Abstract
Background:
The brain vasculature is a key player in neurological manifestations of COVID-19. Infection of brain endothelial cells with SARS-CoV-2 along with circulating cytokines may cause dysfunction of the blood-brain barrier (BBB). Solute carrier transporters (SLCs) in brain endothelial cells regulate substrate transport across the BBB. Here, it was hypothesized that transport functions of SLCs will be impaired by interactions with viral proteins, and subsequently, data-mining studies were performed.
Methods:
Virus-host protein-protein interaction data for SARS-CoV-2 infection were retrieved from the BioGRID database, filtered for SLCs, and then annotated for relevant expression in brain endothelial cells using a mouse brain transcriptomics database. Host SLCs expressed in brain endothelial cells were further explored using publicly available databases and information in the literature. Functional Annotation Clustering was performed using DAVID, and Enrichr served for pathway analysis. Substrates were retrieved from NCBI Gene. Links to monogenic disorders were retrieved from Online Mendelian Inheritance in Man™ and screened for disorders of the nervous system. Interactome data for viral proteins of SARS-CoV-2 were retrieved from BioGRID. Reports for host SLCs in viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle were explored in databases (VThunter) and literature. ATP-binding cassette transporters (ABCs) were studied in parallel.
Results:
N = 80 host SLCs showed relevant expression in brain endothelial cells whereby amino acid transporter stood out. N = 24/80 host SLCs were linked to monogenic disorders of the nervous system. N = 9/29 SARS-CoV-2 viral proteins had strong links to SLCs and key functions in viral infection (e.g., interferon response). SLCs serving as viral receptors and with closely associated functions were significantly enriched among all known listed viral receptors (chi-square test, p = 0.001). Literature searches for host SLCs revealed involvement of a subset of SLCs in infection mechanisms for SARS-CoV-2 and more broadly for other viruses. N = 17 host ABCs were found in brain endothelial cells where they may serve as efflux transporters.
Discussion:
This hypothesis-generating work proposes a set of N = 80 host SLCs expressed in endothelial cells as contributors to BBB impairment after SARS-CoV-2 infection. Theoretically, persistent dysfunction of SLCs at the BBB, in particular insufficient transport of amino acids, could be one of many reasons for cognitive changes in long-COVID. Functions of SLCs in viral entry and associated roles deserve close attention.
Introduction
The coronavirus, SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2), caused a pandemic from early 2020 through mid 2023 and is now considered an endemic virus (1–3). The clinical manifestations of SARS-CoV-2 infection have been named COVID-19 (Coronavirus Disease of 2019), and pulmonary involvement is the leading cause for severe disease and mortality (4, 5). Neurological symptoms and structural damage of the brain (“Neuro-COVID”) were found in a subset of patients acutely affected by COVID-19 (6–10). The mechanisms of brain involvement have been extensively reviewed (9, 11–14). The overall conclusion of these reviews is that viral RNA or proteins can be variably found in the brain, indicating infection of neurons and other cells, but there is limited evidence for replication in neurons or other cells. Little (if any) evidence exists for virus-induced neuronal loss. Neuroinflammation has been also found that does not closely correlate with the detection of viral RNA or protein in the brain, suggesting systemic effects of high levels of cytokines. Ischemic and hypoxic changes are variably found, reflecting systemic effects and local vascular pathology. Functional changes known as Long COVID also appear to affect the brain (15, 16). Thus, SARS-CoV-2 infection may exert functional effects on the brain without leading to overt damage. The nature of these functional effects remains to be defined.
Several lines of evidence converge on the brain vascular system as a key player in COVID-19 (17–26). SARS-CoV-2 may affect endothelial cells through cellular infection (i.e., “endotheliopathy”) or more indirectly through excessive cytokine signaling (or both). Involvement of brain endothelial cells and pericytes may lead to disturbances of the blood brain barrier (BBB) (27–33). Thereby, the invasion of the virus into the brain after the breakdown of the BBB (i.e., hematogenic spread) has attracted most of the attention, while other aspects of the BBB such as controlled substrate influx and efflux are also important.
Solute carrier transporters (SLCs) are a group of membrane transport proteins that carry specific substrates across biological membranes (34–36). SLCs expressed in brain endothelial cells are part of the BBB and selectively transport substrates that are essential for neurons and glial cells (37). Among these are amino acids serving as metabolites, precursors for protein synthesis, and neurotransmitters. The effects of viral infection on SLCs in endothelial cells and BBB could be functional if no cell death occurs in the vasculature. This concept is of particular interest in relation to Long COVID, which can manifest as neurological symptoms, including fatigue and brain fog (15, 16, 38). Several studies have demonstrated BBB changes in Long COVID (14, 29, 39). In addition, studies of various viruses (different from coronavirus) have identified specific SLCs as primary viral receptors or closely associated proteins (40). SLCs for amino acid transport (41, 42) or glucose transport (43) have been identified as viral receptors. Most recently, SLC1A5 has been experimentally shown to participate in SARS-CoV-2 infection (44). Therefore, SLCs could be also involved in the infection process of brain vascular cells. Notably, two amino acid transporters, SLC6A19 and SLC6A20, interact physiologically with ACE2 (Angiotensin Converting Enzyme 2), the key receptor for SARS-CoV-2, in the intestine (45). SLC6A20 has been genetically associated with the clinical course of COVID-19 in multiple GWAS (Genome-wide association studies) (46) and is expressed in brain vasculature (47). While SLCs at the BBB have been predominantly associated with transport into the brain, several ATP-binding cassette transporters (ABCs) in brain endothelial cells move substrates and drugs from the brain across the BBB and into the blood (48). Such efflux could become important if toxins are produced in the brain during infection.
In order to gain a better understanding of the pathophysiology of COVID-19 and to develop drug therapies, major efforts have been undertaken to analyze the interactions between SARS-CoV-2 and host proteins (49). Many viruses can infect the brain whereby viral proteins target host proteins (in neurons or glial cells) to inactivate them or redirect their functions (50). While the focus has been on replication mechanisms, other cellular functions such as substrate transport and protein synthesis are also affected whereby host SLCs could play a role. A large amount of “omics” data collected for SARS-CoV-2 infection has been made publicly available for data-mining (51–53). Proteomics data for virus-host protein interactions in SARS-CoV-2 cellular infection models are available in the public domain through the “SARS-CoV-2 and Coronavirus-Related Interactions” database in BioGRID (Biological General Repository for Interaction Datasets) (54). Combining this resource with other data collections, we carried out a data-mining study and generated a collection of SLCs expressed at the BBB that could be impaired by specific virus-host interactions during SARS-CoV-2 infection in brain endothelial cells. In the first step, information for SLCs as host proteins in SARS-CoV-2 infection was extracted from BioGRID (54). In the second step, expression of these putative virus-interacting SLCs was determined in brain endothelial cells using single cell RNA sequencing data (scRNA seq) as reported by Rosenberg et al. (47) for young mice. The list of virus-interacting host SLCs was further screened in literature and databases to consolidate expression and function in brain endothelial cells and BBB. Subsequently, the list of virus-interacting host SLCs expressed in endothelial cells was screened against various datasets and literature information for infection mechanisms and brain involvement involving seven steps. (1) The DAVID analysis tool was used for Functional Annotation Clustering, resulting in the quantitative classification of groups of SLCs according to their functions. (2) Substrates transported by host SLCs were retrieved from databases. (3) Interactome data were generated for all individual viral proteins of SARS-CoV-2, host SLCs identified, and tested for preferential interaction of SLCs with specific viral proteins. (4) To identify host SLCs critical for brain function, it was assumed that inactivation of host SLCs by viral proteins may have similar effects as disease-causing mutations of the same host proteins, and to this end, host SLCs were annotated for monogenic nervous system disorders using the OMIM™ (Online Mendelian Inheritance in Man) database. (5) Datasets reported for CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) screening of SARS-CoV-2 infected cell lines were screened for reporting of critical host SLCs. (6) Following a lead from the DAVID analysis, roles of SLCs in viral infection processes, including viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle, were explored in databases and literature, first specifically for SARS-CoV-2, and then more generally for other viruses. (7) Finally, SLCs were compared to ABC transporters expressed in endothelial cells.
Materials and methods
The workflow of this study has been outlined in Figure 1.
Figure 1

Flow chart of the data-mining process. Solute carrier proteins (SLCs) interacting with Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral proteins were identified in the virus-host protein–protein interaction (VH-PPI) data. Several analyses were conducted, including a Functional Annotation Clustering analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID); substrate classification using the National Center for Biotechnology Information (NCBI) Gene database; viral protein interactome identification using Biological General Repository for Interaction Datasets (BioGRID); diseases of the brain using Online Mendelian Inheritance in Man (OMIM™); Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) studies; roles of infection based on current literature; a comparison to ATP-binding cassette (ABC) proteins, and an expanded analysis to astrocytes and mural cells.
Selection of host SLCs interacting with SARS-CoV-2 viral proteins
The BioGRID database for proteomics data (54) provides the COVID-19 Coronavirus Curation Project, including the “SARS-CoV-2 and Coronavirus-Related Interactions” database (55). The file called “BIOGRID-PROJECT-covid19_coronavirus_project-LATEST.zip” was downloaded from BioGRID containing the September 1st, 2024 program update. The list was filtered so that “Organism Name Interactor A” and “Organism Name Interactor B” were limited to SARS-CoV-2 and/or Homo sapiens; this list of interaction data of SARS-CoV-2 viral proteins with human host proteins will be named virus-host protein–protein interaction (VH-PPI). The list was further filtered by using the first three letters “SLC” of the gene/protein symbols that have been uniformly assigned to denote solute carrier proteins as a large protein family, including “SLCO”; a few scaffold and adaptor proteins were retained. Validity of the gene/protein symbol was verified using the PANTHER (Protein ANalysis THrough Evolutionary Relationships) database (56), which includes the UniProtKG number and symbol. The number of reported listings was recorded for each extracted SLC protein. After removing duplicates, a final list of host SLCs was generated. To estimate possible enrichment within datasets, the total number of known SLCs was taken as n = 400 from a key review (34). The number of human proteins interrogated by proteomics techniques and appearing in the combined interactome studies was estimated at 12 k (out of 20 k protein-coding genes). It was assumed that the database of VH-PPI is strongly dominated by infection studies using the original strain and major variants circulating in 2020–21.
Annotation of SLCs interacting with SARS-CoV-2 viral proteins for endothelial cell expression
Single cell RNA sequencing data (scRNA seq) for cell classes of young mouse brain (including endothelial cells) were downloaded from a database that employed the split-pool ligation based transcriptomics sequencing (47). Data for n = 382 SLCs were available in this transcriptomics database. Next, the list of SLCs extracted from VH-PPI was merged with the list of n = 382 SLCs in the transcriptomics database and shared genes/proteins determined; human gene/protein symbols will be used during the subsequent reporting. A few host SLC proteins without expression information in the transcriptomics database were removed from the analysis. The selection of SLCs (serving as host proteins) with expression in mouse brain was named “SLC-H.” In the next step, SLCs with relevant expression in brain endothelial cells were determined using the scRNA seq data (column 64 of Table S5, “endothelia”) (47). To this end, a low threshold was set an expression level of n = 10 TPMs (Transcripts per Million), and the set of selected genes was named “SLC-H-E.” For comparison, the total number of SLCs obtained with the same threshold for endothelial cell expression (without selection for VH-PPI) was also determined. Literature searches for known roles in brain were carried out to confirm the vascular expression and functional roles in the BBB.
Functional annotation clustering
The data extracted for SLCs was further analyzed using DAVID (Database for Annotation and Integrated Discovery) (57, 58). This bioinformatics platform provides analysis tools for investigation and annotation of large gene lists for various categories including GO terms and UniprotKB keywords. The list of SLC-H-E was entered using “Official Gene Symbol,” “Homo sapiens” as background, and “Gene List” as selectors. The list of genes assigned with DAVID IDs was analyzed using the Functional Annotation Clustering tool, and clusters were provided with an enrichment score. Each cluster then reported a set of preset terms related to a specific functional annotation, such as GO terms with statistical information. Listed terms with a statistically significant enrichment following Bonferroni correction (at p < 0.05) were considered for the purpose of this study with a focus on GO terms. This approach allowed for the classification of different SLCs according to substrates, and in addition, uncovered functions of SLCs in virus entry. For a second approach, the Enrichr database was used (59–61). The list of SLC-H-E (n = 80) was entered, and the KEGG 2021 Human Pathway option was selected. SLCs were grouped according to functional category, and a Fisher Exact Test with adjusted p-value per Benjamini-Hochberg method and odds ratios were generated. As a third approach, the information under PANTHER Protein Class was compiled, and the number of SLCs in different protein classes was determined.
Substrates transported by selected host SLCs interacting with SARS-CoV-2 viral proteins in brain endothelial cells
Following the detection of several classes of SLCs in the DAVID analysis, a more detailed annotation of individual genes in SLC-H-E for the transported substrates was performed using the summary information provided by NCBI Gene (National Center for Biotechnology Information) (62). The outcome of this analysis was then checked against a specific database named “SLC Tables” at bioparadigms.org (63) that lists detailed information for SLCs, including aliases, transport type and substrates, along with references (mostly dated 2013). Thereby, it must be recognized that substrates remain uncertain or even unknown for a small number of SLCs; these were excluded from the tabulation and graphic presentation. SLCs were grouped by substrates, and expression levels in endothelial cells for each group were compared to “all” other SLCs within SLC-H-E, using unpaired t-test (p < 0.05; EXCEL Data Analysis tool).
