- 1Jiangsu Key Laboratory for Aquatic Crustacean Diseases, College of Marine Science and Engineering, Nanjing Normal University, Nanjing, China
- 2College of Life Sciences, Nanjing Normal University, Nanjing, China
- 3Marine Science and Engineering College, Qingdao Agricultural University, Qingdao, China
- 4Co-innovation Center for Marine Bio-Industry Technology of Jiangsu Province, Lianyungang, China
Spiroplasma eriocheiris is a crustacean pathogen, without a cell wall, that causes enormous economic loss. Macrobrachium rosenbergii hemocytes are the major targets during S. eriocheiris infection. As wall-less bacteria, S. eriocheiris, its membrane protein should interact with host membrane protein directly and firstly when invaded in host cell. In this investigation, six potential hemocyte receptor proteins were identified firstly that mediate interaction between S. eriocheiris and M. rosenbergii. Among these proteins, lipopolysaccharide and β-1, 3-glucan binding protein (MrLGBP) demonstrated to bind to S. eriocheiris using bacterial binding assays and confocal microscopy. Four spiroplasma ligand proteins for MrLGBP were isolated and identified. But, competitive assessment demonstrated that only enolase of S. eriocheiris (SeEnolase) could be a candidate ligand for MrLGBP. Subsequently, the interaction between MrLGBP and SeEnolase was confirmed by co-immunoprecipitation and co-localization in vitro. After the interaction between MrLGBP and SeEnolase was inhibited by antibody neutralization test, the virulence ability of S. eriocheiris was effectively reduced. The quantity of S. eriocheiris decreased in Drosophila S2 cells after overexpression of MrLGBP, compared with the controls. In addition, RNA interference (RNAi) knockdown of MrLGBP made M. rosenbergii more sensitive to S. eriocheiris infection. Further studies found that the immune genes, including MrLGBP and prophenoloxidase (MrproPO), MrRab7A, and Mrintegrin α1 were significantly up-regulated by SeEnolase stimulation. After SeEnolase pre-stimulation, the ability of M. rosenbergii resistance to S. eriocheiris was significantly improved. Collectively, this investigation demonstrated that MrLGBP and pathogen SeEnolase involved in mediating S. eriocheiris invasion into M. rosenbergii hemocytes.
Introduction
Host-pathogen contact is a prerequisite for bacterial invasion and colonization (1, 2). Spiroplasma, a wall-less bacterium (3), undoubtedly depends upon protein interactions between the bacterium and cells of the host. As a novel spiroplasma, Spiroplasma eriocheiris is the causative agent of tremor-disease (4). Previous investigations have demonstrated Macrobrachium rosenbergii hemocytes to be the main cellular targets of S. eriocheiris, from which infections disseminate into the prawn body (5). It is likely that proteins mediate the interaction between S. eriocheiris and hemocytes, permitting entry of spiroplasma into hemocytes.
Lipopolysaccharide and β-1, 3-glucan binding protein (LGBP), a pattern recognition protein (PRP), recognizes and binds to common epitopes on the pathogen surface (6). Subsequent to recognition (7), LGBP activates distinctly a series of immune responses, including phagocytosis, nodule formation, clotting cascade, the synthesis of a wide array of antimicrobial peptides, and the prophenoloxidase system (proPO) (8–10). Our previous research (11) has shown that in M. rosenbergii hemocytes infected with S. eriocheiris, 69 differentially expressed proteins including LGBP were identified compared with control group injected R2 medium [8% sucrose, 2.5% heart infusion broth (HIB), 15% fetal bovine serum (FBS), 100 U ml−1 penicillin, and pH 7.20–7.40]. These results suggested that LGBP play an important role in pathogen invasion into host. However, the specific mechanism by which LGBP facilitates S. eriocheiris entry into M. rosenbergii has not been elucidated.
In order to bind and penetrate target cells, attachment organelles very likely contain specialized receptors similar to those of human mycoplasmas (12, 13). Several spiroplasma adhesins have been identified. These include adhesin-like protein (ALP) (14) and spiralin (15), both of which are involved in spiroplasma transmission. Spiroplasma citri colonization of insect cells is promoted by the interaction of phosphoglycerate kinase (PGK) with actin (16). Further, enolase, a key cytoplasmic glycolytic enzyme, is found on the cell surface of Mycoplasma fermentans (17) as well as Streptococcus pneumoniae (18). Enolase and its receptor protein, plasminogen, are known to promote bacterial binding to host cells. However, no investigations have identification proteins that mediate the interaction of S. eriocheiris with M. rosenbergii hemocytes. Herein, identification of bacterial-host interaction proteins that play a complex and important role in the process of S. eriocheiris entry into M. rosenbergii hemocytes.
Materials and Methods
Spiroplasma Strain, Freshwater Prawns, and Primary Hemocyte Culture
Spiroplasma eriocheiris, obtained from a naturally infected M. rosenbergii, was derived from a livestock farm in Gaoyou, Jiangsu province of China (5) and cultured in R2 liquid medium (8% sucrose, 2.5% HIB, 15% FBS, 100 U ml−1 penicillin, and pH 7.20–7.40) at 30°C (4).
Healthy M. rosenbergii freshwater prawns were obtained from a commercial farm in Nanjing, Jiangsu province of China, and reared in tanks at 28°C with freshwater and an aeration system. A polymerase chain reaction (PCR) was conducted to guarantee that the prawns were free of spiroplasma (19). Prawns were fed daily for 2 weeks with a commercial diet before hemocytes were withdrawn.
Macrobrachium rosenbergii primary hemocytes (20) were cultured at 28°C in Leibovitz-15 (L-15) growth medium (pH 7.2–7.4) supplemented with 15% FBS, 0.1% glucose, 0.5% NaCl, and antibiotics (100 U ml−1 penicillin, 100 U ml−1 streptomycin, and 1 μg ml−1 amphotericin b).
Identification of Receptor Proteins
This experiment was conducted based on the methods of Labroussaa et al. (21), with modifications. Hemocytes were withdrawn from the second abdominal segment of healthy M. rosenbergii using a 1 ml sterile syringe containing 500 μL modified phosphate buffer saline (PBS) (0.9 g/L Na2HPO4, 0.27 g/L KH2PO4, 0.6 g/L KCl, 25.5 g/L NaCl, and 1.0 g/L glucose, pH 7.2) as anticoagulant. The diluted hemocytes were centrifuged for 5 min at 3,800 × g to collect cells and resuspended in Common Lysis Buffer (Generay, China) containing 1 mM phenylmethanesulfonyl fluoride (PMSF). After the mixture was centrifuged for 3 min at 10,000 × g, a bicinchoninic acid (BCA) procedure was used to assess protein concentration. Aliquots of supernatant (20 μg) were separated by electrophoresis in a 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel, and then transferred from the gel to a polyvinylidene difluoride (PVDF) membrane. After transfer, membrane was blocked in 10 ml of tris-buffered saline with Tween (TBST) with 10% bovine serum albumin (BSA) overnight at 4°C. Then, membranes were incubated with 10 ml TBST containing 5% BSA and 5 μg/ml formaldehyde-killed spiroplasma (22) at room temperature (RT) for 1 h. For the control experiment, spiroplasma were not included. After incubation, membrane was washed three times with TBST and incubated in 10 ml of TBST containing 5% BSA with purified polyclonal antibodies (0.5 μg/ml) reactive with S. eriocheiris at a dilution of 1:2,000 at RT for 1 h. After three washings with TBST, membrane was incubated with peroxidase-conjugated goat anti-rabbit IgGs (Transgen Biotech, China) at a 1:20,000 dilution in 5% BSA at RT for 1 h. Detection of the bands was by incubation with an enhanced chemiluminescence (ECL) substrate solution (E411-01/02) according to the manufacturer's instructions (Vazyme Biotech, China). Membranes was washed three times with TBST and then exposed to X-ray film. Based on the method of Killiny et al. (23), proteins of interest (11) were excised from stained gels and digested with trypsin. Peptide mass spectrometry (MS) and tandem mass spectrometry (MS/MS) were performed by BIO-TECH (China) using an ABI 5800 MALDI-TOF/TOF Plus mass spectrometer (Applied Biosystems, USA). Proteins of interest were successfully identified at 95% or higher confidence using the MASCOT V2.3 search engine (Matrix Science Ltd., London, U.K.).
Binding of Recombinant Proteins to Spiroplasma
A pair of primers, MrLGBP-F/-R (Table S1), were used to amplify a 1101-bp open reading frame (ORF) encoding a mature protein. PCR fragments were digested with BamH I and Not I restriction enzymes and cloned into a pGEX-4T1 plasmid. The recombinant plasmids pGEX-4T1-MrLGBP were transformed into Escherichia coli Transetta (DE3) cells for isopropyl-b-D-thio-galactoside (IPTG) (final IPTG concentration of 0.5 mM) induced glutathione S-transferase (GST)-tagged recombinant expression. MrLGBP was purified by Glutathione Sepharose 4 Fast Flow (GE Healthcare).
The bacteria binding assay was conducted as described previously (24). Briefly, spiroplasma were cultured overnight in R2 medium and during exponential growth, collected by centrifugation at 9,000 × g for 5 min at RT. Bacteria were washed three times with PBS and thoroughly resuspended in PBS to an OD600 of 1.0. The purified recombinant MrLGBP was incubated with 100 μL of S. eriocheiris (2 × 108 cells/ml) at RT with gentle rotation for 2 h, washed four times with PBS, and spiroplasma eluted with 100 μL of 8 M urea at RT for 30 min. After centrifugation at 9,000 × g for 5 min, supernatant and precipitate were loaded onto a 12.5% SDS-PAGE gel. A western blotting experiment using anti-GST monoclonal antibodies was conducted to detect the direct binding of recombinant proteins to spiroplasma. Bacteria cells were subjected to the same treatment and incubated with the GST tag as a control.
Co-location of MrLGBP and Spiroplasma by Confocal Microscopy
Primary M. rosenbergii hemocytes cultures were established based on the method of Du et al. (20). After 5 h of seeding, the hemocytes were infected with 100 μL of spiroplasma (108 cells/ml) for 16 h at 28°C. Unbound bacteria were removed by washing three times with PBS. Hemocytes were immersed in fixative (4% paraformaldehyde in PBS) for 30 min at RT. Fixed hemocytes were rinsed three times with PBS and permeabilized with PBS containing 0.5% Triton X-100 for 10 min at RT, and then incubated in blocking buffer (PBS plus 5% BSA) at RT for 1 h.
For MrLGBP and spiroplasma co-localization, hemocytes were incubated at RT for 1 h with rabbit anti-S. eriocheiris (diluted 1:5,000) and mouse anti-MrLGBP (diluted 1:4,000) polyclonal antibodies (prepared by Vazyme Biotech, China) in PBS containing 1% BSA. After three washings, the nuclei was stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) and MrLGBP was stained by Alexa Flour 555 donkey anti-mouse IgGs (Beyotime, China) at a 1:7,000 dilution. Spiroplasma was stained at RT for 1 h by Alexa Flour 488 goat anti-rabbit IgGs (Beyotime, China) at a 1:10,000 dilution in PBS containing 1% BSA. After washing, immunofluorescent samples were visualized with a confocal laser scanning microscope (Nikon TI-E-A1R, Japan).
Identification of Ligand Proteins
Spiroplasma eriocheiris was collected by centrifugation (14,000 × g, 10 min) and then washed three times with PBS. Washed spiroplasma was suspended in Common Lysis Buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM PMSF, and 2 mM ethylene diamine tetraacetic acid (EDTA)] and treated at 50% duty cycles and an intensity of 400 W for 20 min at 4°C. After the mixture was centrifuged for 5 min at 10,000 × g, the supernatant was fractionated by SDS-PAGE, and then transferred from the gel to a PVDF membrane. After blocking, membrane was incubated for 2 h with purified recombinant MrLGBP or the GST tag at a final concentration of 2 μg/ml in TBST containing 5% BSA. The protocol was similar to that described in the “Identification of Receptor Proteins” section except that anti-GST monoclonal antibody at a dilution of 1:2,000 (Transgen Biotech, China) was used instead of spiroplasma-reactive polyclonal antibody.
