- 1Department of Neurology, Goethe University/University Hospital Frankfurt, Frankfurt am Main, Germany
- 2Functional Proteomics, Institute of Cardiovascular Physiology, Faculty of Medicine, Goethe University, Frankfurt am Main, Germany
Background: A reliable distinction between ischemic stroke (IS) and intracerebral hemorrhage (ICH) is required for diagnosis-specific treatment and effective secondary prevention in patients with stroke. However, in resource-limited settings brain imaging, which is the current diagnostic gold standard for this purpose, is not always available in time. Hence, an easily accessible and broadly applicable blood biomarker-based diagnostic test differing stroke subtypes would be desirable. Using an explorative proteomics approach, this pilot study aimed to identify novel blood biomarker candidates for distinguishing IS from ICH.
Material and Methods: Plasma samples from patients with IS and ICH were drawn during hospitalization and were analyzed by using liquid chromatography/mass spectrometry. Proteins were identified using the human reference proteome database UniProtKB, and label-free quantification (LFQ) data were further analyzed using bioinformatic tools.
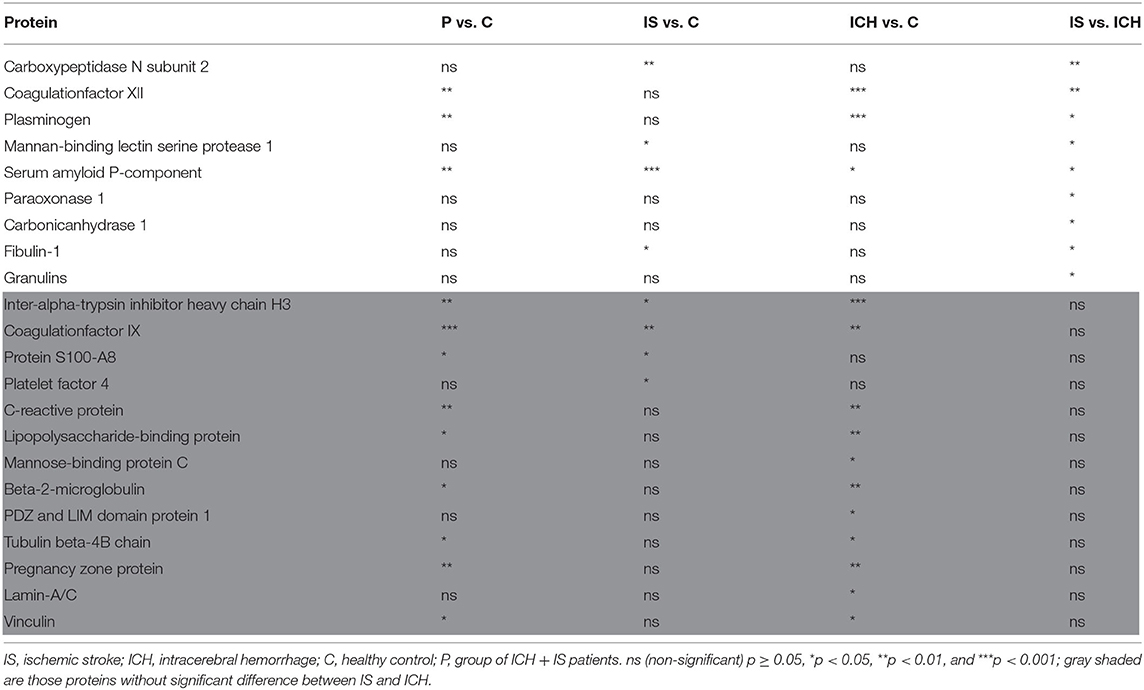
Results: Plasma specimens of three patients with IS and four patients with ICH with a median National Institute of Health Stroke Scale (NIHSS) of 12 [interquartile range (IQR) 10.5–18.5] as well as serum samples from two healthy volunteers were analyzed. Among 495 identified protein groups, a total of 368 protein groups exhibited enough data points to be entered into quantitative analysis. Of the remaining 22 top-listed proteins, a significant difference between IS and ICH was found for Carboxypeptidase N subunit 2 (CPN2), Coagulation factor XII (FXII), Plasminogen, Mannan-binding lectin serine protease 1, Serum amyloid P-component, Paraoxonase 1, Carbonic anhydrase 1, Fibulin-1, and Granulins.
Discussion: In this exploratory proteomics-based pilot study, nine candidate biomarkers for differentiation of IS and ICH were identified. The proteins belong to the immune system, the coagulation cascade, and the apoptosis system, respectively. Further investigations in larger cohorts of patients with stroke using additional biochemical analysis methods, such as ELISA or Western Blotting are now necessary to validate these markers, and to characterize diagnostic accuracy with regard to the development of a point-of-care-system for use in resource-limited areas.
Introduction
In recent years, treatment options for patients with ischemic stroke (IS) have largely expanded. Subsequent to the broad implementation of intravenous thrombolysis, mechanical thrombectomy has now become the standard of care for patients with intracranial large vessel occlusion (1–5). Moreover, multimodal CT- and MR-imaging techniques allow the application of recanalizing treatment strategies even in extended time windows (6–9). In metropolitan areas, mobile stroke units have been released to apply thrombolysis already in the preclinical setting with the shortest possible delays after symptom onset (10, 11).
In contrast to these “high-tech” advances to stand by in many high-income countries, low- to middle-income countries still face immense shortcomings in medical resources. This weighs heavily as these countries have to carry the majority of the global burden of stroke (12). Regarding brain imaging, some countries have only one CT unit available per 1 million inhabitants (13). Hence, the stratification into IS and intracerebral hemorrhage (ICH) is not possible at all or only after long transports and transfer delays (14). This prevents acute target-orientated stroke treatment, but also from the timely initiation of diagnosis-specific secondary prevention (i.e., platelet inhibitors in patients with IS). However, the effect in reducing recurrent stroke for platelet inhibition is highest within the first weeks after the initial event (15).
Thus, in resource-limited areas, an inexpensive and easy-to-use stratification tool to substitute CT-imaging before the initiation of secondary prevention would be desirable. Here recent research on blood-based brain biomarkers has revealed interesting results. Glial fibrillary acidic protein (GFAP) has been characterized in several prospective studies as a biomarker of ICH. However, it reliably distinguishes ICH from IS only within 6 h of symptom onset. Its potential use has been demonstrated in an Indian trial, too (16–21). On the other side, comparable markers of IS have not been identified so far (22–24).
This pilot study aimed to identify candidate biomarkers suitable to differentiate IS and ICH within the first days after symptom onset. Hence, in an exploratory approach, the entire plasma/serum proteome was screened through mass spectrometry (MS) techniques. Ensuing, an extensive literature search was performed, to identify the relevant publications focusing on the diagnostic value of the candidate markers in acute stroke.
Materials and Methods
Study Design
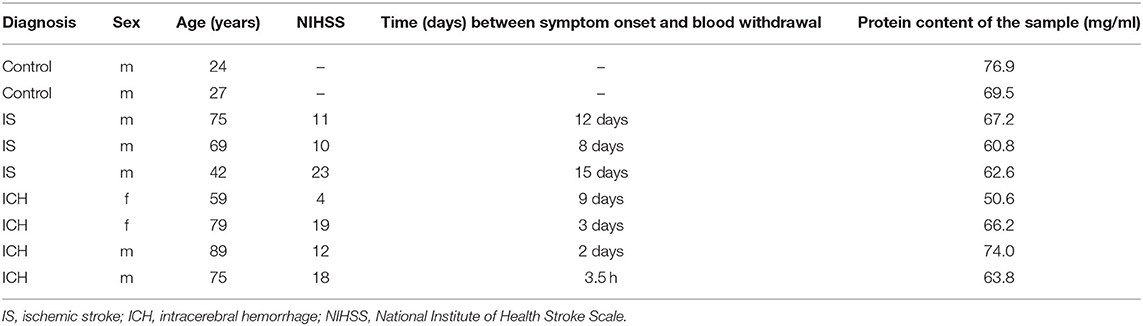
For this explorative pilot study, we targeted to compare the two “prototypes” of stroke, i.e., patients with IS in the middle cerebral artery (MCA) territory and patients with ICH in the basal ganglia or the thalamus (“deep”) as well as in the parietal or temporal lobes (“lobar”). Both conditions typically present as a classical stroke syndrome, and differentiation between the entities solely based on clinical examination alone is usually not possible. To presume a considerable amount of brain tissue damage with release of brain proteins in the bloodstream, only patients with infarctions affecting at least one-third of MCA territory and only patients with hematoma volumes higher than 20 ml were included. For doing so, we screened plasma samples collected in the context of a prior prospective study on GFAP levels in neurological diseases performed in our center for these criteria (25). In total, plasma samples of three patients with IS and of four patients with ICH, who met the above criteria, were randomly chosen among the available samples. Ultimately, the cohort was enriched by serum samples from two healthy controls.
The Ethics Committee of the Goethe University Frankfurt am Main, Germany approved the protocols of the previous and the current study. The studies were conducted according to the principles of the Declaration of Helsinki. Written informed consent was obtained from each patient, or if applicable of the next-of-a-kin.
