- 1Laboratory for Reproductive Health, Institute of Biomedicine and Biotechnology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences (CAS), Shenzhen, China
- 2Chinese Academy of Sciences Key Laboratory of Insect Developmental and Evolutionary Biology, Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN, United States
- 5Institute of Medicine and Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, China
Cilia and flagella are conserved subcellular organelles, which arise from centrioles and play critical roles in development and reproduction of eukaryotes. Dysfunction of cilia leads to life-threatening ciliopathies. HYLS1 is an evolutionarily conserved centriole protein, which is critical for ciliogenesis, and its mutation causes ciliopathy–hydrolethalus syndrome. However, the molecular function of HYLS1 remains elusive. Here, we investigated the function of HYLS1 in cilia formation using the Drosophila model. We demonstrated that Drosophila HYLS1 is a conserved centriole and basal body protein. Deletion of HYLS1 led to sensory cilia dysfunction and spermatogenesis abnormality. Importantly, we found that Drosophila HYLS1 is essential for giant centriole/basal body elongation in spermatocytes and is required for spermatocyte centriole to efficiently recruit pericentriolar material and for spermatids to assemble the proximal centriole-like structure (the precursor of the second centriole for zygote division). Hence, by taking advantage of the giant centriole/basal body of Drosophila spermatocyte, we uncover previously uncharacterized roles of HYLS1 in centriole elongation and assembly.
Introduction
Centrioles are small cylindrical cellular organelles typically composed of ninefold symmetric triplet microtubules, which perform two important functions, building centrosome and templating cilia (Bettencourt-Dias and Glover, 2007; Nigg and Stearns, 2011; Conduit et al., 2015). Centrioles recruit pericentriolar material (PCM) to form centrosomes that function as the microtubule-organizing center to organize the mitotic/meiotic spindles. In quiescent cells, centrioles dock to the cell membrane to function as basal bodies to initiate the biogenesis of cilia. Cilia are critical sensory or motile organelles that regulate development and reproduction of eukaryotes (Goetz and Anderson, 2010; Bettencourt-Dias et al., 2011; Malicki and Johnson, 2017; Anvarian et al., 2019). Dysfunction of cilia results in dozens of human genetic diseases, collectively termed ciliopathies (Hildebrandt et al., 2011; Reiter and Leroux, 2017).
In vertebrates, only the mother centriole, but not the daughter centriole, has the ability to form cilia, because only the mother centriole has distal appendage (DA) structures, which is critical for ciliogenesis initiation by mediating centrioles membrane docking. During ciliogenesis, DAs are converted into transition fibers (TFs), one of the conspicuous structures at the cilia base. In invertebrates, such as Caenorhabditis elegans (C. elegans) and Drosophila, centrioles lack DAs, but TFs, at least functional homologous structures, are formed during ciliogenesis. Transition fibers, together with the transition zone (TZ), act as the ciliary gate to control the ciliary protein entry in the context of cilia (Reiter et al., 2012; Takao and Verhey, 2016; Garcia-Gonzalo and Reiter, 2017). The TZ functions as a diffusion barrier to gate the ciliary compartment (Reiter et al., 2012; Garcia-Gonzalo and Reiter, 2017), whereas the TF promotes the import of ciliary proteins. FBF1 is the key TF protein that mediates the entry of ciliary proteins, including intraflagellar transport (IFT) complexes, which are essential for ciliary axonemal elongation (Wei et al., 2013, 2015; Yang et al., 2018). Despite that great progress has been made in understanding the molecular components of DA/TF and their function in ciliogenesis, how such appendages are uniquely assembled from the distal end of mother centriole remains largely as an unsolved mystery (Tanos et al., 2013; Ye et al., 2014; Yang et al., 2018).
Hydrolethalus syndrome (HLS) is a rare recessive lethal inherited disorder that causes serious defects in fetal development and results in birth defects, with its causal gene HYLS1 first identified in 2005 (Mee et al., 2005). Its function was first studied in detail using C. elegans as a model organism. Dammermann et al. (2009) found that HYLS1 is a centriole protein that is recruited to the outer centriole wall via direct interaction with the core centriolar protein SAS-4/CPAP. Unexpectedly, HYLS1 is dispensable for centriole assembly, cell division, and embryonic viability in worm, but it is specifically involved in cilia formation in C. elegans (Dammermann et al., 2009). Subsequently, we demonstrated that HYLS1 mediates ciliogenesis by regulating the formation of the ciliary gate, plays a major role in recruiting the TF protein FBF1, and plays a minor role in TZ assembly (Wei et al., 2016).
Because of the degeneration of basal bodies after ciliogenesis in C. elegans (Sharma et al., 2016), our understanding of how HYLS1 regulates the ciliary gate is limited. Here we sought to address this question using the Drosophila model organism. We showed that HYLS1 is a conserved centriole and basal body protein, which is required for ciliogenesis and spermatogenesis in Drosophila. Deletion of HYLS1 compromised the localization of ciliary gate proteins in both sensory cilia and spermatocyte cilia. Importantly, by taking advantage of the giant centriole (GC)/basal body in spermatocytes, an excellent model to visualize the dynamic change from centriole to basal body conversion, we found that HYLS1 is critical for the elongation of the GC. In addition, we found that fly HYLS1 is required for efficient PCM components recruitment in spermatocyte and the proximal centriole-like structure (PCL) formation in spermatids. Our findings reveal novel roles of HYLS1 in centriole elongation and assembly during Drosophila spermatogenesis, suggesting that the abnormal ciliary gating associated with hyls1 mutation may arise as a consequential outcome from HYLS1-related distal centriole assembly defects.
Results
Drosophila HYLS1 Is a Conserved Centriole and Basal Body Protein
Reciprocal protein homology queries using BLAST identified CG42231 as the only annotated gene in Drosophila genome with significant homology comparing with mammalian HYLS1 (Supplementary Figure S1). Because CG42231 has never been studied, we named it as HYLS1 hereafter and used genetic manipulations to study its function.
To determine if the centriole/basal body localization of HYLS1 is conserved in Drosophila, we created transgenic flies expressing a green fluorescent protein (GFP) tagged HYLS1 transgene and generated the anti-HYLS1 antibody. The antibody was validated by the following facts: anti-HYLS1 staining showed similar localization pattern as HYLS1-GFP (Figure 1 and Supplementary Figure S2); the staining signal was completely lost in hyls1 deletion mutants (Figure 3D). As demonstrated by both HYLS1-GFP and anti-HYLS1 staining, HYLS1 colocalized with the centrosome marker γ-tubulin or ANA1 in early syncytial stage embryos of Drosophila (Figure 1A and Supplementary Figure S2A), indicating that it is indeed a centriole protein. In Drosophila, only Type I monodendritic sensory neurons of the peripheral nervous system (PNS) and sperm cells are ciliated. Type I sensory neurons include external sensory (Es) and chordotonal (Ch) neurons (Jarman, 2002; Boekhoff-Falk, 2005; Kernan, 2007). In both types of ciliated neurons, co-staining of HYLS1-GFP or anti-HYLS1 with the ciliary marker 21A6/EYS (Vieillard et al., 2015) showed that HYLS1 localized to the cilia base (Figure 1B and Supplementary Figure S2B). By colabeling ectopic HYLS1-GFP with the centriole marker ANA1, we observed that HYLS1 completely overlapped with ANA1 in both the distal centriole and the proximal centriole (Figure 1B).
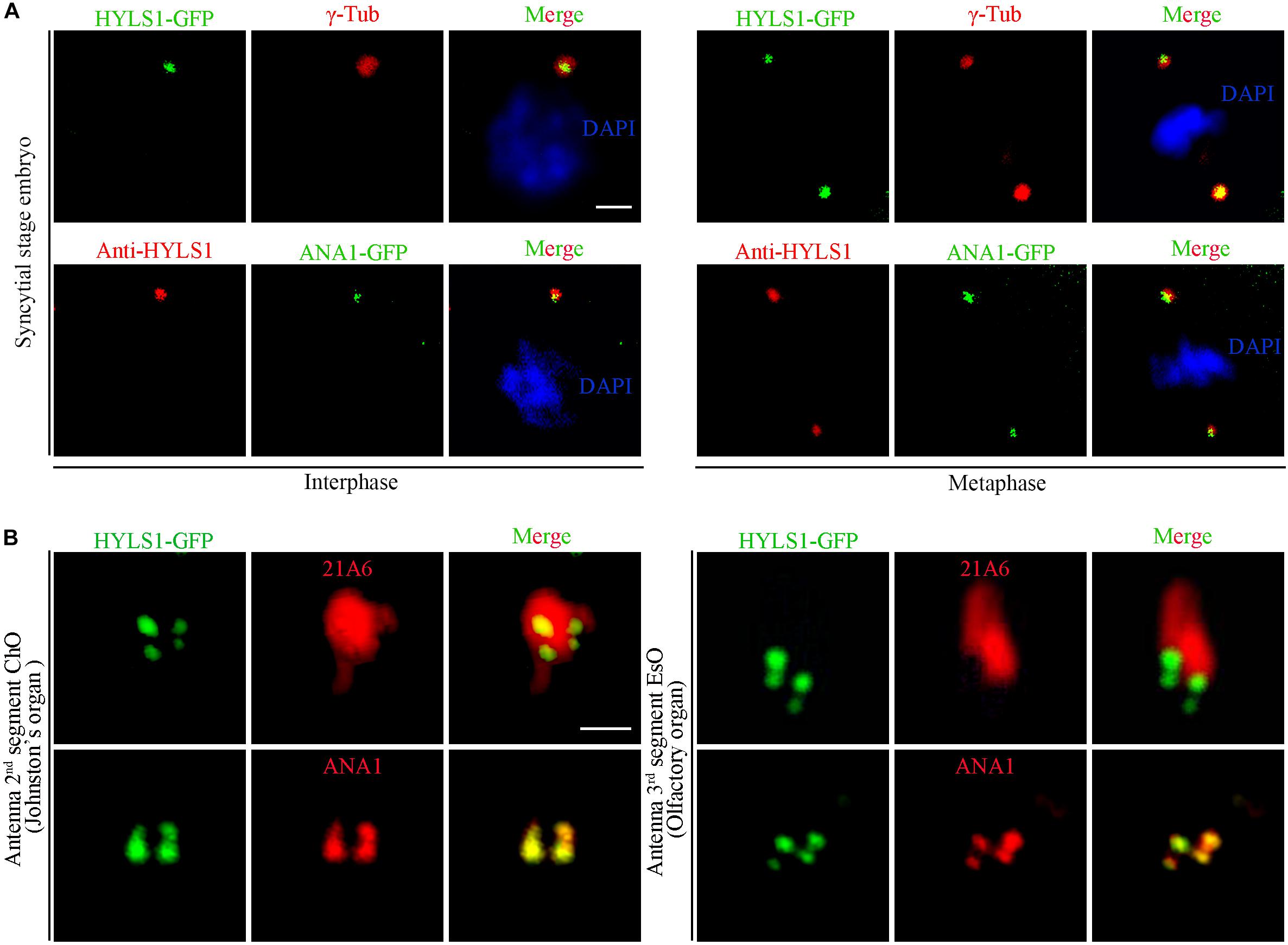
Figure 1. Drosophila HYLS1 localizes to centrioles in embryos and basal bodies in sensory cilia. (A) In early syncytial stage embryos, GFP-tagged HYLS1 localizes to centrosome labeled by γ-tubulin and centriolar protein ANA1 in both interphase and metaphase. Blue channel in the merged panel represents DNA stained by DAPI. Scale bars: 2 μm. (B) Representative images illustrate that HYLS1 is localized to the ciliary base in sensory neurons of both chordotonal organ (ChO) and external sensory organ (EsO). ANA1 is a centriolar protein; 21A6/Eys indicates the base of the sensory cilia. Scale bars: 1 μm.
