Abstract
Sea cucumber has emerged as a crucial economic species in aquaculture in China because of its remarkable nutritional and medicinal value. However, wild sea cucumber populations have experienced a decline due to overfishing and environmental factors, underscoring the urgent need for genetic resource conservation and biotechnology innovation within the sea cucumber aquaculture and breeding industry. The development of the sea cucumber industry is still impeded by challenges and difficulties. Nevertheless, significant progress has been made through the utilization of molecular markers, which have effectively addressed a number of fisheries and aquaculture issues. In recent years, diverse types of molecular markers including mitochondrial DNA, microsatellites, and SNP markers have been developed and extensively applied in various aspects of sea cucumber research. These markers play vital roles in genetic sex identification, germplasm resource evaluation, population structure assessment, as well as marker-assisted breeding in marine ranching and sea cucumber aquaculture and breeding industry. This review provides an overview of the fundamental principles, functions, and characteristics associated with various markers employed across various sea cucumber species while also discussing their applications within different aspects of the sea cucumber fisheries, aquaculture, and breeding industry.
1 Introduction
Sea cucumber is a marine invertebrate belonging to the class Holothuroidea with the phylum Echinodermata. In general, sea cucumbers appear in a cylindrical, highly contractile body with numerous papillae on the dorsal and tube feet on the ventral side. Sea cucumbers live in a wide variety of different habitats, including seaweed ecosystems and coral reefs, ranging from shallow water to depths of more than 3,000 meters (Sulardiono et al., 2022). They feed on microalgae, organic debris, and material in sediments (Madduppa et al., 2017). There are more than 1,700 species of sea cucumber in the world, of which about 50–60 are edible (Elvevoll et al., 2022). Total sea cucumber fisheries increased from 26,000 tonnes of live weight in 2007–2016 to 48,000 tonnes in 2018–2019, with a slight decline to 43,000 tonnes in 2020 (FAO). Most cultured sea cucumber species belong to the order Aspidochirotida, including two sister groups, Holothuriidae and Stichopodidae (Wen et al., 2010). In Asian countries, sea cucumber is a commercially important aquaculture species due to its high nutritional and medicinal value. The wild sea cucumber populations have declined because of overfishing and environmental factors (Peng et al., 2012).
To increase the yield of sea cucumber, the sea cucumber breeding industry is rising gradually. Since the 19th century, sea cucumber farming in most countries has experienced boom-and-bust cycles, with the exception of China, where the majority of sea cucumbers have come from the aquaculture industry since the late 1980s (Liu, 2016). Even though sea cucumber aquaculture has been developed in the past decades, a series of problems occurred that could affect the larva development and juvenile growth, mainly related to disease control, germplasm conservation, and large-scale production in sea cucumber industry (Du et al., 2012). Among these, the slow growth rate is one of the most important issues in the sea cucumber industry (Cui et al., 2021). To meet the increasing demand for sea cucumber in the market, sea cucumber aquaculture and management using molecular methods or based genomic methods are quite important.
The sea cucumber breeding industry requires the application of traditional and novel molecular methods to benefit production and ensure sustainable development. Genetic breeding has been widely used in the breeding of aquaculture species, which focuses on genetic variations among populations and individuals to improve different types of performance traits. Molecular markers are commonly used molecular methods for detecting biological diversity at the DNA level, which is a powerful informatic molecular tool in genetic breeding and population genetics. Commonly used molecular markers include mitochondrial DNA (mtDNA), restricted fragments length polymorphism (RFLP), random amplification polymorphic DNA (RAPD), microsatellite (SSR), and single nucleic polymorphism (SNP), which have been widely used for species identification, population genetics, genetic breeding, marker-assisted selection, stock enhancement, and aquaculture management (Maqsood, 2017).
At the end of the 20th century, with the emergence of restriction enzymes and polymerase chain reaction (PCR), many molecular marker technologies have been developed and widely used. RFLP marker is a highly polymorphic and co-dominant marker that can be used for species identification, which is rarely used at present due to its complex process (Wen et al., 2010). RAPD marker is a simple technique without molecular hybridization and radioautography, which can be used to evaluate the geographical origin of species and genetic diversity analysis (Yun et al., 2017). Microsatellite marker is abundant and widely distributed throughout the genome (Oliveira et al., 2006), which is a simple and fast marker that have high mutation rates and reliable results, which can identify sea cucumber species, which will benefit germplasm conservation and management (Zhan et al., 2007). Subsequently, the advent of single nucleic polymorphism (SNP) molecular marker technology made it possible to perform genetic analysis of the genome at a precise level (Oliveira and Azevedo, 2022). Microsatellite marker and SNP marker are ideal molecular markers because of their high polymorphism and co-dominant inheritance (Maqsood, 2017), which have been widely used in population structure analysis (Kanno et al., 2006), genetic linkage map construction (Yan et al., 2013), QTL mapping (Chen and Li, 2007), marker-assisted selection (MAS) (Wang et al., 2009). Mitochondrial marker is a simple, fast, reliable molecular marker that is inherited from maternal lineage, which have high mutation rates and can be used in species identification, population genetic structure analysis, and phylogenetic analysis (Sulardiono et al., 2022). The advantages and disadvantages of various types of molecular markers are summarized in Table 1. At present, molecular markers are quite useful in the sea cucumber aquaculture and breeding industry. For instance, using sex-specific tags and SNPs to identify the sex can develop and breed a single-sex population, which could improve breeding efficiency, and lay a molecular foundation for studying the sex determination mechanism of sea cucumber (Cui et al., 2021); The high-density genetic linkage map of sea cucumber was constructed based on SNPs, which laid a foundation for mapping and analyzing QTL for performance traits like growth rate (Wei et al., 2021). This review introduces and summarizes the development and application of different types of molecular markers and their based genomic methods in sea cucumbers (Figure 1, Table 2) and examines the present status and prospects in fisheries, aquaculture, and industry.
Table 1
| Molecular marker | Description | Advantage | Disadvantage | Application |
|---|---|---|---|---|
| RFLP | Changes in the length of DNA fragments produced by cutting relevant DNA molecules using specific endonuclease enzymes | High polymorphism; Co-dominant marker; High reproducibility | Require target DNA sequence information for the design of amplification primers; Complicated process | Species identification; Product mislabeling correction |
| RAPD | Genomic DNA is amplified by PCR using a series of single random primers (usually 10 nucleotides), and the polymorphism of PCR products is detected by electrophoresis | Random primers; Simple technique without molecular hybridization and radioautography; Require low amount DNA; Low cost | Poor experimental repeatability; unreliable results; Dominant marker | Genome mapping; Geographical origin and genetic diversity analysis |
| AFLP | A PCR-based molecular technique that uses selective amplification of a subset of digested DNA fragments | Large amount of markers with relatively little lab effort; No prior information about genome; Genome wide coverage; Small amount of DNA needed; Low cost; High experimental repeatability; reliable results | Dominant marker; Low reproducibility | Determine genetic differences among individuals, populations, and species; Investigate population structure and phylogenetic relationships based on genetic distances; Evaluate gene flow and dispersal, outcrossing, introgression, and hybridization |
| SSR | non-coding repetitive DNA regions composed of small motifs of 1 to 6 nucleotides repeated in tandem | Abundant, widely distributed throughout the genome; High mutation rate; Co-Dominant marker; High experimental repeatability; reliable results | Difficult to develop; High cost | Construction of genetic maps; Analyze genetic structure; Species identification; Genetic breeding and assessment of genetic variation |
| SNP | DNA sequence diversity caused by single nucleotide variation at the genomic level | Co-dominant markers; Abundance; Low genotyping cost; Typically biallelic; High-throughput genotyping; Lower mutation rate; Genotyping error rates | Difficult detection; Large workload | Construction of high-dense maps; Quantitative trait loci mapping; Assessment of genetic diversity; Parentage studies; Marker-assisted breeding |
| Mitochondrial DNA markers | Inherited from maternal lineage and efficiently identify and classify species based on a short DNA sequence in population and evolutionary biology | High mutation rate; Maternal inheritance; Simple, fast, and reliable | Limited information compared with nuclear genes | Species identification; Examine population genetic structure; Examine phylogenetic relationships |
Characteristics, advantages, and disadvantages of various types of molecular markers.
Figure 1
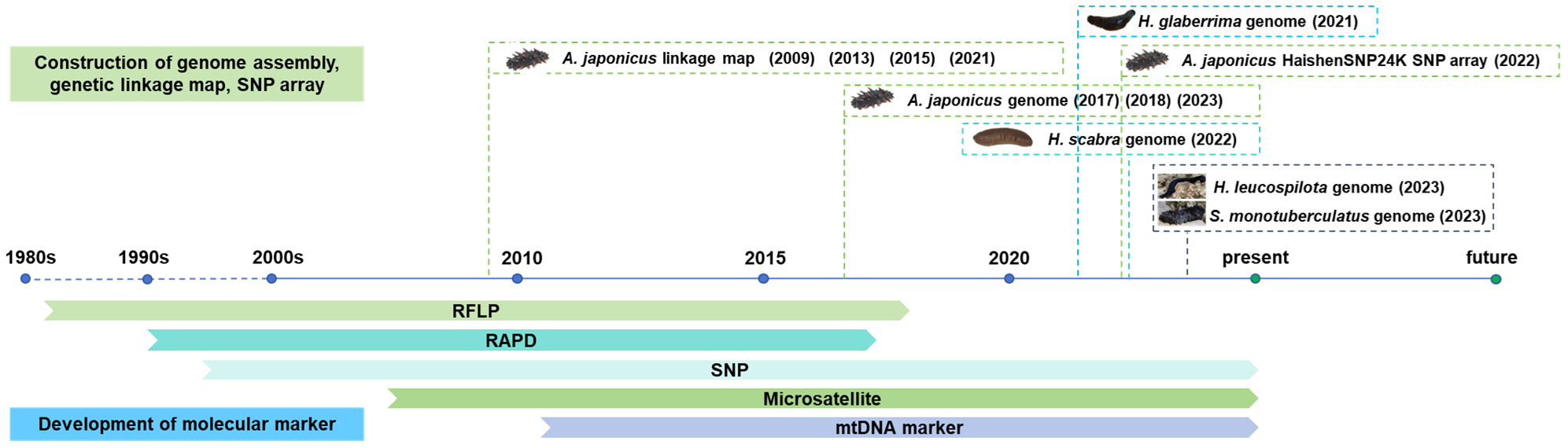
Time history of molecular markers and their based genomic methods applied in sea cucumbers.
