- 1Department of Pathology, Division of Microbiology and Immunology, University of Utah, Salt Lake City, UT, United States
- 2Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, United States
Macrophages are key cells of the innate immune system with functional roles in both homeostatic maintenance of self-tissues and inflammatory responses to external stimuli, including infectious agents. Recent advances in metabolic research have revealed that macrophage functions rely upon coordinated metabolic programs to regulate gene expression, inflammation, and other important cellular processes. Polarized macrophages adjust their use of nutrients such as glucose and amino acids to meet their changing metabolic needs, and this in turn supports the functions of the activated macrophage. Metabolic and inflammatory processes have been widely studied, and a crucial role for their regulation at the post-transcriptional level by microRNAs (miRNAs) has been identified. miRNAs govern many facets of macrophage biology, including direct targeting of metabolic regulators and inflammatory pathways. This review will integrate emerging data that support an interplay between miRNAs and metabolism during macrophage inflammatory responses, highlighting critical miRNAs and miRNA families. Additionally, we will address the implications of these networks for human disease and discuss emerging areas of research in this field.
Introduction
The immune system consists of a vast array of innate and adaptive immune cells necessary for defense against pathogen invasion. Macrophages are key constituents of the innate immune system and are an integral part of the first response to infection through their recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) that include toll-like receptors (TLRs), and NOD-like receptors (NLRs), among others (1–3). Following detection of microbes, macrophages take on a number of roles such as directly killing invading pathogens, phagocytizing dead and dying cells, responding to parasite invasion through cytokine release, and priming the adaptive immune system to generate antigen-specific responses against infectious agents (4, 5).
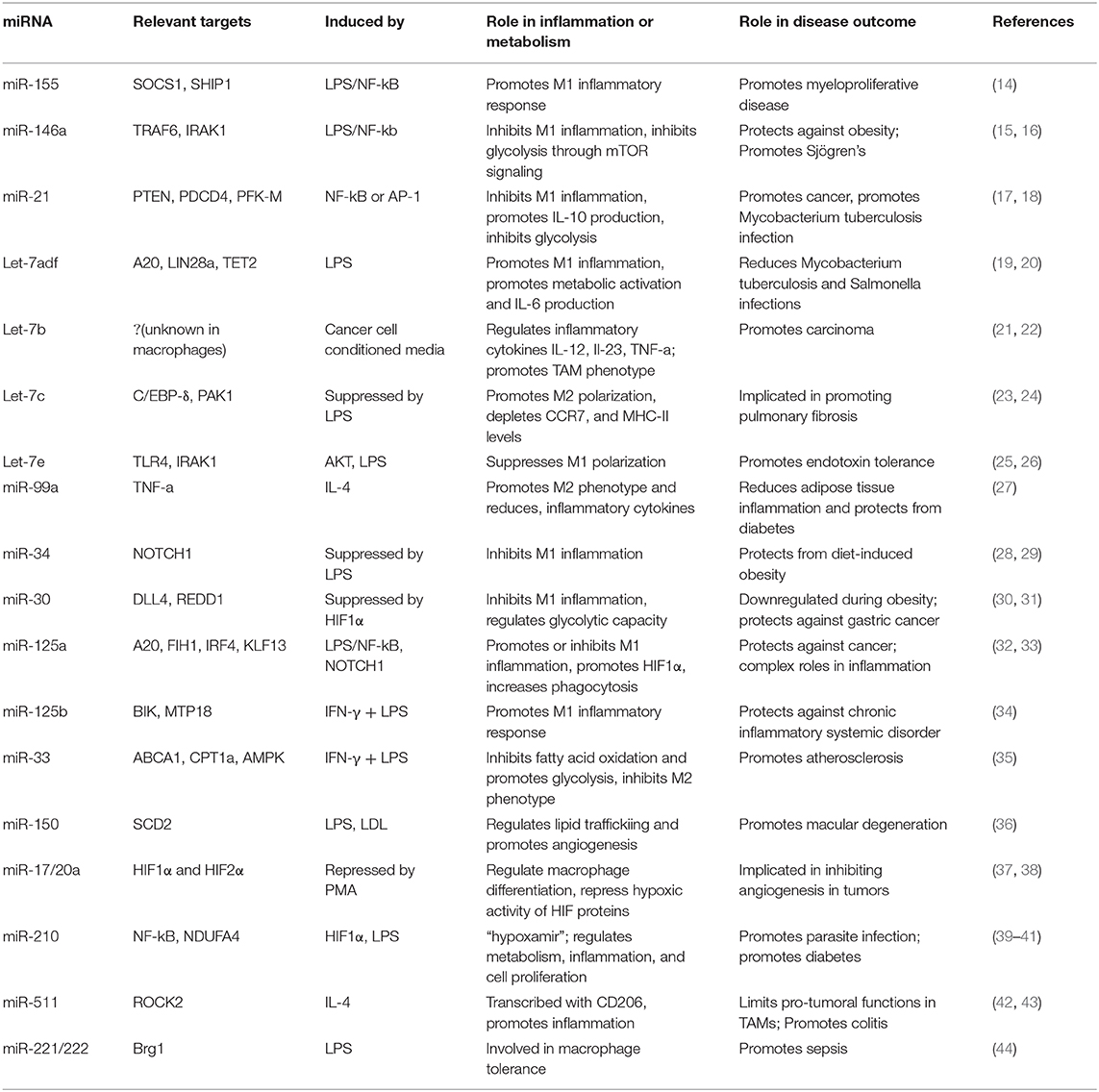
Macrophages are widely distributed throughout the body, residing in nearly every tissue (6). These tissue resident macrophages emerge from distinct sources—either originating from the prenatal yolk sac and seeding into tissues during embryonic development (7), or arising from the bone marrow as monocytes and circulating through the bloodstream prior to injury or infection, prompting differentiation and tissue infiltration (8). Tissue macrophages respond to cytokines, microbial signals, fatty acids, or growth factors in their environment (8), which leads to changes in transcription, cell surface protein expression, and cellular function. Considerable heterogeneity exists among activated macrophage populations (6, 9), and these phenotypic changes are generally described as one of two main polarization subtypes: classical (“M1”) and alternative (“M2”). M1-polarized macrophages are typically activated by inflammatory cytokines such as interferon gamma (IFN-γ) and TLR ligands such as lipopolysaccharide (LPS), and promote pathogen clearance through inflammation and microbicidal activity (10). M2-polarized macrophages, on the other hand, usually respond to Interleukin-4 (IL-4) and IL-13 and promote parasite clearance, inflammatory resolution, and tissue repair (11). The M1/M2 classification system provides a simple paradigm to describe macrophage phenotypes based on their more prominent defining features during activation, yet it simplifies the plastic nature of the macrophage response spectrum, which is subject to an array of activating signals, and which has been extensively discussed in other reviews (12, 13). This review provides a list of relevant activating signals in Table 1 to provide a more in-depth perspective on macrophage activation and polarization.
Macrophages are important for normal homeostatic maintenance and contribute to self-limited inflammation during infection; however, these beneficial functions can be superseded by prolonged activation signals leading to dysregulated macrophage activity, and this can have pathological consequences (5). For instance, unrestrained or chronic inflammation from macrophages can drive inflammatory conditions such as metabolic disease, cytokine storm, and sepsis (45–47). On the other hand, macrophages sometimes fail to mount a sufficient inflammatory response, especially within tumors where they are referred to as tumor-associated macrophages (TAMs). TAMs often resemble M2 macrophages and promote angiogenesis, decrease antigen presentation, dampen inflammatory cytokine production, and impede T cell recruitment into the tumor (48). Research in this field has uncovered many mechanisms that have evolved to regulate macrophage activity and aid in controlling the inflammatory response.
In recent years, the field of immunometabolism has expanded rapidly. It is now widely accepted that leukocyte metabolism is an essential part of a coordinated immune response. Macrophage metabolism in particular has been widely studied, and it has become clear that metabolism plays a considerably larger role in macrophages than simply driving energy production for the cell. Rather, it is a dynamic process with direct and context-specific roles in driving inflammatory signaling and other macrophage effector functions (49, 50). It has also become evident that metabolic programs are responsive to many cues beyond nutrient availability, including cytokine-mediated polarization signals (51, 52). Under polarizing conditions, M1 and M2 macrophages differentially utilize metabolic pathways to promote their inflammatory or anti-inflammatory functions. For instance, M1 macrophages rely on aerobic glycolysis for their energetic needs, and this polarized metabolic programming drives the pentose phosphate pathway (PPP), itaconate production, and nitric oxide (NO) synthesis, all of which support the functions of classically activated macrophages. Conversely, M2 macrophages use oxidative phosphorylation (OXPHOS) for their energetic needs, driving glycosylation of cell surface receptors and producing ornithine to support wound healing functions of alternatively activated macrophages (52). The metabolic activity of macrophages has been an exciting area of research in recent years and has greatly expanded our understanding of how macrophage responses are regulated.
