Abstract
Purpose:
This study aimed to investigate the diagnostic potential of neutrophil extracellular traps (NETs)-related genes in acute ischemic stroke (AIS) through comprehensive bioinformatics analysis.
Methods:
Two GEO datasets (GSE37587 and GSE16561) were integrated to identify differentially expressed genes (DEGs) between AIS patients and healthy controls. Gene Set Enrichment Analysis (GSEA) was performed to explore functional pathways, while single-sample GSEA (ssGSEA) was used to evaluate immune cell infiltration patterns. NETs-related DEGs (NDEGs) were identified by intersecting the DEGs with previously reported NETS-related genes. Functional enrichment of NDEGs was performed using Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Key genes were identified via machine learning algorithms, including least absolute shrinkage and selection operator (LASSO) and random forest (RF). A diagnostic model was constructed based on the identified hub genes and validated using an independent dataset (GSE58294). Potential regulatory miRNAs and candidate therapeutic compounds were predicted using the TargetScan and DSigDB databases, respectively.
Results:
The discovery dataset included 73 AIS patients and 24 healthy controls, revealed 551 DEGs (225 upregulated, 326 downregulated). The analysis of ssGSEA revealed notable immune dysregulation in AIS patients, characterized by increased neutrophil infiltration and decreased level of Th17, Th1, and TFH cells. GSEA indicated that DEGs were enriched in neutrophil degranulation and innate immune system. NDEGs were significantly enriched in immune regulation and leukocyte apoptosis (GO) and NETs formation pathway (KEGG). Four hub genes—SRC, TLR8, FCAR, and HIF1A—were identified using LASSO and RF algorithms. A diagnostic model based on these genes yielded area under the curve (AUC) values of 0.880 in the training dataset and 0.936 in the validation dataset. Furthermore, three regulatory miRNAs (miR-146a-5p, miR-155-5p, and miR-21-5p) and 23 candidate therapeutic drugs were predicted.
Conclusion:
To our knowledge, this represents the first comprehensive investigation of NETs-related gene signatures in AIS patients compared with healthy controls. These findings deepen our understanding of immune cell infiltration and the underlying molecular mechanisms involved in stroke, offering novel insights that may enhance diagnostic accuracy and therapeutic strategies for AIS.
Introduction
Ischemic stroke (IS) is a neurological disorder characterized by an inadequate supply of blood and oxygen to brain tissue, resulting from the obstruction or narrowing of cerebral blood vessels (1). As a major global health burden, stroke affects approximately 26 million individuals annually, making it the second leading cause of mortality worldwide (2). Among the various types of cerebrovascular diseases, ischemic stroke is the most prevalent, accounting for nearly 67% of all stroke cases (3). Acute ischemic stroke (AIS) accounts for approximately 70% of all newly diagnosed stroke (4). Individuals with a history of stroke face a significantly elevated risk of recurrence, long-term disability, and mortality (5). At present, the diagnosis of IS relies primarily on clinical evaluation and neuroimaging techniques (6). However, the absence of reliable blood-based biomarkers remains a major challenge, particularly in cases with atypical symptoms or inconclusive imaging findings (7, 8).
Neutrophil extracellular traps (NETs) are web-like extracellular structures released by neutrophils in response to various stimuli, including bacteria, fungi, inflammatory factors, chemokines, and activated platelets (9). Emerging research suggests that NETs play a critical role in the pathophysiology of stroke by exacerbating blood-brain barrier disruption, promoting thrombosis, inducing resistance to fibrinolytic agents, damaging vascular structures, and participating in revascularization processes (10). Furthermore, bioinformatics studies have identified the diagnostic potential of NETs-related genes (NRGs) in ischemia/reperfusion injury (11). Nevertheless, comprehensive investigations into the relationship between IS and NRGs remain limited.
To address this knowledge gap, we conducted a comprehensive bioinformatics analysis to elucidate the role of neutrophil extracellular traps (NETs) in ischemic stroke (IS), focusing on their associated genes and biological functions. Our study not only advances the understanding of the NETs-IS relationship but also investigates the mechanistic basis of IS pathogenesis. Furthermore, by identifying 4 key hub genes, we developed a predictive model for IS and identified potential regulatory miRNAs and therapeutic agents targeting these genes. These findings provide a foundation for future mechanistic studies and offer promising translational applications for stroke management.
Methods
Data selection and preparation
Gene expression profiles were retrieved from the GEO database1 using the search term “ischemic stroke.” Candidate datasets were screened according to the following criteria: (1) expression data derived from microarray platforms; (2) inclusion of at least 20 IS patients per dataset; and (3) whole blood samples collected within 48 h of symptom onset. For individuals with multiple blood collection time points, only the most recent sample was included in the analysis. Based on these criteria, three datasets were selected: GSE37587 (34 IS patients), GSE16561 (39 IS patients and 24 healthy controls), and GSE58294 (23 IS patients and 23 healthy controls). All subsequent bioinformatics analyses were conducted using R software (version 4.4.2).
