- 1Precision Medicine Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Key Laboratory of Clinical Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Several lines of evidence have confirmed the magnitude of crosstalk between HGF/c-Met axis (hepatocyte growth factor and its high-affinity receptor c-mesenchymal-epithelial transition factor) and non-coding RNAs (ncRNAs) in tumorigenesis. Through activating canonical or non-canonical signaling pathways, the HGF/c-Met axis mediates a range of oncogenic processes such as cell proliferation, invasion, apoptosis, and angiogenesis and is increasingly becoming a promising target for cancer therapy. Meanwhile, ncRNAs are a cluster of functional RNA molecules that perform their biological roles at the RNA level and are essential regulators of gene expression. The expression of ncRNAs is cell/tissue/tumor-specific, which makes them excellent candidates for cancer research. Many studies have revealed that ncRNAs play a crucial role in cancer initiation and progression by regulating different downstream genes or signal transduction pathways, including HGF/c-Met axis. In this review, we discuss the regulatory association between ncRNAs and the HGF/c-Met axis by providing a comprehensive understanding of their potential mechanisms and roles in cancer development. These findings could reveal their possible clinical applications as biomarkers for therapeutic interventions.
Introduction
Cancers are initiated by genetic preconditions and epigenetic alterations, accompanied by various mechanisms, most of which are still unclear (Bach and Lee, 2018). According to Hanahan and Weinberg, these mechanisms are characterized by eight hallmark capabilities which include: sustaining proliferative signaling, evading growth suppressors, activating invasion and metastasis, enabling replicative immortality, inducing angiogenesis, resisting cell death, avoiding immune destruction, and dysregulating cellular energetics (Hanahan and Weinberg, 2011). Despite recent advancements in early diagnosis and personalized treatments, cancer incidence is increasing steadily, and the overall survival rate of patients remains poor (Balani et al., 2017). Therefore, it is crucial to develop a comprehensive understanding of the molecular mechanisms underlying tumor development and progression.
Hepatocyte growth factor (HGF)/c-mesenchymal-epithelial transition factor (c-Met) axis is an essential mediation axis that regulates cellular biological events including cell proliferation, migration and morphogenesis and tumor biological processes such as angiogenesis and drug resistance (Ebens et al., 1996; Arnold et al., 2017). The molecular mechanism of this activation involves gene amplification, overexpression of c-Met and HGF proteins, incremental crosstalk between c-Met and other tyrosine kinases, and MET gene mutation (Zhang et al., 2018). Furthermore, this axis has been generally reported to deregulate in tumor tissues. And its constitutive over-activation, with canonical signaling pathways containing effector molecules such as STAT3/5, PI3K-AKT, and RAS-MAPK activating, which induce malignant phenotype and promoting tumorigenesis, has been implicated in the progression of multifarious cancers (Fukushima et al., 2018). For example, Han et al. found that HGF treatment-induced epithelial-mesenchymal transition (EMT) -like changes and increased the invasive potential of PC-3 cells in human prostate cancer through ERK/MAPK (extracellular-signal-regulated kinase/mitogen-activated protein kinase) and zinc finger E-box binding homeobox 1 (Zeb-1) signaling pathways (Han et al., 2016).
Non-coding RNAs are a cluster of RNA that have non-protein-coding transcripts and are the most common RNA species in eukaryotic cells, including rRNA, tRNA, microRNA, snoRNA, piwi-interacting RNA (piRNA), circRNA, lncRNA, and so on (Slack and Chinnaiyan, 2019). Since the last decade, increasing studies have confirmed that ncRNAs play pivotal roles in various tumors development by targeting diversified downstream genes and signaling pathways and serve as tumor suppressors or stimulators (Higashio et al., 1990; Martianov et al., 2007; De Smet et al., 2015). Among them, miRNAs (small 20- to 25-nucleotide-long RNAs) and lncRNAs (larger than 200 nucleotides long) are the most common molecules that are investigated worldwide. In the recent past, a significant interaction network between HGF/c-Met axis and ncRNAs has been observed in a considerable number of tumors, stressing further the importance of the HGF/c-Met axis in tumorigenesis.
In this review, the crosstalk between the HGF/c-Met axis and ncRNAs (mostly miRNAs and lncRNAs) in common human cancers are summarized. Also, a comprehensive understanding of their potential mechanisms and roles in cancer development are provided. These findings could open a broader horizon for investigators to engage in exploring the molecular mechanisms in cancer development and progression.
An Overview of HGF/c-Met Axis
HGF, also known as scatter factor, is located on chromosome 7q21 and is a member of the peptidase S1 family of serine proteases, but is short of peptidase activity (Garcia-Vilas and Medina, 2018). It was initially identified as a molecule with the ability to stimulate hepatocyte growth and liver regeneration (Ebens et al., 1996). It is a large multi-domain heterodimeric protein comprising of α-chain (69 kD) and β-chain (34 kD) subunits and is mainly secreted by mesenchymal cells. The α-chain subunit contains an N-terminal hairpin loop and four kringle domains (K1–K4), which are responsible for the high-affinity binding to the c-Met receptor (Higashio et al., 1990; Organ and Tsao, 2011; Chen C. T. et al., 2012). Through cleaving at the Arg494-Val495 bond, it can transform from an inactive single-chain precursor (pro-HGF) to mature HGF after activation by extracellular proteases such as cellular type II transmembrane serine proteases (Fukushima et al., 2018).
C-Met is located on chromosome 7q21-31 and belongs to the family of receptor tyrosine kinases (Zhang et al., 2018). It is a heterodimer composed of a highly glycosylated extracellular 50 kDa α-chain and a transmembrane 140 kDa β-chain, linked together by disulfide bonds (Baldanzi and Graziani, 2014). As a natural HGF receptor, it is expressed on the surfaces of various epithelial cells and is known for its roles in promoting tumorigenesis. After binding to the HGF, the two subunits are dimerized, leading to the autophosphorylation of two tyrosine residues (Tyr1234 and 1235). The autophosphorylation of the tyrosine residues results in an activated multifunctional docking site composed of Src homology-2 (SH2) domain, phosphotyrosine binding (PTB) domain, and Met binding domain (MDB), which recruit adapter molecules (Furge et al., 2000). For example, Grb-2, which is a vital element in HGF/c-Met axis, interacts with Y1356 of c-Met and several essential signaling pathway-related proteins such as Ras, SOS, and Gab1 and eventually activates them. Besides, these residues also recruit PI3K, STAT3, PLCγ, and SHP2 thereby linking oncogenes and many receptors which are crucial for cellular functions (Garcia-Vilas and Medina, 2018). Moreover, it has been revealed that c-Met could be activated by non-canonical pathways. Non-canonical pathways are related to c-Met amplification, drug-resistant cancers and malignant tumor features. Several membrane surface proteins (EGFR, β-catenin, MUC-1, CD44, VEGFA, Plexin B, FAK, HER, Integrin α6β4, Fas, etc.) have been confirmed to interact with c-Met and contribute to dynamic c-Met biological responses (Migliore and Giordano, 2008; Scagliotti et al., 2013). The molecular network of the HGF/c-Met signaling is shown in Figure 1.
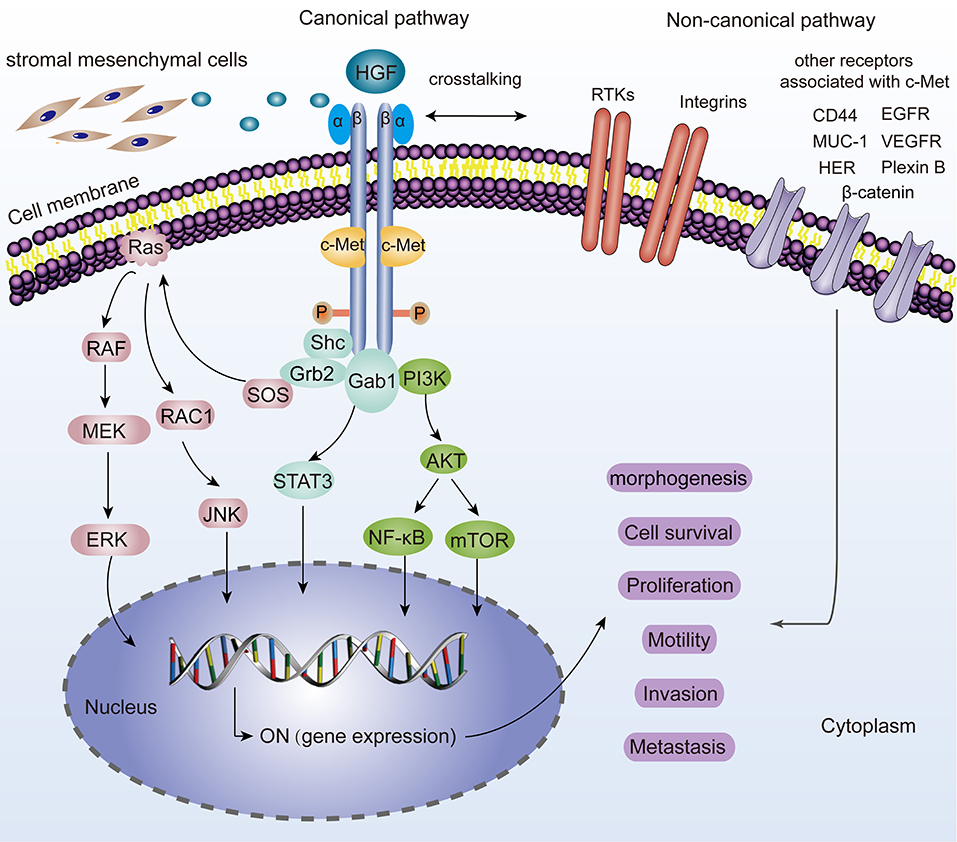
Figure 1. Representation of the HGF/c-Met canonical and non-canonical pathways. For canonical pathways, the binding of HGF, induces two c-Met molecules dimerization, thereby leading to the autophosphorylation of tyrosine residues and subsequent activation of many downstream signaling pathways such as MAPK/ERK, STAT3, PI3K/AKT signaling. JNK is also phosphorylated and activates a variety of downstream substrates, including transcription factors such as AP-1 and apoptosis-related Bcl-2, Bax, etc. All these hereby basically drive a plethora of cell phenotypes such as morphogenesis, survival, proliferation, motility, invasion, and metastasis. “ON” means the activation for gene expression. Non-canonical pathways are activated when c-Met binds to other receptors including EGFR, MUC-1, VEGFR, CD44, Plexin B1, HER, Integrin α6β4, β-catenin, and so on.
Crosstalk Between HGF/c-Met Axis and ncRNAs in Cancers
This chapter summarizes the interaction between the HGF/c-Met axis and ncRNAs in common cancers. The outline is shown in Figure 2, Tables 1, 2.
