- St. Anna Children's Cancer Research Institute, Innovative Cancer Models, Vienna, Austria
The zebrafish (Danio rerio) is a popular vertebrate model organism to investigate molecular mechanisms driving development and disease. Due to its transparency at embryonic and larval stages, investigations in the living organism are possible with subcellular resolution using intravital microscopy. The beneficial optical characteristics of zebrafish not only allow for passive observation, but also active manipulation of proteins and cells by light using optogenetic tools. Initially, photosensitive ion channels have been applied for neurobiological studies in zebrafish to dissect complex behaviors on a cellular level. More recently, exciting non-neural optogenetic tools have been established to control gene expression or protein localization and activity, allowing for unprecedented non-invasive and precise manipulation of various aspects of cellular physiology. Zebrafish will likely be a vertebrate model organism at the forefront of in vivo application of non-neural optogenetic tools and pioneering work has already been performed. In this review, we provide an overview of non-neuromodulatory optogenetic tools successfully applied in zebrafish to control gene expression, protein localization, cell signaling, migration and cell ablation.
Introduction
Experimental control over protein function is an invaluable asset to dissect cellular processes on the molecular level. Conventional means of conditionally inducing protein activity, e.g., by small molecules or by heatshock, provide temporal control, but are typically limited in their spatial resolution. Optogenetic techniques emerged as a highly precise way to establish spatiotemporal control over protein activity by light.
Optogenetics was initially applied in neurobiology by ectopic expression of channelrhodopsins (ChRs), light-sensitive ion-channels, in neuronal cells (Boyden et al., 2005). ChRs depolarize neurons upon illumination and thereby modulate neuronal activity. Since then, the field has evolved to include non-neural applications. At the core of the new toolkit are light-sensitive proteins or protein-domains of bacterial, fungal and plant origin, such as phytochromes (e.g., PHYB-PIF), blue light using flavin (BLUF) domain proteins, cryptochromes (e.g., CRY2-CIB1) and light oxygen voltage (LOV) domains (Figure 1) (Ni et al., 1999; Harper et al., 2003; Liu et al., 2008; Yuan and Bauer, 2008). Irradiation with light of an appropriate wavelength causes conformational changes, commonly resulting in dimerization or oligomerization of the proteins. Careful engineering of photosensitive domains into enzymes, transcription factors or other proteins of interest endowed light-mediated control over the conformation of these proteins and resulted in light-activatable genetic tools for a variety of applications such as control over gene expression, genome editing, and protein relocalization (Beyer et al., 2015; Buckley et al., 2016; Polesskaya et al., 2018).
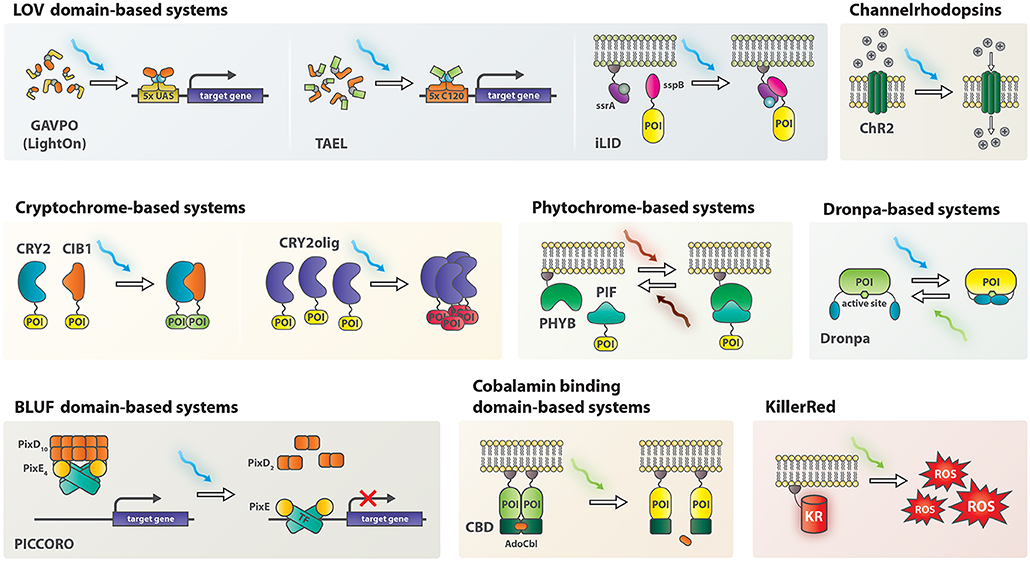
Figure 1. Schematic representation of non-neuromodulatory optogenetic systems applied in zebrafish. AdoCbl, 5′-deoxyadenosylcobalamin; CBD, cobalamin binding domain; ChR2, channelrhodopsin 2; CIB1, cryptochrome interacting bHLH 1; CRY2, cryptochrome 2; KR, KillerRed; PHYB, Phytochrome B; PIF, phytochrome interacting factor; POI, protein of interest; ROS, reactive oxygen species; TF, transcription factor.
