- 1Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Kunming, China
- 2Yunnan Key Laboratory of Vaccine Research and Development on Severe Infectious Disease, Kunming, China
Aim: The objective of this study was to evaluate the association of the human leukocyte antigen (HLA) class II genes HLA-DRB1, HLA-DPB1, and HLA-DQB1 with the humoral immune response elicited by inactivated Japanese encephalitis (JE) vaccine (IJEV).
Methods: A total of 373 individuals aged 3–12 years in the Inner Mongolia Autonomous Region in China, who received two doses of IJEV at 0 and 7 days, were enrolled in the current study. Based on the individuals' specific JE virus (JEV)-neutralizing antibodies (NAbs), they were divided into a seropositive and a seronegative group. HLA-DRB1, HLA-DPB1, and HLA-DQB1 were genotyped using a sequencing-based typing method. Next, the association of the HLA class II genes and their haplotypes with antibody response was evaluated.
Results: Based on NAbs, a total of 161 individuals were classified as seropositive and 212 as seronegative. DQB1*02:01 was significantly associated with JEV seropositivity (P < 0.001, OR = 0.364, 95% CI: 0.221–0.600), while DQB1*02:02 was significantly associated with JEV seronegativity (P = 5.03 × 10−6, OR = 7.341, 95% CI: 2.876–18.736). The haplotypes DRB1*07:01-DPB1*04:01-DQB1* 02:01, DRB1*15:01-DPB1*02:01-DQB1*06:02, DRB1*07:01-DQB1*02:01, and DPB1*02:01-DQB1*06:02 were very frequent in the seropositive group, while DRB1*07:01-DPB1*17:01-DQB1*02:02, DRB1*07:01-DQB1*02:02, and DPB1*17:01-DQB1*02:02 were very frequent in the seronegative group. The presence of DRB1*01:01, DRB1*04:05, DRB1*09:01, DRB1*12:02, DRB1*13:02, and DRB1*14:01 was associated with a higher geometric mean titer (GMT) of NAbs than that of DRB1*11:01 at the DRB1 locus (P < 0.05). At the DPB1 locus, the presence of DPB1*05:01 was associated with higher GMTs than that of DPB1*02:01 and DPB1*13:01 (P < 0.05), and the presence of DPB1*04:01 and DPB1*09:01 was associated with higher GMTs than that of DPB1*13:01 (P < 0.05).
Conclusions: The present study suggests that HLA class II genes may influence the antibody response to IJEV.
Introduction
Japanese encephalitis (JE) is one of the most serious mosquito-borne infectious diseases, with approximately 67,900 individuals being infected by the JE virus (JEV) annually (1). Approximately 75% of these individuals are under 14 years of age, and 50% of the infections occur in China (2, 3).
Vaccination is an efficient method of controlling JEV infection. Four different types of JE vaccine are available in affected countries, namely inactivated mouse brain-derived, live attenuated cell culture-derived, inactivated cell culture-derived, and genetically engineered live attenuated chimeric vaccine. The Vero cell-derived inactivated JE vaccine (IJEV) has been widely used in China, Japan, the US, Europe, Canada, Australia, Hong Kong, Switzerland, and India (4–6). A JEV-neutralizing antibody (NAb) titer of at least 10 has been established as a correlate for protection against JEV, while positive serum conversion rate and geometric mean titer (GMT) have been used as alternative markers of efficacy of JE vaccines (7, 8). After immunization with the JE vaccine, the positive serum conversion rate ranges from 60 to 100% (1, 9). Vaccine efficacy may be influenced by factors such as the type of vaccine and the vaccinated person's age, gender, and nutritional status (6, 10). Several studies have reported that the efficacy of attenuated JE vaccine has reached 85–99.26% in Chinese, South Korean, and Nepalese children; however, it exhibited only 67.2% efficacy in Indians after primary immunization (11–14). These results indicate that different genetic backgrounds of hosts could play an important role in the efficacy of JE vaccines.
As one of the key immune gene complexes, the human leukocyte antigen (HLA) genes play an important role in the adaptive immune response to viruses and vaccines. HLA molecules are divided into three classes: class I, II, and III. Among them, HLA class II molecules (HLA-DR, -DQ, and -DP) bind to extracellular viral antigen peptides and display them on the surface of antigen-presenting cells to CD4+ cells to stimulate their multiplication, which, in turn, stimulate antibody-producing B cells to produce specific antibodies (15, 16). HLA genes exhibit extraordinary polymorphisms, and different alleles can affect the peptide-binding properties of the HLA molecular pocket, which subsequently influences the immune response to a vaccine. In 2005 and 2006, Ovsyannikova et al. (17, 18) observed that HLA-DPB1*03:01, HLA-DPB1*04:01, and HLA-DPB1*15:01 are associated with rubella vaccine-induced antibodies. On the other hand, the HLA-DRB1*15/16-DQB1*06-DPB1*13 haplotype has been associated with high levels of measles antibody response, but low levels of rubella antibody response.
In order to evaluate the association of HLA class II genes HLA-DRB1, HLA-DPB1, and HLA-DQB1 and JEV-NAbs with the humoral immune response to IJEV, this study examined Mongolian Chinese individuals who had been administered IJEV.