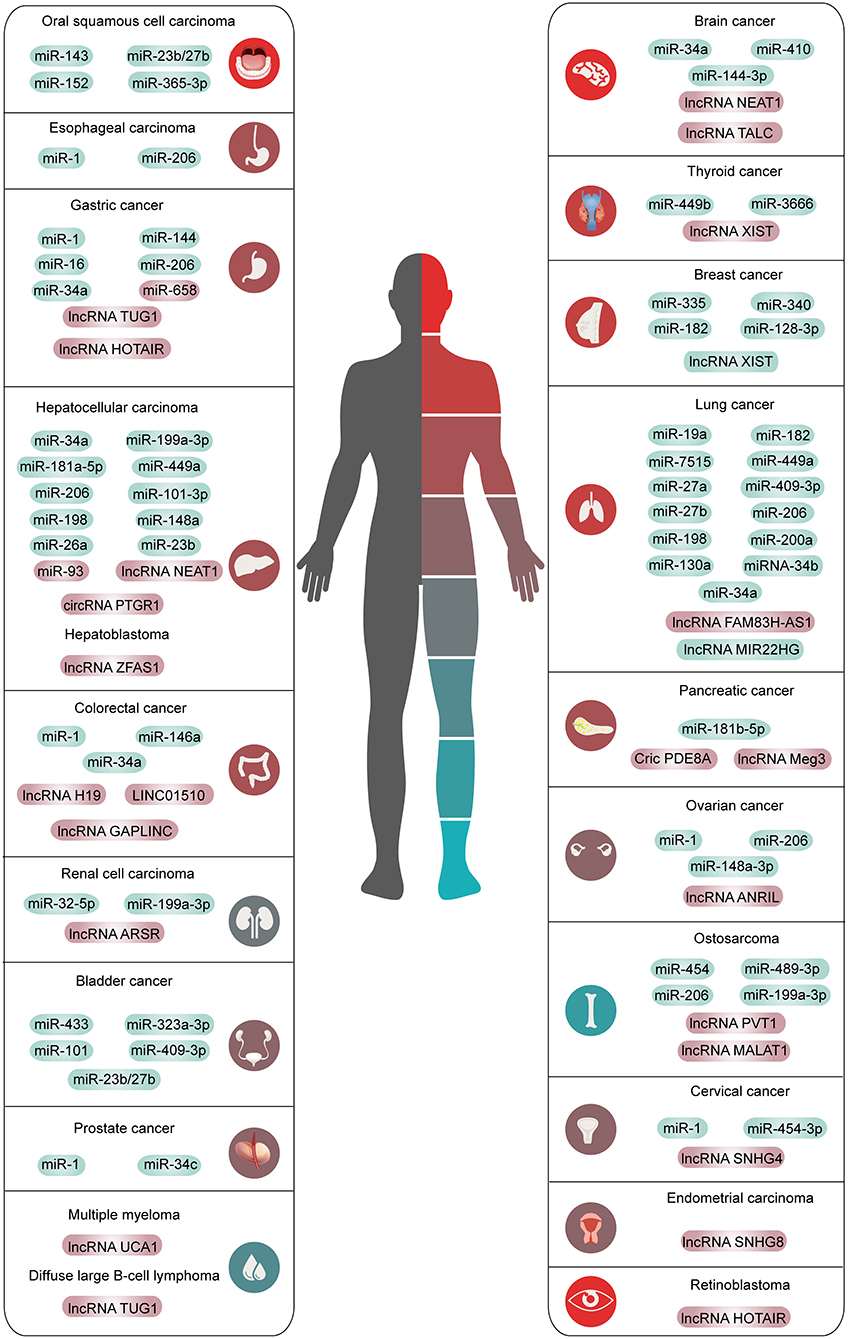
Figure 2. The overview of ncRNAs interacted with HGF/c-Met axis in human common malignancies. The red means high expression and the green means low expression.
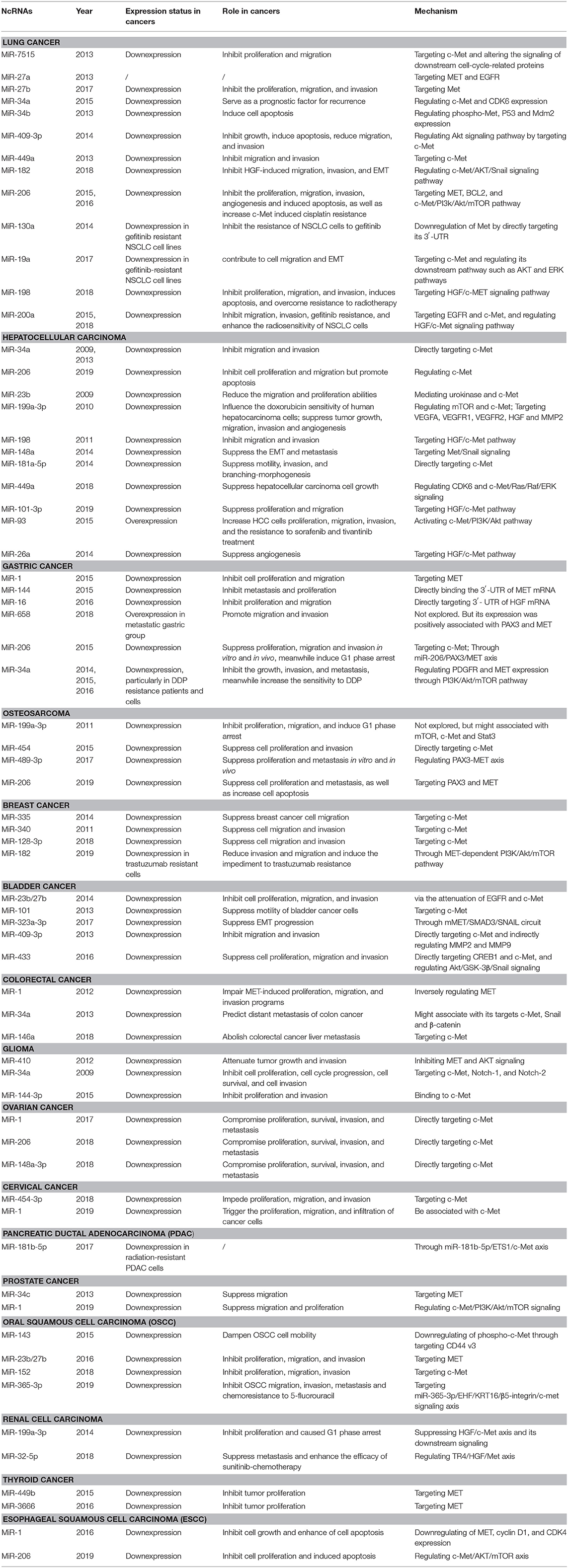
Lung Cancer
Lung cancer is one of the most frequent cancers globally. In 2018, it contributed to 11.6% of all the new cancer cases worldwide (Bray et al., 2018). The disease can be classified into two primary subtypes: non-small cell lung cancer (NSCLC) and small-cell lung cancer, representing 85 and 15% of all lung cancer cases, separately. Lung cancer is very aggressive, more than 50% of the patients die within 1 year after diagnosis, and the 5-year survival rate is below 18% (Sun R. C. et al., 2019). It is still difficult to disclose the indisposition at an early primary phase for possible therapeutic and surgical treatments.
Lee et al. were the first to identify and characterize a novel miR-7515 from lung cancer cells. The miRNA has been shown to inhibit tumor proliferation and migration by directly repressing c-Met expression and altering the signaling of downstream cell cycle-related proteins (Lee et al., 2013). Subsequently, several miRNAs have been reported to regulate c-Met in NSCLC. For example, studies demonstrated that suppression of miR-27a (Acunzo et al., 2013), miR-27b (Zhou et al., 2017), miR-34a (Hong et al., 2015), and miR-34b (Wang et al., 2013) impairs NSCLC progression by regulating c-Met. Another tumor suppressor, miR-409-3p, negatively correlates with c-Met and has been shown to undermine tumor growth, migration, and invasion. Also, it induced apoptosis in vitro via the Akt signaling pathway by targeting c-Met (Wan et al., 2014). Luo et al. revealed that upregulation of miR-449a was significantly associated with less lymph node metastasis and better prognosis in NSCLC via regulation of the target gene c-Met (Luo et al., 2013). Consistently, miR-182 was found to inhibit HGF-induced migration, invasion, and EMT by regulating c-Met/AKT/Snail signaling pathway in NSCLC (Li Y. et al., 2018). Furthermore, Chen et al. showed that miR-206 could inhibit HGF-induced EMT and angiogenesis and induce cisplatin resistance by regulating c-Met/PI3K/Akt/mTOR pathway in lung cancer (Chen Q. Y. et al., 2015; Chen et al., 2016a,b).
The HGF/c-Met axis also contributes to drug resistance in lung cancer. Zhou et al. were the first to verify that miR-130a is highly expressed in gefitinib-sensitive NSCLC cell lines, but not gefitinib-resistant NSCLC cells. They also demonstrated that elevated miR-130a could enhance the sensitivity of NSCLC cells to gefitinib and promote apoptosis by directly reducing c-Met levels (Zhou et al., 2014). Similarly, decreased miR-19a expression in gefitinib-resistant NSCLC cell lines contributes to cell migration, EMT, and gefitinib resistance by negatively regulating c-Met expression and blocking its downstream pathways (such as PI3K-AKT and RAS-ERK pathways) (Cao et al., 2017). Zhu et al. demonstrated that miR-198 could inhibit proliferation, migration, and invasion, induce apoptosis, and overcome resistance to radiotherapy via HGF/c-Met axis. It was downregulated in NSCLC cell lines but overexpressed in radiotherapy sensitive patients (Zhu et al., 2018). Studies also reported that miR-200a plays an active tumor-suppressing role in non-small cell lung cancer, and could also inhibit gefitinib resistance and enhance the radio-sensitivity by deactivating the HGF/c-Met pathway (Zhen et al., 2015; Du et al., 2019).
Moreover, lncRNA FAM83H-AS1 was suggested as a potential therapeutic target that could accelerate cell proliferation, invasion, and migration by activating MET/EGFR signaling pathway in lung adenocarcinoma (Zhang et al., 2017). Su et al. also identified the anticancer effect of lncRNA MIR22HG and there was a significant association between lncRNA MIR22HG and MET expression (Su et al., 2018). These studies provided new insights into prognostic diagnosis and therapeutic strategies for patients with lung cancer. Collectively, the HGF/c-Met axis and ncRNAs play an essential role in the formation and development of lung cancer.
Hepatocellular Carcinoma (HCC)
HCC is one of the ordinary cancers with the highest morbidity and mortality globally. Till now, the underlying molecular mechanisms for pathogenesis and development of HCC remain mostly unknown because of multiple etiologies (Bray et al., 2018). Therefore, studies on the molecular checkpoints involved in HCC development and aggressiveness are crucial.
In 2009, Li et al. were the first to demonstrate that miR-34a is less expressed in HCC tissues than in corresponding normal tissues. In the same study, it was revealed that miR-34a HCC reduces malignant phenotype by downregulating c-Met expression and decreasing the phosphorylation level of ERK1/2 (Li N. et al., 2009). Later, miR-206 (Wang Y. et al., 2019) and miR-23b (Salvi et al., 2009) were found to attenuate cell viability, migration, and invasion by blocking the c-Met activation in HCC. The study concluded that miR-199a-3p is closely associated with the formation and progression of HCCs. In the same study, a significant increase in the G1-phase population following the restoration of miR-199a-3p expression was observed. Further investigation of the mechanism showed that miR-199a-3p contributed to the cell cycle modulation by silencing both mTOR and c-Met (Fornari et al., 2010; Ghosh et al., 2017). MiR-198 bound to the 3′ UTR of c-Met mRNA and blocked p44/42 MAPK activity through HGF/c-Met pathway, leading to the inhibition to migration and invasion of HCC cells (Tan et al., 2011). Consistently, miR-148a was reported to impede EMT and metastasis by targeting HGF/Met/Snail signaling in HCC (Zhang et al., 2014). Besides invasion, miR-181-5p suppressed branching-morphogenesis by directly regulating c-Met (Korhan et al., 2014). Many studies have shown that miR-449a impairs the EMT of tumors by targeting MET, but the precise single target of miR-449a has not been elucidated in HCC (Sun et al., 2017). Cheng et al. revealed that miR-449a suppressed HCC growth by targeting CDK6 and impairing c-Met/Ras/Raf/ERK signaling pathway with an iTRAQ proteomics approach. Their study linked miR-449a, cell cycle, and c-Met/Ras/Raf/ERK signaling pathway in HCC and could also explain the abnormal growth characteristics of HCC cells (Cheng et al., 2018). Besides, in vitro and in vivo studies have demonstrated that miR-101-3p inhibits HCC progression by targeting HGF and it could be regarded as another potential therapeutic tool for regulating cell proliferation and metastasis (Liu et al., 2019). However, miR-93 was identified as HCC stimulator, which increased HCC cell proliferation, migration, and invasion by activating c-Met/PI3K/Akt pathway (Ohta et al., 2015). Thus, HGF/c-Met axis might be of high interest in regulating tumor progression in HCC.
