Abstract
Synechococcus elongatus is a model cyanobacterium with remarkable adaptability to diverse environmental stresses, making it a promising candidate for the photoautotrophic conversion of carbon dioxide into valuable chemicals. This review explores the adaptive mechanisms that allow S. elongatus to survive under various abiotic stresses, such as changes in CO2 levels, heavy metals, and light conditions. We also highlight recent advancements in synthetic biology that have enabled the engineering of S. elongatus to produce biofuels and other value-added compounds, including fatty acids, alcohols, and carotenoids. Additionally, we discuss the applications of modern omics techniques to elucidate the genetic basis of stress tolerance and metabolic regulation. Despite the promising potential of S. elongatus for industrial applications, challenges remain in scaling up production, enhancing genetic stability, and optimizing bioreactor systems. Finally, we provide insights into future directions, including the integration of genome engineering, system-level modeling, and co-culture strategies, to improve the efficiency of cyanobacterial cell factories for sustainable biotechnology applications.
1 Introduction
Cyanobacteria, also known as blue-green algae, are gram-negative bacteria. Synechococcus elongatus, a model species of cyanobacteria, has been widely studied for its fast photoautotrophic growth (Zouni et al., 2001). Bibliometrics analysis indicates a rising trend of global research interest in S. elongatus as relevant publications have increased significantly over the last three decades (Supplementary Figure 1).
The cyanobacterium S. elongatus possessed strong adaptability, endowing their outstanding survival ability in ocean and freshwater environments (Lai et al., 2024). It has an efficient photosynthesis system, rapid reproduction (Table 1), strong carbon sequestration capacity, and good tolerance to extreme environments. Moreover, S. elongatus has a smaller genome (Table 1), with efficient molecular biology tools for gene editing and genetic engineering (Yu et al., 2015). Physiological studies could provide a theoretical basis for cultivating different stress-resistant varieties, and several subspecies of S. elongatus have been used as model strains for various applications (Ungerer et al., 2018a). However, the subspecies’ growth characteristics vary, so it is essential to make selections before conducting specific experiments (Yu et al., 2015; Jaiswal et al., 2020). Compared with other typical microbial cell factories, the growth rate of S. elongatus is slower than that of Escherichia coli (Table 2). However, S. elongatus has a significant advantage among photosynthetic autotrophic cells, with its fastest doubling time being only 1.9 hours (Table 2). Towards a sustainable society, S. elongatus can produce renewable products such as biochemicals and fuels (Table 2).
Table 1
| Subspecies | Genome information | Doubling time | Characteristics |
|---|---|---|---|
| PCC 7942 | 2.7 Mb (Holtman et al., 2005). | 7–10 h (Yu et al., 2013) | The first cyanobacterial strain to be reliably transformed by exogenous DNA (Shestakov and Khyen, 1970); a model organism for studying the circadian rhythm of cyanobacteria (Holtman et al., 2005). |
| PCC 6301 | The homology to PCC 7942 was 99.86% (Sugita et al., 2007). | 3.4 h (Binder and Chisholm, 1990) | The genes for two-component signal transduction systems are only 37 genes (Sugita et al., 2007). |
| PCC 11801 | The homology to PCC 7942 was 83% (Jaiswal et al., 2018). | 2.3 h (Jaiswal et al., 2018) | A high growth rate (Jaiswal et al., 2018). |
| PCC 11802 | The homology to PCC 11801 was 97% (Jaiswal et al., 2020). | 2.8 h (Jaiswal et al., 2020) | Key enzymes of the Calvin cycle are not repressed under elevated CO2 (Jaiswal et al., 2020). |
| UTEX 2973 | There is a difference of 53 SNPs, a 7.5-kb deletion, and a 188-kb inversion compared with PCC 7942 (Yu et al., 2015). | 1.9 h (Yu et al., 2015) | With high light resistance and photosynthetic rate, the biomass productivity was three times that of PCC 7942 (Ungerer et al., 2018b). |
| BDU 130911 | Not available | Not available | High efficacy of uranium adsorption (Rashmi et al., 2013). |
Physiological characteristics of different subspecies of S. elongatus.
Table 2
| Species | Doubling time (h) | Common medium | Cell size (µm) | Representative product |
|---|---|---|---|---|
| Escherichia coli | 0.3 ~1 (Cooper and Helmstetter, 1968) | Luria-Bertani (LB) | 0.5 ~ 3 | Insulin (Baeshen et al., 2014) |
| Saccharomyces cerevisiae | 1.5 (Kaeberlein et al., 2005) | Yeast extract peptone dextrose | 3 ~ 6 | Fuels, chemicals, pharmaceuticals (Hong and Nielsen, 2012) |
| Chlamydomonas reinhardtii | 2.5 (Lien and Knutsen, 1979) | Tris-acetate-phosphate (TAP) | 7 ~ 10 | High quality mammalian proteins (León-Bañares et al., 2004) |
| Synechocystis PCC6803 | 4.3 (Van et al., 2018) | BG-11 | NA | Renewable biofuels and chemicals (Liu et al., 2012) |
| Synechococcus elongatus | 1.9 (Yu et al., 2015) | BG-11 | 2 | Renewable chemicals and fuels (Vayenos et al., 2020) |
Comparison of microbial cell factories.
NA, not available.
Metabolic engineering, developed in the late 20th century, employs genetic modifications (e.g., gene knockout, promoter engineering) to optimize metabolic networks for enhanced product synthesis (Hong and Nielsen, 2012; Keasling, 2012). Emerging in the 21st century, synthetic biology utilizes standardized biological parts (e.g., gene circuits, clustered regularly interspaced short palindromic repeats (CRISPR) technology) to construct novel biological systems (Stephanopoulos, 2012; Baltes and Voytas, 2015). These disciplines synergistically advance microbial cell factories: synthetic biology designs new pathways while metabolic engineering refines their efficiency (Lin et al., 2015; Jarboe et al., 2010).
In this review, we delve into the intricate physiological and biochemical responses exhibited by S. elongatus when subjected to diverse environmental stresses. The aim is to provide a nuanced understanding of the adaptive mechanisms employed by this model cyanobacterium to cope with adverse conditions. To this end, we review the latest research on how S. elongatus responds to various stressors such as CO2 levels, pollutants, and high-light conditions (Figure 1).