Materials and Methods
Subjects and Vaccination
A randomized, double-blinded, positive-control, non-inferiority IJEV trial was implemented in the Inner Mongolia Autonomous Region of China from August 2012 to September 2013. The IJEV (lot: 20101201) was manufactured in a GMP-accredited facility of the Institute of Medical Biology at the Chinese Academy of Medical Sciences (IMBCAMS) and verified by the National Institute for Food and Drug Control (China, approval no. 2010L02035). Briefly, JEV P3 strains were grown on Vero cell microcarriers in a 75 L bioreactor. The virus suspension was harvested, inactivated with ultra-concentrated formalin, and purified by Sepharose 6FF and DEAE Sepharose FF. The resulting vaccine contained 0.5 mL per dose with ≥ 0.6 IU/mL JEV antigens. The clinical study procedure was approved by the Ethics Committee of the Inner Mongolia Autonomous Region Center for Disease Control and Prevention. The IJEV control (lot: 201012B02-1) was manufactured by Liaoning Chengda Biotechnology (Shenyang, China), containing the same concentration of antigens as the vaccine made by IMBCAMS. A total of 1,200 individuals aged 8 months−12 years in the Inner Mongolia Autonomous Region were enrolled to receive two doses of IJEV at 0 and 7 days. They were vaccinated with either the IJEV made by IMBCAMS or the IJEV control at a 1:1 ratio. The inclusion criteria were that the individual was in good health, was not infected by JEV, had not been inoculated with other vaccines within 7 days, and had not been inoculated with attenuated JE vaccine within 1 month. The peripheral blood samples were collected before vaccine administration and 30 days after the second dose received for the detection of neutralization antibody. Considering the limited blood sample volume and the consistency of the test, only individuals of 3–12 years of age, who were negative for NAbs before vaccination, were selected for further HLA genotyping. Finally, after vaccination, 212 individuals negative for NAbs were included in the seronegative group, and 161 individuals positive for NAbs were randomly selected and included in the seropositive group.
Japanese Encephalitis Vaccine Neutralization Antibody Detection
JEV-specific NAbs were determined by the National Institute for Food and Drug Control using the 50% plaque-reduction neutralization test according to the requirement of the Pharmacopeia of the People's Republic of China (19). Briefly, BHK-21 cells were initially inoculated at 106 cells/well in 24-well tissue culture plates and propagated for 48 h at 37°C in a CO2 incubator. The serum samples were inactivated for 30 min in a 56°C water bath, diluted 10-fold, and then serially diluted 2-fold from 1:10 to 1:1280 in Minimum Essential Medium (GIBCO, Grand Island, NY, USA) containing 2% fetal bovine serum and 1% penicillin/streptomycin. The diluted serum and the positive conference serum were mixed with an equal volume of diluted challenge virus (P3 strain, lot 20151102, 500 PFU/mL). The suspensions were kept in a 37°C water bath for 30 min. Afterwards, 0.1 mL aliquots of the virus-serum mixtures were dispensed separately into each well of the 24-well microplates with the BHK-21 cells. The cells were overlaid with medium containing 1% methylcellulose. The cells in the wells were stained after inoculation at 37°C for 5 days in a 5% CO2 incubator, and the plaques were counted. The NAb titer was defined as the reciprocal value of the last serum dilution that showed 50% or greater plaque reduction compared with the plaque counts in the virus-only control wells. NAbs at 50% plaque reduction neutralization titer (PRNT50) < 10 or PRNT50 increased <4-fold after vaccination were considered as negative seroconversion, while PRNT50 > 10, or at least a 4-fold increase after vaccination, was considered to be positive seroconversion. The antibody titers were determined by calculating the GMT as follows: GMT = Log−1 [(LogX1 + LogX2 + … LogXn)/n].
HLA-DRB1, HLA-DPB1, and HLA-DQB1 Genotyping
Genomic DNA was extracted from peripheral lymphocytes using the QIAamp Blood Kit (Qiagen, Hilden, Germany). HLA-DRB1, HLA-DPB1, and HLA-DQB1 were genotyped using a high-resolution sequencing-based typing method (Applied Biosystems, Foster City, CA, USA). Briefly, exons 2 and 3 of DRB1 and DQB1 as well as all exons of DPB1 were amplified, and the PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Finally, the sequence was analyzed with the 3730xl DNA Analyzer (Applied Biosystems), and the HLA alleles were identified using the SBTengine (Applied Biosystems).
Statistical Analysis
The differences in age and sex between the seropositive and seronegative group were determined using Student's t-test or a χ2 test. The HLA-DRB1, -DPB1 and -DQB1 allele frequencies were calculated using the PyPop or PyHLA software based on the genotyping results (20–22). The Hardy-Weinberg equilibrium was assessed using the Guo and Thompson method (23). The haplotypes were constructed based on the genotyping results using the expectation-maximization algorithm (20–22). The χ2 test was used to determine differences in allele and haplotype frequencies between the seropositive and seronegative group. The odds ratios (ORs) and associated 95% confidence intervals (CIs) were also calculated for allele-specific risks. False discovery rate (FDR) correction was used for the multiple comparisons (20). For each gene, the amino acid sequences for all alleles were aligned together. If there was more than one amino acid at one position, a test was performed for each amino acid to examine whether it is distributed differently in the seropositive and seronegative group using PyHLA software (20). Fisher's exact test was used to analyze the association, and the odds ratio was calculated with Haldane's correction of Woolf's method (20). The association between HLA-DRB1, -DPB1, -DQB1 alleles and antibody levels was analyzed through the analysis of variance using GraphPad Prism 7.0. P-values of < 0.05 were considered statistically significant.
Results
Characteristics of Subjects
Table 1 lists the characteristics of the enrolled subjects. They were randomly selected from the two vaccination groups, which had no difference in seroconversion or GMT (P > 0.05). All subjects were negative for NAbs before vaccination. After vaccination, 161 individuals with PRNT50 > 10 were included in the seropositive group, while 212 individuals with PRNT50 < 10 were included in the seronegative group. Table 2 shows that there were no age or gender differences between the seropositive and seronegative group (P > 0.05). In addition, there was no difference in NAbs titers according to age and gender (P > 0.05) in the seropositive group (data not shown). Moreover, there was no difference in HLA allele distribution according to gender (P > 0.05).
