- Department of Tropical Diseases, Naval Medical University, Shanghai, China
Schistosomiasis remains a serious parasitic disease, which is characterized by granulomatous inflammation and hepatic fibrosis. MicroRNAs derived from parasites can regulate host genes and cell phenotype. Here, we showed that a miRNA derived from S. japonicum (Sja-miR-1) exists in the hepatic stellate cells (HSCs) of mice infected with the parasite and up-regulates the expression of collagens and α-SMA by targeting secreted frizzled-related protein 1 (SFRP1). A vector-mediated delivery of Sja-miR-1 into naive mice led to hepatic fibrogenesis in the mice. Accordingly, inhibition of Sja-miR-1 in the infected mice led to reduction of the parasite-induced hepatic fibrosis. The mechanism behind the Sja-miR-1-mediated activation of HSC could be through targeting SFRP1 to regulate the Wnt/β-catenin pathway. These findings reveal that parasite-derived small non-coding RNAs are implicated in cross-species regulation of host pathological process and persistent inhibition of Sja-miR-1 may provide a therapeutic potential for the parasite diseases.
Introduction
At present, schistosomiasis is still a serious parasitic disease, which infects more than 240 million people across 78 countries (Colley et al., 2014). Schistosoma mansoni and Schistosoma japonicum are the major species that cause liver disease in humans. Followed by infecting humans or animals, schistosome cercariae migrate to the host portal-mesenteric vein system where the female worm lay a large number of eggs that, at a part, are transported to the liver through portal veins, causing egg granuloma and hepatic fibrosis, eventually form cirrhosis which is the main cause of mortality of schistosomiasis.
Schistosomiasis is actually an immune pathological disease (Pearce and Macdonald, 2002). At the early stage of schistosome infection, Th1 immune response characterized by elevated IFN-γ is moderately activated. When the eggs deposit in the tissue, the immune response is quickly polarized to Th2 response marked by elevation of IL-13 and IL-4 (Pearce and Macdonald, 2002; Wilson et al., 2007). The Th2-related cytokines are important mediators for regulation of schistosomiasis hepatic fibrosis and progression of the disease (Wilson et al., 2007). Hepatic HSCs have been proven to be the main effector cells of the hepatic fibrosis (Bartley et al., 2006). The eggs trapped in liver induce activation of quiescent HSCs to the proliferative and fibrogenic myofibroblasts, which generate excessive extracellular matrix (ECM), leading to hepatic fibrosis (Burke et al., 2009). The inhibition of the activated HSCs provides a possibility of developing tools for prevention and treatment of the fibrosis (Friedman, 2008).
MicroRNAs (miRNAs), a highly-conserved and endogenous non-coding RNA, widely exist in animals and plants that regulate post-transcriptional gene expression and play an important role in progression of many diseases (Bartel, 2004; Lu et al., 2008). Some miRNAs of plants and parasites have been found in human or animal body fluids and even entry into host cells to modulate host genes and their phenotype in a cross-species manner (Zhang et al., 2012; Hu et al., 2019). MiR-168a abundant in rice can regulate the expression of the gene encoding low-density lipoprotein receptor adapter protein 1 in mammals (Zhang et al., 2012). Small RNAs of fungal, can silence plant genes related to host immunity (Wang et al., 2016). Parasite-derived miRNAs can be absorbed by the intestinal epithelial cells in mice and inhibit host type 2 innate immunity (Buck et al., 2014). Recently, several miRNAs of Schistosoma japonicum were reported to be involved in the modulation of host macrophage proliferation and pathogenesis of schistosomiasis by targeting relative genes of host (He et al., 2019; Liu et al., 2019).
In schistosomiasis, a granulomatous reaction occurs resulting from eggs trapped in the liver tissues, surrounded with immunocytes and other hepatic mesenchymal cells including HSCs (Wilson et al., 2007; Anthony et al., 2012). Exosomes are well-verified to play an important role in pathogen-host interaction or cell-cell communication. Recent studies showed that S. japonicum eggs could release exosomes containing miRNAs that could be transported into recipient cells (Zhu L. et al., 2016; Zhu S. et al., 2016). In this study, we showed a miRNA derived from Schistosoma japonicum exists in HSCs of mice infected with the parasite and contributed to the HSCs activation resulting in hepatic fibrosis. We demonstrate that the miRNA exerts the fibrosis-promoting effect through targeting the gene encoding SFRP1 of the host.
Materials and Methods
Ethics Statement
All the animal experiments were conducted consistent with the Guidelines of the Care and Use of Laboratory Animals of the National Institutes of Health, and the study received the approval of the Animal Ethics Committee of Naval Medical University (access number FYXK [Shanghai] 2014-0003).
Mice and Parasite Infections
Six-week-old male BALB/c mice which were all bought from the experimental animal center of the Naval Medical University (Shanghai, China) were raised under a specific pathogen-free conditions, padded, controlled temperature (23 ± 2°C) and photoperiods (1:1 light-dark cycle). In order to establish a schistosomiasis mouse model, mice were percutaneously exposed to 16 S. japonicum cercariae shed from the lab-cultured snails (Oncomelania hupensis) obtained from the National Institute of Parasitic Disease, Chinese Center for Disease Control and Prevention (Shanghai, China).
RNA Preparation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from the liver tissue or isolated HSCs with the Trizol reagent (Sigma-Aldrich, #T9424) and specific procedure was performed according to manufacturer's protocol. The expression of sja-miR-1, Col1α1, Col3α1, α-Sma, Sfrp1 were detected with a SYBR Green Master Mix kit (Roche, #04913914001). U6 snRNA and β-Actin were used as endogenous controls to normalize the real-time PCR data, and the relative expression was calculated by the 2−ΔΔCt method. The primers used in this study are shown in Table S1.
Isolation and Culture of Primary HSCs
We isolated primary HSCs from mouse liver using the previous protocol (He et al., 2015). Briefly, the livers were initially perfused with pronase/collagenase type IV through the hepatic portal vein, and then the digested liver tissues were triturated and suspended in cold DMEM with 4% fetal bovine serum and antibiotics under a sterilized condition. The hepatocytes were isolated from the collected cell suspension by two runs of centrifugation at 50 g for 4 min each and hepatocytes were pelleted. Then the remaining supernatant was centrifuged at 500 g for 5 min to collect the non-parenchymal cells. Further, the sediment was resuspended in DMEM and added to 11.5% iodixanol gradient (Axis-Shield, #AS1114542) by centrifugation at 1,400 g for 20 min. The HSCs that enriched at the interface of iodixanol gradient and DMEM were resuspended in PBS and centrifugated at 800 g for 7 min. In order to exclude Kupfer cells, the HSCs were further purified by negative selection in the help of magnetic CD11b antibody beads (Miltenyi, Germany). The HSCs were then plated on plastic dishes in DMEM containing 10% fetal bovine serum (Gibco, #10099-141), 1% penicillin-streptomycin (100 mg/ml) and 4 mM L-glutamine (100 IU/ml). Cells were cultured in an incubator (37°C, 5% CO2, 95% humidity).
