- 1Department of Radiation Oncology, Radiotherapy & OncoImmunology Laboratory, Radboud Institute for Molecular Life Sciences, Radboud University Medical Center, Nijmegen, Netherlands
- 2School of Life Science and Technology, Tokyo Institute of Technology, Yokohama, Japan
- 3Department of Medical Biochemistry, Academic Medical Center, Amsterdam Cardiovascular Sciences, Amsterdam, Netherlands
Dendritic cells (DCs) are the professional antigen-presenting cells of the immune system. Proper function of DCs is crucial to elicit an effective immune response against pathogens and to induce antitumor immunity. Different members of the nuclear receptor (NR) family of transcription factors have been reported to affect proper function of immune cells. Nur77 is a member of the NR4A subfamily of orphan NRs that is expressed and has a function within the immune system. We now show that Nur77 is expressed in different murine DCs subsets in vitro and ex vivo, in human monocyte-derived DCs (moDCs) and in freshly isolated human BDCA1+ DCs, but its expression is dispensable for DC development in the spleen and lymph nodes. We show, by siRNA-mediated knockdown of Nur77 in human moDCs and by using Nur77−/− murine DCs, that Nur77-deficient DCs have enhanced inflammatory responses leading to increased T cell proliferation. Treatment of human moDCs with 6-mercaptopurine, an activator of Nur77, leads to diminished DC activation resulting in an impaired capacity to induce IFNγ production by allogeneic T cells. Altogether, our data show a yet unexplored role for Nur77 in modifying the activation status of murine and human DCs. Ultimately, targeting Nur77 may prove to be efficacious in boosting or diminishing the activation status of DCs and may lead to the development of improved DC-based immunotherapies in, respectively, cancer treatment or treatment of autoimmune diseases.
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells. An important function of DCs is to instruct T cells to elicit immunity or tolerance (1, 2). Many factors contribute to the way DCs are shaped to elicit this function. Important factors are the type of pathogens that DCs encounter, such as bacteria or viruses, but also different microenvironmental factors in the tissues they reside in play a crucial role. DCs can be subdivided into classical or conventional DCs (cDC), interferon-producing plasmacytoid DCs (pDC), and monocyte-derived DCs (moDC) each with their own specialized function (3–5). Because of their crucial role in the immune system, different subsets of DCs are exploited in immune therapy (6–15). So far, treatment success is limited and functional knowledge on how DCs initiate and stably steer antitumor responses in vivo is important (13–15). Identification of transcription factors that control DC function in both immunity and tolerance is highly relevant, as these factors may serve as targets to modulate DC activity and function for the development of more successful DC-based immunotherapies.
Different members of the nuclear receptor (NR) family of transcription factors and their ligands have been shown to affect immune cells, including DCs (16–20). NRs are ligand inducible transcription factors having among others, steroid hormones or cellular metabolites as ligands. Several members have been well studied and were shown to play an immune modulatory role in DCs. Another group of NRs are so called “orphan” NRs for which no natural ligand has been identified yet, and the existence of ligands is disputed. The NR4A subfamily of orphan receptors comprises three members, namely, Nur77 (NR4A1/TR3/NGFI-B), Nurr1 (NR4A2/NOT/TINUR), and NOR-1 (NR4A3/TEC/MINOR). Their activity appears to be primarily regulated at the expression level. The expression of the NR4As can be induced by a diverse range of signals, including fatty acids, stress, growth factors, cytokines, peptide hormones, and physical stimuli (21). Hallmark of this subfamily is to respond quickly to such changes in cellular environments and regulate gene expression in a ligand-independent manner.
Members of this subfamily have been shown to be involved in a wide variety of pathological conditions. They have been shown to be dysregulated in multiple cancer types and promote or suppress tumors depending on specific cellular and tissue context, subcellular localization, external stimuli, protein–protein interactions, and post-translational modifications in cancer cells [reviewed in Ref. (22)]. In addition, there is also increasing evidence that the NR4As play a role in neurodegenerative disorders such as Alzheimer’s and Parkinson’s disease by contributing to neuronal cell death via modulating mitochondrial function and ER stress by controlling intracellular levels of ROS and Ca2+ and regulating cellular autophagy (23–26). Also in autoimmune-driven central nervous system (CNS) inflammation, the NR4A NRs have been shown to play an important role (27, 28).
NR4A receptors have emerged to play an important role within the immune balance by transcriptional regulation of cytokines and growth factors in macrophages (29, 30). In addition, they have been shown to be involved in the negative selection of self-reactive T cell clones in the thymus (31, 32) and are essential for thymic regulatory T cell development (33). Studies in Nur77−/− mice imply that Nur77 functions as a master regulator in the differentiation and survival of Ly-6C− monocytes (34, 35). Ly-6C+ and Ly-6C− monocytes that do express Nur77 do not develop into moDCs (36). Thus, Nur77 expression is not required for the development into moDCs but is for differentiation of Ly-6C+ monocytes into Ly-6C− “patrolling” monocytes (34, 36). Moreover, Nur77 has been shown to be involved in the polarization of macrophages toward an inflammatory phenotype important in atherosclerosis (37, 38).
We and others have recently reported expression of Nur77, Nurr1, and NOR-1 in murine DCs (39–43). Nurr1 has been shown to restrict the immunogenicity of bone marrow derived DCs (BMDCs) (43) and NOR-1 leads to activation-induced cell death in DCs (39), is important in DC migration (42), and is involved in TLR-mediated activation and gene expression of DCs (44). However, so far, the role of Nur77 expression in DCs remains elusive. We here set out to assess the expression kinetics and function of Nur77 in multiple subsets of murine and human DCs and its subsequent effect on inducing T cell activation, revealing a function as activation modulator for Nur77 in DCs. Knowledge regarding the possibilities in altering the activation status of DCs may prove to be beneficial in improving DC-based vaccination strategies.
Materials and Methods
Mice
6- to 16-week-old C57BL/6J and Balb/C mice (Charles River), Nur77−/− mice (45) on a C57BL/6 background, and Nur77GFP mice [016607; C57BL/6-Tg(Nr4a1-EGFP/cre)820Khog/J; Jackson Laboratory] were housed under specific pathogen-free conditions in individually ventilated cage units at the Central Animal Laboratory (Nijmegen, The Netherlands). Standard laboratory chow and sterile drinking water were provided ad libitum. All animal experiments were approved by the Radboud University’s Animal Welfare Body (Instantie voor Dierenwelzijn IvD) and the Animal Experiment Committee (DierExperimentenCommissie, RUDEC) that is recognized by the CCD (Central Authority for Scientific Procedures on Animals). The experiments were performed according to institutional, national, and European guidelines as stipulated in the Wet op de dierproeven and in the Dierproevenbesluit.
In Vitro Generation of Murine DCs
DCs were generated from murine BM isolated from the femur/tibia of the mice. To obtain pDCs and cDCs, cells were cultured for 8–10 days (37°C, 10% CO2) in RPMI 1640 supplemented with 10% fetal calf serum (Gibco-BRL Life Technologies), 0.5% antibiotic–antimycotic (Gibco/Invitrogen), 1% ultra-glutamine (Lonza), 50 µM β-mercaptoethanol (Sigma-Aldrich), and 200 ng/ml human rFlt3L (PeproTech). Pure cell populations were isolated by labeling single cell suspensions with anti-SiglecH-FITC (eBiosciences) and anti-CD11c-APC antibodies for pDCs and cDCs, respectively. pDCs were positively sorted with anti-FITC microbeads, the negative fraction was subjected to positive selection with anti-APC microbeads (both Miltenyi Biotec, Germany) to obtain cDCs as described previously (40). CD103+ murine DCs were generated by culturing BM cells in RPMI 1640 supplemented with 10% FCS, 0.5% antibiotic–antimycotic, 1% ultra-glutamine, 50 µM β-mercaptoethanol, 5 ng/ml mGM-CSF, and 200 ng/ml human rFlt3L, fresh medium was added at day 6, and cells were replated in fresh medium at day 9. Cells were harvested and used for experiments at day 14. The purity of the isolated DC subsets was ensured by flow cytometry.
Tumor Induction
The transgenic cell line 9464D was derived from spontaneous tumors from TH-MYCN transgenic mice on C57BL/6 background and were a kind gift from Dr. Orentas (NIH, Bethesda, MD, USA). 9464D cells were cultured in DMEM containing 10% fetal calf serum, 1% non-essential amino acids, 0.5% antibiotic–antimycotic, and 50 µM β-mercaptoethanol. For induction of tumors, 1 × 10e6 9464D cells were injected s.c. in 100 µl PBS on the right flank of the mice. Tumor growth was measured every 3–4 days using calipers. Spleen and lymph nodes (LNs) of the mice were taken when the tumor was more than 5 mm in diameter.
Flow Cytometry
To obtain single cells for flow cytometric staining, murine spleen was passaged over a 100 µm cell strainer, and murine LNs were incubated in serum-free medium containing collagenase (Worthington) and DNAseI (Roche), later supplemented with 1 mM EDTA. In vitro generated human and murine DCs, and ex vivo isolated murine spleen and LN cells were stained using standard antibody staining protocols with antibodies listed in Table S1 in Supplementary Material. Cell viability was assessed by staining with fixable viability dye eFluor™ 450 (eBioscience). Samples were acquired on a FACS Verse (BD Bioscience), and data were analyzed with FlowJo software (Tree Star).
ELISA
Human and mouse IL-6, TNFα, IL-12p70, and human IFNγ present in the supernatant of DC cultures was measured using the ELISA kit (Thermo Fisher) according to the manufacturers protocol.
Murine Type I IFN Bioassay
Type I IFN activity in the supernatant of murine pDCs was measured using L929 cells transfected with an interferon-sensitive luciferase construct (ISRE-L929) (46) with reference to a recombinant mouse IFN-β standard (Sigma). In short, pDC culture supernatants were added to ISRE-L929 IFN reporter cells and incubated for 4–6 h. Then, the cells were lysed in Passive Lysis Buffer (Promega), mixed with firefly luciferin substrate (Promega), and measured on a Victor3 Luminometer.
