- 1College of Animal Science and Technology, Northwest A&F University, Yangling, China
- 2School of Life Sciences, Technical University of Munich, Freising, Germany
Milk casein and triglyceride content are important production traits in goats. Studies on mechanisms in milk casein secretion and mammary gland development is essential for milk goat breeding. miRNAs play an important role in goat lactation. While novel-miR-3880 is highly expressed at goat peak lactation stage, its molecular mechanism has not been studied. The purpose of the present study was to explore the relationship between novel-miR-3880 and lactation, as well as to construct a network among novel-miR-3880, ciRNA13761, and E74 like ETS transcription factor 2 (ELF2), thus further exploring their potential roles in milk components and mammary gland development. ELF2 was previously proven to be important in cell survival and proliferation, and 3′-UTR of ELF2 was predicted to have binding sites of novel-miR-3880. Our study found that the overexpression of novel-miR-3880 exerted anti-apoptotic and proliferative roles in GMEC, induced a boost in triglyceride synthesis, and caused a decrease in α s1-, α s2-, and β-casein, but an increase in κ-casein secretion. Furthermore, treatment in mice indicated that novel-miR-3880 could promote mammary gland development and extend the lactation period, while novel-miR-3880 expression was found to be suppressed by ciRNA13761 as a miRNA sponge. The present study explores a mechanism of triglyceride synthesis and casein secretion, and reveals a crosstalk between ciRNA13761/novel-miR-3880/ELF2 axis and PI3K/AKT/mTOR/S6K1 pathway, to gain a better understanding of lactation traits in dairy goats.
Introduction
Goat’s milk is regarded as a hypoallergenic milk type with therapeutic functions and is accepted as a major functional food (Roncada et al., 2002) when casein, including κ-, β-, αs1,- and αs2-casein, is the main protein in goat’s milk (Singh and Singh, 1980; Chatchatee et al., 2001). MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs of approximately 23 nt in length. They are reported to regulate gene expression by repressing translation or facilitating mRNA degradation (Ambros, 2004) and play an important role in goat lactation (Ji et al., 2017). Novel-miR-3880 was highly expressed during goat peak lactation period based on our previous sequencing data (Hou et al., 2017). The novel-miR-3880 expression data was selected from Supplementary File S1 (Hou et al., 2017). Our study explored effects of novel-miR-3880-related molecules on mammary gland development in vivo and in vitro, thus providing a theoretical molecular basis for breeding and breast care.
Our study found that the overexpression of novel-miR-3880 induced anti-apoptotic and proliferative effects, promoted triglyceride synthesis, and caused a decrease in α s1-, α s2-, and β-casein but an increase in κ-casein secretion. Further, the regulation pattern was studied. Firstly, negative control (NC) and novel-miR-3880 were transfected into goat mammary epithelial cell (MEC) and RT-qPCR was applied to detect the expression of miRNAs, thus ensuring the efficiency of RNA sequencing. Differentially expressed genes (DEGs) of novel-miR-3880 were screened, and 67 downstream genes were found. E74 like ETS transcription factor 2 (ELF2), which is proven to be important in cell survival and proliferation (Qiu et al., 2008), might be essential for mammary gland development and lactation, and was found in the downregulated DEG list of sequencing results.
CircRNAs are another class of endogenous non-coding RNAs from non-classical alternative splicing that can expropriate miRNAs as a sponge to block miRNAs from binding to target genes (Hansen et al., 2013; Vicens and Westhof, 2014; Meng et al., 2017). To explore how novel-miR-3880 expression was regulated, circRNAs that have binding sites of novel-miR-3880 were screened according to circRNA sequencing data our laboratory acquired before. CircRNA13761 (ciRNA13761), whose structure is shown in Supplementary File S2, was selected and verified as a novel-miR-3880 sponge, while its source gene DOCK1 is reported to be involved in mammary gland involution (Bagci et al., 2014). Then, the network among ciRNA13761, novel-miR-3880, and ELF2 was constructed, and the role of DOCK1 in the network was detected.
As is known, mTOR is a central modulator in protein/lipid synthesis and cell growth processes and plays important roles in milk production (Osorio et al., 2016; Saxton and Sabatini, 2017). It serves as a crucial downstream signal of PI3K/AKT pathway to form a functional compound (Yang et al., 2014), which participates in lactation initiation (Chen et al., 2012). S6K1, a downstream effector of mTOR, is also critical for promoting protein and lipid synthesis (Yang et al., 2014); its activation relies on phosphorylation mediated by mTOR (Magnuson et al., 2012). Our study explored whether and how novel-miR-3880 regulates PI3K/AKT/mTOR/S6K1 pathway and participates in MEC biological processes and mammary gland development. In addition, MEC anti-apoptosis signaling was evaluated by the protein expression ratio of Bcl-2 and Bax, which is regarded as a cell survival signal (Basu and Haldar, 1998).
In this study, roles of ciRNA13761, novel-miR-3880, and ELF2 on mammary gland development and lactation traits were studied to provide a basis for molecular breeding of dairy goats. More precisely, the interaction among ciRNA13761, novel-miR-3880, and ELF2, and their effects on MEC triglyceride synthesis, lipid formation, casein secretion, viability, proliferation, and apoptosis were explored, as well as their participation in PI3K/AKT/mTOR/S6K1 pathway and Bcl-2/Bax pathway. Novel-miR-3880 and siELF2 were injected into C57BL/6 mice through the tail vein to examine the participation of PI3K/AKT/mTOR/S6K1 pathway in vivo and to observe the development of mammary glands affected by novel-miR-3880 and siELF2 with ultramicroscopic technique, to judge the availability of the molecular experiments and provide a theoretical basis for practice in dairy goat breeding and breast care.
Materials and Methods
Animals and Ethics
Three-year old female Guanzhong dairy goats 90-day postpartum (peak lactation period) in a research-animal-keeping farm near Northwest A&F University of Shaanxi province in China were selected and anesthetized. Then, one cubic centimeter of mammary gland tissue was removed to PBS with penicillin/streptomycin (100 U/mL, Harbin Pharmaceutical Group, China) from the middle part of the mammary gland with a scalpel. The mammary gland tissue was used to isolate MECs. The wound was sewn and sterilized immediately and animals recovered after the surgical line was removed a week later. C57BL/6 mice used in this study were of a similar age, weight, parity, and litter size, and delivered newborns on the same day. The mice were raised in an SPF environment with natural drink and food in separate nests. Each group had six nests of mice. Injective novel-miR-3880 agomir and siELF2 with 2′OMe and 5′Chol in vivo modification were bought from Ribobio (Guangzhou, China). All surgical procedures conformed to institutional and national guidelines and were approved by the Animal Care and Use Committee of the Northwest A&F University (China).