Interactome of SARS-CoV-2 viral proteins interacting with host SLCs in brain endothelial cells
While most of the attention has been on the spike protein (S) of SARS-CoV-2, multiple viral proteins play a major role in the infection process. It was asked whether SLCs interacted preferentially with specific viral proteins that in turn have specific functions in controlling the immune response of host cells. BioGRID provided specific PPIs for each of the n = 29 viral proteins of SARS-CoV-2 (E, M, N, nsp1-nsp16, ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, ORF10, ORF14, S); definitions and functions are found in recent reviews (64, 65). Following the removal of virus-virus interactions, the total number of interacting host proteins was determined for each viral protein, and then matches for the list of SLC-H-E identified. The relative contribution of SLCs to all proteins was calculated for each viral protein and expressed as %. The random chance was estimated as n = 400 SLC proteins in a total of 12 k proteins (measured by proteomics), or 3.3%. Fisher’s exact tests were then carried out for each viral protein to estimate whether SLCs from the SLC-H-E list were randomly found, and viral proteins with significant underrepresentation (p < 0.002, corrected for multiple comparisons) excluded. Next, the selected viral proteins linked to SLC-H-E were examined for known roles in virus-mediated interference with host antiviral defenses (including interferon response). To this end, a recent review of the innate immune evasion strategies of SARS-CoV-2 was consulted (65). Textual information for n = 23 SARS-CoV-2 viral proteins (provided in Table 1 of the publication) was extracted, including the classification of “activities” of viral proteins. After combining and sorting, the three top “activities” were selected (“Blocks recognition by host sensors,” “Minimizes or masks inflammatory RNA,” “Blocks IFN signaling”) and then tabulated for the viral proteins linked to SLC-H-E.
Analysis of OMIM™ database for monogenic disease of the brain related to host SLCs interacting with SARS-CoV-2 viral proteins
To further prioritize interacting SLCs in relation to brain disease mechanisms, it was hypothesized that interactions with viral proteins may have similar inactivating effects on host SLCs as known mutations. Accordingly, SLC-H-E were screened for known mutations causing (monogenic) brain disorders using the OMIM™ database (66). Some genes affecting the peripheral nervous system and sensory organs were included. Most genes causing nervous system disorders were readily recognized by their “official name.” For some syndromes more detailed information for nervous system symptoms was retrieved from the “Clinical Synopsis” page associated with the entry for the disease. The analysis was then extended to all genes from SLC-H. Literature searches were used to supplement the analysis.
SLCs listed in CRISPR screening studies of SARS-CoV-2 infection
CRISPR techniques using array formats allow to knock-down host genes which can result in the identification of novel pro- or antiviral functions of host proteins (67). Accordingly, the literature reporting data for CRISPR approaches to SARS-CoV-2 infection was surveyed for reports of SLCs. Two studies reviewing previous results were analyzed first (67, 68) and then individual articles were scanned. The data were highly heterogenous across different publications due to different cell types used, variation of the CRISPR tools, and selection or presentation of significant hits, and therefore only few SLCs will be reported here.
Roles of SLCs in infection mechanisms for SARS-CoV-2 and other viruses
Based on the annotation of SLCs for virus entry detected by the DAVID bioinformatics analysis (reported below under Results), a broader search was carried out for links between selected SLCs and viruses. First, a collection of viral receptors in the “VThunter” database (40, 69) was examined for SLCs broadly involved in viral infections. Then, each SLC-H-E was screened using PubMed abstract searches that combined the search term “virus” with the gene symbol for each SLC. Known roles in viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle were identified. The initial search was directed at experimental studies of SARS-CoV-2, and it was then extended more broadly to other viruses.
Analysis of ABC transporters expressed in brain endothelial cells and interacting with SARS-CoV-2 proteins
Several ABC (ATP-binding cassette) transporters in brain endothelial cells move substrates and drugs from the brain over the BBB and into the blood (48). Theoretically, such efflux transporters may become relevant when viral infections lead to a build-up of toxic levels of small molecules within the brain. Accordingly, the list of VH-PPI (as generated before) was screened for host ABC transporters that interact with viral proteins. To this end, the letters “ABC” of the gene/protein symbol shared by this protein family were used for selection. PANTHER (56) was used to verify the gene/protein symbols. Subsequently, expression of selected ABC transporters in mouse brain endothelial cells was determined using the process described above for SLCs, including a cut-off at 10 TPM. A published data collection for n = 50 ABCs expressed at the BBB was retrieved and used to confirm the selections based on the transcriptomics data (48). Two-sample t-tests assuming equal variances were conducted for the number of entries of ABCs (using a cut-off at 10 TPM) compared to those for SLC-H-E; and for the expression levels in endothelial cells in both sets.
Expanded analysis of datasets including astrocytes and mural cells
The BBB function also involves astrocytes and mural cells (e.g., vascular leptomeningeal cells, VMLC), and SLCs expressed by these cell types may also be affected by viral proteins. Thereby, it is of interest to determine which SLCs defined for brain endothelial cells (SLC-H-E) are shared with these other cells and which ones are specific. To this end, datasets for astrocytes (column 70, “Astro GFAP” of table S5 (47)) and mural cells (column 66, “VLMC Slc6a13”) were downloaded, selected for presence on the SLC-H list, and categorized at an expression level of n = 10 TPMSs, using the same approach as for brain endothelial cells. The datasets for selected SLCs for the three cell types were then overlapped with shared genes determined and visualized in a Venn diagram. In addition, the viral proteins interacting with different host SLCs from this analysis were determined (as already described above for the analysis of endothelial cells) and tabulated for each cell type. ANOVA was used to examine the difference in the number of interactions between viral proteins and SLCs across the three cell types. Viral proteins interacting with SLCs in all three cell types were defined as targets of high interest.
Results
Host SLCs interacting with SARS-CoV-2 viral proteins
The file “BIOGRID-PROJECT-covid19_coronavirus_project-LATEST.zip” was downloaded from BioGRID and filtered for SARS-CoV-2 and human studies; the selection of SARS-CoV-2 viral proteins interacting with human host proteins led to n = 39,854 total entries and n = 6,993 singular host proteins. N = 134 SLCs were found among the host proteins by filtering with the first three letters of the gene/protein symbol. The number of listings (or “entries”) per SLC in the VH-PPI was determined, which may serve as a proxy for the relative importance of different SLCs. N = 45 SLCs had 10 or more interactions with the viral proteins listed. It is estimated that with about n = 400 SLCs in total described in the literature (34), one third of all SLCs have been listed as host proteins for SARS-CoV-2 (in VH-PPI). Assuming that 12 k “measurable” proteins were captured by the proteomics studies in the database and that host proteins in the VH-PPI (6.9 k) are about half of these, no relative enrichment for SLCs could be found among host proteins. To obtain independent confirmation for the validity of the SLC nomenclature, the gene list selected from BioGRID was entered into PANTHER for verification, including UniProt KB numbers and symbols; all selections made in BioGRID were matched in PANTHER (Supplementary material 1).
Host SLCs interacting with SARS-CoV-2 viral proteins annotated for endothelial cell expression
Subsequently, the database of Rosenberg et al. (47) for young mouse brain reporting scRNA seq data for n = 382 SLC genes was used for annotation. N = 134/382 genes (or 35%) were on the list of SLCs generated from the VH-PPI. Four genes (SLC18A3, SLC25A52, SLC25A6, SLCO6A1) were not in the mouse transcriptomics study and removed. The remainder of n = 130 genes/proteins will be named “SLC-H.” Next, the host SLCs expressed in mouse brain endothelial cells were determined. It was decided to limit further analyses to an expression level of n = 10 TPMs or higher in brain endothelial cells which resulted in n = 80 genes/proteins named “SLC-H-E.” The dataset SLC-H-E (n = 80) was analyzed in a number of subsequent studies; for some analyses, the additional genes in SLC-H were also considered as specified below. Table 1 lists the findings, with SLC-H-E marked in bold.
Table 1
| SLC1A1 | SLC4A1AP | SLC7A7 | SLC12A7 | SLC20A1 | SLC25A19 | SLC26A11 | SLC31A1 | SLC37A4 | SLC44A1 |
| SLC1A3 | SLC5A2 | SLC7A11 | SLC12A9 | SLC20A2 | SLC25A20 | SLC27A2 | SLC33A1 | SLC38A1 | SLC44A2 |
| SLC1A4 | SLC5A3 | SLC9A1 | SLC15A4 | SLC22A3 | SLC25A21 | SLC27A3 | SLC35A1 | SLC38A2 | SLC44A5 |
| SLC1A5 | SLC5A6 | SLC9A3R1 | SLC16A1 | SLC22A5 | SLC25A22 | SLC27A4 | SLC35A2 | SLC38A9 | SLC45A1 |
| SLC2A1 | SLC6A6 | SLC9A3R2 | SLC16A3 | SLC23A2 | SLC25A23 | SLC29A1 | SLC35B1 | SLC38A10 | SLC45A4 |
| SLC2A3 | SLC6A8 | SLC9A6 | SLC16A4 | SLC25A1 | SLC25A24 | SLC29A2 | SLC35B2 | SLC39A6 | SLC46A1 |
| SLC2A6 | SLC6A9 | SLC9A7 | SLC16A5 | SLC25A2 | SLC25A3 | SLC29A3 | SLC35C2 | SLC39A7 | SLC47A1 |
| SLC2A13 | SLC6A15 | SLC9A8 | SLC16A6 | SLC25A10 | SLC25A4 | SLC30A1 | SLC35D1 | SLC39A10 | SLC47A2 |
| SLC3A2 | SLC6A17 | SLC9B2 | SLC16A10 | SLC25A11 | SLC25A5 | SLC30A4 | SLC35E1 | SLC39A11 | SLC51B |
| SLC4A2 | SLC7A1 | SLC10A1 | SLC17A5 | SLC25A13 | SLC25A46 | SLC30A5 | SLC35F2 | SLC39A14 | SLC52A2 |
| SLC4A4 | SLC7A2 | SLC12A2 | SLC18B1 | SLC25A15 | SLC26A2 | SLC30A6 | SLC35F5 | SLC41A2 | SLCO3A1 |
| SLC4A7 | SLC7A5 | SLC12A4 | SLC19A1 | SLC25A16 | SLC26A4 | SLC30A7 | SLC35F6 | SLC41A3 | SLCO4A1 |
| SLC4A10 | SLC7A6 | SLC12A6 | SLC19A2 | SLC25A17 | SLC26A6 | SLC30A9 | SLC35G2 | SLC43A1 | SLCO4C1 |
Solute carrier proteins in virus-host protein–protein interactions.
Host solute carrier proteins (in total n = 130; SLC-H) found in the virus-host protein–protein interaction (VH-PPI) data are displayed in numerical order. The subset of host solute carrier proteins found at relevant levels in the transcriptome of mouse brain endothelial cells (n = 80; SLC-H-E) is indicated in bold.
Two tests were carried out to look for bias that could arise from the selection based on expression levels. First, proteins with high expression levels may have more chances to interact with other proteins and, hence, may show more entries in VH-PPI. To probe for such bias, the mRNA expression levels in endothelial cells (assuming proportional protein expression) were plotted against the counts for each SLC-H-E in the VH-PPI, and a very low correlation was found (r = 0.2186; Figure 2). Second, the expression levels of SLC-H-E were compared to those of SLCs not in the VH-PPI using the same threshold for endothelial cell expression (at 10 TPM); no difference in expression between these two gene sets was found (t-test, p = 0.58).
Figure 2
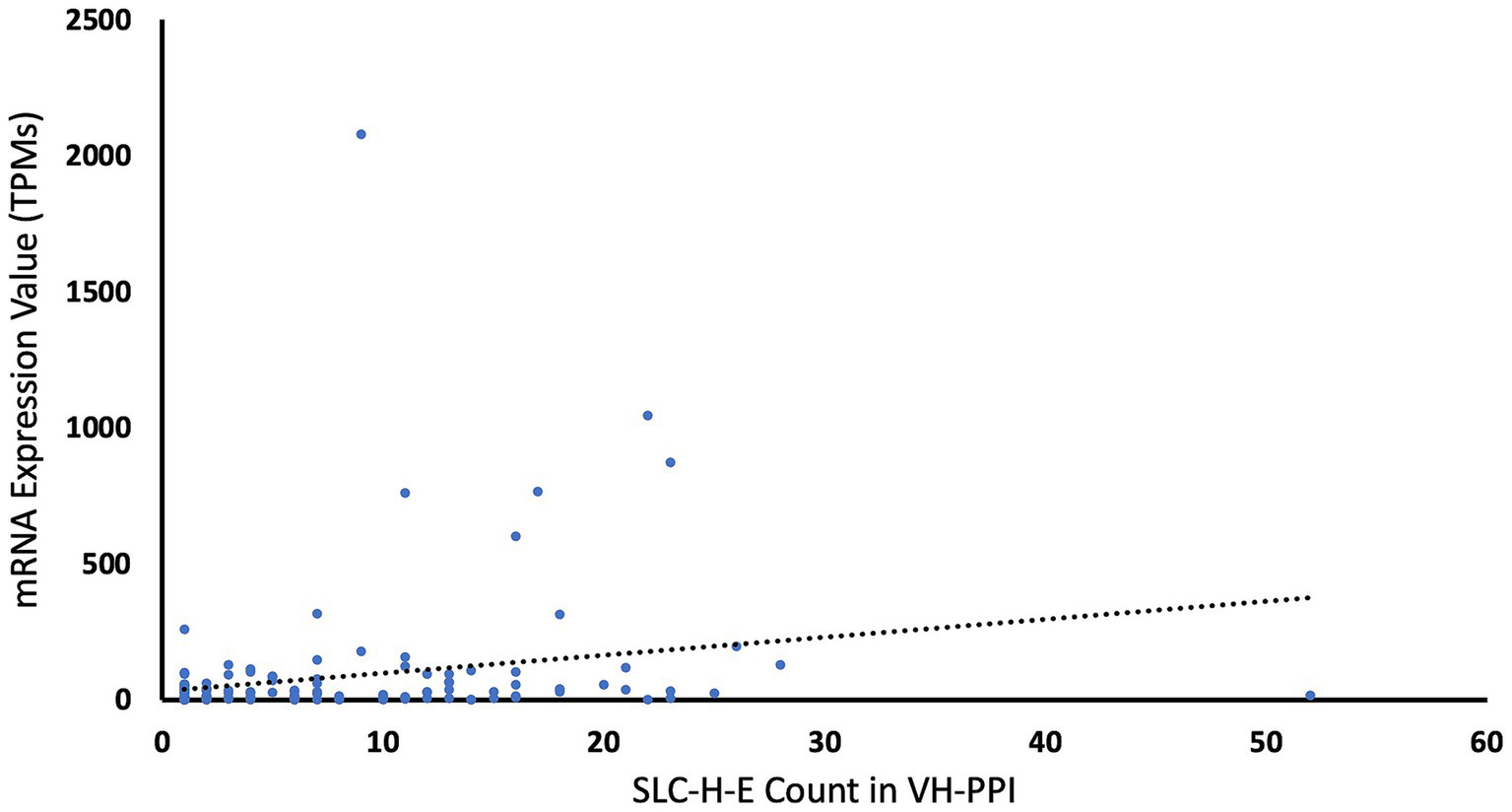
Counts of SLCs in virus-host protein–protein interaction data versus mRNA expression values. The chart displays the counts of each SLC-H-E in the virus-host protein–protein interaction data plotted against its brain endothelial expression value. The dotted line for linear fit indicates low correlation (r = 0.2186).