Association of HLA Alleles With Neutralizing Antibody Seroconversion of Inactivated Japanese Encephalitis Vaccine
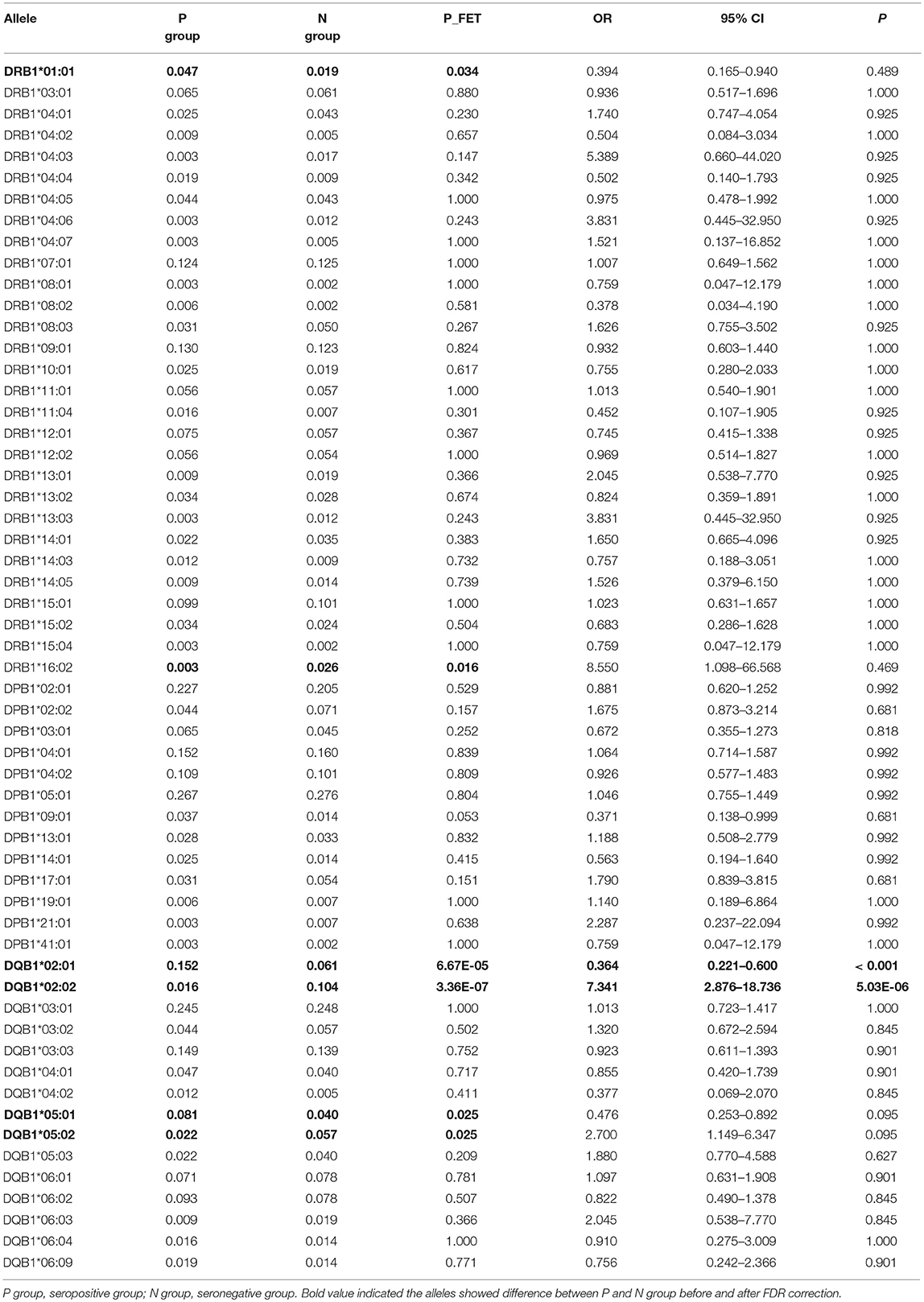
The frequencies of HLA-DRB1, -DPB1, and -DQB1 were in Hardy-Weinberg equilibrium in both the seropositive and seronegative groups (P > 0.05). At the HLA-DRB1 locus, the frequencies of DRB1*01:01 and DRB1*16:02 were different between the seropositive and seronegative group; however, after FDR correction, the difference was not considered significant (Table 3). At the HLA-DPB1 locus, there was no significant difference between the seropositive and seronegative group. At the HLA-DQB1 locus, the frequency of DQB1*02:01 was higher in the seropositive group (0.152) than in the seronegative group (0.061) (P < 0.001; OR = 0.364; 95% CI: 0.221–0.600), while the frequency of DQB1*02:02 was lower in the seropositive group (0.016) than in the seronegative group (0.104) (P = 5.03 × 10−6; OR = 7.341; 95% CI: 2.876–18.736) (Table 3). DQB1*05:01 and DQB1*05:02 frequencies were also different between the groups, but the difference was not significant after FDR correction (Table 3).
Further analysis of HLA residue levels showed that some HLA residues were associated with JEV antibody seroconversion (Supplementary Table 1). At the HLA-DPB1 locus, residues A56, R96, T170, and V265 were associated with seronegative JEV-NAbs. At the HLA-DQB1 locus, the residues S57, V116, A125, G135, and P146 were associated with seronegative NAbs, while V89 was associated with seropositive NAbs. At the HLA-DRB1 locus, D28 and Y30 were associated with seronegative, while L11, K12, F13, L26, C30, I31, and Y32 were associated with seropositive NAbs. Residues containing HLA-DQB1*02:02 were associated with seropositive JEV-NAbs.
