- 1Department of Medicine, University of Washington, Seattle, WA, United States
- 2Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Seattle, WA, United States
- 3Department of Laboratory Medicine, University of Washington, Seattle, WA, United States
- 4Department of Global Health, University of Washington, Seattle, WA, United States
- 5Benaroya Research Institute, Seattle, WA, United States
Human herpesvirus (HHV) 6 is thought to remain clinically latent in most individuals after primary infection and to reactivate to cause disease in persons with severe immunosuppression. In allogeneic hematopoietic stem cell transplant recipients, reactivation of HHV-6 species B is a considerable cause of morbidity and mortality. HHV-6B reactivation is the most frequent cause of infectious meningoencephalitis in this setting and has been associated with a variety of other complications such as graft rejection and acute graft versus host disease. This has inspired efforts to develop HHV-6-targeted immunotherapies. Basic knowledge of HHV-6-specific adaptive immunity is crucial for these endeavors, but remains incomplete. Many studies have focused on specific HHV-6 antigens extrapolated from research on human cytomegalovirus, a genetically related betaherpesvirus. Challenges to the study of HHV-6-specific T-cell immunity include the very low frequency of HHV-6-specific memory T cells in chronically infected humans, the large genome size of HHV-6, and the lack of an animal model. This review will focus on emerging techniques and methodological improvements that are beginning to overcome these barriers. Population-prevalent antigens are now becoming clear for the CD4+ T-cell response, while definition and ranking of CD8+ T-cell antigens and epitopes is at an earlier stage. This review will discuss current knowledge of the T-cell response to HHV-6, new research approaches, and translation to clinical practice.
Introduction
Human herpesvirus 6 was discovered in 1986 and later found to exist as two closely related species, HHV-6A and HHV-6B, in the Betaherpesvirinae subfamily and Roseolovirus genus (1). Hereafter, “HHV-6” refers to both species unless specific data are available to differentiate between species. The two species have genomes roughly 162 kb long with 88–90% sequence identity, but have distinct tropisms and epidemiology (1). The other betaherpesviruses known to infect humans are HHV-7 and human cytomegalovirus (HCMV). About 1% of humans have inherited chromosomally integrated HHV-6 (ici-HHV-6) (2). Interestingly, immune tolerance has not been demonstrated and persons with ici-HHV-6 appear to maintain anti-HHV-6 cell-mediated immunity (CMI) (3). Primary infection with HHV-6B usually occurs once maternal antibodies have waned in early life (4, 5). The clinical syndrome roseola consists of fever and rash, although seizures can occur. The epidemiology of HHV-6A is less well understood, related to difficulties with species-specific serodiagnosis. Like other herpesviruses, HHV-6 establishes lifelong latent infection, usually asymptomatic. Transmission is probably via saliva, as HHV-6 DNA is frequently detectable in oral specimens.
Human herpesvirus-6 reactivation events are thought to occur periodically in healthy carriers and to be subclinical due to intact immune surveillance. Natural killer cells appear to have anti-HHV-6 function (6), as implied by their activity in the acute febrile phase of primary infection (7, 8) and cytotoxicity against HHV-6-infected cells (9) in an interleukin-15-dependent manner (10). There is little evidence that antibody deficiency disorders increase risk of complications from infection by these viruses (11), and B cell deficiency does not increase lethality of murine roseolovirus (MRV), a betaherpesvirus related to HHV-6, in neonatal mice (12).
Compared to other herpesviruses, HHV-6-specific cell-mediated response is delayed in primary infection (8). This correlates with, and could be mechanistically related to, HHV-6 lymphotropism (13–15), since activated HHV-6-responsive T cells may be differentially susceptible to destructive viral infection. HHV-6 also has immunosuppressive mechanisms targeting T cell function (16–20). The T-cell response is considered critical for control of HHV-6B infection since reactivation commonly occurs in cases of T-cell lymphopenia, e.g., in AIDS patients (21) or after bone marrow transplantation (22–28). Moreover, greater overall survival in these posttransplant patients is associated with at least 200 CD3+ T cells/μL in blood at the time of HHV-6B reactivation (29).
The relative importance of different T-cell subsets in HHV-6B immunity is still not well established. In pediatric hematopoietic cell transplant (HCT) patients, increased proportions of perforin-expressing CD8+ T cells have been temporally associated with HHV-6 clearance (30). HHV-6-specific CD8+ T cells with proliferative capacity were more readily detectable in patients after viral reactivation but not in those without (31). Moreover, MRV is lethal to CD8 knockout mice but not to wild-type mice (12). Nevertheless, like other herpesviruses (32–38), HHV-6 can evade CD8+ T cells by downregulating class I MHC molecules (39), which may account for challenges in detecting HHV-6B-specific CD8+ T cells (40, 41).
CD4+ T cells are now considered to exert their own direct antiviral effector functions and to be crucial in controlling herpesvirus infections (42–47), although less is known about their importance for HHV-6B control. Some observers consider it plausible that HHV-6B-induced de novo surface expression of class II MHC molecules (48)—similar to HCMV (49, 50)—could promote recognition of infected cells by CD4+ T lymphocytes. Moreover, HHV-6A-specific CD4+ T-cell lines can produce IFNγ and degranulate (measured by surface CD107a/b) when presented with whole virus or peptide antigen, suggesting HHV-6A-specific cytotoxicity (51). These in vitro studies suggest the importance of Th1 cytotoxic CD4+ T cells in immunity to HHV-6. However, lack of an animal model, the multifaceted nature of human immunodeficiency states such as transplantation and HIV infection, and a paucity of data from direct ex vivo methods to measure expression of cytotoxic machinery in HHV-6-specific CD4+ T cells precludes strong conclusions at present.
Medical Importance of HHV-6
The strongest evidence supporting clinically significant consequences of HHV-6 infection is in immunocompromised patients, particularly recipients of allogeneic HCT and solid organ transplants (SOT). Detection of HHV-6B DNA in blood occurs in 40–50% of HCT recipients at a median of approximately 3 weeks after transplant, corresponding with the time period of lowest lymphocyte and neutrophil counts (25–28, 52, 53). HHV-6B accounts for ≥98% of HHV-6 detection after allogeneic HCT. Given pre-existing seropositivity, most viral detection is likely due to viral reactivation. The factors most prominently associated with HHV-6B reactivation include the use of umbilical cord blood stem cells as the donor source, an HLA mismatched or unrelated donor, receipt of depleting anti-T-cell antibodies, development of acute graft-versus-host disease, and treatment with glucocorticoids (26, 28, 54–59).
Human herpesvirus-6B is the most frequent infectious cause of encephalitis after allogeneic HCT, and this occurs in approximately 1% of all HCT recipients (26, 27, 60–62). HHV-6B encephalitis results in significant morbidity and mortality despite antiviral treatment (26, 61, 63, 64). HHV-6B reactivation has been detected in many other conditions in allogeneic HCT and SOT recipients, although its causal role is less clearly defined. These include myelosuppression, development of acute GVHD (26, 28, 65, 66), increased risk for cytomegalovirus reactivation and disease (28, 67–71), and solid organ allograft dysfunction (72–74).
