- 1Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR, United States
- 2Arkansas Children's Nutrition Center, Little Rock, AR, United States
- 3Department of Pediatrics and Pediatric Infectious Diseases, Institute of Child's Health, Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
- 4Inflammation, Repair and Development Section, Faculty of Medicine, Imperial College London, National Heart and Lung Institute, London, United Kingdom
- 5Research and Clinical Center for Neuropsychiatry, Moscow, Russia
- 6Arkansas Children's Research Institute, Little Rock, AR, United States
Exclusive human milk feeding of the newborn is recommended during the first 6 months of life to promote optimal health outcomes during early life and beyond. Human milk contains a variety of bioactive factors such as hormones, cytokines, leukocytes, immunoglobulins, lactoferrin, lysozyme, stem cells, human milk oligosaccharides (HMOs), microbiota, and microRNAs. Recent findings highlighted the potential importance of adding HMOs into infant formula for their roles in enhancing host defense mechanisms in neonates. Therefore, understanding the roles of human milk bioactive factors on immune function is critical to build the scientific evidence base around breastfeeding recommendations, and to enhance positive health outcomes in formula fed infants through modifications to formulas. However, there are still knowledge gaps concerning the roles of different milk components, the interactions between the different components, and the mechanisms behind health outcomes are poorly understood. This review aims to show the current knowledge about HMOs, milk microbiota, immunoglobulins, lactoferrin, and milk microRNAs (miRNAs) and how these could have similar mechanisms of regulating gut and microbiota function. It will also highlight the knowledge gaps for future research.
Introduction
The immune system is the primary line of defense against environmental exposures such as allergens, bacteria, and viruses. The infant's immune system, often mischaracterized as “immature,” is simply naïve to its new extra-uterine environment (1). Normally it undergoes a series of pre-programmed events during early life in response to exposures that occur primarily through the respiratory tract and gastrointestinal tract (GIT) mucosa (2). The infant's immune system at birth has limited anti-oxidant and anti-inflammatory activity in the respiratory and GIT, underdeveloped physical barriers (e.g., tight junctions), limited GIT acidity (chemical barrier), delayed T-cell function and decreased secretion of immunoglobulins [specifically secretory immunoglobulin A (IgA)] (3–5). Early life in humans (from the fetal stage to early months of life) is associated with developmental milestones and human milk provides a medium for inducing both tolerances to antigens and development of a robust immune defense against harmful pathogens. Human milk feeding has been demonstrated to provide healthy GIT mucosal stimuli, impact gut microbiota composition, and promote the infant's developing immune system likely by human milk bioactives (i.e., HMOs, milk microbiota, miRNA, antibodies, lactoferrin, immunoglobulins, cytokines, and hormones) (6, 7). Careful cultivation of a healthy immune system includes not only protective responses to harmful organisms and antigens (e.g., bacteria, viruses, toxins) but moderating the response to non-harmful antigens in the environment (e.g., food antigens or beneficial commensal organisms) in the form of immune tolerance. The current review focus is on lactoferrin, immunoglobulins, HMOs, milk microbiota, and miRNAs components of human milk and their role in infants' gut microbiota colonization, gut health and immune system modulation.
Lactoferrin
Lactoferrin (LF) membrane structure, membrane receptors and transport have been reviewed elsewhere (8). This section will describe the antimicrobial and immune modulatory properties of lactoferrin as well as ongoing clinical studies of formulas supplemented with lactoferrin. Lactoferrin is an iron-binding glycoprotein that exhibits immunomodulatory, anti-inflammatory, antibacterial, antifungal, and antiviral function (Figure 1A) (9–11). Human lactoferrin levels change as milk matures with colostrum having higher concentrations in both term and preterm milk (12), however, preterm milk tends to maintain higher levels of lactoferrin over time (12–14). A recent study of Chinese women reported that lactoferrin concentration was 3.16 and 1.73 g/L in colostrum and milk, respectively (15). LF binds free iron which is an essential nutrient for bacterial growth, thus leading to a bacteriostatic effect (16). Also, LF promotes the growth of low iron requiring bacteria thought to be beneficial to humans such as Lactobacillus and Bifidobacterium (17). Early studies on LF showed a fungistatic effect through iron sequestration (18, 19). Other studies have shown a more direct fungicidal interaction between lactoferrin and the fungal cell surface that is not dependent on iron (20, 21). Furthermore, in vitro studies in which skim human milk and bovine milk were incubated with lactoferrin, iron, and fungi (Candida albicans) demonstrated that skim human milk inhibits fungal growth while bovine milk did not show a fungistatic effect (22). Additionally, another in vitro study showed that human milk LF had higher effect in preventing bacterial growth relative to bovine LF (23) suggesting human milk LF has a superior effect over bovine milk LF. Unfortunately, not all mothers can provide breastmilk for their infants and human milk LF is difficult to obtain for research. Since human and bovine milk LF are highly similar in sequence homology and structure (24, 25), and share similar antimicrobial and immunomodulatory properties (26–29), bovine LF is used more commonly in research.
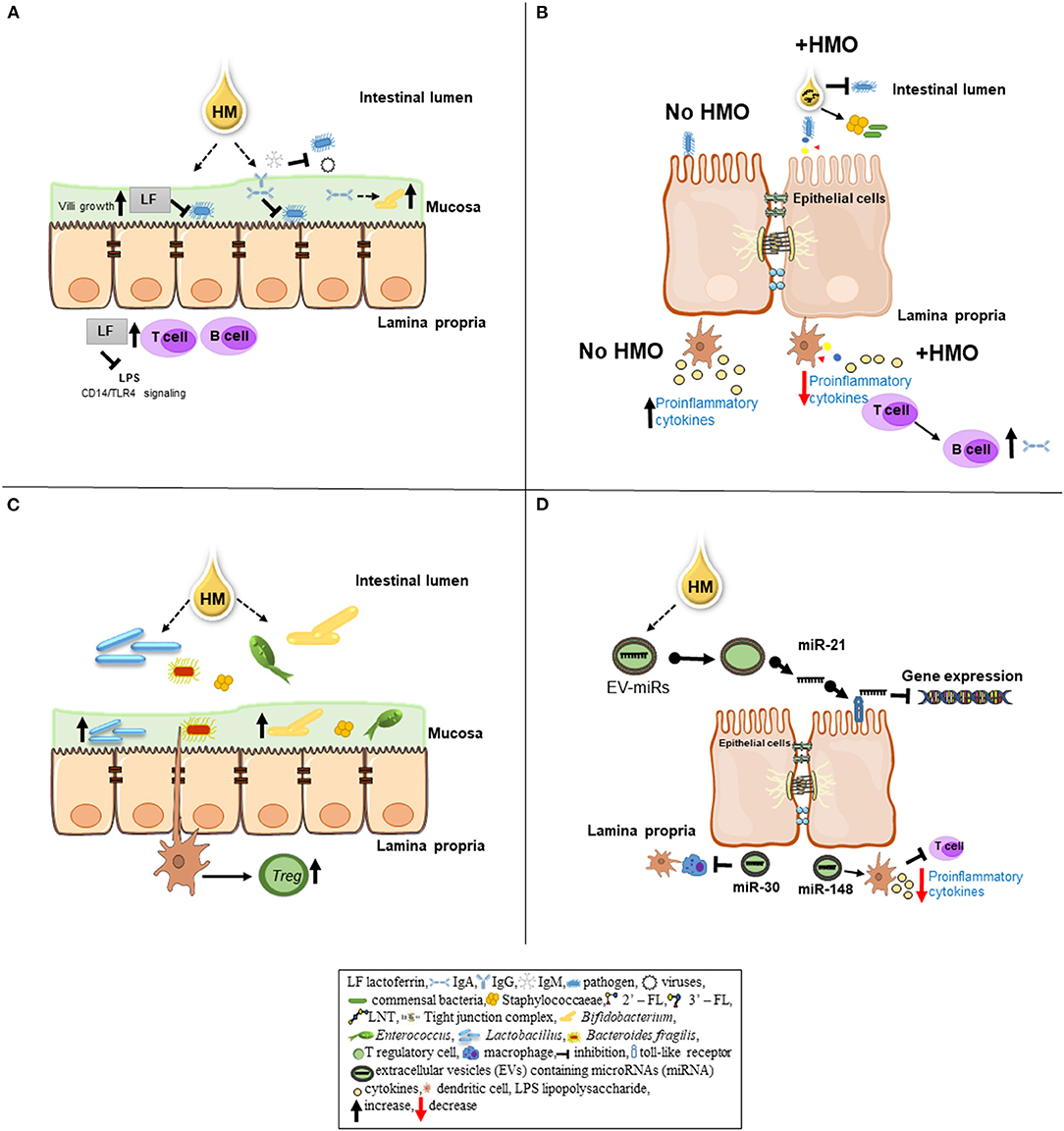
Figure 1. Schematic overview of specific bioactive components of human milk (HM) and their role in immunomodulation. (A) An iron-binding antimicrobial protein lactoferrin (LF) inhibits a number of pathogenic bacteria (i.e., Escherichia coli) from adhering to epithelial cell. LF can promote the growth of intestinal villi. After pathogenic bacteria invasion into the lamina propria of the epithelial gut cells, LF can inhibit the signal between lipopolysaccharide (LPS) released by gram-negative bacteria and the CD14—TLR complex (macrophage signaling). LF can enhance the maturation of B and T cells to improve the immune response. Immunoglobulins IgA, IgM, and IgG present in HM provide passive immunity to the newborn. IgA and IgG can bind to pathogenic bacteria and prevent them from adhering to the epithelial cells in the gut mucosa. Also, IgA can serve as a substrate to obligate anaerobes (i.e., Bacteroides) promoting a healthy microbiota colonization. IgM inhibits enteric bacterial and viral infections by opsonizing the antigen for complement fixation and destruction. (B) In the lumen, human milk oligosaccharides (HMO) inhibit bacterial binding to cell receptors by directly binding to the pathogens. HMOs can stimulate the growth of commensal bacteria by serving as substrates. On epithelial cells, HMOs can prevent pathogen binding by acting as binding decoy receptors. Metabolites of HMOs including short-chain fatty acids can influence epithelial cell maturation and intestinal barrier (i.e., tight junctions) function. HMOs can interact with dendritic cells present in the lamina propria leading to T-cell proliferation, subsequently, T/B cell interaction resulting in increased production of antibodies in order to keep the immune system homeostasis. In the absence of HMOs (no HMO) pathogenic bacteria binding to the epithelial cells increase cytokine production in the lamina propria as a pro-inflammatory response. (C) Bifidobacterium and Lactobacillus, commensal bacteria found in HM, can adhere to intestinal cells, resulting in greater beneficial microbiota colonization. Furthermore, Bacteroides fragilis can interact with dendritic cells, resulting in suppression of inflammation by inducing T regulatory cell (Treg) production. (D) The extracellular vesicles (EVs) contain cargos such as microRNAs (miRNAs). EV-miRNAs likely have immunological and microbial impact on the gastrointestinal tract of neonates. Human milk miRNAs such as miR-21 can regulate gene expression by binding to toll-like receptors 7 and 8 (TLR7/TLR8). Other milk miRNAs (i.e., miR-148 and miR-30) may play a role in gut immune response by decreasing cytokine production via T-cell inhibition and preventing antigen presentation by dendritic cells and macrophages, respectively.
Lactoferrin has been shown to exhibit immunomodulatory properties in several animal models. For example, mice infected with Mycobacterium tuberculosis and supplemented with bovine lactoferrin had decreased levels of M. tuberculosis in their lungs as well as decreased inflammation and increased CD4+ and CD8+ cells (30). A porcine model evaluating the impact of lactoferrin on the immune system showed higher levels of natural killer (NK) cells in mesenteric lymph nodes (MLN), peripheral blood monocytes (PBMC), and in the spleen of piglets fed LF supplemented-formula compared to those fed sow milk and standard formula (31). NK cells are part of the innate immune system and provide protection to the neonate against infections as well as release cytokines that activate other immune cells (32, 33). Piglets fed formula supplemented with bovine lactoferrin had increased crypt cell proliferation and serum immunoglobulin G (IgG) compared to piglets fed formula alone (34, 35). Additionally, piglets that received bovine lactoferrin supplemented formula had greater IL-10 and TNF-α production by splenic cells when compared to the control group (35). Collectively, lactoferrin likely plays a key role in the immune response in neonates. Due to these antimicrobial and immunomodulatory properties of lactoferrin, lactoferrin supplementation in preterm infants has been attempted to decrease late-onset sepsis and necrotizing enterocolitis (36). Moreover, the antifungal property of LF is quite important as premature infants are much more susceptible to fungal infections. Thus, several studies of formulas supplemented with bovine LF to support infants' growth and development have occurred. For example, infant formulas supplemented with bovine LF at 0.6 and 1.0 g/L (range of LF concentration found in mature human milk) were compared to a standard cow's milk formula evaluating growth and tolerance in healthy term infants from 12-days old to 12 months of age. This study reported no growth rate difference between formulas, however the bovine LF supplemented formulas had softer stool consistency relative to the infants fed standard formula (37). Several studies have investigated the addition of bovine LF to neonatal diet (breastmilk, donor milk, and/or formula) in premature infants and have not found significant differences in late onset sepsis outcomes (38–40). Future studies are needed to determine the beneficial effect of enteral LF and LF addition to formulas to enhance the anti-pathogenic effects and immune response in term as well as in preterm infants.