The discovery dataset consisted of two gene expression datasets—GSE37587 and GSE16561—which were merged based on their shared microarray platform (GPL6883). The third dataset, GSE58294, was used as an independent validation dataset. Raw data underwent preprocessing, including background correction, log2 transformation, and normalization using the “limma” package (10). To mitigate batch effects between the merged datasets, we employed the ComBat algorithm from the “sva” package (11). Differentially expressed genes (DEGs) between IS patients and controls were identified using the “limma” package, with significance thresholds set at an adjusted p-value < 0.05 (Benjamini–Hochberg false discovery rate) and an absolute log2 fold change (|logFC|) > 0.05.
Immune cell infiltration analysis
Single-sample Gene Set Enrichment Analysis (ssGSEA) was conducted to quantify the relative abundance of 24 infiltrating immune cell types, using a signature set derived from published literature (12). The analysis was performed using the R package “gsva” (13). For comparisons of immune cell enrichment scores between groups, the Wilcoxon rank-sum test was applied. Differential patterns of immune cell infiltration between the stroke and control cohorts were visualized using the R packages “ggplot2” and “ComplexHeatmap.
NDEGs identification and functional enrichment analysis
NETs-related genes (NRGs) were obtained from a previously published study (14). To identify NETs-related differentially expressed genes (NDEGs), we intersected the differentially expressed genes (DEGs) with the NRGs. The resulting NDEGs were visualized using Venn diagrams, volcano plots, and heatmaps, generated using the R packages “VennDiagram,” “ggplot2,” and “pheatmap,” respectively. To explore the biological roles of the NDEGs, functional enrichment analysis was conducted, focusing on biological processes (BP), cellular components (CC), molecular functions (MF), and associated pathways. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the R package “clusterProfiler,” with p < 0.05 considered statistically significant (15). In addition, gene set enrichment analysis (GSEA) was performed based on the MSigDB database using the “clusterProfiler” package.
Screening of key genes
To identify key genes among the NDEGs, machine learning approaches were applied. First, Least Absolute Shrinkage and Selection Operator (LASSO) regression was used to reduce dimensionality and select the most relevant genes (16). The LASSO model was optimized via a grid search with 10-fold cross-validation (CV) to identify the optimal regularization parameter, ensuring that the selected genes were strongly associated with the outcome while minimizing overfitting. Model performance was evaluated by calculating the mean squared error (MSE) across the cross-validation folds. Next, a Random Forest (RF) algorithm was applied to further refine gene selection based on importance scores (17). The RF model was optimized by performing a grid search with 5-fold cross-validation. Overfitting was mitigated by evaluating model performance on an independent validation dataset and monitoring the out-of-bag (OOB) error rate. The final gene selection was based on the importance ranking derived from the RF model. Both LASSO and RF analyses were implemented using the R packages “glmnet” and “randomForest,” respectively.
Construction and validation of the diagnostic model
A diagnostic model for ischemic stroke was constructed using logistic regression based on the selected key genes, implemented via the R package “rms.” The model’s performance was assessed by receiver operating characteristic (ROC) curve analysis using the “pROC” package, with the area under the curve (AUC) calculated to evaluate diagnostic accuracy. Differential expression of the key genes and the diagnostic efficacy were further validated using the GSE58294 dataset.
Prediction of pivotal miRNAs and candidate drugs
To explore the regulatory mechanisms of the identified key genes, pivotal miRNAs targeting these genes were predicted using the TargetScan database. In parallel, candidate drugs potentially capable of modulating the expression of these genes were identified using DSigDB database. Both analyses were conducted via the Enrichr platform.2 Candidate miRNAs and drugs were screened using a significance threshold of p < 0.05. All resulting networks were visualized using Cytoscape software (version 3.10.3).
Results
DRGs between stroke patients and healthy controls
The merged discovery dataset included 73 ischemic stroke patients and 24 healthy controls, identifying 225 upregulated genes and 326 downregulated genes. Principal component analysis (PCA) demonstrated that the two original datasets, which were initially distributed in distinct clusters, were successfully integrated into a unified distribution following batch effect correction (Supplementary Figures S1A,B). Box plots further confirmed consistent normalization across datasets (Supplementary Figures S1C,D). Single-sample Gene Set Enrichment Analysis (ssGSEA) was performed to assess immune cell infiltration, revealing a significant increase in neutrophil levels and a decrease in Th17, Th1, and T follicular helper (TFH) cells in stroke patients compared to healthy controls. Correlation analysis across 21 immune cell types demonstrated that neutrophils were positively associated with eosinophils and macrophages, but strongly negatively correlated with B cells, cytotoxic cells, T cells, and Th1 cells (Figure 1).
