- 1Departments of Pathology-Immunology and Pediatrics, World Health Organization Collaborating Center for Vaccine Immunology, University of Geneva, Geneva, Switzerland
- 2Department of Clinical and Toxicological Analyses, School of Pharmaceutical Sciences, University of São Paulo, São Paulo, Brazil
- 3Laboratory of Immunology, School of Medicine, Heart Institute, University of São Paulo, São Paulo, Brazil
T follicular helper (Tfh) cells have emerged as a critical limiting factor for controlling the magnitude of neonatal germinal center (GC) reactions and primary vaccine antibody responses. We compared the functional attributes of neonatal and adult Tfh cells at the transcriptomic level and demonstrated that the Tfh cell program is well-initiated in neonates although the Tfh gene-expression pattern (i.e., CXCR5, IL-21, BCL6, TBK1, STAT4, ASCL2, and c-MAF) is largely underrepresented as compared to adult Tfh cells. Importantly, we identified a TH2-bias of neonatal Tfh cells, with preferential differentiation toward short-lived pre-Tfh effector cells. Remarkably, adjuvantation with CpG-ODNs redirect neonatal pre-Tfh cells toward committed GC-Tfh cells, as illustrated by increased expression of Tfh signature genes and reduced expression of TH2-related genes.
Introduction
Neonates and young infants share a high vulnerability to infectious diseases. Inducing efficient and sustained B-cell responses remains challenging in this age group (1, 2). Numerous factors concur to limit primary antibody responses, including delayed follicular dendritic cell maturation (3), the limited development and expansion of T follicular helper (Tfh) cells and, as a result, that of germinal center (GC) B cells and plasma cells (4, 5).
Tfh cell differentiation is a multifactorial, multistep process as illustrated by the extensive list of transcription factors [including BCL6 (6–8), ASCL2 (9), LEF-1 (10, 11), TCF-1 (10, 11), BATF (12), STAT3 (13–15), NFAT (16), IRF4 (17), and c-MAF (18)] playing critical and non-redundant roles in driving Tfh cell differentiation, from the initiation of their development to their maintenance [reviewed in (19)]. The expression of CXC chemokine receptor 5 (CXCR5) through regulation of KLF2 (20, 21) dictates their spatiotemporal distribution, allowing them to migrate in the B cell zone toward CXCL-13 and entering the B cell follicles (22). Along with CXCR5, co-expression of ICOS (23, 24), PD-1 and IL-21 (25, 26) orchestrates Tfh cell differentiation and function. Notably, ICOSL/ICOS signaling plays an important role early in the Tfh cell differentiation program by down-regulating negative regulator molecules (20, 23, 27, 28), such as Blimp-1 (29), T-bet (30), and CCR7 (31). Within the follicles, Tfh cell development could then be negatively regulated by IL-2 (32) and CTLA-4 (33, 34). Once these checkpoints are crossed, cognate Tfh-B cell interactions take place for completing Tfh cell differentiation.
Over the past years, the specific role of Tfh cells as the main providers of B cell help has been unveiled, highlighting a critical role of Tfh cells in vaccine elicited immune responses. We previously demonstrated that Tfh cell development limits early life GC reactions and resulting primary vaccine antibody responses (4). Notably, adjuvantation of a vaccine with CpG-ODNs was sufficient to partially enhance neonatal antibody responses (4). In this study, we compared the transcriptional profile of neonatal and adult Tfh cells and demonstrated that the preferential neonatal polarization toward TH2 is also observed among Tfh cells, with increased expression of IL-13 and other TH2-related factors, which may represent an additional negative regulation checkpoint confining activated neonatal CD4+ T cells at a pre-Tfh stage. Importantly, we showed that adjuvantation with CpG-ODNs reduced the expression of IL-13 and other TH2-related genes and sufficiently strengthened the levels of Tfh cell-associated signature molecules to drive the full completion of GC-Tfh differentiation.
Materials and Methods
Mice
C57BL/6 mice were purchased from Charles River (L'Arbresle, France), bred, and kept in pathogen-free animal facilities in accordance with local guidelines. Mice were used at 1 week (neonates) or 6–8 weeks (adults) of age. All animal experiments were approved by the Geneva Veterinary Office and conducted under relevant Swiss and European guidelines.
Antigens, Adjuvants, Immunization
Groups of C57BL/6 mice (5–16 mice/group) were immunized intramuscularly with Tetanus Toxoid (TT; 1 limit of flocculation (Lf); gift from Berna Biotech, Bern, Switzerland) adsorbed to aluminum hydroxide [AlOH; Former Novartis Vaccines, Siena, Italy (a GSK Company)] or, when indicated, AlOH plus addition of CpG1826 (CpG-ODNs) oligonucleotides (Eurofins MWG Operon). The adult dose of AlOH was weight adjusted to 0.3 mg/adult or 0.15 mg for immunization of 1 week-old mice. The dose of CpG was weight-adjusted as previously described (3) to 50 μg/adult or 3 μg for 1 week-old mice.
Semiquantitative Real-Time PCR
Total cellular RNA was isolated by RNeasy microkit (Qiagen). cDNA was synthesized from 0.5 μg of total RNA using a mix of random hexamers–oligo d(T) primers and PrimerScript reverse transcriptase enzyme (Takara bio inc. Kit) and pre-amplification was performed with TaqMan® PreAmp Master Mix following supplier's instructions (Applied Biosystems). PCR reactions (10 μl volume) contained diluted cDNA, 2× Power SYBR Green Master Mix (Applied Biosystems), 300 nM of forward and reverse primers. RT-PCRs were performed on a SDS 7900 HT instrument (Applied Biosystems). Each reaction was performed in three replicates, with EEf1, GusB, and MmRPS9 as internal controls genes for data normalization. Raw Ct values obtained with SDS 2.2 (Applied Biosystems) were imported in Excel and normalization factor and fold changes were calculated using the GeNorm method (35). Primer sequences are as follows: EEf1 sense, 5′-TCCACTTGGTCGCTTTGCT-3′; anti-sense, 5′-CTTCTTGTCCACAGCTTTGATGA-3′, gusB sense, 5′-ACGGGATTGTGGTCATCGA-3′; anti-sense, 5′-TGACTCGTTGCCAAAACTCTGA-3′, MmRPS9 sense, 5′-GACCAGGAGCTAAAGTTGATTGGA-3′; anti-sense, 5′-TCTTGGCCAGGGTAAACTTGA-3′, s1pr2 sense: 5′-TAACTCCCGTGCAGTGGTTTG; anti-sense: 5′-AGAGCGTGATGAAGGCGG-3′, IL-13 sense: 5′-ACAGGACCCAGAGGATATTGCA; antisense: 5′-GGGAGGCTGGAGACCGTAGT-3′, RXRA sense: 5′-AACACAAGTACCCTGAGCAGCC-3′; antisense: 5′-AGGCGGAGCAGCAGCTT-3′, CCR2 sense: 5′-AGAATTGAACTTGAATCATCTGCAA-3′ antisense: 5′-TGTCTTCCATTTCCTTTGATTTGTT-3′, IL7R sense: 5′-AAATGCCCAGGATGGAGACC-3′; antisense: 5′-AAGGAGTGATCGTCCGCGT-3′, and Bcl6 (36), CXCR5 (37), IL-21 (38), Ascl2 (9), c-maf (39), Pou6f1 (40), and PPAR-γ (41) as stated previously.