Cell Culture and Cell Treatment
A previous method (Wang et al., 2010) was used to isolate goat MECs. Mammary epithelial cells were cultured in a basic DMEM/F12 medium (Hyclone, United States) with 10% bovine serum albumin (Gibco, United States) and penicillin/streptomycin, and incubated in 5% CO2 at 37°C in a humid atmosphere. Novel-miR-3880, siELF2, siDOCK1, and si-ciRNA13761 were synthesized at GenePharma Corporation (Shanghai, China), and the sequences were GGUCCCGCCGCCGCCGCC, CCUAC CUGCUUGAGAGAUU, GCUUCGUACAUCCAUCUUA, and CCUGCACAAGGAAUGUGAU. mammary epithelial cells transfection was performed with Lipofectamine 2000 reagent (Invitrogen, United States). PI3K inhibitor (TGX-221, Selleck, Shanghai, China) in 20 μM, AKT (GDC-0068, Selleck, Shanghai, China) inhibitor in 50 μM, mTOR (Everolimus RAD001, Selleck, Shanghai, China) inhibitor in 0.5 nM, and S6K1 inhibitor (WAY-600, Selleck, Shanghai, China) in 50 μM applied to treat MEC were dissolved in DMSO, and equal DMSO was applied in the control group. The concentration was selected as suggested.
Isolation and Analysis of RNA
Total RNA was isolated from MECs by Trizol Reagent (Invitrogen, United States) according to the manufacturer’s protocol. miRcute Plus miRNA First-Strand cDNA Kit (Tiangen, Beijing, China) was used to perform reverse transcription of novel-miR-3880. The PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan) was used to acquire cDNA. Oligo dT primer was applied for mRNA reverse transcription; for cDNA of ciRNA, total RNA was dealt with RNase, and then random primer was used for reverse transcription. RT-qPCR was conducted using SYBR Green qPCR Master Mix (Takara, Japan) to analyze the mRNA, miRNA, or ciRNA expression levels. Primers used in RT-qPCR are shown below. ELF2: TGTGGCGGTTCAGTCAGTTA (forward), CAGTAGAGGCTGGCATCACA (reverse); Dock1: CGGGA CTCAGAACTCATCGG (forward), CCACCACATCGGTCC TCATC (reverse); ciRNA13761: GAAGTGGTCGGAGGACGTG (forward); TTGTGCAGGTCACAGAGCTT (reverse); β-actin: GATCTGGCACCACACCTTCT (forward); GGGTCATCTTC TCACGGTTG (reverse); novel-miR-3880: TATATAGCCGCCG CCGCC (forward); U6: CTCGCTTCGGCAGCACA (forward); AACGCTTCACGAATTTGCGT (reverse).
RNA-Sequencing
Goat MEC RNA quality was analyzed with an Agilent bioanalyzer 2100, and mRNA was captured using the Poly (A) mRNA Magnetic Isolation Module (NEBNext, United States). The library was constructed using an Ultra RNA Library Prep Kit for Illumina (NEBNext, United States). After the library was purified by Agencourt AMPure XP beads (Beckman, United States), the quality of the library was detected again using an Agilent Bioanalyzer 2100 and Qubit. In addition, before Illumina HiSeq, cBOT automatic clustering was conducted using the TruSeq PE Cluster Kit v4.
Vector Construction
PsiCHECK-2 empty vector was purchased from Promega (Madison, United States). ELF2-1 3′UTR and ELF2-2 3′UTR containing the seed site were amplified and inserted into psiCHECK-2 vector as ELF2-1 and ELF2-2 wild type (Wt) vectors. Primers for ELF2-1 3′UTR were CTAGGGTGTTAGTGCCGGTC (forward) and CGACAAGATCACCCATCCCA (reverse); for ELF2-2 3′UTR were TTCCCAACTGCTGCGTGAA (forward) and GAGTTACAGGACCTAGTTTGGTGT (reverse). The vectors sequencing documents are shown in Supplementarys Files S3, S4. ELF2-1 Mutant (Mu) and ciRNA13761 Wt/Mu psiCHECK-2 vectors were provided by Tsingke Biological Technology Company (Beijing, China).
The complete CDS region of ELF2 was amplified with primers ATGACATCAGCAGTGGTTGAC (forward) and TCATTTCTCACACGCTACCAG (reverse) and inserted into pcDNA3.1(+)vector. The sequencing result of pcDNA3.1-ELF2 vector is provided in Supplementary File S5. In addition, ciRNA13761-pcDNA3.1(+) CircRNA Mini Vector was provided by Tsingke Biological Technology Company (Beijing, China) and marked as pcDNA3.1#-ciRNA13761.
Dual-Luciferase Reporter Assays
Luciferase activities were measured in accordance with the protocol of Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States). The ratio of hRluc and hluc+ activities was used to calculate the relative luciferase activity.
Western Blot Analysis
Equal amounts of each protein sample was loaded to detect protein expression. The following primary antibodies were used in this experiment: β-actin (Beyotime, AA128, Shanghai, China), ELF2 (Proteintech, 12499-1-AP, United States), Bax (BBI, D220073, Shanghai, China), Bcl-2 (BBI, D260117, Shanghai, China), PI3K p110 beta (Bioss, bs-6423R, Beijing, China), p-PI3K p110 beta (Bioss, bs-6417R, Beijing, China), AKT (Cell Signaling Technology, 4685, United States), p-AKT (Cell Signaling Technology, 4060, United States), mTOR (Boster Biological Technology, BM4182, United States), and p-mTOR (Boster Biological Technology, BM4840, United States). Horse Radish Peroxidase-conjugated goat anti-rabbit IgG secondary antibodies were purchased from Beyotime (Shanghai, China).
Immunohistochemistry
Slides cut from paraffin-embedded tissues underwent drying, rehydration, antigen retrieval, and permeation before the samples were blocked in goat serum for 20 min and incubated in the primary antibody then in the second antibody. Color development was performed with a DAB Substrate kit (Solarbio, DA1010, Beijing, China) and counterstained with Hematoxylin (Solarbio, H8070, Beijing, China).
Flow Cytometry
MEC apoptotic rate was evaluated using the Annexin V-FITC/PI apoptosis kit (7Sea Biotech, Shanghai, China) and MEC cell cycle was measured by Cell cycle staining Kit (MultiSciences, Hangzhou, China) according to the manufacturer’s instructions.
Triglyceride Content Detection and Oil Red O Staining
Triglyceride content was measured by Tissue/Cell triglyceride assay kit (Applygen, Beijing, China) and normalized to protein concentration, which was tested by BCA protein assay kit. Oil red O used in staining was purchased from Solarbio (Beijing, China). Relative density of lipid drops was analyzed by Image-Pro Plus 6.0 Software.
ELISA
MEC cultivator was acquired to detect the contents of α s1-, α s2-, β-, and κ-casein respectively using casein ELISA Kits specialized for goats (mlbio, Shanghai).