Blood Sampling
According to the study protocol, 1 ml of ethylenediaminetetraacetic acid (EDTA)-plasma was collected during hospitalization at variable time points after stroke symptom onset and transferred into an Eppendorf tube (25). Within 60 min after blood draw, the samples were centrifugated at 10,000 g for 4 min, and the supernatant was immediately frozen and stored at −25°C; for long-term storing, the samples were transferred to −80°C freezers. Processing and storage of the serum samples of the two healthy controls were done in the same way.
Mass Spectrometry
The protein content was determined by using the method of Lowry (26). For this, 200 μg of plasma/serum proteins were diluted to a final volume of 20 μl with 6 M GdmCl, 50 mM tris(hydroxymethyl)aminomethane (TRIS)/HCl, pH 8.5, 10 mM tris-carboxyethylphosphine (TCEP), and incubated at 95°C for 5 min. Reduced thiols were alkylated with 40 mM chloroacetamide and the samples were diluted with 25 mM TRIS/HCl, pH 8.5, 10% acetonitrile to obtain a final GdmCl concentration of 0.6 M. The proteins were digested with 2 μg trypsin (sequencing grade, Promega, WI, USA) overnight at 37°C under gentle agitation. Digestion was stopped by adding trifluoroacetic acid to a final concentration of 0.5%. The tryptic peptides were cleaned through reversed phase chromatography with C18 material (3M Empore™ SPE Extraction Disks) (27), dried in microtiter plates, and resolved in 1% acetonitrile and 0.1% formic acid before peptide identification.
Liquid chromatography/mass spectrometry (LC/MS) was performed on Thermo Scientific™ Q Exactive Plus equipped with an ultra-high-performance liquid chromatography unit (Thermo Scientific Dionex Ultimate 3000, Thermo Fisher Scientific, MA, USA) and a Nanospray Flex Ion-Source (Thermo Fisher Scientific, MA, USA). The peptides were eluted from the trap column by a continuously increasing concentration of organic solvent (4–50% acetonitrile and 0.1% formic acid) over 90 min at a flow rate of 250 nl/min and then, separated on an analytical column (with 2.4 μm Reprosil C18 resin from Dr. Maisch GmbH in-house packed picotip emitter tip with diameter 100 μm, 15 cm from New Objectives). The peptides were then ionized (2.6 kV) in the ion source and sprayed into the mass spectrometer. MS data were recorded by data dependent acquisition. The full MS scan range was 300–2,000 m/z with a resolution of 70,000, and an automatic gain control (AGC) value of 3 × 106 total ion counts with a maximal ion injection time of 160 ms. Only higher charged ions (2+) were selected for MS/MS scans with a resolution of 17,500, an isolation window of 2 m/z, and an AGC value set to 105 ions with a maximal ion injection time of 150 ms. MS-Data were acquired in profile mode, MS/MS data in Centroid mode. Each patient was measured one time. The two control donors were measured in technical triplicates and quadruplicates, respectively. All samples were measured consecutively with the same instrumental setup (identical analytical column, buffers, and mass calibration).
MS Data Analysis
Mass spectrometry data were analyzed by MaxQuant (Max-Planck-Institute of Biochemistry, Martinsried, Germany) (v1.5.3.30) using default settings (28). Proteins were identified using the human reference proteome database UniProtKB with 71,567 entries, released in July 2017. The enzyme specificity was set to Trypsin. Acetylation (+42.01) at N-terminus, deamidation of N and Q (+0.98), and oxidation of methionine (+15.99) were selected as variable modifications and carbamidomethylation (+57.02) as a fixed modification on cysteines. False discovery rate (FDR) was calculated using the reverse decoy database implemented in MaxQuant. FDR was 1% for the identification of protein and peptides. Label-free quantification (LFQ) data were further analyzed using the bioinformatics tool Perseus (Max-Planck-Institute of Biochemistry, Martinsried, Germany) (v1.5.6.0) (29).
Contaminants from the internal MaxQuant list, only identified by site and reverse hits were removed from the initial protein ID list. The patients were grouped into ICH (n = 4) and IS (n = 3), the control group contains all the replicates (n = 7) of the two donors. Identified proteins were filtered to at least three valid values in one group. Missing values were replaced by the lowest value of the data set. A two-tailed Student's t-test was used to examine the levels of significance.
In addition, the statistics, correlations, and heat maps were created with Perseus as well. Other diagrams were created by using GraphPad Prism 8 (v.8.0.2) and an online Webtool from Bioinformatics and Evolutionary Genomics (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Review of the Literature
After identifying the biomarker candidates, we performed a structured literature search to identify relevant publications focusing on the diagnostic value of these markers in acute stroke. For doing so, a PubMed search was performed with the following search terms: the protein's name, its aliases, and its abbreviations according to UniProtKB (release 2021_01) linked (“AND”) to “stroke, apoplexy, ischemic stroke, ischaemic stroke, intracerebral hemorrhage or intracerebral haemorrhage.” Only studies published before August 2021 were included. Identified reviews were screened for primary sources. The results were filtered by the first author (DM) of this manuscript after a review of the title and abstract of the manuscripts. Publications relevant to the context of the present investigation were finally selected for evaluation. Please see the flow diagram (Figure 3) illustrating the database search for the review of the literature. The exact search terms are listed in the Supplementary Materials (Supplementary Table 5).
Results
Mass spectrometry analysis comprised plasma samples of three patients with MCA infarction, four patients with ICH, and serum samples of two healthy controls (as outlined above in more detail). The baseline characteristics of the study population are depicted in Table 1.
Among the 495 identified protein groups, a total of 368 protein groups exhibited enough data points to be entered into quantitative analysis. Noticeably more proteins are expected to be present in human blood samples, however, proteins with low expression are strongly underrepresented in such analyses. Albumin, immunoglobulins, and transport proteins make up a large part and are increasingly represented in peptide analysis. Therefore, the tryptic peptides were separated by cation exchange chromatography. The serum/plasma amount of thus identified proteins is comparable with those reported in the literature (30). To validate the quality of data, the total results of all proteins were compared with those of all samples (Supplementary Figure 1). The determined correlation coefficients (> 0.88) show that the mass spectrometric analysis provides very homogeneous data (Supplementary Figure 2).
Taking into account these 368 proteins, we then compared the amount of each protein in the patients group (P) as a whole (IS and ICH together) with the healthy (C) controls. Here, 91 proteins could be identified with significantly different amounts (P vs. C) (as shown in Supplementary Table 1; Supplementary Figure 3). In the next step, we compared the number of proteins in each disease entity, IS respectively ICH, with the healthy controls (C) (as shown in Supplementary Tables 2, 3). To create a top list of potential promising biomarkers, only proteins with significant differences with a fold change of at least ±2 between the groups (IS vs. C and ICH vs. C) were selected. Here, 25 proteins could be found both in the IS as well as in the ICH group with significant differences in abundance to the healthy controls, 18 individual proteins were differentially expressed only in the patients with IS, and 29 individual proteins only in the patients with ICH (as shown in Figure 1, Venn diagram).
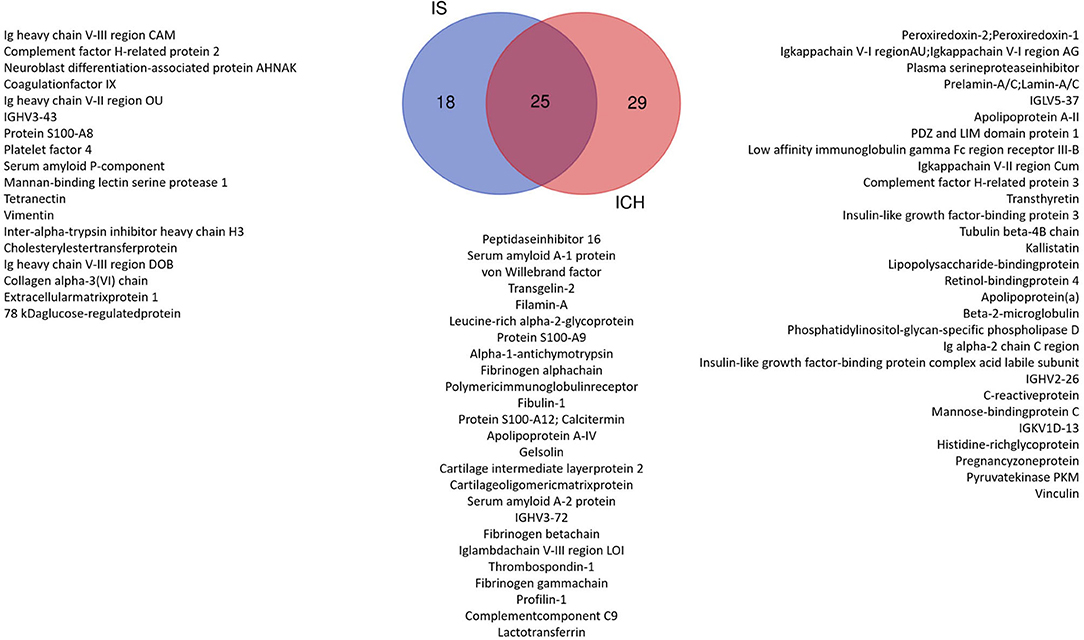
Figure 1. Venn diagram: significant proteins with at least 2-fold change in the patient group ICH vs. Control and IS vs. Control. We identified 25 proteins that were significantly changed in both the patient groups compared with Control. In total, 29 proteins were significantly different only in the ICH group and 18 only in the IS group.
