- 1Department of Genetics, Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo, Brazil
- 2Department of Morphology, Federal University of Sergipe, Aracaju, Brazil
Treatment of fungal infections is difficult due to several reasons, such as side effects of drugs, emergence of resistant strains, and limited number of molecular targets for the drug compounds. In fungi, heat shock proteins (Hsps) have been implicated in several processes with the conserved molecular chaperone Hsp90 emerging as a potential target for antifungal therapy. It plays key cellular roles by eliciting molecular response to environmental changes, morphogenesis, antifungal resistance, and fungal pathogenicity. Here, we evaluated the transcription profiles of hsp genes of the most prevalent dermatophyte Trichophyton rubrum in response to different environmental challenges including nutrient availability, interaction with cells and molecules of the host tissue, and drug exposure. The results suggest that each Hsp responds to a specific stress condition and that the cohort of Hsps facilitates fungal survival under various environmental challenges. Chemical inhibition of Hsp90 resulted in increased susceptibility of the fungus to itraconazole and micafungin, and decreased its growth in human nails in vitro. Moreover, some hsp and related genes were modulated by Hsp90 at the transcriptional level. We are suggesting a role of Hsp90 in the pathogenicity and drug susceptibility of T. rubrum as well as the regulation of other Hsps. The synergism observed between the inhibition of Hsp90 and the effect of itraconazole or micafungin in reducing the fungal growth is of great interest as a novel and potential strategy to treat dermatophytoses.
Introduction
Dermatophytes are pathogenic fungi and primary causative agents of superficial mycoses in humans (Brown et al., 2012). These fungi are keratinolytic and infect keratinized structures such as skin, nails, and hair in the host, giving rise to diseases (also known as dermatophytoses or tineas) such as athlete’s foot, onychomycosis, ringworm, and jock itch. Among the species belonging to this group of filamentous fungi, Trichophyton rubrum is the leading cause of human skin and nail mycoses and has high prevalence worldwide (Havlickova et al., 2008; Seebacher et al., 2008; Nenoff et al., 2014). Although rare, disseminated or deep dermatophytoses have been reported in immunocompromised or immunosuppressed patients (Gong et al., 2007; Marconi et al., 2010; Lanternier et al., 2013). Depending on the amplitude and the site of infection, dermatophytoses can be difficult to cure and often relapse post-treatment, even in immunocompetent individuals (Gupta and Cooper, 2008; Ghannoum and Isham, 2014).
Although a reasonable number of antifungal drugs are commercially available, majority of the clinical drugs act on the ergosterol biosynthesis pathway, thus restricting the number of cellular targets. Besides, resistance to commonly used antifungal drugs has been reported in dermatophytes and other human pathogens, rendering the choice of drug challenging and exacerbating the prospect of successful treatment (Martinez-Rossi et al., 2008; Pfaller, 2012). Therefore, novel antifungal targets have become the prime focus of several researchers in the field of medical mycology. Drug combinations and synergism have been proposed as therapeutically desirable approaches to decrease the development of resistance (Kontoyiannis and Lewis, 2002).
Evaluation of the interconnection among drug resistance, stress response, and the signaling pathways activated in these processes has been revealing key elements or the core circuitry as targets for antifungal therapy (Cowen and Steinbach, 2008; Shapiro et al., 2011). One such promising cellular candidate is the heat shock protein 90 (Hsp90) (Wirk, 2011), a molecular chaperone belonging to the highly conserved family of heat shock proteins (Hsps). These proteins rapidly accumulate in the cytosol in response to heat and environmental challenges such as antifungal drugs, oxidative stress, and heavy metal exposure among others. The heat shock response (HSR) is considered a rescue mechanism that enables the cells to cope under stressful conditions and protects from severe damage. The primary role of Hsps is to sense and assist proper protein folding and refolding, and direct them for degradation in case of misfolding, thereby assuring proteome integrity and homeostasis (Lindquist and Craig, 1988). Hsps act as molecular chaperones or transcriptional regulators in a myriad of physiological functions. These proteins are classified into several families based on their function and molecular weight, which ranges from 9 to 110 kDa. Hsps are also found in all organisms (Lindquist and Craig, 1988; De Maio et al., 2012), and are involved in the assembly of protein complexes, transport and sorting of proteins into the proper cellular compartments, cell-cycle control, and protein fate, among other functions. In fungi, Hsps have been implicated in several processes, including pathogenicity, phase transition in dimorphic fungi, and antifungal drug resistance. Hsps are synthesized as an adaptive response to stress that contributes to the survival of pathogenic microorganisms in the host (Burnie et al., 2006; Brown et al., 2010).
Heat shock protein 90 is highly abundant in cells even in non-stressful state and increases further in response to different forms of stress. However, some eukaryotes present two hsp90 genes, one inducible and the other constitutively expressed (Taipale et al., 2010). Hsp90 can associate with several proteins involved in signaling, metabolism, cell growth, transcription, protein trafficking, chromatin remodeling, and stress response, among others (Leach et al., 2012b). It is an ATP-dependent chaperone and functions as a dimer. Each monomer presents an amino-terminal domain (NTD) that binds ATP and hydrolyzes upon association with the target proteins, a middle domain (MD) crucial for the interaction with the target proteins, and a carboxyl-terminal domain (CTD) responsible for dimerization. The energy produced by the hydrolysis of ATP is used by Hsp90 to fold the target proteins to their active conformations (Taipale et al., 2010). By chaperoning the target proteins, Hsp90 can modulate several downstream processes and regulatory cascades, thus controlling the responses to dynamic environments (Shapiro et al., 2011; Leach et al., 2012b).
Inhibitors of Hsp90 have been thoroughly searched for and some natural compounds produced by microorganisms have been isolated. These include geldanamycin, which is a benzoquinone ansamycin derived from actinomycetes, and a resorcyclic acid lactone called radicicol produced by certain fungal species (Piper and Millson, 2012). Some Hsp90 inhibitors are in clinical trial for cancer therapy and derivatives of natural compounds have been synthesized to increase efficacy and decrease side effects and toxicity (Gorska et al., 2012). In general, these inhibitors act as ATP competitors and interfere with the ATP-binding domain, which turns Hsp90 non-functional and leads to the ubiquitination and proteasome degradation of target proteins because of their aberrant conformation (Wirk, 2011). Besides their therapeutic potential, geldanamycin and its derivatives have been used to characterize the role of Hsp90 in fungal adaptation to host environment and antifungal resistance, as well as to understand their synergism with other antifungal drugs. A promising and interesting consequence of Hsp90 inhibition was that the emergence of resistance to azoles and echinocandins were reduced in vitro in human pathogens Candida albicans and Aspergillus fumigatus, respectively, thus validating the efficiency of these antifungal drugs in experimental infection models (Cowen, 2008; Cowen et al., 2009). Compromising the Hsp90 function was also effective against C. albicans and A. fumigatus biofilms, which are highly drug-resistant recalcitrant structures and an important cause of mortality. Targeting Hsp90 with chemical inhibitors increased the susceptibility of C. albicans biofilms to azoles in vitro and in an animal infection model, and the efficacy of azoles and echinocandins against A. fumigatus biofilms (Robbins et al., 2011). In C. albicans, inhibition of Hsp90 also affected cell wall biogenesis by disrupting the signaling pathways involved in cell wall remodeling (Leach et al., 2012a). Additionally, this also impaired the Hsf1–Hsp90 auto-regulatory circuit in C. albicans. The heat shock transcription factor Hsf1 governs the HSR and is a target of Hsp90. Thus, Hsp90 inhibition affected the Hsf1 regulon, consequently the regulation of HSPs and the resistance to proteotoxic stress (Leach et al., 2012a).