Association of HLA-DRB1, -DPB1, and -DQB1 Haplotypes With Neutralizing Antibody Seroconversion of Inactivated Japanese Encephalitis Vaccine
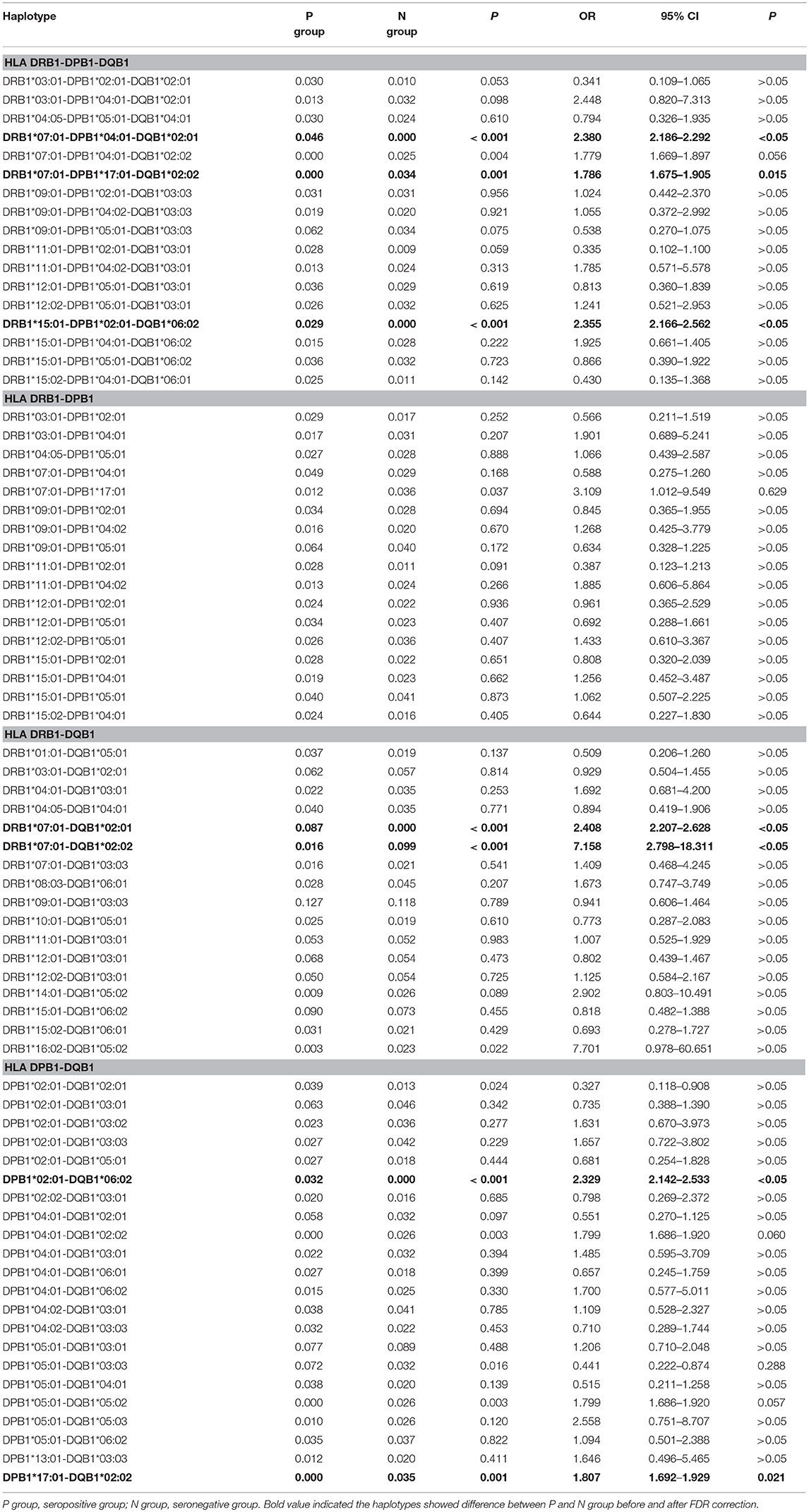
The HLA-DRB1, -DPB1, and -DQB1 alleles with frequencies higher than 0.020 in either the seropositive or seronegative group are listed in Table 4. At the level of the three loci, the frequency of the haplotypes DRB1*07:01-DPB1*04:01-DQB1*02:01 and DRB1*15:01-DPB1*02:01-DQB1*06:02 was higher in the seropositive group than in the seronegative group (P < 0.05), while the frequency of the haplotype DRB1*07:01-DPB1*17:01-DQB1*02:02 was higher in the seronegative group than in the seropositive group (P < 0.05). At the level of the two loci, the frequency of the haplotypes DRB1*07:01-DQB1*02:01 and DPB1*02:01-DQB1*06:02 was higher in the seropositive group (P < 0.05), while that of the haplotypes DRB1*07:01-DQB1*02:02 and DPB1*17:01-DQB1*02:02 was higher in the seronegative group (P < 0.05).
Association of HLA Alleles With Neutralizing Antibody GMTs of Inactivated Japanese Encephalitis Vaccine in Seropositive Group
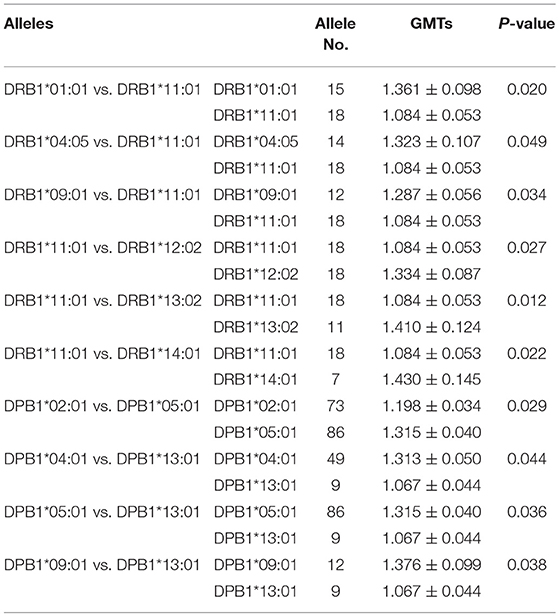
To analyze the association of HLA alleles with JEV-specific NAb GMTs, the 161 individuals in the seropositive group were examined. At the DRB1 locus, the highest GMTs were in subjects with the DRB1*14:03 (1.452 ± 0.174), DRB1*14:01 (1.430 ± 0.383), and DRB1*13:02 (1.410 ± 0.410) alleles, while the lowest were in those with DRB1*01:01 (1.000 ± 0), DRB1*11:04 (1.060 ± 0.135), and DRB1*11:01 (1.084 ± 0.226) alleles. At the DPB1 locus, subjects with DPB1*09:01 (1.376 ± 0.343), DPB1*05:01 (1.315 ± 0.369), and DPB1*04:01 (1.313 ± 0.347) alleles had higher GMTs than those with DPB1*13:01 (1.067 ± 0.133), DPB1*19:01 (1.151 ± 0.213), and DPB1*02:01 (1.198 ± 0.289) alleles (P < 0.05). At the DQB1 locus, the highest GMTs were in subjects with the DQB1*05:03 (1.473 ± 0.383), DQB1*06:09 (1.452 ± 0.369), and DQB1*06:03 (1.401 ± 0.460) alleles, while the lowest were in those with DQB1*05:04 (1.000 ± 0), DQB1*04:02 (1.151 ± 0.301), and DQB1*06:04 (1.181 ± 0.404) alleles (Figure 1).
