- 1Small Molecule Drugs Sichuan Key Laboratory, Institute of Materia Medica, Chengdu Medical College, Chengdu, China
- 2Non-coding RNA and Drug Discovery Laboratory, Chengdu Medical College, Chengdu, China
- 3Clinical Laboratory, The First Affiliated Hospital, Chengdu Medical College, Chengdu, China
- 4Institute of Antibiotics, The Fifth Affiliated Hospital, Wenzhou Medical University, Lishui, China
Carbapenem-resistant Acinetobacter baumannii (CRAB) presents a serious therapeutic and infection control challenge. In this study, we investigated the epidemiological and molecular differences of CRAB and the threatening factors for contributing to increased CRAB infections at a hospital in western China. A total of 110 clinical isolates of A. baumannii, collected in a recent 2-year period, were tested for carbapenem antibiotic susceptibility, followed by a molecular analysis of carbapenemase genes. Genetic relatedness of the isolates was characterized by multilocus sequence typing. Sixty-seven of the 110 isolates (60.9%) were resistant to carbapenems, 80.60% (54/67) of which carried the blaOXA-23 gene. Most of these CRAB isolates (77.62%) were classified as clone complex 92 (CC92), and sequence type (ST) 92 was the most prevalent STs, followed by ST195, ST136, ST843, and ST75. One CRAB isolate of ST195 harbored plasmid pAB52 from a Chinese patient without travel history. This plasmid contains toxin–antitoxin elements related to adaptation for growth, which might have emerged as a common vehicle indirectly mediating the spread of OXA-23 in CRAB. Thus, CC92 A. baumannii carrying OXA-23 is a major drug-resistant strain spreading in China. Our findings indicate that rational application of antibiotics is indispensable for minimizing widespread of drug resistance.
Introduction
Acintobacter baumannii is an opportunistic Gram-negative pathogen, which has recently been successfully spreading worldwide nosocomial infections and causing outbreaks of hospital-acquired infections, primarily due to its prominent ability to acquire antibiotic resistance (Peleg et al., 2008; Kempf and Rolain, 2012). A. baumannii is developing into multidrug resistant (MDR), extensively drug resistant (XDR), and pandrug resistant (PDR) bacteria, and its adaptation to the environment with drug resistance has previously been reported (Durante-Mangoni and Zarrilli, 2011). Carbapenems were regarded as the most powerful antibiotics because of its extremely effective antibacterial activity and low toxicity, but the emergence of carbapenem resistance in A. baumannii has become a global concern recently (Peleg et al., 2008; Tiwari et al., 2012a,b). Resistance of A. baumannii to expanded-spectrum cephalosporins and carbapenems is rapidly growing over the years (Bradford, 2001; Peleg et al., 2008). For example, surveillance by the CHINET project in China revealed that the rate of resistance of A. baumannii to the carbapenem, such as imipenem, doubled from 30.1% in 2006 to 62.8% in 2013 (Wang, 2008; Fupin et al., 2014).
Various types of β-lactam antibiotics, for example carbapenems, contain β-lactam rings in their structures and can be inactivated by β-lactamase enzymes. The β-lactamases are classified into four different molecular groups, Ambler classes A through D, according to amino acid sequence identities (Bush et al., 1995). Class A, C, and D (OXA enzymes) of β-lactamases contain a catalytically active serine residue that cleaves the lactam ring of antibiotics (Bush et al., 1995; Tiwari and Moganty, 2014). Class B of β-lactamases is a metallo-enzyme that requires zinc for their catalytic activity, and therefore, has a completely different mechanism for enzyme activity (Tiwari and Moganty, 2013). Carbapenems, such as imipenem and meropenem, have an exceedingly broad spectrum of activity and are able to resist hydrolysis by most of the β-lactamases, including extended-spectrum and derepressed class C chromosomal AmpC β-lactamases (Bush et al., 1995). However, most of the metallo-β-lactamases and a number of class A and D β-lactamases are able to hydrolyse broad spectrum carbapenems, such as imipenem and meropenem (Walther-Rasmussen and Hoiby, 2006; Evans and Amyes, 2014). Thus, outbreaks of OXA-23-producing A. baumannii have been reported from various regions of the world (Carvalho et al., 2009). It is generally believed that OXA-23 is responsible for carbapenem antibiotic resistance. Previously, Liu et al. (2015) reported the dissemination of MDR OXA-23-producing A. baumannii clones throughout multiple cities in China, but little is known about the molecular mechanisms of resistance to carbapenems in western China.
The multilocus sequence typing (MLST) has been widely applied in genotyping of bacteria, including A. baumannii (Bartual et al., 2005). Molecular epidemiological research of A. baumannii indicates that CC92 has played an important role in nosocomial infection outbreak and spread nationwide (Runnegar et al., 2010).
This study aimed to report the dissemination of A. baumannii harboring carbapenemase genes throughout a university hospital in western China, then identify the risk factors for carbapenem-resistant Acinetobacter baumannii (CRAB) infections, and finally perform a comprehensive evaluation and comparison of their genetic diversity.
Materials and Methods
Bacteria Isolates
A total of 110 consecutive and non-duplicated A. baumannii clinical isolates were collected from different departments [intensive care unit (ICU), gastroenterology, respiratory, neurosurgery and other wards] at the First Affiliated Hospital of Chengdu Medical College, Chengdu, Sichuan, China from 2012 to 2013. Isolates were identified by standard laboratory methods and ATB New (bioMérieux, France). A. baumannii was further verified when two PCR products were yielded as reported: a 425-bp internal control amplicon corresponding to the recA gene of Acinetobacter spp. and the 208-bp fragment of the 16S rRNA (Chiang et al., 2011) intergenic spacer region of A. baumannii (Table 1). All strains were stored at –80°C, and bacteria were grown on tryptose agar or Mueller–Hinton broth or agar (Oxoid, England).