Zebrafish are an excellent model for optogenetic in vivo applications, since they are transparent at embryonic and larval stages and develop extra-uterine, providing light accessibility to the entire organism (Simmich et al., 2012). This optical clarity has been exploited to directly monitor fluorescently tagged cells and proteins in the living organism, hereby gaining great insights into cellular and subcellular processes during development and disease. In addition to this passive observation, novel optogenetic tools now enable manipulation of biological processes in vivo with the possibility of a direct readout of the effects. In this review, we summarize non-neuromodulatory optogenetic tools, which have recently been applied in zebrafish to control gene expression, cell migration, protein localization, signaling pathway activity and cell death (Table 1). Many of them can be readily combined with existing genetic systems like Gal4/UAS and established transgenic strains, making them a powerful addition to the genetic toolbox in zebrafish.
Gene Expression
Control over gene expression is beneficial to interrogate gene function and to model diseases including neurodegeneration and cancer. Temporal control is typically achieved in zebrafish by using either compound-activated systems like tamoxifen for estrogen receptor fusions or heatshock promoters (Mayrhofer and Mione, 2016). Optogenetic gene expression systems additionally offer spatial control and here we present their current applications in zebrafish.
Cryptochrome 2 (CRY2-CIB1)
The Arabidopsis thaliana photoreceptor protein cryptochrome 2 (CRY2) heterodimerizes with the CRY-interacting basic-helix-loop-helix (bHLH) transcription factor 1 (CIB1) upon blue light illumination (Liu et al., 2008). This heterodimerization was exploited to create a light-inducible Gal4 system by fusing the Gal4 DNA binding domain to CRY2 and the Gal4 activation domain containing VP16 to CIB1. A luciferase-based readout revealed that blue-light illumination for 2 h, but not red light, induced luciferase expression in zebrafish with this system (Liu et al., 2012).
LOV Domains
EL222/C120 and TAEL
EL222 is a LOV domain-containing light-inducible transcription factor from Erythrobacter litoralis. Blue light induces homodimerization of EL222 and subsequent binding to its regulatory element C120 activates transcription of downstream genes. EL222 was adapted for eukaryotic use by adding a nuclear localization sequence and the herpes simplex virus-derived VP16 transcriptional activation domain resulting in VP-EL222 (Motta-Mena et al., 2014). Injection of VP-EL222 mRNA together with a C120:mCherry reporter into zebrafish demonstrated that blue light effectively mediates gene expression in this system. As some toxicity was observed with VP-EL222 it was further optimized for applications in zebrafish by exchanging the VP16 transcriptional activation domain with TA4, which is better tolerated in zebrafish, resulting in TA4-EL222 (TAEL) (Distel et al., 2009; Reade et al., 2017).
The TAEL system was used to control the expression of several genes, including sox32 to convert ectoderm to endoderm, lefty to modulate Nodal signaling and Cas9 for light-induced mosaic gene knock out. Importantly, selective illumination using either an epifluorescene, a confocal or light sheet microscope or a digital mirror device (DMD) demonstrated that TAEL endows temporal and spatial control over gene expression in zebrafish (Reade et al., 2017).