In our review, the potential roles of the HGF/c-Met axis in other hallmarks of HCC, such as interactions with tumor microenvironment and drug resistance, are also highlighted. VEGFA/VEGFR2 signaling is a crucial downstream pathway of HGF/c-Met axis and plays a vital role in tumor angiogenesis. VEGFA secreted from cancer cells binds to VEGFR2 on endothelial cells, phosphorylates and activates VEGFR2, which then phosphorylates downstream extracellular signal-regulated kinase (ERK1/2) thus promoting angiogenesis (Ferracini et al., 1995; Fischer et al., 2004; Testini et al., 2019). Recently, Yang et al. confirmed that miR-26a could inhibit tumor proliferation and metastasis of HCC through IL-6-Stat3 signaling pathway. Notably, a negative correlation between miR-26a and VEGFA or microvessel density (MVD), two well-known proangiogenic factors, was observed. MiR-26a significantly impaired in vivo tumor angiogenesis by inhibiting VEGFA production through HGF/c-Met axis in HCC cells. Moreover, miR-26a also impaired VEGFR2 signaling in endothelial cells to exert its antiangiogenesis function (Yang et al., 2014). Accordingly, miR-199a-3p showed important inhibitory effect on angiogenesis by targeting HGF and subsequently blocking downstream signaling pathways. Ghosh et al. demonstrated that miR-199a-3p reduced VEGF secretion in HCC cells and inhibited expression of VEGFR1 and VEGFR2 receptors on endothelial cells. Besides, MMP2 signaling was abrogated by miR-199a-3p (Ghosh et al., 2017). Furthermore, HGF/c-Met axis was reportedly involved in drug resistance. MiR-199a-3p could regulate the doxorubicin sensitivity of human HCC cells through c-Met and mTOR (Fornari et al., 2010). Katsuya revealed that overexpression of miR-93 could improve the resistance of HCC cells against sorafenib and tivantinib treatment through c-Met/PI3K/Akt pathway (Ohta et al., 2015).
With regards to lncRNAs, Chen et al. demonstrated that lncRNA NEAT1 could suppress the sorafenib sensitivity of HCC cells by regulating miR-335/c-Met pathway (Chen and Xia, 2019). In addition, exosomes derived from highly metastatic cells (LM3) with a high abundance of circPTGR1 might enhance the metastatic potential of lower metastatic cells (97L and HepG2) by inhibiting miR-449a-MET interaction, resulting in destruction on tumor microenvironment and thereby promoting HCC development (Wang G. et al., 2019). Collectively, the results of these studies show that HGF/c-Met axis is an effective multifaceted tumor regulator, which could have pivotal implications on the understanding of HCC mechanisms and the improvement of therapeutics.
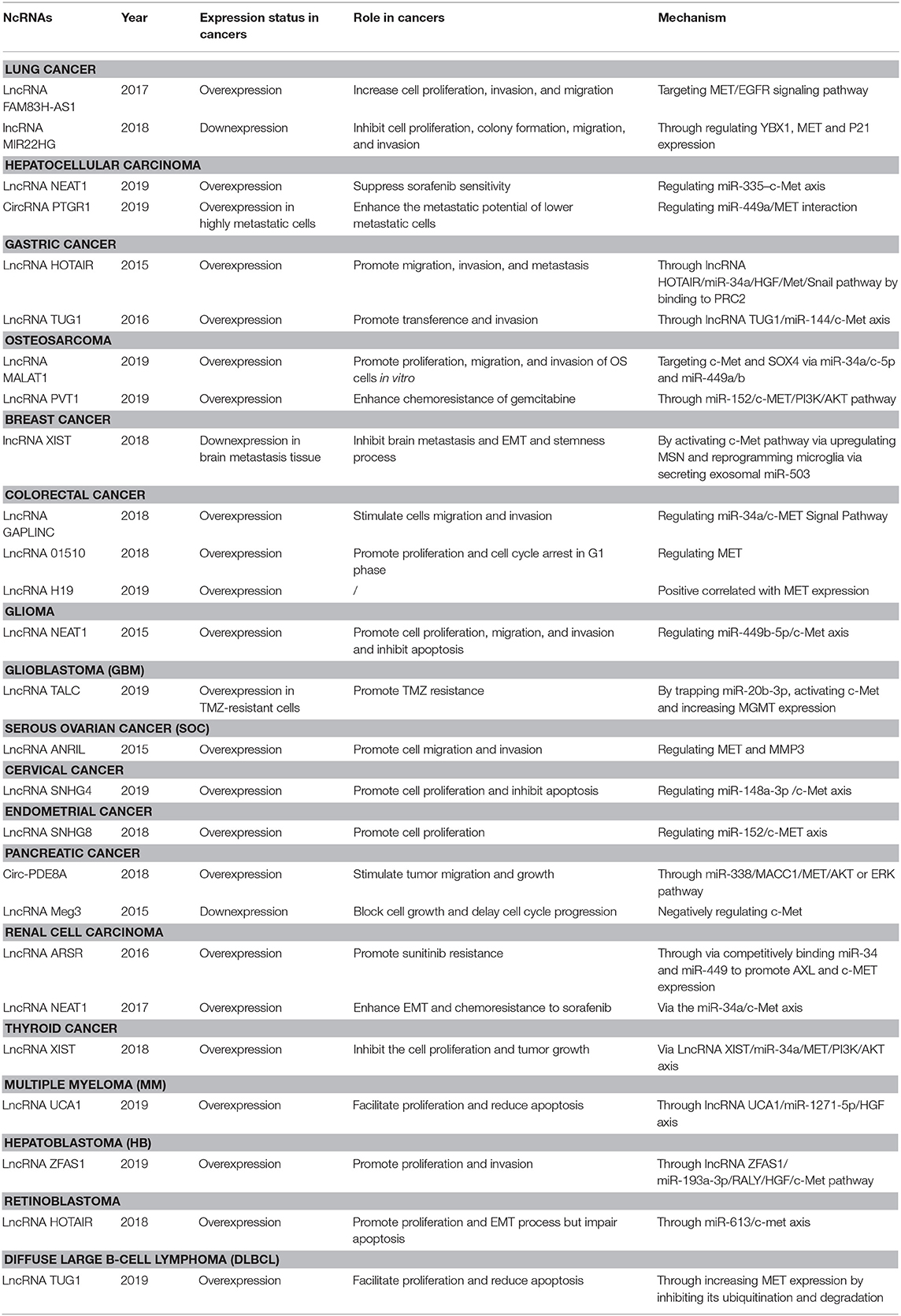
Gastric Cancer (GC)
GC is one of the most ordinary neoplasms with a high incidence, especially in Eastern Asia. Metastasis is the main reason for its which results in high mortality of the patients (Bray et al., 2018). Nonetheless, the precise molecular mechanisms underlying GC metastasis have not uncovered.
The HGF/c-Met axis has been implicated as a crucial modulator of metastasis-associated signal transduction pathways in GC progression. Several miRNAs have been reported to participate in the GC proliferation and metastasis by directly or indirectly regulating the expression of c-Met. MiR-1 (Han et al., 2015) and miR-144 (Liu J. et al., 2015) are downregulated in GC tissues and directly target the 3′-UTR of MET mRNA, thus, leading to the repression of GC progression. Besides, downregulation of miR-16 suppresses HGF expression by binding to its 3′-UTR (Li et al., 2016). In a previous study, the expression of miR-658 was elevated in distant GC cells, which increased the levels of serum miR-658 significantly, thus promoting cell migration and invasion. Besides, a strong positive relation between serum level of miR-658 and mRNA PAX3 and MET expression has been observed (Wu et al., 2018). However, the precise role of miR-658/PAX3/MET axis needs further exploration.
MiR-206, a famous tumor suppressor, is thought to enhance tumor metastasis by regulating diverse mRNA expression at a post-transcription level in some forms of cancer such as breast and colon cancers. In a study by Zhang et al., the expression of miR-206 was weaker in GC cell lines, especially in high metastatic cell lines. Transwell assay confirmed that the ectopic expression of miR-206 inhibited the migration and invasion ability of GC cells by regulating PAX3-MET pathways. In vivo mouse experiments demonstrated that miR-206 overexpression resulted in significant decrease in the incidence of lung metastasis and the number of metastatic lung nodules (Zhang et al., 2015). The study from Zheng et al. also supported the conclusion that miR-206/MET axis modulates the migration and invasion of GC. Besides, suppression of tumor growth was observed after miR-206 mimics transfection, which indicated downregulation of c-Met and cell cycle-related proteins in xenograft mouse models and in vitro (Zheng et al., 2015).
MiR-34a also has tumor suppression ability. A study reported that miR-34a, which was weakly expressed in GC tissues, attenuated the malignant behavior of GC cells by directly targeting PDGFR and MET expression and subsequently regulating the phosphorylation of Akt through PI3K/Akt/mTOR pathway (Peng et al., 2014; Wei et al., 2015). In addition to these findings, Zhang et al. revealed that miR-34a could modulate human GC cells cisplatin (DDP) sensitivity by regulating cell proliferation and apoptosis by targeting MET. Specifically, they found that miR-34a expression was significantly less in DDP resistance human GC tissues and cells than in normal GC tissues and cells. The overexpression of miR-34a enhanced the DDP sensitivity of SGC7901/DDP cells by inhibiting cell proliferation and inducing cell apoptosis. Therefore, its downregulation could weaken the sensitivity to DDP. A subsequent study confirmed that miR-34a modulates DDP resistance by directly repressing MET (Zhang et al., 2016). Collectively, miR-34a could contribute to the development of new therapeutic strategies for gastric cancer.
Furthermore, competing endogenous RNAs (ceRNAs) as essential miRNA-regulators have emerged as therapeutics scavenging oncogenic miRNAs. Recently, they have exposed exciting insights into tumorigenesis. LncRNA HOTAIR was found to be overexpression in GC tissue and cells, particularly in diffuse-type GC. In addition, lncRNA HOTAIR knockdown remarkably impaired migration, invasion and metastasis both in vitro and in vivo and reversed the EMT in GC cells by epigenetically inhibiting miR34a through recruiting and binding to PRC2, mechanically (Liu Y. W. et al., 2015). Similarly, Ji et al. elucidated that overexpression of lncRNA-TUG1 led to a significant proliferation of GC tissue (most likely) by inhibiting miR-144. Also, lncRNA-TUG1 indirectly activated the expression of c-Met, thus promoting the metastasis of GC cells via lncRNA-TUG1/miR-144/c-Met axis (Ji et al., 2016).
Osteosarcoma (OS)
OS predominantly originates from mesenchymal cells of long bones and has a high prevalence in children and young adults. It usually starts as a monoclonal disease and quickly progresses into a polyclonal disease. It is regarded as one of the most complicated cancers in terms of molecular aberration (Kansara and Thomson, 2019). Further elucidation of its exact molecular mechanism could be useful in the identification of its associated markers, which might assist in prognosis and therefore improve its treatment.
