamani et al., 2019) |
| Inhibition of pathogenic fungi | |||||||
| Nirvana beach, Padang, West Sumatra, Indonesia |
Padina sp. | Trichoderma harzianum | NR | Ethyl acetate | NR | Candida albicans | (Handayani et al., 2019) |
| NT9 | |||||||
| Kovalam coast, Trivandrum, Kerala, India | Hypnea musiformis | Aspergillus terreus | MH469513 | Fungal crude extract | NR |
Aspergillus fumigatus
A. flavus Candida albicans C. tropicalis C. duobushaemulonii |
(Sarasan et al., 2020) |
| Chaetomorpha antennina | Aspergillus versicolor | MH469523 MH469532 | |||||
| Sargassum wightii and Hypnea musiformis | Hortea werneckii | MH469501 | |||||
| Rocky coastlines of Praia Dura, Ubatuba, São Paulo, Brazil | Bostrychia tenella |
Penicillium waksmaniia
Penicillium decaturenseb |
NR | n-hexanea Ethyl acetateb |
NR |
Cladosporium cladosporioide
a,b
Cladosporium sphaerospermum a,b |
(De Felício et al., 2015) |
| L’Etete, New Brunswick, Canadá | Devaleraea ramentacea | Penicillium decumbens | KF572138 | Methanol | NR | Candida albicans | (Flewelling et al., 2013) |
| Inhibition of pathogenic protozoa | |||||||
| Scotland and France |
Ascophyllum nodosum
Pelvetia canaliculata Laminaria digitata Saccharina latissima |
Penicillium janczewskii a | MH397623-MH397663 | Ethyl acetate | Pyrenocines (A y B), (S) and (E)b |
Eurychasma dicksonii
a,b,c,d,e,f
Anisolpidium ectocarpii a,b,c,d,e Maullinia ectocarpii a,b,c,d,e |
(Vallet et al., 2018) |
| Phaeosphaeria sp.b | |||||||
| Chaetomium globosum c | |||||||
| Chaetomium globosum d | |||||||
| Phoma exigua e | |||||||
| Paradendryphiella arenaria f | |||||||
Fungi associated with macroalgae and their inhibition capacity against pathogenic microorganisms.
*NR, Not reported; NE, No extract; NT, Not tested; a,b,c,d,e,f, Endophytic species related to the solvent, bioactive compound, and pathogen.
3.1.1 Bioactive compounds and mechanism of action
E. coli, S. aureus, P. aeruginosa, X. campestris, B. subtilis, and B. megaterium were inhibited by C. globosum and A. unguis, and some compounds such as azelaic acid, azetidine, and the furo-pyrans were identified in A. unguis (Kamat et al., 2020). In another study, azelaic acid showed antibiotic action on Propionibacterium acnes (Park et al., 2018); however, its mechanisms of action at the cellular level are unknown. Azitidine, as an antibacterial compound, could cause inhibitory action on protein synthesis and bacterial 50S RNA target molecules (O’Dowd et al., 2008). Furo-pyrans could act by binding to the phosphate group (negative ions) of the phospholipids of the membrane and cell wall, causing an inhibition of lysosomal phospholipase activities due to its neutralization of negative charges (Magesh et al., 2004).
Harikrishnan et al. (2021) showed that P. aeruginosa was inhibited from ethyl acetate extracts of Cystobasidium minutum, Candida railenensis, Rigidoporus vinctus, Grammothele fuligo, and Pichia kudriavzevii, in which Phenol, 2,4-bis(1,1-dimethylethyl)- may be one of the causes of the decrease in reactive oxygen species (ROS) by pumping intracellular protons, acidifying the medium and causing the inhibition of pathogens (Romero-Correa et al., 2014). It could also act, hindering the production of biofilms (Padmavathi et al., 2014) by a bacterial signaling pathway that controls physiological mechanisms called quorum sensing (Samanta et al., 2011). In addition, silicic acid (diethyl bis(trimethylsilyl) ester) identified by Harikrishnan et al. (2021) in a phospholipid compound could act by interacting with the negatively charged compounds of the membranes of pathogenic microbes, altering their permeability and inhibiting their cell wall, nucleic acids, proteins, and enzymes (Abd El Baky et al., 2014); while with arsenous acid (tris(trimethylsilyl) ester) and butylated hydroxytoluene identified by Harikrishnan et al. (2021), an inhibitory mechanism at the cellular level is not reported.
S. aureus and Klebsiella pneumoniaea were inhibited by Penicillium waksmanii, Acremonium implicatum, and Xylaria sp. Cytochalasin D identified in Xylaria sp. (De Felício et al., 2015) probably intervenes in the transduction of signals by the protein tyrosine kinase, and it can affect the actin filaments that form part of the structure of the bacterial cytoskeleton (Akeda et al., 1997) and a decrease in DNA synthesis (Rubtsova et al., 1998). In another study, this compound can inhibit the polymerization of the formed actin contractile rings to be formed on the fungal wall (Gabriel et al., 1998).
Rajamani et al. (2019) showed that A. elegans inhibited E. coli, S. aureus, and B. subtilis, possibly due to chemical modifications in its structure, synthesizing biocompatibility derivatives such as antimicrobial properties, causing a reduction in the formation of biofilms, and generating susceptibility to antibiotics (Roscetto et al., 2020), due to (+)-Epi-Epoformin identified by Rajamani et al. (2019). Additionally, this compound inhibits the germination of Puccinia sp. and Uromyces sp., preventing its penetration into the host (Barilli et al., 2017).
Vallet et al. (2018) showed that pyrenocin A and B; E; and S compounds from C. globosum inhibited E. dicksonii, A. ectocarpii, and M. ectocarpii; E. dicksonii and M. ectocarpii; and E. dicksonii and A. ectocarpii, respectively. Pyrenokines A and B cause an apoptotic process without permeabilization of the plasma membrane, generating a depolarization of the mitochondrial electrochemical gradient (Chan-Bacab et al., 2021). Regarding pyrenocines E and S, their inhibitory mechanism is not clear.
It has been shown that the production of some antifungal compounds were effective when these strains were co-cultivated with marine bacteria (Cueto et al., 2001). Although bioactive compounds are now being sought to act solely as antifungal agents without any detrimental effect on bacteria (or vice versa), they could become a promising source of specific antimicrobial substances (Sarasan et al., 2020) (Figure 3).