Figure 1
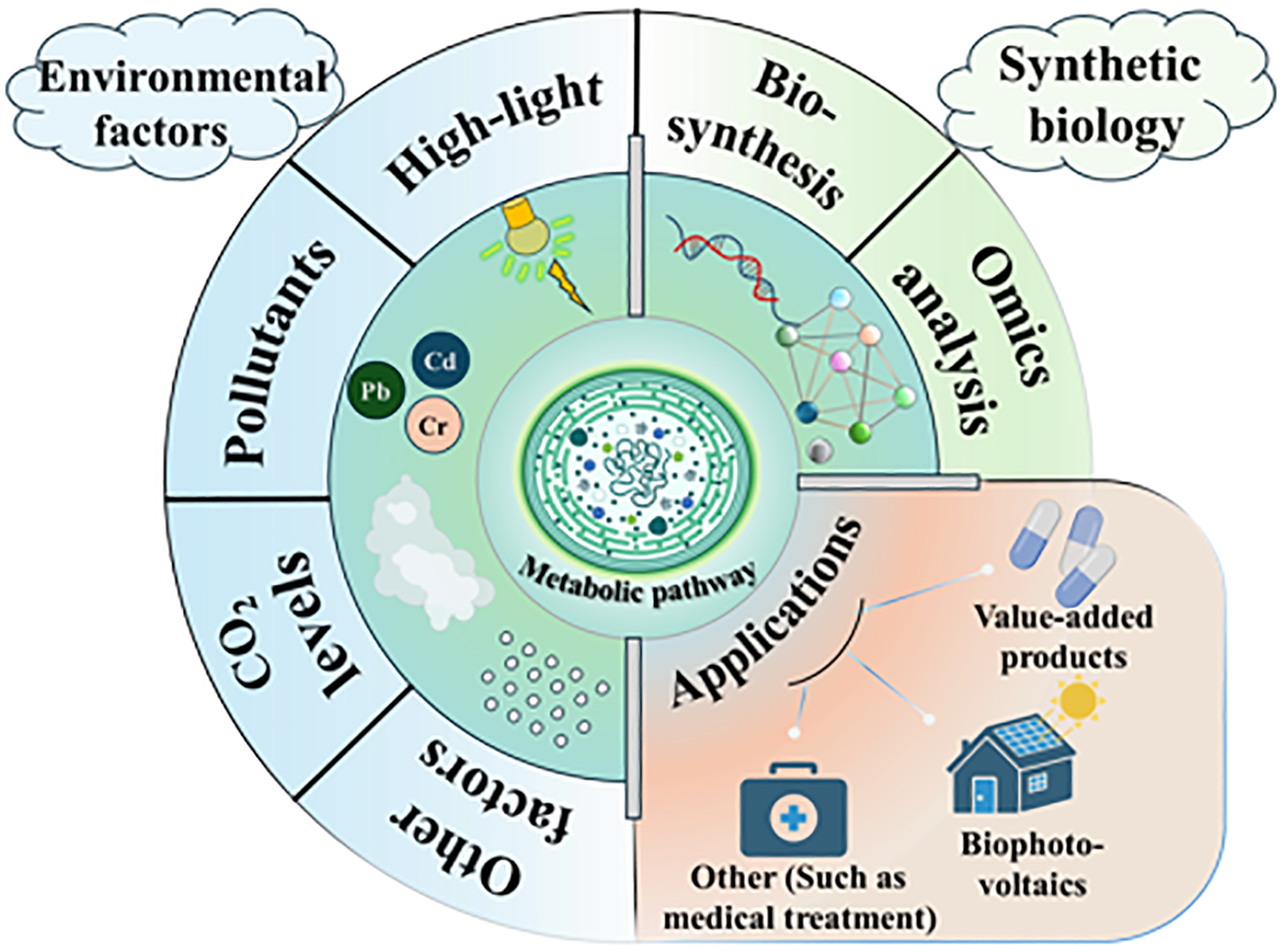
A diagram showing the concepts and applications of cyanobacterial cell factories.
Furthermore, we explore the biosynthetic capabilities of S. elongatus, focusing on both naturally occurring and heterologous bioactive compounds. We summarize the potential applications of S. elongatus in synthetic biology, discussing how its unique characteristics can be harnessed to design novel cell factories to produce high-value chemicals and materials. A timeline of the progress in physiological adaptations and synthetic biology of S. elongatus is shown in Figure 2. We also acknowledge the challenges encountered during the industrialization of S. elongatus, such as scalability, stability, and economic feasibility. By discussing the bottlenecks in algal engineering development, we aim to provide insights that can inform the design of photosynthetic cell factories and promote the practical applications of cyanobacteria.
Figure 2
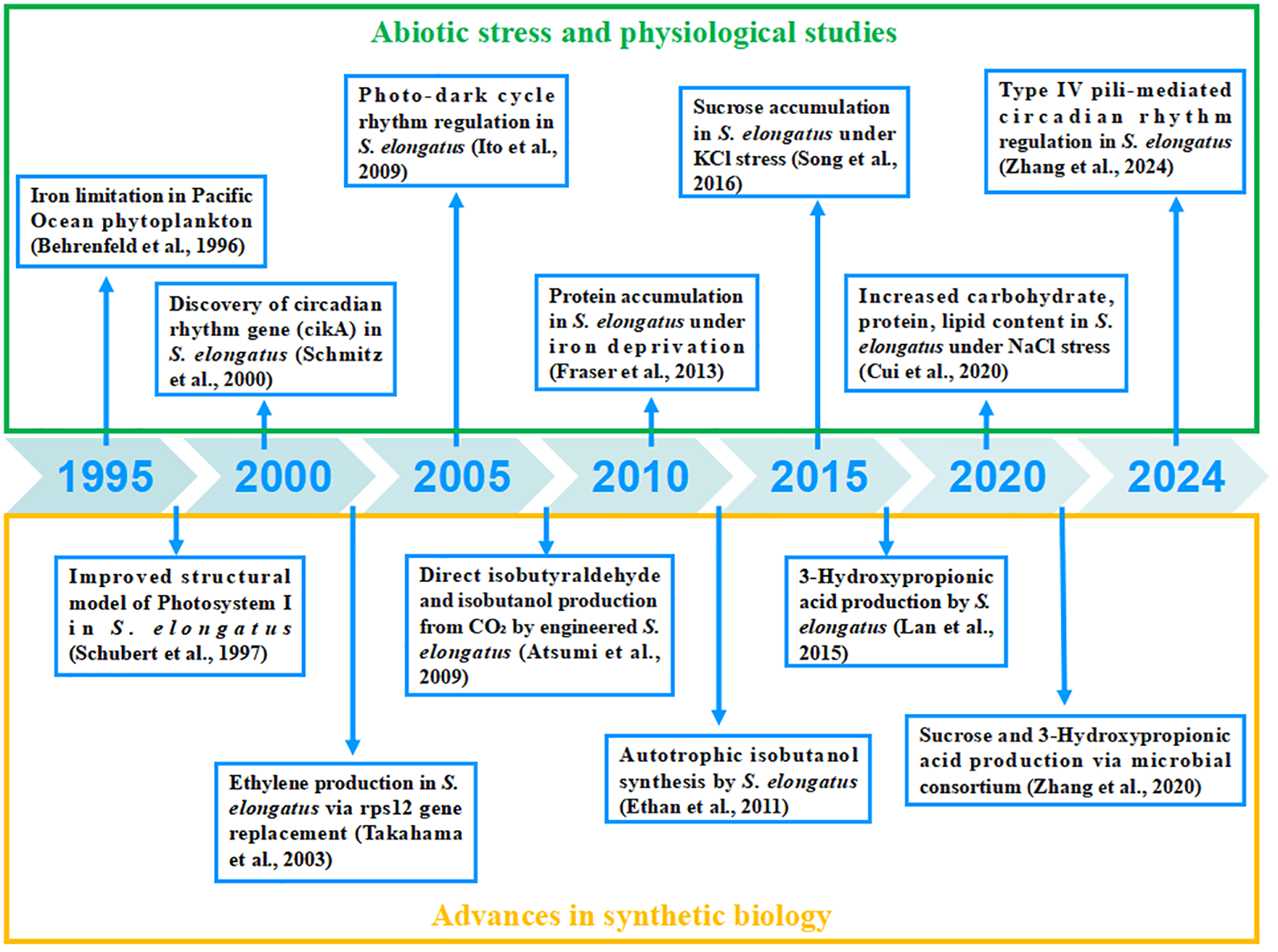
Advances in physiological adaptation and synthetic biology of S. elongatus. References cited: Atsumi et al. (2009), Behrenfeld and Bale (1996), Ito et al. (2009), Schmitz (2000), Schubert et al. (1997), Song et al. (2016), Takahama et al. (2003), Zhang et al. (2024), Fraser et al. (2013) and Cui et al. (2020).
2 Different environmental factors, related abiotic stresses, and adaptive strategies for S. elongatus survival
Advances in omics technologies such as genomics, transcriptomics, and proteomics enable scientists to conduct in-depth research on the molecular mechanisms and genetic basis of environmental resistance in S. elongatus. It is possible to discover a series of genes and proteins related to environmental resistance with crucial roles in cellular stress response, metabolic regulation, and cell repair. Systematic analysis of S. elongatus’ transcriptional regulatory network (TRN) was conducted through machine learning methods, revealing its gene regulatory mechanisms in key biological processes such as photosynthesis, carbon fixation, and nitrogen metabolism (Yuan et al., 2024). Studies on the environmental resistance of S. elongatus have deepened the understanding of the survival strategies of aquatic organisms and also provided novel insights into bioengineering and biotechnology development of microalgae, which could pave the way to produce valuable bioproducts under harsh conditions.