Competitive Assay of SeEnolase in Hemocyte
In the “Identification of Ligand Proteins” assay, SeEnolase, transketolase (TK), and acetaldehyde dehydrogenase (ALDH) were successfully identified. To determine whether ligand proteins participate in the infection process of S. eriocheiris, the gene coding for ligand proteins were amplified and cloned into a pEASY-BluntE1 expression vector (Transgen Biotech, China). Since, TGA is read as a tryptophan codon and not as a termination signal in most Mollicute species, the TGA codons were mutated to TGG codons by the Fast Mutagenesis System (Transgen Biotech, China) according to the instructions. Amplification and mutagenesis primers are listed in Table S1. The recombinant plasmids were transformed into E. coli Transetta (DE3) for protein expression. Recombinant proteins were purified via Ni Sepharose 6 Fast Flow (GE Healthcare). Samples were analyzed by SDS-PAGE.
Primary M. rosenbergii hemocytes were incubated with different ligand proteins at a final concentration of 20 μg/ml, and infected by adding 10 μL of spiroplasma (108 cells/ml) for 16 h at 28°C. Uninfected cells served as positive control, and cells incubated with BSA as a negative control. Following incubation, the hemocytes were then washed three times with PBS to remove spiroplasma that were not attached. Cell viability was determined with cell counting kit-8 (CCK-8) reagent (Beyotime, China) using 10 μL/well for 2 h, and measured the optical density (OD) at 450 nm using a microplate reader (25). To maintain consistency, all data were reported as relative cell viability as the mean ± S.E. Statistical significance was determined by one-way analysis of variance (ANOVA) and by post-hoc Duncan multiple range tests.
Localization of SeEnolase
According to our previous research methods (26), the rabbit poly-antibody of SeEnolase was successfully prepared, namely anti-SeEnolase serum. The proteins from S. eriocheiris and purified recombinant SeEnolase were analyzed by western blot using the anti-SeEnolase serum.
Cytoplasm proteins and outer membrane proteins from S. eriocheiris were obtained as previously described (27, 28), with modification. Briefly, spiroplasma was collected by centrifugation at 12,000 × g for 20 min, washed three times, and resuspended in Tris-HCl (0.02 mol/L, pH 7.5) followed by sonication in an ultrasonic disintegrator (200 W). The sonicate was ultra-centrifuged at 34,000 × g for 30 min, and the supernatant (cytoplasmic proteins) and the pellet (membrane proteins) were collected. Membrane proteins were re-suspended in PBS with a proteinase inhibitor. Protein samples were fractionated by electrophoresis by SDS-PAGE. For western blotting, anti-SeEnolase serum (anti-adhesin serum and anti-arginine deiminase serum were used as controls) was used as the primary antibody (1:2,000) and goat anti-rabbit IgG (whole-molecule) peroxidase conjugate (Sigma) as the secondary antibody (1:5,000). Blots were developed with a 3,3′,5,5′-Tetramethylbenzidine Liquid MB Substrate Kit (Promega, USA).
Antibody Neutralization Assay
An antibody neutralization test was used to investigate the effect of anti-SeEnolase serum on S. eriocheiris infection of M. rosenbergii. The bacteria were pretreated by incubating with anti-SeEnolase serum, pre-immune serum, and PBS, respectively, for 1 h at 30°C (29). Healthy prawns (average 4–5 g, n = 50 for each group) were randomly divided into three groups, anti-SeEnolase serum + S. eriocheiris, pre-immune serum + S. eriocheiris, PBS + S. eriocheiris group. In the anti-SeEnolase serum + S. eriocheiris group, 20 μL pretreated bacteria with anti-SeEnolase serum were injected into prawns. Meanwhile, 20 μL of pretreated bacteria with pre-immune serum or PBS were injected for the pre-immune serum + S. eriocheiris or PBS + S. eriocheiris group. Five prawns were randomly selected from three groups for analysis of S. eriocheiris copies at 1, 3, 5, 7, and 9 days, respectively. The number of S. eriocheiris copies in M. rosenbergii hemocytes were determined by a real-time PCR using primers Se-QF and Se-QR (Table S1) (30).
To further test prawn mortality, 50 μL of pretreated bacteria with anti-SeEnolase serum, pre-immune serum, or PBS were injected for the anti-SeEnolase serum + S. eriocheiris, pre-immune serum + S. eriocheiris, or PBS + S. eriocheiris group. At the same time, three another groups were injected with the anti-SeEnolase serum, pre-immune serum, and PBS, respectively. The cumulative mortality of prawns was recorded daily.
Co-immunoprecipitation and Co-localization Assay
To generate pAc-enolase-V5 and pAc-enolase-RFP plasmids for expression of V5-tagged and RFP-tagged SeEnolase protein, the ORF of SeEnolase was cloned into pAc5.1-V5 and pAc5.1-RFP vectors (31, 32) at Kpn I and Apa I sites using the gene-specific primers pAc-enolase-F and pAc-enolase-R (Table S1). Similarly, ORF of MrLGBP was cloned into pAc5.1-GFP vectors using the primers pAc-MrLGBP-F and pAc-MrLGBP-R (Table S1).
For the co-immunoprecipitation assay, pAc-enolase-V5 was co-transfected with pAc-MrLGBP-GFP or pAc5.1-GFP (as a control) into Drosophila S2 cells. Forty-eight hours after transfection, cells were harvested and washed with ice-cold PBS three times, and then lysed with NP-40 lysis buffer (Beyotime, China) with 1 mM PMSF protease inhibitor (Solarbio, China), and incubated with 2 μg of anti-GFP mouse antibody (Transgen Biotech, China) or anti-V5 mouse antibody (Transgen Biotech, China) overnight at 4°C. Antibodies were precipitated with 40 μL of protein G resin beads (Transgen Biotech, China) for 3 h at 4°C. Beads were then washed three times in ice-cold NP-40 lysis buffer and samples subjected to SDS-PAGE. Western blotting was performed with anti-GFP rabbit antibody (1:2,000) or anti-V5 rabbit antibody (1:2,000), and peroxidase-conjugated goat anti-rabbit secondary antibody at a dilution of 1:5,000 (Transgen Biotech, China). A standardized aliquot (10%) of each total input cell lysate was also examined as a control.
Drosophila S2 cells were seeded onto cover-glass bottom dishes with ~60% confluence and then co-transfected with 2 μg pAc-enolase-RFP and 2 μg pAc-MrLGBP-GFP using the FuGENE HD Transfection Reagent (Promega, USA). At 48 h post-transfection, following three ice-cold washes with PBS, cells were incubated with DAPI (Beyotime, China) for 10 min at RT. The fluorescent images were visualized with a confocal laser scanning microscope (Nikon TI-E-A1R, Japan).
Methods of culture, immobilization, permeability, and blocking of primary M. rosenbergii hemocytes have been described in the section “Co-location of MrLGBP and Spiroplasma by Confocal Microscopy.” Then, hemocytes were incubated with 1 ml PBS containing 1% BSA and 2.5 μg/ml SeEnolase protein at RT for 1 h. For the control experiment, SeEnolase protein was not included. After three washings, hemocytes were incubated at RT for 1 h with rabbit anti-SeEnolase (diluted 1:1,000) and mouse anti-MrLGBP (diluted 1:4,000) polyclonal antibodies (prepared by Vazyme Biotech, China) in PBS containing 1% BSA. Hemocytes nuclei was stained with DAPI. MrLGBP was stained by Alexa Flour 488 goat anti-Mouse IgGs (Beyotime, China) at a 1:7,000 dilution. SeEnolase were stained at RT for 1 h by PE-labeled Goat Anti-Rabbit IgG (Transgen Biotech, China) at a 1:5,000 dilution in PBS containing 1% BSA. After washing, immunofluorescent samples were visualized with a confocal laser scanning microscope (Nikon TI-E-A1R, Japan).
Assay for Over-expression of MrLGBP
Using the FuGENE HD Transfection Reagent (Promega, USA), Drosophila S2 cells from each dish were transfected with 2 μg pAc5.1-MrLGBP-GFP and pAc5.1-GFP plasmids, respectively. Twenty-four hours later, the Drosophila S2 cells were infected with S. eriocheiris (108 cells/ml). Drosophila S2 cells were divided into three groups, S. eriocheiris only, S. eriocheiris + GFP, and S. eriocheiris + LGBP-GFP. At 48 h, after S. eriocheiris infection, samples were washed three times with PBS and fixed with Immunol Staining Fix Solution (Beyotime, China). Fixed Drosophila S2 cells were rinsed three times with PBS and permeabilized with PBS containing 0.2% TritonX-100 for 30 min, and then incubated in blocking buffer (PBS plus 3% BSA) for 30 min. The cells were incubated with S. eriocheiris polyclonal antibody with 1% BSA in PBS overnight. Then, the cells were incubated with PE-labeled Goat anti-Rabbit IgG (Transgen Biotech, China) and examined using a confocal laser scanning microscope (Nikon TI-E-A1R, Japan). The methods for transfection and infection of Drosophila S2 cells were as described above. To quantify the copy number of S. eriocheiris, Drosophila S2 cells were collected from cell culture dishes from each treatment group at 48 h after S. eriocheiris infection and subjected to real-time PCR using the primers Se-QF and Se-QR (Table S1) (30). To confirm MrLGBP over-expression, total protein was extracted with lysis buffer (Beyotime, China) containing 1 mM PMSF and 2 mM EDTA on ice. After sonication and centrifugation (13,000 × g, 15 min, 4°C), the supernatants were collected for protein concentration measurement using the bicinchoninic acid assay (BCA). Thirty micrograms of protein was analyzed using 12% SDS-PAGE and western blotting using anti-GFP (Transgen Biotech, China) and HRP-conjugated Goat Anti-Mouse IgG (Transgen Biotech, China). The bands were visualized using ECL (Vazyme, China).
Drosophila S2 cells were seeded into a 96-well plate with a final ~60% confluence. The methods for transfection of Drosophila S2 cells were as described above. Drosophila S2 cells were divided into four groups, R2 medium, S. eriocheiris only, S. eriocheiris + GFP, and S. eriocheiris + LGBP-GFP. At 48 h after S. eriocheiris infection, the viability of Drosophila S2 cells from 12 wells for each treatment was determined by CCK-8 according to the manufacturer's instructions. All experiments were performed in triplicate.
RNA Interference Assay
Using an in vitro transcription T7 kit for dsRNA synthesis (Takara, Japan), double-stranded RNAs (dsRNAs) targeting the MrLGBP and GFP (as control) genes were synthesized. The DNA template for the MrLGBP dsRNA preparation was generated by PCR using the gene-specific primers dsRNA-MrLGBP-F and dsRNA-MrLGBP-R (Table S1). For preparation of GFP dsRNA, the primers dsRNA-GFP-F and dsRNA-GFP-R were used (Table S1).
To investigate the RNA interference (RNAi) efficiency, healthy M. rosenbergii were cultured in two groups at room temperature. The prawn injected with 20 μg MrLGBP dsRNA as the experimental group and 20 μg GFP dsRNA as the control group, respectively, at the second abdominal segment. After 24 h, the prawns were injected again with the same amount dsRNA. Five prawns were prepared and analyzed by Semi-quantitative PCR using primer pairs MrLGBP-qF/MrLGBP-qR (Table S1) at 48, 72, and 96 h after dsRNA injection, respectively.