In this work, we have chemically inhibited Hsp90 in the dermatophyte T. rubrum and analyzed the effects to assess the roles played by this molecular chaperone in response to antifungal drugs, fungal pathogenicity, and regulation of other genes. To analyze drug susceptibility, three molecules with different modes of action were tested; itraconazole (ITRA), 5-Fluorocytosine (5-FC), and micafungin (MCFG) act on ergosterol biosynthesis, nucleic acids, and glucan synthesis, respectively. The antifungal effects of these drugs in synergy with the Hsp90 inhibitor against the growth of T. rubrum were evaluated. In order to analyze the role of Hsp90 in pathogenicity, the ability of T. rubrum to colonize human skin and nail in the presence of Hsp90 inhibitor was tested in an ex vivo model of infection. The influence of Hsp90 in the regulation of hsp genes and other related genes such as those encoding for the heat shock factor Hsf1 and the pH responsive regulator PacC was evaluated by transcription profile analyses in response to nutritional sources. Finally, in order to assess the adaptive response to various stress conditions, the transcriptional profile of Hsps, including Hsp90, was evaluated after exposure of T. rubrum to antifungal drugs and substrates present in the host.
Materials and Methods
Trichophyton rubrum Strain
Trichophyton rubrum strain CBS118892 (CBS-KNAW Fungal Biodiversity Centre) was cultivated in malt extract agar (MEA: 2% peptone, 2% glucose, 2% agar, pH 5.7) at 28°C. To prepare the conidia suspension, 15-day-old plates were flooded with sterile 0.9% NaCl and the suspension filtered through fiber glass to remove mycelia debris. The conidia concentration in the filtrate was estimated using a Neubauer chamber, as previously described (Persinoti et al., 2014).
Antifungal Drug Susceptibility Test
The synergistic effect between chemical inhibition of Hsp90 and antifungal agents was tested by the following method: T. rubrum conidia (1 × 106 per plate) were spread on the surface of MEA containing 10, 100, and 300 μM of the inhibitor 17-AAG (17-allylamino-17-demethoxygeldanamycin; InvivoGen, San Diego, CA). E-test (AB Biodisk, Solna, Sweden) gradient strips of ITRA, 5-Fluorocytosine (5-FC), or MCFG were then placed on these plates. Gradient concentration of the tested antifungal drugs ranged from 0.002 to 32 μg/mL and the results were observed after incubation at 28°C for 5 days. Plates without antifungal agents were used to assess fungal development in the presence of 17-AAG. Three independent experiments were conducted.
Ex vivo Pathogenicity Test
The ex vivo nail and skin interaction assays were performed as described here. Autoclaved small pieces of human nail obtained from healthy donors were infected with 1 × 104 T. rubrum conidia and incubated at 28°C for 5 days, in the absence or presence of 50, 100, or 200 μM of Hsp90 inhibitor 17-AAG. After incubation, nail fragments were observed under a light microscope to evaluate the hyphal development and fungal morphology.
Small pieces of human skin were obtained from patients who underwent abdominal surgery at the University Hospital of Ribeirão Preto Medical School, University of São Paulo, Brazil (HC-FMRP-USP). After removal of the adipose tissue, the small pieces of human skin were infected with 1 × 104 T. rubrum conidia in the absence or presence of 200 μM 17-AAG and incubated at 28°C for 5 days. Infected skin fragments were maintained in skin graft fluid (SGF; Duek et al., 2004) supplemented with or without 200 μM 17-AAG. Scanning electron microscopy (SEM) was employed to visualize hyphal development. For this purpose, the skin fragments were fixed with 3% glutaraldehyde in 0.1% phosphate buffer (pH 7.2) at 4°C for 2 h, rinsed with 0.1% phosphate buffer (pH 7.2), and post-fixed with 1% osmium tetroxide for 2 h. Samples were dehydrated by a graded ethanol series and sputter-coated with gold to obtain a layer of approximately 200 μm thickness. The samples were viewed under a Jeol JSM -6610 LV scanning electron microscope at an acceleration voltage of 25 kV.
Growth Conditions for Gene Expression Assays
T. rubrum conidia (1 × 106) were inoculated in 100 mL of malt extract (ME) medium (pH 5.0) or keratin medium (KM: 2.5 g/L keratin powder, MP Biomedicals, suspended in water, pH 5.0). After shaking at 28°C for 96 h, the resultant mycelia were filtered, frozen in liquid nitrogen, and stored at -80°C for expression studies. For the Hsp90 chemical inhibition assay, fresh mycelia grown for 96 h at 28°C in ME or KM were incubated for 30 or 90 min at 28°C with 100- or 300 μM 17-AAG. For antifungal drug response assays mycelia grown in ME at 28°C for 96 h were aseptically transferred to RPMI 1640 (Life Biotechnologies—buffered with 0.167M MOPS, pH 7.0) in the absence (control) and presence of sub-inhibitory concentrations of acriflavine (ACR; 1.75 μg/mL) or terbinafine (TRB; 0.2 μg/mL), and incubated for 3 h at 28°C.
For the interaction assays, the small pieces of human skin were cleaned, infected with 1 × 104 T. rubrum conidia, and incubated at 28°C for 96 h. The fungus was then harvested and used for total RNA extraction. For the nail interaction assay, each human nail fragment was exposed to 1 × 104 T. rubrum conidia and incubated at 28°C for 96 h. The infected nail fragments were vortexed to release fungal mycelia for total RNA extraction and the nails discarded. The interaction assays were approved by the local Ethics Committee (Protocol No. 046/2009). Three independent experiments were conducted for each growth condition and interaction assay.
Gene Expression Analysis
Total RNA was isolated from frozen mycelia using Illustra RNAspin Mini RNA Isolation Kit (GE Healthcare). First-strand cDNA was synthesized using the SuperScriptIII First-Strand Synthesis Super Mix for qRT-PCR kit (Invitrogen). Both RNA extraction and cDNA synthesis were performed according to the manufacturer’s recommendations. An intron flanking region of the β-tubulin gene was used as positive control to verify DNA contamination and proper cDNA synthesis, as previously described (Jacob et al., 2012).