Inflammatory and metabolic gene expression must be tightly controlled during macrophage activation for a proper macrophage response to occur. Cellular mechanisms exist to ensure this, with one of these being post-transcriptional gene regulation by miRNAs. miRNAs are small, ~22 nucleotide non-coding RNAs that are fundamental in coordinating expression of a diverse array of genes in immune populations (53). These small RNAs are endogenously expressed and are generated first as long primary transcripts which undergo a series of processing steps to reach their final mature state (54). Once formed, the mature miRNA is loaded into the RISC complex and acts as a guide to complementary mRNA targets. The miRNA-RISC complex binds to the target mRNA, leading to either degradation or translational inhibition of the mRNA transcript. miRNAs are best known for acting intracellularly, but they can also be loaded into exosomes for transport to distant cells (55). A single miRNA can target multiple genes; likewise, a single gene can be repressed by numerous miRNAs, and it is thought that at least 60% of all protein-coding genes are under miRNA regulation (56). miRNAs are numbered sequentially by discovery date and classified into families based on sequence homology, structure, or functional similarities (57).
These small but critically important molecules have been shown to regulate gene expression during each stage of macrophage development, from myelopoiesis all the way through polarization and during effector function (58), and have been identified as a regulatory link between polarization signals and metabolic function (59). miRNAs thus ensure the ability to properly gauge key macrophage processes and inflammatory programs.
miRNAs have distinct roles in regulating macrophage activities to restrain macrophage-associated diseases; meanwhile, miRNA dysregulation is associated with poor prognosis in many of these diseases. Recent work in this field has identified specific miRNAs and their mechanistic functions in macrophages, playing a part in both health and disease. In this review, we will discuss macrophage metabolic and inflammatory programming and the important functional consequences of miRNAs in controlling these processes. As we examine the existing research in this field of study, it should be mentioned that some differences exist between human and mouse macrophages, including distinct metabolic and miRNA variations (60); however, these molecules and processes are also conserved in many cases.
Metabolic Programming in Polarized Macrophages Is Essential for Function
Macrophage activation relies heavily on the metabolic activities of the cell to fuel a specific and timely response. Polarized macrophages rely on specific metabolic pathways to promote the dynamic activities of pro- or anti-inflammatory responses, and metabolic gene expression levels are often used as markers of classically or alternatively activated macrophage populations (13). In this section we will discuss the metabolic changes that occur in macrophages during polarization to promote macrophage functions.
Prior to polarization, macrophages exhibit a resting metabolic state that relies on OXPHOS fueled by the tricarboxylic acid (TCA) cycle for energy needs. When classical activation is initiated by extracellular molecules such as IFN-γ and LPS, murine macrophages undergo a drastic increase in energy demand and induce heightened glucose flux to meet these energetic needs. Macrophage polarizing signals initiate master transcription factors such as nuclear factor-κB (NF-κB) and hypoxia-inducible factor 1α (HIF1α), among others, to carry out changes in inflammatory gene expression and metabolic programming (61, 62). During polarization, inflammatory macrophages switch their metabolic programming to favor aerobic glycolysis over OXPHOS (51, 63, 64). Although aerobic glycolysis, also termed the Warburg Effect, is less efficient at generating ATP than is OXPHOS, the heightened glucose flux into the cell allows for rapid energy production and is used to generate other biosynthetic intermediates as well (65). This includes the pentose phosphate pathway, which supplies NADPH for reactive oxygen species (ROS) and NO production, and which helps maintain the cellular redox state of the cell (66). The pentose phosphate pathway also generates intermediates necessary for nucleotide synthesis, although the need for nucleotide production is unclear since activated macrophages do not proliferate (67). Finally, aerobic glycolysis results in pyruvate being reduced to lactate, which maintains the glycolytic flux in activated macrophages (68).
A hallmark of inflammatory macrophage metabolism is a defective tricarboxylic acid (TCA) cycle. The TCA cycle, which is primarily fed by the products of glycolysis and glutamine metabolism, is broken in two spots in inflammatory murine macrophages, resulting in a buildup of TCA intermediates (64, 69, 70). These breakpoints occur at isocitrate dehydrogenase (IDH), leading to citrate accumulation, and at succinate dehydrogenase (SDH), leading to succinate accumulation (64, 71, 72). These accumulated metabolites are then used for other metabolic and immune signaling functions. First, accumulated citrate is used to drive fatty acid synthesis and NO production to support a classically activated macrophage phenotype (68). Citrate can also be converted into the antimicrobial metabolite itaconate (73, 74), which has multiple roles, including antimicrobial protection against intracellular pathogens and inhibition of SDH to promote succinate buildup (75). Second, accumulated succinate stabilizes HIF1α and leads to sustained IL-1β and ROS production (64). IL-1β is among the inflammatory cytokines characteristically produced by IFN-γ-induced macrophages, along with TNF-α, IL-6, and IL-12 (12). HIF1α is a key transcription factor in regulating both the glycolytic and inflammatory capacities in macrophages, and its absence results in impaired motility, defective bactericidal activity, and poor macrophage accumulation (76). The function of itaconate and succinate in driving antimicrobial programs and pro-inflammatory cytokine expression, respectively, are prime examples of the direct role of metabolic products on macrophage inflammatory function.
This central role for metabolism is further illustrated by seminal findings in the early 1990s demonstrating that murine macrophage subtypes differentially metabolize the amino acid arginine (77–79). These studies showed that inflammatory polarization induces expression of the enzyme NOS2, which converts arginine to the microbicidal molecule NO. NO is cytotoxic to pathogens and thus aids in the antimicrobial macrophage response. In contrast, wound healing macrophages hydrolyze arginine to the tissue-remodeling amino acid ornithine through expression of the enzyme ARG1. Ornithine promotes wound healing by aiding in collagen formation and inhibiting inflammatory cytokine expression (80–83). These distinct metabolic activities in pro- or anti-inflammatory macrophages are driven by macrophage polarization signals, and in return the products of arginine metabolism promote the specific functions of polarized macrophages.
While the metabolic profile undergoes dramatic changes upon pro-inflammatory polarization, the anti-inflammatory macrophage triggered by IL-4 and IL-13 more closely resembles metabolism seen in non-polarized macrophages. Alternatively activated murine macrophages use glucose and glutamine to feed the TCA cycle, and also rely on fatty acid oxidation (FAO) to fuel OXPHOS (84). Important transcription factors and enzymes are involved in alternative macrophage activation and are distinct from inflammatory macrophages. Among these transcription factors, IRF4 is responsive to IL-4 stimulation and supports the roles of inflammatory abatement, wound repair, and angiogenesis, and it contributes to metabolic reprogramming in alternatively activated macrophages (85). Additional molecules are important in M2 macrophages, such as the kinase AMP-activated protein kinase (AMPK), which is induced in response to anti-inflammatory cytokines in murine macrophages and plays a role in stimulating fatty acid oxidation (FAO) (84, 86). Further, the coenzyme UDP-GlcNAc, a product of glucose and glutamine metabolism, is a metabolite produced during alternative macrophage activation which glycosylates receptors, such as the mannose receptors CD206 and CD301. The loss of this necessary enzyme diminishes alternative polarization and cell surface receptor expression (11, 13, 74, 87). Finally, anti-inflammatory macrophages characteristically produce cytokines such as IL-10 and TGF-β, and these cytokines promote alternative activation through multiple mechanisms, including repressing metabolic remodeling in inflammatory macrophages (88). Overall, metabolic programs in polarized macrophages are distinct from one another but promote the inflammatory or reparative functions of the specific macrophage responses. These functions are summarized in Figure 1.
