- 1Department of Medical Microbiology and Infectious Diseases, Canisius Wilhelmina Hospital (CWZ), Nijmegen, Netherlands
- 2Centre of Expertise in Mycology, Radboudumc/CWZ, Nijmegen, Netherlands
- 3Department of Medical Microbiology, Radboudumc, Nijmegen, Netherlands
More than a decade ago a short tandem repeat-based typing method was developed for the fungus Aspergillus fumigatus. This STRAf assay is based on the analysis of nine short tandem repeat markers. Interpretation of this STRAf assay is complicated when there are only one or two differences in tandem repeat markers between isolates, as the stability of these markers is unknown. To determine the stability of these nine markers, a STRAf assay was performed on 73–100 successive generations of five clonally expanded A. fumigatus isolates. In a total of 473 generations we found five times an increase of one tandem repeat unit. Three changes were found in the trinucleotide repeat marker STRAf 3A, while the other two were found in the trinucleotide repeat marker STRAf 3C. The di- or tetranucleotide repeats were not altered. The altered STRAf markers 3A and 3C demonstrated the highest number of repeat units (≥50) as compared to the other markers (≤26). Altogether, we demonstrated that 7 of 9 STRAf markers remain stable for 473 generations and that the frequency of alterations in tandem repeats is positively correlated with the number of repeats. The potential low level instability of STRAf markers 3A and 3C should be taken into account when interpreting STRAf data during an outbreak.
Introduction
Tandem repeats are repetitive DNA sequences of 1 or more nucleotides, which are abundantly present in eukaryotic genomes, both in coding and non-coding regions (Gemayel et al., 2010). They are characterized by their highly polymorphic nature, as the number of repeated sequences often varies within species (Genovese et al., 2018). This polymorphic nature plays an important role in adaptation toward environmental changes by affecting cellular processes like cell surface variability, plasticity in body morphology and tuning of the circadian rhythm. On the other hand it can also cause disease, like Huntington disease (Gemayel et al., 2010).
Tandem repeats are classified according to the length of their repeated sequence. Repeats consisting of 1–9 nucleotides are generally known as microsatellites, or short tandem repeats (STRs), while longer repeats are known as minisatellites (Gemayel et al., 2010). The variation in the number of tandem repeats in both micro- and minisatellites results from strand-slippage by the DNA polymerase during replication, in which a mismatch of one of more repeat units occurs after the transient dissociation of the template and nascent DNA strand. When the template strand produces a loop, one or more repeat units will be deleted from the new DNA strand, however when this occurs with the nascent strand, one or more repeat units will be inserted (Gemayel et al., 2010; Genovese et al., 2018). In addition to strand-slippage, variation in micro- and minisatellites is also caused by events during recombination, like unequal crossing over and gene conversion (Amos, 2010; Ananda et al., 2013). Tandem repeat mutation rates vary from 1:100 to 1:100.000 per locus per generation, which is at least a factor 10 more frequent than point mutations (Brinkmann et al., 1998; Legendre et al., 2007; Jansen et al., 2012; Molnar et al., 2012; Chapuis et al., 2015). This STR mutation rate is dependent on different characteristics of the tandem repeat, like the number of repeated units, unit length and repeat structure and purity (Brinkmann et al., 1998; Legendre et al., 2007; Amos, 2010).
The high variability of STRs and the relative ease of detection of these polymorphisms via PCR amplification make them ideally suited for microbial genotyping. For the fungus A. fumigatus a robust typing method based on nine STRs was developed (STRAf assay) (de Valk et al., 2005, 2007). A. fumigatus is the most common fungus, which can cause invasive aspergillosis in immunocompromised patients with often fatal consequences (Chowdhary et al., 2017). Previously, the high discriminatory power of the STRAf assay was demonstrated by distinguishing 96 genotypes within 99 presumably unrelated isolates (de Valk et al., 2005). Furthermore, its utility in epidemiological surveys was established as the epidemiological concordance among isolates from 6 outbreaks was identified (Balajee et al., 2008). Here, epidemiological related isolates with differences of only one repeat unit in a single STR marker were called micro-variation. It is unknown how to deal with these variations, since no guidelines for the interpretation of STR data have been postulated.