Immunoglobulins
Immunoglobulins (Igs) are glycoprotein molecules produced by plasma cells. They have been shown to provide passive immunity to infants via transfer across the placenta and during breastfeeding. There are five different types of Igs—IgA, IgG, IgM, IgE, and IgD; however, only IgG, crosses the placenta with the majority being transferred in the 3rd trimester (41, 42). All types of Igs have been found in human milk with the most predominant being secretory IgA (sIgA) followed by sIgG (43). sIgA protects against toxins, bacteria, and viruses by preventing binding to the host or directly neutralizing, and serves as the first line of defense in the intestines (Figure 1A) (44–46). sIgA in milk is only partially digested in the stomach of both preterm and term infants while the remainder survives to provide immunity to the lower GI tract (47). Levels in human milk decrease over the first 12 weeks post-partum, most significantly over the first week (48, 49). Although it does decrease, infants rely on human milk sIgA initially, as the cells that produce sIgA in the neonatal gut are low at birth and increase by 10–20 times over the first 6 months of life (50). This correlates with a study comparing fecal sIgA levels in breastfed and formula-fed infants which noted that in the first month of life, sIgA levels were much higher in the breastfed group but were more similar between breastfed and formula fed infants at 6 months of age (51). In mothers immunized with the Neisseria meningococcal vaccine, IgA antibodies specific for Neisseria meningitidis have been shown in human milk for up to 6 months post-partum (52). Interestingly, mother's health status appears to impact sIgA levels in the human milk. sIgA levels have been reported to be lower in the mature milk of mothers with gestational diabetes (53) and in mothers with post-partum stress, anxiety, and depression (54). IgA and IgG levels are lower in the colostrum of mothers with gestational diabetes compared to normo-glycemic women (55, 56). Overall, data suggests that mothers' health condition, vaccination status and lactation period impacts IgA levels in human milk.
IgG is the main immunoglobulin found in serum and is associated with long-term immunity. It not only activates the complement cascade to remove pathogens, but has also been shown to protect against viral infections at the mucosal level through neutralization (57, 58). IgG levels in human milk are low, but increase over time (59). Interestingly, the concentration of IgG is higher in the human milk of exclusively breastfeeding mothers compared to those that are non-exclusive breastfeeding (59). In a mouse model, pathogen-specific IgG was shown to be transferred in milk and protect the pups by coating the pathogen and reducing intestinal colonization (60). Kazimbaya et al. (61) collected human milk samples from mothers prior to their infant receiving the live rotavirus vaccine. For each sample, whole milk, purified IgA, purified IgG, and IgA/IgG depleted milk were isolated. MA104 cells inoculated with the live rotavirus vaccine were exposed to different dilutions of whole milk, purified IgA, purified IgG, and IgA/IgG depleted milk. Interestingly, whole milk and purified IgA and IgG inhibited viral replication suggesting that human milk IgA and IgG can protect against rotavirus infections (61). These studies suggest that human milk IgG plays a role in decreasing infections in infants.
IgM is also transferred to infants via human milk. IgM levels do not vary in human milk in exclusive breastfeeding mothers compared to non-exclusive breastfeeding mothers (59). However, IgM is partially digested by term infants while it is not digested by preterm infants (62). Nevertheless, IgM antibodies protect against bacterial and viral infections by opsonizing the antigen for complement fixation and destruction (63, 64). Serum IgE is associated with a reduction in allergic reactions and parasitic infections. It has also been shown to protect against viruses such as parvovirus B19 (65) and progression of human immunodeficiency virus 1 (HIV-1) (66, 67). Anti-parvovirus B19 IgE antibodies have been found in human milk (68), which might help protect breastfed infants from infection with parvovirus B19. Allergen-specific IgG and IgE antibodies are present in both maternal blood and human milk which may sensitize infants to similar allergens (69). IgD is expressed on mature B cells and it has been shown to bind to certain bacteria resulting in B cell stimulation and activation (70, 71).
Of note, IgA, sIgA, IgM, and IgG concentrations are significantly higher in fresh human milk compared to donor milk (62), which is important to infants in the neonatal intensive care unit due to frequent use of donor milk. This is not unexpected as most donor milk is from mothers of infants that are at least 6 months of age and these samples undergo Holder pasteurization. IgM and IgG are more sensitive to Holder pasteurization than IgA (62, 72, 73). Overall, Igs play a role in reducing pathogenic infections, allergies and likely gut maturation in combination with other components of human milk.
Human Milk Oligosaccharides Promote Beneficial Microbiota Growth, Protect From Inflammation, and Prevent Pathogen Invasion
Human milk oligosaccharides (HMOs) are unconjugated lactose-based carbohydrate structures (74, 75) with concentrations between 7 and 14 g/L in mature milk and 20–24 g/L in colostrum, making HMOs the third most abundant solid component in human milk after lactose and lipids (74, 76). The milk oligosaccharide profile in human milk is more diverse than that of other mammals. For example, the concentration of oligosaccharides in bovine milk is 100 mg/L, and only 50 oligosaccharides structures have been identified in bovine milk (77). However, more than 200 distinct HMO structures have been identified in human milk (74, 75, 78, 79). The structure of HMOs has been reviewed previously (80). The HMOs profile among individual women varies due to differences in the expression of the secretor (Se) and Lewis (Le) genes in the mammary gland. The Se gene encodes for α1,2-fucosyltransferase 2 (FUT2) while the Le gene encodes α1-3/4-fucosyltransferase 3 (81, 82). A systematic review to determine the most abundant HMOs comparing both term and preterm milk reported that for secretor mothers, term milk is most abundant with the neutral HMOs 2′-fucosyllactose (2′FL), difucosyllacto-N-hexaose II (DF-LNH II), Trifucosyllacto-N-hexaose (TF-LNH), and Lacto-N-Fucopentaose I (LNFP-I) and the acidic HMOs 6′-sialyllactose (6′SL), Disialyllacto-N-Tetraose (DS-LNT), and fucosyllacto-N-neohexaose I (FS-LNnH I). For secretor mothers, preterm milk is most abundant with the neutral HMOs 2′FL, DF-LNH II, LNFP-I, and tetrasaccharides lacto-N-tetraose (LNT) and acidic DS-LNT, 6′SL, sialyllacto-N-tetraose c (LST c). Non-secretor milk does not contain α1-2-fucosylated HMOs (83). Additionally, this study revealed that non-secretor term milk is most abundant with neutral DF-LNH II, LNT, and lacto-N-neotetraose (LNnT) and acidic 6′SL. Non-secretor pre-term milk is most abundant for neutral DF-LNH II, LNT, and LNFP II and acidic DS-LNT, LSTc, and 6′SL (83). Erney et al. (84) evaluated 435 women from 10 countries and showed a significant variance in expression of HMOs. In particular, European and Latin American mothers had higher 2′FL expression than those in the US or Asia (84). An in-depth evaluation of regional variation in HMO composition evaluating 410 women from 11 different regions in Europe, North and South America, and sub-Saharan Africa showed variation in secretor status based on regions and self-identified ethnicity (85). It also noted variation in total HMO concentration as well as concentrations of all HMO types except LNFP-I. In addition, several HMO concentrations varied based on environment (rural vs. urban Gambia) including higher LNnT and DSLNT in the rural cohort (85). In addition, HMO composition is likely impacted by exercise. For example, recently Harris et al. (86) demonstrated that exercise induces an increase in 3-SL in human and mice during lactation. In conclusion, HMO composition is impacted by geographic location, likely diet, the secretor status of the mother, term vs. preterm milk and exercise. Thus, future studies need to determine how combination of these factors can optimize HMO synthesis and protect neonates during the infancy period.
HMOs Promote Growth of Healthy Gut Microbiota and Exhibit Protection Against Infections
HMOs have been shown to have a prebiotic effect as they are not digested in the gut and reach the large intestine intact where they are utilized by gut microbiota. HMOs have been shown to stimulate gut microbiota growth and composition. Bifidobacterium, specifically Bifidobacterium longum subsp. infantis and its interaction with HMOs has been well-studied. B. infantis has greater growth when HMOs, not glucose, are the sole source of carbohydrates (87). Its genome has been shown to contain gene clusters dedicated to HMO metabolism and utilization (88). This ability to grow and metabolize HMOs is not present across all bacteria, but seen in B. infantis as well as Bifidobacterium bifidum, Bacteroides fragilis, and Bacteroides vulgatus (89–91). Many bacteria, Lactobacillus gasseri and Enterococcus, for example, do not grow well, or at all, in just the presence of HMOs (87, 91). In a recent animal study, healthy rats were supplemented daily with 2′-FL from days 2 to 16 of life. At day 8, supplemented animals were noted to have increased villus heights as well as higher Lactobacillus proportions in cecal samples. At day 16, animals had higher plasma IgA and IgG as well as more T-cell subsets in their mesenteric lymph nodes (92). This study shows that 2′FL supplementation early in life has a prebiotic effect as well as promotes intestinal growth and immune system maturation.
HMOs not only promote a healthy gut microbiota composition, but also have antimicrobial properties. For instance, α1,2-fucosylated oligosaccharides inhibited Campylobacter jejuni infection in mice (93). In addition, 2′FL percentage in milk has been shown to be inversely proportional to rates of C. jejuni diarrhea (94). HMOs have also recently been shown to have antimicrobial properties against Streptococcus agalactiae [Group B Strep (GBS)], Staphylococcus aureus, and Acinetobacter baumannii (95, 96) by increasing the sensitivity of such bacteria to several antibiotics, particularly antibiotics to which they are not usually susceptible (97). Overall, HMOs provide some protection to infants against bacterial pathogens.
HMOs protect infants from pathogen invasion by various mechanisms (Figure 1B). Several in vitro and in vivo studies highlighted the antiviral properties against different viruses including rotavirus, norovirus, HIV, and influenza. Rotavirus is the most common cause of severe diarrhea worldwide and accounts for 5% of all deaths among children <5 years of age (98). In vitro, 2′FL, 3′SL, 6′SL, and galacto-oligoasccharide reduce infectivity of human rotavirus in MA104 cells, mainly through effects on the virus (99). In experimental settings, 2′FL, LNnT, 3′SL, and 6′SL supplementation in piglets acutely infected with rotavirus downregulated the viral non-structural protein-4 (NSP-4) mRNA expression in the ileum, indicating HMOs inhibit rotavirus replication in the gut (100). Other animal studies in both rats and piglets show that HMOs, in addition to prebiotics, can reduce the length of diarrhea caused by rotavirus (101, 102). HMOs have also been shown to protect against norovirus, the most common cause of acute gastroenteritis outbreaks. Norovirus has been shown to interact with histo-blood group antigens differently with type O having higher susceptibility and B having lower susceptibly to the infection (103, 104). Non-secretors have also been shown to have lower susceptibility to norovirus infections. However, milk from non-secretor mothers does not inhibit attachment of norovirus while milk from secretors does (105). This is likely due to 2′FL binding to the virus and blocking attachment to the gastrointestinal tract (106, 107). 3′FL has also been shown to bind norovirus and block its attachment. Both 2′FL and 3′FL do so by binding to the HBGA pockets on the norovirus capsule, thus, they act as soluble decoy receptors to block pathogens (106). Human milk with higher LDFH-I levels is associated with protection against norovirus as well (94). In both of these gastrointestinal viruses, HMOs have been shown to improve outcomes.
It is estimated that over 38 million people are living with HIV and the rates of transmission from mother to child are as high as 45% (108). In the western world, HIV is considered a contraindication to breastfeeding (109), however, in other countries where access to clean water is unavailable, it is deemed to be the safest option for infant feeding due to lack of nutritional alternatives (110). While breastfeeding is the main post-natal transmission route, many breastfed infants do not become infected. HMOs have been shown to bind the HIV surface glycoprotein, gp120 and decrease binding to dendritic cells (111). HIV infected mothers, particularly those with higher concentrations of LNnT are less likely to transmit HIV to their infants. Mothers with higher concentrations of 3′SL are noted to have higher transmission rates to their offspring as well as a higher viral load and lower CD4 count (112, 113). Higher concentrations of fucosylated HMOs are also associated with decreased mortality in non-infected infants whose mothers are HIV positive (114). Another viral infection that can be ameliorated with HMOs is influenza. Influenza infects more than 3 million people yearly worldwide and causes over 300,000 deaths (115). An in vitro study using pretreated respiratory epithelial cells (Calu-3, 16HBE lines) and PBMCs challenged with either respiratory syncytial virus or influenza and incubated with various concentrations of 6′SL, 3′SL, 2′FL, and LNnT for 24 h showed that 6′SL and LNnT significantly decreased influenza viral load in both airway epithelial cell lines (116). In addition, modified versions of 3′SL and 6′SL have been shown to block hemagglutination and prevent infectivity of influenza viruses (117, 118). HMOs have been shown to improve outcomes in viral gastroenteritis and influenza as well as impact transmission of HIV.