Cell Culture and Transfection
Mouse primary HSCs and human LX-2 cells were cultured in endotoxin-free DMEM mixed with 10% fetal bovine serum, 1% antibiotics and 4 mM L-glutamine described above in an incubator (37°C, 5% CO2, 95% humidity). When the cells density reached 70% confluent in the 12-well plate, the cells were transfected with 1.5 μg/ml pAV-pri-miR-1 plasmid, 80 nM sja-miR-1 mimics (GenePhama, China), 80 nM sja-miR-1 inhibitor (GenePhama, China), 80 nM Sfrp1 siRNA (GenePhama, China) or corresponding negative controls with Lipofectamine 3000 (Life Technology, #L3000-008) according to the recommended protocol. For exosome application 10 μg/mL isolated exosomes were directly added into 12-well plates for 2 days, and then cells were harvested for further detection. 293T cells were cultured in the 24-well plate with endotoxin-free DMEM mixed with 10% fetal bovine serum. When 293T cells density reached 70% confluent, the cells were transfected with plasmids or mimics accordingly. The detailed sequences of the mimics, inhibitor and siRNA are shown in Table S2.
Isolation of Schistosomal Egg Exosomes
Parasite eggs were obtained from the livers of infected mice at 6 weeks after infection using the previous method (Cai et al., 2013). The isolated eggs were examined under a light microscope and incubated in serum-free culture medium for 24 h. Following incubation, supernatant was collected, centrifuged at 3,000 g for 15 min to eliminate remaining eggs and pellets. Then, an exoEasy Maxi Kit (QIAGEN, #76064) was applied to isolate egg exosomes according to the recommended protocol. The final purified exosomes were resuspended in 50 μl sterile PBS and kept in −80°C refrigerator for further study.
Uptake of Schistosomal Egg Exosomes by HSCs
To investigate if the isolated egg exosomes can be internalized by the host HSCs, the egg exosomes were stained with the fluorescent lipid dye PKH67 (Sigma-Aldrich, #MINI67-1KT) according to a previous protocol with minor modification (Lässer et al., 2011). Briefly, 20 μg of the PKH67-labeled exosomes were washed and concentrated using 100 kDa ultra centrifugal filters (Millipore, #UFC810024) in order to remove redundant lipid dye. The HSCs were seeded in a 96-well plate for 6 h and the labeled exosomes were then added to the HSCs culture medium and incubated together for 1 h. At the same time, equal volume of PBS was labeled with PKH67 and added to the HSCs in a parallel experiment to exclude the non-specific labeling. Following 1 h incubation, removed the culture medium and washed the HSCs twice with PBS. Then, fixed the cells in 4% paraformaldehyde for 20 min and washed them twice with PBS. The cell nucleuses were stained using 4′6-diamidino-2-phenylindole (DAPI; Beyotime Biotechnology, China, #C1005). Finally, whether the isolated egg exosomes can be internalized by the host HSCs was observed using confocal fluorescence microscope (Zeiss, Germany).
Pathologic Features of the Liver
After mice were anesthetized, the right liver lobes were isolated and fixed with 4% paraformaldehyde. The processed liver sections were then subjected to hematoxylin-eosin (HE) and Masson's trichrome staining to evaluate the granuloma size and hepatic fibrosis severity, which can be quantified by measuring the fibrosis grade of liver tissue as previously reported (Chiaramonte et al., 1999). For immunohistochemical staining, the liver sections were incubated with primary antibody against α-SMA (CST, #19245). Five different fields on each section were randomly selected to determine the α-SMA expression degree, the examination of which were blindly performed by two independent researchers.
Hydroxyprolin Content Analysis
A Hydroxyproline Content Assay Kit was used to detect the hydroxyproline content of the liver tissues according to the kit's instruction (Nanjing Jiancheng, China, #A030-2). The final data were shown as Hyp (μg) /liver weight (g).
Western Blot
Culture cells or freshly isolated cells were rinsed with pre-cooled PBS three times. Total cell protein extraction was performed using RIPA lysis buffer supplemented with protease and phosphatase inhibitors, while a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, China, #P0027) was applied to extract nuclear and cytoplasmic protein. Then the protein was quantified by the BCA method and boiled at 100°C for 5 min. 30–60 μg total protein per lane was added to the 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Once the electrophoresis was finished, the protein was then transferred to a nitrocellulose membrane (PALL, #P-N66485) according to the wet-transferring method, and non-specific binding sites were sealed for 2 h with 5% skimmed milk powder at room temperature with shaking. Blots were probed with primary antibodies, including anti-SFRP1 (Sigma Aldrich, #AV09053), anti-β-catenin (CST, #8480), anti-GAPDH (Abcam, #ab181602) and anti-Histone H3 (CST, #4499), overnight at 4°C. After that, the membrane was washed 3 times with PBS, followed by incubation with secondary antibody for 2 h at room temperature. The rabbit anti-GAPDH antibody and anti-Histone H3 antibody (CST, #4499) were treated as internal references, while IRDye goat anti-IgG antibody (LI-COR, #926-32211) was treated as secondary antibody. After the membrane was washed 3 times with PBS, the band densities were analyzed.
3′UTR Luciferase Reporter Constructs
According to the online bioinformatic analysis software (miRDB), the seed sequence of sja-miR-1 was predicted to be complementary with the 3'UTR of Sfrp1. To detect if Sfrp1 was a target of sja-miR-1, wild-type or mutant 3'UTRs of Sfrp1 were chemically synthesized (Genomics) and then cloned into the pmirGLO luciferase plasmid (Promega, #E1330). The 293T cells were seeded in a 24-well plate (3 × 105 cells/well). When the cells density reached up to 70%, 40 nM sja-miR-1 mimics or negative control (NC) mimics, together with 0.5 μg wild-type Sfrp1 3'UTR plasmid or mutant Sfrp1 3'UTR plasmid, were transfected into the 293T cells using lipofectamine 3000. Subsequently, the cells were cultured for 24 h and then collected. A Dual-luciferase Reporter Assay Kit (Promega, #E1901) was used to detect the effect of sja-miR-1 on the luciferase activity of the Sfrp1 3'UTR plasmid.
Construction of rAAV8 Vectors
The coding sequence of sja-pri-miR-1 was obtained by PCR using the genome of adult worms as template. The PCR fragment was digested with restriction enzyme AscI and MluI and cloned between the appropriate restriction sites of the pAV-CMV-eGFP plasmid, resulting in the pAV-pri-miR-1 plasmid. To explore the possible effect of sja-miR-1 in vivo, the anti-sja-miR-1 sponge sequence (GACCATACTTGCGACATTCCA) was designed, synthesized and cloned into the pAV-CMV-eGFP plasmid according to a previous study (Ebert et al., 2007). Both the constructed plasmids were verified by DNA sequencing. The AAV8 vectors which were involved in this research were produced and tittered by a professional company (Vigene, China).
AAV8 Vector Injection
For over-expression of sja-miR-1 in mice, 200 μl of rAAV8-pri-miR-1 vectors in PBS, at dose of 2 × 1011 particles/animal, were injected by tail vein into BALB/c mice. Similarly, rAAV8-anti-miR-1 vectors with titer of 1012 genome copies were given through the tail vein into mice infected percutaneously with S. japonicum cercariae. Equal rAAV8-SCR vectors and PBS were used as controls in this study.
Statistical Analysis
Data were presented as Means ± SD. All results were analyzed using the GraphPad Prism 7.0 software. Student's t-test was applied to assess the differences between two groups, and one-way ANOVA followed by Tukey post-test was used among three or more groups. When p < 0.05, differences were considered to indicate statistical significance.