Figure 3
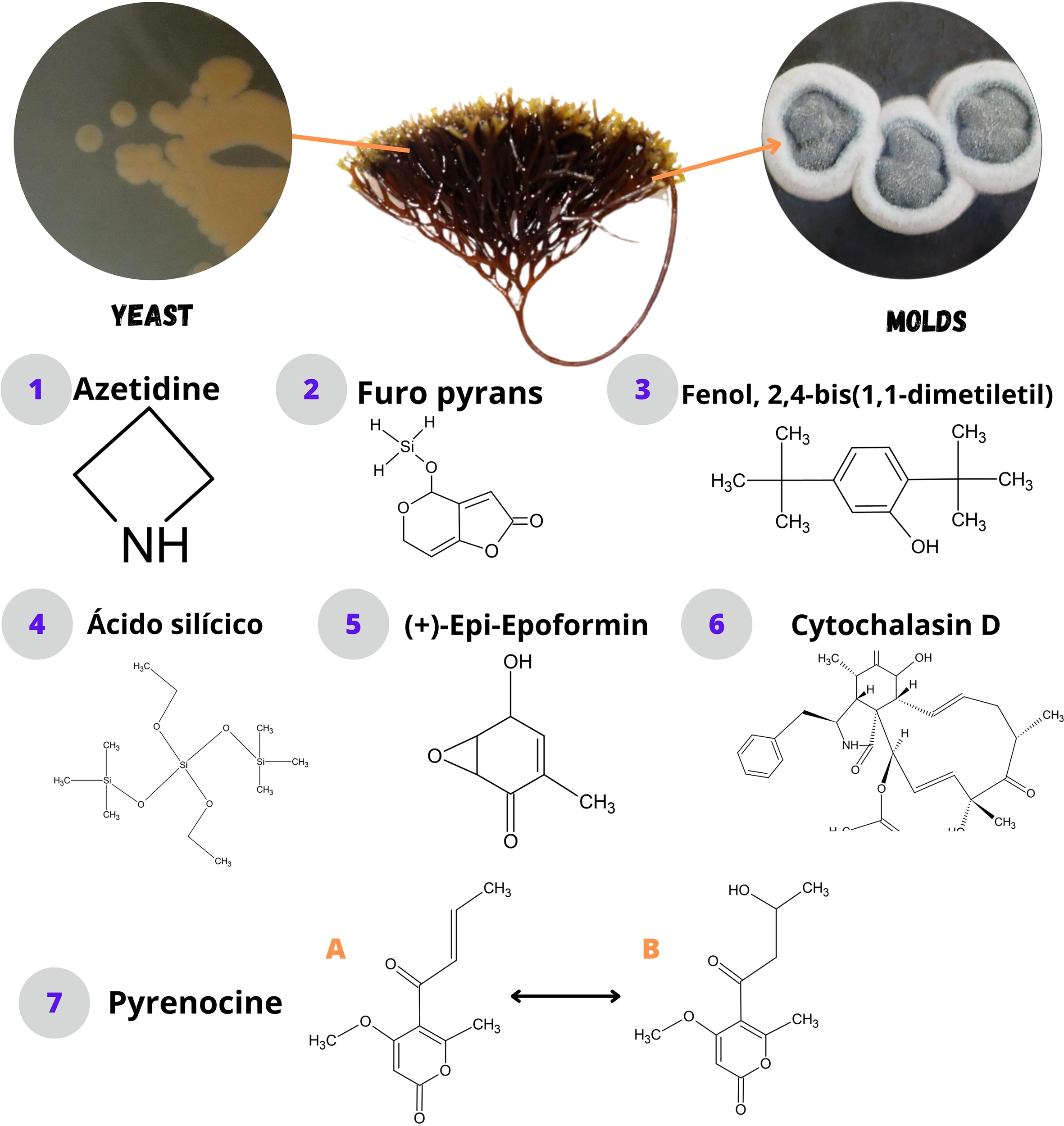
Bioactive compounds synthesized by fungi associated with macroalgae.
Moreover, it was found that a high percentage of fungi associated with macroalgae have the ability to inhibit pathogenic microbes; however there were genera that did not have the expected effect, possibly because these pathogens were resistant due to modifications of the target site, enzymatic activation, or decreased intracellular drug accumulations (Mathan et al., 2013). Even though there are more reports of studies on fungi associated with macroalgae, the mechanisms of action at cellular level of the identified bioactive compounds are still not clear.
It is likely that antimicrobial mechanisms associated to endophytic organisms could be linked to an inhibition of cell wall synthesis, alteration of plasma membrane function (Xu et al., 2019; Hashem et al., 2022), disruption of mitochondrial function (Xu et al., 2019), interference with nucleic acid or protein synthesis, or induction of oxidative stress, highlighting that each mechanism may depend on the synthesized metabolite (Tijith K. et al., 2019) (Figure 4).
Figure 4
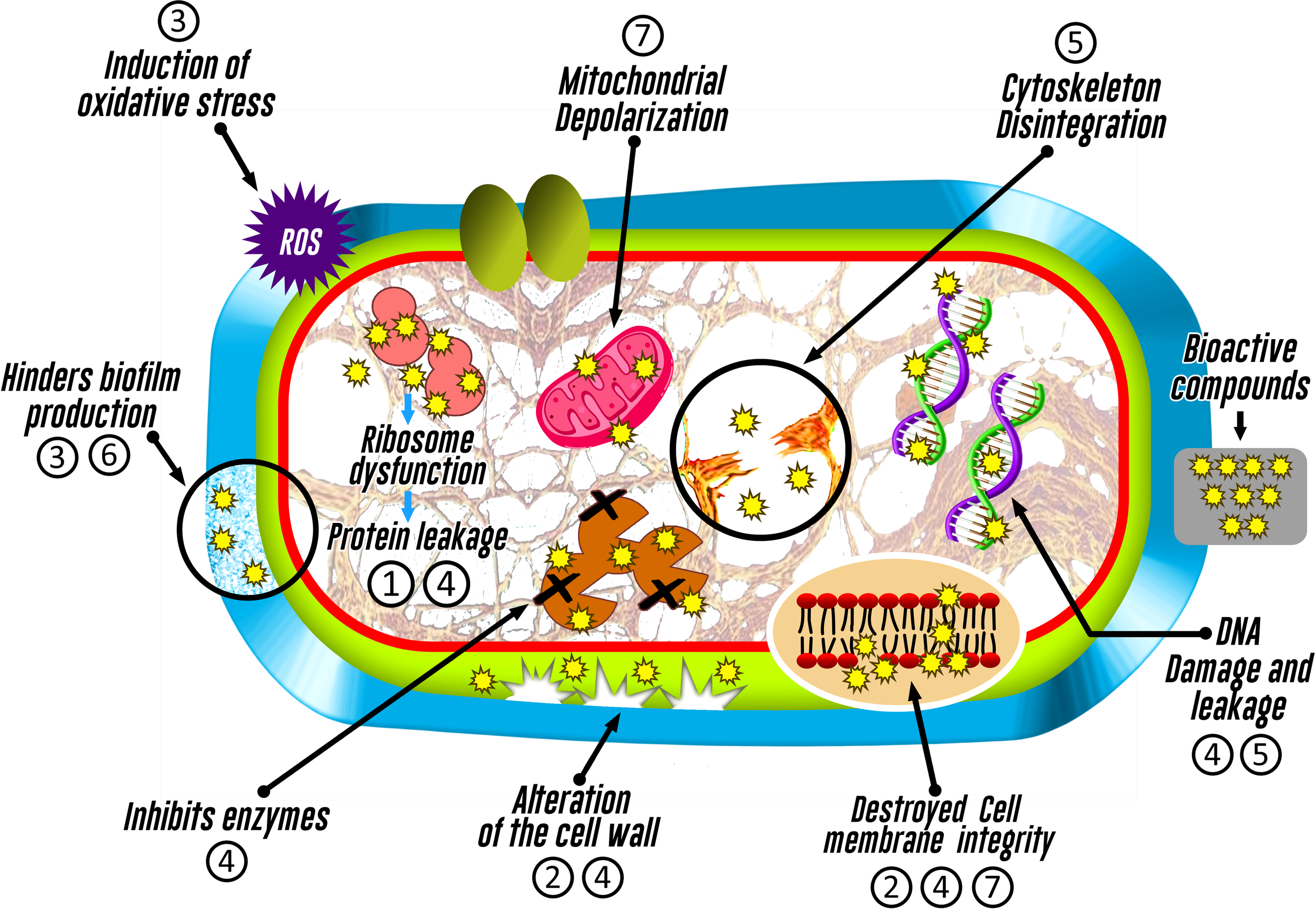
Inhibitory action mechanisms of bioactive compounds identified in fungi associated with macroalgae on pathogenic microorganisms. The numbering represents the bioactive compound (1: Azetidine; 2: Furo-pyrans; 3: Phenol, 2,4-bis(1,1-dimethylethyl)-; 4: Silicic acid; 5: (+)-Epi-Epoformin; 6: Cytochalasin D; and 7: Pyrenocine).
