Abstract
Deep learning has massive potential in predicting phenotype from different omics profiles. However, deep neural networks are viewed as black boxes, providing predictions without explanation. Therefore, the requirements for these models to become interpretable are increasing, especially in the medical field. Here we propose a computational framework that takes the gene expression profile of any primary cancer sample and predicts whether patients’ samples are primary (localized) or metastasized to the brain, bone, lung, or liver based on deep learning architecture. Specifically, we first constructed an AutoEncoder framework to learn the non-linear relationship between genes, and then DeepLIFT was applied to calculate genes’ importance scores. Next, to mine the top essential genes that can distinguish the primary and metastasized tumors, we iteratively added ten top-ranked genes based upon their importance score to train a DNN model. Then we trained a final multi-class DNN that uses the output from the previous part as an input and predicts whether samples are primary or metastasized to the brain, bone, lung, or liver. The prediction performances ranged from AUC of 0.93–0.82. We further designed the model’s workflow to provide a second functionality beyond metastasis site prediction, i.e., to identify the biological functions that the DL model uses to perform the prediction. To our knowledge, this is the first multi-class DNN model developed for the generic prediction of metastasis to various sites.
1 Introduction
Precision medicine is a path that could profoundly change and improve medical practices. This idea proposes using genetic data of individual patients to enhance clinical decision-making, and “omics” technologies now provide a means to acquire such patient data, making precision medicine feasible. Clinical decision-making includes diagnosis, prognosis, choosing the most appropriate treatment, etc. One avenue pursued to support clinical decision-making is building classifiers using gene expression profiles that can function as forms of artificial intelligence (AI).
Many machine learning methods, including support vector machines, random forest, and boosting, are among the primary tools currently being used to make biological discoveries from the vast amount of available gene expression data (Libbrecht and Noble, 2015). However, deep learning (DL) is emerging as a more powerful machine learning method (Goodfellow et al., 2016), although the primary DL application domain is image recognition and speech recognition. Nonetheless, DL is showing promise in many other fields of science, especially in precision medicine and genomics data analysis (Grapov et al., 2018; Martorell-Marugán et al., 2019), as DL can extract intricate structures in high-dimensional data (Najafabadi et al., 2015). However, DL is still new in the bioinformatics community; thus, only a few published works show its application to gene expression-based models (Daoud and Mayo, 2019). Furthermore, unlike images or text data, gene expression data has no clear structure that we can exploit in a neural network architecture. Thus, many new architectures are surfacing for metastasis prediction from gene expression data, such as multilayer perceptron architecture (Albaradei et al., 2019; Albaradei et al., 2021a; Albaradei et al., 2021b) , autoencoder architectures (Sharifi-Noghabi et al., et al.; Albaradei et al., 2021c; Fakoor et al., 2013) and Graph deep learning (Xu et al., 2021). Most of these proposed models try to solve a binary classification problem that classifies samples as metastatic or non-metastatic (Albaradei et al., 2021a). However no generic computational framework based on DL that accepts raw gene expression data to predict whether cancer is primary or has spread to various metastasis sites exists.
The main concern of DL used in medical applications is the lack of interpretability. The reason being, DL networks can be viewed as black boxes that form an input layer (wherein we place the gene expression profile of patients) and an output layer (offering predictions without interpretability). Suppose we do not meet this interpretability criterion at a good standard. In that case, physicians will not be able to trust the decision of the neural network, as they need interpretable data to ensure patients’ safety. Specifically, they need data about neurons, genes, and related biological processes involved in the prediction and the decision-making process to make informed decisions. Thus, researchers are now attempting to make the DL networks more interpretable.
In this work, we attempted to develop an AI method that could translate into a tool that supports clinical decision-making with regard to identifying metastasis and pinpointing the metastasis site (Figure 1). In this process, we also show the biological functions that the model uses to perform the prediction. That is, current work that interprets DL models identifies the genes that impact the prediction. Here, we propose interpreting the hidden neurons by linking the neurons to the enriched biological functions. In this work, we developed such a DL model. The DL framework takes as input raw gene expression data for a sample and predicts whether it is primary or metastasized to the brain, bone, lung, or liver. In the first phase, we used AutoEncoder (AE) to reduce the dimension of the expression data. Then, we applied DeepLIFT to compute an importance score (i.e., the impact of each input layer neuron on the latent layer neurons) used to rank the genes. Finally, to mine the genes that can distinguish the primary and tumor samples metastasized to different sites, we iteratively fed ten top-ranked genes (based upon the importance score) to the DNN model for training. In the second phase, we trained and evaluated a final multi-class DNN model to make the metastasis site predictions. Here, we also used the DeepLIFT approach to identify the essential neurons that lead to the prediction and the set of genes that activate these critical neurons. Then, we linked these critical genes to Gene Ontology (GO). We also provided analyses using Molecular Signatures Database (MSigDB) and the Disease Gene Network (DisGeNet) to support and increase the biology extracted from the essential neurons’ list of genes.
FIGURE 1

General overview of the proposed computational framework that takes the gene expression profile of any primary cancer sample and predicts whether patients’ samples are primary (localized) or had been metastasized to brain, bone, lung, or liver based on deep learning architecture.
2 Method and materials
2.1 Gene expression datasets
We searched for gene expression datasets in Gene Expression Omnibus (GEO) (Edgar et al., 2002) using the following query: “metastas* AND (bone OR brain OR lung OR liver) AND Homo sapiens” filtered by “Expression profiling by array” in September 2021. We retrieved 837 entries which we sifted through and found microarray gene expression data for primary tumors (breast, colorectal, kidney, liver, lung, pancreatic, and prostate cancer samples), and tumors metastasized from these primary tumors to the bone, brain, lung, or liver. Table 1 provides the GEO accession numbers of the samples used in this study, along with the sample statistics. Similar to the approach used in (Chereda et al., 2019), we used the RMA probe-summary algorithm (Irizarry et al., 2003) to process each dataset, after which they were combined based on the HG-U133A array probe names, and quantile normalization was applied across all datasets. In cases where multiple probes were mapped to one gene, the probe with the highest average value was taken. Finally, we used the integrated datasets for each of the four sites as input for the DL models. However, before we fed the data to the DL model, we used the synthetic minority oversampling technique (SMOTE) to oversample the minority class using the imbalanced-learn python library (Chawla et al., 2002), as the number of samples is imbalanced between the primary and metastasized group.
TABLE 1
| Bone | Brain | Lung | Liver | |
|---|---|---|---|---|
| Breast | 220 Primary, | 27 Primary, | 47 Primary, | 28 Primary, |
| 72 Metastasized [GSE 2034, GSE137842] | 65 Metastasized [GSE12276, GSE125989, GSE46928, GSE18549] | 18 Metastasized [GSE16554, GSE5327] | 16 Metastasized [GSE18549] | |
| Colorectal | 0 | 10 Primary, | 186 Primary, | 219 Primary, |
| 23 Metastasized [GSE14108] | 47 Metastasized [GSE18549, GSE41258] | 86 Metastasized [GSE41258, GSE18549, GSE6605] | ||
| Kidney | 0 | 0 | 10 Primary, | 0 |
| 10 Metastasized [GSE22541] | ||||
| Liver | 0 | 0 | 31 Primary, | 0 |
| 31 Metastasized [GSE141016] | ||||
| Lung | 14 Primary, | 15 Primary, | 0 | 0 |
| 19 Metastasized [GSE10096] | 23 Metastasized [GSE18549] | |||
| Pancreas | 0 | 0 | 0 | 15 Primary, |
| 14 Metastasized [GSE19279] | ||||
| Prostate | 16 Primary, | 0 | 0 | 0 |
| 17 Metastasized [GSE18549, GSE43332] |
The gene expression datasets from GEO with the number of primary and metastasized samples for each site.
2.2 Deep learning framework
The first part of our model’s framework comprises three key components, namely the AE (Hinton and Salakhutdinov, 2006), DeepLIFT (Shrikumar et al., 2017), and the deep neural network (DNN) (Svozil et al., 1997) (Figure 2).
FIGURE 2

The workflow represents the first part of our model’s framework. (A) The architecture of AutoEncoder, (B) Applying DeepLIFT to compute the importance scores in the Encoder network, (C) Using DNN as a baseline method to perform the metastasis prediction.
First, the AE-based component is an unsupervised deep neural network with multiple stacked hidden layers composed of two parts, an encoder, and a decoder. The encoder maps the original (high-dimensional) data to a reduced representation (100 dimensions) through the bottleneck layer. The purpose of the decoder is to reconstruct the original data from the low-dimensional representation by minimizing the difference between and . In this manner, the AE extracts features that differ from the original features and functions as a feature extraction method. We used the Python Keras library (https://github.com/fchollet/keras) to implement an AE consisting of three fully connected hidden layers containing 500, 100, and 500 neurons. For each layer, we used “relu” as the activation function. Given m samples, each has a gene expression profile containing n genes; the input vector is reconstructed through a series of matrix transformations of multiple network layers. Training an AE involves finding parameters that minimize a specific loss function; we used mean absolute error (MAE) as the loss function. In addition, we added an L2 regularization penalty to control overfitting and used the early stopping technique. Finally, we trained the AE using the Adam (Kingma and Ba, 2014) optimization algorithm with 500 epochs and a 10% dropout.
Second, the DeepLIFT-based component is a feature scoring algorithm to calculate the contribution scores of each neuron. In our computing framework, we used DeepLIFT to calculate a contribution score for every gene of each input sample. The obtained contribution scores express the importance of the corresponding genes for the compression features of the low-dimensional representation (bottleneck) layer. Then, we ranked the genes based on their importance scores.
