- 1Department of Plant Pathology and Crop Physiology, Louisiana State University Agricultural Center, Baton Rouge, LA, USA
- 2Department of Plant Pathology and Weed Research, The Volcani Center-ARO, Bet-Dagan, Israel
- 3Department of Plant Sciences, North Dakota State University, Fargo, ND, USA
Introduction
Common bean (Phaseolus vulgaris L.) is the most important grain legume for direct human consumption worldwide and represents a rich source of protein, vitamins, minerals, and fiber (Broughton et al., 2003). The recent sequencing of the common bean genome, together with the availability of genomic and transcriptomic data have provided useful information to common bean breeders that will help in the development of genotypes with desirable characteristics (Schmutz et al., 2014; Vlasova et al., 2016).
Endornaviruses are persistent viruses with a non-encapsidated RNA genome that ranges from 9.8 to 17.6 kb, infect plants, fungi, and oomycetes, are transmitted only via gametes, and do not cause apparent symptoms (Stielow et al., 2011; Fukuhara and Gibbs, 2012). Although endornaviruses have been reported in several economically important plant species, little is known about the effect they have on their hosts. One of the major obstacles to study their effect to the host is the lack of a transmission method. In plants, endornaviruses do not move from cell to cell and spread only during cell division.
Recently, Khankhum et al. (2015) reported that most common bean genotypes of Mesoamerican origin are double-infected with Phaseolus vulgaris endornavirus 1 (PvEV1) and Phaseolus endornavirus 2 (PvEV2); in contrast, genotypes of Andean origin are often endornavirus-free. Black Turtle Soup (BTS), a cultivar of Mesoamerican origin has been reported to be double-infected by these two endornaviruses (Okada et al., 2013). A BTS endornavirus-free selection (BTS−), obtained from an endornavirus-infected BTS (BTS+) seed lot has been reported by Okada et al. (2013). To establish the bases for future research on the role that endornaviruses play in the common bean plant, and the effect these viruses have on the host gene expression, we conducted RNAseq on two BTS lines: one endornavirus-infected and the other endornavirus-free.
Value of the Data
Currently, there are no sources of gene annotation for any organism infected with endornaviruses. This information will be helpful in determining the nature of the symbiotic interaction between endornaviruses and their host; more specifically between Mesoamerican common bean and PvEV1 and PvEV2.
These data may help to identify relevant genes in common bean that are differentially expressed under endornavirus infections.
Materials and Methods
Library Preparation and Transcriptome Sequencing
Seeds from the BTS− selection and seeds from a BTS+ plant obtained in previous investigations (Okada et al., 2013) were increased at least three generations by self-pollination. Crosses using the BTS+ selection as male and the BTS− as female were conducted in the greenhouse facilities of the Department of Plant Sciences, North Dakota State University, Fargo, ND. From the F1 generation, a plant double-infected with PvEV1 and PvEV2 designated BTS+ 3 was selected and increased two generations. The original BTS− line was increased two generations and designated BTS− 4. For the detection of the two viruses in the plants selected for the RNAseq, we used two methods reported in previous investigations, electrophoretic analysis of extracted viral dsRNA and RT-PCR using specific primers for each virus (Khankhum et al., 2015, 2016). Seeds of each line were planted under controlled temperature (25°C) and light (16 h photoperiod) conditions. Three weeks after planting, 100 mg of leaf tissue (trifoliate leaves) was collected, placed in a 1.5 ml nuclease-free microcentrifuge tube, and immediately submerged in liquid nitrogen. Samples were kept at −70°C until ready for RNA extraction. Total RNA was extracted following the extraction procedure of the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO). Collected leaf tissues were ground in liquid nitrogen using a micro-pestle. To eliminate residual DNA contamination, the RNA was DNase treated using the On-Spin Column DNase I Kit (MO BIO Laboratory, Inc., Carlsbad, CA) following the manufacturers' directions. Total RNA was eluted out from the column using nuclease-free water. The quantity and quality of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples were placed in RNAstable® (Biomatrica Inc., San Diego, CA) tubes and shipped for sequencing. RNA sequencing was conducted by SeqMatic (SeqMatic, Fremont, CA). A total of six RNA libraries, three from individual plants of BTS− 4 and three from individual plants of BTS+ 3, were prepared using Illumina TruSeq Stranded Total RNA Library Prep Kit (Illumina, Diego, CA) and sequenced using the Illumina Hiseq2500 platform to generating 50 bp single-end reads.