LightOn/GAVPO
The LightOn gene expression system is based on a synthetic protein termed GAVPO (Wang et al., 2012). GAVPO contains a Gal4 DNA binding domain, a p65 transactivation domain and the small fungal protein Vivid including its LOV domain for light-mediated dimerization (Schwerdtfeger and Linden, 2003). As Gal4 binds to UAS sites as a homodimer, its transcriptional activity can be controlled by light using the LightOn system (Wang et al., 2012). Illumination with blue light triggers homodimerization of GAVPO. The dimer then binds to UAS sites to induce expression of a gene of interest. The LightOn system was applied in zebrafish to gain spatiotemporal control over expression of two cell ablation systems, nitroreductase and the cytotoxic viral ion channel M2H37A (Mruk et al., 2019). GAVPO-mediated nitroreductase expression induced apoptosis in embryos when treated with metronidazole and irradiated with blue light. In addition, blue-light induced GAVPO-triggered expression of M2H37A leading to necrotic cell death and developmental defects in zebrafish embryos, which could be rescued by addition of the antiviral channel blocker rimantadine. Furthermore, a transgenic GAVPO strain (elavl3:GAVPO) was successfully established, demonstrating that GAVPO expression can be tolerated in zebrafish, although a previous study reported toxicity of GAVPO (Reade et al., 2017).
PICCORO
The BLUF domain-based “PixD complex dependent control of transcription” (PICCORO) system was engineered to directly control transcription factor activity by protein localization (Masuda et al., 2013). PixD is a bacterial blue-light photoreceptor that interacts with the response regulator-like protein PixE and forms PixD10-PixE4 oligomers in the dark (Masuda and Tanaka, 2016). Illumination with blue light dissociates the large oligomeric complexes into PixD dimers and PixE monomers, triggered by changes in protein conformation (Yuan and Bauer, 2008) (Figure 1).
The PICCORO system was applied in zebrafish for light-mediated control of an engineered transcriptionally repressive version of the transcription factor No Tail (Ntl), which is important for tail tissue formation (Masuda et al., 2013). For this, the DNA binding domain of Ntl and the repressor domain of Engrailed were fused, resulting in the transcriptional repressor Ntl-EnR. When expressed in zebrafish, Ntl-EnR led to the typical “ntl” phenotype with missing tail tissues. To achieve light-dependent transcriptional repression, the N-terminus of PixE was fused to Ntl-EnR and the construct was named NtlPixE. When NtlPixE was expressed in a transgenic strain also ubiquitously expressing PixD (tg(EF1α:PixD)), transcriptional repression and the resulting “ntl” phenotype was ameliorated in the dark, as PixD10-NtlPixE4 oligomers would form and sequester NtlPixE away from its DNA binding site. Blue light illumination could restore the “ntl” phenotype, revealing direct light-mediated control of a chimeric transcription factor in zebrafish.
Cell Migration
The cytoskeleton is a major contributor to cell migration and morphology changes. Several optogenetic tools have been established to control cytoskeleton dynamics in a precise manner, offering possibilities to investigate the mechanisms underlying migration.
Photoactivatable Rac
In an effort to investigate phosphoinositide 3-kinase (PI3K)-dependent movement of neutrophils in zebrafish, photo-activatable Rac (PA-Rac) was employed (Yoo et al., 2010). PA-Rac was engineered by fusion of a phototropin-derived LOV domain to Rac1 to sterically inhibit Rac function. Illumination with blue-light restores Rac activity and induces directed cell movement in vitro (Wu et al., 2009). In zebrafish embryos expressing PA-Rac in neutrophils, precise regulation of directional neutrophil migration was demonstrated using laser illumination. By this means, neutrophils could even be diverted away from wounds, opening up new possibilities to investigate cellular functions in inflammation (Yoo et al., 2010).
Channelrhodopsin (ChR)
In addition to controlling neuronal activity, ChRs were also applied in modulating cell migration. Through tissue-specific expression of ChR2, a blue-light-reactive cation channel protein, zebrafish pigmentation patterns were temporarily disarranged by illumination due to induced migration of pigment melanophores (Aramaki and Kondo, 2019).
Protein Localization
Tight regulation of protein localization in cellular compartments is crucial for many biological processes, and its dysregulation can lead to disease. Studying these processes by manipulation of protein localization requires tools, which act rapidly and with high spatial resolution. Several optogenetic systems have been developed to shuttle proteins between specific subcellular compartments or to form clusters among each other.