Third, the DNN-based component is a neural network with three hidden layers with 64, 32, and 8 neurons, respectively, and uses “relu” as the activation function. We used the Python Keras library to design the DNN model to predict if a sample is primary or metastasized. Finally, we iteratively added ten top-ranked genes (based on the importance scores) to train the DNN model.
The second part of our model uses the output from the first part, i.e., the most important genes for all sites, as an input to the final multi-class DNN model (Figure 3). This multi-class DNN consists of three hidden layers, each with 100 neurons, and uses a “relu” activation layer followed by an output layer with five output neurons (one for each class: primary, and metastasized to bone, brain, lung, or liver) that use the soft-max function to do the prediction. We then used the DeepLIFT to identify the most relevant neurons in each hidden layer for each of the five predictions (see 2.3 for details). Finally, the model was implemented in Python v.3.6 scripting language (https://www.python.org/), using the Keras deep learning and DeepLIFT frameworks (Figures 2B,C). Concerning time complexity, the time needed to train the model was 20.4 min for around 100 epochs for all samples using a workstation with Linux Ubuntu 18.04.5 LTS Intel Xeon Platinum 8,176, 64-bit OS and two GPUs: Quadro and Titan, with CUDA version 11.0.
FIGURE 3

The workflow represents the second part of our model’s framework, which determines the significant neurons in the network to predict metastasis status.
2.3 Identifying the biological functions that the DL model uses to perform the prediction
We interpreted the prediction for each class by first computing the relevance scores through the DL network and identifying the most essential neurons that allow predicting the class. Then, we connected each important neuron with the list of the input genes affecting the neuron activation. In this manner, we associated biological functions with each layer based on its essential neurons.
The first step is to identify the neurons that most influence the predictions for each class (Bach et al., 2015; Hanczar et al., 2020). For this, we computed the relevance scores R of all neurons using the Deep-LIFT approach for each predicted class at each layer. Next, we used the mean of these relevance scores to obtain the average relevance of neuron i in layer L, representing this neuron’s influence on the DL network to predict the class. The relevance score for neuron i in layer L is defined as the sum of incoming scores from each neuron j in layer L+1.
Finally, we ranked the neurons according to their average relevance scores and chose the most essential ones. Similar to (Hanczar et al., 2020), assuming that the average relevance scores follow a Gaussian distribution, we used the two-side t-test (p-value at 0.05 ) to determine each class’s most essential neurons in each layer.
For a given important neuron in layer L, its activation is back propagated using the Deep-LIFT approach to compute the relevance score of each input gene. We then identified the most critical inputs that have an impact on the activation of the neuron. Similar to identifying the essential neurons, we used a two-sided t-test to select the essential input genes.
The second step is to connect each essential neuron to biological functions from GO, signature gene-set from MSigDB, and diseases from DisGeNET. Finally, we used an R interface to the Enrichr database EnricherR (Kuleshov et al., 2016) to identify the over-represented functions in the list of genes connected with each important neuron.
3 Results and discussion
3.1 Determining the gene set that provides optimal prediction performance
In the first part of our model’s framework, we used GEO samples to train an AE and applied DeepLIFT to calculate importance scores for each gene for ranking. Then, ten top-ranked genes (based on the importance scores) were iteratively fed to the DNN model to identify the gene set that provides maximum performance when determining if a sample is primary or metastasized. The DNN reaches its maximum performance when including 60, 80, 20, and 30 top-ranked genes in metastasis to bone, brain, lung, and liver data, respectively (Figure 4). For the metastasis to bone, lung, and liver samples, the DNN achieved an AUC of 1.0. However, the DNN could only achieve an AUC of 0.9597 for the metastasis to brain samples. This might result from the brain samples having less than 50 primary samples, while the metastasis to bone, lung, and liver samples were analyzed using more than 200 primary samples. Adding weight to this suggestion is the number of metastasized samples used for each site being relatively the same (about 100 samples each).
FIGURE 4

AUC is based on different numbers of featured genes using DNN for bone, brain, lung, and liver sites. AUC is indicated in blue, while error rate is shown in red.
3.2 Cross-site generalization analysis
After removing the duplicates, the 190 essential genes for all sites identified in the part of the model’s framework were reduced to 184 genes. The 184 genes were used as an input to the final multi-class DNN model. This model takes these 184 genes and predicts if the input samples are primary or metastasized to the bone, brain, lung, or liver site. Figure 5 provides the prediction performance for the final multi-class DNN model. The best prediction performance was achieved for the primary samples (AUC of 0.93), followed by the metastasis to bone and lung samples (AUC of 0.88). The metastasis to liver and brain samples achieved lower prediction performances with an AUC of 0.84 and 0.82, respectively. Here, we expected the prediction performance for metastasis to the brain to be the lowest, based on the maximum performance the DNN achieved in Figure 3. However, the final multi-class DNN model achieved a more than acceptable prediction performance in all categories.
FIGURE 5
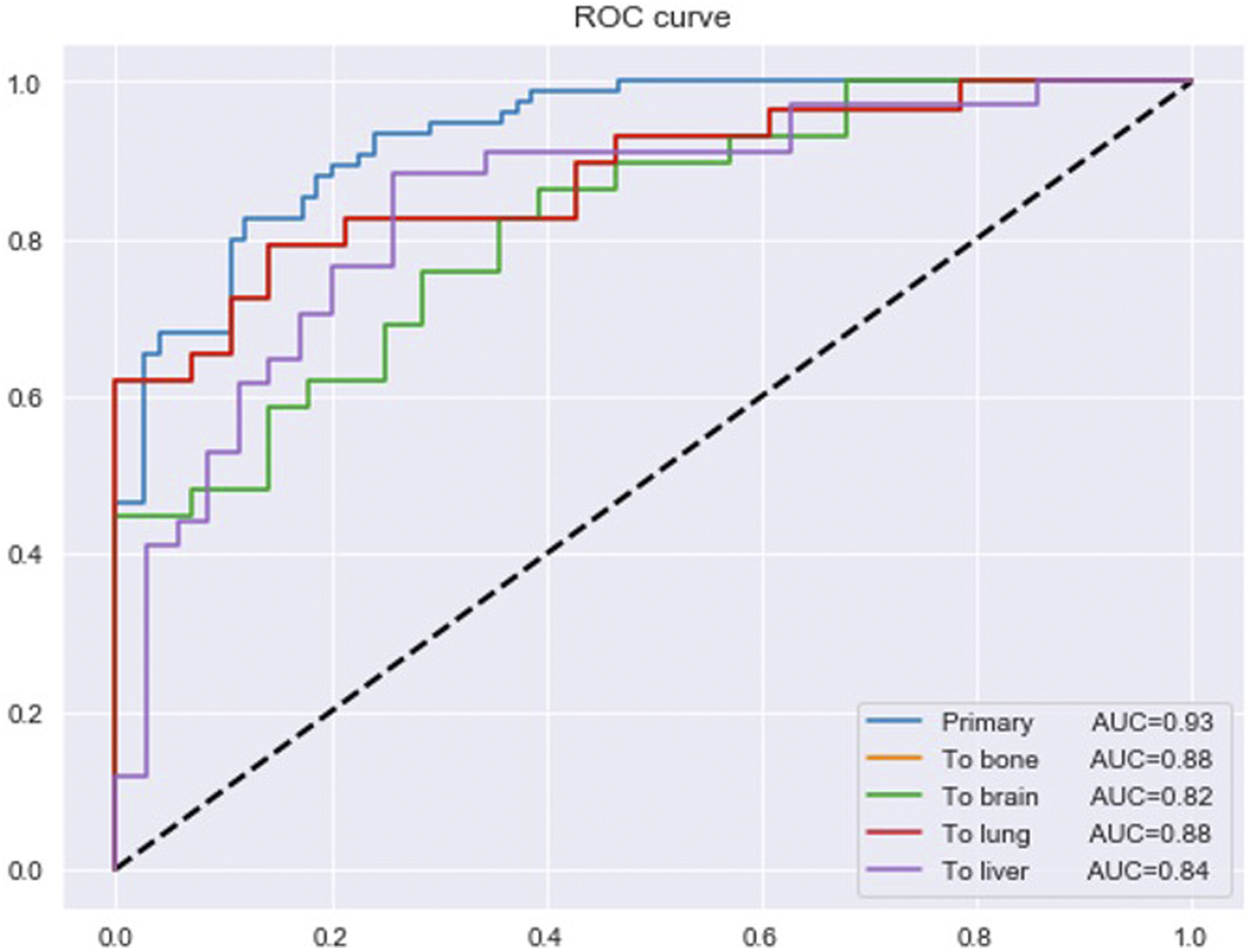
The prediction performance of the final multi-class DNN model.
3.2.1 Testing the robustness of the final DNN model
The final multi-class DNN model achieved a good prediction performance; however, the prediction performance does not indicate the robustness of the final DNN model. Thus, we further evaluated the model’s performance using external testing data from the TCGA datasets (Figure 6) and using a population-based cohort (Figure 7). Also, by using external datasets as a validation technique to show how accurately our predictive model will perform in practice, we eliminate any concerns about over/under -fitting. First, the external set was extracted from the human cancer metastasis database (HCMDB) (Zheng et al., 2018), where we found 378 samples, 250 primary, and 21,2, 44, and 61 were metastasized to bone, brain, lung, and liver (see the complete list of TCGA IDs in Supplementary Table S1), respectively. In addition, we used the gene expression profiles of fresh breast cancer tissue of 45 (21 primary and 24 metastasized) Saudi-Arabian subjects deposited on GSE36295 to test the performance of our model on a population-based cohort (real data).