Cell Counting Kit-8 (CCK-8) Assay
Ten microliter of CCK-8 solution (7Sea Biotech, Shanghai, China) was added into MECs seeded in 96-well plates and incubated at 37°C for 4 h to evaluate MEC viability. Then, the optical density value was measured at 450 nm with a Microplate Reader (Bio Tek, United States).
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
EdU fluorescence labeling was achieved by EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime, Shanghai, China), and cell nucleuses were dyed with DAPI (Beyotime, Shanghai, China).
Statistical Analysis
Sequencing data quality was evaluated using FastQC v0.10.1 and data filtering was performed with Cutadapt 1.9.1. Short read alignment was conducted using Hisat v2.0.14. SNV and InDel information was acquired from Samtools v0.1.18 analysis. The weight of female offspring in every nest was averaged and the data in six nests were set as one group. The acquired data of three groups were applied to analyze. Experiments in this study were conducted in three independent repeats. SPSS 22.0 was used to conduct one-way ANOVA and Student t-test. ∗p < 0.05, which was considered significant, and ∗∗p < 0.01, which was considered highly significant.
Results
Novel-miR-3880 Directly Suppresses the Expression of ELF2
After novel-miR-3880 was transfected into MECs with NC as control, RNA sequencing was conducted and DEGs are shown in Supplementary File S6. There were 57 downregulated DEGs and 10 upregulated DEGs in novel-miR-3880 group (Figure 1A). ELF2 selected from DEGs of RNA-sequencing was verified via RT-qPCR and western blotting to evaluate the sequencing accuracy (Figure 1B).
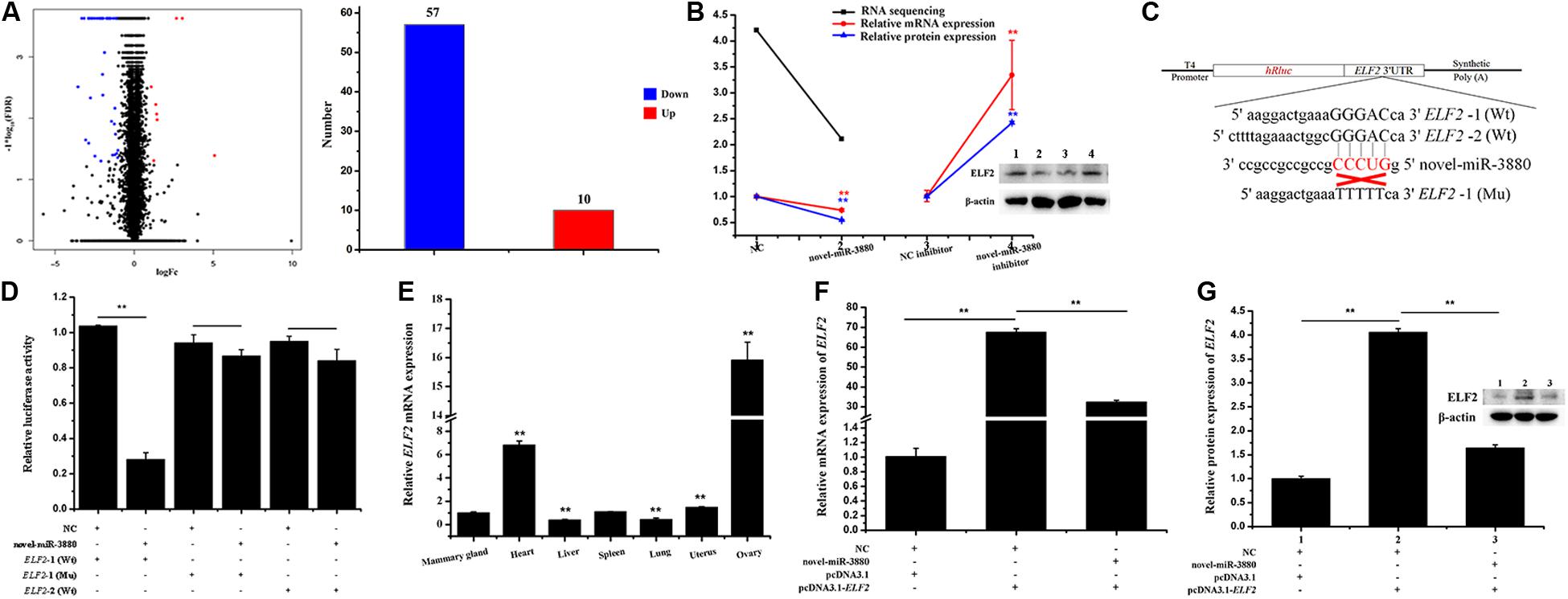
Figure 1. Confirmation of the relationship between novel-miR-3880 and ELF2. (A) DEG number acquired from RNA sequencing. Dots in region logFc < 0 represent downregulated genes, otherwise, in region logFc > 0 means upregulated genes. In total, there are 57 downregulated DEGs and 10 upregulated DEGs. (B) Novel-miR-3880 suppressed ELF2 expression in RNA sequencing, mRNA, and protein levels. When novel-miR-3880 was inhibited, mRNA and protein levels were improved. (C) Schematic diagram explaining the design of ELF2-1 (Wt), ELF2-2 (Wt) and ELF2-1 (Mu) luciferase reporter vectors. (D) ELF2-1 (Wt), ELF2-2 (Wt) and ELF2-1 (Mu) reporter vectors were co-transfected with novel-miR-3880 or NC into MEC, and luciferase assay was performed at 48 h after transfection. (E) Goat tissue expression profile of ELF2. (F,G) ELF2 overexpression vector pcDNA3.1-ELF2 effectiveness was verified in mRNA and protein levels. Novel-miR-3880 significantly decreased the efficiency of pcDNA3.1-ELF2 vector.
A schematic diagram of dual luciferase reporter vectors was described in Figure 1C. It is shown in Figure 1D that the relative luciferase activity was lowered when novel-miR-3880 was co-transfected with psiCHECK2-ELF2-1 3′-UTR Wt vector. However, the relative luciferase activity stayed the same level when either NC or novel-miR-3880 were co-transfected with ELF2-2 3′-UTR Wt vector or psiCHECK2-ELF2-1 3′-UTR Mu vector (Figure 1D). In addition, the expression of ELF2 in different goat tissues was detected (Figure 1E) and pcDNA3.1-ELF2 overexpression vector efficiency is provided in Figures 1F,G.