Several available antiviral agents demonstrate activity against HHV-6A and HHV-6B, including foscarnet, ganciclovir, and cidofovir (25, 75–78), but clinical use has been limited by the toxicities of available antiviral agents and lack of proven efficacy in preventing end-organ disease (79–81). New small molecules with activity against HHV-6 species are in development (77, 82, 83). Given these shortcomings, adoptive immunotherapy using virus-specific T cells (VSTs) is an exciting new therapeutic approach that appears to be safe and to reduce HHV-6 DNA levels, as well as end-organ disease symptoms, in small, uncontrolled studies (84–87). More research is needed to identify HHV-6 epitopes that can be used to generate high affinity T-cell lines to advance adoptive immunotherapeutic strategies.
Challenges and Opportunities for T-Cell Epitope Discovery in HHV-6
Several technical challenges confront investigators seeking to identify T-cell epitopes in HHV-6 and study the roles of T cells in pathogenesis and immunity. First, non-infected humans to serve as negative controls are rare. Second, an affordable small animal model for HHV-6 infection is not available for in vivo studies. Pig tailed macaques (88), marmosets (89), cynomolgus macaques, and African green monkeys (90) are susceptible to human roseolovirus strains and have been used as models in a few studies (91, 92). Endogenous roseoloviruses have recently been described in chimpanzees (93) and rhesus macaques (94). Once we understand more about the natural history of these infections in their natural hosts and how well they mimic HHV-6 in humans, it may become rational to use them as informative model systems. Recently, the genomic sequence of a mouse herpesvirus, MRV, was revealed and appears phylogenetically closer to HHV-6 than to murine cytomegalovirus. It is considered a homolog of HHV-6 and HHV-7 with some potential as an in vivo model (95).
T-cell antigen identification and ranking in animals using animal homologs of HHV-6 may not translate to the HHV-6 infection setting in humans; the optimal human specimens for understanding the in vivo reality have not been defined. Samples from immunocompetent humans with primary or reactivated HHV-6 infection (ideally taken at the peak of virus-specific T cell expansion) might have a high enough frequency of HHV-6-specific T cells to allow direct ex vivo analysis of antigen specificity. This could provide the most true-to-life ranking of HHV-6 antigens with regard to population prevalence and within-patient immunodominance. Samples from such patients have been used in studies of HHV-6 viral loads and clinical associations (96–99), but we are not aware of their use for T cell antigen discovery so far. In addition, study designs in which serial blood samples are obtained from patients with primary or reactivated HHV-6B who spontaneously control infection may help to understand the effective versus bystander T-cell responses. Identification of HHV-6B antigens that activate T cells in this setting may give clues as to the nature of an effective T cell response.
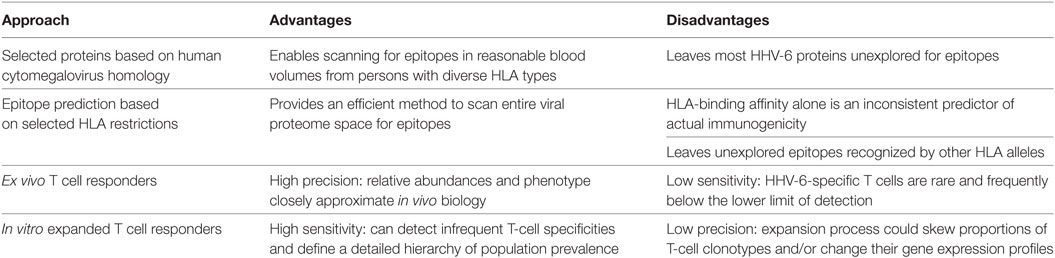
Third, HHV-6A and -6B are quite evasive, with mechanisms including downregulation of CD3 (88), induction of interleukin-10-producing T-regulatory type 1 cells (18), and inhibition of interferon beta (19), interleukin 2 (20), interleukin 12 (100, 101), and MHC class I (6, 39, 102, 103). Perhaps this is why HHV-6-specific T cells in healthy persons, as measured by IFNγ production, are quite rare; between 0.01 and 0.1% of CD4+ T cells respond to whole virus (51) while CD8+ T cells specific for HHV-6 peptide-loaded pentamers are often less than 0.01% of circulating CD8+ T cells and sometimes below the limit of detection (104, 105). The overall frequency of CD8+ T cells specific for whole HHV-6 virus has not been established. By contrast, up to 20% of memory CD4 and CD8 T cells are specific for HCMV in seropositive persons (106). Ex vivo testing of HHV-6B CD4+ T-cell abundance in PBMC by standard methods, such as ELISPOT or intracellular cytokine secretion (ICS), is reliably quantitative if enough cells are studied, but not sensitive enough to meaningfully characterize fine epitope specificity. Researchers, therefore, use in vitro expansion to enrich HHV-6-reactive T cells for detailed epitope identification and definition of epitope breadth and population prevalence (40, 41, 51, 104, 105, 107, 108). However, this process may introduce changes in T-cell expression profiles, and differential proliferation rates could result in skewed T-cell clonotype proportions in the expanded product, introducing challenges into the larger goals of studying the phenotype of HHV-6-specific T cells and measuring the immunodominance hierarchy across a spectrum of epitopes. Overall, in vitro expansion of VSTs offers enhanced sensitivity but decreased quantitative precision (Table 1).
Finally, the HHV-6B genome contains roughly 100 open reading frames totaling tens of thousands of potential T-cell epitopes—a large potential epitope space. Investigators are faced with either down-selecting to a limited number of open reading frames, or using high-throughput methods requiring considerable time and expense.
Approaches to T-Cell Epitope Discovery in HHV-6
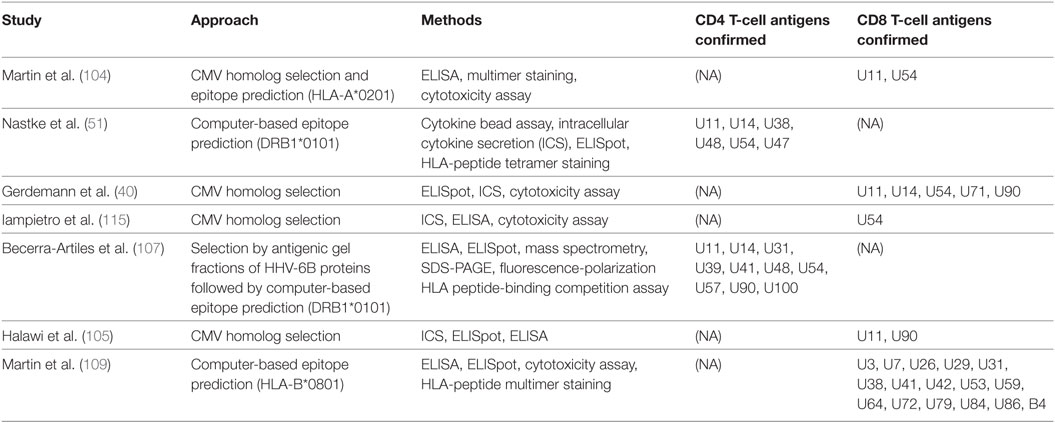
Approaches to discovery of T-cell epitopes in HHV-6B can be categorized by the methods they use to address these challenges. One system focuses on selected HHV-6B proteins and scans cells from persons with a range of HLA haplotypes for induction of T-cell activation. Using the better-characterized HCMV as a springboard, early studies of HHV-6B focused on proteins, which have HCMV homologs (17, 40, 104, 105). The rationale is not possible cross-reactivity due to homolog sequence identity; for example, the identity of U54 to its HCMV homolog UL83 (encoding pp65) is only 20% (104). Rather, it was hypothesized that T-cell antigenicity may be related to biological functional, viral mRNA and protein expression kinetics, virion abundance, and other factors, predisposing certain betaherpesvirus proteins to higher antigenicity.
