Abstract
Teleosts are the most varied vertebrates. They inhabit various environments and are crucial to global fisheries, making them a focus of research using advanced omics approaches. These studies provide insights into the genetic factors, environmental adaptability, disease resistance, and metabolic processes, aiding aquaculture sustainability. Acclimation to salinity stress is complex, influenced by genetics and the environment. Although some species tolerate varying salinity levels, rapid shifts beyond their optimal tolerance cause stress. Euryhaline species experience stress at extreme salinities, whereas stenohaline species are sensitive to minor changes. Osmoregulation maintains homeostasis at varying salinities through acclimation in the intestine, kidney, and gills, ensuring survival in changing environments. Studies on gut microbiota and metabolomics have revealed how teleosts cope with salinity stress. This review delves into the acclimatization processes through transcriptomic, metabolomic, and gut microbiome analyses, which have shed light on the complex mechanisms that teleosts have evolved to cope with salinity stress. Transcriptomic analyses have identified key ion transport, osmoregulation, and stress response genes essential for adaptation, facilitating cellular adjustments and maintaining osmotic balance across habitats. Studies have revealed significant metabolite changes in energy production and osmolyte synthesis during stress, indicating metabolic reorganization for osmoregulation. Gut microbiota analysis highlights microbial diversity in regulating osmoregulatory functions, emphasizing microbiota’s role in resilience. Although research on interactions between salinity, growth conditions, and gut microbiota in teleosts is limited, findings suggest a vital relationship that warrants further study. Understanding these mechanisms is essential for improving fish health and enabling sustainable aquaculture management under environmental fluctuations.
1 Introduction
The increasing global population has led to a higher demand for aquatic products, particularly in terms of the daily consumption of aquatic species (Cooney et al., 2021; Emerenciano et al., 2021; McLean et al., 2020; Yohana et al., 2023; Zarzar et al., 2023). Fish farming is the fastest-growing sector in the aquaculture industry, playing a crucial role in boosting food production, fostering local economic growth, and improving livelihoods (Chang et al., 2020; Mkulo et al., 2024; Yohana et al., 2024). However, wild and cultured aquatic species face stress from pollution, temperature, fluctuations in salinity, and elevated ammonia levels (Zarantoniello et al., 2021). Salinity fluctuations, often caused by extreme weather events, can involve changes of 5–10 ppt or more and rise to be harmful to many marine and freshwater species (Gonzalez, 2012b; Lee et al., 2022b; Röthig et al., 2023). Similarly, sudden temperature shifts exceeding 2-3°C can induce stress, particularly in species with narrow thermal tolerance ranges (Mugwanya et al., 2022). To exist in such environments, fish must actively regulate the balance of ions and water to maintain osmotic homeostasis, as their body fluids and external environments have distinct ionic and osmotic pressures (Ruiz-Jarabo et al., 2017; Soengas et al., 2019). Generally, to maintain osmotic equilibrium, fish rely on both active ion transport, which requires energy, and passive water movement driven by osmotic gradients across epithelial tissues, such as the gills, kidneys, and gut. Additionally, hormonal regulation and physiological adaptations contribute to maintaining a proper balance of salt and water in the body (Arjona et al., 2009; Evans, 2011; Gregório et al., 2013; Takei and Hwang, 2016). Therefore, fish performance can be significantly affected if they cannot efficiently acclimate to osmotic stress (Evans, 2010; Kültz, 2015; Tseng et al., 2022a). Understanding osmoregulatory mechanisms is increasingly essential in the context of the impact of changing climates and increasing salinity variations in aquatic ecosystems (Fridman, 2020; Tseng et al., 2022a; Tresguerres et al., 2023). Recent advances in molecular biology, particularly transcriptomics, metabolomics, and microbiota profiling, have greatly enhanced our knowledge of osmoregulation in response to environmental changes (Kim and Kültz, 2020; Mundy et al., 2020; Escobar-Sierra et al., 2024). Molecular techniques have been widely used to investigate the effects of how salinity changes have influenced fish physiology, metabolic pathways, and gene expression (Houde et al., 2019; Devlin et al., 2020; Rahi et al., 2021).
Teleosts are the most diverse vertebrates and serve as interesting models for osmoregulatory plasticity studies because of their broad tolerance to environmental changes (Takei et al., 2014; Perry et al., 2020). These fish have developed various morphological and physiological adaptations that allow them to thrive in changing environments within their habitats (Harrison and Whitfield, 2006; Christensen et al., 2019). The species that have been most extensively studied in terms of salinity acclimation belong to several families. These include the Cyprinodontidae, such as the sheepshead minnow Cyprinodon variegatus, mummichog Fundulus heteroclitus, and Arabian killifish Aphanius dispar; Cichlidae, such as black-chinned tilapia Sarotherodon melanotheron and Mozambique tilapia Oreochromis mossambicus; and Poecilidae, such as the sailfin molly Poecilia latipinna, on fish salinity adaptations (Gonzalez, 2012a). The intestine is crucial for osmoregulation, as it modulates ion transport, regulates gene expression, interacts with microbes, and facilitates water absorption to assist in adapting to changes in salinity (Chen et al., 2023b; Su et al., 2023a; Takei, 2021a). One crucial function of the gut is to regulate the immune system and create a healthy environment for fish by interacting with the gut microbiota. This interaction is essential for maintaining the overall health and well-being of fish (Diwan et al., 2023; Perry et al., 2020; Xia et al., 2022a; Gyan et al., 2024). Meanwhile, metabolomics explores the complete set of low molecular weight metabolites present in a cell or organism, providing insights into physiological and biochemical responses to stress (Nicholson et al., 1999; Cuykx et al., 2018). Transcriptomics, gut microbiota analysis, and metabolomics have offered valuable insights into the response of teleost fish species such as Danio rerio (Guh et al., 2015), Salmo salar (Tipsmark et al., 2002), and Oreochromis mossambicus (Richards et al., 2003) to salinity stress. Most studies have primarily examined short-term fluctuations in salinity, rather than long-term acclimation. The connection between changes in the gut microbiota and osmoregulatory mechanisms remains unclear, raising the question of whether microbial changes are adaptive responses or play a direct role in osmoregulation. This review explores the osmoregulatory responses of teleost fish to salinity changes, as well as the roles of gut microbiota, metabolomics, and transcriptomics in these responses. By integrating various approaches, we sought to enhance our understanding of how fish adapt to shifting aquatic environments. This review identifies key gaps in the current knowledge and proposes innovative research directions to advance scientific understanding in this field.
2 Interactive effects of salinity, temperature, pH, and oxygen on fish physiology
The interactive effects of environmental stressors, such as salinity, temperature, pH, and oxygen levels, significantly impact fish physiology by disrupting ion regulation, acid-base balance, hormone circulation, metabolic pathways, growth, and survival (Kaushal et al., 2010; Franklin and Edward, 2019; Wang et al., 2019; Mariu et al., 2023). Temperatures exceeding 30°C can impose severe stress and potentially lead to mortality, particularly in cold-water fish species (Jain et al., 2013; Mariu et al., 2023). Extremely high salinity and temperatures, above 35°C, can be lethal to most fish species within hours (He et al., 2017; Mariu et al., 2023; Vadboncoeur et al., 2023; Yang et al., 2023). Temperatures below 5°C can cause a decrease in metabolic rates and activity. Subzero temperatures can lead to bodily fluids freezing, which can result in tissue damage and hinder growth and reproduction (Mariu et al., 2023). In environments with low pH and salinity, aquatic organisms tend to allocate less energy to primary physiological functions to focus on osmoregulation (Lin et al., 2013). A study was conducted to analyze the effects of salinity and temperature on cortisol and glucose concentrations in three different fish species: Cottus bairdii, Catostomus platyrhynchus, and Oncorhynchus clarki pleuriticus. The study findings indicated that temperature had the most significant impact, with the influence of salinity being influenced by temperature. Extended exposure to high salinity levels was seen to lower the baseline cortisol and glucose levels in the fish, leading to an increased stress response when exposed to high temperatures. This ultimately resulted in physiological suppression in the fish. These results highlighted the need for conservation strategies to mitigate the effects of temperature-dependent salinity stress on freshwater fish species, especially considering the expected increase in freshwater salinization and temperature levels (Walker et al., 2020).
The optimal pH range for most freshwater fish species falls between 6.5 and 9.0, promoting maximum growth and reproduction (McCormick and Bradshaw, 2006; Laycock and Meeran, 2012; McCormick et al., 2020). For saltwater species, the ideal pH range is 6.5–8.5, beyond which stress-related effects may become apparent. Fluctuations in pH levels can disrupt ion regulation, enzyme function, and metabolic processes, leading to a disturbance in homeostasis and impairing oxygen transport and enzyme activity. Deviations from optimal pH levels have the potential to alter enzyme configuration and decrease catalytic efficiency (Poff and Zimmerman, 2010; Mariu et al., 2023). Oxygen levels are critical for adaptation to salinity. Research on Perca fluviatilis shows oxygen uptake is minimized in brackish water, where osmotic stress is reduced, compared to freshwater and seawater (Ern et al., 2014). A study of six stenohaline Channel catfish Ictalurus punctatus, and goldfish Carassius auratus and euryhaline rainbow trout Oncorhynchus mykiss, brown trout Salmo trutta, striped bass Morone saxatilis, and Gulf sturgeon Acipenser oxyrinchus fish species examined variations in oxygen consumption across low salinity, highlighting differences in osmoregulatory strategies (Altinok and Grizzle, 2003). In Oreochromis niloticus, it has been observed that as salinity levels increase, there is a decrease in oxygen consumption and ammonia excretion rates. This may reflect a reduction in the energy demand for osmoregulation rather than a limitation in energy availability. The highest survival rates were found at a salinity level of 12 g/L, with the most optimal growth occurring between 6–12 g/L. Salinity levels and duration significantly impact metabolic oxygen consumption rates (MO2), total ammonia excretion (Tamm), ammonia quotient (AQ), and oxygen-to-nitrogen ratio (O: N). Protein metabolism was found to contribute less than 14.74% of the total energy required for osmoregulation (Kombat et al., 2021). A study conducted on Epinephelus malabaricus revealed that fish exposed to a salinity level of 11 psu exhibited lower Na+/K+-ATPase transcript levels, decreased oxygen consumption, and increased growth rates. At 11 psu, the GH-IGF axis was upregulated, leading to improved food conversion efficiency and growth (Zhu et al., 2023b). These findings underscore the importance of taking a comprehensive approach when evaluating fish acclimation, highlighting the interconnected nature of these physiological processes and the effects of salinity, temperature, pH, and oxygen availability on the resilience of aquatic species.
3 Osmoregulation mechanisms in teleost fish: the role of the gill, kidney, and intestine.
3.1 Gill
The gill is a remarkable organ that plays a crucial role in fish life and is a highly distinctive structure with multiple functions. Gills are essential respiratory structures in fish, responsible for respiratory gas exchange, ion and water transfer, filter feeding, ammonia nitrogen excretion, and osmoregulation (Figure 1) (Chen et al., 2023a). Gills have evolved complex morphological structures to perform their vital physiological functions. These structures, which are part of the branchial chamber to which the gills belong, include the gill operculum, filaments, arch, and rakers. Each of these components plays a crucial role in the respiratory process of aquatic organisms (Fiedler et al., 2020; Alsafy et al., 2023). These structures, except gill rakers, undergo morphological and functional modifications in response to variations in water flow, temperature, ion concentration, and salinity within the aquatic environment. One way in which organisms adapt to environmental stressors is by altering the length and surface area of their gill filaments. This adjustment allows for optimal gas exchange and osmoregulatory functions (Evans et al., 2005a; Foyle et al., 2020; Alsafy et al., 2023). Fish have strong ionic/osmotic gradients in their aquatic surroundings, and the methods by which they maintain internal homeostasis are more demanding than those of terrestrial vertebrates (Hwang et al., 2011; Tseng et al., 2022a). Ionocytes are specialized, mitochondria-rich epithelial cells (MR cells) involved in ion regulation and homeostasis in various tissues and organisms. They are primarily responsible for maintaining ionic equilibrium and regulating pH through active ion transport mechanisms. These cells are found in diverse locations, such as the gills of fish, the airway epithelium in mammals, and other organs like the kidney and salivary glands (Griffith, 2017; Guh et al., 2015; Tseng et al., 2022a). A substantial body of literature has proposed models for examining iono-osmoregulatory mechanisms in fish gills. Nevertheless, these models frequently present conflicting or unresolved issues due to variations in species-specific metabolic pathways, differences in experimental methodologies, and inherent limitations of current analytical techniques (Hwang et al., 2011). Recent research has shed light on the challenges of ion regulation and osmoregulation in teleost fish by utilizing cutting-edge molecular and cellular physiological techniques such as whole-body oxygen consumption measurements, ion flux assays, electrophysiology, and immunohistochemistry, as well as animal models like zebrafish (Danio rerio), rainbow trout (Oncorhynchus mykiss), and tilapia (Oreochromis niloticus). Freshwater Teleost gills contain a combination of ionocytes and pavement cells (PNA−) (Shih et al., 2023). Pavement cells, which make up approximately 90% of the gill surface, serve as the main connection between the gills and water (Kovac and Goss, 2024). Ionocytes, primarily situated at the junction of gill filaments and lamellae, play a vital role in regulating ion transport to uphold osmotic balance. Their distribution is subject to alteration in response to environmental conditions; for instance, in fish inhabiting soft water environments, there may be an increase in the density of ionocytes within the lamellae. The apical surfaces of ionocytes vary among different species and environmental conditions, displaying features such as microvilli, a smooth finish, or a spongy texture (Masroor et al., 2019). In some species, ionocytes are situated within the epithelium, with their apical surfaces being partially obscured by pavement cells. The study conducted by (Shih et al., 2022). focused on freshwater teleost fish, particularly medaka (Oryzias latipes) and discovered and characterized the several types of ionocytes and ion transporters involved in the mechanisms of NaCl secretion, Na+ uptake/acid secretion/NH4+ excretion, Ca2+ uptake, and Cl− uptake/base secretion. Additionally, this study also identified the key regulators involved in these processes, including Na+/K+-ATPase (NKA) located in the basolateral membrane, which establishes the primary electrochemical gradient by actively transporting Na+ out of the cell and K+ into the cell. This gradient drives the movement of other ions through various transporters and co-transporters, the cystic fibrosis transmembrane conductance regulator (CFTR), the Na+/K+/2Cl- cotransporter (NKCC1), tight junction proteins such as claudins, and hormonal regulators like cortisol and prolactin, which collectively facilitate ion transport and osmoregulatory adaptations in teleost fish. Marine teleosts and seawater-adapted euryhaline fish regulate ionic balance by actively excreting Na+ and Cl-, along with other minerals, through their gills (Figure 1). This process is facilitated by specialized mitochondrion-rich (MR) cells, also known as chloride cells. Several studies (Kolbadinezhad et al., 2018; Huang et al., 2020; Takvam et al., 2021a) have demonstrated that active trans epithelial Cl- transport drives this process. Active chloride transport is accompanied by the passive flow of sodium ions across the paracellular space and tight junctions (Laverty and Skadhauge, 2012; Saint-Criq and Gray, 2017). The cellular process of chloride (Cl−) secretion is comparable to that observed in several tissues, such as mammals’ intestinal and airway epithelia (Rottgen et al., 2018; Shah et al., 2022; Becker and Seidler, 2024). Cl− ions are initially transported across the basolateral membrane using the sodium-potassium-2 chloride (NKCC) transporter. This transporter depends on the Na+/K+-ATPase to maintain a beneficial inward gradient for Na+ ions. The overall impact of these transporters is to increase the concentration of intracellular Cl− above electrochemical equilibrium (Janoš and Magistrato, 2021; Yurinskaya and Vereninov, 2021; Tseng et al., 2022a). Chloride ions are transported out of the cell by a specific channel called the cystic fibrosis transmembrane conductance regulator (CFTR) channel homolog. This process has been studied and documented by (Ferreira-Martins et al., 2021; Hanssens et al., 2021; Farinha et al., 2024). Several studies have explored the changes in the expression and function of transport components in response to osmoregulatory adaptations. In freshwater teleosts, water absorption and ion leakage through diffusion occur in the gills (Figure 1A). Nevertheless, occludin and claudin proteins restrict the permeability of junctions between gill cells in these fish (Fridman, 2020). These junctions also contain negatively charged amino acid residues that bind to Ca2+ ions. Furthermore, mucosal cells located in the gills secrete mucus that contains charged glycoproteins and mucins. These substances can attract ions to the surface of the gills, aiding in the establishment of localized ionic gradients. This process effectively reduces passive ion loss through diffusion (Wilson and Laurent, 2002; Evans et al., 2005a).
Figure 1
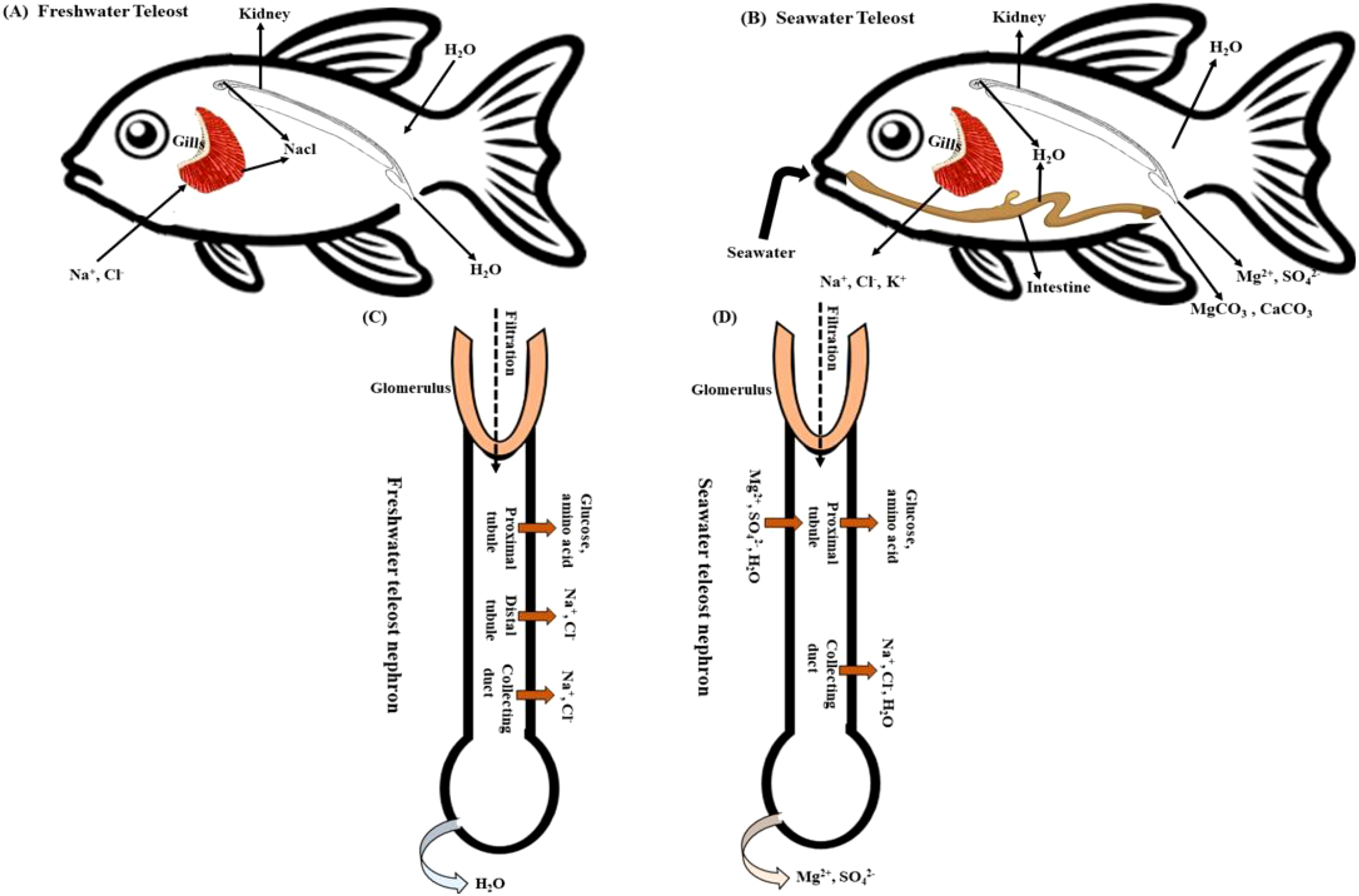
The acclimation process of teleost fishes to freshwater and seawater is crucial for regulating organ functions. This process involves the passage of ions and water through epithelial cells of the gill, kidney, and gut in both freshwater and seawater teleosts, referred to as (A, B). Additionally, the transport of ions and water in the nephrons of freshwater and marine teleosts is a significant aspect of acclimation (C, D). This process helps teleost fishes adapt to their environment and maintain organ function in varying salinity levels.