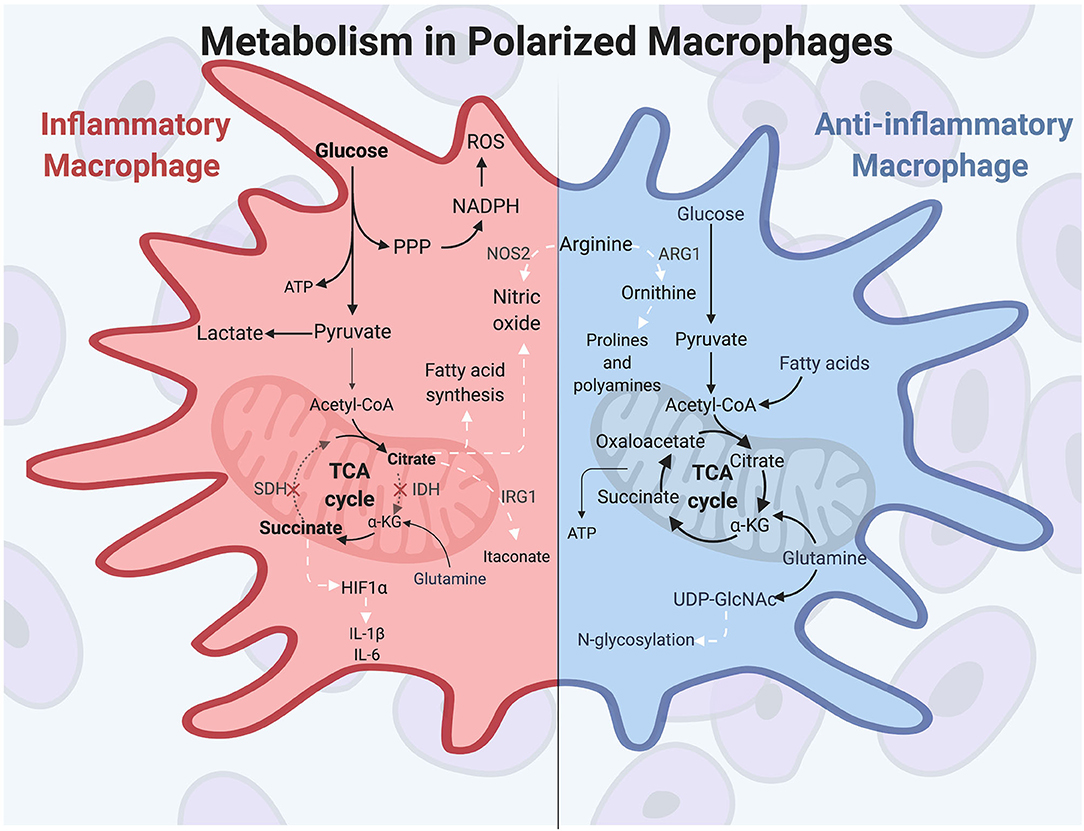
Figure 1. Polarized macrophages have differential metabolic programming. Inflammatory macrophages characteristically increase glucose uptake to fuel aerobic glycolysis, producing increased lactate and ATP production. The pentose phosphate pathway feeds off of the increased flux of glycolytic intermediates, resulting in heightened nucleotide and amino acid synthesis. Classically activated macrophages have a disrupted TCA cycle in two locations: at isocitrate dehydrogenase (IDH) and at succinate dehydrogenase (SDH). As a result, citrate and succinate accumulate and drive such functions as fatty acid and NO synthesis, antimicrobial itaconate production, and HIF1α activity to promote further glycolysis and inflammatory cytokine production. Arginine is differentially metabolized in polarized murine macrophages, producing the antimicrobial molecule NO in inflammatory macrophages. Anti-inflammatory macrophages are not as metabolically active compared to classically activated macrophages, and they utilize glutamine, glucose, and fatty acids to fuel the TCA cycle and OXPHOS. In alternatively activated macrophages, glutamine uptake drives UDP-GlcNAc production, which is important for N-glycosylation of cell surface receptors, such as CD206 and CD301. In anti-inflammatory murine macrophages, arginine is metabolized to ornithine, which promotes tissue repair through production of prolines and polyamines. White arrows indicate metabolic intermediates driving M1 or M2 activities. α-KG, alpha-ketoglutarate; ARG1, arginase; HIF1α, hypoxia-inducible factor 1α; IDH, isocitrate dehydrogenase; IRG1, immune-responsive gene 1; NOS2, nitric oxide synthase; PPP, pentose phosphate pathway; ROS, reactive oxygen species; SDH, succinate dehydrogenase.
MiRNA-Mediated Regulation of Macrophage Function
miRNAs that regulate distinct aspects of macrophage responses have been identified, including those that either promote or mute inflammation in response to polarizing signals. More recently, a growing body of literature has emerged demonstrating the role for miRNAs in regulating metabolic functions to impact macrophage responses (89). While regulation of innate immune metabolism by miRNAs is still up-and-coming, the literature available in this area suggests a strong link between miRNA function and metabolic proficiency in macrophages, with aberrant miRNA expression being linked to deficient antimicrobial responses, metabolic syndromes, and cancer. Here we will review miRNAs that regulate inflammation or metabolism, miRNAs that have been identified to regulate both, and we will discuss how these miRNAs impact macrophage function (Figure 2 and Table 1).
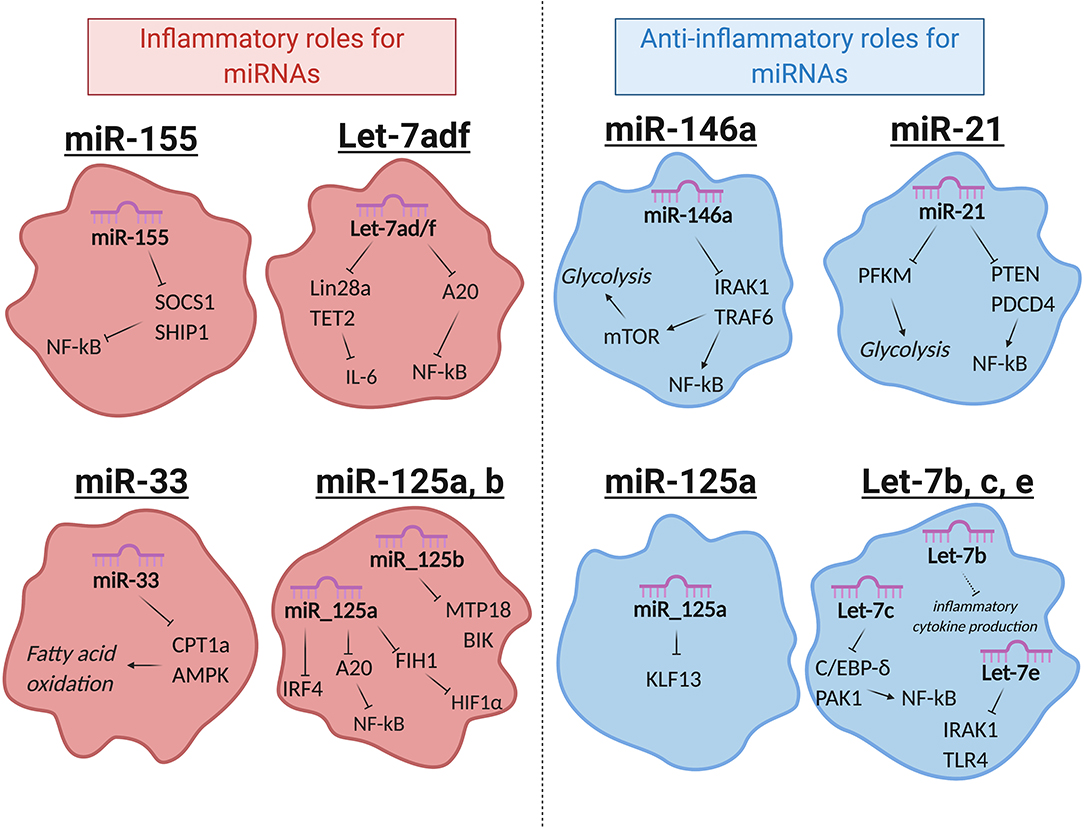
Figure 2. Selected miRNAs that regulate macrophage metabolic and inflammatory pathways. miRNAs are involved in regulating a number of important processes in macrophages. Certain miRNAs, highlighted here, target inflammatory or metabolic pathways to regulate macrophage activities. Red (left): miRNAs that promote inflammatory polarization. Blue (right): miRNAs that promote anti-inflammatory polarization. MiR-125a has noted roles in promoting both inflammatory and anti-inflammatory macrophages, as indicated. Let-7e is part of the miR-99b~Let-7e~miR-125a cluster and acts coordinately with miRNAs in its cluster to have the functions shown.
miR-155
miR-155 was one of the first miRNAs found to regulate the immune system and is one of the most widely studied miRNAs in immunology. It is expressed in both lymphoid and myeloid lineages and is highly induced in macrophages in response to the key inflammatory transcription factor NF-κB (90–93). miR-155 promotes the inflammatory response in humans and mice by targeting SOCS1 and SHIP1, which are negative regulators of inflammation. miR-155 promotes inflammatory cytokine production, PI3K/AKT activity, type I IFN signaling, and further NF-κB function through silencing its anti-inflammatory targets (25, 94–97), and loss of miR-155 attenuates the macrophage antiviral response, highlighting its role in driving inflammatory macrophage function (98, 99).