In 2011, Duan et al. were the first to reveal the negative association between the expression of miR-199a-3p and mTOR, c-Met and Stat3 (Duan et al., 2011). But the molecular mechanism behind the tumor suppressor role of miR-199a-3p in human OS has not been fully elucidated. MiR-454 was found to be downregulated in OS clinical samples and cell lines. And this suppressed proliferation and metastasis of OS cells by inhibiting c-Met expression (Niu et al., 2015). Studies conducted by Liu et al. suggested that miR-489-3p expression could inhibit the proliferation and metastasis of OS tissues and cells, particularly in high metastatic potential cells by negative regulation of PAX3-MET axis. Also, xenografts study results showed smaller cancer nodules and fewer incidences of lung metastases in LV-miR-489-3p group compared with LV-control group, indicating the apparent inhibition of OS metastasis in vivo (Liu Q. et al., 2017). Similarly, miR-206 was reported to repress OS progression by targeting PAX3-MET axis (Zhan et al., 2019).
ceRNAs also play critical roles in the development of OS. Sun et al. demonstrated that serum level of long non-coding RNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (lncRNA MALAT1) was higher in metastatic OS tissues than non-metastasis OS tissues or corresponding normal tissues. In the same study, elevated levels of serum lncRNA MALAT1 predicted worse clinical outcomes in OS patients. The silencing of lncRNA MALAT1 was shown to inhibit proliferation, migration, and invasion of OS cells. Bioinformatic and molecular analyses revealed that lncRNA MALAT1 served as a ceRNA of c-Met and SOX4 by directly targeting miR-34a/c-5p and miR-449a/b (Sun Z. et al., 2019). The function of ceRNAs in drug resistance has also been elucidated. LncRNA PVT1 played critical roles in chemoresistance of OS to gemcitabine (GEM) and was upregulated in OS drug-resistant cells. Functional studies have demonstrated that lncRNA PVT1 could reduce GEM-reduced apoptosis and attenuate GEM-induced tumor growth inhibition in vitro and in vivo. A study on lncRNA PVT1 mechanism revealed that lncRNA PVT1 targeted miR-152, which promoted chemoresistance of OS by activating c-MET/PI3K/AKT pathway (Sun Z. Y. et al., 2019). These findings revealed a novel interaction network that could eventually lead to new therapeutic strategies for OS patients based on ncRNAs.
Breast Cancer
Breast cancer is the leading cause of mortality in women worldwide (Colditz and Bohlke, 2014). It occurs as a consequence of various signaling pathways in mammary epithelial cells, including the HGF/c-Met axis. Many studies have shown that the interaction between ncRNAs and HGF/c-Met axis exerts regulatory effects on breast cancer progression and clinical therapy. Recent studies have suggested that miR-335 could be upregulated in breast cancer tissues. Overexpression of miR-335 has been shown to suppress HGF-induced phosphorylation of c-Met, which could subsequently inhibit the HGF role of prompting breast cancer cell migration in a c-Met-dependent manner by targeting c-Met (Gao et al., 2015). Wu and Breunig confirmed the hypothesis of post-transcriptional regulation of MET by revealing that miR-340 and miR-128-3p negatively regulate MET expression, thus inhibiting the invasion and migration of breast cancer cells (Wu et al., 2011; Breunig et al., 2018).
Concerning drug resistance, Yue et al. revealed that miR-182 binds to MET in breast cancer cells and suppresses the cell's ability to tolerate trastuzumab, therefore reducing the invasion and migration capability of trastuzumab-resistant cells through MET-dependent PI3K/AKT/mTOR signaling pathways (Yue and Qin, 2019). This study highlighted the importance of the interaction between ncRNAs and HGF/c-Met axis in cancer development and therapy.
For lncRNAs, in 2018, Xing et al. profiled lncRNAs in breast cancer tissue with brain metastasis and confirmed that the X-inactive–specific transcript (XIST) was remarkably downregulated. Further in vitro and in vivo experiments revealed that knockdown lncRNA XIST could activate c-Met pathway via upregulating MSN to promote EMT and stemness. Meanwhile, lncRNA XISTlow cells could reprogram microglia in the brain via secreting exosomal miR-503. And the prometastatic effect was inhibited by fludarabine (Xing et al., 2018). These indicated that lncRNA XIST might become an effective target for curing brain metastasis.
Bladder Cancer (BCa)
BCa is the most frequent cancer of the urinary tract (Grayson, 2017). Chlyomaru et al. were the first to reveal the function of miRNA-23b/27b cluster in bladder cancer. In their study, the capacities of cell proliferation, migration, and invasion of bladder cancer cells were repressed after restoring miR-23b and miR-27b expression by attenuation of EGFR and c-Met (Chiyomaru et al., 2015). Hu et al. reported a similar mechanism in miR-101 (Hu et al., 2013). MiR-323a-3p is a member of the miRNA cluster in DLK1-DIO3 genomic region, which plays a role in several pathologic processes of various cancers, especially bladder cancer. Li et al. showed that the methylation of DLK1-MEG3 intergenic DMR contributed to the decrease in levels of serum miR-323a-3p in bladder cancer. The progression of EMT was suppressed through miR-323a-3p/MET/SMAD3/SNAIL circuit (Li et al., 2017). A study elucidated the existence of mutual regulation of BCa progression involving miR-323a-3p/miR-433/miR-409 and MET. The study investigated the role of miR-433 and miR-409 in the development and progression of BCa. The results showed that miR-409-3p could serve as metastasis-suppressor gene by directly targeting c-Met and indirectly regulating MMP2 and MMP9 (Xu et al., 2013). CAMP response element-binding protein1 (CREB1) and c-Met could take part in miR-433-mediated inhibition of the EMT by regulating Akt/GSK-3β/Snail signaling. Also, a reciprocal regulation network between miR-433/miR-409-3p and c-Met was revealed (Xu et al., 2016). These findings might be useful in the search for effective and promising therapies against BCa.
Colorectal Cancer (CRC)
CRC is a public health concern and accounts for over 10.2% of all cancer cases, representing 1.8 million new cases per year (Bray et al., 2018). Despite widespread awareness in the general population regarding CRC, the condition remains a significant cause of mortality and morbidity on account of recurrence and metastasis.
Studies have confirmed that the expression of c-Met in CRC correlates with the presence of local and distal metastasis and poor prognosis. Congruently, Migliore et al. showed that miR-1 is downexpressed in colon cancer cells, and is significantly associated with MET overexpression, especially in metastatic tumors. Further analysis revealed that concomitant MACC1 (MET transcriptional activator) upregulation and miR-1 downregulation are required to elicit the highest level of MET expression. Induced miR-1 expression reduces MET levels and impairs MET-induced proliferation, migration and invasion processes. Notably, a feedback loop between miR-1 and MET has also been revealed. Most likely, MET targeting could result in both abrogations of MET-dependent signaling and some recovery of miR-1 level (Migliore et al., 2012). Siemens verified that the generation of distant metastases is correlated with epigenetic silencing of miR-34a in primary tumors. In their study, they reported that miR-34a expression was inversely correlated with CpG methylation and its targets, that is, c-Met, Snail, and β-catenin, and were associated with distant metastases. In the same study, a multivariate regression model containing miR-34a methylation, high c-Met, and β-catenin levels indicated high prognostic value of CRC cells' metastasis to the liver (Siemens et al., 2013). In 2018, Bleau et al. identified c-Met as a prometastatic gene and miR-146a as a negative regulator of c-Met in highly metastatic variants derived from MC38 cells. Further, they firstly confirmed that miR-146a has a crucial inhibitory role in liver metastasis and this suppressive effect is probably because of targeting several protumorigenic genes, including c-Met (Bleau et al., 2018).
Further, the ceRNA network has also been reported to play a crucial role in CRC progression. The study results showed that lncRNA GAPLINC was upregulated in CRC tissues and thus stimulated CRC cell migration and invasion by regulating miR-34a/c-MET signal pathway (Luo et al., 2018). Analogously, lncRNA 01510 was reported that its overexpression was associated with advanced clinicopathological features. Meanwhile, Cen et al. also found that silencing LINC01510 could restrain cell proliferation and induce G0/G1-phase arrest via a MET-dependent manner (Cen et al., 2018). In 2019, Zhong et al. constructed the lncRNA/pseudogene–miRNA–mRNA ceRNA network using The Cancer Genome Atlas database and verified that lncRNA H19, which was upregulated in CRC tissue and correlated with poor prognosis, was positive related to MET expression. However, the specific molecular mechanism and function were not explored (Zhong et al., 2019). These findings provided substantial evidence that c-Met has excellent potential as an effective agent for both the prevention and treatment of colorectal cancer.
Brain Cancer
Glioma is the most common malignancy of the central nervous system and is associated with a poor prognosis. Several studies suggest that the condition could occur as a result of complicated gene interaction and molecular modulation network (Sturm et al., 2017). In vitro and xenograft model experiments have verified that miR-410 repression attenuates tumor growth and invasion by inhibiting MET and AKT signaling. Also, MET overexpression could significantly abrogate miR-410 dependent effects on glioma proliferation and invasion (Chen L. et al., 2012). With regard to lncRNA, Li et al. found that lncRNA NEAT1was overexpressed in glioma tissue and cell lines and served as a ceRNA to promote glioma pathogenesis. Moreover, lncRNA NEAT1's oncogenic activity was enhanced through inverse regulation of miR-449b-5p and c-Met modulation in glioma cells (Zhen et al., 2016). This was the first report on the role and function of lncRNA NEAT1 in glioma.
Concerning glioblastoma (GBM), it has been reported that transient transfection of miR-34a mimics into glioma and medulloblastoma cell lines remarkably inhibits cell proliferation, invasion, survival, and cell cycle progression by targeting c-Met, Notch-1, and Notch-2 (Li Y. et al., 2009). MiR-144-3p was downregulated in GBM tissue, and this decrease was significantly correlated with ascending grades and poorer overall survival. Exogenetic miR-144-3p expression compromised the malignant biological properties of GBM cells independent of PTEN and resulted in enhancement of radiation and temozolomide (TMZ) sensitivity by binding to c-Met and thus impairing the activity of downstream signaling (Lan et al., 2015). As for lncRNAs, Wu et al. firstly identified that lncRNA TALC, located on the AL358975 locus and consisted of two exons with a full length of 418 nt, had elevated expression in temozolomide-resistant GBM cells. Further mechanism exploration revealed that lncRNA TALC induced TMZ resistance in GBM through trapping miR-20b-3p and activating c-Met. Meanwhile, lncRNA TALC also upregulated MGMT expression by refitting the acetylation of H3K9, H3K27, and H3K36 in MGMT promoter regions via c-Met/Stat3/p300 axis (Wu et al., 2019). Despite the identification of many new biomarkers for brain cancer, improvement in treatment options is yet to be achieved.
Female Reproductive Cancers
One of the most prevalent cancers of the female reproductive system is ovarian cancer (Eisenhauer, 2017). Substantial evidence shows that some miRNAs (such as miR-1, miR-206, and miR-148a-3p) can suppress the proliferation, survival, invasion, and metastasis of ovarian cancer cells by directly targeting c-Met, and subsequently regulating downstream signaling pathway to serve as tumor suppressors such as PI3K/Akt/mTOR and ERK1/2 pathways (Qu et al., 2017; Dai et al., 2018; Wang W. et al., 2018). As for lncRNAs, Qiu et al. highlighted the role of lncRNA antisense non-coding RNA in the INK4 locus (ANRIL) in serous ovarian cancer (SOC). They demonstrated its elevated level in SOC tissue and cell lines, particularly in high metastatic cell lines. Meanwhile, there was significant association between lncRNA ANRIL expression and advanced FIGO stage, high histological grade, lymph node metastasis, and poor prognosis. Moreover, MET and MMP3 were verified as key downstream genes of ANRIL involved in SOC cell migration/invasion by microarray analysis and Western Blotting (Qiu et al., 2015). Therefore, a breakthrough in ovarian cancer treatment could be achieved by a combination of therapy involving both ncRNAs and c-Met inhibitors.
Cervical cancer is a frequent aggressive malignancy and the fourth leading cause of cancer-related deaths among females worldwide, particularly in China. MiR-454-3p mimics treatment has been shown to suppress the proliferation, migration, and invasion ability of cervical tumor cells in vitro by targeting c-Met (Guo et al., 2018). The downregulation of miR-1 in cervical cancer tissues is correlated with high c-Met expression. Also, c-Met upregulation inhibits E-cadherin expression, which triggers the proliferation, migration, and infiltration of cancer cells, thus lowering the survival rates of the patient (Cheng Y. et al., 2019). In addition, ceRNA also has been reported in cervical cancer. LncRNA SNHG4 was overexpressed in cervical cancer tissue and cells. And this upregulation could facilitate proliferation and reduce apoptosis by binding to miR-148a-3pand ultimately activating c-Met (Li et al., 2019). These findings indicate that c-Met isan attractive therapeutic target for cervical cancer.