Mixed Leukocyte Reaction (MLR) Murine DCs
After 16–24 h of stimulation with 1 µg/ml CpGB (1668, Sigma-Aldrich) or 4 µg/ml R848, pDCs, cDCs, or CD103+ DCs were washed and co-incubated with carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled allogeneic BalB/C T cells. T cells were isolated using the T cell isolation kit (EasySep). The cells were co-incubated for 3 days in round-bottom 96-well cluster plates (Corning). T cell proliferation was measured by CFSE dilution by FACS.
Generation of Human DCs
DCs were generated from cells isolated from buffy coats obtained from healthy volunteers (Sanquin, Nijmegen, The Netherlands) after written informed consent as per the Declaration of Helsinki. The study was approved by the Institutional Review Board of the Radboud University Nijmegen Medical Center, Commissie Mensgebonden Onderzoek. Peripheral blood mononuclear cells (PBMCs) were purified via Ficoll density gradient centrifugation (Lucron Bioproducts). moDCs were cultured as described previously (47). In short, plastic-adherent monocytes were cultured for 6 days in RPMI 1640 medium with 1% ultra-glutamine, 0.5% antibiotic–antimycotic, 10% (v/v) fetal calf serum, 300 U/ml IL4, and 450 U/ml GM-CSF (both Cellgenix). IL4 and GM-CSF were added again at day 3. To obtain fresh human myeloid dendritic cells (mDCs), CD14+ cells were depleted from the PBMCs followed by BDCA1+ DC isolation using the CD1c (BDCA1)+ Dendritic Cell Isolation Kit (Miltenyi Biotec). Purity of the freshly isolated mDCs was ensured by flow cytometry.
Small Interfering RNA-Mediated Knockdown
For Nur77 silencing in human moDCs, the ON-TARGETplus SMARTpool NR4A1 (Dharmacon) containing four different Nur77 targeting siRNA oligos each 19 nt long was used. The irrelevant siRNA ON-TARGETplus Non-Targeting siRNA#1 (Dharmacon) was used as control. moDCs were electroporated at day 4 as described before (47). Electroporated DCs were stimulated with 1 µg/ml LPS (Sigma) or 4 µg/ml R848 (Enzo Life Sciences) at day 6. Supernatant was taken 24 h later.
RNA Isolation and Quantitative PCR
Total RNA was isolated and cDNA was synthesized as described before (47). mRNA levels for the genes of interest were determined with a CFX96 sequence detection system (Bio-Rad) using the Faststart SYBR green mastermix (Roche) with SYBR Green as the fluorophore and gene-specific oligonucleotide primers. The primers for human porphobilinogen deaminase (PBGD), IL-6, TNFα, and IL-12 (47) and murine PBGD, TLR7, and TLR9 (40) were described previously. Other primers used (forward and reverse) are listed in Table S2 in Supplementary Material. Reaction mixtures and program conditions were used that were recommended by the manufacturer (Bio-Rad). Quantitative PCR data were analyzed with the CFX Manager V1.6.541.1028 software (Bio-Rad) and checked for correct amplification and dissociation of the products. As we described previously for human and murine DCs, mRNA levels of the genes of interest were normalized to mRNA levels of the housekeeping gene PBGD (19, 20, 40, 47, 48) and were calculated according to the cycle threshold method (49).
Human MLR
Human day 6 moDCs were pretreated with 1 or 10 µM 6-mercaptopurine (6-MP) (Sigma) or vehicle control (DMSO) for 8 h, before o/n stimulation with 4 µg/ml R848. At day 7, the medium was replaced with fresh DC medium, and allogeneic peripheral blood lymphocytes were added to the DCs, in a ratio of 1:10 (DCs:T cells) and cocultured for 144 h. Supernatant was taken for IFNγ measurements.
Statistical Analysis
In each experiment, at least three mice or human donors were used to be able to perform statistical testing. Each legend contains the information of the number of mice or human donors used including the statistics that was used to calculate significance. Statistical testing was performed using GraphPad Prism 5 software (GraphPad, La Jolla, CA, USA). A P < 0.05 was considered significant.
Results
Nur77 Expression and Function in Murine DCs
As NR4A NRs are typical early response genes induced upon stimulation (29), we tested Nur77 expression in cDCs and pDCs 3 h after stimulation with a combination of the TLR7/8 ligand R848 and the TLR9 ligand CpG. DCs were differentiated from murine BM in vitro with FLT3L as this reflects physiologic DC development and gives rise to a mixture of both cDCs and pDCs (50). pDCs were detected as CD11cposB220posSiglecHpos and cDCs were defined as CD11cposB220negSiglecHneg and were sorted and stimulated as described before (40) (Figure S1A in Supplementary Material). In agreement with its classification as early response gene, Nur77 mRNA levels were strongly upregulated after 3 h of stimulation in murine pDCs as well as cDCs compared with freshly sorted cells (0 h) (Figure 1A). To further assess Nur77 expression kinetics, we used BM cells from transgenic Nur77 reporter mice, where the induction of the Nur77 promoter drives GFP expression (Nur77GFP) (51). In line with its mRNA expression, cDCs and to a lesser extend pDCs up regulate Nur77GFP already after 3 h of stimulation with CpG (Figure 1B). In addition to FLT3L-derived BMDCs, we tested Nur77GFP in BMDCs differentiated into Batf3-dependent CD103+ DCs (CD11cposB220negCD103pos) when cultured with GM-CSF and FLT3L (52) (for gating strategy see Figure S1B in Supplementary Material). In CD103+ DCs, there is also already prominent expression of Nur77GFP after 3 h stimulation with CpG (Figure 1B). Our data further show that the expression in cDCs was highest after CpG and LPS stimulation, whereas the expression was less pronounced in response to R848. pDC and CD103+ DCs revealed highest expression of Nur77GFP after stimulation with CpG, compared with LPS and R848 (Figure 1C). These data indicate that in different types of in vitro generated DCs, Nur77 expression is quickly induced upon stimulation with inflammatory ligands, and that the expression in response to TLR-specific agonists varies in different DC subsets.
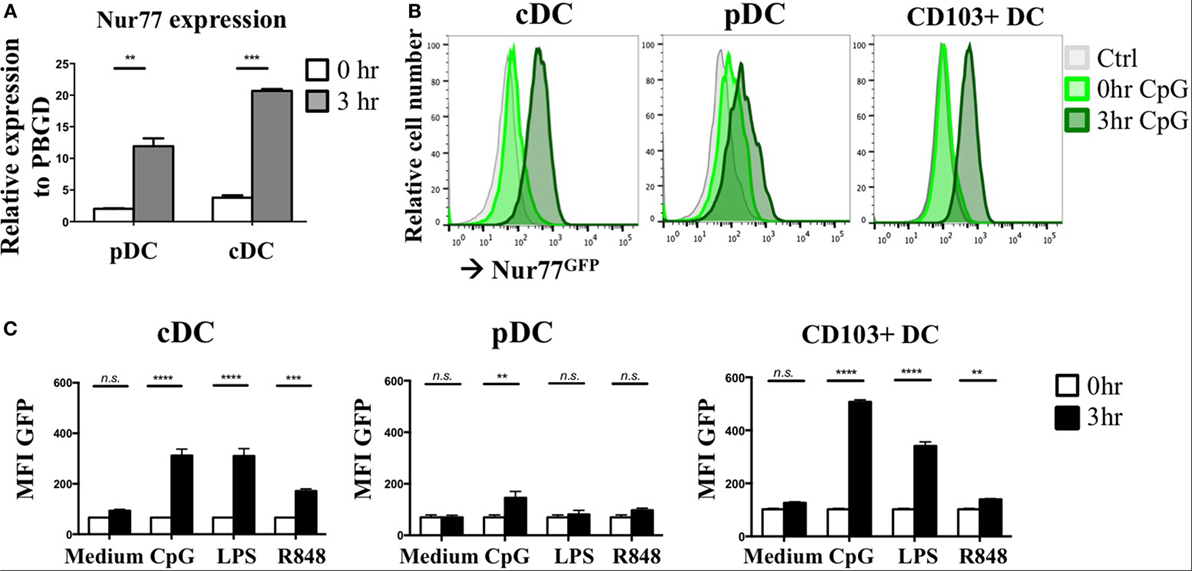
Figure 1. Nur77 expression level in in vitro generated murine dendritic cells (DCs). (A) Plasmacytoid DCs (pDCs) and conventional DCs (cDCs) were sorted from FLT3L bone marrow cultures pooled from three mice per experiment and immediately lysed for RNA isolation (0 h) or stimulated for 3 h with a combination of R848 and CpG. mRNA expression levels of Nur77 were detected by qPCR analysis. Data shown are the mean of three independent experiments ± SEM, two-tailed unpaired t-test: *P < 0.05; **P < 0.01; and ***P < 0.001. (B) In vitro generated cDCs, pDCs, or CD103+ DCs from Nur77GFP or control mice were stimulated for the indicated times with CpG, and GFP expression was determined by flow cytometry. Shown are the representative data of three mice. (C) Quantification of Nur77GFP expression in cDC, pDC, or CD103+ DC stimulated with CpG, LPS, or R848 for 0 or 3 h, presented as the geometric mean (MFI) ± SEM, two-way ANOVA with Sidak’s multiple comparisons test (n = 3). n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Nur77 Does Not Have a Major Impact on the Development of Murine DCs in Spleen and LNs
To test whether Nur77 expression is required for the development of DCs, we investigated the presence of different DC subsets in spleen and LNs of WT and Nur77−/− mice. We observed a small but significant increase in the percentage of total CD11chiMHCIIhi DCs and in CD11b+ DCs (CD11chiMHCIIhiSirpαposCD24negCD115negCD4pos) of the spleen of Nur77−/− mice relative to WT mice (Figure 2A; Figure S2A in Supplementary Material). The number of CD8α+ spleen DCs (CD11chiMHCIIhiSirpαnegCD24posFLT3pos) was similar between WT and Nur77−/− mice. Also in the LNs, the presence of resident (CD11chiMHCII+) and migratory DCs (CD11c+MHCIIhi) was comparable (Figure 2B; Figure S2B in Supplementary Material). Also in a transplantable autologous TH-MYCN 9464D mouse model of neuroblastoma (53), we did not observe differences in the presence of the different subsets of DCs in the spleen or (non)draining LN (Figures S3A,B in Supplementary Material). These data indicate that Nur77 is dispensable for DC development. Next, we tested the expression level of Nur77 in different DC subsets by analyzing DCs from the spleen and LNs from transgenic Nur77 reporter mice that express GFP upon activation of the Nur77 promoter. Nur77GFP was clearly expressed in CD11b+ spleen DCs. The expression in CD8α+ spleen DCs was less well defined and consisted of a population expressing Nur77GFP at a very low level and a population expressing Nur77GFP to a similar level as the CD11b+ DCs (Figures 3A,B). Resident DCs of inguinal and axillary LN expressed clear levels of Nur77GFP, in contrast to significantly lower expression in migratory DCs of these LN (Figures 3C,D). Mice bearing a neuroblastoma tumor showed a similar Nur77GFP expression pattern in DCs (Figures S3C,D in Supplementary Material) as in naïve mice. These data indicate that Nur77 expression does not have a major impact on the development and presence of different DC subsets in the spleen and LNs and that Nur77 is most abundantly expressed in CD11b+ spleen DCs and resident DCs of different LNs.