Figure 1

Differentially infiltrated immune cells in IS patients and healthy controls. (A) Heatmap of differential immune cells. (B) Correlation matrix of immune cells. (C) Histogram of differential immune cells.
Identification of NDEGs
NRGs were obtained from a previously published study, with details provided in Supplementary Table S1. Intersection of NRGs with DEGs yielded five NDEGs: SRC, TLR8, FCAR, HIF1A, and MAPK1. All these five genes were significantly upregulated in ischemic stroke patients compared to healthy controls, as illustrated in the volcano plot and heatmap (Figure 2).
Figure 2

Identification of NDEGs. (A) Venn diagram of the intersection of NETS and DEGs. (B) Volcano plots of DE-GS distribution. (C) Heatmap of NDEGs distribution.
Functional enrichment analysis
Gene Set Enrichment Analysis (GSEA) of the DEGs revealed significant enrichment in pathways related to neutrophil degranulation and the innate immune system (Figure 3A). After identifying the NDEGs, GO and KEGG enrichment analyses were conducted to explore their biological roles. GO results indicated that these NDEGs were primarily involved in immune response regulation and leukocyte apoptosis. KEGG pathway analysis further revealed that the NDEGs were significantly enriched in the pathway associated with NETs formation (Figure 3B). In conclusion, these findings highlight the pivotal role of inflammation and neutrophil-mediated immune responses—particularly NETs—in the pathogenesis of ischemic stroke.
Figure 3

Functional enrichment analysis. (A) GSEA analysis of DEGs between stroke patients and healthy controls. (B) GO/KEGG analysis of differential NDEGs.
Identification of key genes and construction of the diagnostic model
Using LASSO regression, the initial set of NDEGs was narrowed down to four diagnostic biomarkers: SRC, TLR8, FCAR, and HIF1A (Figure 4A). These genes were further validated using RF algorithms, which confirmed their importance based on feature rankings (Figure 4B). A nomogram incorporating the four key genes was then developed to predict the likelihood of ischemic stroke (Figure 5E). The model’s diagnostic performance was assessed using ROC curve analysis, resulting in an AUC of 0.880, indicating high predictive accuracy for ischemic stroke diagnosis (Figure 5F).
Figure 4

Screening for potential diagnostic biomarkers by machine learning. (A) LASSO. (B) Random forest.
Figure 5
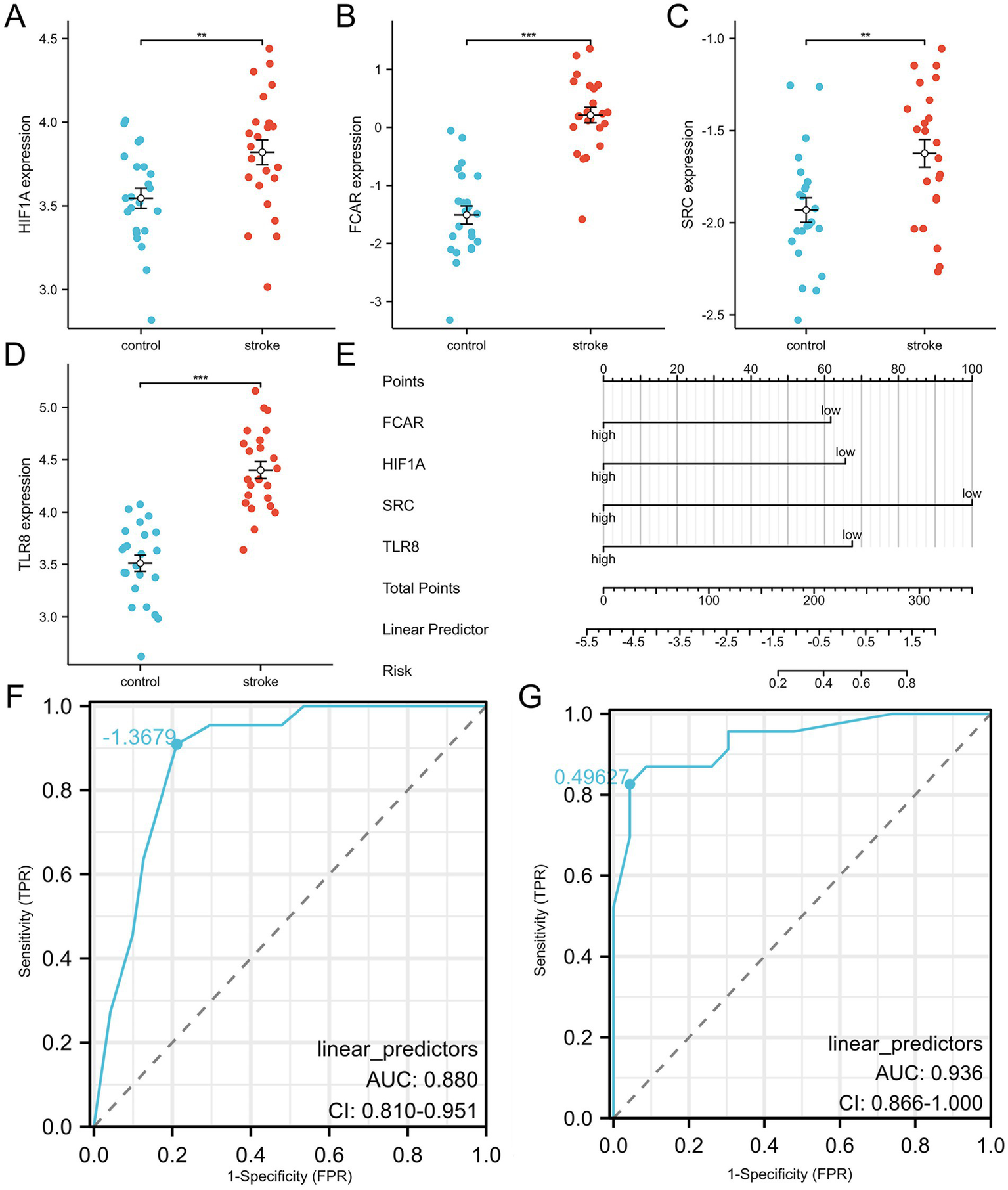
Diagnostic model construction and validation. (A–D) HIFIA, FCAR, SRC and TLR8 expression levels in validation dataset. (E) Construction of a nomogram model with 4 feature genes. (F,G) ROC curve for evaluating and validating the diagnostic model’s performance.