Phytochrome B (PHYB-PIF)
The photoreactive Arabidopsis proteins phytochrome B (PHYB) and the bHLH transcription factor phytochrome interaction partner (PIF) heterodimerize upon red light illumination in the presence of the chromophore phycocyanobilin (PCB) (Ni et al., 1999). The interaction remains stable in the dark for hours after activation. Furthermore, illumination with far-red light dissociates the dimers.
In an elegant study, PHYB-PIF was applied to control nuclear localization of a synthetic transcription factor. Here, PHYB was fused to a VP16 activation domain and a heterodimerizing antiparallel leucine zipper. The tetracycline repressor (TetR) DNA binding protein was attached to PHYB-VP16 via a corresponding leucine zipper. PIF3, which possesses a nuclear localization signal, was used to translocate this chimeric transcription factor into the nucleus in zebrafish upon red-light illumination, hereby controlling the expression of luciferase. PCB was added to the medium in this study (Beyer et al., 2015). The PHYB-PIF system was further optimized for application in zebrafish by truncation of PHYB for efficient expression and injection of PCB to deliver the chromophore into cells beyond the first layer of tissue (Buckley et al., 2016). After these adjustments, PHYB was linked to CAAX for cell-membrane binding, while PIF6 was fused to enhanced green fluorescent protein (EGFP) or Pard3 (Buckley et al., 2016). Red light illumination of a region of interest triggered protein shuttling of PIF6-EGFP or PIF6-Pard3 to the cell membrane, respectively. Activation and binding of the association partners were reported to occur in a matter of seconds, with unbinding kinetics of less than a minute after far-red light illumination.
CRY2olig
CRY2 self-clusters when illuminated, which is an undesirable feature for its function in the CRY2/CIB1 heterodimer but was found useful to induce protein relocalization, leading to the development of the CRY2olig protein clustering system (Bugaj et al., 2013; Lee et al., 2014). Functionality of this system in zebrafish was demonstrated by expression of mRFP1-tagged CRY2olig, which clustered in vivo upon blue-light stimulation (Asakawa et al., 2019). To study the effects of transactivation response element DNA-binding protein 43 (TDP-43) oligomerization in amyotrophic lateral sclerosis in zebrafish, the zebrafish tardp gene was inserted into the mRFP1-CRY2olig construct and named opTDP-43z. Physiological function of opTDP-43z was unaffected by the CRY2olig-fusion compared to wildtype tardp. In dark-state, opTDP-43z was detected mainly in cell nuclei, but started relocalizing to and aggregating in the cytosol after a blue-light stimulus. Light-induced clustering of opTDP-43z expressed in neurons perturbed axonal outgrowth, increased myofiber denervation frequency and seeded non-optogenetically induced delocalization of TDP-43 (Asakawa et al., 2019).
CRY2olig promises to be a general tool to investigate protein-clustering related diseases or to control protein activity by delocalization.
Improved Light-Inducible Dimer (iLID)
The improved light-inducible dimer (iLID) system consisting of the E.coli peptides SspB and SsrA fused to the Avena sativa LOV2 domain was engineered for a high change in affinity for their respective binding partner upon blue light stimulation (Guntas et al., 2015). The iLID system offers rapid activation and deactivation kinetics and lacks homo-monomerization. iLID was applied in zebrafish to regulate mitophagy (D'Acunzo et al., 2019). To achieve this, SsrA was fused to Venus and actin assembly inducing protein (ActA). SspB was fused to RFP and autophagy and beclin-1 regulator 1 (AMBRA1). Venus-ssrA-ActA was tethered to mitochondrial outer membranes while AMBRA1-RFP-sspB was present in the cytosol in the dark. Blue-light illumination triggered association of the two proteins, resulting in relocalization of AMBRA1 to the mitochondrial membrane, inducing mitophagy and leading to a loss of mitochondrial mass (D'Acunzo et al., 2019).
Controlling Cell Signaling and Protein Activity
Precise spatiotemporal control over signaling pathway activity allows for detailed investigation of their role in cellular processes. In contrast to studying the function of proteins and pathways by transient or permanent changes in gene expression, manipulation of protein activity results in an immediate effect. Light is an ideal activator for fast-acting interference in signaling and multiple optogenetic proteins have been engineered to modulate major pathways like Wnt, Nodal or FGF in zebrafish.