Results
Sja-miR-1 Activates HSCs in vitro
The HSCs activation represents a key feature of hepatic fibrosis and is characterized by elevated expression of the genes encoding α-SMA and collagens (Seki and Schwabe, 2015). Once liver injury occurs, quiescent HSCs undergo activation to become the major source of myofibroblasts, which highly express α-SMA and collagens (Kisseleva et al., 2012). α-SMA is regarded as a marker of HSCs activation, while accumulated collagens, especially COL1α1 and COL3α1, contribute to excessive ECM deposition (Böttcher and Pinzani, 2017). The physical properties of ECM, in addition to their chemical composition, affect HSCs activation (Friedman, 2010). To investigate the effect of sja-miR-1 on HSCs activation, we first prepared a construct expressing sja-pri-miR-1 (designated as “pAV-pri-miR-1”) (Figure S1A). To determine whether the schistosome sja-pri-miR-1 can be processed into mature sja-miR-1 in mammal cells, we cloned three perfect sja-miR-1 complementary binding sites into pmirGLO luciferase plasmid to generate a sja-miR-1 sensor plasmid (designated as “pmirGLO-miR-1”), which could regulate the expression of genes encoding firefly luciferase (Fluc) and renilla luciferase (Rluc) reporters (Figure S1A). The two constructs were transiently co-transfected into 293T cells, and the firefly luciferase activity was normalized to renilla luciferase activity. The dual-luciferase reporter assay demonstrated that the presence of the plasmid expressing Sja-pri-miR-1 resulted in the decrease of Fluc activity, indicating that the mature sja-miR-1 was generated in 293T cells and this miRNA suppressed expression of the Fluc through the sites of this miRNA (Figure S1B). Next, we transfected the Sja-pri-miR-1 expression plasmid into primary mouse HSCs and LX-2 cells respectively, and sja-miR-1 significantly increased the expression of α-Sma Col1α1 and Col3α1 mRNA in both cells, which illustrated that sja-miR-1 promoted activation of HSCs in vitro (Figure 1A).
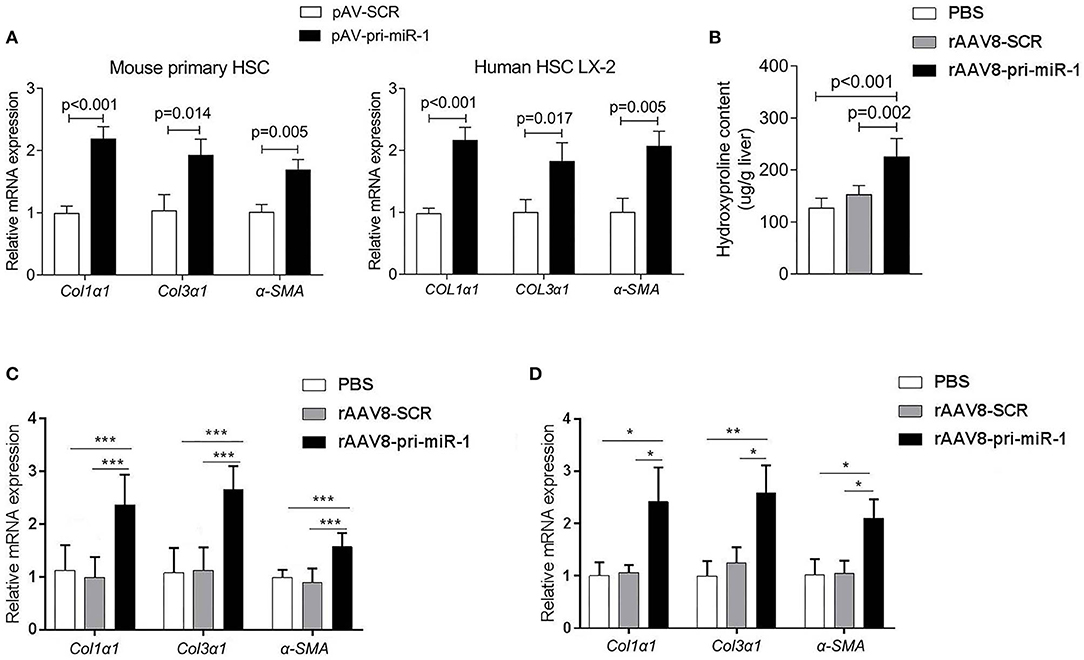
Figure 1. Sja-miR-1 activates HSCs and promotes the hepatic fibrosis in naive mice. (A) Mouse primary HSCs or LX-2 cells were transfected with pAV-pri-miR-1 plasmid and cultivated for 48 h. The level of Col1α1, Col3α1, and α-Sma were detected using qPCR (n = 3). (B,C) Mice were injected with the rAAV8-pri-miR-1 vector or rAAV8-SCR (negative control) at a dose of 2 × 1011 genomes or PBS via the tail vein, and liver tissues were collected at day 50 post-infection (n = 6). The extent of hepatic fibrosis was measured based on the hydroxyproline content (B) or the expression of fibrosis-related genes (C). (D) The expression of Col1α1, Col3α1, and α-Sma in primary HSCs isolated from mice injected with various vectors at day 50 after injection were evaluated (n = 3). Data are presented as the Mean ± SD from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001.
Sja-miR-1 Promotes Hepatic Fibrosis in Naive Mice
To investigate if sja-miR-1 can promote hepatic fibrosis in mice, we constructed recombinant adeno-associated viral serotype 8 (rAAV8) vectors to express pri-sja-miR-1 or scramble miRNA (SCR control) under the control of CMV promoter. AAV8 has been shown to transduce up to 100% liver when injected intravenously in mice (Nakai et al., 2015). Adult BALB/c male mice were injected with the vectors or PBS through the tail vein. The hydroxyproline content and expression of fibrosis-related mRNAs (Col1α1, Col3α1, α-Sma) were significantly elevated in mice injected with rAAV8-pri-miR-1 vector compared with SCR control or PBS at day 50 post injection (Figures 1B,C). This result was also validated by extension of observation time at day 100 after injection (Figures 1C,D). Moreover, the primary HSCs were isolated to measure Col1α1, Col3α1, and α-Sma expression. We found elevated expression of all the genes in mice injected with rAAV8-pri-miR-1 compared with the two control groups (Figure 1D). All these data indicated that sja-miR-1 modulated the expression of fibrosis-related genes and promoted liver fibrogenesis in vivo.
Sja-miR-1 Contributes to Schistosomiasis Hepatic Fibrosis
To evaluate if sja-miR-1 was involved in the parasite-induced liver fibrosis during the parasite infection, we generated a rAAV8-anti-miR-1-sponge vector for knockdown of sja-miR-1 according to previous study (Ebert et al., 2007) (Figure S2A). The 293T cells were co-transfected with sja-miR-1 sensor plasmid, sja-miR-1 expression plasmid and sponge vector to examine whether the sponge vector could inhibit sja-miR-1 function. The dual-luciferase reporter assay showed that the sponge vector specifically inhibited sja-miR-1 function in the 293T cells (Figure S2B). BALB/c mice were infected with the parasite cercariae, followed by intravenously injection with the vectors or PBS at 10 days after infection. The liver tissues were obtained at 8 weeks after injection of the vectors. Mice injected with rAAV8-anti-miR-1 sponge vector revealed a significant reduction in ECM production compared with the control mice, characterized by hydroxyproline quantification (Figure 2A) and Masson's trichrome staining (Figures 2B,C), indicating the attenuation of hepatic fibrosis. Moreover, a significant reduction of the size of hepatic granulomas was visualized by H&E staining (Figures 2B,C). This alleviation of liver fibrosis was also validated by quantification of fibrosis-related genes revealing the expression of Col1α1, Col3α1, and α-Sma were dramatically decreased in livers of mice injected with rAAV8-anti-miR-1 sponge vector compared with the controls (Figure 2D). Moreover, immunohistochemical (IHC) staining showed that the sponge vector also reduced the protein level of α-SMA compared with the controls (Figures 3A,B). The rAAV8 vector, as expected, had no effect on the parasite eggs production in mice (Figure 3C).