Validation of key genes
To confirm the reliability and generalizability of the diagnostic model, the expression levels of the four key genes were validated in the independent dataset GSE58294 (Figures 5A–D). All four biomarkers (SRC, TLR8, FCAR, and HIF1A) were significantly upregulated in ischemic stroke patients compared to healthy controls. The diagnostic model’s performance was further validated in this external cohort, achieving an AUC of 0.936 in ROC analysis, which demonstrates the model’s robust predictive capability (Figure 5G).
Identification of pivotal miRNAs and candidate drugs
Potential miRNAs targeting the four key genes were predicted using the TargetScan database, applying a significance threshold of p < 0.05. The analysis revealed that SRC was targeted by three miRNAs: hsa-miR-4669, hsa-miR-3619-3P, and hsa-miR-4537. FCAR was predicted to interact with hsa-miR-4669 and hsa-miR-4537, while TLR8 was associated solely with hsa-miR-3619-3P.
Additionally, candidate drugs modulating the expression of the key genes were identified using the DSigDB database. The top 10 drug candidates for each gene were selected based on a significance threshold of p < 0.05. Notably, SRC and HIF1A shared the same top 10 candidate compounds, among which “172889-27-9 TTD 00000391” had the highest combined score (17891). For FCAR, actinomycin D (CTD 00005748) yielded the highest score (794). In contrast, only three drug candidates (Arsenenous acid CTD 00000922, IMIQUIMOD BOSS and QUINOLINE CTD 00001749) were identified for TLR8. These findings offer novel insights into the post-transcriptional regulation and therapeutic targeting of ischemic stroke (Figure 6).
Figure 6

(A) Potential miRNAs targeting 4 key genes. (B–E) candidate drugs targeting 4 key genes.
Discussion
To our knowledge, this represents the first comprehensive investigation of NETs-related gene signatures in AIS patients compared with healthy controls. Our findings demonstrated that NETs-related genes are involved in the immune response processes underlying AIS. Using machine learning approaches, we identified four key genes—SRC, TLR8, FCAR, and HIF1A—that exhibit strong diagnostic potential. A diagnostic model based on these genes showed high predictive accuracy, and subsequent analyses identified putative miRNAs and candidate drugs targeting these genes. Collectively, these findings provide novel insights that may contribute to the advancement of diagnostic and therapeutic strategies for AIS.