Rhodopsin-Based Opto-Fz7 to Study Non-canonical Wnt-Fz/PCP Signaling
The non-canonical Wnt receptor frizzled 7 (Fz7) was engineered to be activated by light independent of ligand binding. To achieve this, the intracellular loop 3 and the C-terminus of the light sensitive receptor rhodopsin were replaced by the corresponding intracellular domains of Fz7 to create Opto-Fz7 (Capek et al., 2019). Applying Opto-Fz7 revealed that non-canonical Wnt-Frizzled(Fz)/planar cell polarity (PCP) signaling provides a permissive signal for directed migration of mesenchymal cells during zebrafish gastrulation.
LOV-Domain-Mediated Optogenetic Control Over Nodal Signaling
In order to control Nodal signaling and study its temporal role in mesendoderm induction in zebrafish, light-sensitive Nodal receptors were engineered by fusing the LOV domain of aureochrome 1 from Vaucheria frigida with the C-terminal intracellular domains of Nodal receptors Acvr1b and Acvr2b (Sako et al., 2016). Blue light controlled the dimerization of Opto-Acvr1b and 2b, upon which Nodal signaling was activated resulting in phosphorylation of Smad2 and expression of downstream target genes like goosecoid (Sako et al., 2016).
Cobalamin Binding Domains (CBDs) to Interrogate FGF Signaling
In contrast to the LOV domain based Nodal receptors Opto-Acvr1b and 2b, which dimerize upon blue light illumination, a green light-mediated de-dimerization system was used to inactivate FGF signaling. This system is based on cobalamin binding domains (CBDs) of bacterial CarH transcription factors, which are present as dimers in the dark but dissociate upon illumination with green light (Jost et al., 2015). For CBD-dimer assembly in the dark, 5′-deoxyadenosylcobalamin (AdoCbl) is required, which is cleaved from the protein upon green light illumination. A constitutively active FGF receptor, which can be inactivated by green light, was created by fusing murine mFGFR1 to the CBD from M. xanthus (MxCBD) (Kainrath et al., 2017). Injection of mFGFR1-MxCBD into zebrafish embryos together with AdoCbl led to severe malformations when kept in the dark consistent with hyperactive FGF signaling. However, green light illumination after injection of mFGFR1-MxCBD and AdoCbl completely rescued the phenotype, demonstrating that excessive FGF signaling could be inactivated in vivo (Kainrath et al., 2017).
DRONPA-Caged MEK1 to Control ERK Activity
In order to render MEK1 light-inducible, photo-dimerizable Dronpa (pdDronpa) was used to cage MEK1's active site in a single chain construct termed psMEK1tight (Zhou et al., 2017). Upon illumination with 500 nm Dronpa de-dimerizes and allows psMEK1 to bind ERK, whereas 400 nm light re-establishes the caged conformation. When applied in zebrafish psMEK1tight showed surprisingly no effect during early development (Patel et al., 2019). However, once optimized by introduction of an activating mutation found in cancer (E203K), a non-leaky light-controlled MEK1 (psMEKE203K) for in vivo investigation of MAPK signaling in zebrafish could be generated.
Optical Cell Ablation Systems
Methods to ablate distinct cells are beneficial to study cell function or regeneration. Current chemical/ genetically encoded systems are limited in their spatial resolution, but optogenetic tools offer unique selectivity of target cells.
KillerRed
KillerRed (KR) is a fluorescent protein, which produces large doses of reactive oxygen species (ROS) upon green light illumination, leading to apoptosis (Bulina et al., 2006). KR has been implemented in zebrafish for ROS-induced cell ablation in vivo. Transgenic fish lines with membrane-tethered KR expressed either in the heart or kidney were generated (Teh et al., 2010; Buckley et al., 2017). ROS levels could be controlled to inflict damage to cell membranes or cause death of illuminated KR-positive cells. Additionally, KR-activation in the heart closely resembled heart failure in humans (Teh et al., 2010). For studying neurodegeneration, a transgenic zebrafish model expressing KR in spinal cord neurons was generated, providing single cell control over cell ablation (Formella et al., 2018).
The possibility to control area, intensity and duration of ROS expression makes KR transgenic models a useful tool, not only for directed cell ablation, but also to study effects of free radicals on different tissues.
