Abstract
Early diagnosis of Alzheimer's disease is critical for effective therapeutic intervention. The progressive nature of cognitive decline requires precise computational methods to detect subtle neuroanatomical changes in prodromal stages. Current multi-scale neural networks have limited cross-scale feature integration capabilities, which constrain their effectiveness in identifying early neurodegenerative markers. This paper presents an Efficient Cross-Scale Feature-Aware Self-Attention Network (X-FASNet) designed to address these limitations through systematic hierarchical representation learning. The proposed architecture implements a dual-pathway multi-scale feature extraction approach to identify discriminative neuroanatomical patterns across various spatial resolutions, while integrating a novel cross-scale feature-aware self-attention module that enhances inter-scale information exchange and captures long-range dependencies. Quantitative evaluations on the DPC-SF dataset demonstrate that X-FASNet achieves superior performance with 93.7% accuracy and 0.973 F1-score, outperforming CONVADD by 10.8 percentage points in accuracy and 0.118 in F1-score, while also surpassing EfficientB2 on key performance metrics. Comprehensive experimentation across multiple neuroimaging datasets confirms that X-FASNet provides an effective computational framework for neurodegeneration assessment, characterized by enhanced detection of subtle anatomical variations and improved pathological pattern recognition.
1 Introduction
Alzheimer's disease (AD), the most prevalent neurodegenerative disorder worldwide, is characterized by progressive deterioration of cognitive functions and memory systems (1). Clinically manifested through impaired executive function, visuospatial deficits, and eventual loss of autonomy, AD pathogenesis involves complex interactions between amyloid-beta deposition, tau pathology, and neuroinflammation (2). The disease trajectory typically progresses through three clinically defined stages: cognitively normal (CN), mild cognitive impairment (MCI), and full dementia (AD), with MCI representing a critical window for therapeutic intervention (3, 4).
The insidious nature of AD progression underscores the imperative for early detection methodologies. Current diagnostic paradigms, which rely on neuropsychological assessments (e.g., MMSE, ADAS-Cog), exhibit limited sensitivity to pre-clinical stages. At the same time, cerebrospinal fluid biomarkers remain invasive and cost-prohibitive for population screening (5). Neuroimaging modalities, particularly structural MRI, have emerged as crucial tools for identifying early patterns of neurodegeneration. Automated analysis of MRI-derived biomarkers, including hippocampal atrophy, cortical thinning, and white matter hyperintensities, provides quantifiable metrics that correlate with cognitive decline trajectories (6).
Deep learning architectures have demonstrated remarkable success in decoding complex neuroimaging signatures of Alzheimer's disease (AD) progression (7, 8). Convolutional neural networks (CNNs) excel at capturing hierarchical representations of neurodegeneration patterns through multi-scale feature learning. However, current implementations face three fundamental limitations: (1) inadequate integration of cross-scale contextual information, leading to suboptimal utilization of complementary features across spatial resolutions; (2) limited capacity to model long-range dependencies between distributed neural substrates affected in AD; (3) insufficient attention to neuroanatomically plausible regions of interest, compromising model interpretability (9).
Recent advances in multi-scale architectures and attention mechanisms offer promising solutions to these challenges. While residual connections and dense blocks enhance feature reuse (10), and self-attention modules improve global context modeling (11), existing implementations often neglect the hierarchical nature of neurodegenerative processes. The progressive spatial expansion of AD pathology—from medial temporal lobe structures to association cortices—demands architectures capable of adaptively weighting local and global neurodegeneration signatures (12).
Despite their promising capabilities, current multi-scale convolutional neural networks face two critical limitations (13, 14). First, these architectures struggle to effectively integrate information across different spatial scales during feature fusion processes. This integration challenge frequently results in feature redundancy or information loss, ultimately compromising classification performance. Second, conventional convolutional operations are fundamentally constrained by their local receptive fields, preventing the capture of long-range dependencies between anatomically distant but functionally related regions—a limitation that significantly reduces feature representation capacity.
To address these challenges, we propose the Cross-Scale Feature-Aware Self-Attention Network (X-FASNet). Our architecture implements two complementary innovations: (1) a multi-scale feature extraction framework specifically designed to capture comprehensive neuroanatomical information from Alzheimer's neuroimaging data, and (2) a cross-scale feature-aware self-attention mechanism that facilitates effective information fusion across different scales. This integrated approach optimizes cross-scale feature interactions while simultaneously modeling long-range dependencies, substantially enhancing the network's representational capabilities and diagnostic accuracy.
The main contributions of this paper are summarized as follows:
-
We design a dual-pathway multi-scale feature extraction module by employing parallel 5 × 5 and 7 × 7 convolutional pathways with subsequent depth-separable convolutions. Our approach extracts both fine-grained local features critical for subtle pathological changes and broader contextual patterns that characterize disease progression. Ablation studies demonstrate this dual-pathway architecture improves classification accuracy by 4.7% compared to single-scale approaches.
-
We introduce a cross-scale feature-aware self-attention mechanism that enables dynamic feature integration across spatial scales while modeling long-range dependencies between anatomically distant brain regions affected by AD pathology. This mechanism enhances representation capacity as evidenced by a 6.5% improvement in multi-class classification accuracy over conventional feature fusion methods, with visualization analyses confirming focus on neuroanatomically relevant regions.
-
We propose X-FASNet, an integrated architecture that combines these contributions to address the limitations of conventional multi-scale networks for Alzheimer's disease diagnosis. Extensive evaluations demonstrate state-of-the-art performance across multiple datasets: on DPC-SF, X-FASNet achieves 93.7% accuracy and 0.973 F1-score, outperforming CONVADD (15) by 10.8% in accuracy; on DPC-Pre, it attains 95.5% accuracy with an AUC of 0.997, exceeding current leading models while maintaining clinically relevant interpretability.