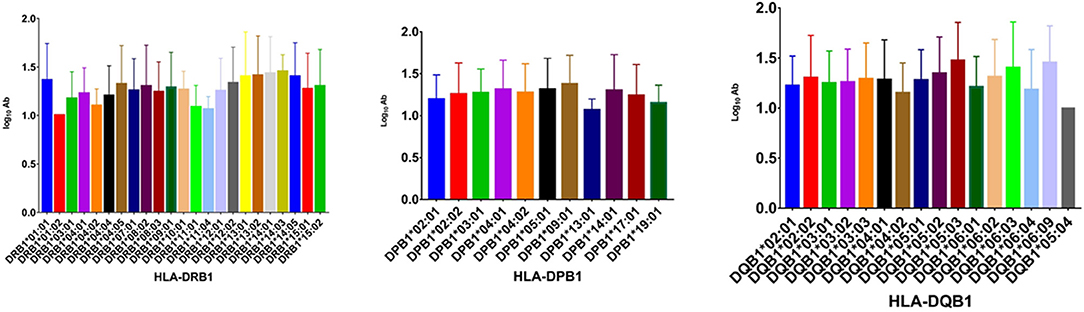
Figure 1. GMTs associated with different HLA-DRB1, -DPB1, and -DQB1 alleles in the IJEV NAb-positive group.
When the alleles were compared one by one, some were associated with higher GMTs (Table 5). The presence of DRB1*01:01, DRB1*04:05, DRB1*09:01, DRB1*12:02, DRB1*13:02, and DRB1*14:01 was associated with higher NAb GMTs than the presence of DRB1*11:01 (P < 0.05) at the DRB1 locus. At the DPB1 locus, the presence of DPB1*05:01 was associated with higher GMTs than the presence of DPB1*02:01 and DPB1*13:01 (P < 0.05), and the presence of DPB1*04:01 and DPB1*09:01 was associated with higher GMTs than the presence of DPB1*13:01 (P < 0.05). There was no significant difference in the GMT between the different DQB1 alleles (P > 0.05).
Discussion
Vaccines are one of the greatest advances in controlling infectious diseases in the past 300 years. The humoral immune response induced by a vaccine produces NAbs, so the seroconversion rate and GMT are widely used to evaluate vaccine efficacy. In the present study, we examined the association of HLA class II genes with the IJEV antibody response to reveal the role of the genetic variation in the HLA class II genes in the IJEV immune response.
HLA class II molecules present viral antigens in the form of peptides derived from the extracellular processing of vaccine peptides, which plays an important role in the humoral immune response to vaccines (24, 25). In an inactivated vaccine, the extracellular vaccine antigens are degraded into smaller peptides and integrated with the HLA class II molecule to constitute the HLA class II peptide complex, which plays a major role in stimulating the differentiation of CD4+ T cells into Th1 and Th2 cells; in turn, the Th2 cells can interact with B cells to promote differentiation into antibody-secreting plasma cells, thus secreting a specific antibody against the vaccine antigen (18).
To date, many studies have reported that HLA class II genes are associated with the vaccine antibody response (17, 26–30). In 1999, McDermott et al. reported that DQB1*02:02 was associated with a negative antibody response to hepatitis B virus (HBV) vaccination in a population in England (31). In 2005, Ovsyannikova et al. performed a study of the association between HLA and the humoral immune response to measles-mumps-rubella vaccination, finding that DQB1*02:02 was negatively associated with rubella-specific lymphoproliferation (17). In the present study, DQB1*02:02 was significantly negatively associated with the IJEV response (P = 5.03 × 10−6; OR = 7.341; 95% CI: 2.876–18.736). However, contrary to previous studies on the HBV, measles, rubella, influenza, and serogroup C meningococcus vaccines, which showed that DQB1*02:01 was negatively associated with vaccine-induced antibody response (32–34), in the present study, DQB1*02:01 had a significantly positive association with IJEV seropositivity (P < 0.05; OR = 0.364; 95% CI: 0.221–0.600). Interestingly, DQB1*02:01 is reportedly associated with high Th1 IFN-γ secretion, while DQB1*02:02 is associated with a low measles-specific Th2 cytokine response (35). There is only one amino-acid difference between DQB1*02:01 and DQB1*02:02, namely at position 135 in the peptide binding groove, where DQB1*02:01 contains aspartic acid and DQB1*02:02 contains glycine. In 2018, Yang et al. predicted the 3D ribbon models of the HLA proteins and indicated that the amino acid position 135 of HLA-DQB1 was located on the junction point of two β-sheet structures and lies on the β2 domain of protein belonging to Ig protein superfamily (36). The domain is expressed on the extracellular part of the antigen presenting cells and could integrate with CD4+ T cells during the antigen presenting process (36). Thus, we deduced that the amino acid change from a negatively charged Asp in DQB1*02:01 to an uncharged polar Gly in DQB1*02:02 could influence the JEV antigen presentation process, in consequence, the inducing of JEV-specific NAb. The further studies on the exact role of how HLA-DQB1*02:01 and HLA-DQB1*02:02 in the progression of JEV-antibody needs to be elucidated in the future.