Endometrial cancer (EC) is the most epidemic neoplasm of the female genital tract worldwide. Despite the great development in early identification and treatment, an abundant number of cases of advanced ECs are still diagnosed (Herrero et al., 2019). LncRNA SNHG8, which are located on Chr4, has been verified to participate in the progression and drug resistance of multiple malignancies, such as esophageal squamous cell carcinoma (Song et al., 2019), gastric cancer (Zhang P. et al., 2019), pancreatic adenocarcinoma (Song et al., 2018), and hepatocellular carcinoma (Dong et al., 2018). In 2018, Yang et al. firstly verified the positive association between lncRNA SNHG8 and c-Met in EC. They found silencing lncRNA SNHG8 resulted in weaker c-MET expression and less proliferation. Meanwhile, this reduction could be reversed by addition of miR-152 inhibitor (Yang C. H. et al., 2018). This study connected ncRNA with HGF/c-Met axis together in EC and demonstrated the significant importance of HGF/c-Met axis in EC.
Pancreatic Cancer
Pancreatic cancer is one of the most aggressive malignancies, and is the seventh cause of cancer-related mortality worldwide, with a 5-year survival rate of about 5% (Bray et al., 2018). Tomihara et al. were the first to confirm the association between preoperative chemoradiation therapy and c-Met expression in pancreatic ductal adenocarcinoma (PDAC). They successfully confirmed the association between irradiation-induced c-Met expression and activated c-Met pathways through miR-181b-5p/ETS1/c-Met axis in PDAC cells (Tomihara et al., 2017). Recently, circRNA and exosomes were shown to play essential roles in various tumors, particularly PDAC. Circ-PDE8A, a highly contained and stable circular RNA screened out from tumor exosomes, is overexpressed in PDAC tissues and is an independent risk factor for PDAC survival. Its upregulation could stimulate tumor migration and growth through miR-338/MACC1/MET/AKT or ERK pathway by sponging miR-338. Further, they found that circ-PDE8A excreted by tumor could be released into blood circulation through exosome transportation. Also, plasma exosomal circ-PDE8A was correlated with PDAC invasion and prognosis (Li Z. et al., 2018). Modali et al. demonstrated the importance of HGF/c-Met axis in development of pancreatic neuroendocrine tumors. They revealed that menin could activate lncRNA Meg3 through H3K4me3 and CpG hypomethylation. And lncRNA Meg3 was shown to have tumor suppressor activity because of its role in suppression of cell growth and cycle progression by negatively targeting c-Met (Modali et al., 2015). All these findings provided a strong basis for considering c-Met suppression as novel therapeutic approach for the treatment of pancreatic cancer through prevention of metastasis and irradiation resistance.
Prostate Cancer
Prostate cancer accounts for 3.8% of cancer-related deaths annually, and this has economic implications on the public health sector worldwide (Bray et al., 2018). Treatment of the condition using exogenous miR-34c and miR-1 has been shown to suppress MET expression by preventing its binding to 3′-UTR and thereby inhibiting prostate cancer's aggressive phenotype (Hagman et al., 2013; Gao et al., 2019). Further studies have confirmed that miR-1 can suppress c-Met/PI3K/Akt/mTOR signaling and impair tumor progression (Gao et al., 2019). However, the specific mechanism of miR-34c is still unclear.
Other Cancers
Oral squamous cell carcinoma (OSCC) represents a unique and major health concern all over the world. A study by Xu et al. indicated that miR-143 inhibited OSCC cell mobility by suppressing phospho-c-Met expression through targeting CD44 v3 (Xu et al., 2015). The miR-23b/27b cluster was also shown to inversely regulate MET in OSCC and bladder cancers (Fukumoto et al., 2016). Analogously, miR-152 served as a tumor suppressor by directly targeting c-Met (Li M. et al., 2018). Moreover, Huang et al. unveiled a novel c-Met regulating mechanism that might be applied as a modality for OSCC therapy. They demonstrated that miR-365-3p/ETS homologous factor (EHF, a KRT16transcription factor) axis could reduce OSCC cell migration, invasion, metastasis, and chemoresistance to 5-fluorouracil through the suppression of KRT16. In the same study, depletion of KRT16 resulted in ample protein degradation of β5-integrin and c-Met via a lysosomal pathway, subsequently led to the repression of downstream Src/STAT3/FAK/ERK signaling in OSCC cells (Huang et al., 2019). Generally, c-Met plays a crucial role in OSCC progression and could be a potential target in OSCC therapy.
In renal cell carcinoma (RCC), miR-199a-3p is known as a tumor attenuator. Huang et al. revealed that miR-199a-3p was downregulated in RCC primary tumor and cell lines, and this loss was associated with advanced tumor-lymph node-metastasis (TNM) stage and Fuhrman grade. Reintroducing miR-199a-3p in RCC cell lines inhibited proliferation and led to the arrest ofG1 phase by negatively regulating c-Met, thus suppressing HGF/c-Met axis and it's downstream signaling such as Akt, ERK1/2 and mTOR (Huang et al., 2014). These provided a potential target for RCC. Wang et al. reported that miR-32-5p functions by altering the TR4/HGF/Met/MMP2-MMP9 signaling to suppress the clear cell renal cell carcinoma (ccRCC) metastasis in vitro and in vivo. Also, they demonstrated that targeting miR-32-5p/TR4/HGF/Met signaling could enhance the efficacy of current sunitinib-chemotherapy for ccRCC suppression (Wang M. et al., 2018). For lncRNA, Qu et al. first identified a novel lncRNA, called lncARSR (lncRNA Activated in RCC with Sunitinib Resistance), which could promote sunitinib resistance and predict poor response of RCC patients. Moreover, they found that intercellular transfer of lncARSR by exosomes disseminated sunitinib resistance. Mechanically, lncARSR functioned as a ceRNA for miR-34 and miR-449 to promote AXL and c-MET expression (Qu et al., 2016). In addition, overexpression of lncRNA NEAT1 was also observed in RCC cells, and its elevated levels were associated with poor prognosis. The silencing of lncRNA NEAT1 has been shown to inhibit RCC cell proliferation, migration, and invasion by inhibiting cell cycle progression and EMT phenotype. Down-regulation of lncRNA NEAT1 enhanced the sensitivity of RCC cells to sorafenib in vitro. Mechanistic analysis revealed that lncRNA NEAT1 acts as a competitive sponge for miR-34a, therefore preventing inhibition of c-Met (Liu F. et al., 2017). Therefore, ncRNAs and c-Met might serve as a predictor and a potential therapeutic target for RCC.
In thyroid cancer (TC), the crosstalk between ncRNAs and HGF/c-Met axis has a significant effect on tumor growth and progression. Several reports have shown that some miRNAs (such as miR-449b and miRNA-3666) act as tumor suppressors that inhibit TC growth and reduce apoptosis by decreasing the levels of MET protein rather than its transcription (Chen L. et al., 2015; Wang et al., 2016). Moreover, ceRNA plays a role in thyroid cancer progression. Liu et al. also observed a significant increase in lncRNA XIST expression in thyroid cancer tissues and cell lines. Currently, lncRNA XIST is used as an important prognostic predictor for TC patients. Further investigation involving in vitro and in vivo experiments, online datasets, and online predicting tools has revealed that lncRNA XIST serves as a ceRNA for miR-34a by sponging miR-34a, and competing with MET for miR-34a binding, hence modulated thyroid cancer cell proliferation and tumor growth (Liu et al., 2018). These findings provided a foundation for TC therapy improvement based on lncRNA-miRNA-mRNA interaction.
Esophageal squamous cell carcinoma (ESCC) is one of the major histological subtypes of esophageal cancer in China. Currently, there are limited therapeutic options for the disease because of limited understanding of its molecular mechanism. MiR-1 functions as a crucial tumor inhibitor in ESCC (same as its role in HCC, GC, and ovarian cancer). Its overexpression is associated with a decrease in acquisition of MET, cyclin D1, and CDK4 (oncogene and cell cycle-related proteins), which eventually lead to are duction in cell growth and enhanced cell apoptosis in vitro and in vivo (Jiang et al., 2016). The downregulation of miR-206 is associated with ESCC of lymph node metastasis, advanced TNM stage, and overall survival. Furthermore, luciferase reporter gene assay revealed that c-Met is a direct target of miR-206, and ectopic miR-206 expression inhibits ESCC cell proliferation and induces apoptosis by inhibiting c-Met/AKT/mTOR pathway (Zhang J. et al., 2019). Collectively, these findings confirm the role of c-Met in ESCC progression.
Multiple myeloma (MM) is the second most general hematological cancer in globe with poor survival on account of high recurrence (Chen et al., 2019). In 2019, Yang and his team demonstrated that HGF/c-Met axis played an essential role in MM development. They first verified the overexpression of lncRNA UCA1 in MM and clarified its tumor-promoting effect (mainly facilitating proliferation and reducing apoptosis) could be regulated by miR-1271-5p/HGF axis (Yang and Chen, 2019). These indicated that RNA regulation might be a new target for the cure of MM.
Hepatoblastoma (HB) is a highly invasive neoplasm in childhood. In 2019, Cui et al. revealed a vital role of crosstalks between ncRNAs and HGF/c-Met axis in HB. LncRNA ZFAS1 is a snoRNA host gene locating in chromosome 20q13.13. They found that lncRNA ZFAS1 was overexpression in HB tissue and correlated with malignant pathological features and poor prognosis through tissue microarray analysis. Furthermore, they also showed that lncRNA ZFAS1 served as a ceRNA to regulate RALY by sponging miR-193a-3p and played an oncogenic role during HB progression via HGF/c-Met pathway during HB development (Cui et al., 2019). This study expounded a specific molecular mechanism of HB development involving ZFAS1/miR-193a-3p/RALY axis, which could be functioned as a potential biomarker and therapeutic target for HB.
As for retinoblastoma, Yang and his team found that there was a negative correlation between the expression of lncRNA HOTAIR and miR-613 in retinoblastoma tissue compared with normal tissue. And lncRNA HOTAIR could stimulate progression and aggravation of retinoblastoma by miR-613/c-Met axis (Yang G. et al., 2018). Although this study provided great evidences for identifying potential diagnostic and therapeutic target in retinoblastoma, further exploration was in demand.
Diffuse large B-cell lymphoma (DLBCL) is the most frequent lymphoid malignancy all over the world. MET was reported to be upregulated in DLBCL and predicted poor outcomes. Cheng et al. investigated that elevated lncRNA TUG1 expression could facilitate cell proliferation and decrease apoptosis in vitro and in vivo by increasing MET expression through inhibiting its ubiquitination and degradation (Cheng H. et al., 2019). In summary, this study provided a novel insight for DLBCL treatment.
Conclusions and Future Perspectives
In this review, we have summarized the crosstalk between HGF/c-Met axis and ncRNAs in common multiple human cancers, which has improved our understanding on the pathogenetic mechanism of tumor occurrence and development. However, further studies are required to fully explore the interaction between ncRNAs and HGF/c-Met axis in cancer. Despite the evidence presented here, the precise interactions between ncRNAs and HGF/c-Met axis, and how these interactions affect tumorigenesis and progression of cancers are still largely unknown. This implies that additional controlled and large-scale clinical studies are required before cancer-specific ncRNAs and HGF/c-Met inhibitors can be recommended for clinical diagnosis and treatment.
Taken together, deeper understanding of HGF/c-Met axis will reveal beneficial avenues and generate new hypotheses regarding cancer pathogenesis and treatment.