In larval zebrafish, Na+ uptake is mediated by Na+/H+ exchanger 3b (Nhe3b), electro genic uptake driven by H+-ATPase in H+-ATPase-rich (HR) cells, and Na+-Cl- cotransporter (Ncc). However, when slc9a3.2 (encoding Nhe3b) was knocked out using CRISPR/Cas9, Na+ uptake remained unaffected, even in low Na+ environments, indicating compensatory mechanisms. Neither H+-ATPase knockdown nor chloride absence influenced Na+ uptake, highlighting zebrafish’s adaptability and revealing gaps in understanding these processes (Zimmer et al., 2020). Prolactin (PRL), crucial for osmoregulation, was examined in zebrafish mutants deficient in PRL due to TALEN-induced mutations. These mutants couldn’t survive in freshwater due to Na+/K+/Cl- uptake defects but thrived in brackish water, emphasizing PRL’s vital role in ion balance (Shu et al., 2016). Investigation into carbonic anhydrase (Ca17a) demonstrated its significance in ion and acid-base equilibrium. CRISPR/Cas9 knockout of ca17a led to death by 19 days post-fertilization, with mutants showing increased Na+ uptake and decreased Cl- uptake, although overall ion content remained stable. Morpholino knockdown and pharmacological inhibition confirmed Ca17a’s involvement in Cl- uptake, yet the exact cause of lethality remains unknown (Zimmer et al., 2021). These findings highlight the complexity of ion transport mechanisms in zebrafish, uncovering compensatory pathways and regulatory networks crucial for osmoregulation. Future research combining genetic and physiological methods will be essential to unravel these complex processes in freshwater and seawater teleosts. The zebrafish (Danio rerio) possesses five distinct ionocyte types: HR, NaR, NCC, SLC26, and KS cells, each with specific roles in ion transport. These regulatory mechanisms are governed by hormonal signaling pathways, which are mediated by isotocin, prolactin, cortisol, vitamin D, and calcitonin (Guh et al., 2015). These ionocytes play a crucial role in maintaining the osmotic balance within the fish. One key component of these ionocytes is the H+ ATPase (V-ATPase), located on the apical membrane (Davidson, 2024). This enzyme works to expel H+ ions, creating an electric gradient that facilitates the entry of Na+ through apical Na+ channels (Davidson, 2024). This process is essential for regulating the fish’s internal environment. Euryhaline teleosts have been extensively studied for their remarkable ability to adapt their ion and water regulation systems to withstand varying osmotic pressures (Agarwal et al., 2024). One critical system involved in these adaptations is the insulin-like growth factor (IGF) system. IGF-1 in particular plays a significant role in regulating myogenic cell proliferation and differentiation, as well as influencing osmoregulatory functions in fish gills by modulating plasma osmolality (Malone et al., 2015; McCormick and Regish, 2018).
Furthermore, Growth hormone (GH) has been found to enhance salinity tolerance in species like rainbow trout, Atlantic salmon, and killifish by stimulating gill Na+/K+-ATPase (NKA) activity (Yada et al., 2012). This hormone plays a crucial role in helping these fish adapt to changing environmental conditions. Recent Genomic advances in salmonids have identified key transporters in ion-coupled fluid regulation, including NKA, Na+/H+ exchangers (NHEs), carbonic anhydrases, V-type H+-ATPase (V-ATPase), Na+:HCO3- co-transporters (NBCs), FXYDs, claudins, aquaporins (AQPs), Na+:K+:2Cl- co-transporters (NKCCs), Na+:Cl- co-transporters (NCCs), and Cl-/HCO-3 exchangers (SLC26A6) (Madsen, 2011). Among these transporters, Claudins maintain epithelial barrier integrity. For example, Zebrafish express claudin-15, while Atlantic salmon use claudin-30 to reduce sodium permeability in gill epithelia (Bagnat et al., 2007; Rosenthal et al., 2010; Amasheh et al., 2011; Engelund and Madsen, 2011; Lingaraju et al., 2015). Identifying claudin-15 and claudin-25b in the salmon intestine has shed light on their specific osmoregulatory functions (Tipsmark, 2008; Tipsmark et al., 2010). Furthermore, in zebrafish, it has been studied that ion homeostasis is maintained through gill ionocytes expressing Na+/K+-ATPase (NKA) and Na+/Cl- cotransporters (NCC) (Hwang and Chou, 2013). Under extreme ion deficiency, zebrafish increase expression of an NKA α-subunit (zatp1a1a.5), altering ATP hydrolysis efficiency (Esbaugh et al., 2019). Anadromous species such as Atlantic salmon undergo significant physiological changes during smoltification, enabling them to survive hypertonic seawater (Christensen et al., 2018; Morera et al., 2021; Morales-Rivera et al., 2022; Silva-Marrero et al., 2025). In freshwater environments, salmon gills exhibit high levels of NCC to absorb Na+ and Cl-. However, when transitioning to seawater, the expression of NCC decreases, while CFTR chloride channels and NKCCs increase. This shift promotes active salt excretion, which is essential for preventing dehydration (Yada et al., 2012; Lema et al., 2019; Inokuchi et al., 2022; Tümmler, 2023). These discoveries shed light on the intricate molecular and physiological adaptations that contribute to salinity tolerance in teleost fish. Understanding these mechanisms is crucial for grasping their ecological resilience and potential applications in aquaculture.
3.2 Kidney
Fish kidneys are essential for regulating ions and water balance in freshwater (FW) (Figure 1A) and seawater (SW) (Figure 1B) environments. Glomerular filtration rate (GFR) as a key indicator of kidney function, representing the rate at which kidneys filter blood, is crucial in determining urine flow (Beyenbach, 2004; Takvam et al., 2023). However, the GFR of teleost fish exhibits considerable variability and is affected by factors such as glomerular intermittency, ambient salinity, renal perfusion pressure, and certain hormones including prolactin, atrial natriuretic peptide (ANP), cortisol, renin-angiotensin system (RAS) and arginine vasotocin (AVT) (Brown et al., 1990; Greenwell et al., 2003; McCormick, 2011). Hickman and Trump examined teleost kidneys’ evolutionary and anatomical features through microscopic observations and investigations of the isolated tubules, analyzing nephron components in euryhaline fish, including the glomerulus, aglomerular fish, proximal tubule, and collecting duct (Hickman and Trump, 1969). The morphological and regulatory characteristics of these kidney structures, such as the glomerulus, proximal tubule, and collecting duct, may vary depending on the acclimation of the fish to salinity or salinity fluctuations. Focusing on previous research conducted on aglomerular fish, which lack glomeruli or the distal tubule, resulting in limited urine dilution, researchers have studied aglomerular toadfish, a species with a lifespan of only three weeks in laboratory freshwater but can survive for months in 10% seawater. Toadfish experience increased metabolic activity when transitioning from seawater to a 10% solution, leading to a double increase in urine flow rate and a decrease in osmotic pressure. The kidneys excrete sodium, chloride, and sulfur at a ratio of 5:1:3, causing a decrease in plasma osmotic pressure (Lahlou et al., 1969; Baustian et al., 1997). To thrive in hypotonic environments, aglomerular toadfish must precisely regulate solute uptake through their branchial and intestinal cells while minimizing solute loss through their kidneys, as they are unable to produce dilute urine (Figures 1C, D) (Baustian et al., 1997).
The concentration of ions was notably low in the (FW) glomerulus of zebra fish and Nile tilapia. The glomerulus has evolved as an expression of the requirement for water excretion in freshwater animals. Dilute urine is regularly released because of the non-permeability of the distal tubule and downstream tissues, such as the collecting tubule/duct and bladder (Takvam et al., 2021a, 2023). The findings indicate that significant ions such as Na+, Mg2+, SO42-, Ca2+, Cl-, K+,and HCO3- undergo reabsorption, whereas the osmotic reabsorption of water, which is present in combination with these ions, is restricted (Bates et al., 2018). Other reviews have provided evidence of this (Evans et al., 2005a; Hwang and Lee, 2007; Takvam et al., 2021a; Huang et al., 2023). Consequently, stenohaline (FW) and euryhaline (when in FW) fish continuously absorb water via their gills and skin while simultaneously losing significant ions through diffusion. To alleviate this phenomenon, the kidneys filter blood within the glomeruli (Loretz et al., 2009). The glomerular filtration rate (GFR) and urine flow rate (UFR) are consistently high, ranging from 4 to 16 ml/kg/h and 1 to 6 ml/kg/h, typically ranging from 20 to 50 mOsm/L (Hickman and Trump, 1969). Consequently, the organism displayed a significant release of diluted urine. This demonstrates the effectiveness of glomerular filtration and epithelial reabsorption of salt in maintaining osmotic balance in freshwater fish (Foskett et al., 1983). Generally, in (SW) conditions, where water conservation is of utmost importance, the renal system has a lower glomerular filtration rate (GFR) than the kidneys in freshwater (FW) environments (Nishimura and Imai, 1982; Ortiz et al., 2002). A low GFR causes the kidneys to filter blood at a lower rate, which preserves water and avoids excessive loss of vital ions. This adaptation facilitates the ability of organisms inhabiting salty habitats to acclimate to their surroundings and uphold internal homeostasis in the face of the difficulties presented by elevated salt concentration in the water. The greater part of sodium (99%), potassium (98%), and chloride (93%) obtained from ingesting seawater are excreted through extrarenal pathways (mainly the gills, but also rectal fluid). The kidney has little or no role in osmoregulation (Evans, 2023).
3.3 Intestine
The intestine serves crucial roles in both digestion and osmoregulatory functions (Grosell, 2011; Rønnestad et al., 2017; de Oliveira et al., 2022) (Figure 1B). Specifically, the intestine plays a vital role in regulating the acid-base balance in marine and euryhaline teleosts, essential for maintaining osmoregulation. Efficient fluid absorption in the intestine is crucial for counteracting water loss in hypertonic environments (Carvalho et al., 2012; Whittamore, 2012). This absorption process is heavily influenced by osmotic gradients, particularly for sodium chloride (NaCl) absorption. The Na+/K+ pump, also known as NKA, plays a crucial role in the absorption of salts and water (Barany et al., 2021). Vertebrates, including fish, possess an impressive ability to absorb water through their digestive systems, effectively reclaiming fluids secreted by the stomach, small intestine, pancreas, and gall bladder. In fish, fluid absorption primarily takes place across the intestinal epithelium, aiding in osmoregulation and the regulation of internal water balance, particularly in hyperosmotic environments. Conversely, in freshwater environments, hydration primarily occurs through the gills (MacKay and Janicki, 1979; Ando, 1980; Ciccotti et al., 1993; Evans et al., 2005a; Takei, 2021a; Ciavoni et al., 2024). The intestine is integral to maintaining osmotic homeostasis in fish, particularly under conditions of salinity stress. This study examines the intricate mechanisms of osmoregulation in teleost fish, with a specific focus on the vital functions of the gill, kidney, and intestine. By analyzing the roles of these essential organs, we aim to advance our understanding of how teleost fish sustain their internal equilibrium of water and salts.
4 Effects of salinity stress on the gut microbiota of teleost fish
The fish intestine is a complex organ crucial for nutrient absorption, immune defense, and osmotic balance (Ciavoni et al., 2024). Along with the gills and kidneys, the gastrointestinal tract is vital for osmoregulation in teleost fish, managing water and electrolyte balance as they transition between FW and SW (Kolbadinezhad et al., 2018; Takvam et al., 2021a). Under hyperosmotic conditions, fish increase their water intake to counteract water loss, causing physiological changes in the intestine, including alterations in stomach acidity, digestive enzymes, and bile salts (Hieu et al., 2022). Environmental factors such as salinity and pollution significantly impact the gastrointestinal microbiota (GM), with salinity changes driving shifts in microbial community structure (Tolas et al., 2025). Microbial species adapted to low-salinity freshwater environments dominate the gut microbiota. Under hypersaline conditions, freshwater microbes with low salinity tolerance are eliminated, whereas marine microbes with higher salinity tolerance thrive (Wang et al., 2018, 2021). The microbial community may also be affected by habitat-generalist bacteria, which possess the ability to thrive across a range of salinities and often become dominant in environments characterized by fluctuations. This phenomenon presents a compelling and significant area of investigation for researchers (Kivistik et al., 2020).Research on anadromous and euryhaline fish has shown that salinity changes shift gut microbiota, impacting microbial diversity (Lai et al., 2020). The properties of water influence the microbiota of tilapia larvae and are correlated with the microbiota present in water (Giatsis et al., 2015). Studies on stenohaline species gut microbiota, such as silver carp (Hypophthalmichthys molitrix), grass carp (Ctenopharyngodon idella), bighead carp (Hypophthalmichthys nobilis), and goldfish (Carassius auratus), which thrive in narrow salinity ranges, show restricted microbial changes under salinity stress. In contrast, euryhaline fish, such as Oncorhynchus mykiss (rainbow trout) and Asian sea bass, showed a strong correlation between gut and water microbiota when compared to the water microbiota of their natural habitats (Dehler et al., 2017; Zeng et al., 2020; Iehata et al., 2021; Lorgen-Ritchie et al., 2021; Morshed et al., 2023), suggesting that waterborne microbes may directly affect the gut microbiota.
Researchers have hypothesized that hypotonic stress leads to changes in the gill and gut microbiota of marine medaka (Oryzias melastigma), aiding salinity acclimation (Lai et al., 2022b) and decreasing bacterial diversity and increasing pathogenic bacteria in the yellowfin seabream (Acanthopagrus latus) (Lin et al., 2020). On the other hand, salinity stress in Nile tilapia (Oreochromis niloticus) increased opportunistic bacteria and decreased beneficial bacteria (Zhang et al., 2016). However, a study on Poecilia mexicana (Mexican mollies) found no such interaction, as key operational taxonomic units (OTUs) in the fish gut were absent in water, indicating that microbial colonization might be influenced by other factors such as host-specific mechanisms (Schmidt et al., 2015; Morshed et al., 2023). While many studies have shown that salinity changes influence the abundance and composition of gut microbiota in teleosts (Sullam et al., 2012; Lorgen-Ritchie et al., 2021; Morales-Rivera et al., 2022), others have reported minimal or no microbial shifts across salinity gradients (Schmidt et al., 2015; Sylvain et al., 2016). Understanding how teleost fish acclimate to extreme salinity is crucial for maintaining physiological homeostasis and ensuring successful osmoregulation. This insight is particularly crucial for enhancing the resilience of aquaculture and facilitating ecological adaptation.
Gut microbiota composition contributes to osmoregulation, affecting host metabolism and ion transport (Minich et al., 2020; Engevik and Engevik, 2021). In freshwater-adapted zebrafish, the microbiota primarily consists of Aeromonas, Pseudomonas, and Vibrio, which are members of the Proteobacteria phylum that contribute to development, immune responses, nutrient absorption, and ion homeostasis (Table 1) (Semova et al., 2012; Flores et al., 2020; Xia et al., 2022a). The gut microbiota of salmon undergoes significant changes during the transition to seawater, characterized by an increase in bacterial load and an increase in Firmicutes, whereas Actinobacteria and Proteobacteria decrease. Research has shown that fluctuations in salinity have a profound effect on the richness, diversity, and taxonomic composition of salmon gut microbiota. Specifically, the study revealed that GSC and FD treatments led to an increase in microbiota diversity in salmon smolts. This suggests a potential correlation between the intestinal microbial community and the overall health of the fish during seawater transfer. These findings have important implications for monitoring the microbiome in smolt fish production, which could ultimately lead to improvements in salmon performance during transfer. By better understanding the relationship between intestinal microbiota and fish health, researchers and fish farmers can optimize the conditions for salmon production (Morales-Rivera et al., 2022). Additionally, core microbiota like Lactobacillus and Clostridium persist across salinity gradients, potentially aiding in osmoregulatory stability (Dehler et al., 2017; Bowman, 2024). Changes in microbial composition play a crucial role in enhancing ion transport mechanisms across the intestinal epithelia, highlighting their significance in maintaining homeostasis during salinity stress. These findings emphasize the complex interplay between the expression of ion transporters, the gut microbiota, and osmoregulatory functions in teleost fish.