Since the primary aim of the study was to identify differentially expressed proteins in patients with symptoms of stroke having either ICH or IS (and not necessarily in comparison with controls), we then screened for differences between ICH and IS, irrespective of differences to the control group and independent of the fold change, starting again from the original 368 proteins with enough data points (Supplementary Table 4). Here, 21 additional proteins could be identified (not part of the Venn diagram). After eliminating general structural and functional proteins, such as transport proteins, myosin chains, hemoglobin subunits, and components of immunoglobulins which are unsuitable as biomarkers due to their ubiquitous abundance, according to the literature (31, 32), 22 proteins remained and listed in Table 2.
Candidate Proteins for Differentiating IS and ICH
Focusing on significant differences between the patients with ICH and IS, Figure 2 shows the nine most promising candidate proteins from the top list. Carboxypeptidase N subunit 2 (CPN2) and the coagulation factor XII (FXII) showed strongly increased (p < 0.01) protein amount in the patients with IS as compared with the patients with ICH. Both proteins play a role in the kinin-kallikrein-system and are involved in thrombus formation (33–36). Significantly increased (p < 0.05) protein amount in IS as compared to ICH patients were found for plasminogen (PLG), a central regulator in the fibrinolytic system, and Mannan-binding lectin serine protease 1 (MASP1), which was attributed a role in the blood clotting system by its thrombin-like activity (37, 38). Other proteins with a considerably increased amount of protein in IS in comparison to ICH patients, whose functionality is mainly not related to the blood coagulation system, were Amyloid P-component (APCS), Paraoxonase 1 (PON1), and Carbonic anhydrase 1 (CA1). Vice versa, only two proteins showed higher values in ICH patients as compared to IS patients [Fibulin-1 (FBLN1) and Granulines (GRN)].
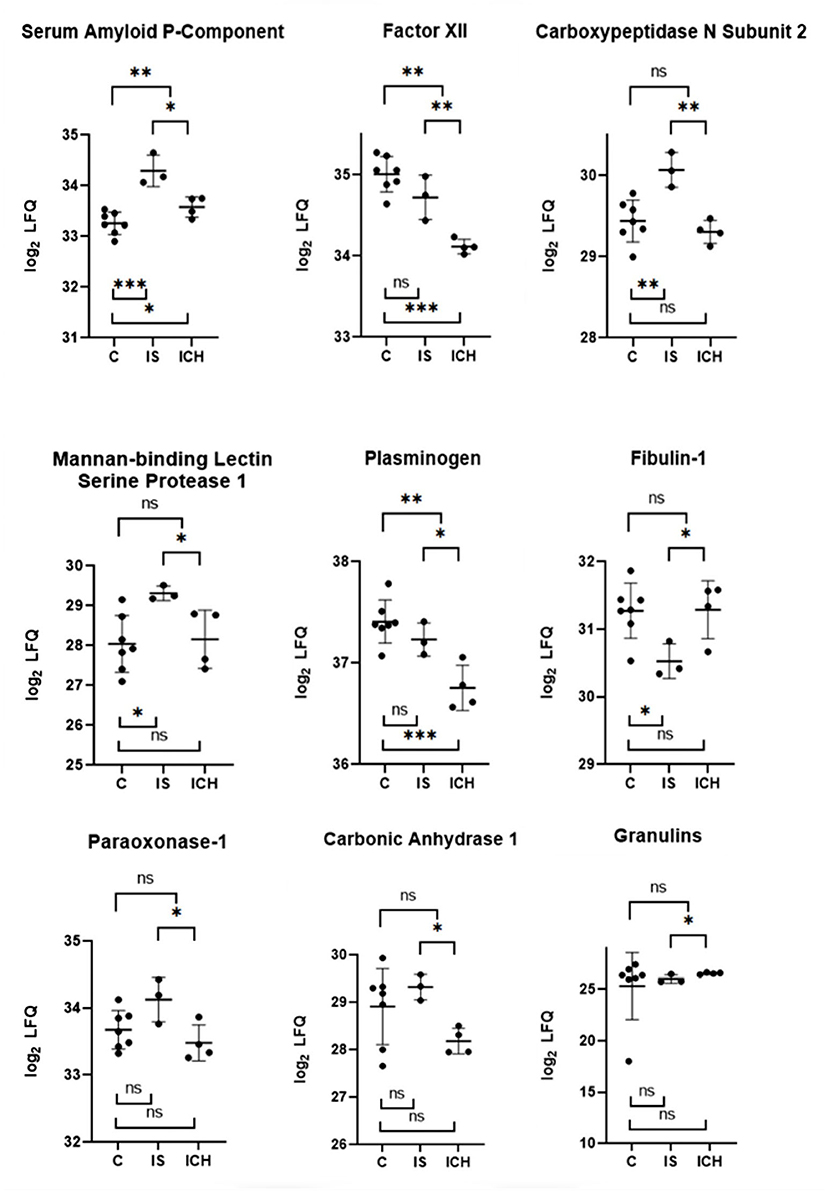
Figure 2. Selected proteins from the “top list” with the most significant differences within each group. The y-axis is transformed logarithmically. The scatter plots show the measured values with mean value and SD. Above the scatter plots, the differences between IS and ICH, respectively between the patients and control group are shown. Below the scatter plots, the differences between IS and control group are shown, as well as between ICH and control group. ns p ≥ 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001.
Review of the Literature and Integrative Evaluation
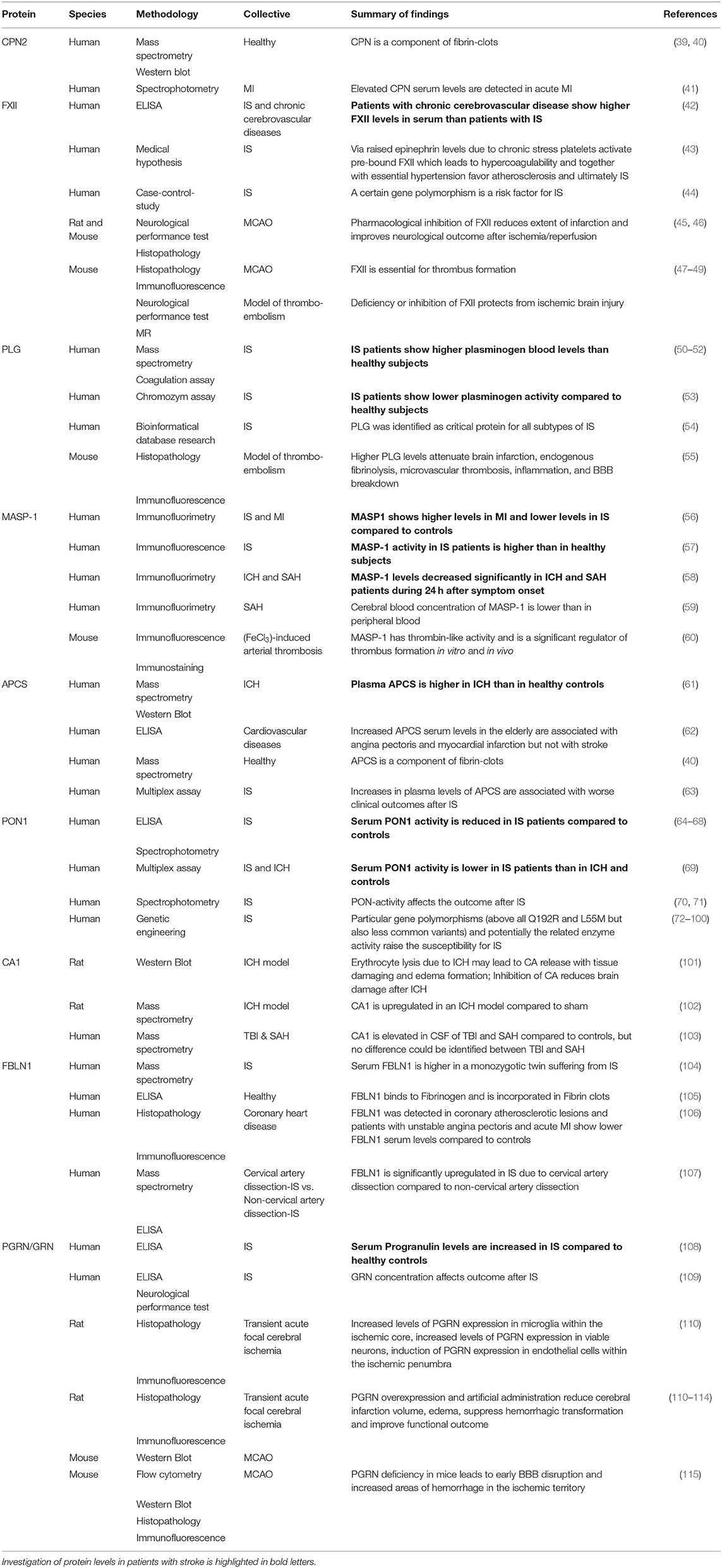
After identifying the above outlined biomarker candidates, we reviewed the literature to figure out which proteins have already been investigated in human or animal studies relating to stroke and to integrate the current knowledge with our findings (as shown in Table 3; Figure 3). Overall, these studies analyzed patients with IS and ICH compared with controls, but not IS and ICH patients compared with each other. Moreover, the largest proportion of the available and identified studies analyzed serum and not plasma samples.