A second approach is to focus on HHV-6B peptides predicted to bind specific HLA allelic variants of interest. Nastke et al. used two predictive algorithms to assess all 42,838 possible HHV-6B nonamers for predicted binding affinity to DRB1*0101 and selected 322 candidates, of which 12 were confirmed as CD4+ T-cell epitopes (51). More recently, Martin et al. took a similar approach aimed at CD8+ T-cell epitopes restricted by HLA-B*0801 and confirmed 16 epitopes by cytolytic functional assays (109). Another study prefaced epitope prediction with a preliminary assay step, by first separating HHV-6 proteins into molecular weight fractions from either HHV-6B virions or infected cell lysates, and testing these fractions for PBMC IFNγ responses (107). Fractions eliciting significant cytokine response were analyzed by mass spectrometry to identify candidate HHV-6B antigenic proteins. Within these, the top 463 predicted HLA DR1-restricted epitopes were then tested using synthetic peptides and 107 were confirmed as epitopes.
Martin et al. used a hybrid of the two approaches, focusing on predicted HLA-A*0201 epitopes within HHV-6B proteins U11, U54, and U90 (homologs of HCMV antigens pp150, pp65, and IE1, respectively) (104). To enrich virus-specific CD8+ T cells, PBMCs were incubated with 12 HHV-6B peptides of interest for 10–14 days, and then re-stimulated with peptide-loaded CD40-activated autologous B cells and IL-2. This study showed CD8+ T-cell clones restricted to three peptides derived from U11 or U54 could recognize HHV-6B-infected cells and produce IFNγ, TNFα, and granzyme B. A summary of these studies is in Table 2.
A High-Throughput Approach to T-Cell Epitope Discovery in HHV-6B
The above approaches have limitations; use of selected ORFs leaves most HHV-6 proteins unexplored while algorithms are imperfect and do not address the full range of possible HLA restrictions. We are thus left with an incomplete view of T-cell specificity for this virus, which could be improved by an approach covering all HHV-6 proteins and diverse HLA haplotypes. Our lab has developed a high throughput, HLA-agnostic method to characterize T-cell immunity to large-genome viruses. Donor PBMCs are stimulated with whole virus, and activated T cells are sorted by FACS and expanded in vitro to produce a polyclonal cell line enriched typically a hundredfold for VSTs above the starting PBMC. Cross-presentation by autologous dendritic cells is used for CD8+ T cells, while addition of UV-inactivated pathogen to PBMC suffices to re-stimulate memory CD4+ responses. Each viral ORF is cloned and expressed via in vitro transcription and translation for CD4+ T-cell work or prepared for transfection of COS-7 artificial antigen-presenting cells (APC) for CD8+ T-cell studies. The bulk expanded T cells are then assayed with suitable APC and each individual viral protein, as documented for vaccinia, HSV-1, HSV-2, VZV, and Mycobacterium tuberculosis (110–114). This approach is now being applied in our lab to study CD4+ T-cell responses to HHV-6B and methods are under development to apply it to CD8+ T cells.
Leading HHV-6 T-Cell Antigens
So far, studies in roseolovirus antigen discovery have investigated HHV-6B. None have explicitly addressed HHV-6A, although cross-reactivity has been described between the two species using bulk expanded cell lines (51) or T-cell clones (108). Using HHV-6B peptides whose HHV-6A homologs differed in one or more amino acid position, Nastke et al. also found that HHV-6A-specific T-cell lines produced IFNγ in response to individual HHV-6B peptides, and vice versa, confirming cross-reactivity at the epitope level (51). Two published studies on CD4+ T cells have both focused on DRB1*0101-restricted HHV-6B epitopes (51, 107); the ORF products identified in both of these studies are U11, U14, U48, U54, and U57, most of which are virion proteins. Becerra-Artiles et al. also correlated HLA-DRB1*0101 peptide-binding affinity with the proportion of responding donors and with the magnitude of T-cell responses as measured by ELISpot (107). Studies on CD8+ T cells have been limited to specific ORF products, except for the recent HLA-B*0801-restricted genome-wide screen performed by Martin et al. mentioned above (109). ORF products described by at least two studies as CD8+ T-cell antigens are U11, U54, and U90 (40, 105, 115). Similarly, studies of VSTs for HHV-6 treatment have used U11, U14, and U90 as antigens for creating VST cell lines (86, 87).
Germline integration of a viral genome presents an interesting question: are viral ORFs at the integration site transcribed and translated, and if so, are they targeted by CMI? Integrated HHV-6 genome sequences are divergent from non-integrated HHV-6 genomes but have all genes fully intact, suggesting replication competence (116). Indeed, PBMCs of iciHHV-6A donors can be induced in vitro to produce virions, which can then infect non-integrated HHV-6A-negative cells, so, in vivo antigen expression from iciHHV-6 ORFs seems plausible (117). Presumably, T cells educated in thymi of people with iciHHV-6 would be tolerized to HHV-6 antigens; however, iciHHV-6 individuals actually have high frequencies of CD8+ T cells specific for products of U54 (118) and U90 (3), suggesting active immune surveillance. More research is needed to compare the T-cell response to HHV-6 in people with and without chromosomal integration.
Future Directions
Research on HHV-6 T-cell epitopes to date has largely shown a selection bias toward viral proteins that are homologs of known HCMV antigenic proteins, and it is not clear where these fit into the overall pattern of immunodominance and population prevalence of T-cell epitopes for this virus. This field is entering a new phase of using high-throughput methods to solve this question. Other methods similar to the one described above would also be useful and informative. For example, instead of using in vitro expressed proteins to screen for viral antigens, one could use peptide mixes for each protein, as has been done for HCMV and HHV-8. This process could be leveraged by focusing on peptides predicted to bind any of a given donor’s HLA alleles; this would reduce cost, making it feasible to study many more donors and identify antigens and epitopes immunoprevalent across a wide range of HLA alleles.