Author Contributions
ZY, ZR, and RS proposed the study. XL, JC, LL, XC, SS, and GC performed the research and wrote the first draft. ZR and ZY are the guarantors. All authors contributed to interpretation of the study and to further drafts.
Funding
This study was supported by the National S&T Major Project of China (2018ZX10301201-008), Natural Science Foundation of China (81702757, 81702346, 81702927, and 81600506), National Key Research and Development Program of China (2018YFC2000500), China Postdoctoral Science Foundation (2017M610463 and 2018M632814). National Engineering Laboratory for Internet Medical System and Application open fund project (NELIMSA2018P03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to all our colleagues and authors whose studies we could not include into this review.
Abbreviations
HGF, hepatocyte growth factor; c-Met, c-mesenchymal-epithelial transition factor; EMT, epithelial-mesenchymal transition; ncRNAs, non-coding RNAs; NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; GC, gastric cancer; DDP, cisplatin; ceRNAs, competing endogenous RNAs; OS, osteosarcoma; BCa, bladder cancer; CRC, colorectal cancer; GBM, glioblastoma; TMZ, temozolomide; SOC, serous ovarian cancer; EC, endometrial cancer; OSCC, oral squamous cell carcinoma; RCC, renal cell carcinoma; ccRCC, clear cell renal cell carcinoma; PDAC, pancreatic ductal adenocarcinoma; PNET, pancreatic neuroendocrine tumors; TC, thyroid cancer; ESCC, esophageal squamous cell carcinoma; MM, multiple myeloma; HB, hepatoblastoma; DLBCL, diffuse large B-cell lymphoma.
References
Acunzo, M., Romano, G., Palmieri, D., Lagana, A., Garofalo, M., Balatti, V., et al. (2013). Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc. Natl. Acad. Sci. U.S.A. 110, 8573–8578. doi: 10.1073/pnas.1302107110
Arnold, L., Enders, J., and Thomas, S. M. (2017). Activated HGF-c-Met axis in head and neck cancer. Cancers 9:E169. doi: 10.3390/cancers9120169
Bach, D. H., and Lee, S. K. (2018). Long noncoding RNAs in cancer cells. Cancer Lett. 419, 152–166. doi: 10.1016/j.canlet.2018.01.053
Balani, S., Nguyen, L. V., and Eaves, C. J. (2017). Modeling the process of human tumorigenesis. Nat. Commun. 8:15422. doi: 10.1038/ncomms15422
Baldanzi, G., and Graziani, A. (2014). Physiological signaling and structure of the HGF receptor MET. Biomedicines 3, 1–31. doi: 10.3390/biomedicines3010001
Bleau, A. M., Redrado, M., Nistal-Villan, E., Villalba, M., Exposito, F., Redin, E., et al. (2018). miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 414, 257–267. doi: 10.1016/j.canlet.2017.11.008
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Breunig, C., Erdem, N., Bott, A., Greiwe, J. F., Reinz, E., Bernhardt, S., et al. (2018). TGF β1 regulates HGF-induced cell migration and hepatocyte growth factor receptor MET expression via C-ets-1 and miR-128-3p in basal-like breast cancer. Mol. Oncol. 12, 1447–1463. doi: 10.1002/1878-0261.12355
Cao, X., Lai, S., Hu, F., Li, G., Wang, G., Luo, X., et al. (2017). miR-19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Sci. Rep. 7:2939. doi: 10.1038/s41598-017-01153-0
Cen, C., Li, J., Liu, J., Yang, M., Zhang, T., Zuo, Y., et al. (2018). Long noncoding RNA LINC01510 promotes the growth of colorectal cancer cells by modulating MET expression. Cancer Cell Int. 18:45. doi: 10.1186/s12935-018-0503-5
Chen, C. T., Kim, H., Liska, D., Gao, S., Christensen, J. G., and Weiser, M. R. (2012). MET activation mediates resistance to lapatinib inhibition of HER2-amplified gastric cancer cells. Mol. Cancer Ther. 11, 660–669. doi: 10.1158/1535-7163.MCT-11-0754
Chen, L., Xu, L., and Wang, G. (2015). Regulation of MET-mediated proliferation of thyroid carcinoma cells by miR-449b. Tumour Biol. 36, 8653–8660. doi: 10.1007/s13277-015-3619-4
Chen, L., Zhang, J., Feng, Y., Li, R., Sun, X., Du, W., et al. (2012). MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem. Cell Biol. 44, 1711–1717. doi: 10.1016/j.biocel.2012.06.027
Chen, Q. Y., Jiao, D. M., Wang, J., Hu, H., Tang, X., Chen, J., et al. (2016a). miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget 7, 24510–24526. doi: 10.18632/oncotarget.8229
Chen, Q. Y., Jiao, D. M., Wu, Y. Q., Chen, J., Wang, J., Tang, X. L., et al. (2016b). MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met /PI3k/Akt/mTOR pathway. Oncotarget 7, 18247–18261. doi: 10.18632/oncotarget.7570
Chen, Q. Y., Jiao, D. M., Yan, L., Wu, Y. Q., Hu, H. Z., Song, J., et al. (2015). Comprehensive gene and microRNA expression profiling reveals miR-206 inhibits MET in lung cancer metastasis. Mol. Biosyst. 11, 2290–2302. doi: 10.1039/c4mb00734d
Chen, R., Zhang, X., and Wang, C. (2019). LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J. Cell. Biochem. doi: 10.1002/jcb.29573
Chen, S., and Xia, X. (2019). Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. J. Cell. Physiol. 234, 14999–15009. doi: 10.1002/jcp.27567
Cheng, H., Yan, Z., Wang, X., Cao, J., Chen, W., Qi, K., et al. (2019). Downregulation of long non-coding RNA TUG1 suppresses tumor growth by promoting ubiquitination of MET in diffuse large B-cell lymphoma. Mol. Cell. Biochem. 461, 47–56. doi: 10.1007/s11010-019-03588-7
Cheng, J., Wu, L. M., Deng, X. S., Wu, J., Lv, Z., Zhao, H. F., et al. (2018). MicroRNA-449a suppresses hepatocellular carcinoma cell growth via G1 phase arrest and the HGF/MET c-Met pathway. Hepatobiliary Pancreat. Dis. Int. 17, 336–344. doi: 10.1016/j.hbpd.2018.07.006
Cheng, Y., Yang, M., and Peng, J. (2019). Correlation the between the regulation of miRNA-1 in c-Met-induced EMT and cervical cancer progression. Oncol. Lett. 17, 3341–3349. doi: 10.3892/ol.2019.9971
Chiyomaru, T., Seki, N., Inoguchi, S., Ishihara, T., Mataki, H., Matsushita, R., et al. (2015). Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int. J. Oncol. 46, 487–496. doi: 10.3892/ijo.2014.2752
Colditz, G. A., and Bohlke, K. (2014). Priorities for the primary prevention of breast cancer. CA Cancer J. Clin. 64, 186–194. doi: 10.3322/caac.21225
Cui, X., Wang, Z., Liu, L., Liu, X., Zhang, D., Li, J., et al. (2019). The Long Non-coding RNA ZFAS1 sponges miR-193a-3p to modulate hepatoblastoma growth by targeting RALY via HGF/c-Met pathway. Front. Cell Dev. Biol. 7:271. doi: 10.3389/fcell.2019.00271
Dai, C., Xie, Y., Zhuang, X., and Yuan, Z. (2018). MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed. Pharmacother. 104, 763–770. doi: 10.1016/j.biopha.2018.05.077
De Smet, E. G., Mestdagh, P., Vandesompele, J., Brusselle, G. G., and Bracke, K. R. (2015). Non-coding RNAs in the pathogenesis of COPD. Thorax 70, 782–791. doi: 10.1136/thoraxjnl-2014-206560
Dong, J., Teng, F., Guo, W., Yang, J., Ding, G., and Fu, Z. (2018). lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell. Physiol. Biochem. 51, 2262–2274. doi: 10.1159/000495871
Du, M., Wang, J., Chen, H., Wang, S., Chen, L., Xu, Y., et al. (2019). MicroRNA200a suppresses migration and invasion and enhances the radiosensitivity of NSCLC cells by inhibiting the HGF/cMet signaling pathway. Oncol. Rep. 41, 1497–1508. doi: 10.3892/or.2018.6925
Duan, Z., Choy, E., Harmon, D., Liu, X., Susa, M., Mankin, H., et al. (2011). MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol. Cancer Ther. 10, 1337–1345. doi: 10.1158/1535-7163.mct-11-0096
Ebens, A., Brose, K., Leonardo, E. D., Hanson, M. G. Jr., Bladt, F., Birchmeier, C., et al. (1996). Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron 17, 1157–1172. doi: 10.1016/s0896-6273(00)80247-0
Eisenhauer, E. A. (2017). Real-world evidence in the treatment of ovarian cancer. Ann. Oncol. 28(Suppl. 8), viii61–viii65. doi: 10.1093/annonc/mdx443
Ferracini, R., Di Renzo, M. F., Scotlandi, K., Baldini, N., Olivero, M., Lollini, P., et al. (1995). The Met/HGF receptor is over-expressed in human osteosarcomas and is activated by either a paracrine or an autocrine circuit. Oncogene 10, 739–749.