FIGURE 6
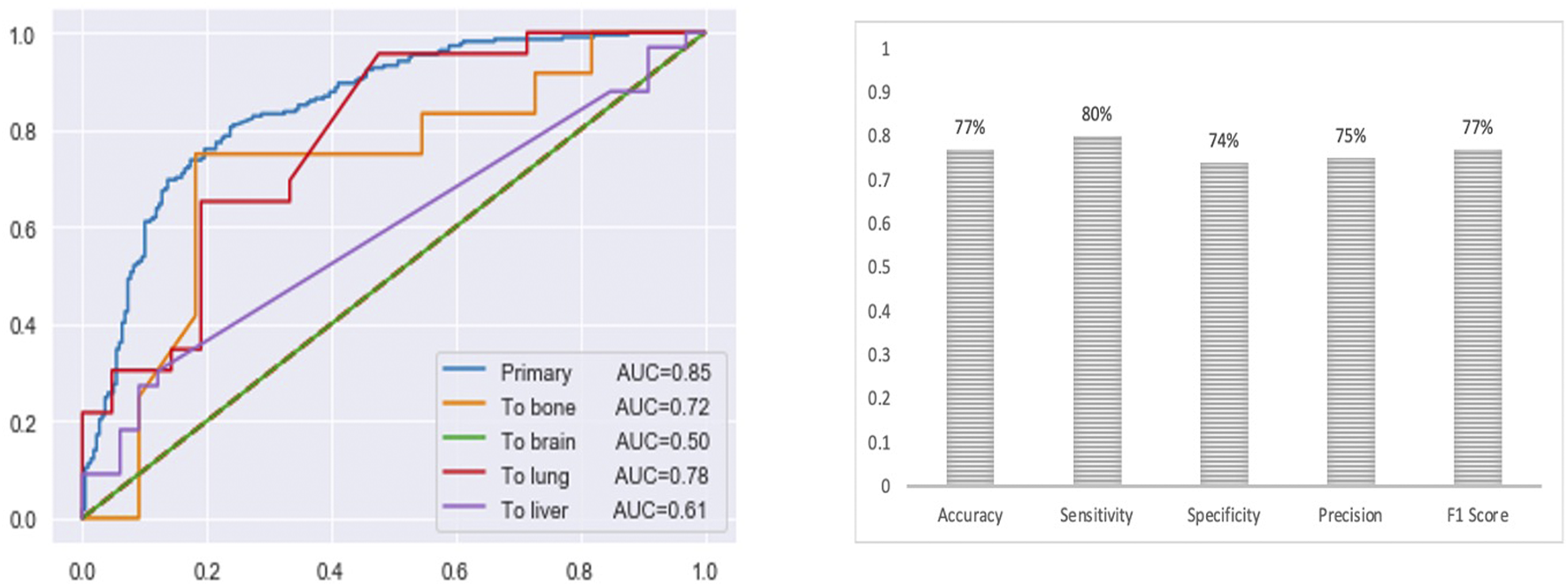
The prediction performance of the final multi-class DNN model using external testing data from the TCGA datasets. Note, for the brain there are only 2 samples in the test set).
FIGURE 7

The prediction performance of the final multi-class DNN model using a specific population-based cohort.
Figure 6 provides the prediction performance using the external set in terms of the area under the ROC curve and shows several other metrics, including accuracy, sensitivity, specificity, precision, and F1 score, ranging between 74%–80%. The prediction performance using the external data followed the same trend with the highest prediction performance achieved for the primary (AUC of 0.85) samples followed by the metastasis to lung (AUC of 0.78), bone (AUC of 0.72), liver (AUC of 0.61) and brain (AUC of 0.50) samples, respectively. This result shows that the multi-class model exhibits robustness concerning the three categories: the primary and metastasis to lung and bone samples. However, the prediction performance for the metastasis to the brain and liver samples dropped by 32% and 23 %, respectively. This suggests that we may have to re-establish the gene set that provides maximum performance using a larger cohort of samples (when the samples become available). Beyond that, here it should also be taken into consideration that for the brain we only had two samples in the test set.
Nonetheless, the prediction performance using samples from a population-based cohort shows that the multi-class DNN model achieved good prediction performance based on area under the ROC curve (AUC of 0.72), when distinguishing between the primary and metastatic samples (Figure 6), and shows several other metrics, including accuracy, sensitivity, specificity, precision, and F1 score, ranging between 73%–85%. This result gives an indication of the potential of our model to accurately predict metastasis sites.
3.2.2 The biological functions associated with the genes used by the DL model to perform the prediction
We further designed the model’s workflow to provide a second functionality beyond metastasis site prediction, i.e., to identify the biological functions that the DL model uses to perform the prediction (Figure 8).
FIGURE 8

The biological interpretation of our deep neural network approach.
We achieved this through the biological interpretation of the neural network predicting the metastasis. That is, for each class, the essential neurons of each layer are selected based on the mean of the relevance scores by using the method described in Section 2.3. We identified 89, 56, 41, 16, and 53 essential neurons in hidden layer one for primary, metastasized to bone, brain, lung, and liver, respectively. We also identified 36, 40, 99, 11, and 18 essential neurons in hidden layer two for primary, metastasized to bone, brain, lung, and liver, respectively. Finally, we identified 54, 48, 22, 84, and 35 essential neurons in hidden layer three for primary, metastasized to bone, brain, lung, and liver, respectively.
For each essential neuron’s list of genes, we determined GO biological functions based on a p-value < 0.05. Figure 9 provides the GO biological functions associated with the list of genes used to differentiate the primary samples from the metastasized ones. The critical neurons in each layer can be grouped depending on the functions enriched among the significant genes they contain. Overall, the enriched functions in layer one belonged to five main categories: “Metabolic process,” “Cellular process,” “Immune response,” “Transport”, and “Cell cycle.” The enriched functions from the essential neurons of layer two included “Adaptive thermogenesis,” “Extracellular matrix disassembly,” “Regulation of transport,” and “Regulation of cell motility.” The enriched functions from the essential neurons of layer three belonged to “Cell-cell adhesion,” “Ion transport,” “Apoptotic process.” The first layer exhibits more general function categories but more specific functions are appearing in subsequent neural network layers.
FIGURE 9
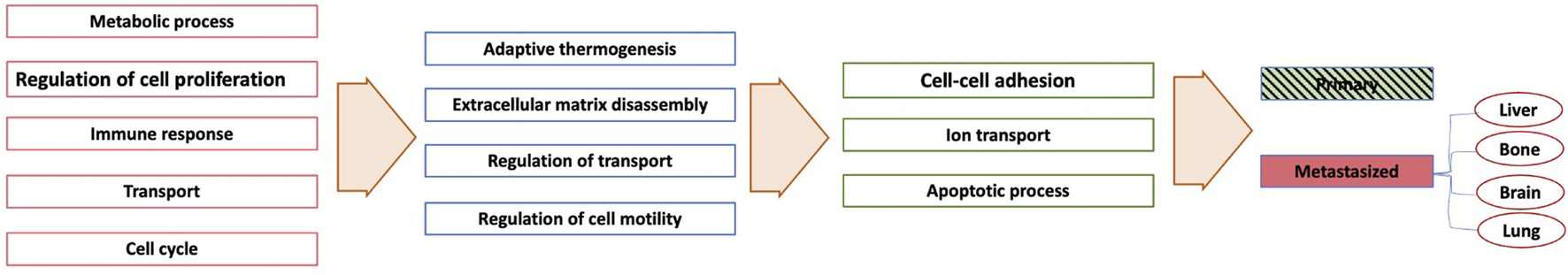
A simplified network showing each layer’s enriched pathways based only on the metastasis sites.
To support and increase the biological insights extracted from the essential neurons’ list of genes, we also performed MSigDB enrichment. In this analysis, we only considered MSigDB enrichments significant to at least three metastasis sites. Only three enriched categories were significant to all four sites, namely “Epithelial Mesenchymal Transition,” “Apoptosis,” and “IL-2/STAT5 Signaling” (Table 2). The apoptosis category is enriched based on the interpretation of the neural network (Figure 9) and the MSigDB enrichment. This is interesting as metastasis cells are subjected to various apoptotic stimuli and epithelial-mesenchymal transition (EMT) (which also features in the MSigDB enrichment) allows a polarized epithelial cell to undergo several biochemical changes to become a mesenchymal cell phenotype with enhanced resistance to apoptosis and increased migratory capacity and invasiveness and production of ECM components (Jason et al., 2003; Kalluri and Weinberg, 2009). Specifically, extracellular matrix disassembly (a GO biological function highlighted by the neural network) enzymes facilitates the remodeling of the extracellular matrix to create a microenvironment in the distant organ that promotes metastasis (Scheau et al., 2019; Winkler et al., 2020). Cell-cell adhesion, another GO biological function highlighted by the neural network, is also a key element of metastasis. For example, it has been shown that S100A8/A9 from tumor cells bind to RAGE on myeloid-derived suppressor cells (MDSCs) and promotes the migration and accumulation of MDSC, while periostin from MDSCs participates in pre-metastatic niche (PMN) formation through promoting extracellular matrix remodeling to facilitate the metastatic colonization of disseminated tumor cells (Cheng et al., 2008; Sinha et al., 2008; Wang et al., 2016). Overall, these biological functions suggest the gene lists used in the DL model to perform the prediction are to a large extent metastasis-specific and can be used to retrieve metastasis-specific biological functions beyond its metastasis site prediction capabilities (Sinha et al., 2008).