Minimal Inhibitory Concentration (MIC)
The minimal inhibitory concentration (MIC) of carbapenems including imipenem and meropenem for A. baumannii were determined by the agar dilution method as previously described in the guidelines from the Clinical and Laboratory Standards Institute (CLSI, 2014). Escherichia coli ATCC25922 and A. baumannii ATCC19606 were used as quality control strain. The results were interpreted according to the CLSI guidelines (CLSI, 2014), i.e., CRAB was defined as an A. baumannii isolate that was resistant to both imipenem and meropenem (i.e., ≥8 μg/ml as resistant), whereas carbapenem-susceptible A. baumannii (CSAB) possessed carbapenem MIC of ≤2 μg/ml and carbapenem-intermediate A. baumannii (CIAB) has MIC of 4 μg/ml).
PCR Experiments
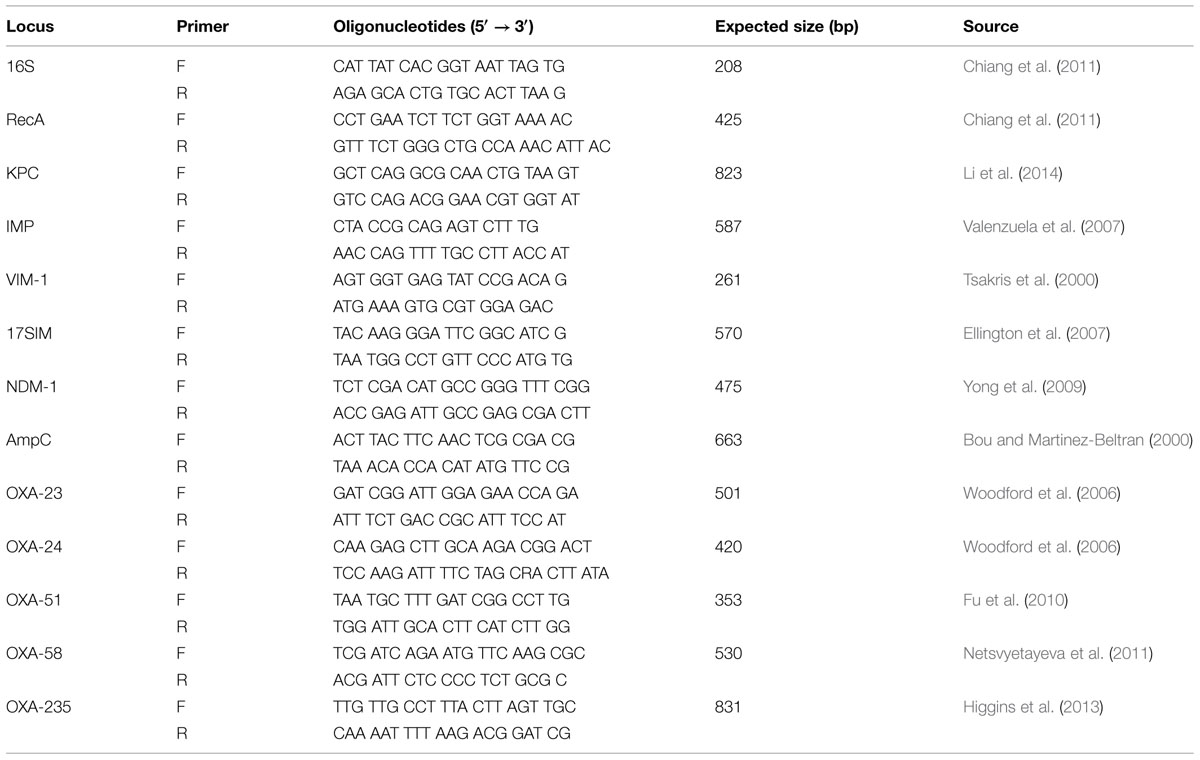
The genes encoding carbapenemases class A [e.g., Klebsiella pneumoniae carbapenemase gene, blaKPC (Li et al., 2014)], class B [e.g., the metallo-β-lactamases, blaIMP (Valenzuela et al., 2007), blaV IM-1 (Tsakris et al., 2000), blaSIM (Ellington et al., 2007), and blaNDM-1 (Yong et al., 2009)], class C [e.g., blaAmpC (Bou and Martinez-Beltran, 2000)], and class D [e.g., blaOXA-23 (Woodford et al., 2006), blaOXA-24 (Woodford et al., 2006), blaOXA-51 (Fu et al., 2010), blaOXA-58 (Netsvyetayeva et al., 2011), and blaOXA-235 (Higgins et al., 2013)] were investigated by polymerase chain reaction (PCR).
To prepare DNA templates, each isolate was grown on tryptose agar (Oxoid, England) plate overnight. Two to three colonies were suspended in 100 μl of sterile distilled water in a 1.5-ml Eppendorf tube, and the suspension was heated at 100°C for 15 min, followed by centrifugation at 12,000 g for 10 min to pellet the debris. The resultant supernatant was used as the DNA template in the PCRs, which were carried out in a 50-μl volume containing 0.2 mM each deoxynucleotide, 0.5 μM each primer, 1.25 U of Taq polymerase, and 5 μl of 10× buffer (Thermo, China). Primers (Table 1) were synthesized by Sangon Company (Sangon, China). Reaction conditions of PCR were 94°C for 5 min and 30 cycles of 94°C for 30 s, 55°C for 30 s (57°C for blaAmpC amplification), and 72°C for 30 s, followed by a final extension at 72°C for 5 min.
Multilocus Sequence Typing
MLST was used to describe the genetic backgrounds of CRAB and CSAB in all clinical isolates (Diancourt et al., 2010; Hamouda et al., 2010; Pournaras et al., 2014). eBURST was performed to cluster sequence types (STs) into clonal complexes (CCs) and infer evolutionary descent. MLST was carried out as described by Bartual et al. (2005) for all isolates. In brief, internal fragments of seven housekeeping genes, gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD, were amplified, purified, and sequenced (Fu et al., 2010; Ruan et al., 2013). And eBURST (version 3, http://eburst.mlst.net/) was used to assign STs to CCs and define the genetic relatedness of STs with the most stringent definition of the groups by sharing the same alleles at ≥6 of 7 loci (Feil et al., 2004).