CiRNA13761 Sponges Novel-miR-3880 and Induces a Regulation in ciRNA13761/Novel-miR-3880/ELF2 Network
Relative luciferase activity shown in Figure 2A illustrates that the relative luciferase activity of Wt psiCHECK2-ciRNA13761 vector was reduced, but that of the Mu vector was not changed, which indicates novel-miR-3880 was absorbed by ciRNA13761. To explore the regulatory pattern of ciRNA13761, the network among ciRNA13761, novel-miR-3880, and ELF2 was established, and the role of ciRNA13761 source gene-DOCK1 in the network was determined. The efficiency of si- and pcDNA3.1#-ciRNA13761 is clarified in Figures 2B,C; it could be seen in Figures 2C,D that novel-miR-3880 decreased the expression of ciRNA13761. Combined with the effect of ciRNA13761 on novel-miR-3880 shown in Figure 2E, it showed a mutual inhibition between novel-miR-3880 and ciRNA13761. Furthermore, ciRNA13761 induced an increase in ELF2 expression, while si-ciRNA13761 decreased ELF2 expression (Figures 2F,G). To determine the function of ELF2 in the network, the efficiency of siELF2 was verified (Figures 2H,I). It is evident that ELF2 inhibited novel-miR-3880 (Figure 2J), but promoted ciRNA13761 and DOCK1 expression (Figures 2K,L). Otherwise, novel-miR-3880 improved but ciRNA13761 restrained the expression of DOCK1 (Figures 2M,N). Interestingly, as DOCK1 was knocked down (Figure 2O), novel-miR-3880 and ciRNA13761 expression were enhanced while ELF2 expression was not affected (Figures 2P,R).
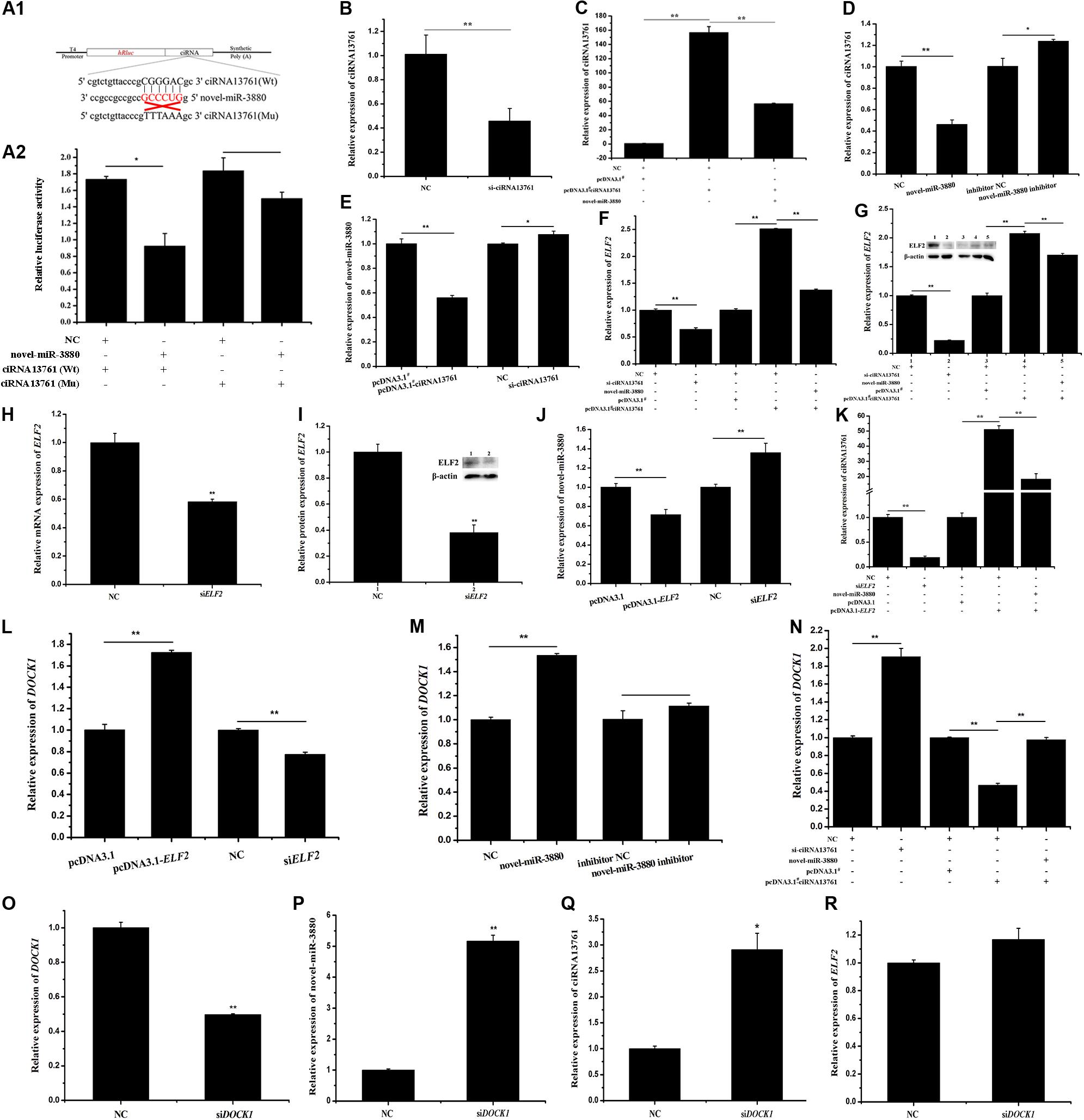
Figure 2. Network construction of ciRNA13761/DOCK1/novel-miR-3880/ELF2 axis. (A) Information of Wt and Mu psiCHECK2-ciRNA13761 Wt and Mu vectors design and relative luciferase activity when vectors were co-transfected with novel-miR-3880 or NC. (B) The efficiency of si-ciRNA13761. (C) The efficiency of pcDNA3.1#-ciRNA13761 and effects of novel-miR-3880 on pcDNA3.1#- ciRNA13761 efficiency. (D) Effects of novel-miR-3880 on ciRNA13761 expression. (E) The regulation of ciRNA13761 to novel-miR-3880 expression. (F,G) The regulation of ciRNA13761 and novel-miR-3880 to ELF2 mRNA and protein. (H,I) The efficiency of siELF2 at mRNA and protein levels. (J,K) Effects of ELF2 on ciRNA13761 and novel-miR-3880 expression; the existence of novel-miR-3880 could impair the expression of ciRNA13761 enhanced by ELF2 overexpression. (L–N) Effects of ELF2, novel-miR-3880 and ciRNA13761 on DOCK1 expression. (O) The efficiency of siDOCK1. (P–R) The regulation of siDOCK1 to ELF2, novel-miR-3880 and ciRNA13761.