Such approaches will provide a much fuller understanding of HHV-6 T-cell epitopes, which will in turn inform development of treatment modalities for immunocompromised patients suffering from reactivation. VST immunotherapy for SCT recipients is a nascent field that has garnered much interest in recent years as a promising alternative to antiviral drugs that have associated toxicities or are altogether ineffective (particularly for adenovirus and BK virus) (3, 40, 86, 87, 119, 120). Results of early clinical trials are promising but still anecdotal. Deeper knowledge of T cell epitopes and antigens could help optimize VST products for broad applicability across HLA alleles. In addition, since HHV-6B infection has been associated with various autoimmune disorders (48, 89, 121) and drug reaction with eosinophilia and systemic symptoms (DRESS) (122–124), VSTs lacking specificity for HHV-6 epitopes implicated in these disorders could be preferable. Finally, although a vaccine for HHV-6B is not a current public health priority, future development may benefit from thorough knowledge of HHV-6B T-cell epitopes gained from ongoing research.
Author Contributions
All authors wrote and edited the manuscript.
Conflict of Interest Statement
JH has served as a consultant for Chimerix, Inc. and Nohla Therapeutics, Inc. and has received research support from Chimerix, Inc. and Shire. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported by NIH T32AI118690 to DH, the NIAID/NIH K23AI119133 to JH, the HHV-6 Foundation’s Dharam Ablashi Research Fund Pilot Grant to DK, and NIH 5P50GM115305-04 to DK.
References
1. Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol (2014) 159(5):863–70. doi:10.1007/s00705-013-1902-5
2. Wood ML, Royle NJ. Chromosomally integrated human herpesvirus 6: models of viral genome release from the telomere and impacts on human health. Viruses (2017) 9(7):184. doi:10.3390/v9070184
3. Barozzi P, Riva G, Vallerini D, Quadrelli C, Lagreca I, Eccheli R, et al. Circulating functional T cells specific to human herpes virus 6 (HHV6) antigens in individuals with chromosomally integrated HHV6. Clin Microbiol Infect (2016) 22(10):893–5. doi:10.1016/j.cmi.2016.07.006
4. Okuno T, Takahashi K, Balachandra K, Shiraki K, Yamanishi K, Takahashi M, et al. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J Clin Microbiol (1989) 27(4):651–3.
5. Cermelli C, Fabio G, Montorsi M, Sabbatini AM, Portolani M. Prevalence of antibodies to human herpesviruses 6 and 7 in early infancy and age at primary infection. New Microbiol (1996) 19(1):1–8.
6. Eliassen E, Di Luca D, Rizzo R, Barao I. The interplay between natural killer cells and human herpesvirus-6. Viruses (2017) 9(12):367. doi:10.3390/v9120367
7. Takahashi K, Segal E, Kondo T, Mukai T, Moriyama M, Takahashi M, et al. Interferon and natural killer cell activity in patients with exanthem subitum. Pediatr Infect Dis J (1992) 11(5):369–73. doi:10.1097/00006454-199205000-00006
8. Kumagai T, Yoshikawa T, Yoshida M, Okui T, Ihira M, Nagata N, et al. Time course characteristics of human herpesvirus 6 specific cellular immune response and natural killer cell activity in patients with exanthema subitum. J Med Virol (2006) 78(6):792–9. doi:10.1002/jmv.20625
9. Malnati MS, Lusso P, Ciccone E, Moretta A, Moretta L, Long EO. Recognition of virus-infected cells by natural killer cell clones is controlled by polymorphic target cell elements. J Exp Med (1993) 178(3):961–9. doi:10.1084/jem.178.3.961
10. Flamand L, Stefanescu I, Menezes J. Human herpesvirus-6 enhances natural killer cell cytotoxicity via IL-15. J Clin Invest (1996) 97(6):1373–81. doi:10.1172/JCI118557
11. Vuorinen T, Kotilainen P, Lautenschlager I, Kujari H, Krogerus L, Oksi J. Interstitial pneumonitis and coinfection of human herpesvirus 6 and Pneumocystis carinii in a patient with hypogammaglobulinemia. J Clin Microbiol (2004) 42(11):5415–8. doi:10.1128/JCM.42.11.5415-5418.2004
12. Patel SJ, Yokoyama WM. CD8(+) T cells prevent lethality from neonatal murine roseolovirus infection. J Immunol (2017) 199(9):3212–21. doi:10.4049/jimmunol.1700982
13. Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell (1999) 99(7):817–27. doi:10.1016/S0092-8674(00)81678-5
14. Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, et al. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol (1989) 63(7):3161–3.
15. Tang H, Serada S, Kawabata A, Ota M, Hayashi E, Naka T, et al. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc Natl Acad Sci U S A (2013) 110(22):9096–9. doi:10.1073/pnas.1305187110
16. Dagna L, Pritchett JC, Lusso P. Immunomodulation and immunosuppression by human herpesvirus 6A and 6B. Future Virol (2013) 8(3):273–87. doi:10.2217/fvl.13.7
17. Iampietro M, Morissette G, Gravel A, Flamand L. Inhibition of interleukin-2 gene expression by human herpesvirus 6B U54 tegument protein. J Virol (2014) 88(21):12452–63. doi:10.1128/JVI.02030-14
18. Wang F, Yao K, Yin QZ, Zhou F, Ding CL, Peng GY, et al. Human herpesvirus-6-specific interleukin 10-producing CD4+ T cells suppress the CD4+ T-cell response in infected individuals. Microbiol Immunol (2006) 50(10):787–803. doi:10.1111/j.1348-0421.2006.tb03855.x
19. Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol (2007) 81(11):5737–48. doi:10.1128/JVI.02443-06
20. Flamand L, Gosselin J, Stefanescu I, Ablashi D, Menezes J. Immunosuppressive effect of human herpesvirus 6 on T-cell functions: suppression of interleukin-2 synthesis and cell proliferation. Blood (1995) 85(5):1263–71.
21. Clark DA, Ait-Khaled M, Wheeler AC, Kidd IM, McLaughlin JE, Johnson MA, et al. Quantification of human herpesvirus 6 in immunocompetent persons and post-mortem tissues from AIDS patients by PCR. J Gen Virol (1996) 77(Pt 9):2271–5. doi:10.1099/0022-1317-77-9-2271
22. Kadakia MP, Rybka WB, Stewart JA, Patton JL, Stamey FR, Elsawy M, et al. Human herpesvirus 6: infection and disease following autologous and allogeneic bone marrow transplantation. Blood (1996) 87(12):5341–54.
