- Cancer Section, Developmental Biology and Cancer Programme, Great Ormond Street Institute of Child Health, University College London, London, United Kingdom
The treatment failure rates of acute leukemia with rearrangements of the Mixed Lineage Leukemia (MLL) gene highlight the need for novel therapeutic approaches. Taking into consideration the limitations of the current therapies and the advantages of novel strategies for drug discovery, drug repurposing offers valuable opportunities to identify treatments and develop therapeutic approaches quickly and effectively for acute leukemia with MLL-rearrangements. These approaches are complimentary to de novo drug discovery and have taken advantage of increased knowledge of the mechanistic basis of MLL-fusion protein complex function as well as refined drug repurposing screens. Despite the vast number of different leukemia associated MLL-rearrangements, the existence of common core oncogenic pathways holds the promise that many such therapies will be broadly applicable to MLL-rearranged leukemia as a whole.
Introduction
Drug repurposing or drug repositioning refers to the use of an already clinically approved or experimental drug to a condition different from the one that the drug was originally developed for (Ashburn and Thor, 2004). It is not surprising that more than one molecular target can potentially be affected by most small molecules. Drug repurposing tackles two main problems of the research development process in the pharmaceutical industry: cost and time. De novo drug discovery and development of a drug for cancer takes approximately 10–17 years, with almost half of this time elapsing before the start of phase I clinical trials (Zhang et al., 2020). As important as time, the cost of bringing new drugs into the market is estimated to be between 2 and 3 billion USD on average. Even though the cost has increased with time, and scientific and technological factors ought to make the process of drug development faster and more efficient, the number of drugs approved every year per USD spent on development has halved roughly every 9 years since the 1950s (Munos, 2009; Scannell et al., 2012; Nosengo, 2016). In addition to this, the possibly most important advantage of drug repurposing is the lower risk of failure due to adverse effects. The safety of a drug that is being repurposed is already known through its clinical application, therefore it is much less likely for it to fail due to unexpected toxicity. The methodologies used to repurpose different therapeutic agents can be hypothesis driven or explorative and they include both computational and experimental approaches. In silico approaches are used to analyze different large-scale datasets, such as genome wide association studies (GWAS), signature matching, computational molecular docking and pathway or network mapping (Pushpakom et al., 2019). In parallel, phenotypic screening and binding assays such as affinity chromatography and mass spectrometry are used to experimentally identify the targets and off-targets of drugs in the lab (Martinez Molina et al., 2013; Moffat et al., 2017). Drug repurposing has already had several success stories. One of the most impressive examples in the field is the repurposing of thalidomide, a sedative that was given to pregnant women suffering from morning sickness with tragic skeletal birth defects. Thalidomide is now used for erythema nodosum leprosum and has achieved excellent results against multiple myeloma (Ashburn and Thor, 2004; Palumbo et al., 2008). In preventive care, the repurposing of aspirin, originally administered as an analgetic, is now helping prevent cardiovascular disease and colorectal cancer (Ishida et al., 2016). More recently, the SARS-CoV-2 emergency created the need for treatments for patients suffering from COVID-19. To this end, many labs screened for agents with the potential to be repurposed for COVID-19 patients and identified among others broad-spectrum antiviral agents (BSAAs) originally approved for Influenza or HIV and immunomodulatory and anti-inflammatory agents, such as the human IL-6 receptor (IL-6R) inhibitor tocilizumab as potential treatments for COVID-19 (Scavone et al., 2020; Singh et al., 2020).
In acute leukemia, 11q23 chromosomal rearrangements, affecting the MLL (KMT2A) gene, are a genetic hallmark and one of the major driver mutations (Meyer et al., 2018). This class of abnormalities is associated with 70–80% of infant acute leukemias, 15–20% of pediatric AML and is also found less frequently in adult patients (Hilden et al., 2006; Pieters et al., 2007; Harrison et al., 2010). The prognosis for these patients is poor. For example, MLL-rearranged infant ALL 5-years event-free survival (EFS) is only 20–50%, in comparison to over 60% for those with wild-type MLL (Hilden et al., 2006; Pieters et al., 2019). For young adult ALL patients, despite very high initial remission rates of 80–90%, unfortunately 50% of these patients will eventually relapse. Similarly, in older patients, initial remission is achieved in 40–90% of the patients with only 10–20% of them reaching a 5-years overall survival rate (Guru Murthy et al., 2015; Winters and Bernt, 2017). MLL-rearranged leukemias are not treated under a distinct protocol, but mainly on regimens based on the disease lineage (AML or ALL) and the age of the patient. The standard treatment typically consists of induction chemotherapy, followed by intensive chemotherapy followed by additional therapy. Over 130 different MLL translocations with numerous different fusion partners have been described, although only a fraction of these are recurrent (Meyer et al., 2018). Development of novel therapies for MLL-rearranged leukemia should take this oncogenic diversity into account and ensure broad applicability by targeting core elements of this disease.
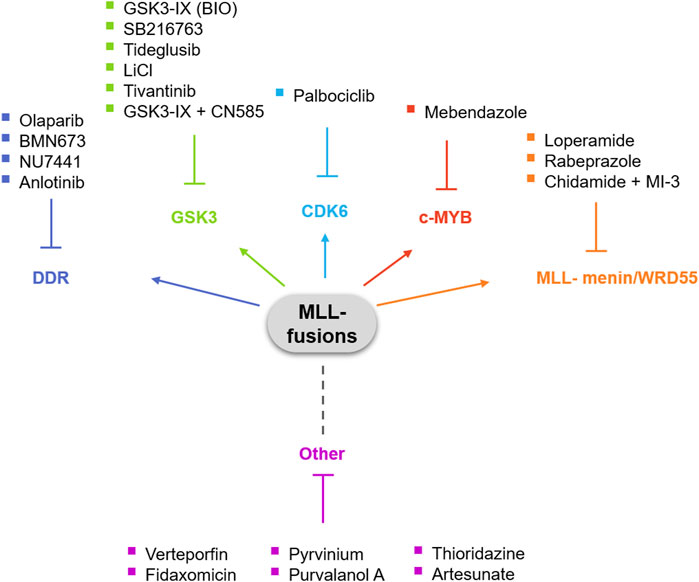
The significant rate of failure of the treatments currently used against acute leukemias with MLL-rearrangements is very frustrating and emphasizes the need for novel therapies that specifically target MLL fusion proteins and/or the downstream pathways these fusions dysregulate. In this review, we aim to highlight the therapeutic opportunities of drug repurposing for acute leukemias with MLL-rearrangements (Figure 1).
Hypothesis Driven and Target-Based Strategies for Drug Repurposing in MLL-Rearranged Leukemia
The understanding of the molecular mechanisms and main targets of MLL fusion proteins can lead us to form hypotheses for the direction of therapeutic intervention. Besides developing de novo drugs acting on the pathways that are known to be dysregulated or implicated in the phenotype of leukemia cells with MLL-rearrangements, we can also get inspired and redirect drugs already targeting those pathways in other conditions towards leukemia treatment. Selective inhibitors can be repositioned to different biological systems based on their activity on their target or even because of their off-target effects. This provides an opportunity to take advantage of the knowledge we already have of MLL-fusion function and quickly test compounds and drugs available in the lab or the clinic.
GSK3 Inhibitors
Glycogen synthase kinase 3 (GSK3) is a serine/threonine kinase with multiple roles in several signaling pathways in physiological and pathological conditions. GSK3 is involved in mRNA transcription, cell cycle, apoptosis, cell fate determination and stem cell maintenance (Beurel et al., 2015). As several of these pathways are implicated in disease pathogenesis, it comes as no surprise that GSK3 specific inhibitors have been in development for years. In hematopoiesis, GSK3 is important for the activity of hematopoietic stem cells and mice treated with GSK3 inhibitors show enhanced hematopoietic stem cell repopulation following bone marrow transplantation (Trowbridge et al., 2006). Importantly, inhibition of GSK3 showed potential as a treatment for MLL-rearranged leukemia, as in a small-scale screen aiming to identify compounds that could induce the growth arrest of genetically defined subsets of leukemia cells, AML and ALL cell lines that expressed MLL-AF4 or MLL-AF5 showed enhanced sensitivity to GSK3-IX (also known as BIO), a GSK3 inhibitor that also targets cyclin-dependent kinases (CDKs) and has been extensively used for the maintenance of pluripotency in human and murine embryonic stem cells (ESCs) (Tseng et al., 2006; Wang et al., 2008). Cell cycle analysis of AML and ALL MLL-rearranged cells treated with SB216763, another GSK3 inhibitor with therapeutic properties for pulmonary inflammation and fibrosis (Gurrieri et al., 2010), revealed reduction of G1-S phase progression. The response of MLL-rearranged cells to GSK3 inhibitors is mediated by p27Kip1, since the knockdown of the protein rescued the effects of GSK3 inhibition on cell cycle progression (Wang et al., 2008). The specificity of GSK3 inhibition in cells with MLL-rearrangements but not wild type cells is supported by studies that have shown that in MLL-rearranged cells, GSK3 regulates the expression of Homeobox (HOX) genes via CREB phosphorylation, leading to the induction of MEIS1-related gene expression and the subsequent maintenance of a stem cell-like phenotype in MLL-rearranged cells (Wang et al., 2010; Yeung et al., 2010). In addition, although MLL-rearranged leukemic stem cells (LSCs) were found to be resistant to GSK3 inhibition by lithium chloride (LiCl), an inhibitor that is approved by the Food and Drug Administration (FDA) for the treatment of epilepsy and bipolar disorder (Abu-Baker et al., 2013), they could be sensitized by β-catenin knockdown, indicating possible avenues for novel combination therapies (Yeung et al., 2010).
DNA Damage Response Inhibitors
In MLL-rearranged leukemia, the DNA damage response (DDR) machinery is perturbed. Some MLL-fusion proteins are suggested to predispose leukemia cells to DNA damage and ultimately cause short latency (Eguchi et al., 2006). Also, although p53, a protein usually activated to protect the cells in response to DNA damage by inducing cell cycle arrest and apoptosis (Aylon and Oren, 2007) is not often mutated in MLL-rearranged leukemia, MLL fusion proteins have been shown to suppress p53 and the p53-mediated response to DNA damage (Wiederschain et al., 2005). Importantly, the DDR machinery is activated during MLL-ENL-oncogene-induced leukemic transformation and is important for the maintenance of stem cell-like properties (Takacova et al., 2012). Finally, in B-ALL with MLL-rearrangements, DDR is required for cells with oncogene-driven replicative stress (Chu et al., 2017). Therefore, it appears that the pharmacologic targeting of the DDR pathway has the potential of inducing specific synthetic lethality in leukemic cells with MLL-rearrangements.