TABLE 2
| p-value | ||||
|---|---|---|---|---|
| MSigDB | Bone | Brain | Lung | Liver |
| Allograft rejection | 2.60E-02 | 5.27E-03 | Na | 3.60E-03 |
| Apoptosis | 2.15E-02 | 2.00E-03 | 2.09E-02 | 2.15E-02 |
| Coagulation | 1.41E-02 | na | 1.82E-02 | 1.88E-02 |
| DNA repair | 1.53E-02 | na | 1.27E-02 | 2.02E-02 |
| Epithelial mesenchymal transition | 2.60E-02 | 1.98E-02 | 1.66E-02 | 2.60E-02 |
| Glycolysis | 2.60E-02 | na | 2.53E-02 | 3.11E-02 |
| IL-2/STAT5 signaling | 2.59E-02 | 2.02E-03 | 2.52E-02 | 2.59E-02 |
| Interferon alpha response | na | 1.39E-03 | 8.68E-04 | 1.39E-03 |
| mTORC1 signaling | 2.60E-02 | na | 1.66E-02 | 2.60E-02 |
| Oxidative phosphorylation | 3.60E-03 | na | 1.37E-03 | 3.28E-04 |
| UV response up | 2.12E-02 | 1.63E-02 | 2.06E-02 | Na |
MSigDB enrichment analysis.
Enrichment associations that are significant to at least three sites
We also performed DisGenNET enrichment. In this analysis, we only considered DisGenNET disease enrichments significant to at least two metastasis sites. Only eight enriched disease categories were significantly associated with at least two metastasis sites, namely Autoimmune Diseases, Carcinoma breast stage IV, Cirrhosis, Dermatomyositis, Giant Cell Tumors, Leukemia, Metastatic malignant neoplasm to brain, and Rheumatoid Arthritis (Table 3). Four disease categories are associated with cancer, and noteworthy is the late-stage and metastasized cancer that is being picked up. Beyond this, Dermatomyositis (Luu et al., 2015), Rheumatoid Arthritis (Racanelli et al., 2008), and Autoimmune Diseases (Milkiewicz et al., 1999) are recognized paraneoplastic syndromes, which are symptoms that occur at sites distant from a tumor or its metastasis site (Pelosof and Gerber, 2010). In addition, several of our differentially expressed genes, including HLA-DMA, SOCS1, HLA-C, CTNNB1, KRAS, MET, and CD244, are associated with Liver Cirrhosis (Knouse et al., 2019), CD79A, HLA-DMA, SOCS1, HLA-B, HLA-C, IFI35, CD68, MET, PTHLH, CD244, and C2 with Rheumatoid Arthritis (Roy et al., 2011), HLA-B, HPRT1, and C2 with dermatomyositis (Bonnetblanc et al., 1990), and RB1, HLA-DMA, CXCR4, and CTGF with Cirrhosis (Shah and Casciola-Rosen, 2015). We also have several genes, including FTO, HLA-DMA, GAP43, SCN8A, HLA-C, CD68, and CDR2, associated with Multiple Sclerosis (Plantone et al., 2015), which suggest Multiple Sclerosis and Cirrhosis may possibly be a paraneoplastic syndrome that arises with metastasis.
TABLE 3
| p-value | ||||
|---|---|---|---|---|
| DisGeNET | Bone | Brain | Lung | Liver |
| Autoimmune Diseases | 5.12E-04 | na | 6.43E-05 | Na |
| Carcinoma breast stage IV | 2.68E-05 | na | 4.90E-04 | Na |
| Cirrhosis | 3.80E-04 | na | 0.00E+00 | 1.37E-02 |
| Dermatomyositis | 2.57E-05 | 2.57E-05 | Na | Na |
| Giant Cell Tumors | 5.69E-06 | na | 7.84E-08 | Na |
| leukemia | 7.13E-05 | 7.13E-05 | Na | Na |
| Metastatic malignant neoplasm to brain | 1.84E-04 | na | 3.43E-07 | Na |
| Rheumatoid Arthritis | 4.84E-05 | na | 2.71E-06 | Na |
DisGeNET enrichment analysis.
Disease associations that are significant to at least two sites
We further determined the overlapping genes between the primary and metastasis samples for the four sites. This analysis includes only the genes used by the DL to perform the classification. If we only considered genes common to at least three sites, we found the products of two genes, HIP1 and LARP4, with expression levels downregulated in the primary samples but upregulated in the metastasis samples. HIP1 was used by the DL to predict metastasis to the bone, brain, and lung, while LARP4 was used to predict metastasis to the brain, lung, and liver. This is interesting as HIP1 is one of the essential proteins involved in clathrin-mediated endocytosis (CME) (Chang et al., 2015), and crosstalk between CLCb/Dyn1-mediated adaptive CME and epidermal growth factor receptor (EGFR) signaling increases metastasis (Chen et al., 2017). Also, LARP4, a known RNA-binding protein (RBP) (Yang et al., 2011; Chothani et al., 2019) that repress or activate the translation of target genes, change the cell shape (which has been correlated with metastatic potential) and LARP4 depletion increases cell migration and invasion (Lyons et al., 2016; Seetharaman et al., 2016). Other proteins also upregulated and common to at least three sites (but do not appear in the primary samples gene list) include CC2D1A (Kumar et al., 2019), CD68 (Huang et al., 2018), EFCAB1 (Fagone et al., 2017), HLA-DMA (Li et al., 2020a), PRAME (Huang et al., 2016; Al-Khadairi et al., 2019), and ULBP2 (Paschen et al., 2009), all of which was linked to metastasis in previous studies. In fact, 87 % of the essential genes are associated with metastasis-related functions based on the current literature (Table 4).
TABLE 4
Literature linking the genes used by the DL to metastasis.
4 Concluding remarks
Metastasis remains the leading cause of cancer-related deaths worldwide, and our inability to identify the tumor cells colonizing distant sites means that the physician cannot treat the metastasized tumors. Here, we developed a DL model that can be fed raw gene expression data to predict whether a sample is primary or metastasized to the brain, bone, lung, or liver. The final multi-class DNN model achieved more than acceptable prediction performance in all categories. We achieved the best prediction performance for the primary samples (AUC of 0.93), followed by the metastasis to bone and lung samples (AUC of 0.88). On the other hand, the metastasis to liver and brain samples achieved lower prediction performance with an AUC of 0.84 and 0.82, respectively. We observed the same trend when evaluating the prediction performance using external data, i.e., the highest prediction performance for the primary (AUC of 0.85) samples followed by the metastasis to lung (AUC of 0.78), bone (AUC of 0.72), liver (AUC of 0.61) and brain (AUC of 0.50) samples, respectively. However, the prediction performance for the metastasis to the brain and liver samples dropped by 32% and 23 %, respectively.
Many factors may contribute to the result we obtained for the brain samples, as this data had the highest number of DEGs and required the highest amount of top-ranked genes to be included in the model, indicating biological complexity associated with the metastasis to the brain. Additionally, the brain samples had less than 50 primary samples. In contrast, we analyzed the metastasis to bone, lung, and liver samples using more than 200 primary samples (the number of metastasized samples used for each site was similar, about 100 samples each). Beyond that, the brain only had two samples in the test set for the external data that exhibited the massive drop in prediction performance. Having this lower number of brain samples may also be contributing to the much lower prediction performance achieved with it. Thus, in the future, we will re-establish the gene set that provides maximum performance using a larger cohort of samples (when the data become available). Nonetheless, we further evaluated the prediction performance using samples from a population-based cohort to show that the multi-class DNN model achieved good prediction performance (AUC of 0.72) when distinguishing between the primary and metastatic samples, which shows the potential of our model.
We further designed the model’s workflow to provide a second functionality beyond metastasis site prediction, i.e., to identify the biological functions that the DL model uses to perform the prediction. We achieved this by associating GO biological functions (p-value < 0.05) with the neuron’s list of genes that differentiate the primary samples from the metastasized ones in the DL model. The critical neurons in each layer are grouped depending on the functions enriched. Thus, the first layer exhibits more general function categories, but more specific functions appear in subsequent neural network layers. Finally, we compared the enrichments retrieved through the DL model neuron interpretations with the MSigDB enrichment analysis. We found only a few functional categories common to both analyses but several inter-related categories. For example, the literature shows “Epithelial Mesenchymal Transition’ involves ‘Ion transport,” and “Extracellular matrix disassembly,” and it is linked to “Cell-cell adhesion,” “regulation of cell motility” and “apoptosis process” (Jason et al., 2003; Cheng et al., 2008; Sinha et al., 2008; Kalluri and Weinberg, 2009; Wang et al., 2016; Scheau et al., 2019; Winkler et al., 2020). Overall, these biological functions suggest that the gene lists used in the DL model to perform the prediction are to a large extent metastasis-specific, which is further supported by literature showing 87% of the genes used by the DL have already been linked to metastasis. These results clearly suggest that our DL model can be used to retrieve metastasis-specific biological functions beyond its metastasis site prediction capabilities.
