- 1Beijing Key Laboratory of Ecological Function Assessment and Regulation Technology of Green Space, Beijing Academy of Forestry and Landscape Architecture, Beijing, China
- 2Beijing Academy of Forestry and Landscape Architecture, Beijing Municipal Forestry and Parks Bureau, Beijing, China
- 3Beijing Academy of Forestry and Landscape Architecture, Beijing Huiwen Middle School, Beijing, China
- 4Beijing Academy of Forestry and Landscape Architecture, Miyun District Forestry and Parks Bureau, Beijing, China
Urban forests constitute vital ecological interfaces between built environments and natural systems, yet the mechanisms driving soil microbial community assembly in these ecosystems remain poorly understood. Through an integrated analysis of five dominant forest types (Populus tomentosa, Salix matsudana, Robinia pseudoacacia, Eucommia ulmoides, and Ailanthus altissima) in Beijing’s plain ecological forests, we reveal hierarchical environmental controls over bacterial diversity and network structure. High-throughput sequencing and co-occurrence network analyses demonstrated that Salix matsudana forest harbored the highest microbial diversity (Shannon index = 5.82 ± 0.14), with Proteobacteria abundance significantly elevated compared to other forest types (P < 0.01). Structural equation modeling (SEM) identified soil total nitrogen (TN) as the principal direct suppressor of bacterial diversity (path coefficient = -0.33, P < 0.001), while forest structural traits—particularly diameter at breast height—emerged as critical mediators of community composition through nutrient modulation (R² = 0.502). Notably, microbial networks exhibited forest-type-specific topologies: Populus tomentosa forest stands showed exceptional connectivity (edge density = 0.29), whereas Robinia pseudoacacia forest developed modular architectures (modularity = 2.30) enhancing ecological resilience. These findings establish a mechanistic framework linking forest management practices to microbial-mediated ecosystem functions, with direct implications for urban green space optimization under accelerating anthropogenic pressures.
1 Introduction
In the context of accelerating global urbanization, urban forests have emerged as a crucial bridge between urban and natural ecosystems, with their ecological functions becoming increasingly significant (1, 2). Soil microorganisms, as fundamental components of these urban forest ecosystems, play a vital role in the regeneration of understory forest and the processes of ecological restoration (3, 4). They contribute to sustaining ecosystem stability through their involvement in key ecological processes, including the soil fertility enhancement, organic matter decomposition, and carbon sequestration (5–7). However, soil microbial communities are highly sensitive to environmental changes, especially those associated with variations in forest types and soil physicochemical properties, further emphasizing the importance of exploring their response mechanisms. Recent studies have revealed the complex effects of shifts in forest types on the diversity and composition of soil microbial communities (8, 9). Therefore, investigating how soil microbial communities respond to environmental changes driven by different forest types in urban plain forests is of profound significance for guiding the scientific management of these forests.
The assembly of soil microbial communities is influenced by a multitude of environmental factors, rendering it a highly intricate process (10, 11). Variations in forest types and soil physicochemical properties, such as bulk density, pH, nutrient content, and enzyme activity, can significantly affect the structure of microbial community and further regulate soil nutrient cycling and forest growth conditions (8). For example, soil nutrients, particularly soil carbon content, constitute the principal energy source for microbial growth and metabolic processes. Nitrogen is an essential element for constituting microbial cellular biomacromolecules, including proteins and nucleic acids, thereby affecting the structure of microbial communities (12). Meanwhile, soil enzymes are also key factors affecting microbial community structure. For example, cellulase facilitates the decomposition of cellulose in soil, thereby providing carbon sources for microorganisms. The activity level of cellulase is directly associated with the rates of soil carbon cycling and the composition of microbial communities (13, 14). Similarly, β-glucosidase, a crucial enzyme in soil carbon cycling, exerts a significant influence on the structure and function of microbial communities (15). Moreover, appropriate soil moisture content creates a favorable metabolic environment for microorganisms, thereby enhancing their activity and facilitating the decomposition and transformation of soil nutrients (16). However, in the practice of urban plain afforestation, there is still a lack of deep understanding of the specific mechanisms by which established forest types and soil physicochemical properties synergistically drive the construction of soil microbial communities.
Soil microorganisms do not respond to environmental conditions in isolation, but interact as integral components of complex microbial communities. These communities constitute cohesive ecological assemblages, with microorganisms sharing similar environmental adaptability and resource utilization preferences. The interrelationships among these microorganisms can be examined through co-occurrence ecological networks, where microbial taxa are considered as network nodes and their relationships as network connections (17). Ecological networks provide a powerful supplement to traditional soil microbial community quantification methods, such as alpha and beta diversity (18). In recent years, ecological network analysis has emerged as a crucial tool for analyzing the structural characteristics of soil microbial communities and microbial co-abundance patterns (19, 20). Empirical studies indicate that the conversion of Populus tomentosa forests to Robinia pseudoacacia forests may strengthen symbiotic relationships within soil microbial networks (21). The nitrogen fixation capability of R. pseudoacacia can provide nitrogen sources for other microorganisms, promoting their growth and reproduction (22). Simultaneously, new competitive relationships may also emerge, as increased nitrogen may trigger intense competition among different microorganisms for nitrogen sources. However, how forest type shifts specifically affect inter-microbial interactions remains insufficient understanding.
In this study, we selected five representative forest types within the plain ecological forests of Beijing: Populus tomentosa Carrière (PT), Salix matsudana Koidz (SM), Robinia pseudoacacia (RP), Eucommia ulmoides Oliver (EU), and Ailanthus altissima (Mill.) Swingle (AA). The species chosen for this study exhibit contrasting ecological strategies: PT and SM, fast-growing riparian species; RP, an N-fixing pioneer; EU, a medicinal tree with deep roots; and AA, a pollution-tolerant invasive species. Collectively, these species account for the main part of the tree biomass in Beijing’s plain afforestation project, enabling an evaluation of how dominant management choices influence soil microbial communities. We conducted an analysis of the soil physicochemical properties and microbial community structures, as well as their interrelationships, across these five forest types. The objective of this study was to reveal the mechanisms through which different forest types influence soil microbial community structures. We hope that this study will provide scientific guidance for ecological management and biodiversity conservation.
2 Materials and methods
2.1 Study site and soil sampling
The study area is located in the Lucheng Collective Forest Farm, Tongzhou District, Beijing (39.84°N, 116.81°E) (Supplementary Figure S1). The terrain of the study area is flat, with loam soil predominant. The climate is warm temperate, featuring a semi-humid and semi-arid continental monsoon pattern, with hot rainy summers and cold, dry winters. Annual precipitation averages 400–600 mm, concentrated mainly from June and August. The soil is neutral in pH. The study area is part of the million-acre afforestation and greening project implemented in the plain areas of Beijing in 2013, with forest stands established as part of the project, each approximately 11 years old. Over the past decade, more than 3 to 5 million trees of species such as Populus tomentosa Carrière (PT), Salix matsudana Koidz (SM), Robinia pseudoacacia (RP), Eucommia ulmoides Oliver (EU), and Ailanthus altissima (Mill.) Swingle (AA) have been planted and were selected for our study. Currently, the forest stands are single-layered, differing significantly from the ideal state of mixed, multi-layered, multi-aged, and multifunctional forest communities. Three spatially independent sampling sites were established for each forest type within an area exceeding 1 hectare. At each site, we randomly established three 20 m × 20 m plots. We recorded geographical coordinates, elevation slope, and aspect using GPS and a compass (23). Tree density (TD) was obtained by counting all trees within each plot, diameter at breast height (DBH) was measured with a vernier caliper, and tree height (TH) was measured with a Blume-Leiss height meter. Crown width in both east-west and north-south directions were also measured to assess tree canopy (TC) size. Understory vegetation (herbs and shrubs < 2 m in height) was surveyed by recording species composition, coverage, and average height within five 2 m × 2 m randomly placed subplots inside each 20 m × 20 m plot. The 0–10 cm soil depth was selected due to its high density of fine roots, microbial biomass, and enzymatic activity in urban forests (24). This layer serves as the primary zone for litter decomposition and nutrient cycling, thus playing a critical role in assessing forest-microbe linkages. Six to nine soil cores (5 cm diameter, 0–10 cm depth) were randomly collected from each site and pooled to form a composited sample. For each site, three composite samples were obtained. Each composite sample was then homogenized and subdivided into four subsamples for different analyses: (1) (1) determination of soil water content (SWC) and bulk density (BD), with soil packed in a ring knife; (2) analysis of soil physicochemical properties, after air-drying and sieving through a 2 mm mesh using a ball mill; (3) molecular analysis, with fresh soil transported in a portable refrigerator at −20 °C and stored at −80 °C until DNA extraction (25, 26); and (4) measurement of soil ammonium nitrogen (NH4+-N), nitrate nitrogen (NO3–N), cellulase (CEL) and β-glucosidase (BGL) activities with fresh soil kept at 4°C.