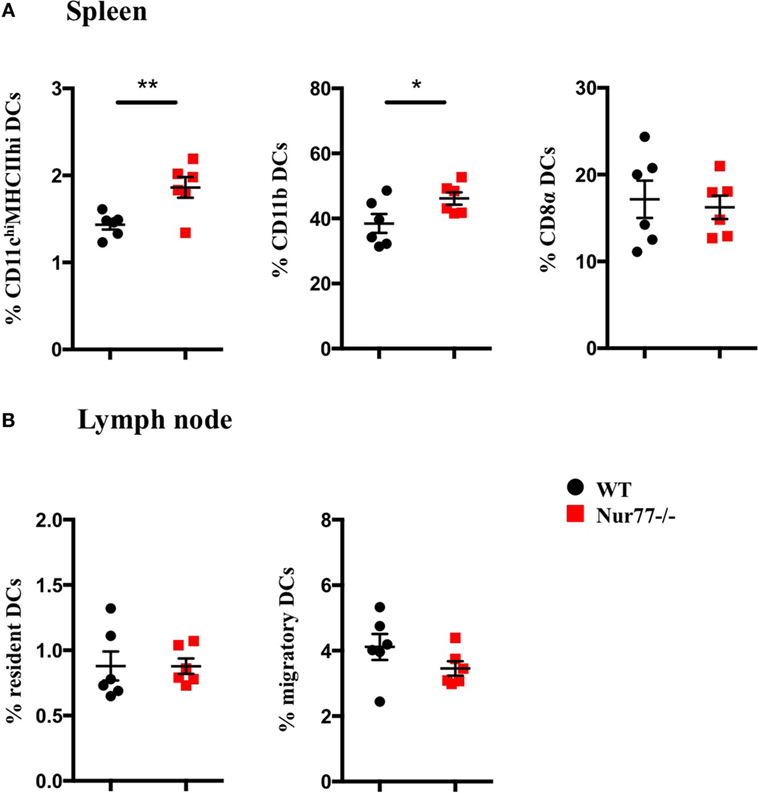
Figure 2. Absence of Nur77 expression does not have a major impact on dendritic cell (DC) development in the spleen and lymph nodes (LNs). % of DCs in (A) spleen and (B) inguinal LNs of WT or Nur77−/− mice. Shown are the means of pooled data of two independent experiments ± SEM for WT or Nur77−/− mice (n = 3). Two-tailed unpaired t-test: *P < 0.05; **P < 0.01; and ***P < 0.001.
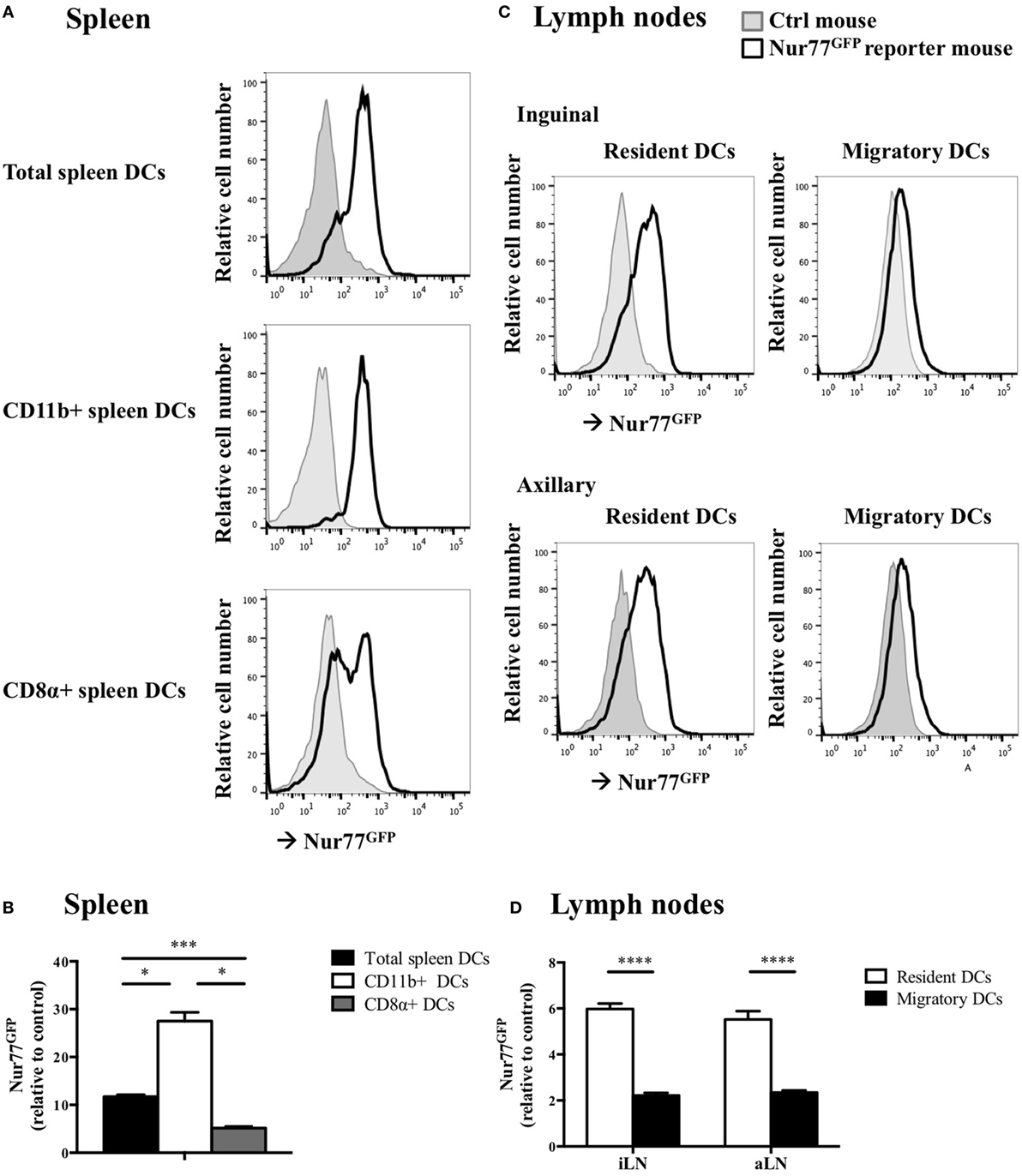
Figure 3. Nur77 expression level in murine dendritic cells (DCs) ex vivo. (A) Ex vivo Nur77GFP expression in the different DC subsets in spleen, shown are the representative data of one out of three transgenic Nur77 reporter mice, where the induction of the Nur77 promoter drives GFP expression (Nur77GFP) mice. (B) Quantification of Nur77GFP expression in the different DC subsets in the spleen, presented as the relative Nur77GFP expression to control mice ± SEM, one-way ANOVA with Sidak’s multiple comparisons test (n = 3). (C) Ex vivo Nur77GFP expression in the resident and migratory DC subsets of the inguinal and axillary lymph nodes (aLN), shown are the representative data of one out of three transgenic Nur77 reporter mice. (D) Quantification of Nur77GFP expression in the resident and migratory DC subsets of the inguinal (iLN) and aLN, presented as the relative Nur77GFP expression to control mice ± SEM, two-way ANOVA with Sidak’s multiple comparisons test (n = 3): *P < 0.05; ***P < 0.001; and ****P < 0.0001.