Table 1
| Intestinal Bacteria Function | The biological function | Mechanism at the molecular level | References |
|---|---|---|---|
| Aeromonas, Pseudomonas, Firmicutes, Actinobacteria, Proteobacteria, Clostridia and lactobacillus | Osmoregulation | Aeromonas and Pseudomonas in the gut microbiota of freshwater zebrafish may be associated with osmoregulation, assisting the fish in maintaining appropriate ion balance in their bodily fluids may also be associated with osmoregulation affecting the survival and growth of fish in the Atlantic salmon | (Flores et al., 2020; Xia et al., 2022a; Tolas et al., 2025) |
| Pseudomonas fluorescens, Aeromonas veroniibiovar sobria | Development | Using specific host recognition mechanisms to promote intestinal epithelial maturation | (Bates et al., 2007; Butt and Volkoff, 2019) |
|
Vibrio
Monospora and Enterobacteriaceae, Firmicutes, Actinobacteria and Proteobacteria |
In larval zebrafish, excessive Vibrio species proliferation prevents the establishment of adaptive immunity and affects the survival and growth of Atlantic salmon fish under salinity fluctuation | (Brugman et al., 2014; Tolas et al., 2025) | |
| Vibrio cholerae: GFP ZWU0020, Aeromonas veronii: dTomato HM21 | Essential for typical neurobehavioral development throughout the early life stages of zebrafish | (Cheesman et al., 2011; Phelps et al., 2017) | |
| Chryseobacterium sp. ZOR0023, Pseudomonas sp. ZWU0006, Exiguobacterium sp. ZWU0009, | Metabolic Activity | Enhancing fatty acid absorption and lipid droplet deposition in intestinal epithelium and hepatic tissue | (Semova et al., 2012) |
| Pseudomonas aeruginosa strain PAK | Immunity | The regulation of the dynamic temporal and spatial activation of NF-кB transcription and subsequent up-regulation of target genes within the gastrointestinal tract. | (Kanther et al., 2011) |
| Aeromonas veronii biovar sobria, Pseudomonas fluorescens, Streptococcus, Staphylococcus, and Vibrio | Regulating host immunity via the control of Myd88 and TNF receptor | (Cheesman et al., 2011; Brugman et al., 2014) |
The biological role of zebrafish and salmon fish gut microbiota.
As climate change continues to alter salinity gradients, it is essential to further investigate the adaptive mechanisms in these species using genomics, metabolomics, and microbiome profiling. Recent advancements in deep sequencing technology have revolutionized our understanding of the fish microbiota, enabling the study of microbial communities without the need for culturing. Overall, the relationship between microbial composition and ion transport mechanisms in teleosts is a critical area of research that can provide valuable insights into how these species adapt to changing environmental conditions (Tian et al., 2020). Quantitative real-time PCR (qPCR), denaturing gradient gel electrophoresis (DGGE), fluorescence in situ hybridization (FISH), temporal temperature gradient electrophoresis (TTGE), marker gene amplification and sequencing (e.g. ITS for fungi, 16S rRNA for bacteria and archaea), and metagenomics have improved our knowledge of fish gut microbiota composition, structure, and diversity (Ou et al., 2021). A metagenomic study of Nile tilapia exposed to acute high-salinity stress revealed significant changes in microbial communities, notably a shift from Actinobacteria to Proteobacteria, while fungal and phage communities remained stable. Functionally, intestinal bacteria show reduced activity in digestion and the nervous system, with increased energy metabolism. Key microbial genes, such as glutathione S-transferase, myo-inositol-1-monophosphatase, and glycine betaine/proline transporters, as well as specific carbohydrate-active enzyme families (GT4, GT2), were upregulated, whereas others (GH15, GH23) were downregulated (Gong et al., 2024). Studies using gnotobiotic fish models suggest that microbiota-derived short-chain fatty acids (SCFAs) influence ion transporter gene expression, affecting epithelial permeability and ion absorption (Lee et al., 2021; Xia et al., 2022a). These findings highlight the complex interactions between the host and the microbiota in osmoregulation. Future research using gene knockout models and microbial colonization assays could provide deeper insights into the role of the microbiota in teleost salinity adaptation (Quan et al., 2021).
These advancements highlight the importance of the gut microbiota in the osmoregulation of euryhaline fish, where microbial changes are linked to osmoregulatory and energy metabolism alterations (Hu et al., 2017; Tseng et al., 2022a). However, much remains to be learned about microbial interactions and physiological changes that occur during salinity acclimation. The interaction between salinity stress and the gut microbiota in teleost fish is intricate and varies by species. Stenohaline species frequently encounter microbial instability when exposed to changes in salinity because their microbiota may experience significant fluctuations or a loss of stability. Conversely, euryhaline fish demonstrate more gradual and adaptive shifts in their microbiota, with alterations in the microbial community composition that facilitate osmoregulation. Gut microbiota responses to salinity stress highlight the balance between the host physiology and environmental factors. Future studies should use multi-omics approaches to clarify the role of gut microbes in salinity adaptation. Comparative research across teleost species will reveal broader microbial patterns under stress, which is crucial for optimizing aquaculture, fish health, and microbiota stability under changing salinity.
5 Influence of salinity stress on metabolomics in teleost fish
Metabolomics, which involves studying all the low-molecular-weight metabolites present in a cell or organism, is known as metabolomics (Nicholson et al., 1999; Cuykx et al., 2018). By integrating metabolomics and transcriptomics, valuable insights can be gained into the effects of various stressors on fish, such as stickleback (Figure 2) (Divino, 2016; Abid et al., 2018; Meador et al., 2020; Iehata et al., 2021; Santos et al., 2023). Understanding the impact of environmental stress on fish can be achieved through comprehensive metabolomic analyses. Investigating the molecular mechanisms controlling teleost fish exposed to high salinity can greatly benefit metabolic analysis (Qin et al., 2022). The gills of euryhaline fish, which are highly oxidative tissues, regulate a variety of ion transporters and enzymes in response to changing salinities in an efficient and timely manner, requiring significant energy (Liu et al., 2023). Since fish ions and osmoregulation processes depend on energy availability for proper operation, this suggests that most metabolic and energy-related genes are expressed more when salinity is higher (Djiba et al., 2021). Hence, the study of Li et al. demonstrated how varying salinity concentrations affect gene expression and energy metabolism. Glycolysis and gluconeogenesis are crucial for generating energy and regulating glucose levels (Li et al., 2023). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a glycolytic enzyme that plays a crucial role in carbon and energy metabolisms. Notably, studies have indicated that a decrease in GAPDH protein levels may facilitate the preservation of other glycolytic enzymes and redirect glucose flux through alternative metabolic pathways. This redirection potentially enhances glucose utilization under specific physiological conditions (Baumgarner et al., 2012; Lazarev et al., 2020). Upon the transfer of fish to seawater, the increased energy requirements in the gills may lead to enhanced reliance on anaerobic metabolism, specifically the conversion of pyruvate to lactate. Lactate dehydrogenase A (LDHA) plays a pivotal role in facilitating this metabolic pathway (Vijayan et al., 1996). Advancements in metabolomics have opened new opportunities for understanding the complex physiological responses of teleost fish to changes in salinity (Ganguly et al., 2020). This emerging discipline focuses on the comprehensive characterization of metabolites, offering detailed insights into the biochemical changes and regulatory processes that enable teleost fish to thrive in variable environments (Alfaro and Young, 2018; Goode et al., 2020). Enhancing genetic traits or regulating nutrition through osmoregulation can significantly improve the ability of fish to adapt to different salinity levels. Metabolomic research on salinity adaptation in teleost fish is needed; however only a limited number of studies have been conducted on various species. Most fish species do not possess a strong ability to adjust to changes in salinity. Exposure to high salinity in their habitat has negative consequences such as stunted growth, susceptibility to diseases, and even mortality (Yamaguchi et al., 2018; Kujawa and Piech, 2022). This has impeded progress in aquaculture and poses a threat to fish biodiversity conservation. This offers a notable opportunity for scientists to further investigate metabolic pathways and identify crucial biomarkers associated with osmoregulation (Liu et al., 2023). Using metabolomics, researchers have been able to identify metabolites from the intestine, hepatopancreas, and muscle of Gymnocypris przewalskii using LC-MS/MS, revealing 5,745 differentially expressed genes. Processes such as gluconeogenesis and long-chain fatty acid metabolism play crucial roles in maintaining glucose homeostasis and providing energy in cold-stressed fishes. Therefore, through the application of metabolomics, scientists can gain a comprehensive understanding of the molecular mechanisms by which teleost fish regulate ion balance, generate energy, and respond to stress (Liu et al., 2023).
Figure 2
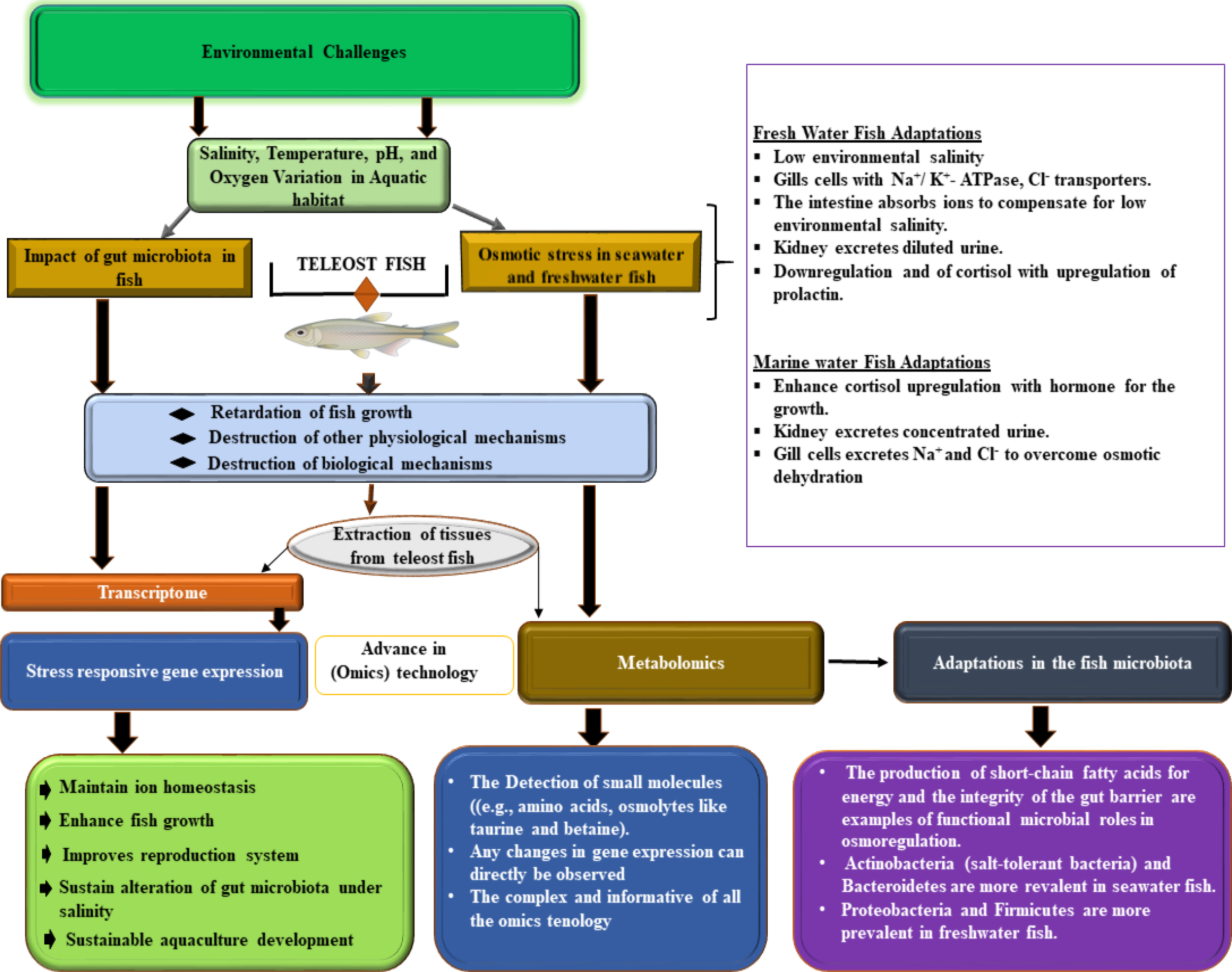
Displays salinity adaptation in teleost fish, covering environmental stressors, physiological osmoregulatory responses, and omics insights. The upper panel shows aquatic ecosystems and stressors, the central panel illustrates cellular osmoregulatory mechanisms in key organs, and the lower panel consolidates omics data, highlighting transcriptomic, metabolomic, and microbiome changes under different salinity conditions. The right panel summarizes the physiological adaptations, offering insights into the salinity management of teleost fish and its implications for aquaculture and conservation.
Acquiring knowledge about the metabolites linked to optimal salinity adaptation is crucial for enhancing resilience and growth of fish in aquaculture settings (Figure 2). By identifying these key metabolites, specialized feeds and breeding programs can be developed to cultivate more resilient fish breeds (Raposo de Magalhães et al., 2022). In a recent study, researchers examined the stress proteome and metabolome of Sparus aurata, a fish species, in response to aquaculture challenges, including factors such as overcrowding, repetitive net handling, air exposure, and hypoxia. Their analysis revealed disrupted pathways in the fish liver, revealing a complex network of regulatory elements that impact cellular stress pathways. This study provides valuable insights into fish welfare (Raposo de Magalhães et al., 2022). Recent research has enhanced our understanding of teleost responses to saline-alkaline stress. Ding et al. examined the physiological and metabolic changes in Crucian carp (Carassius auratus) subjected to varying NaHCO3 concentrations, identifying significant renal impairments such as glomerular atrophy, tubular degranulation, and renal cell proliferation (Ding et al., 2023). Furthermore, the study underscored disruptions in antioxidant systems, energy metabolism, and protein catabolism, indicating that C. auratus exhibits notable sensitivity to saline-alkali exposure. Conversely, comprehensive analyses of the annexin (GpANN) family in naked carp (Gymnocypris przewalskii), a species well-adapted to high-salinity and alkaline environments, revealed critical molecular adaptations. GpANNs displayed diverse functional motifs and tissue-specific expression profiles, with GpANN2 and GpANN3 demonstrating significant transcriptional and translational responses in kidney tissues under stress (Linlin et al., 2024). These findings suggest that, while C. auratus experiences stress-induced damage, G. przewalskii possesses evolved molecular mechanisms that enhance its resilience, offering a comparative model for understanding salinity adaptation in freshwater teleosts.
Metabolomics plays a vital role in promptly identifying stress and diseases in fish, allowing timely interventions to enhance their welfare (Low et al., 2017). By integrating metabolomic data with other omics approaches, we can gain a comprehensive understanding of adaptation mechanisms in fish (Jendoubi, 2021). Given the growing global demand for sustainable fish farming practices, it is essential to prioritize metabolomic research on teleosts fish is essential. Studying their adaptive abilities in response to salinity variations will not only deepen our knowledge, but also inspire innovative solutions for sustainable aquaculture techniques. This will ultimately help the aquaculture business grow and become more resilient in the face of environmental challenges.
6 Influence of salinity stress on transcriptomics in teleost fish
Living species require adequate sensations, reactions, and adaptation to varying salinity conditions for development, reproduction, and survival. In recent decades, aquatic ecosystems have been facing elevated salinity stress due to road de-icing salts, rising sea levels, saltwater intrusion in coastal areas, and increased global temperature changes, affecting their biodiversity directly and indirectly (Cunillera-Montcusí et al., 2022). Salinization has been shown to affect ecological processes, trophic networks, functional trait variety, and community composition in several habitats (Cañedo-Argüelles et al., 2013; Hintz and Relyea, 2019; Tweedley et al., 2019; Röthig et al., 2023). The energy expenses associated with osmoregulation in fish, particularly the transfer of ions across cell membranes against a concentration gradient, can range from a few (‰) to up to 30 (‰) in salinities that differ from their body fluids (Figure 1) (Bœuf and Payan, 2001; Urbina and Glover, 2015). While teleost fish can temporarily adjust to changes in salinity circumstances (Jiang et al., 2019), it has been observed that excessive salinity levels hinder their growth (Zhao et al., 2022) and disrupt their regular metabolism (Jiang et al., 2019). Several physiological studies have been conducted on the osmoregulatory systems involved in the adaptation of fish to different salinity levels, such as freshwater or seawater, which can range from 0 to 38 ‰ (Evans et al., 2005a; Hwang and Lee, 2007; Lee et al., 2022a). The primary focus of their description was on osmoregulatory capabilities within this range of salinity. They provide information on various changes, such as alterations in electrolyte concentration in bodily fluids, remodeling of the epithelium, standard metabolic rates, hormonal regulation, expression patterns of specific genes involved in ion transport and osmoregulation, such as Na+/K+-ATPase [NKA], Na+/K+/2Cl− cotransporter [NKCC], cystic fibrosis transmembrane conductance regulator [CFTR], aquaporins, and the use of high-throughput methods, such as RNA sequencing, to study osmoregulatory tissues (Figure 2). Studies that provide results for more than 40‰ salinity in fish are still uncommon (Lam et al., 2014; Su et al., 2022). The function of ion-transporters has been extensively studied in fish exposed to seawater challenges. The transfer of salmon, specifically Onchorhynchus kisutch or Salmo salar, and brown trout, known as Salmo trutta, in seawater led to a gradual acclimation of Na+/K+-ATPase and Na+/K+/2Cl− co-transporter expression in the cells that make up the gills (Tipsmark et al., 2002). The mRNA levels of certain subunits of Na+/K+-ATPase increased in response to changes in ambient salinity in the tissues of the tilapia Oreochromis mossambicus and rainbow trout Onchorhynchus mykiss. However, the mRNA levels of the other subunits decreased (Richards et al., 2003). In addition, studies have has investigated the responses of fish to the challenges of freshwater environments by assessing and analyzing the expression of prolactin, a hormone that plays a role in osmoregulation and adaptation to freshwater conditions. Prolactin is a key factor in the process of osmoregulation in freshwater fishes. It controls the movement of water and salt through the gills and kidneys by influencing changes in membrane permeability and creation of chloride cells (Forsyth and Wallis, 2002).
Previous studies have attempted to identify molecular processes, such as transcriptome analysis, to provide scientists with a useful set of data for developing specific interventions and management strategies that promote the survival and adaptation of fish populations in environments with different salinity levels (Table 2). Transcriptomics, which involves studying the expression of all genes at a specific time, has been utilized for more than ten years to understand the connections between the environment, genotype, and phenotype in natural populations (Li et al., 2024a). Transcriptomic profiling in animals lacking whole-genome sequencing data is a powerful approach for investigating genomes and discovering functional genes (Escobar-Sierra et al., 2024). Transcriptomic analysis is a widely used technique to gain insights into the functional genomic components and reveal molecular mechanisms in cells and tissues of teleost fish (Figure 2). The transcriptome, consisting of both protein-coding messenger RNA (mRNA) and non-coding RNA (ncRNA), demonstrates heterogeneity in its response to developmental stage, physiological conditions, and external environment (Sun et al., 2020; Yi et al., 2021; Harshini et al., 2022; Ru et al., 2023). High-throughput sequencing technologies offer an opportunity to study changes in gene expression in organisms exposed to various environmental conditions. Transcriptome sequencing allows for comprehensive analysis of several physiological systems, such as metabolism, proteostasis, and osmoregulation (Vij et al., 2020; Escobar-Sierra et al., 2024; Li et al., 2024a).