2.2 Environmental variable selection
We selected environmental variables known to influence the soil microbial community in Beijing’s plain forests, grouping them into forest structure and soil factors (23, 27). Forest structure variables included TD, DBH, TH, and TC. Soil properties included pH, SWC, BD, soil organic carbon (SOC), total nitrogen (TN), carbon-to-nitrogen ratio (C/N), total phosphorus and (TP), and dissolved organic carbon (DOC). Soil pH was measured using a METTLER TOLEDO pH electrode with a 1:2.5 soil/water ratio. BD and SWC were determined using the ring knife method: fresh soil samples were oven-dried at 105 °C ± 2 °C for 12–24 hours to obtain dry weight. SOC was determined using the Potassium dichromate external heating method, while TN was quantified with an elemental analyzer (2400 Series II, Perkin Elmer, Boston, MA, USA). TP was measured by the molybdenum antimony anti-colorimetric method, and DOC was extracted with water, filtered through a 0.45 μm membrane, and analyzed using the potassium permanganate-sulfuric acid heating method. NH4+-N and NO3–N were determined by the indophenol blue colorimetric method and salicylic acid colorimetric method, respectively, following extraction with 2 M KCl. CEL and BGL activities were assayed using 3,5-dinitrosalicylic acid and p-nitrophenol methods, respectively (28, 29).
2.3 DNA extraction sequencing and bioinformation analysis
We extracted genomic DNA from 0.25 g soil samples using kits from Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China). The integrity and purity of the extracted DNA were initially verified by 1% agarose gel electrophoresis, Quantus Fluorometer, and NanoDrop2000. Subsequently, DNA concentration and purity were further assessed using Qubit 4.0 and NanoDrop One instruments (Thermo Fisher Scientific, Waltham, USA). Qualified DNA samples underwent library preparation using the ALFA-SEQ DNA Library Prep Kit. This process included DNA fragmentation, end repair, 3’ adenylate addition, adapter ligation, fragment selection and purification, PCR amplification, and a final purification step. This size distribution of prepared libraries was evaluated using the Qsep400 High-Throughput Nucleic Acid-Protein Analysis System (Hangzhou Houze Biological Technology Co., Ltd., China), and their concentrations were measured with the Qubit 4.0. Finally, sequencing was performed on either the Illumina or MGI platform using PE150 sequencing (Guangdong Magigene Biotechnology Co., Ltd.).
Raw data were processed using Fastp software (parameters: -5-W -5 -W 5 -M 20 -q 15 -u 40 -l 50 –dedup) to obtain clean data (30). The MEGAHIT assembler (V1.2.9) was used for sequence assembly (https://github.com/voutcn/megahit), and Diamond software (https://github.com/bbuchfink/diamond/) was employed for alignments against the NCBI NR database (31). The LCA algorithm in the MEGAN software system was used for classification, and species annotation was determined based on an E value threshold of 1e-10 (32). A table containing gene average depth (AVG depth) and abundance information at various classification levels (kingdom to species) was generated based on LCA annotation and gene AVG depth or abundance tables (33). Species abundance within a sample was calculated as the sum of annotated gene abundances, while gene AVG depth for a species was the sum of non-zero abundance gene AVG depths.
2.4 Microbial network analysis
We constructed ecological association networks for each sampling site based on plot-level estimated bacterial OTUs (34). To ensure the reliability of our correlations, we adopted a rigorous filtering approach (35, 36), retaining only OTUs present in more than 25% of soil samples and with relative abundances exceeding >0.001%. For robust network construction, we utilized Random Matrix Theory (RMT)-based correlations (34), which involved four key steps: majority selection, similarity matrix calculation, RMT-based cutoff determination, and network generation. The network construction and analysis were performed using the integrated Network Analysis Pipeline (iNAP) (accessible at: https://inap.denglab.org.cn/). Visualization of the networks was achieved with the Gephi interactive platform (available at: http://gephi.github.io/) (37).
2.5 Statistical analysis
We evaluated differences in environmental properties (forest structure and soil factors), alpha diversity of the microbial community, and network properties using one-way ANOVA with Tukey HSD tests at p < 0.05. Nonmetric Multidimensional Scaling (NMDS) ordination, based on taxonomic abundance tables, quantified differences in soil microbial community structures, with inter-group differences tested using Adonis and Anosim analyses. To ensure comparability, all predictors and response variables were standardized using Z-scores before conducting Mantel Test (MT), Random Forest (RF), and Generalized Linear Models (GLMs). When necessary, predictors were log-transformed (38). To address potential multicollinearity among soil physiochemical properties, Spearman’s correlation was used, confirming no issue of collinearity (Supplementary Table S1). In ecological prioritization, correlated variables with comparable Variance Inflation Factors (VIFs) were considered, such as the preference for retaining DOC over SOC due to its direct microbial bioavailability. We employed Principle-Component Analysis (PCA) to create a composite variable describing network complexity, encompassing total links, edge density, avgK, APL, avgCC, degree centrality, and closeness centrality. The first component captured 65.21% of the variation (Supplementary Table S2). MT was used to analyze relationships between forest types, climate factors, forest structure, soil characteristics, and microbial diversity (community structure). RF analysis determined the influence of these environmental factors on microbial community assembly. We constructed a Structural Equation Model (SEM) to examine the pathways through which environmental factors affect the microbial diversity. To assess the model’s adequacy, we utilized several goodness-of-fit indices, including the Chi-square test (χ²), Root Mean Square Error of Approximation (RMSEA), Standardized Root Mean Square Residual (SRMR), and Comparative Fit Index (CFI). For evaluating model fit in the presence of non-normally distributed variables, we employed the Bollen-Stine bootstrap test, considering bootstrap P-values ranging from 0.10 to 1.00 as indicative of an acceptable fit. All statistical analyses were performed using RStudio (R version 3.6.1).
3 Results
3.1 Microbial diversity and composition under different forest types
The Shannon diversity index of soil bacteria exhibited significant differences across all study sites (Figure 1). Specifically, the Shannon index was highest in the SM forest, where it was significantly (P<0.05) higher than in the other four forest sites. The AA forest site followed closely, with a Shannon index that was significantly (P<0.05) higher than those in the RP and EU forest sites. Conversely, no significant (P>0.05) differences were observed in the Chao1 and ACE indices across the PT, SM, RP, EU, and AA forest sites. NMDS analysis based on Bray-Curtis distances showed an acceptable model fit (stress = 0.126) and effectively captured the distance relationships within the microbial community data (Supplementary Figure S3). Adonis and Anosim analysis showed that P values for comparisons such as RP versus EU, RP versus SM, EU versus SM, EU versus AA, and EU versus PT were 0.003, 0.001, 0.001, 0.013, and 0.001, respectively, with statistically significant differences in microbial community structures between the compared forest sites.
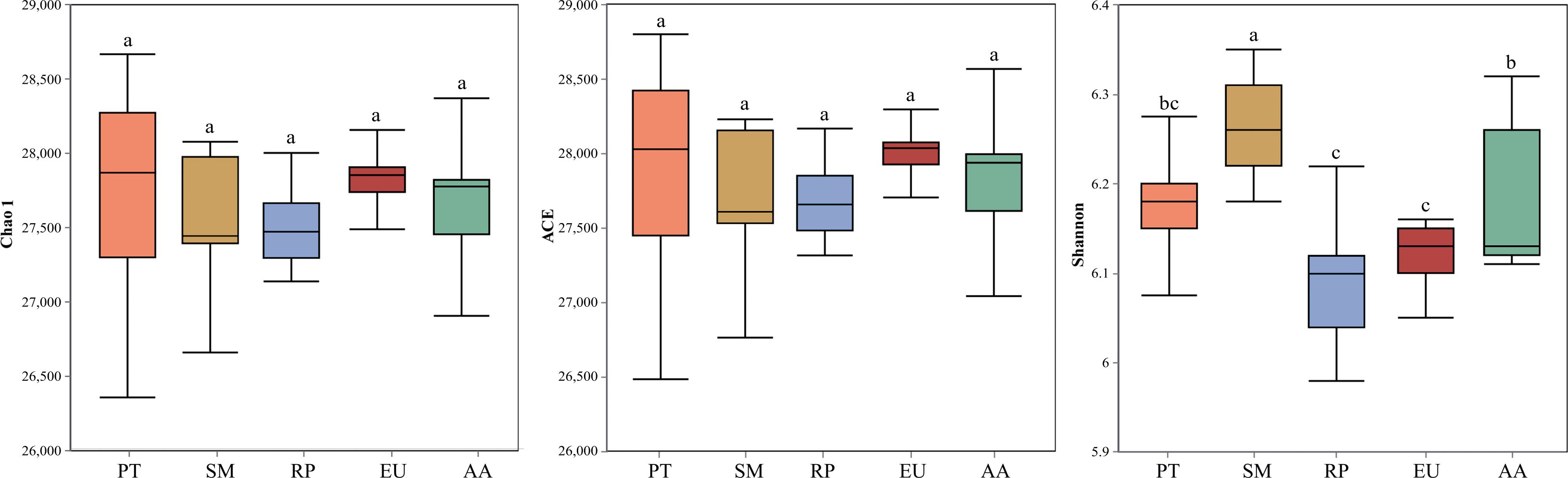
Figure 1. The Chao1, ACE and Shannon indexes of soil bacterial community in the PT, SM, RP, EU and AA forest. Values (mean ± SE) with different letters indicate significant differences at the p < 0.05 level. PT, Populus tomentosa Carrière; SM, Salix matsudana; RP, Robinia pseudoacacia; EU, Eucommia ulmoides; AA, Ailanthus altissima, the same below.