We compared the previously reported HLA-DQA1 and -DQB1 haplotypes to assess whether there is any preference for DQB1*02:01 or DQB1*02:02 over DQA1 and found that DQB1*02:01 and DQB1*02:02 are either in linkage disequilibrium with the same DQA1 alleles, namely DQA1*02:01, DQA1*05:01, and DQA1*03:01 (http://www.allelefrequencies.net/). We then predicted DQB1*02:01 and DQB1*02:02 heterodimers with DQA1 using the E protein sequence of JEV by NetMHCIIpan (http://www.cbs.dtu.dk/services/NetMHCIIpan/logos.php). The predicted peptides of DQA1*03:01-DQB1*02:01 and DQA1*03:01-DQB1*02:02 showed no difference. The identification of the actual DQA1-DQB1 haplotypes existing in the Mongolian population would help for validation of the DQA1-DQB1 molecular binding with specific JEV epitopes in the future.
In 2012, Schillie et al. found that DRB1*13:01 and DRB1*13:02, with an allele difference at position 86, showed contrary roles in the HBV antibody response (37). In the present study, DQB1*05:01 showed a positive response association, and DQB1*05:02 showed a negative response association with IJEV, though the association was not significant after FDR correction. Further HLA residue association study indicated that the DQB1 residue S57, present in DQB1*05:02 and DQB1*05:04, showed an opposite JEV-NAb response from residue V57, present in DQB1*05:01, DQB1*06:04, and DQB1*06:09. The DQ peptide prediction suggested that the peptide FLVHREWFHDLALPW showed strong binding in both DQA1*01:01-DQB1*05:01 and DQA1*01:01-DQB1*05:02, while the peptides HREWFHDLALPWTPP and RNRELLMEFEEAHAT showed strong binding in subjects with DQA1*01:01-DQB1*05:02, but not in DQA1*01:01-DQB1*05:01 (Supplementary Table 2). This finding indicates that DQB1*05:01 and DQB1*05:02 may produce different JEV peptides. Moreover, these data indicate that allele differences may change the binding groove of the antigen-HLA complex, in turn influencing T cell receptors expressed on inactivated JEV-specific CD4+ T cells and, finally, playing different roles in the antibody response (27, 37).
In addition to DQB1*02:01 and DQB1*02:02, other HLA class II genes are reportedly associated with the antibody response to vaccines. For example, Jafarzadeh et al. (27) reported that DRB1*01:01, DRB1*13:01, DRB1:15:01, and DQB1*04:01 were positively associated with HBV antibody response, while DRB1*03:01, DRB1*07:01, and DQB1*02:01 were negatively associated. However, other than DQB1*02:01, no HLA alleles have been associated with IJEV in the present study. One of the reasons for different HLA alleles being associated with antibody responses could be a distinct immune response to different vaccines or pathogens. Most previous association studies have been performed with attenuated vaccines (mumps, measles, rubella vaccine, etc.) or virus-like particle-based vaccines (HBV), which could induce both HLA class I- and class II-mediated immune response to generate an immune response. However, in an inactivated vaccine like IJEV, the humoral immune response mediated by HLA class II molecules would be key in generating JEV-NAbs. Thus, the difference in the immune response mechanism between the inactivated vaccine and the attenuated vaccine may be caused by the association with different HLA alleles (18, 38). Another reason could be a population-specific difference in HLA distribution, as with HLA genes and their motifs, even if the populations were administered the same vaccine. For example, in 2015, Jafarzadeh et al. (27) reported that non-responsiveness to HBV is associated with HLA-A1, -B15, and -B40 in Indians, HLA-A1, -A2, and -B8 in Caucasians, HLA-B54 in Chinese, and HLA-A10 and -Cw4 in Turkish people. In addition, DQA1*05:01-DQB1*02:01 is predominant in Europe, Southwest Asia, and North Africa with frequencies of 19.1, 17.2, and 17.0%, respectively, while its frequency is only 2.7% in North America, and it has not been identified in South America. DQA1*02:01-DQB1*02:01 is predominant in North Africa, common in European and Southwest Asia, and rare in North Africa and South America (39). To the best of our knowledge, this HLA allele diversity was generated in the long evolutionary interaction between hosts and pathogens, which makes it encode adequate products to generate immune responses against different pathogens. Thus, different HLA alleles were formed as an outcome of specific pathogen infections and are therefore associated with different infectious diseases (40–42). As such, the mechanism of the immune response to different vaccine antigens could vary based on different vaccines.
In addition to the seroconversion rate, GMT is an important factor in evaluating vaccine efficacy. In the present study, we evaluated the relationship between GMTs and HLA alleles. Interestingly, we found that the HLA alleles associated with an antibody response were different from the HLA alleles associated with GMTs. These results indicate that HLA alleles have different roles in the host immune response.
In summary, we investigated the association between HLA class II genes and antibody response after IJEV administration, determining that HLA-DQB1*02:01 and HLA-DQB1*02:02 were associated with NAb seroconversion. Furthermore, certain HLA-DRB1 and -DPB1 alleles were associated with higher GMTs than others. The present study suggests that HLA class II genes may influence the antibody response to IJEV. However, as only 161 individuals were examined in the present study, future studies should comprehensively analyze larger samples.
Ethics Statement
This study was carried out in accordance with the recommendations of the ethical standards of the Responsible Committee on Human Experimentation of the Ethics Committee of the Guangxi Centre for Disease Control and Prevention, with written informed consent obtained from all subjects in accordance with the Declaration of Helsinki. The protocol was approved by the Inner Mongolia Autonomous Region Center for Disease Control and Prevention.