In sperm cells, centrioles and basal bodies undergo dynamic changes during spermatogenesis (Fabian and Brill, 2012; Jana et al., 2016; Vieillard et al., 2016; Lattao et al., 2017; Figure 2A). In spermatogonia, centrioles are devoid of appendage structures required for ciliogenesis. In early spermatocytes, centrioles start to accumulate components required for ciliogenesis at their tips. During spermatocyte maturation, paired centrioles dock to the plasma membrane and convert to basal bodies, and then cilium-like structures arise from both the mother and daughter centrioles. Of note, at this stage, paired centrioles/basal bodies elongate dramatically and form GCs/basal bodies with a characteristic V-shaped structure; therefore, it is an attractive model to study the centriole to basal body conversion. In round spermatids, flagellar axonemes start to elongate; cilium-like structures together with ring centriole progressively migrate away from basal bodies. We observed that HYLS1 localized to the centriole in spermatogonia and the basal body in spermatocytes and spermatids, as demonstrated by both HYLS1-GFP and anti-HYLS1 staining (Figure 2B and Supplementary Figure S2C). In spermatocytes, HYLS1 colocalized with γ-tubulin along the whole elongated basal body. In spermatids, HYLS1 presented only at the basal body; no HYLS1 can be detected at the migrated ring centriole marked by UNC (the putative homolog of centriole distal end protein OFD1). Three-dimensional structured illumination microscopy (3D-SIM) showed that HYLS1 localized in a distinctive domain below the TF protein CG5964/FBF1 and the TZ protein MKS1 (Figure 2C). Taken together, we conclude that HYLS1 is a conserved centriole and basal body protein in Drosophila.
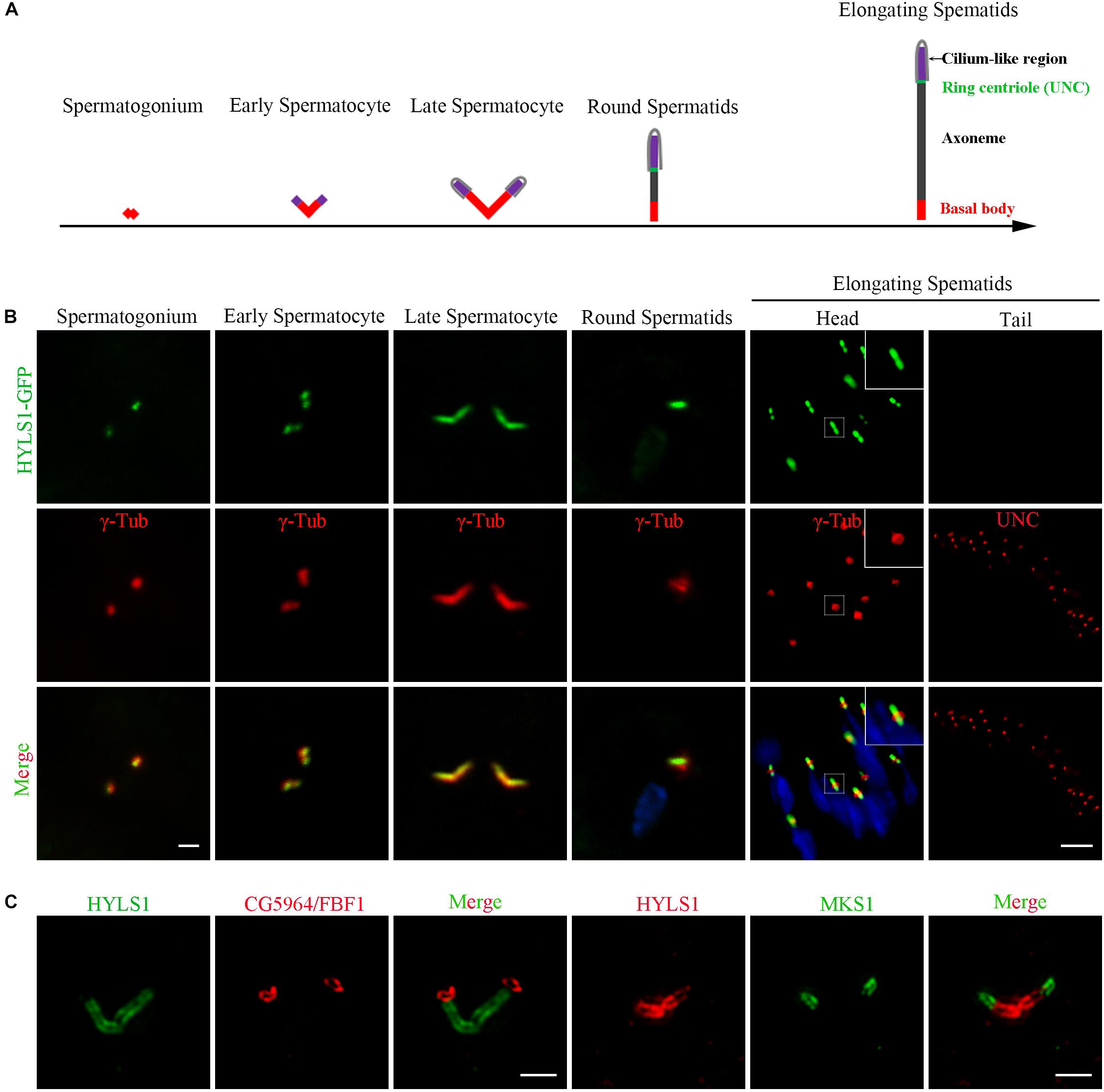
Figure 2. Drosophila HYLS1 is a conserved centriole and basal body protein during spermatogenesis. (A) Schematic illustration of the dynamic behavior of centrioles during spermatogenesis (see text for details). (B) GFP-tagged HYLS1 colocalizes with the centrosome marker γ-tubulin in both spermatogonium and spermatocyte stages. Scale bar: 1 μm. In elongating spermatids, HYLS1 is remained as a basal body protein (magnified region from a head of elongating spermatids, bar: 5 μm), and no HYLS1 migrates to the ring centriole labeled by UNC (bar: 10 μm). Blue channel in the merged panel represents DNA from mature spermatids. (C) 3D-SIM images show that HYLS1 distributes along the entire centrioles and below the transition fiber marker FBF1/CG5964 (left) and the transition zone marker MKS1 (right), indicating that HYLS1 is not a TF or TZ component. Scale bars: 1 μm.
Drosophila HYLS1 Is Required for Sensory Responses
To understand the function of HYLS1 in Drosophila, we employed the CRISPR/Cas9-based genome editing technique to delete most of the second exon of HYLS1 and generated an hyls1 deletion mutant hyls156 (Figures 3A,B). We identified this allele by genomic polymerase chain reaction (PCR) (Figure 3C), and subsequent sequence analysis revealed it bears a 206 bp deletion in the second exon of hyls1 gene, leading to a predicted reading frame shift. Because only the first 38 amino acids (aa) of the very N-terminus of HYLS1 are predicted to be translated followed shortly by a de novo stop codon (Figure 3B), hyls156 is likely a null allele. Such assertion was confirmed by a complete loss of endogenous HYLS1 immunofluorescence signal from the basal body in hyls1 mutants (Figure 3D).