5 Availability
We also developed a web server that the scientific community can access. The web-based tool, MetastaSite https://www.cbrc.kaust.edu.sa/metastasite/, provides a means to implement the final multi-class DNN model developed in the current study. It allows the users to predict the metastasis site (primary, metastasized to bone, brain, lung, or liver). The user needs to provide the raw gene expression for every sample.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
SA, ME, and XG conceived and designed the study; SA performed the experiments; SA, MT, AsA, AbA, SA, and ME analyzed the results; MU designed the web tool; SA, AsA, AbA, SA, MU, MT, TG, ME, and XG contributed to writing and reviewing the manuscript. All authors read and approved the final manuscript.
Funding
The research reported in this publication was supported by King Abdullah University of Science and Technology (KAUST) through grant awards Nos. BAS/1/1059-01-01, BAS/1/1624-01-01, FCC/1/1976-20-01, FCC/1/1976-26-01, URF/1/3450-01-01, REI/1/4216-01-01, REI/1/4437-01-01, REI/1/4473-01-01, and URF/1/4098-01-01.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Al-Khadairi G. Naik A. Thomas R. Al-Sulaiti B. Rizly S. Decock J. (2019). PRAME promotes epithelial-to-mesenchymal transition in triple negative breast cancer. J. Transl. Med.17 (1), 9. 10.1186/s12967-018-1757-3
2
Albaradei S. Napolitano F. Thafar M. A. Gojobori T. Essack M. Gao X. (2021). MetaCancer: A deep learning-based pan-cancer metastasis prediction model developed using multi-omics data. Comput. Struct. Biotechnol. J.19, 4404–4411. 10.1016/j.csbj.2021.08.006
3
Albaradei S. Thafar M. A. Van Neste C. Essack M. (2019). “Metastatic state of colorectal cancer can be accurately predicted with methylome,” in Proceedings of the 2019 6th International Conference on Bioinformatics Research and Applications, Seoul Republic of Korea, December 19 - 21, 2019.
4
Albaradei S. Thafar M. Alsaedi A. Van Neste C. Gojobori T. Essack M. et al (2021). Machine learning and deep learning methods that use omics data for metastasis prediction. Comput. Struct. Biotechnol. J.19, 5008–5018. 10.1016/j.csbj.2021.09.001
5
Albaradei S. Uludag M. Thafar M. A. Gojobori T. Essack M. Gao X. (2021). Predicting bone metastasis using gene expression-based machine learning models. Front. Genet.12, 771092. 10.3389/fgene.2021.771092
6
Bach S. Binder A. Montavon G. Klauschen F. Muller K. R. Samek W. (2015). On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS One10 (7), e0130140. 10.1371/journal.pone.0130140
7
Baik I. H. Jo G. H. Seo D. Ko M. J. Cho C. H. Lee M. G. et al (2016). Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-κB signaling axis. Int. J. Oncol.49 (5), 1953–1962. 10.3892/ijo.2016.3688
8
Balamurugan K. Luu V. D. Kaufmann M. R. Hofmann V. S. Boysen G. Barth S. et al (2009). Onconeuronal cerebellar degeneration-related antigen, Cdr2, is strongly expressed in papillary renal cell carcinoma and leads to attenuated hypoxic response. Oncogene28 (37), 3274–3285. 10.1038/onc.2009.186
9
Bonnetblanc J. M. Bernard P. Fayol J. (1990). Dermatomyositis and malignancy. Dermatology180 (4), 212–216. 10.1159/000248032
10
Boutin A. T. Liao W. T. Wang M. Hwang S. S. Karpinets T. V. Cheung H. et al (2017). Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev.31 (4), 370–382. 10.1101/gad.293449.116
11
Chang P. M.-H. Yeh Y. C. Chen T. C. Wu Y. C. Lu P. J. Cheng H. C. et al (2013). High expression of CHRNA1 is associated with reduced survival in early stage lung adenocarcinoma after complete resection. Ann. Surg. Oncol.20 (11), 3648–3654. 10.1245/s10434-013-3034-2
12
Chang Y.-H. Chen C. M. Chen H. Y. Yang P. C. (2015). Pathway-based gene signatures predicting clinical outcome of lung adenocarcinoma. Sci. Rep.5, 10979. 10.1038/srep10979
13
Chawla N. V. Bowyer K. W. Hall L. O. Kegelmeyer W. P. (2002). Smote: Synthetic minority over-sampling technique. J. Artif. Intell. Res.16, 321–357. 10.1613/jair.953
14
Chen P.-H. Bendris N. Hsiao Y. J. Reis C. R. Mettlen M. Chen H. Y. et al (2017). Crosstalk between CLCb/dyn1-mediated adaptive clathrin-mediated endocytosis and epidermal growth factor receptor signaling increases metastasis. Dev. Cell40 (3), 278–288. 10.1016/j.devcel.2017.01.007
15
Cheng F. Liu J. Zhang Y. You Q. Chen B. Cheng J. et al (2021). Long non-coding RNA UBA6-AS1 promotes the malignant properties of glioblastoma by competitively binding to microRNA-760 and enhancing homeobox A2 expression. Cancer Manag. Res.13, 379–392. 10.2147/CMAR.S287676
16
Cheng P. Corzo C. A. Luetteke N. Yu B. Nagaraj S. Bui M. M. et al (2008). Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med.205 (10), 2235–2249. 10.1084/jem.20080132
17
Cheon S. Lee J. H. Park S. Bang S. I. Lee W. J. Yoon D. Y. et al (2011). Overexpression of IL-32alpha increases natural killer cell-mediated killing through up-regulation of Fas and UL16-binding protein 2 (ULBP2) expression in human chronic myeloid leukemia cells. J. Biol. Chem.286 (14), 12049–12055. 10.1074/jbc.M110.159756
18
Chereda H. Bleckmann A. Kramer F. Leha A. Beissbarth T. (2019). Utilizing molecular network information via Graph convolutional neural networks to predict metastatic event in breast cancer. Stud. Health Technol. Inf.267, 181–186. 10.3233/SHTI190824
19
Chothani S. Schafer S. Adami E. Viswanathan S. Widjaja A. A. Langley S. R. et al (2019). Widespread translational control of fibrosis in the human heart by RNA-binding proteins. Circulation140 (11), 937–951. 10.1161/CIRCULATIONAHA.119.039596
20
Cordon-Cardo C. Fuks Z. DrobnjakM. Moreno C. Eisenbach L. FeldManM. (1991). Expression of HLA-A, B, C antigens on primary and metastatic tumor cell populations of human carcinomas. Cancer Res.51 (23 Pt 1), 6372–6380.
21
Daoud M. Mayo M. (2019). A survey of neural network-based cancer prediction models from microarray data. Artif. Intell. Med.97, 204–214. 10.1016/j.artmed.2019.01.006
22
David M. Naudin C. Letourneur M. Polrot M. Renoir J. M. Lazar V. et al (2014). Suppressor of cytokine signaling 1 modulates invasion and metastatic potential of colorectal cancer cells. Mol. Oncol.8 (5), 942–955. 10.1016/j.molonc.2014.03.014
23
Di Giacomo V. Tian T. V. MAs A. PecoraroM. BatLLe-Morera L. Noya L. et al (2017). ΔNp63α promotes adhesion of metastatic prostate cancer cells to the bone through regulation of CD82. Oncogene36 (31), 4381–4392. 10.1038/onc.2017.42
24
Ding Y. Qi N. Wang K. Huang Y. Liao J. Wang H. et al (2020). FTO facilitates lung adenocarcinoma cell progression by activating cell migration through mRNA demethylation. Onco. Targets. Ther.13, 1461–1470. 10.2147/OTT.S231914
25
Ebright R. Y. Lee S. Wittner B. S. Niederhoffer K. L. Nicholson B. T. Bardia A. et al (2020). Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science367 (6485), 1468–1473. 10.1126/science.aay0939
26
Edgar R. Domrachev M. Lash A. E. (2002). Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res.30 (1), 207–210. 10.1093/nar/30.1.207
27
Egiz M. Usui T. Ishibashi M. Zhang X. Shigeta S. Toyoshima M. et al (2019). La-related protein 4 as a suppressor for motility of ovarian cancer cells. Tohoku J. Exp. Med.247 (1), 59–67. 10.1620/tjem.247.59
28
Ewendt F. Feger M. Föller M. (2020). Role of fibroblast growth factor 23 (FGF23) and αKlotho in cancer. Front. Cell Dev. Biol.8, 601006. 10.3389/fcell.2020.601006
29
Fagone P. Caltabiano R. Russo A. Lupo G. Anfuso C. D. Basile M. S. et al (2017). Identification of novel chemotherapeutic strategies for metastatic uveal melanoma. Sci. Rep.7, 44564. 10.1038/srep44564
30
Fakoor R. Ladhak F. Nazi A. Huber M. (2013). “Using deep learning to enhance cancer diagnosis and classification,” in Proceedings of the international conference on machine learning, New York, USA, 18-24 July 2021.
