- 1Microbial Culture Collection, National Centre for Cell Science, Pune, India
- 2Institute of Bioinformatics and Biotechnology, Savitribai Phule Pune University, Pune, India
Soda lake is hyper alkaline and saline habitat located in closed craters with high evaporation rate. In current study fungal diversity from water and sediment samples of a soda lake (Lonar lake) located in Buldhana district of Maharashtra, India was investigated using extensive culturomics approach and mimicking the natural conditions of Lonar lake in culture media. A total of 104 diverse isolates of extremophilic fungi were recovered from this study and phylogenetically characterized by internal transcribed spacer (ITS) region sequencing. In addition, due to important role of phenol oxidase, and peroxidase in degradation of toxic phenol, lignin, etc., all isolated pure cultures were also screened for extracellular phenol oxidase and peroxidase production potential. Diversity analysis indicated that different groups of extremophilic fungi are present in the water and sediment samples of Lonar lake. A total of 38 species of fungi belonging to 18-different genera were recovered. Out of 104 isolates 32 showed ≤97% sequences similarity, which were morphologically different and could be potential novel isolates of extremophilic fungi. However, out of 104 isolates only 14 showed the extracellular phenol oxidase production potentials at alkaline pH. Curvularia sp. strain MEF018 showed highest phenol oxidase production at alkaline condition and had low sequence similarity with previously characterized species (96% with Curvularia pseudorobusta). Taxonomic characterization (morphological and physiological) and multi locus sequence analysis (MLSA) using combined alignment of ITS-LSU-gpd of strain MEF018 showed that it is a novel species of the genus Curvularia and hence proposed as Curvularia lonarensis sp. nov.
Introduction
Due to immense biotechnological applications of extremophilic enzymes, study of microbial diversity of extreme habitats like soda lake, hot springs, Arctic, and Antarctic polar regions, acid mine drainage and thermal vents are of current interest among the microbiologists (Vargas et al., 2004; Jayani et al., 2005; Calvez et al., 2009; Das et al., 2009; Burgaud et al., 2010; Brown et al., 2015; Chaput et al., 2015). Similar to Bacteria and Archaea, fungi as saprophytes or mutualistic symbionts also provide valuable ecosystem services including degradation of organic materials and mineralization and mobilization of nutrients. Therefore, cultivation and characterization of novel extremophilic fungi from unusual habitats and study of their physiology, genetics and biotechnological potential are equally important from ecological and industrial perspective. According to Ostergaard and Olsen (2011), 75% of the industrial enzymes come from only five genera of fungi which reveal that maximum fungi remain industrially unutilized or un-explored. Moreover, novel fungi are not explored for their potential uses. Despite the immense importance of fungal enzymes in biogeochemical cycling of materials and industrial applications, unlike bacteria little attention has been given on cultivation and characterization of extremophilic fungi from extreme habitats like soda lakes. Although, fungi prefer acidic to neutral pH range for growth, reports on alkaliphilic, and halophilic fungal species from soda lakes like Magadi lake of Kenya, Natron lake of Tanzania (pH 11–12), and Dead sea of Israel are available in literature (Oren and Gunde-Cimerman, 2012; Grum-Grzhimaylo et al., 2013a,b, 2016). These relatively recent contributions are important in light of scarcity of similar studies on fungi and potential of fungi isolated from these unusual habitats.
Phenol oxidase and peroxidase are the enzymes which are important in lignin degradation, humification, carbon mineralization, and dissolved carbon export (Sinsabaugh, 2010). In fungi polyphenol oxidases, particularly laccases play role in lignin degradation, fungal spore formation, pigmentation, detoxification of toxic compounds, pathogenesis, and fungal morphogenesis. Phenol oxidase also play important role in depletion and accumulation of soil organic matter. It has been observed that level of soil organic matter decrease with increasing activity of phenol oxidase, while low level of phenol oxidase promotes its accumulation in soil. In addition, it also degrades toxic phenolic compounds, protects microbial cells from their harmful effects, and play an important role in management of plant residue with high lignin content. Phenol oxidases have widespread applications in pollutant degradation, effluent decolouration, pulp bleaching, removal of phenolics from wines, oxidation of dye, enzymatic conversion of chemical intermediates, biofuel production, etc., Most of the polluted habitats like industrial effluents, leachates, hospital wastes etc. show extreme condition in one or the other aspects. Therefore, organism with potential to produce enzyme in extreme condition has special significance for enzymatic bioremediation of pollutants in comparison to their mesophilic counterparts. Hence, considering the importance of phenol oxidase in biotechnology and carbon cycling of natural ecosystem we screened isolated strains for phenol oxidase production. The chemistry, function and biotechnological use of laccases have recently been reviewed (Baldrian, 2006).
In the current study we cultivated wide range of alkaliphilic and halophilic fungi from water and sediment samples of hyperalkaline and saline Lonar lake using extensive culturomic approaches i.e., use of different media and culture conditions. In addition, we also screened all the isolated fungi for production of phenol oxidase and peroxidase in alkaline condition. Identification of all isolates was done by sequencing internal transcribed spacer (ITS) region. Finally, we did the taxonomical characterization of a novel species of alkaliphilic and halophilic Curvularia which showed efficient phenol oxidase production potential at alkaline pH and proposed it as Curvularia lonarensis sp. nov. Multi locus sequence analysis (MLSA) was used to ascertain its position within the genus Curvularia. To the best of our knowledge, this is the first report on study of fungal diversity and their phenol oxidase producing potential from Lonar lake.
Materials and Methods
Collection of Samples and Geochemical Characterization of Sampling Site
Lonar lake is a hyper-saline and alkaline soda lake located in Buldhana district of Maharashtra, India (19°58′36″N 76°30′30″E) and created by meteor impact during Pleistocene Epoch. The diameter of lake is approximately 1.8 km with 135 m of slope. Due to its unique ecological and geological features it is a site of interest to microbiologists for cultivation of extremophilic microbes. Photographic and tabular representation of lake and its geochemistry are presented in Figure 1A, Table 1 respectively. In current study sediment and water samples were collected from Lonar lake for cultivation of alkaliphilic and halophilic fungi. Water and sediment samples were collected in 50 ml capacity pre-sterilized Falcon tubes (Falcon, USA). Sampling was done from four different collection points located at equal distance (500 m) on circumference of the lake from a reference point. Samples were collected from shore line as well as from 3 m inside the lake from the periphery with a water column depth of approximately 30 cm. Three samples were collected from each point and mixed to form a compound sample. Thus, a total 16 compound samples of water and sediment were collected. Diagram of sampling strategy is given in Figure 1B. Collected samples were stored on crushed ice and transferred to the laboratory and stored at 4°C until further processing.
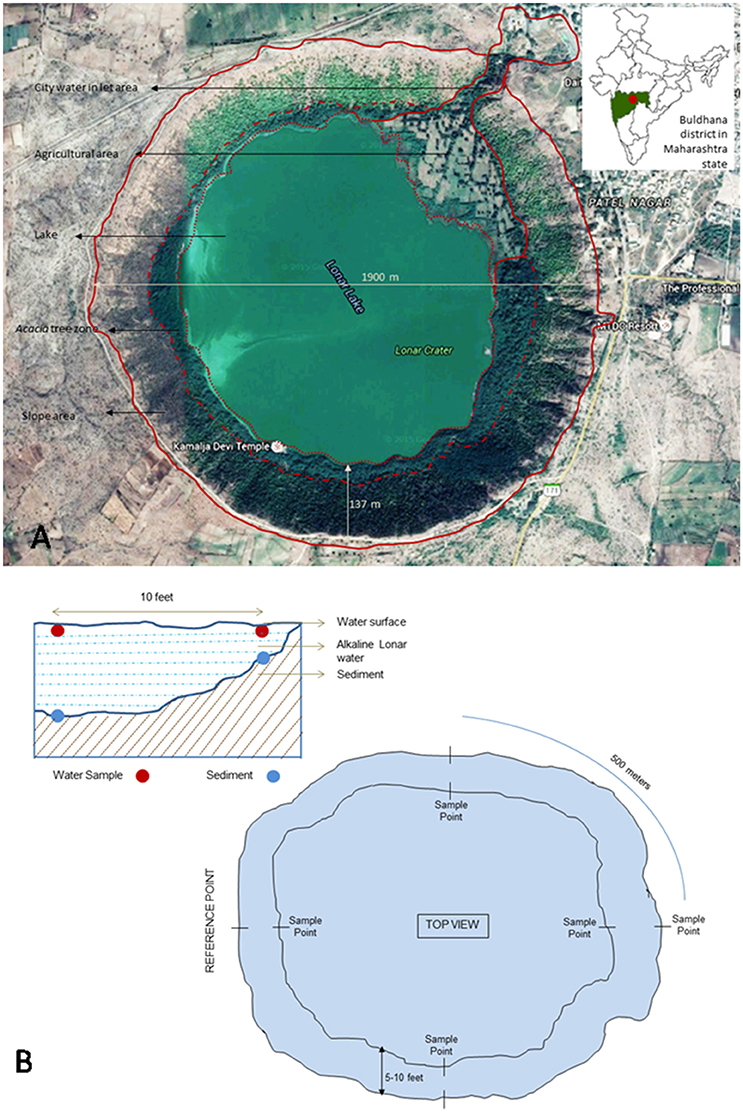
Figure 1. (A) Google image of Lonar lake and its location on Indian map (modified from google maps) (Map data: Google, DigitalGlobe). (B) Schematic representation of lake and details of sampling strategy.