2 Related work
2.1 Machine learning based diagnostics for Alzheimer's detection
Considering that traditional diagnostic methods cannot meet the demand for early detection of Alzheimer's disease. In recent years, machine learning has garnered significant attention in Alzheimer's disease (AD) diagnosis. First, feature mining of medical images is performed by locating key regions through feature extraction algorithms, and then, based on the classification model, features are recognized. Compared to traditional diagnostic methods, machine learning based diagnostic methods are relatively objective and more suitable for large-scale screening. Change et al. (9) summarized several novel biomarkers and, using machine learning algorithms and multivariate analysis, distinguished between patients with AD and healthy individuals. Experiments have demonstrated that combining machine learning algorithms with multivariate analysis can improve the accuracy of Alzheimer's disease (AD) diagnosis. Fan et al. (16) employed a support vector machine (SVM) model to classify and predict the various stages of Alzheimer's disease, utilizing MRI imaging data to facilitate an efficient diagnosis of the disease. Alghamedy et al. (17) proposed a multi-model approach based on machine learning for medical imaging studies, aiming to classify and detect Alzheimer's disease. Neffati et al. (18) proposed a method based on downsized kernel principal component analysis (DKPCA) and multi-class support vector machines for classifying MRI images in different states of Alzheimer's disease. Beheshti et al. (6) developed a feature selection method based on whole-brain voxel analysis of MRI data for the computer-aided diagnosis (CAD) framework. Dong et al. (19) proposed a machine learning method based on functional connectivity of whole-brain connectome for probing the network substrate of cognitive deficits in Alzheimer's disease. Alatrany et al. (20) proposed a machine learning model with interpretability oriented toward early diagnosis of Alzheimer's disease (AD). Specifically, the method introduces Class Association Rules (CAR) and Stable and Interpretable Rule Set for classification (SIRUS) model to enhance the interpretability of the method. Although machine learning based methods have demonstrated better performance in early diagnosis, they usually rely on manually extracted features, which tend to lead to an insufficient generalization ability of the model when facing physiological differences among individual patients.
2.2 Deep learning based diagnostics for Alzheimer's detection
In recent years, deep learning methods have become a significant focus of research in the field of early Alzheimer's disease diagnosis. Using deep learning methods, researchers can automatically extract deep features from various types of medical images to enhance the accuracy and robustness of diagnoses (21). AbdulAzeem et al. (22) proposed a convolutional neural network-based end-to-end image classification framework for Alzheimer's disease, achieving promising results. Al Shehri (23) proposed a deep learning based solution to diagnose and classify Alzheimer's disease by using the CNN architecture of DenseNet-169 and ResNet-50. Yang et al. (12) evaluated the performance of deep learning algorithms in differentiating between patients diagnosed with Alzheimer's disease using baseline MRI brain data. They also combined the features extracted from the neural network with other baseline biomarkers to create prognostic markers for Alzheimer's disease. Liang et al. (24) proposed a distillation multi-residual network (DMRNet) with dilated classifiers and self-distillation for the early diagnosis of Alzheimer's disease (AD), aiming to explore the hidden knowledge between each feature. Khan et al. (25) proposed a deep learning-based multi-class classification method to distinguish the stages of early Alzheimer's disease diagnosis. Through extensive experimental validation, it is shown that the proposed method has good classification performance. Wu et al. (26) proposed a three-dimensional (3D) transfer learning network based on two-dimensional (2D) transfer learning to classify Alzheimer's disease (AD) and normal groups from MRI images. Jiang et al. (27) proposed a new method for classifying images of Alzheimer's disease based on the external-attention mechanism. This approach utilizes the external-attention mechanism to classify images of Alzheimer's disease. To further improve representational capacity, several studies have explored multi-scale feature extraction strategies. Song et al. (28) proposed a three-dimensional multi-scale CNN model for feature extraction, which achieves better overall performance. Tu et al. (29) proposed a multi-modal feature transformation approach for AD diagnosis, which extracts complementary features from multiple modalities to enhance the representation of Alzheimer's-related patterns. Although these approaches attempt to extract multi-scale information, they still rely heavily on convolutional architectures, which are limited in modeling long-range spatial dependencies. In summary, although existing deep learning methods have achieved encouraging progress in the early diagnosis of Alzheimer's disease, many of these methods primarily focus on either local or short-range features. Consequently, there are still deficiencies in multi-scale feature modeling and long-range dependency capture, which limit the ability to extract highly discriminative information of Alzheimer's disease.
3 X-FASNet for Alzheimer's disease detection
3.1 Method overview
Although the Alzheimer's disease state recognition model based on multi-scale convolutional neural networks has made significant progress, it utilizes different sizes of convolutional kernels to extract various parts of the image, effectively capturing both global and local feature information. However, the process of fusing different-scale features is often accompanied by feature information imbalance, which makes the recognition model suffer from poor recognition performance. Furthermore, the absence of long-range dependencies means that the fused features often fail to capture the complex associations in the brain, which in turn affects the accurate diagnosis of early-stage conditions. For this reason, we propose an early recognition model for Alzheimer's disease based on X-FASNet. First, to capture key information at different scales, we constructed multi-scale features of the image using convolution with various kernel sizes. Second, to efficiently fuse multi-scale features, we propose the Cross-Scale Feature-Aware Self-Attention. On the one hand, multiscale features are merged through cross-scale feature fusion, and on the other hand, the long-range dependencies of the fused features are extracted using the self-attention mechanism. Finally, the extracted features are fed into the convolutional and pooling layers to obtain the final classification results. The structure of the overall model is shown in Figure 1.
Figure 1

The overall model structure of X-FASNet. (a) Overall Framework; (b) Multi-scale Feature Extraction; (c) Cross-scale Feature-aware Self-attention (X-FAS) Block; (d) Classifier.