In order to interpret the relationship between isolates with differences in the STR markers, it is necessary to understand the dynamics of the individual markers. In a previous study we demonstrated that 30 subcultures of one A. fumigatus isolate did not affect any of the markers (de Valk et al., 2005). To expand our knowledge on the stability of STR markers in A. fumigatus, we investigated the stability of 9 highly polymorphic microsatellite markers in 100 successive generations of 5 clonally expanded A. fumigatus isolates with randomly chosen genotypes.
Materials and Methods
Generations
Four randomly chosen unrelated clinical A. fumigatus isolates and one reference strain (CBS 487.65) were clonally expanded up to 100 generations. All isolates were subcultured onto Sabouraud agars and incubated at 30°C until sporulation. Successive generations were grown from a single spore using the following procedure: A suspension of A. fumigatus spores was made in aqua dest with 0.05% Tween 40 (Merck Nederland B.V., Amsterdam, The Netherlands). The transmission of this suspensions was measured at a wavelength of 530 nm and adjusted to 80–82%, which corresponds to approximately 1.5 × 106 CFU/ml. This suspension was diluted to a concentration of 3 × 102 CFU/ml. About 100 μL of this suspension was plated onto a Sabouraud agar and incubated at 30°C and examined daily. A single non-sporulating colony was then picked and subcultured on a new Sabouraud agar and incubated at 30°C until sporulation. Subsequent generations were made accordingly.
STRAf Assay
DNA of all generations was extracted as previously described (de Valk et al., 2005). All DNA samples were analyzed using the 9 loci of the STRAf panel consisting of three di-, tri-, and tetra-nucleotide repeats. PCR primers, fluorescent labels and amplification conditions were exactly as previous described (de Valk et al., 2005). Assignment of repeat numbers in each marker was performed using Fragment Profiler 1.2 software (GE Healthcare, Diegem, Belgium) and checked by visual interpretation of the electropherograms.
Results
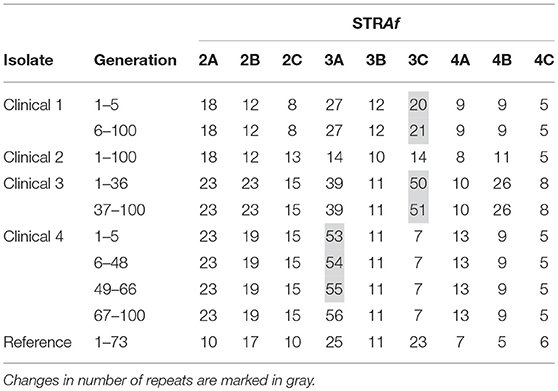
Four unrelated randomly chosen A. fumigatus isolates and one reference isolate were clonally expanded. Four isolates yielded 100 generations and one slow-sporulating isolate yielded 73 generations. In total, 473 generations were grown and analyzed with the nine STR markers of the STRAf assay. This yielded a total of 4,257 marker generations. Results are shown in Table 1.
In isolate #2 and the reference isolate no change in any of the nine STRAf markers was observed. In isolates #1 and #3 one alteration was found, which both took place in the STRAf 3C marker. After five generations of isolate #1, the number of repeat units increased from 20 to 21, while for isolate #3 it increased from 50 to 51 in the 36th generation. Most changes were observed in isolate #4, where in the 5th, 48th, and 66th generation the number of repeat units in the STRAf 3A marker gradually increased from 53 to 56. Altogether, five changes in the number of repeat units were observed, which all involved an increase of one repeat unit in a trinucleotide repeat marker.