2.1 CO2 levels
CO2, the greatest greenhouse gas, is a critical environmental factor and also the major carbon source that affects microalgal growth. It was reported that S. elongatus PCC 7942 achieved optimal growth when supplied with 5% CO2 (Kuan et al., 2015). However, an excessively elevated CO2 concentration reduced the pH value dramatically and inhibited the growth of cyanobacteria (Mortezaeikia et al., 2016). For S. elongatus, the CO2 concentration influences biomass productivity and reduces its ability to absorb CO2 (Hashemi et al., 2020). The CO2 response mainly depended on the autoregulation of the cmpR gene (encoding the DNA-binding transcription factor) in S. elongatus PCC 7942, as the transcription factor CmpR activates the cmpABCD operon under low CO2 conditions while repressing its promoter (Pan et al., 2016). The cmpABCD operon encodes subunits of an ABC-type high-affinity HCO3- transporter, which is activated under low CO2 conditions and repressed under high CO2 conditions (Pan et al., 2016). Since atmospheric CO2 levels are insufficient to saturate Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase), O2 competes as an alternative substrate for Rubisco, impairing carboxylation reaction. Cyanobacteria mitigate this issue through CO2 concentrating mechanisms (CCMs), with the cmpABCD operon playing a key role in enhancing photosynthetic efficiency and environmental adaptation (Pan et al., 2016). Proteomic data revealed that elevated CO2 conditions in S. elongatus PCC 11801 led to the downregulation of photoprotection and redox-related genes while shifting from TCA cycle-dependence to a photosynthesis-dominated NADPH/ATP supply mode (Mehta et al., 2019). These studies suggest that the CO2 stress response in S. elongatus is a complex physiological process.
2.2 Pollutants
Pollutants such as nitrogen and phosphorus are major factors that cause frequent algal blooms. Under eutrophication conditions, S. elongatus has shown potential in water treatment, with its phosphorus and nitrogen removal rates reaching 85.1% and 87.4% respectively (Pishbin et al., 2020). S. elongatus has been used to treat wastewater from dairy and other industries (Ruiz-Güereca and Sánchez-Saavedra, 2016; Samiotis et al., 2021; Usai et al., 2024). In addition, S. elongatus PCC 7492 showed the capability to remove nitrogen from wastewater under different salinities (Samiotis et al., 2022).
Sulfur is a common pollutant in the environment and also an essential element for algae growth, and the use of S. elongatus to treat sulfur-containing wastewater is widely studied (Yang et al., 2015). Recently, the pollution of heavy metals is becoming a serious environmental issue. The survival of cyanobacteria could be affected by heavy metal stress. The heavy metal stress of Cd2+ or Ni2+ prevented S. elongatus PCC 7942 cells from properly entering the chlorosis process under nitrogen starvation (Selim and Haffner, 2020). As a potential carcinogenic pollutant, 2,4-dinitrotoluene is classified as “possibly carcinogenic to humans” (Group 2B) by the International Agency for Research on Cancer with environmental persistence and health risks (Oh et al., 2011). Compared to physical adsorption and chemical oxidation, bioremediation shows potential in degrading 2,4-dinitrotoluene. For example, S. elongatus PCC 7942 could degrade the nitro groups of 2,4-dinitrotoluene, demonstrating its application potential in biological treatment (Fedeson et al., 2020).
2.3 High-light stress
Light is the primary energy to support cyanobacterial growth and development, which affects the physiology of cyanobacteria through light intensity and composition. High-density culture could be achieved through high-light conditions (Moronta-Barrios et al., 2012). S. elongatus UTEX 2973, the fastest-growing cyanobacterial strain, was studied under high-light conditions, and comparative genomics analysis revealed that hltA is a key factor for high-light tolerance as HltA senses environmental signals under high-light conditions and activates stress pathways, thereby helping cyanobacteria avoid photoinhibition and oxidative damage (Walker and Pakrasi, 2022). High-light stress can rapidly decrease the ratio of phosphorylated RpaB to non-phosphorylated RpaB, indicating that RpaB plays a crucial role in high-light signal transduction (Moronta-Barrios et al., 2012). Furthermore, high-light conditions mitigate the growth inhibition caused by salt stress in S. elongatus, which is more severe under low-light conditions. This alleviation likely occurs because high light counteracts the salt-induced suppression of photosynthetic pigment accumulation (Kumar et al., 2021).
2.4 Other environmental factors
There are a large number of emerging environmental pollutants due to human activities. In recent years, there have been many studies on the effects of other emerging environmental stresses on S. elongatus. For example, the discharge of aquaculture wastewater is an essential cause of the anti-growth surge in the water environment. Low-concentration kanamycin enhances the biofilm formation of S. elongatus by upregulating photosynthesis and carbonic anhydrase genes (Tan et al., 2016). The toxic effects of Micro- and nano-sized polystyrene particles on S. elongatus have been demonstrated, resulting in damage to the integrity of the cell membrane (Feng et al., 2019). The zinc oxide used in sunscreen enters domestic wastewater through washing, and improper treatment may lead to water pollution. Zinc oxide could induce oxidative stress, leading to lipid peroxidation and DNA damage in S. elongatus, and genes involved in the photosynthetic system, oxidative phosphorylation, and transcription/translation were down-regulated (Vicente et al., 2019). Due to improper agricultural application, glyphosate can accumulate significantly in soil. Through surface runoff and rainwater erosion, it can enter water bodies, easily causing water pollution. Glyphosate may exert an inhibitory effect on S. elongatus, leading to a notable reduction in its growth rate (Moraes et al., 2021). In summary, emerging environmental pollutants markedly influence the physiology of S. elongatus, which demonstrates strong potential as a candidate for industrial wastewater treatment.
3 Synthetic biology and biotechnology applications
As a photosynthetic microbial cell factory, S. elongatus exhibits remarkable competitiveness in the fields of synthetic biology and biomanufacturing. Beyond its rapid growth rate (Table 2), S. elongatus holds promise for sustainable bioeconomies due to its photoautotrophic metabolism, genetic tractability, and robust metabolic plasticity. Moreover, its inherent capacity to synthesize diverse natural products—such as glycogen, pigments, and lipids—provides essential precursors for metabolic engineering. These attributes have facilitated numerous successful heterologous expression cases (Figure 3), underscoring its potential for high-value compound production.
Figure 3
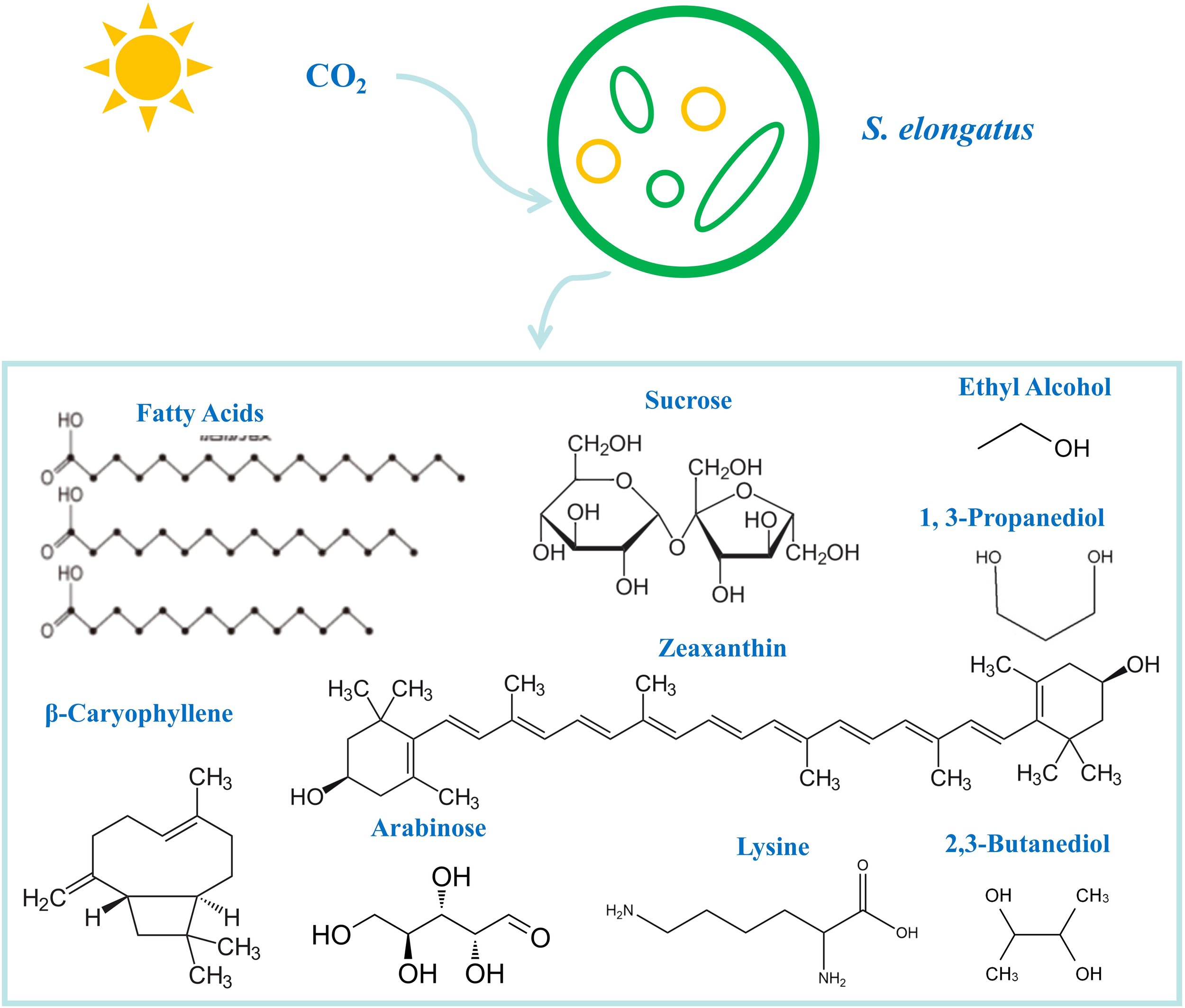
Cell factories of S. elongatus to produce biochemicals such as fatty acids and ethyl alcohol.