Novel-miR-3880 Improves MEC Viability and Suppresses Apoptosis via ELF2
As CCK-8 results revealed, si-ciRNA13761, novel-miR-3880, and siELF2 improved MEC viability (Supplementary File S7a) while novel-miR-3880 recovered MEC viability reduction caused by ciRNA13761 and ELF2 overexpression (Supplementary Files S7b,c). Meanwhile, late apoptotic rates of MEC were analyzed and found the apoptosis of MEC was restrained by si-ciRNA13761, novel-miR-3880, and siELF2, while rising the apoptotic rate induced by ciRNA13761. ELF2 overexpression was balanced by novel-miR-3880 (Supplementary Files S7d–f). MEC cell cycle did not change significantly when the expression of novel-miR-3880 was overexpressed (Supplementary File S7g).
CiRNA13761/Novel-miR-3880/ELF2 Axis Activates PI3K/AKT/mTOR/S6K1 Pathway
C57BL/6 mice during the lactation period were injected with novel-miR-3880 or siELF2 and a schematic diagram about the operational details is shown in Figure 3A. Postpartum mice were injected with 10 nmol novel-miR-3880 or siELF2 per individual with six mice in a group, while an interval of three days and four days was applied alternately. Mammary gland tissue and blood samples were collected at day 22. The phosphorylation levels of PI3K, AKT, mTOR, and S6K1 in mammary gland tissue were shown in Figures 3B,C, which showed a higher phosphorylation of PI3K/AKT/mTOR/S6K1 in novel-miR-3880 and siELF2 groups. In MEC, novel-miR-3880 and siELF2 enhanced the phosphorylation of PI3K, AKT, mTOR, and S6K1, and novel-miR-3880 balanced the reduction of phosphorylation caused by ELF2 overexpression (Figures 3D,E). Then PI3K, AKT, mTOR, and S6K1 inhibitors dissolved in DMSO were applied to demonstrate the role of novel-miR-3880 and siELF2 in PI3K/AKT/mTOR/S6K1 pathway. In Figure 3F, PI3K, AKT, mTOR, and S6K1 inhibitors show a negative effect to the phosphorylation of each other, and the level of Bcl2/Bax was decreased when PI3K, AKT, mTOR, and S6K1 were inhibited. The results indicated that novel-miR-3880 and siELF2 could alleviate the reduction of phosphorylated-PI3K/AKT/mTOR/S6K1 and Bcl2/Bax levels (Figures 3G–J). However, Figures 3K,L show that ciRNA13761 played a negative role in PI3K/AKT/mTOR/S6K1 pathway activation, but PI3K/AKT/mTOR/S6K1 pathway was reactivated when novel-miR-3880 was co-transfected or ciRNA13761/DOCK1 was knocked down.
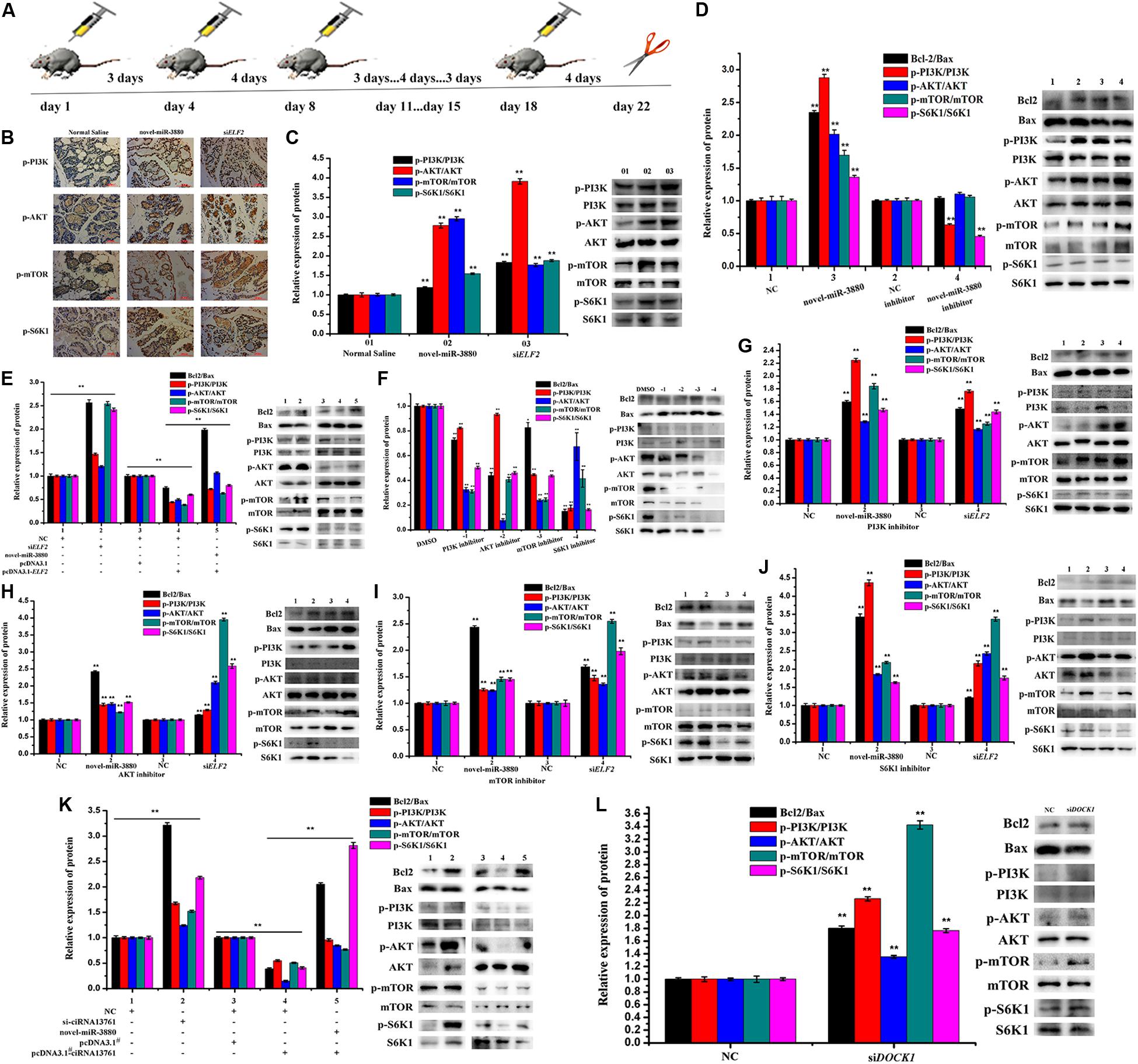
Figure 3. Relationship between ciRNA13761/novel-miR-3880/ELF2 axis and PI3K/AKT/mTOR/S6K1 pathway. (A) Schematic diagram of animal treatment. C57BL/6 mice were injected with novel-miR-3880 or siELF2 in an interval of three days and four days alternatively. Samples were harvested at day 22. (B) Immunohistochemistry of mouse mammary gland for p-PI3K, p-AKT, p-mTOR and p-S6K1 in Normal Saline, novel-miR-3880 and siELF2 groups. (C) Protein phosphorylation level of PI3K, AKT, mTOR and S6K1 in mouse mammary gland. (D,E) Effects of novel-miR-3880 and ELF2 on Bcl2/Bax pathway and protein phosphorylation level of PI3K, AKT, mTOR and S6K1 in MEC. (F) PI3K, AKT, mTOR and S6K1 inhibitors suppressed the phosphorylation of PI3K, AKT, mTOR and S6K1 in MEC. (G–J) The role of novel-miR-3880 and siELF2 in Bcl2/Bax and protein phosphorylation level of PI3K, AKT, mTOR and S6K1 in MEC with PI3K, AKT, mTOR or S6K1 inhibited. (K) Regulation of ciRNA13761 on Bcl2/Bax and PI3K, AKT, mTOR and S6K1 phosphorylation, and the balance effects of novel-miR-3880. (L) Effects of siDOCK1 on Bcl2/Bax and PI3K, AKT, mTOR and S6K1 phosphorylation.
