- Bacterial Genomics and Evolution Laboratory, CSIR-Institute of Microbial Technology, Chandigarh, India
Background
Endophytic bacteria inhabit within plant tissues without causing any evident damage to the host and play crucial roles in plant growth, development, fitness, and protection (Farrar et al., 2014; Truyens et al., 2015). These endophytic bacteria spend a portion of their life cycle inside plants and normally resides on intercellular spaces and gain carbohydrates, amino acids, and inorganic nutrients from plants (Bacon and Hinton, 2007). Despite their beneficial effects on plant growth and development, seed-borne endophytic bacteria are still largely unexplored. Recent developments in high-throughput technologies, such as next-generation sequencing (NGS), permit the investigation of endophytic microbiomes, facilitate sequencing of a larger number of bacteria and encourage in depth analyses of bacterial communities from taxonomy, phylogeny, and evolutionary studies (Kaul et al., 2016).
Genomes of endophytic bacteria encode all the information that is required for an organism to grow under a range of both favorable and unfavorable conditions depending on the plant habitats. Along with the house-keeping machinery, these bacterial genomes also encode genes that are required for their endophytic life style and plant beneficial properties (Hardoim et al., 2015; Sheibani-Tezerji et al., 2015). In the present study, whole genome sequencing of 21 rice seed endophytic bacterial species belonging to the phylum Firmicutes was performed to ascertain their phylogenetic position and to get clue of the genomic signatures for their adaptation to endophytic lifestyle. Genomic dataset of endophytic bacterial strains are valuable pool of information that provides insights into the diversity, distribution, and lifestyle associated genes of these endophytes associated with plants.
Methods
Isolation of Bacteria from Surface Sterilized Rice Seeds
Rice seeds (Variety: Pusa Basmati 1121) were collected from rice field located in Kharar, Punjab (30.6755° N, 76.6723° E) for 3 years and processed in three independent batches for seed sterilization and isolation of endophytic bacteria. Endophytic bacteria were isolated from surface sterilized rice seeds by following the process: The hulls were removed from rice seeds using sterilized forceps, and the seeds (5 g) were put in sterile falcon tubes and washed with sterilized water for 1 min and then with 1% sodium hypochlorite solution for 5 min. The seeds were again washed with 75% ethanol for 1 min. After another wash with sterilized water five times, the surface sterilized rice seeds were crushed in sterile mortar and pestle and suspended in sterile saline solution (0.85% NaCl). The seeds suspension was incubated for 2 h at 28°C under shaking condition. After that, 100 μl of each of Direct, 10−1,10−2, 10−3, and 10−4 dilution in sterile saline was plated in duplicates onto Nutrient agar (NA); King's medium B (KMB); Glucose yeast chalk agar (GYCA); Tryptic soy agar (TSA); Peptone sucrose agar (PSA) supplemented with 0.01% cycloheximide. The confirmation of surface sterilization was conducted by spreading the last water wash as well as placing the washed rice seeds onto different media plates. Further, the isolates were identified by 16S rRNA gene PCR using 16S rRNA universal primers (27F and 1492R) and 16S rRNA gene sequence was used for characterization using EzTaxon server (http://www.ezbiocloud.net/eztaxon; Kim et al., 2012) prior to whole genome sequencing. For preservation, 15% glycerol stocks of the pure culture of each isolate was prepared and maintained at −80°C.
Whole Genome Sequencing (WGS) and Data Collection
Endophytic bacteria belong to Firmicutes phylum were revived from −80°C stocks onto NA plates and genomic DNA was isolated using Zymo ZR- Fungal/bacterial DNA isolation kit as per the instruction manual. The quality of the genomic DNA was assessed using agarose gel electrophoresis. DNA concentrations were estimated using a Nanodrop spectrophotometer ND-100 (Thermo Fisher Scientific, USA) and Qubit 2.0 fluorometer (Invitrogen, USA), with a double-stranded DNA High Sensitivity (dsDNA HS) Assay Kit. Both the ratio A260/280 and gel electrophoresis were used to ascertain the quality and purity of DNA samples. The input of 1 μg of genomic DNA from each sample was taken and standard protocol for the Nextera XT DNA (Illumina, San Diego, CA) sample preparation kit was used for library construction. The purified fragmented DNA was used as a template for a limited cycle PCR using Nextera primers and index adaptors. Cluster generation and sequencing of libraries were performed on the Illumina MiSeq platform (Illumina, San Diego, CA) with a 2 × 250 paired-end run.
Genome Assembly and Annotation
Demultiplexing, FASTQ generation and adapter trimming in raw sequence reads were automatically performed by Illumina- MiSeq software. The paired-end raw reads containing FASTQ files were assembled into contigs. Total number of library reads, number of contigs, genome size, G+C content, and total coverage were analyzed using CLC Genomics Workbench software version 7.0.3. Complete 16S rRNA sequences were extracted from the assembled genomes using RNAmmer 1.2 server (Lagesen et al., 2007) and characterized by EzTaxon server. tRNA was calculated by tRNAScan-SE. Sequences were annotated using NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/) and Rapid Annotation System Technology (RAST) pipeline (Aziz et al., 2008).