3.2 Multi-scale feature extraction
To extract suitable features from Alzheimer's images more efficiently, we employ a multi-scale feature extraction strategy based on a two-way network. First, we target the image for shallow feature extraction using both large and small kernel convolutions. Among them, the use of large kernel convolution aims to extract global features and obtain more spatial information from the image. The small kernel convolution focuses more on the local detailed features of the image. Subsequently, we increased the number of channels by applying a 1 × 1 convolution, which enabled the better extraction of specificity information contained in the data during subsequent feature extraction. Then, to reduce the overall number of parameters in the network, we employ depth-separable convolution and shrink the number of channels through a 1 × 1 convolution to enhance the expressive ability of feature information. Not only that, for each way branch, we use residual concatenation to enhance the performance of the model. After obtaining the features of two-way branches, we combine these features to form multi-scale features. Let the alzheimer's image be X∈RH×W×C, and the following equation can be described as:
Where μB represents the mean of the batch, represents the variance of the batch, and ε is a small positive number guaranteed to be computationally safe. α and β are a set of parameters that are constantly updated during network training. Conv represents the convolution operation, BN represents the batch normalization.
3.3 Cross-scale feature-aware self-attention
Multi-scale features extracted from medical images often contain rich local and global information. To enhance the characterization of features, we designed the Cross-Scale Feature-Aware Self-Attention module. The module is divided into two parts: cross-scale feature fusion and self-attention mechanism. Multi-scale features contain information at multiple levels, both locally and globally. To enhance the characterization ability of features at different scales, we introduce a cross-scale feature fusion module. The module adopts a two-branch structure. In one of the branches, we use a 1 × 1 convolution to adjust the number of channels, thereby realizing residual connectivity while maintaining the original information. In the other branch, the same 1 × 1 convolution is introduced to unify the channel dimensions, followed by three RepVGG blocks (30) for deep feature extraction. As a structural reparameterization unit, it can effectively improve the expression ability of features. Finally, the results of the two branches are summed element by element to realize the residual fusion of multi-scale features. This will provide stable and discriminative feature inputs for subsequent dependency mining. The process can be represented as:
Where XCross−scale is the feature after cross-scale fusion, Conv represents the convolution operation. RepVGG3 represents the operation of RepVGG Block performed three times.
Although cross-scale feature fusion efficiently integrates features at different scales, the dependencies between features are not fully utilized. For this reason, we introduce the multi-head self-attention mechanism. Compared to the traditional mechanism, the spatial dimensions of K and V are first reduced using a depth-separable convolution with a kernel size of k×k in k steps. Secondly, a bias term is added for learning (denoted as B). The process can be represented as:
where XFeature is the output feature, dk is the dimension of the key vector K, Wo, , , and are all parameter matrices, and LN stands for layer normalization.
3.4 Classifier
After the high-level semantic feature extraction is completed, we design a structured classifier module for the final discrimination of Alzheimer's disease (AD). First, to reduce feature dimensionality, the input features are processed using average pooling. Second, the two-dimensional features are converted to one-dimensional using the Flatten operation. Finally, we utilize a compact multilayer perceptron to generate the prediction results through nonlinear mapping. The process can be represented as:
Where O is the final output and AvgPooling is the average pooling operation.
4 Experiments
4.1 Dataset
Three public datasets are used to test our proposed model: the Kaggle Alzheimer's classification (KAC) dataset1 and the preliminary and semi-final rounds of the Disease Prediction Challenge of Alzheimer's Disease (DPC-Pre and DPC-SF).2 The KAC dataset comprises 6,400 MRI samples, categorized into four types of Alzheimer's disease images: Non-Demented, Very Mildly Demented, Mildly Demented, and Moderately Demented. DPC-Pre is a binary classification task that distinguishes between normal individuals (CN) and patients with Alzheimer's disease (AD), using 1,000 MRI images per class. DPC-SF extends the task to three classes—normal(CN), mild cognitive impairment (MCI), and AD, comprising 3,000, 3,000, and 4,000 samples, respectively. The detailed description of the three datasets is shown in Tables 1, 2.
Table 1
| Stage | Total samples |
|---|---|
| Non-demented (ND) | 2,400 |
| Very-mild-demented (VMD) | 2,240 |
| Mild-demented (MD) | 896 |
| Moderate-demented (MoD) | 64 |
Sample distribution across different stages in the KAC dataset.
Table 2
| Name | Stage | Total samples |
|---|---|---|
| DPC-Pre | CN | 1,000 |
| AD | 1,000 | |
| DPC-SF | CN | 3,000 |
| MCI | 3,000 | |
| AD | 4,000 |
Sample distribution across different stages in the DPC-Pre and DPC-SF.
After image pre-processing, all images are resized to a fixed resolution of 168 × 168 pixels for model input. For the KAC dataset, we randomly split the data into 70% for training and 30% for evaluation. For the DPC datasets, both DPC-Pre and DPC-SF, 90% of the data are used to train the classification models, and the remaining 10% are reserved for evaluation.
4.2 Evaluation indicators
To comprehensively assess the performance of the classification models, we employ the following standard evaluation metrics: accuracy, precision, recall, F1-score, and the area under the ROC curve (AUC). These metrics are defined as:
Here, TP, TN, FP, and FN denote true positives, true negatives, false positives, and false negatives, respectively. AUC is the area under the Receiver Operating Characteristic (ROC) curve, which plots the TP rate against the false positive rate. A higher AUC value (closer to 1) indicates better classification performance.
4.3 Performance evaluation
To validate the effectiveness and generalization ability of our proposed model, we conduct comprehensive experiments on three Alzheimer's disease classification datasets: DPC-Pre, DPC-SF, and KAC. The proposed method is compared with a series of representative classifiers, including conventional deep convolution and attention-based models (10, 11, 30–39). The classification results of KAC are shown in Table 3. The classification results of DPC-Pre and DPC-SF are presented in Tables 4, 5, respectively.