Microarray and Analysis
One week-old (16 mice/group) and adult C57BL/6 mice (5 mice/group) were immunized i.m. as described above. Ten days post-vaccination, inguinal draining LNs (dLNs) were pooled per mouse and per group to have a sufficient number of cells. Tfh cell populations were isolated by flow-cytometry cell sorting using a MoFlo® Astrios™ flow cytometer (Beckman Coulter). Six independent experiments have been performed to obtain three independent samples per age group. Total RNA was labeled and hybridized on Agilent Whole Mouse Genome Oligo Microarrays 8 × 60 K at Miltenyi Biotec (Germany) and according to the manufacturer's protocol. Arrays were scanned with the Agilent microarray scanner and raw intensities were extracted with Feature Extraction v10.6 software. Raw intensities were integrated, background corrected and log transformed, following the quantile normalization between arrays. Intensities with detection p-values <0.01 were arbitrarily discarded. Differentially expressed genes (DEGs) were identified by the ANOVA with Tukey post-hoc test considering adjusted p-value ≤ 0.05 and fold-change (FC) ≥ 2. Protein-protein interaction networks were built with DEGs using the NetworkAnalyst program (42) and the InnateDB PPIs as database (43). Enrichment analyses were performed with the program Gene Set Enrichment Analysis (GSEA) (44), using customs gene sets of upregulated genes from CD4+ Tfh effector cells [GSE43863 (45)] and from Bcl6+ Tfh cells or Bcl6− Tfh cells [GSE40068 (8)]. First, raw expression data from GSE43863 and GSE40068 studies were normalized by RMA using the affy R/Bioconductor package (46), and submitted to quality control with the arrayQualityMetrics R/Bioconductor package (47). For both studies, the up-regulated genes were identified using the R/Bioconductor LIMMA package (48). The Tfh effector signature (GSE43863) was generated by comparing CD4+ Tfh effector cells compared with naïve and TH1 CD4+ T cells (adjusted p-value < 0.005 and FC ≥ 1.5), while the Tfh Bcl6+ and Bcl6− signatures (GSE40068) were generated by comparing CD4+ CXCR5+ Bcl6+ and CD4+ CXCR5+ Bcl6− T cells with CD4+ CXCR5− T cells (adjusted p-value < 0.05 and FC ≥ 2). Co-expression modules were identified with the CEMiTool R/Bioconductor package (49) using variance filter p-value < 0.05 and ORA p-value < 0.2. CEMiTool package is available at Bioconductor (https://bioconductor.org/packages/release/bioc/html/CEMiTool.html) (49). This package unifies the discovery and the analysis of coexpression gene modules, evaluating whether modules contain genes that are over-represented by specific pathways or that are altered in a specific sample group. Biological and functional enrichment analyses were also performed with the program GSEA using the REACTOME gene sets (50). Finally, unsupervised hierarchical clustering of the samples was carried out via multiscale bootstrap resampling with the PVCLUST R package (51).
Results
Transcriptional Profile of Neonatal Tfh Cells
We (4, 52) and others (5) have shown that neonatal Tfh cells elicited by aluminum (AlOH)—based adjuvanted vaccines are few and functionally altered compared to adult cells. We therefore investigated the functional attributes of neonatal and adult CD4+ CXCR5highPD-1high Tfh cells at the transcriptomic level. CD4+ CXCR5highPD-1high Tfh and CD4+ CXCR5−PD-1− T (non-Tfh) cells were FACS sorted from the draining lymph nodes (LNs) at the previously identified peak (day 10) of the primary germinal center (GC) reaction induced by TT/AlOH (4) for comparative transcription profile analysis (Figure 1A). To visualize the global gene expression patterns of the various subsets, we first performed a principal component analysis (PCA), retaining the top 2,000 genes that contributed most to the total variance (Figure 1B). The projection of the data variance onto the principal components plane efficiently discriminated Tfh cells from non-Tfh cells in both age groups (Figure 1B), while clustering adult and neonatal Tfh cells together. This was confirmed by unsupervised hierarchical analysis, which grouped Tfh cells from both age groups (Figure S1). Thus, when successful the Tfh differentiation process essentially follows a similar path in early as in adult life.
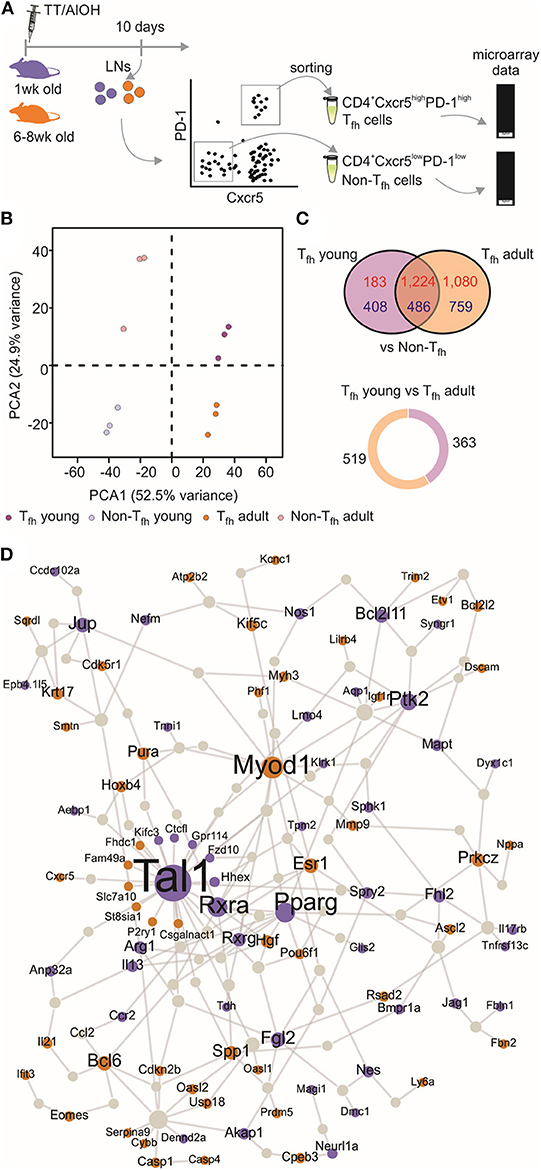
Figure 1. Transcriptional profile of neonatal Tfh cells. (A) One week-old and adult C57BL/6 mice (5–16 mice/group) were immunized i.m. with TT/AlOH. Ten days post-vaccination the two draining inguinal LNs were collected and pooled to simultaneously isolate CD4+ CXCR5highPD-1high Tfh cells and CD4+ CXCR5−PD-1− T cells (non-Tfh) by flow-cytometry cell sorting. The cells obtained from the inguinal draining LNs of either 16 neonates/group or 5 adults/group per experiment were pooled before sorting to recover a sufficient number of cells for experimentation. (B) Principal Component Analysis, based on the top 2,000 genes with highest variance, showing (dis)similarities in gene expression across all samples; 1 week (purple, n = 3 independent experiments including pools of 16 mice) or adult (brown, n = 3 independent experiments including pools of five mice) CD4+ CXCR5highPD-1high Tfh cells and respective controls CD4+ CXCR5−PD-1− T (non-Tfh) cells (light colors). (C) Venn diagram illustrating the overlap of differentially expressed genes between Tfh vs. non-Tfh in young and adult immunized mice. Up-regulated genes in Tfh cells are shown in red and down-regulated genes in blue. Pie chart shows the proportion of genes differentially up-regulated by 1 week-old Tfh cells (purple) when compared to adults, and in brown genes differentially up-regulated by adult Tfh cells compared to 1 week-old. (D) Protein-protein interaction network constructed with the differentially expressed genes in 1 week-old Tfh cells as compared to adults. Up-regulated genes in neonates are illustrated in purple while brown indicates genes up-regulated in adults.
Nevertheless, the gene expression profiles of neonatal and adult Tfh samples differed, revealing functionally differently programmed Tfh cells (Figure 1C). Comparing Tfh cells from neonatal and adult mice with the corresponding age-matched non-Tfh cells identified 2,301 and 3,549 differentially expressed genes, respectively. Overlap comparison showed that 1,710 genes were differentially expressed in Tfh cells of both neonatal and adult immunized mice, 591 genes were exclusively differentially expressed in neonatal Tfh cells, and 1,839 genes were exclusively differentially expressed in adult Tfh cells (Figure 1C).
To get more insight into the key genes leading to functionally differently programmed Tfh cells in early or adult life, PPI networks were generated from the differentially up-regulated genes between neonatal and adult Tfh cells (Figure 1D). The network derived from the genes differentially expressed in adult Tfh cells showed that most of the up-regulated proteins have an established role in Tfh biology and function (Bcl6, Ascl2, Pou6f1, IL-21, and Cxcr5). In accordance with the preferential TH2 polarization of early life responses, IL-13 was strongly enriched in neonates vs. adults (Figure 1D). Unexpectedly, three cancer related-pathways genes (Tal1, PPAR-γ, and RXRA) were identified as hub genes in neonates (Figure 1D). Tal1 is expressed early, in hematopoietic stem cells and progenitor cells (53, 54), and subsequently silenced during T-cell development [reviewed in (55)]. It forms a large transcriptional complex with E proteins, LMO family proteins, LDB1, GATA2, and GATA3 (56–58). TAL1, GATA3, and RUNX1 coordinately regulate the expression of downstream target genes. PPAR-γ is a member of the peroxisome proliferator-activated receptor family and forms heterodimer with RXRs to promote their downstream effects, i.e., suppress the transcription of target genes (59). Remarkably, in adults both PPAR-γ (41) and RXRA (60) negatively regulate T cell activation to prevent Tfh cell formation. These hub genes may thus play an essential role to functionally alter neonatal Tfh cell differentiation.