Nur77-Deficient Murine DCs Have Altered Cytokine Production and T Cell Stimulatory Capacity
To assess the functional role of Nur77 in different murine DC subsets, we investigated cytokine production by murine Nur77−/− BMDCs after stimulation with different inflammatory stimuli. We found that Nur77−/− cDCs produced significantly more IL-6, TNFα, and IL-12 upon CpG and R848 stimulation (Figure 4A). Nur77−/− pDCs showed increased production of IL-6 and IL-12 upon R848 stimulation, whereas TNFα production was not affected. After stimulation with CpG, type I IFN production was much higher in Nur77−/− pDCs compared with WT pDCs (Figure 4B). CD103+ DCs showed a stronger response to CpG than to R848, revealing increased production of IL-6, TNFα, and IL-12 (Figure 4C). To rule out the possibility that the increase in cytokine production was (partly) mediated by enhanced TLR expression, we profiled TLR7 and TLR9 expression in these cells. TLR7 and TLR9 expression was similar in WT and Nur77−/− cDCs, whereas TLR7 expression was reduced in Nur77−/− pDCs (Figure S4 in Supplementary Material). In addition to cytokine production, we investigated the T-cell stimulatory capacities for Nur77-deficient DCs. To this end, control, CpG, or R848 stimulated DCs were added to an allogeneic MLR. All Nur77-deficient DC subsets were significantly more potent in inducing T cell proliferation than WT DCs upon stimulation with CpG (Figure 4D). These data indicate that Nur77 deficiency in DCs leads to enhanced cytokine production and subsequent increased T cell proliferation.
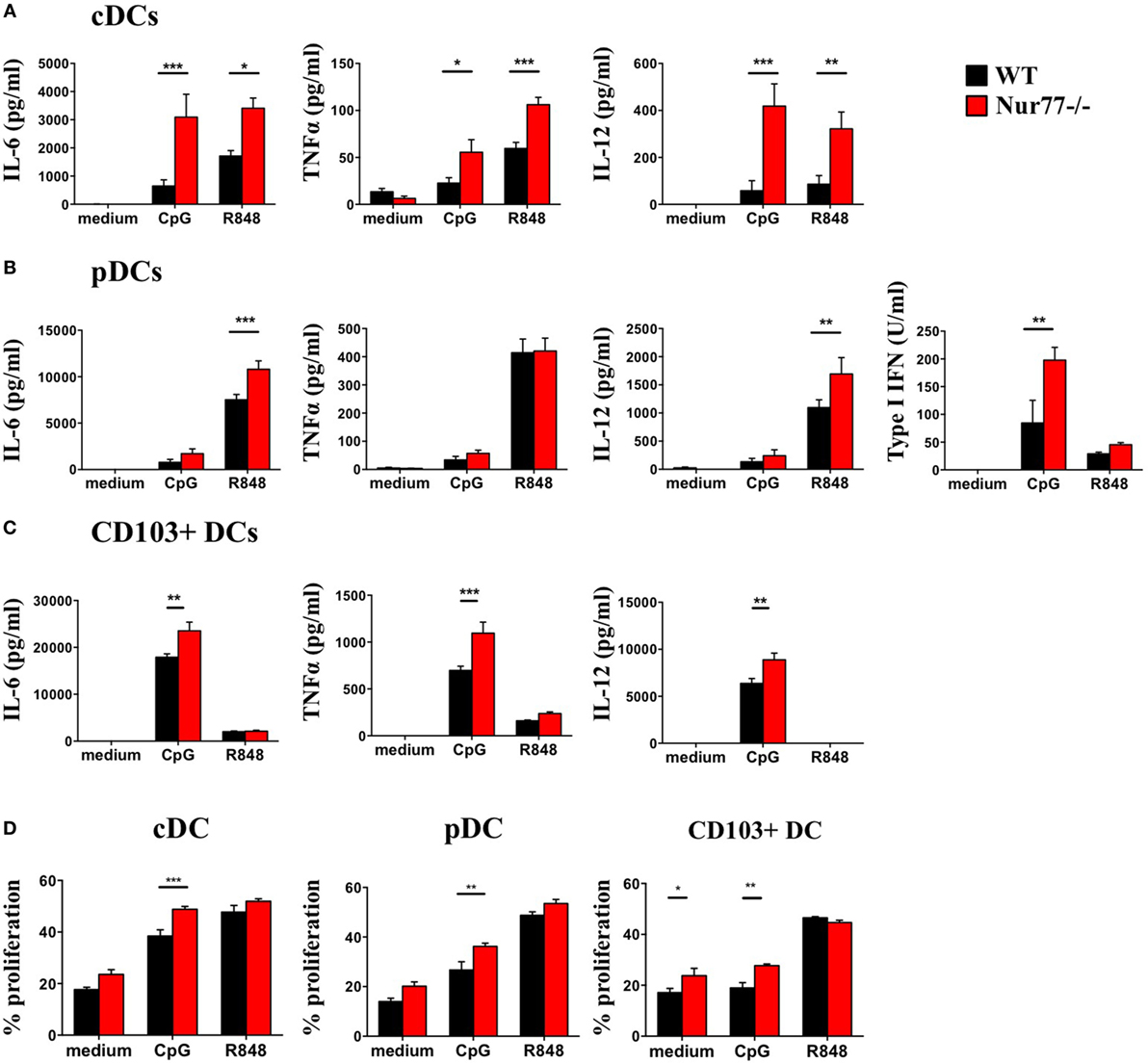
Figure 4. Cytokine production and T cell proliferation by murine Nur77−/− dendritic cells (DCs) are increased compared with WT DCs. Conventional DCs (cDCs) (A) and plasmacytoid DCs (pDCs) (B) were sorted from FLT3L bone marrow cultures or CD103+ DCs (C) derived from FLT3L/GM-CSF cultures were stimulated with CpG or R848, cytokine production was measured with ELISA. T cell proliferation (D) was measured after coculture of CpG or R848 stimulated cDCs, pDCs, or CD103+ DCs with BalB/C T cells and was measured as indicated by CFSE dilution on day 3. Data are shown as the mean ± SEM, two-way ANOVA with Bonferroni posttest (n = 3–6 different mice) (*P < 0.05; **P < 0.01; and ***P < 0.001).
Nur77 Expression and Function in Human DCs
In addition to defining its expression and function in murine DCs we profiled Nur77 mRNA expression in human moDCs after stimulation with LPS and R848. In accordance with murine DCs, human moDCs quickly upregulated Nur77 mRNA expression and the expression remained stable for 24 h after stimulation with either LPS or R848 (Figure 5A). To investigate Nur77 expression in freshly isolated BDCA1+ blood myeloid DCs, purified BDCA1+ DCs were stimulated for different time periods with LPS or R848 (Figure 5B). Compared with moDCs, freshly isolated BDCA1+ DCs had much higher expression levels of Nur77 expression under resting conditions. Stimulation with R848 led to a further increase of Nur77 expression, which diminished to lower levels 16 h after stimulation. These data indicate that in different subsets of human DCs Nur77 is expressed with varying expression levels.
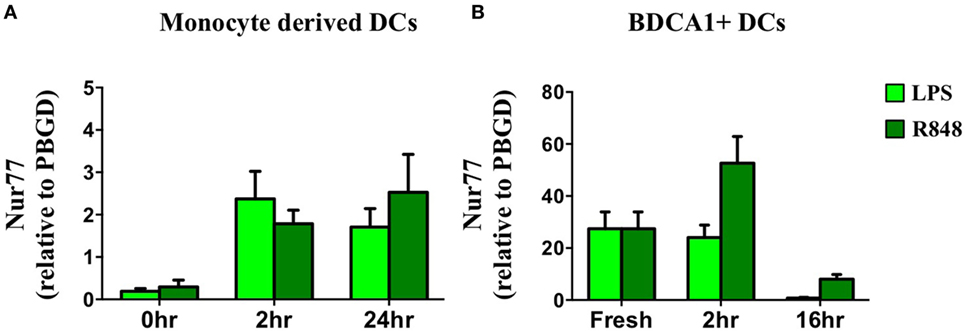
Figure 5. Nur77 mRNA expression in human dendritic cells (DCs). Human day 6 monocyte-derived dendritic cells (A) or freshly isolated BDCA1+ DCs (B) were stimulated for different time periods with LPS or R848. mRNA expression levels of Nur77 were detected by qPCR analysis. Expression levels shown are related to the housekeeping gene porphobilinogen deaminase (PBGD). Data shown are the mean ± SEM (n = 3–5 different donors).
Human Nur77-Modified DCs Have Altered Cytokine Production and T Cell Stimulatory Capacity
To test Nur77 function in human DCs, we silenced Nur77 expression in moDCs using a siRNA smartpool. Nur77 expression in moDCs decreased by 60–70% using siNur77 compared with control siRNA (siCTRL) (Figure 6A). These siNur77 targeted DCs had increased mRNA and protein expression of IL-6 and TNFα compared with siCTRL-treated DCs (Figures 6B,C), especially after R848 stimulation. Nur77-deficient DCs also showed enhanced IL-12 protein production. Profiling of TLR4, TLR7, and TLR8 expression (Figure S5 in Supplementary Material), revealed no change in TLR expression, indicating that the effect on cytokine production is not mediated via altered TLR expression. As NR4A family members have been reported to crosstalk with the NF-κB pathway (54), we investigated whether the enhanced cytokine production was dependent on NF-κB signaling. Blocking NF-κB signaling with the NF-κB inhibitor BAY11-7082 inhibited IL-6 and TNFα production in siNur77 DCs and siCTRL DCs to the same level (Figure 6D), indicating that the enhanced expression of IL-6 and TNFα was indeed dependent on NF-κB signaling. We next determined the expression of CD40, CD86, and CCR7 in siNur77 DCs. While siNur77 and siCTRL DCs show similar expression of the co-stimulatory markers CD40 and CD86, a significantly lower percentage of CCR7+ DCs were present in siNur77 DCs (Figure 6E). To further substantiate these data, we treated DCs with 6-MP, an activator of Nur77 (55–59). Treating DCs with 6-MP before stimulation with R848, led to a dose-dependent decrease of IL-6 and IL-12 production, while TNFα levels were not altered (Figure 6F). No effect of 6-MP on cell viability could be detected (Figure S6 in Supplementary Material). In line with decreased IL-6 and IL-12 production, DCs pretreated with 6-MP were less capable of inducing IFNγ production by T cells in an allogeneic MLR (Figure 6G). These data show that human Nur77-modified moDCs have altered NF-κB-dependent inflammatory responses that are important in inducing T cell activation.