Bioinformatics Analysis
The reference genome of common bean (P. vulgaris) version 1.0 (Schmutz et al., 2014) was downloaded from the Phytozome website (Goodstein et al., 2012). Six RNAseq libraries of BTS common bean, three double-infected with PvEV1 and PvEV2 and three endornavirus-free were mapped to the reference genome using bowtie software (Langmead and Salzberg, 2012). Quantification of the transcript expression was conducted using RSEM method (RNA-Seq by Expectation Maximization) (Li and Dewey, 2011). Differential expression analysis was done using R bioconductor package edgeR (Robinson et al., 2010). To associate sequences and gene expression data with biological functions, gene ontology (GO) distribution analysis was conducted using Blast2GO (Conesa et al., 2005).
Results
Differential expression analysis of RNAseq data revealed that a total of 132 genes were differentially expressed. In the endornavirus-infected line 84 genes were down-regulated while 48 genes up-regulated (Supplementary Tables 1, 2). Figures 1A,B shows a visual reference of the differentially expressed gene vs. samples heatmap and Pearson correlation heatmap. GO distribution data on up-regulated and down-regulated genes is provided as excel files in Data Sheets 1 and 2 respectively in Supplementary Material. Gene ontology distribution show that oxidation-reduction processes were the main process associated with endornavirus infection. Reduction–oxidation (redox) changes have been reported to be associated with plant response to pathogen infection (Frederickson Matika and Loake, 2014), environmental stresses, development, and acclimation (Dietz, 2014; Dietz et al., 2016; Carmody et al., 2016). Data Sheets 3–5 contain excel files with expression levels, p-values, and FPKM (fragments per kilobase of transcript per million mapped reads) values respectively for all genes of the virus-infected and virus-free plants.
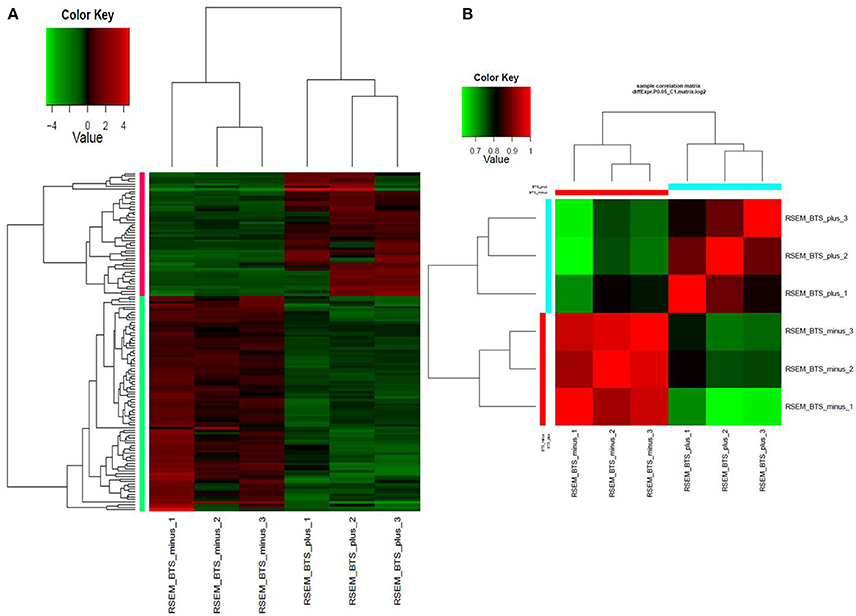
Figure 1. (A) Heatmap of normalized expression matrix of the six RNAseq libraries. Highly expressed genes are in red while low expressed genes are in green. (B) Heatmap of the Pearson correlation between the expression levels of differentially expressed genes. Red color marks highly correlated samples while green color marks low correlation. BTS_minus, endornavirus-free sample; BTS_plus, endornavirus-infected sample.
Direct Link to Deposited Data and Information to Users
Raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/srp090495) under accession SRP090495.
Authors Contributions
RV: conceived the idea and selected the original endornavirus-free BTS plant. SK: conducted the experiments; NS: performed the bioinformatics analysis of the data; JO: conducted the BTS crosses and contributed to the idea; RV, NS: wrote the manuscript. All authors contributed to the review of the manuscript.
Funding
This research was conducted with partial support from research grant No. US-4725-14 F from BARD, the United States—Israel Binational Agricultural Research and Development Fund.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to acknowledge the financial support to RV and NS from BARD and the USDA National Institute of Food and Agriculture and the Louisiana Soybean and Grain Research and Promotion Board for partial support to RV.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01905/full#supplementary-material
Abbreviations
BARD, Binational Agricultural Research and Development; BTS, Black Turtle Soup; BTS−, Black Turtle Soup endornavirus-free; BTS+, Black Turtle Soup endornavirus-infected; FPKM, Fragments per kilobase of transcript per million mapped reads; GO, Gene ontology; PvEV1, Phaseolus vulgaris endornavirus 1; PvEV2, Phaseolus vulgaris endornavirus 2.