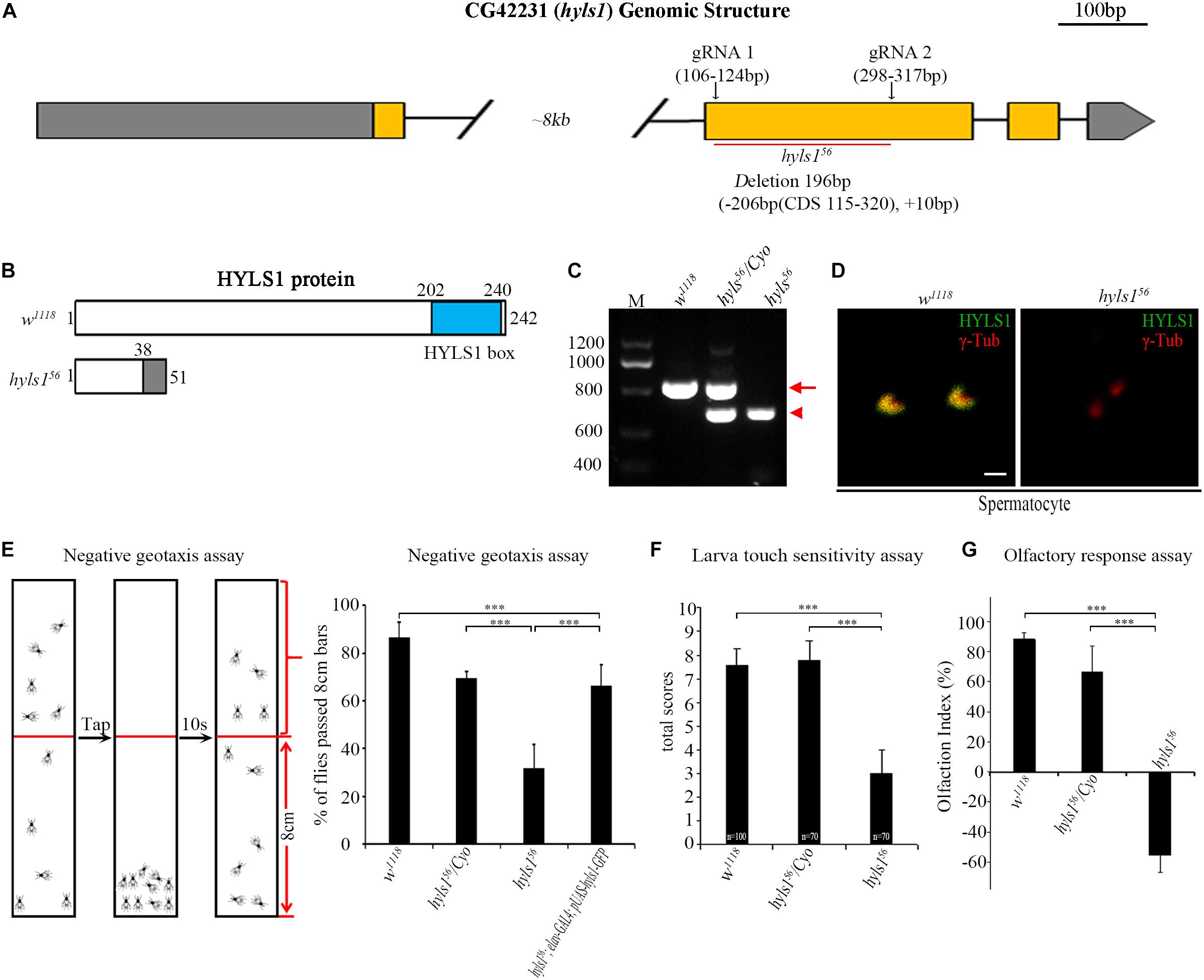
Figure 3. Drosophila HYLS1 is required for sensory responses. (A) Generation of an hyls1 null mutant. Diagram shows the genomic region of Drosophila hyls1 (CG42231). CRISPR/Cas9 system was used to create the null mutant hyls156. hyls156 has a 206-bp nucleotides deletion and an extra 10-bp nucleotides insertion between the targeting sites of gRNA1 and gRNA2 in the second exon. (B) Diagram compares the wild-type HYLS1 protein versus the predicted truncated protein. HYLS1 is a 242-residue-long protein, containing a conserved HYLS1 box at its C terminus. hyls156 encodes a small, C-terminus truncated protein. (C) Genotyping of hyls1 mutants by PCR, primers are listed in Supplementary Table S1. The size of amplicon from hyls1wt is 768 bp (arrow), whereas from hyls156 is only of 572 bp (arrowhead). (D) Immunostaining by anti-HYLS1 and anti–γ-tubulin confirmed that HYLS1 is lost in the spermatocyte centrosome of hyls156 mutants. (E) hyls156 mutants show reduced climbing ability. The percentage of hyls156 flies that passed the 8-cm bars is significantly lower than that in both wild-type and heterozygous hyls156/Cyo flies. Expression of HYLS1 in PNS neurons with elav-GAL4 rescued the defective phenotype. (F) Compared with controls, hyls156 homozygous larvae present touch sensitivity defects. Numbers of tested larvae are indicated in the plot. (G) Mutation in hyls1 causes defective olfactory response. Error bars represent mean with SEM. n.s., p > 0.05; ***p ≤ 0.001 (Student’s t-test). Scale bar: 1 μm.
Of note, another gene, chiffon, which is involved in DNA replication and histone acetylation (Landis and Tower, 1999; Torres-Zelada et al., 2019), is in the intron between the exon 1 and exon 2 of hyls1. chiffon has four annotated isoforms: RB and RD have no shared exons with hyls1 and encode the same 1,695-aa protein; RE encodes the shortest protein of 576 aa residing within RB/RD; RA encodes a 1,711-aa protein. The extra 30-aa C-terminus of RA is encoded by a quite short exon shared with part of exon 2 of hyls1, which is deleted in our hyls156 mutants (Supplementary Figure S3). As the 1,695-aa protein is fully functional (Torres-Zelada et al., 2019), our mutant should not affect the function of chiffon. Such conclusion was further supported by the fact that our hyls156 mutant flies are viable and females are fertile, whereas deletion of CHIFFON results in female infertility and adult lethality (Landis and Tower, 1999; Torres-Zelada et al., 2019).
hyls156 mutants show moderately uncoordinated phenotype (Supplementary Videos S1, S2), a phenotype usually associated with cilia mutants. Interestingly, the uncoordinated phenotype was fully rescued by expression of a WT hyls1 transgene in neurons driven by elav-GAL4, confirming that HYLS1 is the gene responsible for behavior defects in hyls156 mutants (Supplementary Video S3).
First, we examined the cilium ultrastructure of JO in hyls1 mutants using transmission electron microscopy. JO is composed of a various number of scolopidia. A scolopidium usually consists of two neurons with cilia residing on the dendrite tip. In WT, two axonemes are present in the cross section of a scolopidium. However, in hyls1 mutants, missing axoneme was observed (Supplementary Figure S4), indicating that a subset of cilia is truncated in hyls1 mutants.
As cilia are essential for sensory response in neurons, we then examined the sensory abilities of hyls1 mutants with a series of behavioral assays. Negative geotaxis has been well characterized in flies, governed by ciliated sensory organs, which are responsible for gravity perception and locomotor coordination (Kernan, 2007). Wild-type flies climb upward against gravity, after tapping down to the bottom of a tube; most of them move back toward the top in a few seconds. On the contrary, most hyls1 mutants stayed at the bottom of the testing tube after tapping, indicating that negative geotaxis behavior is significantly affected in hyls1 mutants (Figure 3E). Accordingly, climbing defect was effectively rescued by expression of a WT hyls1 transgene in neurons driven by elav-GAL4, confirming that HYLS1 is indeed responsible for negative geotaxis defect in mutant flies. To test the mechanosensory capabilities of hyls1 mutants, we performed touch sensitivity assay and found the response to touch in larva of hyls1 mutants was also severely impaired (Figure 3F). Moreover, we conducted the Y-maze assay to assess the olfactory sensitivity of hyls1 mutants. As expected, hyls1 is required for normal chemical sensation because hyls1 mutants have serious defects in attraction by grape juice. Instead, it seems that hyls1 mutants reject grape juice (Figure 3G). This result indicates that not only the normal olfactory response is disturbed, but also the response pathway also undergoes some changes, which may be due to simultaneous defects in the transport of certain membrane proteins and/or signal molecules that promote or inhibit olfactory function. Altogether, these behavioral assays demonstrate that HYLS1 is required for neuron sensory response in Drosophila.
Drosophila HYLS1 Is Required for Protein Trafficking in Sensory Cilia
It has been reported that HYLS1 is required for the function and formation of ciliary gate (Wei et al., 2016). Therefore, we first examined the localization of ciliary soluble proteins (IFT components) and membrane protein (IAV) in cilia of hyls156 mutants. Axonemes of auditory cilia in the second antennal segment are longer, which are more suitable for cilia study. As expected, we observed that compared with WT, the ciliary level of IFT-B components IFT52 and NOMPB (the ortholog of human IFT88) and the IFT-A components REMPA (the ortholog of human IFT140) was significantly reduced in auditory cilia of hyls156 mutants (Figure 4B). These observations were further confirmed by our quantitative analysis of the signal intensity of these IFT proteins or the ratio of IFT protein intensity to the actin intensity in the associated scolopidia (Figure 4C and Supplementary Figure S5). Accordingly, the amount of ciliary membrane receptor IAV was also dramatically reduced in sensory cilia of hyls156 mutants (Figures 4B,C). These results indicate that deletion of HYLS1 compromised the entry of ciliary proteins.
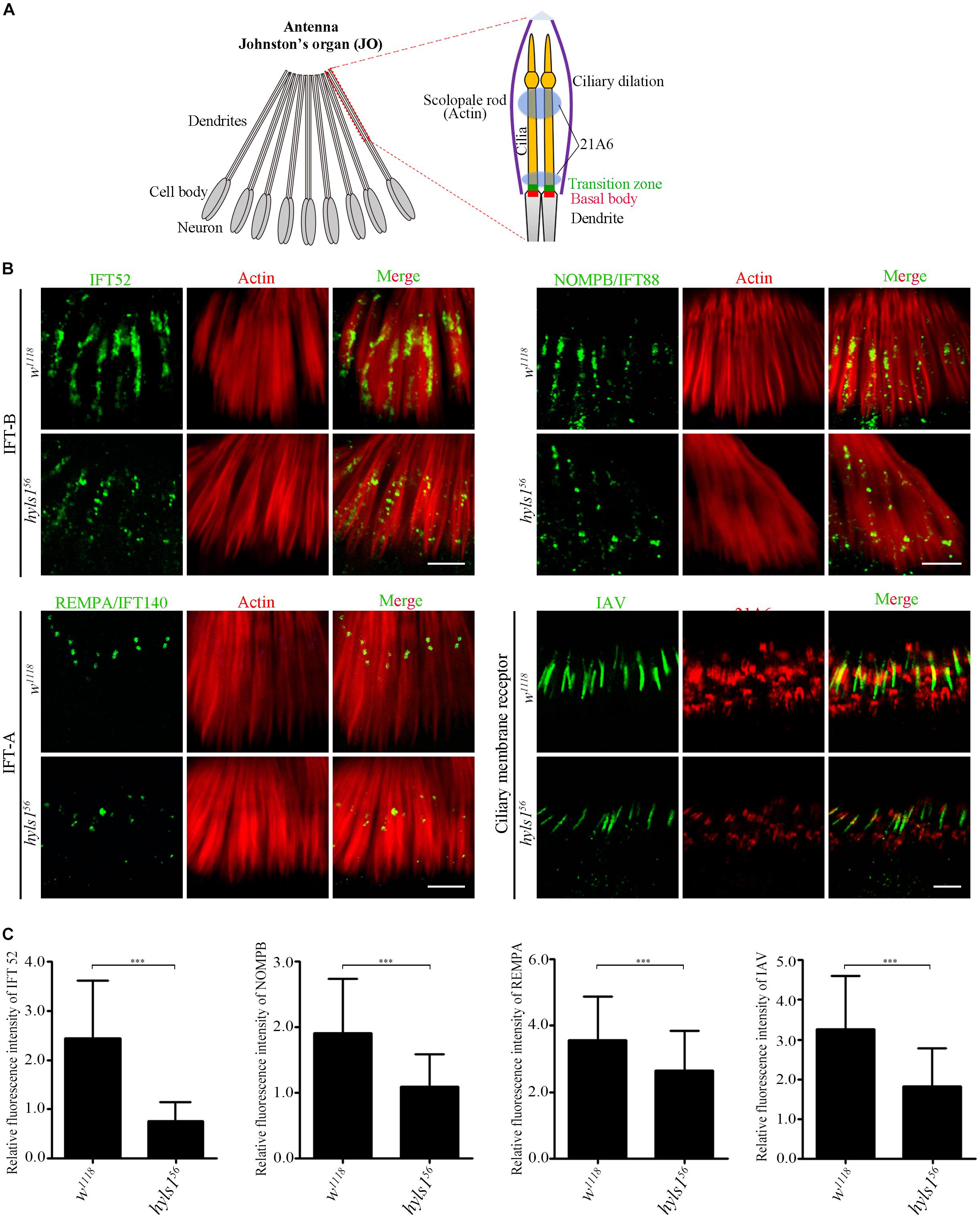
Figure 4. HYLS1 is required for the entry of ciliary proteins in Drosophila. (A) Schematic view of the Johnston’s organ (JO) at the second antennae segment. JO is a specialized chordotonal organ (ChO), which is composed of a various of scolopidia. A scolopidia consists of two Ch neurons and several accessory cells. Ch neurons bear cilia from their dendrite tip; details of the ciliary region of a single scolopidia are illustrated on the right panel. Actin marks scolopale rods, which enclosed the cilia, and 21A6 labels the scolopale lumen around the proximal end of the cilia and the region below the ciliary dilation. (B) HYLS1 is required for the ciliary entry of IFT components in Drosophila. The ciliary signal intensities of IFT-B components IFT52 and NOMPB/IFT88, IFT-A protein REMPA/IFT140, and ciliary membrane protein IAV are significantly reduced in hyls1 mutants. Bars: 5 μm. (C) Quantifications of relative fluorescence intensities of IFT52 (control n = 89; hyls156 n = 85), NOMPB (control n = 91; hyls156 n = 53), REMPA (control n = 148; hyls156 n = 125), and IAV (control n = 132; hyls156 n = 132) in WT and hyls1 mutants. Error bars represent ± s.d., n.s., p > 0.05; ***p ≤ 0.001 (Student’s t-test). Scale bars: 2 μm. “n” is the number of cilia examined.