The dominant bacterial phyla across were Acidobacteria (28.52-29.46%) and Proteobacteria (14.69-17.80%), followed by Candidatus_Rokubacteria (3.37-3.83%), Gemmatimonadetes (2.72-3.62%), Actinobacteria (2.43-3.28%), and Chloroflexi (2.01-2.71%) (Figure 2). The relative abundance of Proteobacteria in the SM site was significantly higher than that in the RP, EU, and AA forests, while the relative abundance of Acidobacteria remained consistent across the PT, SM, RP, EU, and AA forest sites. At the family level, Acidobacteriaceae (1.16-1.38%) and Sphingomonadaceae (0.66-1.08%) were the most abundant, followed by Bradyrhizobiaceae (0.45-0.68%) and Steroidobacteraceae (0.41-0.64%). Notably, the relative abundances of Acidobacteriaceae and Sphingomonadaceae in the SM and AA forests were significantly (P<0.05) higher than those in the PT and RP forests.
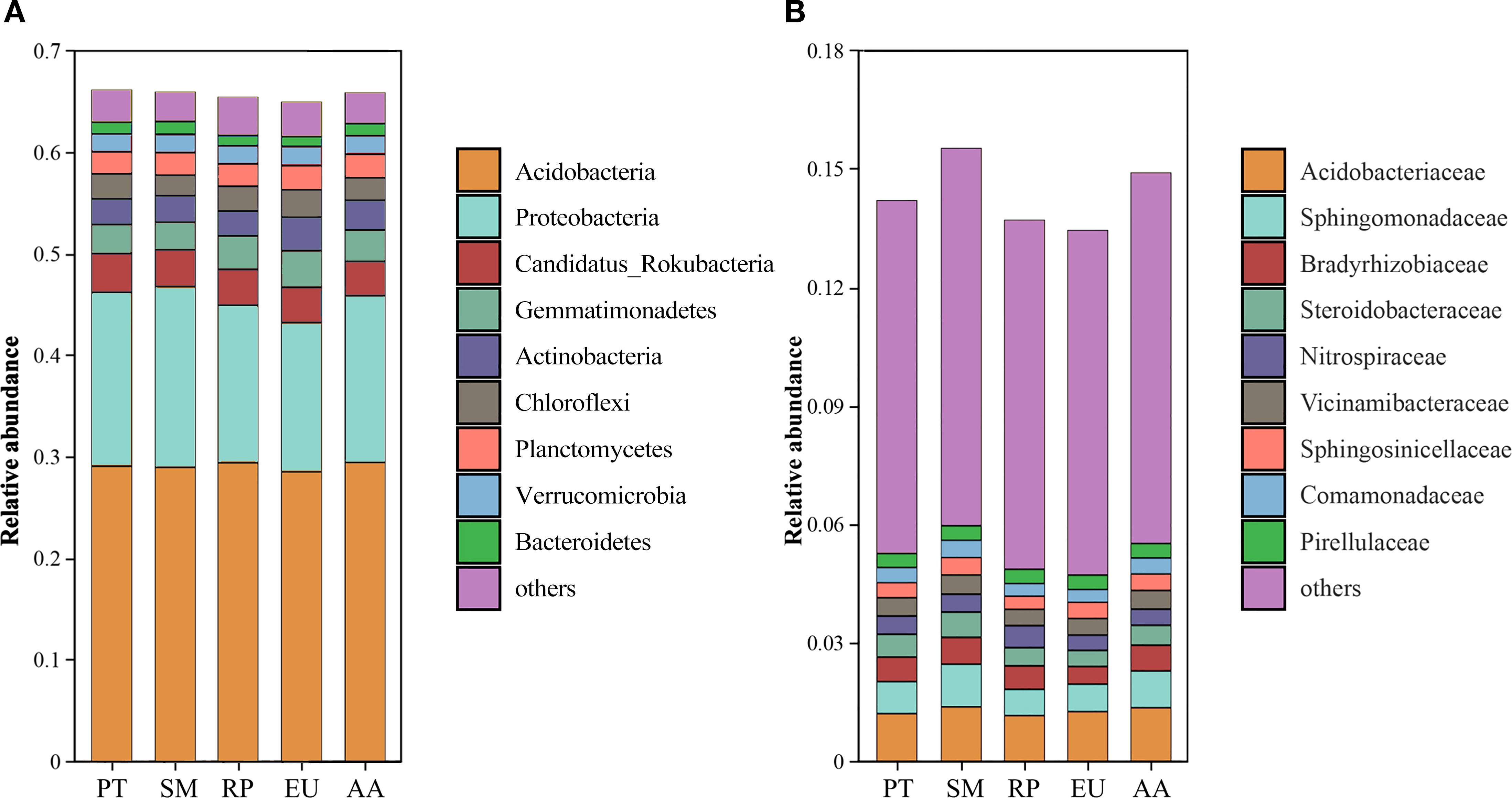
Figure 2. The relative abundances of dominant phyla (A) and family (B) for soil bacterial community in the PT, SM, RP, EU and AA forest.
3.2 Co-occurrence patterns of soil bacterial community
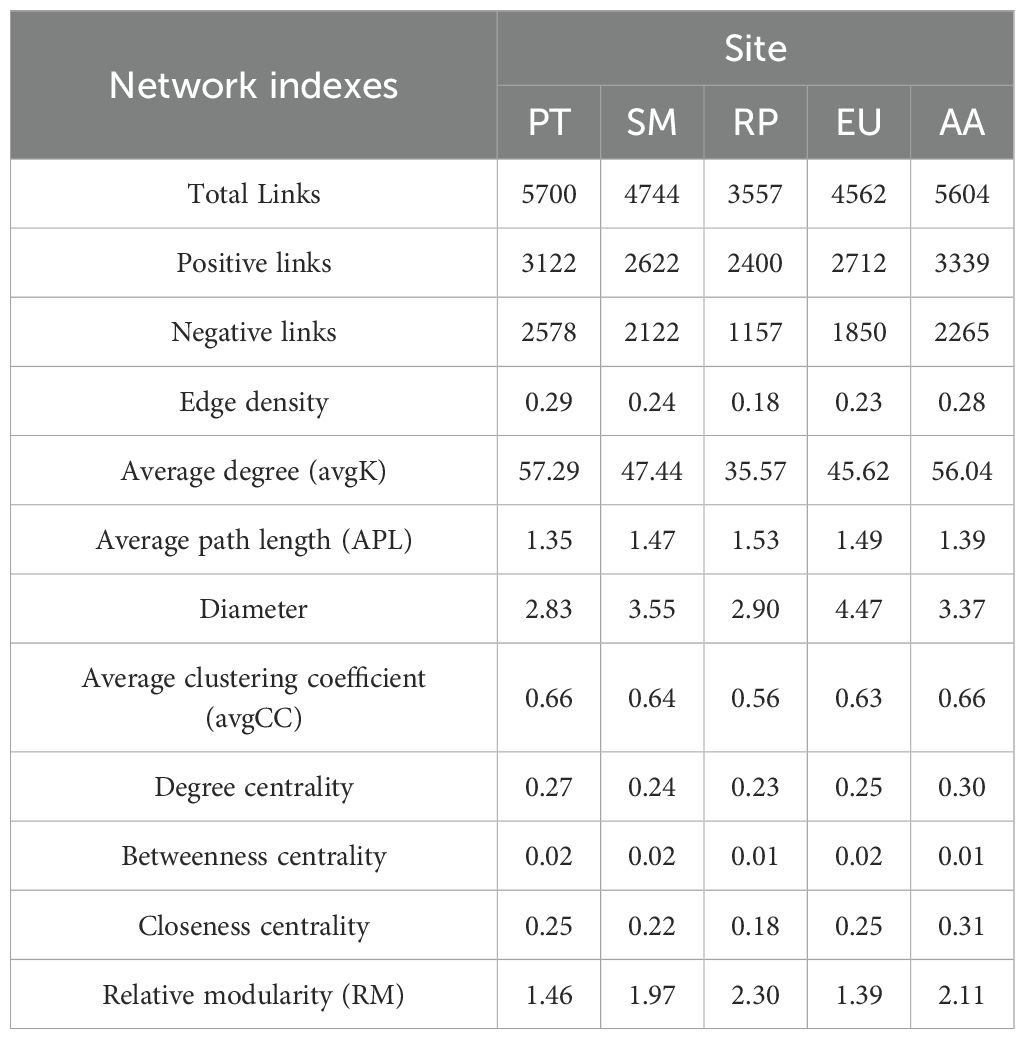
To explore the effects of forest types on microbial communities, five RMT-based correlations were constructed, and the topological properties of networks are shown (Table 1; Supplementary Figure S4). The PT forest has an edge density of 0.29 and an avgK of 57.29, highlighting the network’s exceptional connectivity and compactness. Notably, the APL of PT forest is 1.35, indicating short inter-node distances that facilitate rapid information or material transfer. In contrast, while the RP forest exhibits modest values across most network indexes, its relative modularity reaches up to 2.30, revealing a unique modular architecture that may contribute to network stability. The AA forest shares numerous similarities with the PT forest in network properties, including edge density, APL, and avgCC. Moreover, the degree centrality and closeness centrality in the AA forest are both the highest, highlighting the significance of key nodes and the preferable accessibility among nodes. Meanwhile, the diameter of EU forest is 4.47, suggesting that information or material transfer may involve more intermediary nodes, yet this property also reflects the network with greater robustness.