3.2 Hydrolytic enzymes
3.2.1 Cellulolytic activity
Aspergillus proliferans, Epicoccum sorghinum, Cladosporium sp., Penicillium corylophilum, Penicillium sp., Penicillium fagi, and Mucor circinelloides originating from Ecklonia radiata had a high degradation of cellulose a week after being evaluated; for Truncospora tephropora the degradation only occurred in the third week, decreasing its activity after 6 weeks under oxic conditions (Perkins et al., 2021). Sarasan et al. (2021) also reported that of the 133 endophytic fungi evaluated and isolated from different algae (Sargassum wightii, Caulerpa peltata, U. fasciata, Dictyota dichotoma, Halymenia venusta, Hypnea musiformis, Grateloupia lithophila, Enteromorpha compressa, Ceramium diaphanum, Padina tetrastromatica, and Chaetomorpha antennina), only 37.6% of the endophytes presented cellulolytic activity.
In spite of its wide application in the paper industry, waste plant matter degradation (Balabanova et al., 2018), and the food industry (Kumar et al., 2018), is it is clear that marine fungi have a low cellulolytic capacity (Balabanova et al., 2018) (Patyshakuliyeva et al., 2020) compared to terrestrial strains, possibly due to the 2–10% content in cellulose and hemicellulose in seaweed (Balabanova et al., 2018).
3.2.2 Lignin degradation
E. sorghinum, Cladosporium sp., T. tephropora, Ganoderma sessile, and Pleurotus ostreatus isolated from E. radiata showed high degradation of lignin in the third week of evaluation under oxic conditions. However, M. circinelloides is one of the endophytes that produce more lignin-degrading enzymes under anoxic conditions (Perkins et al., 2021).
3.2.3 Chitinases production
Chitin (N-acetyl-D-glucosamine repeating units linked to β (1→4)) is the second most abundant biopolymer in nature, constituting part of insects, crustaceans, and fungi (Suryanarayanan et al., 2012). Venkatachalam et al. (2015) found that endophytic fungi Aspergillus nidulans, Penicillium sp., and Penicillium sp. isolated from macroalgae Halymenia sp., Halimeda macroloba, and Caulerpa scalpelliformis produced enzymes that acted on chitin and three chitosan substrates (56%, 38%, or 1.6% degree of acetylation) (Venkatachalam et al., 2015).
Similarly, Sarasan et al. (2021), found that 14% of endophytes isolated from different algae (Sargassum wightii, Caulerpa peltata, Ulva fasciata, Dictyota dichotoma, Halymenia venusta, Hypnea musiformis, Grateloupia lithophila, Enteromorpha compressa, Ceramium diaphanum, Padina tetrastromatica, and Chaetomorpha antennina) degraded chitin, with Aspergillus nonius standing out from the 133 endophytic fungi evaluated.
3.2.4 Other enzymes
Of the five magroalgae sampled (Sargassum sp., U. fasciata, Caulerpa sp., Hypnea valentiae, and H. venusta), 29 endophytic fungi were isolated from the Vizhinjam coast, Kerala, India, varying for each algal host, verifying that 90% of all fungal isolates presented amylolytic capacity, 86% lipolytic, 69% gelatinase and asparaginase action, and 62% glutaminase (Manomi et al., 2015). In addition, Sarasan et al. (2021) isolated 133 endophytic fungi from 11 macroalgae (S. wightii, C. peltata, U. fasciata, D. dichotoma, and H. venusta from the coast of Kovalam, Trivandrum, India, and H. musiformis, G. lithophila, E. compressa, C. diaphanum, P. tetrastromatica, and C/antennina from Palakkulam Beach, Calicut, Kerala, India), in which 89.5% have amylolytic activity, 79% gelatinous activity, and 88% lipolytic activity.
In addition, Patyshakuliyeva et al. (2020), showed that Epicoccum nigrum, Penicillium brevicompactum, and Trichoderma paraviridescens isolated from Fucus vesiculosus from the Scheveningen Beach (the Netherlands), had a higher xylanase production, but T. paraviridescens had endoglucanase, xylanase, and glucosidase activities after 120 hours.
3.3 Production of other bioactive compounds
Teixeira et al. (2019b) showed in the mycelia of endophytic fungus A. flavus, Penicillium echinulatum, Penicillium purpurogenum, and Microascus croci isolated from macroalgae Ascoseira mirabilis, Adenocystis utricularis, Phaeurus antarcticus, and Desmarestia anceps the presence of palmitic, linoleic, oleic, and stearic acids. Squalene and ergosterol (ergosta-5,7,9(11) and 22-tetraen-3β-ol) concentrations of about 16% were obtained in P. echinulatum.
Some bacteria like Bacillus pumilus and Bacillus licheniformis isolated from Gracilaria dura produced 445.5 and 335 µgmL-1 of indole acetic acid (IAA) and 12.51 and 10.14 mM mL-1 of ammonia, demonstrating that these compounds induced the growth of new buds in G. dura (Singh et al., 2011) and can be used to improve the production of macroalgae of biotechnological interest.
4 Application of antimicrobials and hydrolytic enzymes produced by bacteria and fungi associated with macroalgae
4.1 Antimicrobial application
Bioactive compounds synthesized by bacteria and fungi associated with macroalgae with antimicrobial potential against pathogens and food spoilers can be applied in the food industry as preservatives for perishable foods (Yong et al., 2007; Bouarab-Chibane et al., 2019; Rani et al., 2021), selection of starter cultures in fermentation processes (Sánchez-Maldonado et al., 2011), disinfectant active compounds (Soni and Brightwell, 2022), water treatment, and pulp processing. In other fields, bioactive compounds can replace synthetic drugs in the treatment of infectious diseases (Danquah et al., 2022) and be used in paper manufacturing, mastics, adhesives, and as disinfectants for textiles, plastics, and cosmetics (Farr et al., 2003).
4.2 Hydrolytic enzyme application
Cellulase, together with hemicelluloses and pectinases, is an important group of enzymes, where its application in food increases its quality and antioxidants (Kumar et al., 2018), and reduces browning (Sims and Bates, 1994), rancidity, and residuality (Kumar et al., 2018). Furthermore, the use of cellulases allows polymers to be degraded, reducing their effect during the baking process, as they are in their pure form, or reducing yield during juice production.