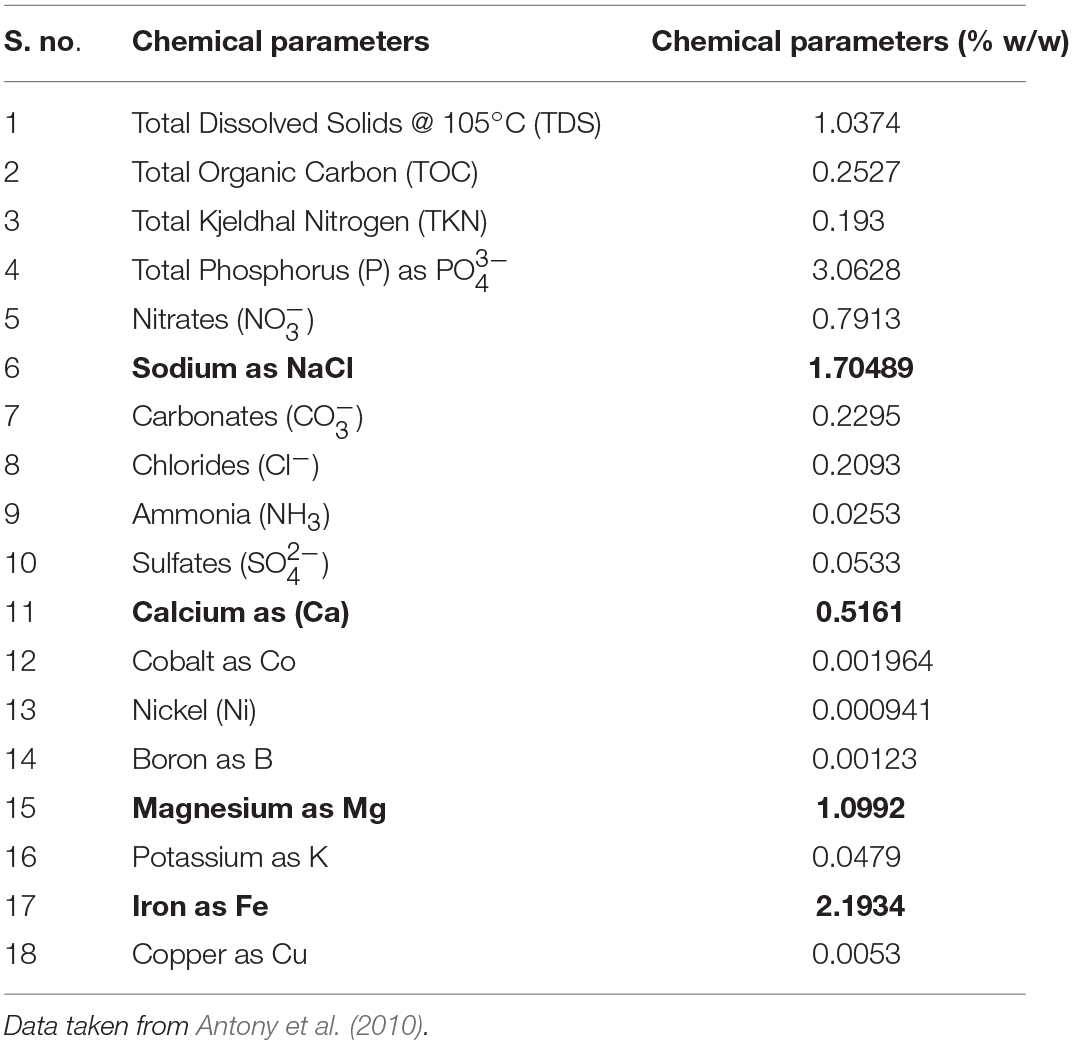
Table 1. Quantification of chemical parameters of Lonar lake sediments (figures in bold shows values of important parameters).
Isolation of Fungi
In order to isolate diverse range of fungi, an exhaustive culturomics approach with wide range of culturing conditions (culture media and temperature) were used. To mimic the natural conditions of salt and pH all the media used for isolation were prepared in Lonar lake water and adjusted to pH 10. Serial dilutions (10−1–10−10) of sediment and water were prepared in filter sterilized Lonar lake water and 100 μl of serially diluted sediment and water samples were plated on the surface of different fungal media. Based on the growth and diversity of fungi captured during preliminary screening on 32-different media (available in the laboratory), only 9 media viz., PDA, CDA, HK Medium 8A, HK Medium 13A, HK Medium 16A, HK Medium 19A, HK Medium 21A, HK Medium 22A, HK Medium 28A (Hi-Media, India) were selected for actual isolation purpose. The compositions of different HK media used for isolation purpose are given in Table ST1. Plates were incubated at two temperatures 28 ± 1°C and 35 ± 1°C up to 21 days. Morphologically different colonies were picked and purified on potato dextrose agar (PDA) plates (pH 10) using hyphal tiping method from the margin of actively growing colonies. After purification, basic morphological characteristics of the isolated fungi were recorded. Purified fungal colonies were preserved with 10% glycerol in liquid nitrogen (−196°C) and in deep freezer (−80°C) at Microbial Culture Collection division of National Centre for Cell Science, Pune India as discussed in Prakash et al. (2013).
Genomic DNA Extraction and Internal Transcribed Spacer (ITS) Region Amplification
Genomic DNA was extracted using the CTAB method as discussed in Voigt et al. (1999). In brief, small amount of fungal mycelia was suspended in 200 μl of extraction buffer containing 10 μl of proteinase K, 5 μl- β-mercaptoethanol, and glass beads in Eppendorf tube and crushed by pestle. After that, 300 μl extraction buffer was added again and vortexed for 5 min. Content was incubated for 2 h at 60°C on a dry bath. After incubation, content was mixed with 140 μl of 5 M NaCl and 64 μl of 10% CTAB and vortexed for 5 min, and incubated at 65°C for 1 h. Total content was mixed with equal volume of chloroform: isoamyl alcohol and incubated for 30 min on ice. Post precipitation, content was centrifuged at 10,000 rpm for 10 min, and aqueous phase was transferred in a fresh eppendorf tube. DNA was precipitated with 1/10th volume of 3 M sodium and washed with 70% ethanol, air dried, and suspended in 100 μl of Tris EDTA buffer. Quantity and quality of DNA was checked by NanoDrop spectrophotometer (Thermo Scientific, USA) and 1% agarose gel electrophoresis.
Fungal ITS region was amplified using universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (word5′-TCCTCCGCTTATTGATATGC-3′) as discussed in White et al. (1990). The D1/D2 region of large subunit (LSU) was amplified using primers LROR (word5′-ACCCGCTGAACTTAAGC-3′) and LR5 (5′-TCCTGAGGGAAACTTCG-3′) as per Vilgalys and Hester (1990). Standard PCR protocol and PCR cycle parameters were used for both. However, the glyceraldehyde-3-phosphate dehydrogenase (gpd) gene was amplified by primer set gpd1 (word5′-CAACGGCTTCGGTCGCATTG-3′) and gpd2 (5′-GCCAAGCAGTTGGTTGTGC-3′) as per the PCR conditions given by Berbee et al. (1999). PCR reaction was performed using 2720 thermal cycler (Applied Biosystems, US). Amplified product was checked on 1.2% agarose gel and purified by PEG- NaCl method as mentioned in Sambrook et al. (1989).