Table 2
| Types | Sea cucumber species | Location | Method | Population number | Description | Reference |
|---|---|---|---|---|---|---|
| 16S rRNA gene region | Stichopus hermanni, Stichopus chloronotus, Thelenota ananas, Thelenota anax, Bohadschia argus, Holothuria fuscopunctata, Holothuria leucospilota, Holothuria scabra, Actinopyga echinites | Sanya, China | species-specific PCR method | 15 individuals per sample | a simple, fast, and reliable protocol for the accurate identification of sea cucumber species | Wen et al., 2012a |
| mitochondrial COI | Holothuria atra | Hawaiian Islands, Line Islands, Marshal Islands, Bonin Islands, and Ryukyu Islands | Sanger sequencing | 385 | examining population genetic structure to aid coral reef management | Skillings et al., 2011 |
| mitochondrial COI and 16S ribosomal RNA (16S), and nuclear histone (H3) | Holothuria edulis | Okinawa Main Island in Ryukyu Archipelago (Motobu, Oyama, Ryugu, Sunabe, Philippine Sea and Awase) | Sanger sequencing | 438 | Investigating the metapopulation structure to provide valuable data to help management in vulnerable locations of Okinawa | Soliman et al., 2016 |
| COI DNA barcoding | 4 species belonged to the Stichopodidae (Stichopus herrmanni, Stichopus ocellatus, Stichopus horrens, Stichopus monotuberculatus), and 3 species to the Holothuridae (Bohadschia bivittata, Actinopyga lecanora and Holothuria leucospilota) | Kepulauan Seribu reefs, Indonesia |
phylogenetic analysis | 96 | species identification | Madduppa et al., 2017 |
| COI DNA barcoding | 23 sea cucumber species | Not applicable | Sanger sequencing and next-generation DNA sequencing | 23 | screening of commercially processed sea cucumber products for clearer labeling | Xing et al., 2021 |
| COI and ITS loci DNA barcoding | Stichopus | Karimunjawa Island, Central Java province, Indonesia | Not applicable | 100 individuals consisting of three sea cucumber types (30 Crengek gamete, 36 Pace gamete, and 34 Kuning gamete) | genetic diversity, species identification, and examine phylogenetic relationships of the Stichopus genus | Sulardiono et al., 2022 |
| RAPD | Apostichopus japonicus | Dalian and Weihai, China | RAPD-PCR | 26 | geographical origin and genetic diversity analysis | Yun et al., 2017 |
| RFLP | Aspidochirotida: Stichopodidae | Sanya, Dalian, and Guangzhou, China | PCR-RFLP and FINS | 70 | identify six species belonging to family Stichopodidae in commercial products | Wen et al., 2010 |
| RFLPs | Holothuria mammata Grube, 1840 and H. sanctori Delle Chiaje, 1823 | The Azores in the north-east Atlantic, nine volcanic oceanic islands and several islets | PCR-RFLPs | Not applicable | quick identification of two species of sea cucumber | Madeira et al., 2018 |
| PCR-RFLP, 16Sar/16Sbr primers | 16 commercial sea cucumber species | Sanya and Dalian, China | PCR | 6 individuals for each species |
identify 16 types of sea cucumber in raw materials and processed foods | Zeng et al., 2018 |
| EST-SSRs | Apostichopus japonicus | Rongcheng, Shandong Province, China | EST database | 48 | population structure analysis, phylogenetic and comparative genomics studies and genetic linkage map construction | Peng et al., 2009 |
| Microsatellite | Stichopus japonicus | Onagawa Bay in Miyagi Prefecture, Japan | Not applicable | Not applicable | tools for genetic analyses, investigate the genetic structure of S. japonicus and to detect reproductive isolation among color types through mating experiments | Kanno et al., 2005 |
| Microsatellite | Apostichopus Japonicus | Aomori, Hokkaido, Miyagi, Oita, Japan; Qingdao, China | Not applicable | 403 | population identity of sympatric green and black, difference in the population structure, and further ecological and genetic research | Kanno et al., 2006 |
| Microsatellite | Apostichopus japonicus | Not applicable | screening from SSR enriched library and EST database | 45 | population structure and the stock enhancement in fishery, the species/hybrid identification, and the linkage map construction | Zhan et al., 2007 |
| Microsatellite | Stichopus japonicus | Penglai, China | expressed sequence tags | 11 | examining genetic population structure, parentage analysis and mapping studies | Chen and Li, 2007 |
| Microsatellite | Apostichopus japonicus | Not applicable | Not applicable | Not applicable | an efficient procedure for isolating microsatellite DNA | Hu et al., 2007 |
| Microsatellite | Apostichopus japonicus | Dalian, China | combining microsatellite DNA polymorphism and its relationship with body weight | 208 | molecular breeding, marker-assisted selection | Wang et al., 2009 |
| Microsatellite | Apostichopus japonicus, Parastichopus parvimensis, and Pathallus mollis warty sea cucumber (Parastichopus parvimensis), and black sea cucumber (Pathallus mollis) | Qingdao, China; United States; Peru | Not applicable | 60 | future genetic breeding and the assessment of genetic variation within sea cucumbers | Liao et al., 2011 |
| Microsatellite | Apostichopus japonicus | Jiaonan, Shandong, China. | expressed sequence tags | 30 | provide sufficient polymorphism for population genetic studies and genome mapping | Jiang et al., 2011 |
| Microsatellite | Stichopus chloronotus | Okinawa Archipelago, Japan | Not applicable | 60 | detailing the genetic structure and gene flow | Taquet et al., 2011 |
| Microsatellite | Apostichopus japonicus | Rongcheng, China | enrichment-colony hybridization protocol | 48 | population genetic studies and molecular marker-assisted breeding | Peng et al., 2012 |
| Microsatellite | Apostichopus japonicus | Noto, Shinminato, and Uozu around Toyama Bay | PCR | 236 wild individuals (146 red and 90 green variants) | clarify larval dispersal among populations by describing the levels of distinctiveness and gene flow among red and green variant populations, provide useful information for genetic conservation and fisheries management | Soliman et al., 2012 |
| Microsatellite | Stichopus japonicus | Jeju island, Korea | multiplex assays with eight highly polymorphic microsatellite loci | 250 | reveals the genetic composition and significant genetic differentiation between wild and hatchery-produced red sea cucumber samples in Korea | An et al., 2013 |
| Microsatellite | Apostichopus japonicus | Liaoning, China | transcriptome sequencing | 20 | linkage mapping, comparative genome analysis and genetic breeding | Chen et al., 2013 |
| Microsatellite | Holothuria scabra | Croker Island, Northern Territory, Australia | next-generation sequencing | 50 | population structure and mating systems, investigation of population structure, levels of gene flow and mating systems | Fitch et al., 2013 |
| Microsatellite | Holothuria leucospilota | Hainan Island, China | amplified fragment length polymorphism | 30 | germplasm conservation, genetic diversity, population structure, and conservation strategy design | Dai et al., 2015 |
| Microsatellite | Holothuria scabra | Hainan Island, China | amplified fragment length polymorphism | 30 | genetic diversity and its effective conservation strategy, population structure | Li et al., 2015a |
| Microsatellite | Stichopus horrens | Hainan Island, China | fast isolation by amplified fragment length | 30 | studying population structure and conservation strategy design, genetic diversity | Li et al., 2015b |
| Microsatellite | Holothuria leucospilota | Not applicable | amplified fragment length polymorphism | 30 | genetic diversity, and the design of conservation strategies, population structure studies and cultivation | Shangguan et al., 2015a |
| Microsatellite | Stichopus horrens (Selenka) | Sanya, Hainan, China | fast isolation method with amplified fragment length | 35 | genetic structure, population conservation, and breeding of wild S. horrens | Shangguan et al., 2015b |
| Microsatellite | Holothuria mammata | Atlantic and Mediterranean Sea | SSR enriched library on a Roche 454 platform | 60 | fisheries management including identification of stocks, assessment of genetic diversity, estimation of gene flow and monitoring the fishery effects on exploited populations | Henriques et al., 2016 |
| Microsatellite | Holothuria grisea | Bitupitá beach, Barroquinha, Ceará, Brazil | Illumina paired-end reads of whole genome shotgun sequencing | 30 | proper conservation of the species and development of sustainable fishery and aquaculture | Pereira et al., 2018 |
| Microsatellite | Apostichopus japonicus Selenka, 1867 | Peter the Great Gulf, Japan | Not applicable | 159 | genetic variability and population structure | Yagodina et al., 2022 |
| SNP | Apostichopus japonicus | Dalian, Yantai, Qingdao, and Wendeng, China | Not applicable | 39 | delineating population structure in the sea cucumber | Sun et al., 2010 |
| SNP | Stichopus monotuberculatus | Xidao Island, Sanya, China | gene-based | 80 | future quantitative trait loci (QTL) analysis, and to facilitate marker-assisted selection (MAS) | Du et al., 2012 |
| SNP | Apostichopus japonicus | Wendeng, China | mining EST | 32 | genetic diversity assessment, genome mapping, reproductive ecology analysis and SNP-based analysis in aquaculture practice | Yang et al., 2012 |
| SNP | Apostichopus japonicus | Dalian, China | SNP markers associated with defense mechanism | 50 | useful for the construction of genetic linkage map and comparative genome analysis | Gao et al., 2013 |
| SNP and SSRs | Apostichopus japonicus | Dalian, China; Vladivostok, Russia; Rajin, North Korea | transcriptome analysis of tube foot | 45 | provide abundant genetics and molecular ecology resources | Zhou et al., 2016 |
| SNPs | Apostichopus japonicus | Changdao, Yantai, Jiaonan, Wendeng, Penglai, China; Oita, Aomori, Japan | EST sequences | 40 | guides selective breeding | Dong et al., 2016 |
| SNPs and SSR | Apostichopus japonicus | Yantai, China | transcriptome sequencing | 150 | genes discovery and functional genomic studies of the sea cucumber | Zhou et al., 2013 |
| SNPs | Holothuria (Metriatyla) scabra | Six locations throughout Fiji (Namuka district, Lakeba island, Tavuki district, Serua island, Raviravi district, Nacula island) | Illumina sequencing | 211 | investigate genetic structure, diversity, signatures of selec-tion, relatedness and connectivity in wild populations | Brown et al., 2022 |
| SNPs | Holothuria leucospilota | Islands from Northern Vietnam, Central Vietnam, and Southern Vietnam; Darwin, Northern Australia and Mooloolaba, Eastern Australia | restricted site associated DNA sequencing | 180 | investigate population genetics to provide basic genetic information, design restocking management plans, genetic conservation initiatives, and sea ranching programs | Chieu et al., 2023 |
Development and application of various types of molecular markers in sea cucumbers.