3.3 Relationship between environmental factors and the soil bacterial community
The Mantel test showed that the bacterial Shannon’s diversity index was significantly correlated with DBH, NH4+-N, and BGL across the five forest types (Figure 3). DBH emerged as the dominant factor influencing the composition of the bacterial community, whereas TN was significantly associated with the co-occurrence network of bacteria. Random forest analysis identified DBH and DOC as the key predictors of the soil bacterial community Shannon index, followed by SOC, TN, and BGL (Figure 4). Notably, DBH alone served as the principal predictor of soil bacterial community composition. TH and TN were the primary factors affecting network complexity, with DBH, TP, and SWC also contributing. These results confirmed that, in addition to soil physicochemical properties, forest structure also played a vital role in shaping microbial community assembly.
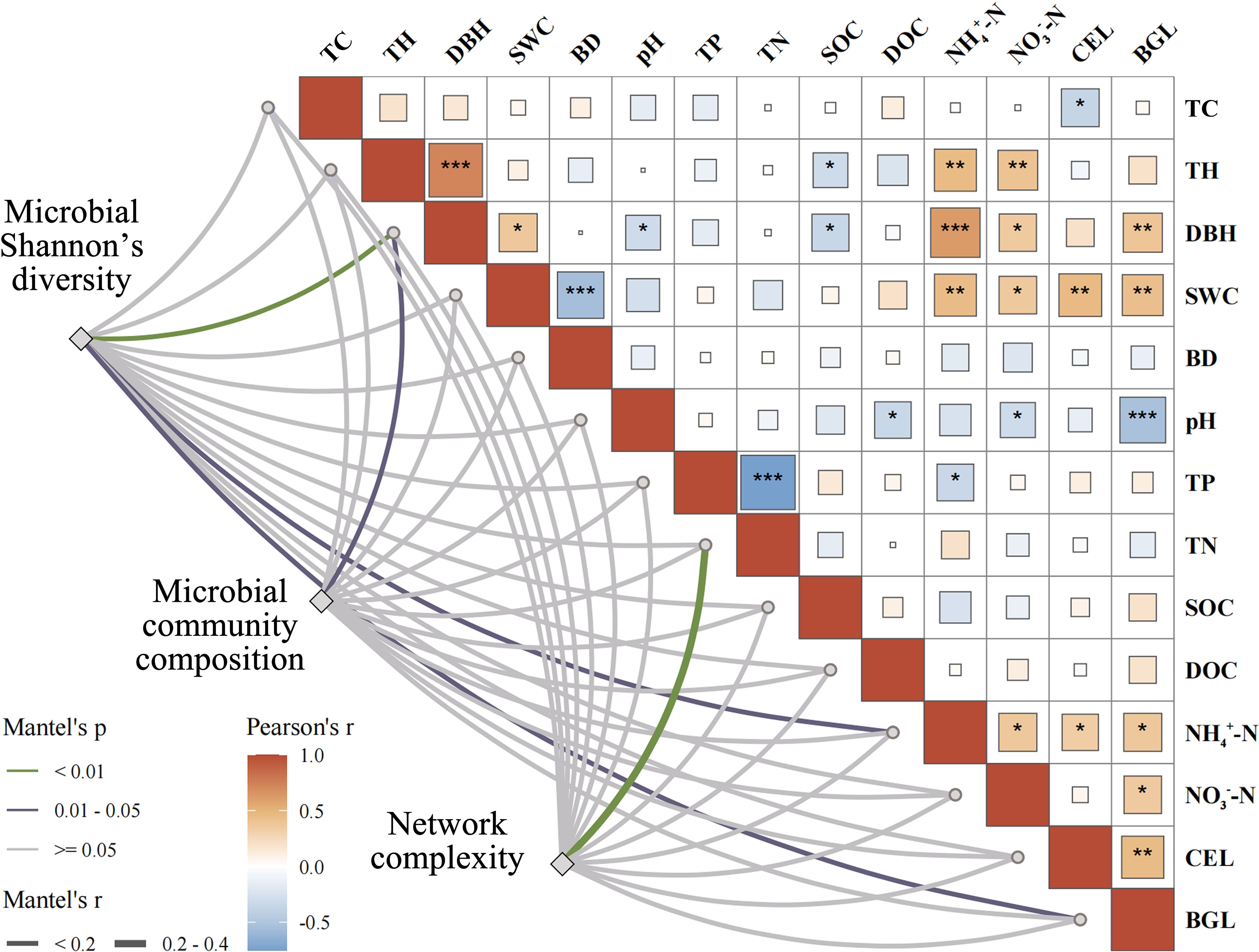
Figure 3. Correlations between soil bacterial community Shannon and composition and network complexity and environmental factors. Mantel test (MT) analyzed the relationship between environmental variables and microbial diversity, structure, co-occurrence networks of soil bacterial community. TC, tree canopy size; TH, tree height; DBH, diameter at breast height; SWC, soil water content; BD, bulk density; TP, total phosphorus; TN, total nitrogen; SOC, soil organic carbon; DOC, dissolved organic carbon; NH4+-N, ammonium nitrogen; NO3–N, nitrate nitrogen; CEL, cellulase; BGL, β-Glucosidase, the same below. *P<0.05, **P<0.01, ***P<0.001.
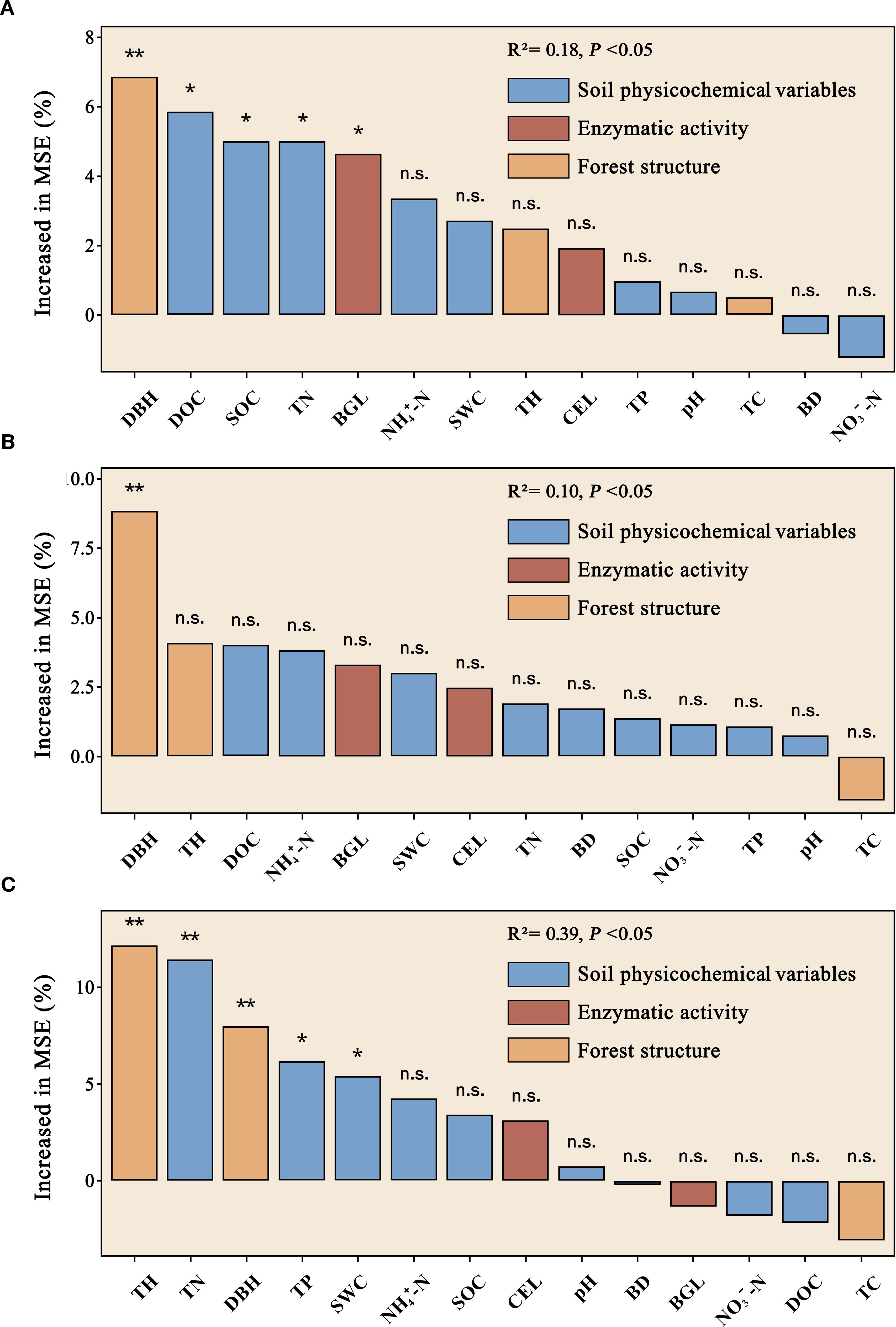
Figure 4. Random Forest (RF) determined the role of environmental variables in microbial diversity (A), structure (B), and co-occurrence networks (C). The thickness of the lines represents the partial Mantel’s r statistic, while the color denotes the correlation between influencing factors. * means P < 0.05, ** means P < 0.01, ns means not significant. Significance levels denoted by asterisks (*P < 0.05; **P < 0.01; n.s. means not significant).