Author Contributions
MS and LiS: conceived and designed the experiments. YY, HY, LeS, SL, and CL: performed the experiments. YY, JC, and ZZ: data analysis. MS and LiS: manuscript writing.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31570918) and the Special Funds for High-Level Health Talents of Yunnan Province (D-201669 and L-201615). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the participation of the patients and control subjects in this study. Appreciation is also owed to the physicians and nursing staff at the Center for Disease Control and Prevention of the Inner Mongolia Autonomous Region.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00428/full#supplementary-material
References
1. Hegde NR, Gore MM. Japanese encephalitis vaccines: Immunogenicity, protective efficacy, effectiveness, and impact on the burden of disease. Hum Vaccin Immunother. (2017) 13:1–18. doi: 10.1080/21645515.2017.1285472
2. Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, et al. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ. (2011) 89:766–74. doi: 10.2471/BLT.10.085233
3. C Centers for Disease and Prevention. Japanese encephalitis surveillance and immunization–Asia and the Western Pacific, 2012. MMWR Morb Mortal Wkly Rep. (2013) 62:658–62.
4. McArthur MA, Holbrook MR. Japanese encephalitis vaccines. J Bioterror Biodef. (2011) S 1:2. doi: 10.4172/2157-2526.S1-002
5. Jelinek T. Ixiaro: a new vaccine against Japanese encephalitis. Expert Rev Vaccines. (2009) 8:1501–11. doi: 10.1586/erv.09.112
6. Yun SI, Lee YM. Japanese encephalitis: the virus and vaccines. Hum Vaccin Immunother. (2014) 10:263–79. doi: 10.4161/hv.26902
7. Markoff L. Points to consider in the development of a surrogate for efficacy of novel Japanese encephalitis virus vaccines. Vaccine. (2000) 18 Suppl 2:26–32. doi: 10.1016/S0264-410X(00)00038-4
8. Van Gessel Y, Klade CS, Putnak R, Formica A, Krasaesub S, Spruth M, et al. Correlation of protection against Japanese encephalitis virus and JE vaccine (IXIARO®;) induced neutralizing antibody titers. Vaccine. (2011) 29:5925–31. doi: 10.1016/j.vaccine.2011.06.062
9. Gao X, Li X, Li M, Fu S, Wang H, Lu Z, et al. Vaccine strategies for the control and prevention of Japanese encephalitis in Mainland China, 1951-2011. PLoS Negl Trop Dis. (2014) 8:e3015. doi: 10.1371/journal.pntd.0003015
10. Kollaritsch H, Paulke-Korinek M, Dubischar-Kastner K. IC51 Japanese encephalitis vaccine. Expert Opin Biol Ther. (2009) 9:921–31. doi: 10.1517/14712590903042282
11. Luo D, Yin H, Xili L, Song J, Wang Z. The efficacy of Japanese encephalitis vaccine in Henan, China: a case-control study. Southeast Asian J Trop Med Public Health. (1994) 25:643–6.
12. Sohn YM, Park MS, Rho HO, Chandler LJ, Shope RE, Tsai TF. Primary and booster immune responses to SA14-14-2 Japanese encephalitis vaccine in Korean infants. Vaccine. (1999) 17:2259–64. doi: 10.1016/S0264-410X(99)00006-7
13. Ohrr H, Tandan JB, Sohn YM, Shin SH, Pradhan DP, Halstead SB. Effect of single dose of SA 14-14-2 vaccine 1 year after immunisation in Nepalese children with Japanese encephalitis: a case-control study. Lancet. (2005) 366:1375–8. doi: 10.1016/S0140-6736(05)67567-8
14. Vashishtha VM, Choudhury P, Kalra A, Bose A, Thacker N, Yewale VN, et al. Indian Academy of Pediatrics (IAP) recommended immunization schedule for children aged 0 through 18 years, India, 2013 and updates on immunization. Indian Pediatr. (2013) 50:1095–108. doi: 10.1007/s13312-013-0292-9
15. Khalil-Daher I, Boisgerault F, Feugeas JP, Tieng V, Toubert A, Charron D. Naturally processed peptides from HLA-DQ7 (alpha1*0501-beta1*0301): influence of both alpha and beta chain polymorphism in the HLA-DQ peptide binding specificity. Eur J Immunol. (1998) 28:3840–9. doi: 10.1002/(SICI)1521-4141(199811)28:11<3840::AID-IMMU3840>3.0.CO;2-T
16. Klein J, Sato A. The HLA system. First of two parts. N Engl J Med. (2000) 343:702–9. doi: 10.1056/NEJM200009073431006
17. Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, Poland GA. Human leukocyte antigen class II alleles and rubella-specific humoral and cell-mediated immunity following measles-mumps-rubella-II vaccination. J Infect Dis. (2005) 191:515–9. doi: 10.1086/427558
18. Ovsyannikova IG, Dhiman N, Jacobson RM, Poland GA. Human leukocyte antigen polymorphisms: variable humoral immune responses to viral vaccines. Expert Rev Vaccines. (2006) 5:33–43. doi: 10.1586/14760584.5.1.33
19. C. P. Commission. Pharmacopoeia of the People's Republic of China. In: Japanese Encephalitis Vaccine(Vero Cell), Inactivated, Freeze-dried. Ed C. P. Commission. Bejing: China Medical Science Press (2015), 170–5.