2 Development of various molecular markers in sea cucumber
2.1 Mitochondrial DNA markers
The DNA barcoding using mitochondrial COI (cytochrome C oxidase subunit I) and ITS (internal transcribed spacers) can be used for identifying species for Stichopus genus and analyzing genetic diversity and phylogenetic relationships (Madduppa et al., 2017; Sulardiono et al., 2022). More specifically, the DNA barcode mitochondrial COI has been used for identifying sea cucumber species in two main families, including Stichopodidae (Stichopus herrmanni, Stichopus ocellatus, Stichopus horrens, Stichopus monotuberculatus) and Holothuridae (Bohadschia bivittata, Actinopyga lecanora, and Holothuria leucospilota), which will benefit germplasm conservation and management (Madduppa et al., 2017).
Using the mitochondrial DNA marker, three local sea cucumber species in Indonesia, Crengek Gamete, Pace gamete, and Kuning gamete, were close relatives to Stichopus monotuberculatus (Sulardiono et al., 2022). A next-generation sequencing-based DNA mini-barcoding using COI has been developed to screen various commercial sea cucumber products and correct mislabeled for clearer labeling guidelines (Xing et al., 2021). The complete mitochondrial genome and cox1 DNA barcoding will be helpful for the sea cucumber breeding industry and studies on population genetic diversity and molecular ecology resources (Fan et al., 2012). In addition, a species-specific PCR of 16S rRNA has been used to authenticate 11 sea cucumber species from processed products, providing a rapid and useful protocol for correct labeling of commercial sea cucumber products (Wen et al., 2012a). By calculating the genetic distance of COI gene among different sea cucumber, Lu et al. proved the feasibility of COI as a DNA barcode for species identification, and the probes designed based on COI gene were used to identify four species of sea cucumber (Lv et al., 2011). Hu et al. used COI gene and 16S rRNA gene as target genes to identify sea cucumber species based on DNA barcoding technology, and the results showed that COI gene or 16S RNA gene could identify most species of sea cucumber (Hu et al., 2019). According to the specific primers for cox1 gene of Holothuria scabra, eDNA technology was used to investigate Holothuria resources in Weizhou Island, and the results showed that the eDNA monitoring method was more suitable and reliable than traditional methods (E et al., 2023).
2.2 Traditional molecular markers
A PCR-RFLP method was developed to identify six species of sea cucumbers in commercial products, which can authenticate species and detect fraudulent labeling (Wen et al., 2010). Another application of the PCR-RFLP method was to identify 16 commercial sea cucumber species from food products to correct the mislabeling (Zeng et al., 2018). Moreover, a PCR-RFLP method using restriction nuclease Sau3AI on 16S rRNA fragments has been developed to rapidly and inexpensively identify holothurian species with no need for taxonomical or genetic expertise (Madeira et al., 2018). DNA polymorphisms identified by RAPD-PCR technology were used to evaluate the geographical origin of A. japonicus and genetic diversity analysis (Yun et al., 2017). EST databases of sea cucumber species have been built, including Holothuria glaberrima during intestinal regeneration (Rojas-Cartagena et al., 2007), multiple tissues (body wall, intestine, respiratory tree) in Apostichopus japonicus (Yang et al., 2009), which were submitted to NCBI Genbank. EST databases have provided a rich resource for the development of molecular markers of sea cucumber species, especially microsatellite markers. SSR can be identified from EST data using screening programs, for example, SSRHUNTER (Chen and Li, 2007; Zhan et al., 2007; Peng et al., 2009; Jiang et al., 2011). In addition, SNP markers were characterized by mining EST data of A. japonicus and using a homogeneous Tm- (melting temperature) genotyping method (Yang et al., 2012; Dong et al., 2016).
2.3 Microsatellite markers
The majority of microsatellite markers in sea cucumber species were from an aquaculture species in the coastal area of the Northwest Pacific, Stichopus (Apostichopus) japonicus. The earliest 20 microsatellite markers of sea cucumber Apostichopus japonicus were developed by Kanno et al. in 2004, which could be excellent molecular markers to facilitate genetic structure analysis (Kanno et al., 2005). Eleven microsatellite markers were used to identify A. japonicus population from green and black color variants for further genetic population structure analysis and ecological research (Kanno et al., 2006). In sea cucumber Apostichopus japonicus, Hu et al. developed an efficient procedure for isolating a reasonable number of microsatellites for genetic analysis (Hu et al., 2007). Forty-five informative microsatellite markers (43 genomic SSRs and 2 EST-SSRs) were developed based on screening SSR-enriched library and EST library, which may contribute to population structure analysis, linkage map construction, stock enhancement, and species identification (Zhan et al., 2007). Eleven microsatellite markers derived from expressed sequence tags were developed to investigate population structure and conduct parentage analysis and QTL mapping (Chen and Li, 2007). Microsatellite markers associated with body weight were developed for not only molecular breeding but also marker-assisted selection (MAS) (Wang et al., 2009). Jiang et al. have developed 43 EST-SSR markers for future studies in population genetics and genomics (Jiang et al., 2011). Twenty novel microsatellite markers from Apostichopus japonicus were used for cross-species application in Parastichopus parvimensis from the United States and Pathallus mollis from Peru, which could facilitate genetic variation assessment and genetic resource management among Holothuroidea populations (Liao et al., 2011). A larger number of ~70 microsatellite markers were developed for the implementation of marker-assisted selection and population genetic studies (Peng et al., 2012). Furthermore, 45 gene-derived microsatellite markers were identified from transcriptome sequencing, which could be useful for comparative genomics studies, genetic linkage mapping, and molecular breeding like MAS (Chen et al., 2013). A total of 8 highly polymorphic microsatellite markers were used for genetic structure studies between wild and hatchery populations and implications in breeding programs and stocking strategies (An et al., 2013). Ten microsatellite markers have revealed the genetic variation and population structure of A. japonicus in Peter the Great Gulf, Japan. In studying microsatellite markers, the detected high heterogeneity among populations might be due to several factors: stochastic larval settlement at a location, and selection pressures on QTL (Yagodina et al., 2022).
Ten microsatellite markers were developed in a tropical sea cucumber species Stichopus chloronotus to investigate the genetic structure and gene flow among populations (Taquet et al., 2011). Eighteen microsatellite markers from another sea cucumber species Holothuria scabra were characterized, which were beneficial for genetic structure and gene flow studies for sea cucumber aquaculture and management (Fitch et al., 2013). A total of another 9 microsatellite markers of H. scabra were developed for conservation strategy and investigation of genetic diversity and population structure (Li et al., 2015a). Eight microsatellite markers were identified in a tropical species Holothuria leucospilota, which could be beneficial for genetic structure, population diversity, and germplasm conservation (Dai et al., 2015). Shangguan et al. identified 16 additional microsatellite markers from H. leucospilota that could be used for studies in genetic diversity and population structure and application in cultivation and conservation strategies (Shangguan et al., 2015a). Li et al. identified 9 microsatellite markers in another tropical sea cucumber Stichopus horrens (Selenka) that could be used as markers to study its conservation and breeding, population structure, phylogeny and evolutionary studies of Holothuroidea (Li et al., 2015b). 9 more microsatellite markers have been characterized in S. horrens, which could be helpful for conservation strategy and management, population structure and genetic diversity of S. horrens (Shangguan et al., 2015b). 9 novel microsatellite markers have been identified from Holothuria mammata, an exploited sea cucumber species in Mediterranean and North-Eastern Atlantic, which could be used for aquaculture and conservation management, gene flow, and genetic diversity assessment (Henriques et al., 2016). Furthermore, eight microsatellite markers in the West Atlantic sea cucumber species Holothuria grisea have been developed and used as markers for proper conservation and advancement in sustainable aquaculture of this species (Alves Pereira et al., 2018). Despite the popularity of SSR in diverse aspects of sea cucumber research, it is gradually replaced by genome-wide molecular markers such as SNPs.