3.1 Synthetic biology for improved biomass production and carbon fixation
Biomass as a renewable resource, has huge potential for developing sustainable feedstock. Due to the increase in global population and the shortage of resource supply, increasing biomass production has become a challenge that needs to be addressed. Knocking out two glucokinase genes caused glucose accumulation and a spontaneous mutation in the genome of S. elongatus PCC 7942, which resulted in direct glucose secretion (Zhang et al., 2023). Genetically engineering S. elongatus PCC 7942 for expressing heterologous hexose transporter gene to perform mixotrophy under natural light is also a scheme to increase biomass yield and productivity (Sarnaik et al., 2017). In addition, heterotrophic partners have a significant growth promotion effect on cyanobacteria, resulting in an 80% increase in growth rate and enhanced photosynthetic capacity (Kratzl et al., 2024). These findings could provide new insights into improving biomass production and carbon sequestration in the future.
3.2 Synthetic biology for value-added products in S. elongatus
3.2.1 Bioenergy sources
Bioenergy is fuel derived from biological sources, also known as biofuels (Voshol, 2015). Currently, there are still some limitations in bioenergy development and applications, such as higher manufacturing costs and lower energy content than fossil fuels. However, bioenergy does have distinct advantages, such as being the only alternative energy source that could replace vehicle fuel without major modifications to vehicle engines and being renewable and relatively simple to process (Kaygusuz, 2009). Different types of bioenergy are summarized below.
3.2.1.1 Fatty acids
Cyanobacteria obtain energy from sunlight and convert carbon dioxide into free fatty acids (FFAs) through photosynthesis. FFAs can be utilized as feedstock and precursors for renewable biodiesel production, and therefore, the production of FFA has attracted much attention (Wijffels et al., 2013). There is rapid progress in the biosynthesis of FFA. For instance, S. elongatus PCC 7942 was engineered to produce free FFAs via gene knockout of the FFA-recycling acyl-ACP synthetase gene and expression of a thioesterase for FFA release, which provided the basis for large-scale FFA production (Ruffing A et al., 2012). However, the final FFA concentration in S. elongatus PCC 7942 was lower compared with other algal strains (Liu et al., 2011). To address this issue, an engineered S. elongatus strain achieved similar FFA secretion rates as other productive cyanobacterial species by modulating the expression level of the acyl-acyl carrier protein thioesterase and increasing the light intensity during cultivation (Kato et al., 2016). To remove FFAs from the medium during cultivation, an aqueous-organic two-phase culture system was developed that provides a basis for S. elongatus to produce FFA industrially (Kato et al., 2017). By covering the aqueous medium with isopropyl myristate (IM), FFA is effectively extracted from the medium into the organic phase, thereby reducing the accumulation of intracellular FFA and avoiding cell death due to FFA toxicity (Kato et al., 2017).
3.2.1.2 Alcohols
Alcohols produced from cyanobacteria have great potential as sustainable biofuels. Recently, a high-yielding strain of 1-butanol was constructed through metabolomics-assisted strain engineering (Fathima et al., 2020). 1-Butanol has the advantage of high energy density, with a heat value close to that of gasoline and superior to ethanol. The development of genetic modifications has increased the production of ethanol and butanediol from S. elongatus (Velmurugan and Incharoensakdi, 2020; Oliver et al., 2013). The synthesis of isopropanol has been the focus of research on biofuels, and S. elongatus has also shown great potential in this direction. Through the construction of synthetic pathways, genetic modification (Hirokawa et al., 2017), and growth optimization (Chandra and Mallick, 2022), it is feasible to increase productivity with reduced costs.
3.2.1.3 Other energy materials
Sugar represents a promising renewable feedstock for biofuel production, with sucrose being the most commonly utilized substrate. S. elongatus has demonstrated efficacy in sucrose synthesis (Ducat et al., 2012). Notably, genetic modifications in S. elongatus PCC 7942 enhanced intracellular sucrose accumulation, significantly improving yield (Vayenos et al., 2020). To further reduce production costs, co-culture fermentation systems have been employed. For instance, a synthetic microbial consortium comprising E. coli and S. elongatus UTEX 2973 was developed to directly convert CO2 into sucrose (Zhang et al., 2020). Additionally, the introduction of the L-arabinose metabolic pathway into S. elongatus boosted biomass productivity under phototrophic conditions (Cao et al., 2017).
In addition to sugars, cyanobacteria can synthesize energy-dense hydrocarbons such as alkanes. For example, the CRISPR-Cpf1 system (derived from Prevotella and Francisella 1) was employed to engineer S. elongatus PCC 11801, enabling regulated expression of ethylene-forming enzyme and high-efficiency ethylene production (Sengupta et al., 2020). Heterologous expression of cyanobacterial genes in fungal hosts has also facilitated the production of pentacene and heptadecane alkenes (Sinha et al., 2017). Moreover, pathway engineering and synthase optimization in S. elongatus UTEX 2973 achieved the highest reported β-caryophyllene yield in a cyanobacterial chassis (Li et al., 2020).
3.2.2 Feed additive
Carotenoids are industrially significant fine chemicals commonly used in the food, pharmaceutical, and healthcare industries (Maoka, 2011). Common carotenoids in the market include β-carotene, astaxanthin, and zeaxanthin, among others (Saini et al., 2018). The CrtR (β-carotene oxygenase) gene was cloned from S. elongatus PCC 7002 by homologous recombination, and then PCC 7942 was genetically modified to enhance β-carotene flux towards zeaxanthin synthesis (Sarnaik et al., 2018).
Cyanobacteria can also synthesize key amino acids for bioplastic production. For instance, engineered S. elongatus UTEX 2973 overproduces lysine, enabling concurrent cadaverine and glutamate biosynthesis for bioplastic production (Dookeran and Nielsen, 2021). Specifically, cadaverine can be used to synthesize biopolyamides (such as polynylon-5,10), while glutamate can be used to synthesize polyesters and other biobased plastics.