31
Feng J. Lan R. Cai G. Lin J. Wang X. Lin J. et al (2016). Verification of TREX1 as a promising indicator of judging the prognosis of osteosarcoma. J. Orthop. Surg. Res.11 (1), 150. 10.1186/s13018-016-0487-6
32
Ge X. Liu W. Zhao W. Feng S. Duan A. Ji C. et al (2020). Exosomal transfer of LCP1 promotes osteosarcoma cell tumorigenesis and metastasis by activating the JAK2/STAT3 signaling pathway. Mol. Ther. Nucleic Acids21, 900–915. 10.1016/j.omtn.2020.07.025
33
Giorello M. B. Matas A. Marenco P. Davies K. M. Borzone F. R. Calcagno M. d. L. et al (2021). CD1a- and CD83-positive dendritic cells as prognostic markers of metastasis development in early breast cancer patients. Breast Cancer28 (6), 1328–1339. 10.1007/s12282-021-01270-9
34
Gonzalez-Avila G. Sommer B. Mendoza-Posada D. A. Ramos C. Garcia-Hernandez A. A. Falfan-Valencia R. (2019). Corrigendum to "Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer" [Crit. Rev. Oncol. Hematol. 137, May (2019) 57-83]. Crit. Rev. Oncol. Hematol.138, 172. 10.1016/j.critrevonc.2019.04.017
35
Goodfellow I. Bengio Y. Courville A. (2016). Deep learning. Cambridge, Massachusetts, United States: MIT Press.
36
Grapov D. Fahrmann J. Wanichthanarak K. Khoomrung S. (2018). Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. OMICS A J. Integr. Biol.22 (10), 630–636. 10.1089/omi.2018.0097
37
Han W. Hu C. Fan Z. J. Shen G. L. (2021). Transcript levels of keratin 1/5/6/14/15/16/17 as potential prognostic indicators in melanoma patients. Sci. Rep.11 (1), 1023. 10.1038/s41598-020-80336-8
38
Hanczar B. Zehraoui F. Issa T. Arles M. (2020). Biological interpretation of deep neural network for phenotype prediction based on gene expression. BMC Bioinforma.21 (1), 501. 10.1186/s12859-020-03836-4
39
Harrison E. B. Porrello A. Bowman B. M. Belanger A. R. Yacovone G. Azam S. H. et al (2020). A circle RNA regulatory Axis promotes lung squamous metastasis via CDR1-mediated regulation of golgi trafficking. Cancer Res.80 (22), 4972–4985. 10.1158/0008-5472.CAN-20-1162
40
Hartung F. Wang Y. Aronow B. Weber G. F. (2017). A core program of gene expression characterizes cancer metastases. Oncotarget8 (60), 102161–102175. 10.18632/oncotarget.22240
41
He W.-P. Zhou J. Cai M. Y. Xiao X. S. Liao Y. J. Kung H. F. et al (2012). CHD1L protein is overexpressed in human ovarian carcinomas and is a novel predictive biomarker for patients survival. BMC Cancer12, 437. 10.1186/1471-2407-12-437
42
Hinton G. E. Salakhutdinov R. R. (2006). Reducing the dimensionality of data with neural networks. Science313 (5786), 504–507. 10.1126/science.1127647
43
Honeyman J. N. Simon E. P. Robine N. Chiaroni-Clarke R. Darcy D. G. Lim I. I. P. et al (2014). Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science343 (6174), 1010–1014. 10.1126/science.1249484
44
Hu M. Fu X. Si Z. Li C. Sun J. Du X. et al (2019). Identification of differently expressed genes associated with prognosis and growth in colon adenocarcinoma based on integrated bioinformatics analysis. Front. Genet.10, 1245. 10.3389/fgene.2019.01245
45
Hu Y.-H. Lu Y. X. Zhang Z. Y. Zhang J. M. Zhang W. J. Zheng L. et al (2019). SSH3 facilitates colorectal cancer cell invasion and metastasis by affecting signaling cascades involving LIMK1/Rac1. Am. J. Cancer Res.9 (5), 1061–1073.
46
Hu Y. Wang B. Yi K. Lei Q. Wang G. Xu X. (2021). IFI35 is involved in the regulation of the radiosensitivity of colorectal cancer cells. Cancer Cell Int.21 (1), 290. 10.1186/s12935-021-01997-7
47
Huang Q. Wei H. Wu Z. Li L. Yao L. Sun Z. et al (2016). Preferentially expressed antigen of melanoma prevents lung cancer metastasis. PLoS One11 (7), e0149640. 10.1371/journal.pone.0149640
48
Huang Y. Pan L. Helou K. Xia Q. Parris T. Z. Li H. et al (2018). Mechanical ventilation promotes lung metastasis in experimental 4T1 breast cancer lung-metastasized models. Cancer Manag. Res.10, 545–555. 10.2147/CMAR.S142650
49
Hwang P. H. Yi H. K. Kim D. S. Nam S. Y. Kim J. S. Lee D. Y. (2001). Suppression of tumorigenicity and metastasis in B16F10 cells by PTEN/MMAC1/TEP1 gene. Cancer Lett.172 (1), 83–91. 10.1016/s0304-3835(01)00632-2
50
Iacobas D. A. Tuli N. Y. Iacobas S. Rasamny J. K. Moscatello A. Geliebter J. et al (2018). Gene master regulators of papillary and anaplastic thyroid cancers. Oncotarget9 (2), 2410–2424. 10.18632/oncotarget.23417
51
Irizarry R. A. Hobbs B. Collin F. Beazer-Barclay Y. D. Antonellis K. J. Scherf U. et al (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics4 (2), 249–264. 10.1093/biostatistics/4.2.249
52
J Sedano M. I Ramos E. Choudhari R. L Harrison A. Subramani R. Lakshmanaswamy R. et al (2020). Hypoxanthine phosphoribosyl transferase 1 is upregulated, predicts clinical outcome and controls gene expression in breast cancer. Cancers12 (6), E1522. 10.3390/cancers12061522
53
Jason L. T. George N. N. Ann F. C. (2003). The role of apoptosis in tumor progression and metastasis. Curr. Mol. Med.3 (7), 631–642. 10.2174/1566524033479483
54
Jiang Y.-Z. Li Q. H. Zhao J. Q. Lv J. J. (2014). Identification of a novel fusion gene (HLA-E and HLA-B) by RNA-seq analysis in esophageal squamous cell carcinoma. Asian pac. J. Cancer Prev.15 (5), 2309–2312. 10.7314/apjcp.2014.15.5.2309
55
Johnson L. A. Vaidya S. V. Goldfarb R. H. Mathew P. A. (2003). 2B4(CD244)-mediated activation of NK cells reduces metastases of B16F10 melanoma in mice. Anticancer Res.23 (5A), 3651–3655.
56
Ju J. A. Godet I. DiGiacomo J. W. Gilkes D. M. (2020). RhoB is regulated by hypoxia and modulates metastasis in breast cancer. Cancer Rep.3 (1), e1164. 10.1002/cnr2.1164
57
Kalluri R. Weinberg R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest.119 (6), 1420–1428. 10.1172/JCI39104
58
Kannathasan T. Kuo W. W. Chen M. C. Viswanadha V. P. Shen C. Y. Tu C. C. et al (2020). Chemoresistance-associated silencing of miR-4454 promotes colorectal cancer aggression through the GNL3L and NF-κB pathway. Cancers12 (5), E1231. 10.3390/cancers12051231
59
Kingma D. P. Ba J. (2014). Adam: A method for stochastic optimization. arXiv [cs.LG].