miR-146a
In contrast to miR-155, miR-146a represses the inflammatory response by targeting transcripts of TRAF6 and IRAK1, two proteins that promote NF-κB activity in the TLR signaling pathway (100). Overexpression of miR-146a diminishes inflammatory cytokine production, while deletion or inhibition of miR-146a leads to elevated inflammatory markers in both human and mouse macrophages, illustrating its role in negatively regulating inflammation (101–103). Recently, our group identified an additional function for miR-146a, showing that it has the ability to regulate metabolic pathways in murine macrophages (15). We found that miR-146a does this by impeding mTOR signaling through its target, Traf6. This restrains glycolysis; further, genetic knockout of miR-146a leads to highly glycolytic and hyperinflammatory macrophages and gives rise to diet-induced inflammation and obesity in mice. These findings demonstrate a dual role for miR-146a in promoting both metabolic and inflammatory abatement.
miR-21
miR-21 is another miRNA exhibiting synchronous roles in both inflammation and metabolism. miR-21 is an endotoxin-responsive miRNA, shown to be induced by either NF-κB or AP-1 (104, 105). This miRNA promotes an anti-inflammatory macrophage phenotype and inflammatory resolution in mouse and human cells by targeting transcripts of the inflammatory proteins PTEN and PDCD4. This leads to decreased NF-κB signaling and increased anti-inflammatory IL-10 production (106, 107). Consistent with this, miR-21 expression in macrophages promotes TAMs which support tumor growth, and its suppression leads to an improved anti-tumor response (17). In addition to targeting inflammatory genes, miR-21 also targets the glycolytic pathway enzyme PFK-M for downregulation (18). This enzyme catalyzes an early step in glycolysis, and its inhibition negatively regulates glycolytic flux (108). Hackett et al. found that miR-21 induction by Mycobacterium tuberculosis during infection limits the switch to a glycolytic state, thus inhibiting inflammatory macrophage polarization. M. tuberculosis is known to promote its own survival by inducing host anti-inflammatory or anti-glycolytic factors (109), and thus sustained miR-21 expression in macrophages is tolerogenic to bacterial growth. miR-21 exhibits roles in restraining pro-inflammatory macrophage activation by targeting both inflammatory and metabolic factors.
Let-7
The Let-7 family of miRNAs contains 10 mature miRNAs in humans, many of which are relevant in macrophage metabolism and have functions in the immune response to pathogens (21, 23, 110). Let-7 was one of the first discovered miRNAs, originally found in C. elegans but with high conservation across species (53). These miRNAs have been extensively studied and a number of them will be highlighted here. First, Let-7f promotes inflammatory polarization and cytokine production in murine macrophages by targeting the negative regulator of NF-κB, A20. During M. tuberculosis infection, Let-7f expression is downregulated and A20 levels rise, leading to diminished NF-κB activity and a poor inflammatory macrophage response (19). This study further showed that Let-7f is sufficient to increase inflammatory NF-κB expression in macrophages and concordantly regulate bacterial growth, as M. tuberculosis survival was diminished after transfection of Let-7f mimics.
Let-7f can also regulate immune metabolism. Let-7f is part of the let-7a-1/let-7d/let-7f-1 (let7-adf) cluster of coordinately transcribed miRNAs, which has been extensively studied for its role in regulating metabolism in multiple immune populations (20, 111). In LPS-activated macrophages, Let-7adf stimulates proinflammatory IL-6 cytokine expression and promotes inflammation through metabolic targets. It does this by targeting two genes in the succinate pathway (20). First, let-7adf regulates one of its well-established targets LIN28a, which directly binds to and enhances SDH activity. By diminishing LIN28a expression, the let-7adf cluster dampens SDH activity and promotes succinate accumulation. Second, let-7adf also inhibits TET2, which normally acts to repress IL-6 downstream of succinate. Inhibition of these two genes by let-7adf promotes succinate accumulation and IL-6 expression.
While Let-7f promotes inflammatory macrophage activity, a number of Let-7 family members instead promote inflammatory resolution. Among these are Let-7b, which promotes anti-inflammatory phenotypes by regulating the inflammatory cytokines IL-12, IL-23, and TNF-α and stimulating angiogenesis in human TAMs (21). Let-7c, another Let-7 family member, promotes wound healing phenotypes by targeting and repressing C/EBP-δ and PAK1, a transcription factor and protein kinase, resulting in reduced NF-κB and AP-1 activity (23, 24). Let-7c overexpression leads to diminished proinflammatory cytokine expression, depleted CCR7 levels, and attenuated MHC-II surface expression in murine macrophages, all indicative of M2 polarization. Finally, let-7e, which falls within the coordinately transcribed let-7e~miR-99b~miR-125a cluster of miRNAs, acts with its cluster members miR-99b and miR-125a to target a number of genes to suppress inflammatory polarization, including TLR4 and IRAK1 (25, 26, 112).
miR-125
As mentioned above, miR-125a is part of the let-7e~miR-99b~miR-125a cluster of miRNAs, though this miRNA may have complex and independent roles from the rest of its cluster in macrophage metabolism and activation. miR-125a promotes pro-inflammatory macrophage responses by targeting FIH1, a negative regulator of HIF1α, and by suppressing the transcription factor IRF4 (32). Additionally, miR-125a has been shown to target A20 to promote NF-kB function (33). In contrast, miR-125a has been reported to have anti-inflammatory roles in macrophages by targeting KLF13, a transcription factor important for inflammation and T lymphocyte activation (113). In this latter context, miR-125a initiation occurs late relative to LPS stimulation. While it has not been shown experimentally, the let-7e~miR-99b~miR-125a cluster may rely on transcriptional timing or relative abundance of distinct miRNAs within this cluster to dictate whether it will favor inflammation or inflammatory resolution since miRNAs in this cluster seem to have opposing roles (114). miR-125a has complex functions in macrophage polarization, but the cluster as a whole has been shown to favor an anti-inflammatory phenotype (26), leading to the idea that cellular context and abundance of individual miRNAs could promote different macrophage programs.
miR-125b is part of the miR-125 family, and it has functions in both inflammation and metabolism to promote an inflammatory response in human and mouse macrophages (34, 115). IFN-γ and LPS stimulation induce miR-125b, which in turn regulates the anti-inflammatory transcription factor IRF4 (115). Further, this miRNA targets genes associated with OXPHOS to promote glycolysis in human monocytes. miR-125b targets BIK, a proapoptotic protein that promotes oxygen consumption, and MTP18, a mitochondria-localized protein involved in mitochondrial fission (34, 116). By targeting these genes, miR-125b abrogates the oxidative capacity of the mitochondria and supports pro-inflammatory activation.
miR-33
A number of publications have focused on the functions of miR-33a and miR-33b, which are co-transcribed with their host genes, SREBF1 and SREBF2 (117–119). These host genes are cholesterol and fatty acid-regulating transcription factors, and studies revealed that miR-33a and b promote the functions of their host genes by targeting genes such as ABCA1 to dampen cholesterol efflux and CPT1a to regulate FAO. Furthermore, it was shown in mouse macrophages that miR-33 targets AMPK, which is a protein kinase important for FAO. miR-33 dampens FAO and promotes glycolytic programming, and thus drives inflammatory polarization (35). miR-33 deletion in macrophages results in M2 polarization that is atheroprotective in hypercholesterolemic mice, demonstrating a physiological role for the activity of this miRNA in a disease driven by inflammatory macrophages.
Additional miRNAs
While this list highlights many crucial miRNAs that regulate macrophage immune responses, a number of other miRNAs have important roles in macrophage function as well. Additional miRNAs are known to dampen inflammation, such as miR-99a, which targets TNF-α (27), and miR-34 and miR-30, which regulate NOTCH signaling to restrain the inflammatory response (28, 30). Conversely, additional miRNAs are known to promote inflammation, such as miR-27a which targets IL-10 to support an inflammatory immune response (120). Other miRNAs have also been noted for their relevance in regulating different aspects of macrophage metabolism. Among these, miR-150 targets SCD2 to regulate lipid metabolism and angiogenesis, promoting macular degeneration in mice (36). miR-17 and miR-20a, which are transcribed as part of the miR-17-92 cluster, regulate HIF1α and HIF2α expression, playing a role in macrophage gene regulation in differentiating monocytes and in TAMs (37, 38). It has become clear from the research in this field that miRNAs are integral for regulating important metabolic processes in macrophages and that their activities promote immune responses. As this field continues to expand, more miRNAs will likely be identified with crucial metabolic roles, and these findings will help us to better understand and ultimately treat diseases.
Looking Ahead: Could Metabolism Play a Role in Regulating miRNAs?
