- Shandong Provincial Hospital for Skin Diseases & Shandong Provincial Institute of Dermatology and Venereology, Shandong First Medical University & Shandong Academy of Medical Sciences, Jinan, China
Leprosy, a disease caused by the intracellular parasite Mycobacterium leprae or Mycobacterium lepromatosis, has affected humans for more than 4,000 years and is a stigmatized disease even now. Since clinical manifestations of leprosy patients present as an immune-related spectrum, leprosy is regarded as an ideal model for studying the interaction between host immune response and infection; in fact, the landscape of leprosy immune responses has been extensively investigated. Meanwhile, leprosy is to some extent a genetic disease because the genetic factors of hosts have long been considered major contributors to this disease. Many immune-related genes have been discovered to be associated with leprosy. However, immunological and genetic findings have rarely been studied and discussed together, and as a result, the effects of gene variants on leprosy immune responses and the molecular mechanisms of leprosy pathogenesis are largely unknown. In this context, we summarized advances in both the immunology and genetics of leprosy and discussed the perspective of the combination of immunological and genetic approaches in studying the molecular mechanism of leprosy pathogenesis. In our opinion, the integrating of immunological and genetic approaches in the future may be promising to elucidate the molecular mechanism of leprosy onset and how leprosy develops into different types of leprosy.
Introduction
Leprosy is an ancient disease caused by Mycobacterium leprae or Mycobacterium lepromatosis infection, which mainly impairs skin and peripheral nerves and can even result in disability (1). The pathogens of leprosy have accompanied and affected humans for more than 4,000 years (2, 3), and over 200,000 new cases of leprosy are still reported each year worldwide (4) despite the application of multidrug therapy by the Word Health Organization. Because of the severe consequences caused by leprosy, including appearance changes and disabilities, leprosy is still a significant public health issue, especially in countries such as Brazil, India, and Indonesia, where the disease is still prevalent.
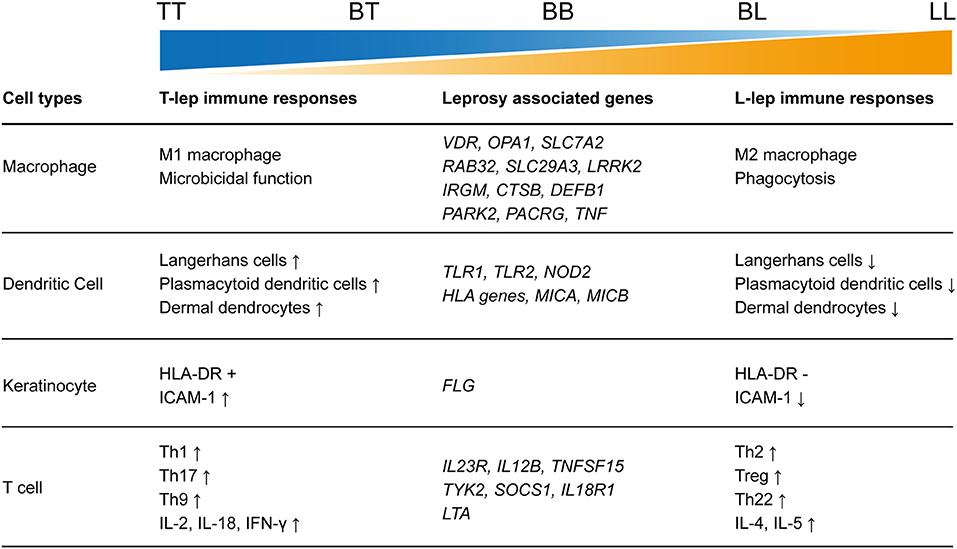
As the clinical manifestation of leprosy presents as a spectrum, it has long been considered an attractive model by immunologists to study the interaction between immune response and infection. Based on the different immune responses observed in patient lesions, leprosy can be categorized into five groups: tuberculoid (TT), borderline tuberculoid (BT), borderline borderline (BB), borderline lepromatous (BL), and lepromatous (LL) (5). The World Health Organization classifies leprosy clinically as multibacillary and paucibacillary, according to the number of skin lesions and nerve involvement (6). During the chronic infectious course, the immune-mediated acute inflammatory episodes called leprosy reactions frequently occurred. Leprosy reactions can be classified into two major types: type 1 reaction (T1R) or reversal reaction occurring mostly in unstable borderline patients (BT, BB, BL) and LL patients and type 2 reaction (T2R) or erythema nodosum leprosum (ENL) occurring mostly in BL and LL patients (7, 8). Therefore, leprosy is considered as an ideal disease model by immunologists to investigate the interrelation between pathogen load in infection and the differential immune responses of the host. Nevertheless, the pathogenesis of leprosy remains ambiguous due to the lack of an ideal animal model for this disease.
Leprosy is one of the most stigmatized diseases worldwide and was once thought to be a punishment from God on sinful persons due to the fact that only specific people developed this disease. In fact, for many infectious diseases, a common feature is that only a portion of the individuals who are exposed to the pathogens are actually infected and develop clinical symptoms, and genetic factors of the hosts have long been considered a major contributor to variances in susceptibility (9). In the case of leprosy, it has been estimated that just 5% of exposed individuals are successfully infected, of whom only 20% actually develop leprosy (10). Since the late 1900s, studies have shown that it is the genetic background and not God's punishment that makes infected individuals develop leprosy (11, 12). In the 21st century, studies using modern genetic approaches such as candidate gene association studies (CGASs) and genome-wide association studies (GWASs) have gradually confirmed that the host genetic background contributes greatly to the development of leprosy, and many leprosy-associated variants or genes have been reported. Most leprosy-associated genes are immune related, which is consistent with the finding that leprosy is caused by infection with pathogens.
Now it is clear that both the genetic background and the immune response of the host play essential roles in the development and manifestation of leprosy. Here, we firstly review studies on the responses of innate and adaptive immune cells in leprosy. We then summarize the leprosy-associated genes discovered by candidate gene and GWASs. Additionally, we proposed the combination of immunological and genetic studies to elucidate the underlying mechanisms of leprosy onset and development.
Leprosy as an Infectious Disease
Innate Immune Responses in Leprosy
Macrophages
As a key part of innate immunity and the major host of leprosy pathogens, macrophages have long been the focus of leprosy research. Macrophages in skin lesions of leprosy were dichotomously categorized into two types, the M1 type and M2 type. Epithelioid macrophages exhibiting M1 phenotype (CD68+CD163-) predominantly present in granulomas of TT patients, whereas macrophages in LL granulomas are foamy and mainly exhibit the M2 phenotype (CD68+CD163+) (13). Recently, de Sousa et al. (14) also characterized the presence of M4 macrophages in TT and LL lesions using double staining with markers of CD68 and MRP8. The expressions of both markers were stronger in LL than in TT lesions (14).
Although the differential polarization of macrophages in leprosy lesions is well-characterized, the intrinsic mechanisms of macrophage divergence in leprosy lesions are largely unknown. Interleukin (IL)-10, a key cytokine present in LL lesions, was shown to induce the phagocytosis program in human peripheral blood mononuclear cell (PBMC)-derived macrophages, whereas IL-15, which is abundant in TT lesions, triggered the vitamin D-dependent antimicrobial pathway (15). Using a co-culture system consisting of endothelial cells and monocytes, Kibbie et al. (16) found that unstimulated endothelial cells could trigger monocytes to become M2 macrophages, while endothelial cells stimulated by interferon (IFN)-γ or certain drugs induced the differentiation of monocytes to M1 macrophages in a Jagged1 (JAG1)-dependent manner (16). This study not only showed how macrophage divergence occurred at the site of infection but also provided new clues for intervening in intracellular infections. Moreover, a recent study focusing on miRNAome expression in leprosy physiopathology found that miR-34a-5p which controlled JAG1 expression was upregulated in lepromatous leprosy (L-lep) lesions (17). Thus, downregulation of JAG1 by miR-34a-5p allows the differentiation of M2 macrophage in L-lep, which is consistent with the discovery of a JAG1-dependent M1 macrophage differentiation (16). These studies demonstrate that cytokines, microRNA, and the microenvironment at the site of infection may play roles in macrophage divergence. Nonetheless, the reason these cytokines are differentially expressed in different types of leprosy remains mysterious, despite the exciting results of these two studies.
By comparing the expression of genes in polar forms of leprosy, three macrophage-related antimicrobial pathways have been found to function in the innate immune response to leprosy. As mentioned above, the vitamin D-dependent antimicrobial pathway has been characterized in TT leprosy (15). Differential expression of the miRNAome between TT and LL lesions found that microRNA hsa-mir-21 was mostly upregulated in LL lesions (18). Further analysis indicated that hsa-mir-21 could inhibit the expression of two vitamin D-dependent antimicrobial peptides by downregulating the Toll-like receptor 2/1 (TLR 2/1) pathway and upregulating IL-10 (18), which clearly demonstrated the interaction between leprosy pathogens and macrophage antimicrobial activity. In addition, other miRNAs targeting TLR4 and IL15R, which also regulated the vitamin D-dependent antimicrobial pathway, were also observed to be upregulated in L-lep, indicating the inhibition of vitamin D-dependent antimicrobial pathway (17). These studies repeatedly demonstrated the role of vitamin D-dependent antimicrobial pathway in the control of M. leprae infection.
Silva et al. (19) systematically analyzed the expression of autophagy genes in tuberculoid leprosy (T-lep) and L-lep lesions. Autophagy genes were found to be significantly upregulated in T leprosy, whereas the autophagic flux was impaired in L-lep but could be restored by IFN-γ or rapamycin (19). This study suggested that autophagy was an innate response of macrophages to control M. leprae. Even more, autophagy was also suggested to play an important role in T1R in multibacillary leprosy. In a study of multibacillary leprosy (BL and LL), the researchers found that autophagy was downregulated in patients who developed T1R in the future compared to patients who did not develop T1R (20). And the authors also demonstrated a significantly higher level of IL-1β in the serum of T1R group months before T1R onset, suggesting IL-1β as a potential marker for T1R prediction (20).
Another canonical antimicrobial pathway, the nitric oxide (NO) antimicrobial pathway, was also studied by a quantitative analysis of inducible NO synthase (iNOS) expression in polar forms of leprosy (21). These results showed that the expression of iNOS in the LL form was significantly higher than that in the TT form, and a linear correlation was also observed between iNOS and CD68 (21). Additionally, an increased expression of iNOS was also observed in T1R patients compared to non-reactional patients, indicating the activation of macrophage in T1R (22, 23). It is possible that M. leprae could induce the expression of iNOS, but the NO antimicrobial pathway alone cannot control M. leprae. This view was confirmed by a report of milestone significance (24). Using a zebrafish model, the authors clearly showed that phenolic glycolipid I (PGL-I) of M. leprae could induce the expression of NOS and increase the production of reactive nitrogen species, which then injured axons by impairing mitochondria and inducing demyelination (24). Thus, the NO antimicrobial pathway cannot control M. leprae and is in fact potentially a key inducement of nerve damage in leprosy. Most recently, studies found that phenolic glycolipid I (PGL-I) shaped the innate immune response not only in macrophages but also in polymorphonuclear neutrophils (PMNs) and dendritic cells (DCs) (25). The interaction of PGL-I with CR3 promoted the invasion of the bacteria into these innate cells and selectively increased the production of IL-2 by DCs, IL-10 by PMNs, and IL-1β by macrophages, respectively, through the CR3–Syk–NFATc axis (25). These studies demonstrated how the virulence factor of M. leprae PGL-I shaped the innate immune responses of innate cells, which may eventually effect the clinical symptoms of leprosy.