HMOs Improve Gut Barrier Function and Optimize Immune Function
Necrotizing enterocolitis (NEC), a common intestinal disease among premature infants, can cause significant morbidity and mortality [reviewed by Neu and Walker (119)], and is far less common in human milk fed vs. formula fed infants (120). Enteral feeding, including breast- and formula-feeding, impacts the gut maturation of neonates by increasing or decreasing intestinal permeability (121, 122). Decreased intestinal permeability is associated with gut maturation while elevated permeability makes neonates more susceptible to enteric infections and inflammation such as NEC (123, 124). Several studies in animals and humans demonstrated that HMOs may contribute to breastfed infants' lower rates of NEC. In a NEC induction model using neonatal mice, HMO supplemented formula-fed pups had increased mucin expression and decreased intestinal permeability (125). In another rat model of NEC, pups fed HMO supplemented formula had improved survival and the HMO disialyllacto-N-tetraose (DSLNT) was noted to be protective (126). Formulas supplemented with 2′FL have been associated with decreased NEC rates in both mice and rat models (127, 128). However, animal models using preterm pigs have shown only minor effects of HMO supplemented formula on gut microbiota (129) and no effects on gut permeability (130). In addition, several studies have found that milk with lower levels of DSLNT is associated with higher rates of NEC (113, 128). In breastfeeding or pumping mothers, decreased diversity of HMOs, specifically lower concentrations of LNDFH-I during the first month of life is associated with a higher risk for NEC development in preterm infants (131). Clinical trials reported an association of breastfeeding with decreased intestinal permeability at 7 and 14 days of life in preterm infants compared to those that were formula fed (122). In preterm infants, decreased intestinal permeability was associated with increased abundance of Clostridium and Bifidobacterium during the first 2 weeks of life (132). However, which components of human milk are providing these effects and interactions remains to be determined. Overall, HMOs have been shown to decrease pro-inflammatory cytokine expression, pathogenic bacteria penetration, and intestinal permeability in the gut (125, 133, 134). These findings suggest that not just HMOs alone, but rather HMOs in combination with maternal and/or host microbiota might regulate the intestinal barrier function.
HMOs play an important role in the enhancement of the immune system both locally and systemically. HMOs enhance the functions of human dendritic cells (135), an antigen-presenting cell that plays a pivotal role in the regulation and development of the immature immune system in neonates through the recruitment of functional regulatory T-cells (136). For instance, an in vitro approach showed that 0.8, 2 and 5 mg/mL of an HMO mixture upregulated interleukin production (IL-10, IL-27, and IL-6) in dendritic cells (135). Furthermore, HMOs at these concentrations protected dendritic cells against the inflammatory impact of 5 mg/mL lipopolysaccharide (LPS) (135). In a recent mouse model, neutral HMO fractions stimulated the immune response in peritoneal macrophage cells by upregulating the release of nitric oxide (NO), prostaglandin E2 (PGE2), reactive oxygen species (ROS), TNF-α and interleukins such as IL-1β, IL-2, IL-6, and IL-10 (137). Therefore, it is reasonable to hypothesize that certain HMOs can inhibit the pro-inflammatory responses in breastfed infants. In a mouse model, 2′FL supplementation with a dose range of 0.25–5% (w/w) 2 weeks before the primary and booster vaccinations enhanced humoral and cellular immune response to vaccines (138). Mice that received 2′FL had increased levels of vaccine-specific IgG1 and IgG2a in the serum that were 2′FL dose dependent and increased CD27 expression in splenic B-cells. When stimulated ex vivo, spleen cells from 2′FL mice had increased interferon-γ production and proliferation of CD8+ and CD4+ T-cells (138). In addition, mice that were fed the 2′FL containing food had increased activation of B-cells, T1-helper cells, and regulatory T-cells in their MLN (135). In a porcine model, piglets that received formula supplemented with HMOs were shown to have increased circulating NK cells and mesenteric lymph node memory T-cells compared to those that only received formula (139). These studies show that HMOs improve immune response to both infections and vaccines.
HMOs have been shown to play a role in toll-like receptors (TLRs) expression. TLRs are a family of pattern recognition receptors that play a key role in the recognition of invading pathogens and initiate host defense (140–142). Studies have reported structure-dependent effects of HMOs on TLR functions. For example, Asakuma et al. (143) showed that 3′SL, 6′SL, and 6′GL increased expression of both TLR2 and TLR4 while LNFP-I upregulated TLR4 in intestinal cell line HT-29 (143). In another in vitro study, Cheng et al. (144) reported that 3′-FL activated TLR2 whereas LNT activated several TLRs in THP1 macrophages. They also found inhibitory effects for HMOs on TLRs in vitro. For instance, 6′SL, 2′FL, and LNnT inhibited TLR5 and TLR7 whereas 3′FL inhibited TLR5, TLR7, and TLR8 (144). A recently published study fed mice and premature piglets with 2′FL, 6′SL or lactose supplemented formula. Those fed 2′FL and/or 6′SL were noted to have decreased signs of NEC. 2′FL and 6′FL inhibited TLR4 signaling in vivo in cultured IEC-6 enterocytes, in human intestinal explants from NEC patients, and in mouse derived enteroids (145). These studies indicate some role for HMOs in modulating TLRs, however, comparisons are difficult due to differences in studies conducted. The complex effects of different HMOs in modulating TLRs need to be investigated through in vivo models. This will enable us to determine the different mechanisms involved in immune modulation by HMOs. Overall, HMOs appear to have a protective effect in reducing inflammation and inducing stronger immune response.
HMOs as Supplements to Boost Immune Function
HMOs and bovine milk oligosaccharides (BMOs) are currently being studied for their ability to improve immune response in infants. Bovine milk serves as a source of simple and complex oligosaccharides that resemble HMOs (146). It is substantially lower in overall total oligosaccharide concentration compared to human milk, however, there are some similarities in the oligosaccharide profile (147). Bovine milk has a much larger proportion of acidic oligosaccharides including 3′SL and 6′SL as well as neutral LNnT, which are identical to the HMOs with the same name (148). Fucosylated structures such as 2′FL have also been isolated from bovine milk, though in far lower concentrations than human milk (146, 148). BMOs have been demonstrated to elicit similar biological functions to those of HMOs including inhibition of pathogen adhesion to intestinal enterocytes, diminished gut permeability, decreased inflammatory markers, and correction of gut dysbiosis (149). Charbonneau et al. (150) investigated breastfed infants' growth parameters and differences in human milk oligosaccharide composition in Malawi (150). This study demonstrated that the human milk of mothers whose infants had poor growth had lower levels of sialylated HMOs and overall lower concentrations of HMOs (150). Based on this data, a germ-free mouse and piglet model was then used to investigate the impact of sialylated HMOs on stunting phenotype. Animals were gavaged with bacterial strains from feces of infants with growth failure and fed a typical Malawian diet. Some of the animals were supplemented with sialylated BMO's (S-BMO) as well. Those that received S-BMO had improved lean body mass gains, improved metabolism, and elevated levels of N-acetylneuraminic acid (150), suggesting sialylated oligosaccharides are involved in infant growth.
Addition of synthesized oligosaccharides to infant formulas is an evolving field. 2′FL is one of the most abundant and well-studied of the human oligosaccharides as previously mentioned. It has been successfully synthesized and shown to be structurally similar to 2′FL found in human milk samples (151). In a neonatal piglet model, enzymatically synthetized 3′SL and 6′SL sodium salt supplemented bovine based formulas were investigated (152, 153). Piglets were fed either a control diet or concentrations of 140, 200 or 500 mg/L 3′SL, and 300, 600, and 1,200 mg/L for 6′SL. These studies showed that the synthesized HMOs are safe and maintain similar growth in supplemented piglets compared to control diet (152, 153). Several clinical studies have evaluated the addition of 2′FL to formula. 2′FL formula fed infants were compared to breastfed infants and all infants had appropriate growth (154). An evaluation of the cytokine profiles in breastfed infants, 2′FL supplemented formula fed infants, and standard dairy-based formula fed infants demonstrated that 2′FL supplemented formula fed infants had lower plasma concentrations of IL-1α, IL-1β, IL-6, TNF-α, and IL-1rα than the standard formula fed infants, and were similar to those that were breastfed (155). 2′FL supplemented formulas have been approved and are being marketed in Europe (156) and the US, however, the supplementation is at much lower concentrations of 2′FL than what is found in human milk. Sialic acid concentrations have also been evaluated in human milk from mothers with term and preterm infants and compared to several infant formulas (157). The highest concentration was noted in colostrum and then decreased over the next 3 months. Milk from mothers with preterm infants had higher levels of sialic acid. Formulas, however, had a much lower sialic acid content, <25% of what was found in human milk (157). Sialic acid is integral to neonatal brain development and childhood malnutrition, specifically decreased sialic acid intake, has been linked to persistent cognitive deficits (158, 159). Thus, future studies of formulas supplemented with sialic acid would need to be tested for the cognitive function in infants and HMO supplementation to formula is an avenue to pursue in the near future.
Human Milk Microbiota Impacts Colonization of Gut Microbiota and Likely Immune System During Neonatal Period
Different maternal factors including pathologies of the breast, intrapartum antibiotics, maternal health, body mass index (BMI), parity, gestational age, and geographic location of the mothers can contribute to shaping the milk microbiota (160–166). The early establishment of infant microbiota relies on maternal microbiota and plays a key role in the formation of the gut barrier and the maturation of the immune system (Figure 1C) (167). Human milk contains a complex community of bacteria (161, 168) which includes, but is not limited to, multiple genera from Bifidobacterium and Lactobacillus spp, Streptococcus, Staphylococcus, Ralstonia, Bacteroides, Enterobacter, and Enterococcus (161, 167, 169–171). Hunt et al. (172) showed that while there are common genera found in milk, there is variation overtime and between mothers. While most studies have focused on human milk bacterial content, several recent studies have noted fungi present in human milk (173–177). These studies are observational and further investigation is required to evaluate fungal population variance between mothers, the functions of milk mycobiome in infant gut development, and its interactions with other milk microbiota/bioactives and infant immune system. Due to this constraint, this review will focus on human milk and infant microbiota.
Human milk microbiota likely establishes a healthy profile of intestinal bacteria, leading to the maturation of the innate and adaptive immune systems in infants. For instance, intestinal bacteria promote the development of B-cells in Peyer's Patches and increase the release of mucosal IgA, which acts as the first line of defense (178, 179). Human milk bacteria can also improve the activity against infections through the induction of cytotoxic Th1 cells maturation in vitro (180). Interestingly, Lactobacillus in the human milk may enhance the release of Th1 cytokines and TNF-α, and activate NK cells, CD4+, and CD8+ T-cells and regulatory T-cells (181). In addition, commensal bacterial in human milk such as Lactobacillus gasseri and Lactobacillus crispatus have adhesion capacity to the intestinal cells, indicating greater colonization for beneficial bacteria in the gut in breastfed infants (182). In a recent study, Damaceno et al. (182) reported that Bifidobacterium breve, Lactobacillus gasseri and Streptococcus salivarius, limit pathogen adhesion to intestinal epithelial cells ex vivo (182). The microbial species identified in human milk have pathogen inhibition and improving immune function properties. Many studies compare human milk bacterial content to stool content of infants. Human milk microbiota composition is also dependent on pumped vs. directly breast fed. Recently, Moossavi et al. (161) noted that providing pumped milk was associated with higher levels of potential pathogens (i.e., Enterobacteriaceae and Enterococcaceae). Infants fed pumped milk had a lower amount of Bifidobacterium in their stool. In addition, Fehr et al. (183) noted that exclusively breastfed infants have a different microbiome than those that are fed pumped milk. The fact that direct breastfeeding vs. pumped milk feeding results in a different gut microbiome in infants needs to be investigated further. It is possible that some of the variations are due to variability in pump hygiene, mothers skin microbiota, and contribution from environment.
Commensal bacteria in human milk may play protective roles against gastrointestinal infections during infancy. Malago et al. (184) found that Lactobacillus casei, Lactococcus lactis and Bifidobacterium infantis suppressed the release of IL-8 in Caco-2 intestinal cell line incubated with pathogenic Salmonella, supporting the notion that human milk bacteria could protect the infant intestine against epithelial damage. In a recent study, higher abundance of Bifidobacterium at 1 week of life was associated with higher levels of IL-13, IL-5, IL-6, TNF, and IL-1β at 36 months of age compared to children with lower abundance of Bifidobacterium at the same time point (185). Bacteroides might also play a key role to support the immune system in infants during the early stages of life. In particular, the surface of Bacteroides fragilis has polysaccharide A which increases FOXP3 T-cells in the lamina propria resulting in suppression of inflammation (186). In a mouse model, Donaldson et al. (187) showed that Bacteroides binds IgA which allows it to colonize the gastrointestinal tract. In conclusion, milk microbiota likely is one of the first things to colonize the infant gut, promote growth of beneficial microbiota, and in turn impact the immune system in infants.