Fischer, O. M., Giordano, S., Comoglio, P. M., and Ullrich, A. (2004). Reactive oxygen species mediate Met receptor transactivation by G protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J. Biol. Chem. 279, 28970–28978. doi: 10.1074/jbc.M402508200
Fornari, F., Milazzo, M., Chieco, P., Negrini, M., Calin, G. A., Grazi, G. L., et al. (2010). MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 70, 5184–5193. doi: 10.1158/0008-5472.can-10-0145
Fukumoto, I., Koshizuka, K., Hanazawa, T., Kikkawa, N., Matsushita, R., Kurozumi, A., et al. (2016). The tumor-suppressive microRNA-23b/27b cluster regulates the MET oncogene in oral squamous cell carcinoma. Int. J. Oncol. 49, 1119–1129. doi: 10.3892/ijo.2016.3602
Fukushima, T., Uchiyama, S., Tanaka, H., and Kataoka, H. (2018). Hepatocyte growth factor activator: a proteinase linking tissue injury with repair. Int. J. Mol. Sci. 19:E3435. doi: 10.3390/ijms19113435
Furge, K. A., Zhang, Y. W., and Vande Woude, G. F. (2000). Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene 19, 5582–5589. doi: 10.1038/sj.onc.1203859
Gao, S., Zhao, Z., Wu, R., Wu, L., Tian, X., and Zhang, Z. (2019). MiR-1 inhibits prostate cancer PC3 cells proliferation through the Akt/mTOR signaling pathway by binding to c-Met. Biomed. Pharmacother. 109, 1406–1410. doi: 10.1016/j.biopha.2018.10.098
Gao, Y., Zeng, F., Wu, J. Y., Li, H. Y., Fan, J. J., Mai, L., et al. (2015). MiR-335 inhibits migration of breast cancer cells through targeting oncoprotein c-Met. Tumour Biol. 36, 2875–2883. doi: 10.1007/s13277-014-2917-6
Garcia-Vilas, J. A., and Medina, M. A. (2018). Updates on the hepatocyte growth factor/c-Met axis in hepatocellular carcinoma and its therapeutic implications. World J. Gastroenterol. 24, 3695–3708. doi: 10.3748/wjg.v24.i33.3695
Ghosh, A., Dasgupta, D., Ghosh, A., Roychoudhury, S., Kumar, D., Gorain, M., et al. (2017). MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 8:e2706. doi: 10.1038/cddis.2017.123
Guo, Y., Tao, M., and Jiang, M. (2018). MicroRNA-454-3p inhibits cervical cancer cell invasion and migration by targeting c-Met. Exp. Ther. Med. 15, 2301–2306. doi: 10.3892/etm.2018.5714
Hagman, Z., Haflidadottir, B. S., Ansari, M., Persson, M., Bjartell, A., Edsjo, A., et al. (2013). The tumour suppressor miR-34c targets MET in prostate cancer cells. Br. J. Cancer 109, 1271–1278. doi: 10.1038/bjc.2013.449
Han, C., Zhou, Y., An, Q., Li, F., Li, D., Zhang, X., et al. (2015). MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol. 36, 6715–6723. doi: 10.1007/s13277-015-3358-6
Han, Y., Luo, Y., Wang, Y., Chen, Y., Li, M., and Jiang, Y. (2016). Hepatocyte growth factor increases the invasive potential of PC-3 human prostate cancer cells via an ERK/MAPK and Zeb-1 signaling pathway. Oncol. Lett. 11, 753–759. doi: 10.3892/ol.2015.3943
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Herrero, C., de la Fuente, A., Casas-Arozamena, C., and Sebastian, V. (2019). Extracellular vesicles-based biomarkers represent a promising liquid biopsy in endometrial cancer. Cancers 11:E2000. doi: 10.3390/cancers11122000
Higashio, K., Shima, N., Goto, M., Itagaki, Y., Nagao, M., Yasuda, H., et al. (1990). Identity of a tumor cytotoxic factor from human fibroblasts and hepatocyte growth factor. Biochem. Biophys. Res. Commun. 170, 397–404. doi: 10.1016/0006-291x(90)91287-3
Hong, J. H., Roh, K. S., Suh, S. S., Lee, S., Sung, S. W., Park, J. K., et al. (2015). The expression of microRNA-34a is inversely correlated with c-MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients. Tumour Biol. 36, 9327–9337. doi: 10.1007/s13277-015-3428-9
Hu, Z., Lin, Y., Chen, H., Mao, Y., Wu, J., Zhu, Y., et al. (2013). MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem. Biophys. Res. Commun. 435, 82–87. doi: 10.1016/j.bbrc.2013.04.042
Huang, J., Dong, B., Zhang, J., Kong, W., Chen, Y., Xue, W., et al. (2014). miR-199a-3p inhibits hepatocyte growth factor/c-Met signaling in renal cancer carcinoma. Tumour Biol. 35, 5833–5843. doi: 10.1007/s13277-014-1774-7
Huang, W. C., Jang, T. H., Tung, S. L., Yen, T. C., Chan, S. H., and Wang, L. H. (2019). A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing beta5-integrin/c-met signaling pathway. J. Exp. Clin. Cancer Res. 38:89. doi: 10.1186/s13046-019-1091-5
Ji, T. T., Huang, X., Jin, J., Pan, S. H., and Zhuge, X. J. (2016). Inhibition of long non-coding RNA TUG1 on gastric cancer cell transference and invasion through regulating and controlling the expression of miR-144/c-Met axis. Asian Pac. J. Trop. Med. 9, 508–512. doi: 10.1016/j.apjtm.2016.03.026
Jiang, S., Zhao, C., Yang, X., Li, X., Pan, Q., Huang, H., et al. (2016). miR-1 suppresses the growth of esophageal squamous cell carcinoma in vivo and in vitro through the downregulation of MET, cyclin D1 and CDK4 expression. Int. J. Mol. Med. 38, 113–122. doi: 10.3892/ijmm.2016.2619
Kansara, M., and Thomson, K. (2019). Infiltrating myeloid cells drive osteosarcoma progression via GRM4 regulation of IL23. 9, 1511–1519. doi: 10.1158/2159-8290.cd-19-0154
Korhan, P., Erdal, E., and Atabey, N. (2014). MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-Met. Biochem. Biophys. Res. Commun. 450, 1304–1312. doi: 10.1016/j.bbrc.2014.06.142
Lan, F., Yu, H., Hu, M., Xia, T., and Yue, X. (2015). miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J. Neurochem. 135, 274–286. doi: 10.1111/jnc.13272
Lee, J. M., Yoo, J. K., Yoo, H., Jung, H. Y., Lee, D. R., Jeong, H. C., et al. (2013). The novel miR-7515 decreases the proliferation and migration of human lung cancer cells by targeting c-Met. Mol. Cancer Res. 11, 43–53. doi: 10.1158/1541-7786.MCR-12-0355
Li, H., Hong, J., and Wijayakulathilaka, W. (2019). Long non-coding RNA SNHG4 promotes cervical cancer progression through regulating c-Met via targeting miR-148a-3p. Cell Cycle 18, 3313–3324. doi: 10.1080/15384101.2019.1674071
Li, J., Xu, X., Meng, S., Liang, Z., Wang, X., Xu, M., et al. (2017). MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial-mesenchymal transition progression in bladder cancer. Cell Death Dis. 8:e3010. doi: 10.1038/cddis.2017.331
Li, M., Li, Z., Wang, X., Wang, Y., Zhao, C., and Wang, L. (2018). Function of miR152 as tumor suppressor in oral squamous cell carcinoma cells by targeting cMET. Oncol. Rep. 39, 1173–1180. doi: 10.3892/or.2017.6157
Li, N., Fu, H., Tie, Y., Hu, Z., Kong, W., Wu, Y., et al. (2009). miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 275, 44–53. doi: 10.1016/j.canlet.2008.09.035
Li, S., Zhang, H., Wang, X., Qu, Y., Duan, J., Liu, R., et al. (2016). Direct targeting of HGF by miR-16 regulates proliferation and migration in gastric cancer. Tumour Biol. 37, 15175–15183. doi: 10.1007/s13277-016-5390-6
Li, Y., Guessous, F., Zhang, Y., Dipierro, C., Kefas, B., Johnson, E., et al. (2009). MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576. doi: 10.1158/0008-5472.CAN-09-0529
Li, Y., Zhang, H., Li, Y., Zhao, C., Fan, Y., Liu, J., et al. (2018). MiR-182 inhibits the epithelial to mesenchymal transition and metastasis of lung cancer cells by targeting the Met gene. Mol. Carcinog. 57, 125–136. doi: 10.1002/mc.22741
Li, Z., Yanfang, W., Li, J., Jiang, P., Peng, T., Chen, K., et al. (2018). Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 432, 237–250. doi: 10.1016/j.canlet.2018.04.035
Liu, F., Chen, N., Gong, Y., Xiao, R., Wang, W., and Pan, Z. (2017). The long non-coding RNA NEAT1 enhances epithelial-to-mesenchymal transition and chemoresistance via the miR-34a/c-Met axis in renal cell carcinoma. Oncotarget 8, 62927–62938. doi: 10.18632/oncotarget.17757
Liu, H., Deng, H., Zhao, Y., Li, C., and Liang, Y. (2018). LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 37, 279. doi: 10.1186/s13046-018-0950-9
Liu, J., Xue, H., Zhang, J., Suo, T., Xiang, Y., Zhang, W., et al. (2015). MicroRNA-144 inhibits the metastasis of gastric cancer by targeting MET expression. J. Exp. Clin. Cancer Res. 34:35. doi: 10.1186/s13046-015-0154-5
Liu, Q., Yang, G., and Qian, Y. (2017). Loss of microRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol. Carcinog. 56, 1312–1321. doi: 10.1002/mc.22593
Liu, Y., Tan, J., Ou, S., Chen, J., and Chen, L. (2019). MicroRNA-101-3p suppresses proliferation and migration in hepatocellular carcinoma by targeting the HGF/c-Met pathway. Invest. New Drugs. doi: 10.1007/s10637-019-00766-8
Liu, Y. W., Sun, M., Xia, R., Zhang, E. B., Liu, X. H., Zhang, Z. H., et al. (2015). LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 6:e1802. doi: 10.1038/cddis.2015.150
Luo, W., Huang, B., Li, Z., Li, H., Sun, L., Zhang, Q., et al. (2013). MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS ONE 8:e64759. doi: 10.1371/journal.pone.0064759
Luo, Y., Ouyang, J., Zhou, D., Zhong, S., Wen, M., Ou, W., et al. (2018). Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Dig. Dis. Sci. 63, 890–899. doi: 10.1007/s10620-018-4915-9
Martianov, I., Ramadass, A., Serra Barros, A., Chow, N., and Akoulitchev, A. (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670. doi: 10.1038/nature05519
Migliore, C., and Giordano, S. (2008). Molecular cancer therapy: can our expectation be MET? Eur J Cancer 44, 641–651. doi: 10.1016/j.ejca.2008.01.022
Migliore, C., Martin, V., Leoni, V. P., Restivo, A., Atzori, L., Petrelli, A., et al. (2012). MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin. Cancer Res. 18, 737–747. doi: 10.1158/1078-0432.CCR-11-1699
Modali, S. D., Parekh, V. I., Kebebew, E., and Agarwal, S. K. (2015). Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol. Endocrinol. 29, 224–237. doi: 10.1210/me.2014-1304
Niu, G., Li, B., Sun, J., and Sun, L. (2015). miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 48, 348–355. doi: 10.1111/cpr.12187
Ohta, K., Hoshino, H., Wang, J., Ono, S., Iida, Y., Hata, K., et al. (2015). MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget 6, 3211–3224. doi: 10.18632/oncotarget.3085
Organ, S. L., and Tsao, M. S. (2011). An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 3, S7–s19. doi: 10.1177/1758834011422556
Peng, Y., Guo, J. J., Liu, Y. M., and Wu, X. L. (2014). MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci. Rep. 34:e00112. doi: 10.1042/BSR20140020
Qiu, J. J., Lin, Y. Y., Ding, J. X., Feng, W. W., Jin, H. Y., and Hua, K. Q. (2015). Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int. J. Oncol. 46, 2497–2505. doi: 10.3892/ijo.2015.2943
Qu, L., Ding, J., Chen, C., Wu, Z. J., Liu, B., Gao, Y., et al. (2016). Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668. doi: 10.1016/j.ccell.2016.03.004
Qu, W., Chen, X., Wang, J., Lv, J., and Yan, D. (2017). MicroRNA-1 inhibits ovarian cancer cell proliferation and migration through c-Met pathway. Clin. Chim. Acta 473, 237–244. doi: 10.1016/j.cca.2017.07.008
Salvi, A., Sabelli, C., Moncini, S., Venturin, M., Arici, B., Riva, P., et al. (2009). MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 276, 2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x
Scagliotti, G. V., Novello, S., and von Pawel, J. (2013). The emerging role of MET/HGF inhibitors in oncology. Cancer Treat. Rev. 39, 793–801. doi: 10.1016/j.ctrv.2013.02.001