References
Broughton, W. J., Hernández, G., Blair, M., Beebe, S., Gepts, P., and Vanderleyden, J. (2003). Beans (Phaseolus spp.)-model food legumes. Plant Soil 252, 55–128. doi: 10.1023/A:1024146710611
Carmody, M., Waszczak, C., Idänheimo, N., Saarinen, T., and Kangasjärvi, J. (2016). ROS signalling in a destabilised world: a molecular understanding of climate change. J. Plant Physiol. 203, 69–83. doi: 10.1016/j.jplph.2016.06.008
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dietz, K. J. (2014). Redox regulation of transcription factors in plant stress acclimation and development. Antioxid. Redox Signal. 21, 1356–1372. doi: 10.1089/ars.2013.5672
Dietz, K. J., Mittler, R., and Noctor, G. (2016). Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 171, 1535–1539. doi: 10.1104/pp.16.00938
Frederickson Matika, D. E., and Loake, G. J. (2014). Redox regulation in plant immune function. Antioxid. Redox Signal. 21, 1373–1378. doi: 10.1089/ars.2013.5679
Fukuhara, T., and Gibbs, M. J. (2012). “Family endornaviridae,” in Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomyof Viruses, eds A. M. Q. King, M. J. Adams, E. B. Carstens, and E. J. Lefkowitz (London: Elsevier Academic Press), 519–521.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Khankhum, S., Escalante, C., Rodrigues de Souto, E., and Valverde, R. A. (2016). Extraction and electrophoretic analysis of large dsRNAs from desiccated plant tissues infected with plant viruses and biotrophic fungi. Eur. J. Plant Pathol. doi: 10.1007/s10658-016-1014-7. [Epub ahead of print].
Khankhum, S., Valverde, R. A., Pastor-Corrales, M., Osorno, J. M., and Sabanadzovic, S. (2015). Two endornaviruses show differential infection patterns between gene pools of Phaseolus vulgaris. Arch. Virol. 160, 1131–1137. doi: 10.1007/s00705-015-2335-0
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Okada, R., Young, C. K., Valverde, R. A., Sabanadzovic, S., Aoki, N., Hotate, S., et al. (2013). Molecular characterization of two evolutionally distinct endornaviruses co-infecting common bean (Phaseolus vulgaris). J. Gen. Virol. 94, 220–229. doi: 10.1099/vir.0.044487-0
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). EdgeR: a Bioeonductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713. doi: 10.1038/ng.3008
Stielow, B., Klenk, H. P., and Menzel, W. (2011). Complete genome sequence of the first endornavirus from the ascocarp of the ectomycorrhizal fungus Tuber aestivum Vittad. Arch. Virol. 156, 343–345. doi: 10.1007/s00705-011-0998-8
Vlasova, A., Capella-Gutiérrez, S., Rendón-Anaya, M., Hernández-Oñate, M., Minoche, A. E., Erb, I., et al. (2016). Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 17:32. doi: 10.1186/s13059-016-0883-6
Keywords: common bean, endornavirus, RNA sequencing, Phaseolus vulgaris endornavirus 1, Phaseolus vulgaris endornavirus 2
Citation: Khankhum S, Sela N, Osorno JM and Valverde RA (2016) RNAseq Analysis of Endornavirus-Infected vs. Endornavirus-Free Common Bean (Phaseolus vulgaris) Cultivar Black Turtle Soup. Front. Microbiol. 7:1905. doi: 10.3389/fmicb.2016.01905
Received: 10 October 2016; Accepted: 15 November 2016;
Published: 29 November 2016.
Edited by:
Ricardo Flores, Polytechnic University of Valencia, SpainReviewed by:
Jesus Navas-Castillo, Instituto de Hortofruticultura Subtropical y Mediterránea La Mayora (IHSM-UMA-CSIC), SpainWon Kyong Cho, Seoul National University, Korea
Copyright © 2016 Khankhum, Sela, Osorno and Valverde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rodrigo A. Valverde, cnZhbHZlcmRlQGFnY2VudGVyLmxzdS5lZHU=
 Surasak Khankhum
Surasak Khankhum Noa Sela
Noa Sela Juan M. Osorno3
Juan M. Osorno3 Rodrigo A. Valverde
Rodrigo A. Valverde