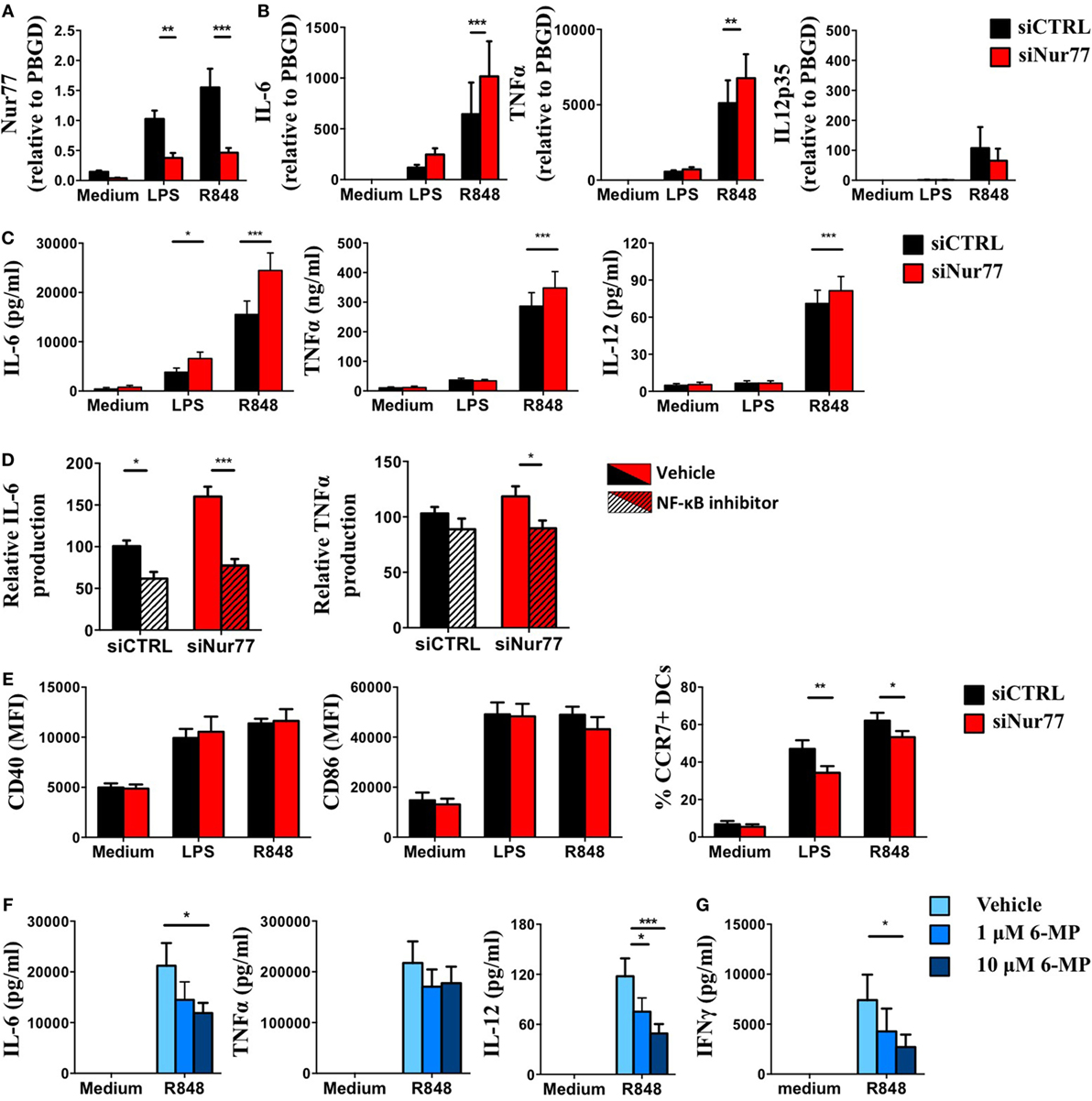
Figure 6. Knockdown or activation of Nur77 in human dendritic cells (DCs) alters DC function and T cell activation. Day 4 monocyte-derived dendritic cells (moDCs) were electroporated with a smartpool siRNA targeting Nur77 (siNur77) or a control siRNA (siCTRL). At day 6, cells were stimulated for 8 h with LPS, and Nur77 mRNA expression was detected by qPCR analysis (A), cytokine mRNA expression after 8 h of stimulation was measured by qPCR analysis (B), and cytokine levels were measured 24 h after stimulation with ELISA. (C) Electroporated moDCs were pretreated with Bay11-7082 and then stimulated with R848 for 24 h. Cytokine levels were measured by ELISA. (D) CD40, CD86, and CCR7 expression was determined by FACS analysis. (E) moDCs were pretreated with 6-mercaptopurine (6-MP) and then stimulated with TLRL for 24 h. Cytokine levels were measured by ELISA. (F) moDCs were pretreated with 6-MP and subsequently stimulated with R848, T cell stimulatory capacity was measured in an allogeneic mixed leukocyte reaction by measuring IFNγ production by ELISA (G). Data shown are the mean ± SEM. Two-way ANOVA with Bonferroni posttest (n = 3–12 independent donors): *P < 0.05; **P < 0.01; and ***P < 0.001.
Discussion
Nuclear receptors have been shown to play a critical role in immune cell function, including members of the NR4A subgroup. However, the expression and function of Nur77 in different DC subsets has not been studied so far. We now show that Nur77 is expressed in different human as well as murine DC subsets. Its expression is rapidly upregulated upon stimulation with different TLR ligands. Deficiency of Nur77 leads to enhanced NF-κB dependent cytokine production and T cell stimulatory capacity of DCs, while stimulation with the Nur77 activator 6-MP limits cytokine production by DCs and its capacity to stimulate allogeneic T cells.
Nur77 expression has been shown to be essential in the differentiation and survival of Ly-6C− monocytes (34, 35), in the polarization of macrophages (37, 38, 60) and in the function and negative selection of T cells (31, 32). This NR is also expressed in infiltrating monocytes and monocyte-derived macrophages of the CNS that are important in experimental autoimmune encephalomyelitis (28) and in patrolling monocytes that control metastasis to the lung (61). We now show, in line with its classification as early response gene, that Nur77 expression is quickly upregulated in different human and murine DC subsets after stimulation with distinct TLR ligands in vitro. However, the expression levels in the different DC subsets and level of response toward diverse stimuli vary. We also found Nur77 expression in different subsets of DCs in the spleen and LNs directly in naïve and in tumor-bearing mice ex vivo. Expression was more pronounced in the CD11b+ DCs of the spleen compared with CD8α+ DCs and higher in the resident than in the migratory DCs of the LNs. Previously, it has been shown that Nur77 is not required for the differentiation of Ly-6Chi monocytes into moDCs (36). We now also show that Nur77 deficiency does not have a major impact on the presence of different DC subsets in the spleen and LN at steady state conditions as well as mice bearing a neuroblastoma tumor. This confirms that in contrast to its expression in Ly-6C− monocytes, Nur77 expression is dispensable for the development of spleen and LN DCs.
Although most studies have reported that Nur77 has an anti-inflammatory role in monocytes and macrophages (37, 38, 62), it has been shown that its overexpression in murine macrophages can lead to a pro-inflammatory response (63). Our data point towards an anti-inflammatory role in human and murine DC subsets. Nur77 deficiency in DCs leads to enhanced production of IL-6, TNFα, and IL-12 and subsequent enhanced T cell proliferation, while Nur77 activation leads to reduced IL-6 and IL-12 production and reduced T cell activation. It has been hypothesized that Nur77 acts to resolve inflammation in macrophages (38, 64) and based on our data we now suggest a similar role for Nur77 in DCs.
All NR4A family members, including Nur77, have been shown to modulate immune cell function via crosstalk to NF-κB (30, 38, 54, 65). Our data show that also in human DCs, Nur77 affects cytokine production by modulating the NF-κB pathway. It has been shown that Nur77 can affect the NF-κB pathway signaling in numerous ways (38, 63, 65–68). Besides modulating phosphorylation of p65 Ser536 and Ser529 in macrophages (38, 69), Nur77 has also been shown to directly interact with the p65 subunit of NF-κB (65, 66) and block p65 binding to DNA (65). Moreover, Nur77 can regulate TRAF6 auto-ubiquitination (67), important for NF-κB signal transduction (70–72). Future studies should reveal which mechanism underlies Nur77-mediated modulation of NF-κB signaling in DCs and whether different DC subsets or different inflammatory conditions involve specific ways of regulating NF-κB signaling.
While Nur77-deficient DCs show enhanced inflammatory responses, pretreating human DCs with 6-MP led to reduced inflammatory responses and a diminished capacity to induce IFNγ production by T cells in an allogeneic MLR. 6-MP is a nucleic acid analog and has been shown to enhance Nur77 transcriptional activity (55–59). Currently, it is being applied as an immunosuppressive drug for the treatment of several chronic inflammatory diseases such as inflammatory bowel disease, systemic lupus erythematosus, acute lymphoblastic leukemia of childhood, inflammatory myopathies, and rheumatoid arthritis and to prevent acute rejection in organ transplant patients (73–75). It has been shown that besides activating Nur77 (55, 56, 76) 6-MP can also activate the NR4A members Nurr1 (77) and NOR-1 (76) and inhibit the GTPase proteins Rac1 and Rac2 (78, 79). Therefore, the effect observed in moDCs may be a combined effect of 6-MP on the function of either of these proteins. In addition to 6-MP many other pharmacological compounds have been generated to modulate Nur77 function. Among them are different C-DIMs [synthetic 1,1-bis(3′-indolyl)-1-(substituted phenyl)methane analogs] (80), cytosporone B and its structural analogs (81, 82), and TMPA (ethyl 2-[2,3,4-trimethoxy-6-(1-octanoyl)phenyl]acetate) (83). They have been shown to regulate Nur77 function by modulating Nur77-dependent transactivation, influencing its expression levels, inducing nuclear export of Nur77 or affecting binding to other proteins (80–88). Many of these compounds have, as also shown for 6-MP, also Nur77-independent actions (85, 89, 90). In cancer cells, neuronal cells, as well as different immune cells, it has been shown that Nur77 function depends on tissue context, subcellular localization, external stimuli, protein–protein interactions, or post-translational modifications (22–26, 31, 32, 34, 35, 37, 38, 60). How Nur77 function in DCs is exactly regulated upon specific immune stimuli and whether that is different in different DC subsets is currently unknown. Future studies should aim at fully elucidating whether specific stimuli in different subsets of DCs and under specific (pathological) conditions affect Nur77 activation and thereby modulate DC function. More knowledge regarding the exact mechanism(s) of Nur77 activation in DCs will help to choose the best pharmacological compound targeting specific actions of Nur77 in DCs. This will not only be important in optimizing current DC-based immunotherapies but also when more generally targeting Nur77 in different cell types and pathological conditions.