Table 2
| Teleosts Species | Salinity environments | Discoveries from Transcriptome Analysis | References |
|---|---|---|---|
|
Fundulus heteroclitus
(killifish) |
Freshwater to Saltwater | Ion transporters and osmosensory signaling genes are differentially expressed in osmoregulation. | (Scott et al., 2004; Whitehead et al., 2012) |
|
Lates calcarifer
(Asian seabass) |
Freshwater to Seawater | An increase in the expression of genes associated with energy production, metabolism, osmoregulation, and the response to oxidative stress was observed. | (Xia et al., 2013; Vij et al., 2020) |
|
Oreochromis niloticus
(Nile tilapia) |
Freshwater to hypersaline (above 35ppt) | The genes responsible for ion transport, osmotic regulation, and stress response are being upregulated. | (Ronkin et al., 2015; Liu et al., 2018; Zhao et al., 2020) |
| Oncorhynchus mykiss (Rainbow trout) | Freshwater to Seawater transition | Alterations in the transcriptome underscore the importance of ion channels and transporters in osmoregulatory adaptation. | (Leguen et al., 2010; Xiong et al., 2020; Liu et al., 2024; Pino-Martinez et al., 2024; Zuloaga et al., 2024) |
| Chinese Sea Bass (Lateolabrax maculatus) | freshwater (FW) to seawater (SW) but moderately adapt to highly alkaline water (AW) | Transcriptomic analysis reveals the unique cellular response in the gills when exposed to fluctuations in salinity and alkalinity. | (Zhang et al., 2017; Li et al., 2022a; Zhu et al., 2023a) |
| Oreochromis mossambicus (Mozambique tilapia) | Variable salinity (freshwater to seawater) | Various genes related to osmoregulation, stress response, prolactin receptors, and ion transport exhibit differential expression. | (Seale et al., 2014; Ronkin et al., 2015; Su et al., 2020; Inokuchi et al., 2021) |
| Acanthopagrus schlegelii (Black Sea bream) | Hypo- and hyper-saline conditions | Significant changes have been observed in the expression of genes related to metabolism, cellular stress, and ion transport. | (Chang et al., 2007; Li et al., 2022b; Nagarajan et al., 2023) |
|
Cyprinodon variegatus
(Sheepshead minnow) |
freshwater to seawater) | Research has identified genes associated with immune response, stress response, and osmotic balance that exhibit changes in their transcriptomes. | (Simning et al., 2019) |
| Dicentrarchus labrax (European sea bass) | Euryhaline species with a Salinity gradient tolerating from FW up to 70 ppt | Differences were observed in the expression of genes associated with immune response, energy metabolism, and ion transport. These variances were crucial for maintaining distinct ecotypes during the early stages of reproductive isolation. | (Boutet et al., 2006; Masroor et al., 2019) |
| Danio rerio (Zebrafish) | Freshwater fish | The stress response triggers alterations in gene expression, particularly in genes that contribute to developmental abnormalities in zebrafish. Furthermore, the research also pinpointed changes in gene expression associated with the regulation of post-embryonic development in response to salinity stress. | (Breves et al., 2013; Thompson et al., 2022; Seli et al., 2024) |
| Gasterosteus aculeatus (Three-spined stickleback) | Has a wide salinity tolerance from seawater to freshwater environments | Genes associated with energy metabolism, oxidative stress, and ion transport exhibit varying expression patterns. | (Jones et al., 2012; Taugbøl et al., 2022) |
Provides an overview of studies that have conducted transcriptome analyses on teleost fish under salinity stress conditions.
In the past years, there has been notable progress in understanding the transcript expression profile during salinity adaptation of various euryhaline teleost species using RNA-Seq, such as Nile tilapia (Oreochromis niloticus), Mozambique tilapia (Oreochromis mossambicus) (Table 2) (Ronkin et al., 2015), medaka (Oryzias melastigma) (Lai et al., 2015), striped catfish (Pangasianodon hypophthalmus) (Thanh et al., 2015; Pasquier et al., 2016), and Asian seabass (Lates carifer) (Xia et al., 2013). Multiple studies have demonstrated alterations in ion transporters, channels, and stress-related proteins, elucidating the physiological mechanisms by which fish can maintain a consistent internal environment under varying salinities. Transcriptomic analyses have recognized many genes involved in ion transport, water channels (aquaporins) (Riera Romo et al., 2016), and stress response pathways (Table 2) (Cutler et al., 2007). Tenualosa ilisha (Hilsa shad), an anadromous fish, was examined in fresh, brackish, and marine water. The analysis revealed 3277 genes that were expressed differently (DEGs), with 232 being shared between marine and freshwater environments. Out of the 54 KEGG Pathways, the focal adhesion, adherens junction, tight junction, and PI3K-Akt signaling pathways were identified as the most significant. Various settings exhibited distinct expression patterns for 24 osmoregulatory genes that were differentially expressed in diverse habitats (Mohindra et al., 2023).
The gill transcriptome of Odontesthes bonariensis, a stenohaline freshwater species, demonstrates distinct gene expression patterns when exposed to high salinity, in contrast to Odontesthes argentinensis, a closely related euryhaline species capable of tolerating a broad range of salinities. This disparity underscores the unique molecular mechanisms that underpin salinity adaptation in species with differing osmoregulatory capacities (Hughes et al., 2017). An analysis was conducted on the transcriptomic data of the livers of juvenile Scatophagus argus following a sudden shift in salinity. The results revealed that this alteration led to the generation of 474 differentially expressed genes. The pathways associated with immune defense showed enrichment, including ‘Antigen processing and presentation’ and ‘Phagosome’, suggesting that S. argus may strengthen the immunological defense system. Low-salinity-induced oxidative stress led to the upregulation of several genes that encode antioxidant enzymes, such as HAO, Trx, and PHGPx. Downregulation of Cul3 could potentially enhance the activation of Nrf2, leading to the upregulation of antioxidant enzyme genes. The results suggested that S. argus could be used to confirm the effectiveness of the transcriptome as a molecular mechanism for resisting low salt stress (Sun et al., 2022). Furthermore, Lai et al. study investigates genetic responses to hypotonic conditions and gill microbiota dynamics in marine medaka following freshwater transfer. Using transcriptome and 16S rRNA sequencing, the study reveals 1,034 genes with altered expression and documents a microbial shift from Vibrio to Pseudomonas and Cetobacterium. The identification of overlapping pathways associated with glycosaminoglycan and chitin suggests significant host-microbiota interaction in gill adaptation, offering novel insights into osmoregulation during osmotic stress (Lai et al., 2022b).
7 Strengthening cross-omics correlation study
Variations in salinity levels stress aquatic organisms, disrupting homeostasis, metabolism, reproduction, and immune function (Leprêtre et al., 2025). Investigating teleost fish adaptation using high-throughput cross-optimal correlation analysis, including gut microbiota composition, metabolomics, and transcriptomics, elucidated salinity adaptation mechanisms (Figure 2). A challenge in gut microbiota studies is the contamination of metagenomic and metatranscriptomic data by host material and the overlap between host and microbial metabolites (Ou et al., 2021). Despite this, integrative omics approaches have advanced our understanding of host-microbiota interactions under stress. This review compiled research on microbial taxa, metabolic pathways, and gene expression interactions in response to salinity stress, offering insights into the adaptive strategies of teleost fish.
7.1 Gut microbiota and metabolomic interactions
Recent advancements in high-throughput chemical fingerprinting methods, such as metabolomics based on nuclear magnetic resonance (NMR), have provided scientists with tools to delve deeper into the intricate interactions between the gut microbiota and host metabolism (van Ravenzwaay et al., 2007; Wei et al., 2018). Gut microbiota is a dynamic entity that evolves alongside the host and is influenced by many factors, including genetics, diet, and environment (Nicholson et al., 2012). Environmental fluctuations, particularly salinity variations, can profoundly affect the gut microbiome, which contains trillions of microorganisms and is considered to play a critical role in host digestion and metabolic homeostasis (Budd et al., 2020; Lai et al., 2022a; Liu et al., 2022). For example, a study on the wild yellowfin goby (Acanthogobius flavimanus) revealed seasonal and latitudinal changes in the metabolome, ionome, and microbiome, with salinity acting as a key regulator of gut microbial communities and homeostasis (Wei et al., 2018). Similarly, research on wild black Amur bream (Megalobrama terminalis) in China’s drainage areas found higher alpha diversity in mainland populations than in those on Hainan Island. Geographic isolation and seasonal variations were found to have a significant effect on the gut microbiome, with distinct regulatory patterns observed in each population. This suggests that environmental factors and genotypes play a crucial role in shaping microbiome diversity in wild M. terminalis (Liu et al., 2022). In juvenile M. salmoides, exposure to 5‰ salinity increased superoxide dismutase activity, while 15‰ salinity elevated levels of aspartate aminotransferase, ALT, acid phosphatase, AKP activity, and total antioxidant capacity. Interestingly, the control group exhibited the highest levels of catalase and glutathione peroxidase activity, suggesting that a salinity of 5‰ enhances immune function. Analysis of paraffin sections revealed a decrease in villus length and an increase in epithelial cell expansion with increasing salinity levels. Furthermore, microbiota analysis showed shifts in dominant taxa, with an increase in Bacillus abundance at a salinity of 10‰. Salinity stress was found to affect lipid and amino acid metabolism, with the 15‰ salinity group showing enriched replication, recombination, and repair processes, as well as salinity stress response mechanisms (Sun et al., 2023). Additionally, different salinity levels were observed to affect the gut microbiota of Atlantic salmon, leading to alterations in microbial communities and an increase in specific taxa, such as Vibrio, Pseudomonas, Acinetobacter, Corynebacterium, Alteromonas, Flavobacterium, and Micrococcus (Ou et al., 2021). Furthermore, metabolomic profiling has demonstrated changes in key metabolites, particularly osmolytes such as taurine and betaine, as well as lipid derivatives (Tian et al., 2022; Li et al., 2024b). Metabolites are linked to osmoregulation and stress adaptation because taurine and betaine serve as compatible solutes that contribute to the stabilization of intracellular osmotic pressure, protection of cellular structures, and maintenance of enzyme activity under hyperosmotic conditions. Concurrently, alterations in lipid metabolism indicate the necessity for membrane remodeling and energy reallocation during salinity stress, both of which are crucial for sustaining homeostasis and physiological functions in dynamic environments. Restructuring of gut microbiota has also been observed in other teleost species, including golden pompano (Trachinotus ovatus) and wild Arctic charr (Salvelinus alpinus), where salinity-induced changes affect amino acid, lipid, and carbohydrate metabolism, influencing host adaptation (Hamilton et al., 2019; Liu et al., 2019). This suggests that the gut microbiota plays a crucial role in maintaining the metabolic equilibrium of the host organism. Overall, these findings highlight the importance of the gut microbiota in maintaining metabolic equilibrium in the host organisms.
7.2 Metabolomics and transcriptomic integration
Through metabolomic and transcriptomic analyses, researchers have linked differentially expressed genes (DEGs) related to lipid metabolism, energy balance, and osmoregulation to metabolic alterations in response to salinity stress (Figure 2) (Kanika et al., 2025). A study of genetically improved farmed tilapia (GIFT) Oreochromis niloticus, identified osmoregulatory genes, such as aquaporin 3, Na+/K+-ATPase, Potassium channel subfamily K member, chloride channel 2, and solute carrier (SLC) transporters, showing differential expression across salinity conditions. The coordinated response between gene regulation and metabolic adaptation is demonstrated by the alignment of over-represented pathways, including UDP-N-acetyl-glucosamine synthesis, N-glycan biosynthesis, and lipid metabolism, all of which play a role in supporting osmoregulatory function. This coordinated response is further underscored by the metabolomic changes observed in osmolytes, such as 12-hydroxyeicosatetraenoic acid, choline, and adenine. These shifts in pathways and metabolites suggest a close connection between the activation of osmoregulatory genes and the metabolic adjustments required to maintain osmotic balance under salinity stress (Qin et al., 2022). This intricate relationship highlights the complex mechanisms at play in the cellular response to environmental challenges, shedding light on the adaptive strategies employed by organisms to survive and thrive in changing conditions. Transcriptomic analyses across various fish species have elucidated critical molecular adaptations to salinity stress, demonstrating how environmental changes influence gene expression and metabolic processes. In Tenualosa ilisha, key pathways related to cell adhesion, ion transport, and osmoregulation have been identified with KEGG pathways such as focal adhesion, tight junctions, and PI3K-Akt signaling being significantly enriched. Notably, the habitat-specific expression patterns of genes from the slc16 and slc2 families, as well as claudin genes (cldn11, cldn10), suggest that these genes play pivotal roles in acclimating to varying salinity conditions. Furthermore, protein-protein interaction (PPI) network analysis revealed that fn1 interacts with genes involved in muscle structure development, underscoring the cellular and structural adjustments necessary for osmoregulation under stress (Mohindra et al., 2023). Similarly, in Lates calcarifer, the differential expression of osmoregulatory genes involved in tissue remodeling, circadian rhythms, and growth regulation indicates that salinity fluctuations not only drive acclimation to freshwater and seawater but also influence physiological behaviors such as migration (Vij et al., 2020). Additionally, in Scatophagus argus, transcriptome profiling of brain tissue under high- and low-salinity conditions revealed metabolic reprogramming, with genes (lipa, sqle, acc, fas, bhmt, mpst, dnmt3a, mtr, hao2, LOC111225351, hmgcs1, hmgcr, and soat1) related to lipid metabolism, steroid biosynthesis, and methylation pathways being significantly altered. These findings suggest that salinity stress affects both metabolic reprogramming and gene expression in the brain, particularly in processes that are vital for osmoregulation and stress adaptation (Lin et al., 2024). Collectively, these studies highlight the complex molecular responses to salinity stress, revealing how different fish species utilize distinct, yet interconnected, gene networks and metabolic pathways to adapt to their respective saline environments.
Alternative splicing (AS) has emerged as a critical post-transcriptional regulatory mechanism for salinity acclimation in teleosts. RNA-Seq analyses of Mozambique tilapia (Oreochromis mossambicus) and rainbow trout revealed significant results. In Mozambique tilapia, the process of differential alternative splicing (DAS) of genes associated with spliceosome assembly, RNA binding, and post-transcriptional processing reveals a highly regulated mechanism of gene expression in response to high-salinity conditions. This intricate regulation suggests that fish have evolved precise mechanisms to acclimate to environmental changes. The enrichment of DAS genes in mitochondrial energy metabolism, ribosomal protein synthesis, and cytoplasmic signal transduction indicates that fish may require rapid and energy-efficient cellular responses to cope with salinity stress, particularly in osmoregulatory tissues such as the gills (Huang et al., 2025). These acclimations likely help fish maintain ionic and osmotic balance when faced with fluctuations in salinity levels. Similarly, in rainbow trout, changes in gene expression related to taurine and glutamine metabolism have been observed, with shifts in metabolite concentrations confirmed using LC-MS/MS analysis. This highlights the importance of organic osmolytes in fish osmoregulatory processes, allowing them to stabilize cellular osmotic pressure without compromising essential cellular functions (Tian et al., 2022). Overall, these findings underscore the critical role of post-transcriptional regulation and compatible solute metabolism in the response of teleosts to salinity stress. By understanding these molecular mechanisms, we can gain insight into how these fish maintain their physiological resilience in challenging environments. Research on Nile tilapia and tongue sole has highlighted the role of amino acid accumulation in salinity adaptation (Jiang et al., 2019; Su et al., 2023b; Wang et al., 2024). KEGG pathway enrichment identified glycerophospholipid metabolism, bile acid biosynthesis, and amino acid metabolism as crucial for salinity adaptation, providing insights into fish osmoregulation. Integrating gut microbiota profiling, metabolomics, and transcriptomics enhances our understanding of salinity stress acclimation in teleosts. Salinity-induced gut microbial changes contribute to metabolic reprogramming, affecting the energy balance, osmoregulation, and immune responses. Transcriptomic analyses have highlighted differentially expressed genes, alternative splicing, and regulatory pathways involved in physiological adaptations. Future research should validate omics predictions, characterize candidate genes, and elucidate host-microbiota interactions to improve the prediction and mitigation of environmental stress effects on aquatic organisms, thereby supporting sustainable fisheries and aquaculture.
We acknowledge the limitations of current research on salinity acclimation in teleost fish. The focus on short-term salinity stress studies has constrained our understanding of long-term acclimation and progressive physiological and microbiota changes. The causal relationship between gut microbiota alterations and osmoregulatory mechanisms remains ambiguous, requiring further functional validation to determine whether these changes are adaptive responses or active regulators. Multi-omics data integration presents technical challenges that affect functional interpretation. Many studies generalize findings across teleost species, overlooking the species-specific osmoregulatory strategies. Although transcriptomics and metabolomics provide valuable insights, experimental validation through in vivo functional studies is limited. Addressing these gaps will enhance our mechanistic understanding and broaden the applicability of our findings in aquaculture and ecological conservation.
8 Conclusion
Teleost fish are known for their ability to thrive in environments with varying salinity levels, requiring them to make precise physiological and molecular adjustments to maintain homeostasis. This review delves into the key osmoregulatory mechanisms found in the gills, kidneys, and intestinal tissues of these fish, highlighting the importance of transcriptomics, metabolomics, and gut microbiota in their ability to acclimate to changes in salinity. Transcriptomic analyses have revealed changes in gene expression related to ion transport, energy metabolism, and signaling pathways, providing insights into how teleost fish respond to salinity stress. Metabolomic studies have identified osmolytes and metabolites that play roles in maintaining cellular stability under these conditions. Furthermore, the gut microbiota of these fish affects their metabolic function and ability to acclimate to salinity fluctuations. By utilizing a combination of these multi-omics approaches, researchers have gained a comprehensive understanding of how teleost fish respond to changes in salinity levels. While these findings have the potential to improve aquaculture practices by enhancing stress resilience and health management, they also contribute to our broader understanding of fish physiology and environmental acclimatization.
Future perspectives
Future research should integrate transcriptomics, gut microbiota, and metabolomics to gain insights into the acclimatization of teleost fish to changes in salinity. Long-term studies will reveal the lasting effects and adaptations to fluctuating salinities. Understanding these mechanisms can improve fisheries, enhance fish health, and optimize species-specific management strategies for aquaculture.