Plasmid Conjugation, Extraction, Sequencing, and Analysis
Plasmid conjugations were performed by using meropenem-resistant A. baumannii as donors and an azide-resistant E. coli J53 as the recipient (MIC > 200 μg/ml; Bogaerts et al., 2006). Matings were performed as described previously (Wang et al., 2003), and transconjugants were recovered on L-agar plates containing meropenem (8 μg/ml) and azide (150 μg/ml).
Plasmid DNA was extracted using a Plasmid Midi Kit (Omega, USA) according to the manufacturer’s instructions. The plasmid was cut by EcoRI, then the fragments were ligated to the vector pET21a. The complete sequencing work was performed by the ABI-3730 XL system (Biosune Biotechnology Company, Shanghai, China) and sequences were assembled by Life Technologies (BigDye V3.1, four fluorescence reagent kit). The sequenced plasmid was annotated by the RAST server (Aziz et al., 2008), then all of the predicted proteins were further compared against the NCBI non-redundant protein database using the BLASTP program. In addition, the DNAMAN software (Lynnon Corporation, USA) was used to generate a circular map of plasmid pAB52.
Nucleotide Sequence Accession Numbers
The complete nucleotide sequences (Supplementary Table S1) of plasmid pAB52 in this study were submitted to GenBank with assigned accession number KR030046.
Results
Characterization of Clinical Isolates
According to our previous study, 67 of the total 110 A. baumannii isolates exhibited resistance to carbapenems, imipenem and meropenem, with MIC values of ≥8 μg/ml. These CRAB isolates were from different types of specimens including sputum (n = 49), throat swabs (n = 5), blood (n = 1), douche (n = 4), and others (n = 8). Epidemiological analysis of the 67 patients with CRAB revealed that 27 were ≥70 years old, 15 were between 60 and 70 years old, 24 were between 21 and 60 years old, and one was ≤20 years old. Among 40 males (59.7%) and 27 females (40.3%), 24 isolates were collected from the ICU, and 17 isolates were collected from the respiratory department. Clinical characteristics of the 67 patients with CRAB are summarized in Table 2.
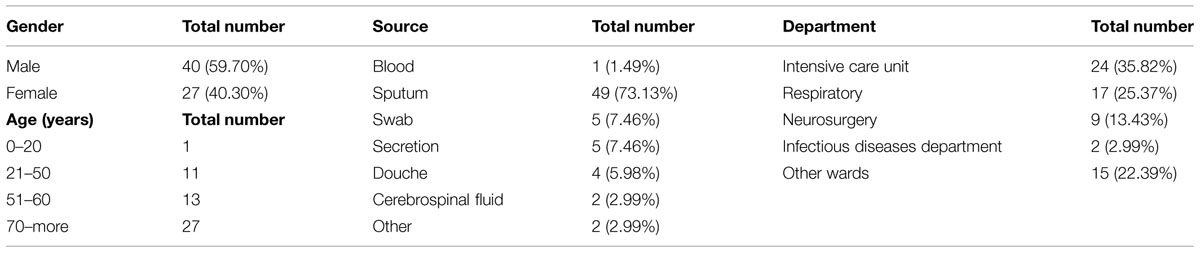
TABLE 2. Clinical characteristics of 67 patients with Carbapenem-resistant Acinetobacter baumannii (CRAB).
Expression of Carbapenemase Genes in A. baumannii Isolates
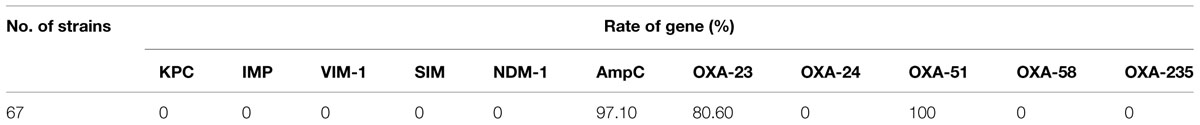
All isolates were screened for the presence of β-lactamases genes (Table 3). In the 67 CRAB isolates, all of them were positive to recA, 16S rRNA conservative region and blaOXA-51. The major carbapenemase gene blaOXA-23 was detected in 80.60% of all isolates. In addition, 65 isolates (97.10%) were found to carry blaAmpC. Other β–lactamase genes including blaKPC, blaIMP, blaV IM-1, blaSIM, blaNDM-1, blaOXA-24, blaOXA-58, and blaOXA-235 were undetectable in all isolates. Thus, the expression of blaOXA-23 is still the dominant carbapenem resistance mechanism in A. baumannii among the isolates investigated aside from the inherent blaOXA-51 gene. One susceptible isolate carried blaOXA-72.
Genetic Analysis of STs
To investigate whether the isolates were genetically related, MLST was performed to characterize the CRAB, CIAB and CSAB (Figure 1). Each ST was represented by a dot that was proportionally sized to the number of A. baumannii containing that ST. The MLST analysis revealed a total of 32 different STs in 67 CRAB, including 6 existing STs and 26 novel STs. 77.62% of the 67 CRAB isolates were classified to CC92, ST92 and its 4 novel single-locus variants (SLVs), ST195, ST136, ST843, and ST75, were the predominant STs, found in 34.33, 7.46, 5.97, 4.48, and 2.99% of isolates, respectively. Among these five predominant STs, 82.35% of carbapenem-resistant isolates carried the blaOXA-23 gene, in which ST92 is one of the most widespread STs across the world and belongs to the CC92 in A. baumannii MLST databases (Wang et al., 2013). Seventeen of the 32 CRAB STs belong to CC92 (Figure 1A). In this study, the number of polymorphic sites in gpi (n = 8), rpoD (n = 4), cpn60 (n = 3), recA (n = 2), and the other three loci (gdhB, gyrB, and gltA) indicated that all were the same in CC92. Seven of the eight CIAB were CC92 (Figure 1B), but only 3 of the 35 CSAB were CC92 (Figure 1C). All these suggested that CC92 represents the most widely distributed carbapenem-resistant CC identified in the hospital.