Tfh Cell Differentiation Is Initiated in Neonates but Tfh Cells Remain Lodged in a pre-Tfh Stage
We then selected the Tfh signature genes from published data sets to perform gene set-enrichment analyses (GSEA) with our data. This confirmed that both neonatal and adult cells were enriched for the Tfh lineage gene set (GEO accession code GSE43863) (Figure 2A), indicating that the Tfh cell differentiation program may succeed in neonates. However, neonatal Tfh Bcl6+ cells exhibited reduced gene expression signatures compared to adults, while using another GSEA (accession code GSE40068) indicated increased Tfh Bcl6− signatures (Figure 2A). Interestingly, Liu et al. demonstrated (8) that CXCR5+ Bcl6low cells develop before CXCR5+ Bcl6high cells and exhibit a non-polarized gene expression pattern. These “intermediate” Tfh cells then further mature into CXCR5+Bcl6high Tfh cells with the help of cognate B cells (8). This suggests that most neonatal Tfh cells are arrested at an early/intermediate stage of Tfh development, only a fraction of activated T cells fully up-regulating their expression of key Tfh genes, while maintaining their expression of IL-13, one of the preferentially expressed neonatal TH2-related cytokine gene.
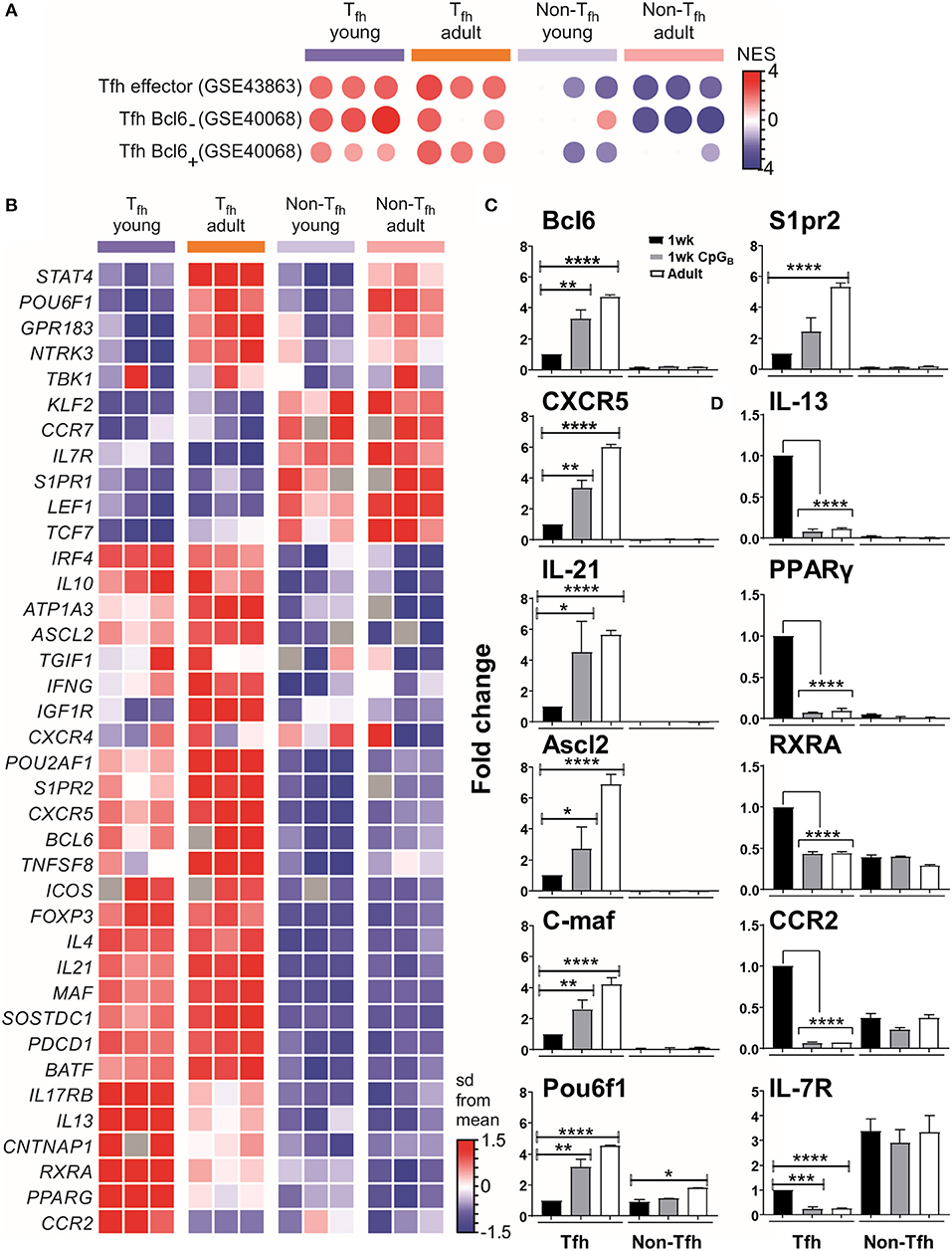
Figure 2. Tfh cell differentiation is initiated in neonates but most of the generated cells are lodged in a pre-Tfh stage. (A) Enrichment analysis of neonatal vs. adult Tfh cells with the gene list from Tfh effector cells or Bcl6 + and Bcl6− Tfh cells. (B) Sample z-score heatmap of significantly differentially expressed genes (row) in Tfh cells and their respective controls from young and adult mice (columns). Semi-quantitative RT-PCR analysis of selected Tfh cell-related genes (C) or TH2 cell-related genes in sorted cells (D), normalized to results obtained for the control genes (EEF1, GusB, RPS9). The graph display mean ± SEM. Cumulative data from adult [TT/AlOH (n = 10)] and 1 week-old [TT/AlOH (n = 64), TT/AlOH+ CpG1826 (n = 32)] mice from at least two independent experiments. Statistical analysis was performed with Prism software (Version 7, GraphPad), using unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further analyze the transcriptional differences among early life and adult Tfh cells, we examined the expression of a set of genes described as up- or down- regulated in Tfh cells compared to non-Tfh CD4+ helper T cells (8, 12, 13, 61, 62). The heatmap of the differentially expressed genes (DEGs) (Figure 2B) confirmed that the gene expression of neonatal and adult Tfh samples differed from the control CD4+ CXCR5−PD-1− samples. Although the overall Tfh signature was again present in neonatal Tfh cells, the expression levels of Cxcr5, Batf, c-maf , and Il-21 were lower than in adult Tfh cells, strengthening our previous observations (4). Compared to adult Tfh cells, neonatal Tfh cells exhibited much lower expression of Bcl6 (8, 29, 63, 64) and achaete-scute homolog 2 (Ascl2) (9), which controls CXCR5 expression and thus the follicular positioning of pre-Tfh cells (9). The TBK1 kinase, which controls the maintenance of Bcl6 expression and is thus required for the commitment to the GC-Tfh program (27), was also less strongly expressed in neonatal Tfh cells.
Bcl6 can bind to promoters and enhancers of genes that encode proteins that control T cell-migration, promoting non-follicular positioning of T cells (7). IL-7R is one of the most repressed Tfh-relevant genes by Bcl6 and its suppression is critical in Tfh cell differentiation (65). Although IL-7R is down-regulated in neonatal Tfh cells, its inhibition is almost two times weaker than in adult Tfh cells (expression level change of 1.89-fold greater in neonates compared to adults). Liu et al. (65) recently demonstrated that IL-7R expression was inversely correlated with Tfh commitment, more precisely with the expression of classical Tfh markers: PD-1, CXCR5, and Bcl6. A limited follicular positioning of neonatal pre-Tfh cells is also supported by the decreased expression (expression level change of −1.92-fold in neonates compared to adults) of S1pr2, known to suppress CXC12/CXCL13-mediated migration, thus restricting premature egress of Tfh cells out of GC (66). Thus, numerous transcription factors contribute to prevent the follicular positioning of neonatal Tfh cells, depriving them from interacting with follicular dendritic cells (FDCs) and germinal center B (GC B) cells.