2.4 Single nucleic polymorphism (SNP)
In A. japonicus, all identified 13 SNPs have two alleles and their heterozygosity could be used as molecular markers in analyzing population structure, population genetics, and QTL mapping (Sun et al., 2010). Fifteen more SNP markers were identified from ESTs and provided a complement to available molecular markers of A. japonicus (Yang et al., 2012). These SNPs may be implied in studies on genetic diversity, QTL mapping, and ecology analyses of A. japonicus (Yang et al., 2012). 26 SNP markers associated with defense mechanisms were developed by mutation scanning from unigene sequences in A. japonicus transcriptome, which may provide insights into genetic maps and comparative genomic studies (Gao et al., 2013). 51 gene-derived SNPs from ESTs using high-resolution melting can be used for MAS, genetic improvement, and aquaculture management (Dong et al., 2016). High throughput transcriptome analysis provided benefit to large-scale marker discovery of A. japonicus. 142,511 SNP markers and 6,417 microsatellite markers with high quality have been identified through transcriptome analysis, which can be used for gene discovery and functional genomic research (Zhou et al., 2014). A total of 219,860 SNP markers have been identified from tube foot transcriptome, providing plentiful resources in genetic and molecular ecology studies (Zhou et al., 2016).
3 Construction of genome assembly, genetic linkage map, and SNP array
Since 2017, the genome assemblies of sea cucumber species have been constructed using next-generation sequencing and third-generation sequencing, including one northern sea cucumber species A. japonicus, and three tropical sea cucumber species, H. scabra, H. leucospilota, H. glaberrima and S. monotuberculatus (Table 3). As the most common commercial sea cucumber species, the A. japonicus genome assembly have been constructed in 2017 (Zhang et al., 2017) and 2018 (Li et al., 2018) and updated in 2023 (Sun et al., 2023). The construction of genome assembly has been used to understand the molecular mechanisms on morphological evolution, visceral regeneration, saponin biosynthesis, aestivation regulation, providing a referable resource for understanding evolution and biodiversity of sea cucumber. In 2022, De novo genome assembly of H. scabra has been built using nanopore sequencing, which provides excellent genetic resource for studying genetic, phylogenetic, molecular biology, and breeding studies (Luo et al., 2022). In 2023, a draft genome of S. monotuberculatus was assembled using Nanopore sequencing, which provides insight into critical genes in fucosylated chondroitin sulfates biosynthetic pathway (Zhong et al., 2023). The high-resolution genome assembly of H. leucospilota was also built in 2023 using Illumina sequencing, Pacbio sequencing, BGI sequencing platform, which provides insights into molecular underpinnings of sacrificial organ expulsion in holothurian species (Chen et al., 2023).
Table 3
| Types | Sea cucumber species | Location | Method | Population number | Description | Reference |
|---|---|---|---|---|---|---|
| genome | Apostichopus japonicus | Laoshan, Qingdao, China | Illumina+PacBio (CLR) | NA | enables exploring key genetic mechanisms behind the unique biological characters, phylogenetic and comparative genomic analyses | Zhang et al., 2017 |
| genome | Apostichopus japonicus | Shandong Province, China | Illumina+PacBio (CLR) | NA | reveals novel genomic features and molecular variations that contribute to the evolutionary adaptation, provides insights into saponin synthesis and aestivation regulation. |
Li et al., 2018 |
| genome | Apostichopus japonicus | Rushan, Shandong Province, China | Illumina+Pacbio (CCS) | NA | an excellent tool to better investigate the mechanisms that drive evolution and biodiversity | Sun et al., 2023 |
| genome | Holothuria glaberrima | Puerto Rico, USA | Illumina | NA | a critical resource for future studies in animal regeneration | Medina-Feliciano et al., 2021 |
| genome | Holothuria scabra | Guangxi Province, China | Nanopore | NA | provides a referable resource for research on genetic, phylogenetic, molecular biology, and breeding studies | Luo et al., 2022 |
| genome | Stichopus monotuberculatus | Guangxi Province, China | Nanopore | NA | provides a genomic approach to investigate the structural diversity, as well as novel perspectives into evolutionary adaptation of critical genes in holothurian fucosylated chondroitin sulfates biosynthesis pathways | Zhong et al., 2023 |
| genome | Holothuria leucospilota | Nanhai, China | Illumina+PacBio+BGI | NA | provides insights into molecular underpinnings of sacrificial organ expulsion in holothurian species | Chen et al., 2023 |
| SNP array | Apostichopus japonicus | Liaoning and Shandong, China; Russia | a high-throughput 24 K SNP genotyping array | 210 | minmax concave penalty (MCP) regularization for sparse deep neural networks (DNN-MCP) high-throughput genotyping platform | Lv et al., 2022 |
| Genetic linkage map | Apostichopus japonicus | Not applicable | AFLP and microsatellite markers | 88 | application of a marker-assisted selection breeding strategy, basis for mapping of the functional genes and quantitative trait loci, mapping QTL for growth heterosis and locating expressed genes in aquaculture. | Li et al., 2009 |
| Genetic linkage map | Apostichopus japonicus | Shandong Province, China | microsatellites and SNPs | 144 | facilitate further sea cucumber genetic studies such as quantitative trait loci (QTL) mapping and comparative genomic analysis | Yan et al., 2013 |
| Genetic map | Apostichopus japonicus | Penglai, China | genotyping-by-sequencing (GBS) methods, 2b-restriction site-associated DNA (2b-RAD) method | 100 | a powerful tool for research involving both fine-scale QTL mapping and marker assisted selection (MAS) | Tian et al., 2015 |
Construction of genome assembly and molecular markers-based genomic methods in sea cucumbers.
NA, not applicable.
Genetic linkage maps are constructed based on the gene linkage and genetic recombination rates to infer the relative positions of genes or genetic markers on chromosomes (Cui et al., 2021). Up to date, a total of four genetic linkage maps of Apostichopus japonicus based on molecular markers have been developed for QTL mapping and marker-assisted selection breeding since 2009 (Table 3). The first genetic linkage map was built on 484 polymorphic AFLP and microsatellite markers, which provides the application of molecular breeding and understanding the genetic basis of economic traits in sea cucumber industry (Li et al., 2009). In 2013, a medium-density genetic linkage map using co-dominant microsatellite and SNP markers was built to benefit QTL mapping, marker-assisted selection, and comparative genomic studies (Yan et al., 2013). A high-density and high-resolution genetic map of 7,839 markers based on 2b-restriction site-associated DNA (2b-RAD) was constructed in 2015, which facilitates fine-scale QTL mapping, precise marker-assisted selection, and chromosome assignment for whole genome assembly (Tian et al., 2015). A 6,144 SNP-based high-density genetic map using genotyping-by-sequencing was built, which provides an important genomic resource for marker-assisted selection with high performance traits like growth traits (Sun et al., 2021). With the development of whole genome sequencing and new molecular techniques, genome-wide molecular markers like SNPs in an SNP array chip have been used as an efficient genotyping tool for genetic research and genome breeding programs in aquaculture. In 2022, a high-throughput 24 K SNP array genotyping platform, HaishenSNP24K, was developed for genetic studies and application in selection and breeding programs with minmax concave penalty regularized deep neural networks (DNN-MCP) (Lv et al., 2022). The new SNP array has significantly improved the genomic assessment of sea cucumber populations for quantitative performance traits, including wet or dry weight, and survival time (Lv et al., 2022).
4 Application of molecular markers and based genomic methods in fisheries, aquaculture, and industry
The molecular marker technology facilitates germplasm resource evaluation, assessment of genetic diversity and population structure, sex identification for live animal, and selection of desirable traits, which has been extensively applied in various aspects of the sea cucumber aquaculture industry (Figure 2). These applications play a pivotal role in enhancing the efficiency of industrialized aquaculture and breeding programs for sea cucumbers, enabling precise breeding strategies and conservation of germplasm resources.