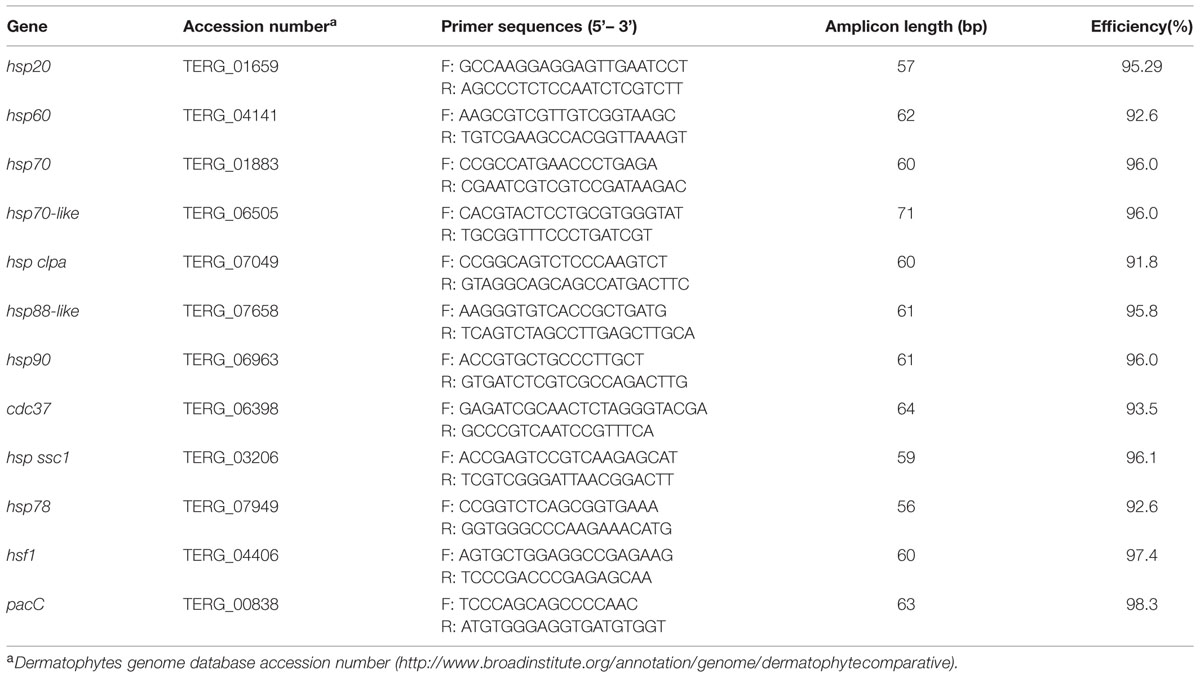
Specific primer pairs for each T. rubrum gene were designed using the Primer Express v.3 software (Life Technologies) and are listed in Table 1. qRT-PCR reactions were carried out in a final volume of 12.5 μL, containing 6.25 μL Power SYBRGreen PCR Master Mix (Life Technologies), 1.0 μL of each primer (hsp20, 250 nM; hsp60, 500 nM; hsp70, 450 nM; hsp70-like, 350 nM; hsp clpa, 300 nM; hsp88-like, 350 nM; hsp90, 300 nM; cdc37, 400 nM; hsp ssc1, 400 nM; hsp78, 350 nM; hsf1, 350 nM; pacC, 350 nM), 2.0-μL template cDNA (50 ng), and 3.25-μL ultra-pure water. Thermal conditions for qRT-PCR were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were performed in triplicate in 96-well reaction plates using the StepOnePlus Real-Time PCR System (Life Technologies). A melting curve for each gene was obtained and 2% agarose gel electrophoresis was performed to confirm the amplification of the unique product of expected size for each hsp gene. To determine PCR efficiency, standard curves were generated using cDNA sample at five-point, twofold dilutions and measured in triplicates. The reference genes rpb2 and actin were used for data normalization as previously described (Jacob et al., 2012). Relative expression was calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001). Statistical significance was evaluated by one-way ANOVA followed by the Tukey’s ad hoc test, using the GraphPad Prism v 5.1 Software.
Results
Inhibition of Hsp90 by 17-AAG
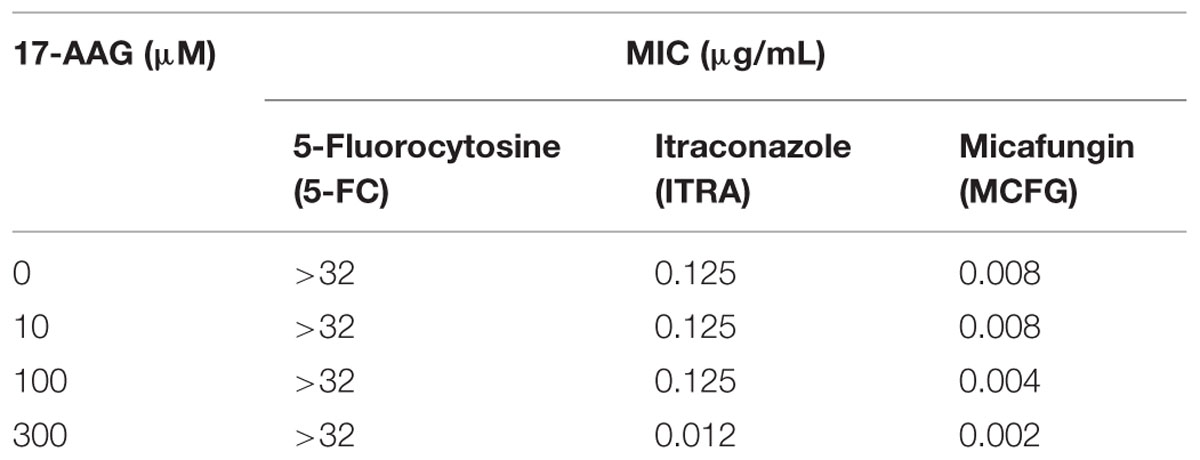
In order to evaluate the synergism of Hsp90 with other antifungal drugs, as well as its role in T. rubrum pathogenicity, 17-AAG was used to chemically inhibit Hsp90 expression in T. rubrum. Although inhibition of Hsp90 using 300 μM of 17-AAG had no effect on T. rubrum growth in MEA, an increase in fungal susceptibility to ITRA and MCFG was observed (Supplementary Figure S1). This was demonstrated by 10- and fourfold decreases in minimal inhibitory concentration (MIC) values, respectively. Moreover, there was no effect when the fungus was challenged with 5-FC (Table 2). The role of Hsp90 in T. rubrum pathogenicity was also analyzed using an ex vivo nail interaction assay. Inhibition of Hsp90 decreased T. rubrum growth on human nail in vitro. This decrease was dependent on the concentration of 17-AAG used, and at 200 μM, T. rubrum growth was almost entirely inhibited (Figure 1), indicating the attenuation of fungal virulence. However, SEM of ex vivo human skin inoculated with T. rubrum conidia showed no significant difference in fungal growth when Hsp90 was inhibited at the same 17-AAG concentration. (Supplementary Figure S2). This suggests that other virulence factors or incomplete inhibition of Hsp90 activity might foster fungal growth in the ex vivo skin model.