Then, we determined ciliary gate formation in hyls156 mutants by examining the localization of ciliary gate proteins. The TZ, together with TFs, forms the ciliary gate (Reiter et al., 2012; Takao and Verhey, 2016; Garcia-Gonzalo and Reiter, 2017). As shown in Figure 5, the signals of TZ protein MKS1 and MKS6 were dramatically reduced in hyls156 mutants compared to WT, although they were still able to target to the basal body. Transition fiber structures have been observed at the base of sensory cilia in Drosophila (Jana et al., 2018). FBF1 is the key TF protein mediating the ciliary protein entry in nematodes and mammals. Its Drosophila homolog is encoded by CG5964, and we have characterized CG5964 as a functional TF protein in Drosophila (Y.H., unpublished results). Interestingly, consistent with what we have reported in C. elegans, CG5964 was completely lost or only minimally retained in sensory cilia in hyls1 mutant flies (Figure 5). Taken together, our results suggest that HYLS1 is involved in the function and formation of the ciliary gate in Drosophila.
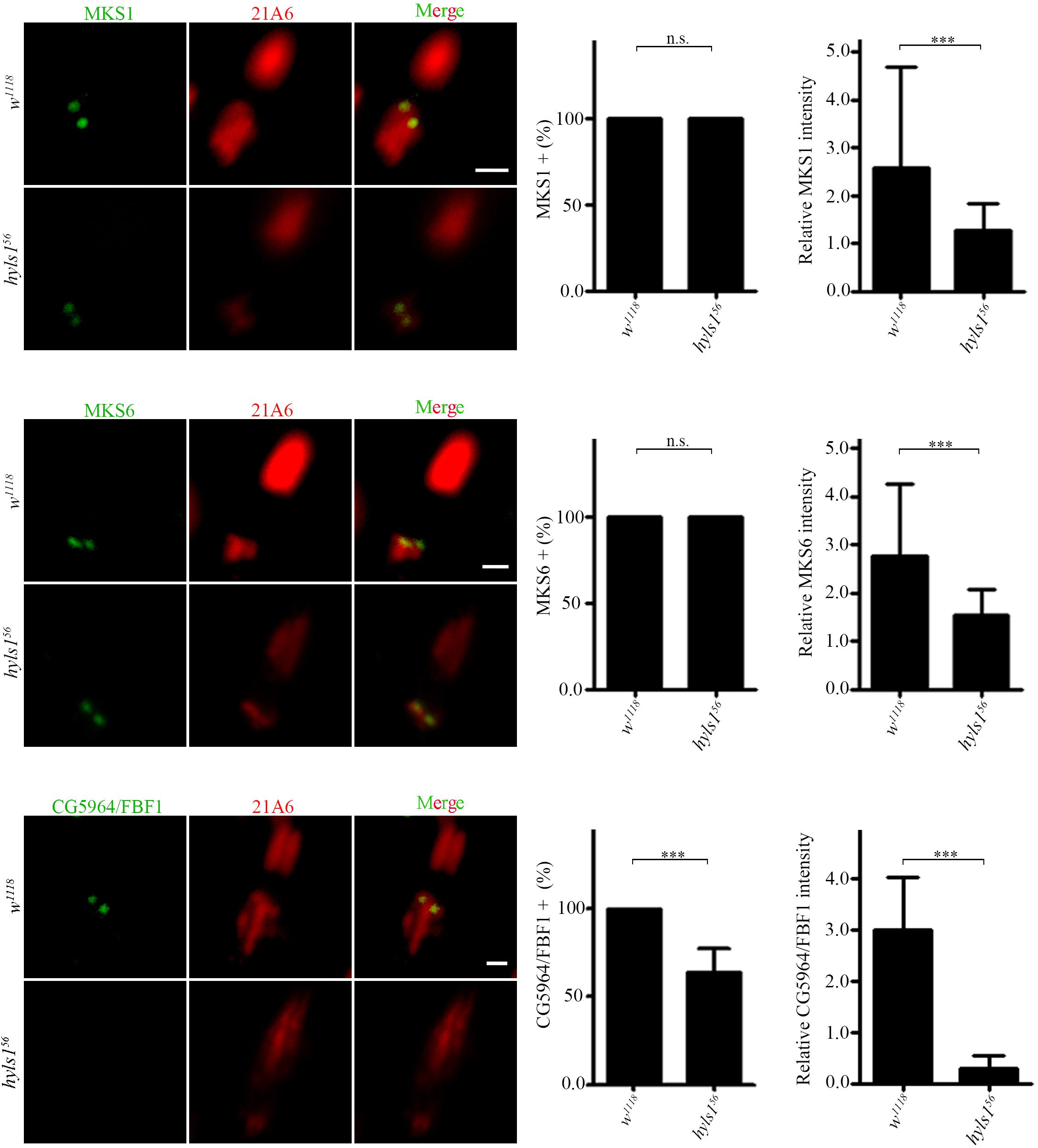
Figure 5. HYLS1 is required for the proper localization of ciliary gate proteins in sensory cilia in Drosophila. Representative images of ciliary gate proteins MKS1 (control n = 115; hyls156 n = 70), MKS6 (control n = 69; hyls156 n = 105), and CG5964 (control n = 98; hyls156 n = 59) in WT and hyls1 mutants. Corresponding quantifications of relative fluorescence intensities were shown in the right panel. Error bars represent ± s.d., n.s., p > 0.05; ***p ≤ 0.001 (Student’s t-test). Scale bars: 2 μm. “n” is the number of cilia examined.
HYLS1 Is Essential for Giant Centriole Elongation in Spermatocytes
Next, we examined the role of HYLS1 in spermatocyte cilia, which have no IFT and only contain conserved TZ proteins (Han et al., 2003; Sarpal et al., 2003; Williams et al., 2011). Although TF structures have not been observed in spermatocyte cilia (Riparbelli et al., 2012; Jana et al., 2018), CG5964 is indeed localized to a distinct region corresponding to the TF, at the tip of centriole and below the TZ. Consistent with what we observed in sensory cilia, the signal intensities of CG5964 and TZ proteins MKS1 and MKS6 were significantly reduced in spermatocyte cilia of hyls1 mutants compared to WT (Figure 6A). And again, CG5964 is more sensitive to HYLS1 deletion. In approximately 30% of cilia, CG5964 was completely lost.
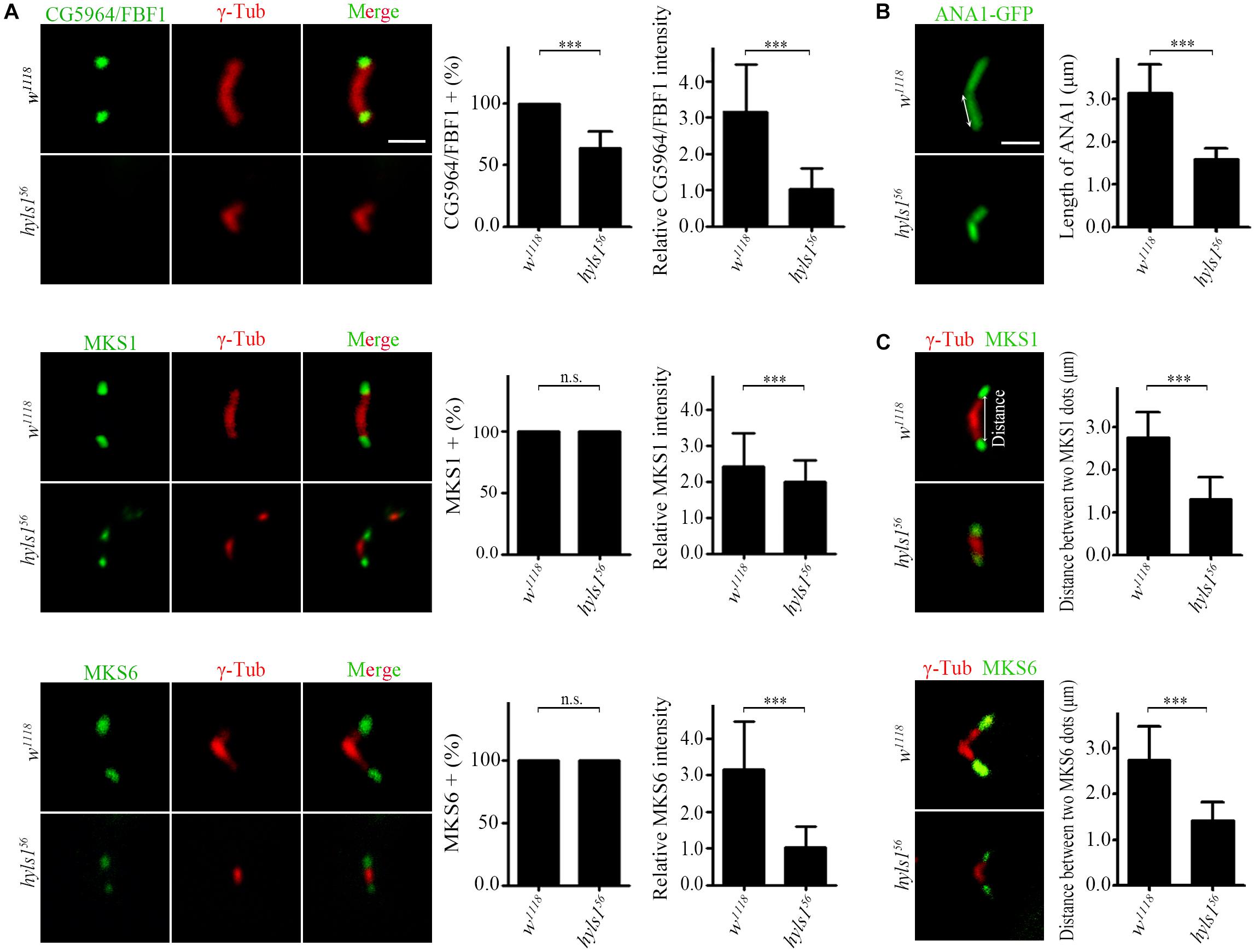
Figure 6. HYLS1 is essential for giant centriole elongation in spermatocytes. (A) In hyls1, CG5964 partially failed to be recruited to centriole tips in spermatocytes (control n = 87; hyls156 n = 57; upper), and reduced signal intensities of MKS1 (n = 143; hyls156 n = 76; middle) and MKS6 (control n = 103; hyls156 n = 98; lower) were observed. (B) Representative micrographs of meiosis II centrioles stained by ANA1-GFP (right) in testes from control and hyls1 mutants. Quantification of centriole length was shown in the right panel. (C) Deletion of HYLS1 leads to shorter distance between two transition zone dots associated with V-shaped centrioles (upper, MKS1; lower, MKS6). The relative distance between two signal dots (double-headed arrow) was quantified in the right panel. Scale bars: 2 μm. For all charts, error bars represent ± s.d.; statistical analysis was done with paired Student’s t-test, n.s., p > 0.05; ***p ≤ 0.001.