Figure 2
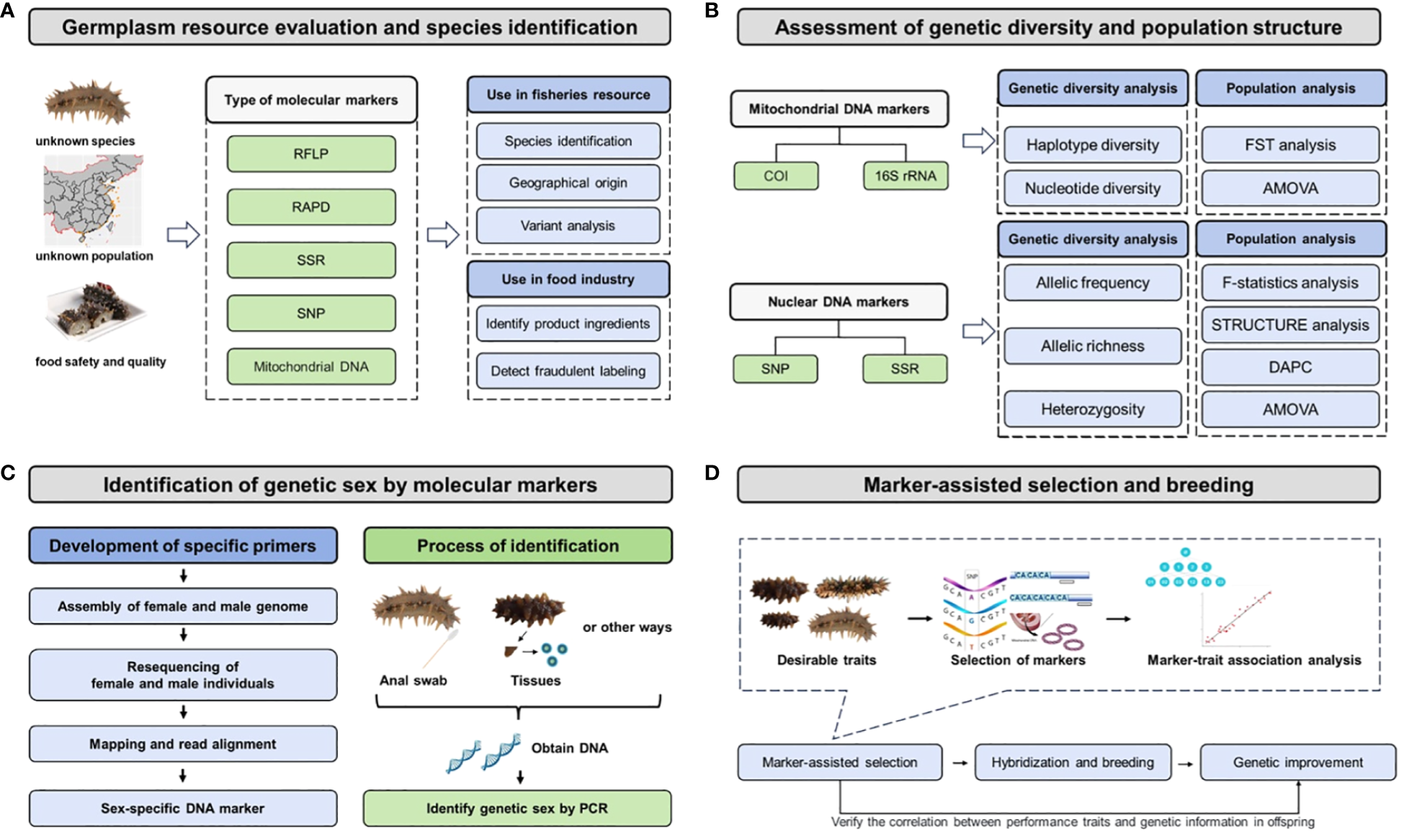
Flow chart of molecular markers or based genomic methods in sea cucumber fisheries, aquaculture, and industry. (A) Germplasm resource evaluation and species identification; (B) Assessment of genetic diversity and population structure; (C) Genetic sex identification in aquaculture; (D) Marker-assisted selection and breeding.
4.1 Germplasm resource evaluation and species identification
Germplasm resource evaluation mainly includes population resource information, ecological factors, habitat conditions, environmental variables, external disturbance factors, and strain identification. Germplasm resource identification has significant implications not only in the discovery and development of new germplasm but also in hybrid and cryptic species identification and the utilization of hybrid advantages (Ding and Chang, 2020). Specific sequence analysis plays an important role in natural hybridization and gene flow between populations. For example, skeletal characteristics could not distinguish H. scabra, colour morphs of H. S. var.versicolor, and intermediate phenotype hybrids in the species boundary study, as their color patterns, morphology, and small-scale distribution patterns were different (Uthicke et al., 2005). Both of population genetics and 16S mtDNA sequence analysis were used to identify new species H. S. var.versicolor and individuals with intermediate phenotypes represented F1 hybrids, demonstrating that the opportunity for introgression exists (Uthicke et al., 2005). The conventional method based on their morphology or external characteristics for species identification have gradually developed towards molecular markers with high accuracy and polymorphism, such as RFLP, RAPD, SSR, and SNP (Lv, 2012). In general, germplasm resource evaluation by molecular markers includes four important steps: sample collection, DNA extraction, PCR amplification using molecular markers, gel electrophoresis or DNA sequencing. For instance, PCR-RFLP was used to develop a method of DNA barcoding-based sea cucumber species identification, and mitochondrial COI gene was used as a DNA barcode to achieve germplasm identification of five sea cucumber species (Lv, 2012). Wen et al. successfully identified 19 commercial sea cucumber species using PCR-RFLP method (Wen et al., 2012b). With the completion of mitochondrial genome sequencing of Apostichopus japonicus and other sea cucumber species at the National Center for Biotechnology Information (NCBI), the basic characteristics of the mitochondrial genome of the taxonomic class Holothuroidea have been revealed, which could benefit germplasm resource evaluation using molecular methods (Shen et al., 2011). The use of all these molecular markers in fisheries resource evaluation include species identification, geographical origin, and variant analysis. In sea cucumber industry, molecular methods could identify product ingredients and detect fraudulent labeling for food safety and better quality.
4.2 Assessment of genetic diversity and population structure
Molecular marker technology has been widely applied in the assessment of population genetic structure and genetic diversity, providing guidance for the sea cucumber aquaculture industry and germplasm resource protection. Bottom-culture farming in marine ranching is an important means of germplasm resource protection, and the evaluation of genetic diversity and population structure of its natural geographical populations is crucial. In general, this includes four important steps: sample collection, DNA extraction, genotyping or sequencing, and data analysis. Numerous sea cucumber studies have utilized mitochondrial DNA markers (e.g. COI, 16S) and nuclear DNA markers (e.g. SNP, SSR) for this purpose. Mitochondrial DNA markers are used to conduct haplotype diversity (h) and nucleotide diversity (π) for genetic diversity analysis, as well as FST (fixation index) and AMOVA (Analysis of molecular variance) for population analysis. For instance, Yan (2006) used COI molecular marker methods to investigate the genetic diversity and population genetic structure of A. japonicus from five geographic subgroups in the Dalian Sea area and revealed that these five geographical subgroups had high genetic diversity. A mitochondrial COI gene analysis for the population genetic structure of Holothuria atra in the waters surrounding the Hawaiian Islands in order to aid coral reef management (Skillings et al., 2011). Additionally, nuclear DNA markers have proven to be an effective method for evaluating genetic diversity, including allelic frequency, allelic richness, and heterozygosity, as well as population analysis including F-statistics analysis (Fixation indices), STRUCTURE analysis, DAPC (discriminant analysis of principal components), AMOVA. For instance, Wang et al. conducted genetic diversity and population structure assessments of 18 geographical populations of A. japonicus from sea ranching in Shandong, Hebei, and Liaoning provinces using specific SNP loci, thereby enabling continuous genetic monitoring of the northern natural habitats of sea cucumbers in China (Wang et al., 2023). Whole-genome SNP data was used to identify three genetically distinct subgroups within populations of Holothuria (Metriatyla) scabra from Fiji, revealing a model of genetic structure isolated by distance (Brown et al., 2022). An investigation of population genetics between Australia and Vietnam using SNP markers in H. Leucospilota was conducted to compare gene flow and genetic similarities among populations and analyzing population genetic structure (Chieu et al., 2023). Now land et al. used population genetic analysis based on microsatellite sequences to assess the population genetic structure of H. scabra in Papua New Guinea and a broader region of northern Australia, in order to determine the genetic diversity within subpopulations and investigate the genetic impact of environmental changes on wild populations (Nowland et al., 2017). Eight polymorphic microsatellite loci were successfully developed in H. leucospilota, which provided crucial molecular markers for studying population genetic diversity and conservation strategies in sea cucumbers (Dai et al., 2015). Liao et al. utilized SSR fingerprinting technology to analyze the genetic diversity and construct fingerprinting profiles of sea cucumber populations from different regions in China, South Korea, and Russia (Liao et al., 2021). The constructed fingerprinting profiles successfully differentiated eight populations and provided technical support for the protection of sea cucumber germplasm resources and the identification of different geographical populations (Liao et al., 2021). Furthermore, genetic diversity and structure assessments of wild populations and hatchery-produced H. scabra in New Caledonia were achieved using nine polymorphic microsatellite markers, aiming to develop an appropriate breeding plan for restocking (Riquet et al., 2022).
4.3 Identification of genetic sex
Due to the lack of visible sexual dimorphism in the external appearance of most sea cucumber species, the development and application of a method based on molecular markers is useful to identify the sex of several sea cucumber species (Table 4). Generally, the development of a sex-specific marker is realized through the assembly of female and male genome sequences, resequencing of female and male individuals, mapping and read alignment, identification of sex-specific region, and design and verification of sex-specific marker. The verification of an accurate and stable sex-specific marker is usually completed by wild populations in multiple geographical regions. Tested samples can be collected by anal swab or tissue dissection, and then genomic DNA can be extracted by multiple protocols. The genetic sex of sea cucumber individual is identified by PCR amplification using sex-specific marker. The utilization of sex-specific molecular markers facilitates the determination of individual gender and enhances the efficiency of artificial breeding in large scale aquaculture. In A. japonicus, a sex-specific marker was developed utilizing 2b-RAD technology, which holds significant implications for genetic breeding (Wei et al., 2021). In addition, a loop-mediated isothermal amplification (LAMP) method was established based on male-specific sequences, which could rapidly identify the genetic sex of A. japonicus (Cong et al., 2024). Furthermore, a rapid and non-destructive method was devised for sex identification in aquaculture practices involving another tropical sea cucumber species (Stichopus monotuberculatus) (Wu et al., 2022). A non-invasive and rapid method based on anal swab sampling using a LAMP system (PCR and loop-mediated isothermal amplification) was employed for sex identification in tropical sea cucumber Holothuria scabra (E et al., 2023).