The infant diet also impacts the microbiome of the gastrointestinal tract and immune system in both animal models and clinical studies. In a rhesus macaques model, formula fed infants were noted to have a different gut microbiome including more Ruminococcus and less Lactobacillus. They also had an increase in pro-inflammatory cytokines TNFα, IFN-γ, IL-1β, and IL-8 (as well as several others) at 1 month of life that decreased overtime (188). Mothers milk fed rhesus macaques are noted to have more memory T-cells as well as T-helper 17 cells compared to formula fed which persists even 6 months after weaning (189). A study of juvenile rhesus macaques noted continued differences, in particular, higher CD8+ T-cell activation (190). These studies show that in rhesus macaques, mothers milk improves immune response while formula changes the microbiome and increases inflammation. There are also several studies carried out with a piglet model that explore diet and its effect on microbiome and the immune system. While many piglet models use sow-fed piglets, this leads to confounding factors due to housing environment, sow milk microbiota, and the maternal environment. Studies from our team housed piglets in the vivarium and fed a regulated diet to eliminate the confounding factors associated with a sow-fed piglet model. Piglets were fed either donor human milk or formula and monitored closely for growth and immune responses. Those fed human milk had a stronger immune response to vaccination in comparison to those fed formula. The piglets who received human milk had lower genera diversity at day 50. At day 21, those fed human milk had higher levels of Bacteroides than those fed formula (191, 192). The human milk fed group also had higher levels of T-cell proliferation (191, 192). These results were similar in comparison to infants fed human milk suggesting the strength of the model. For example, in a small comparative study, fecal samples were collected during the first 20 days of life from 6 breastfed and 6 formula fed infants. In breastfed infants, Bifidobacterium became the most common gut bacteria while in formula fed infants, Bacteroides and Bifidobacterium were found in similar amounts (193). Several other studies have found that in early life, stool Bifidobacterium amount varies in healthy breastfed infants (194–197). Although the reason is unclear, environment may play a role in this. A recent study found three distinct infant gut microbiota, one low in Bifidobacterium but with higher amounts of Streptococcus, one with high amounts of both Bifidobacterium and Bacteroides, and one with higher amounts of Bifidobacterium. Overtime, infant stool transitioned from the profile low in Bifidobacterium to a profiler higher in Bifidobacterium (197). The CHILD cohort has published several studies on infant diet and its impact on microbiome. At 3 months of age, formula fed infants had higher richness and increased Lachnospiraceae. Infants who were breastfed but briefly supplemented with formula had lower levels of Bifidobacteriaceae and higher levels of Enterobacteriaceae at 3 months of age compared to those who did not receive any formula (198). A smaller subset from this cohort noted that formula fed infants had increased richness at 4 months and higher amounts of Clostridium difficile were noted (195). A 2-year study of infant diet and microbiome revealed that formula feeding in the first 3 months of life is associated with decreased diversity and richness at 12–24 months of life. It is also associated with altered beta diversity (199). Andersson et al. (200) compared infants fed 3 different types of formula to breastfed infants and evaluated immune response through 6 months of age. The breastfed group had an increase in leukocyte count, particularly an increase in neutrophils. Formula fed infants had a decrease in the relative amount of NK cells and an increase in CD4+ αβT-cells. Formula fed infants also had a higher ratio of CD4–CD8 cells (200). Data from these studies indicate that human milk feeding is optimal for microbial colonization, promoting robust immune response and decreasing inflammation in early life.
Extracellular Vesicles and Microrna Cargo Role in Immune Function
Extracellular vesicles is a broad term used to describe vesicles released from many cell types. Readers are referred to O'Reilly et al. (201). for a detailed review of human milk extracellular vesicles (EVs) and their role on infant health. The different methodologies (ultracentrifugation, Exoquick) used to isolate EVs indicate the existence of two subsets such as exosomes (30–100 nm) (202–204) and microvesicles (100–1,000 nm) (205, 206). EVs have been reported to contain various molecules (i.e., proteins, microRNA, metabolites) (207–215). It is yet to be determined whether both exosomes and microvesicles contain miRNAs as most of the methods used so far enrich exosomes. Interestingly, milk seems to contain the highest level of miRNAs compared to its volume. The mechanisms involved in loading the miRNAs to EVs in human milk are still unclear and future research is needed. For a more detailed review of EV biogenesis and cargo composition readers are referred to Spencer and Yeruva (216). The focus of this subsection is to describe EV-microRNA cargo role on infant health.
miRNA are small non-coding RNA (~22 nucleotides) that regulate post-transcriptional expression of genes and have biological activities in humans (217–219). Human milk contains several miRNAs (218, 220), and these miRNAs survive in the acidic environment in the GI tract and can be absorbed (221). Infant formulas, however, have a significantly lower amount of miRNAs compared with human milk (218, 222). The origin of these miRNAs is still under debate. However, based on the current knowledge on the composition of the EV proteins, breast cell lines, and miRNA profile of mammary gland cells, these miRNAs are likely from immune-related and mammary gland cells (223–225). The literature review of several studies on miRNA profile suggests that miR-148a-3p, miR-22-3p, miR-200a-3p, miR-146b-5p, miR-30d-5p, let-7a-5p, miR-30a-5p, let-7f-5p, let-7b-5p, and miR-21-5p (226–231) were the most abundant in human milk. In vitro studies suggest that milk miRNAs are taken up by intestinal, immune, and cancer cell lines (218, 220, 232–236). Future animal models and clinical studies under controlled conditions are needed to determine the bioavailability of these miRNAs.
Few studies have been conducted so far on various factors impacting milk miRNA composition. For example, in mice fed high-fat diet, changes in milk miRNA expression was observed (237). Target prediction analysis of these miRNAs in the high-fat diet group impacted developmental process and transcription. Most recently, Carney et al. demonstrated changes in miRNA profile based on delivery status (preterm vs. term) that appear to influence metabolism and lipid biosynthesis. This suggests gestational age likely plays a role in milk miRNA composition and miRNAs appear to directly influence neonatal health and metabolism. This is an area for future studies to determine the underlying mechanisms involved in milk miRNA composition.
The biological impact of human milk EV-miRNAs on infant health is important to address before supplementing formulas. Previous studies using target prediction analysis of human milk miRNAs provided initial evidence that the majority of these miRNAs are likely impacting the immune system. Also, experimental evidence from in vitro and in vivo studies using infection and inflammation models suggest that milk miRNAs could impact the immune system. For example, miR-148, present in pre-term and term human milk but significantly lower in formula (218, 226), appears to be the most abundant in human milk. It is shown to regulate the innate immune response in several ways including limiting cytokine production (238). miR-148 also inhibits T-cell proliferation initiated by the presentation of antigens by dendritic cells in a mouse model (238). Let-7 functions to regulate the innate immune system; it limits B-cell activation, affects T-cell differentiation, and regulates TLR4 signaling and macrophage activation (239, 240). miR-30 is important for intestinal epithelial cell homeostasis (241) and the immune response to Mycobacterium tuberculosis (242) and influenza infections (243). miR-30 also inhibits antigen processing and presentation by dendritic cells and macrophages (244). Other studies identified miR-181 in human milk (220) which induces B- and T-cell differentiation and development (245, 246) and plays a role in inflammation by downregulating TNF-α production in Brucella abortus infections (247). In addition, porcine milk miRNAs were recently shown to reduce LPS-induced apoptosis by preventing TLR4 in intestinal epithelial cells (248). Thus, it is possible that milk miRNAs protect infants from infection, reduces inflammation, and boosts the immune response by various mechanisms (Figure 1D).
The potential for human milk miRNAs acting as TLR7 ligand is a novel concept that we put forth in this review. We hypothesize that GU rich motif (GU or GUUG) of human milk miRNAs activates TLR7/TLR8 and could have an adjuvant effect on immune response during vaccination in breastfed infants. For example, milk miR-21, let-7a, and let-7b have a GU rich region and can bind to TLR7/TLR8 receptors (249–252). Thus, milk miRNAs could have dual functions such as TLR7/TLR8 receptors and/or regulatory role by inhibiting gene expression. Mechanistic studies are needed to determine the specific role of milk miRNAs. In addition, whether miRNAs have direct or indirect effects via microbiota on the infant gut and the immune system is not fully understood. However, the evidence so far suggests that miRNAs could change microbiota composition. Recently, exosome/RNA depleted diet (based on bovine milk exosomes) fed C57Bl6 mice showed changes in the composition of microbiota with relative abundances reported < 1% at family taxonomic level in comparison to exosome/RNA sufficient diet fed mice (253). This study does not show the direct role of miRNAs from bovine milk, nor does it indicate which components of exosomes altered the microbiota composition. However, in a different study it has been demonstrated that bacterial growth is promoted in the presence of certain miRNAs and that endogenous miRNA produced by intestinal epithelial cells alter gut microbial diversity. The increased growth was observed in co-culture of Mission® miRNA mimics and Fusobacterium nucleatum (ATCC® 10953) and E.coli (ATCC® 47016) (254). Results from this study suggest that miRNAs modulate the gut microbiota; to date, however, no studies investigating the effect of exogenous miRNAs from human milk on neonatal microbiota have been conducted. If miRNAs do indeed promote the survival and growth of gut bacteria, these may serve as a novel component to supplement the infant diet.
Perspective and Conclusions
Human milk remains the gold standard for infant nutrition. This review summarized several bioactive components of human milk and their impact on infant microbiome and gut/immune function. Human milk oligosaccharides have been shown to have a prebiotic effect, decrease infectivity as pathogen decoys, and enhance the immune system. Milk microbiota appears to help infants' gut and immune system and protect from pathogens. However, several questions remain unanswered that could ultimately improve term and preterm infant outcomes including decreased infection and improved gut and immune function. Mechanistic studies involving animal models in association with clinical trials are needed. While large animal models (piglet and monkey) are advantageous due to the similarities with infant gut physiology (189, 255), they have multiple limitations. These include a low cost-benefit ratio to generate germ-free animal models due to the specialized facilities required, difficulty and expense of knock-out models, issues obtaining species specific reagents and ethical constraints. Animal models have shown differences in offspring gut microbiome and immune response based on diet. Clinical data, while extremely relevant, only allows for association data due to confounding factors. Thus, alternative models such as germ-free mice could be explored to understand the mechanistic questions about milk bioactives. Determining how different human milk bioactives individually and in combination will impact infants' health needs to be pursued.
Future Research
While many questions relating to human milk bioactives have been addressed, there are areas of research that requires future studies. The questions that remain unanswered are: (1) what combination of HMOs or their derivatives should be added to standard formula? (2) should HMOs be added to formula for premature infants? (3) what are the direct and indirect effects of HMOs on infant immune function? (4) how does maternal microbiota transfer into milk and further shape the milk microbiome? (5) does out-of-body bacteria, including skin bacteria, infant oral bacteria, or bacteria from the environment enter the mammary gland and alter milk microbiota? (6) does milk microbiome affect composition of other milk components such as HMOs and miRNAs? (7) how does milk microbiota affect TLRs in the infant gut and does this impact colonization with commensal bacteria and protection from invading pathogens? (8) does the gut milieu (microbiota and mycobiota) interact and how does the interplay impact overall infant health? and (9) how does the addition of different human milk components to formula impact the gut colonization patterns, and in turn, longitudinal infant health? All these questions need further investigation using preclinical and clinical studies. microRNAs are a newer field of study, thus, many questions remain pertaining to how miRNAs interact with the infant gut microbiome and immune system. In conclusion, determining how different human milk bioactives individually and in combination will promote infants' health needs to be pursued.
Author Contributions
LC, AE, FR, MV, and LY conceived and wrote the paper. FR made the figures. DM and KM edited the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Funding
LC and LY were supported by USDA-ARS Project 6026-51000-012-06S and LY was also supported by NIH 1R21AI146521.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Gervassi AL, Horton H. Is infant immunity actively suppressed or immature? Virology. (2014) 2014:1–9. doi: 10.4137/VRT.S12248
2. Torow N, Marsland BJ, Hornef MW, Gollwitzer ES. Neonatal mucosal immunology. Mucosal Immunol. (2017) 10:5–17. doi: 10.1038/mi.2016.81
3. Medzhitov R, Janeway C Jr. Innate immunity. N Engl J Med. (2000) 343:338–44. doi: 10.1056/NEJM200008033430506
4. Newburg DS, Walker WA. Protection of the neonate by the innate immune system of developing gut and of human milk. Pediatr Res. (2007) 61:2–8. doi: 10.1203/01.pdr.0000250274.68571.18
5. Yu JC, Khodadadi H, Malik A, Davidson B, Salles E, Bhatia J, et al. Innate immunity of neonates and infants. Front Immunol. (2018) 9:1759. doi: 10.3389/fimmu.2018.01759
6. Duijts L, Jaddoe VW, Hofman A, Moll HA. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics. (2010) 126:e18–25. doi: 10.1542/peds.2008-3256
7. Iwasaki, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. (2015) 16:343–53. doi: 10.1038/ni.3123
8. Kell DB, Heyden EL, Pretorius E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. (2020) 11:1221. doi: 10.3389/fimmu.2020.01221
9. Arnold RR, Brewer M, Gauthier JJ. Bactericidal activity of human lactoferrin: sensitivity of a variety of microorganisms. Infect Immun. (1980) 28:893–8.