Table 3
| Name | Accuracy | Recall | Precision | F1-Score | AUC |
|---|---|---|---|---|---|
| Binary classification | |||||
| ResNet18 (10) | 0.7834 | 0.7952 | 0.7759 | 0.7854 | 0.8820 |
| ResNet34 (10) | 0.7771 | 0.7824 | 0.7639 | 0.7730 | 0.8783 |
| DenseNet (32) | 0.8122 | 0.8299 | 0.8128 | 0.8212 | 0.7452 |
| SENet_18 (36) | 0.7628 | 0.7673 | 0.7478 | 0.7574 | 0.8678 |
| ECANet_18 (37) | 0.7951 | 0.7799 | 0.7648 | 0.7723 | 0.8680 |
| HRNet (39) | 0.7628 | 0.7664 | 0.7543 | 0.7603 | 0.8499 |
| GhostNet (35) | 0.7538 | 0.7469 | 0.7311 | 0.7389 | 0.8393 |
| RepVGG (30) | 0.8104 | 0.8149 | 0.7950 | 0.8048 | 0.9158 |
| Vision Transformer (11) | 0.7340 | 0.6943 | 0.7034 | 0.6988 | 0.7465 |
| CONVADD (15) | 0.6827 | 0.6470 | 0.6527 | 0.6498 | 0.6848 |
| Efficient B2 (41) | 0.7333 | 0.6605 | 0.6846 | 0.6723 | 0.7452 |
| Our proposed | 0.8431 | 0.8667 | 0.8486 | 0.8575 | 0.9357 |
| Multi classification | |||||
| ResNet18 (10) | 0.7549 | 0.7976 | 0.8186 | 0.8080 | 0.9353 |
| ResNet34 (10) | 0.7712 | 0.8220 | 0.8485 | 0.8349 | 0.9589 |
| DenseNet (32) | 0.6454 | 0.6685 | 0.7289 | 0.6974 | 0.8235 |
| SENet_18 (36) | 0.5767 | 0.5940 | 0.6434 | 0.6177 | 0.7666 |
| ECANet_18 (37) | 0.6650 | 0.6895 | 0.7305 | 0.7094 | 0.8785 |
| HRNet (39) | 0.7320 | 0.7658 | 0.7940 | 0.7796 | 0.9226 |
| GhostNet (35) | 0.5898 | 0.6225 | 0.6913 | 0.6551 | 0.8184 |
| RepVGG (30) | 0.7173 | 0.7459 | 0.7717 | 0.7586 | 0.8872 |
| Vision Transformer (11) | 0.4771 | 0.4696 | 0.5421 | 0.5034 | 0.6256 |
| CONVADD (15) | 0.7216 | 0.5926 | 0.6268 | 0.6092 | 0.8339 |
| Efficient B2 (41) | 0.6862 | 0.7207 | 0.7869 | 0.7524 | 0.9114 |
| Our proposed | 0.7924 | 0.8422 | 0.8596 | 0.8508 | 0.9635 |
Performance comparison of different models in KAC.
Table 4
| Name | Accuracy (%) | Recall (%) | Precision (%) | F1-Score | AUC |
|---|---|---|---|---|---|
| VGGNet (42) | 48.0 | 50.1 | 49.5 | 0.498 | 0.539 |
| ResNet18 (10) | 90.4 | 93.2 | 93.2 | 0.933 | 0.987 |
| ResNet34 (10) | 92.4 | 96.3 | 95.4 | 0.959 | 0.995 |
| ResNet50 (10) | 87.4 | 90.1 | 90.1 | 0.901 | 0.963 |
| ResNet101 (10) | 85.9 | 88.5 | 88.8 | 0.887 | 0.945 |
| DenseNet (32) | 92.9 | 95.8 | 95.9 | 0.959 | 0.995 |
| SENet_18 (36) | 85.9 | 86.6 | 88.5 | 0.876 | 0.937 |
| ShuffleNet (34) | 89.9 | 92.6 | 92.8 | 0.927 | 0.976 |
| MobileNet V2 (33) | 93.9 | 96.8 | 96.8 | 0.969 | 0.995 |
| ECANet_18 (37) | 94.2 | 97.1 | 97.4 | 0.974 | 0.994 |
| HRNet (39) | 81.3 | 83.9 | 83.9 | 0.839 | 0.940 |
| GhostNet (35) | 88.4 | 91.2 | 91.1 | 0.911 | 0.973 |
| RepVGG (30) | 88.4 | 91.1 | 92.2 | 0.917 | 0.987 |
| Vision Transformer (11) | 82.5 | 86.5 | 80.9 | 0.836 | 0.864 |
| CONVADD (15) | 92.9 | 95.9 | 95.9 | 0.959 | 0.989 |
| Efficient B2 (41) | 87.3 | 90.1 | 90.1 | 0.901 | 0.968 |
| Our Proposed | 95.5 | 97.9 | 98.0 | 0.979 | 0.997 |
Performance comparison of different models in DPC-Pre.
Table 5
| Name | Accuracy (%) | Recall (%) | Precision (%) | F1-Score | AUC |
|---|---|---|---|---|---|
| VGGNet (42) | 38.9 | 33.4 | 40.4 | 0.365 | 0.478 |
| ResNet18 (10) | 87.5 | 91.3 | 90.8 | 0.910 | 0.967 |
| ResNet34 (10) | 74.5 | 77.6 | 78.3 | 0.779 | 0.902 |
| ResNet50 (10) | 76.5 | 80.0 | 79.6 | 0.797 | 0.912 |
| ResNet101 (10) | 79.8 | 83.2 | 83.3 | 0.832 | 0.931 |
| DenseNet (32) | 87.5 | 90.9 | 91.0 | 0.909 | 0.971 |
| SENet_18 (36) | 78.5 | 81.4 | 82.2 | 0.817 | 0.912 |
| ShuffleNet (34) | 71.7 | 75.1 | 74.5 | 0.747 | 0.889 |
| MobileNet V2 (33) | 89.7 | 93.1 | 93.2 | 0.931 | 0.978 |
| ECANet_18 (37) | 86.9 | 90.3 | 90.5 | 0.903 | 0.967 |
| HRNet (39) | 86.8 | 90.0 | 90.2 | 0.901 | 0.968 |
| GhostNet (35) | 86.6 | 89.9 | 90.1 | 0.899 | 0.971 |
| RepVGG (30) | 89.8 | 81.6 | 94.8 | 0.877 | 0.973 |
| Vision transformer (11) | 83.5 | 79.7 | 77.5 | 0.785 | 0.806 |
| CONVADD (15) | 82.9 | 85.5 | 85.6 | 0.855 | 0.946 |
| Efficient B2 (41) | 87.3 | 91.1 | 90.6 | 0.908 | 0.990 |
| Our proposed | 93.7 | 97.3 | 97.3 | 0.973 | 0.998 |
Performance comparison of different models in DPC-SF.