60
Kitamura M. Wada N. Nagata S. Iizuka N. Jin Y. F. Tomoeda M. et al (2010). Malignant peripheral nerve sheath tumor associated with neurofibromatosis type 1, with metastasis to the heart: A case report. Diagn. Pathol.5 (1), 2. 10.1186/1746-1596-5-2
61
Knouse P. Hancock C. Iwaz S. Kaiser P. (2019). Metastatic carcinomatosis Cirrhosis: A rare pattern of metastasis. Cureus11 (1), e3876. 10.7759/cureus.3876
62
Ku S. Y. Rosario S. Wang Y. Mu P. Seshadri M. Goodrich Z. W. et al (2017). Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science355 (6320), 78–83. 10.1126/science.aah4199
63
Kuleshov M. V. Jones M. R. Rouillard A. D. Fernandez N. F. Duan Q. Wang Z. et al (2016). Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res.44 (W1), W90–W97. 10.1093/nar/gkw377
64
Kumar S. Oien D. B. Khurana A. Cliby W. Hartmann L. Chien J. et al (2019). Coiled-coil and C2 domain-containing protein 1A (CC2D1A) promotes chemotherapy resistance in ovarian cancer. Front. Oncol.9, 986. 10.3389/fonc.2019.00986
65
Li C. Ge M. Chen D. Sun T. Jiang H. Xie Y. et al (2020). RPL21 siRNA blocks proliferation in pancreatic cancer cells by inhibiting DNA replication and inducing G1 arrest and apoptosis. Front. Oncol.10, 1730. 10.3389/fonc.2020.01730
66
Li F. Zhang C. Fu L. (2019). PRR14 overexpression promotes cell growth, epithelial to mesenchymal transition and metastasis of colon cancer via the AKT pathway. PLoS One14 (10), e0218839. 10.1371/journal.pone.0218839
67
Li H. Cao Y. Ma J. Luo L. Ma B. (2021). Expression and prognosis analysis of GINS subunits in human breast cancer. Medicine100 (11), e24827. 10.1097/MD.0000000000024827
68
Li M. Jin X. Li H. Wu G. Wang S. Yang C. et al (2020). Key genes with prognostic values in suppression of osteosarcoma metastasis using comprehensive analysis. BMC Cancer20 (1), 65. 10.1186/s12885-020-6542-z
69
Li M. Wang L. Zhan Y. Zeng T. Zhang X. Guan X. Y. et al (2019). Membrane metalloendopeptidase (MME) suppresses metastasis of esophageal squamous cell carcinoma (ESCC) by inhibiting FAK-RhoA signaling Axis. Am. J. Pathol.189 (7), 1462–1472. 10.1016/j.ajpath.2019.04.007
70
Libbrecht M. W. Noble W. S. (2015). Machine learning applications in genetics and genomics. Nat. Rev. Genet.16 (6), 321–332. 10.1038/nrg3920
71
Ling L.-J. Lu C. Zhou G. P. Wang S. (2010). Ectopic expression of RhoBTB2 inhibits migration and invasion of human breast cancer cells. Cancer Biol. Ther.10 (11), 1115–1122. 10.4161/cbt.10.11.13431
72
Liu H. Liu M. You H. Li X. Li X. (2020). Oncogenic network and hub genes for natural killer/T-cell lymphoma utilizing WGCNA. Front. Oncol.10, 223. 10.3389/fonc.2020.00223
73
Liu H. Zhou Y. Qiu H. Zhuang R. Han Y. Liu X. et al (2021). Rab26 suppresses migration and invasion of breast cancer cells through mediating autophagic degradation of phosphorylated Src. Cell Death Dis.12 (4), 284. 10.1038/s41419-021-03561-7
74
Liu J. Wang D. Zhang C. Zhang Z. Chen X. Lian J. et al (2018). Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling. Chin. J. Cancer Res.30 (6), 633–646. 10.21147/j.issn.1000-9604.2018.06.08
75
Liu K. Tang Z. Huang A. Chen P. Liu P. Yang J. et al (2017). Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. Int. J. Oncol.50 (1), 252–262. 10.3892/ijo.2016.3774
76
Liu Y. Wang D. Lei M. Gao J. Cui Y. Jin X. et al (2021). GABARAP suppresses EMT and breast cancer progression via the AKT/mTOR signaling pathway. Aging13 (4), 5858–5874. 10.18632/aging.202510
77
Lopez-Charcas O. Espinosa A. M. Alfaro A. Herrera-Carrillo Z. Ramirez-Cordero B. E. Cortes-Reynosa P. et al (2018). The invasiveness of human cervical cancer associated to the function of NaV1.6 channels is mediated by MMP-2 activity. Sci. Rep.8 (1), 12995. 10.1038/s41598-018-31364-y
78
Luger D. Yang Y. A. Raviv A. Weinberg D. Banerjee S. Lee M. J. et al (2013). Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One8 (10), e76115. 10.1371/journal.pone.0076115
79
Luu X. Leonard S. Joseph K.-A. (2015). Dermatomyositis presenting as a paraneoplastic syndrome with resolution of symptoms following surgical management of underlying breast malignancy. J. Surg. Case Rep.2015 (7), rjv075. 10.1093/jscr/rjv075
80
Lv Y. Wang X. Li X. Xu G. Bai Y. Wu J. et al (2020). Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway. PLoS Biol.18 (11), e3000872. 10.1371/journal.pbio.3000872
81
Lyons S. M. Alizadeh E. Mannheimer J. Schuamberg K. Castle J. Schroder B. et al (2016). Changes in cell shape are correlated with metastatic potential in murine and human osteosarcomas. Biol. Open5 (3), 289–299. 10.1242/bio.013409
82
Madreiter-Sokolowski C. T. Gottschalk B. Sokolowski A. A. Malli R. Graier W. F. (2021). Dynamic control of mitochondrial Ca2+ levels as a survival strategy of cancer cells. Front. Cell Dev. Biol.9, 614668. 10.3389/fcell.2021.614668
83
Mao-De L. Jing X. (2007). Ribosomal proteins and colorectal cancer. Curr. Genomics8 (1), 43–49. 10.2174/138920207780076938
84
Marsh T. Debnath J. (2020). Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy16 (6), 1164–1165. 10.1080/15548627.2020.1753001
85
Martorell-Marugán J. et al (2019). “Deep learning in omics data analysis and precision medicine,” in Computational biology. Editor HusiH. (Brisbane (AU): Codon Publications).
86
McLaren S. University of Western Australia S. (2009). School of, mitochondrial ATP Synthase and associated bioenergetic proteins: Can expression predict Response to Chemotherapy in locally advanced breast cancer?Australia: University Of Western Australia, 382.
87
Medina-Ramirez C. M. Goswami S. Smirnova T. Bamira D. Benson B. Ferrick N. et al (2011). Apoptosis inhibitor ARC promotes breast tumorigenesis, metastasis, and chemoresistance. Cancer Res.71 (24), 7705–7715. 10.1158/0008-5472.CAN-11-2192
88
Milkiewicz P. Mutimer D. Hubscher S. G. Elias E. (1999). Autoimmune liver disease in patients with neoplastic diseases. Eur. J. Gastroenterol. Hepatol.11 (5), 569–573. 10.1097/00042737-199905000-00018
89
Najafabadi M. M. Villanustre F. Khoshgoftaar T. M. Seliya N. Wald R. Muharemagic E. (2015). Deep learning applications and challenges in big data analytics. J. Big Data2 (1), 1–21. 10.1186/s40537-014-0007-7
90
Nałęcz K. A. (2020). Amino acid transporter SLC6A14 (ATB0, +) - a target in combined anti-cancer therapy. Front. Cell Dev. Biol.8, 594464. 10.3389/fcell.2020.594464
91
Ohtaki S. Wanibuchi M. Kataoka-Sasaki Y. Sasaki M. Oka S. Noshiro S. et al (2017). ACTC1 as an invasion and prognosis marker in glioma. J. Neurosurg.126 (2), 467–475. 10.3171/2016.1.JNS152075
92
Okusha Y. Eguchi T. Tran M. T. Sogawa C. Yoshida K. Itagaki M. et al (2020). Extracellular vesicles enriched with moonlighting metalloproteinase are highly transmissive, pro-tumorigenic, and trans-activates cellular communication network factor (CCN2/CTGF): CRISPR against cancer. Cancers12 (4), E881. 10.3390/cancers12040881
93
Paschen A. Sucker A. Hill B. Moll I. Zapatka M. Nguyen X. D. et al (2009). Differential clinical significance of individual NKG2D ligands in melanoma: Soluble ULBP2 as an indicator of poor prognosis superior to S100B. Clin. Cancer Res.15 (16), 5208–5215. 10.1158/1078-0432.CCR-09-0886
94
Pelosof L. C. Gerber D. E. (2010). Paraneoplastic syndromes: An approach to diagnosis and treatment. Mayo Clin. Proc.85 (9), 838–854. 10.4065/mcp.2010.0099
95
Pitarresi J. R. Norgard R. J. Chiarella A. M. Suzuki K. Bakir B. Sahu V. et al (2021). PTHrP drives pancreatic cancer growth and metastasis and reveals a new therapeutic vulnerability. Cancer Discov.11 (7), 1774–1791. 10.1158/2159-8290.CD-20-1098
96
Plantone D. Renna R. Sbardella E. Koudriavtseva T. (2015). Concurrence of multiple sclerosis and brain tumors. Front. Neurol.6, 40. 10.3389/fneur.2015.00040
97
Racanelli V. Prete M. Minoia C. Favoino E. Perosa F. (2008). Rheumatic disorders as paraneoplastic syndromes. Autoimmun. Rev.7 (5), 352–358. 10.1016/j.autrev.2008.02.001
98
Rae D. T. Hocum J. D. Bii V. Deeg H. J. Trobridge G. D. (2015). A novel retroviral mutagenesis screen identifies prognostic genes in RUNX1 mediated myeloid leukemogenesis. Oncotarget6 (31), 30664–30674. 10.18632/oncotarget.5133
99
Roy L. D. Ghosh S. Pathangey L. B. Tinder T. L. Gruber H. E. Mukherjee P. (2011). Collagen induced arthritis increases secondary metastasis in MMTV-PyV MT mouse model of mammary cancer. BMC Cancer11, 365. 10.1186/1471-2407-11-365
100
Scheau C. Badarau I. A. Costache R. Caruntu C. Mihai G. L. Didilescu A. C. et al (2019). The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal. Cell. Pathol.2019, 9423907. 10.1155/2019/9423907
101
Schulten H.-J. Bangash M. Karim S. Dallol A. Hussein D. Merdad A. et al (2017). Comprehensive molecular biomarker identification in breast cancer brain metastases. J. Transl. Med.15 (1), 269. 10.1186/s12967-017-1370-x
102
Seetharaman S. Flemyng E. Shen J. Conte M. R. Ridley A. J. (2016). The RNA-binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton73 (11), 680–690. 10.1002/cm.21336
103
Seguin L. Chaffanet M. Sabatier R. Jose A. Garnier S. Carbuccia N. et al (2018). A major response to carboplatin in a metastatic triple-negative breast cancer patient with somatic mutation of BRCA1 and RAD51B: When chemotherapy meets precision medicine. Ann. Oncol.29, vi29. 10.1093/annonc/mdy314.030
104
Shah A. A. Casciola-Rosen L. (2015). Cancer and scleroderma: A paraneoplastic disease with implications for malignancy screening. Curr. Opin. Rheumatol.27 (6), 563–570. 10.1097/BOR.0000000000000222
105
Sharifi-Noghabi H. Liu Y. Erho N. Shrestha R. Alshalalfa M. Davicioni E. et al Deep Genomic Signature for early metastasis prediction in prostate cancer. bioRxiv.
106
Shrikumar A. Greenside P. Kundaje A. (2017). “Learning important features through propagating activation differences,” in Proceedings of the 34th International Conference on Machine Learning-Volume 70, Sydney NSW Australia, August 6 - 11, 2017.