Molecular Identification and Phylogenetic Study
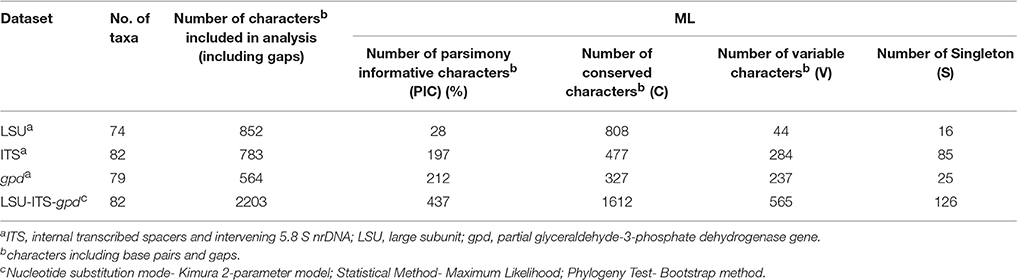
Purified PCR products were sequenced using the ABI BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA) as per manufacturer's instructions and ABI 3730xl (Applied Biosystem, USA) automated DNA sequencer. The quality of the raw sequences were checked and edited using software ChromasPro version 1.34 (Technelysium Pvt. Ltd., Tewantin, Queensland, Australia) and Sequence Scanner version 1.0 (Applied Biosystems, US). Similarity search was carried by BLASTn search with the available ITS sequences in GenBank database (Zhang et al., 2000). Taxonomic affiliations for known species were obtained by a threshold cut-off of ~97% using ITS sequences (Blaalid et al., 2013). The ITS sequences showing <97% similarity were considered as belonging to probable novel taxa. For phylogenetic analysis, published sequences of closely related organisms were retrieved in FASTA format and aligned in CLUSTAL-W. Sequence alignment and phylogenetic analysis was performed with MEGA v.5 computer program (Tamura et al., 2011). For MEF018, the assembled consensus sequences of ITS, LSU, and gpd gene were aligned separately in CLUSTAL-W using MEGA v.5. A concatenated alignment of three regions ITS-LSU-gpd was generated using MEGA v.5 and phylogenetic analysis was conducted using maximum likelihood (ML) and accuracy of the methods was assessed using 1000- bootstrap replicates. Evolutionary distances were computed using the Kimura 2-parameter method (Kimura, 1980) and are in the units of the number of base substitutions per site. Bootstrap confidence intervals were set at 50% (Saitou and Nei, 1987). The neighbor joining (NJ) and maximum parsimony (MP) analysis was also done which yielded similar topologies. All ITS region rRNA gene sequences generated from this study were submitted in NCBI GenBank database under accession numbers KT315397 - KT315422, KT315424 - KT315430, KT315432 - KT315451, KT315453 - KT315503. The D1/D2 region sequence of LSU and gpd gene sequence has been submitted to NCBI [KY007019 (gpd); KY007018 (LSU)]. The GenBank accession numbers strain ID or culture collection numbers and source of the isolates used in the phylogenetic study of genus Curvularia is compiled in Table ST2.
Screening of Strains for Phenol Oxidase Production
All the selected strains were screened for extracellular phenol oxidase production using ABTS [2, 2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid)] as substrate (Floch et al., 2007). For that, PDA medium was supplemented with 10 mM ABTS, sterilized by autoclaving and poured into Petri plates. After solidification, 5 mm diameter fungal bits were cut from actively growing pure cultures of isolated fungi using sterile Cork-borer and inoculated in the center of the plates. Plates were incubated in the dark and observed for the development of colored zone around fungal colonies (Figure S1). Colonies showing positive result for phenol oxidase production were selected for comparative study. Efficiency of the extracellular phenol oxidase production was determined using the phenol oxidase assay as discussed previously (Floch et al., 2007; Sinsabaugh, 2010). In brief, pure culture of selected fungi were raised in liquid medium and amount of phenol oxidase (unit of enzyme production / ml culture broth) was assayed at different time point using the culture broth as crude source of extracellular phenol oxidase and ABTS as substrate. Optima and range of growth at different temperatures, pH and NaCl concentrations of phenol oxidase producing fungi were tested as discussed in Lin et al. (2012). In addition the range and optima of enzyme production at different pH and temperatures was also evaluated by inoculating equal amount of inoculum in optimum medium with different pH and incubation at different temperatures. To determine the correlation between mycelial biomass and enzyme production, mycelial dry weight was also recorded. For that equal amount of fungal bits were inoculated in Erlenmeyer flask in replicate of three. Mycelia was harvested at different time interval on pre-weighted Whatman® Cellulose Filter Paper and incubated at 60°C for 24 h. Cellular biomass was recorded by reducing the weight of filter paper.
Taxonomic Characterization of Curvularia lonarenesis sp. nov. (Morphological and Physiological Study)
Macro-morphological characters like colony morphology, sporulation pattern, surface structure, type, and rate of mycelial growth, colony size, margin, pigmentation, zonation, exudation under different media, and temperature conditions were recorded as discussed in Sharma et al. (2015). Micro-morphological features were observed by mounting the fungus on lactophenol-cotton blue (Hi-Media, India) and observing under light microscope Olympus BX53 (Olympus, Japan). Photomicrographs were taken by ProgRes C5 camera (Jenoptik, USA) attached to the microscope. For Differential Interference Contrast (DIC) microscopy, the slide was observed and images captured on a fully automated upright fluorescence microscope coupled with monochrome and color CCD cameras (Olympus, Japan). DIC was performed at Indian Institute of Science Education and Research, Pune, India. In-depth taxonomical characterization of Curvularia sp. strain MEF018 was conducted as discussed in Madrid et al. (2014). Optimal growth medium was tested by screening strain MEF018 on different range of fungal media like, oat meal agar (OA), Saboraud's dextrose agar (SDA), malt extract agar (MEA), potato dextrose agar (PDA), and Czapek dextrose agar (CDA). The best medium for growth was then subjected to pH study (5–14). After keeping both these factors standard, temperature range was tested (5–40°C). As the salinity of Lonar lake is 3%, the salt tolerance was checked by growing the fungus on a range of salt concentrations (3–12%).
Results
A total of 104 isolates from different groups of fungi were cultivated from water and sediment samples of Lonar lake. Data related with percent identity and query coverage based on ITS sequences (GenBank database) along with key features of closely related fungal strains are presented in Table 2. Sequence similarity data indicated that isolates from Lonar lake showed similarity with members of 18 genera and 38 different fungal species. Except 2 isolates (Coprinopsis calospora of Basidiomycota belonged to family Psathyrellaceae), all other strains belonged to members of phylum Ascomycota. Diversity data from this study indicated that the closest relatives of our isolates showed similarity with fungal species isolated from diverse habitats and most of them are associated with alkaline and saline habitats or soda lakes (Table 2). Phylogenetic analysis using sequences from ITS and 5.8 S region of rRNA gene showed 8 ascomycetous lineages and one basidiomycetous lineage (Figure 2). In total 32 isolates from different genera including Cladorrhinum, Cladosporium, and others showed ≤97% sequence similarity with previously described fungal species and are potential novel genera or species according to the sequence based species delineation in fungal taxonomy (Blaalid et al., 2013). In addition, phylogenetic study also represented two separate clades belonging to genera Cladosporium and Cladorrhinum which contained large number of isolates. Four isolates belonged to genus Curvularia, but none of them showed close relatedness with existing members which indicated their taxonomical novelty.

Table 2. List of all the isolated fungi from Lonar lake with their closest relative in GenBank database and their specific feature reported in literature (figures in bold shows less sequence similarity of probable novel isolates).