A literature survey showed that several SLC-H-E genes/proteins are already known to have brain endothelial cell expression and major BBB functions or they relate to vascular mechanisms relevant to COVID-19. For example, SLC5A6 is involved in pantothenic acid uptake by brain endothelial cells (70). SLC7A5/LAT1 (large neutral amino acids transporter small subunit) (1) is involved in L-leucine transport at the BBB and also serves as a major drug transporter, including transport of L-DOPA (71). SLC9A1 is key to regulating intracellular pH in endothelial cells, and subsequently, for the integrity of the BBB (72). SLC44A1 and SLC44A2 are implied in choline transport through the BBB (73). GWAS have suggested a role for SLC44A2 in regulating thrombosis, which involves mitochondria in platelets (74); this is of particular interest as formation of microthrombosis in cerebral vessels is considered a major cause of brain pathology in COVID-19. In addition, expression at the protein level could be confirmed for n = 34/80 SLC-H-E in a proteomics study of adult mouse brain capillaries (75).
Functional annotation clustering identifies classes of SLCs
The Functional Annotation Clustering tool provided multiple hits for SLC-H-E and terms of interest with significant enrichment after Bonferroni correction (p < 0.05) as shown in Table 2. In the category of GO terms, significant results were related to amino acid transmembrane transporters, zinc ion transport, and regulation of intracellular pH. N = 8 SLCs were classified under the GO term “neurotransmitter transport.” INTERPRO annotation was found for choline transport. The annotation term “Host cell receptor for virus entry” (“UP_KW_MOLECULAR FUNCTION,” derived from UniProt) listed SLC1A5, SLC3A2 and SLC20A2 (Bonferroni, p = 0.005). The related GO term “virus receptor activity” listed SLC1A5, SLC3A2, SLC7A1 and SLC20A2, for which basic enrichment analysis was listed as significant at p = 0.004 but did not pass Bonferroni correction (p = 0.6). The results of the Enrichr analysis are provided in Supplementary material 2. Notable findings by Enrichr (59–61) were “protein digestion and absorption,” “choline metabolism in cancer,” and “glutamatergic synapse,” which corresponds to annotations using DAVID. In addition, the qualitative information for PANTHER (56) protein classes was compiled (Supplementary material 1): secondary carrier transporter (n = 29); transporter (n = 25), primary active transporter (n = 7); amino acid transporter (n = 6), and mitochondrial carrier proteins (n = 3). While this classification is different from the ones provided using GO terms in DAVID, it introduces the new aspect of secondary transport, which depends on gradients produced by energy-dependent ion pumps.
Table 2
| Terms | Count | % | p-value | Fold enrichment | Bonferroni |
|---|---|---|---|---|---|
| GO:0003333 ~ amino acid transmembrane transport | 10 | 12.66 | 8.36E-16 | 90.36 | 5.05E-13 |
| GO:0015179 ~ L-amino acid transmembrane transporter activity | 8 | 10.13 | 9.03E-13 | 98.99 | 2.07E-10 |
| GO:0005283 ~ amino acid: sodium symporter activity | 3 | 3.80 | 1.56E-04 | 148.49 | 0.035059 |
| GO:0032328 ~ alanine transport | 5 | 6.33 | 7.43E-09 | 180.71 | 4.23E-06 |
| GO:0061459 ~ L-arginine transmembrane transporter activity | 5 | 6.33 | 1.13E-07 | 103.12 | 2.59E-05 |
| GO:1903826 ~ L-arginine transmembrane transport | 5 | 6.33 | 2.83E-07 | 84.33 | 1.61E-04 |
| GO:0015173 ~ aromatic amino acid transmembrane transporter activity | 3 | 3.80 | 9.37E-05 | 185.62 | 0.021239 |
| GO:0140009 ~ L-aspartate import across plasma membrane | 4 | 5.06 | 1.13E-06 | 168.67 | 6.44E-04 |
| GO:0015183 ~ L-aspartate transmembrane transporter activity | 4 | 5.06 | 5.04E-06 | 109.99 | 0.001153 |
| GO:0015174 ~ basic amino acid transmembrane transporter activity | 4 | 5.06 | 3.37E-06 | 123.74 | 7.71E-04 |
| GO:0015813 ~ L-glutamate transmembrane transport | 5 | 6.33 | 1.22E-06 | 60.24 | 6.93E-04 |
| GO:0006868 ~ glutamine transport | 5 | 6.33 | 2.66E-08 | 140.56 | 1.51E-05 |
| GO:0015186 ~ L-glutamine transmembrane transporter activity | 5 | 6.33 | 2.90E-08 | 137.49 | 6.65E-06 |
| GO:0015804 ~ neutral amino acid transport | 10 | 12.66 | 6.15E-17 | 115.00 | 6.32E-14 |
| GO:0015823 ~ phenylalanine transport | 3 | 3.80 | 8.97E-05 | 189.75 | 0.049767 |
| GO:0015828 ~ tyrosine transport | 3 | 3.80 | 4.50E-05 | 253.00 | 0.025263 |
| KW-0029 ~ Amino-acid transport | 17 | 21.52 | 7.99E-23 | 45.37 | 2.08E-21 |
| GO:0071577 ~ zinc ion transmembrane transport | 9 | 11.39 | 2.58E-14 | 94.88 | 1.47E-11 |
| GO:0005385 ~ zinc ion transmembrane transporter activity | 9 | 11.39 | 6.50E-14 | 85.67 | 1.49E-11 |
| GO:0071578 ~ zinc ion import across plasma membrane | 4 | 5.06 | 6.72E-06 | 101.20 | 0.003817 |
| GO:0006829 ~ zinc ion transport | 4 | 5.06 | 6.72E-06 | 101.20 | 0.003817 |
| GO:1904257 ~ zinc ion import into Golgi lumen | 3 | 3.80 | 4.50E-05 | 253.00 | 0.025263 |
| GO:0006882 ~ intracellular zinc ion homeostasis | 7 | 8.86 | 4.29E-09 | 50.60 | 2.44E-06 |
| IPR007603: Choline_transptr-like | 3 | 3.80 | 1.40E-04 | 156.51 | 0.016018 |
| GO:0008519 ~ ammonium channel activity | 4 | 5.06 | 1.31E-05 | 82.50 | 0.002991 |
| GO:0140157 ~ ammonium import across plasma membrane | 3 | 3.80 | 8.97E-05 | 189.75 | 0.049767 |
| GO:0015701 ~ bicarbonate transport | 7 | 8.86 | 6.11E-09 | 47.86 | 3.48E-06 |
| GO:1902476 ~ chloride transmembrane transport | 11 | 13.92 | 1.40E-11 | 25.53 | 7.95E-09 |
| GO:0055064 ~ chloride ion homeostasis | 4 | 5.06 | 9.21E-06 | 92.00 | 0.00523 |
| GO:0008509 ~ monoatomic anion transmembrane transporter activity | 4 | 5.06 | 7.18E-06 | 98.99 | 0.001642 |
| GO:0006811 ~ monoatomic ion transport | 16 | 20.25 | 1.42E-18 | 31.87 | 8.06E-16 |
| GO:0071805 ~ potassium ion transmembrane transport | 7 | 8.86 | 1.72E-05 | 12.74 | 0.009758 |
| GO:0055075 ~ potassium ion homeostasis | 4 | 5.06 | 6.21E-05 | 50.60 | 0.034705 |
| GO:0035725 ~ sodium ion transmembrane transport | 13 | 16.46 | 5.73E-14 | 26.52 | 3.26E-11 |
| GO:0098719 ~ sodium ion import across plasma membrane | 5 | 6.33 | 2.55E-06 | 50.60 | 0.001448 |
| GO:0051453 ~ regulation of intracellular pH | 7 | 8.86 | 1.28E-09 | 61.07 | 7.28E-07 |
| GO:0015718 ~ monocarboxylic acid transport | 4 | 5.06 | 3.73E-05 | 59.53 | 0.021023 |
| GO:0008028 ~ monocarboxylic acid transmembrane transporter activity | 4 | 5.06 | 4.77E-05 | 55.00 | 0.010862 |
| GO:0015866 ~ ADP transport | 4 | 5.06 | 4.72E-06 | 112.44 | 0.002681 |
| GO:0000295 ~ adenine nucleotide transmembrane transporter activity | 3 | 3.80 | 9.37E-05 | 185.62 | 0.021239 |
| GO:1990544 ~ mitochondrial ATP transmembrane transport | 4 | 5.06 | 4.72E-06 | 112.44 | 0.002681 |
| GO:0006839 ~ mitochondrial transport | 4 | 5.06 | 2.02E-05 | 72.29 | 0.011404 |
| GO:0006836 ~ neurotransmitter transport | 8 | 10.13 | 2.52E-10 | 48.19 | 1.43E-07 |
| GO:0031902 ~ late endosome membrane | 6 | 7.59 | 2.47E-04 | 10.58 | 0.033093 |
| KW-1183 ~ Host cell receptor for virus entry | 3 | 3.80 | 7.45E-04 | 61.92 | 0.005202 |
List of results of Functional Annotation Clustering.
The table displays the results for host SLCs expressed in endothelial cells (SLC-H-E) obtained by Functional Annotation Clustering analysis using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) tool. “Terms” are mainly from GO (Gene Ontology) categories, whereby counts, % and p-value are provided for each category, along with fold enrichment and the p-value for Bonferroni test (p < 0.05).
Important substrates are transported by selected host SLCs interacting with SARS-CoV-2 viral proteins in brain endothelial cells
In the following, the working hypothesis is that the interaction of a viral protein will negatively affect a related host SLC-H-E by reducing its availability for physiological protein interactions in the infected brain endothelial cells. Information for the substrates transported by n = 73/80 SLC-H-E was found in the summary information provided in NCBI Gene. Several classes of SLCs emerged, whereby n = 21/80 were determined to transport amino acids, n = 3/80 choline, and n = 9/80 zinc (Table 3; Figure 3). The substrates listed in the specific database named “SLC Tables” (at bioparadigms.org) largely coincided with the information provided under NCBI Gene, and inspection of the annotations identified by DAVID bioinformatics also indicated strong overlap. When comparing the expression levels in endothelial cells, the n = 21 SLCs grouped to amino acid transport had significantly higher expression values than all other SLCs (n = 59) within the SLC-H-E set (unpaired t-test, p = 0.006). This was not observed for SLCs related to zinc transport (p = 0.9). An additional analysis was carried out for the five SLC-H-E not annotated with a substrate in NCBI Gene (setting aside SLC4A1AP and SLC9A3R2). Using abstract searches in PubMed, four new annotations were found; SLC18B1 as a polyamine transporter (76); SLC25A46 as a mitochondrial carrier protein (77); SLC35E1 as a regulator of zinc concentration (78); and SLC35F5 as nucleotide sugar transporter (79). Following this, only SLC35F2 remained as an orphan transporter in the SLC-H-E analysis. Interestingly, a detailed study of SLC35F2 in the BBB has appeared (80).