20. Fan Y, Song YQ. PyHLA: tests for the association between HLA alleles and diseases. BMC Bioinformatics. (2017) 18:90. doi: 10.1186/s12859-017-1496-0
21. Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G. PyPop update–a software pipeline for large-scale multilocus population genomics. Tissue Antigens. (2007) 69 (Suppl. 1):192–7. doi: 10.1111/j.1399-0039.2006.00769.x
22. Lancaster A, Nelson MP, Meyer D, Thomson G, Single RM. PyPop: a software framework for population genomics: analyzing large-scale multi-locus genotype data. Pac Symp Biocomput. (2003) 8:514–25. doi: 10.1142/9789812776303_0048
23. Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. (1992) 48:361–72. doi: 10.2307/2532296
24. Poland GA, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Jacobson RM. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. OMICS. (2011) 15:625–36. doi: 10.1089/omi.2011.0032
25. Pulendran B. Systems vaccinology: probing humanity's diverse immune systems with vaccines. Proc Natl Acad Sci USA. (2014) 111:12300–6. doi: 10.1073/pnas.1400476111
26. Nielsen CM, Vekemans J, Lievens M, Kester KE, Regules JA, Ockenhouse CF. RTS,S malaria vaccine efficacy and immunogenicity during Plasmodium falciparum. challenge is associated with HLA genotype. Vaccine. (2018) 36:1637–42. doi: 10.1016/j.vaccine.2018.01.069
27. Jafarzadeh A, Bagheri-Jamebozorgi M, Nemati M, Golsaz-Shirazi F, Shokri F. Human leukocyte antigens influence the antibody response to hepatitis B vaccine. Iran J Allergy Asthma Immunol. (2015) 14:233–45.
28. Xu B, Zhu D, Bi Y, Wang Y, Hu Y, Zhou YH. Minimal association of alleles of human leukocyte antigen class II gene and long-term antibody response to hepatitis B vaccine vaccinated during infancy. Vaccine. (2017) 35:2457–62. doi: 10.1016/j.vaccine.2017.03.021
29. Ovsyannikova IG, Schaid DJ, Larrabee BR, Haralambieva IH, Kennedy RB, Poland GA. A large population-based association study between HLA and KIR genotypes and measles vaccine antibody responses. PLoS ONE. (2017) 12:e0171261. doi: 10.1371/journal.pone.0171261
30. Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. (2012) 30:2146–52. doi: 10.1016/j.vaccine.2012.01.038
31. McDermott AB, Cohen SB, Zuckerman JN, Madrigal JA. Human leukocyte antigens influence the immune response to a pre-S/S hepatitis B vaccine. Vaccine. (1999) 17:330–9. doi: 10.1016/S0264-410X(98)00203-5
32. Poland GA, Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, et al. Identification of an association between HLA class II alleles and low antibody levels after measles immunization. Vaccine. (2001) 20:430–8. doi: 10.1016/S0264-410X(01)00346-2
33. Desombere I, Willems A, Leroux-Roels G. Response to hepatitis B vaccine: multiple HLA genes are involved. Tissue Antigens. (1998) 51:593–604. doi: 10.1111/j.1399-0039.1998.tb03001.x
34. Posteraro B, Pastorino R, Di Giannantonio P, Ianuale C, Amore R, Ricciardi W, et al. The link between genetic variation and variability in vaccine responses: systematic review and meta-analyses. Vaccine. (2014) 32:1661–9. doi: 10.1016/j.vaccine.2014.01.057
35. Ovsyannikova IG, Jacobson RM, Ryan JE, Vierkant RA, Pankratz VS, Jacobsen SJ, et al. HLA class II alleles and measles virus-specific cytokine immune response following two doses of measles vaccine. Immunogenetics. (2005) 56:798–807. doi: 10.1007/s00251-004-0756-0
36. Yang C, Wu J, Zhang X, Wen L, Sun J, Cheng Y, et al. Fine-mapping analysis of the MHC region for vitiligo based on a new Han-MHC reference panel. Gene. (2018) 648:76–81. doi: 10.1016/j.gene.2018.01.053
37. Schillie SF, Spradling PR, Murphy TV. Immune response of hepatitis B vaccine among persons with diabetes: a systematic review of the literature. Diabetes Care. (2012) 35:2690–7. doi: 10.2337/dc12-0312
38. Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. (2011) 11:823–36. doi: 10.1038/nri3084
39. Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, et al. Divergent motifs but overlapping binding repertoires of six HLA-DQ molecules frequently expressed in the worldwide human population. J Immunol. (2010) 185:4189–98. doi: 10.4049/jimmunol.1001006
40. Martin MP, Carrington M. Immunogenetics of viral infections. Curr Opin Immunol. (2005) 17:510–6. doi: 10.1016/j.coi.2005.07.012
41. Thorsby E. On the future of HLA. Tissue Antigens. (2011) 78:229–40. doi: 10.1111/j.1399-0039.2011.01770.x
Keywords: human leukocyte antigen class II genes, HLA haplotype, inactivated Japanese encephalitis virus vaccine, antibody immune response, association
Citation: Yao Y, Yang H, Shi L, Liu S, Li C, Chen J, Zhou Z, Sun M and Shi L (2019) HLA Class II Genes HLA-DRB1, HLA-DPB1, and HLA-DQB1 Are Associated With the Antibody Response to Inactivated Japanese Encephalitis Vaccine. Front. Immunol. 10:428. doi: 10.3389/fimmu.2019.00428
Received: 18 July 2018; Accepted: 18 February 2019;
Published: 08 March 2019.
Edited by:
Rino Rappuoli, GlaxoSmithKline, ItalyReviewed by:
Axel T. Lehrer, University of Hawaii at Manoa, United StatesHenry Alexander Stephens, Royal Free London NHS Foundation Trust, United Kingdom
Copyright © 2019 Yao, Yang, Shi, Liu, Li, Chen, Zhou, Sun and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingbo Sun, c21iQGltYmNhbXMuY29tLmNu
Li Shi, c2hpbGkuaW1iQGdtYWlsLmNvbQ==
 Yufeng Yao
Yufeng Yao Huijuan Yang1,2
Huijuan Yang1,2 Chuanying Li
Chuanying Li Jun Chen
Jun Chen Ziyun Zhou
Ziyun Zhou Li Shi
Li Shi