Table 4
| Types | Sea cucumber species | Location | Method | Population number | Description | Reference |
|---|---|---|---|---|---|---|
| sex-specific markers | Apostichopus japonicus | Liaoning, China. | 2b-RAD | 97 | provide a solid basis to reveal the sex determination mechanism and the origin of sex-determining regions; improve the efficiency of genetic and sex control breeding in sea cucumbers | Wei et al., 2021 |
| sex-specific markers | Stichopus monotuberculatus | Xidao Island, Sanya, China | genomic sequencing | 80 | an effective way to identify sex in sea cucumbers | Wu et al., 2022 |
| PCR and LAMP system | Holothuria scabra | Zhanjiang, China | DNA sequencing | 50 | investigate the basis of sex determination in the Holothuriidae family | E et al., 2023 |
| PCR and LAMP system | Apostichopus japonicus | Dalian, China | genomic sequencing | 100 | A rapid LAMP-based method of identifying the genetic sex without complex equipment. | Cong et al., 2024 |
Development and application of sex-specific markers for genetic sex identification in sea cucumbers.
4.4 Marker-assisted selection and breeding
With the continuous development of breeding technologies, there is a gradual shift from traditional selective breeding and hybrid breeding techniques towards the integration of molecular marker-assisted breeding, whole-genome selection breeding, molecular design breeding, sex control, gene transfer, and gene editing (Ding et al., 2021). Molecular marker-assisted breeding is a technique in aquaculture breeding that involves directly selecting individuals with advantageous alleles or genotypes for specific traits using molecular markers closely associated with those traits, which applies the research findings of genomics and molecular biology to the selection and breeding of aquaculture species (Lu et al., 2019). It demonstrated that the construction and use of genetic linkage maps with a high number of molecular markers could connect desirable performance traits with molecular markers, which can benefit marker-assisted selection and breeding and accomplish genetic improvements for sea cucumber aquaculture. The accomplishments of marker-assisted selection and breeding can be evaluated through the correlation between performance traits and genetic information in offspring. In A. japonicus, the insertion/deletion (INDEL) marker of internal transcribed spacer 1 (ITS1) sequence divergence was successfully used to identify the genetic types of different body color varieties, and hybridization experiments showed that this marker could be used to distinguish and track the genetic composition of F1 hybrid larvae, which explored the relationship between body color variation and genetic background and its impact on larval development (Yoshida et al., 2012). The linkage map using AFLP markers serves as a foundation for constructing high-resolution genetic maps and identifying functional and quantitative trait loci (Li et al., 2009). Yan et al. constructed a common genetic map for sea cucumbers based on microsatellite and SNP markers, which was consistent with the haploid chromosome number of sea cucumbers (Yan et al., 2013). Liu et al. used a high-density genetic linkage map of sea cucumbers to preliminarily locate QTL regions associated with five traits: body length, body width, body weight, total number of spines, and survival rate (Liu et al., 2019). The “Sea Cucumber Superior No.1” was successfully bred using molecular marker-assisted breeding, which has strong resistance to Vibrio splendidus infection and fast growth rate (Ding and Chang, 2020). The new germplasm was achieved through population selection, screening and verification of disease-resistant molecular markers that indicate disease-resistant functional genes (Ding and Chang, 2020). In A. japonicus, a new high-density genetic linkage map was constructed based on single nucleotide polymorphism (SNP) molecular markers, and quantitative trait loci (QTL) mapping analysis was performed to explore the genetic mechanism of sea cucumber growth traits, which could be helpful for future marker-assisted breeding (Cui et al., 2021).
Molecular marker technology has diverse applications in fisheries research and offers significant convenience and solutions to numerous challenges encountered in the aquaculture industry. In order to tackle the obstacles faced by the sea cucumber aquaculture and breeding industry, various molecular markers have been developed and implemented for multiple purposes, including germplasm resource evaluation, species identification, Assessment of genetic diversity and population structure, and marker-assisted selection and breeding. With innovations in genomic methods, molecular markers are expected to continue playing important roles in promoting the future development of sea cucumber fisheries, aquaculture, and industry.
Statements
Author contributions
XL: Investigation, Visualization, Data curation, Writing – original draft. XT: Investigation, Visualization, Data curation, Writing – original draft. MC: Supervision, Resources, Conceptualization, Writing – review & editing. GN: Supervision, Resources, Conceptualization, Writing – review & editing. YY: Visualization, Supervision, Resources, Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by National Key Research and Development Program of China (grant number 2022YFD2401305).
Conflict of interest
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Alves Pereira V. Martins Forte J. Vieira Arruda-Júnior J. P. Mendonça Diniz F. Maggioni R. Brito Salmitovanderley C. S. (2018). Identification and characterization of microsatellite loci in West Atlantic sea cucumber Holothuria grisea (Selenka 1867). J. Genet.97, 1363–1369. doi: 10.1007/s12041-018-1022-9
2
An H. S. Lee J. W. Hong S. W. Hong H. N. Park J. Y. Myeong J. I. et al . (2013). Genetic differences between wild and hatchery populations of red sea cucumber (Stichopus japonicus) inferred from microsatellite markers: implications for production and stocking programs design. Genes Genomics35, 709–717. doi: 10.1007/s13258-013-0139-8
3
Brown K. T. Southgate P. C. Hewavitharane C. A. Lal M. M. (2022). Saving the sea cucumbers: Using population genomic tools to inform fishery and conservation management of the Fijian sandfish Holothuria (Metriatyla) scabra. PloS One17. doi: 10.1371/journal.pone.0274245
4
Chen M. Gao L. Zhang W. You H. Sun Q. Chang Y. (2013). Identification of forty-five gene-derived polymorphic microsatellite loci for the sea cucumber, Apostichopus japonicus. J. Genet.92, e31–e35. doi: 10.1007/s12041-013-0234-2
5
Chen L. Li Q. (2007). Identification and characterization of microsatellite markers derived from expressed sequence tags of the sea cucumber Stichopus japonicus. Mol. Ecol. Notes7, 1057–1059. doi: 10.1111/j.1471-8286.2007.01775.x
6
Chen T. Ren C. Wong N.-K. Yan A. Sun C. Fan D. et al . (2023). The Holothuria leucospilota genome elucidates sacrificial organ expulsion and bioadhesive trap enriched with amyloid-patterned proteins. Proc. Natl. Acad. Sci.120, e2213512120. doi: 10.1073/pnas.2213512120
7
Chieu H. D. Premachandra H. K. A. Powell D. Knibb W. (2023). Genome-wide SNP analyses reveal a substantial gene flow and isolated-genetic structure of sea cucumber Holothuria leucospilota populations in Western Central Pacific. Fish Res.264, 106718. doi: 10.1016/j.fishres.2023.106718
8
Cong J. Kong L. Meng W. Wang Z. Sun Z. Chang Y. (2024). Sex differences in the growth of sea cucumber Apostichopus japonicus. Aquaculture582, 740475. doi: 10.1016/j.aquaculture.2023.740475
9
Cui W. Huo D. Liu S. Xing L. Su F. Yang H. et al . (2021). Construction of a high-density genetic linkage map for the mapping of QTL associated with growth-related traits in sea cucumber (Apostichopus japonicus). Biol. (Basel)11, 50. doi: 10.3390/biology11010050
10
Dai G. Li Z. B. Shangguan J. B. Ning Y. F. Deng H. W. Yuan Y. et al . (2015). Development and characterization of polymorphic microsatellite loci in the sea cucumber Holothuria leucospilota. Genet. Mol. Res.14, 538–541. doi: 10.4238/2015.January.26.8
11
Ding J. Chang Y. (2020). Research progress in conservation and utilization of economic echinoderm: a review. J. Dalian Ocean Univ.35, 645–656. doi: 10.1016/j.rser.2021.111313
12
Ding J. Han L. Chang Y. (2021). Application of germplasm innovation technology in sea cucumber and sea urchin genetic breeding. Prog. Fishery Sci.42, 1–16. doi: 10.19663/j.issn2095-9869.20201208003
13
Dong Y. Li Q. Zhong X. Kong L. (2016). Development of gene-derived SNP markers and their application for the assessment of genetic diversity in wild and cultured populations in sea cucumber, apostichopus japonicus. J. World Aquaculture Soc.47, 873–888. doi: 10.1111/jwas.12334
14