The expression of the Pou2af1 and Pou6f1 transcription factors was also reduced. Although their role in Tfh cells remains to be fully investigated, Pou6f1 is expressed in early fate committed Tfh cells (6) and Pou2af1 is highly expressed in early stage GC-Tfh cells (10, 67). A recent report by Stauss et al. (68) established that the Pou2af1 gene promotes Bcl6 expression and Tfh cell development. A general reduction in CXCR5 expression was observed on Pou2af1−/− CD4+ T cells as well as fewer GL7+ Tfh cells in Pou2af1−/− mice (68). Therefore, the POU family transcription factors seems to fine-tune Tfh cell development and their reduced expression in neonates may contribute to the limited expression of CXCR5 and GL7 in neonatal Tfh cells (4).
The lower expression of STAT4 (expression level change of −2.22-fold in neonates compared to adults) in neonates cements that the specific early life environment prevents the differentiation of pre-Tfh cells toward committed GC-Tfh cells: the IL-12-STAT4 pathway indeed contributes to the expression of key Tfh-associated molecules, such as IL-21, CXCR5, and ICOS as well as multiple important transcription factors involved in Tfh-cell generation, such as Bcl6, c-Maf, and Batf (69, 70).
How does the TH2-like preferential polarization of neonatal effector T cells persist in Tfh cells?
Although, TH2 signature genes, including GATA3, IL-4, and IL-5, were not differentially expressed in neonatal Tfh cells (Table S1), we observed significant changes in IL-13 and PPAR-γ. Nobs et al. recently showed that PPAR-γ expression in T cells controls the development of type-2 immunity (71). Therefore, the increased expression of PPAR-γ and of additional genes associated with TH2 polarization, including RXRA, ccr2 (72), il17rb (73–75), and cntnap1 (75), may all play a role in maintaining the default TH2-bias of neonatal Tfh cells.
Semiquantitative RT-PCR analyses confirmed both the reduced transcript abundance of Tfh-cell-associated signature genes in neonates (black bars) compared to adults (open bars) (Figure 2C and Figure S2), and their preferential bias toward TH2, as shown by the higher levels of IL-13, PPAR-γ, RXRA, and CCR2 (Figure 2D).
Adjuvantation With CpG1826 Bypasses the Neonatal TH2-Bias of pre-Tfh Cells and Supports Terminal GC-Tfh Cell Differentiation
We and others have shown that administration of TT/AlOH supplemented with TLR9 agonist CpG1826 enhanced neonatal antibody responses through the induction of higher Tfh and GC B cell numbers (4), as observed in adult mice (76, 77). We thus asked whether neonatal CpG1826 adjuvantation induced transcriptional changes in the genes/factors identified as differing between neonatal and adult Tfh cells. Semiquantitative RT-PCR on FACS-sorted CD4+ CXCR5highPD-1high Tfh cells isolated 10 days after TT/AlOH + CpG1826 immunization showed that CpG adjuvantation increased the transcriptional abundance of Tfh-cell specific signature genes in neonatal Tfh cells (Figure 2C and Figure S2). Flow cytometry analyses confirmed significantly lower CXCR5 expression by neonatal Tfh cells (4) (Figure S3A). Remarkably, CpG adjuvantation significantly enhanced the expression of CXCR5 on neonatal Tfh cells (Figure S3A), although not to adult like levels. The results support a follicular positioning of neonatal Tfh cells, facilitating the Tfh-GC B cell crosstalk required to provide B cell help during the GC reaction. In contrast, the expression of PD-1 was significantly decreased in 1 week-old and adult mice immunized with CpG-ODNs (Figure S3B).
Notably, TH2-related genes (i.e., PPAR-γ, RXRA, IL-13, and CCR2) were significantly reduced in neonatal Tfh cells, reaching similarly low levels as in adult mice immunized without CpG-ODNs (Figure 2D). However, similar to our previous observation (4), IL-4 mRNA transcripts were significantly lower in neonatal Tfh cells and were not affected by CpG adjuvantation (Figure S2B), suggesting that IL-17 transcription would also remain unaffected as previously demonstrated by Debock et al. (5).
Thus, CpG adjuvantation may (1) abrogate the regulation of early life Tfh cell differentiation exerted by the TH2-related genes PPAR-γ and RXRA, (2) facilitate the follicular positioning of neonatal Tfh cells, as mirrored by increased levels of S1pr2 and CXCR5 and by reduced IL-7R, and (3) support neonatal Tfh cell differentiation toward committed GC-Tfh cells. This explains our previous observations demonstrating the increase of Tfh cell numbers, of GL7 expression by neonatal Tfh cells, of GC reactions and thus of Ab titers following neonatal CpG-ODNs adjuvantation (4) (Figures S3C,D).
Neonatal Tfh Cells Preferentially Give Rise to Short-Lived Effector Cells
To better understand the fate of the pre-Tfh cells elicited in early life, we next ran the Co-Expression Molecules identification Tool (CEMiTool) (49) on our data set. CEMiTool is an R package that provides in an automated manner unsupervised gene filtering, automated parameter selection for identifying modules, enrichment and module functional analyses as well as integration with interactome data (49). This modular expression analysis identified 6 different co-expression modules (Figure 3A and Data Sheet S1), of which only module M2 was significantly enriched in neonates (Figure 3B). This module was enriched for genes related to cell cycle (49) (Figure 3C), suggesting the capacity of neonatal Tfh cells to enter the cell cycle more rapidly than their adult counterparts. CEMITool also integrates co-expression analysis with protein-protein interaction data. Expression of important genes associated with cell-cycle progression, including gene encoding E2F1 and TK1, were identified as hubs in module M2 (Figure 3D). This early life characteristic was previously observed (78–80): neonatal T and B lymphocytes have the capacity to enter the cell cycle more quickly and thus efficiently mobilize responses from an otherwise completely naive population—possibly to compensate for the limitations in immune cell function in early life (i.e., lack of immunological memory) (78–80). Yet, rapid cycle entry only gives rise to short-lived effector cells (78–80).
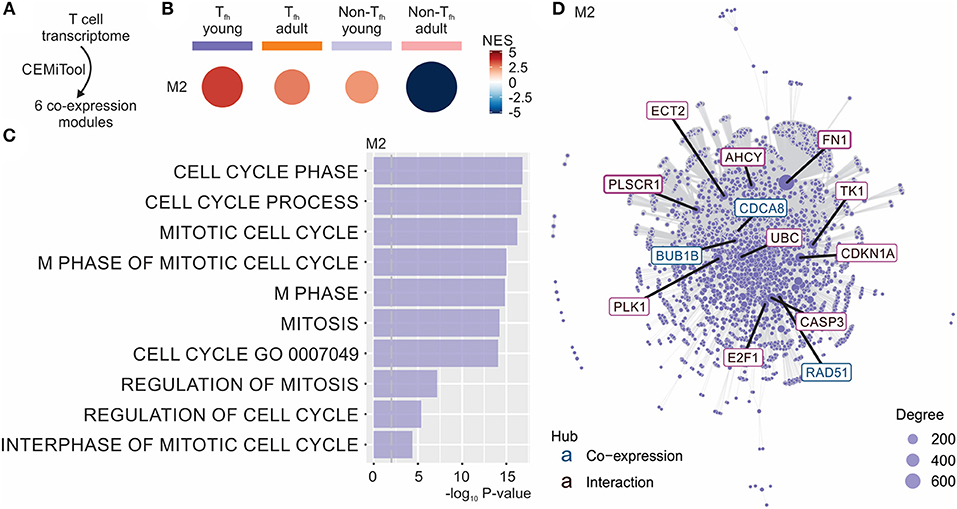
Figure 3. CEMiTool identified co-expression modules in our Tfh dataset. (A) CEMiTool was run on our Tfh dataset and identified 6 co-expression modules. (B) Gene Set Enrichment Analyses showing the M2 module activity on each class of samples. NES, normalized enrichment score. The size and color of the circle represents the normalized enrichment score (NES). (C) Over Representation Analysis of modules M2 using gene sets from the Reactome Pathway database. Bar graph shows the –log10 adjusted P-value of the enrichment between genes in modules and gene sets from Reactome Pathway database. The pathways were ordered by significance as indicated in the x-axis. The vertical dashed gray line indicates an adjusted P-value of 0.01. (D) Interaction plot for M2, with gene nodes highlighted. The nodes represent the 413 genes of M2 plus the genes added by protein-protein interaction information. The genes are connected by co-expression and/or protein-protein interaction. Gene network of module M2 for the most connected genes (hubs) are labeled and colored based on their “origin”: if originally present in the CEMiTool module, they are colored blue; if inserted from the interactions, they are colored red. The size of the node is proportional to its degree.