Table 3
| Substrates | Count | SLC-H-E |
|---|---|---|
| Amino Acids | 21 | SLC1A1, SLC1A3, SLC1A4, SLC1A5, SLC3A2, SLC6A6, SLC6A15, SLC6A17, SLC7A1, SLC7A2, SLC7A5, SLC7A6, SLC7A7, SLC7A11, SLC15A4, SLC16A10, SLC25A13, SLC38A1, SLC38A2, SLC38A9, SLC38A10 |
| Zinc | 9 | SLC30A1, SLC30A4, SLC30A5, SLC30A6, SLC30A7, SLC30A9, SLC39A6, SLC39A10, SLC39A11 |
| Sodium, potassium, chloride, or hydrogen | 8 | SLC9A1, SLC9A6, SLC9A8, SLC12A2, SLC12A4, SLC12A6, SLC12A7, SLCO3A1 |
| ADP/ATP | 4 | SLC25A4, SLC25A17, SLC25A23, SLC25A24 |
| Bicarbonate | 4 | SLC4A2, SLC4A4, SLC4A7, SLC26A6 |
| Monocarboxylates | 3 | SLC16A1, SLC16A4, SLC16A6 |
| Phosphate | 3 | SLC20A1, SLC20A2, SLC25A3 |
| Choline | 3 | SLC44A1, SLC44A2, SLC44A5 |
| Carnitine | 2 | SLC22A5, SLC25A20 |
| Sulfate | 2 | SLC26A2, SLC26A11 |
| Nucleosides | 2 | SLC29A1, SLC29A3 |
| Glucose | 1 | SLC2A1 |
| Myo-inositol | 1 | SLC2A13 |
| Myo-inositol, glucose | 1 | SLC5A3 |
| Biotin | 1 | SLC5A6 |
| Sialic acid | 1 | SLC17A5 |
| Folate | 1 | SLC19A1 |
| Ascorbic Acid | 1 | SLC23A2 |
| Fatty acids | 1 | SLC27A4 |
| Copper | 1 | SLC31A1 |
| Acetyl-coenzyme A | 1 | SLC33A1 |
| CMP-sialic acid | 1 | SLC35A1 |
| UDP-glucuronic acid, UDP-N-acetylgalactosamineT | 1 | SLC35D1 |
List of known substrates of host SLCs expressed in endothelial cells.
The table provides the specific substrates of SLC-H-E as determined by NCBI Gene.
Figure 3
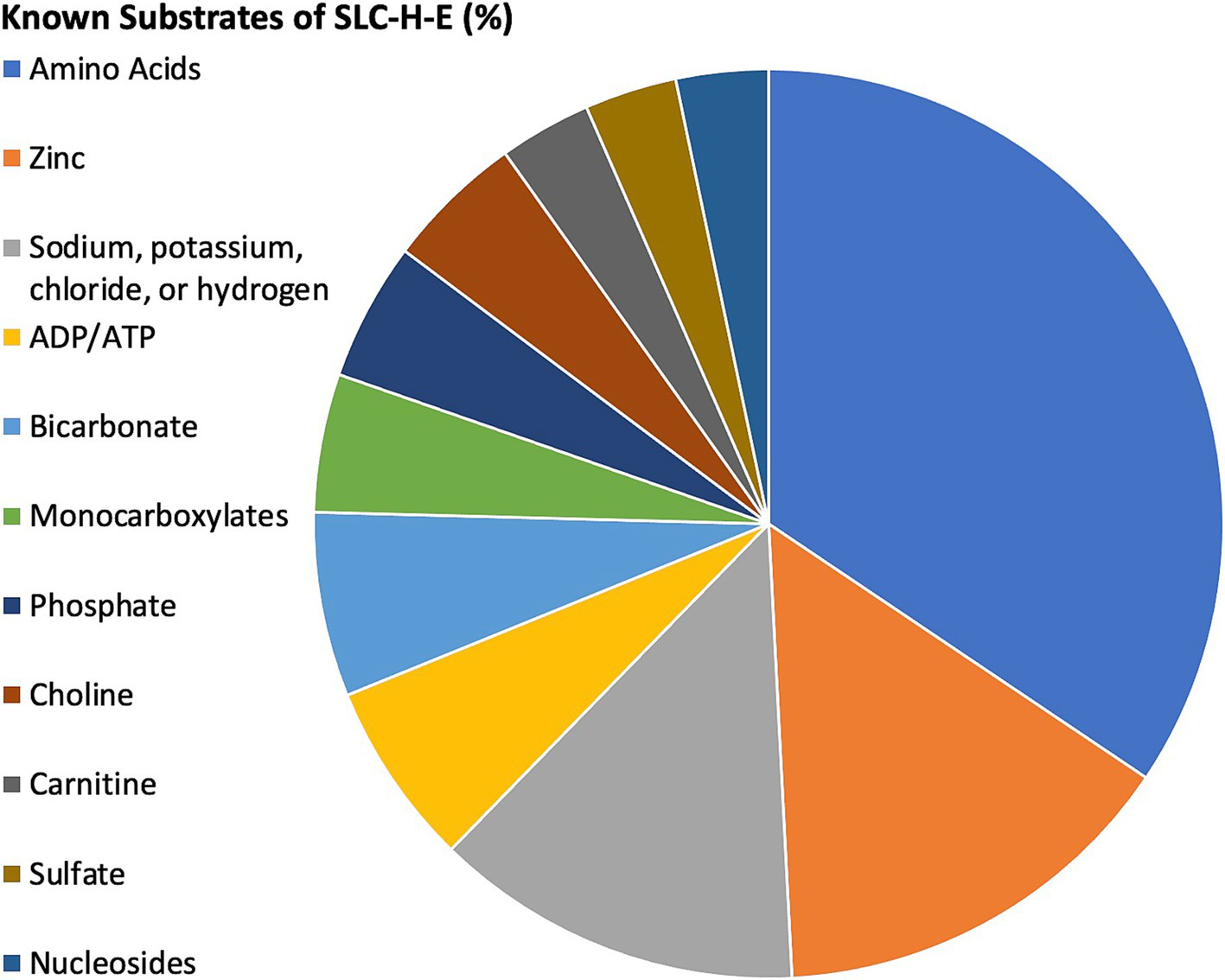
Relative distribution of substrate classes among selected SLCs. The pie chart demonstrates the relative contribution (in percentage) for classes of substrates with at least two members as identified by NCBI Gene for SLC-H-E. SLCs with unknown substrates were excluded. The key to the left shows the color coding for substrate classes. Distribution from highest to lowest were amino acids, zinc, sodium/potassium/chloride/hydrogen, ADP/ATP, bicarbonate, monocarboxylates, phosphate, choline, carnitine, sulfate, and nucleosides. For a complete list, see Table 3.
SARS-CoV-2 viral proteins interact with host SLCs in brain endothelial cells
Subsequently, it was asked whether SLC-H-E interact preferentially with a specific set of viral proteins. PPI information for n = 29 SARS-CoV-2 viral proteins was downloaded from BioGRID.
Following removal of duplicates, the number of host proteins and number of SLC genes from SLC-H-E was determined for each viral protein. Subsequently, the relative contribution of SLC-H-E was calculated and probed using Fisher’s exact test for each viral protein. The distribution according to counts of SLCs from SLC-H-E interacting with each viral protein is shown in Figure 4A, and the relative contribution of SLCs (in %) in Figure 4B. N = 17 viral proteins were excluded due to significant underrepresentation of SLCs; none were enriched. N = 12 viral proteins linked to SLCs remained for further consideration. Information for n = 23 SARS-CoV-2 viral proteins was extracted from a current review including the classification of “activities” of these viral proteins (65). Information for n = 9/12 viral proteins linked to SLCs was available for which the three top activities (“Blocks recognition by host sensors,” “Minimizes or masks inflammatory RNA,” “Blocks IFN signaling”) were assigned as listed in Table 4. A list of the SLCs (including their substrates) interacting with these specific viral proteins is provided in Supplementary material 3. Viral proteins blocking the interferon response stood out (n = 6/9).
Figure 4
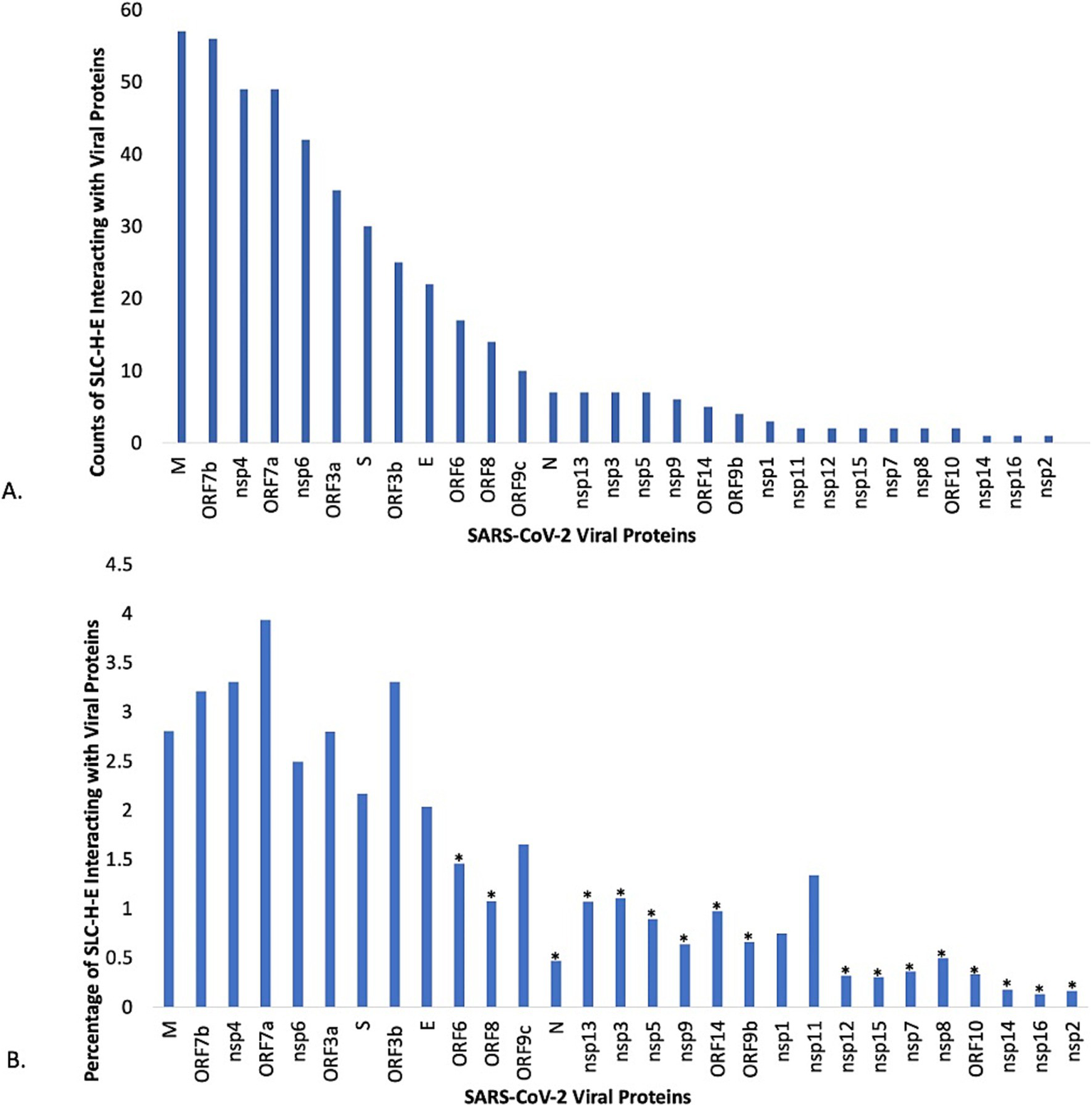
Analysis of individual viral proteins of SARS-CoV-2 for interactions with selected SLCs. (A) The plot displays the counts for host SLCs expressed in endothelial cells (SLC-H-E) after removing duplicates, as found in the interactome for each individual SARS-CoV-2 viral protein. (B) The plot displays the relative contribution of host SLCs expressed in endothelial cells (SLC-H-E) compared to the total count of host proteins in VH-PPI for each SARS-CoV-2 viral protein. Fisher’s exact tests were carried out for over- or underrepresentation of SLCs relative to the total count of host proteins interacting with each viral protein (* p < 0.002, corrected for multiple comparisons).
Table 4
| Viral protein | Blocks recognition by host sensors | Minimizes or masks inflammatory RNA | Blocks IFN signaling |
|---|---|---|---|
| M | X | ||
| ORF7b | X | X | |
| nsp4 | X | ||
| ORF7a | X | X | |
| nsp6 | X | X | X |
| ORF3a | X | ||
| S | X | ||
| ORF3b | X | ||
| nsp1 | X | X |
Functions of selected SARS-CoV-2 viral proteins interacting with host SLCs expressed in endothelial cells.
The table lists nine SARS-CoV-2 viral proteins linked to host SLCs expressed in endothelial cells that matched categories “Blocks recognition by host sensors,” “Minimizes or masks inflammatory RNA,” and “Blocks IFN signaling,” obtained from a recent review (61) listing n = 23 viral proteins related to the innate immune evasion strategies of SARS-CoV-2. More details are found in Supplementary File 3.
A subset of host SLCs interacting with SARS-CoV-2 viral proteins is linked to monogenic diseases of the brain
The assumption was made that interactions with viral proteins may have similar inactivating effects on SLCs as known genomic mutations in the nervous system, and that this allows for classifying specific SLCs as particularly interesting in the context of functional disturbances. Accordingly, sets of SLCs were annotated for known mutations causing nervous system disorders using the OMIM™ database based on the name of the disorder or information under “clinical synopsis.” N = 33/80 SLC-H-E genes were listed for any disorder, and n = 24/80 (or 30%) were classified as nervous system disorders. Endothelial cell expression may contribute to the disease process for these genes while other cells in the brain may also be involved (as discussed below under “Multiple host SLCs are shared between endothelial cells, astrocytes, and mural cells”). Therefore, the search was expanded to the remainder of genes in SLC-H, and n = 18 SLCs were also classified as linked to nervous system disorders. Taken together, n = 42 SLCs among the n = 130 in SLC-H (32%) were related to disorders of the nervous system (mostly affecting the brain) as shown in Supplementary material 4.
CRISPR screening studies of SARS-CoV-2 infection lead to host SLCs
A recent review for CRISPR studies of viral infections was consulted (67) and the genes of interest listed for experimental studies for SARS-CoV-2 retrieved. Among n = 180 genes listed, three SLCs were found, one of which was in SLC-H-E (SLC7A11) and another only in SLC-H (SLC35B2). Another report combined their CRISPR based data with findings from the literature (68) and reported n = 22 SLC genes, whereby n = 5 of these were in SLC-H-E.