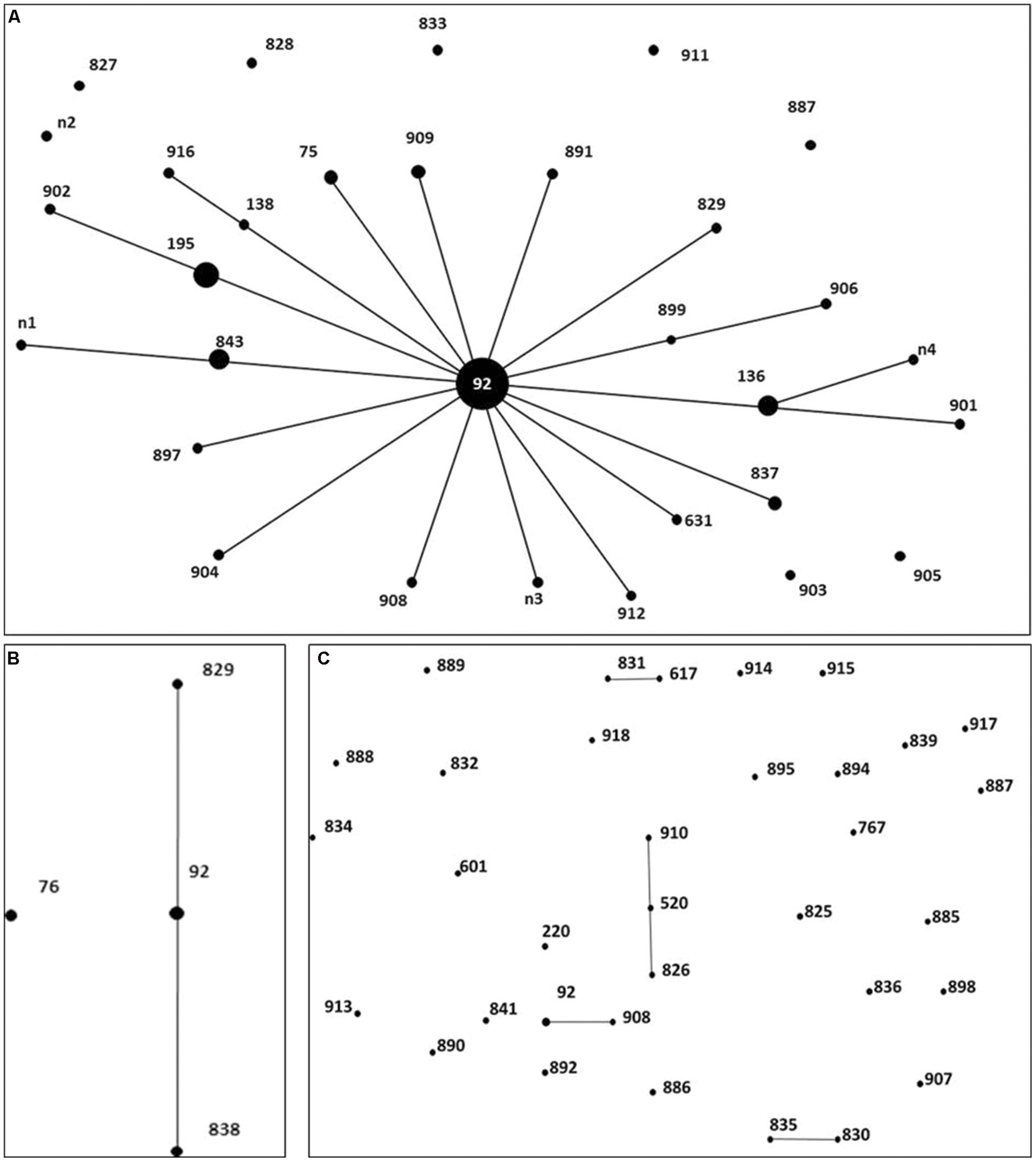
FIGURE 1. Distribution of genes in 67 Carbapenem-resistant Acinetobacter baumannii (CRAB), 8 carbapenem-intermediate A. baumannii (CIAB), and 35 carbapenem-susceptible A. baumannii (CSAB). Minimum spanning tree analysis by eBURST algorithm of CRAB (A), CIAB (B) and CSAB (C) isolates based on MLST data. Each circle represents a specific sequence type (ST). The size of each circle homologizes to a different number of isolates, with larger sizes representing higher frequency of occurrence. The solid lines connecting the circles indicate the relationship between different STs.
Plasmid Analysis
To detect the nosocomial infection mediated by A. baumannii plasmid DNA, a plasmid conjugation assay was performed. However, none of the carbapenem-resistant isolates transferred plasmids from A. baumannii isolates to E. coli J53 successfully. This could be due to multiple reasons. Carbapenem resistance genes were located in the genome, and plasmids were not conjugative or unable to replicate in the E. coli host. Of note, with no. 52 CRAB isolate (which harbored pAB52 plasmid) as the donor strain, a few colonies with azide resistance and displaying a similar morphology as the E. coli J53 recipient were obtained from the conjugation. Yet, these colonies were found to be arisen from A. baumannii based on the amplification and sequence analysis of 16S rRNA and seven housekeeping genes gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD. Likely, the generation of azide-resistant A. baumannii was a result of spontaneous mutation(s). In addition, plasmid extraction assay confirmed no plasmid-containing E. coli J53 colonies, indicating that pAB52 could not be transferred from A. baumannii to E. coli J53.