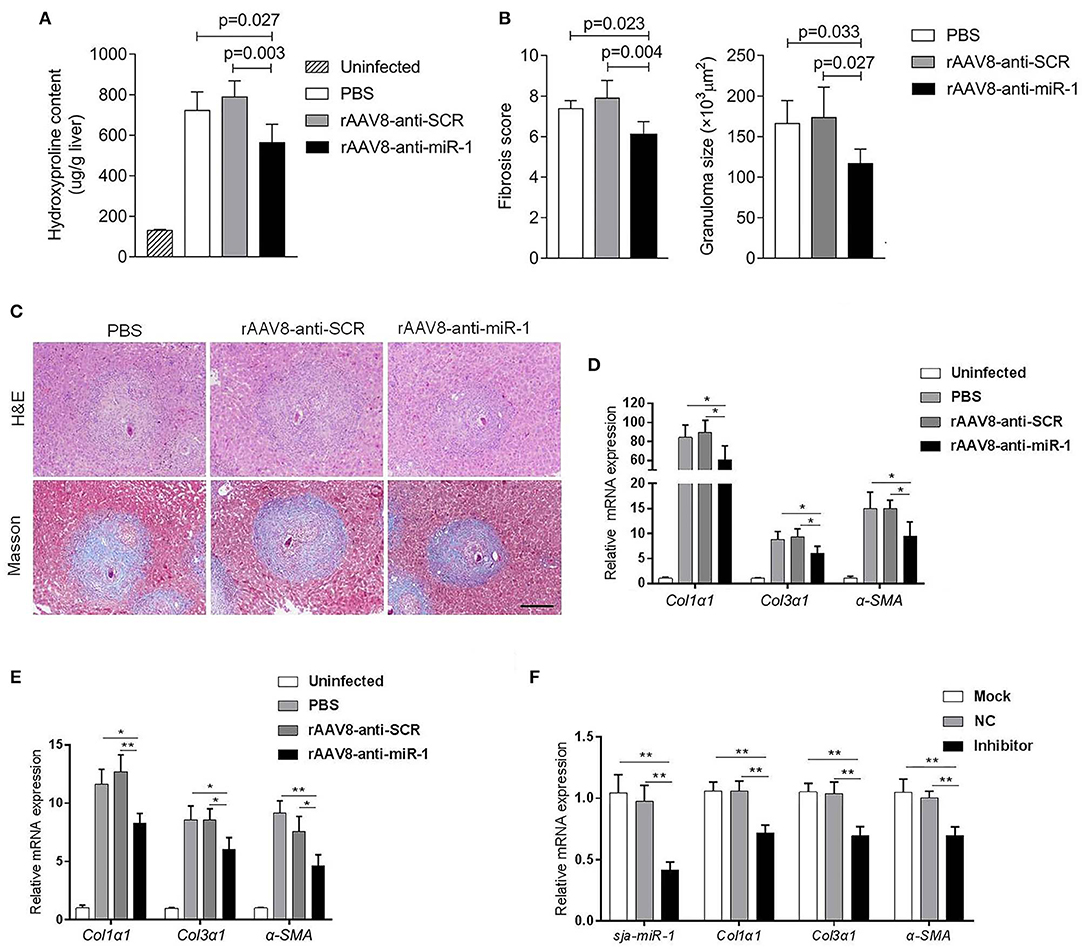
Figure 2. Knockdown of sja-miR-1 alleviates schistosomiasis hepatic fibrosis. (A–E) Mice were infected percutaneously with 16 S. japonicum cercariae at day 0 or remained uninfected. The infected mice received a tail vein-injection of rAAV8-anti-Sja-miR-1-sponge or rAAV8-SCR vectors at a dose of 1 × 1012 virus genomes or PBS via the tail vein. Liver samples were collected at day 56 after the parasite infection (n = 6). (A) Hydroxyproline content in the liver. (B) Fibrosis scores and granuloma size were measured from Masson's trichrome and H&E staining of liver sections. (C) H&E staining and Masson's trichrome staining of liver sections. Scale bar, 250 μm. (D) The expression of Col1α1, Col3α1, and α-Sma in the liver were detected. (E) The expression of Col1α1, Col3α1, and α-Sma in primary HSCs isolated from infected livers were detected (n = 3). (F) Primary HSCs were isolated from the infected mice with the parasite and then transfected with sja-miR-1 inhibitor, negative control (NC) inhibitor or transfection reagent only (Mock) for 48 h. The level of sja-miR-1 and Col1α1, Col3α1, and α-Sma were analyzed (n = 3). Data are presented as the Mean ± SD from three independent experiments, *p < 0.05, **p < 0.01.
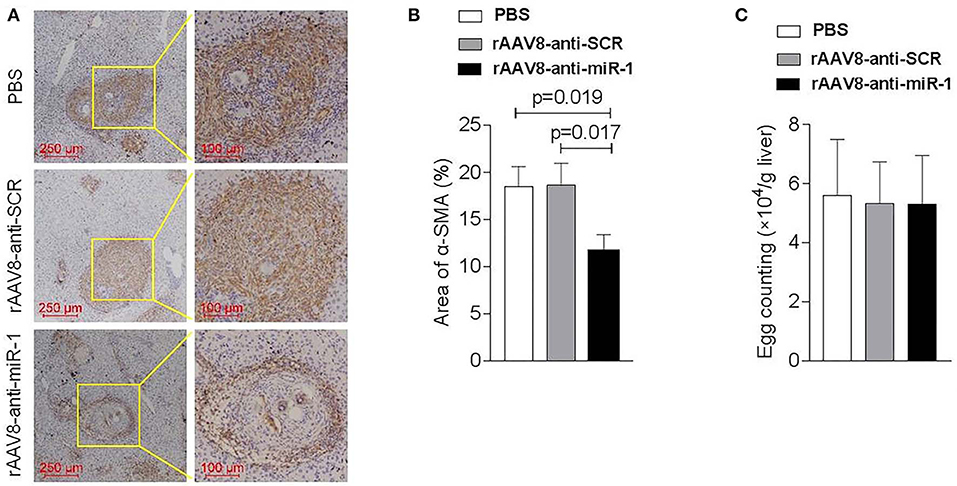
Figure 3. Down-regulation of sja-miR-1 partially blocked the activation of HSCs in schistosomiasis-induced liver fibrosis. Mice were infected percutaneously with 16 S. japonicum cercariae at day 0 or remained uninfected. rAAV8-anti-miR-1 vectors or rAAV8-SCR with titer of 1012 genome copies or PBS were given through the tail vein into the infected mice (n = 3). Liver samples were collected at 56 days post-infection. (A) The expression of HSC marker (α-SMA) in the livers was detected by immunohistochemical staining. (B) The ratio of the α-SMA-stained area to the total area was quantified. (C) Quantification of S. japonicum eggs in the liver. Data are presented as the Mean ± SD from three independent experiments.