Discussion
A better understanding of the epidemiological relation between isolates can be provided by molecular fingerprinting methods. More than 10 years ago a STR-based analysis to type A. fumigatus (STRAf) was developed that consists of three times three di-, tri-, or tetra-nucleotides repeat markers (de Valk et al., 2005). In order to separate related and unrelated isolates within an outbreak, Guinea et al. suggested in 2011 to define isolates with differences in no more than 2 repeat units for a single marker as microvariants and thus genetically related, and isolates with 3 or more repeat units difference as unrelated (Guinea et al., 2011). Information regarding the stability of the STR markers of A. fumigatus was however rather limited. To obtain more insight in the stability of the STRAf markers and improve our understanding whether or not isolates are related during an outbreak, we studied the variation in repeat numbers in successive clonally expanded A. fumigatus isolates by subculturing single spores. In total, 473 generations were analyzed with the STRAf panel. In seven out of nine markers no changes were found. In STRAf 3A and 3C three and two mutations were found, respectively, suggesting that these repeats are potentially less stable. It is known that the variability of some tandem repeats is much higher than others (Bustamante et al., 2013) and that the number of repeat units within a STR can change very rapidly during an outbreak (Hyytiä-Trees et al., 2006; Noller et al., 2006). Importantly, the three mutations in STRAf marker 3A were found in one isolate, demonstrating that the former definition of microvariation for the STRAf assay would not have been valid for this marker if similar variations would have been found in an outbreak. The relative instability of STRAf marker 3A, and to a lesser extent marker 3C, has to be taken into account when interpreting the relationship between epidemiological related A. fumigatus isolates. Another option is the exclusion of the trinucleotide repeat panel from the STRAf analysis, as with the di- and tetra-nucleotide repeats only, the discriminatory power remains very high (Garcia-Rubio et al., 2018).
Four out of five alterations were found in markers with 50 or more repeat units, suggesting that mutation rate may increase with increasing repeat units. Indeed, other studies in various organisms, like primates (Homo sapiens), insects (Drosophila melanogaster), plants (Arabidopsis Thaliana), yeast (Saccharomyces cerevisiae) and bacteria (E. coli), have shown that the variability of repeats is mostly dependent on the number of repeat units (Wierdl et al., 1997; Vogler et al., 2006; Legendre et al., 2007). Therefore, for A. fumigatus, as well as for other organisms, a higher number of repeat units decreases its stability. This means that expansion of one extra repeat unit to a repeat consisting of 10 units is less likely to occur in the same period of time than the expansion of a unit to a repeat that already contain 50 units. For the calculation of relations between isolates with these variations in a marker, this means that a difference of 10 to 11 repeat units in a marker should be given more weight than a change from 50 to 51 repeat units.
All five mutations were found in the trinucleotide repeat markers STRAf 3A and STRAf 3C, while no mutations were present in the di- and tetra-nucleotide repeats. Unit length might be another parameter affecting stability, however we cannot draw any conclusions from our own data, as the STRAf 3A and STRAf 3C markers also demonstrated the highest number of repeat units. There have been other studies though that demonstrated that unit length affects STR stability, however there is no consensus on the direction of the effect. There are studies demonstrating that shorter unit length leads to more instability (Schug et al., 1998; Vigouroux et al., 2002; O'Dushlaine and Shields, 2008; Willems et al., 2014), while other studies found the opposite (O'Dushlaine et al., 2005; Payseur et al., 2011). In the yeast Saccharomyces cerevisiae longer repeat units were more instable, which might be explained by an additive effect of unequal recombination, which is next to strand slippage more frequently observed in longer tandem repeats (Wierdl et al., 1997; Richard and Pâques, 2000). More research is required to demonstrate the potential role of unit length in repeat variability in fungi.
All five mutations were insertions of one repeat unit, while no deletions were found. The rate of insertions vs. deletions of tandem repeats has been studied by others. Some studies found a higher probability for an increase of the number of repeat units (Vigouroux et al., 2002; Noller et al., 2006), while others found no difference or the opposite (Vogler et al., 2006). More recent studies reported that the frequency of insertions or deletions depends on the total length of the tandem repeat, with relative longer repeats have more chance to shorten and relatively shorter repeats more likely to increase in size (Huang et al., 2002; Lai and Sun, 2003). These analyses were based on relative sizes and no absolute threshold at which an insertion is more likely than a deletion or vice versa was established. We found insertions in tri-nucleotide repeats with a length of 53–55 nucleotides, while the A. fumigatus genome contains many tri-nucleotide repeats with a size of >50 repeats (Levdansky et al., 2007). Based on our limited data set we cannot yet determine at which size deletions would be more likely to occur. Finally, we only found insertions of single repeat units, while other studies found that approximately half of all mutations consisted of multiple repeat changes (Bustamante et al., 2013). This might be due to the larger repeat sizes used in these studies, which would more likely lead to involving more repeats, or might be due to the fact that we analyzed every single generation, while other studies only checked a generation after ten passages, possibly leading to a bias of interpreting single additions as one multiple repeat change (Bustamante et al., 2013).