With the recent identification of miRNAs regulating metabolic pathways, it is possible that cross-talk exists between miRNAs and metabolism (Figure 3). Identifying whether metabolism can broadly impact miRNA transcription, biogenesis, or function is a natural next step for this field, owing to the fact that metabolic regulation of protein-coding gene expression and cellular function has been a thriving area of research for many years now.
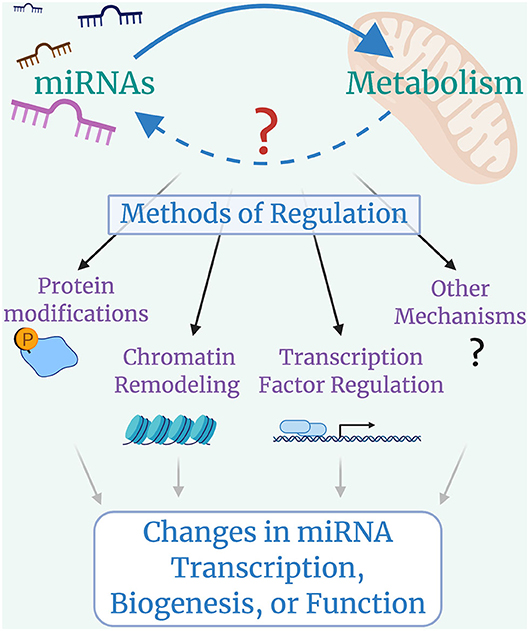
Figure 3. Looking forward: cross-talk between microRNAs and Metabolism. miRNAs control metabolic processes, but it is still largely unclear whether metabolic programs regulate miRNA transcription, biogenesis, or function. Metabolically-induced mechanisms of miRNA regulation could include modifications to miRNA machinery proteins such as DROSHA, DICER, or AGO2, with global impacts on miRNA biogenesis; histone modifications that lead to chromatin remodeling and alterations in miRNA availability; changes in transcription factor activity that lead to up-regulation or down-regulation of specific miRNAs; and other possible mechanisms that have yet to be identified.
The best current examples of metabolically controlled miRNAs are those regulated under hypoxic conditions by the transcription factor HIF1α. A handful of these miRNAs, termed “hypoxamirs,” have been identified, with miR-210 being one of the best-studied. This miRNA is induced by HIF1α in both hypoxic and normoxic conditions, and it in turn regulates metabolism, inflammation, and cell proliferation in macrophages (39, 121–124). Another miRNA, miR-30c, has also been shown to act under HIF1α control to regulate macrophage glycolytic capacity in human macrophages (31). It is important to note that while these examples are of metabolically-regulated miRNAs in macrophages, others have been identified in different cell types as well (125).
Another promising area is in cancer metabolism. Altered metabolism is a hallmark of cancer and it is well-established that miRNAs are dysregulated during cancer, both in immune and non-immune populations (126, 127). Altered metabolic pathways in macrophages impact their function in tumor microenvironments (128, 129), and it is easy to speculate that these metabolic changes could impact miRNAs as part of their regulatory networks. As a result, cancer could be a phenomenal disease model to study how metabolic pathways affect miRNAs to induce differential disease outcomes.
Discussion
In summary, macrophages play a distinct role in maintaining immune system homeostasis and initiating inflammatory responses. We have described how metabolism is an integral part of the macrophage response and how miRNAs are critical mediators of gene expression to either enact or abate inflammatory signals. It is evident that metabolism contributes to the macrophage effector function both by providing needed energy and also through extra-metabolic functions of metabolites to regulate the inflammatory process. miRNAs evidently control both inflammation and metabolism to coordinate a macrophage response. Some of the key miRNAs that control macrophage inflammation and metabolism which we highlighted here include Let-7f, which promotes inflammation, and miR-146a and miR-21, which promote inflammatory resolution. It is certainly possible that additional crucial miRNAs have yet to be described. The fine-tuned effects of miRNAs in macrophages protect the host against disease, and miRNA dysregulation can lead to macrophage-related pathologies.
Despite a robust understanding of the role of miRNAs in macrophage activation and metabolic programming, there are still many open questions. One of the most intriguing new frontiers to address in coming years will be the mechanisms regulating cross-talk between metabolism and miRNAs as we discussed in this review. This question relates not only to specific miRNAs, but can also be applied to global miRNA biogenesis being affected by metabolism (130), which has started to be explored. It was recently shown that miRNA biogenesis processing proteins are regulated in a metabolism-dependent manner, albeit in non-macrophage cells. Drosha, the nuclear protein that processes miRNAs, can be modulated in response to glucose availability, and this has a profound effect on global miRNA biogenesis (131). In addition to this work, other studies have shown that global miRNA regulation greatly affects macrophage function (132–135). Whether and to what extent metabolism regulates miRNA biogenesis in macrophages is a very compelling concept for future study. This will surely be a frontier in immunometabolic research as this exciting area of study moves forward.
Second, miRNAs as extracellular messengers by exosomes-mediated transport is also an intriguing area of research. The full extent of this communication network is still unclear, but some work has revealed that exosomal miRNAs play functional roles on recipient tissues in an endocrine manner and can regulate metabolic processes such as insulin signaling (136). It is still largely unclear what effect macrophage exosomes have on recipient cells and whether or how metabolic functions play a role in exosome biology.
Finally, whether species differences between human and mouse macrophages, including metabolic, inflammatory, and miRNA differences, impact their overall function and phenotype has been a longstanding topic of debate (60). For example, human macrophages induced by LPS do not express NOS2 during arginine metabolism, while NOS2 is a hallmark gene of LPS activation in murine macrophages (60, 137). Limitations in human sampling and variations in commonly used cell sources (for instance, mouse bone marrow-derived macrophages, human peripheral blood macrophages, and mouse or human immortalized cell lines) could contribute to noted phenotypic differences between species. Improving sampling and standardizing methodology for studying macrophage polarization could help improve our understanding of pre-clinical mouse models for human disease research and drug development.
As this field continues to grow, the potential for therapeutic interventions will continue to expand. Clinical trials have shown promise for targeting miRNAs for inhibition (138, 139), and others are exploring miRNA mimics for use in miRNA replacement therapy (140, 141). However, there are still many hurdles to overcome regarding developing and delivering miRNA therapies, and a deeper understanding of miRNA physiology will certainly lend itself to better therapeutic strategies and thus improved clinical outcomes. Furthermore, targeting macrophage metabolism for therapeutic purposes is emerging with the potential to significantly improve cancer, obesity, and autoimmune diseases treatments (142). Taken together, it is clear that there is crosstalk between miRNA, metabolic, and inflammatory pathways in macrophages, and through an improved understanding of these networks, the next generation of targeted therapies will be developed to help fight human disease.
Author Contributions
MN contributed in conceptualization, writing, reviewing, and editing this article. RO'C contributed in conceptualization, writing, reviewing, editing, and supervising this article. All authors contributed to the article and approved the submitted version.
Funding
RO'C was supported by NIH grants R01AI123106-04 and R21AG062923-02.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Kaylyn Bauer and Andrew Ramstead for reviewing drafts of this manuscript and providing useful feedback. Figures were created with BioRender.com.
Abbreviations
A20, negative regulator of NF-kB encoded by TNFAIP3; ABCA1, ATP-Binding Cassette Transporter A1; AKT, also known as PKB, or Protein Kinase B; AMPK, 5′ AMP-activated protein Kinase; AP-1, Activator Protein 1; ARG1, Arginase; BIK, BCL-2 Interacting Killer; CCR7, C-C Motif Chemokine Receptor 7; CD206, Cluster of Differentiation 206, also known as Mannose Receptor; CD301, Cluster of Differentiation 301, also known as CLEC10A; C/EBP-δ, CCAAT/Enhancer-Binding Protein delta; CPT1a, Carnitine Palmitoyltransferase 1a; DLL4, Delta-Like Ligand 4; FIH1, Factor Inhibiting Hif-1; HIF1α, Hypoxia inducible factor 1α; IRAK1, Interleukin-1 Receptor-Associated Kinase 1; IRF4, Interferon Regulatory Factor 4; KLF13, Kruppel-Like Factor 13; LDL, Low-density Lipopolysaccharide; LIN28a, Lin28 homolog A, suppressor of Let-7 and SDH; mTOR, Mammalian Target of Rapamycin; MTP18, Mitochondrial Protein 18; NF-κB, Nuclear factor kappa B; NLR, Nod-like receptor; NO, Nitric Oxide; NOS2, Nitric Oxide Synthase; NOTCH1, Signaling receptor protein in the NOTCH family; OXPHOS, Oxidative Phosphorylation; PAMP, Pathogen associated molecular pattern; PAK1, P21-Activated Kinase 1; PFK-M, Phosphofructokinase, Muscle isoform; PI3K, Phosphatidylinositol 3-Kinase; PMA, Phorbol Myristate Acetate, a monocyte-to-macrophage inducer; PPP, Pentose phosphate pathway; PRR, Pattern recognition receptor; ROCK2, Rho-dependent Kinase 2; SCD2, Stearoyl-CoA Desaturase-2; SHIP1, Src Homology-2 domain-containing Inositol 5-Phosphatase 1; SOCS1, Suppressor of Cytokine Signaling 1; SREBF1, Sterol Regulatory Element-Binding Factor 1; SREBF2, Sterol Regulatory Element-Binding Factor 2; TCA Cycle, Tricarboxylic acid cycle; TET2, TET Methylcytosine Dioxygenase 2; TGF-β, Transforming Growth Factor-β; TLR, Toll-like receptor; TNF-α, Tumor Necrosis Factor-alpha; TRAF6, TNF Receptor Associated Factor 6; UDP-GlcNAc, Uridine Diphosphate N-Acetylglucosamine.