Screening of individual studies using CRISPR-based approaches in cells with SARS-CoV-2 infection led to SLC35B2 (81); SLC35F4 (82); SLC5A1, SLC6A14, SLC6A19 and SLC35B2 (83); SLC1A5 and SLC31A1 (84); and SLC1A12, SLC12A9, SLC62A1 and SLC63A1 (85). Taken together, converging evidence from SLC-H and CRISPR studies points to SLC35B2, which has been also reported for other viruses (86).
SLCs play a role during infection with SARS-CoV-2 and other viruses
Links between SLCs and mechanisms of infection, including viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle, were examined, first for SARS-CoV-2 and then more broadly for other viruses. The database VThunter (40) contained n = 108 genes/proteins as viral receptors, out of which n = 7 were SLCs (SLC10A1, SLC1A5, SLC20A1, SLC20A2, SLC52A1, SLC52A2, SLC7A1). With n = 400 SLCs randomly expected among a total of 20 k genes in the genome, SLCs were estimated as enriched among viral receptors in this database (Chi-square test, GraphPad, two-tailed, p = 0.001). When tested against an estimated total of 12 k proteins measured, marginal significance was found (Chi-square test, one-tailed, p = 0.04). Four of these SLCs listed in VThunter were found on the SLC-H-E list (SLC1A5, SLC20A1, SLC20A2, SLC7A1) and two on list of SLC-H only (SLC10A1, SLC52A2). As reported above, the analysis using the DAVID bioinformatics tool indicated a significant enrichment for “Host cell receptor for virus entry” among the SLC-H-E selection whereby SLC1A5, SLC20A2, and SLC3A2 were listed; weak evidence for SLC7A1 was found.
In the following section, supportive information for links of SLC-H-E to SARS-CoV-2 infections, specifically, and to viral infections (including other viruses), more generally, will be reviewed. Thereby, genes listed in VThunter and/or obtained from DAVID analysis were given high priority, along with SLCs reported in experimental studies of SARS-CoV-2. SLC1A5 is a neutral amino acid transporter listed in VThunter and detected by DAVID analysis. Experimental studies for SARS-CoV-2 uncovered a role for SLC1A5 as a viral entry modulator of ACE2 (44). SLC1A5 was also listed for protein interactions with the SARS-CoV-2 Spike protein (87). SLC1A5 serves as receptor for the RD-114 virus, a feline endogenous retrovirus (42, 88). SLC3A2 transports amino acids and regulates intracellular calcium that was detected by DAVID analysis. An experimental study in SARS-CoV-2 infected cells searched for host proteins by cell surface proximity ligation techniques and tested the effect of chemical inhibition on virion uptake where SLC3A2 was identified as a novel target (89). A role for SLC3A2 had been already described for hepatitis C virus infection (90). SLC6A15 is a neutral amino acid transporter, and it was recently connected to SARS-CoV-2 infection via photocatalytic proximity labeling of the Spike interactome and listed as a candidate auxiliary host entry factor (91). Interestingly, SLC6A15 was grouped with SLC6A19 and SLC6A20 (that are interacting with ACE2) in a recent review of SLCs (45), and it has been linked to the hippocampus and depression (92). SLC7A1 is a transporter for specific amino acids and captured in VThunter as a receptor for bovine leukemia virus (93). SLC20A1 is a sodium-phosphate symporter and featured in VThunter, whereby it serves as receptor for gibbon ape leukemia virus (94). SLC20A2 is an inorganic phosphate transporter, listed in VThunter, emerging from the DAVID analysis and listed as Gibbon Ape Leukemia Retrovirus Receptor 2 (MIM 158378). SLC29A3 is a nucleoside transporter (also known as ENT3) that was experimentally found to be required for SARS-CoV-2 viral genome release (95). SLC38A9 is involved in several transport functions, including amino acids, and it was very recently described to regulate SARS-CoV-2 viral entry (84), whereby the cleavage product S1 of Spike present in the endolysosome lumen may interact with SLC38A9, leading to increased escape of the virus from endolysosomes and enhanced viral entry. Two additional genes from the broader set of SLC-H were also of interest. SLC10A1 (NTPC) is a specific receptor for hepatitis B or D viruses (96). SLC52A2 was captured by DAVID analysis, and it was described as receptor for the Porcine endogenous retrovirus A (97). Extended literature searches for SLC-H-E and viruses in general detected additional links to infection mechanisms for other viruses as shown in Supplementary material 5, illustrating that a variety of substrates can be involved. In summary, evidence from several sources indicates roles of SLCs expressed in brain endothelial cells (as defined by SLC-H-E) in viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle, and several SLCs with viral links are amino acid transporters.
ABC transporters expressed in brain endothelial cells interact with SARS-CoV-2 proteins
Analysis of the VH-PPI for the term “ABC” resulted in n = 169 entries comprised of n = 23 unique ABC genes/proteins (Table 5). Of these n = 17 were expressed in brain endothelial cells based on the transcriptomics data with a cut-off at 10 TPM (bold in Table 5). All selected ABC genes were found listed in a database for brain expression (48). No significant difference was identified between the number of entries in VH-PPI for ABCs (at a cut-off level of 10 TPM) and SLC-H-E (t-test, p = 0.562). No difference was found between the two gene sets for expression levels in brain endothelial cells (p = 0.4).
Table 5
| ABCA1 | ABCB7 | ABCB11 | ABCC4 | ABCD3 | ABCF2 |
| ABCA3 | ABCB8 | ABCC1 | ABCC5 | ABCD4 | ABCF3 |
| ABCB1 | ABCB9 | ABCC2 | ABCC9 | ABCE1 | ABCG2 |
| ABCB6 | ABCB10 | ABCC3 | ABCC10 | ABCF1 |
ABC transporters in virus-host protein–protein interactions.
The table illustrates the ABC transporter proteins identified in VH-PPI (n = 23). ABC proteins with an expression level of n = 10 TPMs or higher in brain endothelial cells are indicated in bold (n = 17).
Multiple host SLCs are shared between endothelial cells, astrocytes, and mural cells
The results of the combined analysis of SLCs interacting with viral proteins in endothelial cells, astrocytes, and mural cells (VLMC) are shown as a Venn diagram in Figure 5. The specific SLCs for each segment of the figure are listed in Supplementary material 6. The total numbers of selected SLCs were well-balanced among cell classes: endothelial cells (n = 80), astrocytes (n = 78), and mural cells (n = 81). The majority of SLCs were shared by all three cell types (n = 51), whereas much fewer SLCs were cell-specific: endothelial cells (n = 6), astrocytes (n = 7), and mural cells (n = 7). Next, the viral proteins (n = 26) were analyzed for interactions with SLCs whereby the number of interactions with SLCs was tabulated for each viral protein and cell type. Nine viral proteins (E, M, nsp4, nsp6, ORF3a, ORF3b, ORF7a, ORF7a, and S) had at least one interaction with SLCs for all three cell types. While the average number of interactions across all plotted viral proteins was highest for host SLCs from astrocytes (2.35), intermediate for mural cells (VLMC, 2.04) and lowest for endothelial cells (1.7), the differences were not significant (ANOVA, F = 0.37, p = 0.69). This analysis suggests that in addition to the central role of SLCs in endothelial cells (reached first by the virus during hematogenic spread), astrocytes and other vascular cells express SLCs relevant to viral infection with SARS-CoV-2. The shared SLCs could be defined as broad targets for interventions.
Figure 5
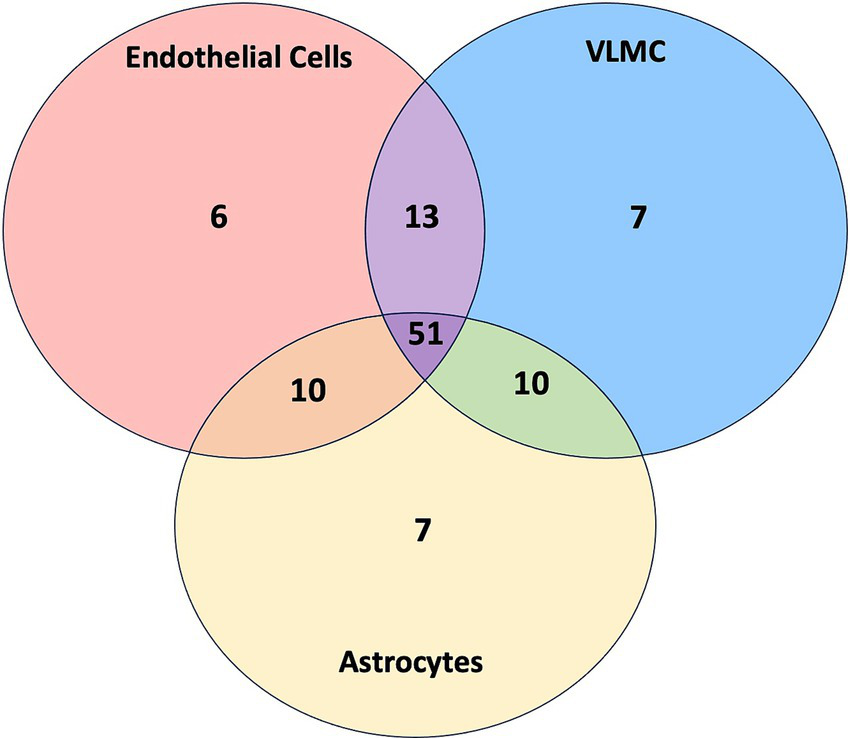
Venn diagram of SLCs in endothelial cells, VLMC, and astrocytes. The Venn diagram displays the number of SLCs belonging to each category, including endothelial cells (red), VLMC (blue), and astrocytes (yellow) as well as SLCs shared between multiple categories. Details are provided in Supplementary File 6.
Therapeutic strategies involving host SLCs
The annotation of SLCs interacting with SARS-CoV-2 viral proteins led to a number of specific transporters with key functions in endothelial cells. Such SLCs could be of interest for therapeutic interventions in COVID-19. SLC2A1 (GLUT1) is a key glucose transporter in brain endothelial cells, and dysfunction of this transporter due to a “block” by viral proteins could impact glucose levels in the brain. As mutations of SCL2A1 cause a clinical disorder, GLUT1 deficiency disorder (as listed under the analysis of OMIM), treatments may be already under development. Most prominently, n = 21 amino acid transporters were identified. Impairment of amino acid transport in brain endothelial cells could affect neuronal function, especially protein synthesis, which in turn is important for memory formation (98). Increasing amino acid delivery across the BBB may be tested as a strategy to overcome chronic, functional neurological changes after COVID-19 infection. Given the complex overlap of transported amino acids amongst different types of transporters and the number of transporters potentially affected, it is difficult to predict which amino acids would be most affected and should be targeted for supplementation. Theoretically, infusion with standard cocktails of amino acids used for parental nutrition could be tested in clinical trials. While this study mostly considered a decrease of transport functions by SLCs targeted by viral proteins, there is a theoretical possibility that viruses utilize host SLC functions to increase amino acid uptake for their own advantage. In this case, supplementation should be avoided. Nine SLCs serving as zinc transporters in brain endothelial cells were found to interact with viral proteins, and zinc has been discussed extensively in relation to COVID-19 (99, 100). Three SLCs serving as choline transporters were identified, and levels of choline in the corpus callosum were shown to be increased in patients affected by COVID-19 when measured with proton MR spectroscopy (101). It remains to be studied whether supplementation with choline would be indicated. The functional annotation tool DAVID returned “neurotransmitter transport” as a significant finding for the list of SLCs expressed in endothelial cells, and similarly Enrichr had “glutamatergic synapse” as a significant result. Transport of amino acids across the BBB may indirectly be connected to the neurotransmitter pools for glutamate and amino acid co-agonists at the NMDA receptor, which in turn is relevant to excitotoxic neuronal damage in viral infections (102, 103). SLC12A2 (NKCC1, Na-K-2Cl cotransporter 1) was detected as a host target shared by all three cell classes. NKCC1 regulates chloride levels important for the function of ionotropic GABA receptors, and NKCC1 is a drug target for neurodevelopmental disorders (104, 105). SLCs are major drug targets, and several compounds already exist to influence SLC functions, including in the brain (71, 105–107). Future studies should directly examine the function of SLCs after infection of brain endothelial cells to identify transport deficits that could be targeted. A caveat is that drugs blocking SLCs used for treatment of CNS disorders may work synergistically with the negative effects of viral proteins and should be critically evaluated. SLCs supporting viral entry and replication may become targets for drug therapy in COVID-19. ABC transporters play important roles in drug movements across the BBB, and abnormalities in ABC functions due to interactions with viral proteins could alter drug levels in the brain during infection. Pharmacogenomics studies have considered genetic variants of ABCs in relation to drug therapy of COVID-19 (108).
Discussion
The rationale for this hypothesis-generating study is based on the critical role the brain vasculature and the BBB play in COVID-19 infection (17–33). Once endothelial cells become infected, viral proteins interacting with SLCs may impair transport of their substrates across the BBB, in particular of amino acids. The downstream effects of impaired transport will be decreased substrate delivery to neurons and glial cells, which impair protein synthesis due to reduced influx of amino acids. In conjunction with the systemic inflammatory response in COVID-19 (11–13), altered transport across the BBB could explain diffuse brain dysfunction, including cognitive deficits (14, 29). Perturbances of transmembrane transport may also affect cellular metabolism of brain endothelial cells themselves, possibly impair their viability, and add to the dysfunction at the BBB. In addition, inflammatory responses may cause abnormal interactions between endothelial cells, thrombocytes, and immune cells, leading to thrombosis followed by local ischemia (9, 39, 109). Disturbances of endothelial cells will be also sensed by perivascular astrocytic processes and microglial cells, contributing to the glial responses (neuroinflammation). Possible roles of selected SLCs as therapeutic targets have been addressed above under “Therapeutic strategies involving host SLCs.”