Genotyping assays based on STRs are a powerful tool for epidemiological studies on a wide range of organisms. The interpretation of data obtained with these assays requires proper understanding of the dynamics of the repeat units in STR markers. A high number of repeat units in a marker seems to have the most influence on the variability of that marker, as was also observed for the STRAf assay. These findings have to be taken into account when interpreting STRAf data during molecular epidemiological analysis.
Author Contributions
TG and JM contributed to the acquisition, the analysis, and the interpretation of the data. Both authors drafted, reviewed, and modified the manuscript.
Conflict of Interest Statement
JM received grants from F2G and Merck. He has been a consultant to Scynexis and Merck and received speaker's fees from Merck, United Medical, TEVA, and Gilead Sciences.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the help of Dr. Hanneke de Valk and Ilse Curfs-Breuker B.Sc. in the initial phases of the experiments.
References
Amos, W. (2010). Mutation biases and mutation rate variation around very short human microsatellites revealed by human-chimpanzee-orangutan genomic sequence alignments. J. Mol. Evol. 71, 192–201. doi: 10.1007/s00239-010-9377-4
Ananda, G., Walsh, E., Jacob, K. D., Krasilnikova, M., Eckert, K. A., Chiaromonte, F., et al. (2013). Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome. Genome Biol. Evol. 5, 606–620. doi: 10.1093/gbe/evs116
Balajee, S. A., de Valk, H. A., Lasker, B. A., Meis, J. F., and Klaassen, C. H. (2008). Utility of a microsatellite assay for identifying clonally related outbreak isolates of Aspergillus fumigatus. J. Microbiol. Methods 73, 252–256. doi: 10.1016/j.mimet.2008.02.011
Brinkmann, B., Klintschar, M., Neuhuber, F., Hühne, J., and Rolf, B. (1998). Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 62, 1408–1415. doi: 10.1086/301869
Bustamante, A. V., Sanso, A. M., Segura, D. O., Parma, A. E., and Lucchesi, P. M. (2013). Dynamic of mutational events in variable number tandem repeats of Escherichia coli O157:H7. Biomed. Res. Int. 2013:390354. doi: 10.1155/2013/390354
Chapuis, M. P., Plantamp, C., Streiff, R., Blondin, L., and Piou, C. (2015). Microsatellite evolutionary rate and pattern in Schistocerca gregaria inferred from direct observation of germline mutations. Mol. Ecol. 24, 6107–6119. doi: 10.1111/mec.13465
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Azole-resistant aspergillosis: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S436–S444. doi: 10.1093/infdis/jix210
de Valk, H. A., Meis, J. F., Curfs, I. M., Muehlethaler, K., Mouton, J. W., and Klaassen, C. H. (2005). Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 43, 4112–4120. doi: 10.1128/JCM.43.8.4112-4120.2005
de Valk, H. A., Meis, J. F., and Klaassen, C. H. (2007). Microsatellite based typing of Aspergillus fumigatus: strengths, pitfalls and solutions. J. Microbiol. Methods 69, 268–272. doi: 10.1016/j.mimet.2007.01.009
Garcia-Rubio, R., Escribano, P., Gomez, A., Guinea, J., and Mellado, E. (2018). Comparison of two highly discriminatory typing methods to analyze Aspergillus fumigatus azole resistance. Front. Microbiol. 9:1626. doi: 10.3389/fmicb.2018.01626
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi: 10.1146/annurev-genet-072610-155046
Genovese, L. M., Geraci, F., Corrado, L., Mangano, E., D'aurizio, R., Bordoni, R., et al. (2018). A census of tandemly repeated polymorphic loci in genic regions through the comparative integration of human genome assemblies. Front. Genet. 9:155. doi: 10.3389/fgene.2018.00155
Guinea, J., García de Viedma, D., Peláez, T., Escribano, P., Muñoz, P., Meis, J. F., et al. (2011). Molecular epidemiology of Aspergillus fumigatus: an in-depth genotypic analysis of isolates involved in an outbreak of invasive aspergillosis. J. Clin. Microbiol. 49, 3498–3503. doi: 10.1128/JCM.01159-11