References
1. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. (1999) 11:443–51. doi: 10.1016/S1074-7613(00)80119-3
2. Schnare M, Holt AC, Takeda K, Akira S, Medzhitov R. Recognition of CpG DNA is mediated by signaling pathways dependent on the adaptor protein MyD88. Curr Biol. (2000) 10:1139–42. doi: 10.1016/S0960-9822(00)00700-4
3. Chen G, Shaw MH, Kim Y-G, Nuñez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. (2009) 4:365–98. doi: 10.1146/annurev.pathol.4.110807.092239
4. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. (2011) 11:723–37. doi: 10.1038/nri3073
5. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. (2013) 496:445–55. doi: 10.1038/nature12034
6. Gordon S, Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. (2017) 15:53. doi: 10.1186/s12915-017-0392-4
7. Perdiguero EG, Geissmann F. The development and maintenance of resident macrophages. Nat Immunol. (2016) 17:2–8. doi: 10.1038/ni.3341
8. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. (2011) 11:762–74. doi: 10.1038/nri3070
9. van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, et al. Yolk Sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity. (2016) 44:755–68. doi: 10.1016/j.immuni.2016.02.017
10. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. (2012) 122:787–95. doi: 10.1172/JCI59643
11. Gordon S. Alternative activation of macrophages. Nat Rev Immunol. (2003) 3:23–35. doi: 10.1038/nri978
12. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. (2008) 8:958–69. doi: 10.1038/nri2448
13. Murray PJ. Macrophage polarization. Annu Rev Physiol. (2017) 79:541–66. doi: 10.1146/annurev-physiol-022516-034339
14. O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. (2008) 205:585–94. doi: 10.1084/jem.20072108
15. Runtsch MC, Nelson MC, Lee SH, Voth W, Alexander M, Hu R, et al. Anti-inflammatory microRNA-146a protects mice from diet-induced metabolic disease. PLoS Genet. (2019) 15:e1007970. doi: 10.1371/journal.pgen.1007970
16. Pauley KM, Stewart CM, Gauna AE, Dupre LC, Kuklani R, Chan AL, et al. Altered miR-146a expression in Sjögren's syndrome and its functional role in innate immunity. Eur J Immunol. (2011) 41:2029–39. doi: 10.1002/eji.201040757
17. Sahraei M, Chaube B, Liu Y, Sun J, Kaplan A, Price NL, et al. Suppressing miR-21 activity in tumor-associated macrophages promotes an antitumor immune response. J Clin Invest. (2019) 129:5518–36. doi: 10.1172/JCI127125
18. Hackett EE, Charles-Messance H, O'Leary SM, Gleeson LE, Munoz-Wolf N, Case S, et al. Mycobacterium tuberculosis limits host glycolysis and IL-1beta by restriction of PFK-M via microRNA-21. Cell Rep. (2020) 30:124–36. e124. doi: 10.1016/j.celrep.2019.12.015
19. Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe. (2015) 17:345–56. doi: 10.1016/j.chom.2015.01.007
20. Jiang S, Yan W, Wang SE, Baltimore D. Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc Natl Acad Sci USA. (2019) 116:12416–21. doi: 10.1073/pnas.1811040116
21. Wang Z, Xu L, Hu Y, Huang Y, Zhang Y, Zheng X, et al. miRNA let-7b modulates macrophage polarization and enhances tumor-associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci Rep. (2016) 6:25602. doi: 10.1038/srep25602
22. Nematian SE, Mamillapalli R, Kadakia TS, Majidi Zolbin M, Moustafa S, Taylor HS. Systemic inflammation induced by microRNAs: endometriosis-derived alterations in circulating microRNA 125b-5p and Let-7b-5p regulate macrophage cytokine production. J Clin Endocrinol Metab. (2018) 103:64–74. doi: 10.1210/jc.2017-01199
23. Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. (2013) 190:6542–9. doi: 10.4049/jimmunol.1202496
24. Zhang W, Liu H, Liu W, Liu Y, Xu J. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-kappaB pathway. Cell Death Differ. (2015) 22:287–97. doi: 10.1038/cdd.2014.142
25. Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. (2009) 31:220–31. doi: 10.1016/j.immuni.2009.06.024
26. Curtale G, Renzi TA, Mirolo M, Drufuca L, Albanese M, De Luca M, et al. Multi-step regulation of the TLR4 pathway by the miR-125a~99b~let-7e cluster. Front Immunol. (2018) 9:2037. doi: 10.3389/fimmu.2018.02037
27. Jaiswal A, Reddy SS, Maurya M, Maurya P, Barthwal MK. MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFalpha. Cell Mol Immunol. (2019) 16:495–507. doi: 10.1038/s41423-018-0038-7
28. Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. (2012) 318:1175–84. doi: 10.1016/j.yexcr.2012.03.018
29. Lavery CA, Kurowska-Stolarska M, Holmes WM, Donnelly I, Caslake M, Collier A, et al. miR-34a(-/-) mice are susceptible to diet-induced obesity. Obesity. (2016) 24:1741–51. doi: 10.1002/oby.21561
30. Miranda K, Yang X, Bam M, Murphy EA, Nagarkatti PS, Nagarkatti M. MicroRNA-30 modulates metabolic inflammation by regulating Notch signaling in adipose tissue macrophages. Int J Obes. (2018) 42:1140–50. doi: 10.1038/s41366-018-0114-1
31. Zhihua Y, Yulin T, Yibo W, Wei D, Yin C, Jiahao X, et al. Hypoxia decreases macrophage glycolysis and M1 percentage by targeting microRNA-30c and mTOR in human gastric cancer. Cancer Sci. (2019) 110:2368–77. doi: 10.1111/cas.14110
32. Zhao JL, Huang F, He F, Gao CC, Liang SQ, Ma PF, et al. Forced activation of notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Res. (2016) 76:1403–15. doi: 10.1158/0008-5472.CAN-15-2019
33. Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J Biol Chem. (2012) 287:21816–25. doi: 10.1074/jbc.M111.327031
34. Duroux-Richard I, Roubert C, Ammari M, Presumey J, Grun JR, Haupl T, et al. miR-125b controls monocyte adaptation to inflammation through mitochondrial metabolism and dynamics. Blood. (2016) 128:3125–36. doi: 10.1182/blood-2016-02-697003
35. Ouimet M, Ediriweera HN, Gundra UM, Sheedy FJ, Ramkhelawon B, Hutchison SB, et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. (2015) 125:4334–48. doi: 10.1172/JCI81676
36. Lin JB, Moolani HV, Sene A, Sidhu R, Kell P, Lin JB, et al. Macrophage microRNA-150 promotes pathological angiogenesis as seen in age-related macular degeneration. JCI Insight. (2018) 3:e120157. doi: 10.1172/jci.insight.120157
37. Poitz DM, Augstein A, Gradehand C, Ende G, Schmeisser A, Strasser RH. Regulation of the Hif-system by micro-RNA 17 and 20a - role during monocyte-to-macrophage differentiation. Mol Immunol. (2013) 56:442–51. doi: 10.1016/j.molimm.2013.06.014
38. Xu Z, Zhao L, Zhu LY, He M, Zheng L, Wu Y. MicroRNA-17, 20a regulates the proangiogenic function of tumor-associated macrophages via targeting hypoxia-inducible factor 2alpha. PLoS ONE. (2013) 8:e77890. doi: 10.1371/journal.pone.0077890
39. Kumar V, Kumar A, Das S, Kumar A, Abhishek K, Verma S, et al. Leishmania donovani activates hypoxia inducible factor-1alpha and miR-210 for survival in macrophages by downregulation of NF-kappaB mediated pro-inflammatory immune response. Front Microbiol. (2018) 9:385. doi: 10.3389/fmicb.2018.00385
40. Qi J, Qiao Y, Wang P, Li S, Zhao W, Gao C. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-kappaB1 in murine macrophages. FEBS Lett. (2012) 586:1201–7. doi: 10.1016/j.febslet.2012.03.011
41. Tian F, Tang P, Sun Z, Zhang R, Zhu D, He J, et al. miR-210 in exosomes derived from macrophages under high glucose promotes mouse diabetic obesity pathogenesis by suppressing NDUFA4 expression. J Diabetes Res. (2020) 2020:6894684. doi: 10.1155/2020/6894684
42. Squadrito ML, Pucci F, Magri L, Moi D, Gilfillan GD, Ranghetti A, et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. (2012) 1:141–54. doi: 10.1016/j.celrep.2011.12.005
43. Heinsbroek SE, Squadrito ML, Schilderink R, Hilbers FW, Verseijden C, Hofmann M, et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. (2016) 9:960–73. doi: 10.1038/mi.2015.113