Genome Based Taxonomy and Phylogenomics
Taxonomic relationship among the endophytic bacterial isolates was deduced using 16S rRNA gene which is considered as the important tool for taxonomic and phylogenetic analysis (Mizrahi-Man et al., 2013). In the era of genome based taxonomy, the 16S rRNA gene is still used for preliminary bacterial typing because it is present in at least one copy in every bacterial genome and its conserved regions enable simple sample identification (Land et al., 2015). Therefore, firstly, the 16S rRNA sequences were used for characterization using EzTaxon server. Further, for genome similarity assessment, BLAST-based average nucleotide identity (ANIb) and Genome to Genome Distance calculator or digital DNA-DNA hybridization (dDDH) values were used. Pairwise ANI was calculated using JSpecies (Richter and Rosselló-Móra, 2009) and dDDH (Auch et al., 2010) was calculated using web tool GGDC 2.0.
Data Deposition
The assembled genome sequences (in FASTA format) of all 21 Rice seed bacterial endophytes is deposited in NCBI GenBank. Assembly statistics and accession numbers of the sequenced genomes are mentioned in Table 1. Figshare link to download genomes in FASTA format for single point access: https://figshare.com/s/6deae778bbdb90c6563c.
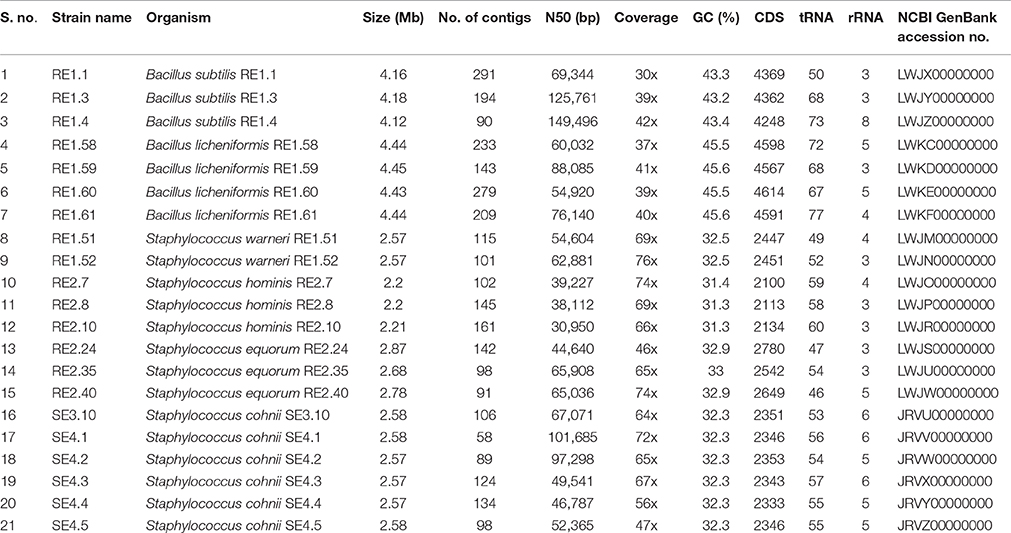
Table 1. Genome assembly statistics and annotation features of bacterial endophytes of rice and their accession numbers.
In silico Mining of Important Genes
RAST annotated genomes was used to search important genes responsible for plant adaptation and plant protection.
Data Interpretation
Isolation and Identification
A total of 21 bacterial endophytes isolated from rice seeds (Variety: Pusa Basmati 1121). The isolates were identified based on 16SrRNA gene for initial characterization prior to whole genome sequencing and their identification using EzTaxon server is summarized in Table S1.