Statements
Author contributions
EMM: Conceptualization, Data curation, Methodology, Writing – original draft. LI: Methodology, Formal analysis, Writing – review & editing. MAY: Investigation, Writing – review & editing. AZ: Investigation, Writing – review & editing. JZ: Investigation, Writing – review & editing. MJ: Investigation, Writing – review & editing. FD: Software, Writing – review & editing. LW: Methodology, Writing – review & editing. HjZ: Validation, Writing – review & editing. BT: Investigation, Writing – review & editing. HZ: Investigation, Writing – review & editing. KA: Visualization, Project administration, Writing – review & editing. JH: Supervision, Writing – review & editing. BW: Resources, Writing – review & editing. ZW: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Guangdong Province Ordinary Colleges and Universities Special Innovative Project (2023KTSCX044); Research on breeding technology of candidate species for Guangdong modern marine ranching (2024-MRB-00-001); Guangdong Province Ordinary Colleges and Universities Key Field Special Project (Science and Technology Services for Rural Revitalization) (2023ZDZX4011); Zhuhai Social Development Field Science and Technology Plan (Key Fields of the “Hundred Counties, Thousand Towns, Ten Thousand Villages Project”) (2320004001603); Guangdong Province Ordinary Colleges and Universities Innovation Team Projects (2021KCXTD026; 2022KCXTD013).
Acknowledgments
We would like to extend our sincere gratitude to our colleagues in the lab for their invaluable assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abid F. Zahid M. A. Abedin Z. U. Nizami S. B. Abid M. J. Kazmi S. Z. H. et al . (2018). “Omics approaches in marine biotechnology: The treasure of ocean for human betterments,” in Omics technologies and bio-engineering. (Academic Press), pp. 47–61. doi: 10.1016/B978-0-12-804659-3.00003-8
2
Agarwal D. Shanmugam S. A. Kathirvelpandian A. Eswaran S. Rather M. A. Rakkannan G. (2024). Unraveling the impact of climate change on fish physiology: A focus on temperature and salinity dynamics. J. Appl. Ichthyol.2024 (1), 5782274. doi: 10.1155/2024/5782274
3
Alfaro A. C. Young T. (2018). Showcasing metabolomic applications in aquaculture: a review. Rev. Aquac.10 (1), 135–152. doi: 10.1111/raq.12152
4
Alsafy M. A. M. El-Gendy S. A. A. El-Bakary N. E. R. Kamal B. M. Derbalah A. Roshdy K. (2023). Morphological comparison of the detailed structure of gill rakers from three different feeding habits of marine fish species. Zoomorphology142, 87–97. doi: 10.1007/s00435-022-00574-4
5
Altinok I. Grizzle J. M. (2003). Effects of low salinities on oxygen consumption of selected euryhaline and stenohaline freshwater fish. J. World Aquac. Soc.34 (1), 113–117. doi: 10.1111/j.1749-7345.2003.tb00046.x
6
Amasheh S. Fromm M. Günzel D. (2011). Claudins of intestine and nephron - a correlation of molecular tight junction structure and barrier function. Acta Physiol.201 (1), 133–140. doi: 10.1111/j.1748-1716.2010.02148.x
7
Ando M. (1980). Chloride-dependent sodium and water transport in the seawater eel intestine. J. Comp. Physiol. B138, 87–91. doi: 10.1007/BF00688739
8
Arjona F. J. Vargas-Chacoff L. Ruiz-Jarabo I. Gonçalves O. Páscoa I. Martín del Río M. P. et al . (2009). Tertiary stress responses in Senegalese sole (Solea Senegalensis Kaup 1858) to osmotic challenge: Implications for osmoregulation, energy metabolism and growth. Aquaculture287 (3-4), 419–426. doi: 10.1016/j.aquaculture.2008.10.047
9
Bagnat M. Cheung I. D. Mostov K. E. Stainier D. Y. R. (2007). Genetic control of single lumen formation in the zebrafish gut. Nat. Cell Biol.9 (8), 954–960. doi: 10.1038/ncb1621
10
Barany A. Shaughnessy C. A. McCormick S. D. (2021). Corticosteroid control of Na+/K+-ATPase in the intestine of the sea lamprey (Petromyzon marinus). Gen. Comp. Endocrinol.307, 113756. doi: 10.1016/j.ygcen.2021.113756
11
Bates J. M. Akerlund J. Mittge E. Guillemin K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe2, 371–382. doi: 10.1016/j.chom.2007.10.010
12
Bates T. Naumann U. Hoppe B. Englert C. (2018). Kidney regeneration in fish. Int. J. Dev. Biol.62 (6-7-8), 419–429. doi: 10.1387/ijdb.170335ce
13
Baumgarner B. L. Riley C. P. Sepulveda M. S. Brown P. B. Meyer J. L. Adamec J. (2012). Increased expression of GAPDH protein is not indicative of nitrosative stress or apoptosis in liver of starved rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem.38, 319–327. doi: 10.1007/s10695-011-9509-9
14
Baustian M. D. Wang S. Q. Beyenbach K. W. (1997). Adaptive responses of aglomerular toadfish to dilute sea water. J. Comp. Physiol. - B Biochem. Syst. Environ. Physiol.167, 61–70. doi: 10.1007/s003600050048
15
Becker H. M. Seidler U. E. (2024). Bicarbonate secretion and acid/base sensing by the intestine. Pflugers Arch. Eur. J. Physiol.476 (4), 593–610. doi: 10.1007/s00424-024-02914-3
16
Beyenbach K. W. (2004). Kidneys sans glomeruli. Am. J. Physiol. - Ren. Physiol.286 (5), F811–F827. doi: 10.1152/ajprenal.00351.2003
17
Bœuf G. Payan P. (2001). How should salinity influence fish growth? Comp. Biochem. Physiol. Part C Toxicol. Pharmacol.130, 411–423. doi: 10.1016/S1532-0456(01)00268-X
18
Boutet I. Long Ky C. L. Bonhomme F. (2006). A transcriptomic approach of salinity response in the euryhaline teleost, Dicentrarchus labrax. Gene379, 40–50. doi: 10.1016/j.gene.2006.04.011
19
Bowman J. P. (2024). Heat stress impacts the gut microbiome of Atlantic salmon by promoting growth of Vibrionaceae and is associated with extensive cast production. bioRxiv, 2024–11. doi: 10.1101/2024.11.23.625002
20
Breves J. P. Serizier S. B. Goffin V. McCormick S. D. Karlstrom R. O. (2013). Prolactin regulates transcription of the ion uptake Na+/Cl– cotransporter (ncc) gene in zebrafish gill. Mol. Cell. Endocrinol.369, 98–106. doi: 10.1016/j.mce.2013.01.021
21
Brown J. A. Gray C. J. Taylor S. M. (1990). The renin-angiotensin system and glomerular function of teleost fish. Prog. Clin. Biol. Res.342, 528–533.
22
Brugman S. Schneeberger K. Witte M. Klein M. R. van den Bogert B. Boekhorst J. et al . (2014). T lymphocytes control microbial composition by regulating the abundance of Vibrio in the zebrafish gut. Gut Microbes5, 737–747. doi: 10.4161/19490976.2014.972228
23
Budd K. Gunn J. C. Finch T. Klymus K. Sitati N. Eggert L. S. (2020). Effects of diet, habitat, and phylogeny on the fecal microbiome of wild African savanna ( Loxodonta africana ) and forest elephants ( L. cyclotis ). Ecol. Evol.10, 5637–5650. doi: 10.1002/ece3.6305
24
Butt R. L. Volkoff H. (2019). Gut microbiota and energy homeostasis in fish. Front. Endocrinol. (Lausanne).10. doi: 10.3389/fendo.2019.00009
25
Cañedo-Argüelles M. Kefford B. J. Piscart C. Prat N. Schäfer R. B. Schulz C.-J. (2013). Salinisation of rivers: An urgent ecological issue. Environ. Pollut.173, 157–167. doi: 10.1016/j.envpol.2012.10.011
26
Carvalho E. S. M. Gregório S. F. Power D. M. Canário A. V. M. Fuentes J. (2012). Water absorption and bicarbonate secretion in the intestine of the sea bream are regulated by transmembrane and soluble adenylyl cyclase stimulation. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol.182. doi: 10.1007/s00360-012-0685-4
27
Chang Y. J. Min B. H. Choi C. Y. (2007). Black porgy (Acanthopagrus schlegeli) prolactin cDNA sequence: mRNA expression and blood physiological responses during freshwater acclimation. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol.147, 122–128. doi: 10.1016/j.cbpb.2007.01.006
28
Chang Z. Q. Neori A. He Y. Y. Li J. T. Qiao L. Preston S. I. et al . (2020). Development and current state of seawater shrimp farming, with an emphasis on integrated multi-trophic pond aquaculture farms, in China – a review. Rev. Aquac.12 (4), 2544–2558. doi: 10.1111/raq.12457
29
Cheesman S. E. Neal J. T. Mittge E. Seredick B. M. Guillemin K. (2011). Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci.108, 4570–4577. doi: 10.1073/pnas.1000072107
30
Chen Z. Huang B. Yan Z. Hong Y. Zhao M. Jin M. et al . (2023b). Genome–wide expression profile analysis of the NHE and NKA gene family in Rachycentron canadum (Linnaeus 1766) and its response to salinity adaptation. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1228933
31
Chen X. Liu S. Ding Q. Teame T. Yang Y. Ran C. et al . (2023a). Research advances in the structure, function, and regulation of the gill barrier in teleost fish. Water Biol. Secur.2 (2), 100139. doi: 10.1016/j.watbs.2023.100139
32
Christensen A. K. Regish A. M. McCormick S. D. (2018). Shifts in the relationship between mRNA and protein abundance of gill ion-transporters during smolt development and seawater acclimation in Atlantic salmon (Salmo salar). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.221, 63–73. doi: 10.1016/j.cbpa.2018.03.020
33
Christensen E. A. F. Stieglitz J. D. Grosell M. Steffensen J. F. (2019). Intra-specific difference in the effect of salinity on physiological performance in european perch (Perca fluviatilis) and its ecological importance for fish in estuaries. Biol. (Basel).8 (4), 89. doi: 10.3390/biology8040089
34
Ciavoni E. Schrama J. W. Radhakrishnan G. Sæle Ø. Prabhu Philip A. J. (2024). Salinity induced changes in the progression of water, ion and nutrient fluxes along the gastrointestinal tract of Atlantic salmon smolt (Salmo salar). Aquaculture580, 740331. doi: 10.1016/j.aquaculture.2023.740331
35
Ciccotti B. E. Macchi E. Rossi A. Cataldi E. Cataudella S. (1993). Glass eel (Anguilla Anguilla) acclimation to freshwater and seawater: morphological changes of the digestive tract. J. Appl. Ichthyol.9 (2), 74–81. doi: 10.1111/j.1439-0426.1993.tb00528.x
36
Cooney R. Wan A. H. L. O’Donncha F. Clifford E. (2021). Designing environmentally efficient aquafeeds through the use of multicriteria decision support tools. Curr. Opin. Environ. Sci. Heal.23, 100276. doi: 10.1016/j.coesh.2021.100276
37
Cunillera-Montcusí D. Beklioğlu M. Cañedo-Argüelles M. Jeppesen E. Ptacnik R. Amorim C. A. et al . (2022). Freshwater salinisation: a research agenda for a saltier world. Trends Ecol. Evol.37 (5), 440–453. doi: 10.1016/j.tree.2021.12.005
38
Cutler C. P. Martinez A. S. Cramb G. (2007). The role of aquaporin 3 in teleost fish. Comp. Biochem. Physiol. - A Mol. Integr. Physiol.148 (1), 82–91. doi: 10.1016/j.cbpa.2006.09.022
39
Cuykx M. Rodrigues R. M. Laukens K. Vanhaecke T. Covaci A. (2018). In vitro assessment of hepatotoxicity by metabolomics: a review. Arch. Toxicol.92. doi: 10.1007/s00204-018-2286-9
40
Davidson S. (2024). “Comparing Ionoregulation and Modes of Nitrogen Excretion Across the Life Cycle of Parasitic and Non-parasitic Lamprey Species”. Theses and Dissertations (Comprehensive), 2637. Available at: https://scholars.wlu.ca/etd/2637.
41
Dehler C. E. Secombes C. J. Martin S. A. M. (2017). Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci. Rep.7, 1–11. doi: 10.1038/s41598-017-13249-8
42
de Oliveira C. G. López-Olmeda J. F. Costa L. S. do Espirito Santo A. H. dos Santos F. A. C. Luz R. K. et al . (2022). Gastrointestinal emptying and daily patterns of activity of proteinolytic enzymes in Nile tilapia (Oreochromis niloticus). Aquaculture546, 737338. doi: 10.1016/j.aquaculture.2021.737338
43
Devlin R. H. Leggatt R. A. Benfey T. J. (2020). Genetic modification of growth in fish species used in aquaculture: Phenotypic and physiological responses. In Fish physiology (Academic Press), 38, 237–272. doi: 10.1016/bs.fp.2020.09.004
44
Ding L. Liu Y. Wei X. Geng C. Liu W. Han L. et al . (2023). Effects of saline-alkaline stress on metabolome, biochemical parameters, and histopathology in the kidney of crucian carp (Carassius auratus). Metabolites13 (2), 159. doi: 10.3390/metabo13020159
45
Divino J. N. (2016). Osmoregulatory Physiology and its Evolution in the Threespine Stickleback (Gasterosteus aculeatus). Doctoral Dissertations. (University of Connecticut - Storrs), 1217. Available at: https://digitalcommons.lib.uconn.edu/dissertations/1217.
46
Diwan A. D. Harke S. N. Panche A. N. (2023). Host-microbiome interaction in fish and shellfish: An overview. Fish Shellfish Immunol. Rep.4, 100091. doi: 10.1016/j.fsirep.2023.100091
47
Djiba P. K. Zhang J. Xu Y. Zhang P. Zhou J. Zhang Y. et al . (2021). Correlation between metabolic rate and salinity tolerance and metabolic response to salinity in grass carp (Ctenopharyngodon idella). Animals11, 3445. doi: 10.3390/ani11123445
48
Emerenciano M. G. C. Miranda-Baeza A. Martínez-Porchas M. Poli M. A. F. do N. (2021). Biofloc technology (BFT) in shrimp farming: past and present shaping the future. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.813091
49
Engelund M. B. Madsen S. S. (2011). The role of aquaporins in the kidney of euryhaline teleosts. Front. Physiol.2. doi: 10.3389/fphys.2011.00051
50
Engevik A. C. Engevik M. A. (2021). Exploring the impact of intestinal ion transport on the gut microbiota. Comput. Struct. Biotechnol. J.19, 134–144. doi: 10.1016/j.csbj.2020.12.008
51
Ern R. Huong D. T. T. Cong N. V. Bayley M. Wang T. (2014). Effect of salinity on oxygen consumption in fishes: A review. J. Fish Biol.84. doi: 10.1111/jfb.12330
52
Esbaugh A. J. Brix K. V. Grosell M. (2019). Na + K + ATPase isoform switching in zebrafish during transition to dilute freshwater habitats. Proc. R. Soc B Biol. Sci.286, 20190630. doi: 10.1098/rspb.2019.0630
53
Escobar-Sierra C. Cañedo-Argüelles M. Vinyoles D. Lampert K. P. (2024). Unraveling the molecular mechanisms of fish physiological response to freshwater salinization: A comparative multi-tissue transcriptomic study in a river polluted by potash mining. Environ. Pollut.357, 124400. doi: 10.1016/j.envpol.2024.124400
54
Evans D. H. (2010). A brief history of the study of fish osmoregulation: The central role of the Mt. Desert Island biological laboratory. Front. Physiol.1. doi: 10.3389/fphys.2010.00013
55
Evans D. H. (2011). Osmotic, ionic and nitrogenous-waste balance|Osmoregulation in Fishes: An Introduction Encyclopedia of Fish Physiology, From Genome to Environment (Reference Module in Life Sciences). (Cambridge, Massachusetts, United States: Academic Press). doi: 10.1016/B978-0-12-374553-8.00210-0
56
Evans R. G. (2023). Evolution of the glomerulus in a marine environment and its implications for renal function in terrestrial vertebrates. Am. J. Physiol. Regul. Integr. Comp. Physiol.324 (2), R143–R151. doi: 10.1152/ajpregu.00210.2022
57
Evans D. H. Piermarini P. M. Choe K. P. (2005a). The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev.85 (1), 97–177. doi: 10.1152/physrev.00050.2003
58
Farinha C. M. Santos L. Ferreira J. F. (2024). Cell type-specific regulation of CFTR trafficking—on the verge of progress. Front. Cell Dev. Biol.12. doi: 10.3389/fcell.2024.1338892
59
Ferreira-Martins D. Wilson J. M. Kelly S. P. Kolosov D. McCormick S. D. (2021). A review of osmoregulation in lamprey. J. Great Lakes Res.47, S59–S71. doi: 10.1016/j.jglr.2021.05.003
60
Fiedler S. Wünnemann H. Hofmann I. Theobalt N. Feuchtinger A. Walch A. et al . (2020). A practical guide to unbiased quantitative morphological analyses of the gills of rainbow trout (Oncorhynchus mykiss) in ecotoxicological studies. PloS One15, e0243462. doi: 10.1371/journal.pone.0243462
61
Flores E. M. Nguyen A. T. Odem M. A. Eisenhoffer G. T. Krachler A. M. (2020). The zebrafish as a model for gastrointestinal tract–microbe interactions. Cell. Microbiol.22 (3), e13152. doi: 10.1111/cmi.13152
62
Forsyth I. A. Wallis M. (2002). Growth hormone and prolactin - Molecular and functional evolution. J. Mammary Gland Biol. Neoplasia7, 291–312. doi: 10.1023/A:1022804817104
63
Foskett J. K. Bern H. A. Machen T. E. Conner M. (1983). Chloride cells and the hormonal control of teleost fish osmoregulation. J. Exp. Biol. VOL.106, 255–281. doi: 10.1242/jeb.106.1.255
64
Foyle K. L. Hess S. Powell M. D. Herbert N. A. (2020). What is gill health and what is its role in marine finfish aquaculture in the face of a changing climate? Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00400
65
Franklin D. A. Edward L. L. (2019). Ammonia toxicity and adaptive response in marine fishes - A review. Indian J. Geo-Marine Sci.48 (3), 273–279. http://nopr.niscpr.res.in/handle/123456789/47054.