This study sought to identify a subset of SARS-CoV-2 viral proteins that interact with SLCs and then related these to known viral protein functions (64, 65). To this end, information for viral proteins was retrieved from a recent review of the innate immune evasion strategies of SARS-CoV-2 (65), and nine viral proteins interacting with host SLCs were categorized for key functions. This observation raises questions about how host SLCs are possibly involved in defense mechanisms through protein interactions, particularly interferon responses. Nsp6 emerged from the present analysis of host SLCs by matching all three categories for immune evasion, and interestingly, nsp6 mutations have been associated with the attenuation of infection seen in Omicron BA.1 variants (110). This study also examined ABC transporters expressed in brain endothelial cells as these proteins may serve as efflux transporters (48). Here, the concept was that inflammation may lead to a buildup of toxic substances within the brain parenchyma that need to be removed from the brain into the blood. Several ABCs were engaged in PPIs with viral proteins, perhaps suggesting decreased removal and intracerebral elevation of toxins. To further strengthen the argument for important functions of SLCs, monogenic disorders of the nervous system involving SLCs were also identified. Thereby, the idea was that mutations of SLCs may allow for predicting the sensitivity of SLCs to inhibitory protein interactions. Several SLCs related to disorders of the nervous system, in particular of the brain, were found. Given the involvement of white matter in COVID-19 (9), it is interesting that some genetic disorders of SLCs can affect the corpus callosum and the optic nerve. Some disorders involving SLCs can be classified as neurodevelopmental (111, 112), which may be of interest when studying the effects of SARS-CoV-2 on the developing brain (113).
The mechanisms of SARS-CoV-2 entry into cells involving the key receptor, ACE2, have been reviewed (114). More recently, several original reports have appeared for roles of SLCs in mechanisms of SARS-CoV-2 infections (44, 84, 87, 89, 91). Thereby, SLC1A5 stands out due to its involvement in other viral infections (44). In the present study, the DAVID analysis and literature evidence indicated that SLCs can be engaged in mechanisms of viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle. This finding suggests a secondary role for SLCs serving in auxiliary functions in SARS-CoV-2 infection that may play out in brain endothelial cells. In fact, literature searches revealed that several SLCs are known to serve as cellular receptors or in closely associated functions for viruses other than coronavirus. Using the data for viral receptors in general from VThunter, it was found that SLCs are enriched among cellular receptors when considered on a genomic basis. It is intriguing that some of these SLCs serving in viral entry for other viruses are expressed in brain endothelial cells and are part of the interactome for SARS-CoV-2 proteins. The significance of this finding is presently unclear. Theoretically, prevention of viral interference could occur if SARS-CoV-2 viral proteins block SLCs used by other viruses as receptors which helps to avoid competing infections. SLCs may be broadly important for a number of viral infections, and some viruses (but not coronaviruses) may have specialized in recruiting specific SLCs as receptors. In the broader context, it is remarkable that ACE2, the key receptor for SARS-CoV-2, is known to interact with two amino acid transporters, SLC6A19 or SLC6A20, in enterocytes (45). These two proteins are not involved in direct interactions with SARS-CoV-2 viral proteins, and therefore, did not emerge from the present search in VH-PPI. Recent studies show that interaction with SLC6A20 may affect ACE2 receptor functions (115).
In the following, six limitations of this study need to be addressed.
-
This research combined data from studies in human cells at the protein level with mRNA data obtained in the mouse brain. The analysis was based on the annotations and listings in BioGRID, which use a unified symbol for the protein and gene based on SLC nomenclature; in addition, gene symbols were confirmed in the PANTHER database. Thereby, the assumption was made that SLCs are expressed in both mouse and human endothelial cells with a similar pattern and that they have closely related functions. The total number of SLCs was assumed to be n = 400 based on a review published in 2015 (34), which is close to the number of SLCs available in the mouse brain database (n = 382) published in 2018 (47). The list of SLCs is growing, in part due to new discoveries and the renaming of genes and proteins (116). The statistical analyses for enrichment of SLCs serving as viral receptors under VThunter depended on the total number used for the Chi-square tests. An estimate with a total number of SLCs of n = 450 showed that the enrichment remained significant (p = 0.0048) when referenced to 20 k genes, but became non-significant (p = 0.08) when referenced to 12 k proteins; this would not change the suggestion that SLCs play a role in viral receptor mechanisms. There is a theoretical possibility that a newly identified solute carrier has been given a protein name but has not yet been assigned a gene symbol beginning with “SLC,” and therefore was not detected. However, given the number of host SLCs with well-known functions in the BBB detected here, this would not change any conclusions.
-
The “SARS-CoV-2 and Coronavirus-Related Interactions” database within BioGRID contains a vast collection of interacting proteins that have been pooled from a large set of experiments using different types of human cell lines and different proteomic techniques. Some of these data are from work at the preprint stage. It is not clear to what extent some techniques have detected weak interactions in the respective assays, leading to an overestimation of the VH-PPI. No virus-host interactome data are available for cell lines closely resembling human brain endothelial cells (to the best of our knowledge). Since new studies with PPI information are regularly being added to the specialized BioGRID database, coverage was assumed to be complete. To test the validity of this assumption, individual studies of virus-host PPIs for SARS-CoV-2 were identified in PubMed abstract searches and successfully located in the BioGRID database. Since the present data analysis was closed by September 2024, newer studies for PPI data were also searched, and only one study was identified (117) in which a small set of SLCs was found, overlapping with the selection used here.
-
Theoretically, inactivation of host SLCs due to interaction with viral proteins may be compensated by the increased transcription of the same SLCs. Several studies have provided transcriptomics data of SARS-CoV-2 infected cells or post-mortem tissues whereby results for SLCs have been listed, e.g., for lungs (118). However, interpretation of such transcriptomics data is difficult, because a change of expression in SLCs, in particular down-regulation, could reflect generalized transcriptional failure and mRNA degradation caused by the acute viral take-over of cellular functions. On the other hand, it will be interesting to study expression levels of SLCs in the chronic state after COVID-19.
-
Only a few SLCs from VH-PPI data were also detected in CRISPR screens. The reason for this discrepancy could be that the read out from CRISPR screens is cell death upon viral infection, whereas no endpoint is used for the protein interaction data. A caveat is that the results from different CRISPR screens are not convergent and depend on the cell type used for screening (67). On the other hand, it will be interesting to test experimentally whether acute knock-down of several SLCs identified here could aggravate cellular deterioration after infection.
-
Several pathophysiological models have been based on the concept that SARS-CoV-2 infects brain endothelial cells and disturbs the BBB (17–33). Accordingly, the present study modeled the interaction of viral proteins with SLCs within infected brain endothelial cells, and the focus was on functional effects of transport across the BBB. It is conceivable that dysfunction of SLCs could also impair metabolism of endothelial cells themselves, which in turn could lead to an increased leakiness of the BBB, including viral entry into the brain parenchyma. Endothelial cells may be also indirectly involved by high levels of cytokines, inflammation and interaction with immune cells, which could impact metabolism and function of endothelial cells. Blood flow regulation could be impaired, leading to intermittent hypoperfusion and reduced oxygen delivery to neurons. To what degree SLCs in brain endothelial cells are affected by specific interactions with viral proteins and (or) by more general factors remains to be studied. It should also be noted that some experimental studies have challenged the concept that infection of brain endothelial cells by SARS-CoV-2 is a major factor (119, 120). Finally, the choroid plexus has been also established as an important site for SARS-CoV-2 infection of the brain (9, 13, 119); multiple SLCs are expressed in the choroid plexus which could be explored in the future using the strategies developed in the present study.
-
This analysis was focused on SLCs as one of the largest protein families with multiple important functions (34), but it should be recognized that many other protein families are involved in SARS-CoV-2 infection. The annotations and correlations presented for SLCs as such do not establish superior importance of these transmembrane transporters for SARS-CoV-2 infections. However, it is notable that SLCs are closely related to receptor functions for several viruses.
In conclusion, given the key role of the brain vasculature in neurological manifestations of COVID-19, the present hypothesis-generating work set out to analyze data for SLCs as important transporters at the BBB. By combining virus-host protein–protein interaction data and transcriptomics data for brain endothelial cells, a set of n = 80 host SLCs was prioritized for analysis. Amino acid transporters formed the largest group of SLCs involved. Information for brain disease associated with mutations of SLCs suggested importance for selected SLCs that may be impaired by viral protein interactions. Specific functions of viral proteins of SARS-CoV-2 interacting with SLCs were defined, including the interferon response. Impairment of SLCs in brain endothelial cells during the acute phase of COVID-19 may reduce substrate supply for neurons and glial cells. Cognitive changes in long-COVID could be theoretically caused by persistent dysfunction of SLCs at the BBB. An additional finding emerging from this study was that some SLCs play key roles in the infection mechanisms for SARS-CoV-2; this observation must be seen in the broader context that SLCs themselves can serve as receptors for other viruses.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants' legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
TF: Writing – original draft. RS-K: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. RS-K was supported by departmental funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2025.1563040/full#supplementary-material
Supplementary Material 1The table displays the results of the PANTHER analysis which confirmed the Gene Name/Gene Symbol and identified the PANTHER Family/Subfamily and Protein Class for SLC-H-E and ABC proteins with an expression level of n = 10 TPMs or higher in brain endothelial cells.
Supplementary Material 2The table displays the results of the Enrichr analysis which classified SLC-H-E into fifty-four functional categories and performed a Fisher Exact test.
Supplementary Material 3The table extends Table 4 by providing the specific SLCs interacting with each viral protein selected based on a recent review (121). For the selection, refer to the text vand legend for Table 4.
Supplementary Material 4The table displays the results of the OMIM™ Analysis which sought to identify SLCs associated with known monogenic disorders of the brain and their corresponding functions per NCBI Gene.
Supplementary Material 5The table illustrates nine SLCs with connections to viruses other than SARS-CoV-2, including the specific virus involved, role during infection, substrates transported, literature source of the SLC-virus interaction, and associated PMID.
Supplementary Material 6Table A illustrates the SLCs in endothelial cells, astrocytes, and VLMC as well as SLCs shared between the three categories. Additionally, Table B displays the number of interactions between SARS-CoV-2 viral proteins and SLCs in endothelial cells, astrocytes, and VLMC.
Abbreviations
ABC, ATP-Binding Cassette; ACE2, Angiotensin Converting Enzyme 2; BBB, Blood-Brain Barrier; BioGRID, Biological General Repository for Interaction Datasets; COVID-19, Coronavirus Disease of 2019; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; DAVID, Database for Annotation, Visualization, and Integrated Discovery; GWAS, Genome-Wide Association Studies; NCBI, National Center for Biotechnology Information; OMIM, Online Mendelian Inheritance in Man; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; scRNA, Single Cell RNA Sequencing Data; SLC, Solute Carrier; SLC-H, SLCs in SARS-CoV-2 VH-PPI and transcriptomics database for mouse brain; SLC-H-E, SLCs in SARS-CoV-2 VH-PPI with brain endothelial expression level of n ≥ 10 TPMs; TPM, Transcripts Per Million; VH-PPI, Virus-Host Protein-Protein Interaction; VLMC, Vascular Leptomeningeal Cells.
References
1.
Hu B Guo H Zhou P Shi ZL . Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. (2021) 19:141–54. doi: 10.1038/s41579-020-00459-7
2.
Carabelli AM Peacock TP Thorne LG Harvey WT Hughes J Peacock SJ et al . SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. (2023) 21:162–77. doi: 10.1038/s41579-022-00841-7
3.
Burki T . WHO ends the COVID-19 public health emergency. Lancet Respir Med. (2023) 11:588. doi: 10.1016/S2213-2600(23)00217-5
4.
Huang C Wang Y Li X Ren L Zhao J Hu Y et al . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:49. doi: 10.1016/S0140-6736(20)30183-5
5.
Zhu N Zhang D Wang W Li X Yang B Song J et al . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
6.
Matschke J Lütgehetmann M Hagel C Sperhake JP Schröder AS Edler C et al . Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. (2020) 19:919–29. doi: 10.1016/S1474-4422(20)30308-2
7.
Schurink B Roos E Radonic T Barbe E Bouman CSC de Boer HH et al . Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. (2020) 1:e290–9. doi: 10.1016/S2666-5247(20)30144-0
8.
Thakur KT Miller EH Glendinning MD Al-Dalahmah O Banu MA Boehme AK et al . COVID-19 neuropathology at Columbia University Irving medical center/New York Presbyterian hospital. Brain. (2021) 144:2696–708. doi: 10.1093/brain/awab148
9.
Meinhardt J Streit S Dittmayer C Manitius RV Radbruch H Heppner FL . The neurobiology of SARS-CoV-2 infection. Nat Rev Neurosci. (2024) 25:30–42. doi: 10.1038/s41583-023-00769-8
10.
Helms J Kremer S Merdji H Clere-Jehl R Schenck M Kummerlen C et al . Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. (2020) 382:2268–70. doi: 10.1056/NEJMc2008597
11.
Iadecola C Anrather J Kamel H . Effects of COVID-19 on the nervous system. Cell. (2020) 183:16–27.e1. doi: 10.1016/j.cell.2020.08.028
12.
Bauer L Laksono BM de Vrij FMS Kushner SA Harschnitz O van Riel D . The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. (2022) 45:358–68. doi: 10.1016/j.tins.2022.02.006
13.