A group of drugs currently redirected for leukemia therapy are the poly (ADP-ribose) polymerase (PARP) inhibitors. Poly (ADP-ribosyl)ation of nuclear proteins supports the survival of cells with low DNA repair capacity. The role of PARP proteins has been well established in BRCA1/2 mutant tumors, that lack the mechanisms for homologous recombination. However, more studies have revealed new roles of PARP for gene transcription, protein stabilization and modulation of chromatin formation (Kraus and Hottiger, 2013; Krietsch et al., 2013). PARP inhibitors regulate critical functions of PARP proteins mainly in DNA-repair mechanisms such as single-strand breaks (SSBs) in base excision repair (BER) and homologous recombination-mediated double-strand break (DSB) repair (Haince et al., 2007; Haince et al., 2008; Bryant et al., 2009). Olaparib, rucaparib, niraparib and talazoparib are four PARP inhibitors that have been approved by the FDA for the treatment of germline BRCA-mutated breast or ovarian cancers (Plummer et al., 2005; Evers et al., 2008; Sandhu et al., 2010; Shen et al., 2013). The repurposing of PARP inhibitors to other malignancies has not been as successful as their use in breast and ovarian cancers. In hematological malignancies, inhibiting the activity of PARP proteins is very relevant, as these proteins regulate components often dysregulated in blood cancers, such as ATM, ATR, CHK1, and RAD51 (Zhao and So, 2016). Importantly, MLL-AF9 expressing leukemia cells depend on mechanisms that prevent the accumulation of DSBs in order to maintain proliferation (Santos et al., 2014; Maifrede et al., 2017). One study has shown that Parp1−/− mouse bone marrow cells (mBMCs) expressing MLL-AF9 show reduced colony formation activity in comparison with Parp1+/+ counterparts. These cells were shown to be modestly sensitive to the PARP1 inhibitor olaparib. In the same study, syngeneic mice transplanted with murine leukemia cells expressing MLL-AF9 were either left untreated or were treated with doxorubicin plus cytarabine, the PARP inhibitor BMN673, or the combination of the two. The results demonstrate a synergistic effect of the PARP1 inhibitor with the cytotoxic drug combination, overcoming the resistance to olaparib (Maifrede et al., 2017). While in leukemia cells with AML1-ETO and PML-RARa fusion proteins inhibition of PARP leads to an accumulation of DNA damage that results in cell differentiation and cell death, leukemia cells expressing MLL fusion proteins exhibit resistance to inhibition of PARP (Yeung et al., 2010). In AML, the expression of MLL fusion proteins results in the expression of the homeodomain transcription factor HOXA9 among others (Krivtsov and Armstrong, 2007; Yip and So, 2013). HOXA9 serves as a prognostic factor associated with poor AML treatment response (Golub et al., 1999) and its suppression has been linked to drug resistance in glioblastoma (Costa et al., 2010). The hypothesis that MLL-AF9-transformed cells show resistance to PARP inhibition possibly via the activation of HOXA9 was tested by Esposito et al. Inhibition of PARP reduced the colony formation ability of MLL-AF9-tranformed cells, but not HOXA9-independent E2A-PBX–transformed control cells while inducing differentiation and senescence (Esposito et al., 2015). Importantly, Hoxa9-deficient MLL-AF9 cells were highly sensitive to olaparib treatment, highlighting the role of HOXA9 in leukemic growth and the resistance of the MLL-rearranged AML cells to PARP inhibition (Esposito et al., 2015). Taking into consideration that HOXA9 is not essential for normal development (So et al., 2004; Lawrence et al., 2005; Smith et al., 2011), PARP inhibitors would appear to have a great therapeutic potential in leukemia with MLL-rearrangements.
ATR and ATM activate the DDR pathway and recognize DNA single or double strand breaks (Hurley and Bunz, 2007). It has been shown that they support cancer cells that are under replicative stress, therefore their inhibition is promising (Weber and Ryan, 2015). Inhibition of ATR in MLL-rearranged leukemia has therapeutic potential, since in mice where ATR expression is reduced, MLL-ENL AML cells growth is affected. Schoppy et al. generated a genetic system where ATR expression was conditionally reduced to 10% of normal levels in adult mice and studied the effects of this reduction in vivo comparing normal tissues and cancer. Although the suppression of ATR did not affect normal bone marrow, the authors reported inhibition of MLL-ENL-driven AML among others (Schoppy et al., 2012). Interestingly, in contrast to its function in overt leukemia the DDR functions as a barrier in the early stages of leukemogenesis. Thus, in a study examining the potential of the inactivation of the DDR barrier in leukemogenesis, Takacova et al. showed that ATM and ATR inhibitors inactivate the DDR barrier and support leukemia progression in a tamoxifen-inducible MLL fusion mouse model (Takacova et al., 2012). At the same time, the genetic ablation of critical regulators of DNA damage response such as MLL4, ATM or BRCA1 in MLL-rearranged cells induces leukemic differentiation (Santos et al., 2014). Importantly, it has been demonstrated that ATR and ATM inhibitors synergize with PARP inhibitors in response to DNA damage (Aguilar-Quesada et al., 2007; Yazinski et al., 2017). Possibly this combination could be successful against MLL-rearranged leukemia as well.
The DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase consisting of a catalytic subunit (DNA-PKcs) and a Ku heterodimer that is made up of the Ku70 and Ku80 subunits and it belongs to the phosphatidylinositol 3-kinase (PI3K)-related kinase protein family (Hartley et al., 1995). DNA-PK is required for repairing DSBs through non-homologous end-joining (NHEJ) (Rassool, 2003; Shrivastav et al., 2008). In addition to its role in NHEJ, DNA-PK also cooperates with ATR and ATM to regulate DNA damage checkpoints and thus, cells that are ATM-deficient, heavily depend on DNA-PK (Meek et al., 2008; Riabinska et al., 2013; Tsuji et al., 2017). In cancers where ATM is dysregulated, inhibition of DNA-PK has a therapeutic potential. For example, CC-115, a selective dual inhibitor of the mammalian target of rapamycin (mTOR) kinase and DNA-PK, has been shown to effectively inhibit proliferation and induce apoptosis in lymphoma, leukemia, breast cancer, hepatocellular carcinoma, head and neck cancer and lung cancer cell lines (Tsuji et al., 2017). In chronic lymphocytic leukemia (CLL) cells, the same inhibitor caused the inhibition of the DNA damage repair pathway and induced apoptosis and is currently under clinical trial for CLL (Thijssen et al., 2016; Tsuji et al., 2017). NU7441 is another DNA-PKi with interesting potential. NU7441 sensitizes cancer cells in B-cell CLL, breast cancer, non-small cell lung carcinoma and nasopharyngeal carcinoma (NPC) cell lines (Elliott et al., 2011; Ciszewski et al., 2014; Yanai et al., 2017; Dong et al., 2018). As we discussed earlier, leukemia cells with MLL-rearrangements also respond to ATM inhibitors. Therefore, the potential synergy between DNA-PK inhibitors and inhibitors of the DDR pathway such as ATMi and PARPi could be interesting.
SET domain-containing protein 1A (SETD1A) is a histone 3 lysine 4 (H3K4) methyltransferase and is one of the six MLL family members found in mammals (MLL1, MLL2, MLL3, MLL4, SETD1A, and SETD1B) (Chan and Chen, 2019). H3K4 methylation is a post-translational modification critical for the regulation and maintenance of gene expression during development and frequently mutated in cancer (Ernst et al., 2004; Kandoth et al., 2013; Bledau et al., 2014; Denissov et al., 2014). In breast cancer, SETD1A controls mitosis by regulating the H3K4 methylation of the promoters of mitotic genes. When SETD1A expression is knocked-down, the cells present severe mitotic defects and senescence (Tajima et al., 2019). In leukemia with MLL-rearrangements, SETD1A is essential for cell proliferation. The non-enzymatic domain of SETD1A interacts with cyclin K to control DNA damage response-related gene expression during the S phase of the cell cycle (Hoshii et al., 2018). In a recent study, Chen J. et al. used anlotinib, an inhibitor originally designed to target receptor tyrosine kinase (RTK) related to tumor vasculogenesis (Sun et al., 2016; Chen et al., 2021). Anlotinib has shown anti-tumor activity in non-small cell lung cancer and hepatocellular carcinoma (Han et al., 2018; He et al., 2018; Shen et al., 2018; Syed, 2018). Anlotinib treatment of MLL-rearranged leukemic cell lines suppressed growth and caused G2/M cell cycle arrest and apoptosis. An RNA sequencing study of cells exposed to anlotinib revealed that anlotinib treatment causes downregulation of genes critical for DDR, such as the genes encoding for the DNA polymerase delts proteins (POLDs) that act as response regulators of the DDR, that strongly correlated with SETD1A and Akt expression, suggesting an anti-leukemic activity of anlotinib by targeting SETD1A- and Akt-mediated DNA damage response in MLL-rearranged leukemia cells (Chen et al., 2021).
These studies highlight the potential of inhibiting molecular players of the DDR pathway in acute leukemias with MLL-rearrangements.