RAST analysis showed that the plasmid pAB52 was 8,893 bp in size and contained 10 open reading frames (ORFs; Figure 2A), with an average G+C content of 34.5%. Three genes with a putative role in virulence were detected in pAB52: a septicolysin-like gene coding for a pore-forming toxin (Rosado et al., 2008; Gan et al., 2012), a TonB-dependent receptor gene coding for an outer membrane protein involved in iron uptake and virulence (Acosta et al., 2011; Gan et al., 2012), and a toxin–antitoxin (TA) gene coding for is a functional TA system and that its toxin, SplT (Gan et al., 2012; Jurenaite et al., 2013), inhibits translation (Figure 2A). One ORF (orf 9) codes for proteins that share similarity to hypothetical proteins encoded by plasmid genes found in other bacteria (Gan et al., 2012), while the predicted products of three others (orf 1, orf 2, and orf 3) do not match any known sequences. orf 9, which is adjacent to the replication initiation protein, is similar to replication protein of other genomes (Figure 2B). Therefore, it might contribute to a replication ability. Further analysis indicated that this plasmid shared most nucleotide identity with previously described isolates of A. baumannii, WM99C, TCDC-AB0715, AB210, AB0057, 1656-2, AYE, UMB001, and ATCC17978, Acinetobacter sp. 6013150 and 6013113 (Figure 2B).
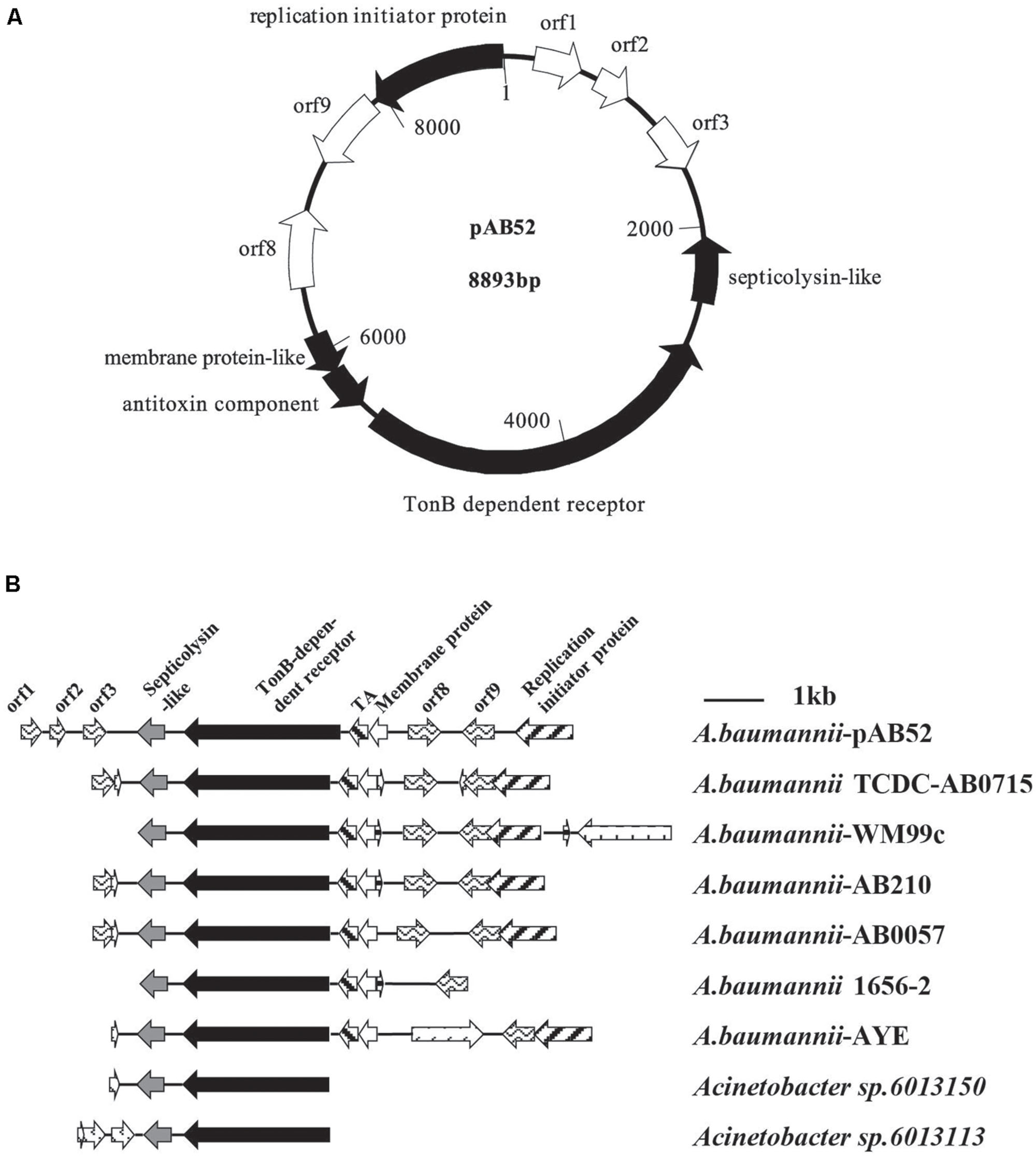
FIGURE 2. (A) Diagram of plasmid pAB52 (GenBank accession number KR03004). The open reading frames (ORFs), represented by arrows pointing in the direction of transcription, are white, except for that for the reported genes, which is black. (B) Comparative analysis of the genetic environment of the TonB-dependent receptor in plasmid DNA by RAST software. Regions with 100% nucleotide sequence identity or similar biological function are filled with the same pattern.
Discussion
Clinical characterization of 67 patients with CRAB indicated that compromised immunity (e.g., elderly patients suffering from respiratory failure, encephalorrhagia, extensive burns, and chronic obstructive pulmonary disease) increases the risk of A. baumannii infection. In addition, males appear to have a greater risk of infection than females.