Jagst M Pottkämper L Gömer A Pitarokoili K Steinmann E . Neuroinvasion and neurotropism of severe acute respiratory syndrome coronavirus 2 infection. Curr Opin Microbiol. (2024) 79:102474. doi: 10.1016/j.mib.2024.102474
14.
Monje M Iwasaki A . The neurobiology of long COVID. Neuron. (2022) 110:3484–96. doi: 10.1016/j.neuron.2022.10.006
15.
Taquet M Geddes JR Husain M Luciano S Harrison PJ . 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry. (2021) 8:416–27. doi: 10.1016/S2215-0366(21)00084-5
16.
Xu E Xie Y Al-Aly Z . Long-term neurologic outcomes of COVID-19. Nat Med. (2022) 28:2406–15. doi: 10.1038/s41591-022-02001-z
17.
Lee MH Perl DP Nair G Li W Maric D Murray H et al . Microvascular injury in the brains of patients with Covid-19. N Engl J Med. (2021) 384:481–3. doi: 10.1056/NEJMc2033369
18.
Lee MH Perl DP Steiner J Pasternack N Li W Maric D et al . Neurovascular injury with complement activation and inflammation in COVID-19. Brain. (2022) 145:2555–68. doi: 10.1093/brain/awac151
19.
Proust A Queval CJ Harvey R Adams L Bennett M Wilkinson RJ . Differential effects of SARS-CoV-2 variants on central nervous system cells and blood-brain barrier functions. J Neuroinflammation. (2023) 20:184. doi: 10.1186/s12974-023-02861-3
20.
Wenzel J Lampe J Müller-Fielitz H Schuster R Zille M Müller K et al . The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat Neurosci. (2021) 24:1522–33. doi: 10.1038/s41593-021-00926-1
21.
Giordo R Paliogiannis P Mangoni AA Pintus G . SARS-CoV-2 and endothelial cell interaction in COVID-19: molecular perspectives. Vasc Biol. (2021) 3:R15–23. doi: 10.1530/VB-20-0017
22.
Yang RC Huang K Zhang HP Li L Zhang YF Tan C et al . SARS-CoV-2 productively infects human brain microvascular endothelial cells. J Neuroinflammation. (2022) 19:149. doi: 10.1186/s12974-022-02514-x
23.
Kakarla V Kaneko N Nour M Khatibi K Elahi F Liebeskind DS et al . Pathophysiologic mechanisms of cerebral endotheliopathy and stroke due to Sars-CoV-2. J Cereb Blood Flow Metab. (2021) 41:1179–92. doi: 10.1177/0271678X20985666
24.
Whitmore HAB Kim LA . Understanding the role of blood vessels in the neurologic manifestations of coronavirus disease 2019 (COVID-19). Am J Pathol. (2021) 191:1946–54. doi: 10.1016/j.ajpath.2021.04.017
25.
Paniz-Mondolfi A Bryce C Grimes Z Gordon RE Reidy J Lednicky J et al . Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J Med Virol. (2020) 92:699–702. doi: 10.1002/jmv.25915
26.
Bernard I Limonta D Mahal LK Hobman TC . Endothelium infection and dysregulation by SARS-CoV-2: evidence and caveats in COVID-19. Viruses. (2021) 13:29. doi: 10.3390/v13010029
27.
Krasemann S Haferkamp U Pfefferle S Woo MS Heinrich F Schweizer M et al . The blood-brain barrier is dysregulated in COVID-19 and serves as a CNS entry route for SARS-CoV-2. Stem Cell Reports. (2022) 17:307–20. doi: 10.1016/j.stemcr.2021.12.011
28.
Yamada S Hashita T Yanagida S Sato H Yasuhiko Y Okabe K et al . SARS-CoV-2 causes dysfunction in human iPSC-derived brain microvascular endothelial cells potentially by modulating the Wnt signaling pathway. Fluids Barriers CNS. (2024) 21:32. doi: 10.1186/s12987-024-00533-9
29.
Greene C Connolly R Brennan D Laffan A O'Keeffe E Zaporojan L et al . Blood-brain barrier disruption and sustained systemic inflammation in individuals with long COVID-associated cognitive impairment. Nat Neurosci. (2024) 27:421–32. doi: 10.1038/s41593-024-01576-9
30.
Hernández-Parra H Reyes-Hernández OD Figueroa-González G González-Del Carmen M González-Torres M Peña-Corona SI et al . Alteration of the blood-brain barrier by COVID-19 and its implication in the permeation of drugs into the brain. Front Cell Neurosci. (2023) 17:1125109. doi: 10.3389/fncel.2023.1125109
31.
Song H Tomasevich A Acheampong KK Schaff DL Shaffer SM Dolle JP et al . Detection of blood-brain barrier disruption in brains of patients with COVID-19, but no evidence of brain penetration by SARS-CoV-2. Acta Neuropathol. (2023) 146:771–5. doi: 10.1007/s00401-023-02624-7
32.
Trevino TN Almousawi AA Robinson KF Fogel AB Class J Minshall RD et al . Caveolin-1 mediates blood-brain barrier permeability, neuroinflammation, and cognitive impairment in SARS-CoV-2 infection. J Neuroimmunol. (2024) 388:578309. doi: 10.1016/j.jneuroim.2024.578309
33.
Martinez TE Mayilsamy K Mohapatra SS Mohapatra S . Modulation of Paracellular permeability in SARS-CoV-2 blood-to-brain Transcytosis. Viruses. (2024) 16:785. doi: 10.3390/v16050785
34.
César-Razquin A Snijder B Frappier-Brinton T Isserlin R Gyimesi G Bai X et al . A call for systematic research on solute carriers. Cell. (2015) 162:478–87. doi: 10.1016/j.cell.2015.07.022
35.
Kandasamy P Gyimesi G Kanai Y Hediger MA . Amino acid transporters revisited: new views in health and disease. Trends Biochem Sci. (2018) 43:752–89. doi: 10.1016/j.tibs.2018.05.003
36.
Pizzagalli MD Bensimon A Superti-Furga G . A guide to plasma membrane solute carrier proteins. FEBS J. (2021) 288:2784–835. doi: 10.1111/febs.15531
37.
Abbott NJ Patabendige AA Dolman DE Yusof SR Begley DJ . Structure and function of the blood-brain barrier. Neurobiol Dis. (2010) 37:13–25. doi: 10.1016/j.nbd.2009.07.030
38.
Mina Y Enose-Akahata Y Hammoud DA Videckis AJ Narpala SR O'Connell SE et al . Deep phenotyping of neurologic Postacute sequelae of SARS-CoV-2 infection. Neurol Neuroimmunol Neuroinflamm. (2023) 10:e200097. doi: 10.1212/NXI.0000000000200097
39.
Kempuraj D Aenlle KK Cohen J Mathew A Isler D Pangeni RP et al . COVID-19 and Long COVID: disruption of the neurovascular unit, blood-brain barrier, and tight junctions. Neuroscientist. (2024) 30:421–39. doi: 10.1177/10738584231194927
40.
Chen D Tan C Ding P Luo L Zhu J Jiang X et al . VThunter: a database for single-cell screening of virus target cells in the animal kingdom. Nucleic Acids Res. (2022) 50:D934–42. doi: 10.1093/nar/gkab894
41.
Kaleeba JA Berger EA . Kaposi's sarcoma-associated herpesvirus fusion-entry receptor: cystine transporter xCT. Science. (2006) 311:1921–4. doi: 10.1126/science.1120878
42.
Rasko JE Battini JL Gottschalk RJ Mazo I Miller AD . The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. (1999) 96:2129–34. doi: 10.1073/pnas.96.5.2129
43.
Manel N Kim FJ Kinet S Taylor N Sitbon M Battini JL . The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell. (2003) 115:449–59. doi: 10.1016/S0092-8674(03)00881-X
44.
Miluzio A Cuomo A Cordiglieri C Donnici L Pesce E Bombaci M et al . Mapping of functional SARS-CoV-2 receptors in human lungs establishes differences in variant binding and SLC1A5 as a viral entry modulator of hACE2. EBioMedicine. (2023) 87:104390. doi: 10.1016/j.ebiom.2022.104390
45.
Kukułowicz J Pietrzak-Lichwa K Klimończyk K Idlin N Bajda M . The SLC6A15-SLC6A20 neutral amino acid transporter subfamily: functions, diseases, and their therapeutic relevance. Pharmacol Rev. (2023) 76:142–93. doi: 10.1124/pharmrev.123.000886
46.
Eshetie S Jullian P Benyamin B Lee SH . Host genetic determinants of COVID-19 susceptibility and severity: a systematic review and meta-analysis. Rev Med Virol. (2023) 33:e2466. doi: 10.1002/rmv.2466
47.
Rosenberg AB Roco CM Muscat RA Kuchina A Sample P Yao Z et al . Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. (2018) 360:176–82. doi: 10.1126/science.aam8999
48.
Zhang W Liu QY Haqqani AS Liu Z Sodja C Leclerc S et al . Differentialexpression of receptors mediating receptor-mediated transcytosis (RMT) in brain microvessels, brain parenchyma and peripheral tissues of the mouse and the human. Fluids Barriers CNS. (2020) 17:47. doi: 10.1186/s12987-020-00209-0
49.
Gordon DE Jang GM Bouhaddou M Xu J Obernier K White KM et al . A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. (2020) 583:459–68. doi: 10.1038/s41586-020-2286-9
50.
Wiley CA . Emergent viral infections of the CNS. J Neuropathol Exp Neurol. (2020) 79:823–42. doi: 10.1093/jnen/nlaa054
51.
Baggen J Vanstreels E Jansen S Daelemans D . Cellular host factors for SARS-CoV-2 infection. Nat Microbiol. (2021) 6:1219–32. doi: 10.1038/s41564-021-00958-0
52.
Aggarwal S Acharjee A Mukherjee A Baker MS Srivastava S . Role of multiomics data to understand host-pathogen interactions in COVID-19 pathogenesis. J Proteome Res. (2021) 20:1107–32. doi: 10.1021/acs.jproteome.0c00771
53.
Kwok AJ Mentzer A Knight JC . Host genetics and infectious disease: new tools, insights and translational opportunities. Nat Rev Genet. (2021) 22:137–53. doi: 10.1038/s41576-020-00297-6
54.
Oughtred R Rust J Chang C Breitkreutz BJ Stark C Willems A et al . The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. (2021) 30:187–200. doi: 10.1002/pro.3978
55.
BioGrid interaction database. Available online at: https://thebiogrid.org. Accessed 14 Sept 2024.
56.
Mi H Muruganujan A Ebert D Huang X Thomas PD . PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. (2019) 47:D419–26. doi: 10.1093/nar/gky1038
57.
DAVID functional annotation bioinformatics microarray analysis. Available online at: https://davidbioinformatics.nih.gov. Accessed 14 Sept 2024.
58.
Sherman BT Hao M Qiu J Jiao X Baseler MW Lane HC et al . DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. (2022) 50:W216–21. doi: 10.1093/nar/gkac194
59.
Enrichr . Available online at: https://maayanlab.cloud/Enrichr/. Accessed 30 March 2025.
60.
Kuleshov MV Jones MR Rouillard AD Fernandez NF Duan Q Wang Z et al . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:W90–7. doi: 10.1093/nar/gkw377
61.
Xie Z Bailey A Kuleshov MV Clarke DJB Evangelista JE Jenkins SL et al . Gene set knowledge discovery with Enrichr. Curr Protoc. (2021) 1:e90. doi: 10.1002/cpz1.90
62.
NCBI gene . Available online at: https://www.ncbi.nlm.nih.gov/gene. Accessed 14 Sept 2024.
63.
BioParadigms . Available online at: https://bioparadigms.org. Accessed 14 Sept 2024.
64.
Justo Arevalo S Castillo-Chávez A Uribe Calampa CS Zapata Sifuentes D Huallpa CJ Landa Bianchi G et al . What do we know about the function of SARS-CoV-2 proteins?Front Immunol. (2023) 14:1249607. doi: 10.3389/fimmu.2023.1249607
65.
Minkoff JM tenOever B . Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. (2023) 21:178–94. doi: 10.1038/s41579-022-00839-1
66.
Online Mendelian Inheritance in Man database . Available online at: https://omim.org. Accessed 14 Sept 2024.
67.
See WR Yousefi M Ooi YS . A review of virus host factor discovery using CRISPR screening. MBio. (2024) 15:e0320523. doi: 10.1128/mbio.03205-23
68.
Synowiec A Jedrysik M Branicki W Klajmon A Lei J Owczarek K et al . Identification of cellular factors required for SARS-CoV-2 replication. Cells. (2021) 10:3159. doi: 10.3390/cells10113159
69.
VThunter . Available online at: https://db.cngb.org/VThunter/. Accessed 14 Sept 2024.
70.
Uchida Y Ito K Ohtsuki S Kubo Y Suzuki T Terasaki T . Major involvement of Na(+) -dependent multivitamin transporter (SLC5A6/SMVT) in uptake of biotin and pantothenic acid by human brain capillary endothelial cells. J Neurochem. (2015) 134:97–112. doi: 10.1111/jnc.13092
71.
Puris E Gynther M Auriola S Huttunen KM . L-type amino acid transporter 1 as a target for drug delivery. Pharm Res. (2020) 37:88. doi: 10.1007/s11095-020-02826-8
72.
Liktor-Busa E Blawn KT Kellohen KL Wiese BM Verkhovsky V Wahl J et al . Functional NHE1 expression is critical to blood brain barrier integrity and sumatriptan blood to brain uptake. PLoS One. (2020) 15:e0227463. doi: 10.1371/journal.pone.0227463
73.
Inazu M . Functional expression of choline transporters in the blood-brain barrier. Nutrients. (2019) 11:2265. doi: 10.3390/nu11102265
74.