Cyclin-Dependent Kinase (CDKs) Inhibitors
The cyclin-dependent kinase 6 (CDK6) is a critical regulator of cell-cycle progression. CDK6 and CDK4 are critical for normal hematopoiesis, as the simultaneous deletion of the two kinases leads to embryonic lethality in mice (Malumbres et al., 2004). In normal hematopoiesis, CDK6 plays a crucial role in regulating the exit of long-term hematopoietic stem cells (HSCs) from quiescence and contributes to maintenance of the HSC pool (Laurenti et al., 2015). Besides its role in cell cycle regulation, CDK6 also regulates transcription by interacting with transcription factors of the STAT and AP-1 family (Kollmann et al., 2013). In ALL and MDS, CDK6 is needed to antagonize p53 responses during transformation (Bellutti et al., 2018). One of the consequences of MLL-rearrangements is the expression of stem cell gene programs that support the leukemia-initiating activity of hematopoietic stem cells (Krivtsov et al., 2006). The importance of CDK6 for leukemia with MLL-rearrangements gained attention when Placke et al. identified CDK6 as selectively implicated in the growth of MLL-AF9, MLL-AF4, and MLL-AF6 rearranged cells compared to wild type controls by using a functional genetic RNA interference screening approach (Placke et al., 2014). It has been shown that in infant MLL-AF4 ALL and MLL-AF9 AML, CDK6 but not CDK4 is a target of the MLL fusion proteins (Placke et al., 2014). MLL-AF9 can bind the CDK6 locus in wild type cells and subsequently increase the levels of CDK6. CDK6 then inhibits myeloid differentiation of AML cells while cell cycle progression is not affected (Placke et al., 2014; Van der Linden et al., 2014). The effect of CDK6 inhibition on myeloid differentiation might also be linked with the fact that CDK6 is a target of miRNA29a, a regulator of myeloid differentiation in hematopoietic progenitors and AML cells (Wang et al., 2013). The catalytic activity of CDK6 appears to be crucial for this effect, as treatment of the cells with the CDK6 inhibitor palbociclib rescues the effect of CDK6 overexpression by inducing myeloid differentiation (Placke et al., 2014). Palbociclib is an FDA-approved CDK4/6 selective inhibitor originally developed for the treatment of hormone receptor (HR)-positive and human epidermal growth factor receptor (HER2)-negative breast cancer patients (Rocca et al., 2014). The study by Placke et al. shows its great potential for reproposing against MLL-rearranged leukemias.
MLL-Menin/WDR5 Interaction Inhibitors
The MLL-menin interaction is considered a requirement for the leukemogenic activity of MLL fusion proteins (Yokoyama et al., 2005; Caslini et al., 2007). Therefore, the de novo drug development of agents able to disrupt this interaction could be a successful strategy in MLL-rearranged leukemia. Li et al. showed that when the MLL-menin interaction is disrupted in mice, only the expression of a small number of genes is affected and importantly, these genes are not the critical downstream effectors of MLL for normal hematopoiesis or B lymphopoiesis. This indicates that menin is not required for the gene expression program regulating development of HSCs and B cells (Li et al., 2013). Therefore, the fact that menin is not required for MLL function during normal hematopoiesis makes drugs that target this interaction safe and selective for MLL-rearranged leukemia (Li et al., 2013; He et al., 2016). In this context, there have been successful de novo designed small molecule inhibitors of the MLL-menin interaction, such as members of the thienopyrimidine class (Shi et al., 2012; Senter et al., 2015; Borkin et al., 2016). The crystal structures of the inhibitors MI-2-2 and MIV-6R were used in scaffold hopping calculations that as a result suggested loperamide, an antidiarrheal drug used in acute and chronic diarrhea as well as in ileostomy, as a possible inhibitor of the interaction between MLL and menin (Yue et al., 2016). Similarly to molecules that disrupt the MLL-menin interaction, histone deacetylase (HDAC) inhibitors also affect the transcription-regulatory machinery governing the epigenetic changes that drive leukemogenesis. It seems, therefore, that there is a potential for synergy between MLL-menin interaction inhibitors and HDAC inhibitors. In a study using MLL-rearranged AML cells, the HDAC inhibitor chidamide, a member of the benzamide class that has been approved by the Chinese FDA for treatment of relapsed or refractory peripheral T cell lymphoma (PTCL), showed synergistic effects with the MLL-menin interaction inhibitor MI-3 on cell growth and apoptosis. The combination of the two agents was also effective in in vivo experiments with a mouse xenograft model established by subcutaneous inoculation with MLL-rearranged MOLM-13 cells (Ye et al., 2019). This study shows the opportunity of combining drugs developed de novo to target MLL with repurposed agents to achieve synergy and optimal results.
MLL-fusion proteins lack the C-terminal SET domain of wild-type MLL. Therefore, wild-type MLL is required for the fusions in order to regulate promoter H3K4 methylation and expression of target genes, such as HOXA/MEIS1 (Milne et al., 2005; Thiel et al., 2010; Chen et al., 2017). Thus, the targeting of the H3K4 methyltransferase activity of MLL can be used to block the function of the MLL-fusion proteins in leukemia with MLL-rearrangements. This can be achieved by blocking the MLL and WDR5 protein-protein interactions (Grembecka et al., 2012; Cao et al., 2014). To this end, Chen et al. performed cell-based screenings with a compound collection to identify new therapeutic agents against MLL-rearranged leukemia cells and identified rabeprazole, a proton pump inhibitor used for heartburn, acid reflux and gastro-esophageal reflux disease (GORD), as a candidate for inhibiting the growth of MLL-rearranged cell lines but not MLL wild type cells (Chen et al., 2020). The treatment with rabeprazole caused the downregulation of the MLL fusion protein targets HOAX9 and MEIS1 and the H3K4me1 and H3K4me2 methylation levels, suggesting a dysregulation of the MLL histone methyltransferase activity. WDR5 is a key structural component of the histone methyltransferase activity of MLL (Dou et al., 2006; Karatas et al., 2013), therefore the authors tested the hypothesis that rabeprazole may exhibit its MLL-specific anti-leukemic effect via disruption of the protein-protein interaction between MLL and WDR5. By tagging the MLL win motif peptide, responsible for binding WDR5, with 5-FAM tracer as probe, they confirmed the inhibition of MLL-WDR5 interaction upon treatment with rabeprazole (Chen et al., 2020). Furthermore, they evaluated the potential of four more proton pump inhibitors for their anti-proliferation activity on leukemia cells and confirmed that proton pump inhibitors selectively inhibit leukemia cells with MLL-rearrangements and not wild type leukemia cells or normal control cells (Chen et al., 2020).
Given the complexity of the MLL recombinome and the large number of different MLL-fusion proteins (Meyer et al., 2018), only drugs targeting regions, pathways or mechanisms that are common to all, e. g., dugs targeting the N-terminus of MLL, will be effective across MLL-rearranged leukemia as a whole. It is also becoming clear that rare cases of non-canonical MLL fusions also occur; for example the recently described fusion incorporating PHD domains (Meyer et al., 2019). This also poses a challenge to therapies targeting conventional MLL-fusion proteins. The success and broad applicability of the later will depend on the effectiveness of them targeting common core pathways and interactions in MLL-rearranged leukemia. Predicting this will depend on further molecular dissection of these different fusions in the laboratory.
Using in Silico Tools and Drug Screens to Identify Novel Drugs With Anti-Leukemic Therapeutic Potential
The gene expression and regulatory profiles that characterize a disease can be used for signature-based strategies for the identification of drugs that can repurposed. Tools such as the Connectivity MAP (CMAP), GWAS and the Library of Integrated Network-based Cellular Signatures (LINCS) can be used among others for signature-based screening (Welter et al., 2014; Koleti et al., 2018). These tools open opportunities for the discovery of new drug-target combinations and significantly expand the potential use of already available therapeutic agents minimizing the time and cost of the research and development phase.
Mebendazole
The transcription factor c-MYB plays a central role in definitive hematopoiesis and the development of multiple hematopoietic lineages (Pattabiraman and Gonda, 2013). In leukemia, c-MYB-regulated transcriptional programs have been demonstrated as essential for the initiation of the disease as well as the self-renewal and maintenance of leukemia cells with MLL-rearrangements (Hess et al., 2006; Somervaille et al., 2009; Zuber et al., 2011). Importantly, reduced c-MYB levels are still compatible with normal hematopoiesis, therefore inhibition of c-MYB appears to have a therapeutic opportunity for MLL-rearranged leukemia (Emambokus et al., 2003; Zuber et al., 2011). However, its nature as a transcription factor makes it difficult to target with small molecule inhibitors and emphasizes the need for alternative targeting approaches. For example, celastrol, a natural compound that targets the interaction of c-MYB with the transcriptional co-activators CBP/P300 has shown promising anti-leukemia activity in AML (Uttarkar et al., 2016). Similarly, peptidomimetics that achieved the inhibition of the assembly of the c-MYB-CBP/P300 complex also caused leukemia cell growth arrest and extended survival of immunodeficient mice engrafted with MLL-rearranged leukemia cells (Ramaswamy et al., 2018). Surprisingly, the treatment of AML cells with mebendazole, an anti-helminth drug routinely used in the clinic for children with parasitic infections, has shown anti-leukemia activity via c-MYB inhibition in both MLL-rearranged and non-rearranged cells (Walf-Vorderwülbecke et al., 2018). Mebendazole was identified as the top hit of in a study that used the CMAP database, that offers a large reference catalogue with the gene expression profiles of various human cells stimulated with different chemicals. The CMAP database was screened with a c-MYB gene expression signature derived from integrating MLL fusion protein specific gene expression changes with a list of previously published direct c-MYB target genes (Walf-Vorderwülbecke et al., 2018). This illustrates the possibility of MLL-rearranged directed drug repurposing approaches leading to the identification of therapeutic approaches that have broad AML applicability. This is supported by recent studies confirming the importance of c-MYB in a broad range of AML subtypes (Armenteros-Monterroso et al., 2019; Takao et al., 2021) (REFs–Takao et al. DOI 10.7554/eLife.65905; Armenteros-Monterroso et al. DOI: 10.1038/s41375-019-0495-8).
Verteporfin
The CMAP database was used in another study for the identification of genes that are exclusively up- or downregulated in LSCs, the self-renewing cell population that is required for the initiation and maintenance of AML (Dick, 2005; Xiu et al., 2018). The screening identified verteporfin as an agent with potential anti-AML activity. Verteporfin has been approved by the FDA in photodynamic therapy to eliminate abnormal blood vessels in macular degeneration. It has also been used in ALL, where its anti-leukemic activity has been studied in respect to its minimal effects on normal hematopoiesis (Gibault et al., 2016; Morishita et al., 2016). Xiu et al., treated AML cells with verteporfin and found that it suppressed cell growth in vitro and delayed the development of the disease in vivo (Xiu et al., 2018). They attributed the effects of verteporfin to the upregulation of the expression of the components of the non-canonical NFκB signaling. This is particularly important for leukemia with MLL-rearrangements, where RUNX1 can interact with MLL and NFκB. This interaction allows the non-canonical signaling component to have an anti-MLL activity by interfering with the RUNX1-MLL complex (Koh et al., 2013).