Interestingly, in tumor cells, the natural steroid Dendrogenin A has been shown to stimulate expression of Nur77 via binding to LXRβ and induce lethal autophagy (91, 92), opening up new perspectives for cancer treatment (93). Moreover, it has been shown that Dendrogenin A, in addition to inducing growth control and improve overall survival in mice, also induces immune cell infiltration, including DCs, in the tumor (94). As LXR has been shown to affect DC differentiation, maturation and migration (95–101), it is tempting to speculate that part of these effects are mediated via regulation of Nur77 expression, especially when DCs are stimulated with Dendrogenin A.
One striking observation is that the percentage of CCR7 expressing human DCs was decreased in siNur77 treated moDCs. Interestingly, another member of the NR4A subfamily, NOR-1, has been shown to affect CCR7-dependent murine CD103+ DC migration from tissues to LNs in vivo (42). Nevertheless, we did not observe a similar effect on CCR7 expression in in vitro cultured murine CD103+ DCs (data not shown). In agreement with Park et al., we did not find differences in the number of migratory murine DCs present in the LN in Nur77−/− mice compared with WT mice, suggesting a less pronounced role for Nur77 in CCR7-dependent DC migration in mice. However, since NR4A family members are highly homologous proteins and can have redundant functions (102–104), it is also possible that the absence of Nur77 is compensated by NOR-1 in murine DCs.
Given that Nur77 modifies DC function with altered inflammatory responses, Nur77 may be an interesting therapeutic target to either boost or diminish the activation status of DCs in DC-based vaccination strategies in cancer or treatment of autoimmune diseases, respectively.
Ethics Statement
All animal experiments were approved by the Radboud University’s Animal Welfare Body (AWB) (Instantie voor Dierenwelzijn IvD) and the Animal Experiment Committee (DierExperimentenCommissie, RUDEC) that is recognized by the CCD (Central Authority for Scientific Procedures on Animals). The experiments were performed according to institutional, national, and European guidelines as stipulated in the Wet op de dierproeven (WOD) and in the Dierproevenbesluit. All experiments involving human material were carried out after obtaining written informed consent from all subjects as per the Declaration of Helsinki. The study was approved by the Institutional Review Board of the Radboud University Nijmegen Medical Center, Commissie Mensgebonden Onderzoek.
Author Contributions
NT-K and MA planned and performed experiments. EK-R and ML performed experiments. HI generated and provided mice. NT-K, CV, and MA contributed to the interpretation of the data. MA wrote the manuscript. NT-K, EK-R, ML, HI, and CV contributed to the review of the manuscript. MA designed the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the constructive discussions and critically reading of the manuscript to Prof. G. J. Adema. This work is supported by project number 016.156.093 of the Netherlands Organisation for Scientific Research (NWO-Veni) (to MA). NT-K is recipient of a PhD grant from the Radboud University Nijmegen Medical Centre. MA is a recipient of a long-term fellowship (BUIT 2012-5347) from the Dutch Cancer Society.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fimmu.2018.01797/full#supplementary-material.
References
1. Granucci F, Foti M, Ricciardi-Castagnoli P. Dendritic cell biology. Adv Immunol (2005) 88:193–233. doi:10.1016/S0065-2776(05)88006-X
2. Adema GJ. Dendritic cells from bench to bedside and back. Immunol Lett (2009) 122(2):128–30. doi:10.1016/j.imlet.2008.11.017
3. Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol (2013) 31:563–604. doi:10.1146/annurev-immunol-020711-074950
4. Granot T, Senda T, Carpenter DJ, Matsuoka N, Weiner J, Gordon CL, et al. Dendritic cells display subset and tissue-specific maturation dynamics over human life. Immunity (2017) 46(3):504–15. doi:10.1016/j.immuni.2017.02.019
5. Pakalniskyte D, Schraml BU. Tissue-specific diversity and functions of conventional dendritic cells. Adv Immunol (2017) 134:89–135. doi:10.1016/bs.ai.2017.01.003
6. Jacobs JF, Punt CJ, Lesterhuis WJ, Sutmuller RP, Brouwer HM, Scharenborg NM, et al. Dendritic cell vaccination in combination with anti-CD25 monoclonal antibody treatment: a phase I/II study in metastatic melanoma patients. Clin Cancer Res (2010) 16(20):5067–78. doi:10.1158/1078-0432.CCR-10-1757
7. Strioga MM, Felzmann T, Powell DJ Jr, Ostapenko V, Dobrovolskiene NT, Matuskova M, et al. Therapeutic dendritic cell-based cancer vaccines: the state of the art. Crit Rev Immunol (2013) 33(6):489–547. doi:10.1615/CritRevImmunol.2013008033
8. Aarntzen EH, De Vries IJ, Lesterhuis WJ, Schuurhuis D, Jacobs JF, Bol K, et al. Targeting CD4(+) T-helper cells improves the induction of antitumor responses in dendritic cell-based vaccination. Cancer Res (2013) 73(1):19–29. doi:10.1158/0008-5472.CAN-12-1127
9. Aarntzen EH, Schreibelt G, Bol K, Lesterhuis WJ, Croockewit AJ, de Wilt JH, et al. Vaccination with mRNA-electroporated dendritic cells induces robust tumor antigen-specific CD4+ and CD8+ T cells responses in stage III and IV melanoma patients. Clin Cancer Res (2012) 18(19):5460–70. doi:10.1158/1078-0432.CCR-11-3368
10. Aarntzen EH, Srinivas M, Schreibelt G, Heerschap A, Punt CJ, Figdor CG, et al. Reducing cell number improves the homing of dendritic cells to lymph nodes upon intradermal vaccination. Oncoimmunology (2013) 2(7):e24661. doi:10.4161/onci.24661
11. Lo J, Clare-Salzler MJ. Dendritic cell subsets and type I diabetes: focus upon DC-based therapy. Autoimmun Rev (2006) 5(6):419–23. doi:10.1016/j.autrev.2005.12.001
12. Dannull J, Haley NR, Archer G, Nair S, Boczkowski D, Harper M, et al. Melanoma immunotherapy using mature DCs expressing the constitutive proteasome. J Clin Invest (2013) 123(7):3135–45. doi:10.1172/JCI67544
13. Sabado RL, Bhardwaj N. Directing dendritic cell immunotherapy towards successful cancer treatment. Immunotherapy (2010) 2(1):37–56. doi:10.2217/imt.09.43
14. Hansen M, Met O, Svane IM, Andersen MH. Cellular based cancer vaccines: type 1 polarization of dendritic cells. Curr Med Chem (2012) 19(25):4239–46. doi:10.2174/092986712802884213
15. Garg AD, Vara Perez M, Schaaf M, Agostinis P, Zitvogel L, Kroemer G, et al. Trial watch: dendritic cell-based anticancer immunotherapy. Oncoimmunology (2017) 6(7):e1328341. doi:10.1080/2162402X.2017.1328341
16. Chow EK, Razani B, Cheng G. Innate immune system regulation of nuclear hormone receptors in metabolic diseases. J Leukoc Biol (2007) 82(2):187–95. doi:10.1189/jlb.1206741
17. Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell (2005) 122(5):707–21. doi:10.1016/j.cell.2005.06.029
18. Chute JP, Ross JR, McDonnell DP. Minireview: nuclear receptors, hematopoiesis, and stem cells. Mol Endocrinol (2010) 24(1):1–10. doi:10.1210/me.2009-0332
19. Hontelez S, Karthaus N, Looman MW, Ansems M, Adema GJ. DC-SCRIPT regulates glucocorticoid receptor function and expression of its target GILZ in dendritic cells. J Immunol (2013) 190(7):3172–9. doi:10.4049/jimmunol.1201776
20. Karthaus N, van Spriel AB, Looman MW, Chen S, Spilgies LM, Lieben L, et al. Vitamin D controls murine and human plasmacytoid dendritic cell function. J Invest Dermatol (2014) 134(5):1255–64. doi:10.1038/jid.2013.501
21. Hamers AA, Hanna RN, Nowyhed H, Hedrick CC, de Vries CJ. NR4A nuclear receptors in immunity and atherosclerosis. Curr Opin Lipidol (2013) 24(5):381–5. doi:10.1097/MOL.0b013e3283643eac
22. Beard JA, Tenga A, Chen T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell Signal (2015) 27(2):257–66. doi:10.1016/j.cellsig.2014.11.009
23. Wei X, Gao H, Zou J, Liu X, Chen D, Liao J, et al. Contra-directional coupling of Nur77 and Nurr1 in neurodegeneration: a novel mechanism for memantine-induced anti-inflammation and anti-mitochondrial impairment. Mol Neurobiol (2016) 53(9):5876–92. doi:10.1007/s12035-015-9477-7
24. Gao H, Chen Z, Fu Y, Yang X, Weng R, Wang R, et al. Nur77 exacerbates PC12 cellular injury in vitro by aggravating mitochondrial impairment and endoplasmic reticulum stress. Sci Rep (2016) 6:34403. doi:10.1038/srep34403
25. Montarolo F, Perga S, Martire S, Navone DN, Marchet A, Leotta D, et al. Altered NR4A subfamily gene expression level in peripheral blood of parkinson’s and alzheimer’s disease patients. Neurotox Res (2016) 30(3):338–44. doi:10.1007/s12640-016-9626-4
26. Zou J, Chen Z, Wei X, Chen Z, Fu Y, Yang X, et al. Cystatin C as a potential therapeutic mediator against Parkinson’s disease via VEGF-induced angiogenesis and enhanced neuronal autophagy in neurovascular units. Cell Death Dis (2017) 8(6):e2854. doi:10.1038/cddis.2017.240
27. Rothe T, Ipseiz N, Faas M, Lang S, Perez-Branguli F, Metzger D, et al. The nuclear receptor Nr4a1 acts as a microglia rheostat and serves as a therapeutic target in autoimmune-driven central nervous system inflammation. J Immunol (2017) 198(10):3878–85. doi:10.4049/jimmunol.1600638