23. Cone RW, Huang ML, Corey L, Zeh J, Ashley R, Bowden R. Human herpesvirus 6 infections after bone marrow transplantation: clinical and virologic manifestations. J Infect Dis (1999) 179(2):311–8. doi:10.1086/314581
24. Wang LR, Dong LJ, Zhang MJ, Lu DP. The impact of human herpesvirus 6B reactivation on early complications following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant (2006) 12(10):1031–7. doi:10.1016/j.bbmt.2006.06.001
25. De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev (2005) 18(1):217–45. doi:10.1128/CMR.18.1.217-245.2005
26. Hill JA, Koo S, Guzman Suarez BB, Ho VT, Cutler C, Koreth J, et al. Cord-blood hematopoietic stem cell transplant confers an increased risk for human herpesvirus-6-associated acute limbic encephalitis: a cohort analysis. Biol Blood Marrow Transplant (2012) 18(11):1638–48. doi:10.1016/j.bbmt.2012.04.016
27. Ogata M, Satou T, Kadota J, Saito N, Yoshida T, Okumura H, et al. Human herpesvirus 6 (HHV-6) reactivation and HHV-6 encephalitis after allogeneic hematopoietic cell transplantation: a multicenter, prospective study. Clin Infect Dis (2013) 57(5):671–81. doi:10.1093/cid/cit358
28. Zerr DM, Boeckh M, Delaney C, Martin PJ, Xie H, Adler AL, et al. HHV-6 reactivation and associated sequelae after hematopoietic cell transplantation. Biol Blood Marrow Transplant (2012) 18(11):1700–8. doi:10.1016/j.bbmt.2012.05.012
29. Greco R, Crucitti L, Noviello M, Racca S, Mannina D, Forcina A, et al. Human herpesvirus 6 infection following haploidentical transplantation: immune recovery and outcome. Biol Blood Marrow Transplant (2016) 22(12):2250–5. doi:10.1016/j.bbmt.2016.09.018
30. de Pagter PJ, Boelens JJ, Jacobi R, Schuurman R, Nanlohy NM, Sanders EA, et al. Increased proportion of perforin-expressing CD8+ T-cells indicates control of herpesvirus reactivation in children after stem cell transplantation. Clin Immunol (2013) 148(1):92–8. doi:10.1016/j.clim.2013.02.023
31. de Pagter AP, Boelens JJ, Scherrenburg J, Vroom-de Blank T, Tesselaar K, Nanlohy N, et al. First analysis of human herpesvirus 6T-cell responses: specific boosting after HHV6 reactivation in stem cell transplantation recipients. Clin Immunol (2012) 144(3):179–89. doi:10.1016/j.clim.2012.06.006
32. Coscoy L, Ganem D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A (2000) 97(14):8051–6. doi:10.1073/pnas.140129797
33. Hudson AW, Blom D, Howley PM, Ploegh HL. The ER-lumenal domain of the HHV-7 immunoevasin U21 directs class I MHC molecules to lysosomes. Traffic (2003) 4(12):824–37. doi:10.1046/j.1398-9219.2003.0137.x
34. Hudson AW, Howley PM, Ploegh HL. A human herpesvirus 7 glycoprotein, U21, diverts major histocompatibility complex class I molecules to lysosomes. J Virol (2001) 75(24):12347–58. doi:10.1128/JVI.75.24.12347-12358.2001
35. Lemmermann NA, Böhm V, Holtappels R, Reddehase MJ. In vivo impact of cytomegalovirus evasion of CD8 T-cell immunity: facts and thoughts based on murine models. Virus Res (2011) 157(2):161–74. doi:10.1016/j.virusres.2010.09.022
36. May NA, Glosson NL, Hudson AW. Human herpesvirus 7 u21 downregulates classical and nonclassical class I major histocompatibility complex molecules from the cell surface. J Virol (2010) 84(8):3738–51. doi:10.1128/JVI.01782-09
37. May NA, Wang Q, Balbo A, Konrad SL, Buchli R, Hildebrand WH, et al. Human herpesvirus 7 U21 tetramerizes to associate with class I major histocompatibility complex molecules. J Virol (2014) 88(6):3298–308. doi:10.1128/JVI.02639-13
38. Zuo J, Currin A, Griffin BD, Shannon-Lowe C, Thomas WA, Ressing ME, et al. The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by targeting MHC class I molecules for degradation. PLoS Pathog (2009) 5(1):e1000255. doi:10.1371/journal.ppat.1000255
39. Glosson NL, Hudson AW. Human herpesvirus-6A and -6B encode viral immunoevasins that downregulate class I MHC molecules. Virology (2007) 365(1):125–35. doi:10.1016/j.virol.2007.03.048
40. Gerdemann U, Keukens L, Keirnan JM, Katari UL, Nguyen CT, de Pagter AP, et al. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood (2013) 121(1):207–18. doi:10.1182/blood-2012-05-430413
41. Yakushijin Y, Yasukawa M, Kobayashi Y. Establishment and functional characterization of human herpesvirus 6-specific CD4+ human T-cell clones. J Virol (1992) 66(5):2773–9.
42. Christensen JP, Cardin RD, Branum KC, Doherty PC. CD4(+) T cell-mediated control of a gamma-herpesvirus in B cell-deficient mice is mediated by IFN-gamma. Proc Natl Acad Sci U S A (1999) 96(9):5135–40. doi:10.1073/pnas.96.9.5135
43. Haque T, Wilkie GM, Jones MM, Higgins CD, Urquhart G, Wingate P, et al. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood (2007) 110(4):1123–31. doi:10.1182/blood-2006-12-063008
44. Sparks-Thissen RL, Braaten DC, Kreher S, Speck SH, Virgin HW IV. An optimized CD4 T-cell response can control productive and latent gammaherpesvirus infection. J Virol (2004) 78(13):6827–35. doi:10.1128/JVI.78.13.6827-6835.2004
45. Stevenson PG, Cardin RD, Christensen JP, Doherty PC. Immunological control of a murine gammaherpesvirus independent of CD8+ T cells. J Gen Virol (1999) 80(Pt 2):477–83. doi:10.1099/0022-1317-80-2-477
46. Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol (2012) 12(2):136–48. doi:10.1038/nri3152
47. Zuo J, Rowe M. Herpesviruses placating the unwilling host: manipulation of the MHC class II antigen presentation pathway. Viruses (2012) 4(8):1335–53. doi:10.3390/v4081335
48. Caselli E, Zatelli MC, Rizzo R, Benedetti S, Martorelli D, Trasforini G, et al. Virologic and immunologic evidence supporting an association between HHV-6 and Hashimoto’s thyroiditis. PLoS Pathog (2012) 8(10):e1002951. doi:10.1371/journal.ppat.1002951
49. Khoury EL, Pereira L, Greenspan FS. Induction of HLA-DR expression on thyroid follicular cells by cytomegalovirus infection in vitro. Evidence for a dual mechanism of induction. Am J Pathol (1991) 138(5):1209–23.