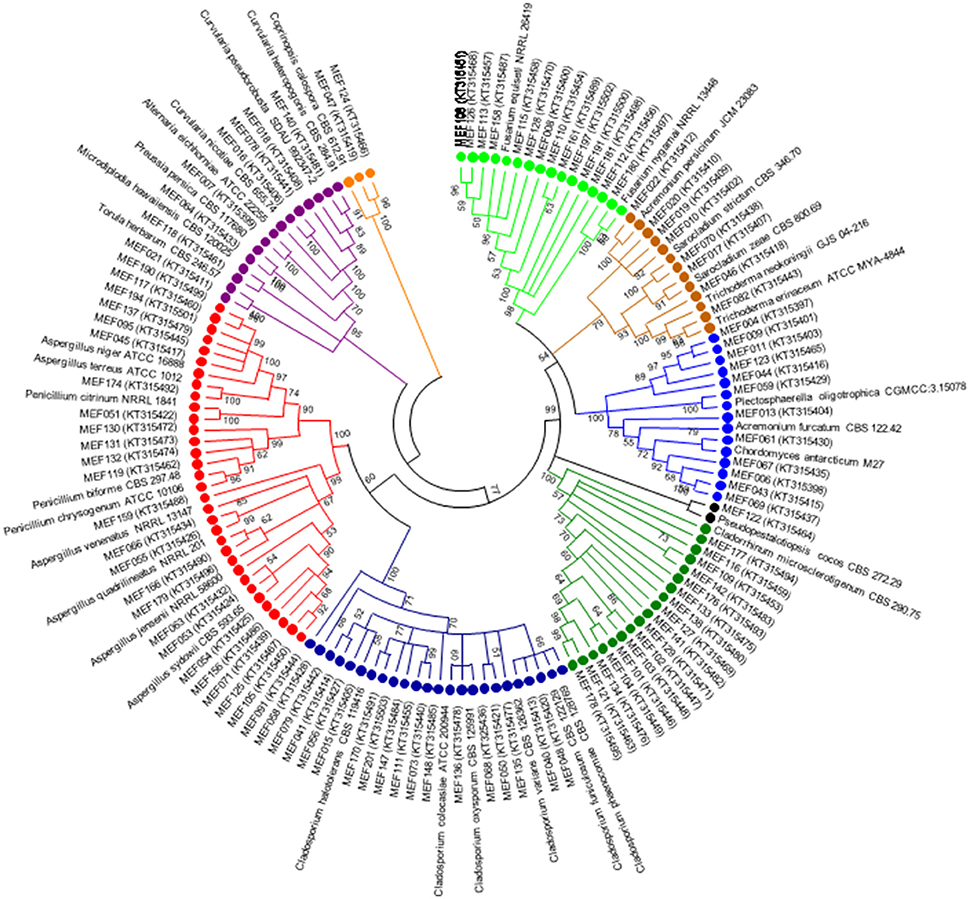
Figure 2. Phylogenetic tree constructed using ITS sequences of 104 isolated strains of fungi along with homologous sequences from type strains of closest match in GenBank. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch length = 3.96668506 is shown. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura, 1980) and are in the units of the number of base substitutions per site. The analysis involved 139 nucleotide sequences. There were a total of 659 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). ( Psathyrellaceae;
Psathyrellaceae;  Pleosporaceae;
Pleosporaceae;  Trichocomaceae;
Trichocomaceae;  Davidiellaceae;
Davidiellaceae;  Lasiosphaeriaceae
Lasiosphaeriaceae  Amphisphaeriaceae;
Amphisphaeriaceae;  Plectosphaerellaceae;
Plectosphaerellaceae;  Hypocreaceae;
Hypocreaceae;  Nectriaceae).
Nectriaceae).
In preliminary screening, 14 different strains from 4-different genera, and 3 classes showed positive result for phenol oxidase production. Result of comparative study of phenol oxidase production indicated that strain MEF018 showed maximum phenol oxidase secretion at pH 12 whereas strain MEF008, MEF109, and MEF135 showed maximum production at pH 10. The effects of salt, pH, and temperature were also studied on growth of these strains and the data are presented in Figures S2A–C. Based on their enzyme production potential at high salinity and pH, Fusarium sp. strain MEF008, Curvularia sp. strain MEF018, Cladorrhinum sp. strain MEF109, and Cladosporium sp. strain MEF135 were selected from the 14 fungal isolates (phenol oxidase producer) for further comparative study of fungal phenol oxidase (Figures S3, S4). Thus, secondary screening was conducted with only 4 strains which were maximum producer in primary screening. Result of enzyme assay indicated that Curvularia sp. strain MEF018 produced high levels of phenol oxidase followed by Cladosporium sp. strain MEF135, while Fusarium sp. strain MEF008 and Cladorrhinum sp. strain MEF109 showed nearly five time less phenol oxidase production efficiency than above two strains (Figures S5, S6). In addition Curvularia sp. strain MEF018 showed optimum phenol oxidase production potential at high pH (pH 12) and at 40°C, which is good for ecological point of view and showed that these potential of strains could be exploited in extreme condition of alkalinity, salinity and temperature (Figures S5, S6). Analysis of data indicated that strain MEF018 and strain MEF109 showed similar amount of cellular biomass at third day but showed big difference in phenol oxidase production while strain MEF135 showed lower cellular biomass than strain MEF109 but produced four to five time more enzymes. Thus, data indicated that quantity of extracellular enzyme production is not related to cellular biomass but it depends on enzyme production potential of the cells. We found that, in addition to efficient phenol oxidase production potential in extreme condition of alkalinity (pH- 12) and at high temperature (40°C) Curvularia sp. strain MEF018 showed low sequence similarity (96%) with previously isolated and characterized Curvularia pseudorobusta. It indicated that strain MEF018 is a novel extremophilic species of genus Curvularia with immense ecological and biotechnological importance and selected for further in-depth taxonomical characterization.
Phylogenetic tree constructed using concatenated alignment of ITS-LSU-gpd sequences from 81 strains of Curvularia and Bipolaris derived from Madrid et al. (2014) and Manamgoda et al. (2014) along with strain MEF018T from this study showed that it clustered with two species of Curvularia, C. hominis and C. muehlenbeckiae (Figure 3). Although the ITS sequence of strain MEF018T showed highest sequence similarity with C. pseudorobusta (96%), the gpd gene sequence showed highest sequence similarity with C. perotidis and C. neerdaardii (93%) and D1/D2 region of LSU showed no differentiation (99% similarity with most species), it formed sister clade with C. muehlenbeckiae CBS 144.63T and C. hominis UTHSC 09-464T. All three species together made separate clade in ML based phylogenetic tree. Even, use of other two methods (NJ and MP) also gave similar tree topologies (tree not included). Overall, the tree showed 8-clades. The strain C. lonarensis MEF018T belongs to clade-V along with C. hominis and C. muehlenbeckaie. The results of phylogenetic analyses of ITS, LSU and gpd gene separately as well as combined dataset along with number of bases, number of parsimony informative characters and other parameters for best substitution model is compiled and presented in Table 3. Thus, the result of phylogenetic study indicated that the strain MEF018 is distantly related with previously cultivated members of the genus Curvularia and is a novel species (Figure 3). Details of morphological, physiological and taxonomic features of the novel fungal species are discussed below.