Surveillance by the CHINET project demonstrated that the A. baumannii rate of imipenem resistance increased over twofold between 2006 and 2013 to 62.8% in China (Fupin et al., 2014). In contrast, the resistance rate of Acinetobacter spp. to cefoperazone/sulbactam was only 36.4% (Fupin et al., 2014), far below the rates of imipenem resistance. Mirroring the national trend, the A. baumannii resistance rate of imipenem and cefoperazone/sulbactamat in our hospital was 60.9 and 14.75%, respectively (Ying et al., 2013).
We observed that OXA-23 was also the major carbapenemase mechanism responsible for the resistance phenotype (Table 3), as the gene was expressed in most CRAB isolates. This result is consistent with previous reports in which the CRAB is often caused due to the expression of OXA enzymes, particularly OXA-23 (Fu et al., 2010; Merino et al., 2014; Zowawi et al., 2015).
In our experiments, we unexpectedly found one particular carbapenem-susceptible isolate, which belonged to ST220 and carried the blaOXA-72 gene, a member of the blaOXA-24 subgroup, but this isolate did not carry any other resistance genes. This is the first time that blaOXA-24 has been detected in western China. Of note, this isolate was not only susceptible to meropenem and imipenem, but also susceptible to other β-lactams, aminoglycosides, tetracyclines, fluoroquinolone, rifampicin, and trimethoprim–sulfamethoxazole (data not shown). Our results are in contrast with previous reports that nosocomial infections were triggered by CRAB-carrying plasmids encoding OXA-72 carbapenemase via plasmid DNA from Lithuanian (Povilonis et al., 2013) and Taiwan (Lu et al., 2009). There are several possible reasons for the different results. For instance, the resistance genes may not get expressed because of different or changed genetic environment. Further investigation is needed why the presence of blaOXA-72 does not produce β-lactam resistance.
MLST is an unambiguous and utility technique that identifies accurate and portable nucleotide sequences of internal fragments from multiple loci housekeeping genes and was used to assess the genetic background of populations in an ocean of bacteria (Maiden et al., 1998). Based on our existing epidemiological data and the eBURST arithmetic, it was shown that CC92 was widespread and this major clone was resistant to carbapenem antibiotics in our hospital. Ruan et al. (2013) explained that CC92 is a widespread variant that has advantages with respect to acquiring resistance determinants and surviving in the nosocomial environment, which renders it preferentially selected under antibiotic pressure. This may be the primary reason for the current epidemiological situation of CC92 in our study. In addition, 82.35% of CRAB from the five predominant STs in CC92 carried the blaOXA-23 gene. We propose that the blaOXA-23 gene is tied to the genetic background of CC92, representing an important setting in which to study the emergence of carbapenem resistance. A novel SLVs of CC92, ST843 was identified in three isolates, and sharing six same alleles with ST92 and n1 (Figure 1A). We could infer that it may be one of the evolutions of A. baumannii to adapt nosocomial environment.
We used eBURST to analyze the distribution of the STs of CSAB, and found it was more dispersive than the distribution of CRAB (Figure 1). This may confirm that CSAB has a more diverse genetic background compared to CRAB. The high genetic diversity in CSAB might be attributed to the decreased survival rate of antibiotic treatment. None of them emerge more than once among 35 carbapenem-susceptible isolates from May 2012 to October 2013 except for 3 isolates belonging to CC92. These data also imply that CSAB may have a more diverse genetic source compared to the CRAB, and suggest CC92 is an evolution for A. baumannii to survive while antibiotics existed.
We also observed that a few A. baumannii isolates could acquire azide resistance after the bacteria were mixed and incubated with E. coli J53 together overnight. This can be totally expected since spontaneous mutation(s) conferring azide resistance could arise and be selected in the presence of azide, similar to those in vitro selections of drug-resistant mutants. This can be considered as a process of adaptation to a selection pressure. Indeed, mutations conferring resistance to azide have been reported in both E. coli (Fortin et al., 1990) and Acinetobacter spp. (Elkeles et al., 1994).
The cryptic plasmid pAB52 from CRAB isolate no. 52 does not carry any carbapenem resistance genes. It matches best with pAC12 which was reported in 2012 in Malaysia (Gan et al., 2012), and the ST of this pAB52 CRAB is ST195 (1-3-3-2-2-96-3) in parallel with A. baumannii AC12. According to the results of pAB52 sequence blast in NCBI (date not shown), all plasmids carry the same genes with over 99.9% identity over the full length, and belong to the A. baumannii plasmid group (GenBank accession number CP008850, CP007535, CP007550, CP001183, CP007578, KJ586856, KJ477078, and CP006964). Thus, plasmid pAB52 is larger with an additional segment of TonB gene (Figure 2B), but these plasmids may have the same function. For example, the septicolysin-like protein encoded in these plasmids suggests that it may be involved in survival of bacteria in the lungs and blood (Acosta et al., 2011). The TA systems have been suggested to mediate bacterial persistence by generating slowly growing cells tolerant to antibiotics and environmental changes (Jurenaite et al., 2013). In particular, the pAB52-containing strain was isolated from the sputum of a 74-year-old male who lived in a rural district in western China and had not traveled to any areas before admission to the hospital’s ICU. This individual was found to have acquired an XDR A. baumannii infection during his hospital stay and eventually, he died from respiratory failure. The observation suggests that probably plasmid-borne A. baumannii TA systems contribute for the evolution of antibiotic resistance in this opportunistic pathogen.
In summary, this investigation confirms that the CC92 producing blaOXA-23 was the leading reason for the dramatic increase in the carbapenem resistance rates in China. Awareness of carbapenem-resistant organisms and their development in hospitals has crucial implications in optimizing infection control practices, establishing antimicrobial stewardship programs within the hospital, and finally establishing active regional surveillance systems (Zowawi et al., 2015).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81373454, 31300659, 31470246, and 31401099), Applied Basic Research Programs of Sichuan Province (grant 2013jy0065), Scientific Research and Innovation Team of Sichuan Province (grant 15TD0025), and Preeminent Youth Fund of Sichuan Province (grant 2015JQO019). We thank Drs. Gang Liu (Xiamen University) and Xian-Zhi Li (Health Canada, Ottawa) for insightful revision of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00910
References
Acosta, J., Merino, M., Viedma, E., Poza, M., Sanz, F., Otero, J. R., et al. (2011). Multidrug-resistant Acinetobacter baumannii Harboring OXA-24 carbapenemase, Spain. Emerg. Infect. Dis. 17, 1064–1067. doi: 10.3201/eid/1706.091866.