Serum levels of CPN2 were not investigated in patients with stroke, but increased concentrations were found in patients with acute myocardial infarction (41). For FXII, studies showed high concentrations in patients with chronic cerebrovascular diseases (42), and FXII inhibition, as well as FXII deficiency improved outcome in experimental IS (45–49). In our study, FXII-levels were decreased in plasma of the patients with ICH compared with both controls and IS. For PLG, increased as well as decreased plasma levels in IS compared with controls have been described in the literature (50–53). We did not find differences between IS patients and controls, but ICH patients had significantly lower PLG protein amounts. MASP1 concentration was increased in myocardial infarction, however, in IS was reported to be either increased or decreased, whereas in ICH and SAH, lower levels were found (56–58). In our study, we detected higher MASP1 protein amount in IS than in healthy controls as well as in ICH. Consistent with our findings, one study showed higher APCS values in ICH than in controls (61). However, we additionally found higher values in IS compared to ICH patients. Among the identified proteins, PON1 seems to be best validated in human stroke patients, especially for several gene polymorphisms leading to higher susceptibility for IS. For PON1, reduced serum levels are described in IS (64–67, 69, 116). We did not find significant differences between IS and controls but within the patient groups elevated PON1 concentrations were found in IS compared to ICH. CA1 was found to be released from erythrocytes due to ICH in animal experiments but studies to evaluate blood levels of CA1 in patients with stroke are not available (101, 102). FBLN1 showed reduced serum concentrations in myocardial infarction (106). Comparable data for IS—except for the certain subgroup of IS caused by cervical artery dissection (107)—could not be identified. GRN was reported to be increased in the serum of patients with IS (108). It is expressed in the microglia in ischemic tissue (110–113). Increased GRN levels have not been confirmed in IS patients in our study.
Most Promising Biomarker Candidates
Based on our data, we suggest that CPN2, FXII, PLG, MASP1, APCS, PON1, and CA1 for IS and FBLN1 and GRN for ICH should be further scientifically pursued as potential stroke blood biomarkers.
Discussion
This exploratory pilot study identified nine proteins by means of mass spectrometry which showed different protein abundance in patients with IS and ICH. These nine proteins alone or in combination may now be evaluated in future prospective studies regarding their diagnostic accuracy to discriminate between those two subtypes of stroke. Moreover, detailed analyses of release kinetics of these markers after IS and ICH onset are mandatory.
At a closer look at the (patho-)physiological function of the identified proteins, it becomes apparent that all proteins, except APCS1 and CA1, are involved in the immune and/or coagulation system. This is not surprising because of previous findings on the pathophysiology of IS described as a “thrombo-inflammatory” disease (36, 117). The individual components of both systems influence each other. Besides its involvement in the coagulation cascade FXIIa activates the kinin-kallikrein system, which mediates, i.e., the activation of PLG and carboxypeptidases (35). Moreover, CPN seems to be involved in the kinin-kallikrein system and is able to reduce the cell binding capacity for PLG (33, 34, 37, 118). However, CPN itself is activated by plasmin as a negative feedback mechanism, ultimately leading to increased antifibrinolytic activity (119, 120). CPN, as well as FBLN1, have also been detected in fibrin clots (39, 106). By interaction of FBLN1 exposed after vascular injury with plasma fibrinogen, a linkage to a platelet integrin is formed and results in the formation of a platelet plug (105). CPN in the fibrin clot seems to act as a fibrinolysis inhibitor and belongs to the same family of zinc metalloproteinases as thrombin-activated fibrinolysis inhibitors (TAFI) (121). In turn, MASP1, traditionally attributed to the complement system, should interact with TAFIs, is conversely activated itself by activated platelets and fibrin and leads to fibrin clot formation and activation of thrombin and platelets, which seems to be essential for obstructive thrombosis at least in a mouse model of arterial injury (38, 60, 122). The formation of reactive oxygen species (ROS) in the context of ischemia/reperfusion processes with the influence of antioxidant components is reflected in the altered activity of associated proteins, such as PON1 (123, 124). Furthermore, the release of pro-inflammatory factors during IS leads to the activation of neuroprotective factors, such as GRN especially in viable neurons and endothelial cells in the ischemic penumbra (110, 112).
APCS and CA1 are not directly involved in coagulation or inflammatory pathways. APCS binds to apoptotic cells, is involved in chromatin degradation, and acts toxic to cerebral neurons (125, 126). CA1 is one of the 14 isoforms of carbonic anhydrases and occurs mainly in the cytosol of erythrocytes. In the context of ICH, erythrocyte lysis occurs around the hematoma, causing the release of iron and CA1 and subsequently increased tissue damage through edema formation and neuronal cell death. In addition, extracellular CA1 should promote the destruction of the blood-brain barrier by activating the kinin-kallikrein system (101, 127).
From a clinical point of view, until now no single biomarker identified in the context of stroke is suitable to certainly distinguish IS from ICH (21–24, 128). The most promising results so far have been published for GFAP. However, the different release kinetics of GFAP in IS and ICH exist only within the first 6 h after symptom onset (18, 129, 130). Thus, this protein is likely not helpful to differentiate strokes at any time point after symptom onset in resource limited settings. Moreover, GFAP release is strongly linked to the extent of damage to astroglial tissue. Thus, smaller ICH or expanding ICH may not always present with increased blood concentrations. More likely for this purpose, a combination of several markers may be favorable (131, 132). However, ischemic stroke is a heterogeneous disease comprising patients with large territorial infarctions and small lacunar strokes, as well as a large diversity of underlying etiologies. This makes it difficult to identify a biomarker panel that copes with all the facets of ischemic stroke. Interestingly, most of the proteins identified in our pilot study play pivotal roles in the immune and coagulation system. Thus, it is likely that they are directly involved in the pathophysiology of stroke and are not just an epiphenomenon. They are interesting candidates that add to the existing portfolio of potential biomarkers in stroke.
A shortcoming of this explorative pilot study is the very limited sample size. Reconfirmation of the core findings in larger patient cohorts is mandatory. In addition, other detection methods, such as ELISA or Western Blotting need to be applied to verify the proteins identified by mass spectrometry.
Furthermore, plasma samples were used for the primary comparison between IS and ICH. The control group, however, consisted of serum samples, thereby increasing the heterogeneity of the study.
Another limitation with the risk of a possible selection bias is the imbalance of baseline variables (such as age and sex) between the diagnosis groups. However, we focused on comparing “prototype” strokes as described above, and other exploratory studies based on the mass spectrometry techniques were designed in a similar way, nevertheless allowing the successful identification of novel biomarker candidates (133–135).
In summary, in this exploratory proteomics-based study, nine candidate blood biomarkers for differentiation of IS and ICH were identified. The proteins belong to the immune system, the coagulation cascade, and the apoptosis system, respectively. Due to the exploratory nature of the study, further investigations in independent, well-matched, and large-scaled cohorts of stroke patients are now necessary to validate these markers, and to characterize diagnostic accuracy with regard to the development of a point-of-care-system differentiating IS and ICH in resource-limited areas.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the Goethe University Frankfurt am Main. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
DM and IW: acquisition, analysis and interpretation of data, and drafting the work or revising it critically for important intellectual content. SL and CF: substantial contributions to the conception or design of the work and drafting the work or revising it critically for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Deutsche Forschungsgesellschaft (DFG) project SFB815/Z1 (IW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2021.713124/full#supplementary-material
Abbreviations
AGC, automatic gain control; APCS, serum amyloid P-component; BBB, blood brain barrier; CA1, carbonic anhydrase 1; CPN2, carboxypeptidase N subunit 2; CT, computed tomography; EDTA, ethylenediaminetetraacetic acid; ELISA, enzyme-linked immunosorbent assay; f, female; FDR, false discovery rate; Fig., figure; FXII, factor XII; FBLN1, fibulin-1; GdmCl, guanidinium hydrochloride; GFAP, glial fibrillary acidic protein; GRN, granulins; HCl, hydrochloric acid; ICH, intracerebral hemorrhage; IQR, interquartile range; IS, ischemic stroke; LC-MS, liquid chromatography mass spectrometry; LFQ, label free quantification; m, male; MASP1, Mannan-binding lectin serine protease 1; MCA, middle cerebral artery; MCAO, middle cerebral artery occlusion; MI, myocardial infarction; MR, magnetic resonance; MS, mass spectrometry; m/z, mass-to-charge ratio; NIHSS, National Institutes of Health Stroke Scale; PLG, plasminogen; PON1, paraoxonase 1; Ref., references; SAH, subarachnoid hemorrhage; Supp., supplementary; TAFI, thrombin-activated fibrinolysis inhibitor; TBI, traumatic brain injury; TCEP, tris-carboxyethylphosphine; TRIS, tris(hydroxymethyl)aminomethane.