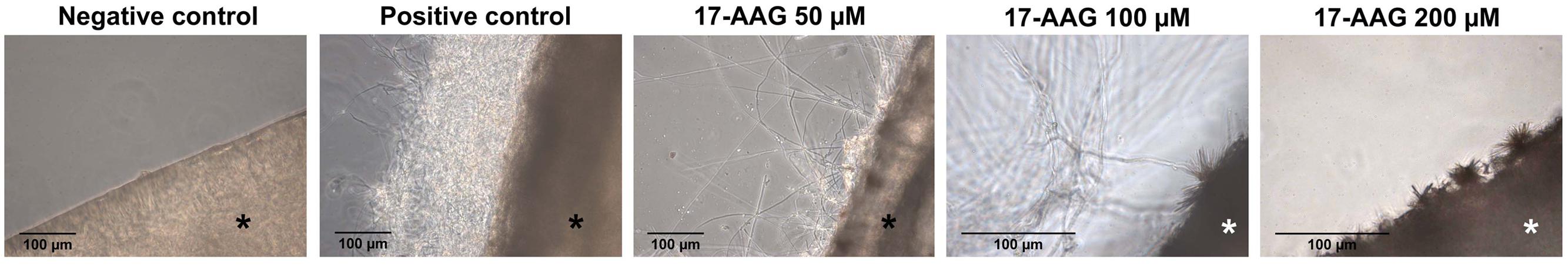
FIGURE 1. Effect of heat shock protein (Hsp90) inhibition on Trichophyton rubrum growth in human nail. Light microscopy was used to analyze hyphal development in human nail fragments infected with T. rubrum conidia in the absence (positive control) or presence of the Hsp90 inhibitor 17-AAG (50, 100, or 200 μM) after 5 days at 28°C. Negative control consists of uninoculated and untreated nails. Asterisks indicate nail fragments.
Expression Profile of hsps and Related Genes
The expression of several hsps and related genes is modulated during the growth of T. rubrum in KM and ME in the presence of Hsp90 inhibitor 17-AAG. For some genes, this effect is time- and concentration-dependent (Figure 2). While the inhibition of Hsp90 led to decreased accumulation of hsp70-like and hsf1 transcripts, regardless of the growth medium, the levels of hsp60 and hsp88-like transcripts were decreased in response to ME and KM, respectively. However, hsp70 and hsp clpa genes were upregulated in the absence of Hsp90 activity, which led to increased accumulation of hsp90 transcripts, perhaps in an attempt to compensate for the chemical inhibition of the protein. Interestingly, pacC was downregulated when Hsp90 was inhibited in the presence of keratin. Moreover, the gene coding for the Hsp90 co-chaperone cdc37 was upregulated in ME but not in KM, whereas hsp ssc1 was upregulated only in KM (Figure 2) upon Hsp90 inhibition. These results suggest a regulatory role for Hsp90 in the expression of pacC and other hsp genes in T. rubrum, depending on the growth medium.
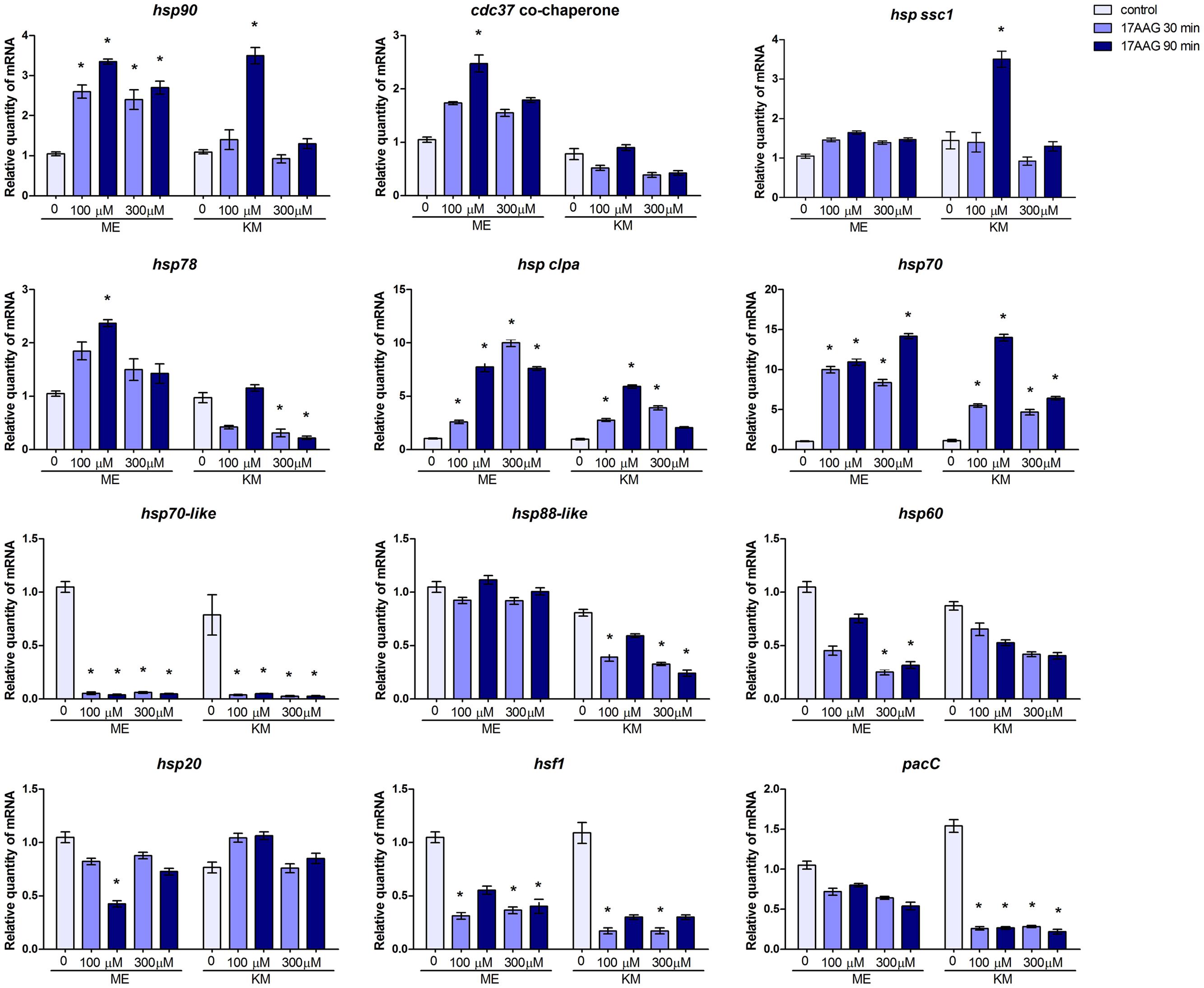
FIGURE 2. Effect of Hsp90 on the expression of hsp and related genes in T. rubrum. qRT-PCR analyses of the hsp and related genes of T. rubrum grown in malt extract (ME) or keratin medium (KM) in the absence or presence of Hsp90 inhibitor. Gene expression levels are represented by the quantities of mRNA in each condition relative to the control (ME). Data are represented as mean ± SD from three independent experiments with reactions performed in triplicate. Tukey’s ad hoc test was used for statistical analysis ; ∗P < 0.05.