The phenol oxidase (PO) activity of hemocytes was determined after MrLGBP silencing. Hemocytes were withdrawn from the ventral sinus of experimental prawns at 48, 72, and 96 h, respectively, after the first dsRNA injection. Protein concentration was measured using a Total protein quantitative assay kit (Nanjing Jiancheng, China). Hemocyte PO activity was detected using L-3,4-dihydroxyphenylalanine (L-dopa) dissolved in water according to Liu et al. (33). Briefly, 2 μg of total hemolymph proteins in 435 μL of Tris-HCl (10 mM, pH 8.0) were mixed with 65 μL of freshly prepared L-dopa (3 mg/ml in water). After incubation at room temperature for 30 min, PO activity was measured by monitoring the absorbance at 490 nm and recorded as OD490 per μg total protein.
For pathogen challenge tests, healthy prawns (average 4–5 g, n = 50 for each group) were cultured at RT and randomly divided into three groups, PBS + S. eriocheiris, dsRNA-GFP + S. eriocheiris, and dsRNA-LGBP + S. eriocheiris. In the PBS + S. eriocheiris group, the prawns were treated with 20 μL of PBS. Meanwhile, 20 μg of GFP dsRNA or MrLGBP dsRNA were injected for the dsRNA-GFP + S. eriocheiris group, or dsRNA-LGBP + S. eriocheiris group. All of the prawns were received an injection of 10 μL S. eriocheiris (108 cells/ml). Five prawns were randomly selected from three groups for analysis of S. eriocheiris copies at 1, 3, 5, 7, and 9 days, respectively. The number of S. eriocheiris copies in M. rosenbergii hemocytes was determined by a real-time PCR using primers Se-QF and Se-QR (Table S1) (30). All samples were assessed three times.
The healthy prawns (average 4–5 g, n = 30 for each group) were cultured at RT and randomly divided into six groups; PBS, dsRNA-GFP, dsRNA-LGBP, PBS + S. eriocheiris, dsRNA-GFP + S. eriocheiris, and dsRNA-LGBP + S. eriocheiris. The prawns of the dsRNA-LGBP group and dsRNA-LGBP + S. eriocheiris group were injected individually with 20 μg of MrLGBP dsRNA. The prawns of the dsRNA-GFP group and the dsRNA-GFP + S. eriocheiris group were injected individually with 20 μg of GFP dsRNA. The prawns of the PBS group and PBS + S. eriocheiris group were injected individually with 20 μL PBS. After 24 h, the prawns were injected again with the same amount dsRNA or PBS. Forty-eight hours after the first injection, the prawns of the PBS + S. eriocheiris group, dsRNA-GFP + S. eriocheiris group, and dsRNA-LGBP + S. eriocheiris group received an injection of 50 μL S. eriocheiris (108 cells/ml). The cumulative mortality of prawns was recorded daily.
SeEnolase Stimulation Assay
One hundred healthy prawns (average 4–5 g, n = 50 for each group) were randomly divided into two groups. Fifty prawns were injected individually with 50 μL SeEnolase protein (1 μg/μL) as an experimental group. For the control group, fifty prawns were injected with 50 μL PBS. The hemocytes were sampled from every five individuals at 0, 2, 4, 6, 8, 12, 24, 36, and 48 h post-injection. After extraction, total RNA was reverse-transcribed into cDNA with a PrimeScript RT reagent Kit (Takara, Japan). Quantitative real-time PCR (qRT-PCR) was conducted using a 2 × SYBR Premix Ex Taq Kit (Takara, Japan). GAPDH was amplified for internal standardization using the primers GAPDH-qF and GAPDH-qR (Table S1). The PCR reaction was performed in a 10 μL volume with a SYBR Premix Ex Taq™ Kit (Takara, Japan), 0.4 μM of each specific primer (Table S1), and 1 μL of cDNA in StepOnePlus™ Real-Time PCR System. The relative expression levels of immune relative genes, including MrLGBP, MrproPO, MrRab7A, and Mrintegrin α1 were calculated according to the 2−ΔΔCT method. All experiments were performed in triplicate. To maintain consistency, all data are given in terms of relative mRNA expression levels as the mean ± S.E. Statistical significance was determined by Student's t-test.
Healthy prawns (average 4–5 g, n = 50 for each group) were cultured at RT and randomly divided into two groups, SeEnolase + S. eriocheiris and PBS + S. eriocheiris group. Fifty prawns were injected individually with 50 μL of SeEnolase protein (1 μg/μL) in SeEnolase + S. eriocheiris group. For PBS + S. eriocheiris group, fifty prawns were injected with 50 μL of PBS. After 12 h stimulation, all of the prawns were received an injection of 10 μL S. eriocheiris (108 cells/ml). The mothed of S. eriocheiris copies analysis was described in the section Antibody Neutralization Assay.
To further test prawn mortality, two groups of prawns were received an injection of 50 μL S. eriocheiris (108 cells/ml) under the same culture conditions and protein stimulation as above. At the same time, two another groups were injected with the SeEnolase and PBS, respectively. The cumulative mortality of prawns was recorded daily.
Results
Identification of Receptor Proteins on M. rosenbergii Hemocytes
Far western blotting was used to detect the proteins interacting with S. eriocheiris. Compared to the control group, eight different bands were found in the experimental group, having apparent molecular masses of 24, 26, 41, 42, 45, 50, 52, and 56 kDa (Figure 1A). All bands were successfully sequenced, except for the 26 kDa band. By use of blastp and the national center for biotechnology information (NCBI) non-redundant protein database, mass spectra matched the following, ras-related nuclear protein (Ran), MrLGBP, beta-Actin, prophenoloxidase (proPO), beta tubulin, and alpha-tubulin (Figure S1). Related outputs obtained by MASCOT are shown in Table S2.
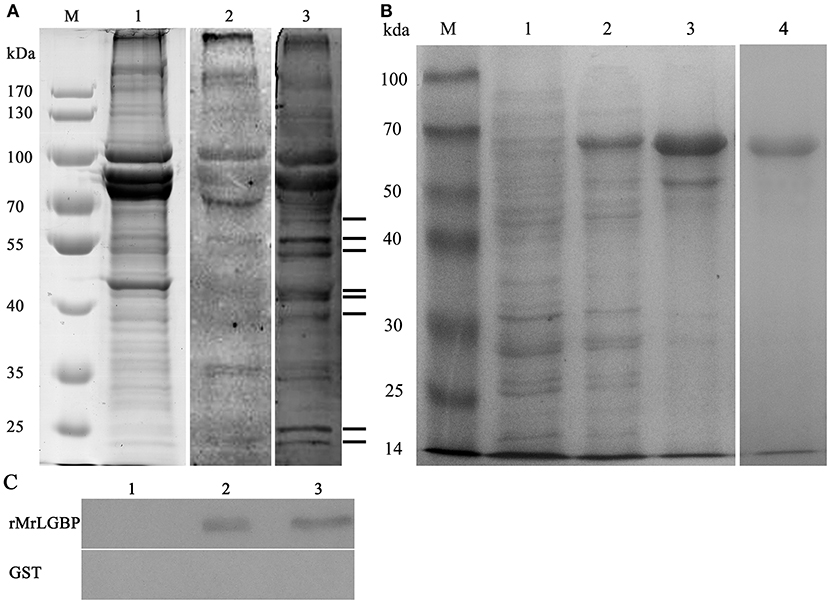
Figure 1. The identified receptor proteins from interactions between M. rosenbergii hemocytes and S. eriocheiris. (A) The far western blotting analysis to identify proteins on hemocytes that could bind to S. eriocheiris. M. rosenbergii hemocytes total proteins were analyzed by SDS-PAGE (Lane M and 1). Hemocyte receptor proteins were detected by far western blotting (Lane 2 and 3). Lane M, molecular weight marker. Lane 1, M. rosenbergii hemocyte proteins (20 μg). Lane 2, control blot not probed with S. eriocheiris bacteria. Lane 3, experimental blot probed with S. eriocheiris bacteria. Black lines on the right indicate the eight significant binding activities. (B) MrLGBP recombinant expression in E. coli analyzed by SDS-PAGE. Lane M, molecular weight marker; lane 1, negative control for MrLGBP (without induction); lane 2, supernatant component after induction by IPTG; lane 3, precipitation component after induction by IPTG; lane 4, purified recombinant MrLGBP protein. (C) direct binding of MrLGBP to spiroplasma using bacteria binding assay through western blot analysis. GST tag was used as a negative control. Lane 1, eluent of the fourth PBS washing; lane 2, supernatant component after treatment with 8 M urea; lane 3, precipitation component after treatment with 8 M urea.
Receptor Protein Expression and Spiroplasma Binding Assay
Recombinant MrLGBP proteins were isolated from supernatants after IPTG induction. Apparent molecular weight was 68 kDa by glutathione Sepharose 4B chromatography. After expression and purification, a clear band was detected by SDS-PAGE (Figure 1B, lanes 4). A direct binding assay suggested that although treated with 8 M urea, MrLGBP bound spiroplasma in vitro (Figure 1C).
As shown in Figure 2 of co-localization results, hemocytes were visualized in bright field (Figures 2a,f), blue fluorescence only (Figures 2b,g), red fluorescence only (Figures 2c,h), green fluorescence only (Figures 2d,i), and bright field merge with all fluorescent molecules (Figures 2e,j), respectively. MrLGBP proteins were visualized located in the membrane and cytoplasm (Figures 2c,e). But, S. eriocheiris mainly remained on the hemocyte membrane, but a few of them entered the cytoplasm (Figure 2i) after infection for 16 h. So, the co-localization of MrLGBP and S. eriocheiris was mainly shown on the membrane, as shown in Figure 2j. These results suggested that S. eriocheiris could bind to MrLGBP.
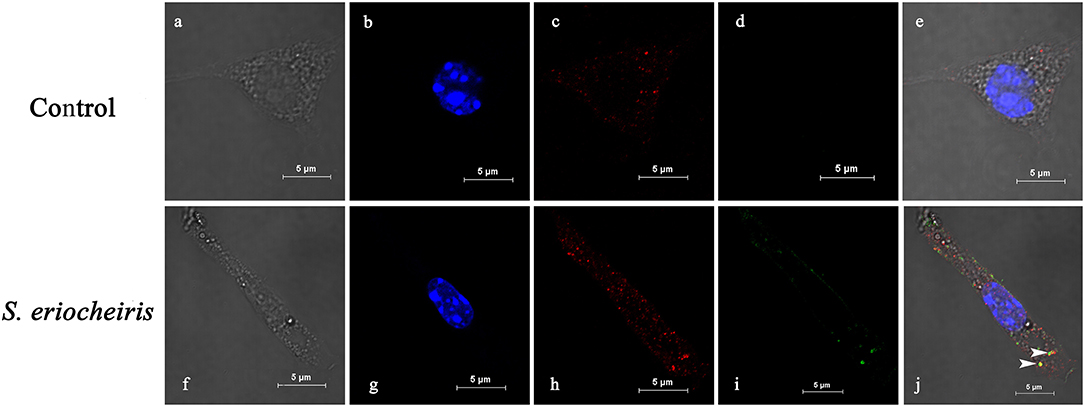
Figure 2. Spiroplasma eriocheiris bind to the MrLGBP protein of M. rosenbergii hemocyte. The co-localization of MrLGBP and S. eriocheiris in prawn hemocytes was analyzed by confocal microscopy. Hemocytes were immobilized after S. eriocheiris injection for 16 h. Spiroplasma were stained by Alexa Flour 488 goat anti-rabbit IgGs (green). MrLGBP were stained used the mouse anti-MrLGBP as primary antibody and donkey anti-mouse IgG Alexa-555 as the second antibody (red). Nuclei were stained with DAPI (blue). Hemocytes were visualized by bright field (a,f), blue fluorescence only (b,g), red fluorescence only (c,h), green fluorescence only (d,i), and by bright field merge with all fluorescent molecules (e,j) in both the non-infected and S. eriocheiris infected group. The white arrows indicate co-localization of MrLGBP and S. eriocheiris. Bars = 5 μm.