Commercial animal feed can also be prepared using cellulase enzymes, due to their low digestion in some mammals and birds (Kumar et al., 2018), and farmers can use the enzymes as supplements (Liu and Kokare, 2017). In addition, cellulase enzymes present a potential utility in biorefineries, where certain estuaries and mangroves with a rich lignocellulosic biomass constitute a feeding zone for cellulose digesters (Barzkar and Sohail, 2020). Additionally, Trivedi et al. (2016) mentioned that microbial cellulases have demonstrated their efficiency in the conversion of cellulosic plant biomass into fermentable sugar, presenting great bioenergetic potential. Ligninase from these marine microbes may be suitable for bioremediation of effluents from the paper industry (Sarasan et al., 2021) due to its ability to degrade lignocellulosic biomass into fermentable sugars (Suryanarayanan et al., 2012) and could be used in the biofuels field (Bonugli-Santos et al., 2015).
Chitin deacetylases and chitosanases could be used to remodel natural chitin or chitosans and can be adapted according to needs (Suryanarayanan et al., 2012) such as functional foods and nutraceuticals for humans or in the preservation of foods from agriculture and livestock (Zhang et al., 2022). In addition, they allow the biodegradation of chitinous waste, reducing environmental risks and generating products of industrial value (Tamadoni Jahromi and Barzkar, 2018).
Although Patyshakuliyeva et al. (2020) evidenced the presence of endoglucanases, xylanases, and glucosidases in T. paraviridescens, the knowledge of the enzymes involved in algal carbon sources (laminaria, alginate, and carrageenan) is still scarce. These results show that amylolytic activities are enzymatic processes that allow the formation of very specific products compared to chemical modifications (hard conditions and a greater number of by-products), providing various starch products with multiple uses for the food industry and with good biochemical characteristics for industrial processes (Farias et al., 2021). Similarly, amylases and gelatinases are enzymes that hydrolyze gelatin into smaller oligopeptides, being important for the production of hydrolysates that could be applied in the production of various foods (desserts and candies) and non-foods (cosmetics) (Mekonnen et al., 2022). Likewise, lipolytic activity mediated by lipases is important, as it allows the degradation of oleaginous substances used in the food industry, releasing fatty acids as health protectors from edible oil converting themselves into good additives of functional food products (Kotogán et al., 2022).
Asparaginases may be an alternative source for anticancer enzyme production (Hatamzadeh et al., 2020) by converting asparagine into aspartic acid, reducing the presence of acrylamide produced at high temperatures in starchy foods and maintaining sensory quality (Xu et al., 2016) and as a source of therapeutic enzymes by being part of functional foods. Regarding glutaminase, it is an enzyme that could improve functional properties such as solubility, emulsification, and foaming properties, and flavor, and could reduce allergenicity. The redesign of protein foods could improve their process performance and consumer acceptance (Liu et al., 2022a).
5 Conclusion
Bacteria and fungi associated with macroalgae have the ability to inhibit pathogenic bacteria, fungi, yeasts, and protozoa that affect aquaculture, public health, and the food industry. Pyrenocins A, B, E, and S obtained and fractionated from Phaeosphaeria sp. inhibited pathogenic protozoa. Other compounds, such as phenol, 3, 5-bis (1, 1-dimethylethyl), heptacosane, 2-methyloctacosane, polyketides, lipopeptides, desferrioxamine B and B2, azelaic acid, azitidine, furo-pyrans, phenol, 2,4-bis(1,1-dimethylethyl)-, silicic acid, cytochalasin D, and (+)-Epi-Epoformin, were only identified, and the mechanisms of action of each compound have not been reported yet, except pyrenocines A and B, which demonstrated inhibitory action. These compounds found in living organisms act by altering the membrane and wall cells, inhibiting and denaturing proteins and enzymes, damaging genetic material, decreasing reactive oxygen species (ROS), and altering the actin filaments of the cytoskeleton, causing mitochondrial electrochemical depolarization or hindering biofilms production.
Independent of antimicrobial activity, the associated microorganisms can synthesize hydrolytic enzymes such as xylanase, amylase, cellulase, pectinase, agarase, lignocellulase, chitinase, gelatinase, asparaginase, glutaminase, and lipase that can be used to reduce enzymatic oxidation and browning, improve digestibility, functionality, and food preservation. Likewise, they could be used for the bioremediation of agricultural waste, industrial effluents, or production of hydrolysates.
Statements
Author contributions
EV-P and MR-C conceived, designed, and wrote the original draft of the manuscript. JV-M and NA-T edited, reviewed, and proposed amendments to the manuscript. LE-E, LM-Q and HC-S supervised, reviewed, and critically commented on and edited the manuscript. All authors contributed to the manuscript and approved the submitted version.
Funding
This research received financial support from the Universidad Nacional de Frontera, Peru, through a Resolution of the Organizing Commission No 014-2022-UNF/CO.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abd El Baky H. H. El Baz F. K. El Baroty G. S. Asker M. M. S. Ibrahim E. A. (2014). Phospholipids of some marine macroalgae: identification, antivirus, anticancer and antimicrobial bioactivities. Der Pharma Chem.6, 370–382.
2
Abdullah A. S. H. Mirghani M. E. S. Jamal P. (2011). Antibacterial activity of Malaysian mango kernel. Afr. J. Biotechnol.10, 18739–18748. doi: 10.5897/AJB11.2746
3
Akeda Y. Nagayama K. Yamamoto K. Honda T. (1997). Invasive phenotype of Vibrio parahaemolyticus. J. Infect. Dis.176, 822–824. doi: 10.1086/517312
4
Aleti G. Sessitsch A. Brader G. (2015). Genome mining: prediction of lipopeptides and polyketides from bacillus and related firmicutes. Comput. Struct. Biotechnol. J.13, 192–203. doi: 10.1016/j.csbj.2015.03.003
5