3.4 Pathways of environmental factors affecting microbial community
Soil physicochemical properties emerged as the dominant predictor, accounting for 80% of the explained variance, followed by forest structural attributes (20%) (Figure 5). Adjusted R² values (0.502) indicate that the model explains approximately half of the observed variation in bacterial diversity. The proportional contribution analysis demonstrated that TN emerged as the predominant driver, accounting for 36.58% of the explained variation (95% CI [-0.25, -0.12], P < 0.001). This was followed by TP (31.39%, 95% CI [-0.15, -0.95], P < 0.01) and DOC (25.31%, 95% CI [-0.11, -0.78], P < 0.01). SEM revealed distinct environmental drivers of bacterial diversity. Soil BD exhibited moderate direct effects (R²=0.044), while NH4+-N displayed stronger associations (R²=0.226) (Figure 6). TN exerted the strongest total effect (-0.33), directly suppressing bacterial diversity (path coefficient=-0.33) and indirectly modulating NH4+-N. TD showed a direct positive effect on NH4+-N (path coefficient=0.29), yet its indirect contributions to microbial diversity were negligible. These findings suggest the predominance of direct pathways—particularly nutrient availability (TN, NH4+-N)—in shaping microbial community structure, with minimal mediation through secondary environmental interactions.
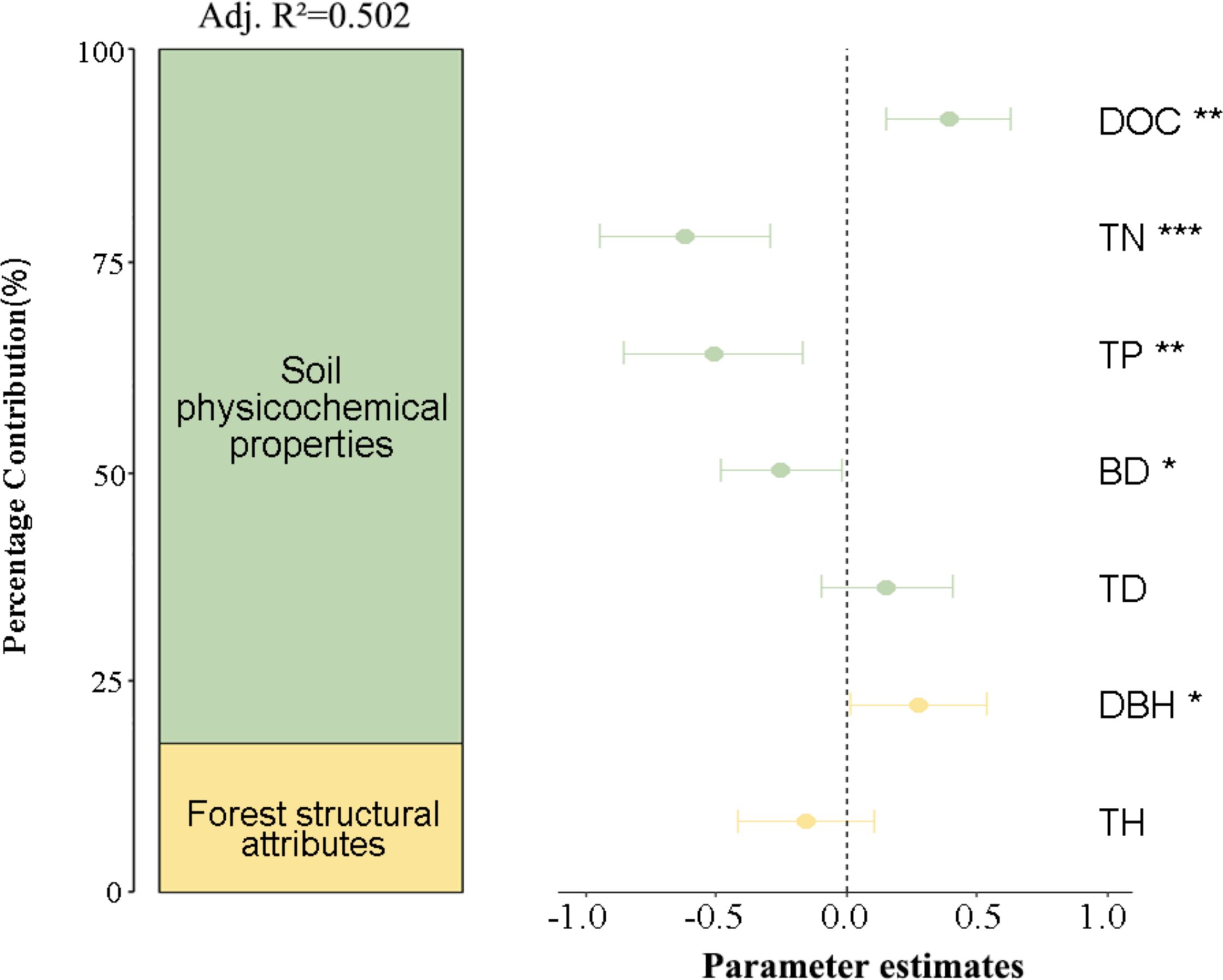
Figure 5. Proportional contributions of environmental drivers to bacterial diversity variation Standardized regression coefficients with 95% CIs demonstrate the effect magnitude hierarchy among predictors. Variable importance is expressed as percentage contribution derived from coefficient ratios. Significance levels denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). TD, tree density; detailed abbreviation are given in Figure 3.
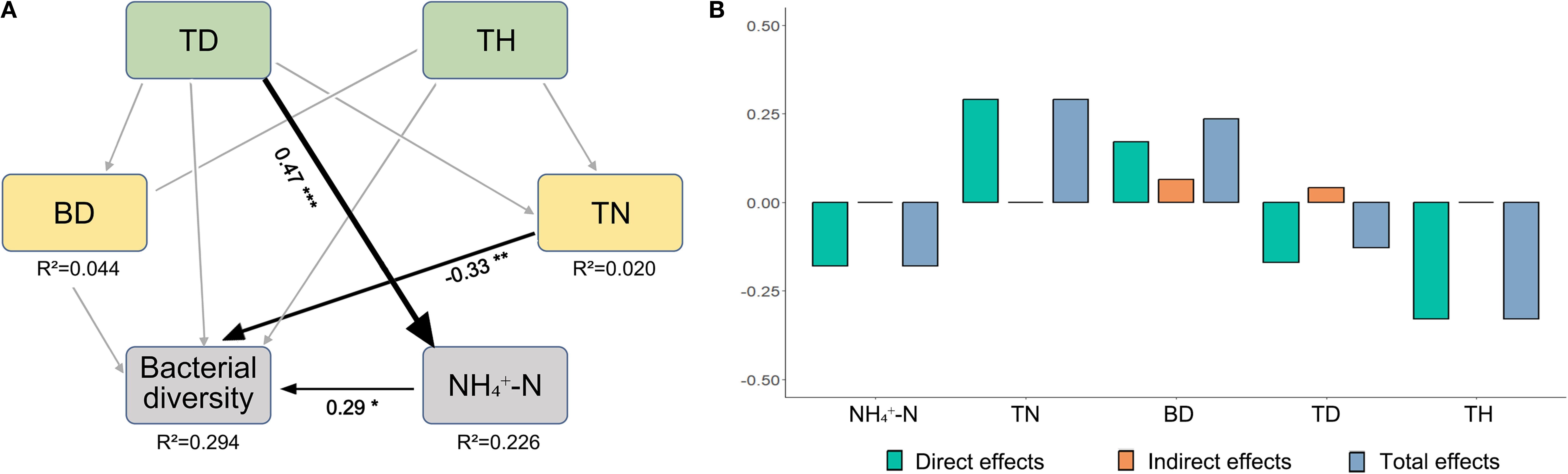
Figure 6. The effects of environmental factors on the soil bacterial diversity using SEMs (A). Line thickness indicates effect size. Bar graphs illustrate the standardized effects of each factor on soil bacterial diversity (B). Significance levels denoted by asterisks (*P < 0.05; **P < 0.01; ***P < 0.001).
4 Discussion
4.1 Soil microbial diversity and composition under different forest types
The observed variations in soil microbial diversity and composition among different forest types in this study are consistent with previous findings that highlight the influence of vegetation type on soil microbiomes (39, 40). The higher Shannon diversity index observed in SM forests, compared to other forest types, suggests the presence of distinct ecological conditions favoring a more diverse bacterial community. The lack of significant differences in the Chao1 and ACE indices across forest types suggests that while dominant taxa shift, the underlying richness of rare species remains conserved. This discrepancy between diversity indices is well-documented and underscores that the Shannon index is more sensitive to changes in abundant taxa, whereas Chao1 and ACE better reflect the rare biosphere (41). Local environmental conditions, such as soil type and forest structure, are known to be strong confounding factors in mediating tree species effects on soil microbes (42, 43), which may explain the specific pattern observed here.