Fidaxomicin
In one study aiming to identify vulnerabilities in HSC-derived human MLL-rearranged AML cells resistant to chemotherapy, Zeisig et al. identified genes differentially expressed in MLL-rearranged HSC and common myeloid progenitor (CMP) cells and compared them with the Genomic Element Associated with drug Resistance (GEAR) database (Wang et al., 2017). The screening came up with two genes associated with resistance to doxorubicin treatment; IL6, encoding for interleukin-6 and ABCC3, a member of the ABC family of organic anion transporters (Zeisig et al., 2021). Intriguingly, ABCC3 overexpression has been associated with chemoresistance and poor prognosis in both pediatric and adult AML (Steinbach et al., 2003; Benderra et al., 2005) and Zeisig et al. confirmed its higher expression in MLL-rearranged HSC cells and thus revealed the translational potential of targeting ABCC3 in MLL-rearranged leukemia (Zeisig et al., 2021). ABCC3 is targeted by fidaxomicin, a drug used for diarrhea associated with Clostridium difficile infection (Golan and Epstein, 2012). Fidaxomicin treatment of MLL-rearranged HSC cells inhibited cell growth in vitro and reduced tumor burden when combined with doxorubicin in an NSG xenograft model transplanted with HSC-derived MLL-AF6 cells (Zeisig et al., 2021). The fact that the combination treatment had significantly higher effects in the reduction of the tumor burden in the animal models that the authors used suggests that the repurposing of fidaxomicin to target ABCC3 in leukemia acts synergistically with the de novo drugs to overcome chemoresistance in MLL-rearranged leukemia.
Pyrvinium
With a different approach using a high-throughput drug library screening methodology, more than 4,000 compounds were screened at various final concentrations on two primary AML patient samples with MLL-AF9 translocation aiming to identify drugs that can achieve inhibition of more than 50% leukemic cell viability. Pyrvinium was the highest-ranked hit at the highest dose tested and achieved elimination of the leukemic cells in both samples, while affecting the viability of non-leukemic bone marrow control samples by 50% (Wander et al., 2021). Pyrvinium is an FDA approved anthelmintic drug used in the clinic for the treatment of pinworms in humans (Beck et al., 1959). Anti-cancer effects of pyrvinium have been reported in the past, with two mechanisms of action usually suggested; inhibition of the canonical Wnt signaling pathway via β-catenin degradation (Xu et al., 2018) and mitochondrial respiration impairment (Xiang et al., 2015; Xiao et al., 2016). Although the authors did not investigate in detail the mechanism of the anti-leukemia activity of pyrvinium in the MLL samples, they showed lack of β-catenin protein in MLL-rearranged AML cells and, at the same time, mitochondrial localization of the drug in the MLL cells. Therefore, they speculate that pyrvinium might cause mitochondrial respiration impairment in leukemia cells with MLL translocations (Wander et al., 2021).
Thioridazine and Artesunate
In a high-throughput drug screening of 1,280 compounds, 104 were identified as active against MLL-AF6 rearranged cell lines, with 20 of them showing activity specifically in MLL-rearranged cells and 10 exclusively on the t (6;11)-rearranged cells (Tregnago et al., 2020). One of those hits was thioridazine, an anti-psychotic agent primarily used for schizophrenia. Thioridazine caused apoptosis and cell cycle arrest exclusively in the t (6;11) AML cells. Importantly, thioridazine sensitized non- t (6;11)-rearranged cells when they were transfected with an MLL-AF6 expressing vector. The anti-leukemia effects of thioridazine were also tested in vivo, where the treatment significantly reduced the progression of tumor growth in t (6;11) SHI-1 cell transplanted mice, but not in mice transplanted with t (5;17) HL60 or t (9;11) THP-1 cells, highlighting the specificity of thioridazine against cells with the MLL-AF6 fusion. To understand the mechanism behind the MLL-AF6 specific anti-leukemia activity of thioridazine, the authors used a large-scale method combining quantitative proteomics with affinity enrichment and identified the calcium binding proteins S100A8 and S100A9, and ANXA6, a component of the annexin family, as targets of thioridazine. These three proteins are known to form a complex associated with cytoskeletal filaments (Bode et al., 2008). Therefore, the authors investigated the hypothesis that thioridazine treatment may induce cytoskeletal changes in the MLL-AF6 expressing cells and confirmed that morphologic changes and accumulation of large F-actin aggregates were induced upon exposure to thioridazine (Tregnago et al., 2020). The treatment with thioridazine caused an influx of external Ca2+ possibly via the cytoskeletal rearrangement caused by the drug. This Ca2+ overload led to reactive oxygen species (ROS) overproduction that was detrimental for the t (6;11) AML cells (Tregnago et al., 2020). Intriguingly, another class of drugs that leads to cellular and mitochondrial ROS accumulation in MLL-rearranged cells are the artemisinins, a family of antimalarial compounds. In their study, Kumar B. et al., treated MLL-rearranged AML cells expressing MLL-AF9 with artesunate and confirmed its cytotoxic activity in vitro and in vivo as well as its synergy with the BCL-2 inhibitor venetoclax. Mechanistically, the authors linked the activity of artesunate with the loss of the mitochondrial membrane potential and the generation of ROS (Kumar et al., 2017).
ABT-737 and Purvalanol A
In a study using an alternative multiplex screening approach, Lappin et al. screened a library of 80 apoptosis-inducing agents for potential combinations with compounds with therapeutic potential for pediatric AML (Lappin et al., 2020). The screening identified ABT-737, a B-cell lymphoma (BCL)-family inhibitor and Purvalanol A, a CDK2 inhibitor, as a combination with therapeutic potential for MLL-rearranged leukemia and internal tandem duplications of the fms-related tyrosine kinase 3 gene (FLT3-ITD). This high-throughput screening approach proved very effective, as it combined the selection of a therapeutically highly relevant drug library as step one, and then the application of an all-pairs testing algorithm for identification of novel partners for combination treatments. This methodology achieves the reduction of consumables and time in the laboratory and opens up opportunities identification of novel combination therapies including the repurposing of therapeutic agents (Lappin et al., 2020).
Conclusion
The understanding of the network of gene expression regulation driven by MLL fusion proteins in acute leukemia has given us a better insight of the molecular mechanisms that these fusion proteins hijack in order to initiate and maintain leukemogenesis. The protein complexes that cooperate with MLL fusion proteins and the subsequent gene expression changes open up numerous opportunities for pharmacological targeting specific to the leukemia cells with MLL-rearrangements. The successful modelling of MLL-rearranged acute leukemia has also provided fruitful experimental strategies in the search for new therapies. In this direction, we can take advantage of the technologies used for drug repurposing and identify therapeutic agents with the potential to specifically target leukemia cells with MLL-rearrangements. Exploring drug repurposing provides a new toolkit to tackle the challenges of clinical translation. The opportunity of applying known therapeutic strategies against new targets by repurposing drugs in the clinic is developing into a core strategy in drug development.
Author Contributions
AT conducted the literature review and wrote the manuscript. OW supervised the review and wrote the manuscript.
Funding
This work was funded by grants from the MRC (MR/S021000/1), GOSH Children’s Charity (W1003, V1305, V2617, V4819) and by the NIHR Great Ormond Street Hospital Biomedical Research Centre.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Baker, A., Laganiere, J., Gaudet, R., Rochefort, D., Brais, B., Neri, C., et al. (2013). Lithium Chloride Attenuates Cell Death in Oculopharyngeal Muscular Dystrophy by Perturbing Wnt/β-Catenin Pathway. Cell Death Dis 4 (10), e821. doi:10.1038/cddis.2013.342
Aguilar-Quesada, R., Muñoz-Gámez, J. A., Martín-Oliva, D., Peralta, A., Valenzuela, M. T., Matínez-Romero, R., et al. (2007). Interaction between ATM and PARP-1 in Response to DNA Damage and Sensitization of ATM Deficient Cells through PARP Inhibition. BMC Mol. Biol. 8 (1), 29. doi:10.1186/1471-2199-8-29
Armenteros-Monterroso, E., Zhao, L., Gasparoli, L., Brooks, T., Pearce, K., Mansour, M. R., et al. (2019). The AAA+ATPase RUVBL2 Is Essential for the Oncogenic Function of C-MYB in Acute Myeloid Leukemia. Leukemia 33 (12), 2817–2829. doi:10.1038/s41375-019-0495-8
Ashburn, T. T., and Thor, K. B. (2004). Drug Repositioning: Identifying and Developing New Uses for Existing Drugs. Nat. Rev. Drug Discov. 3 (8), 673–683. doi:10.1038/nrd1468
Aylon, Y., and Oren, M. (2007). Living with P53, Dying of P53. Cell. 130 (4), 597–600. doi:10.1016/j.cell.2007.08.005
Beck, J. W., Saavedra, D., Antell, G. J., and Tejeiro, B. (1959). The Treatment of Pinworm Infections in Humans (Enterobiasis) with Pyrvinium Chloride and Pyrvinium Pamoate. Am. J. Trop. Med. Hyg. 8 (3), 349–352. doi:10.4269/ajtmh.1959.8.349
Bellutti, F., Tigan, A. S., Nebenfuehr, S., Dolezal, M., Zojer, M., Grausenburger, R., et al. (2018). CDK6 Antagonizes P53-Induced Responses during Tumorigenesis. Cancer Discov. 8 (7), 884–897. doi:10.1158/2159-8290.CD-17-0912
Benderra, Z., Faussat, A. M., Sayada, L., Perrot, J. Y., Tang, R., Chaoui, D., et al. (2005). MRP3, BCRP, and P-Glycoprotein Activities Are Prognostic Factors in Adult Acute Myeloid Leukemia. Clin. Cancer Res. 11 (21), 7764–7772. doi:10.1158/1078-0432.CCR-04-1895
Beurel, E., Grieco, S. F., and Jope, R. S. (2015). Glycogen Synthase Kinase-3 (GSK3): Regulation, Actions, and Diseases. Pharmacol. Ther. 148, 114–131. doi:10.1016/j.pharmthera.2014.11.016
Bledau, A. S., Schmidt, K., Neumann, K., Hill, U., Ciotta, G., Gupta, A., et al. (2014). The H3K4 Methyltransferase Setd1a Is First Required at the Epiblast Stage, whereas Setd1b Becomes Essential after Gastrulation. Development 141 (5), 1022–1035. doi:10.1242/dev.098152
Bode, G., Lüken, A., Kerkhoff, C., Roth, J., Ludwig, S., and Nacken, W. (2008). Interaction between S100A8/A9 and Annexin A6 Is Involved in the Calcium-Induced Cell Surface Exposition of S100A8/A9. J. Biol. Chem. 283 (46), 31776–31784. doi:10.1074/jbc.M803908200
Borkin, D., Pollock, J., Kempinska, K., Purohit, T., Li, X., Wen, B., et al. (2016). Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL). J. Med. Chem. 59 (3), 892–913. doi:10.1021/acs.jmedchem.5b01305
Bryant, H. E., Petermann, E., Schultz, N., Jemth, A. S., Loseva, O., Issaeva, N., et al. (2009). PARP Is Activated at Stalled forks to Mediate Mre11-dependent Replication Restart and Recombination. EMBO J. 28 (17), 2601–2615. doi:10.1038/emboj.2009.206
Cao, F., Townsend, E. C., Karatas, H., Xu, J., Li, L., Lee, S., et al. (2014). Targeting MLL1 H3K4 Methyltransferase Activity in Mixed-Lineage Leukemia. Mol. Cel. 53 (2), 247–261. doi:10.1016/j.molcel.2013.12.001
Caslini, C., Yang, Z., El-Osta, M., Milne, T. A., Slany, R. K., and Hess, J. L. (2007). Interaction of MLL Amino Terminal Sequences with Menin Is Required for Transformation. Cancer Res. 67 (15), 7275–7283. doi:10.1158/0008-5472.CAN-06-2369
Chan, A. K. N., and Chen, C. W. (2019). Rewiring the Epigenetic Networks in MLL-Rearranged Leukemias: Epigenetic Dysregulation and Pharmacological Interventions. Front Cel Dev Biol 7, 81. doi:10.3389/fcell.2019.00081
Chen, J., Feng, J., Fang, Z., Ye, J., Chen, Q., Chen, Q., et al. (2021). Anlotinib Suppresses MLL-Rearranged Acute Myeloid Leukemia Cell Growth by Inhibiting SETD1A/AKT-Mediated DNA Damage Response. Am. J. Transl Res. 13 (3), 1494–1504.