28. Shaked I, Hanna RN, Shaked H, Chodaczek G, Nowyhed HN, Tweet G, et al. Transcription factor Nr4a1 couples sympathetic and inflammatory cues in CNS-recruited macrophages to limit neuroinflammation. Nat Immunol (2015) 16(12):1228–34. doi:10.1038/ni.3321
29. Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal (2006) 4:e002. doi:10.1621/nrs.04002
30. McMorrow JP, Murphy EP. Inflammation: a role for NR4A orphan nuclear receptors? Biochem Soc Trans (2011) 39(2):688–93. doi:10.1042/BST0390688
31. Fassett MS, Jiang W, D’Alise AM, Mathis D, Benoist C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc Natl Acad Sci U S A (2012) 109(10):3891–6. doi:10.1073/pnas.1200090109
32. Zhou T, Cheng J, Yang P, Wang Z, Liu C, Su X, et al. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med (1996) 183(4):1879–92. doi:10.1084/jem.183.4.1879
33. Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol (2013) 14(3):230–7. doi:10.1038/ni.2520
34. Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C-monocytes. Nat Immunol (2011) 12(8):778–85. doi:10.1038/ni.2063
35. Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell (2013) 153(2):362–75. doi:10.1016/j.cell.2013.03.010
36. Briseno CG, Haldar M, Kretzer NM, Wu X, Theisen DJ, Kc W, et al. Distinct transcriptional programs control cross-priming in classical and monocyte-derived dendritic cells. Cell Rep (2016) 15(11):2462–74. doi:10.1016/j.celrep.2016.05.025
37. Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, Ferreira V, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol (2006) 26(10):2288–94. doi:10.1161/01.ATV.0000238346.84458.5d
38. Hanna RN, Shaked I, Hubbeling HG, Punt JA, Wu R, Herrley E, et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ Res (2012) 110(3):416–27. doi:10.1161/CIRCRESAHA.111.253377
39. Wang T, Jiang Q, Chan C, Gorski KS, McCadden E, Kardian D, et al. Inhibition of activation-induced death of dendritic cells and enhancement of vaccine efficacy via blockade of MINOR. Blood (2009) 113(13):2906–13. doi:10.1182/blood-2008-08-176354
40. Karthaus N, Hontelez S, Looman MW, van Spriel AB, Ansems M, Adema GJ. Nuclear receptor expression patterns in murine plasmacytoid and conventional dendritic cells. Mol Immunol (2013) 55(3–4):409–17. doi:10.1016/j.molimm.2013.03.020
41. Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, Kc W, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol (2015) 16(7):708–17. doi:10.1038/ni.3197
42. Park K, Mikulski Z, Seo GY, Andreyev AY, Marcovecchio P, Blatchley A, et al. The transcription factor NR4A3 controls CD103+ dendritic cell migration. J Clin Invest (2016) 126(12):4603–15. doi:10.1172/JCI87081
43. Saini A, Mahajan S, Gupta P. Nuclear receptor expression atlas in BMDCs: Nr4a2 restricts immunogenicity of BMDCs and impedes EAE. Eur J Immunol (2016) 46(8):1842–53. doi:10.1002/eji.201546229
44. Nagaoka M, Yashiro T, Uchida Y, Ando T, Hara M, Arai H, et al. The orphan nuclear receptor NR4A3 is involved in the function of dendritic cells. J Immunol (2017) 199(8):2958–67. doi:10.4049/jimmunol.1601911
45. Sekiya T, Kashiwagi I, Inoue N, Morita R, Hori S, Waldmann H, et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun (2011) 2:269. doi:10.1038/ncomms1272
46. Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol (2005) 6(6):565–70. doi:10.1038/ni1207
47. Hontelez S, Ansems M, Karthaus N, Zuidscherwoude M, Looman MW, Triantis V, et al. Dendritic cell-specific transcript: dendritic cell marker and regulator of TLR-induced cytokine production. J Immunol (2012) 189(1):138–45. doi:10.4049/jimmunol.1103709
48. Sondergaard JN, Poghosyan S, Hontelez S, Louche P, Looman MW, Ansems M, et al. DC-SCRIPT regulates IL-10 production in human dendritic cells by modulating NF-kappaBp65 activation. J Immunol (2015) 195(4):1498–505. doi:10.4049/jimmunol.1402924
49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (2001) 25(4):402–8. doi:10.1006/meth.2001.1262
50. Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, et al. Cutting edge: generation of splenic CD8+ and CD8-dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol (2005) 174(11):6592–7. doi:10.4049/jimmunol.174.11.6592
51. Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med (2011) 208(6):1279–89. doi:10.1084/jem.20110308
52. Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, et al. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood (2014) 124(20):3081–91. doi:10.1182/blood-2013-12-545772
53. Kroesen M, Nierkens S, Ansems M, Wassink M, Orentas RJ, Boon L, et al. A transplantable TH-MYCN transgenic tumor model in C57Bl/6 mice for preclinical immunological studies in neuroblastoma. Int J Cancer (2014) 134(6):1335–45. doi:10.1002/ijc.28463
54. Murphy EP, Crean D. Molecular interactions between NR4A orphan nuclear receptors and NF-kappaB are required for appropriate inflammatory responses and immune cell homeostasis. Biomolecules (2015) 5(3):1302–18. doi:10.3390/biom5031302
55. Yoo YG, Na TY, Yang WK, Kim HJ, Lee IK, Kong G, et al. 6-Mercaptopurine, an activator of Nur77, enhances transcriptional activity of HIF-1alpha resulting in new vessel formation. Oncogene (2007) 26(26):3823–34. doi:10.1038/sj.onc.1210149
56. Pires NM, Pols TW, de Vries MR, van Tiel CM, Bonta PI, Vos M, et al. Activation of nuclear receptor Nur77 by 6-mercaptopurine protects against neointima formation. Circulation (2007) 115(4):493–500. doi:10.1161/CIRCULATIONAHA.106.626838
57. Huang HY, Chang HF, Tsai MJ, Chen JS, Wang MJ. 6-Mercaptopurine attenuates tumor necrosis factor-alpha production in microglia through Nur77-mediated transrepression and PI3K/Akt/mTOR signaling-mediated translational regulation. J Neuroinflammation (2016) 13(1):78. doi:10.1186/s12974-016-0543-5
58. Qin Q, Chen M, Yi B, You X, Yang P, Sun J. Orphan nuclear receptor Nur77 is a novel negative regulator of endothelin-1 expression in vascular endothelial cells. J Mol Cell Cardiol (2014) 77:20–8. doi:10.1016/j.yjmcc.2014.09.027
59. Shao Q, Shen LH, Hu LH, Pu J, Qi MY, Li WQ, et al. Nuclear receptor Nur77 suppresses inflammatory response dependent on COX-2 in macrophages induced by oxLDL. J Mol Cell Cardiol (2010) 49(2):304–11. doi:10.1016/j.yjmcc.2010.03.023
60. Hamers AA, Argmann C, Moerland PD, Koenis DS, Marinkovic G, Sokolovic M, et al. Nur77-deficiency in bone marrow-derived macrophages modulates inflammatory responses, extracellular matrix homeostasis, phagocytosis and tolerance. BMC Genomics (2016) 17:162. doi:10.1186/s12864-016-2469-9
61. Hanna RN, Cekic C, Sag D, Tacke R, Thomas GD, Nowyhed H, et al. Patrolling monocytes control tumor metastasis to the lung. Science (2015) 350(6263):985–90. doi:10.1126/science.aac9407
62. Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, et al. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res (2014) 114(10):1611–22. doi:10.1161/CIRCRESAHA.114.303204
63. Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol (2006) 20(4):786–94. doi:10.1210/me.2005-0331
64. Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem (2005) 280(32):29256–62. doi:10.1074/jbc.M502606200
65. Li L, Liu Y, Chen HZ, Li FW, Wu JF, Zhang HK, et al. Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat Chem Biol (2015) 11(5):339–46. doi:10.1038/nchembio.1788
66. Hong CY, Park JH, Ahn RS, Im SY, Choi HS, Soh J, et al. Molecular mechanism of suppression of testicular steroidogenesis by proinflammatory cytokine tumor necrosis factor alpha. Mol Cell Biol (2004) 24(7):2593–604. doi:10.1128/MCB.24.7.2593-2604.2004
67. Li XM, Zhang S, He XS, Guo PD, Lu XX, Wang JR, et al. Nur77-mediated TRAF6 signalling protects against LPS-induced sepsis in mice. J Inflamm (Lond) (2016) 13:4. doi:10.1186/s12950-016-0112-9
68. Hamers AA, van Dam L, Teixeira Duarte JM, Vos M, Marinkovic G, van Tiel CM, et al. Deficiency of nuclear receptor Nur77 aggravates mouse experimental colitis by increased NFkappaB activity in macrophages. PLoS One (2015) 10(8):e0133598. doi:10.1371/journal.pone.0133598
69. Ipseiz N, Uderhardt S, Scholtysek C, Steffen M, Schabbauer G, Bozec A, et al. The nuclear receptor Nr4a1 mediates anti-inflammatory effects of apoptotic cells. J Immunol (2014) 192(10):4852–8. doi:10.4049/jimmunol.1303377
70. Kobayashi T, Walsh PT, Walsh MC, Speirs KM, Chiffoleau E, King CG, et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity (2003) 19(3):353–63. doi:10.1016/S1074-7613(03)00230-9
71. Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell (2000) 103(2):351–61. doi:10.1016/S0092-8674(00)00126-4
72. Wang Y, Tang Y, Teng L, Wu Y, Zhao X, Pei G. Association of beta-arrestin and TRAF6 negatively regulates toll-like receptor-interleukin 1 receptor signaling. Nat Immunol (2006) 7(2):139–47. doi:10.1038/ni1294
73. Murray JE, Merrill JP, Harrison JH, Wilson RE, Dammin GJ. Prolonged survival of human-kidney homografts by immunosuppressive drug therapy. N Engl J Med (1963) 268:1315–23. doi:10.1056/NEJM196306132682401