Aziz, R. K., Bartels, D., Best, A. A., Dejongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bartual, S. G., Seifert, H., Hippler, C., Luzon, M. A., Wisplinghoff, H., and Rodriguez-Valera, F. (2005). Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43, 4382–4390. doi: 10.1128/jcm.43.9.4382-4390.2005
Bogaerts, P., Naas, T., Wybo, I., Bauraing, C., Soetens, O., Pierard, D., et al. (2006). Outbreak of infection by carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-58 in Belgium. J. Clin. Microbiol. 44, 4189–4192. doi: 10.1128/jcm.00796-06
Bou, G., and Martinez-Beltran, J. (2000). Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44, 428–432. doi: 10.1128/AAC.44.2.428-432.2000
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/cmr.14.4.933-951.2001
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Carvalho, K. R., Carvalho-Assef, A. P., Peirano, G., Santos, L. C., Pereira, M. J., and Asensi, M. D. (2009). Dissemination of multidrug-resistant Acinetobacter baumannii genotypes carrying bla(OXA-23) collected from hospitals in Rio de Janeiro, Brazil. Int. J. Antimicrob. Agents 34, 25–28. doi: 10.1016/j.ijantimicag.2008.12.009
Chiang, M. C., Kuo, S. C., Chen, Y. C., Lee, Y. T., Chen, T. L., and Fung, C. P. (2011). Polymerase chain reaction assay for the detection of Acinetobacter baumannii in endotracheal aspirates from patients in the intensive care unit. J. Microbiol. Immunol. Infect. 44, 106–110. doi: 10.1016/j.jmii.2010.04.003
CLSI. (2014). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute.
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 5:e10034. doi: 10.1371/journal.pone.0010034
Durante-Mangoni, E., and Zarrilli, R. (2011). Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6, 407–422. doi: 10.2217/fmb.11.23
Elkeles, A., Rosenberg, E., and Ron, E. Z. (1994). Azide-resistant mutants in Acinetobacter calcoaceticus A2 are defective in protein secretion. FEMS Microbiol. Lett. 116, 221–224. doi: 10.1111/j.1574-6968.1994.tb06704.x
Ellington, M. J., Kistler, J., Livermore, D. M., and Woodford, N. (2007). Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob Chemother. 59, 321–322. doi: 10.1093/jac/dkl481
Evans, B. A., and Amyes, S. G. (2014). OXA beta-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/cmr.00117-113
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004
Fortin, Y., Phoenix, P., and Drapeau, G. R. (1990). Mutations conferring resistance to azide in Escherichia coli occur primarily in the secA gene. J. Bacteriol. 172, 6607–6610.
Fu, Y., Zhou, J., Zhou, H., Yang, Q., Wei, Z., Yu, Y., et al. (2010). Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65, 644–650. doi: 10.1093/jac/dkq027
Fupin, H., Demei, Z., Fu, W., Xiaofei, J., Ziyong, S., Zhongju, C., et al. (2014). CHINET 2013 surveillance of bacterial resistance in China. Chin. J. Infect. Chemother. 14, 365–374.
Gan, H. M., Lean, S. S., Suhaili, Z., Thong, K. L., and Yeo, C. C. (2012). Genome sequence of Acinetobacter baumannii AC12, a polymyxin-resistant strain isolated from Terengganu, Malaysia. J. Bacteriol. 194, 5979–5980. doi: 10.1128/jb.01466-1412
Hamouda, A., Evans, B. A., Towner, K. J., and Amyes, S. G. (2010). Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J. Clin. Microbiol. 48, 2476–2483. doi: 10.1128/jcm.02431-2439
Higgins, P. G., Perez-Llarena, F. J., Zander, E., Fernandez, A., Bou, G., and Seifert, H. (2013). OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 2121–2126. doi: 10.1128/aac.02413-2412
Jurenaite, M., Markuckas, A., and Suziedeliene, E. (2013). Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 195, 3165–3172. doi: 10.1128/JB.00237-213
Kempf, M., and Rolain, J. M. (2012). Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int. J. Antimicrob. Agents 39, 105–114. doi: 10.1016/j.ijantimicag.2011.10.004
Li, Y., Guo, Q., Wang, P., Zhu, D., Ye, X., Wu, S., et al. (2014). Clonal dissemination of extensively drug-resistant Acinetobacter baumannii producing an OXA-23 beta-lactamase at a teaching hospital in Shanghai, China. J. Microbiol. Immunol. Infect. 48, 101–108. doi: 10.1016/j.jmii.2014.04.005
Liu, L. L., Ji, S. J., Ruan, Z., Fu, Y., Fu, Y. Q., Wang, Y. F., et al. (2015). Dissemination of blaOXA-23 in Acinetobacter spp. in China: main roles of conjugative plasmid pAZJ221 and Transposon Tn2009. Antimicrob. Agents Chemother. 59, 1998–2005. doi: 10.1128/AAC.04574-4514
Lu, P. L., Doumith, M., Livermore, D. M., Chen, T. P., and Woodford, N. (2009). Diversity of carbapenem resistance mechanisms in Acinetobacter baumannii from a Taiwan hospital: spread of plasmid-borne OXA-72 carbapenemase. J. Antimicrob. Chemother. 63, 641–647. doi: 10.1093/jac/dkn553
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Merino, M., Poza, M., Roca, I., Barba, M. J., Sousa, M. D., Vila, J., et al. (2014). Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb. Drug Resist. 20, 259–263. doi: 10.1089/mdr.2013.0127
Netsvyetayeva, I., Sikora, M., Golas, M., Swoboda-Kopec, E., De Walthoffen, S.W., Dembicka, O., et al. (2011). Acinetobacter baumannii multidrug-resistant strain occurrence in liver recipients with reference to other high-risk groups. Transplant Proc. 43, 3116–3120. doi: 10.1016/j.transproceed.2011.07.008