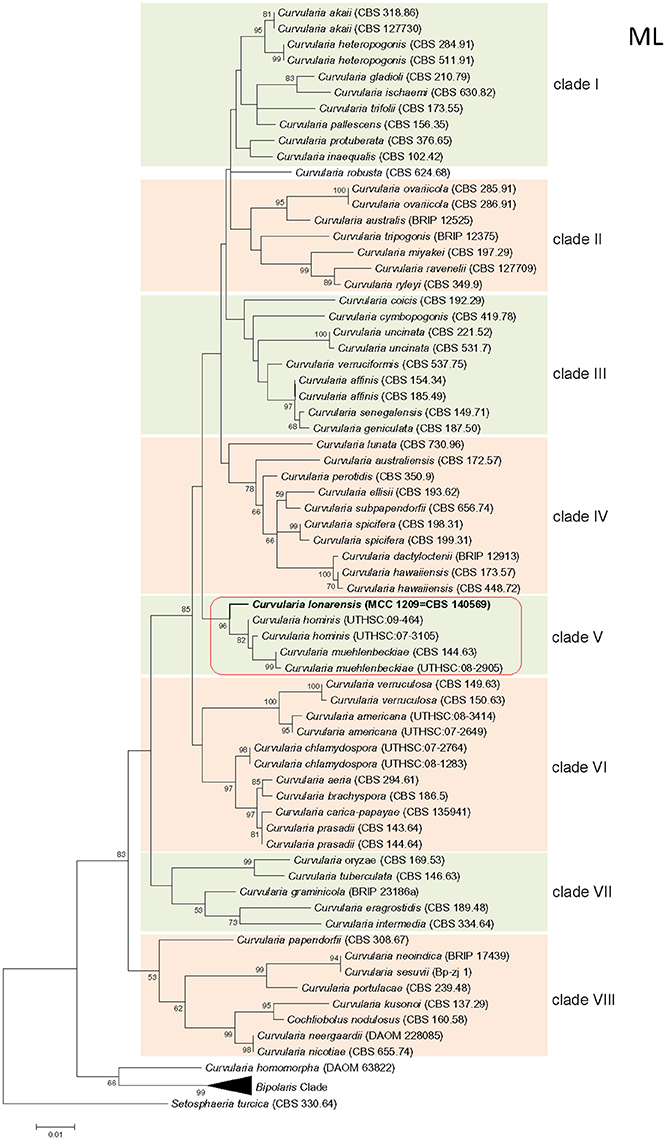
Figure 3. Maximum Likelihood (ML) tree of the Curvularia strains studied. Bootstrap support of branches indicated on the node was obtained using 1000 replicates. Only statistically significant bootstrap values (≥50%) are indicated. Branch lengths are indicated as 0.01 substitutions per positions according to the scale bar underneath the tree. Number on right side of species name denotes the strain number/ culture collection accession number. Number in parentheses denotes accession number of the sequence deposited to online database.
Description of Curvularia lonarensis Rohit Sharma and Rahul Sharma sp. nov.
Curvularia lonarensis Rohit Sharma and Rahul Sharma sp. nov. (Figure 4).

Figure 4. (A) Colony morphology of Curvularia lonarensis strain MEF018 on OA; (B–E) Morphology of conidia as captured by DIC imaging showing ornamentation on all cells of conidia (Scale bars: B–E = 10 μm); (F–K) Structural features of conidia and conidiophores of Curvularia lonarensis strain MEF018 as depicted during light microscopic imaging. (scale bars: F–K = 20 μm).
MycoBank: MB 814557.
Etymology
The epithet lonarensis is derived from the Lonar lake situated in Buldhana district of Maharashtra, India from where the fungus was isolated.
Vegetative hyphae
Vegetative hyphae are septate, branched, hyaline to sub-hyaline, smooth-walled and ranges from 3.2 to 4.9 μm (width). Conidiophores are septate, simple to branched, semi–macronematous, mononematous, straight, or flexuous, geniculate toward the apex, subhyaline to dark brown, smooth to aperculate with cell wall often thicker than those of the vegetative hyphae. Size of Conidiophores ranged from 40–306.5 μm (length) × 4.4–6.2 μm (width), with subnodulose and nodulose intercalary swellings ranging from 3.9 to 6.0 μm (width) which coincide with conidiogenous cells. Conidiogenous cells are subcylindrical to irregularly shaped and integrated with terminal and intercalary conidiophores cells. They are monopolytrectic, proliferating sympodially, and size of intercalary conidiogenous cells are ranging from 4.6–21.0 μm (length) × 3.4–5.7 μm (width) whereas the size of terminal conidiogenous cells range 8.3–14.7 μm (length). Conidia of strain MEF018 are 4–celled, asymmetrical to curve and their size ranged from 19.2–30.7 μm (length) × 10.7–15.0 μm (width). Middle cells are dark brown and usually verruculose while the cells located at terminal ends are paler, and ornamented. Chlamydospores and microconidiation were not observed.
Cultural Characteristics
It forms 66 and 77 mm wide colonies on MEA and OA respectively after 5 days of incubation at 30°C. Colonies are flat, circular, filamentous, greenish-black on OA but grayish-green on MEA with pale colored margin and lavish sporulation. Reverse side of the colonies showed blackish pigmentation with pale colored margin. Colonies on CDA and PDA are 68 and 71 mm wide respectively after 5 days at 30°C. It forms grayish-green colonies on CDA and greenish-black on PDA. Colonies on both the media looks circular, filamentous, flat, with slightly hairy mycelia, spreading, white margins, and with full of sporulation. Reverse view of the colonies have grayish-black spots. Colonies on SDA are 73 mm wide after 5 days of incubation at 30°C, looks grayish-green slightly raised, with pale colored margins, and with abundant aerial mycelia.
Sexual morph: Not observed.
Habitat: Hyper alkaline and saline Lonar lake.
Distribution: Buldhana (Maharashtra, India).
Type: INDIA, Maharashtra, Buldhana, Lonar, from water and sediment of Lonar lake, 01 Oct. 2010, Rohit Sharma (holotype CBS 140569T = MCC 1209T = MEF018T).
Gene sequences ex-holotype: KT315408 (ITS); KY007019 (gpd); KY007018 (LSU).
The genus Curvularia was first time described by Schmidt and Kunze (1817) with Curvularia lunata as type species of the genus. It is characterized by production of transversely septate conidia with dark hila, which is asymmetrically curved from middle cell. Its closest genus is Bipolaris which forms symmetrically swollen central cell (distoseptate) while sensu stricto species of Curvularia lack this feature. Traditionally, both the genera (Bipolaris and Curvularia) were distinguished by conidial features but molecular data have now confirmed their positions in the family Pleosporaceae order Pleosporales, (Zhang et al., 2009, 2012). Manamgoda et al. (2012) reclassified several Bipolaris and Curvularia species based on phylogeny of ITS, LSU, gpd, tef sequences. Following re-classification, some of the plant pathogenic species of Bipolaris were shifted to Curvularia (da Cunha et al., 2013; Madrid et al., 2014). Even the genus Pseudocochliobolus was merged in Curvularia with type species P. nisikadoi now described as Curvularia coicis. Manamgoda et al. (2014) revised the generic boundaries between Bipolaris and Curvularia based on ITS and gpd phylogenetic tree. Similar to the present study, Manamgoda et al. (2012); Manamgoda et al., 2014 found two major groups, one group includes species of Bipolaris and other group include species of Curvularia (having 8 clades). Thus, the phylogenetic analysis (Figure 3) clearly shows that the sequences of ITS-LSU-gpd combined dataset resolves the two genera as well as species within individual genus. Although single name of both morphs have reduced the complexities in fungal taxonomy of many genera, lack of authentic, curated database of sequences had made it difficult for correct identification of species (Sharma, 2012).
According to our observation the strain MEF018 belong to the genus Curvularia because it forms curved conidia with dark enlarged, central cell and double layered wall. As per Manamgoda et al. (2012), conidia of Curvularia can be straight or curved. When curved, the conidia have enormously enlarged intermediate cells contributing to their curvature. Similar to C. americana, C. tuberculata and C. verruculosa the surface of the conidia of the strain MEF018 is rough (Figures 4B–E). The conidia of the strain MEF018 is slightly larger (19–30 × 10–15 μm) than the conidia of C. americana (13–28 × 7–15 μm) but smaller than C. tuberculata (23–52 × 13–20 μm) and C. verruculosa (20–40 × 12–17 μm). Differential features of MEF018 with phylogenetically closest relatives of the genus Curvularia are given in the Table 4.
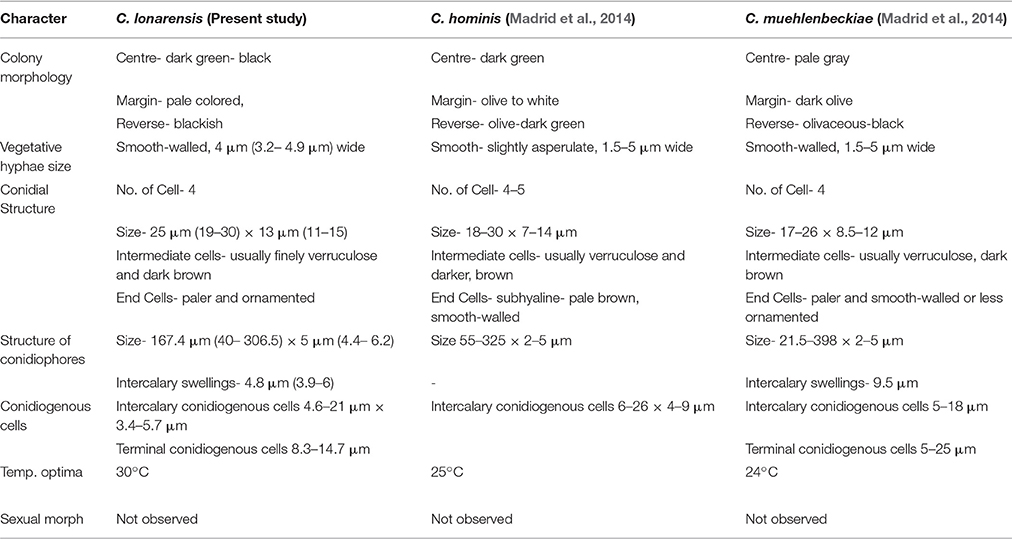
Table 4. Comparison of morphological characters of Curvularia lonarensis, C. hominis, C. muehlenbeckiae.