Novel-miR-3880 Enhances MEC Proliferation Through PI3K/AKT/mTOR/S6K1 and Bcl-2/Bax Pathway via Targeting ELF2
The results show that MEC proliferation would be suppressed when PI3K, AKT, mTOR, or S6K1 were inhibited (Supplementary Files S8a,b), but the participation of novel-miR-3880 and siELF2 played a positive part in freeing MEC to proliferate (Supplementary Files S8a,c–g). In addition, results shown in Supplementary Files S8h–k provide evidence for the restraint of ciRNA13761 and ELF2 overexpression on MEC proliferation and the restraint was alleviated by novel-miR-3880, and further affirmed the modulation of ciRNA13761, novel-miR-3880, and ELF2 on MEC proliferation.
ELF2 Contributes to the Modulation of Novel-miR-3880 to MEC Lipid Formation, Triglyceride Synthesis and Casein Secretion
The Oil Red O staining and triglyceride content test revealed that novel-miR-3880, si-ciRNA13761, and siELF2 enhanced while ciRNA13761 and ELF2 overexpression reduced MEC lipid formation (Supplementary Files S9a–c) and triglyceride synthesis (Supplementary Files S9d–f), with the reduction weakened by novel-miR-3880 (Supplementary Files S9b,c,e,f). The participation of PI3K/AKT/mTOR/S6K1 pathway is illustrated in Supplementary File S9g; it is shown that MEC triglyceride content declined significantly when PI3K, AKT, mTOR, or S6K1 was inhibited. It is worthwhile to mention that novel-miR-3880 and siELF2 improved the triglyceride content in mouse mammary glands but did not change blood triglyceride content (Supplementary Files S9h,i).
As is shown in Supplementary File S10a, novel-miR-3880, si-ciRNA13761, and siELF2 increased MEC κ-casein secretion, but decreased α s1-, α s2-, and β-casein secretion. The ciRNA13761 and ELF2 overexpression and novel-miR-3880 co-transfection experiment demonstrated an interaction among ciRNA13761, novel-miR-3880, and ELF2 in MEC κ-, α s1-, α s2-, and β-casein secretion regulation (Supplementary Files S10b,c).
Novel-miR-3880 Promotes Mammary Gland Development, Extends Lactation Period and Benefits Offspring Growth
The ultrastructure of collected mammary gland tissues was observed. It is significant that lipid droplets became larger but fewer, as the acini cavities turned smaller in the control group (Figure 4A). However, animals injected with novel-miR-3880 or siELF2 were still lactating with plump acini filled with secreta and had more abundant mammary ducts (Figures 4B,C). More images can be found in Supplementary File S11. The weight of all female offspring in six nests of each group was measured, and it was found that newborns fed by mothers in novel-miR-3880 or siELF2 groups gained more weight (Figure 4D).
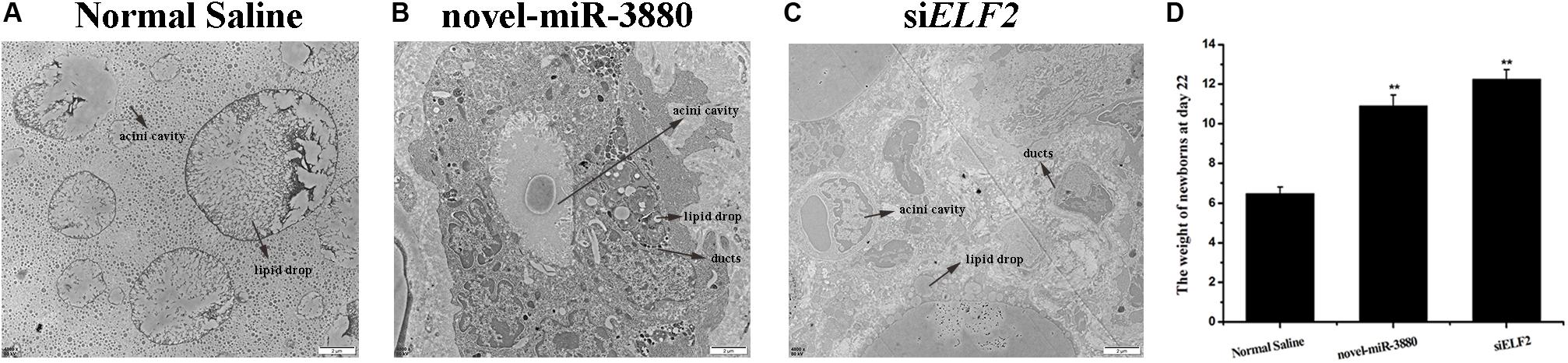
Figure 4. Mammary gland morphology after treatment with Normal Saline, novel-miR-3880, and siELF2 and weight of female offspring in each group. (A–C) Novel-miR-3880 and siELF2 promoted mammary gland development and extended lactation period. (D) Offspring fed by mothers in novel-miR-3880 and siELF2 groups grew faster than normal. There were 28 females (n = 4, 5, 5, 4, 6, 4) weighed in Normal Saline group, 30 females (n = 6, 7, 5, 4, 4, 4) in novel-miR-3880 group and 32 females (n = 4, 6, 5, 5, 6, 6) in siELF2 group.