10. Kruzel ML, Harari Y, Chen CY, Castro GA. Lactoferrin protects gut mucosal integrity during endotoxemia induced by lipopolysaccharide in mice. Inflammation. (2000) 24:33–44. doi: 10.1023/A:1006935908960
11. Håversen L, Ohlsson BG, Hahn-Zoric M, Hanson LA, Mattsby-Baltzer I. Lactoferrin down-regulates the LPS-induced cytokine production in monocytic cells via NF-kappa B. Cell Immunol. (2002) 220:83–95. doi: 10.1016/S0008-8749(03)00006-6
12. Hirai Y, Kawakata N, Satoh K, Ikeda Y, Hisayasu S, Orimo H, et al. Concentrations of lactoferrin and iron in human milk at different stages of lactation. J Nutr Sci Vitaminol. (1990) 36:531–44. doi: 10.3177/jnsv.36.531
13. Ronayne de Ferrer PA, Baroni A, Sambucetti ME, López NE, Ceriani Cernadas JM. Lactoferrin levels in term and preterm milk. J Am Coll Nutr. (2000) 19:370–3. doi: 10.1080/07315724.2000.10718933
14. Hsu YC, Chen CH, Lin MC, Tsai CR, Liang JT, Wang TM. Changes in preterm breast milk nutrient content in the first month. Pediatr Neonatol. (2014) 55:449–54. doi: 10.1016/j.pedneo.2014.03.002
15. Yang Z, Jiang R, Chen Q, Wang J, Duan Y, Pang X, et al. Concentration of lactoferrin in human milk and its variation during lactation in different Chinese populations. Nutrients. (2018) 10:91235. doi: 10.3390/nu10091235
16. Anghel L, Radulescu A, Erhan RV. Structural aspects of human lactoferrin in the iron-binding process studied by molecular dynamics and small-angle neutron scattering. Eur Phys J E Soft Matter. (2018) 41:109. doi: 10.1140/epje/i2018-11720-x
17. Sherman MP, Bennett SH, Hwang FF, Yu C. Neonatal small bowel epithelia: enhancing anti-bacterial defense with lactoferrin and Lactobacillus GG. Biometals. (2004) 17:285–9. doi: 10.1023/B:BIOM.0000027706.51112.62
18. Kirkpatrick CH, Green I, Rich RR, Schade AL. Inhibition of growth of Candida albicans by iron-unsaturated lactoferrin: relation to host-defense mechanisms in chronic mucocutaneous candidiasis. J Infect Dis. (1971) 124:539–44. doi: 10.1093/infdis/124.6.539
19. Al-Sheikh H. Effect of lactoferrin and iron on the growth of human pathogenic Candida species. Pak J Biol Sci. (2009) 12:91–4. doi: 10.3923/pjbs.2009.91.94
20. Kondori N, Baltzer L, Dolphin GT, Mattsby-Baltzer I. Fungicidal activity of human lactoferrin-derived peptides based on the antimicrobial alphabeta region. Int J Antimicrob Agents. (2011) 37:51–7. doi: 10.1016/j.ijantimicag.2010.08.020
21. Nikawa H, Samaranayake LP, Tenovuo J, Pang KM, Hamada T. The fungicidal effect of human lactoferrin on Candida albicans and Candida krusei. Arch Oral Biol. (1993) 38:1057–63. doi: 10.1016/0003-9969(93)90167-K
22. Andersson Y, Lindquist S, Lagerqvist C, Hernell O. Lactoferrin is responsible for the fungistatic effect of human milk. Early Hum Dev. (2000) 59:95–105. doi: 10.1016/S0378-3782(00)00086-4
23. Woodman T, Strunk T, Patole S, Hartmann B, Simmer K, Currie A. Effects of lactoferrin on neonatal pathogens and Bifidobacterium breve in human breast milk. PLoS ONE. (2018) 13:e0201819. doi: 10.1371/journal.pone.0201819
24. Pierce, Colavizza D, Benaissa M, Maes P, Tartar A, Montreuil J, et al. Molecular cloning and sequence analysis of bovine lactotransferrin. Eur J Biochem. (1991) 196:177–84. doi: 10.1111/j.1432-1033.1991.tb15801.x
25. Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, et al. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. (2012) 11:2912–24. doi: 10.1021/pr300008u
26. Puddu P, Latorre D, Carollo M, Catizone A, Ricci G, Valenti P, et al. Bovine lactoferrin counteracts Toll-like receptor mediated activation signals in antigen presenting cells. PLoS ONE. (2011) 6:e22504. doi: 10.1371/journal.pone.0022504
27. Velusamy SK, Markowitz K, Fine DH, Velliyagounder K. Human lactoferrin protects against Streptococcus mutans-induced caries in mice. Oral Dis. (2016) 22:148–54. doi: 10.1111/odi.12401
28. Perdijk O, van Neerven RJJ, van den Brink E, Savelkoul HFJ, Brugman S. Bovine lactoferrin modulates dendritic cell differentiation and function. Nutrients. (2018) 10:848. doi: 10.3390/nu10070848
29. Velliyagounder K, Rozario SD, Fine DH. The effects of human lactoferrin in experimentally induced systemic candidiasis. J Med Microbiol. (2019) 68:1802–12. doi: 10.1099/jmm.0.001098
30. Welsh KJ, Hwang SA, Boyd S, Kruzel ML, Hunter RL, Actor JK. Influence of oral lactoferrin on Mycobacterium tuberculosis induced immunopathology. Tuberculosis. (2011) 1:S105–13. doi: 10.1016/j.tube.2011.10.019
31. Liu KY, Comstock SS, Shunk JM, Monaco MH, Donovan SM. Natural killer cell populations and cytotoxic activity in pigs fed mother's milk, formula, or formula supplemented with bovine lactoferrin. Pediatr Res. (2013) 74:402–7. doi: 10.1038/pr.2013.125
32. Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol. (2007) 7:379–90. doi: 10.1038/nri2075
33. Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. (1999) 17:189–220. doi: 10.1146/annurev.immunol.17.1.189
34. Reznikov EA, Comstock SS, Yi C, Contractor N, Donovan SM. Dietary bovine lactoferrin increases intestinal cell proliferation in neonatal piglets. J Nutri. (2014) 144:1401–8. doi: 10.3945/jn.114.196568
35. Comstock SS, Reznikov EA, Contractor N, Donovan SM. Dietary bovine lactoferrin alters mucosal and systemic immune cell responses in neonatal piglets. J Nutr. (2014) 144:525–32. doi: 10.3945/jn.113.190264
36. Pammi M, Suresh G. Enteral lactoferrin supplementation for prevention of sepsis and necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. (2017) 6:CD007137. doi: 10.1002/14651858.CD007137.pub5
37. Johnston WH, Ashley C, Yeiser M, Harris CL, Stolz SI, Wampler JL, et al. Growth and tolerance of formula with lactoferrin in infants through one year of age: double-blind, randomized, controlled trial. BMC Pediatr. (2015) 15:173. doi: 10.1186/s12887-015-0488-3
38. Griffiths J, Jenkins P, Vargova M, Bowler U, Juszczak E, King A, et al. Enteral lactoferrin to prevent infection for very preterm infants: the ELFIN RCT. Health Technol Assess. (2018) 22:1–60. doi: 10.3310/hta22740
39. The ELFIN trial investigators group. Enteral lactoferrin supplementation for very preterm infants: a randomised placebo-controlled trial. Lancet. (2019) 393:423–33. doi: 10.1016/S0140-6736(18)32221-9
40. Asztalos EV, Barrington K, Lodha A, Tarnow-Mordi W, Martin A. Lactoferrin infant feeding trial_Canada (LIFT_Canada): protocol for a randomized trial of adding lactoferrin to feeds of very-low-birth-weight preterm infants. BMC Pediatr. (2020) 20:40. doi: 10.1186/s12887-020-1938-0
41. Malek, Sager R, Kuhn P, Nicolaides KH, Schneider H. Evolution of maternofetal transport of immunoglobulins during human pregnancy. Am J Reprod Immunol. (1996) 36:248–55. doi: 10.1111/j.1600-0897.1996.tb00172.x
42. Firan M, Bawdon R, Radu C, Ober RJ, Eaken D, Antohe F, et al. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. Int Immunol. (2001) 13:993–1002. doi: 10.1093/intimm/13.8.993
43. Peitersen B, Bohn L, Andersen H. Quantitative determination of immunoglobulins, lysozyme, and certain electrolytes in breast milk during the entire period of lactation, during a 24-hour period, and in milk from the individual mammary gland. Acta Paediatr Scand. (1975) 64:709–17. doi: 10.1111/j.1651-2227.1975.tb03909.x
44. Apter FM, Lencer WI, Finkelstein RA, Mekalanos JJ, Neutra MR. Monoclonal immunoglobulin A antibodies directed against cholera toxin prevent the toxin-induced chloride secretory response and block toxin binding to intestinal epithelial cells in vitro. Infect Immun. (1993) 61:5271–8. doi: 10.1128/IAI.61.12.5271-5278.1993
45. Mantis NJ, Farrant SA, Mehta S. Oligosaccharide side chains on human secretory IgA serve as receptors for ricin. J Immunol. (2004) 172:6838–45. doi: 10.4049/jimmunol.172.11.6838
46. Silvey KJ, Hutchings AB, Vajdy M, Petzke MM, Neutra MR. Role of immunoglobulin A in protection against reovirus entry into Murine Peyer's patches. J Virol. (2001) 75:10870–9. doi: 10.1128/JVI.75.22.10870-10879.2001
47. Demers-Mathieu V, Underwood MA, Beverly RL, Nielsen SD, Dallas DC. Comparison of human milk immunoglobulin survival during gastric digestion between preterm and term infants. Nutrients. (2018) 10:631. doi: 10.3390/nu10050631
48. Kawano, Emori Y. Changes in maternal secretory immunoglobulin a levels in human milk during 12 weeks after parturition. Am J Hum Biol. (2013) 25:399–403. doi: 10.1002/ajhb.22387
49. Ballabio, Bertino E, Coscia A, Fabris C, Fuggetta D, Molfino S, et al. Immunoglobulin-A profile in breast milk from mothers delivering full term and preterm infants. Int J Immunopathol Pharmacol. (2007) 20:119–28. doi: 10.1177/039463200702000114
50. Perkkiö M, Savilahti E. Time of appearance of immunoglobulin-containing cells in the mucosa of the neonatal intestine. Pediatr Res. (1980) 14:953–5. doi: 10.1203/00006450-198008000-00012
51. Kohler H, Donarski S, Stocks B, Parret A, Edwards C, Schroten H. Antibacterial characteristics in the feces of breast-fed and formula-fed infants during the first year of life. J Pediatr Gastroenterol Nutr. (2002) 34:188–93. doi: 10.1097/00005176-200202000-00015
52. Shahid NS, Steinhoff MC, Roy E, Begum T, Thompson CM, Siber GR. Placental and breast transfer of antibodies after maternal immunization with polysaccharide meningococcal vaccine: a randomized, controlled evaluation. Vaccine. (2002) 20:2404–9. doi: 10.1016/S0264-410X(02)00061-0
53. Smilowitz JT, Totten SM, Huang J, Grapov D, Durham HA, Lammi-Keefe CJ, et al. Human milk secretory immunoglobulin a and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J Nutr. (2013) 143:1906–12. doi: 10.3945/jn.113.180695
54. Kawano, Emori Y. The relationship between maternal postpartum psychological state and breast milk secretory immunoglobulin A level. J Am Psychiatr Nurses Assoc. (2015) 21:23–30. doi: 10.1177/1078390314566882
55. Franca EL, Calderon Ide M, Vieira EL, Morceli G, Honorio-Franca AC. Transfer of maternal immunity to newborns of diabetic mothers. Clin Dev Immunol. (2012) 2012:928187. doi: 10.1155/2012/928187
56. Morceli G, Franca EL, Magalhaes VB, Damasceno DC, Calderon IM, Honorio-Franca AC. Diabetes induced immunological and biochemical changes in human colostrum. Acta Paediatr. (2011) 100:550–6. doi: 10.1111/j.1651-2227.2010.02070.x
57. Van Rompay KK, Berardi CJ, Dillard-Telm S, Tarara RP, Canfield DR, Valverde CR, et al. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J Infect Dis. (1998) 177:1247–59. doi: 10.1086/515270
58. Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. (2000) 6:200–6. doi: 10.1038/72309
59. Abuidhail J, Al-Shudiefat AA, Darwish M. Alterations of immunoglobulin G and immunoglobulin M levels in the breast milk of mothers with exclusive breastfeeding compared to mothers with non-exclusive breastfeeding during 6 months postpartum: The Jordanian cohort study. Am J Hum Biol. (2019) 31:e23197. doi: 10.1002/ajhb.23197
60. Caballero-Flores G, Sakamoto K, Zeng MY, Wang Y, Hakim J, Matus-Acuna V, et al. Maternal immunization confers protection to the offspring against an attaching and effacing pathogen through delivery of IgG in breast milk. Cell Host Microbe. (2019) 25:313–23.e4. doi: 10.1016/j.chom.2018.12.015
61. Kazimbaya KM, Chisenga CC, Simuyandi M, Phiri CM, Laban NM, Bosomprah S, et al. In-vitro inhibitory effect of maternal breastmilk components on rotavirus vaccine replication and association with infant seroconversion to live oral rotavirus vaccine. PLoS ONE. (2020) 15:e0240714. doi: 10.1371/journal.pone.0240714
62. Demers-Mathieu V, Huston RK, Markell AM, McCulley EA, Martin RL, Spooner M, et al. Differences in maternal immunoglobulins within mother's own breast milk and donor breast milk and across digestion in preterm infants. Nutrients. (2019) 11:920. doi: 10.3390/nu11040920
63. Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J Exp Med. (2000) 192:271–80. doi: 10.1084/jem.192.2.271
64. Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J Exp Med. (1998) 188:2381–6. doi: 10.1084/jem.188.12.2381
65. Bluth MH, Norowitz KB, Chice S, Shah VN, Nowakowski M, Josephson AS, et al. Detection of IgE anti-parvovirus B19 and increased CD23+ B cells in parvovirus B19 infection: relation to Th2 cytokines. Clin Immunol. (2003) 108:152–8. doi: 10.1016/S1521-6616(03)00098-6
66. Pellegrino MG, Bluth MH, Smith-Norowitz T, Fikrig S, Volsky DJ, Moallem H, et al. HIV type 1-specific IgE in serum of long-term surviving children inhibits HIV type 1 production in vitro. AIDS Res Hum Retroviruses. (2002) 18:363–72. doi: 10.1089/088922202753519142
67. Seroogy CM, Wara DW, Bluth MH, Dorenbaum A, White C, Durkin HG, et al. Cytokine profile of a long-term pediatric HIV survivor with hyper-IgE syndrome and a normal CD4 T-cell count. J Allergy Clin Immunol. (1999) 104:1045–51. doi: 10.1016/S0091-6749(99)70087-4
68. Smith-Norowitz TA, Drew H, Norowitz HM, Nowakowski M, Bluth EF, Durkin HG, et al. Detection of IgE anti-parvovirus antibodies in human breast milk. Ann Clin Lab Sci. (2008) 38:168–73.