Phylogenetic analysis of combined dataset dissects the available species of the genus Curvularia in to eight main clades (Figure 3). Study of tree topology indicated that strain MEF018 showed distant relationship with other existing members of the genus Curvularia and clustered with C. hominis and C. muehlenbeckiae with strong bootstrap support (96%). All the three species possess warted or verruculose conidia. The species of Curvularia with warted or verruculose conidia appear in different clades outside C. hominis-clade (C. hominis, C. muehlenbeckiae, strain MEF018), which suggests polyphyletic origin of conidial ornamentation in genus Curvularia (Madrid et al., 2014). The C. hominis and C. muehlenbeckiae species are isolated from human and leaf of Muehlenbeckia plant respectively growing optimally at 24°C. Whereas, the strain MEF018 is only isolate among the clade which is isolated from a soda lake and grows at high pH and 30°C temperature. The close clustering of 3 Curvularia species belonging to different habitat indicates that ecological gradient may not be a factor in differentiating these species, hence not correlated with fungal diversity. We also observed that the clustering pattern of the phylogenetic tree supports the morphological data of the conidia in the genus Curvularia. The clade -IV contains mostly 4-celled, conspicuously distoseptate conidia with darker middle cell. They are mostly curved at the third cell from the base, and larger in size than conidia of the members of the others clade. Morphologically, strain MEF018 is closer to clade IV (consisting of C. lunata) as it forms curved conidia with darker third cell. Hence, in the present study, phylogeny of combined dataset along with morphological details gives good resolution to distinguish the strain MEF018 from its close relatives and proves that strain MEF018 is a novel species of the genus Curvularia.
Strain MEF018 showed optimum growth on OA among the tested fungal growth media. It tolerated 10% NaCl concentration in the medium but showed optimum growth at 1% (Figure S2a). It showed positive growth between pH 5–14 with optimal mycelial growth at pH- 11 (Figure S7). Evaluation of the pH of the medium after fungal growth indicated that strain MEF018 secreted some metabolites which shift the pH of the growth medium toward its optima (pH 10–11). Temperature range for the growth was between 10 and 40°C with optimum growth at 30°C. Thus, in conclusion based on morphological, physiological and phylogenetic details strain MEF018 sufficiently delineates with existing members of the genus Curvularia and proposed as Curvularia lonarensis Rohit Sharma & Rahul Sharma sp. nov. In conclusion, the above described novel fungus is an important finding of current study because till date no Curvularia species with phenol oxidase producing potential has been reported from hyper alkaline and saline habitats.
Discussion
Lonar lake is a soda lake located in Buldhana district of Maharashtra, India but understudied in terms of fungal diversity. Available literature indicated that except a single report on keratinophillic fungi from the soil of slope by Deshmukh and Verekar (2006) very little or no information on the soda lake fungi from Lonar lake are available. Generally, fungi prefer acidic, or neutral pH, and very few data are available on extremophilic fungi especially from soda lakes. Therefore, isolation of alkaliphilic fungi from Lonar lake and other such habitats would be beneficial to improve the database as well as for their future commercial exploitation. Due to commercial and physiological importance of extremophiles fungi several other groups are working on this aspect from various soda lakes across the world (Table 5). Our study also demonstrated that a wide range of fungal diversity inhabit in the hyper alkaline and saline habitat of Lonar lake. Furthermore, all the fungal isolates recovered from Lonar lake are members of the Dikarya, most of them (98%) belong to Ascomycota, and are distributed throughout the sub-phylum Pezizomycotina (Figure 5). Phylogenetic analyses reveals that alkaliphilic trait is widely distributed among the various sub-phyla of Ascomycota suggesting that diverse groups of fungi have the potential to adopt themselves in extremophilic conditions of the various soda lakes. While, phylum Basidiomycota was represented by a single member Coprinopsis sp. (2 strains) belonging to family Psathyrellaceae. The species of Coprinopsis generally inhabits terrestrial habitat growing on either coprophilous or lignicolous substrate. Strains MEF047 and MEF124 showed closest similarity with Coprinopsis calospora (≡ Coprinopsis calosporus) which was isolated from a stem in flowerpot from Netherlands. In Lonar lake it is possible that they were associated with some dead wood debris inside the lake. Sequence similarity data from present study indicated that most of the strains isolated showed similarity with previously characterized strains isolated from alkaline, saline, or other extreme environments (Table 2) and confirm that the isolated strains are native to the lake samples and not contaminant of isolation procedures. In addition, out of 38 species reported from current study, 12 are putative novel (involving 32 strains) based on the current criteria (≤97% sequence similarity with closest relative) set for fungal species delineation by Blaalid et al. (2013), which suggest that Lonar lake is an important reservoir for the ecologically and economically important fungi and need further investigation in terms of physiology and genetics to explore their role in biogeochemical cycling and to get the valuable products of industrial importance.
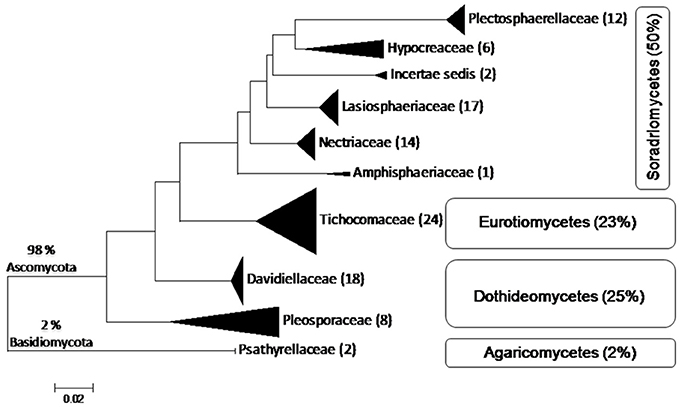
Figure 5. Schematic representation of placement of 104 isolates belonging to 38 species derived from ITS sequences of fungi from Lonar lake, Buldhana, Maharashtra, India. Classification follows Hibbett et al. (2007). All isolates belong to Ascomycota except 2% belonging to Basidiomycota. The isolates of Ascomycota are distributed among three classes, Sordariomycetes, Dothideomycetes, and Eurotiomycetes. Percentages indicate the total number of isolates out of the 104 isolated fungi.
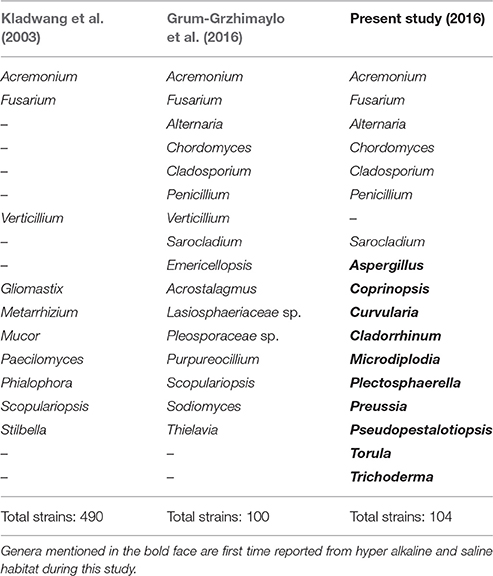
The Table 6 shows diversity of fungal species isolated from various hyper alkaline and hyper saline environments across the world along with data on the habitats of isolation. Among the isolated strains in present study, members of Acremonium, Alternaria, Chordomyces, Cladosporium, Fusarium, Penicillium have also been isolated from other soda lakes supported the findings of our study. In addition a total of 10 genera including Aspergillus, Cladorrhinum, Coprinopsis, Curvularia, Microdiplodia, Plectosphaerella, Preussia, Pseudopestalotiopsis, Torula, and Trichoderma were unique to this study and reported from Lonar lake only (Table 5). We also observed that Cladorrhinum with 17 different isolates dominate among the isolated strain which is not reported from any soda lake in earlier studies (Tables 5, 6). Furthermore, studies conducted on soda lakes from other part of the world reported lesser diversity than reported in current study (Kladwang et al., 2003; Grum-Grzhimaylo et al., 2016). Thus, our diversity data indicated that Lonar lake harbor wide range of unique fungal diversity and also indicated that these strains are indigenous to alkaline habitat of Lonar lake.