In a genome-wide study of mRNA expression in leprosy, mRNA of AKR1B10 was observed to be overexpressed in T2R lesions (26). Further immunohistochemistry investigation confirmed the overexpression of AKR1B10 in T2R and showed that AKR1B10 was principally expressed by macrophage in leprosy lesions (27). Due to the unknown function of AKR1B10 in leprosy, the authors could not determine whether AKR1B10 overexpression was an event accompanying T2R or contributed to T2R development. Nonetheless, AKR1B10 expression was a potential biomarker of T2R.
Dendritic Cells
DCs are professional antigen-presenting cells and can process and present antigens to T cells. Considering the essential roles of DCs in cell-mediated immunity, DCs were thought to play roles in leprosy pathogenesis and different types of DCs have been examined in leprosy by several studies (Table 1).
Langerhans cells (LCs) are resident DCs located in the epidermis, which express the lipid-presenting molecules, CD1a and CD207 (Langerin). A number of studies have shown similar results, indicating that the number of LCs in the epidermis of T-lep lesions was significantly larger than in L-lep lesions (28, 29, 31). Since LCs gathered antigens and drove T cell responses through antigen presentation in the draining lymph node (35), these findings are consistent with the fact that cell-mediated immune responses dominate in T-lep, whereas L-lep is characterized by a humoral immune response. For indeterminate leprosy, no significant difference in LC expression was observed by the authors comparing indeterminate leprosy lesions and normal skin (30). But an increase in the numbers of LCs was observed in both T1R and T2R compared to non-reactional leprosy, which was consistent with the acute inflammatory reactions in leprosy reactions (36, 37).
Leprosy lesions and granulomas appear mainly in the dermis of patients, so DCs in the dermis of leprosy lesions have also received attention. Plasmacytoid DCs (pDCs), which express CD123 and dermal dendrocytes (DDs) characterized by the FXIIIa marker were studied in polar forms of leprosy. Similar to LCs, the presence of a relatively large number of pDCs and DDs was observed in T-lep lesions, but much fewer of these DCs were found in L-lep lesions (29–34). The expression of CD123 marker was also evaluated in T1R and corresponding BL and LL lesions. Using immunohistochemistry, real-time polymerase chain reaction (RT-PCR), and flow cytometry methods, the authors clearly showed that CD123 was significantly more abundant in T1R compared to BL and LL lesions (32).
CD207+ DCs were evaluated in both the epidermis and dermis in polar forms of leprosy by Hirai et al. (34). Compared to L-lep, the authors found a larger number of CD207+ DCs not only in the epidermis but also in the inflammation area of the dermis of T-lep lesions (34). However, the authors were not sure whether these Langerin-positive DCs in the dermis of leprosy lesions were LCs migrating from the epidermis or were another type of dermis-resident DCs (34). DC-specific intercellular adhesion molecule (ICAM)-grabbing non-integrin (DC-SIGN) is a C-type lectin expressed by subsets of DCs and macrophages and is an entry receptor for pathogens (38). Different from LCs, pDCs, and DDs, more DC-SIGN+ cells were found in L-lep other than T-lep (39, 40). Moreover, CD11c+ cells in PBMC of L-lep patients also showed higher expression of DC-SIGN (41), and peripheral monocyte of L-lep patients differentiated into DC-SIGN+ macrophages but not CD1b+ DCs with efficient antigen presentation function after TLR2/1 activation (39).
In contrast to the characterization of different subsets of DCs in leprosy lesions, fewer studies have focused on the specific roles of DCs in leprosy pathogenesis. CD1+ DCs induced by M. leprae antigens have been demonstrated to be efficient antigen-presenting cells for T cells (33, 42). LCs isolated from the epidermis of healthy volunteers also showed better efficiency than monocyte-derived DCs in presenting non-peptide antigens of M. leprae to T cells (43). Conversely, the recognition of M. leprae by DC-SIGN+ DCs showed immunosuppressive function by inducing IL-10 (44). However, the ability to present M. leprae antigens and the effect of M. leprae on DC differentiation were not investigated in pDCs or DDs. Moreover, the roles of DCs in leprosy pathogenesis are not restricted to antigen presentation, as DCs were also suggested to contribute to granuloma formation (45). Gene MMP12, part of the tissue remodeling network, was found to be connected to DCs in T-lep lesions using a cell type deconvolution-based gene expression analysis of leprosy lesions, which suggested the involvement of DCs in leprosy granuloma formation and/or maintenance (45).
Keratinocytes
Although the lesions of leprosy and the causal pathogens appear mainly in the dermis, the presence of M. leprae in the pilosebaceous unit and epidermis in BL and LL patients were repeatedly reported (46–48). Keratinocytes in the epidermis also show several immune responses in leprosy lesions or against M. leprae infection. Immunohistological studies have found that keratinocytes are strongly positive for HLA-DR antigens overlying T-lep lesions, but anti HLA-DR reactivity was negative for keratinocytes in L-lep lesions (49, 50). Further investigation on the interplay between keratinocytes and T cells indicated that HLA-DR+ keratinoc ytes could present M. leprae antigens to CD4+ Th1-like cells in a human leukocyte antigen (HLA) class II-restricted manner (51), consistent with the observation that HLA-DR-positive keratinocytes were abundantly overlying T-lep lesions. The intracellular adhesion between keratinocytes and lymphocytes, which is important for efficient antigen presenting, was also discovered in leprosy (52). In lesions of T-lep, as well as in reversal reactions and Mitsuda reactions, keratinocytes were found to express pronounced levels of ICAM-1, and lymphocytes contained within these lesions were positive for the ICAM-1 ligand lymphocyte function-associated antigen-1 (LFA) (52).
In addition to acting as effective antigen-presenting cells, keratinocytes were also found to defend against M. leprae directly using antimicrobial means. Compared to normal skin, strong expression of nitrotyrosine and iNOS were observed in keratinocytes in granulomas from borderline leprosy patients (53). Higher expression of human beta-defensin 3 was found in lesions from patients displaying the type 1 reaction compared to leprosy patients negative for the type 1 reaction (54). Further studies indicated that keratinocytes, rather than macrophages, upregulated human beta-defensin 2 and human beta-defensin 3 in response to M. leprae stimulation (54). The in vitro phagocytosis of M. leprae by keratinocytes was shown for the first time by Lyrio et al. (55), who also demonstrated that keratinocytes infected by M. leprae increased the expression of cathelicidin and tumor necrosis factor (TNF)-α.
Adaptive Immune Responses in Leprosy
The Th1/Th2 Paradigm
Although the role of innate immune cells in leprosy pathogenesis cannot be neglected, it appears that the responses of T cells determine the outcome in leprosy development. In a murine model, Th1 cells that produced IL-2 and IFN-γ could prime macrophages to the microbicidal M1 polar state and produce a restricted form of the disease. In contrast, Th2 cells that produced IL-4 and IL-5 inhibited the microbicidal function of macrophages, resulting in the progressive form of the disease (56). In tuberculoid patient lesions, it was found that cytokines IL-2 and IFN-γ showed remarkably higher expression while IL-4, IL-5, and IL-10 were more abundant in lepromatous lesions (56). This cytokine pattern in leprosy lesions is very similar to the murine Th1/Th2 model. In addition, cytokine profiles of PBMCs from leprosy patients also showed a polar pattern according to leprosy type (57, 58). Although half of PBMCs from subjects showed non-discriminating Th0 responses upon M. leprae antigen stimulation, the remaining L-lep presented Th2 responses while T-lep showed Th1 cytokine responses (57). But the Th1 or Th2 responses is not irreversible in patients, for example, the shifting from a Th2 profile to a Th1 profile with the increased production of IFN-γ and CXCL10 is a prominent character of T1R (59–61).
The Th1/Th2 paradigm can explain the manifestations and histopathology of the two polar forms of leprosy. In localized TT, the cell-mediated immune response is strong and the bacilli are rarely observed, whereas in disseminated LL, the humoral response dominates and the bacilli load is high. In addition, during the past few decades, several other smaller lymphocyte subsets have been described in leprosy lesions, which may be involved in leprosy pathogenesis, especially in the shaping of host immune responses to infection.
The Reciprocal Relationship Between Regulatory T Cells and Th17 Subsets
Regulatory T cells (Tregs) and IL-17-producing Th17 cells are functionally and developmentally reciprocal to each other (62–64). Naive T cells develop into Tregs in the presence of transforming growth factor (TGF)-β, while the combination of TGF-β and IL-6/IL-21 introduce naive T cells into Th17 (62–64). Tregs with tolerance/immunosuppression functions are the primary mediators maintaining peripheral tolerance and are essential for the prevention of autoimmune diseases and chronic inflammatory diseases (65). However, Tregs may also suppress the appropriate host immune responses against infections (65). IL-17-producing Th17 cells with immunity/inflammation functions are a recently discovered and characterized subset of effector T helper cells, which have a reciprocal relationship with Tregs in subsets of developmental programs (63).
IL-10 producing CD4+CD25+FoxP3+ Tregs were analyzed in LL/BL, BT/TT, and healthy controls by Kumar et al. (66) who found that the expression of Tregs was higher in patients than in healthy controls and that LL/BL patients showed the highest expression of Tregs, which was consistent with the anergy of T cell responses in L-lep (66). Similarly, a high frequency of TGF-β secreting CD4+CD25+ FOXP3+ Tregs was also observed in L-lep (67). IL-35 is another suppressive cytokine and was shown to increase significantly in CD4+CD25+ Tregs of leprosy patients as compared to healthy controls (68). In contrast, Th17-related cytokines, chemokines, transcription factors, and Th17 cells showed higher expression in T-lep compared to L-lep (69, 70). The reciprocal relationship between Treg and Th17 cells has also been observed in leprosy (71). After M. leprae antigen stimulation, a higher frequency of Tregs was found in PBMCs of BL/LL patients, while a conversely higher frequency of Th17 cells was found in PBMCs from BT/TT patients (71). But the reciprocal relationship between Treg and Th17 was not irreversible, a recent study has shown that Tregs from leprosy patients could be converted to IL-17 producing Th17-like cells by rIL-23, suggesting a new way to overcome the immunosuppression in leprosy patients, especially in L-lep (72).
However, the reciprocal relationship between Treg and Th17 may be not applicable in leprosy reactions, especially for T1R. Since T1R and T2R are both acute inflammation reactions, it is reasonable to discover a higher frequency of Th17 in both T1R and T2R compared to non-reactional leprosy considering the inflammation functions of Th17 cells (73–75). Comparing to other forms of leprosy and healthy controls, T2R showed lowest circulating Treg frequency, which was consistent with the high Th17 frequency in T2R (58, 76). But it is not the case for T1R, several studies have demonstrated that T1R showed a significantly higher frequency of Treg compared to non-reactional leprosy in both PBMC and skin lesions (58, 74, 76–78). One possible explanation for the increase of Treg in T1R is that the increase of Treg in T1R is a self-protection mechanism to reduce the tissue damage caused by the exacerbated cell-mediated immune responses.
Th9 and Th22
In the presence of IL-4 and TGF-β, Th0 lymphocytes differentiate into Th9 lymphocytes, which preferentially produce IL-9, IL-10, and IL-21 (79). The Th22 lineage is characterized by the production of IL-22 and several fibroblast growth factors and also expresses the skin homing receptors CCR4 and CCR10, suggestive of the possible roles of Th22 in skin diseases (80). These two subsets of T helper cells were both studied by quantitative determination of their signature cytokines. IL-9 was found to be expressed more highly in TT lesions compared to LL lesions (81), consistent with the early finding that IL-9 could promote anti-M. leprae cytotoxicity (82). In contrast, the expression of Th22 signatures IL-22 and fibroblast growth factor basic (FGF-b) was higher in LL lesions compared to TT lesions (83).