Given the involvement of Hsps in the pathogenicity of several fungal pathogens and in drug susceptibility, the expression profile of some of these hsp genes was evaluated in T. rubrum in response to other antifungal drugs, such as TRB and ACR, and during its growth on nail and skin fragments (Figure 3). The analysis revealed different expression profiles in response to these environmental challenges. While hsp clpa gene was not modulated in response to any stimuli analyzed (Figures 3A,B), hsp90, hsp88-like, and hsp20 transcripts accumulated in response to drug exposure (Figure 3B). However, hsp60 and hsp78 presented a slightly broad range of modulation with transcripts accumulating during growth on nails and in response to one or both drugs (Figures 3A,B).
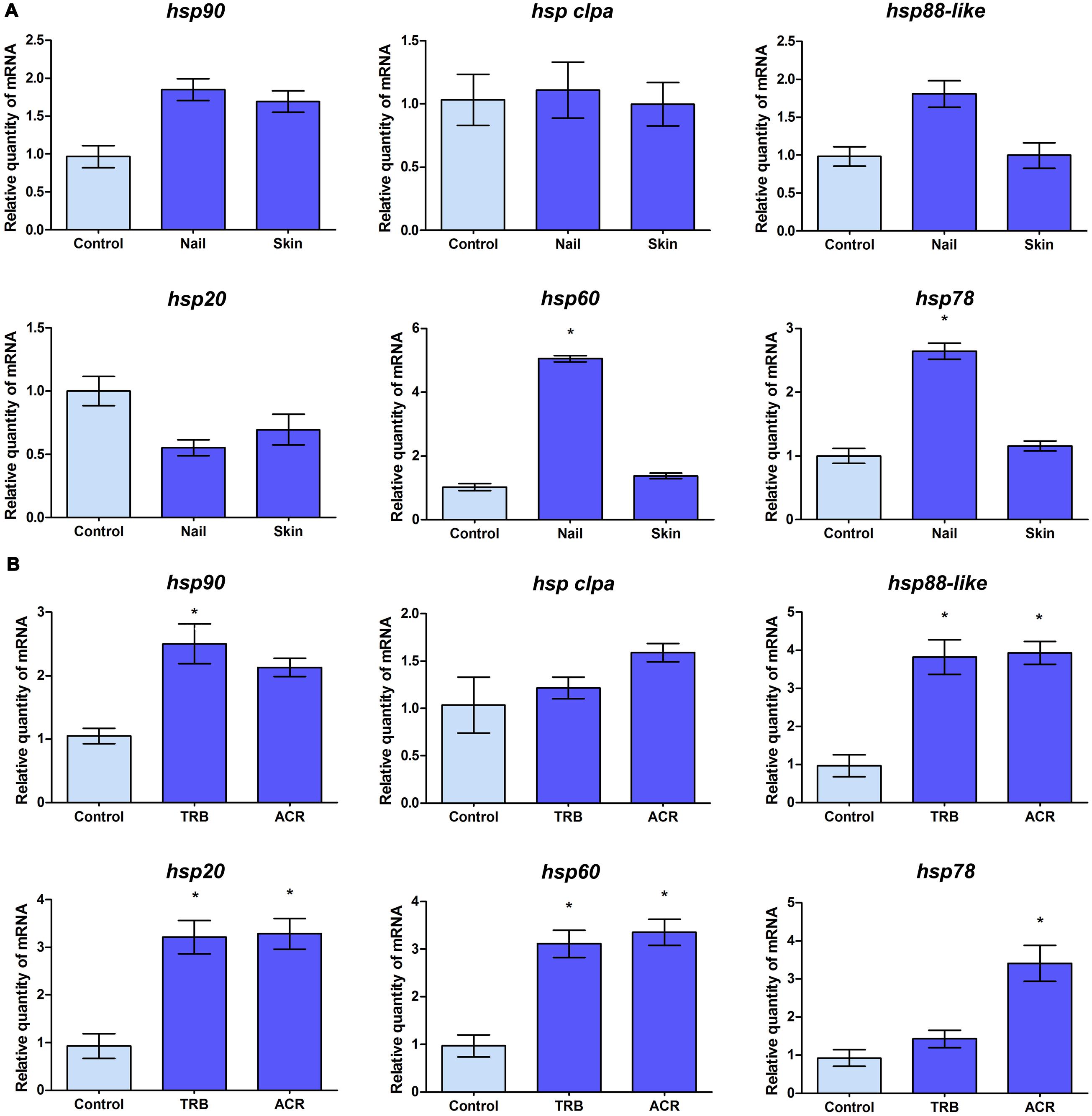
FIGURE 3. Transcript levels of T. rubrum hsp genes in response to environmental challenges. (A) qRT-PCR analyses of the transcript levels of hsp genes of T. rubrum grown in malt extract (control), human nail, or human skin fragments for 96 h. (B) Mycelia were transferred to RPMI medium in the absence (control) or presence of sub-inhibitory doses of terbinafine (TRB) or acriflavine (ACR) for 3 h for assessment of drug response. mRNA quantity in each condition relative to the control are represented as mean ±SD from three independent experiments with reactions performed in triplicate. Tukey’s ad hoc test was used for statistical analysis, ∗P < 0.05.