Surprisingly, we observed that, compared with WT that has a large V-shaped centriole pair, the centrioles in hyls1 mutant spermatocytes were significantly shorter, as indicated by the centriole marker ANA1/CEP295 (Figure 6B). This phenotype is specific to the centriole elongation, because TZ proteins can still be recruited to the tip of growing centrioles throughout spermatocytes development. To further confirm the role of HYLS1 in regulating centriole elongation, we measured the distance between two TZ protein dots associated with a given pair of centrioles. As expected, the results showed that the mean distance between two protein dots in hyls1 mutants was significantly shorter than that in WT (Figure 6C).
As a centriole protein, how could HYLS1 regulate the localization of TF/TZ proteins? One possible explanation is that HYLS1 regulates the elongation/assembly of the distal centriole where TF/TZ are localized. However, no supportive evidence has been reported. Our observations provide the strong evidence for this hypothesis. We suggest that defects in centriole elongation contribute to defects in TZ and TF in hyls1 mutants. Without proper HYLS1-dependent centriole growth and structural remodeling, TF and TZ can be formed but with significant abnormalities (such as decreased localization of TF and TZ proteins).
Drosophila HYLS1 Is Required for PCM Recruitment in Spermatocytes and PCL Formation in Spermatids
In addition to the short centrioles, we observed that the signal of γ-tubulin was dramatically reduced in centrosomes in spermatocytes of hyls1 mutants compared to WT (Figure 7A). Because γ-tubulin is a PCM component, we examined the level of another PCM component CNN. Compared with WT, the amount of CNN was significantly decreased in interphase in hyls1 mutant cells as well (Figure 7B). Pericentriolar material enlarges in meiosis, and its component CNN is robustly recruited to the centrosome. Quantification of the CNN signal revealed that the recruitment of CNN around HYLS1-negative meiotic centrosomes was significantly less robust comparing with WT (Figure 7C). All these results collectively indicated that HYLS1 is involved in PCM recruitment in spermatocytes.
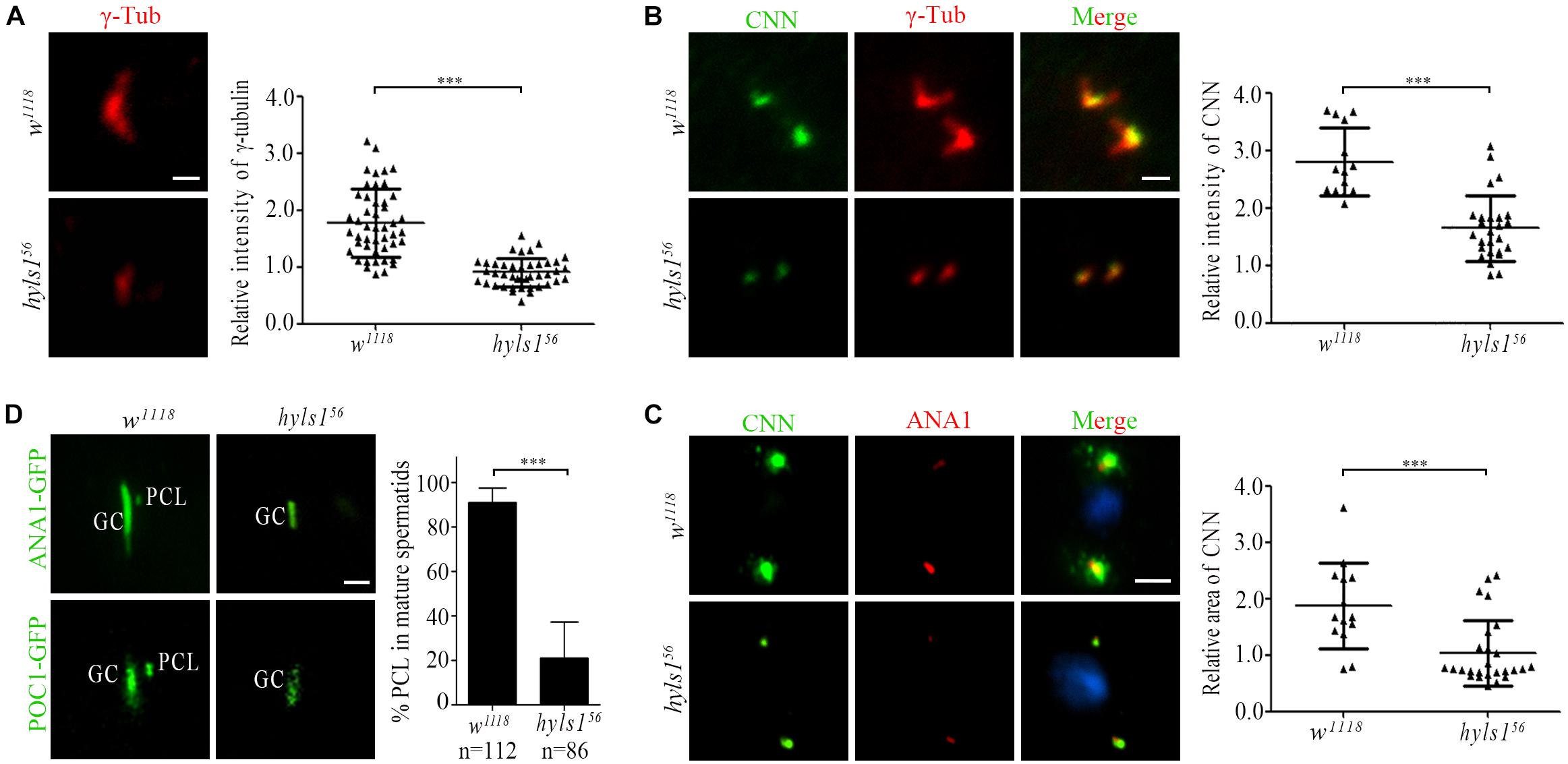
Figure 7. Drosophila HYLS1 is required for PCM recruitment in spermatocytes and PCL formation in spermatids. (A) γ-Tubulin signal was significantly reduced in spermatocyte centriole in hyls156. Quantification data of the relative signal intensity are shown in the right panel. (B) HYLS1 is required for recruitment of PCM protein CNN in interphase of spermatocyte. The right panel shows the quantification of relative centrosome fluorescence intensities of CNN in controls and hyls1 mutants. (C) Images of CNN signal and corresponding quantification data (right panel) in metaphase of meiosis in controls and hyls1 mutants. Blue channel in all panels represents DNA from meiotic chromosomes. (D) Deletion of HYLS1 results in abnormal PCL formation indicated by ANA1-GFP and POC1-GFP. GC, giant centriole; PCL, proximal centriole-like structure. Scale bars: 1 μm (A, B, D), 5 μm (C). Scattered plots with mean and SD are shown. n.s., p > 0.05; ***p ≤ 0.001 (Student’s t-test).
Drosophila spermatids have two centrioles: one is a typical centriole (GC), and the other one is an atypical centriole (PCL) adjacent to the distal part of GC (Blachon et al., 2009). Both centrioles are required for zygote mitosis (Gottardo et al., 2015; Khire et al., 2016). Strikingly, we observed that, in addition to the short typical centriole, PCL was often lost in hyls1 mutant spermatids. We confirmed this result by using two PCL marker proteins ANA1 and POC1 (Figure 7D). It is not likely that HYLS1 is a PCL protein, as we did not observe that HYLS1 localizes to PCL (Figure 2B and Supplementary Figure S2B). Given the fact that PCL is adjacent to the distal part of GC, it is tempting to speculate that PCL formation depends on centriole distal part; loss of PCL in hyls1 mutants may be caused by the defects in centriole elongation.
Taken together, our results indicate that HYLS1 is required for centriole elongation and assembly in Drosophila sperm cells.
HYLS1 Is Required for Spermatogenesis in Drosophila
To determine if HYLS1 is required for spermatogenesis, we examined sperm production. In hyls1 mutants, mature sperms were accumulated in the seminal vesicles and were motile; no visible abnormalities were observed. Colabeling of DNA and α-tubulin in spermatocyte cyst showed that spermatids elongate in hyls1 mutants. Surprisingly, in wild type, 64 flagella tightly associate with each other to form a well-organized bundle, whereas in hyls1 mutants, nuclei were dispersed, and the bundled organization of spermatids was severely disordered (Figure 8A). When examining the sperm flagella axonemal ultrastructure by electron microscopy, we frequently observed incomplete and broken axonemal in hyls1 mutants (32.5%; n = 471; Figure 8B; red arrow), and 8 of 19 (42.15%) cysts had 63 or fewer spermatids, whereas all control cysts possessed 64 spermatids (Figure 8B). Considering that sperm flagella formation is independent of IFT in Drosophila, we speculated that the role of HYLS1 in basal body might directly contribute to defects in spermatogenesis.