References
1. Saver JL, Goyal M, Bonafe A, Diener H-C, Levy EI, Pereira VM, et al. Stent-retriever thrombectomy after intravenous t-PA vs. t-PA alone in stroke. N Engl J Med. (2015) 372:2285–95. doi: 10.1056/NEJMoa1415061
2. Jovin TG, Chamorro A, Cobo E, de Miquel MA, Molina CA, Rovira A, et al. Thrombectomy within 8 hours after symptom onset in ischemic stroke. N Engl J Med. (2015) 372:2296–306. doi: 10.1056/NEJMoa1503780
3. Campbell BCV, Mitchell PJ, Kleinig TJ, Dewey HM, Churilov L, Yassi N, et al. Endovascular therapy for ischemic stroke with perfusion-imaging selection. N Engl J Med. (2015) 372:1009–18. doi: 10.1056/NEJMoa1414792
4. Goyal M, Demchuk AM, Menon BK, Eesa M, Rempel JL, Thornton J, et al. Randomized assessment of rapid endovascular treatment of ischemic stroke. N Engl J Med. (2015) 372:1019–30. doi: 10.1056/NEJMoa1414905
5. Berkhemer OA, Fransen PSS, Beumer D, van den Berg LA, Lingsma HF, Yoo AJ, et al. A randomized trial of intraarterial treatment for acute ischemic stroke. N Engl J Med. (2015) 372:11–20. doi: 10.1056/NEJMoa1411587
6. Silva GS, Nogueira RG. Endovascular treatment of acute ischemic stroke. Continuum. (2020) 26:310–31. doi: 10.1212/CON.0000000000000852
7. Nogueira RG, Jadhav AP, Haussen DC, Bonafe A, Budzik RF, Bhuva P, et al. Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. New England J Med. (2018) 378:11–21. doi: 10.1056/NEJMoa1706442
8. Albers GW, Thijs VN, Wechsler L, Kemp S, Schlaug G, Skalabrin E, et al. Magnetic resonance imaging profiles predict clinical response to early reperfusion: the diffusion and perfusion imaging evaluation for understanding stroke evolution (DEFUSE) study. Ann Neurol. (2006) 60:508–17. doi: 10.1002/ana.20976
9. Li X, Wu L, Xie H, Bao Y, He D, Luo X. Endovascular treatment for ischemic stroke beyond the time window: a meta-analysis. Acta Neurol Scand. (2020) 141:3–13. doi: 10.1111/ane.13161
10. Calderon VJ, Kasturiarachi BM, Lin E, Bansal V, Zaidat OO. Review of the mobile stroke unit experience worldwide. Interv Neurol. (2018) 7:347–58. doi: 10.1159/000487334
11. Czap AL, Singh N, Bowry R, Jagolino-Cole A, Parker SA, Phan K, et al. Mobile stroke unit computed tomography angiography substantially shortens door-to-puncture time. Stroke. (2020) 51:1613–5. doi: 10.1161/STROKEAHA.119.028626
12. Johnson W, Onuma O, Owolabi M, Sachdev S. Stroke: a global response is needed. Bull World Health Organ. (2016) 94:634–634A. doi: 10.2471/BLT.16.181636
13. WHO. Global Atlas of Medical Devices. WHO. Available online at: http://www.who.int/medical_devices/publications/global_atlas_meddev2017/en/ (accessed May 11, 2021)
14. Berkowitz AL. Managing acute stroke in low-resource settings. Bull World Health Organ. (2016) 94:554–6. doi: 10.2471/BLT.15.162610
15. Johnston SC, Easton JD, Farrant M, Barsan W, Conwit RA, Elm JJ, et al. Clinical research collaboration, neurological emergencies treatment trials network, and the POINT investigators. Clopidogrel and aspirin in acute ischemic stroke and high-risk. TIA N Engl J Med. (2018) 379:215–25. doi: 10.1056/NEJMoa1800410
16. Hayakawa T, Ushio Y, Maeda Y, Arita N, Yoshimine T, Taneda M, et al. Astroprotein (GFAP) levels in cerebrospinal fluid of stroke patients. Neurol Med Chir (Tokyo). (1984) 24:13–8. doi: 10.2176/nmc.24.13
17. Luger S, Jæger HS, Dixon J, Bohmann FO, Schaefer J, Richieri SP, et al. Diagnostic accuracy of glial fibrillary acidic protein and ubiquitin carboxy-terminal hydrolase-L1 serum concentrations for differentiating acute intracerebral hemorrhage from ischemic stroke. Neurocrit Care. (2020) 33:39–48. doi: 10.1007/s12028-020-00931-5
18. Foerch C, Niessner M, Back T, Bauerle M, De Marchis GM, Ferbert A, et al. Diagnostic accuracy of plasma glial fibrillary acidic protein for differentiating intracerebral hemorrhage and cerebral ischemia in patients with symptoms of acute stroke. Clin Chem. (2012) 58:237–45. doi: 10.1373/clinchem.2011.172676
19. Perry LA, Lucarelli T, Penny-Dimri JC, McInnes MD, Mondello S, Bustamante A, et al. Glial fibrillary acidic protein for the early diagnosis of intracerebral hemorrhage: systematic review and meta-analysis of diagnostic test accuracy. Int J Stroke. (2019) 14:390–9. doi: 10.1177/1747493018806167
20. Kumar A, Misra S, Yadav AK, Sagar R, Verma B, Grover A, et al. Role of glial fibrillary acidic protein as a biomarker in differentiating intracerebral haemorrhage from ischaemic stroke and stroke mimics: a meta-analysis. Biomarkers. (2020) 25:1–8. doi: 10.1080/1354750X.2019.1691657
21. Bhatia R, Warrier AR, Sreenivas V, Bali P, Sisodia P, Gupta A, et al. Role of blood biomarkers in differentiating ischemic stroke and intracerebral hemorrhage. Neurol India. (2020) 68:824–9. doi: 10.4103/0028-3886.293467
22. Misra S, Montaner J, Ramiro L, Arora R, Talwar P, Nath M, et al. Blood biomarkers for the diagnosis and differentiation of stroke: a systematic review and meta-analysis. Int J Stroke. (2020) 15:704–21. doi: 10.1177/1747493020946157
23. Kamtchum-Tatuene J, Jickling GC. Blood biomarkers for stroke diagnosis and management. Neuromolecular Med. (2019) 21:344–68. doi: 10.1007/s12017-019-08530-0
24. Qin C, Zhao X-L, Ma X-T, Zhou L-Q, Wu L, Shang K, et al. Proteomic profiling of plasma biomarkers in acute ischemic stroke due to large vessel occlusion. J Transl Med. (2019) 17:214. doi: 10.1186/s12967-019-1962-8
25. Mayer CA, Brunkhorst R, Niessner M, Pfeilschifter W, Steinmetz H, Foerch C. Blood levels of glial fibrillary acidic protein (GFAP) in patients with neurological diseases. PLoS ONE. (2013) 8:e62101. doi: 10.1371/journal.pone.0062101
26. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. (1951) 193:265–75.
27. Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. (2007) 2:1896–906. doi: 10.1038/nprot.2007.261
28. Cox J. Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. (2008) 26:1367–72. doi: 10.1038/nbt.1511
29. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. (2016) 13:731–40. doi: 10.1038/nmeth.3901
30. Zhang R, Barker L, Pinchev D, Marshall J, Rasamoelisolo M, Smith C, et al. Mining biomarkers in human sera using proteomic tools. Proteomics. (2004) 4:244–56. doi: 10.1002/pmic.200300495
31. Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. (2017) 13:6297. doi: 10.15252/msb.20156297
32. Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteomics Clin Appl. (2008) 2:1386–402. doi: 10.1002/prca.200780174
33. Matthews KW, Mueller-Ortiz SL, Wetsel RA. Carboxypeptidase N: a pleiotropic regulator of inflammation. Mol Immunol. (2004) 40:785–93. doi: 10.1016/j.molimm.2003.10.002
34. Redlitz A, Tan AK, Eaton DL, Plow EF. Plasma carboxypeptidases as regulators of the plasminogen system. J Clin Invest. (1995) 96:2534–8. doi: 10.1172/JCI118315
35. Renné T, Schmaier AH, Nickel KF, Blombäck M, Maas C. In vivo roles of factor XII. Blood. (2012) 120:4296–303. doi: 10.1182/blood-2012-07-292094
36. Nieswandt B, Kleinschnitz C, Stoll G. Ischaemic stroke: a thrombo-inflammatory disease? J Physiol. (2011) 589:4115–23. doi: 10.1113/jphysiol.2011.212886
37. Herren T, Swaisgood C, Plow EF. Regulation of plasminogen receptors. Front Biosci. (2003) 8:d1–8. doi: 10.2741/916
38. Hess K, Ajjan R, Phoenix F, Dobó J, Gál P, Schroeder V. Effects of MASP-1 of the complement system on activation of coagulation factors and plasma clot formation. PLoS ONE. (2012) 7:e35690. doi: 10.1371/journal.pone.0035690