γδ T Cells
γδ T cells is one of three lymphocyte lineages and only constitutes a very small proportion of lymphocytes as compared to the conventional αβ T cells and B cells (84). But γδ T cells have various immune functions and have been suggested to play roles in infectious diseases including leprosy (84, 85). In the case of leprosy, at least two different roles of γδ T cells have been suggested, namely, contributing to leprosy reactions and immunosuppression. Modlin et al. (86) firstly observed a significant increase of γδ T cell frequency in granulomatous reactions of leprosy. Following studies also confirmed that γδ T cell frequency increased significantly in Mitsuda reaction, a form of delayed hypersensitivity in leprosy (87, 88). But underlying mechanism of the contributions of γδ T cells to leprosy reactions was unknown until recently. Saini et al. (89) found that γδ T cells showed a higher frequency in both T1R and T2R reaction patients as compared to stable patients, and they also showed that these γδ T cells produced a notable amount of IL-17 and IFN-γ, which may explain the mechanism that γδ T cells contribute to leprosy reactions (89). In 2004, Sridevi et al. (90) reported the high level of γδ T cells in L-lep patients. And then the immunosuppressive role of γδ T cells was systematically studied (91). CD4+TCRγδ+FoxP3+ cells were observed to be significantly increased when moved from healthy controls and T-lep to L-lep patients, and the immunosuppressive nature of CD4+TCRγδ+FoxP3+ cells was also evidenced by in vitro experiments (91).
B Regulatory Cell
Humoral immunity in leprosy was suggested to be ineffective in pathogen elimination because M. leprae could survive and multiply in humoral immune-dominated L-lep despite the greater antibody responses in L-lep (92, 93). Therefore, B cell is the least cell population to be considered in leprosy pathogenesis studies, although it is a major immune cell population with antibodies secretory and antigen presentation functions. But recent studies on B cells showed that three subsets of B regulatory cells (Bregs) which showed immunosuppressive functions may play important roles in leprosy pathogenesis (68, 94–96). IL-35 producing Bregs showed a higher frequency in leprosy patients compared to healthy controls, and a positive correlation between IL-35 and bacteriological index was also observed (68). The second reported subset is IL-10 producing Breg, and this subset was also demonstrated to show an increased frequency in PBMCs of leprosy patients as compared to healthy controls (96). Furthermore, the authors showed that IL-10 producing Breg could convert effector T cells to Tregs and enhance the function of Treg (96). Besides IL-10 and IL-35 producing Breg, tissue-like memory B cells with immunosuppressive functions were also reported to be more abundant in L-lep comparing to T2R patients (95). These studies on Breg clearly showed that immunosuppressive functions of Breg may play an important role in the immunopathogenesis of leprosy.
The Hereditability of Leprosy Genetic Risk Factors
Association Between Leprosy and Innate Immune-Related Genes
Skin is the first physical barrier of human hosts against microbial invasion. Filaggrin, encoded by gene FLG, is the main constituent of keratohyalin granules and is indispensable for the proper function of the epidermal barrier. And loss-of-function mutations of FLG have been demonstrated to cause skin barrier deficiency and increase the risk to bacterial infection (97, 98). In an exome-wide association study, rs146466242 (K4022X), a loss-of-function mutation of FLG, was found to be associated with leprosy in Chinese populations (99). This finding raised the speculation that the impaired skin barrier might be an important route of M. leprae and other bacterial infections. But further genetically or biological experiments were needed to confirm this speculation. Genetically, more studies on the association between other infectious skin diseases and loss-of-function mutations of FLG could be performed. Biologically, the invasion of M. leprae and other bacteria into skin could be performed using mouse carrying flg loss-of-function mutations and human epidermal model constructed from keratinocyte with FLG loss-of-function mutations.
Innate immune cells recognize invading pathogens through receptors for pathogen-associated molecular patterns and then initiate specific innate immune responses. Both extracellular and intracellular pattern recognition receptor-coding genes have been found to be associated with leprosy. In a CGAS, functionally relevant coding single-nucleotide polymorphisms (SNPs) of TLR1/TLR2 were studied in 543 Bangladeshi leprosy patients and 842 healthy controls, and the polymorphism N248S was found to be associated with leprosy (100). Also, in the first large-scale GWAS of leprosy, it was discovered that the intracellular pattern recognition receptor gene NOD2 was associated with leprosy in Chinese (101). These findings were replicated in a case-control CGAS study using 933 patients in Nepal (102). The role of NOD2 in leprosy immune response was also functionally confirmed by cell-based experiments. In monocyte, recognition of NOD2 by leprosy muramyl dipeptide could induce the expression of IL-32, which regulates the differentiation from monocyte to CD1b+ DC (42, 103). But knowledge about effect of variants on function of NOD2 was still absent in leprosy.
Autophagy and phagocytosis are important defense mechanisms of the host innate immune system against intracellular pathogens (104, 105). Intron variant rs2275606 in gene RAB32, encoding a critical molecule required for the biogenesis of lysosomal-related organelles (106), was identified as associated with leprosy in a GWAS using Chinese Han population subjects (107). In a Listeria monocytogenes-infected DC model, Li et al. (108) provided direct evidence that Rab32 was part of a complex which encompassed bacteria and controlled the intracellular growth of L. monocytogenes. Additionally, the lysosome and endosome membrane protein-encoding gene SLC29A3 was also discovered by this research group to be a leprosy susceptibility gene (99). These findings indicated the involvement of autophagy or phagocytosis in leprosy pathogenesis. Moreover, two other genes that regulate autophagy, LRRK2 and IRGM, were found to be associated with leprosy (109, 110). In a zebrafish model, the LRRK2 mutant showed a weakened immune response to Mycobacterium marinum infection, functionally confirming the role of LRRK2 in infectious diseases (111).
To directly kill invading pathogens is one important part of innate cell functions. Several genes related to microbicidal functions have been identified as leprosy susceptibility genes. The exonic variant rs13259978 in SLC7A2 was found to be associated with leprosy (112). Since SLC7A2 is an important component of the classic nitric oxide antimicrobial pathway in macrophages, this finding suggested the involvement of the nitric oxide microbicidal pathway in leprosy. Another classic antimicrobial pathway, the vitamin D antimicrobial pathway has also been suggested to be involved in leprosy. Early in 1999, the TaqI polymorphism in the 3′ region of the vitamin D receptor (VDR) gene VDR was reported to be associated with leprosy in Indians (113). Furthermore, a silent change in codon 352 of the VDR gene also showed association with leprosy in Malawians (114). However, consistent results were not obtained in a case-control study including 933 leprosy patients and 101 controls in Nepal, in which the TaqI polymorphism of VDR showed no association with leprosy (115). As explained by the authors, this negative finding might be caused by population heterogeneity, different sample size, or alteration in the virulence of M. leprae in different geographical regions (115). Association between OPA1 common variants and L-lep was observed in Han Chinese of southwest China (116). Gene OPA1 encodes an inner membrane protein of mitochondria, which suggests a third antimicrobial pathway in leprosy, the mitochondrial antimicrobial pathway that functions by generating reactive oxygen species (116). In addition to these three antimicrobial pathways, several genes encoding products that have microbicidal functions were also found to be associated with leprosy, such as the lysosomal cysteine protease encoding gene CTSB (cathepsin B) (117), the antimicrobial peptide encoding gene DEFB1 (beta-defensin 1) (118) and the IFNG gene (119, 120).
HLAs play key roles in the presentation of antigens to T cells and are indispensable for adaptive immune responses. Several members belonging to the classical class I and class II HLA genes have been shown to be associated with leprosy. Leprosy-associated class I HLA genes include HLA-A*28 in Mestizo populations (121) and HLA-B*15 and HLA-C*05 in the Brazilian population (122). Leprosy-associated class II HLA genes include HLA-DQA1 in Brazilians (123), HLA-DQB1*06 and HLA-DQB1*07 in Mestizo populations (121), HLA-DRB1*11 in Brazilians (124), HLA-DRB1*1501 in Chinese and Indians (125, 126), and HLA-DRB1*1501 in Indians (125, 126). In addition, other antigen-processing and presentation-related genes were found to be associated with leprosy, such as the MICA gene (127), MICB gene (127), HLA-G gene (128), and TAP1 gene (129). Interestingly, even in the same population, some HLA genes showed susceptibility to leprosy and others showed resistance to leprosy, which indicated that some HLA molecules could activate T cells by presenting M. leprae antigens, while others are responsible for the T cell anergy in leprosy (121).
The complement system is an important component of the host innate immune system, and several gene members of the complement system were identified as associated with leprosy. In 2007, the association between MBL2 and leprosy was revealed by a study examining polymorphisms at the promoter and exon 1 regions of this gene (130). This group also found that FCN2 was associated with Brazilian leprosy patients (131). The mannose binding lectin encoded by MBL2 and ficolin-2 encoded by FCN2 both have essential roles in the lectin complement pathway. These two findings were subsequently confirmed by a candidate loci association study using Han Chinese, in which the authors found that genetic variants of FCN2 and MBL2 genes conferred susceptibility to leprosy (132). Moreover, this study also found two variants of the CFH gene, which showed significant associations with leprosy, indicating the involvement of the alternative complement pathway in leprosy pathogenesis (132).
Leprosy-Associated Adaptive Immune-Related Genes
Compared to innate immune-related genes, far fewer adaptive immune-related genes have been found to be associated with leprosy. Nevertheless, various genes involved in Th1, Th2, and Th17 differentiation and responses were found to be associated with leprosy. In a multiple-stage candidate susceptibility loci association study, IL18R1 was identified as a leprosy risk gene in a Chinese population. IL18R1 encodes the receptor of IL-18, which can promote Th1 responses to M. leprae (133). Also in a Chinese population, the 590T/C polymorphism of IL4 was found to be associated with leprosy, which indicated Th2 involvement in leprosy since IL-4 is a typical Th2 cytokine. Two IL-23/Th-17 pathway genes, IL12B encoding the heterodimeric subunit of IL-23 and IL23R encoding the IL23 receptor, were identified as leprosy susceptibility genes in different populations (107, 133–136). The Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathway is the downstream pathway of IL-23 signaling, and two key genes of this pathway, TYK2 and SOCS1, were also found to be associated with leprosy (99, 137). These findings strongly suggest the involvement of Th17 responses in leprosy. Moreover, TNFSF15, which encodes a mediator of the switch from Th1 to Th2 phenotype, was also found to be a leprosy susceptibility gene (101, 138).
Roles of Cytokine Gene Polymorphisms in Leprosy
Cytokines are secreted by various immune cells and play essential roles in shaping the immune responses in leprosy and even can drive the conversion between functionally antagonistic cells (Figure 1). Therefore, association between cytokine gene polymorphisms and leprosy has been one of the focuses in leprosy genetic study. In addition to IL4 and IL12B mentioned above, other cytokine genes that may play important roles in leprosy immune responses were also genetically investigated. The tumor necrosis factor (TNF)–lymphotoxin-α (LTA) locus was found to be leprosy-associated by both linkage analysis and SNP scanning of this region (139, 140). This region encodes pleiotropic cytokines TNF and LTA, which have various immune regulatory functions. Moreover, the most frequently involved SNP of TNF in infectious diseases, TNF-308 G>A (rs1800629), has been widely studied in leprosy with different ethnicities, including Nepalese, Brazilians, and Indians (115, 141, 142). However, these studies obtained inconsistent results for the association of TNF-308 G>A with leprosy, which inspired a meta-analysis of 14 studies on this topic (143). The meta-analysis found that no association was observed in the overall population or in Asians, but TNF-308 G>A showed a protective effect against leprosy risk in the Latin American population (143). For LTA of this region, a fine linkage disequilibrium mapping study discovered that LTA+80 A allele was significantly associated with leprosy risk (144). TNF and LTA were also suggested to play essential but different roles in the regulation of leprosy granuloma formation (145).