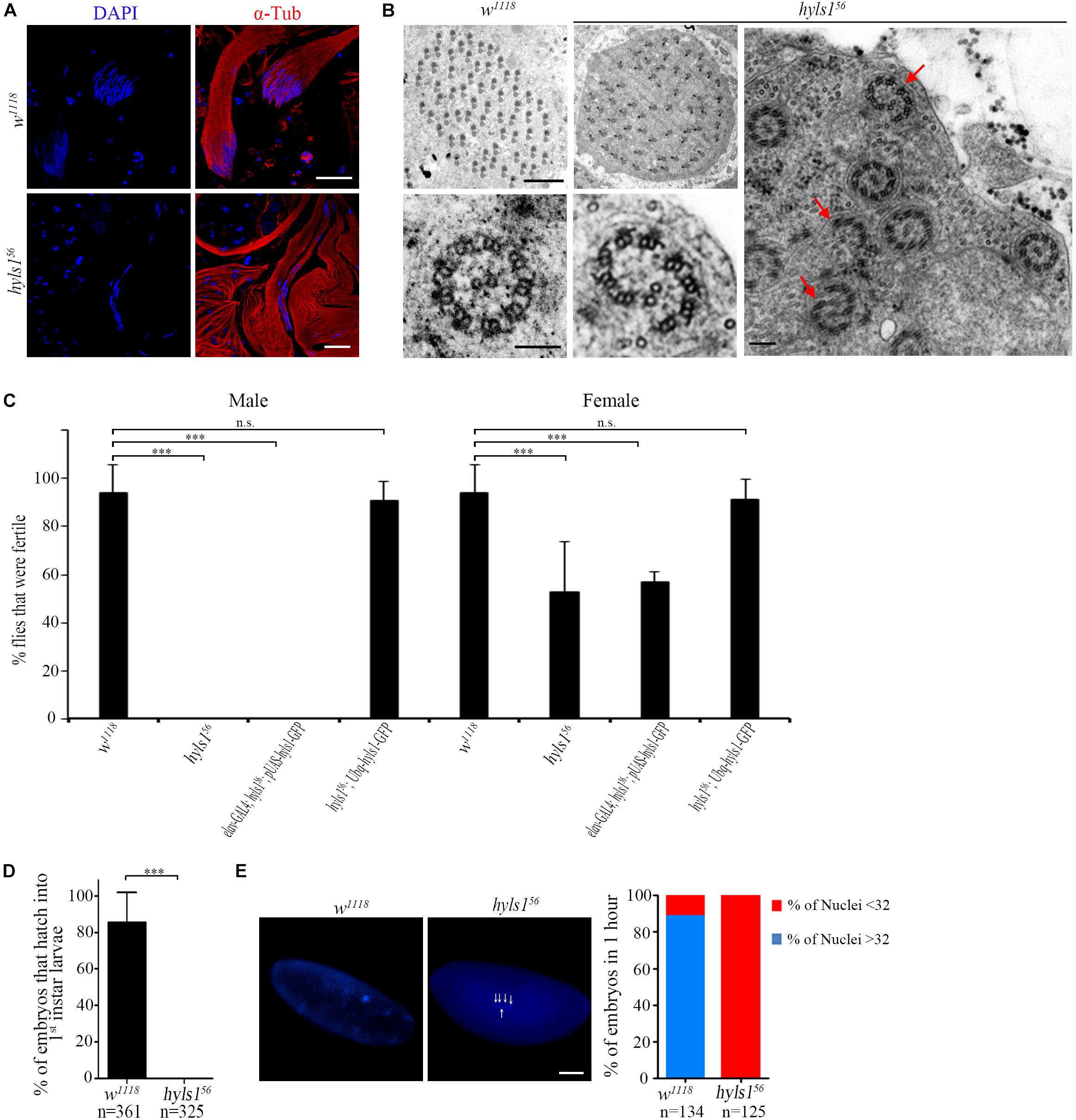
Figure 8. HYLS1 is required for spermatogenesis in Drosophila. (A) Cysts of elongating spermatids in wild-type (upper) and hyls1 mutant (lower) testis stained by nuclei marker DAPI (blue) and sperm flagellar marker α-tubulin (red). Nuclei were clustered in WT cysts but were abnormally dispersed in hyls156 mutants. (B) Ultrastructure of sperm flagellar axoneme of WT and hyls156 mutants showed that the spermatids in the hyls156 cyst were irregularly arranged, and some axonemes were incomplete or broken (red arrows). (C) hyls156 males are completely infertile. Introduction of HYLS1 in neurons driven by elav-Gal4 could not rescue the male fertility. Fertility is fully recovered with global HYSL1 rescue (Ubq-hyls1-GFP). Mutations in hyls1 also affected female fertility. (D,E) HYLS1 is essential for embryogenesis. Embryos fathered by hyls1 mutant failed to develop into larva (D) and were arrested in the early development stage (E). Scale bars: 20 μm in (A), 2 μm in (B left), 100 nm in (B right), 100 μm in (E). Error bars represent ± s.d., n.s., p > 0.05; ***p ≤ 0.001 (paired Student’s t-test).
Although only moderate sperm defects in hyls1 mutants, homozygous hyls1 males are completely infertile (Figure 8C). No zygotes derived from hyls1 mutant males could successfully develop into larvae even when homozygous mutant males were mated to wild-type (w1118) females (Figure 8D). Male fertility can be fully rescued by a ubiquitously expressed WT hyls1 transgene (Ubq-hyls1-GFP), but not by restricted rescue of HYLS1 in neurons (elav-GAL4; pUAS-hyls1-GFP), indicating that infertility is not likely due to the impaired sensory or courtship capacity but owing to defects in spermatogenesis and/or embryogenesis after fertilization (Figure 8C). Further studies revealed that the development of embryos fathered by hyls156 was dramatically delayed and arrested at the early cleavage stage of Drosophila embryos. Nearly 85% embryos have more than 32 nuclei in 1-hour-old control embryos, whereas no embryos with more than 32 nuclei were observed in 1-hour-old hyls1 embryos (Figure 8E). Because both GC and PCL of spermatids are required for zygote mitosis, it is tempting to speculate that defects in GC and PCL may also contribute to embryonic development arrest in paternal hyls1 mutants. Of note, female hyls1 flies were partial fertile, and the phenotype was fully rescued by expressing a Ubq-hyls1-GFP transgene but not a pUAS-hyls1-GFP only in nervous system (Figure 8C), suggesting that maternal HYLS1 may also play a role in embryogenesis. More works will be needed to understand the role of HYLS1 in embryogenesis in the future.
Discussion
Here we investigated the function of HYLS1 in ciliogenesis and spermatogenesis in Drosophila. We demonstrated that the centriole/basal body localization of HYLS1 and its role in ciliary function are highly conserved in Drosophila. More importantly, by taking advantage of the GC in Drosophila spermatocytes, we demonstrated that Drosophila HYLS1 is required for centriole elongation during spermatogenesis.
Although conventional TF structures observed in mammalian cells may not be conserved in C. elegans and Drosophila, the presence of conserved components suggests that, at least, alternative functional homologous structures should exist (Ye et al., 2014; Yang et al., 2018). In both C. elegans and Drosophila, HYLS1 is required for the localization of key TF protein FBF1. As a centriole protein, how could HYLS1 regulate the localization of a TF protein? One highly possible explanation is that HYLS1 regulates the elongation of the distal centriole required for TFs anchoring. However, short or degenerated basal body limited the analysis of centriole elongation during centriole to basal body conversion in sensory cilia in both C. elegans and Drosophila. Remarkably, by taking advantages of the GC in Drosophila spermatocytes, we do observe that HYLS1 promotes centriole elongation. Our observation leads us to propose that HYLS1 is required for distal centriole elongation and recruiting distal centriole proteins to build a specialized construction stage to support the de novo assembly of TFs. Without HYLS1, such stage is compromised, which has direct structural and functional consequences toward the associated appendages made at the distal tip of growing centriole. Although our model is largely based on observations made on the growth of specialized centrioles (Drosophila spermatocytes GCs), we speculate similar HYLS1-dependent distal centriole extension process is a general prerequisite for cilia formation based on the mother centriole. Verification of such generalization calls for further refinement of super-resolution molecular imaging techniques to resolve the full dynamics of cilia formation.
What is the possible role of HYLS1 in centriole elongation or assembly? It has been reported that HYLS1 interacts with the core centriolar protein SAS-4 (Dammermann et al., 2009). Because SAS-4 binds tubulin and is essential for centriole elongation and assembly, it is highly possible that HYSL1 functions through SAS-4 to regulate centriole elongation. In addition, SAS-4 cooperates with CEP120 and SPICE1 to regulate centriole elongation (Comartin et al., 2013; Lin et al., 2013); it is also possible that HYLS1 serves as a critical mediator bridging interactions between SAS-4 and other regulators of centriole elongation. Of note, SAS-4 is also required for PCM assembly (Gopalakrishnan et al., 2011, 2012; Zheng et al., 2014). It is also possible that HYLS1 functions through SAS-4 to regulate PCM recruitment. It will be interesting to test all these hypotheses in the future.
In C. elegans, HYLS1 is not required for centriole assembly and centrosome function (Dammermann et al., 2009). In Drosophila, the development of hyls1 mutant is grossly normal, suggesting that HYLS1 may be also dispensable for centriole and centrosome function in tissues other than sperm cells. In both C. elegans and Drosophila, centrioles are short and lack of appendage structures and probably lack of distal structures found in vertebrate centrioles. Because the distal part of the mammalian centriole is usually not important for centriole duplication and assembly, but is critical for ciliogenesis, it is tempting to speculate that HYLS1 mainly plays a role in the elongation or assembly of the centriole distal part. In conclusion, our finding advances understanding on HYLS1 function and suggests that not only cilia but also the HYLS1-dependent centrosome remodeling may directly contribute to the pathogenesis of human HLS.
Materials and Methods
Fly Stocks
All flies were cultured on standard media at 18°C or 25°C. w1118 was used as wild-type fly. The following flies were obtained from Bloomington Drosophila Stock Center: elav-GAL4 (BS458; BDSC). Mks1-GFP and Mks6-GFP strains were described previously (Vieillard et al., 2016) and were gifts from Bénédicte Durand. IAV-GFP, RempA-YFP, and NompB-GFP were gifts from Seok Jun Moon (Park et al., 2013). All the other following transgenes were generated in the laboratory: Ubq-hyls1-GFP, pUAS-hyls1-GFP, Ubq-ana1-GFP, Ubq-cnn-GFP, Ubq-poc1-GFP, CG5964-GFP, unc-GFP, and ift52-GFP.
Transgenic Drosophila Lines
To generate ubiquitin-driven transgenic lines, promoter of ubiquitin was inserted into the HindIII site of the pJFRC2 vector1 first, and then CDS sequences of HYLS1, POC1B, and CNN-PA were cloned into the NotI–BamHI site of the pJFRC2 plasmid in frame with the downstream GFP sequence. To make CG5964-GFP, unc-GFP and ift52-GFP transgenic lines, their coding, and upstream regulatory sequences were amplified from genomic DNA and then cloned into the HindIII–BamHI site of the pJFRC2 plasmid in frame with the downstream GFP sequence.