The dominance of Acidobacteria and Proteobacteria across these managed forest ecosystems is a common phenomenon observed in various soil types (44, 45). Recent studies, such as Deltedesco et al. (46), have also highlighted the consistent dominance of these phyla in different forest ecosystems, reinforcing their ecological importance. The SM forest exhibited the highest Shannon diversity index, accompanied by a significantly higher relative abundance of Proteobacteria. This finding suggests that the soil environment of the SM forest may be particularly conducive to the growth and proliferation of this phylum. The reason may be that the microbial community in SM forest harbors positive interactions, such as symbiosis or mutualism, that promote the growth of Proteobacteria (47, 48). These interactions could involve nutrient exchange, signaling molecules, or other forms of cooperation that benefit both Proteobacteria and their associated microbes. The stable relative abundance of Acidobacteria across all forest types indicates its broad ecological adaptability, likely due to its extensive metabolic versatility and niche breadth within acidic to neutral soils (49, 50). This underscores that forest type shifts primarily restructure dominant community components rather than alter rare species pools in these managed ecosystems.
4.2 Soil microbial co-occurrence network in different forest types
The high complexity and connectivity of the microbial network in PT forests may be attributed to the high resource availability and diverse ecological niches. As a fast-growing species, Populus tomentosa likely provides abundant carbon and nitrogen sources for soil microbes through root exudates and litterfall, promoting microbial diverse microbial interactions (47, 51–53). This high ecological diversity likely contributes to the high connectivity and short average path length of the microbial network, enhancing network complexity and information transfer efficiency (20). The high modularity of the microbial network in RP forests may be associated with ecological niche differentiation. Modules can act as ‘firewalls’ containing disturbances, enhancing ecological stability (54). In RP forests, different microbial species may form relatively independent modules due to niche differentiation, with microbes within these modules sharing similar ecological requirements and functions, thereby fostering close interactions within modules (55, 56). Furthermore, variations in soil nutrient distribution, light intensity, and other environmental factors in RP forests may further drive the modular distribution of microbial communities. In ecological co-occurrence networks, modularity (Q) values in RP forests typically range from 0.3 to 0.7, with values exceeding 0.4 generally indicating a significant modular structure. The exceptionally high value observed in this study (2.30) strongly implies a hyper-modular architecture, which may substantially enhance network stability through the compartmentalization of ecological interactions (54).
The prominence of key nodes in the microbial network of AA forests is likely related to competitive dominance and niche overlap. In AA forests, certain dominant microbial species may influence the growth and distribution of other microbes by producing inhibitory compounds or competing for resources, thus forming highly connected key nodes (57, 58). These key nodes play pivotal roles in the network, serving as crucial hubs for information transfer, material cycling, or energy flow. Simultaneously, niche overlap and competition among microbes may prompt some species to enhance interactions to consolidate their ecological niches, thereby becoming key nodes in the network (59, 60). The robustness of the microbial network in EU forests may be attributed to its large diameter and redundant paths. EU forests may face severe environmental pressures such as nutrient-poor soils and drought. EU’s large diameter creates redundant pathways for nutrient cycling, allowing microbial functions to persist. These pressures may prompt microbial communities to develop more robust network structures to cope with adverse conditions (61–63). A large diameter implies the existence of multiple redundant paths and alternative routes in the network, ensuring that information or materials can be transmitted through other paths when certain nodes or links are disrupted, thereby maintaining network stability and continuous functioning (64, 65).
4.3 Relationships between environmental factors and microbial communities
The results of Mantel tests and Random Forest analysis highlight the significance of DBH as a primary predictor of soil bacterial community composition. This is consistent with previous studies indicating a positive correlation between DBH and soil bacterial diversity (66, 67). Mature trees, defined as those with a DBH greater than 20 cm, release labile carbon compounds, such as succinic acid and fructose, as well as antimicrobial phenolic substances higher than that of saplings. This differential secretion contributes to the formation of distinct microbial niches. However, several studies suggest that the influence of DBH on soil bacterial communities may be modulated by other environmental factors (52, 66). Despite the conflicting opinions, the majority of these studies support the notion that larger trees provide more stable and diverse habitats for soil microorganisms (68). These studies emphasize how tree size and age indirectly shape bacterial communities by influencing soil microenvironments, including BD, SOC, and TN. Moreover, our study identified significant correlations between NH4+-N, TN, DOC, and bacterial diversity and network complexity, emphasizing the importance of soil nitrogen content on bacterial community structure (69, 70). Nevertheless, concerning the role of TP in contributing to network complexity, our findings differ from those of previous studies, which posited a minimal effect of phosphorus availability on bacterial network structure (71). Our research has underscored the significant roles of nitrogen and carbon, the impact of phosphorus may be contingent upon various ecosystem types, soil conditions, and additional environmental factors.
Our study found a significant correlation between BGL and soil bacterial diversity, emphasizing the crucial role of soil enzymes in driving bacterial community dynamics. The main reason is that soil enzymes, as products of microbial activity and drivers of nutrient cycling, play a vital role in bacterial community dynamics. However, several studies have pointed out that different soil enzymes may have varied impacts on bacterial communities, influenced by soil types (72, 73). In addition, our study reveals that forest structure and soil physicochemical properties jointly shape microbial community assembly and network complexity, which is consistent with previous studies (48, 74). However, some researches have emphasized the dominant role of single factors, such as soil pH, in shaping bacterial communities, contrasting with our findings (75). The formation and maintenance of microbial communities in forest ecosystems are multifactorial processes. Our results emphasize the synergistic effects of forest structure and soil physicochemical properties, but this does not diminish the importance of other single factors. Rather, these studies may reveal differences and complexities in microbial community assembly mechanisms across different ecosystems. Urban-specific pressures, such as nitrogen deposition from traffic, soil compaction due to human activities, and residual heavy metal contamination, collectively intensify the suppression of diversity mediated by TN and alter the structure of microbial networks.
4.4 Environmental drivers and their pathways in shaping microbial communities
Our findings on the pathways through which environmental factors affect microbial community assembly provide valuable insights into the mechanisms underlying microbial dynamics in urban forest ecosystems. The SEM analysis revealed distinct hierarchical pathways through which environmental factors regulate soil bacterial diversity in urban forest ecosystems. The predominance of direct effects, particularly through nutrient availability (TN and NH4+-N), suggests that microbial communities in these managed ecosystems are primarily shaped by bottom-up resource control rather than secondary environmental interactions. This finding is consistent with recent studies demonstrating the central role of nitrogen dynamics in structuring microbial communities in urban green spaces (51, 70). The strong negative direct effect of TN on bacterial diversity (-0.33) contrasts with previous observations in natural forests, potentially reflecting the unique nutrient dynamics of urban ecosystems where nitrogen deposition and management practices may create distinct selective pressures (69).
Moreover, the moderate direct effects of BD on bacterial diversity, coupled with the strong association of NH4+-N with diversity, underscore the complex interplay between physical and chemical soil properties. While BD influences microbial habitat structure and oxygen availability, its relatively weaker effect compared to nutrient factors indicates that chemical properties, particularly nitrogen dynamics, may override physical constraints in shaping microbial communities in these managed systems. This contrasts with observations in natural forests, where physical soil properties often play a more dominant role (68, 75). The negligible indirect contributions of TD through environmental mediation further emphasize the primacy of direct nutrient pathways, suggesting that in urban forests, tree size effects on microbial communities are less mediated by secondary environmental factors. This finding has important implications for urban forest management, suggesting that nutrient management strategies, particularly targeting nitrogen dynamics, could be effective in modulating microbial community assembly and functions in these ecosystems. For instance, they could enable the prediction of network collapse risks under warming scenarios and enhance the optimization of green infrastructure to enhance carbon sequestration rates.
5 Conclusions
This study provides a comprehensive assessment of how different forest types influence soil microbial community structures in plain ecological forests of Beijing, China. Soil nitrogen dynamics emerge as the primary selector of bacterial diversity (TN path coefficient = -0.33), overriding physical constraints like BD, reflecting intensified nutrient cycling in anthropogenic systems. Forest structural attributes—particularly TD and TH—mediate microbial composition through cascading effects on soil chemistry, redefining plant-soil feedback mechanisms in managed landscapes. Crucially, microbial networks exhibit adaptive topological signatures across forest types: Populus stands’ hyperconnectivity (edge density = 0.29) enhances metabolic coordination, while Robinia’s modular architecture (modularity = 2.30) confers resilience against environmental perturbations. These insights advocate for targeted afforestation strategies—prioritizing Salix for diversity enhancement and Robinia for stability—while highlighting network properties as novel bioindicators for urban ecosystem health. Future research must unravel how climate change alters these structure-function hierarchies through long-term experimental manipulations. Implementation of our species-network matching strategy could enhance ecosystem functions in Beijing’s urban forest spaces.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI SRA, accession PRJNA1313477.