Chen, W. L., Li, D. D., Chen, X., Wang, Y. Z., Xu, J. J., Jiang, Z. Y., et al. (2020). Proton Pump Inhibitors Selectively Suppress MLL Rearranged Leukemia Cells via Disrupting MLL1-WDR5 Protein-Protein Interaction. Eur. J. Med. Chem. 188, 112027. doi:10.1016/j.ejmech.2019.112027
Chen, Y., Anastassiadis, K., Kranz, A., Stewart, A. F., Arndt, K., Waskow, C., et al. (2017). MLL2, Not MLL1, Plays a Major Role in Sustaining MLL-Rearranged Acute Myeloid Leukemia. Cancer cell 31 (6), 755–e6. doi:10.1016/j.ccell.2017.05.002
Chu, S. H., Song, E. J., Chabon, J., Minehart, J., Feng, Z., Ross, K., et al. (2017). Targeting the DNA Damage Response Pathway in MLL-Rearranged B-Acute Lymphoblastic Leukemias. Blood 130 (Suppl. 1), 3880. doi:10.1182/blood.V130.Suppl_1.3880.3880
Ciszewski, W. M., Tavecchio, M., Dastych, J., and Curtin, N. J. (2014). DNA-PK Inhibition by NU7441 Sensitizes Breast Cancer Cells to Ionizing Radiation and Doxorubicin. Breast Cancer Res. Treat. 143 (1), 47–55. doi:10.1007/s10549-013-2785-6
Costa, B. M., Smith, J. S., Chen, Y., Chen, J., Phillips, H. S., Aldape, K. D., et al. (2010). Reversing HOXA9 Oncogene Activation by PI3K Inhibition: Epigenetic Mechanism and Prognostic Significance in Human Glioblastoma. Cancer Res. 70 (2), 453–462. doi:10.1158/0008-5472.CAN-09-2189
Denissov, S., Hofemeister, H., Marks, H., Kranz, A., Ciotta, G., Singh, S., et al. (2014). Mll2 Is Required for H3K4 Trimethylation on Bivalent Promoters in Embryonic Stem Cells, whereas Mll1 Is Redundant. Development 141 (3), 526–537. doi:10.1242/dev.102681
Dick, J. E. (2005). Acute Myeloid Leukemia Stem Cells. Ann. N. Y Acad. Sci. 1044 (1), 1–5. doi:10.1196/annals.1349.001
Dong, J., Ren, Y., Zhang, T., Wang, Z., Ling, C. C., Li, G. C., et al. (2018). Inactivation of DNA-PK by Knockdown DNA-PKcs or NU7441 Impairs Non-homologous End-Joining of Radiation-Induced Double Strand Break Repair. Oncol. Rep. 39 (3), 912–920. doi:10.3892/or.2018.6217
Dou, Y., Milne, T. A., Ruthenburg, A. J., Lee, S., Lee, J. W., Verdine, G. L., et al. (2006). Regulation of MLL1 H3K4 Methyltransferase Activity by its Core Components. Nat. Struct. Mol. Biol. 13 (8), 713–719. doi:10.1038/nsmb1128
Eguchi, M., Eguchi-Ishimae, M., Knight, D., Kearney, L., Slany, R., and Greaves, M. (2006). MLL Chimeric Protein Activation Renders Cells Vulnerable to Chromosomal Damage: an Explanation for the Very Short Latency of Infant Leukemia. Genes Chromosomes Cancer 45 (8), 754–760. doi:10.1002/gcc.20338
Elliott, S. L., Crawford, C., Mulligan, E., Summerfield, G., Newton, P., Wallis, J., et al. (2011). Mitoxantrone in Combination with an Inhibitor of DNA-dependent Protein Kinase: a Potential Therapy for High Risk B-Cell Chronic Lymphocytic Leukaemia. Br. J. Haematol. 152 (1), 61–71. doi:10.1111/j.1365-2141.2010.08425.x
Emambokus, N., Vegiopoulos, A., Harman, B., Jenkinson, E., Anderson, G., and Frampton, J. (2003). Progression through Key Stages of Haemopoiesis Is Dependent on Distinct Threshold Levels of C-Myb. EMBO J. 22 (17), 4478–4488. doi:10.1093/emboj/cdg434
Ernst, P., Fisher, J. K., Avery, W., Wade, S., Foy, D., and Korsmeyer, S. J. (2004). Definitive Hematopoiesis Requires the Mixed-Lineage Leukemia Gene. Dev. Cel 6 (3), 437–443. doi:10.1016/s1534-5807(04)00061-9
Esposito, M. T., Zhao, L., Fung, T. K., Rane, J. K., Wilson, A., Martin, N., et al. (2015). Synthetic Lethal Targeting of Oncogenic Transcription Factors in Acute Leukemia by PARP Inhibitors. Nat. Med. 21 (12), 1481–1490. doi:10.1038/nm.3993
Evers, B., Drost, R., Schut, E., de Bruin, M., van der Burg, E., Derksen, P. W., et al. (2008). Selective Inhibition of BRCA2-Deficient Mammary Tumor Cell Growth by AZD2281 and Cisplatin. Clin. Cancer Res. 14 (12), 3916–3925. doi:10.1158/1078-0432.CCR-07-4953
Gibault, F., Corvaisier, M., Bailly, F., Huet, G., Melnyk, P., and Cotelle, P. (2016). Non-photoinduced Biological Properties of Verteporfin. Curr. Med. Chem. 23 (11), 1171–1184. doi:10.2174/0929867323666160316125048
Golan, Y., and Epstein, L. (2012). Safety and Efficacy of Fidaxomicin in the Treatment of Clostridium Difficile-Associated Diarrhea. Therap Adv. Gastroenterol. 5 (6), 395–402. doi:10.1177/1756283X12461294
Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., et al. (1999). Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring. Science 286 (5439), 531–537. doi:10.1126/science.286.5439.531
Grembecka, J., He, S., Shi, A., Purohit, T., Muntean, A. G., Sorenson, R. J., et al. (2012). Menin-MLL Inhibitors Reverse Oncogenic Activity of MLL Fusion Proteins in Leukemia. Nat. Chem. Biol. 8 (3), 277–284. doi:10.1038/nchembio.773
Gurrieri, C., Piazza, F., Gnoato, M., Montini, B., Biasutto, L., Gattazzo, C., et al. (2010). 3-(2,4-dichlorophenyl)-4-(1-methyl-1H-indol-3-yl)-1H-pyrrole-2,5-dione (SB216763), a Glycogen Synthase Kinase-3 Inhibitor, Displays Therapeutic Properties in a Mouse Model of Pulmonary Inflammation and Fibrosis. J. Pharmacol. Exp. Ther. 332 (3), 785–794. doi:10.1124/jpet.109.153049
Guru Murthy, G. S., Venkitachalam, R., and Mehta, P. (2015). Trends in Survival Outcomes of B-Lineage Acute Lymphoblastic Leukemia in Elderly Patients: Analysis of Surveillance, Epidemiology, and End Results Database. Leuk. Lymphoma 56 (8), 2296–2300. doi:10.3109/10428194.2014.991921
Haince, J. F., Kozlov, S., Dawson, V. L., Dawson, T. M., Hendzel, M. J., Lavin, M. F., et al. (2007). Ataxia Telangiectasia Mutated (ATM) Signaling Network Is Modulated by a Novel poly(ADP-ribose)-dependent Pathway in the Early Response to DNA-Damaging Agents. J. Biol. Chem. 282 (22), 16441–16453. doi:10.1074/jbc.M608406200
Haince, J. F., McDonald, D., Rodrigue, A., Déry, U., Masson, J. Y., Hendzel, M. J., et al. (2008). PARP1-dependent Kinetics of Recruitment of MRE11 and NBS1 Proteins to Multiple DNA Damage Sites. J. Biol. Chem. 283 (2), 1197–1208. doi:10.1074/jbc.M706734200
Han, B., Li, K., Wang, Q., Zhang, L., Shi, J., Wang, Z., et al. (2018). Effect of Anlotinib as a Third-Line or Further Treatment on Overall Survival of Patients with Advanced Non-small Cell Lung Cancer: The ALTER 0303 Phase 3 Randomized Clinical Trial. JAMA Oncol. 4 (11), 1569–1575. doi:10.1001/jamaoncol.2018.3039
Harrison, C. J., Hills, R. K., Moorman, A. V., Grimwade, D. J., Hann, I., Webb, D. K., et al. (2010). Cytogenetics of Childhood Acute Myeloid Leukemia: United Kingdom Medical Research Council Treatment Trials AML 10 and 12. J. Clin. Oncol. 28 (16), 2674–2681. doi:10.1200/JCO.2009.24.8997
Hartley, K. O., Gell, D., Smith, G. C., Zhang, H., Divecha, N., Connelly, M. A., et al. (1995). DNA-dependent Protein Kinase Catalytic Subunit: a Relative of Phosphatidylinositol 3-kinase and the Ataxia Telangiectasia Gene Product. Cell. 82 (5), 849–856. doi:10.1016/0092-8674(95)90482-4