Conclusion and Perspectives
Applying optogenetics in zebrafish offers unprecedented spatiotemporal control over protein function in a living vertebrate model organism. We have summarized recent studies using chimeric proteins engineered to endow light-mediated control over their activity in biological processes ranging from gene expression, cell migration, mitophagy and signaling pathways to cell ablation.
So far, LOV and BLUF domain-, pdDronpa-, CRY2-CIB-, PHYB-PIF-, and CBD-based systems have been successfully applied in zebrafish, revealing that zebrafish are generally permissive for various optogenetic systems (Table 1). Some of the systems needed zebrafish-specific optimization to ensure proper expression (e.g., truncation of the PHYB-PIF system Buckley et al., 2016) or to reduce toxicity (e.g., replacement of the transactivation domain of VP-EL222 to generate TAEL Reade et al., 2017).
LOV and BLUF domain-based systems as well as CRY-CIB are activated by blue light, CBD by green light and PHYB-PIF by red light, offering potential orthogonal applications, which have not been realized in zebrafish to date.
An important aspect to be considered is the availability of the co-factor for the respective optogenetic system in zebrafish. Blue-light activated systems typically rely on Flavin mononucleotides (FMN) or Flavin adenine dinucleotides (FAD), which are readily available in zebrafish cells (Table 1). However, cobalamins needed for CBD or PCB for PHYB-PIF have to be supplemented and as absorption from the medium is limited, these cofactors are ideally injected. Injection at the one cell-stage will result in dilution of the cofactor while cells are dividing and will thus limit the application of the system to early developmental stages. Possible ways to apply these systems at later stages are to engineer transgenic zebrafish strains, expressing the enzymes needed to produce the cofactor (e.g., mitochondria-localized cyanobacterial heme oxygenase 1 and PCB:ferredoxin oxidoreductase for PCB). This would greatly enhance the applicability of the PHYB-PIF system with its unique and elegant far-red light off-switch.
Some of the systems like LightOn (GAVPO) can be combined with available UAS strains for light-mediated control of a plethora of transgenes promising widespread use. The ideal optogenetic system for zebrafish still needs to be determined and further rounds of optimization of expression levels, toxicity and increased activity change will likely be required for their easy application. Nevertheless, the possibility to manipulate a biological process and to image the effects using real time reporters as readout, puts zebrafish in a prime position to take full advantage of the optogenetic toolkit and shine light on many biological processes.
Author Contributions
AV and MD wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Benjamin Natha and Michael Grusch for proofreading the manuscript.
Abbreviations
ActA, actin assembly inducing protein; AdoCbl, 5′-deoxyadenosylcobalamin; AMBRA1, autophagy and beclin-1 regulator 1; BLUF, blue light using flavin; bHLH, basic-helix-loop-helix transcription factor; CBD, cobalamin binding domain; ChR, Channel rhodopsin; CIB1, cryptochrome interacting bHLH 1; CRY2, cryptochrome 2; DMD, digital mirror device; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; Fz(7), frizzled (7); iLID, improved light-inducible dimer; KR, KillerRed; LOV, light oxygen voltage; MxCBD, M. xanthus CBD; Ntl, No Tail; PA-Rac, photo-activatable Rac; PCB, phycocyanobilin; PCP, planar cell polarity; PHYB, phytochrome B; PI3K, phosphoinositide 3-kinase; PICCORO, PixD complex dependent control of transcription; PIF, phytochrome interacting factor; RFP, red fluorescent protein; ROS, reactive oxygen species; TAEL, TA4-EL222; TDP-43, transactivation response element DNA-binding protein 43; TetR, tetracycline repressor; UAS, upstream activating sequence.