Because of the key roles of HSCs in schistosome-induced hepatic fibrosis (Burke et al., 2009), we then obtained primary HSCs derived from the infected mice injected with the sponge vector to quantify the expression of genes related to HSCs activation. The results showed a lower level of Col1α1, Col3α1, and α-Sma in the HSCs of mice injected with rAAV8-anti-miR-1 sponge vector compared with the control mice (Figure 2E).
To further validate the pro-fibrotic effect of sja-miR-1 and its existence in HSCs of mice infected with S. japonicum, the primary HSCs isolated from the infected mice at 56 days post-infection were transfected with sja-miR-1 inhibitor or NC inhibitor in vitro. The qPCR analysis showed that the miR-1 inhibitor treatment significantly reduced the expression of the genes encoding α-Sma, Col1α1, and Col3α1 compared with the NC or Mock (Figure 2F). These results suggested that the parasite-derived sja-miR-1 may be a virulence factor contributing to pathogenicity of schistosomiasis.
S. japonicum Egg-Derived Exosomes and Internalization of Sja-miR-1 in HSCs
Exosomes cargo, including miRNAs, play a regulatory role in host-pathogen interactions (Szempruch et al., 2016). In our previous research, we found that sja-miR-1 was abundantly present in host HSCs through RNA sequencing (He et al., 2019). To ascertain whether the S. japonicum egg-derived exosomes can enter host cells, we labeled S. japonicum exosomes with the lipid dye PKH67 and incubated the labeled exosomes in primary HSCs derived from mice and LX-2 cell line in vitro. We revealed efficient internalization of the egg exosomes by both primary HSCs and LX-2 cells using confocal image analysis (Figure S3A). To investigate the roles of S. japonicum egg exosomes in activating HSCs, we incubated them with primary HSCs from naive mice for 24 h and found that sja-miR-1 was detectable in the cells treated with the exosomes (Figure S3B). Crucially, the exosomes promoted activation of HSCs, evidenced by elevated level of Col1α1, Col3α1, and α-Sma. While pretreatment with sja-miR-1 inhibitor abrogated the effects of exosomes on level of Col1α1, Col3α1, and α-Sma production compared with the controls (Figure S3C). These data suggested that sja-miR-1 carried by the egg-derived exosomes could be transferred to the HSCs and regulated fibrosis-related genes in HSCs.
Sfrp1 Is a Target Gene of Sja-miR-1
The online bioinformatic analysis using miRDB software suggested the Sfrp1 as the potential target gene of sja-miR-1. To further validate the target gene, the luciferase reporter constructs were generated that contained 3'UTR of Sfrp1 of either wild type (Sfrp1-3'UTR-WT) or mutant (Sfrp1-3'UTR-Mut) complementary sites of sja-miR-1 (Figure 4A). The luciferase assay indicated a significant decrease in the luciferase activity in the cells transfected with Sfrp1-3'UTR-WT and sja-miR-1 mimics compared with the control groups (Figure 4B). We then analyzed the expression of sja-miR-1 and Sfrp1 in HSCs isolated from the liver samples of infected mice at various time points post infection. The qPCR analysis showed that sja-miR-1 level was significantly up-regulated since day 30 and peaked at day 50 (Figure 4C). In contrast, Sfrp1 mRNA in HSCs was significantly decreased since post infection, which was reversely related to the sja-miR-1 level in HSCs (Figure 4D). Next, we detected both mRNA and protein levels of SFRP1 in primary HSCs transfected with sja-miR-1 mimics in vitro. The results showed that both mRNA and protein level of Sfrp1 were dramatically decreased in the HSCs transfected with sja-miR-1 mimics (Figures 4E,F), while inhibition of sja-miR-1 in the HSCs derived from the infected mice by the sja-miR-1 inhibitor dramatically elevated the levels of both Sfrp1 mRNA and protein (Figures 4G,H). Furthermore, the primary HSCs isolated from the livers of infected mice treated with the sponge vector showed significant up-regulation of the Sfrp1 expression compared with the controls (Figures 4I,J). Thus, our result suggested that Sfrp1 could be a target gene of sja-miR-1 in HSCs.