To get pUAS-hyls1-GFP flies, full-length HYLS1 CDS was cloned into NotI–BamHI digested pJFRC14 vector.
All vectors were injected into Drosophila embryos by the Core Facility of Drosophila Resource and Technology, SIBCB, CAS. The resulting transgenic flies were selected according to standard procedure.
All GFP signals in colabeling immunofluorescence assay were stained with anti-GFP.
All DNA fragments used were amplified from genomic DNA by us. All CDS fragments were amplified from cDNA reverse transcribed from our own extracted total RNA. All primers used are listed in Supplementary Table S1.
Generation of hyls1 Mutant
We used the CRISPR/Cas9 technique to generate the hyls1 knockout mutants. Two gRNAs were selected using Target Finder website2, gRNA 1: 5′-CGCAAGCTCATCAAGCATGAGG-3′ and gRNA 2: 5′-CTTAAGGAGCGCTCCGATGGTGG-3′. gRNA sequences were separately cloned into a modified pEASY-Blunt vector with a U6.3 promoter. Two gRNA plasmids were collectively injected into vasa:Cas9 embryos, and deletion lines were screened with genomic PCR and were outcrossed five times to wild-type flies (w1118).
Immunofluorescence and Microscopy
Embryo Staining
Embryos were dechorionated in 50% bleach for 5 min to remove chorion, and vitelline membranes were gently stripped under the stereomicroscope and then fixed in a mixture of heptane and 4% formaldehyde (1:1) for 30 min with shaking. The fixed embryos were washed in phosphate-buffered saline (PBS) and blocked with 3% bovine serum albumin (BSA) in PBS-T (PBS 1 × + 0.1% Triton X-100) for 1 h and then incubated with primary antibodies for 24 h at 4°C. The samples were then washed three times with PBS-T for 15 min each and then incubated with secondary antibodies for 3 h at room temperature. After washing three times with PBS-T, embryos were then mounted on slides.
Whole-Mount Antennae Staining
Antennae from pupal or 1- or 2-days-old flies were dissected in PBS-T (PBS 1 × + 0.3% Triton) and then fixed in 4% paraformaldehyde in PBS-T for 1 h at room temperature. The fixed antennae were then incubated in blocking solution (PBS 1 × + 0.1% BSA + 0.1% Triton X-100) for 1 h and stained with primary antibody for 48 h and then secondary antibodies for 24 h.
Testis Squashes Staining
Testes from young males (1 or 2 days old) or pupae were dissected in PBS and then were squished between cover glass and microscopy slide. After squashing, the samples were snap-frozen in liquid nitrogen, and then the coverslips were removed off, and the slides were immersed immediately into methanol for 10 min at –20°C and sequentially in acetone for 10 min at –20°C. Slides were washed in PBS-T and blocked with 3% BSA in PBS-T for 1 h and then were incubated with primary antibodies overnight at 4°C and secondary antibodies for 3 h at room temperature. Lastly, testes were mounted and examined using fluorescence microscopy.
Antibodies
The following primary antibodies were used: mouse anti–γ-tubulin (1:500; Sigma-Aldrich, St. Louis, MO, United States), mouse anti–α-tubulin (1:500; Santa Cruz Biotechnology, Dallsa, TX, United States), mouse anti-GFP (1:500; Roche, Basel, Switzerland), rabbit anti-GFP (1:500; Abcam, Cambridge, United Kingdom), mouse 22C10 (1:200; DSHB), mouse 21A6 (1:200; DSHB), iFluorTM 555 Phalloidin (1:200; Yeasen, Shanghai, China). The following secondary antibodies were used: goat anti–mouse Alexa Fluor 488 or Alexa Fluor 594 or goat anti–rabbit Alexa Fluor 488 or Alexa Fluor 594.
Generation of Drosophila HYLS1 Antibody
All the work of generation of HYLS1 antibody is done by YOUKE Biotech, Shanghai, China. Briefly, His-tagged full-length HYLS1 protein was expressed in Escherichia coli and purified as immunogen. Antisera were raised in two rabbits and purified by protein G beads; 1:200 dilution was used for immunofluorescence assay.
Microscopy
Samples were imaged using either the fluorescence microscopy (Nikon Eclipse Ti, Nikon, Japan) or the confocal microscopy (Olympus FV1000a, Olympus, Japan) with a 100 × /NA 1.45 oil immersion objective. For super resolution imaging, samples mounted in ProLongTM Diamond Antifade Mountant (Invitrogen, Carlsbad, CA, United States) were imaged using a 3D-SIM (Delta Vision OMX SR, GE Healthcare, Chicago, IL, United States).
Quantification of Fluorescence Intensity
To minimize the staining variation between mutant and control, both samples were prepared and stained at the same time and mounted on a same slide, and fluorescence images were obtained under the same microscopic conditions and settings. The fluorescence intensity was measured using Nikon NIS-Elements software, and background fluorescence was subtracted.
Transmission Electron Microscopy
Dissected fly antennae and testes were immediately held in 2.5% glutaraldehyde for at least 24 h before postfixation in OsO4. Then samples were dehydrated in alcohol and then embedded in epoxy resin. Sections of (∼70 nm) of samples were cut in Ultracut S ultramicrotome, collected on Formvar-coated copper grids, and stained with uranyl acetate and lead citrate. Samples were imaged with H-7650 electron microscope (Hitachi, Japan) at 80 KV.
Fertility Ratio Assay
For male fertility ratio assay, virgin w1118 females and mutant males are crossed. For female fertility ratio assay, mutant females and w1118 males are crossed. Before testing, virgin females and newly enclosed males were separately collected and held in room temperature for 3–4 days. In each test, a single male was mated individually with a single virgin female for 4 days at 25°C. Crosses with dead flies were eliminated from the test. Fertility ratio was quantified.
24 h Embryo Development Assay and Larval Hatching
Virgins and males of 3–4 days were placed in a mating chamber at 25°C to lay eggs. Parents were removed until more than 80 embryos were laid, and embryos were cultured at room temperature for 3 days. The percent of hatched eggs was counted. Experiments were repeated at least three times.
Negative Geotaxis Assay (Climbing Assay)
Climbing assay was performed as described previously (Chen et al., 2015). Briefly, 10 flies were transferred as a group to a fresh food vial the day before the test. Just before the experiment, flies were transferred into a 20-cm-long clear testing vial. During the test, flies were gently tapped down to the vial bottom; the flies climbing above the 8-cm mark were counted after 10 s. After the test, flies were given 1 min to recover, repeated 10 times. At least five groups of flies were assessed.
Larva Touch Assay
Larva touch assay was performed as previously described (Chen et al., 2015). Briefly, 10 larvae as a group were gently touched one by one on their head segments with a human hair. According to the response of the larva to the touch, a score was assigned: 0 for the larva showed no response; 1 for the larva showed hesitation with ceased movement; 2 for the larva that showed anterior contract; 3 for one full wave of body contraction; and 4 for the larva that showed two or more full waves of body contractions. Each group was tested four times, and the four scores were added up to a total score; at least five groups of larvae were assayed.
Olfactory Response Assay
Olfactory response was tested by using the Y-maze assay as described (Simonnet et al., 2014). Briefly, before testing, 10 flies as a group were starved for 16–18 h at 25°C in a glass tube. Just before the test, a small filter paper with odorant or corresponding solvent was placed into the two trap vials, respectively, and flies were loaded into the loading vial. Olfactory index was calculated after 24 h. At least five groups of flies were tested each experiment.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
QW and YH designed the study and analyzed the data. YH performed the experiments. ZW, YZ, and HC assisted the experiments and performed the SIM imaging. YP, YG, and JH assisted the experiments and provide some reagents. QW, YP, and YH wrote the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31671549 and 31871357) to QW, and National Natural Science Foundation Youth Project of China (31802009) to YH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Bénédicte Durand, Dr. Chul Hoon Kim, the BDSC and Tsinghua Drosophila Stock Center for fly stocks. We thank Wu Wei, Core Facility of Drosophila Resource and Technology, SIBCB, CAS for transgenic Drosophila.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00301/full#supplementary-material
FIGURE S1 | Alignment of human HYLS1, mice HYLS1 and Drosophila CG42231. Conserved amino acids are highlighted, and the conserved HYLS1 box is underlined. Hs: Homo sapiens; Mm: Mus musculus; Dm: Drosophila melanogaster. Red * stands for the mutation site of hydrolethalus syndrome.
FIGURE S2 | HYLS1 is a conserved centriole and basal body protein. (A) HYLS1 localizes to centrosomes in syncytial stage embryos. (B) Endogenous HYLS1 localizes to cilia base in sensory neurons. HYLS1 colocalizes with basal body marker γ-tubulin and is detected at the ciliary base of external sensory organ marked by 21A6. 21A6 indicates the cilium base. (C) Localization of endogenous HYLS1 in centriole and basal body during spermatogenesis. Blue channel in the merged panel represents DNA from mature spermatids. Bars: 2 μm.
FIGURE S3 | Structural map of hyls1 gene and chiffon gene. chiffon gene (CG5813, blue) overlaps with hyls1 gene (CG42231, blue) and shares a common promoter, but differs in their open reading frames and codes different proteins. The coding sequences (black boxes), untranslated nucleotides (gray boxes), and introns (lines) are respectively illustrated to chiffon and hyls1. There are four annotated splice isoforms for chiffon: RA encodes a 1711-aa protein from two exons spanning a ∼1.1 kb intron; RB and RD encode 1695-aa proteins from a single ∼5 kb exon; RE encodes a 576-aa protein from a shorter exon within RB/RD. Deleted regions in hyls1 and chiffon-RA are illustrated by black lines. gRNA editing to hyls1 enabled the deletion of the last C-terminal 30 amino acids of CHIFFON-RA, including the stop codon (from 5043 bp to the 5136 bp).
FIGURE S4 | Ultrastructure of the chordotonal cilia in WT and hyls1 mutants. In WT, two axonemes are usually present in the cross section of a scolopidium. However, in hyls1 mutants, missing axonemes is observed. Red arrow, normal axoneme. Green arrow, abnormal axoneme. Scale bars: 1 μm.
FIGURE S5 | Quantification of IFT protein intensity relative to Actin intensity in the associated scolopidia in WT and hyls1 mutants. Compare to WT, the ratios of IFT52 intensity/Actin intensity (control n = 35; hyls156 n = 35), NOMPB intensity/Actin intensity (control n = 35; hyls156 n = 26) and REMPA intensity/Actin intensity (control n = 31; hyls156 n = 20) were significantly reduced in hyls1 mutants. Error bars represent ± s.d., *p < 0.05; ***p ≤ 0.001 (Student’s t-test). “n” is the number of cilia examined.
TABLE S1 | Primers used in this paper.