References
Aramaki, T., and Kondo, S. (2019). Method for disarranging the pigment pattern of zebrafish by optogenetics. Dev. Biol. 460, 12–19. doi: 10.1016/j.ydbio.2018.12.019
Asakawa, K., Handa, H., and Kawakami, K. (2019). Optogenetic modulation of TDP-43 oligomerization fast-forwards ALS-related pathologies in the spinal motor neurons. Nat Commun. 11:1004. doi: 10.1101/789057
Beyer, H. M., Juillot, S., Herbst, K., Samodelov, S. L., Müller, K., Schamel, W. W., et al. (2015). Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth. Biol. 4, 951–958. doi: 10.1021/acssynbio.5b00004
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., and Deisseroth, K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. doi: 10.1038/nn1525
Buckley, C., Carvalho, M. T., Young, L. K., Rider, S. A., McFadden, C., Berlage, C., et al. (2017). Precise spatio-temporal control of rapid optogenetic cell ablation with mem-killerred in zebrafish. Sci. Rep. 7:5096. doi: 10.1038/s41598-017-05028-2
Buckley, C. E., Moore, R. E., Reade, A., Goldberg, A. R., Weiner, O. D., and Clarke, J. D. W. (2016). Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev. Cell 36, 117–126. doi: 10.1016/j.devcel.2015.12.011
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S., and Schaffer, D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252. doi: 10.1038/nmeth.2360
Bulina, M. E., Chudakov, D. M., Britanova, O. V., Yanushevich, Y. G., Staroverov, D. B., Chepurnykh, T. V., et al. (2006). A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95–99. doi: 10.1038/nbt1175
Capek, D., Smutny, M., Tichy, A. M., Morri, M., Janovjak, H., and Heisenberg, C. P. (2019). Light-activated frizzled7 reveals a permissive role of non-canonical wnt signaling in mesendoderm cell migration. Elife 8:e42093. doi: 10.7554/eLife.42093
D'Acunzo, P., Strappazzon, F., Caruana, I., Meneghetti, G., Di Rita, A., Simula, L., et al. (2019). Reversible induction of mitophagy by an optogenetic bimodular system. Nat. Commun. 10:1533. doi: 10.1038/s41467-019-09487-1
Distel, M., Wullimann, M. F., and Köster, R. W. (2009). Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 106, 13365–13370. doi: 10.1073/pnas.0903060106
Formella, I., Svahn, A. J., Radford, R. A. W., Don, E. K., Cole, N. J., Hogan, A., et al. (2018). Real-time visualization of oxidative stress-mediated neurodegeneration of individual spinal motor neurons in vivo. Redox Biol. 19, 226–234. doi: 10.1016/j.redox.2018.08.011
Guntas, G., Hallett, R. A., Zimmerman, S. P., Williams, T., Yumerefendi, H., Bear, J. E., et al. (2015). Engineering an improved light-induced dimer (ILID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 112–117. doi: 10.1073/pnas.1417910112
Harper, S. M., Neil, L. C., and Gardner, K. H. (2003). Structural basis of a phototropin light switch. Science 301, 1541–1544. doi: 10.1126/science.1086810
Jost, M., Fernández-Zapata, J., Polanco, M. C., Ortiz-Guerrero, J. M., Chen, P. Y., Kang, G., et al. (2015). Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526, 536–541. doi: 10.1038/nature14950
Kainrath, S., Stadler, M., Reichhart, E., Distel, M., and Janovjak, H. (2017). Green-light-induced inactivation of receptor signaling using cobalamin-binding domains. Angew. Chem. Int. Ed. 56, 4608–4611. doi: 10.1002/anie.201611998
Kolar, K., Knobloch, C., Stork, H., Žnidarič, M., and Weber, W. (2018). OptoBase: a web platform for molecular optogenetics. ACS Synth. Biol. 7, 1825–1828. doi: 10.1021/acssynbio.8b00120
Lee, S., Park, H., Kyung, T., Kim, N. Y., Kim, S., Kim, J., et al. (2014). Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods 11, 633–636. doi: 10.1038/nmeth.2940
Liu, H., Gomez, G., Lin, S., Lin, S., and Lin, C. (2012). Optogenetic control of transcription in zebrafish. PLoS ONE 7:e50738. doi: 10.1371/journal.pone.0050738
Liu, H., Yu, X., Li, K., Klejnot, J., Yang, H., Lisiero, D., et al. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in arabidopsis. Science 322, 1535–1539. doi: 10.1126/science.1163927