44. Seeley JJ, Baker RG, Mohamed G, Bruns T, Hayden MS, Deshmukh SD, et al. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature. (2018) 559:114–9. doi: 10.1038/s41586-018-0253-5
45. Evans TJ. The role of macrophages in septic shock. Immunobiology. (1996) 195:655–9. doi: 10.1016/S0171-2985(96)80029-5
46. Snelgrove RJ, Goulding J, Didierlaurent AM, Lyonga D, Vekaria S, Edwards L, et al. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat Immunol. (2008) 9:1074–83. doi: 10.1038/ni.1637
47. Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. (2011) 11:738–49. doi: 10.1038/nri3071
48. Coffelt SB, Hughes R, Lewis CE. Tumor-associated macrophages: effectors of angiogenesis and tumor progression. Biochim Biophys Acta. (2009) 1796:11–8. doi: 10.1016/j.bbcan.2009.02.004
49. O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. (2016) 213:15–23. doi: 10.1084/jem.20151570
50. Noe JT, Mitchell RA. Tricarboxylic acid cycle metabolites in the control of macrophage activation and effector phenotypes. J Leukoc Biol. (2019) 106:359–67. doi: 10.1002/JLB.3RU1218-496R
51. Biswas SK, Mantovani A. Orchestration of metabolism by macrophages. Cell Metab. (2012) 15:432–7. doi: 10.1016/j.cmet.2011.11.013
52. Kelly B, O'Neill LAJ. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. (2015) 25:771–84. doi: 10.1038/cr.2015.68
53. Ambros V. microRNAs: tiny regulators with great potential. Cell. (2001) 107:823–6. doi: 10.1016/S0092-8674(01)00616-X
54. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–297. doi: 10.1016/s0092-8674(04)00045-5
55. Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. (2009) 9:581–93. doi: 10.1038/nri2567
56. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. (2008) 9:102–114. doi: 10.1038/nrg2290
57. Kamanu TK, Radovanovic A, Archer JA, Bajic VB. Exploration of miRNA families for hypotheses generation. Sci Rep. (2013) 3:2940. doi: 10.1038/srep02940
58. Roy S. miRNA in macrophage development and function. Antioxid Redox Signal. (2016) 25:795–804. doi: 10.1089/ars.2016.6728
59. Yao Q, Song Z, Wang B, Zhang JA. Emerging roles of microRNAs in the metabolic control of immune cells. Cancer Lett. (2018) 433:10–7. doi: 10.1016/j.canlet.2018.06.024
60. Murray PJ, Wynn TA. Obstacles and opportunities for understanding macrophage polarization. J Leukocyte Biol. (2011) 89:557–63. doi: 10.1189/jlb.0710409
61. Kawai T, Akira S. TLR signaling. Cell Death Differ. (2006) 13:816–25. doi: 10.1038/sj.cdd.4401850
62. Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. (2012) 4:a006049. doi: 10.1101/cshperspect.a006049
63. Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. (2010) 185:605–14. doi: 10.4049/jimmunol.0901698
64. Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. (2013) 496:238–42. doi: 10.1038/nature11986
65. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. (2011) 27:441–64. doi: 10.1146/annurev-cellbio-092910-154237
66. Viola A, Munari F, Sanchez-Rodriguez R, Scolaro T, Castegna A. The metabolic signature of macrophage responses. Front Immunol. (2019) 10:1462. doi: 10.3389/fimmu.2019.01462
67. Liu L, Lu Y, Martinez J, Bi Y, Lian G, Wang T, et al. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1alpha-dependent. Proc Natl Acad Sci USA. (2016) 113:1564–9. doi: 10.1073/pnas.1518000113
68. O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. (2016) 16:553–65. doi: 10.1038/nri.2016.70
69. O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. (2013) 493:346–55. doi: 10.1038/nature11862
70. Galvan-Pena S, O'Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. (2014) 5:420. doi: 10.3389/fimmu.2014.00420
71. Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFalpha- and IFNgamma-triggered inflammation. Biochim Biophys Acta. (2014) 1839:1217–25. doi: 10.1016/j.bbagrm.2014.07.013
72. Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. (2016) 167:457–70. e413. doi: 10.1016/j.cell.2016.08.064
73. Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci USA. (2013) 110:7820–5. doi: 10.1073/pnas.1218599110
74. Jha AK, Huang SC-C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. (2015) 42:419–30. doi: 10.1016/j.immuni.2015.02.005
75. Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. (2016) 24:158–66. doi: 10.1016/j.cmet.2016.06.004
76. Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. (2003) 112:645–57. doi: 10.1016/S0092-8674(03)00154-5
77. Nathan CF, Hibbs JB Jr. Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr Opin Immunol. (1991) 3:65–70. doi: 10.1016/0952-7915(91)90079-G
78. Mills CD, Shearer J, Evans R, Caldwell MD. Macrophage arginine metabolism and the inhibition or stimulation of cancer. J Immunol. (1992) 149:2709–14.
79. Tripathi P, Tripathi P, Kashyap L, Singh V. The role of nitric oxide in inflammatory reactions. FEMS Immunol Med Microbiol. (2007) 51:443–52. doi: 10.1111/j.1574-695X.2007.00329.x
80. MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. (1997) 15:323–50. doi: 10.1146/annurev.immunol.15.1.323
81. Herbert DR, Orekov T, Roloson A, Ilies M, Perkins C, O'Brien W, et al. Arginase I suppresses IL-12/IL-23p40-driven intestinal inflammation during acute schistosomiasis. J Immunol. (2010) 184:6438–46. doi: 10.4049/jimmunol.0902009
82. Sheldon KE, Shandilya H, Kepka-Lenhart D, Poljakovic M, Ghosh A, Morris SM Jr. Shaping the murine macrophage phenotype: IL-4 and cyclic AMP synergistically activate the arginase I promoter. J Immunol. (2013) 191:2290–8. doi: 10.4049/jimmunol.1202102
83. Rath M, Muller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol. (2014) 5:532. doi: 10.3389/fimmu.2014.00532
84. Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. (2014) 15:846–55. doi: 10.1038/ni.2956
85. Huang SCC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, et al. Metabolic reprogramming mediated by the mTORC2-IRF4 signaling axis is essential for macrophage alternative activation. Immunity. (2016) 45:817–30. doi: 10.1016/j.immuni.2016.09.016
86. Sag D, Carling D, Stout RD, Suttles J. Adenosine 5'-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J Immunol. (2008) 181:8633–41. doi: 10.4049/jimmunol.181.12.8633
87. Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. (2012) 13:270–6. doi: 10.1038/nrm3305
88. Ip WKE, Hoshi N, Shouval DS, Snapper S, Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. (2017) 356:513. doi: 10.1126/science.aal3535
89. Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. (2006) 4:9–12. doi: 10.1016/j.cmet.2006.05.009
90. O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. (2007) 104:1604–9. doi: 10.1073/pnas.0610731104
91. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. (2007) 316:604–8. doi: 10.1126/science.1141229
92. Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. (2008) 9:839–45. doi: 10.1038/ni.f.209
93. Hsin JP, Lu Y, Loeb GB, Leslie CS, Rudensky AY. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol. (2018) 19:1137–45. doi: 10.1038/s41590-018-0208-x
94. An H, Xu H, Zhang M, Zhou J, Feng T, Qian C, et al. Src homology 2 domain-containing inositol-5-phosphatase 1 (SHIP1) negatively regulates TLR4-mediated LPS response primarily through a phosphatase activity- and PI-3K-independent mechanism. Blood. (2005) 105:4685–92. doi: 10.1182/blood-2005-01-0191
95. O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. (2009) 106:7113–8. doi: 10.1073/pnas.0902636106
96. Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. (2011) 108:11193–8. doi: 10.1073/pnas.1019536108
97. Lu LF, Gasteiger G, Yu IS, Chaudhry A, Hsin JP, Lu Y. A single miRNA-mRNA interaction affects the immune response in a context- and cell-type-specific manner. Immunity. (2015) 43:52–64. doi: 10.1016/j.immuni.2015.04.022
98. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. (2007) 316:608–11. doi: 10.1126/science.1139253