VIDEO S1 | Heterozygous hyls156/Cyo presented normal climbing activities.
VIDEO S2 | hyls156 null mutant flies showed climbing defect.
VIDEO S3 | Uncoordinated phenotype in hyls156 was rescued by introduction of a WT hyls1 transgene in neurons driven by elav-GAL4.
Footnotes
References
Anvarian, Z., Mykytyn, K., Mukhopadhyay, S., Pedersen, L. B., and Christensen, S. T. (2019). Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15, 199–219. doi: 10.1038/s41581-019-0116-9
Bettencourt-Dias, M., and Glover, D. M. (2007). Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463. doi: 10.1038/nrm2180
Bettencourt-Dias, M., Hildebrandt, F., Pellman, D., Woods, G., and Godinho, S. A. (2011). Centrosomes and cilia in human disease. Trends Genet. 27, 307–315.
Blachon, S., Cai, X. Y., Roberts, K. A., Yang, K., Polyanovsky, A., Church, A., et al. (2009). A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics 182, 133–144. doi: 10.1534/genetics.109.101709
Boekhoff-Falk, G. (2005). Hearing in Drosophila: development of Johnston’s organ and emerging parallels to vertebrate ear development. Dev. Dyn. 232, 550–558. doi: 10.1002/dvdy.20207
Chen, J. V., Kao, L. R., Jana, S. C., Sivan-Loukianova, E., Mendonca, S., Cabrera, O. A., et al. (2015). Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J. Cell. Biol. 211, 435–453. doi: 10.1083/jcb.201502032
Comartin, D., Gupta, G. D., Fussner, E., Coyaud, E., Hasegan, M., Archinti, M., et al. (2013). CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23, 1360–1366. doi: 10.1016/j.cub.2013.06.002
Conduit, P. T., Wainman, A., and Raff, J. W. (2015). Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell. Biol. 16, 611–624. doi: 10.1038/nrm4062
Dammermann, A., Pemble, H., Mitchell, B. J., McLeod, I., Yates, J. R. III, Kintner, C., et al. (2009). The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 23, 2046–2059. doi: 10.1101/gad.1810409
Fabian, L., and Brill, J. A. (2012). Drosophila spermiogenesis: big things come from little packages. Spermatogenesis 2, 197–212. doi: 10.4161/spmg.21798
Garcia-Gonzalo, F. R., and Reiter, J. F. (2017). Open sesame: how transition fibers and the transition zone control ciliary composition. CSH Perspect. Biol. 9:a028134. doi: 10.1101/cshperspect.a028134
Goetz, S. C., and Anderson, K. V. (2010). The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 11, 331–344. doi: 10.1038/nrg2774
Gopalakrishnan, J., Chim, Y. C., Ha, A., Basiri, M. L., Lerit, D. A., Rusan, N. M., et al. (2012). Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat. Cell Biol. 14, 865–873. doi: 10.1038/ncb2527
Gopalakrishnan, J., Mennella, V., Blachon, S., Zhai, B., Smith, A. H., Megraw, T. L., et al. (2011). Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2:359. doi: 10.1038/ncomms1367
Gottardo, M., Callaini, G., and Riparbelli, M. G. (2015). Structural characterization of procentrioles in Drosophila spermatids. Cytoskeleton 72, 576–584. doi: 10.1002/cm.21260
Han, Y. G., Kwok, B. H., and Kernan, M. J. (2003). Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr. Biol. 13, 1679–1686. doi: 10.1016/j.ydbio.2011.06.028
Hildebrandt, F., Benzing, T., and Katsanis, N. (2011). Ciliopathies. N. Engl. J. Med. 364, 1533–1543.
Jana, S. C., Bettencourt-Dias, M., Durand, B., and Megraw, T. L. (2016). Drosophila melanogaster as a model for basal body research. Cilia 5:22. doi: 10.1186/s13630-016-0041-5
Jana, S. C., Mendonca, S., Machado, P., Werner, S., Rocha, J., Pereira, A., et al. (2018). Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nat. Cell. Biol. 20, 928–941. doi: 10.1038/s41556-018-0132-1
Jarman, A. P. (2002). Studies of mechanosensation using the fly. Hum. Mol. Genet. 11, 1215–1218. doi: 10.1093/hmg/11.10.1215
Kernan, M. J. (2007). Mechanotransduction and auditory transduction in Drosophila. Pflugers Arch. 454, 703–720. doi: 10.1007/s00424-007-0263-x
Khire, A., Jo, K. H., Kong, D., Akhshi, T., Blachon, S., Cekic, A. R., et al. (2016). Centriole remodeling during spermiogenesis in Drosophila. Curr. Biol. 26, 3183–3189. doi: 10.1016/j.cub.2016.07.006
Landis, G., and Tower, J. (1999). The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126, 4281–4293.
Lattao, R., Kovacs, L., and Glover, D. M. (2017). The centrioles, centrosomes, basal bodies, and cilia of Drosophila melanogaster. Genetics 206, 33–53. doi: 10.1534/genetics.116.198168
Lin, Y. N., Wu, C. T., Lin, Y. C., Hsu, W. B., Tang, C. J., Chang, C. W., et al. (2013). CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell. Biol. 202, 211–219. doi: 10.1083/jcb.201212060
Malicki, J. J., and Johnson, C. A. (2017). The cilium: cellular antenna and central processing unit. Trends Cell Biol. 27, 126–140. doi: 10.1016/j.tcb.2016.08.002
Mee, L., Honkala, H., Kopra, O., Vesa, J., Finnila, S., Visapaa, I., et al. (2005). Hydrolethalus syndrome is caused by a missense mutation in a novel gene HYLS1. Hum. Mol. Genet. 14, 1475–1488. doi: 10.1093/hmg/ddi157
Nigg, E. A., and Stearns, T. (2011). The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell. Biol. 13, 1154–1160. doi: 10.1038/ncb2345
Park, J., Lee, J., Shim, J., Han, W., Lee, J., Bae, Y. C., et al. (2013). dTULP, the Drosophila melanogaster homolog of tubby, regulates transient receptor potential channel localization in cilia. PLoS Genet. 9:e1003814. doi: 10.1371/journal.pgen.1003814
Reiter, J. F., Blacque, O. E., and Leroux, M. R. (2012). The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13, 608–618. doi: 10.1038/embor.2012.73
Reiter, J. F., and Leroux, M. R. (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell. Biol. 18, 533–547. doi: 10.1038/nrm.2017.60
Riparbelli, M. G., Callaini, G., and Megraw, T. L. (2012). Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev Cell. 23, 425–432. doi: 10.1016/j.devcel.2012.05.024
Sarpal, R., Todi, S. V., Sivan-Loukianova, E., Shirolikar, S., Subramanian, N., Raff, E. C., et al. (2003). Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 13, 1687–1696. doi: 10.1016/j.cub.2003.09.025
Sharma, A., Aher, A., Dynes, N. J., Frey, D., Katrukha, E. A., Jaussi, R., et al. (2016). Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev Cell. 37, 362–376. doi: 10.1016/j.devcel.2016.04.024
Simonnet, M. M., Berthelot-Grosjean, M., and Grosjean, Y. (2014). Testing Drosophila olfaction with a Y-maze assay. J. Vis. Exp. 88:51241. doi: 10.3791/51241
Takao, D., and Verhey, K. J. (2016). Gated entry into the ciliary compartment. Cell. Mol. Life Sci. 73, 119–127. doi: 10.1007/s00018-015-2058-0
Tanos, B. E., Yang, H. J., Soni, R., Wang, W. J., Macaluso, F. P., Asara, J. M., et al. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168. doi: 10.1101/gad.207043.112
Torres-Zelada, E. F., Stephenson, R. E., Alpsoy, A., Anderson, B. D., Swanson, S. K., Florens, L., et al. (2019). The Drosophila Dbf4 ortholog Chiffon forms a complex with Gcn5 that is necessary for histone acetylation and viability. J. Cell. Sci. 132:jcs214072. doi: 10.1242/jcs.214072
Vieillard, J., Duteyrat, J. L., Cortier, E., and Durand, B. (2015). Imaging cilia in Drosophila melanogaster. Methods Cell Biol. 127, 279–302.
Vieillard, J., Paschaki, M., Duteyrat, J. L., Augiere, C., Cortier, E., Lapart, J. A., et al. (2016). Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J. Cell Biol. 214, 875–889. doi: 10.1083/jcb.201603086
Wei, Q., Ling, K., and Hu, J. (2015). The essential roles of transition fibers in the context of cilia. Curr. Opin. Cell Biol. 35, 98–105. doi: 10.1016/j.ceb.2015.04.015
Wei, Q., Xu, Q. W., Zhang, Y. X., Li, Y. J., Zhang, Q., Hu, Z., et al. (2013). Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat. Commun. 4:2750. doi: 10.1038/ncomms3750
Wei, Q., Zhang, Y., Schouteden, C., Zhang, Y., Zhang, Q., Dong, J., et al. (2016). The hydrolethalus syndrome protein HYLS-1 regulates formation of the ciliary gate. Nat. Commun. 7:12437. doi: 10.1038/ncomms12437
Williams, C. L., Li, C., Kida, K., Inglis, P. N., Mohan, S., Semenec, L., et al. (2011). MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192, 1023–1041. doi: 10.1083/jcb.201012116
Yang, T. T., Chong, W. M., Wang, W. J., Mazo, G., Tanos, B., Chen, Z., et al. (2018). Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023. doi: 10.1038/s41467-018-04469-1
Ye, X., Zeng, H. Q., Ning, G., Reiter, J. F., and Liu, A. M. (2014). C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. U.S.A. 111, 2164–2169. doi: 10.1073/pnas.1318737111
Keywords: HYLS1, ciliogenesis, ciliary gate, PCL, centriole elongation, spermatogenesis
Citation: Hou Y, Wu Z, Zhang Y, Chen H, Hu J, Guo Y, Peng Y and Wei Q (2020) Functional Analysis of Hydrolethalus Syndrome Protein HYLS1 in Ciliogenesis and Spermatogenesis in Drosophila. Front. Cell Dev. Biol. 8:301. doi: 10.3389/fcell.2020.00301
Received: 19 February 2020; Accepted: 07 April 2020;
Published: 21 May 2020.
Edited by:
Tomer Avidor-Reiss, University of Toledo, United StatesReviewed by:
Timothy L. Megraw, Florida State University, United StatesGiuliano Callaini, University of Siena, Italy
Copyright © 2020 Hou, Wu, Zhang, Chen, Hu, Guo, Peng and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Wei, cWluZy53ZWlAc2lhdC5hYy5jbg==
 Yanan Hou
Yanan Hou Zhimao Wu
Zhimao Wu Yingying Zhang
Yingying Zhang Huicheng Chen1,2,3
Huicheng Chen1,2,3 Qing Wei
Qing Wei