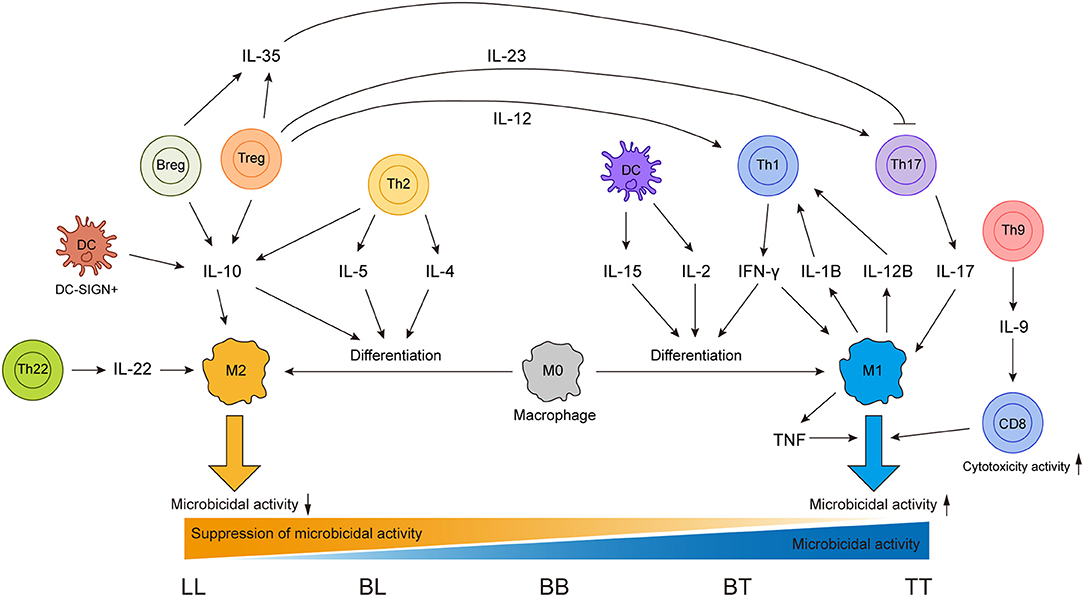
Figure 1. Sources of different cytokines and effects of these cytokines on different immune cells in leprosy immune responses.
IFN-γ can shape the immune responses in leprosy by activating innate immune cells (15); studies focused on the SNP +874 A/T (rs2430561) have been conducted in leprosy. In the Brazilian population, a protective effect of IFNG +874 A/T was observed which was confirmed by the higher level of IFN-γ produced by PBMCs (119). A meta-analysis using data from different populations also found that IFNG +874 A/T showed a protective effect for leprosy (146). IL-10, a cytokine with immunosuppressive properties secreted by monocyte and lymphocyte lineages, has been demonstrated to play roles in the development of L-lep (15, 96). Variant−819 C/T (rs1800871) in the promoter region of gene IL10 was repeatedly found to be associated with leprosy (142, 147). And two meta analyses focusing on IL10 variants both confirmed the significant susceptibility association between−819 C/T (rs1800871) and leprosy (148, 149). These results suggested that these variants of IFNG and IL10 might regulate the immune response and even the disease progression of leprosy.
Genes Associated With Leprosy Reactions
Although immunological studies have made significant progress in the pathogenesis of leprosy reactions, the intrinsic mechanism involved in the onset of leprosy reactions remains largely unknown. And host genetic factors have also been suggested to contribute to the developing of leprosy reactions (7). A comprehensive review on the genetics of leprosy reaction has well summarized the studies on the association between leprosy reactions and genes TLR1, TLR2, NOD2, IL6, C4B, VDR, and SLC11A1 (7). Therefore, here we reviewed several important advances of leprosy reactions genetic studies made in recent years.
TNFSF15 and TNFSF8 are both members of TNF-like molecules. TNFSF15 play roles in the switch from Th1 to Th2 cells (150, 151), while TNFSF8 impact the balance between Th1 and Th2 cells (152). Since immune responses in T1R is characterized by the switch from Th2 to Th1, it is reasonable to study the association between T1R and these two genes. To evaluate the association of these two genes with T1R, the fine mapping of the TNFSF15-TNFSF8 locus was performed separately in Vietnamese and Brazilians populations. And the results indicated that TNFSF8 was genetically associated with T1R in these two geographically and ethnically distinct populations, but not TNFSF15 (153). This study suggested that TNFSF8 might be a mediator of excessive inflammatory responses, which needs to be further confirmed. But the strong associations of TNFSF15/TNFSF8 variants rs6478108 and rs7863183 with T1R in Vietnamese patients were not observed in the Brazilian patients (153). Considering the significant difference in the age of diagnosis between Vietnamese and Brazilian populations, this research group further validated three regulators of the TNFSF8 gene transcription rs3181348, rs6478108, and rs7863183 in additional two Vietnamese and two Brazilian populations (154). They found that the associations with T1R for rs7863183 and rs3181348 were observed in all diagnosis age groups, but for rs6478108 was diagnosis age dependent which suggested that the regulation of TNFSF8 transcription might be age dependent (154).
The involvement of HLA complex in leprosy has been demonstrated immunologically and genetically, but genetic studies of HLA genes in borderline leprosy and leprosy reactions were rarely reported. de Souza-Santana et al. (122) genotyped the HLA class I (A*, B*, and C*) and class II (DRB1* and DQB1*) loci in a cohort consisting of 202 borderline leprosy (BT, BB, BL) patients of which 94 had T1R and 478 healthy controls (122). Their results demonstrated that HLA-C*05, HLA-DRB1*07, and HLA-DQB1*02 were genetically associated with borderline leprosy, while HLA-B*15 showed a significantly high frequency in T1R (122). Since the association between HLA and T1R is rarely reported in the literature, these results need further confirmation in different populations.
T1R is the main cause of nerve damage in leprosy patients (155). Causal genes of Parkinson's LRRK2 and PRKN were found to be associated with T1R in a genewise enrichment analysis including 63 coding variants of seven genes (156). Moreover, gain-of-function mutation LRRK2 R1628P was functionally shown to be protective for T1R by experiments performed in gene-edited RAW cells, and similarly, rare non-synonymous variants of PRKN were also functionally determined to be T1R risk factors (156). This study not only functionally linked the genetic associated mutations with nerve damage in T1R but also provided a good example to follow in the future for studying the pathogenesis of leprosy and T1R-associated variants.
Other Leprosy-Associated Genes
Genetic studies have also discovered other leprosy-associated genes, such as genes related to lipid metabolism, nerve damage, oxidative stress, and ubiquitin-mediated proteolysis. The accumulation of lipids in macrophages is thought to be beneficial for mycobacterium survival, and variants of two lipid metabolism related-genes, ALDH2 and APOE, were shown to confer risk for leprosy (112, 157). Although nerve damage is frequently observed in leprosy patients, the association between leprosy and nerve-related genes has rarely been reported. SYN2 encodes a neuronal phosphoprotein, which is a member of the synapsin gene family. In a large-scale genome-wide association and meta-analysis study, rs6807915 near SYN2 was identified as a susceptibility locus for leprosy. This finding suggested a role for synapsin-2 in the progression of infection from mycobacteria to the nerves (117). Unexpectedly, two oxidative stress-related genes, SOD2 and HIF1A, were also found to be associated with leprosy, which may be explained by the possibility that oxidative stress is a means of defense used by M. leprae-infected cells (158, 159). Early in 2004, a genome-wide linkage scan study reported the association between the 5′ regulatory region of PARK2/PACRG and Vietnamese leprosy, which was confirmed in Brazilian leprosy patients (160). Combining this genetic finding with the expression of PARK2 and PACRG in macrophages and Schwann cells, the authors pointed out that ubiquitin-mediated proteolysis might function in leprosy pathogenesis, but this association signal was not observed in Indians or Chinese (161, 162).
Expression of Human Genes in Leprosy
Plenty of literature have reported the genetic association between human genes and leprosy at DNA level using variants genotyping, while only a few studies have focused on the association between expression of human genes and leprosy. Studies on the expression of human genes in leprosy have been performed using various specimen types from different clinical forms of leprosy patients. A comprehensive genome-wide analysis of human mRNA expression in skin lesions of all forms of leprosy (TT, BT, BB, BL, LL, T1R, and T2R) was performed using microarrays, RT-PCR, and immunohistochemistry, and different forms of leprosy showed some unique differentially expressed genes, such as GPNMB, IL1B, MICAL2, and FOXQ1 in T1R and AKR1B10, FAM180B, FOXQ1, NNMT, NR1D1, PTX3, and TNFRSF25 in T2R (26). Although this finding revealed the complexity of this disease from a molecular point of view with solid evidences, few of discovered genes could be used to explain specific pathogenesis due to that these genes could hardly be linked to a specific cell type of leprosy lesions (26). In a cell-type deconvolution analysis of leprosy lesions transcriptome data, Inkeles et al. (45) found roles of some cell-defined genes in leprosy immunopathology. For example, the authors demonstrated granuloma formation/inflammation roles of MMP12 in DCs of T-Lep/T1R and chemotaxis roles of gene CXCL1, CXCL5, and CCR2 associated with neutrophil in T2R (45). Besides transcriptome profiling of leprosy lesions, transcriptome data of whole blood cells stimulated with M. leprae were also an efficient way to reveal gene expression signature related to leprosy. An elaborated analysis of transcriptome response of whole blood from former T1R and T1R-free patients to M. leprae sonicate identified a T1R gene set signature mainly consisting of innate pro-inflammation genes, such as CCL2, IL1A, and IL1B (163). This study suggested that defect in the regulation of innate pro-inflammation genes contributed to risk of T1R onset (163). A major problem of studies on expression of human genes in leprosy is that in rare cases, genetic associated genes were observed to be up or downregulated in any form of leprosy as compared to corresponding controls. Considering the high complexity in cell types of leprosy lesions, this problem may be caused by that genes per se are heterogeneously expressed in different types of cells.
Discussion
As an infectious disease, the landscape of immune responses in leprosy has been widely studied using skin lesions or PBMCs from leprosy patients. Various innate and adaptive immune cells have been shown to be involved in the immune responses to leprosy (Figure 2). Particularly, immunological studies have revealed remarkable differences between the two polar forms of leprosy (Figure 1). However, due to the lack of an efficient leprosy animal model, immunological studies were unable to delve deep into the gene or molecule level and could not explain why leprosy occurs only in certain populations and how leprosy develops into spectrum-like clinical manifestations. Genetic studies give us another opportunity to uncover the molecular mechanism underlying immune responses. Since most leprosy-associated genes are immune related (Figure 1), it is reasonable to speculate that changes in gene functions caused by leprosy-associated variants may make the immune system of the host unresponsive, disordered, or over-reactive when exposed to M. leprae infection, which contributes to the onset and spectrum development of leprosy. However, many leprosy-associated genes or variants have not been confirmed by biological experiments, leaving these genetic findings unsubstantiated.