66
Fridman S. (2020). Ontogeny of the osmoregulatory capacity of teleosts and the role of ionocytes. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00709
67
Ganguly S. Mitra T. Mahanty A. Mohanty S. Mohanty B. P. (2020). A comparative metabolomics study on anadromous clupeid Tenualosa ilisha for better understanding the influence of habitat on nutritional composition. Metabolomics16, 30. doi: 10.1007/s11306-020-01655-5
68
Giatsis C. Sipkema D. Smidt H. Heilig H. Benvenuti G. Verreth J. et al . (2015). The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci. Rep.5 (1), 18206. doi: 10.1038/srep18206
69
Gong J. Xu F. Li Y. He Y. Liang Z. Chen X. et al . (2024). Metagenomic analysis of intestinal microbial function and key genes responsive to acute high-salinity stress in Nile tilapia (Oreochromis niloticus). Gene913, 148371. doi: 10.1016/j.gene.2024.148371
70
Gonzalez R. J. (2012a). The physiology of hyper-salinity tolerance in teleost fish: a review. J Comp Physiol B.182, 321–329. doi: 10.1007/s00360-011-0624-9
71
Gonzalez R. J. (2012b). The physiology of hyper-salinity tolerance in teleost fish: a review. J. Comp. Physiol. B182, 321–329. doi: 10.1007/s00360-011-0624-9
72
Goode K. L. Dunphy B. J. Parsons D. M. (2020). Environmental metabolomics as an ecological indicator: Metabolite profiles in juvenile fish discriminate sites with different nursery habitat qualities. Ecol. Indic.115, 106361. doi: 10.1016/j.ecolind.2020.106361
73
Greenwell M. G. Sherrill J. Clayton L. A. (2003). Osmoregulation in fish mechanisms and clinical implications. Vet. Clin. North Am. - Exot. Anim. Pract.6, 169–189. doi: 10.1016/S1094-9194(02)00021-X
74
Gregório S. F. Carvalho E. S. M. Encarnação S. Wilson J. M. Power D. M. Canário A. V. M. et al . (2013). Adaptation to different salinities exposes functional specialization in the intestine of the sea bream (Sparus aurata L.). J. Exp. Biol.216 (3), 470–479. doi: 10.1242/jeb.073742
75
Griffith M. B. (2017). Toxicological perspective on the osmoregulation and ionoregulation physiology of major ions by freshwater animals: Teleost fish, crustacea, aquatic insects, and Mollusca. Environ. Toxicol. Chem.36 (3), 576–600. doi: 10.1002/etc.3676
76
Grosell M. (2011). Intestinal anion exchange in marine teleosts is involved in osmoregulation and contributes to the oceanic inorganic carbon cycle. Acta Physiol. (Oxf).202 (3), 421–434. doi: 10.1111/j.1748-1716.2010.02241.x
77
Guh Y. J. Lin C. H. Hwang P. P. (2015). Osmoregulation in zebrafish: Ion transport mechanisms and functional regulation. EXCLI J.14, 627–659. doi: 10.17179/excli2015-246
78
Gyan R. W. Alatwinusa M. Yang Q. Tan B. Chi S. Yi Y. (2024). The effects of dietary L-carnitine supplementation in fish diet containing high corn gluten meal on immunity, lipid metabolism, and metabolomics in juvenile hybrid grouper (♀Epinephelus fuscoguttatus × ♂Epinephelus lanceolatus). Aquacult Int.32, 833–869. doi: 10.1007/s10499-023-01193-6
79
Hamilton E. F. Element G. van Coeverden de Groot P. Engel K. Neufeld J. D. Shah V. et al . (2019). Anadromous arctic char microbiomes: bioprospecting in the high arctic. Front. Bioeng. Biotechnol.7. doi: 10.3389/fbioe.2019.00032
80
Hanssens L. S. Duchateau J. Casimir G. J. (2021). CFTR protein: not just a chloride channel? Cells10, 2844. doi: 10.3390/cells10112844
81
Harrison T. D. Whitfield A. K. (2006). Temperature and salinity as primary determinants influencing the biogeography of fishes in South African estuaries. Estuar. Coast. Shelf Sci.66 (1-2), 335–345. doi: 10.1016/j.ecss.2005.09.010
82
Harshini V. Shukla N. Raval I. Kumar S. Shrivastava V. Patel A. K. et al . (2022). Kidney transcriptome response to salinity adaptation in Labeo rohita. Front. Physiol.13. doi: 10.3389/fphys.2022.991366
83
He Y. F. Wang L. M. Zhu W. B. Dong Z. J. Liu N. (2017). Effects of salinity on cold tolerance of Malaysian red tilapia. Aquac. Int.25, 777–792. doi: 10.1007/s10499-016-0077-y
84
Hickman C. P. Trump B. F. (1969). “The kidney,”. in Fish Physiology eds. HoarW. S.RandallD. J. (Cambridge, CA: Academic), p. 91–239. doi: 10.1016/S1546-5098(08)60083-7
85
Hieu D. Q. Hang B. T. B. Lokesh J. Garigliany M. M. Huong D. T. T. Yen D. T. et al . (2022). al microbiota and gene expression in striped catfish juvenilesSalinity significantly affects intestin. Appl. Microbiol. Biotechnol.106, 3245–3264. doi: 10.1007/s00253-022-11895-1
86
Hintz W. D. Relyea R. A. (2019). A review of the species, community, and ecosystem impacts of road salt salinisation in fresh waters. Freshw. Biol.64, 1081–1097. doi: 10.1111/fwb.13286
87
Houde A. L. S. Akbarzadeh A. Günther O. P. Li S. Patterson D. A. Farrell A. P. et al . (2019). Salmonid gene expression biomarkers indicative of physiological responses to changes in salinity, temperature, but not dissolved oxygen. J. Exp. Biol.222 (13), jeb198036. doi: 10.1242/jeb.198036
88
Hu Y. C. Chu K. F. Yang W. K. Lee T. H. (2017). Na+, K+-ATPase β1 subunit associates with α1 subunit modulating a “higher-NKA-in-hyposmotic media” response in gills of euryhaline milkfish, Chanos chanos. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol.187, 995–1007. doi: 10.1007/s00360-017-1066-9
89
Huang M. Gao Q. Yang X. Jiang W. Hao L. Yu Y. et al . (2023). Free amino acids in response to salinity changes in fishes: relationships to osmoregulation. Fish Physiol. Biochem.49, 1031–1042. doi: 10.1007/s10695-023-01244-y
90
Huang P. C. Liu T. Y. Hu M. Y. Casties I. Tseng Y. C. (2020). Energy and nitrogenous waste from glutamate/glutamine catabolism facilitates acute osmotic adjustment in non-neuroectodermal branchial cells. Sci. Rep.10 (1), 9460. doi: 10.1038/s41598-020-65913-1
91
Huang M. Yu H. Wang B. Jiang W. Xue N. Bao H. et al . (2025). Landscapes of alternative splicing genes/events in the gills of Mozambique tilapia (Oreochromis mossambicus) and their roles in high-salinity adaptation. Aquac. Int.33, 174. doi: 10.1007/s10499-025-01846-8
92
Hughes L. C. Somoza G. M. Nguyen B. N. Bernot J. P. González-Castro M. Díaz de Astarloa J. M. et al . (2017). Transcriptomic differentiation underlying marine-to-freshwater transitions in the South American silversides Odontesthes argentinensis and O. bonariensis (Atheriniformes). Ecol. Evol.7, 5258–5268. doi: 10.1002/ece3.3133
93
Hwang P.-P. Chou M.-Y. (2013). Zebrafish as an animal model to study ion homeostasis. Pflügers Arch. - Eur. J. Physiol.465, 1233–1247. doi: 10.1007/s00424-013-1269-1
94
Hwang P.-P. Lee T.-H. (2007). New insights into fish ion regulation and mitochondrion-rich cells. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.148, 479–497. doi: 10.1016/j.cbpa.2007.06.416
95
Hwang P.-P. Lee T.-H. Lin L.-Y. (2011). Ion regulation in fish gills: recent progress in the cellular and molecular mechanisms. Am. J. Physiol. Integr. Comp. Physiol.301, R28–R47. doi: 10.1152/ajpregu.00047.2011
96
Iehata S. Nosi M. Z. M. Danish-Daniel M. Sharifah N. E. (2021). Gut microbiome associated with cultured Malaysian mahseer tor tambroides. AACL Bioflux14 (1), 46–58.
97
Inokuchi M. Hiroi J. Kaneko T. (2022). Why can Mozambique tilapia acclimate to both freshwater and seawater? Insights from the plasticity of ionocyte functions in the euryhaline teleost. Front. Physiol.13. doi: 10.3389/fphys.2022.914277
98
Inokuchi M. Yamaguchi Y. Moorman B. P. Seale A. P. (2021). Age-dependent decline in salinity tolerance in a euryhaline fish. Front. Aging2. doi: 10.3389/fragi.2021.675395
99
Jain S. Sharma G. Mathur Y. (2013). Effects of temperature variations on fish in lakes. Int. J. Eng. Res. Technol. IJERT2 (10), 2516–2523.
100
Janoš P. Magistrato A. (2021). All-atom simulations uncover the molecular terms of the NKCC1 transport mechanism. J. Chem. Inf. Model.61 (7), 3649–3658. doi: 10.1021/acs.jcim.1c00551
101
Jendoubi T. (2021). Approaches to integrating metabolomics and multi-omics data: A primer. Metabolites11, 184. doi: 10.3390/metabo11030184
102
Jiang W. Tian X. Fang Z. Li L. Dong S. Li H. et al . (2019). Metabolic responses in the gills of tongue sole (Cynoglossus semilaevis) exposed to salinity stress using NMR-based metabolomics. Sci. Total Environ.653, 465–474. doi: 10.1016/j.scitotenv.2018.10.404
103
Jones F. C. Grabherr M. G. Chan Y. F. Russell P. Mauceli E. Johnson J. et al . (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature484, 55–61. doi: 10.1038/nature10944
104
Kanika N. H. Liaqat N. Chen H. Ke J. Lu G. Wang J. et al . (2025). Fish gut microbiome and its application in aquaculture and biological conservation. Front. Microbiol.15. doi: 10.3389/fmicb.2024.1521048
105
Kanther M. Sun X. Mühlbauer M. Mackey L. C. Flynn E. J. Bagnat M. et al . (2011). Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology141, 197–207. doi: 10.1053/j.gastro.2011.03.042
106
Kaushal S. S. Likens G. E. Jaworski N. A. Pace M. L. Sides A. M. Seekell D. et al . (2010). Rising stream and river temperatures in the United States. Front. Ecol. Environ.8. doi: 10.1890/090037
107
Kim C. Kültz D. (2020). An osmolality/salinity-responsive enhancer 1 (OSRE1) in intron 1 promotes salinity induction of tilapia glutamine synthetase. Sci. Rep.10 (1), 12103. doi: 10.1038/s41598-020-69090-z
108
Kivistik C. Knobloch J. Käiro K. Tammert H. Kisand V. Hildebrandt J.-P. et al . (2020). Impact of salinity on the gastrointestinal bacterial community of theodoxus fluviatilis. Front. Microbiol.11. doi: 10.3389/fmicb.2020.00683
109
Kolbadinezhad S. M. Coimbra J. Wilson J. M. (2018). Osmoregulation in the Plotosidae catfish: Role of the salt secreting dendritic organ. Front. Physiol.9. doi: 10.3389/fphys.2018.00761
110
Kombat E. O. Zhao J. L. Abakari G. Owusu-Afriyie G. Birteeb P. T. Alhassan E. H. (2021). Metabolic cost of acute and chronic exposure of Nile tilapia (Oreochromis niloticus) to different levels of salinity. Aquac. Res.52 (12), 6152–6163. doi: 10.1111/are.15477
111
Kovac A. Goss G. G. (2024). Cellular mechanisms of ion and acid-base regulation in teleost gill ionocytes. J. Comp. Physiol. B.194 (5), 645–662. doi: 10.1007/s00360-024-01560-6
112
Kujawa R. Piech P. (2022). Influence of Water Salinity on the Growth and Survivability of Asp Larvae Leuciscus aspius (Linnaeus 1758) under Controlled Conditions. Animals12, 2299. doi: 10.3390/ani12172299
113
Kültz D. (2015). Physiological mechanisms used by fish to cope with salinity stress. J. Exp. Biol.218, 1907–1914. doi: 10.1242/jeb.118695
114
Lahlou B. Henderson I. W. Sawyer W. H. (1969). Renal adaptations by Opsanus tau, a euryhaline aglomerular teleost, to dilute media. Am. J. Physiol.216 (5), 1266–1272. doi: 10.1152/ajplegacy.1969.216.5.1266
115
Lai K. P. Boncan D. A. T. Yang L. Leung C. C. T. Ho J. C. H. Lin X. et al . (2022a). Osmotic gradient is a factor that influences the gill microbiota communities in oryzias melastigma. Biol. (Basel).11, 1528. doi: 10.3390/biology11101528
116
Lai K. P. Li J. W. Wang S. Y. Chiu J. M. Y. Tse A. Lau K. et al . (2015). Tissue-specific transcriptome assemblies of the marine medaka Oryzias melastigma and comparative analysis with the freshwater medaka Oryzias latipes. BMC Genomics16, 1–13. doi: 10.1186/s12864-015-1325-7
117
Lai K. P. Lin X. Tam N. Ho J. C. H. Wong M. K. Gu J. et al . (2020). Osmotic stress induces gut microbiota community shift in fish. Environ. Microbiol.22, 3784–3802. doi: 10.1111/1462-2920.15150
118
Lai K. P. Zhu P. Boncan D. A. T. Yang L. Leung C. C. T. Ho J. C. H. et al . (2022b). Integrated omics approaches revealed the osmotic stress-responsive genes and microbiota in gill of marine medaka. mSystems7 (2), e00047–22. doi: 10.1128/msystems.00047-22
119
Lam S. Lui E. Li Z. Cai S. Sung W.-K. Mathavan S. et al . (2014). Differential transcriptomic analyses revealed genes and signaling pathways involved in iono-osmoregulation and cellular remodeling in the gills of euryhaline Mozambique tilapia, Oreochromis mossambicus. BMC Genomics15, 921. doi: 10.1186/1471-2164-15-921
120
Laverty G. Skadhauge E. (2012). Adaptation of teleosts to very high salinity. Comp. Biochem. Physiol. - A Mol. Integr. Physiol.163, 1–6. doi: 10.1016/j.cbpa.2012.05.203
121
Laycock J. Meeran K. (2012). The endocrine control of salt and water balance [Internet]. In: Integrated endocrinology. John Wiley & Sons, Ltd; 2012 [cited 2021 Aug 3]. pp. 259–73. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118450642.ch12.
122
Lazarev V. F. Guzhova I. V. Margulis B. A. (2020). Glyceraldehyde-3-phosphate dehydrogenase is a multifaceted therapeutic target. Pharmaceutics12, 416. doi: 10.3390/pharmaceutics12050416
123
Lee C. E. Charmantier G. Lorin-Nebel C. (2022a). Mechanisms of Na+ uptake from freshwater habitats in animals. Front. Physiol.13. doi: 10.3389/fphys.2022.1006113
124
Lee J.-G. Cho H.-J. Jeong Y.-M. Lee J.-S. (2021). Genetic approaches using zebrafish to study the microbiota–gut–brain axis in neurological disorders. Cells10, 566. doi: 10.3390/cells10030566
125
Lee C. E. Downey K. Colby R. S. Freire C. A. Nichols S. Burgess M. N. et al . (2022b). Recognizing salinity threats in the climate crisis. Integr. Comp. Biol.62, 441–460. doi: 10.1093/icb/icac069
126
Leguen I. Odjo N. Le Bras Y. Luthringer B. Baron D. Monod G. et al . (2010). Effect of seawater transfer on CYP1A gene expression in rainbow trout gills. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.156, 211–217. doi: 10.1016/j.cbpa.2010.02.002
127
Lema S. C. Washburn E. H. Crowley M. E. Carvalho P. G. Egelston J. N. McCormick S. D. (2019). Evidence for a role of arginine vasotocin receptors in the gill during salinity acclimation by a euryhaline teleost fish. Am. J. Physiol. - Regul. Integr. Comp. Physiol.316 (6), R735–R750. doi: 10.1152/ajpregu.00328.2018
128
Leprêtre M. Hamar J. Urias M. B. Kültz D. (2025). Comparative proteomics of salinity stress responses in fish and aquatic invertebrates. Proteomics, e202400255. doi: 10.1002/pmic.202400255
129
Li Y. Li Y. Lai Q. Yao Z. Zhou K. Niu D. (2024b). Proline metabolism and its osmotic regulatory role in enhancing hypersalinity tolerance of the razor clam (Sinonovacula constricta). Aquaculture589, 741019. doi: 10.1016/j.aquaculture.2024.741019
130
Li X. Liu S. Wang Y. Lu W. Zhang Q. Cheng J. (2022a). Genomic and Transcriptomic Landscape and Evolutionary Dynamics of Heat Shock Proteins in Spotted Sea Bass (Lateolabrax maculatus) under Salinity Change and Alkalinity Stress. Biol. (Basel).11, 353. doi: 10.3390/biology11030353
131
Li N. Shao T. Xu L. Long X. Rengel Z. Zhang Y. (2024a). Transcriptome analysis reveals the molecular mechanisms underlying the enhancement of salt-tolerance in Melia azedarach under salinity stress. Sci. Rep.14, 10981. doi: 10.1038/s41598-024-61907-5
132
Li X. Shen Y. Bao Y. Wu Z. Yang B. Jiao L. et al . (2022b). Physiological responses and adaptive strategies to acute low-salinity environmental stress of the euryhaline marine fish black seabream (Acanthopagrus schlegelii). Aquaculture554, 738117. doi: 10.1016/j.aquaculture.2022.738117
133
Li Y. Ye Y. Li W. Liu X. Zhao Y. Jiang Q. et al . (2023). Effects of salinity stress on histological changes, glucose metabolism index and transcriptomic profile in freshwater shrimp, macrobrachium nipponense. Animals13, 2884. doi: 10.3390/ani13182884
134
Lin T. Lai Q. Yao Z. Lu J. Zhou K. Wang H. (2013). Combined effects of carbonate alkalinity and pH on survival, growth and haemocyte parameters of the Venus clam Cyclina sinensis. Fish Shellfish Immunol.35, 525–531. doi: 10.1016/j.fsi.2013.05.006
135
Lin T. Zhai Y. Tian C. Jiang D. Shi H. Jiang M. et al . (2024). Brain transcriptome analysis reveals metabolic changes adapting to hyperhaline or hypohaline environments in spotted scat (Scatophagus argus). J. Ocean Univ. China23, 755–765. doi: 10.1007/s11802-024-5702-1
136
Lin G. Zheng M. Li S. Xie J. Fang W. Gao D. et al . (2020). Response of gut microbiota and immune function to hypoosmotic stress in the yellowfin seabream (Acanthopagrus latus). Sci. Total Environ.745, 140976. doi: 10.1016/j.scitotenv.2020.140976
137
Linlin A. Zhang Y. Xu B. Zhang H. Li Y. Wang L. et al . (2024). Comprehensive analyses of annexins in naked carp (Gymnocypris przewalskii) unveil their roles in saline-alkaline stress. Aquaculture579, 740175.