To complete our observations, we performed a pathway analysis which revealed cell-intrinsic differences between neonatal and adult Tfh cells (Figure 4): neonatal Tfh cells were enriched in pathways associated with cell proliferation, apoptosis and key metabolic reactions, such as glycolysis, considered to play an important role in T cell activation and differentiation, while adult cells were enriched in mitogen-activated protein kinase (MAPK)-signaling pathways, thus outlining age-associated differences in the maturity and basic function. Interestingly, enrichment of Hedgehog signaling, which predispose T cell differentiation toward the TH2 pathway (81), further supports the TH2-bias of neonatal Tfh cells. Altogether, these transcriptional analyses of neonatal vs. adult Tfh cells reveal the existence of multiple coordinated regulatory mechanisms resulting into the preferential differentiation of neonatal CD4+ T cells toward innate, short-lived pre-Tfh effectors rather than adaptative (GC-derived) immunity defense mechanisms.
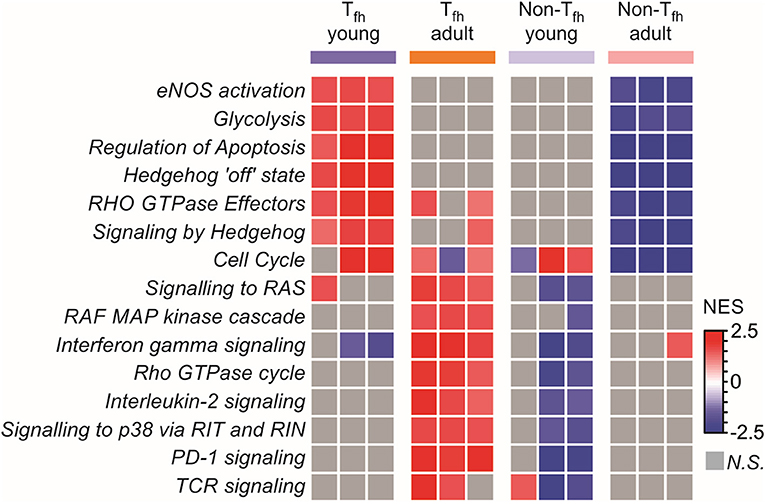
Figure 4. Pathway analysis reveals cell intrinsic differences between neonatal and adult Tfh cells. Gene Set enrichment analysis using Reactome gene sets. Heat map showing the normalized enrichment scores for the pathways consistently induced or repressed in a Tfh young or adult population are illustrated by color.
Discussion
We previously identified the induction of Tfh cells as limiting early life GC and Ab responses elicited by vaccines including aluminum-based adjuvants (4). We now demonstrate that the few Tfh cells elicited in early life retain a preferential bias toward TH2, strongly expressing IL-13, and PPAR-γ and RXRA which negatively regulate Tfh cell differentiation (41, 60), and that numerous transcription factors contribute to restrict activated neonatal CD4+ T cells at a pre-Tfh cell stage of short-lived effectors favoring innate rather than GC-associated adaptive responses. Importantly, we show that this fate is not inevitable as adjuvantation with CpG-ODNs reduced the expression of TH2-related genes and sufficiently strengthened the Tfh cell-associated signature molecules to drive the GC-Tfh differentiation program to its completion and fine tune the GC reaction.
Following immune challenges, neonatal responses are often weak (82). This has been associated with a propensity of neonatal T cells to give rise to short-lived effector cells (78–80) and to produce elevated levels of TH2-type cytokines compared to adults (83–85). We show that these two key neonatal characteristics persist during Tfh cell differentiation, lodging the cells in a pre-Tfh stage characterized by a TH2 bias. Our results suggest that a delicate balance of several signals known to promote TH2 development may contribute in maintaining an optimal environment for the TH2-biased Tfh cell differentiation in neonates, including increased expression of PPAR-γ (71, 86), RXRA (87), ccr2 (72), il17rb (73–75), cntnap1 (75), Hedgehog signaling (81), and lower levels of c-maf mRNA transcripts. A critical role for c-maf in limiting TH2 responses and in driving Tfh cell development was recently unveiled by Andris et al. (18). Further investigations are warranted to delineate whether Tal1 may also play a fundamental role in the generation and persistence of TH2-biased Tfh cells in neonates.
A limitation of our study is that the very few Tfh cells induced in early life (about 2 × 104 Tfh cells from a pool of 8 neonates) and thus the small amount of recovered RNA precluded the analysis of all potentially interesting genes. As the microarray did not reveal significant changes in TH17-related genes, in GATA3 or in IL-5 expression in neonatal Tfh vs. non-Tfh cells (Table S1), we did not compare their mRNA transcript levels to those of adult cells. In our model, both IFN-γ and IL-4 mRNA transcript levels are significantly lower in neonatal Tfh cells—and not affected by CpG adjuvantation (Figure S2B). The similar expression of Foxp3 in neonatal and adult Tfh cells was confirmed by semiquantitative RT-PCR (Figure S2D). Altogether, these results suggest that in our model, neonatal Tfh cells do not exhibit a bias toward TH1, TH17 nor Treg cells. Our attempts to develop validated assays to reliably measure several proteins in the few recovered neonatal Tfh cells did not succeed, and such proteins remained below detection levels when assessed in lymph nodes homogenates (not shown).
PPAR-γ and RXRA were identified as critical hub genes in neonates. Therefore, besides their role in maintaining the overall TH2 bias, PPAR-γ and RXRA may also negatively regulate Tfh cell differentiation. PPAR-γ is known to (71, 86) promote Tregs survival (88–90) and inhibit the formation of Tfh cells and GC reactions via the regulation of Bcl6 and IL-21 (41). That inhibition of Bcl6 expression is illustrated in neonatal Tfh cells by increased expression of the TH2-related gene, IL-13, previously identified as one of the most repressed Bcl6-target gene (65). A role for Tgif1-RXR interaction in the establishment or inhibition of a chronically elevated Tfh cell population was recently computationally predicted and demonstrated by Leber et al. (60); an increase with Tgif1 was associated with an increase in the Tfh response, while an increase in RXR was more closely correlated with the Tfh decline phase (60). Small changes in RXRA, such as 10% change in expression were previously demonstrated to result in a 50% change in activity and significant alteration of downstream transcriptional targets (91). We conclude that the differential expression of TH2-related genes PPAR-γ and RXRA might be involved in the distinct genetic programming of neonatal and adult Tfh cells.
Although the Tfh cell program is well-initiated in neonates, the gene-expression pattern of neonatal Tfh cells underrepresented that of adult Tfh cells, suggesting that T-B interactions fail to elicit appropriate signals and provide efficient help to neonatal pre-Tfh cells to further differentiate into committed GC-Tfh cells. Indeed, Tfh cells differentiation involves a multi-signal process that includes expression of CXCR5, IL-21, Bcl6, TBK1, STAT4, Ascl2, and c-maf which were all expressed to lower levels as compared to adult Tfh cells. Remarkably, adjuvantation with CpG-ODNs, skewed neonatal pre-Tfh cells toward committed GC-Tfh cells, as illustrated by increased expression of Tfh-signature genes (Figure 2C and Figure S2). In parallel, genes associated with follicular positioning of Tfh cells were increased (i.e., s1pr2, Ascl2, and CXCR5), facilitating the cognate Tfh-B cell interactions for completing Tfh cell differentiation (92–96), with concomitant increase in Bcl6 and IL-21 expression (10, 95). Ascl2 directly regulates the localization of Tfh cells via CXCR5 expression and suppression of CCR7 and PSGL1 (9). CXCR5 allows Tfh cells to migrate into the B cell follicles and form stable contacts with antigen-primed B cells (92, 97). These results indicate that a combination of several Tfh-specific signals, in addition to previously described environment factors and CD4+ T cells intrinsic determinants (4), maintain a favorable environment for TH2-biased Tfh cell differentiation, restricting neonatal CD4+ T cells at a pre-Tfh stage of short-lived effector cells. Adjuvantation with CpG-ODNs is sufficient to counteract the TH2-biased response of neonatal Tfh cells, reducing TH2-related genes to adult-like levels, while Tfh signature genes (i.e., Bcl6, CXCR5, IL-21, Ascl2, C-maf, Pou6f1, s1pr2, Batf, CXCR4, and TBK1) are progressively enhanced, resulting in differentiated and GC-committed Tfh cells. As illustrated in Figures 2C,D, the switch from pre-Tfh to mature Tfh cells involves changes in the expression levels of several factors—such that “classical” mechanistic approaches including knockout/knock-in mice were not attempted.