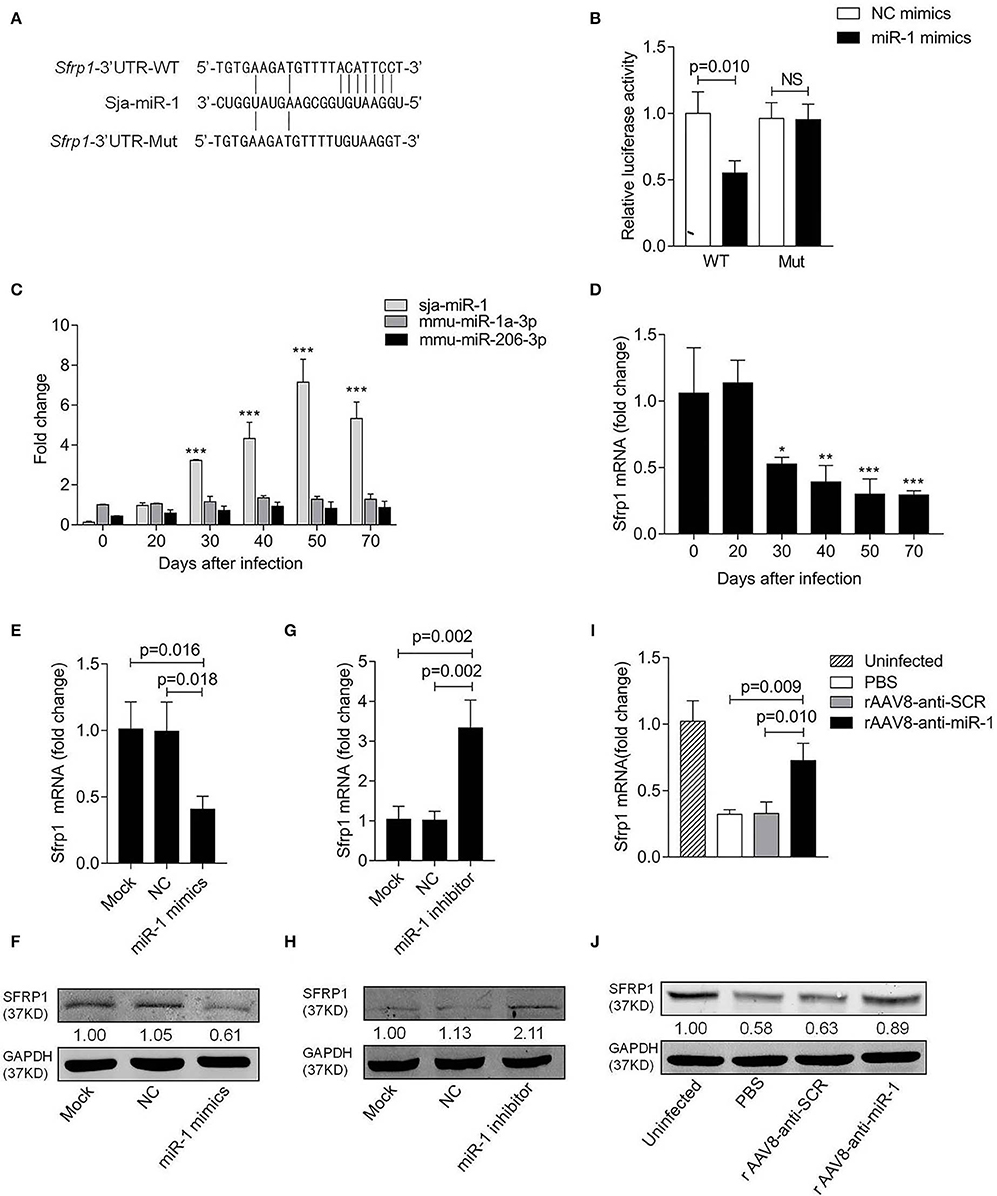
Figure 4. Sfrp1 is the direct target gene of sja-miR-1. (A) Sequence alignment of sja-miR-1 and the target region in the 3'UTR of Sfrp1. The corresponding luciferase reporter constructs were designated as pmirGLO-Sfrp1-3'UTR-WT and pmirGLO-Sfrp1-3'UTR-Mut, respectively. (B) Luciferase reporter assays of 293T cells transfected with pmirGLO-Sfrp1-3'UTR-WT or pmirGLO-Sfrp1-3'UTR-Mut in the absence or presence of sja-miR-1 mimics (n = 3). WT, pmirGLO-Sfrp1-3'UTR-WT; Mut, pmirGLO-Sfrp1-3'UTR-Mut. (C,D) The expression of sja-miR-1, mmu-miR-1a-3p, mmu-miR-206-3p and Sfrp1 in primary HSCs isolated from the infected mice at various time points post-infection. (E,F) Primary HSCs isolated from naive mice were transfected with the sja-miR-1 mimics, a negative control (NC) mimics or transfection reagent only (Mock) for 48 h. The expression of SFRP1 was detected by qPCR or western blot (n = 3). (G,H) Primary HSCs isolated from infected mice were transfected with sja-miR-1 inhibitor, a negative control (NC) inhibitor or transfection reagent only (Mock) for 48 h. The expression of SFRP1 was detected by qPCR or western blot (n = 3). (I,J) The expression of SFRP1 in primary HSCs from the infected mice treated with the sponge vectors at day 56 post-infection was detected using qPCR or western blot (n = 3). Data are presented as the Mean ± SD from three independent experiments, *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
Sja-miR-1 Activates Wnt/β-Catenin Pathway in HSCs by Targeting Sfrp1
It was reported that SFRP1 can inhibit Wnt signaling pathway, which was involved in promoting hepatic fibrosis by enhancing HSCs activation and survival (Myung et al., 2007). To investigate the biological functions of SFRP1 in the HSC activation, the primary HSCs from naive mice were transfected with Sfrp1 siRNA and qPCR analysis showed that Sfrp1 siRNA reduced 70% level of Sfrp1 (Figure 5A), which led to the activation of HSCs, evidenced by significant elevation of Col1α1, Col3α1, and α-Sma compared with the controls (Figure 5B).
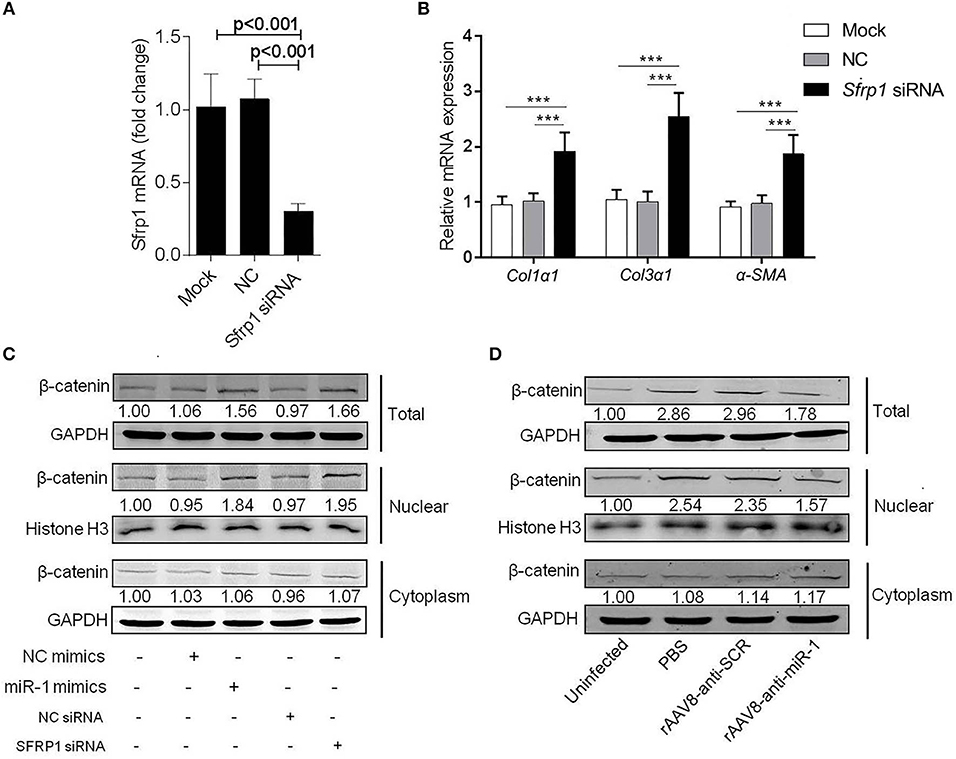
Figure 5. Sja-miR-1 activates HSCs via Wnt/β-catenin signaling pathway. (A,B) Primary HSCs isolated from naive mice were transfected with Sfrp1 siRNA, a negative control siRNA (NC) or transfection reagent only (Mock) for 48 h. The expression of Sfrp1 (A), Col1α1, Col3α1, and α-Sma. (B) were detected (n = 3). (C) The protein level of total, nuclear, and cytoplasm β-catenin in HSCs with various treatments indicated were detected using western blot. (D) The protein level of β-catenin in primary HSCs isolated from infected mice treated with sponge vectors were detected. Data are presented as the Mean ± SD from three independent experiments, ***p < 0.001.
To investigate involvement of the Wnt/β-catenin signaling in the sja-miR-1-mediated activation of HSCs, the protein level of β-catenin was detected in HSCs treated with sja-miR-1 mimics or Sfrp1 siRNA. Consistent with previous reports that SFRP1 functioned as a negative modulator of Wnt/β-catenin pathway (Sklepkiewicz et al., 2015), western blot analysis showed that the nuclear protein level of β-catenin was increased in the HSCs transfected with sja-miR-1 mimics or Sfrp1siRNA compared with the controls, whereas no significant difference was detected in the cytoplasm protein level of β-catenin (Figure 5C). Next, we analyzed β-catenin level in primary HSCs isolated from mice treated with the sponge vector and showed that β-catenin protein was obviously decreased in total HSCs homogenate and the nuclear protein compared with the control vector (Figure 5D). These data indicated that sustained and efficient suppression of sja-miR-1 led to marked elevation of SFRP1 and thereby inhibited the Wnt/β-catenin signaling pathway and collagens production.