In the KAC dataset (shown in Table 3), we conducted comprehensive evaluations of the proposed method's effectiveness by comparing its performance in binary classification and multi-classification tasks. In the binary classification task, our proposed method outperforms the other comparative models in all evaluation metrics, with an accuracy of 0.8431 and an F1-score of 0.8575. It significantly outperforms most deep learning models, such as the DenseNet and ResNet series, and exhibits a strong discriminative ability. In the multi-classification task, although the overall recognition difficulty is higher and the performance of the models decreases, our proposed method still achieves the optimal results. It achieves an accuracy of 0.7924 and an F1-score of 0.8508. Our method, as demonstrated by its recall and precision metrics of 0.8422 and 0.8596, respectively, accurately discriminates between multiple classes. On the DPC-Pre dataset (shown in Table 4), most of the deep networks can achieve high recognition accuracy on this dataset, among which MobileNetV2 (93.9%), ResNet34 (92.4%), and DenseNet (92.9%) perform well. In contrast, however, our proposed method achieves the best performance, with an accuracy of 95.5% and an F1-score of 0.979. On the DPC-SF dataset (see Table 5), the performance of most models degrades due to increased task complexity. However, our proposed method still maintains a significant advantage, outperforming the other compared methods in accuracy (93.7%) and F1-Score (0.973). It still offers substantial improvement in all metrics. In summary, the experimental results on the KAC, DPC-Pre, and DPC-SF datasets demonstrate that our proposed method consistently outperforms existing mainstream models and exhibits superior discriminative ability.
4.4 Ablation study
To further validate the effectiveness of each module in our proposed method, we conducted a series of ablation experiments on the KAC dataset. Specifically, we start from a baseline architecture and progressively extend the proposed modules. The compared models are listed in Table 6. The specific results are shown in Table 7. The basic single-branch structure exhibits limited performance, with accuracies of 0.7810 (Branch I) and 0.7843 (Branch II). Fusing the two into a dual branch results in a significant performance improvement. A slight decrease in performance was observed after adding enhancement modules to Dual Branch, probably due to the introduced structural complexity not working effectively. The further introduction of the attention mechanism improved several metrics, including an accuracy of 0.8319 and an AUC of up to 0.9368. Ultimately, the proposed method is optimal, achieving an accuracy of 0.8431 and an F1 score of 0.8575, which proves the effectiveness of the overall architectural design. In the multi-classification task, the single-branch model performed weakly, with a highest accuracy of 0.6764, while the dual-branch model significantly improved performance, achieving an accuracy of 0.7385. The attention mechanism was equally effective. The final proposed method also achieves the best results in the multi-classification task, with an accuracy of 0.7924 and an F1 score of 0.8508, further validating the adaptability and superiority of the proposed module in complex tasks. In summary, the ablation experiments fully demonstrate the effectiveness and complementarity of the overall design in the classification task, showing that the proposed method has robust generalization ability and high discriminative performance.
Table 6
| Model name | Conv_5 | Conv_7 | Cross-scale feature fusion | Attention |
|---|---|---|---|---|
| Branch I - small | ✓ | |||
| Branch II - large | ✓ | |||
| Dual branch | ✓ | ✓ | ||
| Dual branch enhance | ✓ | ✓ | ✓ | |
| Dual branch attention | ✓ | ✓ | ✓ | |
| Our proposed | ✓ | ✓ | ✓ | ✓ |
The compared models of the ablation experiment.
Table 7
| Name | Accuracy | Recall | Precision | F1-Score | AUC |
|---|---|---|---|---|---|
| Binary classification | |||||
| Branch I - small | 0.7810 | 0.8034 | 0.7856 | 0.7944 | 0.9006 |
| Branch II - large | 0.7843 | 0.7986 | 0.7794 | 0.7889 | 0.8991 |
| Dual branch | 0.8310 | 0.8508 | 0.8290 | 0.8397 | 0.9288 |
| Dual branch enhance | 0.7672 | 0.7627 | 0.7430 | 0.7527 | 0.8752 |
| Dual branch attention | 0.8319 | 0.8566 | 0.8386 | 0.8475 | 0.9368 |
| Our proposed | 0.8431 | 0.8667 | 0.8486 | 0.8575 | 0.9357 |
| Multi classification | |||||
| Branch I - small | 0.6764 | 0.7130 | 0.7477 | 0.7299 | 0.8896 |
| Branch II - large | 0.6960 | 0.7366 | 0.7749 | 0.7553 | 0.9148 |
| Dual branch | 0.7385 | 0.7759 | 0.8026 | 0.7891 | 0.9314 |
| Dual branch enhance | 0.7156 | 0.7635 | 0.7925 | 0.7777 | 0.9264 |
| Dual branch attention | 0.7679 | 0.7586 | 0.7895 | 0.7737 | 0.9248 |
| Our proposed | 0.7924 | 0.8422 | 0.8596 | 0.8508 | 0.9635 |
Ablation study results on the binary classification task.