Amin E. Elwekeel A. Alshariedh N. F. Abdel-Bakky M. S. Hassan M. H. A. (2022). GC-MS analysis and bioactivities of the essential oil of suaeda aegyptiaca. Separations9, 1–13. doi: 10.3390/separations9120439
6
Arshad M. S. Batool S. A. (2017). Natural antimicrobials, their sources and food safety. Food Addit. 87, 87–101. doi: 10.5772/intechopen.70197
7
Aumack C. F. Amsler C. D. McClintock J. B. Baker B. J. (2011). Impacts of mesograzers on epiphyte and endophyte growth associated with chemically defended macroalge from the western antarctic peninsula: a mesocosm experiment. J. Phycol.47, 36–41. doi: 10.1111/j.1529-8817.2010.00927.x
8
Balabanova L. Slepchenko L. Son O. Tekutyeva L. (2018). Biotechnology potential of marine fungi degrading plant and algae polymeric substrates. Front. Microbiol.9. doi: 10.3389/fmicb.2018.01527
9
Bankeu J. J. K. Madjouka S. Feuya G. R. T. Fongang Y. S. F. Siddiqui S. Ali I. et al . (2018). Pobeguinine: a monoterpene indole alkaloid and other bioactive constituents from the stem bark of nauclea pobeguinii. Z. Naturforsch. C.73, 335–344. doi: 10.1515/znc-2017-0127
10
Barilli E. Cimmino A. Masi M. Evidente M. Rubiales D. Evidente A. (2017). Inhibition of early development stages of rust fungi by the two fungal metabolites cyclopaldic acid and epi-epoformin. Pest Manage. Sci.73, 1161–1168. doi: 10.1002/ps.4438
11
Barzkar N. Sohail M. (2020). An overview on marine cellulolytic enzymes and their potential applications. Appl. Microbiol. Biotechnol.104, 6873–6892. doi: 10.1007/s00253-020-10692-y
12
Bogas A. C. Henrique Rodrigues S. Gonçalves M. O. De Assis M. Longo E. Paiva De Sousa C. (2022). Endophytic microorganisms from the tropics as biofactories for the synthesis of metal-based nanoparticles: healthcare applications. Front. Nanotechnol.4. doi: 10.3389/fnano.2022.823236
13
Bonthond G. Barilo A. Allen R. J. Cunliffe M. Krueger-Hadfield S. A. (2022). Fungal endophytes vary by species, tissue type, and life cycle stage in intertidal macroalgae. J. Phycol.58, 330–342. doi: 10.1111/jpy.13237
14
Bonugli-Santos R. C. Vasconcelos dos M. R.S. Passarini M. R.Z. Vieira G. A.L. Lopes V. C.P. Mainardi P. H. et al . (2015). Marine-derived fungi: Diversity of enzymes and biotechnological applications. Front. Microbiol.6. doi: 10.3389/fmicb.2015.00269
15
Bouarab-Chibane L. Forquet V. Lantéri P. Clément Y. Léonard-Akkari L. Oulahal N. et al . (2019). Antibacterial properties of polyphenols: characterization and QSAR (Quantitative structure-activity relationship) models. Front. Microbiol.10. doi: 10.3389/fmicb.2019.00829
16
Campos F. M. Couto J. A. Figueiredo A. R. Tóth I. V. Rangel A. O. S. S. Hogg T. A. (2009). Cell membrane damage induced by phenolic acids on wine lactic acid bacteria. Int. J. Food Microbiol.135, 144–151. doi: 10.1016/j.ijfoodmicro.2009.07.031
17
Cesa-Luna C. Baez A. Quintero-Hernández V. de la Cruz-Enríquez J. Castañeda-Antonio M. D. Muñoz-Rojas J. (2020). The importance of antimicrobial compounds produced by beneficial bacteria on the biocontrol of phytopathogens. Acta Biol. Colomb.25, 140–154. doi: 10.15446/abc.v25n1.76867
18
Cha H. J. Chiang M. W. L. Guo S. Y. Lin S. M. Pang K. L. (2021). Culturable fungal community of pterocladiella capillacea in keelung, taiwan: effects of surface sterilization method and isolation medium. J. Fungi7, 651. doi: 10.3390/jof7080651
19
Chan-Bacab M. J. Reyes-Estebanez M. M. Camacho-Chab J. C. Ortega-Morales B. O. (2021). Microorganisms as a potential source of molecules to control trypanosomatid diseases. Molecules26, 1388. doi: 10.3390/molecules26051388
20
Chen J. Zang Y. Yang Z. Qu T. Sun T. Liang S. et al . (2022). Composition and functional diversity of epiphytic bacterial and fungal communities on marine macrophytes in an intertidal zone. Front. Microbiol.13. doi: 10.3389/fmicb.2022.839465
21
Cooper C. Walker A. K. (2022). Endophytic fungi from marine macroalgae in Nova Scotia. Northeastern Naturalist29, 295–310. doi: 10.1656/045.029.0212
22
Cueto M. Jensen P. R. Kauffman C. Fenical W. Lobkovsky E. Clardy J. (2001). Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod.64, 1444–1446. doi: 10.1021/np0102713
23
Danquah C. A. Minkah P. A. B. Junior I. O. D. Amankwah K. B. Somuah S. O. (2022). Antimicrobial compounds from microorganisms. Antibiotics11, 1–20. doi: 10.3390/antibiotics11030285
24
Darienko T. Rad-Menéndez C. Campbell C. N. Pröschold T. (2021). Molecular phylogeny of unicellular marine coccoid green algae revealed new insights into the systematics of the ulvophyceae (Chlorophyta). Microorganisms9, 1586. doi: 10.3390/microorganisms9081586
25
Deshmukh S. K. Prakash V. Ranjan N. (2018). Marine fungi: a source of potential anticancer compounds. Front. Microbiol.8. doi: 10.3389/fmicb.2017.02536
26
De Felício R. Pavão G. B. De Oliveira A. L.L. Erbert C. Conti R. Pupo M. T. et al . (2015). Antibacterial, antifungal and cytotoxic activities exhibited by endophytic fungi from the Brazilian marine red alga Bostrychia tenella (Ceramiales). Rev. Bras. Farmacogn.25, 641–650. doi: 10.1016/j.bjp.2015.08.003
27
Deutsch Y. Gur L. Frank I. B. Ezra D. (2021). Endophytes from algae , a potential source for new biologically active metabolites for disease management in aquaculture. Frontiers in Marine Science8, 1–13. doi: 10.3389/fmars.2021.636636
28
Dhanya K. I. Swati V. I. Vanka K. S. Osborne W. J. (2016). Antimicrobial activity of ulva reticulata and its endophytes. J. Ocean Univ. China15, 363–369. doi: 10.1007/s11802-016-2803-7
29
Ecevit K. Barros A. A. Silva J. M. Reis R. L. (2022). Preventing microbial infections with natural phenolic compounds. Futur. Pharmacol.2, 460–498. doi: 10.3390/futurepharmacol2040030
30
Espinoza C. Herrera-García C. D. Espinosa-García V. Couttolenc A. Andrade-Torres A. Padrón J. M. et al . (2021). The antiproliferative potential of fungi associated with coral and algae collected from a veracruz reef system, gulf of Mexico. Lat. Am. J. Aquat. Res.49, 843–849. doi: 10.3856/vol49-issue5-fulltext-2704
31
Farias T. C. Kawaguti H. Y. Bello Koblitz M. G. (2021). Microbial amylolytic enzymes in foods: technological importance of the bacillus genus. Biocatal. Agric. Biotechnol.35, 102054. doi: 10.1016/j.bcab.2021.102054
32
Farr J. P. Smith W. L. Steichen D. S. (2003). Pericles_0471238961.