3.2.3 Other value-added products
3-Hydroxypropionic acid (3-HP) is a valuable chemical product used to synthesize polymers and other chemicals such as acrylic acid. However, cyanobacteria do not have a native pathway to synthesize 3-HP. By constructing an alternative pathway in S. elongatus PCC 7942, 3-HP could be synthesized (Lan et al., 2015). However, the yield remains insufficient for industrial production purposes. A microbial complex composed of S. elongatus UTEX 2973 and E. coli was constructed to convert CO2 into sucrose from S. elongatus UTEX 2973, and then sucrose was used as raw material for the production of 3-HP by E. coli (Zhang et al., 2020). The artificial co-culture system could significantly increase the yield of 3-HP and does not require foreign carbon sources (Matson and Atsumi, 2018). Further, the xylose utilization pathway from E. coli was introduced into S. elongatus UTEX 2973 through genetic engineering, which reconstituted the natural glycolytic pathway to transfer more carbon flux from xylose to acetyl-CoA, thereby increasing 3-HP biosynthesis by approximately 4.1-fold (Yao et al., 2022). The genetic engineering method also has great potential for the production of other value-added chemicals that require acetyl-CoA as a precursor.
3.3 Biophotovoltaic platforms
Cyanobacteria have been studied for biopower generation. In photosynthesis, only a small part of the absorbed solar energy is converted into chemical energy, while the rest of the energy is wasted as heat and fluorescence (Yagishita et al., 1997). Therefore, it is doable to harvest solar energy through a biophotovoltaic (BPV) platform to generate electricity. Cyanobacteria exhibit light-dependent electrogenic characteristics in photo-bioelectrochemical cells that generate substantial photocurrents, but the current densities are lower than their photovoltaic counterparts (Logan, 2009). However, by studying the algal biofilms formed on indium tin oxide anodes that were used in the algal biophotovoltaic platforms, it was found that several strains of cyanobacteria had high photosynthetic performance, and their biofilm and power generation capacity had application potential in the BPV platform (Ng et al., 2014). In a cyanobacterium called Nostoc sp. (NOS), it was found that the power generation capacity of NOS could be significantly increased by adding 1, 4-benzoquinone as a redox medium (Sekar et al., 2014). Inspired by the study on NOS, S. elongatus PCC 7942 was genetically modified to express a non-natural redox protein, which significantly improved the bioelectricity production capacity of the cyanobacterium (Sekar et al., 2016). Reduced graphene oxide-based BPV devices were found to produce reduced bioelectricity under dark conditions (Ng et al., 2018). An initial cross-comparison of S. elongatus PCC 7942 with other exoelectrogenic cultures showed a hindered exoelectrogenic capacity (McCormick et al., 2011).
3.4 Other applications
S. elongatus has been studied for medical purposes such as the photosynthetic therapies that protect ischemic tissues and ensure the aerobic metabolism of tissue cells (Williams et al., 2020; Zhu and Woo, 2022). S. elongatus PCC 7942 has shown potential as a new treatment for burn wounds (Yin et al., 2019). However, these applications are still in their experimental stage before clinical practice.
4 Concluding remarks and future perspectives
In this review, we explored the physiological and biochemical responses of S. elongatus to various environmental stresses and its potential applications in synthetic biology and biotechnology. S. elongatus is a highly adaptable organism that can thrive under diverse conditions, making it a promising candidate for industrial-scale applications in renewable energy, wastewater treatment, and bio-based manufacturing. Its rapid growth rate and metabolic versatility position it as an ideal model for advancing cyanobacterial biotechnology.
However, several challenges still impede the industrial application of S. elongatus. Although S. elongatus has good genetic operability, its gene editing tools and expression systems are not as mature as model organisms such as E. coli. Furthermore, light availability remains a critical bottleneck for high-density cultures, and scalability issues make it difficult to translate laboratory success into large-scale production. To address these challenges, advancements in genome editing tools, such as CRISPR-Cas systems specifically tailored for S. elongatus, will enable more precise genetic modifications. Additionally, genome-scale reconstruction and modeling through collaborative research could accelerate metabolic pathway optimization.
On the bioprocess front, optimizing photobioreactor designs is crucial to enhancing light utilization. Strategies such as incorporating light-harvesting technologies, like optical fibers or adjustable light emission strategies, could increase the efficiency of photosynthesis and boost biomass productivity. Furthermore, engineering S. elongatus to enhance carbon fixation, for example, by introducing high-affinity Rubisco variants or optimizing bicarbonate transport, could improve CO2 utilization, especially under suboptimal conditions.
Co-culture systems, where S. elongatus is paired with heterotrophic microorganisms, may also provide solutions for improving overall productivity. These systems can facilitate nutrient recycling and create stable growth environments, helping to reduce costs and improve efficiency in large-scale applications. For instance, it was demonstrated that co-culturing S. elongatus with E. coli can enhance the production of biofuels by leveraging complementary metabolic pathways. However, due to the metabolic byproducts generated during co-cultivation, the complexity of downstream processing is increased.
Downstream processing, including cell harvesting and product extraction, also presents a significant barrier. Current methods are often energy-intensive and costly, particularly for low-value products like biofuels. Advances in cell lysis techniques, such as enzymatic or mechanical disruption, and improved separation technologies, such as membrane filtration or chromatography, are needed to enhance efficiency and reduce costs.
Looking ahead, the integration of genetic engineering, bioreactor optimization, and system-level modeling will be essential to overcoming current challenges. For example, the use of artificial intelligence and machine learning to predict optimal metabolic pathways and cultivation conditions could significantly accelerate the transition from lab-scale to industrial-scale production. Furthermore, policy support, such as government subsidies or tax incentives for sustainable biotechnologies, will play a crucial role in fostering commercialization.
In conclusion, while S. elongatus holds immense potential for industrial applications, realizing this potential will require interdisciplinary innovations and collaborative efforts across academia, industry, and government. By addressing the technical, economic, and policy-related challenges, S. elongatus can be positioned as a key player in sustainable biotechnology, contributing to global efforts to address environmental and energy challenges.
Statements
Author contributions
WM: Visualization, Writing – original draft, Writing – review & editing. MX: Writing – review & editing. JH: Writing – original draft. CW: Writing – original draft. CQ: Writing – review & editing. MZ: Writing – review & editing. WF: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Natural Science Foundation of China Joint Fund Project (Grant No. U23A2034), the Zhejiang Province Leading Geese Plan (Grant No. 2024C03110), and the Science Foundation of Donghai Laboratory (grant no. DH-2022KF0203).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1542670/full#supplementary-material
Supplementary Figure 1The annual publication of S. elongatus-related article based on Google Scholar (https://scholar.google.com) using the keyword “Synechococcus elongatus”. The literature search was completed on December 1st, 2024.