Discussion
Sja-miR-1 was abundant in Schistosoma japonicum and considered playing an important role in schistosome sexual maturation (Cai et al., 2013; Han et al., 2015). This study provides the following evidence that the sja-miR-1 is involved in the occurrence and progression of hepatic fibrosis during schistosome infection: (1) sja-miR-1 was present in HSCs and involved in host HSC activation by targeting the Sfrp1; (2) rAAV8-mediated delivery of sja-miR-1 to the mouse liver promotes hepatic fibrosis in vivo; (3) the sustained inhibition of sja-miR-1 alleviated schistosomiasis hepatic fibrosis via reduction in production of collagens and α-SMA in the infected mice with the parasite; (4) the sja-miR-1 inhibitor reduced the expression of fibrosis-related genes in the HSCs isolated from infected mice or in the egg exosome-activated HSCs in vitro.
Wnt proteins are involved in the modeling and remodeling processes (van Gijn et al., 2002; Moon et al., 2004; Reya and Clevers, 2005; Brade et al., 2006). Wnt proteins are divided into two classes, involving the canonical and non-canonical Wnt pathways. The former stabilizes intracellular β-catenin, which can transfer β-catenin into the nucleus to activate the Wnt-dependent genes via regulating the relationship between the transcription factors and the transcriptional co-repressors (Huelsken and Behrens, 2002; Moon et al., 2004). Altered Wnt signaling is related to various disease progression including liver diseases (Thompson and Monga, 2007). Accumulated studies reveal that the activation of Wnt/β-catenin signaling can promote hepatic fibrosis by enhancing HSCs activation via HSCs proliferation, and ECM accumulation and also play an important role in schistosomiasis-induced hepatic fibrosis (Jiang et al., 2006; Myung et al., 2007). SFRP1 is homologous protein of the extracellular cysteine-rich-domain of the Wnt receptor Frizzled, but lacks the transmembrane and intracellular domain (Yang et al., 2009). SFRP1 protein functions as a negative regulator of Wnt/β-catenin signaling via inhibiting the binding of the Wnt receptor and thereby suppressing ligand-receptor interactions and signal transduction (Jones and Jomary, 2002; Kang et al., 2014). The reduction in production of SFRP1 leads to the activation of Wnt/β-catenin signaling pathways (Sklepkiewicz et al., 2015). It was demonstrated that the expression of fibrosis-related markers was elevated in SFRP1 knock-out mice (Matsuyama et al., 2014; Sklepkiewicz et al., 2015). Here, our study revealed that Sfrp1 gene was a direct target gene of sja-miR-1 both in vitro and in vivo models, and the sja-miR-1 mediated down-regulation of Sfrp1 elevated the expression of collagens and α-SMA and thereby activated the HSCs. Further, sustained and efficient inhibition of sja-miR-1 in vivo led to significant increase in the expression of SFRP1 and reduction in the collagen production. In addition, siRNA-mediated inhibition of Sfrp1 expression remarkably increased intra-nuclear β-catenin level, and consequently caused hepatic fibrosis. Therefore, these findings demonstrate that SFRP1 is a negative regulator of Wnt/β-catenin pathway which is consistent with the previous findings (Jones and Jomary, 2002; Kang et al., 2014).
The hepatic fibrosis is main pathology of schistosomiasis. The previous studies demonstrated the Th2 cytokines such as TGF-β1 and IL-13 were major profibrogenic factors. Recently, we reported that the sja-miR-2162 modulated pathogenicity of schistosomiasis in the infected mice by targeting the transforming growth factor beta receptor III (He et al., 2019). In this study, we demonstrated that sja-miR-1 derived from the parasite also contributed to the schistosomiasis hepatic fibrosis by regulating the fibrosis-related genes in a cross-species manner, which enriched our understanding of the pathogenesis of schistosomiasis. Further, the miRNAs derived from parasites may represent the emerging generic regulators that are involved in the pathogenesis of other parasite diseases, and the rAAV8-mediated sustained suppression of the heterogenous miRNAs may present a promising intervention for the disease therapy.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Naval Medical University (approval number FYXK [Shanghai] 2014-0003).
Author Contributions
YW, XF, and WP designed research and wrote the paper. YW, XF, NL, XW, and XL performed research. XH, YW, DZ, and WP analyzed data. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81972985 and 81430051), and Foundation of Shanghai Municipal Commission of Health and Family Planning (201740099).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staffs of the National Institute of Parasitic Disease, Chinese Center for Disease Control and Prevention for their help with parasite infections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00101/full#supplementary-material
References
Anthony, B. J., Ramm, G. A., and McManus, D. P. (2012). Role of resident liver cells in the pathogenesis of schistosomiasis. Trends Parasitol. 28, 572–579. doi: 10.1016/j.pt.2012.09.005
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 16, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartley, P. B., Ramm, G. A., Jones, M. K., Ruddell, R. G., Li, Y., and McManus, D. P. (2006). A contributory role for activated hepatic stellate cells in the dynamics of Schistosoma japonicum egg-induced fibrosis. Int. J. Parasitol. 36, 993–1001. doi: 10.1016/j.ijpara.2006.04.015
Böttcher, K., and Pinzani, M. (2017). Pathophysiology of liver fibrosis and the methodological barriers to the development of anti-fibrogenic agents. Adv. Drug. Deliv. Rev. 121, 3–8. doi: 10.1016/j.addr.2017.05.016
Brade, T., Manner, J., and Kühl, M. (2006). The role of wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovasc. Res. 72, 198–209. doi: 10.1016/j.cardiores.2006.06.025
Buck, A. H., Coakley, G., Simbari, F., McSorley, H. J., Quintana, J. F., Le Bihan, T., et al. (2014). Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5:5488. doi: 10.1038/ncomms6488
Burke, M. L., Jones, M. K., Gobert, G. N., Li, Y. S., Ellis, M. K., McManus, D., et al. (2009). Immunopathogenesis of human schistosomiasis. Parasite Immunol. 31, 163–176. doi: 10.1111/j.1365-3024.2009.01098.x
Cai, P., Piao, X., Hao, L., Liu, S., Hou, N., Wang, H., et al. (2013). A deep analysis of the small non-coding RNA population in Schistosoma japonicum eggs. PLoS ONE 8:e64003. doi: 10.1371/journal.pone.0064003
Chiaramonte, M. G., Donaldson, D. D., Cheever, A. W., and Wynn, T. A. (1999). An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J. Clin. Invest. 104, 777–785. doi: 10.1172/JCI7325
Colley, D. G., Bustinduy, A. L., Secor, W. E., and King, C. H. (2014). Human schistosomiasis. Lancet 383, 2253–2264. doi: 10.1016/S0140-6736(13)61949-2