Following stroke onset, neutrophils are the earliest immune cells to become activated and infiltrate ischemic brain tissue, with their numbers increasing markedly within hours (18). Activated neutrophils undergo morphological changes, including chromatin condensation, nuclear membrane fragmentation, and the fusion of nucleoplasm with chromatin. Eventually, the cells condense into a spherical shape, and the cell membrane ruptures, releasing intracellular components and forming fibrous structures known as NETs (19). In 2015, Perez-de-Puig et al. first reported histone citrullination in neutrophils of stroke patients, accompanied by extracellular release of DNA and histones (20). Two years later, Laridan et al. further confirmed the critical role of NETs in thrombus formation in patients with stroke (21). In the same year, Valles et al. identified significantly elevated plasma levels of NETs markers—including citrullinated histone 3 (CitH3), cell-free DNA (cfDNA), and nucleosomes—in stroke patients, with increases of 72, 33, and 39%, respectively, compared to healthy controls (22). NETs levels begin to rise within 24 h of stroke onset, peaking around 2–3 days, and remaining elevated for up to 7 days. Accumulating evidence suggests that NETs contribute to the stroke pathology through multiple mechanisms: (1) accelerating blood–brain barrier disruption; (2) promoting thrombus formation; (3) inducing resistance to tissue plasminogen activator (t-PA); and (4) impairing vascular integrity and inhibiting remodeling (10). These findings underscore the central role of NETs in the progression of AIS and support the relevance of NETs-related genes as potential diagnostic and therapeutic targets.
In this study, we identified 551 DEGs from a combined dataset comprising 73 ischemic stroke patients and 24 healthy controls. Immune cell infiltration analysis revealed a notable trend: a significant increase in neutrophil infiltration accompanied by a reduction in lymphocyte populations, including both T cells and B cells. Although neutrophils and lymphocytes are both integral components of the immune response, they displayed markedly divergent patterns in stroke patients. This imbalance may be attributed to the massive infiltration of neutrophils into ischemic brain tissue, which disrupts neuroimmune homeostasis and induces a shift in the peripheral immune system toward a suppressed state (23). This immune suppression is characterized by reduced lymphocyte counts and impaired lymphocyte function (24–26), and increases susceptibility to systemic infections, supporting our earlier hypothesis that neutrophil levels are positively correlated with infection risk in stroke patients (27). By intersecting DEGs with previously reported NETs-related genes, we identified five key genes: SRC, TLR8, FCAR, MAPK1 and HIF1A. Functional enrichment analysis revealed that these genes are significantly involved in immune regulatory pathways and leukocyte apoptosis, reinforcing their connection to NETs-mediated immune dysregulation in stroke. These findings underscore the central role of NETs-related genes in the pathophysiology of stroke and suggest that they may serve as promising diagnostic and therapeutic targets.
Based on the four key genes identified through the machine learning, we constructed a diagnostic nomogram, which demonstrated high predictive performance, with AUC values of 0.880 in the training dataset and 0.936 in the validation dataset. Most of the key genes has been implicated in stroke-related pathogenesis. SRC, a non-receptor protein tyrosine kinase encoded by SRC proto-oncogene, exhibits rapid activation within hours following stroke onset, with expression levels progressively increasing in infarcted regions for up to 24 h (28). It exacerbates stroke damage by promoting M1 microglial polarization, enhancing inflammation, and inducing neuronal apoptosis (29). Toll-like receptor (TLR) are transmembrane pattern recognition receptors encoded by TLR genes, which initiate intracellular signaling cascades upon recognition of pathogen-associated molecular patterns (PAMPs) (30). In AIS, TLR8 exacerbates neuronal damage by promoting apoptosis, mediating T cell-driven inflammation, and has been proposed as a biomarker of poor prognosis (31, 32). HIF1A, a master regulator of cellular oxygen homeostasis, modulates the expression of genes involved in hypoxic adaptation (33). In the context of AIS, it contributes to inflammasome activation, mitochondrial dysfunction, and cell death under severe hypoxia (34).
Finally, we predicted miRNAs and candidate drugs that may regulate the identified key genes. Among them, miR-4669 and miR-4537 were found to simultaneously target FCAR and SRC, while miR-3619-3P was predicted to regulate TLR8 and SRC. Although the roles of these miRNAs in AIS remain unclear, studies in other disease contexts provide preliminary insights. miR-4669 has been reported to enhance tumor invasiveness and contribute to an immunosuppressive tumor microenvironment (35). miR-4537 inhibits tumor cell proliferation, promotes apoptosis, and increases cellular radiosensitivity (36). In contrast, miR-3619-3P facilitates tumor cell migration and invasion (37). These findings suggest that these miRNAs may participate in the regulation of immune and inflammatory pathways in AIS, but their specific functions in stroke pathogenesis warrant further investigation.