He, C., Wu, T., and Hao, Y. (2018). Anlotinib Induces Hepatocellular Carcinoma Apoptosis and Inhibits Proliferation via Erk and Akt Pathway. Biochem. Biophys. Res. Commun. 503 (4), 3093–3099. doi:10.1016/j.bbrc.2018.08.098
He, S., Malik, B., Borkin, D., Miao, H., Shukla, S., Kempinska, K., et al. (2016). Menin-MLL Inhibitors Block Oncogenic Transformation by MLL-Fusion Proteins in a Fusion Partner-independent Manner. Leukemia 30 (2), 508–513. doi:10.1038/leu.2015.144
Hess, J. L., Bittner, C. B., Zeisig, D. T., Bach, C., Fuchs, U., Borkhardt, A., et al. (2006). c-Myb Is an Essential Downstream Target for Homeobox-Mediated Transformation of Hematopoietic Cells. Blood 108 (1), 297–304. doi:10.1182/blood-2005-12-5014
Hilden, J. M., Dinndorf, P. A., Meerbaum, S. O., Sather, H., Villaluna, D., Heerema, N. A., et al. (2006). Analysis of Prognostic Factors of Acute Lymphoblastic Leukemia in Infants: Report on CCG 1953 from the Children's Oncology Group. Blood 108 (2), 441–451. doi:10.1182/blood-2005-07-3011
Hoshii, T., Cifani, P., Feng, Z., Huang, C. H., Koche, R., Chen, C. W., et al. (2018). A Non-catalytic Function of SETD1A Regulates Cyclin K and the DNA Damage Response. Cell. 172 (5), 1007–e17. doi:10.1016/j.cell.2018.01.032
Hurley, P. J., and Bunz, F. (2007). ATM and ATR: Components of an Integrated Circuit. Cell cycle 6 (4), 414–417. doi:10.4161/cc.6.4.3886
Ishida, J., Konishi, M., Ebner, N., and Springer, J. (2016). Repurposing of Approved Cardiovascular Drugs. J. Transl Med. 14 (1), 269. doi:10.1186/s12967-016-1031-5
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational Landscape and Significance across 12 Major Cancer Types. Nature 502 (7471), 333–339. doi:10.1038/nature12634
Karatas, H., Townsend, E. C., Cao, F., Chen, Y., Bernard, D., Liu, L., et al. (2013). High-affinity, Small-Molecule Peptidomimetic Inhibitors of MLL1/WDR5 Protein-Protein Interaction. J. Am. Chem. Soc. 135 (2), 669–682. doi:10.1021/ja306028q
Koh, C. P., Wang, C. Q., Ng, C. E., Ito, Y., Araki, M., Tergaonkar, V., et al. (2013). RUNX1 Meets MLL: Epigenetic Regulation of Hematopoiesis by Two Leukemia Genes. Leukemia 27 (9), 1793–1802. doi:10.1038/leu.2013.200
Koleti, A., Terryn, R., Stathias, V., Chung, C., Cooper, D. J., Turner, J. P., et al. (2018). Data Portal for the Library of Integrated Network-Based Cellular Signatures (LINCS) Program: Integrated Access to Diverse Large-Scale Cellular Perturbation Response Data. Nucleic Acids Res. 46 (D1), D558–D66. doi:10.1093/nar/gkx1063
Kollmann, K., Heller, G., Schneckenleithner, C., Warsch, W., Scheicher, R., Ott, R. G., et al. (2013). A Kinase-independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer cell 24 (2), 167–181. doi:10.1016/j.ccr.2013.07.012
Kraus, W. L., and Hottiger, M. O. (2013). PARP-1 and Gene Regulation: Progress and Puzzles. Mol. Aspects Med. 34 (6), 1109–1123. doi:10.1016/j.mam.2013.01.005
Krietsch, J., Rouleau, M., Pic, É., Ethier, C., Dawson, T. M., Dawson, V. L., et al. (2013). Reprogramming Cellular Events by poly(ADP-Ribose)-Binding Proteins. Mol. Aspects Med. 34 (4), 1066–1087. doi:10.1016/j.mam.2012.12.005
Krivtsov, A. V., and Armstrong, S. A. (2007). MLL Translocations, Histone Modifications and Leukaemia Stem-Cell Development. Nat. Rev. Cancer 7 (11), 823–833. doi:10.1038/nrc2253
Krivtsov, A. V., Twomey, D., Feng, Z., Stubbs, M. C., Wang, Y., Faber, J., et al. (2006). Transformation from Committed Progenitor to Leukaemia Stem Cell Initiated by MLL-AF9. Nature 442 (7104), 818–822. doi:10.1038/nature04980
Kumar, B., Kalvala, A., Chu, S., Rosen, S., Forman, S. J., Marcucci, G., et al. (2017). Antileukemic Activity and Cellular Effects of the Antimalarial Agent Artesunate in Acute Myeloid Leukemia. Leuk. Res. 59, 124–135. doi:10.1016/j.leukres.2017.05.007
Lappin, K. M., Davis, L., Matchett, K. B., Ge, Y., Mills, K. I., and Blayney, J. K. (2020). A Compound Combination Screening Approach with Potential to Identify New Treatment Options for Paediatric Acute Myeloid Leukaemia. Sci. Rep. 10 (1), 18514. doi:10.1038/s41598-020-75453-3
Laurenti, E., Frelin, C., Xie, S., Ferrari, R., Dunant, C. F., Zandi, S., et al. (2015). CDK6 Levels Regulate Quiescence Exit in Human Hematopoietic Stem Cells. Cell Stem Cell 16 (3), 302–313. doi:10.1016/j.stem.2015.01.017
Lawrence, H. J., Christensen, J., Fong, S., Hu, Y. L., Weissman, I., Sauvageau, G., et al. (2005). Loss of Expression of the Hoxa-9 Homeobox Gene Impairs the Proliferation and Repopulating Ability of Hematopoietic Stem Cells. Blood 106 (12), 3988–3994. doi:10.1182/blood-2005-05-2003
Li, B. E., Gan, T., Meyerson, M., Rabbitts, T. H., and Ernst, P. (2013). Distinct Pathways Regulated by Menin and by MLL1 in Hematopoietic Stem Cells and Developing B Cells. Blood 122 (12), 2039–2046. doi:10.1182/blood-2013-03-486647
Maifrede, S., Martinez, E., Nieborowska-Skorska, M., Di Marcantonio, D., Hulse, M., Le, B. V., et al. (2017). MLL-AF9 Leukemias Are Sensitive to PARP1 Inhibitors Combined with Cytotoxic Drugs. Blood Adv. 1 (19), 1467–1472. doi:10.1182/bloodadvances.2017006247
Malumbres, M., Sotillo, R., Santamaría, D., Galán, J., Cerezo, A., Ortega, S., et al. (2004). Mammalian Cells Cycle without the D-type Cyclin-dependent Kinases Cdk4 and Cdk6. Cell. 118 (4), 493–504. doi:10.1016/j.cell.2004.08.002
Martinez Molina, D., Jafari, R., Ignatushchenko, M., Seki, T., Larsson, E. A., Dan, C., et al. (2013). Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular thermal Shift Assay. Science 341 (6141), 84–87. doi:10.1126/science.1233606
Meek, K., Dang, V., and Lees-Miller, S. P. (2008). DNA-PK: the Means to Justify the Ends?. Adv. Immunol. 99, 33–58. doi:10.1016/S0065-2776(08)00602-0
Meyer, C., Burmeister, T., Gröger, D., Tsaur, G., Fechina, L., Renneville, A., et al. (2018). The MLL Recombinome of Acute Leukemias in 2017. Leukemia 32 (2), 273–284. doi:10.1038/leu.2017.213
Meyer, C., Lopes, B. A., Caye-Eude, A., Cavé, H., Arfeuille, C., Cuccuini, W., et al. (2019). Human MLL/KMT2A Gene Exhibits a Second Breakpoint Cluster Region for Recurrent MLL-USP2 Fusions. Leukemia 33 (9), 2306–2340. doi:10.1038/s41375-019-0451-7
Milne, T. A., Martin, M. E., Brock, H. W., Slany, R. K., and Hess, J. L. (2005). Leukemogenic MLL Fusion Proteins Bind across a Broad Region of the Hox A9 Locus, Promoting Transcription and Multiple Histone Modifications. Cancer Res. 65 (24), 11367–11374. doi:10.1158/0008-5472.CAN-05-1041
Moffat, J. G., Vincent, F., Lee, J. A., Eder, J., and Prunotto, M. (2017). Opportunities and Challenges in Phenotypic Drug Discovery: an Industry Perspective. Nat. Rev. Drug Discov. 16 (8), 531–543. doi:10.1038/nrd.2017.111