Author contributions
YZ: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. CH: Data curation, Writing – review & editing. JC: Project administration, Writing – review & editing. YW: Project administration, Writing – review & editing. HH: Data curation, Writing – review & editing. DY: Resources, Writing – review & editing. YSZ: Visualization, Writing – review & editing. XZ: Resources, Writing – review & editing. WD: Visualization, Writing – review & editing. ZL: Investigation, Writing – review & editing. XD: Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Open project of Beijing Academy of Forestry and Landscape Architecture (STBH202401).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsoil.2025.1573531/full#supplementary-material
References
1. Pataki DE, Alig RJ, Fung AS, Golubiewski NE, Kennedy CA, Mcpherson EG, et al. Urban ecosystems and the North American carbon cycle. Glob Change Biol. (2006) 12:2092–102. doi: 10.1111/j.1365-2486.2006.01242.x
2. Njana MA, Mbilinyi B, and Eliakimu Z. The role of forests in the mitigation of global climate change: emprical evidence from Tanzania. Environ Challenges. (2021) 4:100170. doi: 10.1016/j.envc.2021.100170
3. Eisenhauer N. Plant diversity effects on soil microorganisms: spatial and temporal heterogeneity of plant inputs increase soil biodiversity. Pedobiologia. (2016) 59:175–7. doi: 10.1016/j.pedobi.2016.04.004
4. Lu Y, Lyu M, Xiong X, Deng C, Jiang Y, Zeng M, et al. Understory ferns promote the restoration of soil microbial diversity and function in previously degraded lands. Sci Total Environ. (2023) 870:161934. doi: 10.1016/j.scitotenv.2023.161934
5. Van der Heijden MG, Bardgett RD, and Van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett. (2008) 11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x
6. Philippot L, Raaijmakers JM, Lemanceau P, and van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. (2013) 11:789–99. doi: 10.1038/nrmicro3109
7. Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core arabidopsis thaliana root microbiome. Nature. (2012) 488:86–90. doi: 10.1038/nature11237
8. Liu Z, Lyu Y, Liu Y, Wang Y, Xiong M, Tang Y, et al. Differential spatial responses and assembly mechanisms of soil microbial communities across region-scale taiga ecosystems. J Environ Manage. (2024) 370:122653. doi: 10.1016/j.jenvman.2024.122653
9. Kang Y, Shen L, Li C, Huang Y, and Chen L. Effects of vegetation restoration on soil microbial community composition and function in a karst area, Southwest China. J Environ Manage. (2024) 363:121395. doi: 10.1016/j.jenvman.2024.121395
10. Delgado-Baquerizo M, Maestre FT, Reich PB, Jeffries TC, Gaitan JJ, Encinar D, et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat Commun. (2016) 7:10541. doi: 10.1038/ncomms10541
11. Tripathi BM, Stegen JC, Kim M, Dong K, Adams JM, and Lee YK. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. (2018) 12:1072–83. doi: 10.1038/s41396-018-0082-4
12. Craig H, Antwis RE, Cordero I, Ashworth D, Robinson CH, Osborne TZ, et al. Nitrogen addition alters composition, diversity, and functioning of microbial communities in mangrove soils: an incubation experiment. Soil Biol Biochem. (2021) 153:1–12. doi: 10.1016/j.soilbio.2020.108076
13. Xiao W, Chen X, Jing X, and Zhu B. A meta-analysis of soil extracellular enzyme activities in response to global change. Soil Biol Biochem. (2018) 123:21–32. doi: 10.1016/j.soilbio.2018.05.001
14. Trivedi P, Delgado-Baquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, et al. Microbial regulation of the soil carbon cycle: evidence from gene–enzyme relationships. ISME J. (2016) 10:2593–604. doi: 10.1038/ismej.2016.65
15. Bowles TM, Acosta-Martinez V, Calderón F, and Jackson LE. Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol Biochem. (2014) 68:252–63. doi: 10.1016/j.soilbio.2013.10.004
16. Wang GS, Huang WJ, Zhou GY, Mayes M, and Zhou JZ. Modeling the processes of soil moisture in regulating microbial and carbon-nitrogen cycling. J Hydrol. (2020) 585:124777. doi: 10.1016/j.jhydrol.2020.124777
17. Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, et al. Core microbiomes for sustainable agroecosystems. Nat Plants. (2018) 4:247–57. doi: 10.1038/s41477-018-0139-4
18. Tu Q, Yan Q, Deng Y, Michaletz S, Buzzard V, Weiser M, et al. Biogeographic patterns of microbial co-occurrence ecological networks in six American forests. Soil Biol Biochem. (2020) 148:107897. doi: 10.1016/j.soilbio.2020.107897
19. Jiao S, Lu Y, and Wei G. Soil multitrophic network complexity enhances the link between biodiversity and multifunctionality in agricultural systems. Glob Change Biol. (2021) 28:140–53. doi: 10.1111/gcb.15917
20. Yuan MM, Guo X, Wu L, Zhang Y, Xiao N, Ning D, et al. Climate warming enhances microbial network complexity and stability. Nat Clim Change. (2021) 11:343–8. doi: 10.1016/j.scitotenv.2023.168444
21. Wang X, Hong M, Huang Z, Zhao Y, Ou Y, Jia H, et al. Biomechanical properties of plant root systems and their ability to stabilize slopes in geohazard-prone regions. Soil Tillage Res. (2019) 189:148–57. doi: 10.1016/j.still.2019.02.003
22. Moshki A and Lamersdorf N. Symbiotic nitrogen fixation in black locust (Robinia pseudoacacia L.) seedlings from four seed sources. JFR. (2011) 22:689–92. doi: 10.1007/s11676-011-0212-6
23. Li Z, Wang T, Zhu J, Tian H, Yang Y, Jin Y, et al. Scale effect of landscape characteristics on undergrowth vegetation variance with different ecological traits. Eco Frontiers. (2024) 6:1269–79. doi: 10.1016/j.ecofro.2024.08.003
24. Vogt KA and Persson H. Measuring growth and development of roots. FT - Ecophys Tech Appr. (1991) pp:477–501. doi: 10.1007/978-3-540-69321-5-50
25. Angel R, Soares MIM, Ungar ED, and Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. (2010) 4:553–63. doi: 10.1038/ismej.2009.136
26. Pellissier L, Niculita-Hirzel H, Dubuis A, Pagni M, Guex N, Ndiribe C, et al. Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Mol Ecol. (2014) 23:4274–90. doi: 10.1111/mec.12854
27. Chen S, Lin L, Deng Y, Yuan S, and Zhang N. Tree species richness and mycorrhizal types drive soil nitrogen cycling by regulating soil microbial community composition and diversity in tropical forests. For Ecol Manage. (2024) 569:122187. doi: 10.1016/j.foreco.2024.122187
28. Boehme L, Langer U, and Böhme F. Microbial biomass, enzyme activities and microbial community structure in two European long-term field experiments. Agro. Eco. Env. (2005) 109:141–52. doi: 10.1016/j.agee.2005.01.017
29. DeForest JL. The influence of time, storage temperature, and substrate age on potential soil enzyme activity in acidic forest soils using MUB-linked substrates and l-DOPA. Soil Biol Biochem. (2009) 41:1180–6. doi: 10.1016/j.soilbio.2009.02.029
30. Chen S, Zhou Y, Chen Y, and Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. (2018) 34:884–90. doi: 10.1093/bioinformatics/bty560
31. Buchfink B, Reuter K, and Drost HG. Sensitive protein Alignments at Tree-Of-Life Scale Using DIAMOND. Nat Methods. (2021) 18:366–8. doi: 10.1038/s41592-021-01101-x
32. Wooley JC, Godzik A, and Friedberg I. A primer on metagenomics. PloS Comput Biol. (2010) 6:e1000667. doi: 10.1371/journal.pcbi.1000667
33. Peng Y, Leung HCM, Yiu SM, and Chin FYL. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. (2012) 28:1420–8. doi: 10.1093/bioinformatics/bts174
34. Deng Y, Jiang Y, Yang Y, He Z, Luo F, and Zhou J. Molecular ecological network analyses. Bioinformatics. (2012) 13:113. doi: 10.1186/1471-2105-13-113
35. Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. (2018) 359:320–5. doi: 10.1126/science.aap9516
36. Jiao S, Wang J, Wei G, Chen W, and Lu Y. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere. (2019) 235:248–59. doi: 10.1016/j.chemosphere.2019.06.174
37. Bastian M, Heymann S, and Jacomy M. Gephi: an open source software for exploring and manipulating networks. ICWSM. (2009) 3:361–2. doi: 10.1609/icwsm.v3i1.13937
38. García-Palacios P, Gross N, Gaitán J, and Maestre FT. Climate mediates the biodiversity-ecosystem stability relationship globally. Proc Natl Acad Sci. (2018) 115:8400–5. doi: 10.1073/pnas.1800425115
39. Xia Q, Rufty T, and Shi W. Soil microbial diversity and composition: links to soil texture and associated properties. Soil Biol Biochem. (2020) 149:107953. doi: 10.1016/j.soilbio.2020.107953
40. Ding X, Liu G, Fu S, and Chen HYH. Tree species composition and nutrient availability affect soil microbial diversity and composition across forest types in subtropical China. Catena. (2021) 201:105224. doi: 10.1016/j.catena.2021.105224