69. Hochwallner H, Alm J, Lupinek C, Johansson C, Mie A, Scheynius A, et al. Transmission of allergen-specific IgG and IgE from maternal blood into breast milk visualized with microarray technology. J Allergy Clin Immunol. (2014) 134:1213–5. doi: 10.1016/j.jaci.2014.08.041
70. Gjörloff Wingren, Hadzic R, Forsgren A, Riesbeck K. The novel IgD binding protein from Moraxella catarrhalis induces human B lymphocyte activation and Ig secretion in the presence of Th2 cytokines. J Immunol. (2002) 168:5582–8. doi: 10.4049/jimmunol.168.11.5582
71. Chen K, Xu W, Wilson M, He B, Miller NW, Bengtén E, et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat Immunol. (2009) 10:889–98. doi: 10.1038/ni.1748
72. Ford JE, Law BA, Marshall VM, Reiter B. Influence of the heat treatment of human milk on some of its protective constituents. J Pediatr. (1977) 90:29–35. doi: 10.1016/S0022-3476(77)80759-2
73. Adhisivam B, Vishnu Bhat B, Rao K, Kingsley SM, Plakkal N, Palanivel C. Effect of Holder pasteurization on macronutrients and immunoglobulin profile of pooled donor human milk. J Matern Fetal Neonatal Med. (2019) 32:3016–9. doi: 10.1080/14767058.2018.1455089
74. Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, metabolic aspects. Annu Rev Nutr. (2000) 20:699–722. doi: 10.1146/annurev.nutr.20.1.699
75. Coppa GV, Gabrielli O, Pierani P, Catassi C, Carlucci A, Giorgi PL. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. (1993) 91:637–41.
76. Coppa GV, Pierani P, Zampini L, Carloni I, Carlucci A, Gabrielli O. Oligosaccharides in human milk during different phases of lactation. Acta Paediatr Suppl. (1999) 88:89–94. doi: 10.1111/j.1651-2227.1999.tb01307.x
77. Albrecht S, Lane JA, Marino K, Al Busadah KA, Carrington SD, Hickey RM, et al. A comparative study of free oligosaccharides in the milk of domestic animals. Br J Nutr. (2014) 111:1313–28. doi: 10.1017/S0007114513003772
78. Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. (2010) 9:4138–51. doi: 10.1021/pr100362f
79. Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. (2011) 10:856–68. doi: 10.1021/pr101006u
80. Bode L, Jantscher-Krenn E. Structure-function relationships of human milk oligosaccharides. Adv Nutr. (2012) 3:383S−91S. doi: 10.3945/an.111.001404
81. Thurl S, Henker J, Siegel M, Tovar K, Sawatzki G. Detection of four human milk groups with respect to Lewis blood group dependent oligosaccharides. Glycoconj J. (1997) 14:795–9. doi: 10.1023/A:1018529703106
82. Kumazaki T, Yoshida A. Biochemical evidence that secretor gene, Se, is a structural gene encoding a specific fucosyltransferase. Proc Natl Acad Sci USA. (1984) 81:4193–7. doi: 10.1073/pnas.81.13.4193
83. Thurl S, Munzert M, Boehm G, Matthews C, Stahl B. Systematic review of the concentrations of oligosaccharides in human milk. Nutr Rev. (2017) 75:920–33. doi: 10.1093/nutrit/nux044
84. Erney RM, Malone WT, Skelding MB, Marcon AA, Kleman-Leyer KM, O'Ryan ML, et al. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr. (2000) 30:181–92. doi: 10.1097/00005176-200002000-00016
85. McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, et al. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. (2017) 105:1086–100. doi: 10.3945/ajcn.116.139980
86. Harris JE, Pinckard KM, Wright KR, Baer LA, Arts PJ, Abay E, et al. Exercise-induced 3'-sialyllactose in breast milk is a critical mediator to improve metabolic health and cardiac function in mouse offspring. Nat Metab. (2020) 2:678–87. doi: 10.1038/s42255-020-0223-8
87. Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. (2006) 72:4497–9. doi: 10.1128/AEM.02515-05
88. Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. (2008) 105:18964–9. doi: 10.1073/pnas.0809584105
89. Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. (2007) 51:1398–405. doi: 10.1002/mnfr.200700150
90. Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. (2011) 286:34583–92. doi: 10.1074/jbc.M111.248138
91. Marcobal, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. (2010) 58:5334–40. doi: 10.1021/jf9044205
92. Azagra-Boronat, Massot-Cladera M, Mayneris-Perxachs J, Knipping K, Van't Land B, Tims S, et al. Immunomodulatory and prebiotic effects of 2'-fucosyllactose in suckling rats. Front Immunol. (2019) 10:1773. doi: 10.3389/fimmu.2019.01773
93. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. (2003) 278:14112–20. doi: 10.1074/jbc.M207744200
94. Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr. (2004) 145:297–303. doi: 10.1016/j.jpeds.2004.04.054
95. Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B Streptococcus. ACS Infect Dis. (2017) 3:595–605. doi: 10.1021/acsinfecdis.7b00064
96. Ackerman DL, Craft KM, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, et al. Antimicrobial and antibiofilm activity of human milk oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, Acinetobacter baumannii. ACS Infect Dis. (2018) 4:315–324. doi: 10.1021/acsinfecdis.7b00183
97. Craft KM, Gaddy JA, Townsend SD. Human milk oligosaccharides (HMOs) sensitize group B Streptococcus to clindamycin, erythromycin, gentamicin, and minocycline on a strain specific basis. ACS Chem Biol. (2018) 13:2020–6. doi: 10.1021/acschembio.8b00661
98. Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin Infect Dis. (2016) 62:S96–105. doi: 10.1093/cid/civ1013
99. Laucirica DR, Triantis V, Schoemaker R, Estes MK, Ramani S. Milk oligosaccharides inhibit human rotavirus infectivity in MA104 cells. J Nutr. (2017) 147:1709–14. doi: 10.3945/jn.116.246090
100. Hester SN, Chen X, Li M, Monaco MH, Comstock SS, Kuhlenschmidt TB, et al. Human milk oligosaccharides inhibit rotavirus infectivity in vitro and in acutely infected piglets. Br J Nutr. (2013) 110:1233–42. doi: 10.1017/S0007114513000391
101. Li M, Monaco MH, Wang M, Comstock SS, Kuhlenschmidt TB, Fahey GC, et al. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J. (2014) 8:1609–20. doi: 10.1038/ismej.2014.10
102. Azagra-Boronat, Massot-Cladera M, Knipping K, Van't Land B, Stahl B, Garssen J, et al. Supplementation With 2'-FL and scGOS/lcFOS ameliorates rotavirus-induced diarrhea in suckling rats. Front Cell Infect Microbiol. (2018) 8:372. doi: 10.3389/fcimb.2018.00372
103. Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. (2002) 185:1335–7. doi: 10.1086/339883
104. Rockx BH, Vennema H, Hoebe CJ, Duizer E, Koopmans MP. Association of histo-blood group antigens and susceptibility to norovirus infections. J Infect Dis. (2005) 191:749–54. doi: 10.1086/427779
105. Le Pendu J. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv Exp Med Biol. (2004) 554:135–43. doi: 10.1007/978-1-4757-4242-8_13
106. Weichert S, Koromyslova A, Singh BK, Hansman S, Jennewein S, Schroten H, et al. structural basis for norovirus inhibition by human milk oligosaccharides. J Virol. (2016) 90:4843–48. doi: 10.1128/JVI.03223-15
107. Koromyslova, Tripathi S, Morozov V, Schroten H, Hansman GS. Human norovirus inhibition by a human milk oligosaccharide. Virology. (2017) 508:81–9. doi: 10.1016/j.virol.2017.04.032
108. World Health Organization. HIV/AIDS (2020). Available online at: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed December 14, 2020).
109. Committee on Pediatric Aids. Infant feeding and transmission of human immunodeficiency virus in the United States. Pediatrics. (2013) 131:391–6. doi: 10.1542/peds.2012-3543
110. World Health Organization. Guidelines on HIV and Infant Feeding 2010: Principles and Recommendations for Infant Feeding in the Context of HIV and a Summary of Evidence. (2010). Available online at: http://whqlibdoc.who.int/publications/2010/9789241599535_eng.pdf (accessed December 14, 2020).
111. Hong P, Ninonuevo MR, Lee B, Lebrilla C, Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br J Nutr. (2009) 101:482–6. doi: 10.1017/S0007114508025804
112. Bode L, Kuhn L, Kim HY, Hsiao L, Nissan C, Sinkala M, et al. Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. Am J Clin Nutr. (2012) 96:831–9. doi: 10.3945/ajcn.112.039503
113. Van Niekerk E, Autran CA, Nel DG, Kirsten GF, Blaauw R, Bode L. Human milk oligosaccharides differ between HIV-infected and HIV-uninfected mothers and are related to necrotizing enterocolitis incidence in their preterm very-low-birth-weight infants. J Nutr. (2014) 144:1227–33. doi: 10.3945/jn.113.187799
114. Kuhn L, Kim HY, Hsiao L, Nissan C, Kankasa C, Mwiya M, et al. Oligosaccharide composition of breast milk influences survival of uninfected children born to HIV-infected mothers in Lusaka, Zambia. J Nutr. (2015) 145:66–72. doi: 10.3945/jn.114.199794
115. World Health Organization. Influena (Seasonal) (2018). https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal) (accessed December 14, 2020).