Huang, Q. Y., Xu, F. H., Shen, H., Deng, H. Y., Liu, Y. J., Liu, Y. Z., et al. (2002). Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 70, 625–634. doi: 10.1086/338997
Hyytiä-Trees, E., Smole, S. C., Fields, P. A., Swaminathan, B., and Ribot, E. M. (2006). Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3, 118–131. doi: 10.1089/fpd.2006.3.118
Jansen, A., Gemayel, R., and Verstrepen, K. J. (2012). Unstable microsatellite repeats facilitate rapid evolution of coding and regulatory sequences. Genome Dyn. 7, 108–125. doi: 10.1159/000337121
Lai, Y., and Sun, F. (2003). The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20, 2123–2131. doi: 10.1093/molbev/msg228
Legendre, M., Pochet, N., Pak, T., and Verstrepen, K. J. (2007). Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 17, 1787–1796. doi: 10.1101/gr.6554007
Levdansky, E., Romano, J., Shadkchan, Y., Sharon, H., Verstrepen, K. J., Fink, G. R., et al. (2007). Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 6, 1380–1391. doi: 10.1128/EC.00229-06
Molnar, R. I., Witte, H., Dinkelacker, I., Villate, L., and Sommer, R. J. (2012). Tandem-repeat patterns and mutation rates in microsatellites of the nematode model organism Pristionchus pacificus. G3 2, 1027–1034. doi: 10.1534/g3.112.003129
Noller, A. C., Mcellistrem, M. C., Shutt, K. A., and Harrison, L. H. (2006). Locus-specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J. Clin. Microbiol. 44, 374–377. doi: 10.1128/JCM.44.2.374-377.2006
O'Dushlaine, C. T., Edwards, R. J., Park, S. D., and Shields, D. C. (2005). Tandem repeat copy-number variation in protein-coding regions of human genes. Genome Biol. 6:R69. doi: 10.1186/gb-2005-6-8-r69
O'Dushlaine, C. T., and Shields, D. C. (2008). Marked variation in predicted and observed variability of tandem repeat loci across the human genome. BMC Genomics 9:175. doi: 10.1186/1471-2164-9-175
Payseur, B. A., Jing, P., and Haasl, R. J. (2011). A genomic portrait of human microsatellite variation. Mol. Biol. Evol. 28, 303–312. doi: 10.1093/molbev/msq198
Richard, G. F., and Pâques, F. (2000). Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 1, 122–126. doi: 10.1093/embo-reports/kvd031
Schug, M. D., Hutter, C. M., Wetterstrand, K. A., Gaudette, M. S., Mackay, T. F., and Aquadro, C. F. (1998). The mutation rates of di-, tri- and tetranucleotide repeats in Drosophila melanogaster. Mol. Biol. Evol. 15, 1751–1760. doi: 10.1093/oxfordjournals.molbev.a025901
Vigouroux, Y., Jaqueth, J. S., Matsuoka, Y., Smith, O. S., Beavis, W. D., Smith, J. S., et al. (2002). Rate and pattern of mutation at microsatellite loci in maize. Mol. Biol. Evol. 19, 1251–1260. doi: 10.1093/oxfordjournals.molbev.a004186
Vogler, A. J., Keys, C., Nemoto, Y., Colman, R. E., Jay, Z., and Keim, P. (2006). Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7. J. Bacteriol. 188, 4253–4263. doi: 10.1128/JB.00001-06
Wierdl, M., Dominska, M., and Petes, T. D. (1997). Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics 146, 769–779.
Keywords: Aspergillus fumigatus, microsatellites, short tandem repeats, STRAf, stability
Citation: de Groot T and Meis JF (2019) Microsatellite Stability in STR Analysis Aspergillus fumigatus Depends on Number of Repeat Units. Front. Cell. Infect. Microbiol. 9:82. doi: 10.3389/fcimb.2019.00082
Received: 21 December 2018; Accepted: 11 March 2019;
Published: 29 March 2019.
Edited by:
Dea Garcia-Hermoso, Institut Pasteur, FranceReviewed by:
Emilia Mellado, Instituto de Salud Carlos III, SpainJesus Guinea, Hospital General Universitario Gregorio Marañón, Spain
Copyright © 2019 de Groot and Meis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theun de Groot, dGhldW5kZWdyb290N0BnbWFpbC5jb20=
 Theun de Groot
Theun de Groot Jacques F. Meis
Jacques F. Meis