PP2, a reversible ATP-competitive inhibitor of the Src family kinases (SFKs), demonstrates dual-targeting activity against both SRC and HIF1A. In the context of stroke, PP2 has been shown to attenuate blood–brain barrier (BBB) disruption by suppressing Src kinase phosphorylation (38). Similarly, Purvalanol A, a selective cyclin-dependent kinase (CDK) inhibitor, was predicted to target both SRC and HIF1A, with a binding affinity score slightly lower than that of PP2 (14,316 vs. 17,891). Emerging evidence suggests that Purvalanol A reduces ischemia/reperfusion-induced Golgi fragmentation and apoptosis through CDK inhibition (39). Nevertheless, the precise molecular mechanisms and therapeutic efficacy of Purvalanol A in stroke remain to be elucidated through further experimental studies.
The pathological progression of stroke is modulated by multiple regulatory systems, including the autonomic nervous system, hypothalamic–pituitary–adrenal (HPA) axis, and the immune system (40). We hypothesize that NETs may play a pivotal role in this process and directly contribute to stroke pathogenesis. However, the precise regulatory mechanisms underlying this involvement require further investigation. While our findings offer valuable insights, we acknowledge that the study is based entirely on in silico analyses, without experimental validation of the key findings. Moreover, the relatively small sample size and the absence of in vivo or in vitro validation of the predicted miRNAs and therapeutic candidates limit the generalizability and translational potential of our results. Future studies, including experimental validation (e.g., qPCR, Western blot, luciferase reporter assays), along with larger, well-characterized cohorts, are crucial for deepening our understanding of the relationship between these biomarkers and acute ischemic stroke. This will facilitate the development of more accurate diagnostic approaches and therapeutic interventions for stroke management.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
HZ: Funding acquisition, Writing – original draft, Conceptualization. TW: Data curation, Investigation, Software, Formal analysis, Writing – original draft. XL: Data curation, Software, Investigation, Formal analysis, Writing – original draft. SL: Data curation, Investigation, Software, Formal analysis, Writing – original draft. YW: Data curation, Investigation, Software, Formal analysis, Writing – original draft. YC: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was sponsored by Tianjin Health Research Project (TJWJ2023QN019) and Natural Science Foundation of Tianjin (No. 18JCZDJC36000).
Acknowledgments
We acknowledge the Gene Expression Omnibus (GEO) database for providing publicly available IS data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2025.1611776/full#supplementary-material
References
1.
Feske SK . Ischemic Stroke. Am J Med. (2021) 134:1457–64. doi: 10.1016/j.amjmed.2021.07.027
2.
Krishnamurthi RV Feigin VL Forouzanfar MH Mensah GA Connor M Bennett DA et al . Global and regional burden of first-ever ischaemic and haemorrhagic stroke during 1990-2010: findings from the global burden of disease study 2010. Lancet Glob Health. (2013) 1:e259–81. doi: 10.1016/s2214-109x(13)70089-5
3.
Adams HP Jr Bendixen BH Kappelle LJ Biller J Love BB Gordon DL et al . Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of org 10172 in acute stroke treatment. Stroke. (1993) 24:35–41. doi: 10.1161/01.str.24.1.35
4.
Ma Q Li R Wang L Yin P Wang Y Yan C et al . Temporal trend and attributable risk factors of stroke burden in China, 1990-2019: an analysis for the global burden of disease study 2019. Lancet Public Health. (2021) 6:e897–906. doi: 10.1016/S2468-2667(21)00228-0
5.
Tu WJ Chao BH Ma L Yan F Cao L Qiu H et al . Case-fatality, disability and recurrence rates after first-ever stroke: a study from bigdata observatory platform for stroke of China. Brain Res Bull. (2021) 175:130–5. doi: 10.1016/j.brainresbull.2021.07.020
6.
Chalela JA Kidwell CS Nentwich LM Luby M Butman JA Demchuk AM et al . Magnetic resonance imaging and computed tomography in emergency assessment of patients with suspected acute stroke: a prospective comparison. Lancet. (2007) 369:293–8. doi: 10.1016/S0140-6736(07)60151-2
7.
Coutts SB Moreau F Asdaghi N Boulanger JM Camden MC Campbell BCV et al . Diagnosis of uncertain-origin benign transient neurological symptoms study: rate and prognosis of brain ischemia in patients with lower-risk transient or persistent minor neurologic events. JAMA Neurol. (2019) 76:1439–45. doi: 10.1001/jamaneurol.2019.3063
8.
Yew KS Cheng E . Acute stroke diagnosis. Am Fam Physician. (2009) 80:33–40. PMID:
9.
Lipinska-Gediga M . Neutrophils, NETs, NETosis - old or new factors in sepsis and septic shock?Anaesthesiol Intensive Ther. (2017) 49:235–40. doi: 10.5603/AIT.2017.0041
10.