116. Duska-McEwen G, Senft AP, Ruetschilling TL, Barrett EG, Buck R. Human milk oligosaccharides enhance innate immunity to respiratory syncytial virus and influenza in vitro. Food Nutr Sci. (2014) 5:1387–98. doi: 10.4236/fns.2014.514151
117. Zevgiti S, Zabala JG, Darji A, Dietrich U, Panou-Pomonis E, Sakarellos-Daitsiotis M. Sialic acid and sialyl-lactose glyco-conjugates: design, synthesis and binding assays to lectins and swine influenza H1N1 virus. J Pept Sci. (2012) 18:52–8. doi: 10.1002/psc.1415
118. Kwon SJ, Na DH, Kwak JH, Douaisi M, Zhang F, Park EJ, et al. Nanostructured glycan architecture is important in the inhibition of influenza A virus infection. Nat Nanotechnol. (2017) 12:48–54. doi: 10.1038/nnano.2016.181
119. Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med. (2011) 364:255–64. doi: 10.1056/NEJMra1005408
120. Lucas, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet. (1990) 336:1519–23. doi: 10.1016/0140-6736(90)93304-8
121. Weaver LT, Laker MF, Nelson R, Lucas A. Milk feeding and changes in intestinal permeability and morphology in the newborn. J Pediatr Gastroenterol Nutr. (1987) 6:351–8. doi: 10.1097/00005176-198705000-00008
122. Taylor SN, Basile LA, Ebeling M, Wagner CL. Intestinal permeability in preterm infants by feeding type: mother's milk versus formula. Breastfeed Med. (2009) 4:11–5. doi: 10.1089/bfm.2008.0114
123. Moore SA, Nighot P, Reyes C, Rawat M, McKee J, Lemon D, et al. Intestinal barrier dysfunction in human necrotizing enterocolitis. J Pediatr Surg. (2016) 51:1907–13. doi: 10.1016/j.jpedsurg.2016.09.011
124. Ravisankar S, Tatum R, Garg PM, Herco M, Shekhawat PS, Chen YH. Necrotizing enterocolitis leads to disruption of tight junctions and increase in gut permeability in a mouse model. BMC Pediatr. (2018) 18:372. doi: 10.1186/s12887-018-1346-x
125. Wu RY, Li B, Koike Y, Maattanen P, Miyake H, Cadete M, et al. Human milk oligosaccharides increase mucin expression in experimental necrotizing enterocolitis. Mol Nutr Food Res. (2019) 63:e1800658. doi: 10.1002/mnfr.201800658
126. Jantscher-Krenn E, Zherebtsov M, Nissan C, Goth K, Guner YS, Naidu N, et al. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut. (2012) 61:1417–25. doi: 10.1136/gutjnl-2011-301404
127. Good M, Sodhi CP, Yamaguchi Y, Jia H, Lu P, Fulton WB, et al. The human milk oligosaccharide 2'-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr. (2016) 116:1175–87. doi: 10.1017/S0007114516002944
128. Autran CA, Kellman BP, Kim JH, Asztalos E, Blood AB, Spence ECH, et al. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut. (2018) 67:1064–70. doi: 10.1136/gutjnl-2016-312819
129. Cilieborg MS, Bering SB, Ostergaard MV, Jensen ML, Krych L, Newburg DS, et al. Minimal short-term effect of dietary 2'-fucosyllactose on bacterial colonisation, intestinal function and necrotising enterocolitis in preterm pigs. Br J Nutr. (2016) 116:834–41. doi: 10.1017/S0007114516002646
130. Rasmussen SO, Martin L, Ostergaard MV, Rudloff S, Roggenbuck M, Nguyen DN, et al. Human milk oligosaccharide effects on intestinal function and inflammation after preterm birth in pigs. J Nutr Biochem. (2017) 40:141–54. doi: 10.1016/j.jnutbio.2016.10.011
131. Wejryd E, Marti M, Marchini G, Werme A, Jonsson B, Landberg E, et al. Low diversity of human milk oligosaccharides is associated with necrotising enterocolitis in extremely low birth weight infants. Nutrients. (2018) 10:1556. doi: 10.3390/nu10101556
132. Ma B, McComb E, Gajer P, Yang H, Humphrys M, Okogbule-Wonodi AC, et al. Microbial biomarkers of intestinal barrier maturation in preterm infants. Front Microbiol. (2018) 9:2755. doi: 10.3389/fmicb.2018.02755
133. Autran CA, Schoterman MH, Jantscher-Krenn E, Kamerling JP, Bode L. Sialylated galacto-oligosaccharides and 2'-fucosyllactose reduce necrotising enterocolitis in neonatal rats. Br J Nutr. (2016) 116:294–9. doi: 10.1017/S0007114516002038
134. Wang C, Zhang M, Guo H, Yan J, Liu F, Chen J, et al. Human milk oligosaccharides protect against necrotizing enterocolitis by inhibiting intestinal damage via increasing the proliferation of crypt cells. Mol Nutr Food Res. (2019) 63:e1900262. doi: 10.1002/mnfr.201900262
135. Xiao L, van De Worp WR, Stassen R, van Maastrigt C, Kettelarij N, Stahl B. Human milk oligosaccharides promote immune tolerance via direct interactions with human dendritic cells. Eur J Immunol. (2019) 49:1001–14. doi: 10.1002/eji.201847971
136. Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. (2008) 8:435–46. doi: 10.1038/nri2335
137. Zhang W, Yan J, Wu L, Yu Y, Ye RD, Zhang Y, et al. In vitro immunomodulatory effects of human milk oligosaccharides on murine macrophage RAW264.7 cells. Carbohydr Polym. (2019) 207:230–8. doi: 10.1016/j.carbpol.2018.11.039
138. Xiao L, Leusink-Muis T, Kettelarij N, van Ark I, Blijenberg B, Hesen Na. Human milk oligosaccharide 2'-fucosyllactose improves innate and adaptive immunity in an influenza-specific murine vaccination model. Front Immunol. (2018) 9:452. doi: 10.3389/fimmu.2018.00452
139. Comstock SS, Li M, Wang M, Monaco MH, Kuhlenschmidt TB, Kuhlenschmidt MS, et al. Dietary human milk oligosaccharides but not prebiotic oligosaccharides increase circulating natural killer cell and mesenteric lymph node memory T cell populations in noninfected and rotavirus-infected neonatal piglets. J. Nutr. (2017) 147:1041–7. doi: 10.3945/jn.116.243774
140. Christmas P. Toll-Like Receptors: Sensors that Detect Infection. Nat Educ. (2010) 3:85. Available online at: https://www.nature.com/scitable/topicpage/toll-like-receptors-sensors-that-detect-infection-14396559/ (accessed December 14, 2020).
141. El-Zayat SR, Sibaii H, Mannaa FA. Toll-like receptors activation, signaling, and targeting: an overview. Bull Natl Res Centre. (2019) 43:187. doi: 10.1186/s42269-019-0227-2
142. Cheng L, Akkerman R, Kong C, Walvoort MTC, de Vos P. More than sugar in the milk: human milk oligosaccharides as essential bioactive molecules in breast milk and current insight in beneficial effects. Crit Rev Food Sci Nutr. (2020). doi: 10.1080/10408398.2020.1754756. [Epub ahead of print].
143. Asakuma S, Yokoyama T, Kimura K, Watanabe Y, Nakamura T, Fukuda K, et al. Effect of human milk oligosaccharides on messenger ribonucleic acid expression of toll-like receptor 2 and 4, and of MD2 in the intestinal cell line HT-29. J Appl Glycosci. (2010) 57:177–83. doi: 10.5458/jag.57.177
144. Cheng L, Kiewiet MBG, Groeneveld A, Nauta A, de Vos P. Human milk oligosaccharides and its acid hydrolysate LNT2 show immunomodulatory effects via TLRs in a dose and structure-dependent way. J Funct Foods. (2019) 59:174–84. doi: 10.1016/j.jff.2019.05.023
145. Sodhi CP, Wipf P, Yamaguchi Y, Fulton WB, Kovler M, Nino DF, et al. The human milk oligosaccharides 2'-fucosyllactose and 6'-sialyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signaling. Pediatr Res. (2020). doi: 10.1038/s41390-020-0852-3. [Epub ahead of print].
146. Tao N, DePeters EJ, Freeman S, German JB, Grimm R, Lebrilla CB. Bovine milk glycome. J Dairy Sci. (2008) 91:3768–78. doi: 10.3168/jds.2008-1305
147. Barile D, Tao N, Lebrilla CB, Coisson JD, Arlorio M, German JB. Permeate from cheese whey ultrafiltration is a source of milk oligosaccharides. Int Dairy J. (2009) 19:524–30. doi: 10.1016/j.idairyj.2009.03.008
148. Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. (2013) 23:664–76. doi: 10.1093/glycob/cwt007
149. Boudry G, Hamilton MK, Chichlowski M, Wickramasinghe S, Barile D, Kalanetra KM, et al. Bovine milk oligosaccharides decrease gut permeability and improve inflammation and microbial dysbiosis in diet-induced obese mice. J Dairy Sci. (2017) 100:2471–81. doi: 10.3168/jds.2016-11890
150. Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. (2016) 164:859–71. doi: 10.1016/j.cell.2016.01.024
151. Albermann C, Piepersberg W, Wehmeier UF. Synthesis of the milk oligosaccharide 2'-fucosyllactose using recombinant bacterial enzymes. Carbohydr Res. (2001) 334:97–103. doi: 10.1016/S0008-6215(01)00177-X
152. Monaco MH, Gurung RB, Donovan SM. Safety evaluation of 3'-siallylactose sodium salt supplementation on growth and clinical parameters in neonatal piglets. Regul Toxicol Pharmacol. (2019) 101:57–64. doi: 10.1016/j.yrtph.2018.11.008
153. Monaco MH, Kim DH, Gurung RB, Donovan SM. Evaluation of 6'-sialyllactose sodium salt supplementation to formula on growth and clinical parameters in neonatal piglets. Nutrients. (2020) 12:1030. doi: 10.3390/nu12041030
154. Marriage BJ, Buck RH, Goehring KC, Oliver JS, Williams JA. Infants fed a lower calorie formula with 2'FL show growth and 2'FL uptake like breast-fed infants. J Pediatr Gastroenterol Nutr. (2015) 61:649–58. doi: 10.1097/MPG.0000000000000889
155. Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG, Buck RH. Similar to those who are breastfed, infants fed a formula containing 2'-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr. (2016) 146:2559–66. doi: 10.3945/jn.116.236919
156. Commission Implementing Regulation (EU) 2017/2470 of 20 December 2017 Establishing the Union List of Novel Foods in Accordance With Regulation (EU) 2015/2283 of the European Parliament and of the Council on novel foods (2017).
157. Wang B, Brand-Miller J, McVeagh P, Petocz P. Concentration and distribution of sialic acid in human milk and infant formulas. Am J Clin Nutr. (2001) 74:510–5. doi: 10.1093/ajcn/74.4.510
158. Victora CG, Adair L, Fall C, Hallal PC, Martorell R, Richter L, et al. Child undernutrition study, maternal and child undernutrition: consequences for adult health and human capital. Lancet. (2008) 371:340–57. doi: 10.1016/S0140-6736(07)61692-4
159. Wang B, Yu B, Karim M, Hu H, Sun Y, McGreevy P, et al. Dietary sialic acid supplementation improves learning and memory in piglets. Am J Clin Nutr. (2007) 85:561–9. doi: 10.1093/ajcn/85.2.561
160. Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. (2019) 6:4. doi: 10.3389/fnut.2019.00004
161. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. (2019) 25:324–35.e4. doi: 10.1016/j.chom.2019.01.011
162. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. (2016) 4:1. doi: 10.1186/s40168-015-0145-y
163. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. (2012) 96:544–51. doi: 10.3945/ajcn.112.037382
164. Toscano M, De Grandi R, Peroni DG, Grossi E, Facchin V, Comberiati P, et al. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. (2017) 17:205. doi: 10.1186/s12866-017-1109-0
165. Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. (2016) 7:54–60. doi: 10.1017/S2040174415001397
166. Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martinez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. (2014) 34:599–605. doi: 10.1038/jp.2014.47
167. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
168. Zimmermann P, Curtis N. Breast milk microbiota: a review of the factors that influence composition. J Infect. (2020) 81:17–47. doi: 10.1016/j.jinf.2020.01.023
169. Kordy K, Gaufin T, Mwangi M, Li F, Cerini C, Lee DJ, et al. Contributions to human breast milk microbiome and enteromammary transfer of Bifidobacterium breve. PLoS ONE. (2020) 15:e0219633. doi: 10.1371/journal.pone.0219633
170. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. (2014) 16:2891–904. doi: 10.1111/1462-2920.12238
171. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. (2016) 7:1619. doi: 10.3389/fmicb.2016.01619
172. Hunt KM, Foster JA, Forney LJ, Schutte UM, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE. (2011) 6:e21313. doi: 10.1371/journal.pone.0021313
173. Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B. Mycobiome profiles in breast milk from healthy women depend on mode of delivery, geographic location, and interaction with bacteria. Appl Environ Microbiol. (2019) 85:e02994–18. doi: 10.1128/AEM.02994-18
174. Boix-Amoros, Martinez-Costa C, Querol A, Collado MC, Mira A. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci Rep. (2017) 7:13016. doi: 10.1038/s41598-017-13270-x
175. Moossavi S, Fehr K, Derakhshani H, Sbihi H, Robertson B, Bode L, et al. Human milk fungi: environmental determinants and inter-kingdom associations with milk bacteria in the CHILD cohort Study. BMC Microbiol. (2020) 20:146. doi: 10.1186/s12866-020-01829-0
176. Heisel T, Nyaribo L, Sadowsky MJ, Gale CA. Breastmilk and NICU surfaces are potential sources of fungi for infant mycobiomes. Fungal Genet Biol. (2019) 128:29–35. doi: 10.1016/j.fgb.2019.03.008
177. Dinleyici M, Perez-Brocal V, Arslanoglu S, Aydemir O, Ozumut SS, Tekin N, et al. Human milk mycobiota composition: relationship with gestational age, delivery mode, birth weight. Benef Microbes. (2020) 11:151–62. doi: 10.3920/BM2019.0158
178. Hashizume T, Togawa A, Nochi T, Igarashi O, Kweon MN, Kiyono H, et al. Peyer's patches are required for intestinal immunoglobulin A responses to Salmonella spp. Infect Immun. (2008) 76:927–34. doi: 10.1128/IAI.01145-07
179. Kotani Y, Kunisawa J, Suzuki Y, Sato I, Saito T, Toba M, et al. Role of Lactobacillus pentosus Strain b240 and the Toll-like receptor 2 axis in Peyer's patch dendritic cell-mediated immunoglobulin A enhancement. PLoS ONE. (2014) 9:e91857. doi: 10.1371/journal.pone.0091857
180. M'Rabet L, Vos AP, Boehm G, Garssen J. Breast-feeding and its role in early development of the immune system in infants: consequences for health later in life. J Nutr. (2008) 138:1782s–90s. doi: 10.1093/jn/138.9.1782S
181. Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. (2013) 69:1–10. doi: 10.1016/j.phrs.2012.09.001
182. Damaceno QS, Souza JP, Nicoli JR, Paula RL, Assis GB, Figueiredo HC, et al. Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics Antimicrob Proteins. (2017) 9:371–9. doi: 10.1007/s12602-017-9270-1
183. Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, et al. Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers' milk and the infant gut: the child cohort study. Cell Host Microbe. (2020) 28:285–97.e4. doi: 10.1016/j.chom.2020.06.009
184. Malago JJ, Tooten PC, Koninkx JF. Anti-inflammatory properties of probiotic bacteria on Salmonella-induced IL-8 synthesis in enterocyte-like Caco-2 cells. Benef Microbes. (2010) 1:121–30. doi: 10.3920/BM2009.0021
185. Rabe H, Lundell AC, Sjoberg F, Ljung A, Strombeck A, Gio-Batta M, et al. Neonatal gut colonization by Bifidobacterium is associated with higher childhood cytokine responses. Gut Microbes. (2020) 12:1–14. doi: 10.1080/19490976.2020.1847628
186. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. (2010) 107:12204–9. doi: 10.1073/pnas.0909122107
187. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. (2018) 360:795–800. doi: 10.1126/science.aaq0926
188. O'Sullivan, He X, McNiven EM, Haggarty NW, Lonnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res. (2013) 12:2833–45. doi: 10.1021/pr4001702
189. Ardeshir, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. (2014) 6:252ra120. doi: 10.1126/scitranslmed.3008791
190. Narayan NR, Mendez-Lagares G, Ardeshir A, Lu D, Van Rompay KK, Hartigan-O'Connor DJ. Persistent effects of early infant diet and associated microbiota on the juvenile immune system. Gut Microbes. (2015) 6:284–9. doi: 10.1080/19490976.2015.1067743
191. Miklavcic JJ, Badger TM, Bowlin AK, Matazel KS, Cleves MA, LeRoith T, et al. Human breast-milk feeding enhances the humoral and cell-mediated immune response in neonatal piglets. J Nutr. (2018) 148:1860–70. doi: 10.1093/jn/nxy170
192. Brink LR, Matazel K, Piccolo BD, Bowlin AK, Chintapalli SV, Shankar K, et al. Neonatal diet impacts bioregional microbiota composition in piglets fed human breast milk or infant formula. J Nutr. (2019) 149:2236–46. doi: 10.1093/jn/nxz170
193. Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. (2000) 30:61–7. doi: 10.1097/00005176-200001000-00019
194. Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. (2014) 87:280–90. doi: 10.1111/1574-6941.12223
195. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. (2013) 185:385–94. doi: 10.1503/cmaj.121189
196. Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE. (2012) 7:e44595. doi: 10.1371/journal.pone.0044595
197. Borewicz K, Suarez-Diez M, Hechler C, Beijers R, de Weerth C, Arts I, et al. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci Rep. (2019) 9:2434. doi: 10.1038/s41598-018-38268-x
198. Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, et al. Canadian healthy infant longitudinal development study, association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. (2018) 172:e181161. doi: 10.1001/jamapediatrics.2018.1161
199. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. (2016) 8:343ra82. doi: 10.1126/scitranslmed.aad7121
200. Andersson Y, Hammarstrom ML, Lonnerdal B, Graverholt G, Falt H, Hernell O. Formula feeding skews immune cell composition toward adaptive immunity compared to breastfeeding. J Immunol. (2009) 183:4322–8. doi: 10.4049/jimmunol.0900829
201. O'Reilly D, Dorodnykh D, Avdeenko NV, Nekliudov NA, Garssen J, Elolimy AA, et al. Perspective: the role of human breast-milk extracellular vesicles in child health and disease. Adv Nutr. (2020). doi: 10.1093/advances/nmaa094. [Epub ahead of print].