References
1
Atsumi S. Higashide W. Liao J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol.27, 1177–1180. doi: 10.1038/nbt.1586
2
Baeshen N. A. Baeshen M. N. Sheikh A. Bora R. S. Ahmed M. M. M. Ramadan H. A. et al . (2014). Cell factories for insulin production. Microb. Cell Factories13, 1–9. doi: 10.1186/s12934-014-0141-0
3
Baltes N. J. Voytas D. F. (2015). Enabling plant synthetic biology through genome engineering. Trends Biotechnol.33, 120–131. doi: 10.1016/j.tibtech.2014.11.008
4
Behrenfeld M. J. Bale A. J. (1996). Confirmation of iron limitation of phytoplankton photosynthesis in the equatorial Pacific Ocean. Nature. 333(6600):508–511. doi: 10.1126/science.289.5480.765
5
Binder B. J. Chisholm S. W. (1990). Relationship between DNA cycle and growth rate in Synechococcus sp. strain PCC 6301. J. Bacteriol.172, 2313–2319. doi: 10.1128/jb.172.5.2313-2319.1990
6
Cao Y. Q. Li Q. Xia P. F. Wei L. J. Guo N. Li J. W. et al . (2017). AraBAD based toolkit for gene expression and metabolic robustness improvement in Synechococcus elongatus. Sci. Rep.7, 1–10. doi: 10.1038/s41598-017-17035-4
7
Chandra N. Mallick N. (2022). Co-production of bioethanol and commercially important exopolysaccharides from the marine cyanobacterium Synechococcus elongatus BDU 10144 in a novel low-cost seawater-fertilizer-based medium. Int. J. Energy Res. 46(10):13487–13510. doi: 10.1002/er.8069
8
Cooper S. Helmstetter C. E. (1968). Chromosome replication and the division cycle of Escherichia coli Br. J. Mol. Biol.31, 519–540. doi: 10.1016/0022-2836(68)90425-7
9
Cui J. Sun T. Li S. Xie Y. Song X. Wang F. et al (2020). Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing Mrp antiporters. Front. Bioeng. Biotechnol8, 500. doi: 10.3389/fbioe.2020.00500
10
Dookeran Z. A. Nielsen D. R. (2021). Systematic Engineering of Synechococcus elongatus UTEX 2973 for Photosynthetic Production of l-Lysine, Cadaverine, and Glutarate. ACS Synth. Biol.10, 3561–3575. doi: 10.1021/acssynbio.1c00492
11
Ducat D. C. Avelar-Rivas J. A. Way J. C. Silver P. A. (2012). Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol.78, 2660–2668. doi: 10.1128/AEM.07901-11
12
Fathima A. M. Laviña W. A. Putri S. P. Fukusaki E. (2020). Accumulation of sugars and nucleosides in response to high salt and butanol stress in 1-butanol producing Synechococcus elongatus. J. Biosci. Bioeng.129, 177–183. doi: 10.1016/j.jbiosc.2019.08.015
13
Fedeson D. T. Saake P. Calero P. Nikel P. I. Ducat D. C. (2020). Biotransformation of 2, 4-dinitrotoluene in a phototrophic co-culture of engineered Synechococcus elongatus and Pseudomonas putida. Microb. Biotechno.13, 997–1011. doi: 10.1111/1751-7915.13544
14
Feng L. J. Li J. W. Xu E. G. Sun X. D. Zhu F. P. Ding Z. et al . (2019). Short-term exposure to positively charged polystyrene nanoparticles causes oxidative stress and membrane destruction in cyanobacteria. Environ. Sci. Nano6, 3072–3079. doi: 10.1039/c9en00807a
15
Fraser J. M. Tulk S. E. Jeans J. A. Campbell D. A. Bibby T S. Cockshutt A. M. (2013). Photophysiological and photosynthetic complex changes during iron starvation in Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. PLoS One8 (3), e59861. doi: 10.1371/journal.pone.0059861
16
Hashemi A. Pajoum S. F. Sohani E. Azizi S. Hosseinifar S. Z. Delavari A. H. (2020). CO2 biofixation by Synechococcus elongatus from the power plant flue gas under various light–dark cycles. Clean Technol. Environ. Policy22, 1735–1743. doi: 10.1007/s10098-020-01912-0
17
Hirokawa Y. Dempo Y. Fukusaki E. Hanai T. (2017). Metabolic engineering for isopropanol production by an engineered cyanobacterium, Synechococcus elongatus PCC 7942, under photosynthetic conditions. J. Biosci. Bioeng.123, 39–45. doi: 10.1016/j.jbiosc.2016.07.005
18
Holtman C. K. Chen Y. Sandoval P. Gonzales A. Nalty M. S. Thomas T. L. et al . (2005). High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res.12, 103–115. doi: 10.1093/dnares/12.2.103
19
Hong K. K. Nielsen J. (2012). Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell. Mol. Life Sci.69, 2671–2690. doi: 10.1007/s00018-012-0945-1
20
Ito H. Mutsuda M. Murayama Y. Tomita J. Hosokawa N. Terauchi K. et al . (2009). Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci.106, 14168–14173. doi: 10.1073/pnas.0902587106
21
Jaiswal D. Sengupta A. Sengupta S. Madhu S. Pakrasi H. B. Wangikar P. P. (2020). A novel cyanobacterium Synechococcus elongatus PCC 11802 has distinct genomic and metabolomic characteristics compared to its neighbor PCC 11801. Sci. Rep.10, 1–15. doi: 10.1038/s41598-019-57051-0
22
Jaiswal D. Sengupta A. Sohoni S. Sengupta S. Phadnavis A. G. Pakrasi H. B. et al . (2018). Genome features and biochemical characteristics of a robust, fast growing and naturally transformable cyanobacterium Synechococcus elongatus PCC 11801 isolated from India. Sci. Rep.8, 1–13. doi: 10.1038/s41598-018-34872-z
23
Jarboe L. R. Zhang X. Wang X. Moore J. C. Shanmugam K. T. Ingram L. O. (2010). Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J. Biomed. Biotechnol.2010, 1–18. doi: 10.1155/2010/761042
24
Kaeberlein M. Powers R. W. Steffen K. K. Westman E. A. Hu D. Dang N. et al . (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science310, 1193–1196. doi: 10.1126/science.1115535
25
Kato A. Takatani N. Ikeda K. Maeda S. I. Omata T. (2017). Removal of the product from the culture medium strongly enhances free fatty acid production by genetically engineered Synechococcus elongatus. Biotechnol. Biofuels10, 1–8. doi: 10.1186/s13068-017-0831-z
26
Kato A. Takatani N. Ikeda K. Matsuura M. Kojima K. Aichi M. et al . (2016). Modulation of the balance of fatty acid production and secretion is crucial for enhancement of growth and productivity of the engineered mutant of the cyanobacterium Synechococcus elongatus. Biotechnol. Biofuels9, 1–10. doi: 10.1186/s13068-016-0506-1
27
Kaygusuz K. (2009). Bioenergy as a clean and sustainable fuel. Energy Sources Part A31, 1069–1080. doi: 10.1080/15567030801909839
28
Keasling J. D. (2012). Synthetic biology and the development of tools for metabolic engineering. Metab. Eng.14, 189–195. doi: 10.1016/j.ymben.2012.01.004
29
Kratzl F. Urban M. Pandhal J. Shi M. Meng C. Kleigrewe K. et al . (2024). Pseudomonas putida as saviour for troubled Synechococcus elongatus in a synthetic co-culture – interaction studies based on a multi-OMICs approach. Commun. Biol.7(1), 452. doi: 10.1038/s42003-024-06098-5
30
Kuan D. Duff S. Posarac D. Bi X. (2015). Growth optimization of Synechococcus elongatus PCC7942 in lab flasks and a 2-D photobioreactor. Can. J. Chem. Eng.93, 640–647. doi: 10.1002/cjce.22154
31
Kumar V. Mondal S. Gupta A. Maurya P. K. Sinha R. P. Häder D. P. et al . (2021). Light-dependent impact of salinity on the ecophysiology of Synechococcus elongatus PCC 7942: Genetic and comparative protein structure analyses of UV- absorbing mycosporine-like amino acids (MAAs) biosynthesis. Environ. Exp. Bot.191, 104620. doi: 10.1016/j.envexpbot.2021.104620
32
Lai J. L. Li Z. G. Han M. W. Huang Y. Xi H. L. Luo X. J. (2024). Analysis of environmental biological effects and OBT accumulation potential of microalgae in freshwater systems exposed to tritium pollution. Water Res.250, 15. doi: 10.1016/j.watres.2023.121013
33