Morishita, T., Hayakawa, F., Sugimoto, K., Iwase, M., Yamamoto, H., Hirano, D., et al. (2016). The Photosensitizer Verteporfin Has Light-independent Anti-leukemic Activity for Ph-Positive Acute Lymphoblastic Leukemia and Synergistically Works with Dasatinib. Oncotarget 7 (35), 56241–56252. doi:10.18632/oncotarget.11025
Munos, B. (2009). Lessons from 60 Years of Pharmaceutical Innovation. Nat. Rev. Drug Discov. 8 (12), 959–968. doi:10.1038/nrd2961
Nosengo, N. (2016). Can You Teach Old Drugs New Tricks?. Nature 534 (7607), 314–316. doi:10.1038/534314a
Palumbo, A., Facon, T., Sonneveld, P., Bladè, J., Offidani, M., Gay, F., et al. (2008). Thalidomide for Treatment of Multiple Myeloma: 10 Years Later. Blood 111 (8), 3968–3977. doi:10.1182/blood-2007-10-117457
Pattabiraman, D. R., and Gonda, T. J. (2013). Role and Potential for Therapeutic Targeting of MYB in Leukemia. Leukemia 27 (2), 269–277. doi:10.1038/leu.2012.225
Pieters, R., De Lorenzo, P., Ancliffe, P., Aversa, L. A., Brethon, B., Biondi, A., et al. (2019). Outcome of Infants Younger Than 1 Year with Acute Lymphoblastic Leukemia Treated with the Interfant-06 Protocol: Results from an International Phase III Randomized Study. J. Clin. Oncol. 37 (25), 2246–2256. doi:10.1200/JCO.19.00261
Pieters, R., Schrappe, M., De Lorenzo, P., Hann, I., De Rossi, G., Felice, M., et al. (2007). A Treatment Protocol for Infants Younger Than 1 Year with Acute Lymphoblastic Leukaemia (Interfant-99): an Observational Study and a Multicentre Randomised Trial. Lancet 370 (9583), 240–250. doi:10.1016/S0140-6736(07)61126-X
Placke, T., Faber, K., Nonami, A., Putwain, S. L., Salih, H. R., Heidel, F. H., et al. (2014). Requirement for CDK6 in MLL-Rearranged Acute Myeloid Leukemia. Blood 124 (1), 13–23. doi:10.1182/blood-2014-02-558114
Plummer, R., Middleton, M., Wilson, R., Jones, C., Evans, J., Robson, L., et al. (2005). First in Human Phase I Trial of the PARP Inhibitor AG-014699 with Temozolomide (TMZ) in Patients (Pts) with Advanced Solid Tumors. J. Clin. Oncol. 23, 3065. doi:10.1200/jco.2005.23.16_suppl.3065
Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., et al. (2019). Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 18 (1), 41–58. doi:10.1038/nrd.2018.168
Ramaswamy, K., Forbes, L., Minuesa, G., Gindin, T., Brown, F., Kharas, M. G., et al. (2018). Peptidomimetic Blockade of MYB in Acute Myeloid Leukemia. Nat. Commun. 9 (1), 110–113. doi:10.1038/s41467-017-02618-6
Rassool, F. V. (2003). DNA Double Strand Breaks (DSB) and Non-homologous End Joining (NHEJ) Pathways in Human Leukemia. Cancer Lett. 193 (1), 1–9. doi:10.1016/s0304-3835(02)00692-4
Riabinska, A., Daheim, M., Herter-Sprie, G. S., Winkler, J., Fritz, C., Hallek, M., et al. (2013). Therapeutic Targeting of a Robust Non-oncogene Addiction to PRKDC in ATM-Defective Tumors. Sci. Transl Med. 5 (189), 189ra78. doi:10.1126/scitranslmed.3005814
Rocca, A., Farolfi, A., Bravaccini, S., Schirone, A., and Amadori, D. (2014). Palbociclib (PD 0332991) : Targeting the Cell Cycle Machinery in Breast Cancer. Expert Opin. Pharmacother. 15 (3), 407–420. doi:10.1517/14656566.2014.870555
Sandhu, S. K., Wenham, R. M., Wilding, G., McFadden, M., Sun, L., Toniatti, C., et al. (2010). First-in-human Trial of a Poly (ADP-Ribose) Polymerase (PARP) Inhibitor MK-4827 in Advanced Cancer Patients (Pts) with Antitumor Activity in BRCA-Deficient and Sporadic Ovarian Cancers. J. Clin. Oncol. 28, 3001. doi:10.1200/jco.2010.28.15_suppl.3001
Santos, M. A., Faryabi, R. B., Ergen, A. V., Day, A. M., Malhowski, A., Canela, A., et al. (2014). DNA-damage-induced Differentiation of Leukaemic Cells as an Anti-cancer Barrier. Nature 514 (7520), 107–111. doi:10.1038/nature13483
Scannell, J. W., Blanckley, A., Boldon, H., and Warrington, B. (2012). Diagnosing the Decline in Pharmaceutical R&D Efficiency. Nat. Rev. Drug Discov. 11 (3), 191–200. doi:10.1038/nrd3681
Scavone, C., Brusco, S., Bertini, M., Sportiello, L., Rafaniello, C., Zoccoli, A., et al. (2020). Current Pharmacological Treatments for COVID-19: What's Next?. Br. J. Pharmacol. 177 (21), 4813–4824. doi:10.1111/bph.15072
Schoppy, D. W., Ragland, R. L., Gilad, O., Shastri, N., Peters, A. A., Murga, M., et al. (2012). Oncogenic Stress Sensitizes Murine Cancers to Hypomorphic Suppression of ATR. J. Clin. Invest. 122 (1), 241–252. doi:10.1172/JCI58928
Senter, T., Gogliotti, R. D., Han, C., Locuson, C. W., Morrison, R., Daniels, J. S., et al. (2015). Progress towards Small Molecule Menin-Mixed Lineage Leukemia (MLL) Interaction Inhibitors with In Vivo Utility. Bioorg. Med. Chem. Lett. 25 (13), 2720–2725. doi:10.1016/j.bmcl.2015.04.026
Shen, G., Zheng, F., Ren, D., Du, F., Dong, Q., Wang, Z., et al. (2018). Anlotinib: a Novel Multi-Targeting Tyrosine Kinase Inhibitor in Clinical Development. J. Hematol. Oncol. 11 (1), 120. doi:10.1186/s13045-018-0664-7
Shen, Y., Rehman, F. L., Feng, Y., Boshuizen, J., Bajrami, I., Elliott, R., et al. (2013). BMN 673, a Novel and Highly Potent PARP1/2 Inhibitor for the Treatment of Human Cancers with DNA Repair Deficiency. Clin. Cancer Res. 19 (18), 5003–5015. doi:10.1158/1078-0432.CCR-13-1391
Shi, A., Murai, M. J., He, S., Lund, G., Hartley, T., Purohit, T., et al. (2012). Structural Insights into Inhibition of the Bivalent Menin-MLL Interaction by Small Molecules in Leukemia. Blood 120 (23), 4461–4469. doi:10.1182/blood-2012-05-429274
Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008). Regulation of DNA Double-Strand Break Repair Pathway Choice. Cell Res 18 (1), 134–147. doi:10.1038/cr.2007.111
Singh, T. U., Parida, S., Lingaraju, M. C., Kesavan, M., Kumar, D., and Singh, R. K. (2020). Drug Repurposing Approach to Fight COVID-19. Pharmacol. Rep. 72, 1479–1508. doi:10.1007/s43440-020-00155-6
Smith, L. L., Yeung, J., Zeisig, B. B., Popov, N., Huijbers, I., Barnes, J., et al. (2011). Functional Crosstalk between Bmi1 and MLL/Hoxa9 axis in Establishment of normal Hematopoietic and Leukemic Stem Cells. Cell stem cell 8 (6), 649–662. doi:10.1016/j.stem.2011.05.004
So, C. W., Karsunky, H., Wong, P., Weissman, I. L., and Cleary, M. L. (2004). Leukemic Transformation of Hematopoietic Progenitors by MLL-GAS7 in the Absence of Hoxa7 or Hoxa9. Blood 103 (8), 3192–3199. doi:10.1182/blood-2003-10-3722
Somervaille, T. C., Matheny, C. J., Spencer, G. J., Iwasaki, M., Rinn, J. L., Witten, D. M., et al. (2009). Hierarchical Maintenance of MLL Myeloid Leukemia Stem Cells Employs a Transcriptional Program Shared with Embryonic rather Than Adult Stem Cells. Cell stem cell 4 (2), 129–140. doi:10.1016/j.stem.2008.11.015
Steinbach, D., Lengemann, J., Voigt, A., Hermann, J., Zintl, F., and Sauerbrey, A. (2003). Response to Chemotherapy and Expression of the Genes Encoding the Multidrug Resistance-Associated Proteins MRP2, MRP3, MRP4, MRP5, and SMRP in Childhood Acute Myeloid Leukemia. Clin. Cancer Res. 9 (3), 1083–1086.