Bennett JA Mastrangelo MA Ture SK Smith CO Loelius SG Berg RA et al . The choline transporter Slc44a2 controls platelet activation and thrombosis by regulating mitochondrial function. Nat Commun. (2020) 11:3479. doi: 10.1038/s41467-020-17254-w
75.
Hamada Y Ogata S Masuda T Ito S Ohtsuki S . Development of a method for isolating brain capillaries from a single neonatal mouse brain and comparison of proteomic profiles between neonatal and adult brain capillaries. Fluids Barriers CNS. (2023) 20:50. doi: 10.1186/s12987-023-00449-w
76.
Fredriksson R Sreedharan S Nordenankar K Alsiö J Lindberg FA Hutchinson A et al . The polyamine transporter Slc18b1 (VPAT) is important for both short and long time memory and for regulation of polyamine content in the brain. PLoS Genet. (2019) 15:e1008455. doi: 10.1371/journal.pgen.1008455
77.
Bitetto G Malaguti MC Ceravolo R Monfrini E Straniero L Morini A et al . SLC25A46 mutations in patients with Parkinson's disease and optic atrophy. Parkinsonism Relat Disord. (2020) 74:1–5. doi: 10.1016/j.parkreldis.2020.03.018
78.
Huang T Chen S Ding K Zheng B Lv W Wang X et al . SLC35E1 promotes keratinocyte proliferation in psoriasis by regulating zinc homeostasis. Cell Death Dis. (2023) 14:354. doi: 10.1038/s41419-023-05874-1
79.
Hao GJ Ding YH Wen H Li XF Zhang W Su HY et al . Attenuation of deregulated miR-369-3p expression sensitizes non-small cell lung cancer cells to cisplatin via modulation of the nucleotide sugar transporter SLC35F5. Biochem Biophys Res Commun. (2017) 488:501–8. doi: 10.1016/j.bbrc.2017.05.075
80.
Mochizuki T Mizuno T Kurosawa T Yamaguchi T Higuchi K Tega Y et al . Functional investigation of solute carrier family 35, member F2, in three cellular models of the primate blood-brain barrier. Drug Metab Dispos. (2021) 49:3–11. doi: 10.1124/dmd.120.000115
81.
Sunshine S Puschnik AS Replogle JM Laurie MT Liu J Zha BS et al . Systematic functional interrogation of SARS-CoV-2 host factors using perturb-seq. Nat Commun. (2023) 14:6245. doi: 10.1038/s41467-023-41788-4
82.
Israeli M Finkel Y Yahalom-Ronen Y Paran N Chitlaru T Israeli O et al . Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2. Nat Commun. (2022) 13:2237. doi: 10.1038/s41467-022-29896-z
83.
Rebendenne A Roy P Bonaventure B Chaves Valadão AL Desmarets L Arnaud-Arnould M et al . Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat Genet. (2022) 54:1090–102. doi: 10.1038/s41588-022-01110-2
84.
Datta G Rezagholizadeh N Hasler WA Khan N Chen X . SLC38A9 regulates SARS-CoV-2 viral entry. iScience. (2024) 27:110387. doi: 10.1016/j.isci.2024.110387
85.
Ugalde AP Bretones G Rodríguez D Quesada V Llorente F Fernández-Delgado R et al . Autophagy-linked plasma and lysosomal membrane protein PLAC8 is a key host factor for SARS-CoV-2 entry into human cells. EMBO J. (2022) 41:e110727. doi: 10.15252/embj.2022110727
86.
Thamamongood T Aebischer A Wagner V Chang MW Elling R Benner C et al . A genome-wide CRISPR-Cas 9 screen reveals the requirement of host cell Sulfation for Schmallenberg virus infection. J Virol. (2020) 94:e00752–20. doi: 10.1128/JVI.00752-20
87.
Iacobucci I Monaco V Canè L Bibbò F Cioffi V Cozzolino F et al . Spike S1 domain interactome in non-pulmonary systems: a role beyond the receptor recognition. Front Mol Biosci. (2022) 9:975570. doi: 10.3389/fmolb.2022.975570
88.
Miyaho RN Nakagawa S Hashimoto-Gotoh A Nakaya Y Shimode S Sakaguchi S et al . Susceptibility of domestic animals to a pseudotype virus bearing RD-114 virus envelope protein. Gene. (2015) 567:189–95. doi: 10.1016/j.gene.2015.04.079
89.
Williams N Silva F Schmolke M . Harnessing host enhancers of SARS-CoV-2 entry as novel targets for antiviral therapy. Antivir Res. (2024) 228:105951. doi: 10.1016/j.antiviral.2024.105951
90.
Nguyen NNT Lim YS Nguyen LP Tran SC Luong TTD Nguyen TTT et al . Hepatitis C virus modulates solute carrier family 3 member 2 for viral propagation. Sci Rep. (2018) 8:15486. doi: 10.1038/s41598-018-33861-6
91.
Suzuki S Geri JB Knutson SD Bell-Temin H Tamura T Fernández DF et al . Photochemical identification of auxiliary severe acute respiratory syndrome coronavirus 2 host entry factors using μMap. J Am Chem Soc. (2022) 144:16604–11. doi: 10.1021/jacs.2c06806
92.
Santarelli S Namendorf C Anderzhanova E Gerlach T Bedenk B Kaltwasser S et al . The amino acid transporter SLC6A15 is a regulator of hippocampal neurochemistry and behavior. J Psychiatr Res. (2015) 68:261–9. doi: 10.1016/j.jpsychires.2015.07.012
93.
Bai L Sato H Kubo Y Wada S Aida Y . CAT1/SLC7A1 acts as a cellular receptor for bovine leukemia virus infection. FASEB J. (2019) 33:14516–27. doi: 10.1096/fj.201901528R
94.
Farrell KB Tusnady GE Eiden MV . New structural arrangement of the extracellular regions of the phosphate transporter SLC20A1, the receptor for gibbon ape leukemia virus. J Biol Chem. (2009) 284:29979–87. doi: 10.1074/jbc.M109.022566
95.
Hsieh YT Tsai TL Huang SY Heng JW Huang YC Tsai PY et al . IFN-stimulated metabolite transporter ENT3 facilitates viral genome release. EMBO Rep. (2023) 24:e55286. doi: 10.15252/embr.202255286
96.
Ni Y Lempp FA Mehrle S Nkongolo S Kaufman C Fälth M et al . Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. (2014) 146:1070–1083.e6. doi: 10.1053/j.gastro.2013.12.024
97.
Colon-Moran W Argaw T Wilson CA . Three cysteine residues of SLC52A1, a receptor for the porcine endogenous retrovirus-a (PERV-a), play a critical role in cell surface expression and infectivity. Virology. (2017) 507:140–50. doi: 10.1016/j.virol.2017.04.019
98.
Finch CE Morgan DG . RNA and protein metabolism in the aging brain. Annu Rev Neurosci. (1990) 13:75–88. doi: 10.1146/annurev.ne.13.030190.000451
99.
Cereda G Ciappolino V Boscutti A Cantù F Enrico P Oldani L et al . Zinc as a neuroprotective nutrient for COVID-19-related neuropsychiatric manifestations: a literature review. Adv Nutr. (2022) 13:66–79. doi: 10.1093/advances/nmab110
100.
Lahaye C Parant F Haesebaert J Goldet K Bendim'red L Henaff L et al . Minerals and antioxidant micronutrients levels and clinical outcome in older patients hospitalized for COVID-19 during the first wave of the pandemic. Nutrients. (2023) 15:1516. doi: 10.3390/nu15061516
101.
Pajuelo D Dezortova M Hajek M Ibrahimova M Ibrahim I . Metabolic changes assessed by 1H MR spectroscopy in the corpus callosum of post-COVID patients. MAGMA. (2024) 37:937–46. doi: 10.1007/s10334-024-01171-w
102.
Mizuguchi M Yamanouchi H Ichiyama T Shiomi M . Acute encephalopathy associated with influenza and other viral infections. Acta Neurol Scand Suppl. (2007) 186:45–56. doi: 10.1111/j.1600-0404.2007.00809.x
103.
Chaganti J Poudel G Cysique LA Dore GJ Kelleher A Matthews G et al . Blood brain barrier disruption and glutamatergic excitotoxicity in post-acute sequelae of SARS COV-2 infection cognitive impairment: potential biomarkers and a window into pathogenesis. Front Neurol. (2024) 15:1350848. doi: 10.3389/fneur.2024.1350848
104.
Ben-Ari Y . The GABA excitatory/inhibitory developmental sequence: a personal journey. Neuroscience. (2014) 279:187–219. doi: 10.1016/j.neuroscience.2014.08.001
105.
Cherubini E Di Cristo G Avoli M . Dysregulation of GABAergic signaling in neurodevelomental disorders: targeting cation-chloride co-transporters to re-establish a proper E/I balance. Front Cell Neurosci. (2021) 15:813441. doi: 10.3389/fncel.2021.813441
106.
Garibsingh RA Schlessinger A . Advances and challenges in rational drug design for SLCs. Trends Pharmacol Sci. (2019) 40:790–800. doi: 10.1016/j.tips.2019.08.006
107.
Giacomini KM Yee SW Koleske ML Zou L Matsson P Chen EC et al . New and emerging research on solute carrier and ATP binding cassette transporters in drug discovery and development: outlook from the international transporter consortium. Clin Pharmacol Ther. (2022) 112:540–61. doi: 10.1002/cpt.2627
108.
Takahashi T Luzum JA Nicol MR Jacobson PA . Pharmacogenomics of COVID-19 therapies. NPJ Genom Med. (2020) 5:35. doi: 10.1038/s41525-020-00143-y
109.
Ryu JK Yan Z Montano M Sozmen EG Dixit K Suryawanshi RK et al . Fibrin drives thromboinflammation and neuropathology in COVID-19. Nature. (2024) 633:905–13. doi: 10.1038/s41586-024-07873-4
110.
Chen DY Chin CV Kenney D Tavares AH Khan N Conway HL et al . Spike and nsp 6 are key determinants of SARS-CoV-2 omicron BA.1 attenuation. Nature. (2023) 615:143–50. doi: 10.1038/s41586-023-05697-2
111.
McNeill A Iovino E Mansard L Vache C Baux D Bedoukian E et al . SLC12A2 variants cause a neurodevelopmental disorder or cochleovestibular defect. Brain. (2020) 143:2380–7. doi: 10.1093/brain/awaa176
112.
Tărlungeanu DC Deliu E Dotter CP Kara M Janiesch PC Scalise M et al . Impaired amino acid transport at the blood brain barrier is a cause of autism Spectrum disorder. Cell. (2016) 167:1481–94.e18. doi: 10.1016/j.cell.2016.11.013
113.
Dubey H Sharma RK Krishnan S Knickmeyer R . SARS-CoV-2 (COVID-19) as a possible risk factor for neurodevelopmental disorders. Front Neurosci. (2022) 16:1021721. doi: 10.3389/fnins.2022.1021721
114.
Jackson CB Farzan M Chen B Choe H . Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. (2022) 23:3–20. doi: 10.1038/s41580-021-00418-x
115.
Li HZ Pike ACW Lotsaris I Chi G Hansen JS Lee SC et al . Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2. Nat Commun. (2024) 15:5503. doi: 10.1038/s41467-024-48921-x
116.
Gyimesi G Hediger MA . Systematic in silico discovery of novel solute carrier-like proteins from proteomes. PLoS One. (2022) 17:e0271062. doi: 10.1371/journal.pone.0271062
117.
Huuskonen S Liu X Pöhner I Redchuk T Salokas K Lundberg R et al . The comprehensive SARS-CoV-2 'hijackome' knowledge base. Cell Discov. (2024) 10:125. doi: 10.1038/s41421-024-00748-y
118.
Bhargava A Szachnowski U Chazal M Foretek D Caval V Aicher SM et al . Transcriptomic analysis of sorted lung cells revealed a proviral activity of the NF-κB pathway toward SARS-CoV-2. iScience. (2023) 26:108449. doi: 10.1016/j.isci.2023.108449
119.
Stüdle C Nishihara H Wischnewski S Kulsvehagen L Perriot S Ishikawa H et al . SARS-CoV-2 infects epithelial cells of the blood-cerebrospinal fluid barrier rather than endothelial cells or pericytes of the blood-brain barrier. Fluids Barriers CNS. (2023) 20:76. doi: 10.1186/s12987-023-00479-4
120.
Constant O Barthelemy J Bolloré K Tuaillon E Gosselet F Chable-Bessia C et al . SARS-CoV-2 poorly replicates in cells of the human blood-brain barrier without associated deleterious effects. Front Immunol. (2021) 12:697329. doi: 10.3389/fimmu.2021.697329
121.
Minkoff JM tenOever B . Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. (2023) 21:178–94.
Summary
Keywords
SARS-CoV-2, COVID-19, blood-brain barrier, brain endothelial cells, solute carrier proteins, amino acid transport, virus-host interactions, protein-protein interactions
Citation
Fradkin T and Schmidt-Kastner R (2025) A data-mining analysis of host solute carrier family proteins in SARS-CoV-2 infection with reference to brain endothelial cells and the blood-brain barrier in COVID-19. Front. Neurol. 16:1563040. doi: 10.3389/fneur.2025.1563040
Received
18 January 2025
Accepted
03 June 2025
Published
01 July 2025
Volume
16 - 2025
Edited by
Kempuraj Duraisamy, Nova Southeastern University, United States
Reviewed by
Sven Marcel Stefan, University of Oslo, Norway
Deepyaman Das, Raiganj University, India
Updates
Copyright
© 2025 Fradkin and Schmidt-Kastner.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rainald Schmidt-Kastner, schmidtk@health.fau.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.