petras kundrotas
University of Kansas
Lawrence, United States
5,278
Total downloads
26k
Total views and downloads
This Research Topic is closed for submissions.
This collection of articles highlights recent advances in the computational prediction, analysis, and understanding of protein-protein and peptide-protein interactions, which are central to elucidating biological processes and disease mechanisms. The first study focuses on predicting protein-protein interactions (PPIs) between rice and the rice blast fungus, employing multiple genomic and machine learning models to achieve high accuracy and specificity, thereby offering valuable insights for crop disease management. The second article introduces an integrated PPI network and co-expression analysis model (PRNet) to identify and prioritize cancer-associated genes in pancreatic adenocarcinoma, demonstrating its utility in discovering novel disease biomarkers and improving prognostic predictions. The third paper presents InterPepRank, a graph-based deep learning approach that significantly enhances the ranking and scoring of peptide-protein complexes compared to existing methods, thus improving the selection of relevant models for biological studies. Lastly, the fourth article explores the use of residue interaction networks and various centrality measures to pinpoint key binding residues in protein complexes, recommending a combined analytic strategy for optimal prediction accuracy. Collectively, these studies underscore the power of integrating computational and network-based approaches to advance our understanding of complex biological interactions and facilitate translational research in agriculture and medicine.
----
This Research Topic addresses recent methodological advances in computational methodologies for the analysis and identification of protein-protein interactions, structural modelling of protein-protein complexes, and their applications in the fields of life sciences and human health.
Proteins often function by interacting either with copies of themselves or with other proteins. During the last decades tremendous efforts were applied to reconstruct protein-protein interaction (PPI) networks on the level of entire organisms and/or complete biochemical pathways. Those efforts are mainly focused on elucidating which proteins are interacting partners, or in other words predicting the fact of interaction. However, the interactions between proteins occur in a particular biological context and are closely related to the protein characteristics. This makes it difficult to obtain reliable PPIs. Despite the significant challenges of such an undertaking, significant efforts have been devoted to characterizing and cataloging PPIs and major strides have been made over the past decade.
Although the number of protein-protein complexes in PDB is constantly growing, high costs and methodological laboriousness prevent relying only on experimental methods for 3D reconstruction of protein-protein complexes. On the other hand, ongoing proteomics and structural genomics studies routinely produce massive amounts of data that needs to be interpreted at a fast pace. Hence, there has been an increasing effort to develop computational strategies aiming to elucidate protein complexes, such as protein-protein interaction prediction, protein–protein docking, protein interface prediction, and reconstruction of biological pathways. In addition, PPI networks could provide a platform to systematically predict protein functions, pathways, essential proteins and disease genes, which helps to reveal the underlying molecular mechanism of complex phenotypes and illustrate biological processes of health and disease.
This Research Topic welcomes the submission of manuscripts (Original Research, Reviews, Perspective, Methods, Tools, and Databases) covering the following areas:
1) Construction and interpretations of protein-protein interaction networks to inform precision cancer medicine
2) Development of template-based and template-free docking methodologies including:
a. Advances in development of generic and specialized algorithms for fast generation of putative docking models.
b. Development of physics- and knowledge-based generic and specialized functions for ranking of putative docking models
c. Advances in the development of generic and specialized refining protocols for improving quality of near-native docking models.
3) The effects of missense variants on protein-protein interactions
4) Databases and software that can be expected to provide novel insights about protein interactions
5) Generation of generic and specialized high-quality benchmark datasets of protein-protein complexes for testing various aspects of docking methodologies.
6) Application of docking methodologies to specific biological problems
7) Protein-sequence and structure-guided discovery of functional sites or potential causative mutations.
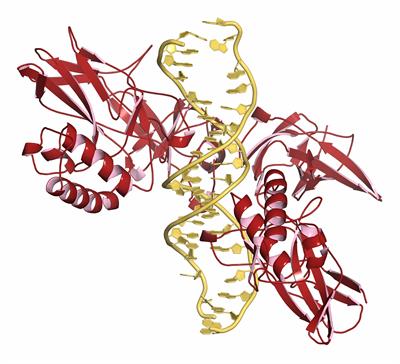
Keywords: Protein-protein interactions, missense mutations, PPI networks, protein–protein docking, binding hot-spots, biomedicine, genotype, phenotype
Important note: All contributions to this Research Topic must be within the scope of the section and journal to which they are submitted, as defined in their mission statements. Frontiers reserves the right to guide an out-of-scope manuscript to a more suitable section or journal at any stage of peer review.
Share on WeChat
Scan with WeChat to share this article