202. Trams EG, Lauter CJ, Salem N Jr, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. (1981) 645:63–70. doi: 10.1016/0005-2736(81)90512-5
203. Bobrie, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. (2012) 1:18397. doi: 10.3402/jev.v1i0.18397
204. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. (2015) 8:83. doi: 10.1186/s13045-015-0181-x
205. Taylor DD, Taylor CG, Jiang CG, Black PH. Characterization of plasma membrane shedding from murine melanoma cells. Int J Cancer. (1988) 41:629–35. doi: 10.1002/ijc.2910410425
206. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. (2015) 25:364–72. doi: 10.1016/j.tcb.2015.01.004
207. Singh PP, Li L, Schorey JS. Exosomal RNA from Mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic. (2015) 16:555–71. doi: 10.1111/tra.12278
208. Lo Cicero, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. (2015) 35:69–77. doi: 10.1016/j.ceb.2015.04.013
209. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. (2016) 126:1208–15. doi: 10.1172/JCI81135
210. Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9
211. Gutierrez-Vazquez, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol Rev. (2013) 251:125–42. doi: 10.1111/imr.12013
212. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. (2012) 1820:940–8. doi: 10.1016/j.bbagen.2012.03.017
213. Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. (2010) 117:1–4. doi: 10.1007/s00702-009-0288-8
214. Guescini M, Guidolin D, Vallorani L, Casadei L, Gioacchini AM, Tibollo P, et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp Cell Res. (2010) 316:1977–84. doi: 10.1016/j.yexcr.2010.04.006
215. Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci USA. (2017) 114:E9066–75. doi: 10.1073/pnas.1704862114
216. Spencer N, Yeruva L. Role of bacterial infections in extracellular vesicles release and impact on immune response. Biomed J. (2020). doi: 10.1016/j.bj.2020.05.006. [Epub ahead of print].
217. Cui J, Zhou B, Ross SA, Zempleni J. Nutrition, microRNAs, human health. Adv Nutr. (2017) 8:105–12. doi: 10.3945/an.116.013839
218. Golan-Gerstl R, Elbaum Shiff Y, Moshayoff V, Schecter D, Leshkowitz D, Reif S. Characterization and biological function of milk-derived miRNAs. Mol Nutr Food Res. (2017) 61:9. doi: 10.1002/mnfr.201700009
219. Zempleni J, Aguilar-Lozano A, Sadri M, Sukreet S, Manca S, Wu D, et al. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J Nutr. (2017) 147:3–10. doi: 10.3945/jn.116.238949
220. Liao Y, Du X, Li J, Lonnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. (2017) 61: doi: 10.1002/mnfr.201700082
221. Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. (2010) 1:7. doi: 10.1186/1758-907X-1-7
222. Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. (2010) 20:1128–37. doi: 10.1038/cr.2010.80
223. Benmoussa, Gotti C, Bourassa S, Gilbert C, Provost P. Identification of protein markers for extracellular vesicle (EV) subsets in cow's milk. J Proteomics. (2019) 192:78–88. doi: 10.1016/j.jprot.2018.08.010
224. Benmoussa, Ly S, Shan ST, Laugier J, Boilard E, Gilbert C, et al. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow's milk. J Extracell Vesicles. (2017) 6:1401897. doi: 10.1080/20013078.2017.1401897
225. Lasser, Shelke GV, Yeri A, Kim DK, Crescitelli R, Raimondo S, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. (2017) 14:58–72. doi: 10.1080/15476286.2016.1249092
226. van Herwijnen MJC, Driedonks TAP, Snoek BL, Kroon AMT, Kleinjan M, Jorritsma R, et al. Abundantly present miRNAs in milk-derived extracellular vesicles are conserved between mammals. Front Nutr. (2018) 5:81. doi: 10.3389/fnut.2018.00081
227. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. (2012) 8:118–23. doi: 10.7150/ijbs.8.118
228. Alsaweed M, Hepworth AR, Lefevre C, Hartmann PE, Geddes DT, Hassiotou F. Human milk MicroRNA and total RNA differ depending on milk fractionation. J Cell Biochem. (2015) 116:2397–407. doi: 10.1002/jcb.25207
229. Hill AF, Pegtel DM, Lambertz U, Leonardi T, O'Driscoll L, Pluchino S, et al. Nolte-'t Hoen, ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles. (2013). doi: 10.3402/jev.v2i0.22859. [Epub ahead of print].
230. Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles. (2014) 3:26913. doi: 10.3402/jev.v3.26913
231. Vaswani K, Koh YQ, Almughlliq FB, Peiris HN, Mitchell MD. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod Biol. (2017) 17:341–8. doi: 10.1016/j.repbio.2017.09.007
232. Kahn S, Liao Y, Du X, Xu W, Li J, Lonnerdal B. Exosomal microRNAs in milk from mothers delivering preterm infants survive in vitro digestion and are taken up by human intestinal cells. Mol Nutr Food Res. (2018) 62:e1701050. doi: 10.1002/mnfr.201701050
233. Shandilya S, Rani P, Onteru SK, Singh D. Small interfering RNA in milk exosomes is resistant to digestion and crosses the intestinal barrier in vitro. J Agric Food Chem. (2017) 65:9506–13. doi: 10.1021/acs.jafc.7b03123
234. Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, mouse livers. J Nutr. (2014) 144:1495–500. doi: 10.3945/jn.114.196436
235. Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. (2012) 95:4831–41. doi: 10.3168/jds.2012-5489
236. Maburutse BE, Park MR, Oh S, Kim Y. Evaluation and characterization of milk-derived microvescicle isolated from bovine colostrum. Korean J Food Sci Anim Resour. (2017) 37:654–62. doi: 10.5851/kosfa.2017.37.5.654
237. Cheng AA, Li W, Hernandez LL. Effect of high-fat diet feeding and associated transcriptome changes in the peak lactation mammary gland in C57BL/6 dams. Physiol Genomics. (2018) 50:1059–70. doi: 10.1152/physiolgenomics.00052.2018
238. Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol. (2010) 185:7244–51. doi: 10.4049/jimmunol.1001573
239. Wells AC, Daniels KA, Angelou CC, Fagerberg E, Burnside AS, Markstein M, et al. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife. (2017) 6:e26398. doi: 10.7554/eLife.26398
240. Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. (2013) 190:6542–9. doi: 10.4049/jimmunol.1202496
241. Peck BC, Sincavage J, Feinstein S, Mah AT, Simmons JG, Lund PK, et al. miR-30 family controls proliferation and differentiation of intestinal epithelial cell models by directing a broad gene expression program that includes SOX9 and the ubiquitin ligase pathway. J Biol Chem. (2016) 291:15975–84. doi: 10.1074/jbc.M116.733733
242. Wu Y, Sun Q, Dai L. Immune regulation of miR-30 on the Mycobacterium tuberculosis-induced TLR/MyD88 signaling pathway in THP-1 cells. Exp Ther Med. (2017) 14:3299–303. doi: 10.3892/etm.2017.4872
243. Lin X, Yu S, Ren P, Sun X, Jin M. Human microRNA-30 inhibits influenza virus infection by suppressing the expression of SOCS1, SOCS3, and NEDD4. Cell Microbiol. (2020) 22:e13150. doi: 10.1111/cmi.13150
244. Naqvi AR, Fordham JB, Ganesh B, Nares S. miR-24, miR-30b and miR-142-3p interfere with antigen processing and presentation by primary macrophages and dendritic cells. Sci Rep. (2016) 6:32925. doi: 10.1038/srep32925
245. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. (2004) 303:83–6. doi: 10.1126/science.1091903
246. Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. (2007) 129:147–61. doi: 10.1016/j.cell.2007.03.008
247. Corsetti PP, de Almeida LA, Goncalves ANA, Gomes MTR, Guimaraes ES, Marques JT, et al. miR-181a-5p Regulates TNF-alpha and miR-21a-5p influences gualynate-binding protein 5 and IL-10 expression in macrophages affecting host control of Brucella abortus infection. Front Immunol. (2018) 9:1331. doi: 10.3389/fimmu.2018.01331
248. Xie MY, Hou LJ, Sun JJ, Zeng B, Xi QY, Luo JY, et al. Porcine milk exosome MiRNAs attenuate LPS-induced apoptosis through inhibiting TLR4/NF-kappaB and p53 pathways in intestinal epithelial cells. J Agric Food Chem. (2019) 67:9477–91. doi: 10.1021/acs.jafc.9b02925
249. Gantier MP, Tong S, Behlke MA, Xu D, Phipps S, Foster PS, et al. TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. (2008) 180:2117–24. doi: 10.4049/jimmunol.180.4.2117
250. Lee EK, Chung KW, Kim YR, Ha S, Kim SD, Kim DH, et al. Small RNAs induce the activation of the pro-inflammatory TLR7 signaling pathway in aged rat kidney. Aging Cell. (2017) 16:1026–34. doi: 10.1111/acel.12629
251. Yelamanchili SV, Lamberty BG, Rennard DA, Morsey BM, Hochfelder CG, Meays BM, et al. Correction: MiR-21 in extracellular vesicles leads to neurotoxicity via TLR7 signaling in SIV neurological disease. PLoS Pathog. (2015) 11:e1005131. doi: 10.1371/journal.ppat.1005131
252. Kim SJ, Chen Z, Essani AB, Elshabrawy HA, Volin MV, Volkov S, et al. Identification of a novel toll-like receptor 7 endogenous ligand in rheumatoid arthritis synovial fluid that can provoke arthritic joint inflammation. Arthritis Rheumatol. (2016) 68:1099–110. doi: 10.1002/art.39544
253. Zhou F, Paz HA, Sadri M, Cui J, Kachman SD, Fernando SC, et al. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am J Physiol Gastrointest Liver Physiol. (2019) 317:G618–24. doi: 10.1152/ajpgi.00160.2019
254. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. (2016) 19:32–43. doi: 10.1016/j.chom.2015.12.005
Keywords: human milk, immunity, infants, neonates, development, breastmilk, immune system, gut
Citation: Carr LE, Virmani MD, Rosa F, Munblit D, Matazel KS, Elolimy AA and Yeruva L (2021) Role of Human Milk Bioactives on Infants' Gut and Immune Health. Front. Immunol. 12:604080. doi: 10.3389/fimmu.2021.604080
Received: 08 September 2020; Accepted: 22 January 2021;
Published: 12 February 2021.
Edited by:
Francisco José Pérez-Cano, University of Barcelona, SpainReviewed by:
Markus Xie, Genentech, United StatesMaciej Chichlowski, Mead Johnson Nutrition Institute, United States
Copyright © 2021 Carr, Virmani, Rosa, Munblit, Matazel, Elolimy and Yeruva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laxmi Yeruva, dmx5ZXJ1dmFAdWFtcy5lZHU=; Laura E. Carr, bGVjYXJyQHVhbXMuZWR1
 Laura E. Carr
Laura E. Carr Misty D. Virmani
Misty D. Virmani Fernanda Rosa
Fernanda Rosa Daniel Munblit
Daniel Munblit Katelin S. Matazel
Katelin S. Matazel Ahmed A. Elolimy
Ahmed A. Elolimy Laxmi Yeruva
Laxmi Yeruva