Identification and Expression of Ligand Proteins of MrLGBP
To determine the S. eriocheiris ligands interacting with MrLGBP, spiroplasma proteins were fractionated by SDS-PAGE (Figure 3A) and incubated with MrLGBP. The ligand proteins were identified by western blotting using anti-GST monoclonal antibodies. As shown in Figure 3A, lane 3, four significant protein bands, located at ~50, 70, 100, and 130 kDa were found to bind MrLGBP. The corresponding protein bands were excised from colloidal blue stained gels and used for MS/MS analysis. All four proteins were successfully sequenced and blasted. The 50 kDa protein was identified as enolase (GenBank accession number: AHF58090), and the 70 kDa protein were identified as transketolase (TK) (GenBank accession number: AHF57705). The 100 kDa protein was identified as ALDH (GenBank accession number: AHF57596), and the 130 kDa protein was identified as DNA-directed RNA polymerase subunit beta (Figure S2). Related outputs obtained by MASCOT are shown in Table S3. Based on previous reports (18, 34, 35), enolase, TK, and ALDH were focused. Enolase, TK and ALDH of S. eriocheiris have five, eight, and five TGA codons, respectively (Figure S3). For expression in E. coli, the TGA codons were mutated to TGG using the Fast Mutagenesis System. SeEnolase, TK, and ALDH proteins were successfully expressed and purified (Figures 3B1–B3, respectively).
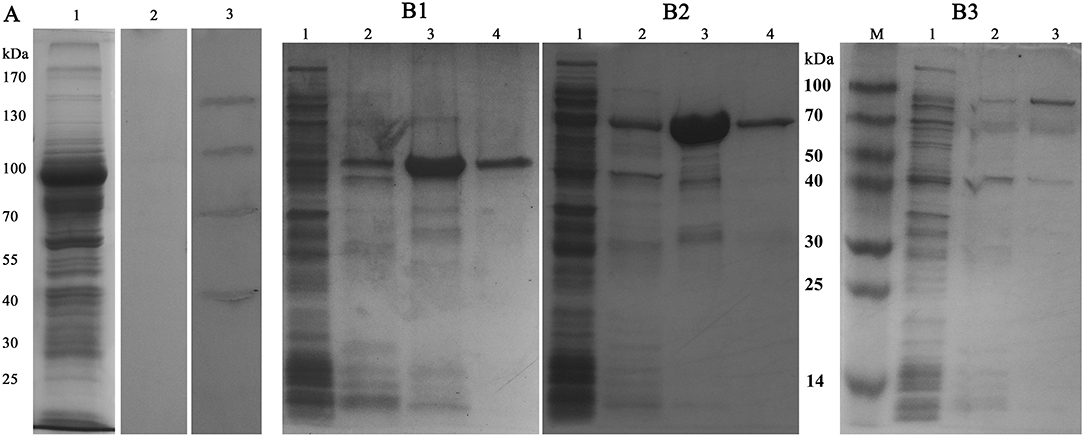
Figure 3. Identification and expression of ligand proteins for MrLGBP. (A) Far western blotting was used to identify proteins on S. eriocheiris that could bind to MrLGBP. S. eriocheiris total proteins were analyzed by SDS-PAGE (Lane 1). S. eriocheiris ligand proteins were detected by far western blotting (Lane 2 and 3). Line 1, S. eriocheiris proteins (20 μg). Line 2, GST tag as a control blot. Line 3, identification of ligand proteins for MrLGBP. (B1–B3) ligand proteins (SeEnolase, TK, and ALDH) recombinant expression in E. coli analyzed by SDS-PAGE. Lane 1, flow-through component; lane 2, eluate with 50 mM imidazole; lane 3, eluate with 100 mM imidazole; lane 4, eluate with 500 mM imidazole.
Influence of Ligand Proteins on Hemocyte Viability and Localization of S. eriocheiris SeEnolase
Once S. eriocheiris enolase, TK and ALDH were demonstrated to be involved in adhesion to prawn hemocyte surfaces, hemocyte surface proteins were identified by competitive binding. Hemocyte viability percentages were determined after infection with S. eriocheiris. The results showed that with SeEnolase incubation (Figure 4A), relative cell viability increased 20% (p < 0.05) compared with the negative control. However, for TK and ALDH, there was little or no effect on relative cell viability (Figures 4B,C).
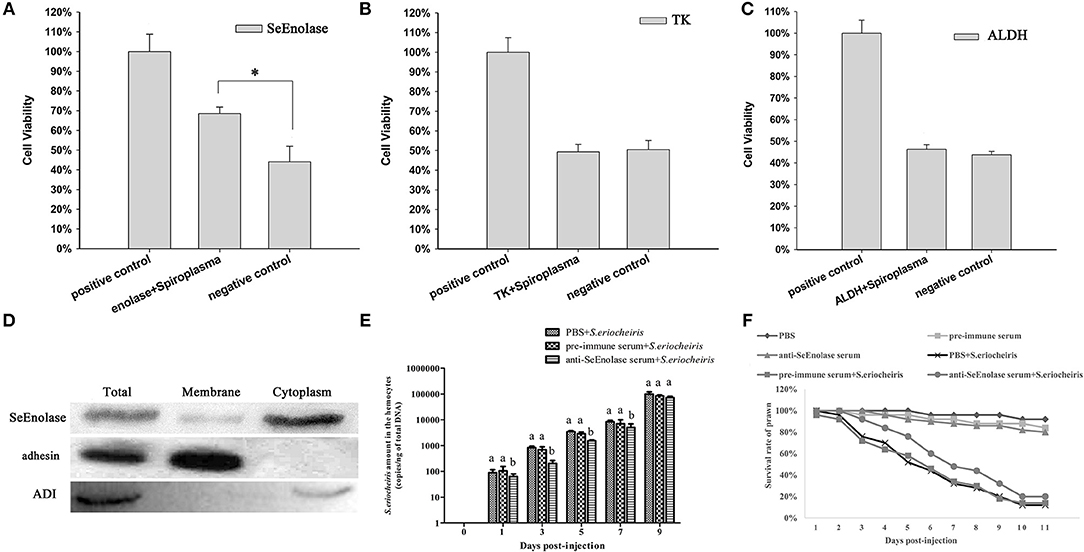
Figure 4. SeEnolase proteins mediate S. eriocheiris invasion into M. rosenbergii hemocytes. (A–C) Show competitive binding experiments with SeEnolase, TK, and ALDH, respectively. M. rosenbergii hemocyte were incubated with different ligand proteins and infected by spiroplasma. Cell viability was determined with CCK-8. Cells used for different treatments are shown on the abscissa, and relative cell viability rate on the ordinate. Vertical bars depict the mean ± S.E (n = 36). Significant differences were analyzed using one-way ANOVA by post-hoc Duncan multiple range tests. *p < 0.05. (D) Subcellular localization of SeEnolase. Western blotting was used to analyze the location of SeEnolase, three lanes are represented: total protein, membrane proteins, and cytoplasmic proteins of S. eriocheiris, respectively. (E) The quantification of S. eriocheiris copies in hemocytes from the three groups detected by real-time PCR at 1, 3, 5, 7, and 9 days, respectively. Differences between each group were analyzed using one-way ANOVA. Different letters indicate statistical significance (p < 0.05) and the same letter indicate no statistical difference (p > 0.05). (F) The survival rate of prawn infected using S. eriocheiris pretreated with antibody. Pretreated bacteria with anti-SeEnolase serum, pre-immune serum, or PBS were injected into prawn for the anti-SeEnolase serum + S. eriocheiris, pre-immune serum + S. eriocheiris, or PBS + S. eriocheiris group. Three another groups were injected with the anti-SeEnolase serum, pre-immune serum, and PBS, respectively. The cumulative mortality of prawns was recorded daily.
Western blot analysis of proteins from S. eriocheiris and purified recombinant SeEnolase using the anti-SeEnolase serum suggested that a protein of 50 kDa was detected (Figure S4). No immunoreactive band was detected in the control group using pre-immune rabbit serum. As shown in Figure 4D, SeEnolase was detected in lanes containing S. eriocheiris total protein, membrane proteins, and cytoplasmic proteins, suggesting that SeEnolase is exposed on the surface of S. eriocheiris. S. eriocheiris adhesin and arginine deiminase (ADI) were previously demonstrated (11) to be S. eriocheiris membrane and cytoplasmic proteins, respectively, as controls.
Enolase Antibody Prevented S. eriocheiris Invasion Into M. rosenbergii Hemocytes
As shown in Figure 4E, the copies of S. eriocheiris in the pre-immune serum + S. eriocheiris or PBS + S. eriocheiris group was significantly increased in the hemocytes from 1 to 7 days compared to the anti-SeEnolase serum + S. eriocheiris group (p < 0.05). These results shown that anti-SeEnolase serum could effectively prevent the S. eriocheiris invasion into M. rosenbergii. And, the survival rate of the pre-immune serum + S. eriocheiris or PBS + S. eriocheiris group was decreased compared with the anti-SeEnolase serum + S. eriocheiris group (Figure 4F). The number of live prawns during this experiment was recorded in Table S4. The results showed that the neutralization of SeEnolase using specific antibody could significantly suppress the S. eriocheiris pathogenicity.
Confirmation of the Interaction of MrLGBP With SeEnolase
The expression of respective constructs was detected a strong co-precipitation of SeEnolase-V5 with MrLGBP-GFP (Figure 5A). In contrast, SeEnolase-V5 did not precipitate with control GFP. These data demonstrate a molecular interaction between MrLGBP and SeEnolase. Recombinant MrLGBP and SeEnolase were assessed in cultured Drosophila S2 cells using a confocal laser scanning microscope. MrLGBP and SeEnolase were found to co-localize in Drosophila S2 cells (Figure 5B). Both co-immunoprecipitation and co-localization demonstrated that SeEnolase binds MrLGBP directly in Drosophila S2 cells. As shown in Figure 5C, MrLGBP protein with green fluorescence and SeEnolase protein with red fluorescence could overlap to produce yellow fluorescence. These results showed that the interaction of MrLGBP and SeEnolase was present not only in Drosophila S2 cells but also in M. rosenbergii hemocytes.