Zhao Z Pan Z Zhang S Ma G Zhang W Song J et al . Neutrophil extracellular traps: a novel target for the treatment of stroke. Pharmacol Ther. (2023) 241:108328. doi: 10.1016/j.pharmthera.2022.108328
11.
Xie M He Z Bin B Wen N Wu J Cai X et al . Bulk and single-cell RNA sequencing analysis with 101 machine learning combinations reveal neutrophil extracellular trap involvement in hepatic ischemia-reperfusion injury and early allograft dysfunction. Int Immunopharmacol. (2024) 131:111874. doi: 10.1016/j.intimp.2024.111874
12.
Bindea G Mlecnik B Tosolini M Kirilovsky A Waldner M Obenauf AC et al . Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. (2013) 39:782–95. doi: 10.1016/j.immuni.2013.10.003
13.
Park TJ Park JH Lee GS Lee JY Shin JH Kim MW et al . Quantitative proteomic analyses reveal that GPX4 downregulation during myocardial infarction contributes to ferroptosis in cardiomyocytes. Cell Death Dis. (2019) 10:835. doi: 10.1038/s41419-019-2061-8
14.
Wu J Zhang F Zheng X Zhang J Cao P Sun Z et al . Identification of renal ischemia reperfusion injury subtypes and predictive strategies for delayed graft function and graft survival based on neutrophil extracellular trap-related genes. Front Immunol. (2022) 13:1047367. doi: 10.3389/fimmu.2022.1047367
15.
Yu G Wang LG Han Y He QY . clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
16.
Wang Q Qiao W Zhang H Liu B Li J Zang C et al . Nomogram established on account of Lasso-cox regression for predicting recurrence in patients with early-stage hepatocellular carcinoma. Front Immunol. (2022) 13:1019638. doi: 10.3389/fimmu.2022.1019638
17.
Chen H Jiang R Huang W Chen K Zeng R Wu H et al . Identification of energy metabolism-related biomarkers for risk prediction of heart failure patients using random forest algorithm. Front Cardiovasc Med. (2022) 9:993142. doi: 10.3389/fcvm.2022.993142
18.
Jickling GC Liu D Ander BP Stamova B Zhan X Sharp FR . Targeting neutrophils in ischemic stroke: translational insights from experimental studies. J Cereb Blood Flow Metab. (2015) 35:888–901. doi: 10.1038/jcbfm.2015.45
19.
Pinegin B Vorobjeva N Pinegin V . Neutrophil extracellular traps and their role in the development of chronic inflammation and autoimmunity. Autoimmun Rev. (2015) 14:633–40. doi: 10.1016/j.autrev.2015.03.002
20.
Perez-de-Puig I Miro-Mur F Ferrer-Ferrer M Gelpi E Pedragosa J Justicia C et al . Neutrophil recruitment to the brain in mouse and human ischemic stroke. Acta Neuropathol. (2015) 129:239–57. doi: 10.1007/s00401-014-1381-0
21.
Laridan E Denorme F Desender L Francois O Andersson T Deckmyn H et al . Neutrophil extracellular traps in ischemic stroke thrombi. Ann Neurol. (2017) 82:223–32. doi: 10.1002/ana.24993
22.
Valles J Lago A Santos MT Latorre AM Tembl JI Salom JB et al . Neutrophil extracellular traps are increased in patients with acute ischemic stroke: prognostic significance. Thromb Haemost. (2017) 117:1919–29. doi: 10.1160/TH17-02-0130
23.
Shi K Wood K Shi FD Wang X Liu Q . Stroke-induced immunosuppression and poststroke infection. Stroke Vasc Neurol. (2018) 3:34–41. doi: 10.1136/svn-2017-000123
24.
Liu Q Jin WN Liu Y Shi K Sun H Zhang F et al . Brain ischemia suppresses immunity in the periphery and brain via different neurogenic innervations. Immunity. (2017) 46:474–87. doi: 10.1016/j.immuni.2017.02.015
25.
Prass K Meisel C Hoflich C Braun J Halle E Wolf T et al . Stroke-induced immunodeficiency promotes spontaneous bacterial infections and is mediated by sympathetic activation reversal by poststroke T helper cell type 1-like immunostimulation. J Exp Med. (2003) 198:725–36. doi: 10.1084/jem.20021098
26.
Vogelgesang A Grunwald U Langner S Jack R Broker BM Kessler C et al . Analysis of lymphocyte subsets in patients with stroke and their influence on infection after stroke. Stroke. (2008) 39:237–41. doi: 10.1161/STROKEAHA.107.493635
27.