39. Talens S, Lebbink JHG, Malfliet JJMC, Demmers JA, Uitte de Willige S, Leebeek FWG, Rijken DC. Binding of carboxypeptidase N to fibrinogen and fibrin. Biochem Biophys Res Commun. (2012) 427:421–5. doi: 10.1016/j.bbrc.2012.09.081
40. Talens S, Leebeek FWG, Demmers JAA, Rijken DC. Identification of fibrin clot-bound plasma proteins. PLoS ONE. (2012) 7:e41966. doi: 10.1371/journal.pone.0041966
41. Zaninotto M, Altinier S, Lachin M, Plebani M. Carboxypeptidase N and creatine kinase-MB isoforms in acute myocardial infarction. Eur J Clin Chem Clin Biochem. (1997) 35:291–5. doi: 10.1515/cclm.1997.35.4.291
42. Kraft P, Drechsler C, Gunreben I, Heuschmann PU, Kleinschnitz C. Regulation of blood coagulation factors XI and XII in patients with acute and chronic cerebrovascular disease: a case-control study. Cerebrovasc Dis. (2014) 38:337–43. doi: 10.1159/000368434
43. Eggers AE. Factor XII (Hageman factor) is a missing link between stress and hypercoagulability and plays an important role in the pathophysiology of ischemic stroke. Med Hypotheses. (2006) 67:1065–71. doi: 10.1016/j.mehy.2006.04.009
44. Santamaría A, Mateo J, Tirado I, Oliver A, Belvís R, Martí-Fábregas J, et al. Homozygosity of the T allele of the 46 C → T polymorphism in the F12 gene is a risk factor for ischemic stroke in the Spanish population. Stroke. (2004) 35:1795–9. doi: 10.1161/01.STR.0000133127.68041.a3
45. Krupka J, May F, Weimer T, Pragst I, Kleinschnitz C, Stoll G, et al. The coagulation factor XIIa inhibitor rHA-infestin-4 improves outcome after cerebral ischemia/reperfusion injury in rats. PLoS ONE. (2016) 11:e0146783. doi: 10.1371/journal.pone.0146783
46. Hagedorn I, Schmidbauer S, Pleines I, Kleinschnitz C, Kronthaler U, Stoll G, et al. Factor XIIa inhibitor recombinant human albumin infestin-4 abolishes occlusive arterial thrombus formation without affecting bleeding. Circulation. (2010) 121:1510–7. doi: 10.1161/CIRCULATIONAHA.109.924761
47. Renné T, Pozgajová M, Grüner S, Schuh K, Pauer H-U, Burfeind P, et al. Defective thrombus formation in mice lacking coagulation factor XII. J Exp Med. (2005) 202:271–81. doi: 10.1084/jem.20050664
48. Kleinschnitz C, Stoll G, Bendszus M, Schuh K, Pauer H-U, Burfeind P, et al. Targeting coagulation factor XII provides protection from pathological thrombosis in cerebral ischemia without interfering with hemostasis. J Exp Med. (2006) 203:513–8. doi: 10.1084/jem.20052458
49. Pham M, Kleinschnitz C, Helluy X, Bartsch AJ, Austinat M, Behr VC, et al. Enhanced cortical reperfusion protects coagulation factor XII-deficient mice from ischemic stroke as revealed by high-field MRI. Neuroimage. (2010) 49:2907–14. doi: 10.1016/j.neuroimage.2009.11.061
50. Lee J, Mun S, Park A, Kim D, Lee Y-J, Kim H-J, et al. Proteomics reveals plasma biomarkers for ischemic stroke related to the coagulation cascade. J Mol Neurosci. (2020) 70:1321–31. doi: 10.1007/s12031-020-01545-4
51. Lee J, Park A, Mun S, Kim H-J, Son H, Choi H, Kim D, Lee SJ, Kim JG, Kang H-G. Proteomics-based identification of diagnostic biomarkers related to risk factors and pathogenesis of ischemic stroke. Diagnostics. (2020) 10:340. doi: 10.3390/diagnostics10050340
52. Fletcher AP, Alkjaersig N, Davies A, Lewis M, Brooks J, Hardin W, et al. Blood coagulation and plasma fibrinolytic enzyme system pathophysiology in stroke. Stroke. (1976) 7:337–48. doi: 10.1161/01.STR.7.4.337
53. Petersen NH, Schmied AB, Zeller JA, Plendl H, Deuschl G, Zunker P. Lp(a) lipoprotein and plasminogen activity in patients with different etiology of ischemic stroke. CED. (2007) 23:188–93. doi: 10.1159/000097640
54. Wei LK, Quan LS. Biomarkers for ischemic stroke subtypes: a protein-protein interaction analysis. Comput Biol Chem. (2019) 83:107116. doi: 10.1016/j.compbiolchem.2019.107116
55. Singh S, Houng AK, Wang D, Reed GL. Physiologic variations in blood plasminogen levels affect outcomes after acute cerebral thromboembolism in mice: a pathophysiologic role for microvascular thrombosis. J Thromb Haemost. (2016) 14:1822–32. doi: 10.1111/jth.13390
56. Frauenknecht V, Thiel S, Storm L, Meier N, Arnold M, Schmid J-P, et al. Plasma levels of Mannan-binding lectin (MBL)-associated serine proteases (MASPs) and MBL-associated protein in cardio- and cerebrovascular diseases. Clin Exp Immunol. (2013) 173:112–20. doi: 10.1111/cei.12093
57. Tsakanova G, Stepanyan A, Nahapetyan K, Sim RB, Arakelyan A, Boyajyan A. Serine proteases of the complement lectin pathway and their genetic variations in ischaemic stroke. J Clin Pathol. (2018) 71:141–7. doi: 10.1136/jclinpath-2017-204403
58. Sandgaard E, Troldborg A, Lauridsen SV, Gyldenholm T, Thiel S, Hvas A-M. Changes in the lectin pathway following intracerebral or spontaneous subarachnoid hemorrhage. Mol Neurobiol. (2019) 56:78–87. doi: 10.1007/s12035-018-1066-0
59. Anker-Møller T, Hvas A, Sunde N, Thiel S, Troldborg A. Proteins of the Lectin Pathway of complement activation at the site of injury in subarachnoid hemorrhage compared with peripheral blood. Brain Behav. (2020) 10:e01728. doi: 10.1002/brb3.1728
60. La Bonte LR, Pavlov VI, Tan YS, Takahashi K, Takahashi M, Banda NK, et al. Mannose-binding lectin-associated serine protease-1 is a significant contributor to coagulation in a murine model of occlusive thrombosis. J Immunol. (2012) 188:885–91. doi: 10.4049/jimmunol.1102916
61. Li G, Zhang L, Yu M, Jia H, Tian T, Wang J, Wang F, Zhou L. Identification of novel biomarker and therapeutic target candidates for acute intracerebral hemorrhage by quantitative plasma proteomics. Clin Proteomics. (2017) 14:14. doi: 10.1186/s12014-017-9149-x
62. Jenny NS, Arnold AM, Kuller LH, Tracy RP, Psaty BM. Serum amyloid P and cardiovascular disease in older men and women: results from the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. (2007) 27:352–8. doi: 10.1161/01.ATV.0000254150.97741.fe
63. Appleton JP, O'Sullivan SE, Hedstrom A, May JA, Donnelly R, Sprigg N, et al. Blood markers in remote ischaemic conditioning for acute ischaemic stroke: data from the REmote ischaemic conditioning after stroke trial. Eur J Neurol. (2021) 28:1225–33. doi: 10.1111/ene.14650
64. Chawhan SS, Mogarekar MR, Wagh RV, Das RR, Pramanik SS, Sonune SM, et al. Relation of paraoxonase1, arylesterase and lipid profile in ischemic stroke patients. J Clin Diagn Res. (2015) 9:BC01–3. doi: 10.7860/JCDR/2015/15345.6707
65. Kotur-Stevuljevic J, Bogavac-Stanojevic N, Jelic-Ivanovic Z, Stefanovic A, Gojkovic T, Joksic J, et al. Oxidative stress and paraoxonase 1 status in acute ischemic stroke patients. Atherosclerosis. (2015) 241:192–8. doi: 10.1016/j.atherosclerosis.2015.05.016
66. Ferretti G, Bacchetti T, Masciangelo S, Nanetti L, Mazzanti L, Silvestrini M, Bartolini M, Provinciali L. Lipid peroxidation in stroke patients. Clinical Chemistry and Laboratory Medicine. (2008) 46: 113–117. doi: 10.1515/CCLM.2008.011
67. Zhu H, Zhao T, Liu J. Role of paraoxonase 1 activity and oxidative/antioxidative stress markers in patients with acute cerebral infarction. Clin Lab. (2018) 64:1049–53. doi: 10.7754/Clin.Lab.2018.180201
68. Gokdemir MT, Karakilcik AZ, Gokdemir GS. Prognostic importance of paraoxonase, arylesterase and mean platelet volume efficiency in acute ischaemic stroke. J Pak Med Assoc. (2017) 67:1679–83.