Table 6. Fungal species isolated from various hyper alkaline and hyper saline environments around world with the data on the habitats they were isolated from.
Being an active decomposer fungi are the crucial component of carbon cycling and play active role in global climate change and carbon sequestration (Thormann, 2006). Fungal extracellular phenol oxidase/peroxidase is an important class of enzyme due to its participation in the degradation of lignin and toxic phenolics. The ABTS is a commonly used substrate to detect the activity of phenol oxidase (PO) in biological samples due to rapid oxidation of ABTS in colored blue-green ABTS+ by phenol oxidase. Although several different species of fungi with extracellular phenol oxidase producing potential have been isolated and characterized from different habitats (Szklarz et al., 1989; Crognale et al., 2012) but study on isolation and characterization of fungi with phenol oxidase producing potential from soda lake habitat are lacking. Isolation and characterization of phenol oxidase producing fungi from hyperalkaline and saline habitats like Lonar lake has special ecological significance because most of the contaminated habitat and industrial effluents have high alkalinity and salinity and fungal agent with survival potential in extreme condition with active phenol oxidase production potential can be used as bioinoculant for bioaugmentation based bioremediation. Furthermore, detection of active enzyme secretion in extreme conditions (high pH and salinity) indicates their probable role in the degradation of complex organics like leaf-litter, plant debris and detoxification of phenolics present in lake ecosystem and contribution in carbon turnover of the lake. Recently, Vavourakis et al. (2016) and Ausec et al. (2011) have shown in metagenomic studies of various soda lake that many uncultured fungi have laccases-like Cu-oxidase encoded which may be involved in degradation of phenolic compounds. Hence, this is the first culture based study on Soda lake showing phenolic compound degradation capacities of fungal isolates from such habitat.
Geographical location of Lonar lake (Figure 1) indicated that it is almost a closed ecosystem with only inflow of water by the surface run-off and discharge from a village effluent by small creek. It contains high levels of dissolved organic matter (DOM), high pH (pH 10), high salinity and high content of iron, magnesium and phosphorus which is generally considered not suitable for fungal growth. Despite unfavorable conditions for fungal growth, occurrence of wide range of fungi from diverse genera with polyphyletic affiliations with upland fungal species indicated that fungal flora of the Lonar lake are not a real habitants of the lake ecosystem. They might have arrived to the lake from outside in the form of spores and fruiting bodies. After that they adopted and established themselves according to the geographical conditions of the lake during the course of evolution and contributing in the functionality of the lake ecosystem. The present study provides first hand information about the diversity of fungi from hyperalkaline and saline Lonar lake. It is the first extensive investigation of the fungal diversity of Lonar lake and their physiological and functional potential in the lake. It will be interesting to study in detail the functional role of these fungi in the ecophysiology of lake habitat and mechanism by which they are able to tolerate and survive the extreme environment. Moreover, in depth sampling of the lake over time and area would help to study the spatial and temporal diversity and their impact on the lake ecology.
Author Contributions
RoS and OP conceived the study and also involved in sample collection, isolation and MS writing. RaS did phylogenetic study, novel species identification, and MS writing. PG, MS, and YN did molecular and enzymatic work.
Funding
This work was supported by the Department of Biotechnology, New Delhi, India (BT/PR10054/NDB/52/94/2007). Senior Research Associateship of RaS is supported by the Council of Scientific and Industrial Research, New Delhi, India (CSIR Pool No. 8766-A).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Mr. Vishal Thite and Mr. Sandeep Walujkar, MCC, Pune are acknowledged for their help during sampling at Lonar lake. Ministry of Environment and Forest (MoEF) is duly acknowledged for sampling of Lonar-Lake, Buldhana.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01847/full#supplementary-material
References
Antony, C. P., Kumaresan, D., Ferrando, L., Boden, R., Moussard, H., Scavino, A. F., et al. (2010). Active methylotrophs in the sediments of Lonar lake, a saline and alkaline ecosystem formed by meteor impact. ISME J. 4, 1470–1480. doi: 10.1038/ismej.2010.70
Ausec, L., Zakrzewski, M., Goesmann, A., Schlüter, A., and Mandic-Mulec, I. (2011). Bioinformatic analysis reveals high diversity of bacterial genes for laccase-like enzymes. PLoS ONE 6:e25724. doi: 10.1371/journal.pone.0025724
Baldrian, P. (2006). Fungal laccases–occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. doi: 10.1111/j.1574-4976.2005.00010.x
Berbee, M. L., Pirseyedi, M., and Hubbard, S. (1999). Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91, 964–977. doi: 10.2307/3761627
Blaalid, R., Kumar, S., Nilsson, R. H., Abarenkov, K., Kirk, P. M., and Kauserud, H. (2013). ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 13, 218–224. doi: 10.1111/1755-0998.12065
Brown, S. P., Olson, B. J. S. C., and Jumpponen, A. (2015). Fungi and Algae Co-Occur in snow: an issue of shared habitat or algal facilitation of heterotrophs? Arct. Antarct. Alp. Res. 47, 729–749. doi: 10.1657/AAAR0014-071
Burgaud, G., Arzur, D., Durand, L., Cambon-Bonavita, M.-A., and Barbier, G. (2010). Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol. Ecol. 73, 121–133. doi: 10.1111/j.1574-6941.2010.00881.x
Calvez, T., Burgaud, G., Mahé, S., Barbier, G., and Vandenkoornhuyse, P. (2009). Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 75, 6415–6421. doi: 10.1128/AEM.00653-09
Chaput, D. L., Hansel, C. M., Burgos, W. D., and Santelli, C. M. (2015). Profiling microbial communities in manganese remediation systems treating coal mine drainage. Appl. Environ. Microbiol. 81, 2189–2198. doi: 10.1128/AEM.03643-14
Crognale, S., Pesciaroli, L., Petruccioli, M., and D'Annibale, A. (2012). Phenoloxidase-producing halotolerant fungi from olive brine wastewater. Process Biochem. 47, 1433–1437. doi: 10.1016/j.procbio.2012.05.014
da Cunha, K. C., Sutton, D. A., Fothergill, A. W., Gené, J., Cano, J., Madrid, H., et al. (2013). In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn. Microbiol. Infect. Dis. 76, 168–174. doi: 10.1016/j.diagmicrobio.2013.02.034
Das, B. K., Roy, A., Koschorreck, M., Mandal, S. M., Wendt-Potthoff, K., and Bhattacharya, J. (2009). Occurrence and role of algae and fungi in acid mine drainage environment with special reference to metals and sulfate immobilization. Water Res. 43, 883–894. doi: 10.1016/j.watres.2008.11.046
Deshmukh, S. K., and Verekar, S. (2006). Keratinophilic fungi from the vicinity of meteorite crater soils of Lonar (India). Mycopathologia 162, 303–306. doi: 10.1007/s11046-006-0044-7
Floch, C., Alarcon-Gutiérrez, E., and Criquet, S. (2007). ABTS assay of phenol oxidase activity in soil. J. Microbiol. Methods 71, 319–324. doi: 10.1016/j.mimet.2007.09.020
Georgieva, M. L., Lebedeva, M. P., and Bilanenko, E. N. (2012). Mycelial fungi in saline soils of the western Transbaikal region. Eurasian Soil Sci. 45, 1159–1168. doi: 10.1134/S1064229312120058
Grum-Grzhimaylo, A. A., Debets, A. J., van Diepeningen, A. D., Georgieva, M. L., and Bilanenko, E. N. (2013a). Sodiomyces alkalinus, a new holomorphic alkaliphilic ascomycete within the Plectosphaerellaceae. Persoonia 31, 147–158. doi: 10.3767/003158513X673080
Grum-Grzhimaylo, A. A., Georgieva, M. L., Bondarenko, S. A., Debets, A. J. M., and Bilanenko, E. N. (2016). On the diversity of fungi from soda soils. Fungal Divers 76, 27–74. doi: 10.1007/s13225-015-0320-2
Grum-Grzhimaylo, A. A., Georgieva, M. L., Debets, A. J., and Bilanenko, E. N. (2013b). Are alkalitolerant fungi of the Emericellopsis lineage (Bionectriaceae) of marine origin? IMA Fungus 4, 213–228. doi: 10.5598/imafungus.2013.04.02.07
Hibbett, D. S., Binder, M., Bischoff, J. F., Blackwell, M., Canon, P. F., Eriksson, O. E., et al. (2007). A higher-level phylogenetic classification of the fungi. Mycol. Res. 111, 509–547. doi: 10.1016/j.mycres.2007.03.004
Horikoshi, K. (1991). “Isolation and classification of alkalophilic microorganisms,” in Microorganisms in Alkaline Environments, ed K. Horikoshi (Tokyo; Kodansha Limited), 15–24.