Discussion
Dermatophytes affect millions of individuals annually and have become an important public health concern because of their refractivity to therapy, which prolongs the duration of treatment especially in aging populations (Coelho et al., 2008; Martinez et al., 2012). Because both the host and the pathogen are eukaryotic organisms, treatment of fungal infections is difficult due to the limited number of antifungal targets available (Martinez-Rossi et al., 2008). Another concern is the emergence of resistance to antifungal drugs currently in clinical use. Thus, it is necessary to identify new strategies for therapy against fungal infections. In this study, we observed that it is possible to increase the efficacy of antifungal drugs, ITRA and MCFG, by targeting the molecular chaperone Hsp90 with 17-AAG, an inhibitor of the Hsp90 ATPase activity. Interestingly, these antifungal agents have different mechanisms of action. While ITRA inhibits the enzyme lanosterol 14α-demethylase, thus preventing the biosynthesis of ergosterol, a key sterol in the fungal membrane (Lupetti et al., 2002; Odds et al., 2003), MCFG inhibits the enzyme 1, 3-β-D-glucan synthase, thereby blocking the biosynthesis of a key linker molecule in the fungal cell wall (Onishi et al., 2000). When challenged with antifungal drugs, several dermatophytes generally react by activating stress responses (Paião et al., 2007; Yu et al., 2007; Zhang et al., 2009; Peres et al., 2010), which often depends on the Hsp90 chaperone. Hsp90 can associate itself with a myriad of target proteins such as its co-chaperones and functional regulators, and modulate the activation and stability of the complex. Thus, functional inhibitors of the Hsp90 that act on its ATPase-coupled conformation (Siligardi et al., 2002; Lotz et al., 2003) disassemble the molecular complex of Hsp90 with co-chaperones and target proteins, thereby abrogating drug resistance and increasing the efficacy of traditional antifungal drugs (Veri and Cowen, 2014). Although Hsp90 is conserved among eukaryotes, it presents some conformational differences in fungi, especially in regions such as the ATP binding and the MDs that could be selectively targeted by the chemical inhibitors (Wider et al., 2009; Shahinas et al., 2015). Alternatively, it is possible to interfere with the Hsp90 targets or functional regulators, thus expanding the possibility to find clearer discrepancies between the pathogen and the host (Veri and Cowen, 2014). Therefore, targeting Hsp90 or other related proteins may be a viable alternative to treat fungal infections caused by T. rubrum and probably by other dermatophytes as well. We have also shown that the Hsp90 chaperone has a role in conferring the fungus with the ability to colonize human nails in vitro. The involvement of this chaperone in the pathogenicity of other pathogens such as C. albicans and C. glabrata has been demonstrated (Noble et al., 2010; Leach et al., 2012b; Singh-Babak et al., 2012). Thus, blocking the action of Hsp90 in dermatophytes becomes a potential strategy that combines this therapy with conventional antifungal drugs, which would enhance the overall outcome of the treatment.
The broad spectrum of Hsp90 functions were confirmed by changes in the expression profile of various hsp and related genes upon chemical inhibition of this chaperone in T. rubrum. Moreover, these changes in gene expression in the fungus were nutrient-dependent (ME and KM medium). The heat shock transcription factor Hsf1 positively regulates the transcription of the hsp90 gene in C. albicans and Saccharomyces cerevisiae; both pharmacological inhibition and genetic depletion of Hsp90 correlate with Hsf1 activation in response to thermal stress (Wu, 1995; Leach et al., 2012b). However, hsf1 transcript levels decreased when T. rubrum was challenged with the Hsp90 inhibitor in MEA or KM, under non-stressful temperature conditions (Figure 2). Additionally, there was an evident increase in hsp90 transcripts in T. rubrum treated with the Hsp90 inhibitor, which was likely to compensate for the absence of Hsp90 function (Figure 2). This suggests a regulatory role for Hsp90 over hsf1 transcript levels or a compensatory mechanism upon Hsp90 inhibition, in which most of the hsf1 transcripts in the cell are efficiently transduced into protein, in turn aiding hsp90 transcription. In T. rubrum, putative DNA-binding sites for Hsf1 in the hsp90 promoter region enable direct regulation. Another interesting observation from this study is the decreased amount of the pacC transcripts during growth in keratin cultures containing 17-AAG (Figure 2). The transcription factor PacC mediates diverse metabolic events, including virulence and keratinolytic activity, in T. rubrum (Ferreira-Nozawa et al., 2006; Silveira et al., 2010; Martinez-Rossi et al., 2012), suggesting a correlation between the pacC and hsp90 genes and fungal virulence. Additionally, we observed an increased accumulation of some hsp transcripts on Hsp90 inhibition, while others, including those belonging to the same family, suffered drastic decline. For example, regardless of the culture condition, transcription of the hsp70-like gene showed a significant reduction, whereas hsp70 transcripts increased significantly and hssc1 (Hsp70 family protein) transcripts practically accumulated to the same amount, suggesting different roles for each Hsp (Figure 2).
In order to evaluate the expression profiles of some hsp genes in ex vivo models, as well as during treatment with antifungal drugs, T. rubrum was cultured in MEA or in nail or skin fragments (Figure 3A) and in RPMI medium containing sub-inhibitory concentrations of TRB or ACR (Figure 3B). hsp90 gene expression increased only in response to TRB, an antifungal used to treat dermatophytosis. Unexpectedly, hsp90 transcript levels were similar to those observed in other experimental conditions tested, including the human nail, in which the fungus was dependent on Hsp90 function when this substrate was its sole nutrient source. It is possible that the fungus depends on similar amounts of the Hsp90 protein to cope with varying culture conditions. The transcription profile of the hsp clpA gene was unchanged in the analyzed conditions, demonstrating constitutive expression in response to various antifungal drugs and growth conditions. However, there was an increase in hsp20, hsp60, hsp78, and hsp88-like gene transcript levels when T. rubrum was challenged with ACR and/or TRB, suggesting their involvement in cellular stress responses. An increase in hsp60 and hsp78 transcripts was also observed when the fungus was grown in nail fragments, suggesting that Hsp60 and Hsp78 proteins act as Hsp90 co-chaperones in nail infections.
Conclusion
Complex formation of Hsp90 and its chaperones depends on an ATPase-coupled conformational cycle, which links ATP binding and hydrolysis. This is a highly conserved mechanism in eukaryotic organisms, including C. albicans and other human pathogenic fungi, since ATP binding and hydrolysis are essential for Hsp90 function. Functional inhibition of ATP binding to Hsp90 disassembles the molecular complex between Hsp90 with target proteins and co-chaperones, consequently abrogating drug resistance and increasing the efficacy of traditional antifungal drugs such as ITRA and MCFG. Blocking the Hsp90 activity drastically decreased the ability of T. rubrum to grow on human nail fragments and interfered with the modulation of some hsp genes and the pacC gene, a regulator involved in T. rubrum virulence. Thus, blocking Hsp90 function in dermatophytes is suggested as a potential strategy for combination of this therapy with traditional antifungal drugs, which would enhance the antifungal efficacy. This is an attractive hypothesis to be explored further probably using in vitro and/or ex vivo models of infection as well as new inhibitors for Hsp90 and other co-chaperones.