69. Walsh KB, Hart K, Roll S, Sperling M, Unruh D, Davidson WS, et al. Apolipoprotein A-I and paraoxonase-1 are potential blood biomarkers for ischemic stroke diagnosis. J Stroke Cerebrovasc Dis. (2016) 25:1360–5. doi: 10.1016/j.jstrokecerebrovasdis.2016.02.027
70. Michalak S, Kazmierski R, Hellmann A, Wysocka E, Kocialkowska-Adamczewska D, Wencel-Warot A, et al. Serum paraoxonase/arylesterase activity affects outcome in ischemic stroke patients. Cerebrovasc Dis. (2011) 32:124–32. doi: 10.1159/000328227
71. Xu Y, Wang K, Wang Q, Ma Y, Liu X. The antioxidant enzyme PON1: a potential prognostic predictor of acute ischemic stroke. Oxid Med Cell Longev. (2021) 2021:1–8. doi: 10.1155/2021/6677111
72. Aydin M, Gencer M, Cetinkaya Y, Ozkok E, Ozbek Z, Kilic G, et al. PON1 55/192 polymorphism, oxidative stress, type, prognosis and severity of stroke. IUBMB Life. (2006) 58:165–72. doi: 10.1080/15216540600688462
73. Banerjee I. Relationship between Paraoxonase 1 (PON1) gene polymorphisms and susceptibility of stroke: a meta-analysis. Eur J Epidemiol. (2010) 25:449–58. doi: 10.1007/s10654-010-9470-4
74. Baum L, Ng HK, Woo KS, Tomlinson B, Rainer TH, Chen X, et al. Paraoxonase 1 gene Q192R polymorphism affects stroke and myocardial infarction risk. Clin Biochem. (2006) 39:191–5. doi: 10.1016/j.clinbiochem.2006.01.010
75. Bhattacharyya T. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. (2008) 299:1265. doi: 10.1001/jama.299.11.1265
76. Can Demirdögen B, Türkanoglu A, Bek S, Sanisoglu Y, Demirkaya S, Vural O, et al. Paraoxonase/arylesterase ratio, PON1 192Q/R polymorphism and PON1 status are associated with increased risk of ischemic stroke. Clin Biochem. (2008) 41:1–9. doi: 10.1016/j.clinbiochem.2007.08.010
77. Cozzi L, Campolo J, Parolini M, De Maria R, Patrosso MC, Marocchi A, et al. Paraoxonase 1 L55M, Q192R and paraoxonase 2 S311C alleles in atherothrombosis. Mol Cell Biochem. (2013) 374:233–8. doi: 10.1007/s11010-012-1525-2
78. Demirdögen BCDemirkaya S, Türkanoglu A, Bek S, Arinç E, Adali O. Analysis of paraoxonase 1 (PON1) genetic polymorphisms and activities as risk factors for ischemic stroke in Turkish population. Cell Biochem Funct. (2009) 27:558–67. doi: 10.1002/cbf.1607
79. Imai Y, Morita H, Kurihara H, Sugiyama T, Kato N, Ebihara A., Oh-hashi Y, et al. Evidence for association between paraoxonase gene polymorphisms and atherosclerotic diseases. Atherosclerosis. (2000) 149:435–42. doi: 10.1016/S0021-9150(99)00340-8
80. Kim NS, Kang BK, Cha MH, Oh S-M, Ko MM, Bang O-S. Association between PON1 5′-regulatory region polymorphisms, PON1 activity and ischemic stroke. Clin Biochem. (2009) 42:857–63. doi: 10.1016/j.clinbiochem.2009.02.008
81. Kim NS, Kang K, Cha MH, Kang B-J, Moon J, Kang BK Yu B-C, et al. Decreased paraoxonase-1 activity is a risk factor for ischemic stroke in Koreans. Biochem Biophys Res Commun. (2007) 364:157–62. doi: 10.1016/j.bbrc.2007.09.119
82. Kim DS, Crosslin DR, Auer PL, Suzuki SM, Marsillach J, Burt AA, et al. Rare coding variation in paraoxonase-1 is associated with ischemic stroke in the NHLBI Exome Sequencing Project. J Lipid Res. (2014) 55:1173–8. doi: 10.1194/jlr.P049247
83. Li X-Q, Ma N, Li X-G, Wang B, Sun S-S, Gao F, et al. Association of PON1, P2Y12 and COX1 with recurrent ischemic events in patients with extracranial or intracranial stenting. PLoS ONE. (2016) 11:e0148891. doi: 10.1371/journal.pone.0148891
84. Liu M-E, Liao Y-C, Lin R-T, Wang Y-S, Hsi E, Lin H-F, et al. functional polymorphism of PON1 interferes with microRNA binding to increase the risk of ischemic stroke and carotid atherosclerosis. Atherosclerosis. (2013) 228:161–7. doi: 10.1016/j.atherosclerosis.2013.01.036
85. Luu HN, Kingah PL, North K, Boerwinkle E, Volcik KA. Interaction of folate intake and the paraoxonase Q192R polymorphism with risk of incident coronary heart disease and ischemic stroke: the atherosclerosis risk in communities study. Ann Epidemiol. (2011) 21:815–23. doi: 10.1016/j.annepidem.2011.08.007
86. Mahrooz A, Gohari G, Hashemi M-B, Zargari M, Musavi H, Abedini M, et al. R-carrying genotypes of serum paraoxonase (PON1) 192 polymorphism and higher activity ratio are related to susceptibility against ischemic stroke. Mol Biol Rep. (2012) 39:11177–85. doi: 10.1007/s11033-012-2027-8
87. Mahrooz A, Alizadeh A, Gohari G. The salt stimulation property of serum paraoxonase (PON1) could be a valuable factor in evaluating the enzyme status in ischemic stroke: the role of activity-determined PON1 192Q/R phenotypes. J Neurol Sci. (2014) 338:197–202. doi: 10.1016/j.jns.2014.01.006
88. Man BL, Baum L, Fu YP, Chan YY, Lam W, Hui CF, et al. Genetic polymorphisms of Chinese patients with ischemic stroke and concurrent stenoses of extracranial and intracranial vessels. J Clin Neurosci. (2010) 17:1244–7. doi: 10.1016/j.jocn.2010.01.050
89. Pan Y, He B, Sun H, Xu T, Pan B, Wang S, et al. Susceptibility of PON1/PON2 genetic variations to ischemic stroke risk in a Chinese HAN population. PGPM. (2020) 13:563–70. doi: 10.2147/PGPM.S275341
90. Ranade K, Kirchgessner TG, Iakoubova OA, Devlin JJ, DelMonte T, Vishnupad P, et al. Evaluation of the paraoxonases as candidate genes for stroke: Gln192Arg polymorphism in the paraoxonase 1 gene is associated with increased risk of stroke. Stroke. (2005) 36:2346–50. doi: 10.1161/01.STR.0000185703.88944.7d
91. Rodríguez-Esparragón F, López-Fernández JC, Buset-Ríos N, García-Bello MA, Hernández-Velazquez E, Cappiello L, et al. Paraoxonase 1 and 2 gene variants and the ischemic stroke risk in Gran Canaria population: an association study and meta-analysis. Int J Neurosci. (2017) 127:191–8. doi: 10.3109/00207454.2016.1165675
92. Schiavon R, Turazzini M, De Fanti E, Battaglia P, Targa L, Del Colle R, et al. PON1 activity and genotype in patients with arterial ischemic stroke and in healthy individuals. Acta Neurol Scand. (2007) 116:26–30. doi: 10.1111/j.1600-0404.2006.00765.x
93. Shenhar-Tsarfaty S, Waiskopf N, Ofek K, Shopin L, Usher S, Berliner S, et al. Atherosclerosis and arteriosclerosis parameters in stroke patients associate with paraoxonase polymorphism and esterase activities. Eur J Neurol. (2013) 20:891–8. doi: 10.1111/ene.12074
94. Sun J, Wang L, Yang Q, Zhou T, Ding X, Yang K, et al. The association of paraoxonase-1 polymorphism with carotid artery stenosis among elderly Chinese population. Oxid Med Cell Longev. (2020) 2020:1–5. doi: 10.1155/2020/3084120
95. Ueno T, Shimazaki E, Matsumoto T, Watanabe H, Tsunemi A, Takahashi Y, et al. Paraoxonase1 polymorphism Leu-Met55 is associated with cerebral infarction in Japanese population. Med Sci Monit. (2003) 9:CR208–212.
96. Voetsch B, Benke KS, Damasceno BP, Siqueira LH, Loscalzo J. Paraoxonase 192 Gln → Arg polymorphism: an independent risk factor for nonfatal arterial ischemic stroke among young adults. Stroke. (2002) 33:1459–64. doi: 10.1161/01.STR.0000016928.60995.BD
97. Voetsch B, Benke KS, Panhuysen CI, Damasceno BP, Loscalzo J. The combined effect of paraoxonase promoter and coding region polymorphisms on the risk of arterial ischemic stroke among young adults. Arch Neurol. (2004) 61:351. doi: 10.1001/archneur.61.3.351
98. Wei LK, Au A, Menon S, Griffiths LR, Kooi CW, Irene L, et al. Polymorphisms of MTHFR, eNOS, ACE, AGT, ApoE, PON1, PDE4D, and ischemic stroke: meta-analysis. J Stroke Cerebrovasc Dis. (2017) 26:2482–93. doi: 10.1016/j.jstrokecerebrovasdis.2017.05.048
99. Zeng Q, Zeng J A. meta-analysis on relationship between paraoxonase 1 polymorphisms and atherosclerotic cardiovascular diseases. Life Sci. (2019) 232:116646. doi: 10.1016/j.lfs.2019.116646
100. Zhang G, Li W, Li Z, Lv H, Ren Y, Ma R, et al. Association between paraoxonase gene and stroke in the Han Chinese population. BMC Med Genet. (2013) 14:16. doi: 10.1186/1471-2350-14-16
101. Guo F, Hua Y, Wang J, Keep RF Xi G. Inhibition of carbonic anhydrase reduces brain injury after intracerebral hemorrhage. Transl Stroke Res. (2012) 3:130–7. doi: 10.1007/s12975-011-0106-0
102. Liu T, Zhou J, Cui H, Li P, Li H, Wang Y, et al. Quantitative proteomic analysis of intracerebral hemorrhage in rats with a focus on brain energy metabolism. Brain Behav. (2018) 8:e01130. doi: 10.1002/brb3.1130
103. Connor DE, Chaitanya GV, Chittiboina P, McCarthy P, Scott LK, Schrott L, et al. Variations in the cerebrospinal fluid proteome following traumatic brain injury and subarachnoid hemorrhage. Pathophysiology. (2017) 24:169–83. doi: 10.1016/j.pathophys.2017.04.003
104. Vadgama N, Lamont D, Hardy J, Nasir J, Lovering RC. Distinct proteomic profiles in monozygotic twins discordant for ischaemic stroke. Mol Cell Biochem. (2019) 456:157–65. doi: 10.1007/s11010-019-03501-2
105. Godyna S, Diaz-Ricart M, Argraves WS. Fibulin-1 mediates platelet adhesion via a bridge of fibrinogen. Blood. (1996) 88:2569–77.