Jayani, R. S., Saxena, S., and Gupta, R. (2005). Microbial pectinolytic enzymes: a review. Process Biochem. 40, 2931–2944. doi: 10.1016/j.procbio.2005.03.026
Johnson, H. W. (1923). Relationships between Hydrogen Ion, Hydroxyl Ion and Salt Concentrations and the Growth of Seven Soil Molds. Research Bulletin No. 76. Ames: Iowa.
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kladwang, W., Bhumirattana, A., and Hywel-Jones, N. (2003). Alkaline-tolerant fungi from Thailand. Fungal Divers 13, 69–84.
Lin, X., Green, S., Tfaily, M. M., Prakash, O., Konstantinidis, K. T., Corbett, J. E., et al. (2012). Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the Glacial lake agassiz peatland. Appl. Environ. Microbiol. 78, 7023. doi: 10.1128/AEM.01750-12
Liu, R., Jiang, X., Mou, H., Guan, H., Hwang, H., and Li, X. (2009). A novel low-temperature resistant alkaline lipase from a soda lake fungus strain Fusarium solani N4-2 for detergent formulation. Biochem. Eng. J. 46, 265–270. doi: 10.1016/j.bej.2009.05.016
Madrid, H., da Cunha, K. C., Gené, J, Dijksterhuis, J., Cano, J., Sutton, D. A., et al. (2014). Novel Curvularia species from clinical specimens. Persoonia 33, 48–60. doi: 10.3767/003158514X683538
Manamgoda, D. S., Cai, L., McKenzie, E. H. C., Crous, P. W., Madrid, H., Chukeatirote, E., et al. (2012). A phylogenetic and taxonomic re-evaluation of the Bipolaris–Cochliobolus–Curvularia complex. Fungal Divers 56, 131–144. doi: 10.1007/s13225-012-0189-2
Manamgoda, D. S., Rossman, A. Y., Castlebury, L. A., Crous, P. W., Madrid, H., Chukeatirote, E., et al. (2014). The genus Bipolaris. Stud. Mycol. 79, 221–288. doi: 10.1016/j.simyco.2014.10.002
Nagai, K., Sakai, T., Rantiatmodjo, R. M., Suzuki, K., Gams, W., and Okada, G. (1995). Studies on the distribution of alkalophilic and alkalitolerant soil fungi I. Mycoscience 36, 247–256. doi: 10.1007/BF02268598
Nagai, K., Suzuki, K., and Okada, G. (1998). Studies on the distribution of alkaliphilic and alkali-tolerant soil fungi II: fungal flora in two limestone caves in Japan. Mycoscience 39, 293–298. doi: 10.1007/BF02464011
Okada, G., Niimura, Y., Sakata, T., Uchimura, T., Ohara, N., Suzuki, H., et al. (1993). Acremonium alcalophilum, a new alkalophilic cellulolytic hyphomycete. Trans. Mycol. Soc. Jpn 34, 171–185.
Oren, A., and Gunde-Cimerman, N. (2012). Fungal life in the Dead Sea. Prog. Mol. Subcell. Biol. 53, 115–132. doi: 10.1007/978-3-642-23342-5_6
Ostergaard, L. H., and Olsen, H. S. (2011). “Industrial applications of fungal enzymes,” in Industrial Applications- Mycota, Vol 10, ed M. Hofrichter (Berlin; Heidelberg: Springer), 269–290.
Prakash, O., Nimonkar, Y., and Shouche, Y. S. (2013). Practice and prospects of microbial preservation. FEMS Microbiol. Lett. 339, 1–9. doi: 10.1111/1574-6968.12034
Saitou, N., and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Sharma, R. (2012). Urgent need for authentic (derived from type or typified material) ITS sequence database for all fungi. Curr. Sci. 103, 1270–1272.
Sharma, R., Polkade, A. V., and Shouche, Y. S. (2015). “Species Concept” in microbial taxonomy and systematics. Curr. Sci. 108, 1804–1814.
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404. doi: 10.1016/j.soilbio.2009.10.014
Szklarz, G. D., Antibus, R. K., Sinsabaugh, R. L., and Linkiins, A. E. (1989). Production of phenol oxidases and peroxidises by wood-rotting fungi. Mycologia 81, 234–240. doi: 10.2307/3759705
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thormann, M. N. (2006). Diversity and function of fungi in peatlands: a carbon cycling perspective. Can. J. Soil Sci. 86, 281–293. doi: 10.4141/S05-082
Vargas, V. A., Delgado, O. D., Hatti-Kaul, R., and Mattiasson, B. (2004). Lipase-producing microorganisms from a Kenyan alkaline soda lake. Biotechnol. Lett. 26, 81–86. doi: 10.1023/B:BILE.0000012898.50608.12
Vavourakis, C. D., Ghai, R., Rodriguez-Valera, F., Sorokin, D. Y., Tringe, S. G., Hugenholtz, P., et al. (2016). Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline Soda Lake brines. Front. Microbiol. 7:211. doi: 10.3389/fmicb.2016.00211
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4239–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Voigt, K., Cigelnik, E., and O'Donnell, K. (1999). Phylogeny and PCR identification of clinically important zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 37, 3957–3964.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA; Academic Press), 315–322.
Zak, J. C., and Wildman, H. G. (2004). “Fungi in stressful environments,” in Biodiversity of Fungi, Inventory, and Monitoring Methods, eds G. M. Mueller, G. F. Bills and M. S. Foster (London: Elsevier/Academic), 303–331.
Zhang, Y., Crous, P. W., Schoch, C. L., and Hyde, K. D. (2012). Pleosporales. Fungal Divers 53, 1–221. doi: 10.1007/s13225-011-0117-x
Zhang, Y., Schoch, C. L., Fournier, J., Crous, P. W., de Gruyter, J., Woudenberg, J. H., et al. (2009). Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Stud. Mycol. 64, 85–102. doi: 10.3114/sim.2009.64.04
Keywords: Curvularia, fungal diversity, extremophilic fungi, lonar lake, phenol oxidase, soda lake
Citation: Sharma R, Prakash O, Sonawane MS, Nimonkar Y, Golellu PB and Sharma R (2016) Diversity and Distribution of Phenol Oxidase Producing Fungi from Soda Lake and Description of Curvularia lonarensis sp. nov. Front. Microbiol. 7:1847. doi: 10.3389/fmicb.2016.01847
Received: 30 August 2016; Accepted: 03 November 2016;
Published: 22 November 2016.
Edited by:
Martin G. Klotz, Queens College (CUNY), USAReviewed by:
Bhim Pratap Singh, Mizoram University, IndiaRavindra Nath Kharwar, Banaras Hindu University, India
Copyright © 2016 Sharma, Prakash, Sonawane, Nimonkar, Golellu and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohit Sharma, cm9oaXRAbmNjcy5yZXMuaW4=
Om Prakash, b21wcmFrYXNoQG5jY3MucmVzLmlu
†These authors have contributed equally to this work.
 Rahul Sharma1†
Rahul Sharma1† Om Prakash
Om Prakash Mahesh S. Sonawane
Mahesh S. Sonawane Yogesh Nimonkar
Yogesh Nimonkar