Du H. Bao Z. Yan J. Tian M. Mu X. Wang S. et al . (2012). Development of 101 gene-based single nucleotide polymorphism markers in sea cucumber, apostichopus japonicus. IJMS13, 7080–7097. doi: 10.3390/ijms13067080
15
E Z. Cheng C. Wu F. Ren C. Chen R. Rao Y. et al . (2023). Nondestructive and rapid method for sex identification of the tropical sea cucumber Holothuria scabra by anal swab sampling. Aquaculture562, 738749. doi: 10.1016/j.aquaculture.2022.738749
16
Elvevoll E. O. James D. Toppe J. Gamarro E. G. Jensen I.-J. (2022). Food safety risks posed by heavy metals and persistent organic pollutants (POPs) related to consumption of sea cucumbers. Foods11, 3992. doi: 10.3390/foods11243992
17
Fan S. Hu C. Zhang L. Sun H. Wen J. Luo P. (2012). Complete mitochondrial genome of the sea cucumber Stichopus sp. and its application in the identification of this species. Aquaculture Res.43, 1306–1316. doi: 10.1111/are.2012.43.issue-9
18
Fitch A. J. Leeworthy G. Li X. Bowman W. Turner L. Gardner M. G. (2013). Isolation and characterisation of eighteen microsatellite markers from the sea cucumber Holothuria scabra (Echinodermata: Holothuriidae). Aust. J. Zoology60, 368–371. doi: 10.1071/zo12114
19
Gao L.-l. Chen M. Chang Y.-Q. Ji N.-J. Li C.-Z. (2013). Development of SNP markers associated with defense mechanism of sea cucumber, Apostichopus japonicas. Conserv. Genet. Resour.5, 587–591. doi: 10.1007/s12686-013-9858-z
20
Henriques F. F. Serrao E. A. González-Wangüemert M. (2016). Novel polymorphic microsatellite loci for a new target species, the sea cucumber Holothuria mammata. Biochem. Systematics Ecol.66, 109–113. doi: 10.1016/j.bse.2016.03.012
21
Hu R. Xing R. Wang N. Ge Y. Chen Y. (2019). Species identification of sea cucumber based on DNA barcode technology. Food Industry Sci. Technol.40, 145–151+157. doi: 10.13386/j.issn1002-0306.2019.10.024
22
Hu J. Zhan A. Lu W. Hu X. Bao Z. (2007). An efficient procedure for isolating microsatellite DNAs from sea cucumber (Apostichopus japonicus). J. Ocean Univ. China. JOUC6, 12. doi: 10.1007/s11802-007-0012-y
23
Jiang Q. Li Q. Yu H. Kong L. (2011). Identification and characterization of 43 microsatellite markers derived from expressed sequence tags of the sea cucumber (Apostichopus japonicus). J. Ocean Univ. China10, 185–190. doi: 10.1007/s11802-011-1814-5
24
Kanno M. Li Q. Kijima A. (2005). Isolation and characterization of twenty microsatellite loci in Japanese sea cucumber (Stichopus japonicus). Mar. Biotechnol. (NY)7, 179–183. doi: 10.1007/s10126-004-0006-3
25
Kanno M. Suyama Y. Li Q. Kijima A. (2006). Microsatellite analysis of Japanese sea cucumber, Stichopus (Apostichopus) japonicus, supports reproductive isolation in color variants. Mar. Biotechnol. (NY)8, 672–685. doi: 10.1007/s10126-006-6014-8
26
Li Q. Chen L. Kong L. (2009). A genetic linkage map of the sea cucumber, Apostichopus japonicus (Selenka), based on AFLP and microsatellite markers. Anim. Genet.40, 678–685. doi: 10.1111/j.1365-2052.2009.01900.x
27
Li Z. B. Dai G. Shangguan J. B. Ning Y. F. Li Y. Y. Chen R. B. et al . (2015a). Isolation and characterization of polymorphic microsatellite loci in the sea cucumber Holothuria scabra. Genet. Mol. Res.14, 6529–6532. doi: 10.4238/2015.June.12.5
28
Li Z. B. Dai G. Shangguan J. B. Ning Y. F. Li Y. Y. Chen R. B. et al . (2015b). Isolation and characterization of microsatellite markers of sea cucumber Stichopus horrens. Genet. Mol. Res.14, 8496–8499. doi: 10.4238/2015.July.28.18
29
Li Y. Wang R. Xun X. Wang J. Bao L. Thimmappa R. et al . (2018). Sea cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discovery4, 1–17. doi: 10.1038/s41421-018-0030-5
30
Liao M. Wang J. Li B. Wang Y. Rong X. Zhang Z. et al . (2021). Genetic structure analysis and fingerprint construction of different geographical populations of sea cucumber based on SSR markers. Prog. Fishery Sci.42, 165–176. doi: 10.19663/j.issn2095-9869.20191125001
31
Liao M. Wang Y. Rong X. Zhang Z. Li B. Wang L. et al . (2011). Development of new microsatellite DNA markers from Apostichopus japonicus and their cross-species application in Parastichopus parvimensis and Pathallus mollis. Int. J. Mol. Sci.12, 5862–5870. doi: 10.3390/ijms12095862
32
Liu D. (2016). A review of sea cucumber aquaculture, ranching, and stock enhancement in China. Rev. Fisheries Sci. Aquaculture24, 326–341. doi: 10.1080/23308249.2016.1193472
33
Liu A. Liao M. Li B. Wang Y. Rong X. Zhang Z. et al . (2019). Validation analysis of SNP markers associated with important economic traits in sea cucumber (Apostichopus japonicus). Prog. Fishery Sci.40, 101–109. doi: 10.19663/j.issn2095-9869.20171217002
34
Lu C. Kuang Y. Zheng X. Li C. Sun X. (2019). Research progress on molecular marker-assisted breeding in aquatic animals. J. Fisheries43, 36–53. doi: 10.11964/jfc.20181211592
35
Luo H. Huang G. Li J. Yang Q. Zhu J. Zhang B. et al . (2022). De novo genome assembly and annotation of Holothuria scabra (Jaeger 1833) from nanopore sequencing reads. Genes Genomics44, 1487–1498. doi: 10.1007/s13258-022-01322-0
36
Lv Y. (2012). Molecular biology methods for identifying sea cucumber species based on DNA barcode. Ocean University of China. doi: 10.7666/d.y2157861
37
Lv J. Wang Y. Ni P. Lin P. Hou H. Ding J. et al . (2022). Development of a high-throughput SNP array for sea cucumber (Apostichopus japonicus) and its application in genomic selection with MCP regularized deep neural networks. Genomics114, 110426. doi: 10.1016/j.ygeno.2022.110426
38
Lv Y. Zuo T. Tang Q. Duan G. Wang C. Li J. et al . (2011). Construction and application of sea cucumber DNA barcodes. J. Fisheries Sci. China18, 782–789. doi: 10.3724/sp.j.1118.2011.00782
39
Madduppa H. Subhan B. Anggraini N. P. Fadillah R. Tarman K. (2017). DNA barcoding reveals vulnerable and not evaluated species of sea cucumbers (Holothuroidea and Stichopodidae) from Kepulauan Seribu reefs, Indonesia. Biodiversitas18, 893–898. doi: 10.13057/biodiv/d180305
40
Madeira P. Stefanni S. Ávila S. (2018). Non-destructive tissue sampling and the use of PCR-RFLPs in two edible sea cucumbers from the north-eastern Atlantic, Holothuria mammata Grube 1840 and H. sanctori Delle Chiaje 1823 (Echinodermata: holothuroidea). Eur. Zoological J.85, 88–93. doi: 10.1080/24750263.2018.1438529
41
Maqsood H. M. (2017). Advances in molecular markers and their applications in aquaculture and fisheries. GenAqua1. doi: 10.4194/2459-1831-v1_1_05
42
Medina-Feliciano J. G. Pirro S. Garcia-Arraras J. E. Mashanov V. Ryan J. F. (2021). Draft genome of the sea cucumber holothuria glaberrima, a model for the study of regeneration. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.603410
43
Nowland S. J. Southgate P. C. Basiita R. K. Jerry D. R. (2017). Elucidation of fine-scale genetic structure of sandfish (Holothuria scabra) populations in Papua New Guinea and northern Australia. Mar. Freshw. Res.68, 1901–1911. doi: 10.1071/MF16223
44
Oliveira M. Azevedo L. (2022). Molecular markers: an overview of data published for fungi over the last ten years. J. Fungi (Basel)8, 803. doi: 10.3390/jof8080803
45
Oliveira E. J. Pádua J. G. Zucchi M. I. Vencovsky R. Vieira M. L. C. (2006). Origin, evolution and genome distribution of microsatellites. Genet. Mol. Biol.29 (2), 294–307. doi: 10.1590/s1415-47572006000200018
46
Peng W. Bao Z. Du H. Dong Y. Lv Y. Zhou Z. et al . (2009). Development and characterization of 38 novel EST-SSRs for the sea cucumber Apostichopus japonicus. Conserv. Genet. Resour.1, 447–450. doi: 10.1007/s12686-009-9103-y
47
Peng W. Bao Z. M. Du H. X. Yan J. J. Zhang L. L. Hu J. J. (2012). Development and characterization of 70 novel microsatellite markers for the sea cucumber (Apostichopus japonicus). Genet. Mol. Res.11, 434–439. doi: 10.4238/2012.February.24.2
48
Pereira V. A. Forte J. M. Arruda-Júnior J. P. V. Diniz F. M. Maggioni R. Salmito-Vanderley C. S. B. (2018). Identification and characterization of microsatellite loci in west atlantic sea cucumber holothuria grisea (Selenka 1867). J. Genet.97 (5), 1363–1369. doi: 10.1007/s12041-018-1022-9
49
Riquet F. Fauvelot C. Fey P. Grulois D. Leopold M. (2022). Hatchery-produced sandfish (Holothuria scabra) show altered genetic diversity in New Caledonia. Fish Res.252, 10. doi: 10.1016/j.fishres.2022.106343
50
Rojas-Cartagena C. Ortíz-Pineda P. Ramírez-Gómez F. Suárez-Castillo E. C. Matos-Cruz V. Rodríguez C. et al . (2007). Distinct profiles of expressed sequence tags during intestinal regeneration in the sea cucumber Holothuria glaberrima. Physiol. Genomics31, 203–215. doi: 10.1152/physiolgenomics.00228.2006
51
Shangguan J. B. Li Z. B. Ning Y. F. Huang Y. S. Yuan Y. Lu J. et al . (2015a). Screening and characterization of novel polymorphic microsatellite markers from sea cucumber Holothuria leucospilota. Genet. Mol. Res.14, 6555–6560. doi: 10.4238/2015.June.12.9