138
Lingaraju A. Long T. M. Wang Y. Austin J. R. Turner J. R. (2015). Conceptual barriers to understanding physical barriers. Semin. Cell Dev. Biol.42, 13–21. doi: 10.1016/j.semcdb.2015.04.008
139
Liu B. Guo H. Zhu K. Guo L. Liu B. Zhang N. et al . (2019). Salinity effect on intestinal microbiota in golden pompano trachinotus ovatus (Linnaeus 1758). Isr. J. Aquac. - Bamidgeh71. doi: 10.46989/001c.20995
140
Liu Y. Li Y. Li J. Zhou Q. Li X. (2022). Gut microbiome analyses of wild migratory freshwater fish (Megalobrama terminalis) through geographic isolation. Front. Microbiol.13. doi: 10.3389/fmicb.2022.858454
141
Liu Y. Li E. Xu C. Su Y. Qin J. G. Chen L. et al . (2018). Brain transcriptome profiling analysis of Nile tilapia (Oreochromis niloticus) under long-term hypersaline stress. Front. Physiol.9. doi: 10.3389/fphys.2018.00219
142
Liu S. Tian F. Qi D. Qi H. Wang Y. Xu S. et al . (2023). Physiological, metabolomic, and transcriptomic reveal metabolic pathway alterations in Gymnocypris przewalskii due to cold exposure. BMC Genomics24, 545. doi: 10.1186/s12864-023-09587-9
143
Liu D. Yu H. Xue N. Bao H. Gao Q. Tian Y. (2024). Alternative splicing patterns of hnrnp genes in gill tissues of rainbow trout (Oncorhynchus mykiss) during salinity changes. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol.271, 110948. doi: 10.1016/j.cbpb.2024.110948
144
Loretz C. A. Pollina C. Hyodo S. Takei Y. (2009). Extracellular calcium-sensing receptor distribution in osmoregulatory and endocrine tissues of the tilapia. Gen. Comp. Endocrinol.161 (2), 216–228. doi: 10.1016/j.ygcen.2008.12.020
145
Lorgen-Ritchie M. Clarkson M. Chalmers L. Taylor J. F. Migaud H. Martin S. A. M. (2021). A temporally dynamic gut microbiome in atlantic salmon during freshwater recirculating aquaculture system (RAS) production and post-seawater transfer. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.711797
146
Low C. Rozaini M. Z. H. Musa N. Syarul Nataqain B. (2017). Current knowledge of metabolomic approach in infectious fish disease studies. J. Fish Dis.40, 1267–1277. doi: 10.1111/jfd.12610
147
MacKay W. C. Janicki R. (1979). Changes in the EEL intestine during seawater adaptation. Comp. Biochem. Physiol. – Part A Physiol.62 (3), 757–761. doi: 10.1016/0300-9629(79)90135-X
148
Madsen S. (2011). Functional characterization of water transport and cellular localization of three aquaporin paralogs in the salmonid intestine. Front. Physiol.2. doi: 10.3389/fphys.2011.00056
149
Malone A. M. Cozzi R. R. F. Marshall W. S. (2015). Cold acclimation allows regulation of chloride secretion in a eurythermic teleost fish Fundulus heteroclitus. Comp. Biochem. Physiol. -Part A Mol. Integr. Physiol.180, 68–74. doi: 10.1016/j.cbpa.2014.11.010
150
Mariu A. Chatha A. M. M. Naz S. Khan M. F. Safdar W. Ashraf I. (2023). Effect of temperature, pH, salinity and dissolved oxygen on fishes. J. Zool. Syst.1, 1–12. doi: 10.56946/jzs.v1i2.198
151
Masroor W. Farcy E. Blondeau-Bidet E. Venn A. Tambutté E. Lorin-Nebel C. (2019). Effect of salinity and temperature on the expression of genes involved in branchial ion transport processes in European sea bass. J. Therm. Biol.85. doi: 10.1016/j.jtherbio.2019.102422
152
McCormick S. D. (2011). The hormonal control of osmoregulation in teleost fish. Life Sciences1, 1466–1473. doi: 10.1016/B978-0-12-374553-8.00212-4
153
McCormick S. D. Bradshaw D. (2006). Hormonal control of salt and water balance in vertebrates. Gen. Comp. Endocrinol.147 (1), 3–8. doi: 10.1016/j.ygcen.2005.12.009
154
McCormick S. D. Regish A. M. (2018). Effects of ocean acidification on salinity tolerance and seawater growth of Atlantic salmon Salmo salar smolts. J. Fish Biol.93 (3), 560–566. doi: 10.1111/jfb.13656
155
McCormick S. D. Taylor M. L. Regish A. M. (2020). Cortisol is an osmoregulatory and glucose-regulating hormone in Atlantic sturgeon, a basal ray-finned fish. J. Exp. Biol.223 (18), jeb220251. doi: 10.1242/jeb.220251
156
McLean E. Barrows F. T. Craig S. R. Alfrey K. Tran L. (2020). Complete replacement of fishmeal by soybean and poultry meals in Pacific whiteleg shrimp feeds: Growth and tolerance to EMS/AHPND and WSSV challenge. Aquaculture527, 735383. doi: 10.1016/j.aquaculture.2020.735383
157
Meador J. P. Bettcher L. F. Ellenberger M. C. Senn T. D. (2020). Metabolomic profiling for juvenile Chinook salmon exposed to contaminants of emerging concern. Sci. Total Environ.747, 141097. doi: 10.1016/j.scitotenv.2020.141097
158
Minich J. J. Poore G. D. Jantawongsri K. Johnston C. Bowie K. Bowman J. et al . (2020). Microbial ecology of atlantic salmon (Salmo salar) hatcheries: impacts of the built environment on fish mucosal microbiota. Appl. Environ. Microbiol.86 (12), e00411–20. doi: 10.1128/AEM.00411-20
159
Mkulo E. M. Wang B. Amoah K. Huang Y. Cai J. Jin X. et al . (2024). The current status and development forecasts of vaccines for aquaculture and its effects on bacterial and viral diseases. Microb. Pathog.196, 106971. doi: 10.1016/j.micpath.2024.106971
160
Mohindra V. Chowdhury L. M. Chauhan N. Paul A. Singh R. K. Kushwaha B. et al . (2023). Transcriptome Analysis Revealed Osmoregulation Related Regulatory Networks and Hub Genes in the Gills of Hilsa shad, Tenualosa ilisha, during the Migratory Osmotic Stress. Mar. Biotechnol.25 (1), 161–173. doi: 10.1007/s10126-022-10190-4
161
Morales-Rivera M. F. Valenzuela-Miranda D. Nuñez-Acuña G. Benavente B. P. Gallardo-Escárate C. Valenzuela-Muñoz V. (2022). Atlantic salmon (Salmo salar) transfer to seawater by gradual salinity changes exhibited an increase in the intestinal microbial abundance and richness. Microorganisms11, 76. doi: 10.3390/microorganisms11010076
162
Morera F. J. Castro-Guarda M. Nualart D. Espinosa G. Muñoz J. L. Vargas-Chacoff L. (2021). The biological basis of smoltification in Atlantic salmon. Austral J. Vet. Sci.53, 73–82. doi: 10.4067/S0719-81322021000100073
163
Morshed S. M. Chen Y. Y. Lin C. H. Chen Y. P. Lee T. H. (2023). Freshwater transfer affected intestinal microbiota with correlation to cytokine gene expression in Asian sea bass. Front. Microbiol.14. doi: 10.3389/fmicb.2023.1097954
164
Mugwanya M. Dawood M. A. O. Kimera F. Sewilam H. (2022). Anthropogenic temperature fluctuations and their effect on aquaculture: A comprehensive review. Aquac. Fish.7, 223–243. doi: 10.1016/j.aaf.2021.12.005
165
Mundy P. C. Jeffries K. M. Fangue N. A. Connon R. E. (2020). Differential regulation of select osmoregulatory genes and Na+/K+-ATPase paralogs may contribute to population differences in salinity tolerance in a semi-anadromous fish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.240, 110584. doi: 10.1016/j.cbpa.2019.110584
166
Nagarajan G. Aruna A. Chang Y.-M. Alkhamis Y. A. Mathew R. T. Chang C.-F. (2023). Effects of Osmotic Stress on the mRNA Expression of prl, prlr, gr, gh, and ghr in the Pituitary and Osmoregulatory Organs of Black Porgy, Acanthopagrus schlegelii. Int. J. Mol. Sci.24, 5318. doi: 10.3390/ijms24065318
167
Nicholson J. K. Lindon J. C. Holmes E. (1999). “Metabonomics”: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica29, 1181–1189. doi: 10.1080/004982599238047
168
Nicholson J. K. Holmes E. Kinross J. Burcelin R. Gibson G. Jia W. et al . (2012). Host-gut microbiota metabolic interactions. Science336, 1262–1267. doi: 10.1126/science.1223813
169
Nishimura H. Imai M. (1982). Control of renal function in freshwater and marine teleosts. Fed. Proc.41 (8), 2355–2360.
170
Ortiz R. M. Wade C. E. Costa D. P. Ortiz C. L. (2002). Renal effects of fresh water-induced hypo-osmolality in a marine adapted seal. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol.172, 297–307. doi: 10.1007/s00360-002-0254-3
171
Ou W. Yu G. Zhang Y. Mai K. (2021). Recent progress in the understanding of the gut microbiota of marine fishes. Mar. Life Sci. Technol.3, 434–448. doi: 10.1007/s42995-021-00094-y
172
Pasquier J. Cabau C. Nguyen T. Jouanno E. Severac D. Braasch I. et al . (2016). Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genomics17, 1–10. doi: 10.1186/s12864-016-2709-z
173
Perry W. B. Lindsay E. Payne C. J. Brodie C. Kazlauskaite R. (2020). The role of the gut microbiome in sustainable teleost aquaculture. Proc. R. Soc B Biol. Sci.287. doi: 10.1098/rspb.2020.0184
174
Phelps D. Brinkman N. E. Keely S. P. Anneken E. M. Catron T. R. Betancourt D. et al . (2017). Microbial colonization is required for normal neurobehavioral development in zebrafish. Sci. Rep.7, 11244. doi: 10.1038/s41598-017-10517-5
175
Pino-Martinez E. Balseiro P. Kvittingen H. F. Pedrosa C. Gorissen M. Handeland S. O. (2024). Influence of salinity on rainbow trout (Oncorhynchus mykiss) smolt development and postsmolt performance. Aquaculture587, 740874. doi: 10.1016/j.aquaculture.2024.740874
176
Poff N. L. Zimmerman J. K. (2010). Ecological responses to altered flow regimes: a literature review to inform the science and management of environmental flows. Freshwater Biology55 (1), 194–205. doi: 10.1111/j.1365-2427.2009.02272.x
177
Qin H. Yu Z. Zhu Z. Lin Y. Xia J. Jia Y. (2022). The integrated analyses of metabolomics and transcriptomics in gill of GIFT tilapia in response to long term salinity challenge. Aquac. Fish.7, 131–139. doi: 10.1016/j.aaf.2021.02.006
178
Quan F. B. Gaillard A. L. Alejevski F. Pézeron G. Tostivint H. (2021). Urotensin II-related peptide (Urp) is expressed in motoneurons in zebrafish, but is dispensable for locomotion in larva. Peptides146, 170675. doi: 10.1016/j.peptides.2021.170675
179
Rahi M. L. Azad K. N. Tabassum M. Irin H. H. Hossain K. S. Aziz D. et al . (2021). Effects of salinity on physiological, biochemical and gene expression parameters of black tiger shrimp (Penaeus monodon): potential for farming in low-salinity environments. Biol. (Basel).10, 1220. doi: 10.3390/biology10121220
180
Raposo de Magalhães C. Farinha A. P. Blackburn G. Whitfield P. D. Carrilho R. Schrama D. et al . (2022). Gilthead seabream liver integrative proteomics and metabolomics analysis reveals regulation by different prosurvival pathways in the metabolic adaptation to stress. Int. J. Mol. Sci.23, 15395. doi: 10.3390/ijms232315395
181
Richards J. G. Semple J. W. Bystriansky J. S. Schulte P. M. (2003). Na+/K+-ATPase α-isoform switching in gills of rainbow trout (Oncorhynchus mykiss) during salinity transfer. J. Exp. Biol.206 (24), 4475–4486. doi: 10.1242/jeb.00701
182
Riera Romo M. Pérez-Martínez D. Castillo Ferrer C. (2016). Innate immunity in vertebrates: An overview. Immunology148 (2), 125–139. doi: 10.1111/imm.12597
183
Ronkin D. Seroussi E. Nitzan T. Doron-Faigenboim A. Cnaani A. (2015). Intestinal transcriptome analysis revealed differential salinity adaptation between two tilapiine species. Comp. Biochem. Physiol. - Part D Genomics Proteomics13, 35–43. doi: 10.1016/j.cbd.2015.01.003
184
Rønnestad I. Gomes A. S. Murashita K. Angotzi R. Jönsson E. Volkoff H. (2017). Appetite-controlling endocrine systems in teleosts. Front. Endocrinol. (Lausanne).8. doi: 10.3389/fendo.2017.00073
185
Rosenthal R. Milatz S. Krug S. M. Oelrich B. Schulzke J. D. Amasheh S. et al . (2010). Claudin-2, a component of the tight junction, forms a paracellular water channel. J. Cell Sci.123 (11), 1913–1921. doi: 10.1242/jcs.060665
186
Röthig T. Trevathan-Tackett S. M. Voolstra C. R. Ross C. Chaffron S. Durack P. J. et al . (2023). Human-induced salinity changes impact marine organisms and ecosystems. Glob. Change Biol.29, 4731–4749. doi: 10.1111/gcb.16859
187
Rottgen T. S. Nickerson A. J. Rajendran V. M. (2018). Calcium-activated cl– channel: insights on the molecular identity in epithelial tissues. Int. J. Mol. Sci.19, 1432. doi: 10.3390/ijms19051432
188
Ru X. Huang Y. Shi H. Peng Y. Hao R. Yang T. et al . (2023). RNA-seq of hypo- and hyper-salinity stress-response transcriptome in the liver of greater amberjack (Seriola dumerili) juveniles. Aquac. Rep.29, 101498. doi: 10.1016/j.aqrep.2023.101498
189
Ruiz-Jarabo I. Barany A. Jerez-Cepa I. Mancera J. M. Fuentes J. (2017). Intestinal response to salinity challenge in the Senegalese sole (Solea Senegalensis). Comp. Biochem. Physiol. -Part A Mol. Integr. Physiol.204, 57–64. doi: 10.1016/j.cbpa.2016.11.009
190
Saint-Criq V. Gray M. A. (2017). Role of CFTR in epithelial physiology. Cell. Mol. Life Sci.74 (1), 93–115. doi: 10.1007/s00018-016-2391-y
191
Santos L. H. Maulvault A. L. Jaén-Gil A. Marques A. Barceló D. Rodríguez-Mozaz S. (2023). Linking chemical exposure and fish metabolome: Discovering new biomarkers of environmental exposure of Argyrosomus regius to the antidepressant venlafaxine. Environ. Toxicol. Pharmacol.98, 104063. doi: 10.1016/j.etap.2023.104063
192
Schmidt V. T. Smith K. F. Melvin D. W. Amaral-Zettler L. A. (2015). Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol.24, 2537–2550. doi: 10.1111/mec.13177
193
Scott G. R. Richards J. G. Forbush B. Isenring P. Schulte P. M. (2004). Changes in gene expression in gills of the euryhaline killifish Fundulus heteroclitus after abrupt salinity transfer. Am. J. Physiol. Physiol.287, C300–C309. doi: 10.1152/ajpcell.00054.2004
194
Seale A. P. Stagg J. J. Yamaguchi Y. Breves J. P. Soma S. Watanabe S. et al . (2014). Effects of salinity and prolactin on gene transcript levels of ion transporters, ion pumps and prolactin receptors in Mozambique tilapia intestine. Gen. Comp. Endocrinol.206, 146–154. doi: 10.1016/j.ygcen.2014.07.020
195
Seli D. A. Prendergast A. Ergun Y. Tyagi A. Taylor H. S. (2024). High naCl concentrations in water are associated with developmental abnormalities and altered gene expression in zebrafish. Int. J. Mol. Sci.25, 4104. doi: 10.3390/ijms25074104
196
Semova I. Carten J. D. Stombaugh J. Mackey L. C. Knight R. Farber S. A. et al . (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe12, 277–288. doi: 10.1016/j.chom.2012.08.003
197
Shah V. S. Chivukula R. R. Lin B. Waghray A. Rajagopal J. (2022). Cystic fibrosis and the cells of the airway epithelium: what are ionocytes and what do they do? Annu. Rev. Pathol. Mech. Dis.17, 23–46. doi: 10.1146/annurev-pathol-042420-094031
198
Shih S. W. Yan J. J. Chou M. Y. Hwang P. P. (2023). Recent progress and debates in molecular physiology of Na+ uptake in teleosts. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1066929
199
Shih S.-W. Yan J.-J. Tsou Y.-L. Lu S.-W. Wang M.-C. Chou M.-Y. et al . (2022). In vivo functional assay in fish gills: exploring branchial acid-excreting mechanisms in zebrafish. Int. J. Mol. Sci.23, 4419. doi: 10.3390/ijms23084419
200
Shu Y. Lou Q. Dai Z. Dai X. He J. Hu W. et al . (2016). The basal function of teleost prolactin as a key regulator on ion uptake identified with zebrafish knockout models. Sci. Rep.6, 18597. doi: 10.1038/srep18597
201
Silva-Marrero J. I. Lai F. Handeland S. O. Pedrosa C. Gelebart V. Balseiro P. et al . (2025). Integration of gill and intestinal osmoregulatory functions to assess the smoltification window in atlantic salmon. Fishes10, 119. doi: 10.3390/fishes10030119
202
Simning D. Sepulveda M. De Guise S. Bosker T. Griffitt R. J. (2019). The combined effects of salinity, hypoxia, and oil exposure on survival and gene expression in developing sheepshead minnows, Cyprinodon variegatus. Aquat. Toxicol.214, 105234. doi: 10.1016/j.aquatox.2019.105234
203
Soengas J. L. Sangiao-Alvarellos S. Laiz-Carrión R. Mancera J. M. (2019). “Energy metabolism and osmotic acclimation in teleost fish,” in Fish Osmoregulation (CRC Press), 277–307. doi: 10.1201/b10994-10
204
Su H. Li Y. Ma D. Fan J. Zhong Z. Zhu H. (2023a). Metabolism responses in the intestine of Oreochromis mossambicus exposed to salinity, alkalinity and salt-alkalinity stress using LC-MS/MS-based metabolomics. Comp. Biochem. Physiol. Part D Genomics Proteomics45, 101044. doi: 10.1016/j.cbd.2022.101044
205
Su M. Liu N. Zhang Z. Zhang J. (2022). Osmoregulatory strategies of estuarine fish Scatophagus argus in response to environmental salinity changes. BMC Genomics23. doi: 10.1186/s12864-022-08784-2
206