Discussion
This research aimed to study the effects of novel-miR-3880 and relative molecules on mammary gland development and lactation traits, thus gaining a better understanding of lactation in dairy goats. A general view of our work is presented in Supplementary File S12. Novel-miR-3880 was selected from our previous study (Hou et al., 2017) for its higher expression in peak lactation stage. The present study reveals that novel-miR-3880 targeted ELF2 and was sponged by ciRNA13761. CiRNA13761, novel-miR-3880, and ELF2 as well as DOCK1 had an interaction with each other, although siDOCK1 did not directly affect the expression of ELF2. It indicates that the molecules we studied have a mutual regulation to keep a steady state in MEC. Novel-miR-3880 combined with ELF2 3′-UTR to suppress the expression of ELF2, while ciRNA13761 sponged novel-miR-3880 and reduced the chance of combination between novel-miR-3880 and ELF2, and ELF2 inhibited the expression of novel-miR-3880 to protect itself from too low expression. Interestingly, novel-miR-3880 also presented resistance to ciRNA13761 to ensure its own function in the network. DOCK1, as the source gene of ciRNA13761, improved both expression of novel-miR-3880 and ciRNA13761 when it was knocked down, while ELF2 exerted a facilitated role in DOCK1 expression. Our findings suggest that the network participates in MEC triglyceride synthesis, casein secretion, cell viability, and proliferation, as well as anti-apoptosis regulation, and plays an important part in PI3K/AKT/mTOR/S6K1 and Bcl-2/Bax pathways.
Novel-miR-3880 improved MEC triglyceride synthesis, casein secretion, cell viability, proliferation, and anti-apoptotic ability via targeting ELF2. Novel-miR-3880 would offset the reduction caused by ciRNA13761 and ELF2 overexpression. This confirms the mutual regulation among ciRNA13761, novel-miR-3880, and ELF2 on MEC. Besides, the results shown in Figure 3 not only demonstrate that ciRNA13761, novel-miR-3880, ELF2, and DOCK1 engaged in PI3K/AKT/mTOR/S6K1 pathway and Bcl2/Bax signaling, but also certify a close relationship among PI3K, AKT, mTOR, and S6K. Once any of them was inhibited, all of them would face a decrease in phosphorylation level as well as a decline in the expression of Bcl2/Bax. Fortunately, novel-miR-3880 and siELF2 could activate PI3K/AKT/mTOR/S6K1 and Bcl2/Bax pathways, and possesses the ability to alleviate the decline of PI3K/AKT/mTOR/S6K1 phosphorylation levels and Bcl2/Bax expression ratio, which might be a reason for the mitigative effects of novel-miR-3880 and siELF2 on reduced MEC proliferation induced by PI3K, AKT, mTOR, or S6K1 inhibitor. It thus appears that the results are consistent with previous research that PI3K/AKT/mTOR/S6K1 pathway activation benefits cell growth (Park et al., 2011; Zhang et al., 2019), and provide sufficient evidence for the participation of novel-miR-3880 and ELF2 in the modulation of PI3K/AKT/mTOR/S6K1 pathway both in vitro and in vivo. In addition, the role of ciRNA13761 and DOCK1 in PI3K/AKT/mTOR/S6K1 pathway and Bcl2/Bax pathway was elaborated, which increases evidence for the regulation of ciRNA13761/novel-miR-3880/ELF2 to PI3K/AKT/mTOR/S6K1 and Bcl2/Bax pathway.
The results provide evidence that novel-miR-3880 is a key molecule in regulating the effects of ciRNA13761 and ELF2 on MEC function, illustrating the role of their mutual regulation in MEC cellular processes. It is worth mentioning that novel-miR-3880 decreased content of most casein (α s1-, α s2-, and β-casein), which makes it closer to human milk protein composition (Kunz and Lonnerdal, 1992; Hao-Feng, 2012), and increased κ-casein content to make milk products easier to process (Brigid et al., 2003).
Importantly, it was confirmed that novel-miR-3880 and siELF2 work well in facilitating mammary gland development, improving triglyceride content in mammary glands but not in blood, extending lactation days, and promoting newborn growth through PI3K/AKT/mTOR/S6K1 pathway. Therefore, novel-miR-3880 application is promising in the goat breeding and milk industry.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Northwest A&F University.
Author Contributions
BC and XA guided the experiment and applied the funds. YZ designed and performed most of the experiment and wrote the manuscript. FC, WL, JL, and XD conducted some of experiments. GN and SJ revised the manuscript. All of the authors have read and approved the submitted version of the manuscript.
Funding
This study was supported by Shaanxi Key Research and Development Plan Project (2020ZDLNY02-01 and 2020ZDLNY02-02), National Natural Science Foundation of China (31601925), Natural Science Foundation of Shaanxi Province (2018JM3006 and 2015JM3087), China Postdoctoral Science Foundation (2016T90954 and 2014M552498) and Shaanxi Science and Technology Innovation Project Plan (2015KTCQ03-08, 2016KTZDNY02-04, 2017ZDXM-NY-081, and 2018ZDCXL-NY-01-04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks for Northwest A&F University’s Life Science Core Services Facility.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00383/full#supplementary-material
FILE S1 | Differentially expressed miRNAs between Colostrum period and peak period acquired from previous sequencing results in our laboratory (Hou et al., 2017).
FILE S2 | Source and structure of ciRNA13761.
FILE S3 | Result of psiCHECK2-ELF2-1 vector sequencing.
FILE S4 | Result of psiCHECK2-ELF2-2 vector sequencing.
FILE S5 | Result of pcDNA3.1-ELF2 vector sequencing.
FILE S6 | Novel-miR-3880 regulated differentially expressed genes in MEC acquired from RNA-sequencing.
FILE S7 | MEC viability and apoptosis regulated by ELF2, novel-miR-3880, and ciRNA13761. (a–c) MEC viability was regulated by novel-miR-3880, ciRNA13761 and ELF2. (d–f) The modulation of novel-miR-3880, ciRNA13761 and ELF2 to MEC apoptosis. (g) The effect of novel-miR-3880 on MEC cycle.
FILE S8 | Roles of PI3K/AKT/mTOR/S6K1 pathway and ciRNA13761/novel- miR-3880/ELF2 axis in MEC proliferation. (a–g) In accordance with the EdU results (a), PI3K, AKT, mTOR, and S6K1 innhibitors restrained MEC proliferation (b), while novel-miR-3880 and siELF2 aroused MEC proliferation (c); and novel-miR-3880 and siELF2 could suppress the inhibition come from PI3K (d), AKT (e), mTOR (f) and S6K1 innhibitors (g). (h,i) Regulation of ciRNA13761 to MEC proliferation and the function of novel-miR-3880 on balancing reduced MEC proliferation caused by ciRNA13761 overexpression. (j,k) Regulation of ELF2 to MEC proliferation and the balance of novel-miR-3880 on reduced MEC proliferation caused by ELF2 overexpression.
FILE S9 | Effects of ciRNA13761, novel-miR-3880, and ELF2 on MEC lipid droplets formation and triglyceride synthesis. (a–c) Oil red O staining illustrating the amount of MEC lipid droplets. (d) MEC triglyceride content was improved by si-ciRNA13761, novel-miR-3880, siELF2. (e,f) ELF2 and ciRNA13761 overexpression restrained triglyceride synthesis while novel-miR-3880 eliminated some restraint. (g) PI3K, AKT, mTOR and S6K1 inhibitors suppressed triglyceride synthesis in MEC. (h) Novel-miR-3880 and siELF2 improved triglyceride content in mouse mammary gland. (i) Novel-miR-3880 and siELF2 did not change triglyceride content in blood of mouse.