The combination of immunological and genetic approaches is a promising way to explain leprosy pathogenesis more clearly and deeply. That is to say, we can fine-map the leprosy-associated gene loci and find the candidate causal variants and then explore the effect of these candidate causal variants on protein function and immune responses. The fine-mapping can be performed using large cohorts by next-generation sequencing, and cells from volunteers who carry candidate variants or gene-edited cells may be subjects for studies on the effects of these candidate variants on protein functions and immune responses; however, analysis may still be difficult because of the lack of an in vivo or in vitro leprosy model. The co-culture of innate and adaptive immune cells may be an option for an in vitro leprosy model, but up to now no such reported in vitro model has been widely accepted and applied. A mouse model for leprosy also failed due to the immune differences between mice and humans. A humanized mouse model may be a solution to this problem. Studies on the effects of disease-associated variants on protein function and immune response is not only important for the elucidation of leprosy pathogenesis but may also provide clues for genetic diagnosis and more precise therapeutic interventions.
Author Contributions
FZ and HL designed this work and revised the manuscript. ZM wrote the manuscript.
Funding
This work was funded by The National Key Research and Development Program of China (2016YFE0201500), the Academic promotion program of Shandong First Medical University (2019LJ002 and 2019RC007), the National Natural Science Foundation of China (81822038), the Key research and development program of Shandong Province (2019JZZY011010), and the Shandong Province Taishan Scholar Project (tspd20150214 and tsqn201812124).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Han XY, Silva FJ. On the age of leprosy. PLoS Negl Trop Dis. (2014) 8:e2544. doi: 10.1371/journal.pntd.0002544
2. Robbins G, Tripathy VM, Misra VN, Mohanty RK, Shinde VS. Gray KM, et al. Ancient skeletal evidence for leprosy in India. (2000 B.C.). PLoS ONE. (2009) 4:e5669. doi: 10.1371/journal.pone.0005669
3. Witas HW, Donoghue HD, Kubiak D, Lewandowska M, Gladykowska-Rzeczycka JJ. Molecular studies on ancient M. tuberculosis and M leprae: methods of pathogen and host DNA analysis. Eur J Clin Microbiol Infect Dis. (2015) 34:1733–49. doi: 10.1007/s10096-015-2427-5
4. WHO. Global leprosy update, 2016: accelerating reduction of disease burden. Wkly Epidemiol Rec. (2017). 92:501–19. http://apps.who.int/iris/bitstream/10665/258841/1/WER9235.pdf
5. Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis. (1966) 34:255–73.
6. Pannikar V. Enhanced global strategy for further reducing the disease burden due to leprosy: 2011-2015. Lepr Rev. (2009) 80:353–4. https://apps.who.int/iris/bitstream/handle/10665/205004/B4304.pdf?sequence=1
7. Fava V, Orlova M, Cobat A, Alcais A, Mira M, Schurr E. Genetics of leprosy reactions: an overview. Mem Inst Oswaldo Cruz. (2012) 107 (Suppl. 1):132–42. doi: 10.1590/S0074-02762012000900020
8. Voorend CG, Post EB. A systematic review on the epidemiological data of erythema nodosum leprosum, a type 2 leprosy reaction. PLoS Negl Trop Dis. (2013) 7:e2440. doi: 10.1371/journal.pntd.0002440
9. Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. (2012) 13:175–88. doi: 10.1038/nrg3114
10. Nunzi E, Massone C. Leprosy: A Practical Guide. New York, NY; Dordrecht: Springer Milan Heidelberg. (2012). doi: 10.1007/978-88-470-2376-5
11. Chakravartti M, Vogel F. A twin study on leprosy. In: Becker PE, Lenz W, Vogel F, Wendt GG, editors. Topics in Human Genetics. Stuttgart: Georg Thieme (1973). p. 1–123.
12. Shields ED, Russell DA, Pericak-Vance MA. Genetic epidemiology of the susceptibility to leprosy. J Clin Invest. (1987) 79:1139–43. doi: 10.1172/JCI112930
13. Fachin LR, Soares CT, Belone AF, Trombone AP, Rosa PS, Guidella CC, et al. Immunohistochemical assessment of cell populations in leprosy-spectrum lesions and reactional forms. Histol Histopathol. (2017) 32:385–96. doi: 10.14670/HH-11-804
14. de Sousa JR, Lucena Neto FD, Sotto MN, Quaresma JAS. Immunohistochemical characterization of the M4 macrophage population in leprosy skin lesions. BMC Infect Dis. (2018) 18:576. doi: 10.1186/s12879-018-3478-x
15. Montoya D, Cruz D, Teles RM, Lee DJ, Ochoa MT, Krutzik SR, et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe. (2009) 6:343–53. doi: 10.1016/j.chom.2009.09.002
16. Kibbie J, Teles RM, Wang Z, Hong P, Montoya D, Krutzik S, et al. Jagged1 instructs macrophage differentiation in leprosy. PLoS Pathog. (2016) 12:e1005808. doi: 10.1371/journal.ppat.1005808
17. Salgado CG, Pinto P, Bouth RC, Gobbo AR, Messias ACC, Sandoval TV, et al. miRNome expression analysis reveals new players on leprosy immune physiopathology. Front Immunol. (2018) 9:463. doi: 10.3389/fimmu.2018.00463
18. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, et al. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med. (2012) 18:267–73. doi: 10.1038/nm.2584
19. Silva BJ, Barbosa MG, Andrade PR, Ferreira H, Nery JA, Corte-Real S, et al. Autophagy is an innate mechanism associated with leprosy polarization. PLoS Pathog. (2017) 13:e1006103. doi: 10.1371/journal.ppat.1006103
20. de Mattos Barbosa MG, de Andrade Silva BJ, Assis TQ, da Silva Prata RB, Ferreira H, Andrade PR, et al. Autophagy impairment is associated with increased inflammasome activation and reversal reaction development in multibacillary leprosy. Front Immunol. (2018) 9:1223. doi: 10.3389/fimmu.2018.01223
21. de Sousa JR, de Sousa RPM, de Souza Aarao TL, Dias LB Jr, Oliveira Carneiro FR, Simoes Quaresma JA. Response of iNOS and its relationship with IL-22 and STAT3 in macrophage activity in the polar forms of leprosy. Acta Trop. (2017) 171:74–9. doi: 10.1016/j.actatropica.2017.03.016
22. Little D, Khanolkar-Young S, Coulthart A, Suneetha S, Lockwood DN. Immunohistochemical analysis of cellular infiltrate and gamma interferon, interleukin-12, and inducible nitric oxide synthase expression in leprosy type 1. (reversal) reactions before and during prednisolone treatment. Infect Immun. (2001) 69:3413–7. doi: 10.1128/IAI.69.5.3413-3417.2001
23. Lockwood DN, Suneetha L, Sagili KD, Chaduvula MV, Mohammed I, van Brakel W, et al. Cytokine and protein markers of leprosy reactions in skin and nerves: baseline results for the North Indian INFIR cohort. PLoS Negl Trop Dis. (2011) 5:e1327. doi: 10.1371/journal.pntd.0001327
24. Madigan CA, Cambier CJ, Kelly-Scumpia KM, Scumpia PO, Cheng TY, Zailaa J, et al. A macrophage response to mycobacterium leprae phenolic glycolipid initiates nerve damage in leprosy. Cell. (2017) 170:973–85 e910. doi: 10.1016/j.cell.2017.07.030
25. Doz-Deblauwe E, Carreras F, Arbues A, Remot A, Epardaud M, Malaga W, et al. CR3 Engaged by PGL-I triggers syk-calcineurin-NFATc to rewire the innate immune response in leprosy. Front Immunol. (2019) 10:2913. doi: 10.3389/fimmu.2019.02913
26. Belone Ade F, Rosa PS, Trombone AP, Fachin LR, Guidella CC, Ura S, et al. Genome-wide screening of mRNA expression in leprosy patients. Front Genet. (2015) 6:334. doi: 10.3389/fgene.2015.00334
27. Soares CT, Fachin LRV, Trombone APF, Rosa PS, Ghidella CC, Belone AFF. Potential of AKR1B10 as a biomarker and therapeutic target in type 2 leprosy reaction. Front Med. (2018) 5:263. doi: 10.3389/fmed.2018.00263
28. Gimenez MF, Gigli I, Tausk FA. Differential expression of Langerhans cells in the epidermis of patients with leprosy. Br J Dermatol. (1989) 121:19–26. doi: 10.1111/j.1365-2133.1989.tb01395.x
29. Simoes Quaresma JA, de Oliveira MF, Ribeiro Guimaraes AC, de Brito EB, de Brito RB, Pagliari C, et al. CD1a and factor XIIIa immunohistochemistry in leprosy: a possible role of dendritic cells in the pathogenesis of Mycobacterium leprae infection. Am J Dermatopathol. (2009) 31:527–31. doi: 10.1097/DAD.0b013e31819f1997
30. de Alvarenga Lira ML, Pagliari C, de Lima Silva AA, de Andrade HF Jr, Duarte MI. Dermal dendrocytes FXIIIa+ are essential antigen-presenting cells in indeterminate leprosy. Am J Dermatopathol. (2015) 37:269–73. doi: 10.1097/DAD.0000000000000238
31. Hirai KE, Aarao TL, Silva LM, de Sousa JR, de Souza J, Dias LB Jr, et al. Langerhans cells. (CD1a and CD207), dermal dendrocytes. (FXIIIa) and plasmacytoid dendritic cells. (CD123) in skin lesions of leprosy patients. Microb Pathog. (2016) 91:18–25. doi: 10.1016/j.micpath.2015.11.013
32. Andrade PR, Amadeu TP, Nery JA, Pinheiro RO, Sarno EN. CD123, the plasmacytoid dendritic cell phenotypic marker, is abundant in leprosy type 1 reaction. Br J Dermatol. (2015) 172:268–71. doi: 10.1111/bjd.13430
33. Sieling PA, Jullien D, Dahlem M, Tedder TF, Rea TH, Modlin RL, et al. CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol. (1999) 162:1851–8.
34. Hirai KE, Silva LM, de Sousa JR, de Souza J, Dias LB Jr, Oliveira Carneiro FR, et al. Langerin. (CD207)-positive cells in leprosy: Possible implications for pathogenesis of the disease with special emphasis on dermal immunoreactivity. Microb Pathog. (2018) 124:1–4. doi: 10.1016/j.micpath.2018.08.020
35. Kashem SW, Haniffa M, Kaplan DH. Antigen-presenting cells in the skin. Annu Rev Immunol. (2017) 35:469–99. doi: 10.1146/annurev-immunol-051116-052215
36. Rea TH, Shen JY, Modlin RL. Epidermal keratinocyte Ia expression, Langerhans cell hyperplasia and lymphocytic infiltration in skin lesions of leprosy. Clin Exp Immunol. (1986) 65:253–9.
37. Thangaraj H, Laal S, Thangaraj I, Nath I. Epidermal changes in reactional leprosy: keratinocyte Ia expression as an indicator of cell-mediated immune responses. Int J Lepr Other Mycobact Dis. (1988) 56:401–7.