Su H. Ma D. Fan J. Zhong Z. Li Y. Zhu H. (2023b). Metabolism response mechanism in the gill of Oreochromis mossambicus under salinity, alkalinity and saline-alkalinity stresses. Ecotoxicol. Environ. Saf.251, 114523. doi: 10.1016/j.ecoenv.2023.114523
207
Su H. Ma D. Zhu H. Liu Z. Gao F. (2020). Transcriptomic response to three osmotic stresses in gills of hybrid tilapia (Oreochromis mossambicus female × O. urolepis hornorum male). BMC Genomics21. doi: 10.1186/s12864-020-6512-5
208
Sullam K. E. Essinger S. D. Lozupone C. A. O’Connor M. P. Rosen G. L. Knight R. et al . (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol.21 (13), 3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x
209
Sun S. Gong C. Deng C. Yu H. Zheng D. Wang L. et al . (2023). Effects of salinity stress on the growth performance, health status, and intestinal microbiota of juvenile Micropterus salmoides. Aquaculture576, 739888. doi: 10.1016/j.aquaculture.2023.739888
210
Sun Z. Lou F. Zhang Y. Song N. (2020). Gill transcriptome sequencing and de novo annotation of acanthogobius ommaturus in response to salinity stress. Genes (Basel).11 (6), 631. doi: 10.3390/genes11060631
211
Sun X. N. Shen C. C. Feng G. P. Liu J. Y. Song C. Yu Y. F. (2022). Transcriptomic response to sudden salinity drop in the liver of juvenile scatophagus argus. Indian J. Anim. Res.1, 6. doi: 10.18805/ijar.bf-1496
212
Sylvain F.-É. Cheaib B. Llewellyn M. Gabriel Correia T. Barros Fagundes D. Luis Val A. et al . (2016). pH drop impacts differentially skin and gut microbiota of the Amazonian fish tambaqui (Colossoma macropomum). Sci. Rep.6, 32032. doi: 10.1038/srep32032
213
Takei Y. (2021a). The digestive tract as an essential organ for water acquisition in marine teleosts: lessons from euryhaline eels. Zool. Lett.7, 10. doi: 10.1186/s40851-021-00175-x
214
Takei Y. Hiroi J. Takahashi H. Sakamoto T. (2014). Diverse mechanisms for body fluid regulation in teleost fishes. Am. J. Physiol. - Regul. Integr. Comp. Physiol.307, R778–R792. doi: 10.1152/ajpregu.00104.2014
215
Takei Y. Hwang P. P. (2016). “Homeostatic responses to osmotic stress,” in Fish physiology. (Academic Press) Vol. 35, pp. 207–249. doi: 10.1016/B978-0-12-802728-8.00006-0
216
Takvam M. Wood C. M. Kryvi H. Nilsen T. O. (2021a). Ion transporters and osmoregulation in the kidney of teleost fishes as a function of salinity. Front. Physiol.12. doi: 10.3389/fphys.2021.664588
217
Takvam M. Wood C. M. Kryvi H. Nilsen T. O. (2023). Role of the kidneys in acid-base regulation and ammonia excretion in freshwater and seawater fish: implications for nephrocalcinosis. Front. Physiol.14. doi: 10.3389/fphys.2023.1226068
218
Taugbøl A. Solbakken M. H. Jakobsen K. S. Vøllestad L. A. (2022). Salinity-induced transcriptome profiles in marine and freshwater threespine stickleback after an abrupt 6-hour exposure. Ecol. Evol.12 (10), e9395. doi: 10.1002/ece3.9395
219
Thanh N. M. Jung H. Lyons R. E. Njaci I. Yoon B. H. Chand V. et al . (2015). Optimizing de novo transcriptome assembly and extending genomic resources for striped catfish (Pangasianodon hypophthalmus). Mar. Genomics23, 87–97. doi: 10.1016/j.margen.2015.05.001
220
Thompson E. Hensley J. Taylor R. (2022). Effect of high glucose and salinity on embryological development of zebrafish, brachyodanio rerio. FASEB J.36. doi: 10.1096/fasebj.2022.36.s1.r4243
221
Tian Y. Gao Q. Yu H. Liu D. Dong S. Zhou Y. et al . (2022). Dynamic transcriptome and LC-MS/MS analysis revealed the important roles of taurine and glutamine metabolism in response to environmental salinity changes in gills of rainbow trout (Oncorhynchus mykiss). Int. J. Biol. Macromol.221, 1545–1557. doi: 10.1016/j.ijbiomac.2022.09.124
222
Tian L. Tan P. Yang L. Zhu W. Xu D. (2020). Effects of salinity on the growth, plasma ion concentrations, osmoregulation, non-specific immunity, and intestinal microbiota of the yellow drum (Nibea albiflora). Aquaculture528. doi: 10.1016/j.aquaculture.2020.735470
223
Tipsmark C. K. (2008). Identification of FXYD protein genes in a teleost: Tissue-specific expression and response to salinity change. Am. J. Physiol. - Regul. Integr. Comp. Physiol.294 (4), R1367–R1378. doi: 10.1152/ajpregu.00454.2007
224
Tipsmark C. K. Madsen S. S. Seidelin M. Christensen A. S. Cutler C. P. Cramb G. (2002). Dynamics of Na+,K+,2Cl- cotransporter and Na+,K+-ATPase expression in the branchial epithelium of brown trout (Salmo trutta) and Atlantic salmon (Salmo salar). J. Exp. Zool.293 (2), 106–118. doi: 10.1002/jez.10118
225
Tipsmark C. K. Sørensen K. J. Madsen S. S. (2010). Aquaporin expression dynamics in osmoregulatory tissues of Atlantic salmon durings moltification and seawater acclimation. J. Exp. Biol.213 (3), 368–379. doi: 10.1242/jeb.034785
226
Tolas I. Zhou Z. Zhang Z. Teame T. Olsen R. E. Ringø E. et al . (2025). A fishy gut feeling – current knowledge on gut microbiota in teleosts. Front. Mar. Sci.11. doi: 10.3389/fmars.2024.1495373
227
Tresguerres M. Kwan G. T. Weinrauch A. (2023). Evolving views of ionic, osmotic and acid base regulation in aquatic animals. J. Exp. Biol.226 (14), jeb245747. doi: 10.1242/jeb.245747
228
Tseng Y.-C. Yan J.-J. Furukawa F. Chen R.-D. Lee J.-R. Tsou Y.-L. et al . (2022a). Teleostean fishes may have developed an efficient Na+ uptake for adaptation to the freshwater system. Front. Physiol.13. doi: 10.3389/fphys.2022.947958
229
Tümmler B. (2023). Puzzle resolved: CFTR mediates chloride homeostasis by segregating absorption and secretion to different cell types. J. Clin. Invest.133 (20). doi: 10.1172/JCI174667
230
Tweedley J. R. Dittmann S. R. Whitfield A. K. Withers K. Hoeksema S. D. Potter I. C. (2019). “Hypersalinity: Global Distribution, Causes, and Present and Future Effects on the Biota of Estuaries and Lagoons,” in Coasts and Estuaries (Elsevier), 523–546. doi: 10.1016/B978-0-12-814003-1.00030-7
231
Urbina M. A. Glover C. N. (2015). Effect of salinity on osmoregulation, metabolism and nitrogen excretion in the amphidromous fish, inanga ( Galaxias maculatus ). J. Exp. Mar. Bio. Ecol.473, 7–15. doi: 10.1016/j.jembe.2015.07.014
232
Vadboncoeur É. Nelson C. Clow K. A. Sandrelli R. M. Brauner C. J. Swanson A. K. et al . (2023). ‘Cold shock’ has few physiological effects on cultured Atlantic Salmon (Salmo salar) acclimated to low temperatures. Aquaculture577, 739900. doi: 10.1016/j.aquaculture.2023.739900
233
van Ravenzwaay B. Cunha G. C.-P. Leibold E. Looser R. Mellert W. Prokoudine A. et al . (2007). The use of metabolomics for the discovery of new biomarkers of effect. Toxicol. Lett.172, 21–28. doi: 10.1016/j.toxlet.2007.05.021
234
Vij S. Purushothaman K. Sridatta P. S. R. Jerry D. R. (2020). Transcriptomic analysis of gill and kidney from asian seabass (Lates calcarifer) acclimated to different salinities reveals pathways involved with euryhalinity. Genes (Basel).11, 733. doi: 10.3390/genes11070733
235
Vijayan M. M. Morgan J. D. Sakamoto T. Grau E. G. Iwama G. K. (1996). Food-deprivation affects seawater acclimation in tilapia: hormonal and metabolic changes. J. Exp. Biol.199, 2467–2475. doi: 10.1242/jeb.199.11.2467
236
Walker R. H. Smith G. D. Hudson S. B. French S. S. Walters A. W. (2020). Warmer temperatures interact with salinity to weaken physiological facilitation to stress in freshwater fishes. Conserv. Physiol.8, 1–18. doi: 10.1093/conphys/coaa107
237
Wang J. Jaramillo-Torres A. Li Y. Kortner T. M. Gajardo K. Brevik Ø. J. et al . (2021). Microbiota in intestinal digesta of Atlantic salmon (Salmo salar), observed from late freshwater stage until one year in seawater, and effects of functional ingredients: a case study from a commercial sized research site in the Arctic region. Anim. Microbiome3, 14. doi: 10.1186/s42523-021-00075-7
238
Wang Z. Lui G. C. S. Burton G. A. Leung K. M. Y. (2019). Thermal extremes can intensify chemical toxicity to freshwater organisms and hence exacerbate their impact to the biological community. Chemosphere224, 256–264. doi: 10.1016/j.chemosphere.2019.02.103
239
Wang A. R. Ran C. Ringø E. Zhou Z. G. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquac.10, 626–640. doi: 10.1111/raq.12191
240
Wang M. Yan Y. Liu W. Fan J. Li E. Chen L. et al . (2024). Proline metabolism is essential for alkaline adaptation of Nile tilapia (Oreochromis niloticus). J. Anim. Sci. Biotechnol.15, 142. doi: 10.1186/s40104-024-01100-w
241
Wei F. Sakata K. Asakura T. Date Y. Kikuchi J. (2018). Systemic homeostasis in metabolome, ionome, and microbiome of wild yellowfin goby in estuarine ecosystem. Sci. Rep.8 (1), 3478. doi: 10.1038/s41598-018-20120-x
242
Whitehead A. Roach J. L. Zhang S. Galvez F. (2012). Salinity- and population-dependent genome regulatory response during osmotic acclimation in the killifish ( Fundulus heteroclitus ) gill. J. Exp. Biol.215, 1293–1305. doi: 10.1242/jeb.062075
243
Whittamore J. M. (2012). Osmoregulation and epithelial water transport: Lessons from the intestine of marine teleost fish. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol.182 (1), 1–39. doi: 10.1007/s00360-011-0601-3
244
Wilson J. M. Laurent P. (2002). Fish gill morphology: inside out. J. Exp. Zool.293, 192–213. doi: 10.1002/jez.10124
245
Xia H. Chen H. Cheng X. Yin M. Yao X. Ma J. et al . (2022a). Zebrafish: an efficient vertebrate model for understanding role of gut microbiota. Mol. Med.28, 161. doi: 10.1186/s10020-022-00579-1
246
Xia J. H. Liu P. Liu F. Lin G. Sun F. Tu R. et al . (2013). Analysis of stress-responsive transcriptome in the intestine of Asian Seabass (Lates calcarifer) using RNA-seq. DNA Res.20 (5), 449–460. doi: 10.1093/dnares/dst022
247
Xiong Y. Dong S. Huang M. Li Y. Wang X. Wang F. et al . (2020). Growth, osmoregulatory response, adenine nucleotide contents, and liver transcriptome analysis of steelhead trout (Oncorhynchus mykiss) under different salinity acclimation methods. Aquaculture520, 734937. doi: 10.1016/j.aquaculture.2020.734937
248
Yada T. McCormick S. D. Hyodo S. (2012). Effects of environmental salinity, biopsy, and GH and IGF-I administration on the expression of immune and osmoregulatory genes in the gills of Atlantic salmon (Salmo salar). Aquaculture362, 177–183. doi: 10.1016/j.aquaculture.2010.12.029
249
Yamaguchi Y. Breves J. P. Haws M. C. Lerner D. T. Grau E. G. Seale A. P. (2018). Acute salinity tolerance and the control of two prolactins and their receptors in the Nile tilapia (Oreochromis niloticus) and Mozambique tilapia (O. mossambicus): A comparative study. Gen. Comp. Endocrinol.257, 168–176. doi: 10.1016/j.ygcen.2017.06.018
250
Yang S. Li D. Feng L. Zhang C. Xi D. Liu H. et al . (2023). Transcriptome analysis reveals the high temperature induced damage is a significant factor affecting the osmotic function of gill tissue in Siberian sturgeon (Acipenser baerii). BMC Genomics24 (1), 2. doi: 10.1186/s12864-022-08969-9
251
Yi H. Chen X. Liu S. Han L. Liang J. Su Y. et al . (2021). Growth, osmoregulatory and hypothalamic–pituitary–somatotropic (HPS) axis response of the juvenile largemouth bass (Micropterus salmoides), reared under different salinities. Aquac. Rep.20, 100727. doi: 10.1016/j.aqrep.2021.100727
252
Yohana M. A. Ray G. W. Qihui Y. Shiyu K. Tan B. Wu J. et al . (2024). Response of gut microbiota, antioxidation, and disease resistance to Pacific shrimp fed distiller’s dried grains with solubles replaced soybean meal. Aquac. Int.32 (6), 7551–7573. doi: 10.1007/s10499-024-01529-w
253
Yohana M. A. Ray G. W. Yang Q. Beiping T. Shuyan C. Junming D. (2023). Effect of corn gluten meal on the replacement of soybean meal on the survival, biochemical and metabolic responses, and disease resistance of Pacific white shrimp (Litopenaeus vannamei). Ann. Anim. Sci.24 (2), 575–591. doi: 10.2478/aoas-2023-0085
254
Yurinskaya V. E. Vereninov A. A. (2021). Cation-chloride cotransporters, na/K pump, and channels in cell water and ion regulation: in silico and experimental studies of the U937 cells under stopping the pump and during regulatory volume decrease. Front. Cell Dev. Biol.9. doi: 10.3389/fcell.2021.736488
255
Zarantoniello M. Bortoletti M. Olivotto I. Ratti S. Poltronieri C. Negrato E. et al . (2021). Article salinity, temperature and ammonia acute stress response in seabream (Sparus aurata) juveniles: A multidisciplinary study. Animals11. doi: 10.3390/ani11010097
256
Zarzar C. A. Fernandes T. J. Cardoso de Oliveira I. R. (2023). Modeling the growth of Pacific white shrimp (Litopenaeus vannamei) using the new Bayesian hierarchical approach based on correcting bias caused by incomplete or limited data. Ecol. Inform.77, 102271. doi: 10.1016/j.ecoinf.2023.102271
257
Zeng A. Tan K. Gong P. Lei P. Guo Z. Wang S. et al . (2020). Correlation of microbiota in the gut of fish species and water. 3 Biotech.10, 472. doi: 10.1007/s13205-020-02461-5
258
Zhang M. Sun Y. Liu Y. Qiao F. Chen L. Liu W.-T. et al . (2016). Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture454, 72–80. doi: 10.1016/j.aquaculture.2015.12.014
259
Zhang X. Wen H. Wang H. Ren Y. Zhao J. Li Y. (2017). RNA-Seq analysis of salinity stress–responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus). PloS One12, e0173238. doi: 10.1371/journal.pone.0173238
260
Zhao H. Wang Q. Zhao H. Chen C. (2022). Transcriptome profiles revealed high- and low-salinity water altered gill homeostasis in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D Genomics Proteomics42, 100989. doi: 10.1016/j.cbd.2022.100989
261
Zhao Y. Zhang C. Zhou H. Song L. Wang J. Zhao J. (2020). Transcriptome changes for Nile tilapia (Oreochromis niloticus) in response to alkalinity stress. Comp. Biochem. Physiol. - Part D Genomics Proteomics33, 100651. doi: 10.1016/j.cbd.2019.100651
262
Zhu Q. Li M. Lu W. Wang Y. Li X. Cheng J. (2023a). Transcriptomic Modulation Reveals the Specific Cellular Response in Chinese Sea Bass (Lateolabrax maculatus) Gills under Salinity Change and Alkalinity Stress. Int. J. Mol. Sci.24, 5877. doi: 10.3390/ijms24065877
263
Zhu Y. Negishi R. Fukunaga K. Udagawa S. Shimabukuro A. Takemura A. (2023b). Activation of the growth–IGF-1 axis, but not appetite, is related to high growth performance in juveniles of the Malabar grouper, Epinephelus malabaricus, under isosmotic condition. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol.283, 111456. doi: 10.1016/j.cbpa.2023.111456
264
Zimmer A. M. Mandic M. Yew H. M. Kunert E. Pan Y. K. Ha J. et al . (2021). Use of a carbonic anhydrase Ca17a knockout to investigate mechanisms of ion uptake in zebrafish ( Danio rerio ). Am. J. Physiol. Integr. Comp. Physiol.320, R55–R68. doi: 10.1152/ajpregu.00215.2020
265
Zimmer A. M. Shir-Mohammadi K. Kwong R. W. M. Perry S. F. (2020). Reassessing the contribution of the Na+/H+ exchanger Nhe3b to Na+ uptake in zebrafish ( Danio rerio ) using CRISPR/Cas9 gene editing. J. Exp. Biol.223 (2), jeb215111. doi: 10.1242/jeb.215111
266
Zuloaga R. Ahumada-Langer L. Aedo J. E. Molina A. Valdés J. A. (2024). 11-deoxycorticosterone (DOC)’s action on the gill osmoregulation of juvenile rainbow trout (Oncorhynchus mykiss). Biol. (Basel).13, 107. doi: 10.3390/biology13020107
Summary
Keywords
salinity adaptation, teleost fish, osmoregulation, gut microbiota, metabolomics
Citation
Mkulo EM, Iddrisu L, Yohana MA, Zheng A, Zhong J, Jin M, Danso F, Wang L, Zhang H, Tang B, Zhou H, Amoah K, Huang J, Wang B and Wang Z (2025) Exploring salinity adaptation in teleost fish, focusing on omics perspectives on osmoregulation and gut microbiota. Front. Mar. Sci. 12:1559871. doi: 10.3389/fmars.2025.1559871
Received
13 January 2025
Accepted
21 April 2025
Published
20 May 2025
Volume
12 - 2025
Edited by
Edward Mager, University of North Texas, United States
Reviewed by
Yaqiu Liu, Chinese Academy of Fishery Sciences, China
Amit Ranjan, Tamil Nadu Fisheries University, India
Updates
Copyright
© 2025 Mkulo, Iddrisu, Yohana, Zheng, Zhong, Jin, Danso, Wang, Zhang, Tang, Zhou, Amoah, Huang, Wang and Wang.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongliang Wang, wangzl@gdou.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.