Lan E. I. Chuang D. S. Shen C. R. Lee A. M. Ro S. Y. Liao J. C. (2015). Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942. Metab. Eng.31, 163–170. doi: 10.1016/j.ymben.2015.08.002
34
León-Bañares R. González-Ballester D. Galván A. Fernández E. (2004). Transgenic microalgae as green cell-factories. Trends Biotechnol.22, 45–52. doi: 10.1016/j.tibtech.2003.11.003
35
Li S. Sun T. Chen L. Zhang W. (2020). Light and carbon dioxide-driven synthesis of high-density fuel in Synechococcus elongates UTEX 2973. Chin. J. Biotechnol.36, 2126–2138. doi: 10.13345/j.cjb.200090
36
Lien T. Knutsen G. (1979). Synchronous growth of Chlamydomonas reinhardtii (chlorophyceae): a review of optimal conditions 1. J. Phycol.15, 191–200. doi: 10.1111/j.1529-8817.1979.tb02984.x
37
Lin Y. F. Lin Z. Q. Huang C. Zhang Y. Wang Z. W. Tang Y. J. et al . (2015). Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 31:13–21. doi: 10.1016/j.ymben.2015.06.006
38
Liu J. Chen L. Wang J. Qiao J. Zhang W. (2012). Proteomic analysis reveals resistance mechanism against biofuel hexane in Synechocystis sp. PCC 6803. Biotechnol. Biofuels5, 1–17. doi: 10.1186/1754-6834-5-68
39
Liu X. Sheng J. Curtiss R. (2011). Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci.108, 6899–6904. doi: 10.1073/pnas.1103014108
40
Logan B. E. (2009). Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol.7, 375–381. doi: 10.1038/nrmicro2113
41
Maoka T. (2011). Carotenoids in marine animals. Mar. Drugs9, 278–293. doi: 10.3390/md9020278
42
Matson M. M. Atsumi S. (2018). Photomixotrophic chemical production in cyanobacteria. Curr. Opin. Biotechnol.50, 65–71. doi: 10.1016/j.copbio.2017.11.008
43
McCormick A. J. Bombelli P. Scott A. M. Philips A. J. Smith A. G. Fisher A. C. et al . (2011). Photosynthetic biofilms in pure culture harness solar energy in a mediatorless bio-photovoltaic cell (BPV) system. Energy Environ. Sci.4, 4699–4709. doi: 10.1039/C1EE01965A
44
Mehta K. Jaiswal D. Nayak M. Prasannan C. B. Wangikar P. P. Srivastava S. (2019). Elevated carbon dioxide levels lead to proteome-wide alterations for optimal growth of a fast-growing cyanobacterium, Synechococcus elongatus PCC 11801. Sci. Rep.9, 1–14. doi: 10.1038/s41598-019-42576-1
45
Moraes J. S. Oliveira T. P. Guimarães P. S. Martins C. M. G. (2021). Short-term effects of glyphosate and Roundup Transorb® formulation on the cyanobacteria Synechococcus elongatus. Ecotoxic. Environ. Contam.16, 57–62. doi: 10.5132/eec.2021.01.07
46
Moronta-Barrios F. Espinosa J. Contreras A. (2012). In vivo features of signal transduction by the essential response regulator RpaB from Synechococcus elongatus PCC 7942. Microbiology158, 1229–1237. doi: 10.1099/mic.0.057679-0
47
Mortezaeikia V. Yegani R. Tavakoli O. (2016). Membrane-sparger vs. membrane contactor as a photobioreactors for carbon dioxide biofixation of Synechococcus elongatus in batch and semi-continuous mode. J. CO2 Util.16, 23–31. doi: 10.1016/j.jcou.2016.05.009
48
Ng F. L. Phang S. M. Periasamy V. Beardall J. Yunus K. Fisher A. C. (2018). Algal biophotovoltaic (BPV) device for generation of bioelectricity using Synechococcus elongatus (Cyanophyta). J. Appl. Phycol.30, 2981–2988. doi: 10.1007/s10811-018-1515-1
49
Ng F. L. Phang S. M. Periasamy V. Yunus K. Fisher A. C. (2014). Evaluation of algal biofilms on indium tin oxide (ITO) for use in biophotovoltaic platforms based on photosynthetic performance. PloS One9, e97643. doi: 10.1371/journal.pone.0097643
50
Oh S. Y. Kang S. G. Kim D. W. Chiu P. C. (2011). Degradation of 2, 4-dinitrotoluene by persulfate activated with iron sulfides. Chem. Eng. J.172, 641–646. doi: 10.1016/j.cej.2011.06.023
51
Oliver J. W. MaChado I. M. Yoneda H. Atsumi S. (2013). Cyanobacterial conversion of carbon dioxide to 2, 3-butanediol. Proc. Natl. Acad. Sci.110, 1249–1254. doi: 10.1073/pnas.1213024110
52
Pan L. L. Onai K. Uesaka K. Ihara K. Natsume T. Takatani N. et al . (2016). Transcriptional regulation of CmpR, the LysR family protein involved in CO2-responsive gene regulation in the cyanobacterium Synechococcus elongatus. Biomed. Genet. Genomics1, 1–6. doi: 10.15761/BGG.1000123
53
Pishbin M. Sarrafzadeh M. H. Faramarzi M. A. (2020). Nitrate and phosphate removal efficiency of Synechococcus elongatus under mixotrophic and heterotrophic conditions for wastewater treatment. Iran. J. Sci. Technol. Trans. Civ. Eng.45 (3), 1831–1843.1–13. doi: 10.1007/s40996-020-00514-6
54
Rashmi V. ShylajaNaciyar M. Rajalakshmi R. D’Souza S. F. Prabaharan D. Uma L. (2013). Siderophore mediated uranium sequestration by marine cyanobacterium Synechococcus elongatus BDU 130911. Bioresour. Technol.130, 204–210. doi: 10.1016/j.biortech.2012.12.016
55
Ruiz-Güereca D. A. Sánchez-Saavedra M. (2016). Growth and phosphorus removal by Synechococcus elongatus co-immobilized in alginate beads with Azospirillum brasilense. J. Appl. Phycol.28, 1501–1507. doi: 10.1007/s10811-015-0728-9
56
Saini D. K. Pabbi S. Shukla P. (2018). Cyanobacterial pigments: Perspectives and biotechnological approaches. Food Chem. Toxicol.120, 616–624. doi: 10.1016/j.fct.2018.08.002
57
Samiotis G. Stamatakis K. Amanatidou E. (2021). Assessment of Synechococcus elongatus PCC 7942 as an option for sustainable wastewater treatment. Water Sci. Technol.84, 1438–1451. doi: 10.2166/wst.2021.319
58
Samiotis G. Stamatakis K. Amanatidou E. (2022). Dimensioning of Synechococcus elongatus PCC 7492 cultivation photobioreactor for valorization of wastewater resources. Chem. Eng. J.435, 134895. doi: 10.1016/j.cej.2022.134895
59
Sarnaik A. Pandit R. Lali A. (2017). Growth engineering of Synechococcus elongatus PCC 7942 for mixotrophy under natural light conditions for improved feedstock production. Biotechnol. Prog.33, 1182–1192. doi: 10.1002/btpr.2490
60
Schmitz O. (2000). CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science289, 765–768. doi: 10.1126/science.289.5480.765
61
Schubert W. D. Klukas O. Krauß N. Saenger W. Fromme P. Witt H. T. (1997). Photosystem I of Synechococcus elongatus at 4 Å resolution: comprehensive structure analysis. J. Mol. Biol.272, 741–769. doi: 10.1006/jmbi.1997.1269
62
Sekar N. Jain R. Yan Y. Ramasamy R. P. (2016). Enhanced photo-bioelectrochemical energy conversion by genetically engineered cyanobacteria. Biotechnol. Bioeng113, 675–679. doi: 10.1002/bit.25829
63
Sekar N. Umasankar Y. Ramasamy R. P. (2014). Photocurrent generation by immobilized cyanobacteria via direct electron transport in photo-bioelectrochemical cells. Phys. Chem. Chem. Phys.16, 7862–7871. doi: 10.1039/c4cp00494a
64
Selim K. A. Haffner M. (2020). Heavy metal stress alters the response of the unicellular cyanobacterium Synechococcus elongatus PCC 7942 to nitrogen starvation. Life10, 275. doi: 10.3390/life10110275
65
Sengupta S. Jaiswal D. Sengupta A. Shah S. Gadagkar S. Wangikar P. P. (2020). Metabolic engineering of a fast-growing cyanobacterium Synechococcus elongatus PCC 11801 for photoautotrophic production of succinic acid. Biotechnol. Biofuels13, 1–18. doi: 10.1186/s13068-020-01727-7
66
Shestakov S. V. Khyen N. T. (1970). Evidence for genetic transformation in blue-green alga Anacystis nidulans. Mol. Gen. Genet. MGG107, 372–375. doi: 10.1007/BF00441199
67