We recently demonstrated that adjuvantation of a vaccine with a liposome including a C-type lectin receptor agonist was able to elicit potent GC reactions in neonates after a single dose (98). Altogether, these results show that immune deficiencies seen in early life can be overcame by providing the right signals, and are in accordance with the current understanding that the neonatal immune system is not deficient but tightly regulated to best adapt to the unique challenge of a rapidly required adaptation from a sterile to a microbial environment (99).
Further studies are necessary to investigate whether abrogating the TH2 bias of Tfh cells in early life is critical for the full commitment of Tfh cell differentiation and the subsequent GC B cell and antibody responses, resulting in effective responses to vaccination in early life.
Data Availability
The data has been deposited at the Gene Expression Omnibus repository—accession number is GSE126843.
Ethics Statement
This study was carried out in accordance with the recommendations of the Geneva Veterinary Office and conducted under relevant Swiss and European guidelines. The protocol was approved by the Geneva Veterinary Office.
Author Contributions
BM-G, P-HL, and C-AS contributed to formulation of theory and prediction. BM-G, P-HL, and C-AS designed the research. BM-G and MV performed the experiments and analyzed and/or interpreted the data. BM-G, MV, and C-AS wrote the manuscript. PG-D, FF, LC, and HN performed the microarray analysis and critically revised the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the Swiss National Science Foundation (grant numbers 310000-111926/1 and 310030-165960 to C-AS) and by research grants of the Center for Vaccinology and Neonatal Immunology. HN was supported by FAPESP (grant numbers: 2012/19278-6 and 2017/50137-3).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Paola Fontannaz, Stéphane Grillet, and Chantal Tougne for their key contribution to the complex experimental work required by this study; Anthony Joubin for excellent assistance with animal care; Anne Rochat for running the real-time PCR; Mylène Docquier and the Genomics Platform for assistance with the real-time PCR; Jutta Kollet for assistance with the Microarray (Miltenyi Biotech GmbH Bioinformatics); Jean-Pierre Aubry-Lachainaye and Cécile Gameiro from the flow cytometry platform.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01845/full#supplementary-material
Figure S1. Unsupervised hierarchical analysis groups together Tfh cells from both age groups. Hierarchical clustering of CD4+ CXCR5highPD-1high Tfh cells and respective controls CD4+ CXCR5−PD-1− T cell samples was obtained by pvclust R package. CD4+ CXCR5highPD-1high Tfh cells from young (purple) and adult (orange) mice and their respective controls (light colors) formed distinct groups.
Figure S2. CpG adjuvantation is sufficient to increase Bcl6 targeted Tfh specific genes, such as Atp1a3 as well as Tfh cell-related genes, including BATF, TBK1, and CXCR4. One week-old and adult C57BL/6 mice (5–8 mice/group) were immunized i.m. with TT/AlOH. Ten days post-vaccination the draining LNs were collected to simultaneously isolate CD4+ CXCR5highPD-1high Tfh cells and CD4+ CXCR5−PD-1− T (non-Tfh) cells by flow-cytometry cell sorting. The cells obtained from the two inguinal draining LNs of either 16 neonates/group or 5 adults/group per experiment were pooled before sorting to recover a sufficient number of cells for experimentation. Semi-quantitative RT-PCR analysis of selected Tfh cell-related genes (A), IL-4 (B), TH1 cell-related genes (C), Foxp3 (D), or Bcl6 targeted Tfh specific genes (E) in sorted cells, normalized to results obtained for the control genes (EEF1, GusB, RPS9). The graph display mean ± SEM. Cumulative data from adult [TT/AlOH (n = 10)] and 1 week-old [TT/AlOH (n = 64), TT/AlOH+ CpG1826 (n = 32)] mice from at least two independent experiments. Fold changes are shown relative to 1 week-old mice immunized with TT/AlOH. Statistical analysis was performed with Prism software (Version 7, GraphPad), using unpaired t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure S3. Progressive increase of TT-specific IgG1 titers and Tfh cell responses in neonates with CpG adjuvantation. C57BL/6 mice were immunized i.m. at 1 week or as adult with TT/AlOH with or without CpG (5–8 mice per group). The adult dose of AlOH was weight adjusted to 0.3 mg/adult or 0.15 mg for immunization of 1 week-old mice. (A) CXCR5 and (B) PD-1 surface marker mean fluorescence intensity (MFI) on Tfh cells from 1 week and adult mice day 10 post-immunization. TT-specific (C) IgG1 or (D) IgG2a titers 10 days post-immunization. Numbers are represented as mean ± SEM. Data are representative of one of three independent experiments. Mann–Whitney U test differences with p > 0.05 were considered to be insignificant. *p-value (0.01–0.05), **p-value (0.001–0.01), ***p-value (<0.001).
Table S1. Summary table of TH1, TH2, TH17, and Treg selected genes. Indicated are the p-values, Median intensity values of all samples, the flag counts and the ProbeID. Fold-change values (columns C–E) which did not pass the selection criteria (Anova P-value ≤ 0.05, Tukey P-value ≤ 0.05, fold-change ≥2 or ≤-2, and reliable detection of the signal (Flag counts) ≤1 in the group with higher expression) are in black color, while up-regulated ones are shown in red and down-regulated ones are shown in green.
Data Sheet s1. CEMiTool output html file for all modules.
References
1. Siegrist CA, Aspinall R. B-cell responses to vaccination at the extremes of age. Nat Rev Immunol. (2009) 9:185–94. doi: 10.1038/nri2508
2. Wood N, Siegrist CA. Neonatal immunization: where do we stand? Curr Opin Infect Dis. (2011) 24:190–5. doi: 10.1097/QCO.0b013e328345d563
3. Pihlgren M, Tougne C, Bozzotti P, Fulurija A, Duchosal MA, Lambert PH, et al. Unresponsiveness to lymphoid-mediated signals at the neonatal follicular dendritic cell precursor level contributes to delayed germinal center induction and limitations of neonatal antibody responses to T-dependent antigens. J Immunol. (2003) 170:2824–32. doi: 10.4049/jimmunol.170.6.2824
4. Mastelic B, Kamath AT, Fontannaz P, Tougne C, Rochat AF, Belnoue E, et al. Environmental and T cell-intrinsic factors limit the expansion of neonatal follicular T helper cells but may be circumvented by specific adjuvants. J Immunol. (2012) 189:5764–72. doi: 10.4049/jimmunol.1201143
5. Debock I, Jaworski K, Chadlaoui H, Delbauve S, Passon N, Twyffels L, et al. Neonatal follicular Th cell responses are impaired and modulated by IL-4. J Immunol. (2013) 191:1231–9. doi: 10.4049/jimmunol.1203288
6. Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. (2013) 190:4014–26. doi: 10.4049/jimmunol.1202963
7. Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med. (2015) 212:539–53. doi: 10.1084/jem.20141380
8. Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. (2012) 209:1841–52, S1–24. doi: 10.1084/jem.20120219
9. Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. (2014) 507:513–8. doi: 10.1038/nature12910
10. Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. (2015) 16:980–90. doi: 10.1038/ni.3226
11. Kubo M. TCF-1 and LEF-1 help launch the T(FH) program. Nat Immunol. (2015) 16:900–1. doi: 10.1038/ni.3254
12. Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. (2011) 12:536–43. doi: 10.1038/ni.2037
13. Eddahri F, Denanglaire S, Bureau F, Spolski R, Leonard WJ, Leo O, et al. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. (2009) 113:2426–33. doi: 10.1182/blood-2008-04-154682
14. Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. (2014) 40:367–77. doi: 10.1016/j.immuni.2014.02.005
15. Wu H, Xu LL, Teuscher P, Liu H, Kaplan MH, Dent AL. An inhibitory role for the transcription factor Stat3 in controlling IL-4 and Bcl6 expression in follicular helper T cells. J Immunol. (2015) 195:2080–9. doi: 10.4049/jimmunol.1500335