99. Wang P, Hou J, Lin L, Wang C, Liu X, Li D, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. (2010) 185:6226–33. doi: 10.4049/jimmunol.1000491
100. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
101. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. (2011) 208:1189–201. doi: 10.1084/jem.20101823
102. Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci USA. (2011) 108:9184–9. doi: 10.1073/pnas.1105398108
103. Luly FR, Lévêque M, Licursi V, Cimino G, Martin-Chouly C, Théret N, et al. MiR-146a is over-expressed and controls IL-6 production in cystic fibrosis macrophages. Sci Rep. (2019) 9:16259. doi: 10.1038/s41598-019-52770-w
104. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. (2008) 378:492–504. doi: 10.1016/j.jmb.2008.03.015
105. Sheedy FJ. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. (2015) 6:19. doi: 10.3389/fimmu.2015.00019
106. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. (2007) 133:647–58. doi: 10.1053/j.gastro.2007.05.022
107. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. (2010) 11:141–7. doi: 10.1038/ni.1828
108. Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb Symp Quant Biol. (2011) 76:211–6. doi: 10.1101/sqb.2011.76.010868
109. Beamer GL, Flaherty DK, Assogba BD, Stromberg P, Gonzalez-Juarrero M, de Waal Malefyt R, et al. Interleukin-10 promotes Mycobacterium tuberculosis disease progression in CBA/J mice. J Immunol. (2008) 181:5545–50. doi: 10.4049/jimmunol.181.8.5545
110. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. (2008) 18:505–16. doi: 10.1016/j.tcb.2008.07.007
111. Jiang S, Yan W, Wang SE, Baltimore D. Let-7 suppresses B cell activation through restricting the availability of necessary nutrients. Cell Metab. (2018) 27:393–403. e394. doi: 10.1016/j.cmet.2017.12.007
112. Muxel SM, Acuna SM, Aoki JI, Zampieri RA, Floeter-Winter LM. Toll-like receptor and miRNA-let-7e expression alter the inflammatory response in leishmania amazonensis-infected macrophages. Front Immunol. (2018) 9:2792. doi: 10.3389/fimmu.2018.02792
113. Banerjee S, Cui H, Xie N, Tan Z, Yang S, Icyuz M, et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. (2013) 288:35428–36. doi: 10.1074/jbc.M112.426866
114. Lu L, McCurdy S, Huang S, Zhu X, Peplowska K, Tiirikainen M, et al. Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci Rep. (2016) 6:37446. doi: 10.1038/srep37446
115. Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O'Connell RM, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. (2011) 187:5062–8. doi: 10.4049/jimmunol.1102001
116. Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. (2008) 27(Suppl. 1):S20–9. doi: 10.1038/onc.2009.40
117. Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. (2010) 107:12228–32. doi: 10.1073/pnas.1005191107
118. Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. (2010) 328:1566–9. doi: 10.1126/science.1189123
119. Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. (2010) 328:1570–3. doi: 10.1126/science.1189862
120. Xie N, Cui H, Banerjee S, Tan Z, Salomao R, Fu M, et al. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol. (2014) 193:327–34. doi: 10.4049/jimmunol.1400203
121. Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: the master hypoxamir. Microcirculation. (2012) 19:215–23. doi: 10.1111/j.1549-8719.2011.00154.x
122. Cicchillitti L, Di Stefano V, Isaia E, Crimaldi L, Fasanaro P, Ambrosino V, et al. Hypoxia-inducible factor 1-alpha induces miR-210 in normoxic differentiating myoblasts. J Biol Chem. (2012) 287:44761–71. doi: 10.1074/jbc.M112.421255
123. Lemaire J, Mkannez G, Guerfali FZ, Gustin C, Attia H, Sghaier RM, et al. MicroRNA expression profile in human macrophages in response to Leishmania major infection. PLoS Negl Trop Dis. (2013) 7:e2478. doi: 10.1371/journal.pntd.0002478
124. Zaccagnini G, Maimone B, Fuschi P, Maselli D, Spinetti G, Gaetano C, et al. Overexpression of miR-210 and its significance in ischemic tissue damage. Sci Rep. (2017) 7:9563. doi: 10.1038/s41598-017-09763-4
125. Jin F, Yang R, Wei Y, Wang D, Zhu Y, Wang X, et al. HIF-1alpha-induced miR-23a approximately 27a approximately 24 cluster promotes colorectal cancer progression via reprogramming metabolism. Cancer Lett. (2019) 440–1:211–22. doi: 10.1016/j.canlet.2018.10.025
126. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. (2014) 20:460–9. doi: 10.1016/j.molmed.2014.06.005
127. Curtale G, Rubino M, Locati M. MicroRNAs as molecular switches in macrophage activation. Front Immunol. (2019) 10:799. doi: 10.3389/fimmu.2019.00799
128. Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. (2014) 513:559–63. doi: 10.1038/nature13490
129. Netea-Maier RT, Smit JWA, Netea MG. Metabolic changes in tumor cells and tumor-associated macrophages: a mutual relationship. Cancer Lett. (2018) 413:102–9. doi: 10.1016/j.canlet.2017.10.037
130. Jiang S. Recent findings regarding let-7 in immunity. Cancer Lett. (2018) 434:130–1. doi: 10.1016/j.canlet.2018.07.027
131. Ye P, Liu Y, Chen C, Tang F, Wu Q, Wang X, et al. An mTORC1-Mdm2-Drosha axis for miRNA biogenesis in response to glucose- and amino acid-deprivation. Mol Cell. (2015) 57:708–20. doi: 10.1016/j.molcel.2014.12.034
132. Baer C, Squadrito ML, Laoui D, Thompson D, Hansen SK, Kiialainen A, et al. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. (2016) 18:790–802. doi: 10.1038/ncb3371
133. Varol D, Mildner A, Blank T, Shemer A, Barashi N, Yona S, et al. Dicer deficiency differentially impacts microglia of the developing and adult brain. Immunity. (2017) 46:1030–44 e1038. doi: 10.1016/j.immuni.2017.05.003
134. Wei Y, Corbalan-Campos J, Gurung R, Natarelli L, Zhu M, Exner N, et al. Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation. (2018) 138:2007–20. doi: 10.1161/CIRCULATIONAHA.117.031589
135. Cho YK, Son Y, Kim SN, Song HD, Kim M, Park JH, et al. MicroRNA-10a-5p regulates macrophage polarization and promotes therapeutic adipose tissue remodeling. Mol Metab. (2019) 29:86–98. doi: 10.1016/j.molmet.2019.08.015
136. Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, et al. Adipose tissue macrophage-derived exosomal mirnas can modulate in vivo and in vitro insulin sensitivity. Cell. (2017) 171:372–84:e312. doi: 10.1016/j.cell.2017.08.035
137. Gross TJ, Kremens K, Powers LS, Brink B, Knutson T, Domann FE, et al. Epigenetic silencing of the human NOS2 gene: rethinking the role of nitric oxide in human macrophage inflammatory responses. J Immunol. (2014) 192:2326–38. doi: 10.4049/jimmunol.1301758
138. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. (2013) 368:1685–94. doi: 10.1056/NEJMoa1209026
139. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
140. Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. (2010) 70:5923–30. doi: 10.1158/0008-5472.CAN-10-0655
141. Su YL, Wang X, Mann M, Adamus TP, Wang D, Moreira DF, et al. Myeloid cell-targeted miR-146a mimic inhibits NF-kappaB-driven inflammation and leukemia progression in vivo. Blood. (2020) 135:167–80. doi: 10.1182/blood.2019002045
Keywords: microRNA, metabolism, macrophage, immunometabolism, inflammation, macrophage polarization
Citation: Nelson MC and O'Connell RM (2020) MicroRNAs: At the Interface of Metabolic Pathways and Inflammatory Responses by Macrophages. Front. Immunol. 11:1797. doi: 10.3389/fimmu.2020.01797
Received: 01 May 2020; Accepted: 06 July 2020;
Published: 14 August 2020.
Edited by:
Luminita Aurelia Stanciu, Imperial College London, United KingdomReviewed by:
Joel W. Graff, The University of Iowa, United StatesGyorgy Fejer, University of Plymouth, United Kingdom
Copyright © 2020 Nelson and O'Connell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan M. O'Connell, cnlhbi5vY29ubmVsbEBwYXRoLnV0YWguZWR1
 Morgan C. Nelson
Morgan C. Nelson Ryan M. O'Connell
Ryan M. O'Connell