41. Cao M, Zheng X, Cui L, Wu F, Gao H, and Jiang J. Soil bacterial communities are more sensitive to short-term nitrogen deposition than fungal communities in subtropical Chinese fir forests. For Ecol Management. (2023) 549:121490. doi: 10.1016/j.foreco.2023.121490
42. Pan J, Wu H, Xiang W, Ouyang S, Chen L, Zeng Y, et al. Soil microbial richness and community composition are primarily mediated by functional trait diversity of fine roots in subtropical forests. Plant Soil. (2023) 497:485–501. doi: 10.1007/s11104-023-06408-6
43. López-Goldar X and Agrawal AA. Ecological interactions, environmental gradients, and gene flow in local adaptation. Trends Plant Sci. (2021) 26:796–809. doi: 10.1016/j.tplants.2021.03.006
44. Giguere AT, Eichorst SA, Meier DV, Herbold CW, Richter A, Greening C, et al. Acidobacteria are active and abundant members of diverse atmospheric H2-oxidizing communities detected in temperate soils. ISME J. (2021) 15:363–76. doi: 10.1038/s41396-020-00750-8
45. Kim HS, Lee SH, Jo HY, Finneran KT, and Kwon MJ. Diversity and composition of soil acidobacteria and proteobacteria communities as a bacterial indicator of past land-use change from forest to farmland. Sci Total Environ. (2021) 797:148944. doi: 10.1016/j.scitotenv.2021.148944
46. Deltedesco E, Keiblinger KM, Piepho H-P, Antonielli L, Pötsch EM, Zechmeister-Boltenstern S, et al. Soil microbial community structure and function mainly respond to indirect effects in a multifactorial climate manipulation experiment. Soil Biol Biochem. (2020) 142:107704. doi: 10.1016/j.soilbio.2020.107704
47. Wang M, Shao Y, Zhang W, Yu B, Shen Z, Fan Z, et al. Secondary succession increases diversity and network complexity of soil microbial communities in subtropical and temperate forests. Catena. (2024) 249:108662. doi: 10.1016/j.catena.2024.108662
48. Huo X, Ren C, Wang D, Wu R, Wang Y, Li Z, et al. Microbial community assembly and its influencing factors of secondary forests in Qinling Mountains. Soil Biol Biochem. (2023) 184:109075. doi: 10.1016/j.soilbio.2023.109075
49. Sikorski J, Baumgartner V, Birkhofer K, Boeddinghaus RS, Bunk B, Fischer M, et al. The evolution of ecological diversity in acidobacteria. Front Microbiol. (2022) 13:715637. doi: 10.3389/fmicb.2022.715637
50. Zhang Y, Cong J, Lu H, Li G, Qu Y, Su X, et al. Community structure and elevational diversity patterns of soil Acidobacteria. J Environ Sci. (2014) 26:1717–24. doi: 10.1016/j.jes.2014.04.014
51. Zhang J, Sáez-Sandino T, Maestre FT, Feng Y, Yu Y, Berdugo M, et al. Global environmental dependences of soil biodiversity and functions are modified by water availability thresholds. Sci Total Environ. (2024) 958:178033. doi: 10.1016/j.scitotenv.2024.178033
52. Bastida F, Eldridge DJ, García C, Png GK, Bardgett RD, and Delgado-Baquerizo M. Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. (2021) 15:2081–91. doi: 10.1038/s41396-021-00906-0
53. Chen Y, Tao S, Ma J, Qu Y, Sun Y, Wang M, et al. New insights into assembly processes and driving factors of urban soil microbial community under environmental stress in Beijing. Sci Total Environ. (2024) 947:174551. doi: 10.1016/j.scitotenv.2024.174551
54. Delgado-Baquerizo M, Reich PB, Trivedi C, Eldridge DJ, Abades S, Alfaro FD, et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat Ecol Evol. (2024) 4:210–20. doi: 10.1038/s41559-019-1084-y
55. Faust K and Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. (2012) 10:538–50. doi: 10.1038/nrmicro2832
56. DeCelis M, Duque J, Marquina D, Salvadó H, Serrano S, Arregui L, et al. Niche differentiation drives microbial community assembly and succession in full-scale activated sludge bioreactors. NPJ Biofilms Microbiomes. (2022) 8:23. doi: 10.1038/s41522-022-00291-2
57. Nizamani MM, Hughes AC, Qureshi S, Zhang Q, Tarafder E, Das D, et al. Microbial biodiversity and plant functional trait interactions in multifunctional ecosystems. Appl Soil Ecol. (2024) 201:105515. doi: 10.1016/j.apsoil.2024.105515
58. Jiao S, Xu Y, Zhang J, Hao X, and Lu Y. Core microbiota in agricultural soils and their potential associations with nutrient cycling. mSystems. (2019) 4:e00313–00318. doi: 10.1128/msystems.00313-18
59. Pastore AI, Barabás G, Bimler MD, Mayfield MM, and Miller TE. The evolution of niche overlap and competitive differences. Nat Ecol Evol. (2021) 5:330–7. doi: 10.1038/s41559-020-01383-y
60. Ashby B, Watkins E, Lourenço J, Gupta S, and Foster KR. Competing species leave many potential niches unfilled. Nat Ecol Evol. (2017) 1:1495–501. doi: 10.1038/s41559-017-0295-3
61. Metze D, Schnecker J, Canarini A, Fuchslueger L, Koch BJ, Stone BW, et al. Microbial growth under drought is confined to distinct taxa and modified by potential future climate conditions. Nat Commun. (2023) 14:5895. doi: 10.1038/s41467-023-41524-y
62. Kost E, Kundel D, Conz RF, Mäder P, Krause H, Six J, et al. Soil microbial resistance and resilience to drought under organic and conventional farming. Eur J Soil Biol. (2024) 123:103690. doi: 10.1016/j.ejsobi.2024.103690
63. Jing X, Chen X, Fang J, Ji C, Shen H, Zheng C, et al. Soil microbial carbon and nutrient constraints are driven more by climate and soil physicochemical properties than by nutrient addition in forest ecosystems. Soil Biol Biochem. (2020) 141:107657. doi: 10.1016/j.soilbio.2019.107657
64. Xu X, Chen A, Xu G, Yang C, and Lam WH. Enhancing network resilience by adding redundancy to road networks. TRE. (2021) 154:102448. doi: 10.1016/j.tre.2021.102448
65. Fan T, Lü L, Shi D, and Zhou T. Characterizing cycle structure in complex networks. Commun Phys. (2021) 4:272. Available online at: https://www.nature.com/articles/s42005-021-00781-3 (Accessed October 3, 2025).
66. Ren C, Zhang W, Zhong Z, Han X, Yang G, Feng Y, et al. Differential responses of soil microbial biomass, diversity, and compositions to altitudinal gradients depend on plant and soil characteristics. Sci Total Environ. (2018) 610-611:750–8. doi: 10.1016/j.scitotenv.2017.08.110
67. Zhou G, Long F, Zu L, Jarvie S, Peng Y, Zang L, et al. Stand spatial structure and microbial diversity are key drivers of soil multifunctionality during secondary succession in degraded karst forests. Sci Total Environ. (2024) 937:173504. doi: 10.1016/j.scitotenv.2024.173504
68. Xu Z, Hu Z, Jiao S, Bell SM, Xu Q, Ma L, et al. Depth-dependent effects of tree species identity on soil microbial community characteristics and multifunctionality. Sci Total Environ. (2023) 878:162972. doi: 10.1016/j.scitotenv.2023.162972
69. Liao X, Tang T, Li J, Wang J, Neher DA, Zhang W, et al. Nitrogen fertilization increases the niche breadth of soil nitrogen-cycling microbes and stabilizes their co-occurrence network in a karst agroecosystem. Agr Ecosyst Environ. (2024) 374:109177. doi: 10.1016/j.agee.2024.109177
70. Wang X, Feng J, Ao G, Qin W, Han M, Shen Y, et al. Globally Nitrogen Addition Alters Soil Microbial Community Structure, but has Minor Effects on Soil Microbial Diversity and Richness. Soil Biol Biochem. (2023) 179:108982. doi: 10.1016/j.soilbio.2023.108982
71. Samaddar S, Chatterjee P, Truu J, Anandham R, Kim S, and Sa T. Long-term phosphorus limitation changes the bacterial community structure and functioning in paddy soils. Appl Soil Ecol. (2018) 134:111–5. doi: 10.1016/j.apsoil.2018.10.016
72. Caldwell BA. Enzyme activities as a component of soil biodiversity: A review. Pedobiologia. (2005) 49:637–44. doi: 10.1016/j.pedobi.2005.06.003
73. Burns RG, DeForest J, Marxsen J, Sinsabaugh R, Stromberger ME, Wallenstein M, et al. Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem. (2022) 58:216–34. doi: 10.1016/j.soilbio.2012.11.009
74. Yan G, Luo X, Huang B, Wang H, Sun X, Gao H, et al. Assembly processes, driving factors, and shifts in soil microbial communities across secondary forest succession. Land Degrad Dev. (2023) 34:3130–43. doi: 10.1002/ldr.4671
Keywords: soil microbial community, plain ecological forests, co-occurrence networks, forest structural traits, forest management practices
Citation: Zheng Y, Han C, Cao J, Wang Y, Hu H, Yan D, Zhang Y, Zhang X, Dai W, Li Z and Ding X (2025) Dominance of forest structural traits in shaping soil bacterial community assembly in Beijing’s urban forests. Front. Soil Sci. 5:1573531. doi: 10.3389/fsoil.2025.1573531
Received: 09 February 2025; Accepted: 25 September 2025;
Published: 13 October 2025.
Edited by:
Rodrigo Gouvea Taketani, Rothamsted Research, United KingdomReviewed by:
Yawei Wei, Shenyang Agricultural University, ChinaWen Zhang, Ecology Institute of Shandong Academy of Sciences, China
Copyright © 2025 Zheng, Han, Cao, Wang, Hu, Yan, Zhang, Zhang, Dai, Li and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Ding, ZGlueGkxMjM0QDE2My5jb20=
 Yi Zheng
Yi Zheng Conghai Han2
Conghai Han2