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/cmr.00058-57
Pournaras, S., Gogou, V., Giannouli, M., Dimitroulia, E., Dafopoulou, K., Tsakris, A., et al. (2014). Single-locus-sequence-based typing of blaOXA-51-like genes for rapid assignment of Acinetobacter baumannii clinical isolates to international clonal lineages. J. Clin. Microbiol. 52, 1653–1657. doi: 10.1128/jcm.03565-3513
Povilonis, J., Seputiene, V., Krasauskas, R., Juskaite, R., Miskinyte, M., Suziedelis, K., et al. (2013). Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68, 1000–1006. doi: 10.1093/jac/dks499
Rosado, C. J., Kondos, S., Bull, T. E., Kuiper, M. J., Law, R. H., Buckle, A. M., et al. (2008). The MACPF/CDC family of pore-forming toxins. Cell Microbiol. 10, 1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x
Ruan, Z., Chen, Y., Jiang, Y., Zhou, H., Zhou, Z., Fu, Y., et al. (2013). Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int. J. Antimicrob. Agents 42, 322–328. doi: 10.1016/j.ijantimicag.2013.06.019
Runnegar, N., Sidjabat, H., Goh, H. M., Nimmo, G. R., Schembri, M. A., and Paterson, D. L. (2010). Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a single institution over a 10-year period. J. Clin. Microbiol. 48, 4051–4056. doi: 10.1128/jcm.01208-1210
Tiwari, V., Kapil, A., and Moganty, R. R. (2012a). Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb. Pathog. 53, 81–86. doi: 10.1016/j.micpath.2012.05.004
Tiwari, V., Vashistt, J., Kapil, A., and Moganty, R. R. (2012b). Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS ONE 7:e39451. doi: 10.1371/journal.pone.0039451
Tiwari, V., and Moganty, R. R. (2013). Structural studies on New Delhi Metallo-beta-lactamase (NDM-2) suggest old beta-lactam, penicillin to be better antibiotic for NDM-2-harbouring Acinetobacter baumanni. J. Biomol. Struct. Dyn. 31, 591–601. doi: 10.1080/07391102.2012.706075
Tiwari, V., and Moganty, R. R. (2014). Conformational stability of OXA-51 beta-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 32, 1406–1420. doi: 10.1080/07391102.2013.819789
Tsakris, A., Pournaras, S., Woodford, N., Palepou, M.-F. I., Babini, G. S., Douboyas, J., et al. (2000). Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38, 1290–1292.
Valenzuela, J. K., Thomas, L., Partridge, S. R., Van Der Reijden, T., Dijkshoorn, L., and Iredell, J. (2007). Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45, 453–460. doi: 10.1128/jcm.01971-1976
Walther-Rasmussen, J., and Hoiby, N. (2006). OXA-type carbapenemases. J. Antimicrob. Chemother. 57, 373–383. doi: 10.1093/jac/dki482
Wang, F. (2008). CHINET 2006 surveillance of bacterial resistance in China. Chin. J. Infect. Chemother. 8, 1–9.
Wang, M., Tran, J. H., Jacoby, G. A., Zhang, Y., Wang, F., and Hooper, D. C. (2003). Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47, 2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003
Wang, X., Qiao, F., Yu, R., Gao, Y., and Zong, Z. (2013). Clonal diversity of Acinetobacter baumannii clinical isolates revealed by a snapshot study. BMC Microbiol. 13:234. doi: 10.1186/1471-2180-13-234
Woodford, N., Ellington, M. J., Coelho, J. M., Turton, J. F., Ward, M. E., Brown, S., et al. (2006). Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27, 351–353. doi: 10.1016/j.ijantimicag.2006.01.004
Ying, X. U., Wang, D., Wang, J. Q., and Ling, B. D. (2013). Drug resistance surveillance of common bacteria in a hospital between 2010 and 2013 years. J. Chengdu Med. College 8, 506–511.
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-779
Zowawi, H. M., Sartor, A. L., Sidjabat, H. E., Balkhy, H. H., Walsh, T. R., Al Johani, S. M., et al. (2015). Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the gulf cooperation council states: dominance of oxa-23-type producers. J. Clin. Microbiol. 53, 896–903. doi: 10.1128/jcm.02784-2714
Keywords: Acinetobacter baumannii, carbapenem resistance, OXA-23, plasmid, CC92
Citation: Chang Y, Luan G, Xu Y, Wang Y, Shen M, Zhang C, Zheng W, Huang J, Yang J, Jia X and Ling B (2015) Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front. Microbiol. 6:910. doi: 10.3389/fmicb.2015.00910
Received: 28 April 2015; Accepted: 19 August 2015;
Published: 01 September 2015.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Benjamin Andrew Evans, Anglia Ruskin University, UKVishvanath Tiwari, Central University of Rajasthan, India
Copyright © 2015 Chang, Luan, Xu, Wang, Shen, Zhang, Zheng, Huang, Yang, Jia and Ling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baodong Ling, Small Molecule Drugs Sichuan Key Laboratory, Institute of Materia Medica, Chengdu Medical College, Chengdu 610500, China,bGluZ2Jhb2RvbmdAY21jLmVkdS5jbg==; Xu Jia, Non-coding RNA and Drug Discovery Laboratory, Chengdu Medical College, Chengdu 610500, China,ZmRqaWF4dUBnbWFpbC5jb20=
†These authors have contributed equally to this work.
 Yaowen Chang
Yaowen Chang Guangxin Luan
Guangxin Luan Ying Xu
Ying Xu Yanhong Wang2
Yanhong Wang2 Xu Jia
Xu Jia