4.5 Confusion matrix
To further investigate the classification performance of each ablation model, we conduct and visualize its confusion matrix. The confusion matrix results of binary and multi-classification are shown in Figures 2, 3. The results demonstrate a significant improvement in classification accuracy as the model structure is gradually enhanced. The initial “Branch I - Small” model had high rates of false positives and false negatives in its predictions. The “Branch I - Large” model improved the prediction accuracy and reduced the false negatives. Then, the classification performance was further enhanced by introducing the “Dual Branch” structure. The model will better recognize Alzheimer's disease, although many false positives will still exist. Ultimately, the proposed model performs well in the classification task by combining all the improvement strategies. This shows that enhancing the model structure and introducing the attention mechanism effectively improve the classification accuracy and optimize the model's generalization ability.
Figure 2

Confusion matrix results of binary classification task in KAC dataset.
Figure 3

Confusion matrix results of multi-classification task in KAC dataset.
4.6 Model interpretability
To further validate the proposed module's ability to improve model discrimination and interpretability, we performed Grad-CAM (40) analysis on the model under various ablation settings. Specifically, we compare the heat maps generated by the whole model with those generated by the model with the key module removed. As shown in Figures 4, 5, the region of attention of the whole model is more focused and highly aligned with the lesion region. In contrast, the model with the modules removed tends to focus on scattered or irrelevant areas. This result shows that the modules we introduced not only bring performance improvements in quantitative metrics but also prompt the model to focus more on medically significant regions in the decision-making process. It enhances the interpretability and reliability of the model in clinical diagnosis.
Figure 4
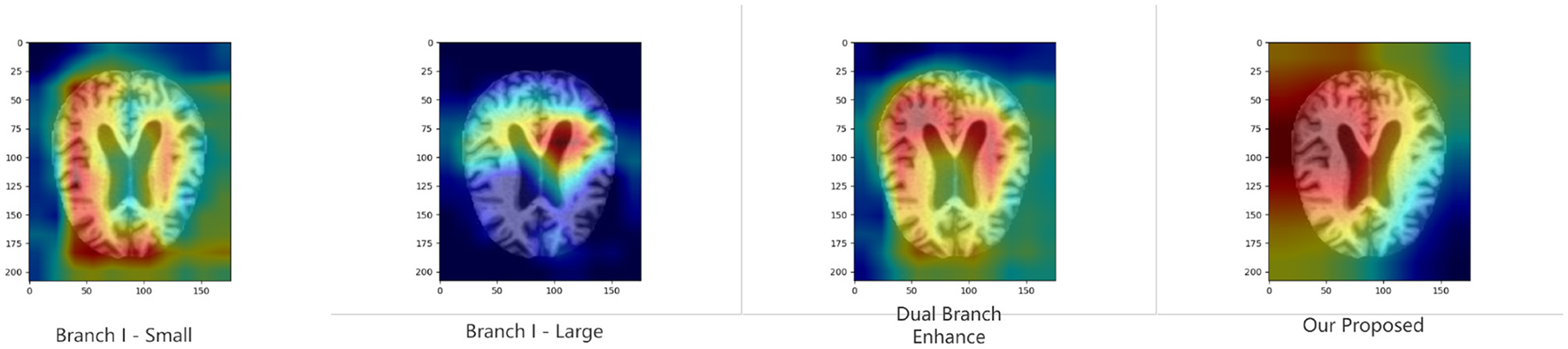
Grad-CAM results of binary classification task in KAC dataset.
Figure 5

Grad-CAM results of multi-classification task in KAC dataset.
5 Discussion
In recent years, the application of deep learning in Alzheimer's disease (AD) image recognition has achieved remarkable results; however, existing methods still face numerous challenges in handling complex brain structural features. Specifically, multi-scale convolutional neural networks often struggle to achieve effective inter-scale interaction when fusing information from different spatial scales. This results in a limited representation of information in key focal regions. Meanwhile, the local receptive fields of traditional convolutional operations hinder the model's ability to effectively capture long-range dependencies between features at different scales. To this end, the X-FASNet proposed in this paper introduces three core strategies at the structural design level. First, the multi-scale feature extraction structure is employed to fully capture image information across different receptive fields, thereby enhancing the model's sensitivity to Alzheimer's disease. Second, the cross-scale feature-aware self-attention is designed to facilitate information interaction and integration between different scales. In the experimental analysis, we further verify the effectiveness of the above structural design through a series of quantitative evaluations and visualization methods. Furthermore, the CAM-based results indicate that the whole model can focus more on typical regions associated with Alzheimer's disease compared to the control model with key modules removed. This suggests that the introduced structural design not only improves the classification performance but also aligns the model's focus mechanism more closely with the actual clinical regions of concern, thereby enhancing the credibility of the model output.
The effectiveness of the proposed structural design is validated through a series of experiments. First, the ablation results (Table 7) show that our model consistently outperforms all baseline variants across multiple evaluation metrics, confirming that the three core strategies make a significant contribution to performance enhancement. Further, the multi-scale feature extraction provides richer information for diagnosis, while the cross-scale feature-aware self-attention strengthens the feature representation capability. In addition, these quantitative improvements are further supported by qualitative evidence from Grad-CAM visualizations (Figures 4, 5). Compared with models using only single-scale branches or lacking attention mechanisms, our proposed model demonstrates more focused, semantically meaningful, and structurally coherent attention regions. The activation patterns are concentrated in brain areas closely associated with AD pathology, indicating stronger clinical consistency and interpretability.
However, the method presented still has some limitations. First, due to the high cost of acquiring high-quality neuro-imaging data related to Alzheimer's disease, the size and diversity of training data are still limited at this stage. In addition, this study utilizes unimodal structural MRI data, which, to some extent, limits the model's ability to comprehensively represent the complex pathological mechanisms of AD. Second, this study was evaluated based on publicly available datasets. Its image acquisition conditions are relatively uniform, and it lacks heterogeneous data validation from different centers, different devices, or different populations. In the future, we will consider combining multi-modal data, such as PET images, to enhance the model's ability to express complex pathological mechanisms. At the same time, data from different hospitals or devices are introduced to construct cross-center validation sets, enhancing the robustness and generalization performance of the model in real-world scenarios. Beyond dataset expansion, we will also consider verifying the practical application effectiveness of the model. Firstly, we will collaborate with clinical institutions to conduct prospective studies and integrate the model into computer-aided diagnosis (CAD) systems for evaluation in practical workflows. Additionally, we aim to test the model's inference efficiency and deployment feasibility on hospital-grade hardware to ensure that it meets the performance requirements for clinical deployment. These efforts will help transition our approach from research validation to practical implementation.