50. Li Y, Yan H, Xue WJ, Tian PX, Ding XM, Pan XM, et al. Allograft rejection-related gene expression in the endothelial cells of renal transplantation recipients after cytomegalovirus infection. J Zhejiang Univ Sci B (2009) 10(11):820–8. doi:10.1631/jzus.B0920115
51. Nastke MD, Becerra A, Yin L, Dominguez-Amorocho O, Gibson L, Stern LJ, et al. Human CD4+ T cell response to human herpesvirus 6. J Virol (2012) 86(9):4776–92. doi:10.1128/JVI.06573-11
52. Dulery R, Salleron J, Dewilde A, Rossignol J, Boyle EM, Gay J, et al. Early human herpesvirus type 6 reactivation after allogeneic stem cell transplantation: a large-scale clinical study. Biol Blood Marrow Transplant (2012) 18(7):1080–9. doi:10.1016/j.bbmt.2011.12.579
53. Olson AL, Dahi PB, Zheng J, Devlin SM, Lubin M, Gonzales AM, et al. Frequent human herpesvirus-6 viremia but low incidence of encephalitis in double-unit cord blood recipients transplanted without antithymocyte globulin. Biol Blood Marrow Transplant (2014) 20(6):787–93. doi:10.1016/j.bbmt.2014.02.010
54. Yoshikawa T, Asano Y, Ihira M, Suzuki K, Ohashi M, Suga S, et al. Human herpesvirus 6 viremia in bone marrow transplant recipients: clinical features and risk factors. J Infect Dis (2002) 185(7):847–53. doi:10.1086/339411
55. Zerr DM, Corey L, Kim HW, Huang ML, Nguy L, Boeckh M. Clinical outcomes of human herpesvirus 6 reactivation after hematopoietic stem cell transplantation. Clin Infect Dis (2005) 40(7):932–40. doi:10.1086/428060
56. Ljungman P, Wang FZ, Clark DA, Emery VC, Remberger M, Ringdén O, et al. High levels of human herpesvirus 6 DNA in peripheral blood leucocytes are correlated to platelet engraftment and disease in allogeneic stem cell transplant patients. Br J Haematol (2000) 111(3):774–81. doi:10.1046/j.1365-2141.2000.02422.x
57. Ogata M, Kikuchi H, Satou T, Kawano R, Ikewaki J, Kohno K, et al. Human herpesvirus 6 DNA in plasma after allogeneic stem cell transplantation: incidence and clinical significance. J Infect Dis (2006) 193(1):68–79. doi:10.1086/498531
58. Yamane A, Mori T, Suzuki S, Mihara A, Yamazaki R, Aisa Y, et al. Risk factors for developing human herpesvirus 6 (HHV-6) reactivation after allogeneic hematopoietic stem cell transplantation and its association with central nervous system disorders. Biol Blood Marrow Transplant (2007) 13(1):100–6. doi:10.1016/j.bbmt.2006.09.003
59. Zerr DM, Gooley TA, Yeung L, Huang ML, Carpenter P, Wade JC, et al. Human herpesvirus 6 reactivation and encephalitis in allogeneic bone marrow transplant recipients. Clin Infect Dis (2001) 33(6):763–71. doi:10.1086/322642
60. Scheurer ME, Pritchett JC, Amirian ES, Zemke NR, Lusso P, Ljungman P. HHV-6 encephalitis in umbilical cord blood transplantation: a systematic review and meta-analysis. Bone Marrow Transplant (2013) 48(4):574–80. doi:10.1038/bmt.2012.180
61. Seeley WW, Marty FM, Holmes TM, Upchurch K, Soiffer RJ, Antin JH, et al. Post-transplant acute limbic encephalitis: clinical features and relationship to HHV6. Neurology (2007) 69(2):156–65. doi:10.1212/01.wnl.0000265591.10200.d7
62. Ward KN. Child and adult forms of human herpesvirus 6 encephalitis: looking back, looking forward. Curr Opin Neurol (2014) 27(3):349–55. doi:10.1097/WCO.0000000000000085
63. Sakai R, Kanamori H, Motohashi K, Yamamoto W, Matsuura S, Fujita A, et al. Long-term outcome of human herpesvirus-6 encephalitis after allogeneic stem cell transplantation. Biol Blood Marrow Transplant (2011) 17(9):1389–94. doi:10.1016/j.bbmt.2011.01.014
64. Shimazu Y, Kondo T, Ishikawa T, Yamashita K, Takaori-Kondo A. Human herpesvirus-6 encephalitis during hematopoietic stem cell transplantation leads to poor prognosis. Transpl Infect Dis (2013) 15(2):195–201. doi:10.1111/tid.12049
65. Gotoh M, Yoshizawa S, Katagiri S, Suguro T, Asano M, Kitahara T, et al. Human herpesvirus 6 reactivation on the 30th day after allogeneic hematopoietic stem cell transplantation can predict grade 2-4 acute graft-versus-host disease. Transpl Infect Dis (2014) 16(3):440–9. doi:10.1111/tid.12229
66. Pichereau C, Desseaux K, Janin A, Scieux C, Peffault de Latour R, Xhaard A, et al. The complex relationship between human herpesvirus 6 and acute graft-versus-host disease. Biol Blood Marrow Transplant (2012) 18(1):141–4. doi:10.1016/j.bbmt.2011.07.018
67. Guardia AC, Stucchi RS, Sampaio AM, Milan A, Costa SC, Pavan CR, et al. Human herpesvirus 6 in donor biopsies associated with the incidence of clinical cytomegalovirus disease and hepatitis C virus recurrence. Int J Infect Dis (2012) 16(2):e124–9. doi:10.1016/j.ijid.2011.10.008
68. Humar A, Malkan G, Moussa G, Greig P, Levy G, Mazzulli T. Human herpesvirus-6 is associated with cytomegalovirus reactivation in liver transplant recipients. J Infect Dis (2000) 181(4):1450–3. doi:10.1086/315391
69. Razonable RR, Brown RA, Humar A, Covington E, Alecock E, Paya CV, et al. Herpesvirus infections in solid organ transplant patients at high risk of primary cytomegalovirus disease. J Infect Dis (2005) 192(8):1331–9. doi:10.1086/466529
70. Tormo N, Solano C, de la Cámara R, Garcia-Noblejas A, Cardeñoso L, Clari MA, et al. An assessment of the effect of human herpesvirus-6 replication on active cytomegalovirus infection after allogeneic stem cell transplantation. Biol Blood Marrow Transplant (2010) 16(5):653–61. doi:10.1016/j.bbmt.2009.12.003
71. Van Leer-Buter CC, Sanders JS, Vroom HE, Riezebos-Brilman A, Niesters HG. Human herpesvirus-6 DNAemia is a sign of impending primary CMV infection in CMV sero-discordant renal transplantations. J Clin Virol (2013) 58(2):422–6. doi:10.1016/j.jcv.2013.07.014
72. Buyse S, Roque-Afonso AM, Vaghefi P, Gigou M, Dussaix E, Duclos-Vallée JC, et al. Acute hepatitis with periportal confluent necrosis associated with human herpesvirus 6 infection in liver transplant patients. Am J Clin Pathol (2013) 140(3):403–9. doi:10.1309/AJCP0FWI2XAHECBJ
73. Dohna-Schwake C, Fiedler M, Gierenz N, Gerner P, Ballauf A, Breddemann A, et al. Primary HHV 6 infection after liver transplantation with acute graft rejection and multi-organ failure: successful treatment with a 2.5-fold dose of cidofovir and reduction of immunosuppression. Pediatr Transplant (2011) 15(6):E126–9. doi:10.1111/j.1399-3046.2010.01310.x
74. Pischke S, Gösling J, Engelmann I, Schlue J, Wölk B, Jäckel E, et al. High intrahepatic HHV-6 virus loads but neither CMV nor EBV are associated with decreased graft survival after diagnosis of graft hepatitis. J Hepatol (2012) 56(5):1063–9. doi:10.1016/j.jhep.2011.12.017
75. Agut H, Aubin JT, Huraux JM. Homogeneous susceptibility of distinct human herpesvirus 6 strains to antivirals in vitro. J Infect Dis (1991) 163(6):1382–3. doi:10.1093/infdis/163.6.1382
76. Akhyani N, Fotheringham J, Yao K, Rashti F, Jacobson S. Efficacy of antiviral compounds in human herpesvirus-6-infected glial cells. J Neurovirol (2006) 12(4):284–93. doi:10.1080/13550280600880772
77. Prichard MN, Whitley RJ. The development of new therapies for human herpesvirus 6. Curr Opin Virol (2014) 9:148–53. doi:10.1016/j.coviro.2014.09.019