16. Vaeth M, Muller G, Stauss D, Dietz L, Klein-Hessling S, Serfling E, et al. Follicular regulatory T cells control humoral autoimmunity via NFAT2-regulated CXCR5 expression. J Exp Med. (2014) 211:545–61. doi: 10.1084/jem.20130604
17. Bollig N, Brustle A, Kellner K, Ackermann W, Abass E, Raifer H, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci USA. (2012) 109:8664–9. doi: 10.1073/pnas.1205834109
18. Andris F, Denanglaire S, Anciaux M, Hercor M, Hussein H, Leo O. The transcription factor c-Maf promotes the differentiation of follicular helper T cells. Front Immunol. (2017) 8:480. doi: 10.3389/fimmu.2017.00480
19. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. (2016) 34:335–68. doi: 10.1146/annurev-immunol-041015-055605
20. Weber JP, Fuhrmann F, Feist RK, Lahmann A, Al Baz MS, Gentz LJ, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. J Exp Med. (2015) 212:217–33. doi: 10.1084/jem.20141432
21. Lee JY, Skon CN, Lee YJ, Oh S, Taylor JJ, Malhotra D, et al. The transcription factor KLF2 restrains CD4(+) T follicular helper cell differentiation. Immunity. (2015) 42:252–64. doi: 10.1016/j.immuni.2015.01.013
22. Chen X, Ma W, Zhang T, Wu L, Qi H. Phenotypic Tfh development promoted by CXCR5-controlled re-localization and IL-6 from radiation-resistant cells. Protein Cell. (2015) 6:825–32. doi: 10.1007/s13238-015-0210-0
23. Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. (2011) 34:932–46. doi: 10.1016/j.immuni.2011.03.023
24. Xu H, Li X, Liu D, Li J, Zhang X, Chen X, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. (2013) 496:523–7. doi: 10.1038/nature12058
25. Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. (2011) 6:e17739. doi: 10.1371/journal.pone.0017739
26. Zotos D, Coquet JM, Zhang Y, Light A, D'Costa K, Kallies A, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. (2010) 207:365–78. doi: 10.1084/jem.20091777
27. Pedros C, Zhang Y, Hu JK, Choi YS, Canonigo-Balancio AJ, Yates JR III, et al. A TRAF-like motif of the inducible costimulator ICOS controls development of germinal center TFH cells via the kinase TBK1. Nat Immunol. (2016) 17:825–33. doi: 10.1038/ni.3463
28. Stone EL, Pepper M, Katayama CD, Kerdiles YM, Lai CY, Emslie E, et al. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. (2015) 42:239–51. doi: 10.1016/j.immuni.2015.01.017
29. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. (2009) 325:1006–10. doi: 10.1126/science.1175870
30. Weinmann AS. Regulatory mechanisms that control T-follicular helper and T-helper 1 cell flexibility. Immunol Cell Biol. (2014) 92:34–9. doi: 10.1038/icb.2013.49
31. Gatto D, Wood K, Brink R. EBI2 operates independently of but in cooperation with CXCR5 and CCR7 to direct B cell migration and organization in follicles and the germinal center. J Immunol. (2011) 187:4621–8. doi: 10.4049/jimmunol.1101542
32. Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. (2012) 36:847–56. doi: 10.1016/j.immuni.2012.02.012
33. Wang CJ, Heuts F, Ovcinnikovs V, Wardzinski L, Bowers C, Schmidt EM, et al. CTLA-4 controls follicular helper T-cell differentiation by regulating the strength of CD28 engagement. Proc Natl Acad Sci USA. (2015) 112:524–9. doi: 10.1073/pnas.1414576112
34. Sage PT, Paterson AM, Lovitch SB, Sharpe AH. The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity. (2014) 41:1026–39. doi: 10.1016/j.immuni.2014.12.005
35. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. (2002) 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
36. Chevrier S, Genton C, Kallies A, Karnowski A, Otten LA, Malissen B, et al. CD93 is required for maintenance of antibody secretion and persistence of plasma cells in the bone marrow niche. Proc Natl Acad Sci USA. (2009) 106:3895–900. doi: 10.1073/pnas.0809736106
37. Nemajerova A, Petrenko O, Trumper L, Palacios G, Moll UM. Loss of p73 promotes dissemination of Myc-induced B cell lymphomas in mice. J Clin Invest. (2010) 120:2070–80. doi: 10.1172/JCI40331
38. Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. (2009) 10:385–93. doi: 10.1038/ni.1715
39. Tanaka Y, So T, Lebedeva S, Croft M, Altman A. Impaired IL-4 and c-Maf expression and enhanced Th1-cell development in Vav1-deficient mice. Blood. (2005) 106:1286–95. doi: 10.1182/blood-2004-10-4074
40. David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, et al. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev. (2006) 209:212–36. doi: 10.1111/j.0105-2896.2006.00355.x
41. Park HJ, Kim DH, Choi JY, Kim WJ, Kim JY, Senejani AG, et al. PPARgamma negatively regulates T cell activation to prevent follicular helper T cells and germinal center formation. PLoS ONE. (2014) 9:e99127. doi: 10.1371/journal.pone.0099127
42. Xia J, Benner MJ, Hancock RE. NetworkAnalyst–integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. (2014) 42:W167–74. doi: 10.1093/nar/gku443
43. Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. (2013) 41:D1228–33. doi: 10.1093/nar/gks1147
44. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
45. Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. (2013) 38:805–17. doi: 10.1016/j.immuni.2013.02.020
46. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of affymetrix GeneChip data at the probe level. Bioinformatics. (2004) 20:307–15. doi: 10.1093/bioinformatics/btg405
47. Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. (2009) 25:415–6. doi: 10.1093/bioinformatics/btn647
48. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
49. Russo PST, Ferreira GR, Cardozo LE, Burger MC, Arias-Carrasco R, Maruyama SR, et al. CEMiTool: a bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinformatics. (2018) 19:56. doi: 10.1186/s12859-018-2053-1
50. Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The reactome pathway knowledgebase. Nucleic Acids Res. (2018) 46:D649–55. doi: 10.1093/nar/gkx1132
51. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. (2001) 98:13784–9. doi: 10.1073/pnas.241500798
52. Mastelic Gavillet B, Eberhardt CS, Auderset F, Castellino F, Seubert A, Tregoning JS, et al. MF59 mediates its B cell adjuvanticity by promoting t follicular helper cells and thus germinal center responses in adult and early life. J Immunol. (2015) 194:4836–45. doi: 10.4049/jimmunol.1402071
53. Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and Pre-t alpha chain expression. Nat Immunol. (2000) 1:138–44. doi: 10.1038/77819
54. Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. (2004) 32:11–24. doi: 10.1016/j.exphem.2003.10.010
55. Sanda T, Leong WZ. TAL1 as a master oncogenic transcription factor in T-cell acute lymphoblastic leukemia. Exp Hematol. (2017) 53:7–15. doi: 10.1016/j.exphem.2017.06.001
56. Hsu HL, Cheng JT, Chen Q, Baer R. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol Cell Biol. (1991) 11:3037–42. doi: 10.1128/MCB.11.6.3037
57. Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. (1997) 16:3145–57. doi: 10.1093/emboj/16.11.3145
58. Rabbitts TH. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. (1998) 12:2651–7. doi: 10.1101/gad.12.17.2651
59. Ravnskjaer K, Boergesen M, Rubi B, Larsen JK, Nielsen T, Fridriksson J, et al. Peroxisome proliferator-activated receptor alpha (PPARalpha) potentiates, whereas PPARgamma attenuates, glucose-stimulated insulin secretion in pancreatic beta-cells. Endocrinology. (2005) 146:3266–76. doi: 10.1210/en.2004-1430
60. Leber A, Abedi V, Hontecillas R, Viladomiu M, Hoops S, Ciupe S, et al. Bistability analyses of CD4+ T follicular helper and regulatory cells during Helicobacter pylori infection. J Theor Biol. (2016) 398:74–84. doi: 10.1016/j.jtbi.2016.02.036
61. Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. (2009) 10:167–75. doi: 10.1038/ni.1690
62. Weinstein JS, Herman EI, Lainez B, Licona-Limon P, Esplugues E, Flavell R, et al. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. (2016) 17:1197–205. doi: 10.1038/ni.3554
63. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science. (2009) 325:1001–5. doi: 10.1126/science.1176676
64. Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. (2009) 31:457–68. doi: 10.1016/j.immuni.2009.07.002
65. Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S, et al. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. (2016) 14:1735–47. doi: 10.1016/j.celrep.2016.01.038
66. Moriyama S, Takahashi N, Green JA, Hori S, Kubo M, Cyster JG, et al. Sphingosine-1-phosphate receptor 2 is critical for follicular helper T cell retention in germinal centers. J Exp Med. (2014) 211:1297–305. doi: 10.1084/jem.20131666
67. Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol. (2015) 16:991–9. doi: 10.1038/ni.3229
68. Stauss D, Brunner C, Berberich-Siebelt F, Hopken UE, Lipp M, Muller G. The transcriptional coactivator Bob1 promotes the development of follicular T helper cells via Bcl6. EMBO J. (2016) 35:881–98. doi: 10.15252/embj.201591459
69. Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. (2011) 35:919–31. doi: 10.1016/j.immuni.2011.11.012
70. Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. (2009) 31:158–69. doi: 10.1016/j.immuni.2009.04.016
71. Nobs SP, Natali S, Pohlmeier L, Okreglicka K, Schneider C, Kurrer M, et al. PPARgamma in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J Exp Med. (2017) 214:3015–35. doi: 10.1084/jem.20162069
72. Luther SA, Cyster JG. Chemokines as regulators of T cell differentiation. Nat Immunol. (2001) 2:102–7. doi: 10.1038/84205
73. Angkasekwinai P, Park H, Wang YH, Wang YH, Chang SH, Corry DB, et al. Interleukin 25 promotes the initiation of proallergic type 2 responses. J Exp Med. (2007) 204:1509–17. doi: 10.1084/jem.20061675
74. Wang YH, Angkasekwinai P, Lu N, Voo KS, Arima K, Hanabuchi S, et al. IL-25 augments type 2 immune responses by enhancing the expansion and functions of TSLP-DC-activated Th2 memory cells. J Exp Med. (2007) 204:1837–47. doi: 10.1084/jem.20070406
75. Ono C, Yu Z, Kasahara Y, Kikuchi Y, Ishii N, Tomita H. Fluorescently activated cell sorting followed by microarray profiling of helper T cell subtypes from human peripheral blood. PLoS ONE. (2014) 9:e111405. doi: 10.1371/journal.pone.0111405
76. Chakarov S, Fazilleau N. Monocyte-derived dendritic cells promote T follicular helper cell differentiation. EMBO Mol Med. (2014) 6:590–603. doi: 10.1002/emmm.201403841
77. Rookhuizen DC, DeFranco AL. Toll-like receptor 9 signaling acts on multiple elements of the germinal center to enhance antibody responses. Proc Natl Acad Sci USA. (2014) 111:E3224–33. doi: 10.1073/pnas.1323985111
78. Adkins B, Williamson T, Guevara P, Bu Y. Murine neonatal lymphocytes show rapid early cell cycle entry and cell division. J Immunol. (2003) 170:4548–56. doi: 10.4049/jimmunol.170.9.4548
79. Smith NL, Wissink E, Wang J, Pinello JF, Davenport MP, Grimson A, et al. Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J Immunol. (2014) 193:177–84. doi: 10.4049/jimmunol.1400553
80. Galindo-Albarran AO, Lopez-Portales OH, Gutierrez-Reyna DY, Rodriguez-Jorge O, Sanchez-Villanueva JA, Ramirez-Pliego O, et al. CD8(+) T cells from human neonates are biased toward an innate immune response. Cell Rep. (2016) 17:2151–60. doi: 10.1016/j.celrep.2016.10.056
81. Furmanski AL, Saldana JI, Ono M, Sahni H, Paschalidis N, D'Acquisto F, et al. Tissue-derived hedgehog proteins modulate Th differentiation and disease. J Immunol. (2013) 190:2641–9. doi: 10.4049/jimmunol.1202541
82. Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol. (2007) 7:379–90. doi: 10.1038/nri2075
83. Chen N, Gao Q, Field EH. Expansion of memory Th2 cells over Th1 cells in neonatal primed mice. Transplantation. (1995) 60:1187–93. doi: 10.1097/00007890-199512000-00002
84. Opiela SJ, Levy RB, Adkins B. Murine neonates develop vigorous in vivo cytotoxic and Th1/Th2 responses upon exposure to low doses of NIMA-like alloantigens. Blood. (2008) 112:1530–8. doi: 10.1182/blood-2007-08-106500
85. Rose S, Lichtenheld M, Foote MR, Adkins B. Murine neonatal CD4+ cells are poised for rapid Th2 effector-like function. J Immunol. (2007) 178:2667–78. doi: 10.4049/jimmunol.178.5.2667
86. Saubermann LJ, Nakajima A, Wada K, Zhao S, Terauchi Y, Kadowaki T, et al. Peroxisome proliferator-activated receptor gamma agonist ligands stimulate a Th2 cytokine response and prevent acute colitis. Inflamm Bowel Dis. (2002) 8:330–9. doi: 10.1097/00054725-200209000-00004
87. Du X, Tabeta K, Mann N, Crozat K, Mudd S, Beutler B. An essential role for Rxr alpha in the development of Th2 responses. Eur J Immunol. (2005) 35:3414–23. doi: 10.1002/eji.200535366
88. Choi JM, Bothwell AL. The nuclear receptor PPARs as important regulators of T-cell functions and autoimmune diseases. Mol Cells. (2012) 33:217–22. doi: 10.1007/s10059-012-2297-y
89. Baratelli F, Lin Y, Zhu L, Yang SC, Heuze-Vourc'h N, Zeng G, et al. Prostaglandin E2 induces FOXP3 gene expression and T regulatory cell function in human CD4+ T cells. J Immunol. (2005) 175:1483–90. doi: 10.4049/jimmunol.175.3.1483
90. Wohlfert EA, Nichols FC, Nevius E, Clark RB. Peroxisome proliferator-activated receptor gamma (PPARgamma) and immunoregulation: enhancement of regulatory T cells through PPARgamma-dependent and -independent mechanisms. J Immunol. (2007) 178:4129–35. doi: 10.4049/jimmunol.178.7.4129
91. Akram A, Schmeidler J, Katsel P, Hof PR, Haroutunian V. Increased expression of RXRalpha in dementia: an early harbinger for the cholesterol dyshomeostasis? Mol Neurodegener. (2010) 5:36. doi: 10.1186/1750-1326-5-36
92. Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. (2007) 179:5099–108. doi: 10.4049/jimmunol.179.8.5099
93. Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. (2011) 34:961–72. doi: 10.1016/j.immuni.2011.03.025
94. Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, et al. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. (2011) 34:947–60. doi: 10.1016/j.immuni.2011.03.024
95. Baumjohann D, Okada T, Ansel KM. Cutting edge: distinct waves of BCL6 expression during T follicular helper cell development. J Immunol. (2011) 187:2089–92. doi: 10.4049/jimmunol.1101393
96. Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, et al. Optimal germinal center responses require a multistage T cell: B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. (2010) 32:253–65. doi: 10.1016/j.immuni.2010.01.010
97. Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. (2000) 192:1545–52. doi: 10.1084/jem.192.11.1545
98. Vono M, Eberhardt CS, Mohr E, Auderset F, Christensen D, Schmolke M, et al. Overcoming the neonatal limitations of inducing germinal centers through liposome-based adjuvants including C-type lectin agonists trehalose dibehenate or curdlan. Front Immunol. (2018) 9:381. doi: 10.3389/fimmu.2018.00381
Keywords: T follicular helper cells, neonates, vaccines, adjuvant, transcriptional profile analysis
Citation: Mastelic-Gavillet B, Vono M, Gonzalez-Dias P, Ferreira FM, Cardozo L, Lambert P-H, Nakaya HI and Siegrist C-A (2019) Neonatal T Follicular Helper Cells Are Lodged in a Pre-T Follicular Helper Stage Favoring Innate Over Adaptive Germinal Center Responses. Front. Immunol. 10:1845. doi: 10.3389/fimmu.2019.01845
Received: 15 February 2019; Accepted: 22 July 2019;
Published: 13 August 2019.
Edited by:
Pedro A. Reche, Complutense University of Madrid, SpainReviewed by:
Roza Nurieva, University of Texas MD Anderson Cancer Center, United StatesAlex Dent, Purdue University Indianapolis, United States
Copyright © 2019 Mastelic-Gavillet, Vono, Gonzalez-Dias, Ferreira, Cardozo, Lambert, Nakaya and Siegrist. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatris Mastelic-Gavillet, YmVhdHJpcy5tYXN0ZWxpYy1nYXZpbGxldEBjaHV2LmNo
†Present address: Beatris Mastelic-Gavillet, Department of Oncology, Center for Experimental Therapeutics, Ludwig Center for Cancer Research, University of Lausanne, Lausanne, Switzerland
 Beatris Mastelic-Gavillet
Beatris Mastelic-Gavillet Maria Vono
Maria Vono Patrícia Gonzalez-Dias
Patrícia Gonzalez-Dias Frederico Moraes Ferreira
Frederico Moraes Ferreira Lucas Cardozo
Lucas Cardozo Paul-Henri Lambert
Paul-Henri Lambert Helder I. Nakaya
Helder I. Nakaya Claire-Anne Siegrist
Claire-Anne Siegrist