106. Argraves WS, Tanaka A, Smith EP, Twal WO, Argraves KM, Fan D, et al. Fibulin-1 and fibrinogen in human atherosclerotic lesions. Histochem Cell Biol. (2009) 132:559–65. doi: 10.1007/s00418-009-0628-7
107. Yang Y, Peng J, Wang S, Huang J, Ran H, Chen K, et al. Serum-based proteomics reveals lipid metabolic and immunoregulatory dysregulation in cervical artery dissection with stroke. Front Neurol. (2020) 11:352. doi: 10.3389/fneur.2020.00352
108. Xie S, Lu L, Liu L, Bi G, Zheng L. Progranulin and short-term outcome in patients with acute ischaemic stroke. Eur J Neurol. (2016) 23:648–55. doi: 10.1111/ene.12920
109. Lasek-Bal A, Jedrzejowska-Szypulka H, Student S, Wianecka A, Zareba K, Puz P, Bal W, Pawletko K, Lewin-Kowalik J. The importance of selected markers of inflammation and blood-brain barrier damage for short-term ischemic stroke prognosis. J Physiol Pharmacol. (2019) 70:209–217. doi: 10.26402/jpp.2019.2.04
110. Kanazawa M, Kawamura K, Takahashi T, Miura M, Tanaka Y, Koyama M, et al. Multiple therapeutic effects of progranulin on experimental acute ischaemic stroke. Brain. (2015) 138:1932–48. doi: 10.1093/brain/awv079
111. Liu Y, Ren J, Kang M, Zhai C, Cheng Q, Li J, et al. Progranulin promotes functional recovery and neurogenesis in the subventricular zone of adult mice after cerebral ischemia. Brain Res. (2021) 1757:147312. doi: 10.1016/j.brainres.2021.147312
112. Tao J, Ji F, Wang F, Liu B, Zhu Y. Neuroprotective effects of progranulin in ischemic mice. Brain Res. (2012) 1436:130–6. doi: 10.1016/j.brainres.2011.11.063
113. Egashira Y, Suzuki Y, Azuma Y, Takagi T, Mishiro K, Sugitani S, et al. The growth factor progranulin attenuates neuronal injury induced by cerebral ischemia-reperfusion through the suppression of neutrophil recruitment. J Neuroinflammation. (2013) 10:884. doi: 10.1186/1742-2094-10-105
114. Li X, Cheng S, Hu H, Zhang X, Xu J, Wang R, et al. Progranulin protects against cerebral ischemia-reperfusion (I/R) injury by inhibiting necroptosis and oxidative stress. Biochem Biophys Res Commun. (2020) 521:569–76. doi: 10.1016/j.bbrc.2019.09.111
115. Jackman K, Kahles T, Lane D, Garcia-Bonilla L, Abe T, Capone C, et al. Progranulin deficiency promotes post-ischemic blood-brain barrier disruption. J Neurosci. (2013) 33:19579–89. doi: 10.1523/JNEUROSCI.4318-13.2013
116. Gokdemir MT, Karakilcik AZ, Gokdemir GS. Prognostic importance of paraoxonase, arylesterase and mean platelet volume efficiency in acute ischaemic stroke. J Pak Med Assoc. (2017) 67:1679–83.
117. De Meyer SF, Denorme F, Langhauser F, Geuss E, Fluri F, Kleinschnitz C. Thromboinflammation in stroke brain damage. Stroke. (2016) 47:1165–72. doi: 10.1161/STROKEAHA.115.011238
118. Skidgel RA, Erdös EG. Structure and function of human plasma carboxypeptidase N, the anaphylatoxin inactivator. Int Immunopharmacol. (2007) 7:1888–99. doi: 10.1016/j.intimp.2007.07.014
119. Quagraine MO, Tan F, Tamei H, Erdös EG, Skidgel RA. Plasmin alters the activity and quaternary structure of human plasma carboxypeptidase N. Biochem J. (2005) 388:81–91. doi: 10.1042/BJ20041471
120. Walker JB, Binette TM, Mackova M, Lambkin GR, Mitchell L, Bajzar L. Proteolytic cleavage of carboxypeptidase N markedly increases its antifibrinolytic activity. J Thromb Haemost. (2008) 6:848–55. doi: 10.1111/j.1538-7836.2008.02912.x
121. Tran H, Tanaka A, Litvinovich SV, Medved LV, Haudenschild CC, Argraves WS. The interaction of fibulin-1 with fibrinogen. A potential role in hemostasis and thrombosis. J Biol Chem. (1995) 270:19458–64. doi: 10.1074/jbc.270.33.19458
122. Sarma JV, Ward PA. The complement system. Cell Tissue Res. (2011) 343:227–35. doi: 10.1007/s00441-010-1034-0
123. Furlong CE, Suzuki SM, Stevens RC, Marsillach J, Richter RJ, Jarvik GP, et al. Human PON1, a biomarker of risk of disease and exposure. Chem Biol Interact. (2010) 187:355–61. doi: 10.1016/j.cbi.2010.03.033
124. Chan PH. Role of oxidants in ischemic brain damage. Stroke. (1996) 27:1124–9. doi: 10.1161/01.str.27.6.1124
125. Hutchinson WL, Hohenester E, Pepys MB. Human serum amyloid P component is a single uncomplexed pentamer in whole serum. Mol Med. (2000) 6:482–93. doi: 10.1007/BF03401789
126. Al-Shawi R, Tennent GA, Millar DJ, Richard-Londt A, Brandner S, Werring DJ, Simons JP, Pepys MB. Pharmacological removal of serum amyloid P component from intracerebral plaques and cerebrovascular Aβ amyloid deposits in vivo. Open Biol. (2016) 6:150202. doi: 10.1098/rsob.150202
127. Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol. (2012) 11:720–31. doi: 10.1016/S1474-4422(12)70104-7
128. Harpaz D, Eltzov E, Seet RCS, Marks RS, Tok AIY. Point-of-care-testing in acute stroke management: an unmet need ripe for technological harvest. Biosensors. (2017) 7:30. doi: 10.3390/bios7030030
129. Luger S, Witsch J, Dietz A, Hamann GF, Minnerup J, Schneider H, et al. Glial fibrillary acidic protein serum levels distinguish between intracerebral hemorrhage and cerebral ischemia in the early phase of stroke. Clin Chem. (2017) 63:377–85. doi: 10.1373/clinchem.2016.263335
130. Dvorak F, Haberer I, Sitzer M, Foerch C. Characterisation of the diagnostic window of serum glial fibrillary acidic protein for the differentiation of intracerebral haemorrhage and ischaemic stroke. Cerebrovasc Dis. (2009) 27:37–41. doi: 10.1159/000172632
131. Jickling GC, Sharp FR. Biomarker panels in ischemic stroke. Stroke. (2015) 46:915–20. doi: 10.1161/STROKEAHA.114.005604
132. Katan M, Elkind MS. The potential role of blood biomarkers in patients with ischemic stroke: an expert opinion. Clin Transl Neurosci. (2018) 2:2514183X18768050. doi: 10.1177/2514183X18768050
133. Fish-Low C-Y, Than LTL, Ling K-H, Lin Q, Sekawi Z. Plasma proteome profiling reveals differentially expressed lipopolysaccharide-binding protein among leptospirosis patients. J Microbiol Immunol Infect. (2020) 53:157–62. doi: 10.1016/j.jmii.2018.12.015
134. Lemańska-Perek A, Lis-Kuberka J, Lepczyński A, Dratwa-Chałupnik A, Tupikowski K, Katnik-Prastowska I, et al. Potential plasma biomarkers of bladder cancer identified by proteomic analysis: a pilot study. Adv Clin Exp Med. (2019) 28:339–46. doi: 10.17219/acem/79296
Keywords: mass spectrometry, blood, biomarker, differentiation, ischemic stroke, intracerebral hemorrhage
Citation: Malicek D, Wittig I, Luger S and Foerch C (2021) Proteomics-Based Approach to Identify Novel Blood Biomarker Candidates for Differentiating Intracerebral Hemorrhage From Ischemic Stroke—A Pilot Study. Front. Neurol. 12:713124. doi: 10.3389/fneur.2021.713124
Received: 21 May 2021; Accepted: 01 November 2021;
Published: 17 December 2021.
Edited by:
Timo Uphaus, Johannes Gutenberg University Mainz, GermanyReviewed by:
Luis Rafael Moscote-Salazar, Latinamerican Council of Neurocritical Care (CLaNi), ColombiaSteffen Tiedt, LMU Munich University Hospital, Germany
Copyright © 2021 Malicek, Wittig, Luger and Foerch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Foerch, Zm9lcmNoQGVtLnVuaS1mcmFua2Z1cnQuZGU=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
 David Malicek
David Malicek Ilka Wittig
Ilka Wittig Sebastian Luger
Sebastian Luger Christian Foerch
Christian Foerch