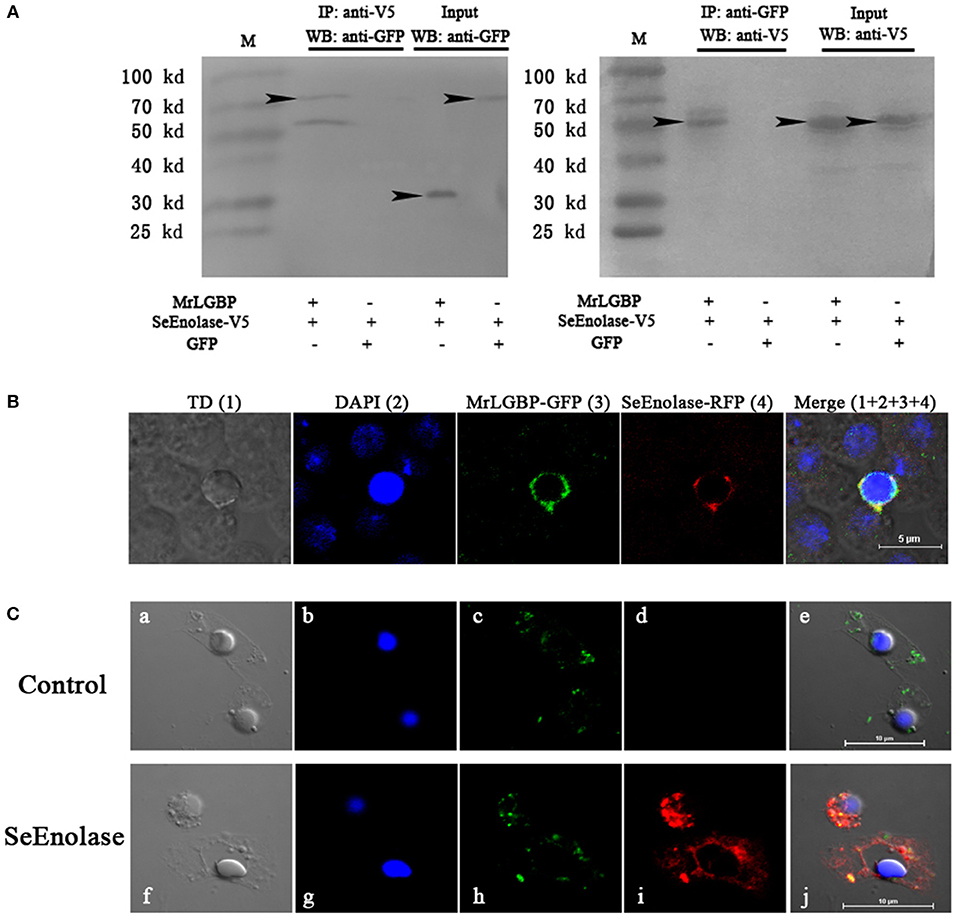
Figure 5. Interaction between MrLGBP and SeEnolase. (A) Co-immunoprecipitation demonstrating that GFP-tagged MrLGBP but not control GFP is co-precipitated with V5-tagged SeEnolase. Immunoprecipitation (IP) and western-blotting (WB) were performed using anti-V5 mouse antibody and anti-GFP rabbit antibodies, respectively. Input: western blotting of the input cell lysates before immunoprecipitation. Approximate molecular sizes: MrLGBP-GFP, ~68 kDa; SeEnolase-V5, ~50 kDa; GFP, ~28 kDa. (B) Subcellular localization of MrLGBP and SeEnolase. Recombinant plasmid of MrLGBP with GFP-tag and SeEnolase with RFP-tag were co-transfected into Drosophila S2 cells. MrLGBP and SeEnolase glowed green and red, respectively. Nuclei were stained with DAPI (blue). Bars = 5 μm. (C) The co-localization of MrLGBP and SeEnolase in prawn hemocytes analyzed by confocal microscopy. Hemocytes were incubated with SeEnolase protein for 1 h. For the control experiment, SeEnolase protein was not included. Hemocytes were incubated with mouse anti-MrLGBP and rabbit anti-SeEnolase as primary antibody, respectively. MrLGBP and SeEnolase stained by Alexa Flour 488 goat anti-Mouse IgG (green) and PE-labeled Goat Anti-Rabbit IgG (red). Hemocytes nuclei were stained with DAPI (blue). Bars = 10 μm. Hemocytes were visualized by bright field (a,f), blue fluorescence only (b,g), red fluorescence only (c,h), green fluorescence only (d,i), and by bright field merge with all fluorescent molecules (e,j) in both the control and SeEnolase group.
Overexpression of MrLGBP Promote Drosophila S2 Cells to Resist S. eriocheiris Infection
To assess the role of MrLGBP protein on S. eriocheiris infection, Drosophila S2 cells were transfected with a pAc5.1-MrLGBP-GFP plasmid and then infected with S. eriocheiris. Results showed that the number of S. eriocheiris was decreased in the S. eriocheiris + LGBP-GFP group compared to the S. eriocheiris + GFP group and the S. eriocheiris only group (Figure 6A). The result of western blot shown that the expression of MrLGBP, GFP-fusion MrLGBP and the GFP-tag were successful (Figure 6B). Real-time PCR results showed that the copy number of S. eriocheiris was 38,869/ng total DNA in the S. eriocheiris + LGBP-GFP group at 48 h post infection, whereas 389,311/ng and 646,956/ng total DNA was found in the S. eriocheiris + GFP group and in the S. eriocheiris only group, respectively (Figure 6C). Based on the CCK-8 assay, relative cell viability of the LGBP-GFP + S. eriocheiris group was significantly higher (p < 0.05) than that in the S. eriocheiris + GFP and S. eriocheiris only groups at 48 h post S. eriocheiris infection. CCK-8 test results showed that relative cell viability was 100% in the R2 medium group, whereas 50.57, 51.65, and 54.37% viability was found in the S. eriocheiris only group, the S. eriocheiris + GFP group, and the LGBP-GFP + S. eriocheiris group, respectively (Figure 6D). In a word, the overexpression of MrLGBP could help Drosophila S2 cells to resist S. eriocheiris infection.
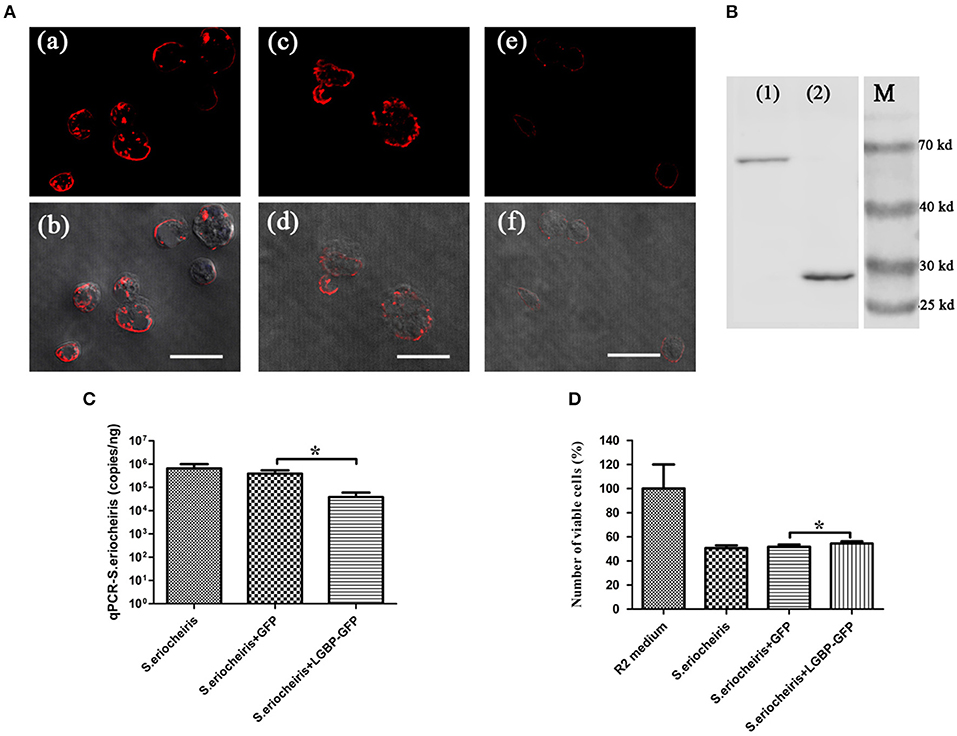
Figure 6. Overexpression of MrLGBP promote Drosophila S2 cells to resist S. eriocheiris infection. (A) Immunocytochemistry analysis S. eriocheiris quantity in Drosophila S2 cells. Drosophila S2 cells were transfected with pAc5.1-MrLGBP-GFP or pAc5.1-GFP plasmids. Drosophila S2 cells were infected with S. eriocheiris for 48 h. Then, the cells were incubated with anti-S. eriocheiris (primary antibody) and PE-labeled Goat anti-Rabbit IgG (second antibody, red), and examined using a confocal laser scanning microscope. (a,b) Represent the S. eriocheiris only group: (a), red fluorescence only, (b), the bright field merge with red fluorescence, (c,d) represents the S. eriocheiris + GFP group: (c), red fluorescence only, (d), bright field merge with red fluorescence, (e,f), represent the S. eriocheiris + LGBP-GFP group: (e), red fluorescence only, (f), bright field merge with red fluorescence, Bars, 10 μm. (B) Western blot of MrLGBP expression in Drosophila S2 cells. Drosophila S2 cells were transfected with pAc5.1-MrLGBP-GFP or pAc5.1-GFP plasmids. Efficiency of MrLGBP overexpression in Drosophila S2 cells, as detected by western blotting with anti-GFP antibody. (1), experimental group; (2), control group; (M), protein marker. Approximate molecular sizes: MrLGBP, ~68 kDa; GFP, ~28 kDa. (C) Real-time PCR analysis of copy number for S. eriocheiris in Drosophila S2 cells. Drosophila S2 cells were transfected with pAc5.1-MrLGBP-GFP or pAc5.1-GFP plasmids. Drosophila S2 cells were infected with S. eriocheiris for 48 h. S. eriocheiris quantity in Drosophila S2 cells. Vertical bars depict the mean ± S.E (n = 9). (D) Cell viability by CCK-8 assay. Drosophila S2 cells were transfected with pAc5.1-MrLGBP-GFP or pAc5.1-GFP plasmids. Drosophila S2 cells were infected with S. eriocheiris for 48 h. Relative cell viability was tested using CCK-8 assay. Cells used for different treatments are shown on the abscissa and relative cell viability rate on the ordinate. The assay was repeated three times. Vertical bars depict the mean ± S.E (n = 36). Significant difference *p < 0.05.
MrLGBP Silencing Reduced the Ability of M. rosenbergii Resistance to S. eriocheiris
In order to assess MrLGBP function during the prawn immune response, MrLGBP was silenced using RNAi during pathogen infection. The data show (Figure 7A) that the MrLGBP transcription was declined dramatically in the dsRNA-MrLGBP group compared to the dsRNA-GFP group, and maintained for 96 h after MrLGBP dsRNA inoculation. Therefore, the interference assay for MrLGBP was high efficiency. After MrLGBP interference, M. rosenbergii hemocyte PO activity was remarkably lower from 72 to 96 h (Figure 7B). The results suggested that MrLGBP silencing might reduce the innate immunity of prawns.
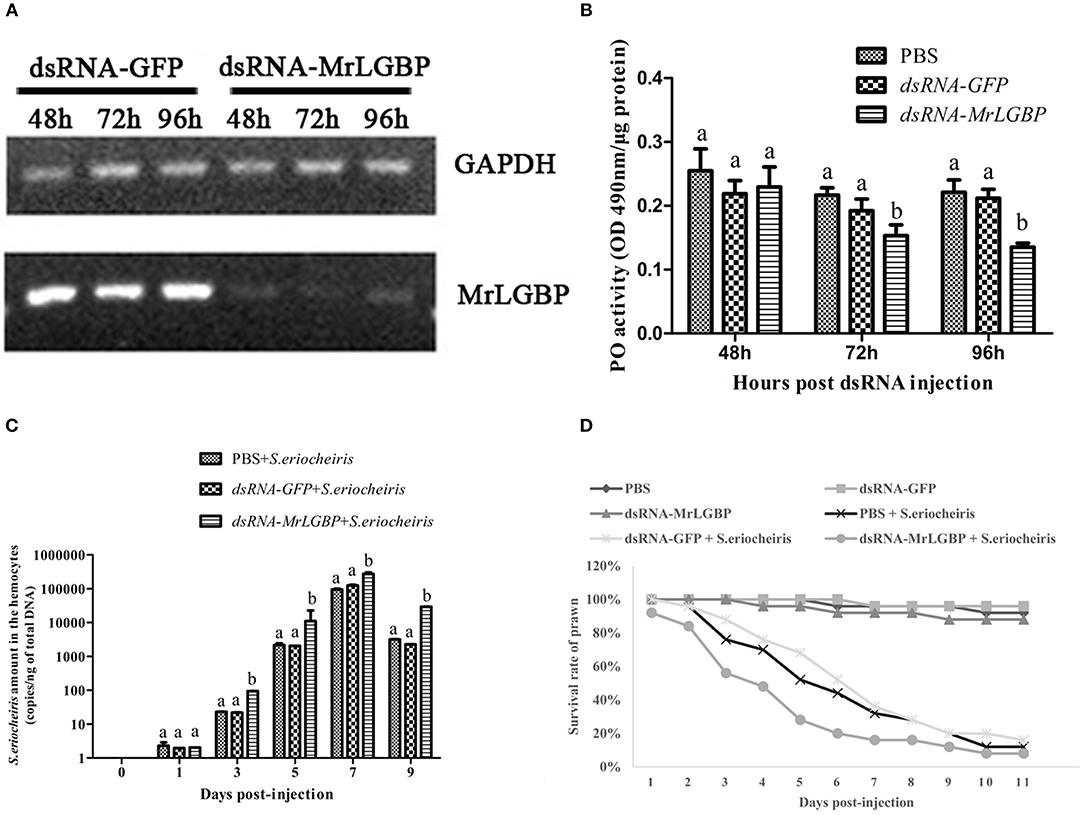
Figure 7. MrLGBP silencing reduced the ability of M. rosenbergii resistance to S. eriocheiris. (A) Efficiency of MrLGBP silencing in hemocytes, as detected by semi-quantitative PCR. (B) The phenol oxidase (PO) activity in hemocytes was determined after MrLGBP silencing. Differences between each group were analyzed using one-way ANOVA. Different letters indicate statistical significance (p < 0.05) and the same letter indicate no statistical difference (p > 0.05). (C) The quantification of S. eriocheiris copies in hemocytes from the three groups detected by real-time PCR at 1, 3, 5, 7, and 9 days, respectively. Statistical significance was determined by Student's t-test. Different letters indicate statistical significance (p < 0.05) and the same letter indicate no statistical difference (p > 0.05). (D) The survival rate of MrLGBP silencing prawn infected with S. eriocheiris. Prawn were divided six groups (50 prawn in each group). The prawns of the dsRNA-LGBP group and dsRNA-LGBP + S. eriocheiris group were injected with MrLGBP dsRNA. The prawns of the dsRNA-GFP group and the dsRNA-GFP + S. eriocheiris group were injected with GFP dsRNA. The prawns of the PBS group and PBS + S. eriocheiris group were injected with PBS. After 24 h, the prawns were injected again with the same amount dsRNA or PBS. Forty-eight hours after the first injection, the prawns of the PBS + S. eriocheiris group, dsRNA-GFP + S. eriocheiris group, and dsRNA-LGBP + S. eriocheiris group received an injection of S. eriocheiris. The cumulative mortality of prawns was recorded daily.