Author Contributions
TJ performed most of the experimental procedures, such as fungal cultivation, RNA extraction, synergism assays, and qPCR. NP participated in the experimental design and procedures, and drafted the manuscript. PS performed computational analyses to identify the genes used in this work, their promoter regions, identifying DNA-binding domains and the putative proteins. He also performed the statistical analyses of qPCR. EL performed some experimental procedures, such as skin infections, image acquisition, and drafted the manuscript. MM performed the nail infections, synergism assays and image acquisition. AR and NM-R designed the project, supervised the research study, and prepared the manuscript. All authors participated in data analysis and interpretation, read, revised critically the manuscript and approved the final version. Also, all authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Brazilian Funding Agencies: São Paulo Research Foundation (FAPESP; Grant No. 2014/03847-7, and Postdoctoral Fellowships No. 2011/08424-9 and 2009/08411-4), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Grants No 305252/2013-5, 304222/2013-5, 150980/2013-2, 150133/2013-8, and 553778/2009-9), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Apoio ao Ensino, Pesquisa e Assistência (FAEPA). We thank V.M. Oliveira, M. Mazucato, and M. D. Martins for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01241
References
Brown, A. J., Leach, M. D., and Nicholls, S. (2010). The relevance of heat shock regulation in fungal pathogens of humans. Virulence 1, 330–332. doi: 10.4161/viru.1.4.12364
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. doi: 10.1126/scitranslmed.3004404
Burnie, J. P., Carter, T. L., Hodgetts, S. J., and Matthews, R. C. (2006). Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 30, 53–88. doi: 10.1111/j.1574-6976.2005.00001.x
Coelho, L. M., Aquino-Ferreira, R., Maffei, C. M. L., and Martinez-Rossi, N. M. (2008). In vitro antifungal drug susceptibilities of dermatophytes microconidia and arthroconidia. J. Antimicrob. Chemother. 62, 758–761. doi: 10.1093/jac/dkn245
Cowen, L. E. (2008). The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6, 187–198. doi: 10.1038/nrmicro1835
Cowen, L. E., Singh, S. D., Kohler, J. R., Collins, C., Zaas, A. K., Schell, W. A., et al. (2009). Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U.S.A. 106, 2818–2823. doi: 10.1073/pnas.0813394106
Cowen, L. E., and Steinbach, W. J. (2008). Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot. Cell 7, 747–764. doi: 10.1128/EC.00041-08
De Maio, A., Santoro, M. G., Tanguay, R. M., and Hightower, L. E. (2012). Ferruccio Ritossa’s scientific legacy 50 years after his discovery of the heat shock response: a new view of biology, a new society, and a new journal. Cell Stress Chaperones 17, 139–143. doi: 10.1007/s12192-012-0320-z
Duek, L., Kaufman, G., Ulman, Y., and Berdicevsky, I. (2004). The pathogenesis of dermatophyte infections in human skin sections. J. Infect. 48, 175–180. doi: 10.1016/j.jinf.2003.09.008
Ferreira-Nozawa, M. S., Silveira, H. C. S., Ono, C. J., Fachin, A. L., Rossi, A., and Martinez-Rossi, N. M. (2006). The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med. Mycol. 44, 641–645. doi: 10.1080/13693780600876553
Ghannoum, M., and Isham, N. (2014). Fungal nail infections (onychomycosis): a never-ending story? PLoS Pathog. 10:e1004105. doi: 10.1371/journal.ppat.1004105
Gong, J. Q., Liu, X. Q., Xu, H. B., Zeng, X. S., Chen, W., and Li, X. F. (2007). Deep dermatophytosis caused by Trichophyton rubrum: report of two cases. Mycoses 50, 102–108. doi: 10.1111/j.1439-0507.2006.01324.x
Gorska, M., Popowska, U., Sielicka-Dudzin, A., Kuban-Jankowska, A., Sawczuk, W., Knap, N., et al. (2012). Geldanamycin and its derivatives as Hsp90 inhibitors. Front. Biosci. (Landmark Ed.) 17:2277. doi: 10.2741/4050
Gupta, A. K., and Cooper, E. A. (2008). Update in antifungal therapy of dermatophytosis. Mycopathologia 166, 353–367. doi: 10.1007/s11046-008-9109-0
Havlickova, B., Czaika, V. A., and Friedrich, M. (2008). Epidemiological trends in skin mycoses worldwide. Mycoses. 51(Suppl. 4), 2–15. doi: 10.1111/j.1439-0507.2008.01606.x
Jacob, T. R., Peres, N. T. A., Persinoti, G. F., Silva, L. G., Mazucato, M., and Rossi, A. (2012). rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 50, 368–377. doi: 10.3109/13693786.2011.616230
Kontoyiannis, D. P., and Lewis, R. E. (2002). Antifungal drug resistance of pathogenic fungi. Lancet 359, 1135–1144. doi: 10.1016/S0140-6736(02)08162-X
Lanternier, F., Cypowyj, S., Picard, C., Bustamante, J., Lortholary, O., Casanova, J. L., et al. (2013). Primary immunodeficiencies underlying fungal infections. Curr. Opin. Pediatr. 25, 736–747. doi: 10.1097/MOP.0000000000000031
Leach, M. D., Budge, S., Walker, L., Munro, C., Cowen, L. E., and Brown, A. J. (2012a). Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 8:e1003069. doi: 10.1371/journal.ppat.1003069
Leach, M. D., Klipp, E., Cowen, L. E., and Brown, A. J. (2012b). Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat. Rev. Microbiol. 10, 693–704. doi: 10.1038/nrmicro2875
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lotz, G. P., Lin, H., Harst, A., and Obermann, W. M. (2003). Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 278, 17228–17235. doi: 10.1074/jbc.M212761200
Lupetti, A., Danesi, R., Campa, M., Del Tacca, M., and Kelly, S. (2002). Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8, 76–81. doi: 10.1016/S1471-4914(02)02280-3
Marconi, V. C., Kradin, R., Marty, F. M., Hospenthal, D. R., and Kotton, C. N. (2010). Disseminated dermatophytosis in a patient with hereditary hemochromatosis and hepatic cirrhosis: case report and review of the literature. Med. Mycol. 48, 518–527. doi: 10.3109/13693780903213512