6 Conclusion
In this study, an X-FASNet is proposed for the early diagnosis task of Alzheimer's disease and validated based on Alzheimer's disease MRI data. The model effectively captures both local and global features in brain images through cross-scale information fusion. Experiments on multiple publicly available datasets demonstrate that X-FASNet outperforms existing methods in terms of classification performance. Meanwhile, the visualization analysis, combined with the model interpretability, shows that the model can focus on clinically significant brain regions stably, providing intuitive support for the model's discrimination process.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants' legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
WC: Methodology, Writing – original draft, Conceptualization. SX: Resources, Writing – review & editing, Formal analysis, Software. YP: Formal analysis, Conceptualization, Writing – original draft, Investigation. HZha: Project administration, Writing – review & editing, Resources. JZ: Resources, Writing – review & editing, Validation. HZhe: Software, Writing – review & editing, Validation. HY: Writing – review & editing, Resources. ZC: Writing – review & editing, Investigation, Methodology, Visualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Natural Science Foundation of Fujian Province (Grant No. 2024J01971), the National Natural Science Foundation of China (Grant No. 62071123), and Fujian Province Marine Economy Development Subsidy Fund Project (FJHJF-L-2019-7).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1.^ https://www.kaggle.com/datasets/borhanitrash/alzheimer-mri-disease-classification-dataset
References
1.
Golovanevsky M Eickhoff C Singh R . Multimodal attention-based deep learning for Alzheimer's disease diagnosis. J Am Med Inform Assoc. (2022) 29:2014–22. 10.1093/jamia/ocac168
2.
Song S Li T Lin W Liu R Zhang Y . Application of artificial intelligence in Alzheimer's disease: a bibliometric analysis. Front Neurosci. (2025) 19:1511350. 10.3389/fnins.2025.1511350
3.
Yang J Wang S Initiative AND . A novel coupling model of physiological degradation and emotional state for prediction of Alzheimer's disease progression. Brain Sci. (2022) 12:1132. 10.3390/brainsci12091132
4.
Bigi A Limbocker R Cecchi C . Promising therapeutic strategies for Alzheimer's disease: a focus on amyloid-β targeting. Front Neurosci. (2024) 18:1415641. 10.3389/fnins.2024.1415641
5.
Frisoni GB Fox NC Jack Jr CR Scheltens P Thompson PM . The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. (2010) 6:67–77. 10.1038/nrneurol.2009.215
6.
Beheshti I Mahdipour Hossein-Abad H Matsuda H Japanese-Alzheimer's Disease Neuroimaging Initiative . Identification of Alzheimer's disease on the basis of a voxel-wise approach. Appl Sci. (2019) 9:3063. 10.3390/app9153063
7.
Dana D Gadhiya SV St Surin LG Li D Naaz F Ali Q et al . Deep learning in drug discovery and medicine; scratching the surface. Molecules. (2018) 23:2384. 10.3390/molecules23092384
8.
Liu Y Wang L Ning X Gao Y Wang D . Enhancing early Alzheimer's disease classification accuracy through the fusion of sMRI and rsMEG data: a deep learning approach. Front Neurosci. (2024) 18:1480871. 10.3389/fnins.2024.1480871
9.
Chang CH Lin CH Lane HY . Machine learning and novel biomarkers for the diagnosis of Alzheimer's disease. Int J Mol Sci. (2021) 22:2761. 10.3390/ijms22052761
10.
He K Zhang X Ren S Sun J . Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2016). p. 770–778. 10.1109/CVPR.2016.90
11.
Dosovitskiy A Beyer L Kolesnikov A Weissenborn D Zhai X Unterthiner T et al . An image is worth 16x16 words: transformers for image recognition at scale. In: International Conference on Learning Representations (ICLR) (2021).
12.
Yang L Wang X Guo Q Gladstein S Wooten D Li T et al . Deep learning based multimodal progression modeling for Alzheimer's disease. Stat Biopharm Res. (2021) 13:337–43. 10.1080/19466315.2021.1884129
13.
Yazdan SA Ahmad R Iqbal N Rizwan A Khan AN Kim DH . An efficient multi-scale convolutional neural network based multi-class brain MRI classification for SaMD. Tomography. (2022) 8:1905–27. 10.3390/tomography8040161
14.
Yin Y Han Z Jian M Wang GG Chen L Wang R . AMSUnet: a neural network using atrous multi-scale convolution for medical image segmentation. Comput Biol Med. (2023) 162:107120. 10.1016/j.compbiomed.2023.107120
15.
Alsubaie MG Luo S Shaukat K . ConvADD: exploring a novel CNN architecture for Alzheimer's disease detection. Pathology. (2024) 15:1–14. 10.14569/IJACSA.2024.0150431
16.
Fan Z Xu F Qi X Li C Yao L . Classification of Alzheimer's disease based on brain MRI and machine learning. Neural Comput Applic. (2020) 32:1927–36. 10.1007/s00521-019-04495-0
17.
Alghamedy FH Shafiq M Liu L Yasin A Khan RA Mohammed HS . Machine learning-based multimodel computing for medical imaging for classification and detection of Alzheimer disease. Comput Intell Neurosci. (2022) 2022:1–14. 10.1155/2022/9211477
18.
Neffati S Ben Abdellafou K Jaffel I Taouali O Bouzrara K . An improved machine learning technique based on downsized KPCA for Alzheimer's disease classification. Int J Imaging Syst Technol. (2019) 29:121–31. 10.1002/ima.22304
19.