33
Flewelling A. J. Ellsworth K. T. Sanford J. Forward E. Johnson J. A. and Gray C. A. (2013). Macroalgal endophytes from the atlantic coast of canada: A potential source of antibiotic natural products? Microorganisms1, 175–187. doi: 10.3390/microorganisms1010175
34
Flewelling A. J. Currie J. Gray C. A. Johnson J. A. (2015). Endophytes from marine macroalgae: promising sources of novel natural products. Curr. Sci.109, 88–111. Available at: https://www.jstor.org/stable/24905694
35
Gabriel M. Horký D. Svoboda A. Kopecká M. (1998). Cytochalasin d interferes with contractile actin ring and septum formation in schizosaccharomyces japonicus var. versatilis. Microbiology144 (Pt 8), 2331–2344. doi: 10.1099/00221287-144-8-2331
36
Gonçalves M. F. M. Vicente T. F. L. Esteves A. C. Alves A. (2020). Novel halotolerant species of emericellopsis and parasarocladium associated with macroalgae in an estuarine environment. Mycologia112, 154–171. doi: 10.1080/00275514.2019.1677448
37
Gray D. A. Wenzel M. (2020). More than a pore: a current perspective on the in vivo mode of action of the lipopeptide antibiotic daptomycin. Antibiotics9, 17. doi: 10.3390/antibiotics9010017
38
Habbu P. Warad V. Shastri R. Savant C. Madagundi S. Kekare P. (2016). In vitro and in vivo antimicrobial activity of ulva lactuca linn. (greer algae) associated endophytic bacterial strains. J. Appl. Pharm. Sci.6, 138–146. doi: 10.7324/JAPS.2016.601019
39
Hagaggi N. S. A. Abdul-Raouf U. M. (2022). Macroalga-associated bacterial endophyte bioactive secondary metabolites twinning: cystoseira myrica and its associated catenococcus thiocycli QCm as a model. World J. Microbiol. Biotechnol.38, 1–11. doi: 10.1007/s11274-022-03394-2
40
Handayani D. Ananda N. Artasasta M. A. Ruslan R. Fadriyanti O. Tallei T. E. (2019). Antimicrobial activity screening of endophytic fungi extracts isolated from brown algae padina sp. J. Appl. Pharm. Sci.9, 9–13. doi: 10.7324/JAPS.2019.90302
41
Harikrishnan M. Prakash P. Jayabaskaran C. Bhat S. G. (2021). Multi-functional bioactive secondary metabolites derived from endophytic fungi of marine algal origin. Curr. Res. Microb. Sci.2, 100037. doi: 10.1016/j.crmicr.2021.100037
42
Hashem A. H. Shehabeldine A. M. Abdelaziz A. M. Amin B. H. Sharaf M. H. (2022). Antifungal activity of endophytic aspergillus terreus extract against some fungi causing mucormycosis: ultrastructural study. Appl. Biochem. Biotechnol.194, 3468–3482. doi: 10.1007/s12010-022-03876-x
43
Hatamzadeh S. Rahnama K. Nasrollahnejad S. Fotouhifar K. B. Hemmati K. White J. F. et al . (2020). Isolation and identification of l-asparaginase-producing endophytic fungi from the asteraceae family plant species of Iran. PeerJ2020, 1–16. doi: 10.7717/peerj.8309
44
Hollants J. Leroux O. Leliaert F. Decleyre H. de Clerck O. Willems A. (2011). Who is in there? exploration of endophytic bacteria within the siphonous green seaweed bryopsis (Bryopsidales, chlorophyta). PloS One6, 1–23. doi: 10.1371/journal.pone.0026458
45
Ismail A. Ktari L. Ahmed M. Bolhuis H. Boudabbous A. Stal L. J. et al . (2016). Antimicrobial activities of bacteria associated with the brown alga padina pavonica. Front. Microbiol.7. doi: 10.3389/fmicb.2016.01072
46
Jakubczyk D. Dussart F. (2020). Selected fungal natural products with antimicrobial properties. Molecules25, 1–18. doi: 10.3390/molecules25040911
47
Kamat S. Kumari M. Taritla S. Jayabaskaran C. (2020). Endophytic fungi of marine alga from konkan coast, India–a rich source of bioactive material. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00031
48
Katif C. Chilczuk T. Sabour B. Belattmania Z. Hilmi A. Niedermeyer T. H. J. et al . (2022). Isolation and structure elucidation of desferrioxamine b and the new desferrioxamine b2 antibiotics from a brown marine macroalga carpodesmia tamariscifolia associated streptomyces isolate. Biointerface Res. Appl. Chem.12, 5647–5662. doi: 10.33263/BRIAC124.56475662
49
Kumar V. A. Kurup S. C. R. Snishamol C. Nagendra Prabhu G. (2018). Role of cellulases in food, feed and beverage industries. Green Bio-processes: Enzymes in Industrial Food Processing, 323–343.
50
Kizhakkekalam V. K. Chakraborty K. (2020). Marine macroalgae-associated heterotrophic firmicutes and gamma-proteobacteria: prospective anti-infective agents against multidrug resistant pathogens. Arch. Microbiol.202, 905–920. doi: 10.1007/s00203-019-01800-2
51
Kjer J. Debbab A. Aly A. H. Proksch P. (2010). Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc.5, 479–490. doi: 10.1038/nprot.2009.233
52
Kotogán A. Furka Z. T. Kovács T. Volford B. Papp D. A. Varga M. et al . (2022). Hydrolysis of edible oils by fungal lipases: an effective tool to produce bioactive extracts with antioxidant and antimicrobial potential. Foods11, 1711. doi: 10.3390/foods11121711
53
Kumar P. Verma A. Sundharam S. S. Ojha A. K. Krishnamurthi S. (2022). Exploring diversity and polymer degrading potential of epiphytic bacteria isolated from marine macroalgae. Microorganisms10, 2513. doi: 10.3390/microorganisms10122513
54
Le P. N. T. Desbois A. P. (2017). Antibacterial effect of eicosapentaenoic acid against bacillus cereus and staphylococcus aureus: killing kinetics, selection for resistance, and potential cellular target. Mar. Drugs15, 334. doi: 10.3390/md15110334
55
Liu X. Kokare C. (2017). “Chapter 11 - Microbial Enzymes of Use in Industry,” in Biotechnol. Microb. Enzym. Prod. Biocatal. Ind. Appl., ed. G. B. T.-B. of M. E. Brahmachari (Academic Press), 267–298. doi: 10.1016/B978-0-12-803725-6.00011-X
56
Liu X. Wang C. Zhang X. Zhang G. Zhou J. Chen J. (2022a). Application prospect of protein-glutaminase in the development of plant-based protein foods. Foods11, 1–11. doi: 10.3390/foods11030440
57
Liu X. Zhao J. Jiang P. (2022b). Easy removal of epiphytic bacteria on ulva (Ulvophyceae, chlorophyta) by vortex with silica sands. Microorganisms10, 1–8. doi: 10.3390/microorganisms10020476
58
Lucini F. de Andrade G. A. K. Victoria F. de C. de Albuquerque M. P. (2022). Impact of the temperature in endophytic ascomycota isolated from Antarctic hair-grass. Life12, 1501. doi: 10.3390/life12101501
59
Magesh C. J. Makesh S. V. Perumal P. T. (2004). Highly diastereoselective inverse electron demand (IED) diels-alder reaction mediated by chiral salen-AlCl complex: the first, target-oriented synthesis of pyranoquinolines as potential antibacterial agents. Bioorganic Med. Chem. Lett.14, 2035–2040. doi: 10.1016/j.bmcl.2004.02.057
60
Magina M. D. A. Dalmarco E. M. Wisniewski A. Simionatto E. L. Dalmarco J. B. Pizzolatti M. G. et al . (2009). Chemical composition and antibacterial activity of essential oils of Eugenia species. J. Nat. Med.63, 345–350. doi: 10.1007/s11418-009-0329-5
61
Manomi S. Neema J. Rosamma P. (2015). Bioactive potential of endophytic fungi from macroalgae. Int. J. Res. Mar. Sci.4, 27–32.
62
Mathan S. Subramanian V. Nagamony S. Ganapathy K. (2013). Isolation of endophytic fungi from marine algae and its bioactivity. Int. J. Res. Pharm. Sci.4, 45–49.