52
Shangguan J. B. Li Z. B. Yuan Y. Huang Y. S. (2015b). Identification and characterization of microsatellite markers from the tropical sea cucumber, Stichopus horrens (Selenka). Genet. Mol. Res.14, 13582–13587. doi: 10.4238/2015.October.28.18
53
Shen X. Tian M. Cheng H. Meng X. Ren J. (2011). Analysis of mitochondrial genome characteristics and discussion on molecular markers in sea cucumbers. Fisheries Sci.30, 400–404. doi: 10.16378/j.cnki.1003-1111.2011.07.007
54
Skillings D. J. Bird C. E. Toonen R. J. (2011). Gateways to hawai‘i: genetic population structure of the tropical sea cucumber holothuria atra. J. Mar. Biol.2011, 783030. doi: 10.1155/2011/783030
55
Soliman T. Kanno M. Kijima A. Yamazaki Y. (2012). Population genetic structure and gene flow in the japanese sea cucumber apostichopus japonicus across toyama bay, japan. Fisheries Sci.78 (4), 775–783. doi: 10.1007/s12562-012-0509-1
56
Soliman T. Fernandez-Silva I. Reimer J. D. (2016). Genetic population structure and low genetic diversity in the over-exploited sea cucumber Holothuria edulis Lesson, 1830 (Echinodermata: Holothuroidea) in Okinawa Island. Conserv. Genet.17 (4), 811–821. doi: 10.1007/s10592-016-0823-88
57
Sulardiono B. Hartoko A. Aini A. N. Wulandari D. Budiharjo A. (2022). Genetic diversity of commercial sea cucumbers Stichopus (Echinoderm: Stichopodidae) based on DNA Barcoding in Karimunjawa, Indonesia. Biodiversitas23, 922–927. doi: 10.13057/biodiv/d230234
58
Sun L. Jiang C. Su F. Cui W. Yang H. (2023). Chromosome-level genome assembly of the sea cucumber Apostichopus japonicus. Sci. Data10, 454. doi: 10.1038/s41597-023-02368-9
59
Sun W.-J. Li Q. Kong L.-F. (2010). Characterization of thirteen single nucleotide polymorphism markers in the sea cucumber (Apostichopus japonicus). Conserv. Genet. Resour.2, 141–144. doi: 10.1007/s12686-010-9270-x
60
Sun Z.-H. Wei J.-L. Cui Z.-P. Han Y.-L. Zhang J. Song J. et al . (2021). Identification and functional characterization of piwi1 gene in sea cucumber, apostichopus japonicas. Comp. Biochem. Physiol. B-Biochemistry Mol. Biol.252. doi: 10.1016/j.cbpb.2020.110536
61
Taquet C. Nagai S. Yasuda N. Nadaoka K. (2011). First report of the development of microsatellite markers for a tropical sea cucumber (Stichopus chloronotus). Conserv. Genet. Resour.3, 201–203. doi: 10.1007/s12686-010-9322-2
62
Tian M. Li Y. Jing J. Mu C. Du H. Dou J. et al . (2015). Construction of a high-density genetic map and quantitative trait locus mapping in the sea cucumber apostichopus japonicus. Sci. Rep.5, 14852. doi: 10.1038/srep14852
63
Uthicke S. Purcell S. Blockmans B. (2005). Natural hybridization does not dissolve species boundaries in commercially important sea cucumbers. Biol. J. Linn. Soc.85, 261–270. doi: 10.1111/bij.2005.85.issue-3
64
Wang X. Shan X. Qiu X. Meng X. Chang Y. (2009). Microsatellite DNA polymorphisms and the relation with body weight in sea cucumber Apostichopus japonicus. Chin. J. Oceanology Limnology27, 331. doi: 10.1007/s00343-009-9162-7
65
Wang Q. L. Yu S. S. Ren H. Tang J. B. Gao J. J. Li X. Y. et al . (2023). Genetic diversity and population structure of the endangered Japanese sea cucumber (Apostichopus japonicus) in natural seas of northern China. Aquacult. Rep.30, 7. doi: 10.1016/j.aqrep.2023.101595
66
Wei J.-L. Cong J.-J. Sun Z.-H. Song J. Zhao C. Chang Y.-Q. (2021). A rapid and reliable method for genetic sex identification in sea cucumber, Apostichopus japonicus. Aquaculture543, 737021. doi: 10.1016/j.aquaculture.2021.737021
67
Wen J. Hu C. Zhang L. Fan S. (2012a). Identification of 11 sea cucumber species by species-specific PCR method. Food control27, 380–384. doi: 10.1016/j.foodcont.2012.04.007
68
Wen J. Hu C. Zhang L. Luo P. Zhao Z. Fan S. et al . (2010). The application of PCR–RFLP and FINS for species identification used in sea cucumbers (Aspidochirotida: Stichopodidae) products from the market. Food Control21, 403–407. doi: 10.1016/j.foodcont.2009.06.014
69
Wen J. Zeng L. Wang Y. Tian L. Zhou X. (2012b). Identification of counterfeit sea cucumber products in the market using morphological and molecular biology methods. J. Zhanjiang Normal Univ.33, 95–99 (in Chinese). doi: 10.1016/j.foodcont.2012.04.007
70
Wu F. Cheng C. Li X. Ren C. Luo P. Jiang X. et al . (2022). Identification of sex-specific molecular markers and development of PCR-based sex detection techniques in tropical sea cucumber (Stichopus monotuberculatus). Aquaculture547, 737458. doi: 10.1016/j.aquaculture.2021.737458
71
Xing R.-R. Hu R.-R. Wang N. Zhang J.-K. Ge Y.-Q. Chen Y. (2021). Authentication of sea cucumber products using NGS-based DNA mini-barcoding. Food Control129, 108199. doi: 10.1016/j.foodcont.2021.108199
72
Yagodina V. D. Bondar E. I. Brykov V. A. (2022). Genetic variability and population structure of the Japanese sea cucumber, Apostichopus japonicus Selenka 1867 revealed by microsatellites in Peter the Great Gulf, Sea of Japan. Mar. Biodiversity52, 40. doi: 10.1007/s12526-022-01278-0
73
Yan H. (2006). Analysis of Genetic Diversity in Different Geographic Populations of Apostichopus japonicus Using ISSR and COI Sequence Markers (Liaoning Normal University).
74
Yan J. Jing J. Mu X. Du H. Tian M. Wang S. et al . (2013). A genetic linkage map of the sea cucumber (Apostichopus japonicus) based on microsatellites and SNPs. Aquaculture404, 1–7. doi: 10.1016/j.aquaculture.2013.04.011
75
Yang A. Sun D. Liu S. Dong Y. Chen Z. Zhou Z. (2012). Characterization of fifteen SNP markers by mining EST in sea cucumber, Apostichopus japonicus. J. Genet.91, e49–e53. doi: 10.1007/s12041-012-0141-y
76
Yang A.-F. Zhou Z.-C. He C.-B. Hu J.-J. Chen Z. Gao X.-G. et al . (2009). Analysis of expressed sequence tags from body wall, intestine and respiratory tree of sea cucumber (Apostichopus japonicus). Aquaculture296, 193–199. doi: 10.1016/j.aquaculture.2009.08.016
77
Yoshida W. Ishida S. Ono K. Izumi S. Hasegawa K. (2012). Developmental styles and larval morphology of hybridized sea cucumbers (Echinodermata: Holothuroidea). Invertebrate Reprod. Dev.56, 249–259. doi: 10.1080/07924259.2011.601140
78
Yun Z. Sun Z. Xu H. Sun Z. Zhang Y. Liu Z. (2017). Identifying the geographical origin of protected sea cucumbers (Apostichopus japonicus) in china using random amplified polymorphic DNA polymerase chain reaction (RAPD-PCR). Food Sci. Biotechnol.26 (2), 357–362. doi: 10.1007/s10068-017-0048-8
79
Zeng L. Wen J. Fan S. Chen Z. Xu Y. Sun Y. et al . (2018). Identification of sea cucumber species in processed food products by PCR-RFLP method. Food Control90, 166–171. doi: 10.1016/j.foodcont.2018.02.048
80
Zhan A. Bao Z. Lu W. Hu X. Peng W. Wang M. et al . (2007). Development and characterization of 45 novel microsatellite markers for sea cucumber (Apostichopus japonicus). Mol. Ecol. Notes7, 1345–1348. doi: 10.1111/j.1471-8286.2007.01876.x
81
Zhang X. Sun L. Yuan J. Sun Y. Gao Y. Zhang L. et al . (2017). The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PloS Biol.15, e2003790. doi: 10.1371/journal.pbio.2003790
82
Zhong S. Ma X. Jiang Y. Liu X. Zeng M. Zhao L. et al . (2023). The draft genome of the tropical sea cucumber Stichopus monotuberculatus (Echinodermata, Stichopodidae) reveals critical genes in fucosylated chondroitin sulfates biosynthetic pathway. Front. Genet.14. doi: 10.3389/fgene.2023.1182002
83
Zhou Z. C. Dong Y. Sun H. J. Yang A. F. Chen Z. Gao S. et al . (2014). Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol. Ecol. Resour14, 127–138. doi: 10.1111/1755-0998.12147
84
Zhou X. Wang H. Cui J. Qiu X. Chang Y. Wang X. (2016). Transcriptome analysis of tube foot and large scale marker discovery in sea cucumber, Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genomics Proteomics20, 41–49. doi: 10.1016/j.cbd.2016.07.005
85
Zhou Z. C. Dong Y. Sun H. J. Yang A. F. Chen Z. Gao S. et al . (2013). Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol. Ecol. Resour.14 (1), 127–138. doi: 10.1111/1755-0998.12147
Summary
Keywords
sea cucumber, molecular marker, aquaculture, germplasm conservation, molecular breeding
Citation
Liu X, Tang X, Chen M, Ni G and Yang Y (2024) Development and application of molecular markers in fisheries, aquaculture, and industry of representative temperate and tropical sea cucumbers: a review. Front. Mar. Sci. 11:1423096. doi: 10.3389/fmars.2024.1423096
Received
25 April 2024
Accepted
06 June 2024
Published
25 June 2024
Volume
11 - 2024
Edited by
Arnold Rakaj, University of Rome Tor Vergata, Italy
Reviewed by
Chryssanthi Antoniadou, Aristotle University of Thessaloniki, Greece
Ting Chen, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2024 Liu, Tang, Chen, Ni and Yang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujia Yang, yangyujia@ouc.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.