The change in S. eriocheiris copies in M. rosenbergii hemocytes was measured by real-time PCR (Figure 7C). Meanwhile, the number of died prawns during this experiment was recorded in Table S5. The copy number of S. eriocheiris in the dsRNA-LGBP + S. eriocheiris group was significantly increased in the hemocytes from 3 to 9 days compared to the dsRNA-GFP + S. eriocheiris group (p < 0.05). The survival rate of MrLGBP dsRNA injected prawns was decreased compared with the dsRNA-GFP + S. eriocheiris group (Figure 7D). The cumulative survival rate of the dsRNA-LGBP + S. eriocheiris group was 8% at 11 day, compared to the dsRNA-GFP + S. eriocheiris group and PBS + S. eriocheiris group, which was 28 and 32%, respectively. These results showed that MrLGBP interference reduced the ability of M. rosenbergii resistance to S. eriocheiris.
SeEnolase Induced Immune Responses and Improved the Ability of M. rosenbergii Resistance to S. eriocheiris
The expression of immune genes mRNA after SeEnolase protein stimulation in hemocytes was detected by qRT-PCR. The MrLGBP mRNA in the SeEnolase stimulation group was significantly up-regulated (p < 0.05) from 4 to 36 h (Figure 8A). And, compared with the PBS group, the expression of gene MrprpPO in the SeEnolase injected group was remarkably up-regulated from 2 to 48 h and demonstrated the highest expression at 12 h (Figure 8B). In addition, MrRab7A and Mrintegrin α1 genes were also obviously up-regulated compared with the control group from 12 to 36 h (Figure 8C) and 8 to 48 h (Figure 8D), respectively. These results indicated that the SeEnolase stimulation triggered the prawn innate immune response.
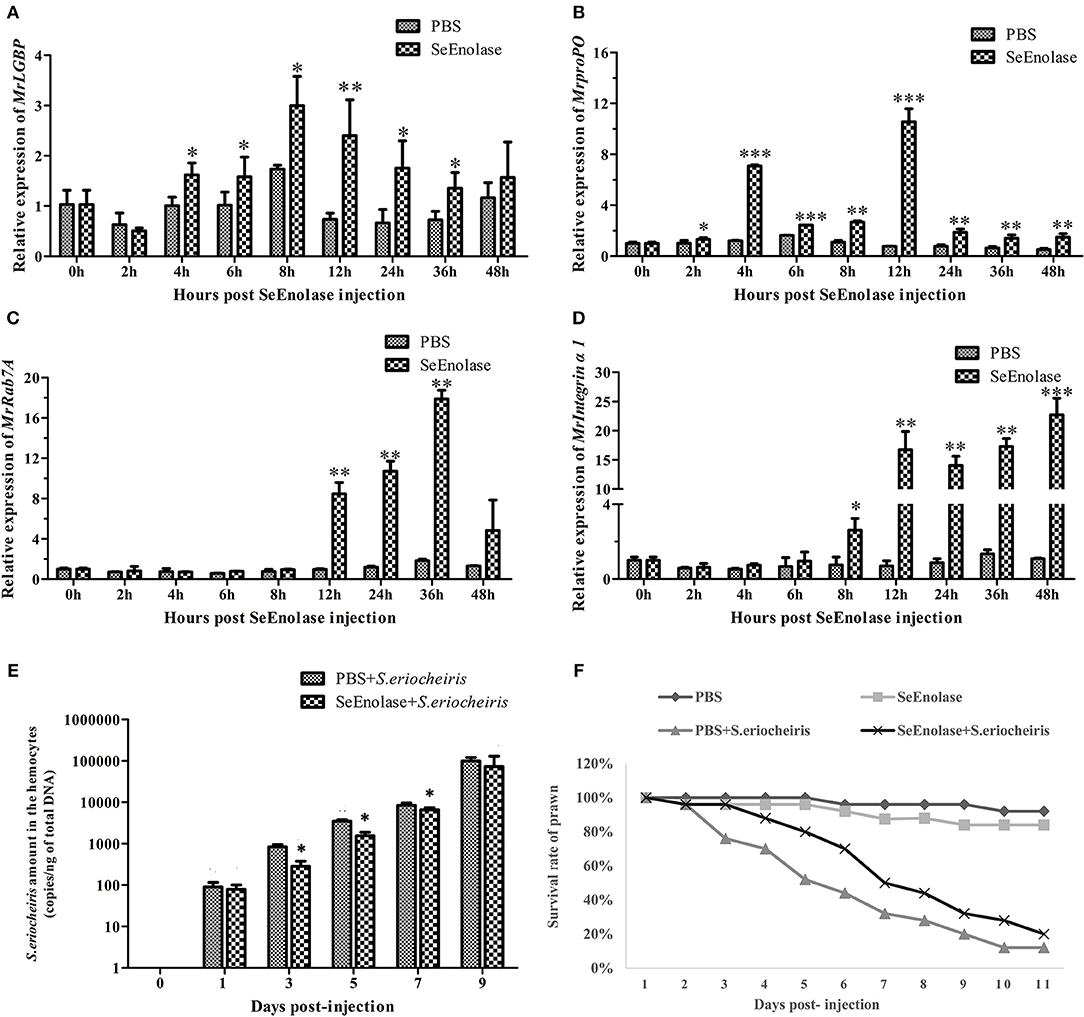
Figure 8. SeEnolase induced immune responses and improved the ability of M. rosenbergii resistance to S. eriocheiris. (A) Transcription levels of MrLGBP in hemocytes after SeEnolase stimulation. Prawns were injected with SeEolase protein as an experimental group and injected with PBS as control group. The hemocytes were sampled from every five individuals at 0, 2, 4, 6, 8, 12, 24, 36, and 48 h post-injection. Transcription levels of MrLGBP in hemocytes were analyzed by QRT-PCR. GAPDH was used as a reference gene. Statistical significance was determined by Student's t-test. Significant differences are indicated: *p < 0.05 and **p < 0.01. (B) QRT-PCR analysis of MrproPO in prawn hemocytes after SeEnolase stimulation. Statistical significance was determined by Student's t-test. Significant differences are indicated: *p < 0.05, **p < 0.01, and ***p < 0.001. (C) QRT-PCR analysis of MrRab7A in prawn hemocytes after SeEnolase stimulation. Statistical significance was determined by Student's t-test. Significant differences are indicated: **p < 0.01. (D) QRT-PCR analysis of Mrintegrin α1 in prawn hemocytes after SeEnolase stimulation. Statistical significance was determined by Student's t-test. Significant differences are indicated: *p < 0.05, **p < 0.01, and ***p < 0.001. (E) the quantification of S. eriocheiris copies in hemocytes from the three groups detected by real-time PCR at 1, 3, 5, 7, and 9 days, respectively. Statistical significance was determined by Student's t-test. Significant differences are indicated: *p < 0.05. (F) The survival rate of SeEnolase stimulation prawn infected with S. eriocheiris. Prawns were injected with SeEnolase protein in SeEnolase + S. eriocheiris group. For PBS + S. eriocheiris group, prawns were injected with PBS. After 12 h stimulation, the prawns were received an injection of S. eriocheiris. Two another groups were injected with the SeEnolase or PBS only. The cumulative mortality of prawns was recorded daily.
After being stimulated for another 12 h, the prawns were injected with S. eriocheiris. As shown in Figure 8E, the copy number of S. eriocheiris in the SeEnolase + S. eriocheiris group was significantly decreased in the hemocytes from 3 to 7 days compared to the PBS + S. eriocheiris group (p < 0.05). Meanwhile, the number of live prawns during this experiment was recorded in Table S6. The survival rate of the SeEnolase + S. eriocheiris group was increased comparison with the PBS + S. eriocheiris group (Figure 8F). The cumulative survival rate of the SeEnolase + S. eriocheiris group was 38% at 8 days, compared to the PBS + S. eriocheiris group, which was 8%. At the same time, cumulative survival rate of SeEnolase group and PBS group was 85 and 88%, respectively. The results showed that SeEnolase induced immune responses and enhanced the ability of M. rosenbergii resistance to S. eriocheiris.
Discussion
Spiroplasma belongs to a mollicute species that lacks cell wall and does not produce external toxins or endotoxins (3). Therefore, proteins that mediate the interaction of spiroplasma with host cells likely play an important role in pathogenesis. Identification of those interactive proteins will provide for a greater understanding of host-pathogen relationships.
Using a far western blotting assay, Killiny et al. (36) demonstrated spiroplasma proteins to have an affinity for seven leafhopper proteins. A leafhopper protein overlay assay on an S. citri protein blot showed that spiralin, which is the most abundant membrane protein of S. citri, displayed insect protein-binding activity. In 2010, five significant binding activities among S. citri proteins and insect host proteins were identified in salivary glands using an in vitro protein overlay assay (21) that identified actin as one of the insect binding proteins. An S. citri actin-binding protein of 44 kDa was isolated by affinity chromatography and identified as PGK. Competitive spiroplasma attachment and internalization assays demonstrated that PGK-FL5-actin interaction is required for the internalization of S. citri (16). In their crustacean hosts, an essential step in the pathogenic spiroplasma's life cycle is hemocyte invasion. In this investigation, six potential receptors were identified, including Ran, LGBP, beta-Actin, proPO, beta tubulin, and alpha-tubulin. These six proteins can be classified into three groups, a cytoskeleton group including beta-Actin, beta tubulin, and alpha-tubulin; an innate immunity group including LGBP and proPO and a signal transduction group including Ran.
As previously described, bacteria binding analysis suggested that the Fenneropenaeus chinensis LGBP (Fc-LGBP) protein was able to strongly bind to Gram-negative bacteria, with little or no binding to Gram-positive bacteria or yeast (37). Herein, the in vitro spiroplasma binding assay and confocal analysis confirmed that MrLGBP also bound spiroplasma, though S. eriocheiris belongs to a mollicute species that lacks a cell wall. Therefore, our point is that MrLGBP could not only bind Gram-negative bacteria, but also the cell wall-less bacteria. In addition, Fc-LGBP hemocyte membrane localization (37) was confirmed by immunohistochemistry as similar as our result. This suggested that transmembrane MrLGBP protein might have a function of recognizing invading microorganisms.