Masuda, S., Nakatani, Y., Ren, S., and Tanaka, M. (2013). Blue light-mediated manipulation of transcription factor activity in vivo. ACS Chem. Biol. 8, 2649–2653. doi: 10.1021/cb400174d
Masuda, S., and Tanaka, M. (2016). PICCORO: a technique for manipulating the activity of transcription factors with blue light. Methods Cell Biol. 135, 289–295. doi: 10.1016/bs.mcb.2016.03.009
Mayrhofer, M., and Mione, M. (2016). The toolbox for conditional zebrafish cancer models. Adv. Exp. Med. Biol. 916, 21–59. doi: 10.1007/978-3-319-30654-4_2
Motta-Mena, L. B., Reade, A., Mallory, M. J., Glantz, S., Weiner, O. D., Lynch, K. W., et al. (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196–202. doi: 10.1038/nchembio.1430
Mruk, K., Ciepla, P., Piza, P. A., Alnaqib, M., and Chen, J. K. (2019). Targeted cell ablation in zebrafish using optogenetic transcriptional control. bioRxiv [Preprint]. doi: 10.1101/730507
Ni, M., Tepperman, J. M., and Quail, P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. doi: 10.1038/23500
Patel, A. L., Yeung, E., McGuire, S. E., Wu, A. Y., Toettcher, J. E., Burdine, R. D., et al. (2019). Optimizing photoswitchable MEK. Proc. Natl. Acad. Sci. U.S.A. 116, 25756–25763. doi: 10.1073/pnas.1912320116
Polesskaya, O., Baranova, A., Bui, S., Kondratev, N., Kananykhina, E., Nazarenko, O., et al. (2018). Optogenetic regulation of transcription. BMC Neurosci. 19(Suppl. 1):12. doi: 10.1186/s12868-018-0411-6
Reade, A., Motta-Mena, L. B., Gardner, K. H., Stainier, D. Y., Weiner, O. D., and Woo, S. (2017). TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development 144, 345–355. doi: 10.1242/dev.139238
Sako, K., Pradhan, S. J., Barone, V., Inglés-Prieto, Á., Müller, P., Ruprecht, V., et al. (2016). Optogenetic control of nodal signaling reveals a temporal pattern of nodal signaling regulating cell fate specification during gastrulation. Cell Rep. 16, 866–877. doi: 10.1016/j.celrep.2016.06.036
Schwerdtfeger, C., and Linden, H. (2003). VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22, 4846–4855. doi: 10.1093/emboj/cdg451
Simmich, J., Staykov, E., and Scott, E. (2012). Zebrafish as an appealing model for optogenetic studies. Prog. Brain Res. 196, 145–162. doi: 10.1016/B978-0-444-59426-6.00008-2
Teh, C., Chudakov, D. M., Poon, K. L., Mamedov, I. Z., Sek, J. Y., Shidlovsky, K., et al. (2010). Optogenetic in vivo cell manipulation in killerred-expressing zebrafish transgenics. BMC Dev. Biol. 10:110. doi: 10.1186/1471-213X-10-110
Wang, X., Chen, X., and Yang, Y. (2012). Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 9, 266–269. doi: 10.1038/nmeth.1892
Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., et al. (2009). A genetically-encoded photoactivatable rac controls the motility of living cells. Nature 461, 104–108. doi: 10.1038/nature08241
Yoo, S. K., Deng, Q., Cavnar, P. J., Wu, Y. I., Hahn, K. M., and Huttenlocher, A. (2010). Differential regulation of protrusion and polarity by PI(3)K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236. doi: 10.1016/j.devcel.2009.11.015
Yuan, H., and Bauer, C. E. (2008). PixE promotes dark oligomerization of the BLUF photoreceptor PixD. Proc. Natl. Acad. Sci. U.S.A. 105, 11715–11719. doi: 10.1073/pnas.0802149105
Keywords: zebrafish, optogenetics, non-neural optogenetics, synthetic biology, gene expression, protein localization, cell signaling
Citation: Varady A and Distel M (2020) Non-neuromodulatory Optogenetic Tools in Zebrafish. Front. Cell Dev. Biol. 8:418. doi: 10.3389/fcell.2020.00418
Received: 19 February 2020; Accepted: 05 May 2020;
Published: 03 June 2020.
Edited by:
Maria Caterina Mione, University of Trento, ItalyReviewed by:
Stephan C. F. Neuhauss, University of Zurich, SwitzerlandGerald Radziwill, University of Freiburg, Germany
Copyright © 2020 Varady and Distel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Distel, bWFydGluLmRpc3RlbEBjY3JpLmF0
 Adam Varady
Adam Varady Martin Distel
Martin Distel