Sun, Y., Niu, W., Du, F., Du, C., Li, S., Wang, J., et al. (2016). Safety, Pharmacokinetics, and Antitumor Properties of Anlotinib, an Oral Multi-Target Tyrosine Kinase Inhibitor, in Patients with Advanced Refractory Solid Tumors. J. Hematol. Oncol. 9 (1), 105–109. doi:10.1186/s13045-016-0332-8
Syed, Y. Y. (2018). Anlotinib: First Global Approval. Drugs 78 (10), 1057–1062. doi:10.1007/s40265-018-0939-x
Tajima, K., Matsuda, S., Yae, T., Drapkin, B. J., Morris, R., Boukhali, M., et al. (2019). SETD1A Protects from Senescence through Regulation of the Mitotic Gene Expression Program. Nat. Commun. 10 (1), 2854. doi:10.1038/s41467-019-10786-w
Takacova, S., Slany, R., Bartkova, J., Stranecky, V., Dolezel, P., Luzna, P., et al. (2012). DNA Damage Response and Inflammatory Signaling Limit the MLL-ENL-Induced Leukemogenesis In Vivo. Cancer cell 21 (4), 517–531. doi:10.1016/j.ccr.2012.01.021
Takao, S. F. L., Uni, M., Cheng, S., Pineda, J. M., Tarumoto, Y., Cifani, P., et al. (2021). Convergent Organization of Aberrant MYB Complex Controls Oncogenic Gene Expression in Acute Myeloid Leukemia. Elife 10, e65905. doi:10.7554/elife.65905
Thiel, A. T., Blessington, P., Zou, T., Feather, D., Wu, X., Yan, J., et al. (2010). MLL-AF9-induced Leukemogenesis Requires Coexpression of the Wild-type Mll Allele. Cancer Cell. 17 (2), 148–159. doi:10.1016/j.ccr.2009.12.034
Thijssen, R., Ter Burg, J., Garrick, B., van Bochove, G. G., Brown, J. R., Fernandes, S. M., et al. (2016). Dual TORK/DNA-PK Inhibition Blocks Critical Signaling Pathways in Chronic Lymphocytic Leukemia. Blood 128 (4), 574–583. doi:10.1182/blood-2016-02-700328
Tregnago, C., Da Ros, A., Porcù, E., Benetton, M., Simonato, M., Simula, L., et al. (2020). Thioridazine Requires Calcium Influx to Induce MLL-AF6-Rearranged AML Cell Death. Blood Adv. 4 (18), 4417–4429. doi:10.1182/bloodadvances.2020002001
Trowbridge, J. J., Xenocostas, A., Moon, R. T., and Bhatia, M. (2006). Glycogen Synthase Kinase-3 Is an In Vivo Regulator of Hematopoietic Stem Cell Repopulation. Nat. Med. 12 (1), 89–98. doi:10.1038/nm1339
Tseng, A. S., Engel, F. B., and Keating, M. T. (2006). The GSK-3 Inhibitor BIO Promotes Proliferation in Mammalian Cardiomyocytes. Chem. Biol. 13 (9), 957–963. doi:10.1016/j.chembiol.2006.08.004
Tsuji, T., Sapinoso, L. M., Tran, T., Gaffney, B., Wong, L., Sankar, S., et al. (2017). CC-115, a Dual Inhibitor of mTOR Kinase and DNA-PK, Blocks DNA Damage Repair Pathways and Selectively Inhibits ATM-Deficient Cell Growth In Vitro. Oncotarget 8 (43), 74688–74702. doi:10.18632/oncotarget.20342
Uttarkar, S., Dassé, E., Coulibaly, A., Steinmann, S., Jakobs, A., Schomburg, C., et al. (2016). Targeting Acute Myeloid Leukemia with a Small Molecule Inhibitor of the Myb/p300 Interaction. Blood 127 (9), 1173–1182. doi:10.1182/blood-2015-09-668632
Van der Linden, M. H., Willekes, M., van Roon, E., Seslija, L., Schneider, P., Pieters, R., et al. (2014). MLL Fusion-Driven Activation of CDK6 Potentiates Proliferation in MLL-Rearranged Infant ALL. Cell cycle 13 (5), 834–844. doi:10.4161/cc.27757
Walf-Vorderwülbecke, V., Pearce, K., Brooks, T., Hubank, M., van den Heuvel-Eibrink, M. M., Zwaan, C. M., et al. (2018). Targeting Acute Myeloid Leukemia by Drug-Induced C-MYB Degradation. Leukemia 32 (4), 882–889. doi:10.1038/leu.2017.317
Wander, P., Arentsen-Peters, S. T. C. J. M., Pinhanҫos, S. S., Koopmans, B., Dolman, M. E. M., Ariese, R., et al. (2021). High-throughput Drug Screening Reveals Pyrvinium Pamoate as Effective Candidate against Pediatric MLL-Rearranged Acute Myeloid Leukemia. Transl Oncol. 14 (5), 101048. doi:10.1016/j.tranon.2021.101048
Wang, J., Zhao, Y., Wang, Y., and Huang, J. (2013). Molecular Dynamics Simulations and Statistical Coupling Analysis Reveal Functional Coevolution Network of Oncogenic Mutations in the CDKN2A-CDK6 Complex. FEBS Lett. 587 (2), 136–141. doi:10.1016/j.febslet.2012.11.001
Wang, Y. Y., Chen, W. H., Xiao, P. P., Xie, W. B., Luo, Q., Bork, P., et al. (2017). GEAR: A Database of Genomic Elements Associated with Drug Resistance. Sci. Rep. 7 (1), 44085–44088. doi:10.1038/srep44085
Wang, Z., Iwasaki, M., Ficara, F., Lin, C., Matheny, C., Wong, S. H., et al. (2010). GSK-3 Promotes Conditional Association of CREB and its Coactivators with MEIS1 to Facilitate HOX-Mediated Transcription and Oncogenesis. Cancer cell 17 (6), 597–608. doi:10.1016/j.ccr.2010.04.024
Wang, Z., Smith, K. S., Murphy, M., Piloto, O., Somervaille, T. C., and Cleary, M. L. (2008). Glycogen Synthase Kinase 3 in MLL Leukaemia Maintenance and Targeted Therapy. Nature 455 (7217), 1205–1209. doi:10.1038/nature07284
Weber, A. M., and Ryan, A. J. (2015). ATM and ATR as Therapeutic Targets in Cancer. Pharmacol. Ther. 149, 124–138. doi:10.1016/j.pharmthera.2014.12.001
Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., et al. (2014). The NHGRI GWAS Catalog, a Curated Resource of SNP-Trait Associations. Nucleic Acids Res. 42 (D1), D1001–D1006. doi:10.1093/nar/gkt1229
Wiederschain, D., Kawai, H., Shilatifard, A., and Yuan, Z. M. (2005). Multiple Mixed Lineage Leukemia (MLL) Fusion Proteins Suppress P53-Mediated Response to DNA Damage. J. Biol. Chem. 280 (26), 24315–24321. doi:10.1074/jbc.M412237200
Winters, A. C., and Bernt, K. M. (2017). MLL-rearranged Leukemias-An Update on Science and Clinical Approaches. Front. Pediatr. 5, 4. doi:10.3389/fped.2017.00004
Xiang, W., Cheong, J. K., Ang, S. H., Teo, B., Xu, P., Asari, K., et al. (2015). Pyrvinium Selectively Targets Blast Phase-Chronic Myeloid Leukemia through Inhibition of Mitochondrial Respiration. Oncotarget 6 (32), 33769–33780. doi:10.18632/oncotarget.5615
Xiao, M., Zhang, L., Zhou, Y., Rajoria, P., and Wang, C. (2016). Pyrvinium Selectively Induces Apoptosis of Lymphoma Cells through Impairing Mitochondrial Functions and JAK2/STAT5. Biochem. Biophys. Res. Commun. 469 (3), 716–722. doi:10.1016/j.bbrc.2015.12.059
Xiu, Y., Dong, Q., Li, Q., Li, F., Borcherding, N., Zhang, W., et al. (2018). Stabilization of NF-Κb-Inducing Kinase Suppresses MLL-AF9-Induced Acute Myeloid Leukemia. Cell Rep 22 (2), 350–358. doi:10.1016/j.celrep.2017.12.055
Xu, F., Zhu, Y., Lu, Y., Yu, Z., Zhong, J., Li, Y., et al. (2018). Anthelmintic Pyrvinium Pamoate Blocks Wnt/β-Catenin and Induces Apoptosis in Multiple Myeloma Cells. Oncol. Lett. 15 (4), 5871–5878. doi:10.3892/ol.2018.8006
Yanai, M., Makino, H., Ping, B., Takeda, K., Tanaka, N., Sakamoto, T., et al. (2017). DNA-PK Inhibition by NU7441 Enhances Chemosensitivity to Topoisomerase Inhibitor in Non-small Cell Lung Carcinoma Cells by Blocking DNA Damage Repair. Yonago Acta Med. 60 (1), 9–15.
Yazinski, S. A., Comaills, V., Buisson, R., Genois, M. M., Nguyen, H. D., Ho, C. K., et al. (2017). ATR Inhibition Disrupts Rewired Homologous Recombination and fork protection Pathways in PARP Inhibitor-Resistant BRCA-Deficient Cancer Cells. Genes Dev. 31 (3), 318–332. doi:10.1101/gad.290957.116
Ye, J., Zha, J., Shi, Y., Li, Y., Yuan, D., Chen, Q., et al. (2019). Co-inhibition of HDAC and MLL-Menin Interaction Targets MLL-Rearranged Acute Myeloid Leukemia Cells via Disruption of DNA Damage Checkpoint and DNA Repair. Clin. Epigenetics 11 (1), 137. doi:10.1186/s13148-019-0723-0
Yeung, J., Esposito, M. T., Gandillet, A., Zeisig, B. B., Griessinger, E., Bonnet, D., et al. (2010). β-Catenin Mediates the Establishment and Drug Resistance of MLL Leukemic Stem Cells. Cancer cell 18 (6), 606–618. doi:10.1016/j.ccr.2010.10.032
Yip, B. H., and So, C. W. (2013). Mixed Lineage Leukemia Protein in normal and Leukemic Stem Cells. Exp. Biol. Med. (Maywood) 238 (3), 315–323. doi:10.1177/1535370213480717
Yokoyama, A., Somervaille, T. C., Smith, K. S., Rozenblatt-Rosen, O., Meyerson, M., and Cleary, M. L. (2005). The Menin Tumor Suppressor Protein Is an Essential Oncogenic Cofactor for MLL-Associated Leukemogenesis. Cell. 123 (2), 207–218. doi:10.1016/j.cell.2005.09.025
Yue, L., Du, J., Ye, F., Chen, Z., Li, L., Lian, F., et al. (2016). Identification of Novel Small-Molecule Inhibitors Targeting Menin-MLL Interaction, Repurposing the Antidiarrheal Loperamide. Org. Biomol. Chem. 14 (36), 8503–8519. doi:10.1039/c6ob01248e
Zeisig, B. B., Fung, T. K., Zarowiecki, M., Tsai, C. T., Luo, H., Stanojevic, B., et al. (2021). Functional Reconstruction of Human AML Reveals Stem Cell Origin and Vulnerability of Treatment-Resistant MLL-Rearranged Leukemia. Sci. Transl Med. 13, eabc4822. doi:10.1126/scitranslmed.abc4822
Zhang, Z., Zhou, L., Xie, N., Nice, E. C., Zhang, T., Cui, Y., et al. (2020). Overcoming Cancer Therapeutic Bottleneck by Drug Repurposing. Signal. Transduct Target. Ther. 5 (1), 113–125. doi:10.1038/s41392-020-00213-8
Zhao, L., and So, C. W. (2016). PARP-inhibitor-induced Synthetic Lethality for Acute Myeloid Leukemia Treatment. Exp. Hematol. 44 (10), 902–907. doi:10.1016/j.exphem.2016.07.007
Keywords: MLL-rearrangements, leukemia, drug repurposing, AML, ALL
Citation: Tsakaneli A and Williams O (2021) Drug Repurposing for Targeting Acute Leukemia With KMT2A (MLL)—Gene Rearrangements. Front. Pharmacol. 12:741413. doi: 10.3389/fphar.2021.741413
Received: 14 July 2021; Accepted: 01 September 2021;
Published: 14 September 2021.
Edited by:
Maria Teresa Esposito, University of Roehampton London, United KingdomReviewed by:
Akihiko Yokoyama, National Cancer Centre, JapanHansen Kosasih, Royal Children’s Hospital, Australia
Copyright © 2021 Tsakaneli and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexia Tsakaneli, a.tsakaneli@ucl.ac.uk; Owen Williams, owen.williams@ucl.ac.uk
 Alexia Tsakaneli
Alexia Tsakaneli Owen Williams
Owen Williams