Siemens, H., Neumann, J., Jackstadt, R., Mansmann, U., Horst, D., Kirchner, T., et al. (2013). Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 19, 710–720. doi: 10.1158/1078-0432.CCR-12-1703
Slack, F. J., and Chinnaiyan, A. M. (2019). The role of non-coding RNAs in oncology. Cell 179, 1033–1055. doi: 10.1016/j.cell.2019.10.017
Song, H., Song, J., Lu, L., and Li, S. (2019). SNHG8 is upregulated in esophageal squamous cell carcinoma and directly sponges microRNA-411 to increase oncogenicity by upregulating KPNA2. Onco. Targets. Ther. 12, 6991–7004. doi: 10.2147/ott.s214881
Song, Y., Zou, L., Li, J., Shen, Z. P., Cai, Y. L., and Wu, X. D. (2018). LncRNA SNHG8 promotes the development and chemo-resistance of pancreatic adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 22, 8161–8168. doi: 10.26355/eurrev_201812_16508
Sturm, D., Pfister, S. M., and Jones, D. T. W. (2017). Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 35, 2370–2377. doi: 10.1200/jco.2017.73.0242
Su, W., Feng, S., Chen, X., Yang, X., Mao, R., Guo, C., et al. (2018). Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 78, 3207–3219. doi: 10.1158/0008-5472.CAN-18-0222
Sun, R. C., Dukhande, V. V., Zhou, Z., Young, L. E. A., Emanuelle, S., Brainson, C. F., et al. (2019). Nuclear glycogenolysis modulates histone acetylation in human non-small cell lung cancers. Cell Metab. 30, 903–916.e7. doi: 10.1016/j.cmet.2019.08.014
Sun, X., Liu, S., Chen, P., Fu, D., Hou, Y., Hu, J., et al. (2017). miR-449a inhibits colorectal cancer progression by targeting SATB2. Oncotarget 8, 100975–100988. doi: 10.18632/oncotarget.10900
Sun, Z., Zhang, T., and Chen, B. (2019). Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes proliferation and metastasis of osteosarcoma cells by targeting c-Met and SOX4 via miR-34a/c-5p and miR-449a/b. Med. Sci. Monit. 25, 1410–1422. doi: 10.12659/msm.912703
Sun, Z. Y., Jian, Y. K., Zhu, H. Y., and Li, B. (2019). lncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to gemcitabine through activating c-MET/PI3K/AKT pathway. Pathol. Res. Pract. 215, 555–563. doi: 10.1016/j.prp.2018.12.013
Tan, S., Li, R., Ding, K., Lobie, P. E., and Zhu, T. (2011). miR-198 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the HGF/c-MET pathway. FEBS Lett. 585, 2229–2234. doi: 10.1016/j.febslet.2011.05.042
Testini, C., Smith, R. O., Jin, Y., Martinsson, P., Sun, Y., Hedlund, M., et al. (2019). Myc-dependent endothelial proliferation is controlled by phosphotyrosine 1212 in VEGF receptor-2. 20:e47845. doi: 10.15252/embr.201947845
Tomihara, H., Yamada, D., Eguchi, H., Iwagami, Y., Noda, T., Asaoka, T., et al. (2017). MicroRNA-181b-5p, ETS1, and the c-Met pathway exacerbate the prognosis of pancreatic ductal adenocarcinoma after radiation therapy. Cancer Sci. 108, 398–407. doi: 10.1111/cas.13159
Wan, L., Zhu, L., Xu, J., Lu, B., Yang, Y., Liu, F., et al. (2014). MicroRNA-409-3p functions as a tumor suppressor in human lung adenocarcinoma by targeting c-Met. Cell. Physiol. Biochem. 34, 1273–1290. doi: 10.1159/000366337
Wang, G., Cai, C., and Chen, L. (2016). MicroRNA-3666 regulates thyroid carcinoma cell proliferation via MET. Cell. Physiol. Biochem. 38, 1030–1039. doi: 10.1159/000443054
Wang, G., Liu, W., Zou, Y., Wang, G., Deng, Y., Luo, J., et al. (2019). Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 40, 432–445. doi: 10.1016/j.ebiom.2018.12.062
Wang, L. G., Ni, Y., Su, B. H., Mu, X. R., Shen, H. C., and Du, J. J. (2013). MicroRNA-34b functions as a tumor suppressor and acts as a nodal point in the feedback loop with Met. Int. J. Oncol. 42, 957–962. doi: 10.3892/ijo.2013.1767
Wang, M., Sun, Y., Xu, J., Lu, J., Wang, K., Yang, D. R., et al. (2018). Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int. J. Cancer 143, 100–112. doi: 10.1002/ijc.31289
Wang, W., Dong, J., Wang, M., Yao, S., Tian, X., Cui, X., et al. (2018). miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol. Lett. 15, 6131–6136. doi: 10.3892/ol.2018.8110
Wang, Y., Tai, Q., Zhang, J., Kang, J., Gao, F., Zhong, F., et al. (2019). MiRNA-206 inhibits hepatocellular carcinoma cell proliferation and migration but promotes apoptosis by modulating cMET expression. Acta Biochim. Biophys. Sin. 51, 243–253. doi: 10.1093/abbs/gmy119
Wei, B., Huang, Q. Y., Huang, S. R., Mai, W., and Zhong, X. G. (2015). MicroRNA34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol. Med. Rep. 12, 5255–5261. doi: 10.3892/mmr.2015.4110
Wu, P., Cai, J., Chen, Q., Han, B., Meng, X., Li, Y., et al. (2019). Lnc-TALC promotes O(6)-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 10:2045. doi: 10.1038/s41467-019-10025-2
Wu, Y., Wan, X., Ji, F., Song, Z., and Fang, X. (2018). Serum miR-658 induces metastasis of gastric cancer by activating PAX3-MET pathway: a population-based study. Cancer Biomark. 22, 111–118. doi: 10.3233/CBM-171045
Wu, Z. S., Wu, Q., Wang, C. Q., Wang, X. N., Huang, J., Zhao, J. J., et al. (2011). miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer 117, 2842–2852. doi: 10.1002/cncr.25860
Xing, F., Liu, Y., Wu, S. Y., Wu, K., Sharma, S., Mo, Y. Y., et al. (2018). Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78, 4316–4330. doi: 10.1158/0008-5472.can-18-1102
Xu, P., Li, Y., Yang, S., Yang, H., Tang, J., and Li, M. (2015). Micro-ribonucleic acid 143 (MiR-143) inhibits oral squamous cell carcinoma (OSCC) cell migration and invasion by downregulation of phospho-c-Met through targeting CD44 v3. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 120, 43–51. doi: 10.1016/j.oooo.2015.02.486
Xu, X., Chen, H., Lin, Y., Hu, Z., Mao, Y., Wu, J., et al. (2013). MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol. Cells 36, 62–68. doi: 10.1007/s10059-013-0044-7
Xu, X., Zhu, Y., Liang, Z., Li, S., Xu, X., Wang, X., et al. (2016). c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3beta/Snail signaling. Cell Death Dis. 7:e2088. doi: 10.1038/cddis.2015.274
Yang, C. H., Zhang, X. Y., Zhou, L. N., Wan, Y., Song, L. L., Gu, W. L., et al. (2018). LncRNA SNHG8 participates in the development of endometrial carcinoma through regulating c-MET expression by miR-152. Eur. Rev. Med. Pharmacol. Sci. 22, 1629–1637. doi: 10.26355/eurrev_201803_14698
Yang, G., Fu, Y., Lu, X., Wang, M., Dong, H., and Li, Q. (2018). LncRNA HOTAIR/miR-613/c-met axis modulated epithelial-mesenchymal transition of retinoblastoma cells. J. Cell. Mol. Med. 22, 5083–5096. doi: 10.1111/jcmm.13796
Yang, X., Zhang, X. F., Lu, X., Jia, H. L., Liang, L., Dong, Q. Z., et al. (2014). MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology 59, 1874–1885. doi: 10.1002/hep.26941
Yang, Y., and Chen, L. (2019). Downregulation of lncRNA UCA1 facilitates apoptosis and reduces proliferation in multiple myeloma via regulation of the miR-1271-5p/HGF axis. J. Chin. Med. Assoc. 82, 699–709. doi: 10.1097/JCMA.0000000000000145
Yue, D., and Qin, X. (2019). miR-182 regulates trastuzumab resistance by targeting MET in breast cancer cells. Cancer Gene Ther. 26, 1–10. doi: 10.1038/s41417-018-0031-4
Zhan, F. B., Zhang, X. W., Feng, S. L., Cheng, J., Zhang, Y., Li, B., et al. (2019). MicroRNA-206 reduces osteosarcoma cell malignancy in vitro by targeting the PAX3-MET axis. Yonsei Med. J. 60, 163–173. doi: 10.3349/ymj.2019.60.2.163
Zhang, J., Fa, X., and Zhang, Q. (2019). MicroRNA206 exerts antioncogenic functions in esophageal squamous cell carcinoma by suppressing the cMet/AKT/mTOR pathway. Mol. Med. Rep. 19, 1491–1500. doi: 10.3892/mmr.2018.9775
Zhang, J., Feng, S., Su, W., Bai, S., Xiao, L., Wang, L., et al. (2017). Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci. Rep. 7:42819. doi: 10.1038/srep42819
Zhang, J. P., Zeng, C., Xu, L., Gong, J., Fang, J. H., and Zhuang, S. M. (2014). MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene 33, 4069–4076. doi: 10.1038/onc.2013.369
Zhang, L., Xia, L., Zhao, L., Chen, Z., Shang, X., Xin, J., et al. (2015). Activation of PAX3-MET pathways due to miR-206 loss promotes gastric cancer metastasis. Carcinogenesis 36, 390–399. doi: 10.1093/carcin/bgv009
Zhang, P., Li, S., Chen, Z., Lu, Y., and Zhang, H. (2019). LncRNA SNHG8 promotes proliferation and invasion of gastric cancer cells by targeting the miR-491/PDGFRA axis. Hum. Cell. 33, 123–130. doi: 10.1007/s13577-019-00290-0
Zhang, Y., Xia, M., Jin, K., Wang, S., Wei, H., Fan, C., et al. (2018). Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol. Cancer 17:45. doi: 10.1186/s12943-018-0796-y
Zhang, Z., Kong, Y., Yang, W., Ma, F., Zhang, Y., Ji, S., et al. (2016). Upregulation of microRNA-34a enhances the DDP sensitivity of gastric cancer cells by modulating proliferation and apoptosis via targeting MET. Oncol. Rep. 36, 2391–2397. doi: 10.3892/or.2016.5016
Zhen, L., Yun-Hui, L., Hong-Yu, D., Jun, M., and Yi-Long, Y. (2016). Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 37, 673–683. doi: 10.1007/s13277-015-3843-y
Zhen, Q., Liu, J., Gao, L., Liu, J., Wang, R., Chu, W., et al. (2015). MicroRNA-200a targets EGFR and c-Met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. Cytogenet. Genome Res. 146, 1–8. doi: 10.1159/000434741
Zheng, Z., Yan, D., Chen, X., Huang, H., Chen, K., Li, G., et al. (2015). MicroRNA-206: effective inhibition of gastric cancer progression through the c-Met pathway. PLoS ONE 10:e0128751. doi: 10.1371/journal.pone.0128751
Zhong, M. E., Chen, Y., Zhang, G., Xu, L., Ge, W., and Wu, B. (2019). LncRNA H19 regulates PI3K-Akt signal pathway by functioning as a ceRNA and predicts poor prognosis in colorectal cancer: integrative analysis of dysregulated ncRNA-associated ceRNA network. Cancer Cell Int. 19:148. doi: 10.1186/s12935-019-0866-2
Zhou, H., Liu, Y., Xiao, L., Hu, Z., and Xia, K. (2017). Overexpression of MicroRNA-27b inhibits proliferation, migration, and invasion via suppression of MET expression. Oncol. Res. 25, 147–154. doi: 10.3727/096504016x14732772150505
Zhou, Y. M., Liu, J., and Sun, W. (2014). MiR-130a overcomes gefitinib resistance by targeting met in non-small cell lung cancer cell lines. Asian Pac. J. Cancer Prev. 15, 1391–1396. doi: 10.7314/apjcp.2014.15.3.1391
Keywords: non-coding RNA, HGF/c-Met axis, cancer, crosstalk, mechanism
Citation: Liu X, Sun R, Chen J, Liu L, Cui X, Shen S, Cui G, Ren Z and Yu Z (2020) Crosstalk Mechanisms Between HGF/c-Met Axis and ncRNAs in Malignancy. Front. Cell Dev. Biol. 8:23. doi: 10.3389/fcell.2020.00023
Received: 20 November 2019; Accepted: 13 January 2020;
Published: 31 January 2020.
Edited by:
Pedro M. Fernández-Salguero, University of Extremadura, SpainReviewed by:
Qiulun Lu, Nanjing Medical University, ChinaWei-Hsiung Yang, Mercer University, United States
Copyright © 2020 Liu, Sun, Chen, Liu, Cui, Shen, Cui, Ren and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Ren, ZmNjcmVuemdAenp1LmVkdS5jbg==; Zujiang Yu, am9obnl1ZW1Aenp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Xin Liu
Xin Liu Ranran Sun
Ranran Sun Jianan Chen1,2†
Jianan Chen1,2† Zhigang Ren
Zhigang Ren Zujiang Yu
Zujiang Yu