78. Zerr DM, Gupta D, Huang ML, Carter R, Corey L. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis (2002) 34(3):309–17. doi:10.1086/338044
79. Ishiyama K, Katagiri T, Hoshino T, Yoshida T, Yamaguchi M, Nakao S. Preemptive therapy of human herpesvirus-6 encephalitis with foscarnet sodium for high-risk patients after hematopoietic SCT. Bone Marrow Transplant (2011) 46(6):863–9. doi:10.1038/bmt.2010.201
80. Ogata M, Satou T, Inoue Y, Takano K, Ikebe T, Ando T, et al. Foscarnet against human herpesvirus (HHV)-6 reactivation after allo-SCT: breakthrough HHV-6 encephalitis following antiviral prophylaxis. Bone Marrow Transplant (2013) 48(2):257–64. doi:10.1038/bmt.2012.121
81. Ogata M, Satou T, Kawano R, Goto K, Ikewaki J, Kohno K, et al. Plasma HHV-6 viral load-guided preemptive therapy against HHV-6 encephalopathy after allogeneic stem cell transplantation: a prospective evaluation. Bone Marrow Transplant (2008) 41(3):279–85. doi:10.1038/sj.bmt.1705907
82. De Bolle L, Andrei G, Snoeck R, Zhang Y, Van Lommel A, Otto M, et al. Potent, selective and cell-mediated inhibition of human herpesvirus 6 at an early stage of viral replication by the non-nucleoside compound CMV423. Biochem Pharmacol (2004) 67(2):325–36. doi:10.1016/j.bcp.2003.08.042
83. Williams-Aziz SL, Hartline CB, Harden EA, Daily SL, Prichard MN, Kushner NL, et al. Comparative activities of lipid esters of cidofovir and cyclic cidofovir against replication of herpesviruses in vitro. Antimicrob Agents Chemother (2005) 49(9):3724–33. doi:10.1128/AAC.49.9.3724-3733.2005
84. Becerra A, Gibson L, Stern LJ, Calvo-Calle JM. Immune response to HHV-6 and implications for immunotherapy. Curr Opin Virol (2014) 9:154–61. doi:10.1016/j.coviro.2014.10.001
85. Gerdemann U, Keirnan JM, Katari UL, Yanagisawa R, Christin AS, Huye LE, et al. Rapidly generated multivirus-specific cytotoxic T lymphocytes for the prophylaxis and treatment of viral infections. Mol Ther (2012) 20(8):1622–32. doi:10.1038/mt.2012.130
86. Papadopoulou A, Gerdemann U, Katari UL, Tzannou I, Liu H, Martinez C, et al. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med (2014) 6(242):242ra83. doi:10.1126/scitranslmed.3008825
87. Tzannou I, Papadopoulou A, Naik S, Leung K, Martinez CA, Ramos CA, et al. Off-the-shelf virus-specific T cells to treat BK virus, human herpesvirus 6, cytomegalovirus, Epstein-Barr virus, and adenovirus infections after allogeneic hematopoietic stem-cell transplantation. J Clin Oncol (2017) 35(31):3547–57. doi:10.1200/JCO.2017.73.0655
88. Lusso P, Crowley RW, Malnati MS, Di Serio C, Ponzoni M, Biancotto A, et al. Human herpesvirus 6A accelerates AIDS progression in macaques. Proc Natl Acad Sci U S A (2007) 104(12):5067–72. doi:10.1073/pnas.0700929104
89. Leibovitch E, Wohler JE, Cummings Macri SM, Motanic K, Harberts E, Gaitán MI, et al. Novel marmoset (Callithrix jacchus) model of human herpesvirus 6A and 6B infections: immunologic, virologic and radiologic characterization. PLoS Pathog (2013) 9(1):e1003138. doi:10.1371/journal.ppat.1003138
90. Yalcin S, Mukai T, Kondo K, Ami Y, Okawa T, Kojima A, et al. Experimental infection of cynomolgus and African green monkeys with human herpesvirus 6. J Gen Virol (1992) 73(Pt 7):1673–7. doi:10.1099/0022-1317-73-7-1673
91. Reynaud JM, Horvat B. Animal models for human herpesvirus 6 infection. Front Microbiol (2013) 4:174. doi:10.3389/fmicb.2013.00174
92. Horvat B, Berges BK, Lusso P. Recent developments in animal models for human herpesvirus 6A and 6B. Curr Opin Virol (2014) 9:97–103. doi:10.1016/j.coviro.2014.09.012
93. Lacoste V, Verschoor EJ, Nerrienet E, Gessain A. A novel homologue of Human herpesvirus 6 in chimpanzees. J Gen Virol (2005) 86(Pt 8):2135–40. doi:10.1099/vir.0.81034-0
94. Staheli JP, Dyen MR, Deutsch GH, Basom RS, Fitzgibbon MP, Lewis P, et al. Complete unique genome sequence, expression profile, and salivary gland tissue tropism of the herpesvirus 7 homolog in pigtailed macaques. J Virol (2016) 90(15):6657–74. doi:10.1128/JVI.00651-16
95. Patel SJ, Zhao G, Penna VR, Park E, Lauron EJ, Harvey IB, et al. A murine herpesvirus closely related to ubiquitous human herpesviruses causes T-cell depletion. J Virol (2017) 91(9):e02463-16. doi:10.1128/JVI.02463-16
96. Jacobs F, Knoop C, Brancart F, Gilot P, Mélot C, Byl B, et al. Human herpesvirus-6 infection after lung and heart-lung transplantation: a prospective longitudinal study. Transplantation (2003) 75(12):1996–2001. doi:10.1097/01.TP.0000058809.42027.66
97. Ward KN, Leong HN, Thiruchelvam AD, Atkinson CE, Clark DA. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J Clin Microbiol (2007) 45(4):1298–304. doi:10.1128/JCM.02115-06
98. Ylinen E, Lehtinen S, Jahnukainen T, Karlsson T, Loginov R, Mannonen L, et al. Human herpes virus 6 infection in pediatric organ transplant patients. Pediatr Transplant (2017) 21(4):1–6. doi:10.1111/petr.12905
99. Nagasaka M, Morioka I, Kawabata A, Yamagishi Y, Iwatani S, Taniguchi-Ikeda M, et al. Comprehensive analysis of serum cytokines/chemokines in febrile children with primary human herpes virus-6B infection. J Infect Chemother (2016) 22(9):593–8. doi:10.1016/j.jiac.2016.05.010
100. Smith A, Santoro F, Di Lullo G, Dagna L, Verani A, Lusso P. Selective suppression of IL-12 production by human herpesvirus 6. Blood (2003) 102(8):2877–84. doi:10.1182/blood-2002-10-3152
101. Smith AP, Paolucci C, Di Lullo G, Burastero SE, Santoro F, Lusso P. Viral replication-independent blockade of dendritic cell maturation and interleukin-12 production by human herpesvirus 6. J Virol (2005) 79(5):2807–13. doi:10.1128/JVI.79.5.2807-2813.2005
102. Hirata Y, Kondo K, Yamanishi K. Human herpesvirus 6 downregulates major histocompatibility complex class I in dendritic cells. J Med Virol (2001) 65(3):576–83. doi:10.1002/jmv.2075
103. Ota M, Serada S, Naka T, Mori Y. MHC class I molecules are incorporated into human herpesvirus-6 viral particles and released into the extracellular environment. Microbiol Immunol (2014) 58(2):119–25. doi:10.1111/1348-0421.12121
104. Martin LK, Schub A, Dillinger S, Moosmann A. Specific CD8(+) T cells recognize human herpesvirus 6B. Eur J Immunol (2012) 42(11):2901–12. doi:10.1002/eji.201242439
105. Halawi M, Khan N, Blake N. Identification of novel CD8+ T cell epitopes in human herpesvirus 6B U11 and U90. Immun Inflamm Dis (2015) 3(2):118–31. doi:10.1002/iid3.55
106. Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med (2005) 202(5):673–85. doi:10.1084/jem.20050882
107. Becerra-Artiles A, Dominguez-Amorocho O, Stern LJ, Calvo-Calle JM. A simple proteomics-based approach to identification of immunodominant antigens from a complex pathogen: application to the CD4 T cell response against human herpesvirus 6B. PLoS One (2015) 10(11):e0142871. doi:10.1371/journal.pone.0142871
108. Yasukawa M, Yakushijin Y, Furukawa M, Fujita S. Specificity analysis of human CD4+ T-cell clones directed against human herpesvirus 6 (HHV-6), HHV-7, and human cytomegalovirus. J Virol (1993) 67(10):6259–64.