Sinha M. Weyda I. Sørensen A. Bruno K. S. Ahring B. K. (2017). Alkane biosynthesis by Aspergillus carbonarius ITEM 5010 through heterologous expression of Synechococcus elongatus acyl-ACP/CoA reductase and aldehyde deformylating oxygenase genes. Amb. Express7, 1–9. doi: 10.1186/s13568-016-0321-x
68
Song K. Tan X. Liang Y. Lu X. (2016). The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl. Microbiol. Biotechnol.100, 7865–7875. doi: 10.1007/s00253-016-7510-z
69
Stephanopoulos G. (2012). Synthetic biology and metabolic engineering. ACS Synth. Biol.1, 514–525. doi: 10.1021/sb300094q
70
Sugita C. Ogata K. Shikata M. Jikuya H. Takano J. Furumichi M. et al . (2007). Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth. Res.93, 55–67. doi: 10.1007/s11120-006-9122-4
71
Takahama K. Matsuoka M. Nagahama K. Ogawa T. (2003). Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng95, 302–305. doi: 10.1016/S1389-1723(03)80034-8
72
Tan L. R. Xia P. F. Sun X. F. Guo N. Song C. Li Q. et al . (2016). Ecological insights into low-level antibiotics interfered biofilms of Synechococcus elongatus. RSC Adv.6, 78132–78135. doi: 10.1039/C6RA15025J
73
Ungerer J. Lin P. C. Chen H. Y. Pakrasi H. B. (2018a). Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio9, e02327–e02317. doi: 10.1128/mBio.02327-17
74
Ungerer J. Wendt K. E. Hendry J. I. Maranas C. D. Pakrasi H. B. (2018b). Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. U.S.A.115, E11761–E11770. doi: 10.1073/pnas.1814912115
75
Usai G. Cordara A. Mazzocchi E. Re A. Fino D. Pirri C. D. et al . (2024). Coupling dairy wastewaters for nutritional balancing and water recycling: sustainable heterologous 2-phenylethanol production by engineered cyanobacteria. Front. Bioeng. Biotechnol.12. doi: 10.3389/fbioe.2024.1359032
76
Van P. Abedini H. Branco F. Hellingwerf K. J. (2018). Increasing the photoautotrophic growth rate of Synechocystis sp. PCC 6803 by identifying the limitations of its cultivation. Biotechnol. J.13, 1700764. doi: 10.1002/biot.201700764
77
Vayenos D. Romanos G. E. Papageorgiou G. C. Stamatakis K. (2020). Synechococcus elongatus PCC7942: a cyanobacterium cell factory for producing useful chemicals and fuels under abiotic stress conditions. Photosynth. Res.146, 235–245. doi: 10.1007/s11120-020-00747-6
78
Velmurugan R. Incharoensakdi A. (2020). Heterologous expression of ethanol synthesis pathway in glycogen deficient Synechococcus elongatus PCC 7942 resulted in enhanced production of ethanol and exopolysaccharides. Front. Plant Sci.11. doi: 10.3389/fpls.2020.00074
79
Vicente A. Sohm B. Flayac J. Rousselle P. Bauda P. Pagnout C. (2019). Toxicity mechanisms of ZnO UV-filters used in sunscreens toward the model cyanobacteria Synechococcus elongatus PCC 7942. Sci. pollut. Res.26, 22450–22463. doi: 10.1007/s11356-019-05057-6
80
Voshol G. (2015). Biodiesel production using blue-green cyanobacterium Synechococcus elongatus PCC 7942. Front. Nutr.11, 2296–861X. doi: 10.1093/femsle/fnx066
81
Walker P. L. Pakrasi H. B. (2022). A ubiquitously conserved cyanobacterial protein phosphatase essential for high light tolerance in a fast-growing cyanobacterium. Microbiol. Spectr.10, e01008–e01022. doi: 10.1128/spectrum.01008-22
82
Wijffels R. H. Kruse O. Hellingwerf K. J. (2013). Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol.24, 405–413. doi: 10.1016/j.copbio.2013.04.004
83
Williams K. M. Wang H. Paulsen M. J. Thakore A. D. Rieck M. Lucian H. J. et al . (2020). Safety of photosynthetic Synechococcus elongatus for in vivo cyanobacteria–mammalian symbiotic therapeutics. Microb. Biotechnol.13, 1780–1792. doi: 10.1111/1751-7915.13596
84
Yagishita T. Sawayama S. Tsukahara K. I. Ogi T. (1997). Effects of intensity of incident light and concentrations of Synechococcus sp. and 2-hydroxy-1, 4-naphthoquinone on the current output of photosynthetic electrochemical cell. Sol. Energy61, 347–353. doi: 10.1016/S0038-092X(97)00069-8
85
Yang C. C. Wen R. Shen C. Yao D. J. (2015). Using a microfluidic gradient generator to characterize BG-11 medium for the growth of cyanobacteria Synechococcus elongatus PCC7942. Micromachines6(11), 1755–1767. doi: 10.3390/mi6111454
86
Yao J. Wang J. Ju Y. Dong Z. Song X. Chen L. et al . (2022). Engineering a xylose-utilizing Synechococcus elongatus UTEX 2973 chassis for 3-hydroxypropionic acid biosynthesis under photomixotrophic conditions. ACS Synth. Biol.11, 678–688. doi: 10.1021/acssynbio.1c00364
87
Yin H. Chen C. Y. Liu Y. W. Tan Y. J. Deng Z. L. Yang F. et al . (2019). Synechococcus elongatus PCC7942 secretes extracellular vesicles to accelerate cutaneous wound healing by promoting angiogenesis. Theranostics9, 2678. doi: 10.7150/thno.31884
88
Yu J. Liberton M. Cliften P. F. Head R. D. Jacobs J. M. Smith R. D. et al . (2015). Synechococcus elongatus UTEX 2973, a fast-growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep.5, 1–10. doi: 10.1038/srep08132
89
Yu Y. You L. Liu D. Hollinshead W. Tang Y. J. Zhang F. (2013). Development of Synechocystis sp. PCC 6803 as a phototrophic cell factory. Mar. Drugs11, 2894–2916. doi: 10.3390/md11082894
90
Yuan Y. Bulushi T. A. Sastry A. V. Sancar C. Szubin R. Golden S. S. et al . (2024). Machine learning reveals the transcriptional regulatory network and circadian dynamics of Synechococcus elongatus PCC 7942. PNAS121, 10. doi: 10.1073/pnas.2410492121
91
Zhang L. Chen L. Diao J. Song X. Shi M. Zhang W. (2020). Construction and analysis of an artificial consortium based on the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce the platform chemical 3-hydroxypropionic acid from CO2. Biotechnol. Biofuels13, 1–14. doi: 10.1186/s13068-020-01720-0
92
Zhang J. Li S. Tao S. Y. Wu Z. Y. Luo Y. F. Wei W. W. et al . (2024). Oscillation of type IV pili regulated by the circadian clock in cyanobacterium Synechococcus elongatus PCC7942. Sci. Adv.10 (4), eadd9485. doi: 10.1126/sciadv.add9485
93
Zhang S. Sun J. Feng D. Sun H. Cui J. Zeng X. et al . (2023). Unlocking the potentials of cyanobacterial photosynthesis for directly converting carbon dioxide into glucose. Nat. Commun.14, 3425. doi: 10.1038/s41467-022-31251-3
94
Zhu Y. Woo Y. J. (2022). Photosynthetic symbiotic therapeutics–An innovative, effective treatment for ischemic cardiovascular diseases. J. Mol. Cell. Cardiol.164, 51–57. doi: 10.1016/j.yjmcc.2021.11.007
95
Zouni A. Witt H. T. Kern J. Krauss N. Saenger W. Orth P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature409, 739–743. doi: 10.1038/35055589
Summary
Keywords
Synechococcus elongatus , cyanobacteria, synthetic biology, cell factory, stress tolerance
Citation
Meng W, Xia M, Hu J, Wang C, Qian C, Zhang M and Fu W (2025) Adaptation and synthetic biology of the model cyanobacterium Synechococcus elongatus for sustainable development: a review. Front. Mar. Sci. 12:1542670. doi: 10.3389/fmars.2025.1542670
Received
10 December 2024
Accepted
05 May 2025
Published
03 June 2025
Volume
12 - 2025
Edited by
María Belén Fernández, Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina
Reviewed by
Giselle Martínez-Noël, CONICET Instituto de Investigaciones en Biodiversidad y Biotecnología (INBIOTEC), Argentina
Xupeng Cao, Chinese Academy of Sciences (CAS), China
Updates
Copyright
© 2025 Meng, Xia, Hu, Wang, Qian, Zhang and Fu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiqi Fu, weiqifu@zju.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.