Ligand proteins of MrLGBP were identified as SeEnolase, TK, ALDH, and DNA-directed RNA polymerase subunit beta. In competitive assays, recombinant proteins, SeEnolase, TK, and ALDH were added to M. rosenbergii primary hemocyte cultures prior to infection with spiroplasma. Significant difference was found with SeEnolase but little or no difference for TK or ALDH. Invasion by S. eriocheiris was decreased after competitive inhibition by SeEnolase. Enolase has been reported to play a role as plasminogen receptor on the surface of several pathogenic bacteria (38, 39), fungi (40), and protozoa (41–43). For example, in Vibrio parahaemolyticus, the glycolytic enzyme enolase, a membrane associated protein located on the cell surface was found to bind plasminogen (28). Similar results were obtained with S. pneumoniae (18) and M. fermentans (17). And, enolases have raised interest as inducers of protective immunity and potential vaccine candidates in two tapeworm infections in recent years (44, 45). In this study, a portion of SeEnolase was located on S. eriocheiris' membrane, where SeEnolase could directly bind to combine with MrLGBP in vitro. The result suggests that S. eriocheiris adhesion to M. rosenbergii hemocytes may be due to interaction between SeEnolase and MrLGBP. From further investigation, the virulence ability of S. eriocheiris was effectively reduced by anti-SeEnolase serum neutralization assay. These results suggest that the loss of SeEnolase function reduces the ability of S. eriocheiris adhesion to host cells. In other words, SeEnolase plays an important role in the process of S. eriocheiris invasion into M. rosenbergii hemocytes.
Pattern recognition proteins (PRPs), including LGBP and lipopolysaccharides, have an ability to bind to pathogen-associated molecular patterns, on the surface of microorganisms, triggering cellular responses to that resist penetration of various pathogens (46). And in general, subsequent to recognition, LGBP induced a series of immune responses including encapsulation, phagocytosis, and the activation of the prophenoloxidase (proPO) system (6, 47) in invertebrates (46). Over-expression of MrLGBP decreased invasion of S. eriocheiris, and increased cellular proliferation. MrLGBP interaction with S. eriocheiris activates the proPO system, which increases the cellular immune response. Meanwhile, RNAi depletion of MrLGBP significantly reduced M. rosenbergii hemocyte PO activity and prawn survival rate, increased S. eriocheiris copies at same time. These results suggest that silencing of MrLGBP increased M. rosenbergii sensitivity to S. eriocheiris. In a word, MrLGBP was involved in the regulation of S. eriocheiris invasion into to M. rosenbergii hemocytes. Herein, MrLGBP, a hemocyte receptor protein, was demonstrated to bind spiroplasma by direct interaction with SeEnolase. Therefore, SeEnolase stimulation might induce the prawn innate immune responses. It is well-known that LGBP and prpPO were important components of proPO system (6). In addition, Rab and integrin proteins could regulate invertebrate hemocytic phagocytosis (48, 49) and encapsulation (50, 51), respectively. Our research showed that the transcription of genes MrLGBP, MrprpPO, MrRab7A, and Mrintegrin α1 were significantly up-regulated after SeEnolase stimulation, which suggested that prawn immune responses, including proPO system, phagocytosis and encapsulation were activated by SeEnolase. Crustaceans could use the innate immune to eliminate pathogens through activation of immune systems by LGBP. In our study, it was found that the activation of three immune systems by enolase stimulation led to an increase in resistibility against S. eriocheiris in M. rosenbergii. The evidence suggested that prawn immune system activation is due to SeEnolase binding to MrLGBP.
In conclusion, this investigation has identified interacting proteins between S. eriocheiris and M. rosenbergii hemocytes. Evidence showed S. eriocheiris SeEnolase, a surface-exposed protein, to promote pathogen-host interaction, involved in colonization and/or invasion of S. eriocheiris into M. rosenbergii hemocytes. In addition, MrLGBP, as a recognition protein, interacted with SeEnolase to defense the pathogen by activate prawn three innate immune systems (Figure 9). In summary, MrLGBP and SeEnolase involved in mediating S. eriocheiris invasion into M. rosenbergii hemocytes.
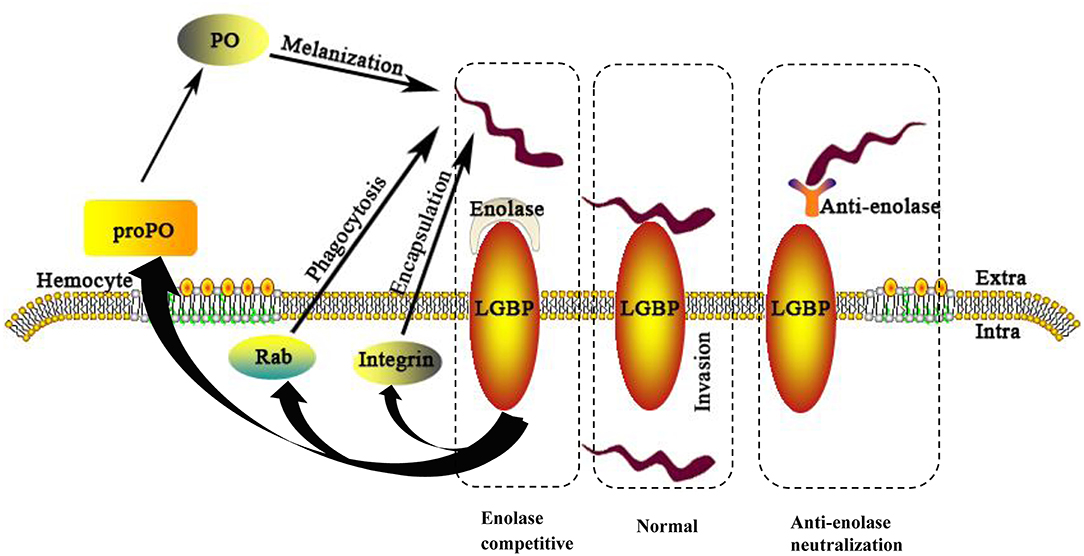
Figure 9. A schematic model of the M. rosenbergii hemocytes immune reaction against S. eriocheiris infected. For abbreviations and explanation see the text.
Ethics Statement
The animal subjects used in the present study are freshwater prawn, which are invertebrates and are exempt from this requirement.
Author Contributions
QM, MN, and YX designed experiments, analyzed experimental results, and wrote the manuscript. MY, JB, LH, WG, and WW conceived the idea, discussed data, and supervised this work.
Funding
The current study was supported by grants from the National Natural Sciences Foundation of China (NSFC Nos. 31570176; 31602198; 31870168), Project for Aquaculture in Jiangsu Province (Grant Nos. D2015-13; Y2016-28), the National Key Research and Development Program of China (Grant No. 2018YFD0900600), the Modern Fisheries Industry Technology System Project of Jiangsu Province (Grant No.JFRS-01), and the project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor O. Roger Anderson for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01852/full#supplementary-material
Figure S1. Mass spectrometry analysis of receptor proteins. Red letters represent successful blast with NCBInr. (A) Ras-related nuclear protein (Ran), similar to Marsupenaeus japonicus Ran with a shared coverage of 18%; (B) lipopolysaccharide and beta-1,3-glucan binding protein (LGBP), similar to M. rosenbergii LGBP with a shared coverage of 8%; (C) beta-actin protein, similar to Litopenaeus vannamei beta-actin with a shared coverage of 37%; (D) prophenoloxidase protein (proPO), similar to M. rosenbergii proPO with a shared coverage of 13%; (E) beta tubulin protein, to Penaeus monodon beta tubulin with a shared coverage of 28%; (F) alpha-tubulin protein, similar to Eriocheir sinensis alpha-tubulin with a shared coverage of 24 %; (G) alpha-tubulin protein, similar to Penaeus monodon alpha-tubulin with a shared coverage of 10%.
Figure S2. Mass spectrometry of ligand proteins. Red letters represent successful blast with NCBInr. (A) 50 kDa band was identified as enolase, shared of 7%; (B) 70 kDa band identified as transketolase (TK), shared coverage of 8%; (C) 100 kDa band was identified as acetaldehyde dehydrogenase (ALDH), shared coverage of 6%; (D) 130 kDa band was identified as DNA-directed RNA polymerase subunit beta, shared coverage of 7%.
Figure S3. Nucleotide and deduced amino acid sequence of enolase, transketolase (TK), and acetaldehyde dehydrogenase (ALDH) from S. eriocheiris. Nucleotide mutagenesis of recombinant plasmids are boxed. (A) The SeEnolase N-terminal domain (I4–L134) is underlined and the C-terminal domain (A139–N448) shaded in gray. (B) The TK dehydrogenases domain is underlined (F87–G244) and the pyrimidine binding domain shaded in gray (Q345–E515). (C) The aldehyde dehydrogenases domain is underlined (M1–R279). Stop codons are indicated by asterisks (*).
Figure S4. Western blotting of S. eriocheiris protein and purified recombinant SeEnolase. M, protein marker. Lane 1, S. eriocheiris; lane 2, purified recombinant SeEnolase, anti-SeEnolase serum as primary antigen. Lane 3, S. eriocheiris; lane 4, purified recombinant SeEnolase, pre-immune serum as the primary antibody.
Table S1. Sequences of primers used in this study.
Table S2. Related outputs obtained from MASCOT analysis.
Table S3. Related outputs obtained from MASCOT analysis.
Table S4. The number of lived prawns.
Table S5. The number of lived prawns.
Table S6. The number of lived prawns.
References
1. Kahlon A, Ojogun N, Ragland SA, Seidman D, Troese MJ, Ottens AK, et al. Anaplasma phagocytophilum asp14 is an invasin that interacts with mammalian host cells via its c terminus to facilitate infection. Infect Immun. (2013) 81:65–79. doi: 10.1128/IAI.00932-12
2. Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, et al. Anaplasma phagocytophilum outer membrane protein a interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun. (2012) 80:3748–60. doi: 10.1128/IAI.00654-12
3. Trachtenberg S. Mollicutes-wall-less bacteria with internal cytoskeletons. J Struct Biol. (1998) 124:244–56. doi: 10.1006/jsbi.1998.4063
4. Wang W, Gu W, Gasparich GE, Bi K, Ou J, Meng Q, et al. Spiroplasma eriocheiris sp. nov., associated with mortality in the Chinese mitten crab, Eriocheir sinensis. Int J Syst Evol Micr. (2011) 61 (Pt 4):703–8. doi: 10.1099/ijs.0.020529-0
5. Liang T, Li X, Du J, Yao W, Sun G, Dong X, et al. Identification and isolation of a spiroplasma pathogen from diseased freshwater prawns, Macrobrachium rosenbergii, in China: a new freshwater crustacean host. Aquaculture. (2011) 318:1–6. doi: 10.1016/j.aquaculture.2011.03.018
6. Cerenius L, Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. (2004) 198:116–26. doi: 10.1111/j.0105-2896.2004.00116.x
7. Lee SY, Söderhäll K. Early events in crustacean innate immunity. Fish Shellfish Immunol. (2002) 12:421–37. doi: 10.1006/fsim.2002.0420
8. Lai X, Kong J, Wang Q, Wang W, Meng X. Cloning and characterization of a β-1,3-glucan-binding protein from shrimp fenneropenaeus chinensis. Mol Biol Rep. (2011) 38:4527–35. doi: 10.1007/s11033-010-0583-3
9. Sritunyalucksana K, Soderhall K. The propo and clotting system in crustaceans. Aquaculture. (2000) 191:53–69. doi: 10.1016/S0044-8486(00)00411-7
10. Hoffmann JA, Reichhart JM, Hetru C. Innate immunity in higher insects. Curr Opin Immunol. (1996) 8:8–13. doi: 10.1016/S0952-7915(96)80098-7
11. Hou L, Xiu Y, Jian W, Liu X, Liu Y, Gu W, et al. iTRAQ-based quantitative proteomic analysis of Macrobrachium rosenbergii hemocytes during Spiroplasma eriocheiris infection. J Proteomics. (2015) 136:112–22. doi: 10.1016/j.jprot.2015.12.026