Ebert, M. S., Neilson, J. R., and Sharp, P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726. doi: 10.1038/nmeth1079
Friedman, S. L. (2008). Mechanisms of hepatic fibrogenesis. Gastroenterology 134, 1655–1669. doi: 10.1053/j.gastro.2008.03.003
Friedman, S. L. (2010). Evolving challenges in hepatic fibrosis. Nat. Rev. Gastroenterol. Hepatol. 7, 425–436. doi: 10.1038/nrgastro.2010.97
Han, H., Peng, J., Hong, Y., Fu, Z., Lu, K., Li, H., et al. (2015). Comparative characterization of microRNAs in Schistosoma japonicum schistosomula from Wistar rats and BALB/c mice. Parasitol. Res. 114, 2639–2647. doi: 10.1007/s00436-015-4468-1
He, X., Wang, Y. G., Fan, X. B., Lei, N. H., Zhang, D. M., and Pan, W. Q. (2019). A schistosome miRNA promotes host hepatic fibrosis by targeting transforming growth factor beta receptor III. J. Hepatol. 19, 30674–30679. doi: 10.1016/j.jhep.2019.10.029
He, X., Xie, J., Zhang, D., Su, Q., Sai, X., Bai, R., et al. (2015). Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology 61, 2008–2017. doi: 10.1002/hep.27671
Hu, C., Zhu, S., Wang, J., Lin, Y., Ma, L., Zhu, L., et al. (2019). Schistosoma japonicum MiRNA-7-5p inhibits the growth and migration of hepatoma cells via cross-species regulation of S-phase kinase-associated protein 2. Front. Oncol. 9:175. doi: 10.3389/fonc.2019.00175
Huelsken, J., and Behrens, J. (2002). The Wnt signalling pathway. J. Cell Sci. 115, 3977–3978. doi: 10.1242/jcs.00089
Jiang, F., Parsons, C. J., and Stefanovic, B. (2006). Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J. Hepatol. 45, 401–409. doi: 10.1016/j.jhep.2006.03.016
Jones, S. E., and Jomary, C. (2002). Secreted frizzled-related proteins: searching for relationships and patterns. Bioessays 24, 811–820. doi: 10.1002/bies.10136
Kang, P., Wan, M., Huang, P., Li, C., Wang, Z., Zhong, X., et al. (2014). The Wnt antagonist sFRP1 as a favorable prognosticator in human biliary tract carcinoma. PLoS ONE 9:e90308. doi: 10.1371/journal.pone.0090308
Kisseleva, T., Cong, M., Paik, Y., Scholten, D., Jiang, C., Benner, C., et al. (2012). Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl. Acad. Sci. U.S.A. 109, 9448–9453. doi: 10.1073/pnas.1201840109
Lässer, C., Alikhani, V. S., Ekström, K., Eldh, M., Paredes, P. T., Bossios, A., et al. (2011). Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 9:9. doi: 10.1186/1479-5876-9-9
Liu, J., Zhu, L., Wang, J., Qiu, L., Chen, Y., Davis, R. E., et al. (2019). Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PLoS Pathog. 15:e1007817. doi: 10.1371/journal.ppat.1007817
Lu, M., Zhang, Q., Deng, M., Miao, J., Guo, Y., Gao, W., et al. (2008). An analysis of human microRNA and disease associations. PLoS ONE 3:e3420. doi: 10.1371/journal.pone.0003420
Matsuyama, M., Nomori, A., Nakakuni, K., Shimono, A., and Fukushima, M. (2014). Secreted frizzled-related Protein 1 (Sfrp1) regulates the progression of renal fibrosis in a mouse model of obstructive nephropathy. J. Biol. Chem. 289, 31526–31533. doi: 10.1074/jbc.M114.584565
Moon, R. T., Kohn, A. D., de Ferrari, G. V., and Kaykas, A. (2004). Wnt and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691–701. doi: 10.1038/nrg1427
Myung, S. J., Yoon, J. H., Gwak, G. Y., Kim, W., Lee, J. H., Kim, K. M., et al. (2007). Wnt signaling enhances the activation and survival of human hepatic stellate cells. FEBS Lett. 581, 2954–2958. doi: 10.1016/j.febslet.2007.05.050
Nakai, H., Fuess, S., Storm, T. A., Muramatsu, S., Nara, Y., and Kay, M. A. (2015). Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 79, 214–224. doi: 10.1128/JVI.79.1.214-224.2005
Pearce, E. J., and Macdonald, A. S. (2002). The immunobiology of schistosomiasis. Nat. Rev Immunol. 2, 499–511. doi: 10.1038/nri843
Reya, T., and Clevers, H. (2005). Wnt signalling in stem cells and cancer. Nature 434, 843–850. doi: 10.1038/nature03319
Seki, E., and Schwabe, R. F. (2015). Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology 61, 1066–1079. doi: 10.1002/hep.27332
Sklepkiewicz, P., Shiomi, T., Kaur, R., Sun, J., Kwon, S., Mercer, B., et al. (2015). Loss of secreted frizzled-related protein-1 leads to deterioration of cardiac function in mice and plays a role in human cardiomyopathy. Circ. Heart Fail. 8, 362–372. doi: 10.1161/CIRCHEARTFAILURE.114.001274
Szempruch, A. J., Dennison, L., Kieft, R., Harrington, J. M., and Hajduk, S. L. (2016). Sending a message: extracellular vesicles of pathogenic protozoan parasites. Nat. Rev. Microbiol. 14, 669–675. doi: 10.1038/nrmicro.2016.110
Thompson, M. D., and Monga, S. P. (2007). WNT/β-catenin signaling in liver health and disease. Hepatology 45, 1298–1305. doi: 10.1002/hep.21651
van Gijn, M. E., Daemen, M. J., Smits, J. F., and Blankesteijn, W. M. (2002). The Wnt-frizzled cascade in cardiovascular disease. Cardiovasc. Res. 55, 16–24. doi: 10.1016/S0008-6363(02)00221-3
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. doi: 10.1038/nplants.2016.151
Wilson, M. S., Mentink-Kane, M. M., Pesce, J. T., Ramalingam, T. R., Thompson, R., and Wynn, T. A. (2007). Immunopathology of schistosomiasis. Immunol. Cell. Biol. 85, 148–154. doi: 10.1038/sj.icb.7100014
Yang, Z. Q., Liu, G., Bollig-Fischer, A., Haddad, R., Tarca, A. L., and Ethier, S. P. (2009). Methylation-associated silencing of SFRP1 with an 8p11-12 amplification inhibits canonical and non-canonical WNT pathways in breast cancers. Int. J. Cancer 125, 1613–1621. doi: 10.1002/ijc.24518
Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., et al. (2012). Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22,107–126 doi: 10.1038/cr.2011.158
Zhu, L., Liu, J., Dao, J., Lu, K., Li, H., Gu, H., et al. (2016). Molecular characterization of S. japonicum exosome-like vesicles reveals their regulatory roles in parasite-host interactions. Sci. Rep. 6:25885. doi: 10.1038/srep25885
Keywords: Schistosoma japonicum, microRNA, hepatic fibrosis, SFRP1, cross-species regulation
Citation: Wang Y, Fan X, Lei N, He X, Wang X, Luo X, Zhang D and Pan W (2020) A MicroRNA Derived From Schistosoma japonicum Promotes Schistosomiasis Hepatic Fibrosis by Targeting Host Secreted Frizzled-Related Protein 1. Front. Cell. Infect. Microbiol. 10:101. doi: 10.3389/fcimb.2020.00101
Received: 20 November 2019; Accepted: 26 February 2020;
Published: 13 March 2020.
Edited by:
Martin Craig Taylor, University of London, United KingdomReviewed by:
César López-Camarillo, Universidad Autónoma de la Ciudad de México, MexicoJingwen Wang, Yale University, United States
Copyright © 2020 Wang, Fan, Lei, He, Wang, Luo, Zhang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongmei Zhang, ZG16aGFuZ2NuQDE2My5jb20=; Weiqing Pan, d3FwYW4wOTEyQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
 Yange Wang
Yange Wang Xiaobin Fan
Xiaobin Fan Nanhang Lei
Nanhang Lei Xing He
Xing He Weiqing Pan
Weiqing Pan