63
Mekonnen M. Girma S. Atnafu A. (2022). Hydrolysis of gelatin from animal hoof using bacterial gelatinase. Int. J. Microbiol. Biotechnol.7, 135–142. doi: 10.11648/j.ijmb.20220703.14
64
Menaa F. Wijesinghe P. A. U. I. Thiripuranathar G. Uzair B. Iqbal H. Khan B. A. et al . (2020). Ecological and industrial implications of dynamic seaweed-associated microbiota interactions. Mar. Drugs18, 1–25. doi: 10.3390/md18120641
65
Ochieng P. Isanda G. Piero M. (2023). Heliyon antibacterial properties and GC-MS analysis of ethyl acetate extracts of xerophyta spekei ( baker ) and grewia tembensis ( fresen ). Heliyon9, e14461. doi: 10.1016/j.heliyon.2023.e14461
66
O’Dowd H. Lewis J. G. Trias J. Asano R. Blais J. Lopez S. L. et al . (2008). Novel antibacterial azetidine lincosamides. Bioorganic Med. Chem. Lett.18, 2645–2648. doi: 10.1016/j.bmcl.2008.03.032
67
Ortiz-Ojeda C. P. de Andrade S. L. Procópio R. E. L . (2020). Antifungal activity of endophytic microorganisms isolated from acmella ciliata (Asteraceae). Genet. Mol. Res.19, 1–14. doi: 10.4238/gmr18570
68
Padmavathi A. R. Abinaya B. Pandian S. K. (2014). Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen serratia marcescens. Biofouling30, 1111–1122. doi: 10.1080/08927014.2014.972386
69
Patyshakuliyeva A. Falkoski D. L. Wiebenga A. Timmermans K. de Vries R. P. (2020). Macroalgae derived fungi have high abilities to degrade algal polymers. Microorganisms8, 1–16. doi: 10.3390/microorganisms8010052
70
Park H. H. Ko S. C. Oh G. W. Jang Y. M. Kim Y. M. Park W. S. et al (2018). Characterization and biological activity of PVA hydrogel containing chitooligosaccharides conjugated with gallic acid. Carbohydr. Polym.198, 197–205. doi: 10.1016/j.carbpol.2018.06.070
71
Pereira L. (2021). Macroalgae. Encyclopedia1, 177–188. doi: 10.3390/encyclopedia1010017
72
Pereira D. T. Simioni C. Filipin E. P. Bouvie F. Ramlov F. Maraschin M. et al . (2017). Effects of salinity on the physiology of the red macroalga, acanthophora spicifera (Rhodophyta, ceramiales). Acta Bot. Brasilica31, 555–565. doi: 10.1590/0102-33062017abb0059
73
Perkins A. K. Rose A. L. Grossart H. P. Rojas-Jimenez K. Barroso Prescott S. K. Oakes J. M. (2021). Oxic and anoxic organic polymer degradation potential of endophytic fungi from the marine macroalga, ecklonia radiata. Front. Microbiol.12. doi: 10.3389/fmicb.2021.726138
74
Purro M. Qiao J. Liu Z. Ashcraft M. Xiong M. P. (2018). Desferrioxamine:gallium-pluronic micelles increase outer membrane permeability and potentiate antibiotic activity against pseudomonas aeruginosa. Chem. Commun.54, 13929–13932. doi: 10.1039/c8cc08134d
75
Rajamani S. Sengodan T. Thangavelu S. Shanmukhan N. K. Radhakrishnan A. (2019). Naringenin-loaded TPGS polymeric nanosuspension: in-vitro and in-vivo anti-inflammatory activity. Indones. J. Pharm.30, 225–232. doi: 10.14499/indonesianjpharm30iss3pp217
76
Rani A. Saini K. C. Bast F. Varjani S. Mehariya S. Bhatia S. K. et al . (2021). A review on microbial products and their perspective application as antimicrobial agents. Biomolecules11, 1860. doi: 10.3390/biom11121860
77
Rego Y. F. Queiroz M. P. Brito T. O. Carvalho P. G. de Queiroz V. T. de Fátima Â. et al . (2018). A review on the development of urease inhibitors as antimicrobial agents against pathogenic bacteria. J. Adv. Res.13, 69–100. doi: 10.1016/j.jare.2018.05.003
78
Res I. J. A. (2018). Manuscript info abstract introduction. Materials Methods6, 525–538. doi: 10.21474/IJAR01/6700
79
Romero-Correa M. T. Villa-Gómez R. Castro-Mercado E. Garcia-Pineda E. (2014). The avocado defense compound phenol-2,4-bis (1,1-dimethylethyl) is induced by arachidonic acid and acts via the inhibition of hydrogen peroxide production by pathogens. Physiol. Mol. Plant Pathol.87, 32–41. doi: 10.1016/j.pmpp.2014.05.003
80
Roscetto E. Masi M. Esposito M. Di Lecce R. Delicato A. Maddau L. et al . (2020). Anti-biofilm activity of the fungal phytotoxin sphaeropsidin a against clinical isolates of antibiotic-resistant bacteria. Toxins (Basel).12, 1–14. doi: 10.3390/toxins12070444
81
Rubtsova S. N. Kondratov R. V. Kopnin P. B. Chumakov P. M. Kopnin B. P. Vasiliev J. M. (1998). Disruption of actin microfilaments by cytochalasin d leads to activation of p53. FEBS Lett.430, 353–357. doi: 10.1016/S0014-5793(98)00692-9
82
Sahoo S. Subban K. Chelliah J. (2021). Diversity of marine macro-algicolous endophytic fungi and cytotoxic potential of biscogniauxia petrensis metabolites against cancer cell lines. Front. Microbiol.12. doi: 10.3389/fmicb.2021.650177
83
Samanta S. Dey P. Gupta N. Mouleeswaran K. S. Nijhawan R. (2011). Micronucleus in atypical squamous cell of undetermined significance. Diagn. Cytopathol.39, 242–244. doi: 10.1002/dc.21368
84
Sánchez Hinojosa V. Asenjo J. Leiva S. (2018). Agarolytic culturable bacteria associated with three antarctic subtidal macroalgae. World J. Microbiol. Biotechnol.34, 73. doi: 10.1007/s11274-018-2456-1
85
Sánchez-Maldonado A. F. Schieber A. Gänzle M. G. (2011). Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol.111, 1176–1184. doi: 10.1111/j.1365-2672.2011.05141.x
86
Sarasan M. Job N. Puthumana J. Ravinesh R. Kachiprath B. Philip R. (2021). Enzyme profiling of macroalgal endophytes: an attempt to uncover the arsenal of novel biocatalysts. J. Mar. Biol. Assoc. India62, 112–116. doi: 10.6024/jmbai.2020.62.2.2183-14
87
Sarasan M. Job N. Puthumana J. Ravinesh R. Prabhakaran. M. P. Thomas L. C. et al . (2020). Exploration and profiling of hidden endophytic mycota of marine macroalgae with potential drug leads. FEMS Microbiol. Lett.367, 1–9. doi: 10.1093/femsle/fnaa078
88
Sarasan M. Puthumana J. Job N. Han J. Lee J.-S. Philip R. (2017). Marine algicolous endophytic fungi-a promising drug resource of the era. J. Microbiol. Biotechnol.27, 1039–1052. doi: 10.4014/jmb.1701.01036
89
Sharon P. P. Oluwafemi A. A. Ezekiel G. (2021). Chemotaxonomic profiling of fungal endophytes of solanum mauritianum (alien weed) using gas chromatography high resolution time-of-flight mass spectrometry (GC-HRTOF-MS). Metabolomics17, 1–13. doi: 10.1007/s11306-021-01790-7
90
Silva A. Silva S. A. Carpena M. Garcia-Oliveira P. Gullón P. Barroso M. F. et al . (2020). Macroalgae as a source of valuable antimicrobial compounds: extraction and applications. Antibiotics9, 1–41. doi: 10.3390/antibiotics9100642
91
Sims C. A. Bates R. P. (1994). Challenges to processing tropical fruit juices: banana as an example. Proc. Florida State Hortic. Soc107, 315–318.