38. Zhang F, Ren S, Zuo Y. DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection. Int Rev Immunol. (2014) 33:54–66. doi: 10.3109/08830185.2013.834897
39. Krutzik SR, Tan B, Li H, Ochoa MT, Liu PT, Sharfstein SE, et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med. (2005) 11:653–60. doi: 10.1038/nm1246
40. Soilleux EJ, Sarno EN, Hernandez MO, Moseley E, Horsley J, Lopes UG, et al. DC-SIGN association with the Th2 environment of lepromatous lesions: cause or effect? J Pathol. (2006) 209:182–9. doi: 10.1002/path.1972
41. Barreiro LB, Quach H, Krahenbuhl J, Khaliq S, Mohyuddin A, Mehdi SQ, et al. DC-SIGN interacts with Mycobacterium leprae but sequence variation in this lectin is not associated with leprosy in the Pakistani population. Hum Immunol. (2006) 67:102–7. doi: 10.1016/j.humimm.2006.02.028
42. Schenk M, Krutzik SR, Sieling PA, Lee DJ, Teles RM, Ochoa MT, et al. NOD2 triggers an interleukin-32-dependent human dendritic cell program in leprosy. Nat Med. (2012) 18:555–63. doi: 10.1038/nm.2650
43. Hunger RE, Sieling PA, Ochoa MT, Sugaya M, Burdick AE, Rea TH, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest. (2004) 113:701–8. doi: 10.1172/JCI200419655
44. Kumar S, Naqvi RA, Bhat AA, Rani R, Ali R, Agnihotri A, et al. IL-10 production from dendritic cells is associated with DC SIGN in human leprosy. Immunobiology. (2013) 218:1488–96. doi: 10.1016/j.imbio.2013.05.004
45. Inkeles MS, Teles RM, Pouldar D, Andrade PR, Madigan CA, Lopez D, et al. Cell-type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight. (2016) 1:e88843. doi: 10.1172/jci.insight.88843
46. Seo VH, Cho W, Choi HY, Hah YM, Cho SN. Mycobacterium leprae in the epidermis: ultrastructural study I. Int J Lepr Other Mycobact Dis. (1995) 63:101–4.
47. Hosokawa A. A clinical and bacteriological examination of Mycobacterium leprae in the epidermis and cutaneous appendages of patients with multibacillary leprosy. J Dermatol. (1999) 26:479–88. doi: 10.1111/j.1346-8138.1999.tb02032.x
48. Satapathy J, Kar BR, Job CK. Presence of Mycobacterium leprae in epidermal cells of lepromatous skin and its significance. Indian J Dermatol Venereol Leprol. (2005) 71:267–9. doi: 10.4103/0378-6323.16620
49. Volc-Platzer B, Stemberger H, Luger T, Radaszkiewicz T, Wiedermann G. Defective intralesional interferon-gamma activity in patients with lepromatous leprosy. Clin Exp Immunol. (1988) 71:235–40.
50. Volc-Platzer B, Kremsner P, Stemberger H, Wiedermann G. Restoration of defective cytokine activity within lepromatous leprosy lesions. Zentralbl Bakteriol. (1990) 272:458–66. doi: 10.1016/S0934-8840(11)80047-2
51. Mutis T, De Bueger M, Bakker A, Ottenhoff TH. HLA class II+ human keratinocytes present Mycobacterium leprae antigens to CD4+ Th1-like cells. Scand J Immunol. (1993) 37:43–51. doi: 10.1111/j.1365-3083.1993.tb01663.x
52. Sullivan L, Sano S, Pirmez C, Salgame P, Mueller C, Hofman F, et al. Expression of adhesion molecules in leprosy lesions. Infect Immun. (1991) 59:4154–60. doi: 10.1128/IAI.59.11.4154-4160.1991
53. Schon T, Hernandez-Pando RH, Negesse Y, Leekassa R, Sundqvist T, Britton S. Expression of inducible nitric oxide synthase and nitrotyrosine in borderline leprosy lesions. Br J Dermatol. (2001) 145:809–15. doi: 10.1046/j.1365-2133.2001.04491.x
54. Cogen AL, Walker SL, Roberts CH, Hagge DA, Neupane KD, Khadge S, et al. Human beta-defensin 3 is up-regulated in cutaneous leprosy type 1 reactions. PLoS Negl Trop Dis. (2012) 6:e1869. doi: 10.1371/journal.pntd.0001869
55. Lyrio EC, Campos-Souza IC, Correa LC, Lechuga GC, Vericimo M, Castro HC, et al. Interaction of Mycobacterium leprae with the HaCaT human keratinocyte cell line: new frontiers in the cellular immunology of leprosy. Exp Dermatol. (2015) 24:536–42. doi: 10.1111/exd.12714
56. Modlin RL. Th1-Th2 paradigm: insights from leprosy. J Invest Dermatol. (1994) 102:828–32. doi: 10.1111/1523-1747.ep12381958
57. Misra N, Murtaza A, Walker B, Narayan NP, Misra RS, Ramesh V, et al. Cytokine profile of circulating T cells of leprosy patients reflects both indiscriminate and polarized T-helper subsets: T-helper phenotype is stable and uninfluenced by related antigens of Mycobacterium leprae. Immunology. (1995) 86:97–103.
58. Abdallah M, Attia EA, Saad AA, El-Khateeb EA, Lotfi RA, El-Shennawy D. Serum Th1/Th2 and macrophage lineage cytokines in leprosy; correlation with circulating CD4(+) CD25(high) FoxP3(+) T-regs cells. Exp Dermatol. (2014) 23:742–7. doi: 10.1111/exd.12529
59. Geluk A, van Meijgaarden KE, Wilson L, Bobosha K, van der Ploeg-van Schip JJ, van den Eeden SJ, et al. Longitudinal immune responses and gene expression profiles in type 1 leprosy reactions. J Clin Immunol. (2014) 34:245–55. doi: 10.1007/s10875-013-9979-x
60. Andrade PR, Pinheiro RO, Sales AM, Illarramendi X, Barbosa MG, Moraes MO, et al. Type 1 reaction in leprosy: a model for a better understanding of tissue immunity under an immunopathological condition. Exp Rev Clin Immunol. (2015) 11:391–407. doi: 10.1586/1744666X.2015.1012501
61. Santos MB, de Oliveira DT, Cazzaniga RA, Varjao CS, Dos Santos PL, Santos MLB, et al. Distinct roles of Th17 and Th1 cells in inflammatory responses associated with the presentation of paucibacillary leprosy and leprosy reactions. Scand J Immunol. (2017) 86:40–9. doi: 10.1111/sji.12558
62. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. (2006) 441:235–8. doi: 10.1038/nature04753
63. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
64. Chen X, Oppenheim JJ. Th17 cells and tregs: unlikely allies. J Leukoc Biol. (2014) 95:723–31. doi: 10.1189/jlb.1213633
65. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. (2008) 8:523–32. doi: 10.1038/nri2343
66. Kumar S, Naqvi RA, Ali R, Rani R, Khanna N, Rao DN. CD4+CD25+ T regs with acetylated FoxP3 are associated with immune suppression in human leprosy. Mol Immunol. (2013) 56:513–20. doi: 10.1016/j.molimm.2013.04.015
67. Saini C, Ramesh V, Nath I. Increase in TGF-beta secreting CD4(+)CD25(+) FOXP3(+) T regulatory cells in anergic lepromatous leprosy patients. PLoS Negl Trop Dis. (2014) 8:e2639. doi: 10.1371/journal.pntd.0002639
68. Tarique M, Saini C, Naqvi RA, Khanna N, Rao DN. Increased IL-35 producing Tregs and CD19(+)IL-35(+) cells are associated with disease progression in leprosy patients. Cytokine. (2017) 91:82–8. doi: 10.1016/j.cyto.2016.12.011
69. Saini C, Ramesh V, Nath I. CD4+ Th17 cells discriminate clinical types and constitute a third subset of non Th1, non Th2 T cells in human leprosy. PLoS Negl Trop Dis. (2013) 7:e2338. doi: 10.1371/journal.pntd.0002338
70. Dang AT, Teles RM, Weiss DI, Parvatiyar K, Sarno EN, Ochoa MT, et al. IL-26 contributes to host defense against intracellular bacteria. J Clin Invest. (2019) 130:1926–39. doi: 10.1172/JCI99550
71. Sadhu S, Khaitan BK, Joshi B, Sengupta U, Nautiyal AK, Mitra DK. Reciprocity between Regulatory T Cells and Th17 Cells: relevance to polarized immunity in leprosy. PLoS Negl Trop Dis. (2016) 10:e0004338. doi: 10.1371/journal.pntd.0004338
72. Tarique M, Saini C, Naqvi RA, Khanna N, Sharma A, Rao DN. IL-12 and IL-23 modulate plasticity of FoxP3(+) regulatory T cells in human leprosy. Mol Immunol. (2017) 83:72–81. doi: 10.1016/j.molimm.2017.01.008
73. Saini C, Siddiqui A, Ramesh V, Nath I. Leprosy reactions show increased Th17 cell activity and reduced FOXP3+ tregs with concomitant decrease in TGF-beta and increase in IL-6. PLoS Negl Trop Dis. (2016) 10:e0004592. doi: 10.1371/journal.pntd.0004592
74. Costa MB, Hungria EM, Freitas AA, Sousa A, Jampietro J, Soares FA, et al. In situ T regulatory cells and Th17 cytokines in paired samples of leprosy type 1 and type 2 reactions. PLoS ONE. (2018) 13:e0196853. doi: 10.1371/journal.pone.0196853
75. Saini C, Srivastava RK, Kumar P, Ramesh V, Sharma A. A distinct double positive IL-17A(+)/F(+) T helper 17 cells induced inflammation leads to IL17 producing neutrophils in type 1 reaction of leprosy patients. Cytokine. (2020) 126:154873. doi: 10.1016/j.cyto.2019.154873
76. Attia EA, Abdallah M, El-Khateeb E, Saad AA, Lotfi RA, El-Shennawy D. Serum Th17 cytokines in leprosy: correlation with circulating CD4(+) CD25. (high)FoxP3. (+) T-regs cells, as well as down regulatory cytokines. Arch Dermatol Res. (2014) 306:793–801. doi: 10.1007/s00403-014-1486-2
77. Massone C, Nunzi E, Ribeiro-Rodrigues R, Talhari C, Talhari S, Schettini AP, et al. T regulatory cells and plasmocytoid dentritic cells in hansen disease: a new insight into pathogenesis? Am J Dermatopathol. (2010) 32:251–6. doi: 10.1097/DAD.0b013e3181b7fc56
78. Parente JN, Talhari C, Schettini AP, Massone C. T regulatory cells. (TREG)(TCD4+CD25+FOXP3+) distribution in the different clinical forms of leprosy and reactional states. An Bras Dermatol. (2015) 90:41–7. doi: 10.1590/abd1806-4841.20153311
79. Schmitt E, Klein M, Bopp T. Th9 cells, new players in adaptive immunity. Trends Immunol. (2014) 35:61–8. doi: 10.1016/j.it.2013.10.004
80. Jia L, Wu C. The biology and functions of Th22 cells. Adv Exp Med Biol. (2014) 841:209–30. doi: 10.1007/978-94-017-9487-9_8
81. de Sousa JR, Pagliari C, de Almeida DSM, Barros LFL, Carneiro FRO, Dias LB Jr, et al. Th9 cytokines response and its possible implications in the immunopathogenesis of leprosy. J Clin Pathol. (2017) 70:521–7. doi: 10.1136/jclinpath-2016-204110
82. Finiasz MR, Franco MC, de la Barrera S, Rutitzky L, Pizzariello G, del Carmen Sasiain M, et al. IL-9 promotes anti-Mycobacterium leprae cytotoxicity: involvement of IFNgamma. Clin Exp Immunol. (2007) 147:139–47. doi: 10.1111/j.1365-2249.2006.03241.x