Dong N Fu C Li R Zhang W Liu M Xiao W et al . Machine learning decomposition of the anatomy of neuropsychological deficit in Alzheimer's disease and mild cognitive impairment. Front Aging Neurosci. (2022) 14:854733. 10.3389/fnagi.2022.854733
20.
Alatrany AS Khan W Hussain A Kolivand H Al-Jumeily D . An explainable machine learning approach for Alzheimer's disease classification. Sci Rep. (2024) 14:2637. 10.1038/s41598-024-51985-w
21.
Yaqoob N Khan MA Masood S Albarakati HM Hamza A Alhayan F et al . Prediction of Alzheimer's disease stages based on ResNet-Self-attention architecture with Bayesian optimization and best features selection. Front Comput Neurosci. (2024) 18:1393849. 10.3389/fncom.2024.1393849
22.
AbdulAzeem Y Bahgat WM Badawy M . A CNN based framework for classification of Alzheimer's disease. Neural Comput Applic. (2021) 33:10415–28. 10.1007/s00521-021-05799-w
23.
Al Shehri W . Alzheimer's disease diagnosis and classification using deep learning techniques. PeerJ Computer Science. (2022) 8:e1177. 10.7717/peerj-cs.1177
24.
Liang X Wang Z Chen Z Song X . Alzheimer's disease classification using distilled multi-residual network. Appl Intell. (2022) 53:11934–11950. 10.1007/s10489-022-04084-0
25.
Khan R Qaisar ZH Mehmood A Ali G Alkhalifah T Alturise F et al . A practical multiclass classification network for the diagnosis of Alzheimer's disease. Appl Sci. (2022) 12:6507. 10.3390/app12136507
26.
Wu H Luo J Lu X Zeng Y . 3D transfer learning network for classification of Alzheimer's disease with MRI. Int J Mach Learn Cybern. (2022) 13:1997–2011. 10.1007/s13042-021-01501-7
27.
Jiang M Yan B Li Y Zhang J Li T Ke W . Image classification of Alzheimer's disease based on external-attention mechanism and fully convolutional network. Brain Sci. (2022) 12:319. 10.3390/brainsci12030319
28.
Song J Zheng J Li P Lu X Zhu G Shen P . An effective multimodal image fusion method using MRI and PET for Alzheimer's disease diagnosis. Front Digital Health. (2021) 3:637386. 10.3389/fdgth.2021.637386
29.
Tu Y Lin S Qiao J Zhuang Y Zhang P . Alzheimer's disease diagnosis via multimodal feature fusion. Comput Biol Med. (2022) 148:105901. 10.1016/j.compbiomed.2022.105901
30.
Ding X Zhang X Ma N Han J Ding G Sun J . RepVGG: making VGG-style ConvNets great again. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2021). p. 13733–13742. 10.1109/CVPR46437.2021.01352
31.
Simonyan K Zisserman A . Very deep convolutional networks for large-scale image recognition. In: International Conference on Learning Representations (ICLR) (2015).
32.
Huang G Liu Z Van Der Maaten L Weinberger KQ . Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017). p. 4700–4708. 10.1109/CVPR.2017.243
33.
Sandler M Howard A Zhu M Zhmoginov A Chen LC . Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018). p. 4510–4520. 10.1109/CVPR.2018.00474
34.
Zhang X Zhou X Lin M Sun J . ShuffleNet: an extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2018). p. 6848–6856. 10.1109/CVPR.2018.00716
35.
Han K Wang Y Tian Q Guo J Xu C Xu C . GhostNet: more features from cheap operations. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2020). p. 1580–1589. 10.1109/CVPR42600.2020.00165
36.
Hu J Shen L Sun G . Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2018). p. 7132–7141. 10.1109/CVPR.2018.00745
37.
Wang Q Wu B Zhu P Li P Zuo W Hu Q . ECA-Net: efficient channel attention for deep convolutional neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2020). p. 11531–11539. 10.1109/CVPR42600.2020.01155
38.
Tan M Le QV . EfficientNetV2: smaller models and faster training. In: Proceedings of the International Conference on Machine Learning (ICML) (2021). p. 10096–10106.
39.
Wang J Sun K Cheng T Jiang B Deng C Zhao Y et al . Deep high-resolution representation learning for visual recognition. In: IEEE Transactions on Pattern Analysis and Machine Intelligence (2020). p. 3349–3364. 10.1109/TPAMI.2020.2983686
40.
Selvaraju RR Cogswell M Das A Vedantam R Parikh D Batra D . Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision (2017). p. 618–626. 10.1109/ICCV.2017.74
41.
Zubair Rahman A Gupta M Aarathi S Mahesh T Vinoth Kumar V Yogesh Kumaran S et al . Advanced AI-driven approach for enhanced brain tumor detection from MRI images utilizing EfficientNetB2 with equalization and homomorphic filtering. BMC Med Inform Decis Mak. (2024) 24:113. 10.1186/s12911-024-02519-x
42.
Simonyan K Zisserman A . Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. (2014).
Summary
Keywords
Alzheimer's disease, multi-scale model, cross-scale feature-aware self-attention, feature fusion, cognitive decline assessment
Citation
Chen W, Xu S, Peng Y, Zhang H, Zhang J, Zheng H, Yan H and Chen Z (2025) X-FASNet: cross-scale feature-aware with self-attention network for cognitive decline assessment in Alzheimer's disease. Front. Neurol. 16:1630838. doi: 10.3389/fneur.2025.1630838
Received
19 May 2025
Accepted
23 July 2025
Published
12 August 2025
Volume
16 - 2025
Edited by
Pei Shang, Mayo Clinic, United States
Reviewed by
Song Qiao, Zhejiang Hospital, China
Xinyuan Yuan, Mayo Clinic, United States
Chenyu Wang, Mayo Clinic, United States
Updates
Copyright
© 2025 Chen, Xu, Peng, Zhang, Zhang, Zheng, Yan and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiran Peng jack97yiran@gmail.comHong Zhang zhangh@fpnu.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.