Martinez, D. A., Oliver, B. G., Graser, Y., Goldberg, J. M., Li, W., Martinez-Rossi, N. M., et al. (2012). Comparative Genome Analysis of Trichophyton rubrum and Related Dermatophytes Reveals Candidate Genes Involved in Infection. MBio 3:e00259–e00312. doi: 10.1128/mBio.00259-12
Martinez-Rossi, N. M., Peres, N. T. A., and Rossi, A. (2008). Antifungal resistance mechanisms in dermatophytes. Mycopathologia 166, 369–383. doi: 10.1007/s11046-008-9110-7
Martinez-Rossi, N. M., Persinoti, G. F., Peres, N. T. A., and Rossi, A. (2012). Role of pH in the pathogenesis of dermatophytoses. Mycoses 55, 381–387. doi: 10.1111/j.1439-0507.2011.02162.x
Nenoff, P., Kruger, C., Ginter-Hanselmayer, G., and Tietz, H. J. (2014). Mycology - an update. Part 1: dermatomycoses: causative agents, epidemiology and pathogenesis. J. Dtsch. Dermatol. Ges. 12, 188–209. doi: 10.1111/ddg.12245
Noble, S. M., French, S., Kohn, L. A., Chen, V., and Johnson, A. D. (2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598. doi: 10.1038/ng.605
Odds, F. C., Brown, A. J., and Gow, N. A. (2003). Antifungal agents: mechanisms of action. Trends Microbiol. 11, 272–279. doi: 10.1016/S0966-842X(03)00117-3
Onishi, J., Meinz, M., Thompson, J., Curotto, J., Dreikorn, S., Rosenbach, M., et al. (2000). Discovery of novel antifungal (1,3)-beta-D-glucan synthase inhibitors. Antimicrob. Agents Chemother. 44, 368–377. doi: 10.1128/AAC.44.2.368-377.2000
Paião, F. G., Segato, F., Cursino-Santos, J. R., Peres, N. T., and Martinez-Rossi, N. M. (2007). Analysis of Trichophyton rubrum gene expression in response to cytotoxic drugs. FEMS Microbiol. Lett. 271, 180–186. doi: 10.1111/j.1574-6968.2007.00710.x
Peres, N. T. A., Sanches, P. R., Falcão, J. P., Silveira, H. C. S., Paião, F. G., Maranhão, F. C. A., et al. (2010). Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 10:39. doi: 10.1186/1471-2180-10-39
Persinoti, G. F., Peres, N. T. A., Jacob, T. R., Rossi, A., Vencio, R. Z., and Martinez-Rossi, N. M. (2014). RNA-seq Trichophyton rubrum transcriptome analysis in response to the cytotoxic drug acriflavine. BMC Genomics 15:S1. doi: 10.1186/1471-2164-15-S7-S1
Pfaller, M. A. (2012). Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125, S3–S13. doi: 10.1016/j.amjmed.2011.11.001
Piper, P. W., and Millson, S. H. (2012). Spotlight on the microbes that produce heat shock protein 90-targeting antibiotics. Open Biol. 2:120138. doi: 10.1098/rsob.120138
Robbins, N., Uppuluri, P., Nett, J., Rajendran, R., Ramage, G., Lopez-Ribot, J. L., et al. (2011). Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7:e1002257. doi: 10.1371/journal.ppat.1002257
Seebacher, C., Bouchara, J. P., and Mignon, B. (2008). Updates on the epidemiology of dermatophyte infections. Mycopathologia 166, 335–352. doi: 10.1007/s11046-008-9100-9
Shahinas, D., Debnath, A., Benedict, C., McKerrow, J. H., and Pillai, D. R. (2015). Heat shock protein 90 inhibitors repurposed against Entamoeba histolytica. Front. Microbiol. 6:368. doi: 10.3389/fmicb.2015.00368
Shapiro, R. S., Robbins, N., and Cowen, L. E. (2011). Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75, 213–267. doi: 10.1128/MMBR.00045-10
Siligardi, G., Panaretou, B., Meyer, P., Singh, S., Woolfson, D. N., Piper, P. W., et al. (2002). Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277, 20151–20159. doi: 10.1074/jbc.M201287200
Silveira, H. C. S., Gras, D. E., Cazzaniga, R. A., Sanches, P. R., Rossi, A., and Martinez-Rossi, N. M. (2010). Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb. Pathog. 48, 91–96. doi: 10.1016/j.micpath.2009.10.006
Singh-Babak, S. D., Babak, T., Diezmann, S., Hill, J. A., Xie, J. L., Chen, Y. L., et al. (2012). Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 8:e1002718. doi: 10.1371/journal.ppat.1002718
Taipale, M., Jarosz, D. F., and Lindquist, S. (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528. doi: 10.1038/nrm2918
Veri, A., and Cowen, L. E. (2014). Progress and prospects for targeting Hsp90 to treat fungal infections. Parasitology 141, 1127–1137. doi: 10.1017/S0031182013002072
Wider, D., Peli-Gulli, M. P., Briand, P. A., Tatu, U., and Picard, D. (2009). The complementation of yeast with human or Plasmodium falciparum Hsp90 confers differential inhibitor sensitivities. Mol. Biochem. Parasitol. 164, 147–152. doi: 10.1016/j.molbiopara.2008.12.011
Wirk, B. (2011). Heat shock protein inhibitors for the treatment of fungal infections. Recent Pat. Antiinfect. Drug Discov. 6, 38–44. doi: 10.2174/157489111794407840
Wu, C. (1995). Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441–469. doi: 10.1146/annurev.cb.11.110195.002301
Yu, L., Zhang, W., Wang, L., Yang, J., Liu, T., Peng, J., et al. (2007). Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum. Antimicrob. Agents Chemother. 51, 144–153. doi: 10.1128/AAC.00755-06
Keywords: Hsp, antifungal therapy, molecular target, drug synergism, itraconazole, micafungin, 17-AAG
Citation: Jacob TR, Peres NTA, Martins MP, Lang EAS, Sanches PR, Rossi A and Martinez-Rossi NM (2015) Heat Shock Protein 90 (Hsp90) as a Molecular Target for the Development of Novel Drugs Against the Dermatophyte Trichophyton rubrum. Front. Microbiol. 6:1241. doi: 10.3389/fmicb.2015.01241
Received: 27 August 2015; Accepted: 26 October 2015;
Published: 10 November 2015.
Edited by:
Chaminda Jayampath Seneviratne, National University of Singapore, SingaporeReviewed by:
Siddharth Kaushal Tripathi, University of Mississippi, USASabine Fillinger, Institut National de la Recherche Agronomique, France
Mehdi Razzaghi-Abyaneh, Pasteur Institute of Iran, Iran
Copyright © 2015 Jacob, Peres, Martins, Lang, Sanches, Rossi and Martinez-Rossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nilce M. Martinez-Rossi, bm1tcm9zc2lAdXNwLmJy
 Tiago R. Jacob
Tiago R. Jacob Nalu T. A. Peres
Nalu T. A. Peres Maíra P. Martins
Maíra P. Martins Elza A. S. Lang
Elza A. S. Lang Pablo R. Sanches
Pablo R. Sanches Antonio Rossi
Antonio Rossi Nilce M. Martinez-Rossi
Nilce M. Martinez-Rossi