83. de Lima Silveira E, de Sousa JR, de Sousa Aarao TL, Fuzii HT, Dias Junior LB, Carneiro FR, et al. New immunologic pathways in the pathogenesis of leprosy: role for Th22 cytokines in the polar forms of the disease. J Am Acad Dermatol. (2015) 72:729–30. doi: 10.1016/j.jaad.2014.11.023
84. Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. (2002) 2:336–45. doi: 10.1038/nri797
85. Saini C, Tarique M, Rai R, Siddiqui A, Khanna N, Sharma A. T helper cells in leprosy: an update. Immunol Lett. (2017) 184:61–6. doi: 10.1016/j.imlet.2017.02.013
86. Modlin RL, Pirmez C, Hofman FM, Torigian V, Uyemura K, Rea TH, et al. Lymphocytes bearing antigen-specific gamma delta T-cell receptors accumulate in human infectious disease lesions. Nature. (1989) 339:544–8. doi: 10.1038/339544a0
87. Uyemura K, Ho CT, Ohmen JD, Rea TH, Modlin RL. Selective expansion of V delta 1 + T cells from leprosy skin lesions. J Invest Dermatol. (1992) 99:848–52. doi: 10.1111/1523-1747.ep12614809
88. Fujita M, Miyachi Y, Nakata K, Imamura S. Appearance of gamma delta T cell receptor-positive cells following alpha beta T cell receptor-positive cells in the lepromin reaction of human skin. Immunol Lett. (1993) 35:39–44. doi: 10.1016/0165-2478(93)90145-R
89. Saini C, Tarique M, Ramesh V, Khanna N, Sharma A. Gammadelta T cells are associated with inflammation and immunopathogenesis of leprosy reactions. Immunol Lett. (2018) 200:55–65. doi: 10.1016/j.imlet.2018.07.005
90. Sridevi K, Neena K, Chitralekha KT, Arif AK, Tomar D, Rao DN. Expression of costimulatory molecules. (CD80, CD86, CD28, CD152), accessory molecules. (TCR alphabeta, TCR gammadelta) and T cell lineage molecules. (CD4+, CD8+) in PBMC of leprosy patients using Mycobacterium leprae antigen. (MLCWA) with murabutide and T cell peptide of Trat protein. Int Immunopharmacol. (2004) 4:1–14. doi: 10.1016/j.intimp.2003.09.001
91. Tarique M, Naqvi RA, Ali R, Khanna N, Rao DN. CD4(+) TCRgammadelta(+) FoxP3(+) cells: an unidentified population of immunosuppressive cells towards disease progression leprosy patients. Exp Dermatol. (2017) 26:946–8. doi: 10.1111/exd.13302
92. Ochoa MT, Teles R, Haas BE, Zaghi D, Li H, Sarno EN, et al. A role for interleukin-5 in promoting increased immunoglobulin M at the site of disease in leprosy. Immunology. (2010) 131:405–14. doi: 10.1111/j.1365-2567.2010.03314.x
93. Montoya DJ, Andrade P, Silva BJA, Teles RMB, Ma F, Bryson B, et al. Dual RNA-Seq of human leprosy lesions identifies bacterial determinants linked to host immune response. Cell Rep. (2019) 26:3574–85 e3573. doi: 10.1016/j.celrep.2019.02.109
94. Mizoguchi A, Bhan AK. A case for regulatory B cells. J Immunol. (2006) 176:705–10. doi: 10.4049/jimmunol.176.2.705
95. Negera E, Walker SL, Bekele Y, Dockrell HM, Lockwood DN. Increased activated memory B-cells in the peripheral blood of patients with erythema nodosum leprosum reactions. PLoS Negl Trop Dis. (2017) 11:e0006121. doi: 10.1371/journal.pntd.0006121
96. Tarique M, Naz H, Kurra SV, Saini C, Naqvi RA, Rai R, et al. Interleukin-10 producing regulatory B cells transformed CD4(+)CD25(-) into tregs and enhanced regulatory T cells function in human leprosy. Front Immunol. (2018) 9:1636. doi: 10.3389/fimmu.2018.01636
97. Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. (2006) 38:337–42. doi: 10.1038/ng1743
98. Cai SC, Chen H, Koh WP, Common JE, van Bever HP, McLean WH, et al. Filaggrin mutations are associated with recurrent skin infection in Singaporean Chinese patients with atopic dermatitis. Br J Dermatol. (2012) 166:200–3. doi: 10.1111/j.1365-2133.2011.10541.x
99. Liu H, Wang Z, Li Y, Yu G, Fu X, Wang C, et al. Genome-wide analysis of protein-coding variants in leprosy. J Invest Dermatol. (2017) 137:2544–51. doi: 10.1016/j.jid.2017.08.004
100. Schuring RP, Hamann L, Faber WR, Pahan D, Richardus JH, Schumann RR, et al. Polymorphism N248S in the human Toll-like receptor 1 gene is related to leprosy and leprosy reactions. J Infect Dis. (2009) 199:1816–9. doi: 10.1086/599121
101. Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, et al. Genomewide association study of leprosy. N Engl J Med. (2009) 361:2609–18. doi: 10.1056/NEJMoa0903753
102. Berrington WR, Macdonald M, Khadge S, Sapkota BR, Janer M, Hagge DA, et al. Common polymorphisms in the NOD2 gene region are associated with leprosy and its reactive states. J Infect Dis. (2010) 201:1422–35. doi: 10.1086/651559
103. Schenk M, Mahapatra S, Le P, Kim HJ, Choi AW, Brennan PJ, et al. Human NOD2 recognizes structurally unique muramyl dipeptides from mycobacterium leprae. Infect Immun. (2016) 84:2429–38. doi: 10.1128/IAI.00334-16
104. Flannagan RS, Jaumouille V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. (2012) 7:61–98. doi: 10.1146/annurev-pathol-011811-132445
105. Lai SC, Devenish RJ. LC3-associated phagocytosis. (LAP): connections with host autophagy. Cells. (2012) 1:396–408. doi: 10.3390/cells1030396
106. Solano-Collado V, Rofe A, Spano S. Rab32 restriction of intracellular bacterial pathogens. Small GTPases. (2018) 9:216–23. doi: 10.1080/21541248.2016.1219207
107. Zhang F, Liu H, Chen S, Low H, Sun L, Cui Y, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet. (2011) 43:1247–51. doi: 10.1038/ng.973
108. Li Y, Wang Y, Zou L, Tang X, Yang Y, Ma L, et al. Analysis of the Rab GTPase interactome in dendritic cells reveals anti-microbial functions of the Rab32 complex in bacterial containment. Immunity. (2016) 44:422–37. doi: 10.1016/j.immuni.2016.01.027
109. Yang D, Chen J, Shi C, Jing Z, Song N. Autophagy gene polymorphism is associated with susceptibility to leprosy by affecting inflammatory cytokines. Inflammation. (2014) 37:593–8. doi: 10.1007/s10753-013-9773-1
110. Wang D, Xu L, Lv L, Su LY, Fan Y, Zhang DF, et al. Association of the LRRK2 genetic polymorphisms with leprosy in Han Chinese from Southwest China. Genes Immun. (2015) 16:112–9. doi: 10.1038/gene.2014.72
111. Sheng D, See K, Hu X, Yu D, Wang Y, Liu Q, et al. Disruption of LRRK2 in Zebrafish leads to hyperactivity and weakened antibacterial response. Biochem Biophys Res Commun. (2018) 497:1104–9. doi: 10.1016/j.bbrc.2018.02.186
112. Wang Z, Mi Z, Wang H, Sun L, Yu G, Fu X, et al. Discovery of 4 exonic and 1 intergenic novel susceptibility loci for leprosy. Clin Genet. (2018) 94:259–63. doi: 10.1111/cge.13376
113. Roy S, Frodsham A, Saha B, Hazra SK, Mascie-Taylor CG, Hill AV. Association of vitamin D receptor genotype with leprosy type. J Infect Dis. (1999) 179:187–91. doi: 10.1086/314536
114. Fitness J, Floyd S, Warndorff DK, Sichali L, Mwaungulu L, Crampin AC, et al. Large-scale candidate gene study of leprosy susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg. (2004) 71:330–40. doi: 10.4269/ajtmh.2004.71.330
115. Sapkota BR, Macdonald M, Berrington WR, Misch EA, Ranjit C, Siddiqui MR, et al. Association of TNF, MBL, and VDR polymorphisms with leprosy phenotypes. Hum Immunol. (2010) 71:992–8. doi: 10.1016/j.humimm.2010.07.001
116. Xiang YL, Zhang DF, Wang D, Li YY, Yao YG. Common variants of OPA1 conferring genetic susceptibility to leprosy in Han Chinese from Southwest China. J Dermatol Sci. (2015) 80:133–41. doi: 10.1016/j.jdermsci.2015.09.001
117. Wang Z, Sun Y, Fu X, Yu G, Wang C, Bao F, et al. A large-scale genome-wide association and meta-analysis identified four novel susceptibility loci for leprosy. Nat Commun. (2016) 7:13760. doi: 10.1038/ncomms13760
118. Prado-Montes de Oca E, Velarde-Felix JS, Rios-Tostado JJ, Picos-Cardenas VJ, Figuera LE. SNP 668C. (-44) alters a NF-kappaB1 putative binding site in non-coding strand of human beta-defensin 1. (DEFB1) and is associated with lepromatous leprosy. Infect Genet Evol. (2009) 9:617–25. doi: 10.1016/j.meegid.2009.03.006
119. Cardoso CC, Pereira AC, Brito-de-Souza VN, Dias-Baptista IM, Maniero VC, Venturini J, et al. IFNG +874 T>A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Genet. (2010) 128:481–90. doi: 10.1007/s00439-010-0872-x
120. Wang D, Feng JQ, Li YY, Zhang DF, Li XA, Li QW, et al. Genetic variants of the MRC1 gene and the IFNG gene are associated with leprosy in Han Chinese from Southwest China. Hum Genet. (2012) 131:1251–60. doi: 10.1007/s00439-012-1153-7
121. Aguilar-Medina M, Escamilla-Tilch M, Frias-Castro LO, Romero-Quintana G, Estrada-Garcia I, Estrada-Parra S, et al. HLA alleles are genetic markers for susceptibility and resistance towards leprosy in a mexican mestizo population. Ann Hum Genet. (2017) 81:35–40. doi: 10.1111/ahg.12183
122. de Souza-Santana FC, Marcos EV, Nogueira ME, Ura S, Tomimori J. Human leukocyte antigen class I and class II alleles are associated with susceptibility and resistance in borderline leprosy patients from Southeast Brazil. BMC Infect Dis. (2015) 15:22. doi: 10.1186/s12879-015-0751-0
123. Wong SH, Gochhait S, Malhotra D, Pettersson FH, Teo YY, Khor CC, et al. Leprosy and the adaptation of human toll-like receptor 1. PLoS Pathog. (2010) 6:e1000979. doi: 10.1371/journal.ppat.1000979
124. Lavado-Valenzuela R, Jose Bravo M, Junqueira-Kipnis AP, Ramos de Souza M, Moreno C, Alonso A, et al. Distribution of the HLA class II frequency alleles in patients with leprosy from the mid-west of Brazil. Int J Immunogenet. (2011) 38:255–8. doi: 10.1111/j.1744-313X.2010.00993.x
125. Mehra NK, Rajalingam R, Mitra DK, Taneja V, Giphart MJ. Variants of HLA-DR2/DR51 group haplotypes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis in Asian Indians. Int J Lepr Other Mycobact Dis. (1995) 63:241–8.