Zhang H Wu T Tian X Lyu P Wang J Cao Y . High neutrophil percentage-to-albumin ratio can predict occurrence of stroke-associated infection. Front Neurol. (2021) 12:705790. doi: 10.3389/fneur.2021.705790
28.
Guldbrandsen HO Staehr C Iversen NK Postnov DD Matchkov VV . Does Src kinase mediated vasoconstriction impair penumbral reperfusion?Stroke. (2021) 52:e250–8. doi: 10.1161/STROKEAHA.120.032737
29.
Zhang X Guo J Liu J Liu J Li Z Chen J et al . Exosomal Src from hypoxic vascular smooth muscle cells exacerbates ischemic brain injury by promoting M1 microglial polarization. Neurochem Int. (2024) 179:105819. doi: 10.1016/j.neuint.2024.105819
30.
Kawai T Akira S . TLR signaling. Semin Immunol. (2007) 19:24–32. doi: 10.1016/j.smim.2006.12.004
31.
Tang SC Yeh SJ Li YI Wang YC Baik SH Santro T et al . Evidence for a detrimental role of TLR8 in ischemic stroke. Exp Neurol. (2013) 250:341–7. doi: 10.1016/j.expneurol.2013.10.012
32.
Brea D Sobrino T Rodriguez-Yanez M Ramos-Cabrer P Agulla J Rodriguez-Gonzalez R et al . Toll-like receptors 7 and 8 expression is associated with poor outcome and greater inflammatory response in acute ischemic stroke. Clin Immunol. (2011) 139:193–8. doi: 10.1016/j.clim.2011.02.001
33.
Amin N Abbasi IN Wu F Shi Z Sundus J Badry A et al . The Janus face of HIF-1alpha in ischemic stroke and the possible associated pathways. Neurochem Int. (2024) 177:105747. doi: 10.1016/j.neuint.2024.105747
34.
Choi YK . Detrimental roles of hypoxia-inducible factor-1alpha in severe hypoxic brain diseases. Int J Mol Sci. (2024) 25. doi: 10.3390/ijms25084465
35.
Nakano T Chen CL Chen IH Tseng HP Chiang KC Lai CY et al . Cheng: overexpression of miR-4669 enhances tumor aggressiveness and generates an immunosuppressive tumor microenvironment in hepatocellular carcinoma: its clinical value as a predictive biomarker. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24097908
36.
Liu J Yan S Hu J Ding D Liu Y Li X et al . MiRNA-4537 functions as a tumor suppressor in gastric cancer and increases the radiosensitivity of gastric cancer cells. Bioengineered. (2021) 12:8457–67. doi: 10.1080/21655979.2021.1982843
37.
Yu S Cao S Hong S Lin X Guan H Chen S et al . miR-3619-3p promotes papillary thyroid carcinoma progression via Wnt/beta-catenin pathway. Ann Transl Med. (2019) 7:643. doi: 10.21037/atm.2019.10.71
38.
Bai Y Xu G Xu M Li Q Qin X . Inhibition of Src phosphorylation reduces damage to the blood-brain barrier following transient focal cerebral ischemia in rats. Int J Mol Med. (2014) 34:1473–82. doi: 10.3892/ijmm.2014.1946
39.
Li Z Zhang W Xu J Mo X . Cdk1 protects against oxygen-glucose deprivation and reperfusion-induced Golgi fragmentation and apoptosis through mediating GM130 phosphorylation. J Mol Histol. (2023) 54:609–19. doi: 10.1007/s10735-023-10164-3
40.
Cui P McCullough LD Hao J . Brain to periphery in acute ischemic stroke: mechanisms and clinical significance. Front Neuroendocrinol. (2021) 63:100932. doi: 10.1016/j.yfrne.2021.100932
Summary
Keywords
ischemic stroke, neutrophil extracellular traps, bioinformatics analysis, biomarker, machine learning
Citation
Zhang H, Wu T, Li X, Liu S, Wang Y and Cao Y (2025) Machine learning identifies neutrophil extracellular traps-related biomarkers for acute ischemic stroke diagnosis. Front. Neurol. 16:1611776. doi: 10.3389/fneur.2025.1611776
Received
19 May 2025
Accepted
11 August 2025
Published
29 August 2025
Volume
16 - 2025
Edited by
Jose Laffita Mesa, Karolinska Institutet (KI), Sweden
Reviewed by
Amanda Alves Marcelino Da Silva, Universidade de Pernambuco, Brazil
Feng Yan, Shandong University, China
Updates
Copyright
© 2025 Zhang, Wu, Li, Liu, Wang and Cao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Cao, ttykcaochen@126.com
†These authors have contributed equally to this work and share first authorship
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.