92
Singh R. P. Bijo A. J. Baghel R. S. Reddy C. R. K. Jha B. (2011). Role of bacterial isolates in enhancing the bud induction in the industrially important red alga gracilaria dura. FEMS Microbiol. Ecol.76, 381–392. doi: 10.1111/j.1574-6941.2011.01057.x
93
Singh R. P. Reddy C. R. K. (2016). Unraveling the functions of the macroalgal microbiome. Front. Microbiol.6. doi: 10.3389/fmicb.2015.01488
94
Soni A. Brightwell G. (2022). Nature-inspired antimicrobial surfaces and their potential applications in food industries. Foods11, 844. doi: 10.3390/foods11060844
95
Suryanarayanan T. S. Thirunavukkarasu N. Govindarajulu M. B. Gopalan V. (2012). Fungal endophytes: an untapped source of biocatalysts. Fungal Divers.54, 19–30. doi: 10.1007/s13225-012-0168-7
96
Tamadoni Jahromi S. Barzkar N. (2018). Marine bacterial chitinase as sources of energy, eco-friendly agent, and industrial biocatalyst. Int. J. Biol. Macromol.120, 2147–2154. doi: 10.1016/j.ijbiomac.2018.09.083
97
Teixeira T. R. Dos Santos G. S. Armstrong L. Colepicolo P. (2019a). Antitumor potential of seaweed derived-endophytic fungi. Antibiotics8, 1–24. doi: 10.3390/antibiotics8040205
98
Teixeira T. R. Santos G. S. Turatti I. C. C. Paziani M. H. von Zeska Kress M. R. Colepicolo P. et al . (2019b). Characterization of the lipid profile of Antarctic brown seaweeds and their endophytic fungi by gas chromatography–mass spectrometry (GC–MS). Polar Biol.42, 1431–1444. doi: 10.1007/s00300-019-02529-w
99
Tijith K. G. Akhil J. Divya K. Jisha M. S. (2019). Microbial pathogenesis In vitro and in silico docking studies of antibacterial compounds derived from endophytic penicillium setosum. Microb. Pthogenes.131, 87–97. doi: 10.1016/j.micpath.2019.03.033
100
Trivedi N. Reddy C. R. K. Lali A. M. (2016). Marine microbes as a potential source of cellulolytic enzymes. Advances in food and nutrition research79, 27–41. doi: 10.1016/bs.afnr.2016.07.002
101
Vallet M. Strittmatter M. Murúa P. Lacoste S. Dupont J. Hubas C. et al . (2018). Chemically-mediated interactions between macroalgae, their fungal endophytes, and protistan pathogens. Front. Microbiol.9. doi: 10.3389/fmicb.2018.03161
102
Venkatachalam A. Govinda Rajulu M. B. Thirunavukkarasu N. Suryanarayanan T. S. (2015). Endophytic fungi of marine algae and seagrasses: a novel source of chitin modifying enzymes. Mycosphere6, 345–355. doi: 10.5943/MYCOSPHERE/6/3/10
103
Villarreal-Gómez L. J. Soria-Mercado I. E. Guerra-Rivas G. Ayala-Sánchez N. E. (2010). Antibacterial and anticancer activity of seaweeds and bacteria associated with their surface. Rev. Biol. Mar. Oceanogr.45, 267–275. doi: 10.4067/s0718-19572010000200008
104
Wichard T. Beemelmanns C. (2018). Role of chemical mediators in aquatic interactions across the prokaryote–eukaryote boundary. J. Chem. Ecol.44, 1008–1021. doi: 10.1007/s10886-018-1004-7
105
Wong Chin J. M. Puchooa D. Bahorun T. Jeewon R. (2021). Molecular characterization of marine fungi associated with haliclona sp. (sponge) and turbinaria conoides and sargassum portierianum (brown algae). Proc. Natl. Acad. Sci. India Sect. B - Biol. Sci.91, 643–656. doi: 10.1007/s40011-021-01229-y
106
Xu T. Cao L. Zeng J. Franco C. M. M. Yang Y. Hu X. (2019). The antifungal action mode of the rice endophyte streptomyces hygroscopicus OsiSh-2 as a potential biocontrol agent against the rice blast pathogen. Pestic. Biochem. Physiol.160, 58–69. doi: 10.1016/j.pestbp.2019.06.015
107
Xu F. Oruna-Concha M. J. Elmore J. S. (2016). The use of asparaginase to reduce acrylamide levels in cooked food. Food Chem.210, 163–171. doi: 10.1016/j.foodchem.2016.04.105
108
Yong S. Bajpai V. K. Ryul H. Chul S. (2007). Antibacterial activity of bioconverted eicosapentaenoic ( EPA ) and docosahexaenoic acid ( DHA ) against foodborne pathogenic bacteria. International Journal of Food Microbiology113, 233–236. doi: 10.1016/j.ijfoodmicro.2006.05.020
109
Zainee N. F. A. Ibrahim N. Hidayah N. Rozaimi M. (2021). Variation in antibacterial properties of endophytic fungi isolated from phaeophytes and rhodophytes of johor, Malaysia. J. Environ. Biol.42, 840–848. doi: 10.22438/JEB/42/3(SI)/JEB-16
110
Zhang R. Zhao Q. Yi Z. Zhang K. Shi J. Zhu L. et al . (2022). Chitin oligosaccharides for the food industry: production and applications. Syst. Microbiol. Biomanufacturing3, 49–74. doi: 10.1007/s43393-022-00127-2
Summary
Keywords
bioactive compounds, hydrolytic enzymes, bacteria, fungus, yeasts
Citation
Vega-Portalatino EJ, Rosales-Cuentas MM, Valdiviezo-Marcelo J, Arana-Torres NM, Espinoza-Espinoza LA, Moreno-Quispe LA and Cornelio-Santiago HP (2023) Antimicrobial and production of hydrolytic enzymes potentials of bacteria and fungi associated with macroalgae and their applications: a review. Front. Mar. Sci. 10:1174569. doi: 10.3389/fmars.2023.1174569
Received
26 February 2023
Accepted
26 April 2023
Published
16 May 2023
Volume
10 - 2023
Edited by
Sophia Letsiou, University of West Attica, Greece
Reviewed by
Satheesh Sathianeson, King Abdulaziz University, Saudi Arabia; Lixia Pan, Guangxi Academy of Sciences, China; Anthimia Batrinou, University of West Attica, Greece
Updates
Copyright
© 2023 Vega-Portalatino, Rosales-Cuentas, Valdiviezo-Marcelo, Arana-Torres, Espinoza-Espinoza, Moreno-Quispe and Cornelio-Santiago.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edwin Jorge Vega-Portalatino, evega@unf.edu.pe
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.