107
Sinha P. Okoro C. Foell D. Freeze H. H. Ostrand-Rosenberg S. Srikrishna G. (2008). Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol.181 (7), 4666–4675. 10.4049/jimmunol.181.7.4666
108
Song S. Jacobson K. N. McDermott K. M. Reddy S. P. Cress A. E. Tang H. et al (2016). ATP promotes cell survival via regulation of cytosolic [Ca2+] and Bcl-2/Bax ratio in lung cancer cells. Am. J. Physiol. Cell Physiol.310 (2), C99–C114. 10.1152/ajpcell.00092.2015
109
Song W. He D. Chen Y. Yeh C. R. Hsu I. Huang Q. et al (2018). Targeting newly identified ERβ/TGF-β1/SMAD3 signals with the FDA-approved anti-estrogen Faslodex or an ERβ selective antagonist in renal cell carcinoma. Mol. Oncol.12 (12), 2055–2071. 10.1002/1878-0261.12377
110
Strömvall K. Sundkvist K. Ljungberg B. Halin Bergstrom S. Bergh A. (2017). Reduced number of CD169+ macrophages in pre-metastatic regional lymph nodes is associated with subsequent metastatic disease in an animal model and with poor outcome in prostate cancer patients. Prostate77 (15), 1468–1477. 10.1002/pros.23407
111
Sun Y. Zhou Y. Xia J. Wen M. Wang X. Zhang J. et al (2021). Abnormally high HIP1 expression is associated with metastatic behaviors and poor prognosis in ESCC. Oncol. Lett.21 (2), 79. 10.3892/ol.2020.12340
112
Svozil D. Kvasnicka V. Pospichal J. í. (1997). Introduction to multi-layer feed-forward neural networks. Chemom. Intelligent Laboratory Syst.39 (1), 43–62. 10.1016/s0169-7439(97)00061-0
113
Wang M. Liu Y. Qian X. Wei N. Tang Y. Yang J. (2018). Downregulation of occludin affects the proliferation, apoptosis and metastatic properties of human lung carcinoma. Oncol. Rep.40 (1), 454–462. 10.3892/or.2018.6408
114
Wang X. Wang L. Xu Y. Zhang G. Wu Y. Chen P. (2018). PIAS1 inhibited the metastasis of gastric cancer cell by epithelial-mesenchymal transition regulation within the inflammatory microenvironment. Oncol. Lett.15 (3), 3828–3837. 10.3892/ol.2018.7811
115
Wang Y. Wu Y. Xiao K. Zhao Y. Lv G. Xu S. et al (2020). RPS24c isoform facilitates tumor angiogenesis via promoting the stability of MVIH in colorectal cancer. Curr. Mol. Med.20 (5), 388–395. 10.2174/1566524019666191203123943
116
Wang Z. Xiong S. Mao Y. Chen M. Ma X. Zhou X. et al (2016). Periostin promotes immunosuppressive premetastatic niche formation to facilitate breast tumour metastasis. J. Pathol.239 (4), 484–495. 10.1002/path.4747
117
Wen J. Min X. Shen M. Hua Q. Han Y. Zhao L. et al (2019). ACLY facilitates colon cancer cell metastasis by CTNNB1. J. Exp. Clin. Cancer Res.38 (1), 401. 10.1186/s13046-019-1391-9
118
Winkler J. Abisoye-Ogunniyan A. Metcalf K. J. Werb Z. (2020). Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun.11 (1), 5120. 10.1038/s41467-020-18794-x
119
Wong H. Y. Wang G. M. Croessmann S. Zabransky D. J. Chu D. Garay J. P. et al (2015). TMSB4Y is a candidate tumor suppressor on the Y chromosome and is deleted in male breast cancer. Oncotarget6 (42), 44927–44940. 10.18632/oncotarget.6743
120
Wu J. Tian B. Yang J. Huo H. Song Z. Yu J. et al (2020). Reduction of Hip2 suppresses gastric cancer cell proliferation, migration, invasion and tumorigenesis. Transl. Cancer Res.9 (2), 774–785. 10.21037/tcr.2019.12.12
121
Xu L. Zhou R. Yuan L. Wang S. Li X. Ma H. et al (2017). IGF1/IGF1R/STAT3 signaling-inducible IFITM2 promotes gastric cancer growth and metastasis. Cancer Lett.393, 76–85. 10.1016/j.canlet.2017.02.014
122
Xu L. Ziegelbauer J. Wang R. Wu W. W. Shen R. F. Juhl H. et al (2016). Distinct profiles for mitochondrial t-RNAs and small nucleolar RNAs in locally invasive and metastatic colorectal cancer. Clin. Cancer Res.22 (3), 773–784. 10.1158/1078-0432.CCR-15-0737
123
Xu Q. Liu X. Chen W. Zhang Z. (2010). Inhibiting adenoid cystic carcinoma cells growth and metastasis by blocking the expression of ADAM 10 using RNA interference. J. Transl. Med.8 (1), 136. 10.1186/1479-5876-8-136
124
Xu Y. Cui X. Wang Y. (2021). Pan-cancer metastasis prediction based on Graph deep learning method. Front. Cell Dev. Biol.9, 675978. 10.3389/fcell.2021.675978
125
Yamaguchi M. Osuka S. Weitzmann M. N. Shoji M. Murata T. (2016). Increased regucalcin gene expression extends survival in breast cancer patients: Overexpression of regucalcin suppresses the proliferation and metastatic bone activity in MDA-MB-231 human breast cancer cells in vitro. Int. J. Oncol.49 (2), 812–822. 10.3892/ijo.2016.3538
126
Yang R. Gaidamakov S. A. Xie J. Lee J. Martino L. Kozlov G. et al (2011). La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol. Cell. Biol.31 (3), 542–556. 10.1128/MCB.01162-10
127
Yano Y. Akiba J. Naito Y. Sadashima E. Cho H. Hishima T. et al (2021). Sulfite oxidase is a novel prognostic biomarker of advanced gastric cancer. Vivo35 (1), 229–237. 10.21873/invivo.12251
128
Yi X. Luk J. M. Lee N. P. Peng J. Leng X. Guan X. Y. et al (2008). Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell. Proteomics7 (2), 315–325. 10.1074/mcp.M700116-MCP200
129
Zeng H. Yuan F. Mi Y. Xian G. Qin C. Zhang D. (2018). As an independent prognostic factor, USP6 promotes the invasion and metastasis of colon cancer. Biochem. Biophys. Res. Commun.505 (3), 816–822. 10.1016/j.bbrc.2018.08.168
130
Zhang B. Li Y. Wu Q. Xie L. Barwick B. Fu C. et al (2021). Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat. Commun.12 (1), 1714. 10.1038/s41467-021-21976-w
131
Zhang C. Li H. Gao J. Cui X. Yang S. Liu Z. (2021). Prognostic significance of ANO1 expression in cancers. Medicine100 (4), e24525. 10.1097/MD.0000000000024525
132
Zhang F. Ying L. Jin J. Feng J. Chen K. Huang M. et al (2018). GAP43, a novel metastasis promoter in non-small cell lung cancer. J. Transl. Med.16 (1), 310. 10.1186/s12967-018-1682-5
133
Zhang J. Huang J. Z. Zhang Y. Q. Zhang X. Zhao L. Y. Li C. G. et al (2020). Microtubule associated protein 9 inhibits liver tumorigenesis by suppressing ERCC3. EBioMedicine53, 102701. 10.1016/j.ebiom.2020.102701
134
Zhang Y. Xia M. Jin K. Wang S. Wei H. Fan C. et al (2018). Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer17 (1), 45. 10.1186/s12943-018-0796-y
135
Zheng G. Ma Y. Zou Y. Yin A. Li W. Dong D. (2018). Hcmdb: The human cancer metastasis database. Nucleic Acids Res.46 (D1), D950–D955. 10.1093/nar/gkx1008
136
Zhou D. Tang W. Zhang Y. An H. X. (2019). JAM3 functions as a novel tumor suppressor and is inactivated by DNA methylation in colorectal cancer. Cancer Manag. Res.11, 2457–2470. 10.2147/CMAR.S189937
137
Zhou Y. Zang Y. Yang Y. Xiang J. Chen Z. (2019). Candidate genes involved in metastasis of colon cancer identified by integrated analysis. Cancer Med.8 (5), 2338–2347. 10.1002/cam4.2071
Summary
Keywords
machine learning, deep learning, artificial intelligence, metastasis, metastasis site, gene expression, clinical decision-making
Citation
Albaradei S, Albaradei A, Alsaedi A, Uludag M, Thafar MA, Gojobori T, Essack M and Gao X (2022) MetastaSite: Predicting metastasis to different sites using deep learning with gene expression data. Front. Mol. Biosci. 9:913602. doi: 10.3389/fmolb.2022.913602
Received
05 April 2022
Accepted
29 June 2022
Published
22 July 2022
Volume
9 - 2022
Edited by
Yong Teng, Emory University, United States
Reviewed by
Abdulrazak Yahya Saleh, Universiti Malaysia Sarawak, Malaysia
Dong Xu, University of Missouri, United States
Updates
Copyright
© 2022 Albaradei, Albaradei, Alsaedi, Uludag, Thafar, Gojobori, Essack and Gao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magbubah Essack, magbubah.essack@kaust.edu.sa; Xin Gao, xin.gao@kaust.edu.sa
This article was submitted to Molecular Diagnostics and Therapeutics, a section of the journal Frontiers in Molecular Biosciences
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.