12. Dallo SF, Chavoya A, Baseman JB. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect Immun. (1990) 58:4163–5.
13. Rottem S. Interaction of mycoplasmas with host cells. Physiol Rev. (2003) 83:417–32. doi: 10.1152/physrev.00030.2002
14. Hou L, Liu Y, Gao Q, Xu X, Ning M, Bi J, et al. Spiroplasma eriocheiris adhesin-like protein (ALP) interacts with epidermal growth factor (EGF) domain proteins to facilitate infection. Front Cell Infect Microbiol. (2017) 7:13. doi: 10.3389/fcimb.2017.00013
15. Duret S, Berho N, Danet JL, Garnier M, Renaudin J. Spiralin is not essential for helicity, motility, or pathogenicity but is required for efficient transmission of Spiroplasma citri by its leafhopper vector Circulifer haematoceps. Appl Environ Microb. (2003) 69:6225–34. doi: 10.1128/AEM.69.10.6225-6234.2003
16. Labroussaa F, Dubrana MP, Arricau-Bouvery N, Béven L, Saillard C. Involvement of a minimal actin-binding region of Spiroplasma citri phosphoglycerate kinase in spiroplasma transmission by its leafhopper vector. PLoS ONE. (2011) 6:e17357. doi: 10.1371/journal.pone.0017357
17. Yavlovich A, Rechnitzer H, Rottem S. α-Enolase resides on the cell surface of Mycoplasma fermentans and binds plasminogen. Infect Immun. (2007) 75:5716–9. doi: 10.1128/IAI.01049-07
18. Bergmann S, Rohde M, Chhatwal GS, Hammerschmidt S. alpha-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol Microbiol. (2001) 40:1273–87. doi: 10.1046/j.1365-2958.2001.02448.x
19. Ding Z, Bi K, Wu T, Gu W, Wang W, Chen J. A simple PCR method for the detection of pathogenic spiroplasmas in crustaceans and environmental samples. Aquaculture. (2007) 265:49–54. doi: 10.1016/j.aquaculture.2007.01.032
20. Du J, Ou J, Li W, Ding Z, Wu T, Meng Q, et al. Primary hemocyte culture of the freshwater prawn Macrobrachium rosenbergii and its susceptibility to the novel pathogen spiroplasma strain MR-1008. Aquaculture. (2012) 330:21–8. doi: 10.1016/j.aquaculture.2011.12.003
21. Labroussaa F, Arricau-Bouvery N, Dubrana MP, Saillard C. Entry of Spiroplasma citri into Circulifer haematoceps cells involves interaction between spiroplasma phosphoglycerate kinase and leafhopper actin. Appl Environ Microbiol. (2010) 76:1879–86. doi: 10.1128/AEM.02384-09
22. Wang J, Huang H, Feng Q, Liang T, Bi K, Gu W, et al. Enzyme-liked immunosorbent assay for the detection of pathogenic spiroplasma in commercially exploited crustaceans from China. Aquaculture. (2009) 292:166–71. doi: 10.1016/j.aquaculture.2009.04.022
23. Killiny N, Batailler B, Foissac X, Saillard C. Identification of a Spiroplasma citri hydrophilic protein associated with insect transmissibility. Microbiology. (2006) 152 (Pt 4):1221–30. doi: 10.1099/mic.0.28602-0
24. Zhang XW, Wang XW, Sun C, Zhao XF, Wang JX. C-type lectin from red swamp crayfish Procambarus clarkii participates in cellular immune response. Arch Insect Biochem Physiol. (2011) 76:168–84. doi: 10.1002/arch.20416
25. Xia H, Liang W, Song Q, Chen X, Chen X, Hong J. The in vitro study of apoptosis in NB4 cell induced by citral. Cytotechnology. (2013) 65:49–57. doi: 10.1007/s10616-012-9453-2
26. Meng Q, Li W, Liang T, Jiang X, Gu W, Wang W. Identification of adhesin-like protein ALP41 from Spiroplasma eriocheiris and induction immune response of Eriocheir sinensis. Fish Shellfish Immunol. (2010) 29:587–93. doi: 10.1016/j.fsi.2010.06.005
27. Kornspan JD, Rottem S. Phospholipase A and glycerophosphodiesterase activities in the cell membrane of Mycoplasma hyorhinis. Fems Microbiol Lett. (2012) 332:34–9. doi: 10.1111/j.1574-6968.2012.02571.x
28. Jiang W, Han X, Wang Q, Li X, Yi L, Liu Y, et al. Vibrio parahaemolyticus enolase is an adhesion-related factor that binds plasminogen and functions as a protective antigen. Appl Microbio Biotechnol. (2014) 98:4937–48. doi: 10.1007/s00253-013-5471-z
29. Zhu H, Liu P, Du J, Wang J, Jing Y, Zhang J, et al. Identification of lysophospholipase protein from Spiroplasma eriocheiris and verification of its function. Microbiology. (2017) 163:175–84. doi: 10.1099/mic.0.000407
30. Ding Z, Tang J, Xue H, Li J, Ren Q, Gu W, et al. Quantitative detection and proliferation dynamics of a novel Spiroplasma eriocheiris pathogen in the freshwater crayfish, Procambarus clarkii. J Invertebr Pathol. (2014) 115: 51–4. doi: 10.1016/j.jip.2013.10.012
31. Li C, Chen YX, Shuang Z, Ling L, Chen YH, Chai J, et al. Identification, characterization, and function analysis of the cactus gene from Litopenaeus vannamei. PLoS ONE. (2012) 7:e49711. doi: 10.1371/journal.pone.0049711
32. Wang S, Qian Z, Li H, Lu K, Xu X, He J, et al. Identification and characterization of MKK7 as an upstream activator of JNK in Litopenaeus vannamei. Fish Shellfish Immunol. (2016) 48:285–94. doi: 10.1016/j.fsi.2015.12.014
33. Liu H, Jiravanichpaisal P, Cerenius L, Lee BL, Soderhall I, Soderhall K. Phenoloxidase is an important component of the defense against Aeromonas hydrophila infection in a crustacean, Pacifastacus leniusculus. J Biol Chem. (2007) 282:33593–8. doi: 10.1074/jbc.M706113200
34. Kochetov GA. Functional flexibility of the transketolase molecule. Biochemistry. (2001) 66:1077–85. doi: 10.1023/A:1012424711871
35. Schenk G, Duggleby RG, Nixon PF. Properties and functions of the thiamin diphosphate dependent enzyme transketolase. Int J Biochem Cell B. (1998) 30:1297–318. doi: 10.1016/S1357-2725(98)00095-8
36. Killiny N, Castroviejo M, Saillard C. Spiroplasma citri spiralin acts in vitro as a lectin binding to glycoproteins from its insect vector Circulifer haematoceps. Phytopathology. (2005) 95:541–8. doi: 10.1094/PHYTO-95-0541
37. Du XJ, Zhao XF, Wang JX. Molecular cloning and characterization of a lipopolysaccharide and beta-1,3-glucan binding protein from fleshy prawn (Fenneropenaeus chinensis). Mol Immunol. (2007) 44:1085–94. doi: 10.1016/j.molimm.2006.07.288
38. Xolalpa W, Vallecillo AJ, Lara M, Mendoza-Hernandez G, Comini M, Spallek R, et al. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics. (2010) 7:3332–41. doi: 10.1002/pmic.200600876
39. Sanderson-Smith ML, De Oliveira DM, Ranson M, McArthur JD. Bacterial plasminogen receptors: mediators of a multifaceted relationship. J Biomed Biotechnol. (2012) 2012:272148. doi: 10.1155/2012/272148
40. Verhamme IM, Panizzi PR, Bock PE. Pathogen activators of plasminogen. J Thromb Haemost. (2015) 13:S106–14. doi: 10.1111/jth.12939
41. Vanegas G, Quinnes V, Carrasco-Lopez C, Concepcion JL, Albericio F, Avilan L. Enolase as a plasminogen binding protein in Leishmania mexicana. Parasitol Res. (2007) 101:1511–16. doi: 10.1007/s00436-007-0668-7
42. Ghosh AK, Jacobs-Lorena M. Surface-expressed enolases of Plasmodium and other pathogens. Mem Inst Oswaldo Cruz. (2011) 106:85–90. doi: 10.1590/S0074-02762011000900011
43. Pancholi V, Fischetti VA. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. (1998) 273:14503–15. doi: 10.1074/jbc.273.23.14503
44. Yang J, Qiu C, Xia Y, Yao L, Fu Z, Yuan C, et al. Molecular cloning and functional characterization of Schistosoma japonicum enolasa: which is highly expressed at the schistosomulum stage. Parasitol Res. (2010) 107:667–77. doi: 10.1007/s00436-010-1913-z
45. Chen N, Yuan ZG, Xu MJ, Zhou DH, Zhang XX, Zhang YZ. Ascaris suum enolasa is a potential vaccine candidate against ascariasis. Vaccine. (2012) 30:3478–82. doi: 10.1016/j.vaccine.2012.02.075
46. Medzhitov R, Janeway C Jr. Innate immune recognition: mechanisms and pathways. Immunol Rev. (2000) 173:89–97. doi: 10.1034/j.1600-065X.2000.917309.x
47. Lee SY, Wang R, Söderhäll K. A lipopolysaccharide- and beta-1,3-glucan-binding protein from hemocytes of the freshwater crayfish Pacifastacus leniusculus. Purification, characterization, and cDNA cloning. J Biol Chem. (2000) 275:1337–43. doi: 10.1074/jbc.275.2.1337
48. Wu W, Zong R, Xu J, Zhang X. Antiviral phagocytosis is regulated by a novel rab-dependent complex in shrimp, Penaeus japonicus. J Proteome Res. (2008) 7:424–31. doi: 10.1021/pr700639t
49. Huang Y, Ren Q. Identification and function of 11 rab gtpases in giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. (2015) 43:120–30. doi: 10.1016/j.fsi.2014.12.021
50. Wang P, Zhuo X, Tang L, Liu X, Wang Y, Wang G, et al. C-type lectin interacting with β-integrin enhances hemocytic encapsulation in the cotton bollworm, Helicoverpa armigera. Insect Biochem Mol. (2017) 86:29–40. doi: 10.1016/j.ibmb.2017.05.005
Keywords: Macrobrachium rosenbergii, Spiroplasma eriocheiris, interactive proteins, innate immunity, infection
Citation: Ning M, Xiu Y, Yuan M, Bi J, Hou L, Gu W, Wang W and Meng Q (2019) Spiroplasma eriocheiris Invasion Into Macrobrachium rosenbergii Hemocytes Is Mediated by Pathogen Enolase and Host Lipopolysaccharide and β-1, 3-Glucan Binding Protein. Front. Immunol. 10:1852. doi: 10.3389/fimmu.2019.01852
Received: 29 August 2018; Accepted: 23 July 2019;
Published: 08 August 2019.
Edited by:
Gerardo R. Vasta, University of Maryland, Baltimore, United StatesReviewed by:
Kunlaya Somboonwiwat, Chulalongkorn University, ThailandHao-Ching Wang, Taipei Medical University, Taiwan
Copyright © 2019 Ning, Xiu, Yuan, Bi, Hou, Gu, Wang and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingguo Meng, bWx6emNsZEBhbGl5dW4uY29t
†These authors have contributed equally to this work
 Mingxiao Ning
Mingxiao Ning Yunji Xiu
Yunji Xiu Meijun Yuan1,2
Meijun Yuan1,2 Jingxiu Bi
Jingxiu Bi Qingguo Meng
Qingguo Meng