74. Kornbluth A, George J, Sachar DB. Immunosuppressive drugs in Crohn’s disease. Gastroenterologist (1994) 2(3):239–46.
75. Aarbakke J, Janka-Schaub G, Elion GB. Thiopurine biology and pharmacology. Trends Pharmacol Sci (1997) 18(1):3–7. doi:10.1016/S0165-6147(96)01007-3
76. Wansa KD, Harris JM, Yan G, Ordentlich P, Muscat GE. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. J Biol Chem (2003) 278(27):24776–90. doi:10.1074/jbc.M300088200
77. Ordentlich P, Yan Y, Zhou S, Heyman RA. Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J Biol Chem (2003) 278(27):24791–9. doi:10.1074/jbc.M302167200
78. Marinkovic G, Hamers AA, de Vries CJ, de Waard V. 6-Mercaptopurine reduces macrophage activation and gut epithelium proliferation through inhibition of GTPase Rac1. Inflamm Bowel Dis (2014) 20(9):1487–95. doi:10.1097/MIB.0000000000000122
79. Marinkovic G, Kroon J, Hoogenboezem M, Hoeben KA, Ruiter MS, Kurakula K, et al. Inhibition of GTPase Rac1 in endothelium by 6-mercaptopurine results in immunosuppression in nonimmune cells: new target for an old drug. J Immunol (2014) 192(9):4370–8. doi:10.4049/jimmunol.1302527
80. Lee SO, Li X, Hedrick E, Jin UH, Tjalkens RB, Backos DS, et al. Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Mol Endocrinol (2014) 28(10):1729–39. doi:10.1210/me.2014-1102
81. Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol (2008) 4(9):548–56. doi:10.1038/nchembio.106
82. Liu JJ, Zeng HN, Zhang LR, Zhan YY, Chen Y, Wang Y, et al. A unique pharmacophore for activation of the nuclear orphan receptor Nur77 in vivo and in vitro. Cancer Res (2010) 70(9):3628–37. doi:10.1158/0008-5472.CAN-09-3160
83. Zhan YY, Chen Y, Zhang Q, Zhuang JJ, Tian M, Chen HZ, et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol (2012) 8(11):897–904. doi:10.1038/nchembio.1069
84. Chintharlapalli S, Burghardt R, Papineni S, Ramaiah S, Yoon K, Safe S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J Biol Chem (2005) 280(26):24903–14. doi:10.1074/jbc.M500107200
85. Cho SD, Lei P, Abdelrahim M, Yoon K, Liu S, Guo J, et al. 1,1-bis(3′-indolyl)-1-(p-methoxyphenyl)methane activates Nur77-independent proapoptotic responses in colon cancer cells. Mol Carcinog (2008) 47(4):252–63. doi:10.1002/mc.20378
86. Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family – orphans no more. J Steroid Biochem Mol Biol (2016) 157:48–60. doi:10.1016/j.jsbmb.2015.04.016
87. Yoon K, Lee SO, Cho SD, Kim K, Khan S, Safe S. Activation of nuclear TR3 (NR4A1) by a diindolylmethane analog induces apoptosis and proapoptotic genes in pancreatic cancer cells and tumors. Carcinogenesis (2011) 32(6):836–42. doi:10.1093/carcin/bgr040
88. Lee SO, Li X, Khan S, Safe S. Targeting NR4A1 (TR3) in cancer cells and tumors. Expert Opin Ther Targets (2011) 15(2):195–206. doi:10.1517/14728222.2011.547481
89. Dawson MI, Ye M, Cao X, Farhana L, Hu QY, Zhao Y, et al. Derivation of a retinoid X receptor scaffold from peroxisome proliferator-activated receptor gamma ligand 1-Di(1H-indol-3-yl)methyl-4-trifluoromethylbenzene. ChemMedChem (2009) 4(7):1106–19. doi:10.1002/cmdc.200800447
90. Cho SD, Yoon K, Chintharlapalli S, Abdelrahim M, Lei P, Hamilton S, et al. Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer Res (2007) 67(2):674–83. doi:10.1158/0008-5472.CAN-06-2907
91. Segala G, David M, de Medina P, Poirot MC, Serhan N, Vergez F, et al. Dendrogenin A drives LXR to trigger lethal autophagy in cancers. Nat Commun (2017) 8(1):1903. doi:10.1038/s41467-017-01948-9
92. Poirot M, Silvente-Poirot S. The tumor-suppressor cholesterol metabolite, dendrogenin A, is a new class of LXR modulator activating lethal autophagy in cancers. Biochem Pharmacol (2018) 153:75–81. doi:10.1016/j.bcp.2018.01.046
93. Silvente-Poirot S, Segala G, Poirot MC, Poirot M. Ligand-dependent transcriptional induction of lethal autophagy: a new perspective for cancer treatment. Autophagy (2018) 14(3):555–7. doi:10.1080/15548627.2018.1425059
94. de Medina P, Paillasse MR, Segala G, Voisin M, Mhamdi L, Dalenc F, et al. Dendrogenin A arises from cholesterol and histamine metabolism and shows cell differentiation and anti-tumour properties. Nat Commun (2013) 4:1840. doi:10.1038/ncomms2835
95. Zhong L, Yang Q, Xie W, Zhou J. Liver X receptor regulates mouse GM-CSF-derived dendritic cell differentiation in vitro. Mol Immunol (2014) 60(1):32–43. doi:10.1016/j.molimm.2014.03.006
96. Canavan M, McCarthy C, Larbi NB, Dowling JK, Collins L, O’Sullivan F, et al. Activation of liver X receptor suppresses the production of the IL-12 family of cytokines by blocking nuclear translocation of NF-kappaBp50. Innate Immun (2014) 20(7):675–87. doi:10.1177/1753425913501915
97. Geyeregger R, Zeyda M, Bauer W, Kriehuber E, Saemann MD, Zlabinger GJ, et al. Liver X receptors regulate dendritic cell phenotype and function through blocked induction of the actin-bundling protein fascin. Blood (2007) 109(10):4288–95. doi:10.1182/blood-2006-08-043422
98. Torocsik D, Barath M, Benko S, Szeles L, Dezso B, Poliska S, et al. Activation of liver X receptor sensitizes human dendritic cells to inflammatory stimuli. J Immunol (2010) 184(10):5456–65. doi:10.4049/jimmunol.0902399
99. Villablanca EJ, Raccosta L, Zhou D, Fontana R, Maggioni D, Negro A, et al. Tumor-mediated liver X receptor-alpha activation inhibits CC chemokine receptor-7 expression on dendritic cells and dampens antitumor responses. Nat Med (2010) 16(1):98–105. doi:10.1038/nm.2074
100. Beceiro S, Pap A, Czimmerer Z, Sallam T, Guillen JA, Gallardo G, et al. LXR nuclear receptors are transcriptional regulators of dendritic cell chemotaxis. Mol Cell Biol (2018) 38(10):e00534–17. doi:10.1128/MCB.00534-17
101. Bruckner M, Dickel D, Singer E, Legler DF. Converse regulation of CCR7-driven human dendritic cell migration by prostaglandin E(2) and liver X receptor activation. Eur J Immunol (2012) 42(11):2949–58. doi:10.1002/eji.201242523
102. Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J (1997) 16(8):1865–75. doi:10.1093/emboj/16.8.1865
103. Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol (1999) 19(11):7549–57. doi:10.1128/MCB.19.11.7549
Keywords: dendritic cells, dendritic cell-based immunotherapy, nuclear receptors, NR4A, Nur77
Citation: Tel-Karthaus N, Kers-Rebel ED, Looman MW, Ichinose H, de Vries CJ and Ansems M (2018) Nuclear Receptor Nur77 Deficiency Alters Dendritic Cell Function. Front. Immunol. 9:1797. doi: 10.3389/fimmu.2018.01797
Received: 01 May 2018; Accepted: 20 July 2018;
Published: 03 August 2018
Edited by:
Silvia Beatriz Boscardin, Universidade de São Paulo, BrazilReviewed by:
Dennis Qing Wang, Zhujiang Hospital of Southern Medical University, ChinaAmanda Jane Gibson, Royal Veterinary College, United Kingdom
Marc Poirot, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright: © 2018 Tel-Karthaus, Kers-Rebel, Looman, Ichinose, de Vries and Ansems. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marleen Ansems, bWFybGVlbi5hbnNlbXNAcmFkYm91ZHVtYy5ubA==
 Nina Tel-Karthaus1
Nina Tel-Karthaus1 Esther D. Kers-Rebel
Esther D. Kers-Rebel Marleen Ansems
Marleen Ansems