109. Martin LK, Hollaus A, Stahuber A, Hübener C, Fraccaroli A, Tischer J, et al. Cross-sectional analysis of CD8 T cell immunity to human herpesvirus 6B. PLoS Pathog (2018) 14(4):e1006991. doi:10.1371/journal.ppat.1006991
110. Jing L, Chong TM, Byrd B, McClurkan CL, Huang J, Story BT, et al. Dominance and diversity in the primary human CD4 T cell response to replication-competent vaccinia virus. J Immunol (2007) 178(10):6374–86. doi:10.4049/jimmunol.178.10.6374
111. Jing L, Haas J, Chong TM, Bruckner JJ, Dann GC, Dong L, et al. Cross-presentation and genome-wide screening reveal candidate T cells antigens for a herpes simplex virus type 1 vaccine. J Clin Invest (2012) 122(2):654–73. doi:10.1172/JCI60556
112. Jing L, Laing KJ, Dong L, Russell RM, Barlow RS, Haas JG, et al. Extensive CD4 and CD8 T cell cross-reactivity between alphaherpesviruses. J Immunol (2016) 196(5):2205–18. doi:10.4049/jimmunol.1502366
113. Jing L, Schiffer JT, Chong TM, Bruckner JJ, Davies DH, Felgner PL, et al. CD4 T-cell memory responses to viral infections of humans show pronounced immunodominance independent of duration or viral persistence. J Virol (2013) 87(5):2617–27. doi:10.1128/JVI.03047-12
114. Nayak K, Jing L, Russell RM, Davies DH, Hermanson G, Molina DM, et al. Identification of novel Mycobacterium tuberculosis CD4 T-cell antigens via high throughput proteome screening. Tuberculosis (Edinb) (2015) 95(3):275–87. doi:10.1016/j.tube.2015.03.001
115. Iampietro M, Morissette G, Gravel A, Dubuc I, Rousseau M, Hasan A, et al. Human herpesvirus 6B immediate-early I protein contains functional HLA-A*02, HLA-A*03, and HLA-B*07 class I restricted CD8(+) T-cell epitopes. Eur J Immunol (2014) 44(12):3573–84. doi:10.1002/eji.201444931
116. Zhang E, Bell AJ, Wilkie GS, Suárez NM, Batini C, Veal CD, et al. Inherited chromosomally integrated human herpesvirus 6 genomes are ancient, intact, and potentially able to reactivate from telomeres. J Virol (2017) 91(22): e01137-17. doi:10.1128/JVI.01137-17
117. Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A (2010) 107(12):5563–8. doi:10.1073/pnas.0913586107
118. Strenger V, Kayser S, Witte KE, Lassner D, Schwinger W, Jahn G, et al. Individuals with inherited chromosomally integrated human herpes virus 6 (ciHHV-6) have functionally active HHV-6 specific T-cell immunity. Clin Microbiol Infect (2016) 22(2):209.e5–209.e8. doi:10.1016/j.cmi.2015.10.006
119. Bollard CM, Heslop HE. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood (2016) 127(26):3331–40. doi:10.1182/blood-2016-01-628982
120. Naik S, Nicholas SK, Martinez CA, Leen AM, Hanley PJ, Gottschalk SM, et al. Adoptive immunotherapy for primary immunodeficiency disorders with virus-specific T lymphocytes. J Allergy Clin Immunol (2016) 137(5):1498–1505.e1. doi:10.1016/j.jaci.2015.12.1311
121. Tejada-Simon MV, Zang YC, Hong J, Rivera VM, Zhang JZ. Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Ann Neurol (2003) 53(2):189–97. doi:10.1002/ana.10425
122. Descamps V, Ranger-Rogez S. DRESS syndrome. Joint Bone Spine (2014) 81(1):15–21. doi:10.1016/j.jbspin.2013.05.002
123. Mine S, Suzuki K, Sato Y, Fukumoto H, Kataoka M, Inoue N, et al. Evidence for human herpesvirus-6B infection of regulatory T-cells in acute systemic lymphadenitis in an immunocompetent adult with the drug reaction with eosinophilia and systemic symptoms syndrome: a case report. J Clin Virol (2014) 61(3):448–52. doi:10.1016/j.jcv.2014.08.025
Keywords: human herpesvirus 6, CD4+ T cell, CD8+ T cell, antigen, epitope
Citation: Hanson DJ, Hill JA and Koelle DM (2018) Advances in the Characterization of the T-Cell Response to Human Herpesvirus-6. Front. Immunol. 9:1454. doi: 10.3389/fimmu.2018.01454
Received: 21 March 2018; Accepted: 12 June 2018;
Published: 25 June 2018
Edited by:
Clemencia Pinilla, Torrey Pines Institute for Molecular Studies, United StatesReviewed by:
Günther H. S. Richter, Technische Universität München, GermanyHui Xiao, Institut Pasteur of Shanghai (CAS), China
Copyright: © 2018 Hanson, Hill and Koelle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David M. Koelle, ZGtvZWxsZUBtZWRpY2luZS53YXNoaW5ndG9uLmVkdQ==
 Derek J. Hanson
Derek J. Hanson Joshua A. Hill
Joshua A. Hill David M. Koelle
David M. Koelle