126. Wang D, Zhang DF, Feng JQ, Li GD, Li XA, Yu XF, et al. Common variants in the PARL and PINK1 genes increase the risk to leprosy in Han Chinese from South China. Sci Rep. (2016) 6:37086. doi: 10.1038/srep37086
127. Tosh K, Ravikumar M, Bell JT, Meisner S, Hill AV, Pitchappan R. Variation in MICA and MICB genes and enhanced susceptibility to paucibacillary leprosy in South India. Hum Mol Genet. (2006) 15:2880–7. doi: 10.1093/hmg/ddl229
128. Lucena-Silva N, Teixeira MA, Ramos Ade L, de Albuquerque RS, Diniz GT, Mendes-Junior CT, et al. The +3187A/G HLA-G polymorphic site is associated with polar forms and reactive reaction in leprosy. Mol Genet Genomic Med. (2013) 1:123–30. doi: 10.1002/mgg3.14
129. Shinde V, Marcinek P, Rani DS, Sunder SR, Arun S, Jain S, et al. Genetic evidence of TAP1 gene variant as a susceptibility factor in Indian leprosy patients. Hum Immunol. (2013) 74:803–7. doi: 10.1016/j.humimm.2013.01.001
130. de Messias-Reason IJ, Boldt AB, Moraes Braga AC, Von Rosen Seeling Stahlke E, Dornelles L, Pereira-Ferrari L, et al. The association between mannan-binding lectin gene polymorphism and clinical leprosy: new insight into an old paradigm. J Infect Dis. (2007) 196:1379–85. doi: 10.1086/521627
131. de Messias-Reason I, Kremsner PG, Kun JF. Functional haplotypes that produce normal ficolin-2 levels protect against clinical leprosy. J Infect Dis. (2009) 199:801–4. doi: 10.1086/597070
132. Zhang DF, Huang XQ, Wang D, Li YY, Yao YG. Genetic variants of complement genes ficolin-2, mannose-binding lectin and complement factor H are associated with leprosy in Han Chinese from Southwest China. Hum Genet. (2013) 132:629–40. doi: 10.1007/s00439-013-1273-8
133. Liu H, Irwanto A, Tian H, Fu X, Yu Y, Yu G, et al. Identification of IL18RAP/IL18R1 and IL12B as leprosy risk genes demonstrates shared pathogenesis between inflammation and infectious diseases. Am J Hum Genet. (2012) 91:935–41. doi: 10.1016/j.ajhg.2012.09.010
134. Alvarado-Navarro A, Montoya-Buelna M, Munoz-Valle JF, Lopez-Roa RI, Guillen-Vargas C, Fafutis-Morris M. The 3'UTR 1188 A/C polymorphism in the interleukin-12p40 gene. (IL-12B) is associated with lepromatous leprosy in the west of Mexico. Immunol Lett. (2008) 118:148–51. doi: 10.1016/j.imlet.2008.03.015
135. Ali S, Srivastava AK, Chopra R, Aggarwal S, Garg VK, Bhattacharya SN, et al. IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J Med Genet. (2013) 50:34–42. doi: 10.1136/jmedgenet-2012-101214
136. Bonefeld CM, Petersen TH, Bandier J, Agerbeck C, Linneberg A, Ross-Hansen K, et al. Epidermal filaggrin deficiency mediates increased systemic T-helper 17 immune response. Br J Dermatol. (2016) 175:706–12. doi: 10.1111/bjd.14570
137. Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, et al. Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet. (2015) 47:267–71. doi: 10.1038/ng.3212
138. Sun Y, Irwanto A, Toyo-Oka L, Hong M, Liu H, Andiappan AK, et al. Fine-mapping analysis revealed complex pleiotropic effect and tissue-specific regulatory mechanism of TNFSF15 in primary biliary cholangitis, Crohn's disease and leprosy. Sci Rep. (2016) 6:31429. doi: 10.1038/srep31429
139. Shaw MA, Donaldson IJ, Collins A, Peacock CS, Lins-Lainson Z, Shaw JJ, et al. Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun. (2001) 2:196–204. doi: 10.1038/sj.gene.6363754
140. Ali S, Chopra R, Aggarwal S, Srivastava AK, Kalaiarasan P, Malhotra D, et al. Association of variants in BAT1-LTA-TNF-BTNL2 genes within 6p21.3 region show graded risk to leprosy in unrelated cohorts of Indian population. Hum Genet. (2012) 131:703–16. doi: 10.1007/s00439-011-1114-6
141. Silva GA, Ramasawmy R, Boechat AL, Morais AC, Carvalho BK, Sousa KB, et al. Association of TNF−1031 C/C as a potential protection marker for leprosy development in Amazonas state patients, Brazil. Hum Immunol. (2015) 76:137–41. doi: 10.1016/j.humimm.2015.01.011
142. Tarique M, Naqvi RA, Santosh KV, Kamal VK, Khanna N, Rao DN. Association of TNF-alpha-(308(GG)), IL-10(-819(TT)), IL-10(-1082(GG)) and IL-1R1(+1970(CC)) genotypes with the susceptibility and progression of leprosy in North Indian population. Cytokine. (2015) 73:61–5. doi: 10.1016/j.cyto.2015.01.014
143. Areeshi MY, Mandal RK, Dar SA, Jawed A, Wahid M, Lohani M, et al. Impact of TNF−308 G>A. (rs1800629) gene polymorphism in modulation of leprosy risk: a reappraise meta-analysis of 14 case-control studies. Biosci Rep. (2017) 37:pii:BSR20170806. doi: 10.1042/BSR20170806
144. Alcais A, Alter A, Antoni G, Orlova M, Nguyen VT, Singh M, et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat Genet. (2007) 39:517–22. doi: 10.1038/ng2000
145. Hagge DA, Saunders BM, Ebenezer GJ, Ray NA, Marks VT, Britton WJ, et al. Lymphotoxin-alpha and TNF have essential but independent roles in the evolution of the granulomatous response in experimental leprosy. Am J Pathol. (2009) 174:1379–89. doi: 10.2353/ajpath.2009.080550
146. Silva GA, Naveca FG, Ramasawmy R, Boechat AL. Association between the IFNG +874A/T gene polymorphism and leprosy resistance: a meta-analysis. Cytokine. (2014) 65:130–3. doi: 10.1016/j.cyto.2013.12.002
147. Cardona-Castro N, Sanchez-Jimenez M, Rojas W, Bedoya-Berrio G. IL-10 gene promoter polymorphisms and leprosy in a Colombian population sample. Biomedica. (2012) 32:71–6. doi: 10.7705/biomedica.v32i1.386
148. Pereira AC, Brito-de-Souza VN, Cardoso CC, Dias-Baptista IM, Parelli FP, Venturini J, et al. Genetic, epidemiological and biological analysis of interleukin-10 promoter single-nucleotide polymorphisms suggests a definitive role for−819C/T in leprosy susceptibility. Genes Immun. (2009) 10:174–80. doi: 10.1038/gene.2008.97
149. Alvarado-Arnez LE, Amaral EP, Sales-Marques C, Duraes SM, Cardoso CC, Nunes Sarno E, et al. Association of IL10 polymorphisms and leprosy: a meta-analysis. PLoS ONE. (2015) 10:e0136282. doi: 10.1371/journal.pone.0136282
150. Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. (2002) 2:933–44. doi: 10.1038/nri954
151. Meylan F, Davidson TS, Kahle E, Kinder M, Acharya K, Jankovic D, et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity. (2008) 29:79–89. doi: 10.1016/j.immuni.2008.04.021
152. Pellegrini P, Berghella AM, Contasta I, Adorno D. CD30 antigen: not a physiological marker for TH2 cells but an important costimulator molecule in the regulation of the balance between TH1/TH2 response. Transpl Immunol. (2003) 12:49–61. doi: 10.1016/S0966-3274(03)00014-5
153. Fava VM, Cobat A, Van Thuc N, Latini AC, Stefani MM, Belone AF, et al. Association of TNFSF8 regulatory variants with excessive inflammatory responses but not leprosy per se. J Infect Dis. (2015) 211:968–77. doi: 10.1093/infdis/jiu566
154. Fava VM, Sales-Marques C, Alcais A, Moraes MO, Schurr E. Age-dependent association of TNFSF15/TNFSF8 variants and leprosy type 1 reaction. Front Immunol. (2017) 8:155. doi: 10.3389/fimmu.2017.00155
155. Naafs B, van Hees CL. Leprosy type 1 reaction. (formerly reversal reaction). Clin Dermatol. (2016) 34:37–50. doi: 10.1016/j.clindermatol.2015.10.006
156. Fava VM, Xu YZ, Lettre G, Van Thuc N, Orlova M, Thai VH, et al. Pleiotropic effects for Parkin and LRRK2 in leprosy type-1 reactions and Parkinson's disease. Proc Natl Acad Sci USA. (2019) 116:15616–24. doi: 10.1073/pnas.1901805116
157. Wang D, Zhang DF, Li GD, Bi R, Fan Y, Wu Y, et al. A pleiotropic effect of the APOE gene: association of APOE polymorphisms with multibacillary leprosy in Han Chinese from Southwest China. Br J Dermatol. (2018) 178:931–9. doi: 10.1111/bjd.16020
158. Ramos GB, Salomao H, Francio AS, Fava VM, Werneck RI, Mira MT. Association analysis suggests SOD2 as a newly identified candidate gene associated with leprosy susceptibility. J Infect Dis. (2016) 214:475–8. doi: 10.1093/infdis/jiw170
159. Wang D, Fan Y, Malhi M, Bi R, Wu Y, Xu M, et al. Missense variants in HIF1A and LACC1 contribute to leprosy risk in Han Chinese. Am J Hum Genet. (2018) 102:794–805. doi: 10.1016/j.ajhg.2018.03.006
160. Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. (2004) 427:636–40. doi: 10.1038/nature02326
161. Malhotra D, Darvishi K, Lohra M, Kumar H, Grover C, Sood S, et al. Association study of major risk single nucleotide polymorphisms in the common regulatory region of PARK2 and PACRG genes with leprosy in an Indian population. Eur J Hum Genet. (2006) 14:438–42. doi: 10.1038/sj.ejhg.5201563
162. Li J, Liu H, Liu J, Fu X, Yu Y, Yu G, et al. Association study of the single nucleotide polymorphisms of PARK2 and PACRG with leprosy susceptibility in Chinese population. Eur J Hum Genet. (2012) 20:488–9. doi: 10.1038/ejhg.2011.190
Keywords: leprosy, innate immunity, adaptive immunity, genetic association, variant
Citation: Mi Z, Liu H and Zhang F (2020) Advances in the Immunology and Genetics of Leprosy. Front. Immunol. 11:567. doi: 10.3389/fimmu.2020.00567
Received: 13 December 2019; Accepted: 12 March 2020;
Published: 16 April 2020.
Edited by:
Juarez Antonio Simões Quaresma, Evandro Chagas Institute, BrazilReviewed by:
Mohd Tarique, University of Miami Health System, United StatesMalcolm Scott Duthie, HDT Biotech Corporation, United States
John S. Spencer, Colorado State University, United States
Cleverson Teixeira Soares, Instituto Lauro de Souza Lima, Brazil
Copyright © 2020 Mi, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Furen Zhang, emhhbmdmdXJlbkBob3RtYWlsLmNvbQ==
 Zihao Mi
Zihao Mi Hong Liu
Hong Liu