FILE S10 | Effects of ciRNA13761, novel-miR-3880, and ELF2 on κ-, α s1-, αs2- and β-casein secretion in MEC. (a) Effects of si-ciRNA13761, novel-miR-3880 and siELF2 on casein secretion. (b) Regulation of overexpressed ciRNA13761 to casein secretion. (c) Regulation of overexpressed ELF2 to casein secretion.
FILE S11 | General view of mutual regulation among ciRNA13761, DOCK1, novel-miR-3880, and ELF2.
References
Bagci, H., Laurin, M., Huber, J., Muller, W. J., and Côté, J. F. (2014). Impaired cell death and mammary gland involution in the absence of Dock1 and Rac1 signaling. Cell Death Dis. 5:e1375. doi: 10.1038/cddis.2014.338
Basu, A., and Haldar, S. (1998). The relationship between BcI2. Bax and p53: consequences for cell cycle progression and cell death. Mol. Hum. Reprod. 4, 1099–1109. doi: 10.1093/molehr/4.12.1099
Brigid, B., Smolenski, G., Wheeler, T., Wells, D., L’Huillier, P., and Laible, G. (2003). Cloned transgenic cattle produce milk with higher levels of beta-casein and kappa-casein. Nat. Biotechnol. 21:157. doi: 10.1038/nbt783
Chatchatee, P., Järvinen, K. M., Bardina, L., Vila, L., Beyer, K., Sampson, H. A., et al. (2001). Identification of IgE and IgG binding epitopes on beta- and kappa-casein in cow’s milk allergic patients. Clin. Exp. Allergy 31, 1256–1262. doi: 10.1046/j.1365-2222.2001.01167.x
Chen, C.-C., Stairs, D. B., Boxer, R. B., Belka, G. K., Horseman, N. D., Alvarez, J. V., et al. (2012). Autocrine prolactin induced by the Pten-Akt pathway is required for lactation initiation and provides a direct link between the Akt and Stat5 pathways. Genes Dev. 26, 2154–2168. doi: 10.1101/gad.197343.112
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hao-Feng, G. U. (2012). Comparison of nutritional components for goat milk, cow milk and human milk. Sci. Technol. Food Indus. 33, 369–373.
Hou, J., An, X., Song, Y., Cao, B., Yang, H., Zhang, Z., et al. (2017). Detection and comparison of microRNAs in the caprine mammary gland tissues of colostrum and common milk stages. BMC Genet. 18:38. doi: 10.1186/s12863-017-0498-2
Ji, Z., Liu, Z., Chao, T., Hou, L., Fan, R., He, R., et al. (2017). Screening of miRNA profiles and construction of regulation networks in early and late lactation of dairy goat mammary glands. Sci. Rep. 7:11933.
Kunz, C., and Lonnerdal, B. (1992). Re-evaluation of the whey protein/casein ratio of human milk. Acta Paediatr. 81, 107–112. doi: 10.1111/j.1651-2227.1992.tb12184.x
Magnuson, B., Ekim, B., and Fingar, D. C. (2012). Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 441, 1–21. doi: 10.1042/bj20110892
Meng, X., Li, X., Zhang, P., Wang, J., Zhou, Y., Chen, M., et al. (2017). Circular RNA: an emerging key player in RNA world. Brief. Bioinform. 18, 547–557.
Osorio, J. S., Lohakare, J., and Bionaz, M. (2016). Biosynthesis of milk fat, protein, and lactose: roles of transcriptional and posttranscriptional regulation. Physiol. Genomics 48, 231–256. doi: 10.1152/physiolgenomics.00016.2015
Park, K. R., Nam, D., Yun, H. M., Lee, S. G., Jang, H. J., Sethi, G., et al. (2011). beta-Caryophyllene oxide inhibits growth and induces apoptosis through the suppression of PI3K/AKT/mTOR/S6K1 pathways and ROS-mediated MAPKs activation. Cancer Lett. 312, 178–188. doi: 10.1016/j.canlet.2011.08.001
Qiu, Y., Morii, E., Zhang, B., Tomita, Y., and Aozasa, K. (2008). E74-like factor 2 transactivates valosin-containing protein gene, a gene involved in cancer growth? Exp. Mol. Pathol. 84, 226–229. doi: 10.1016/j.yexmp.2008.04.004
Roncada, P., Gaviraghi, A., Liberatori, S., Canas, B., Bini, L., and Greppi, G. F. (2002). Identification of caseins in goat milk. Proteomics 2, 723–726. doi: 10.1002/1615-9861(200206)2:6<723::aid-prot723>3.0.co;2-i
Saxton, R. A., and Sabatini, D. M. (2017). mTOR Signaling in Growth. Metabolism, and Disease. Cell 69, 361–371. doi: 10.1016/j.cell.2017.03.035
Singh, V. B., and Singh, S. N. (1980). Total protein, whey protein and casein content milk of four Indian goat breeds during lactation. Int. Goat Sheep Res.
Vicens, Q., and Westhof, E. (2014). Biogenesis of Circular RNAs. Cell 159, 13–14. doi: 10.1016/j.cell.2014.09.005
Wang, Z., Luo, J., Wang, W., Zhao, W., and Lin, X. (2010). [Characterization and culture of isolated primary dairy goat mammary gland epithelial cells]. Sheng Wu Gong Cheng Xue Bao 26, 1123–1127.
Yang, L., Miao, L., Liang, F., Huang, H., Teng, X., Li, S., et al. (2014). The mTORC1 effectors S6K1 and 4E-BP play different roles in CNS axon regeneration. Nat. Commun. 5:5416.
Keywords: ciRNA13761, novel-miR-3880, ELF2, PI3K/AKT/mTOR/S6K1 pathway, mammary gland development
Citation: Zhang Y, Liu J, Li W, Cao F, Niu G, Ji S, Du X, Cao B and An X (2020) A Regulatory Circuit Orchestrated by Novel-miR-3880 Modulates Mammary Gland Development. Front. Cell Dev. Biol. 8:383. doi: 10.3389/fcell.2020.00383
Received: 24 March 2020; Accepted: 28 April 2020;
Published: 16 June 2020.
Edited by:
Dario Krapf, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Zhi Chen, Yangzhou University, ChinaLucas B. Pontel, Instituto de Investigación en Biomedicina de Buenos Aires (CONICET), Argentina
Copyright © 2020 Zhang, Liu, Li, Cao, Niu, Ji, Du, Cao and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binyun Cao, Y2FvYmlueXVuQDEyNi5jb20=; Xiaopeng An, YW54aWFvcGVuZ2RreUAxNjMuY29t
 Yue Zhang1
Yue Zhang1 Binyun Cao
Binyun Cao