Genome Sequencing and Phylogenomic Inference
Sequence data generated for each strain (Table 1) was de novo assembled with coverage ranging from 30x to 76x. Analysis based on complete 16SrRNA gene sequence extracted from the whole genome sequences assigned the bacterial isolates into 2 distinct genera and 6 species (Table S1). To gain better taxonomic resolution, all the 21 bacterial isolates have been validated by species delineation cut-off of >95% ANI values and >70% dDDH values (Richter and Rosselló-Móra, 2009; Auch et al., 2010). Genome sequence of reference strains were taken from NCBI database. The ANI value heatmap and dDDH values of 21 endophytes genomes with Type strain/reference strain genome are depicted in Figure 1 and Table S2, respectively. Description of the strains assigned to 2 different genera is provided as follows:
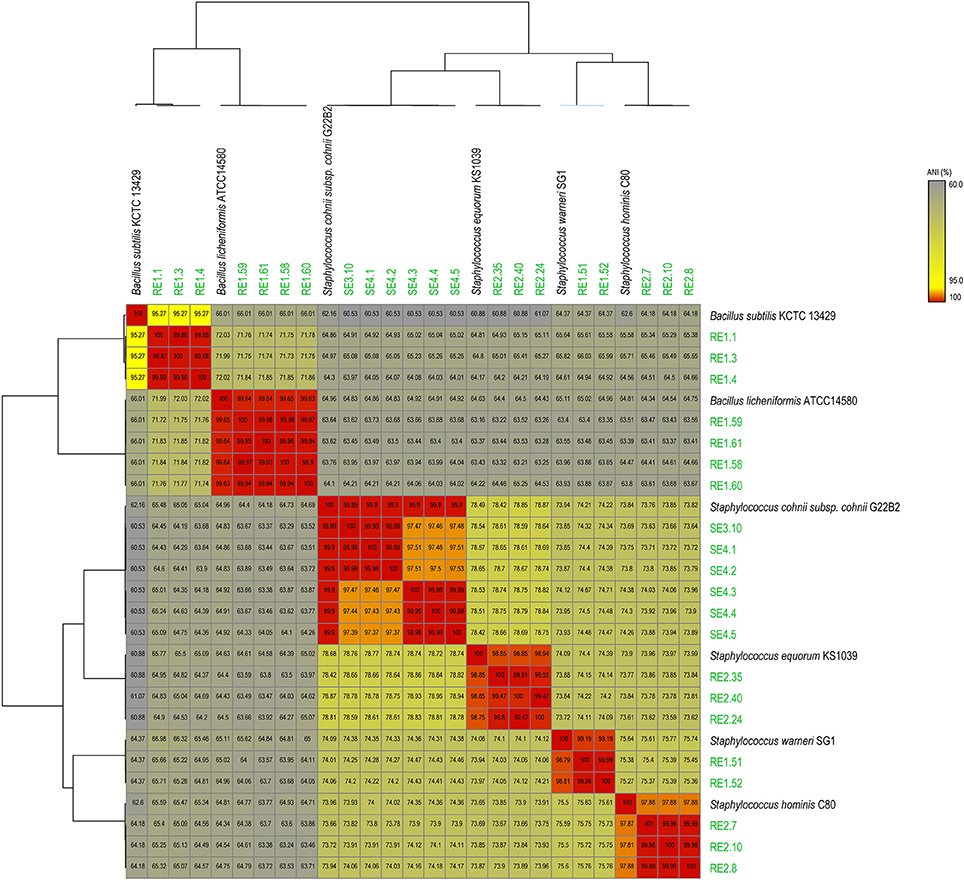
Figure 1. Average Nucleotide Identity (ANI) heat-map showing the 21 rice bacterial endophytes in our dataset, along with genomes of best matches from NCBI genome database, indicating distribution of 6 species based on >95% ANI values cutoff.
Genus: Staphylococcus
Staphylococcus is cocci shaped gram-positive member of phylum Firmicutes and 14 different strains were selected for sequencing from this genus belonging to 3 different year rice seeds sample lots. 16S rRNA sequences have assigned them to four different species with more than 99% identity to Staphylococcus warneri (RE1.51 and RE1.52), Staphylococcus hominis (RE2.7, RE2.8, and RE2.10), Staphylococcus equorum (RE2.24, RE2.35, and RE2.40), and Staphylococcus cohnii (SE3.10, SE4.1, SE4.2, SE4.3, SE4.4, and SE4.5). Moreover, ANI values also supported the species distinction between the four groups as they are above the cut-off (>95%) for species delineation (Figure 1). Further dDDH values of all the strains were more than 70% which further confirms their species identity (Table S2). Both ANI and dDDH resolved Staphylococcus species up to subspecies/strain level which were not possible with 16S rRNA gene sequences based analysis. This suggests that atleast 4 species of Staphylococcus inhabits rice seeds.
Genus: Bacillus
Bacillus is a genus of gram-positive, rod-shaped bacteria and a member of the phylum Firmicutes. Seven isolates from different years of rice seed lots belong to two different species, Bacillus subtilis (RE1.1, RE1.3, and RE1.4), and Bacillus licheniformis (RE1.58, RE1.59, RE1.60, and RE1.61) with more than 99% identity of 16SrRNA gene sequence identity. ANI and dDDH values of RE1.58, RE1.59, RE1.60, and RE1.61 of >99 and >97% with type strain genomes unequivocally establish their species identity as B. licheniformis (Figure 1; Table S2). However, in case of RE1.1, RE1.3, and RE1.4, ANI value with type strain of B. subtilis was 95.2% that is close to borderline of species demarcation, and dDDH values of 48.9% also showed that it is closely related to B. subtilis and may belongs to separate species.
Genome Mining of Plant Beneficial Traits
From RAST analysis, it was observed that more than 100 genes in the genomes of B. subtilis and B. licheniformis strains whereas more than 70 genes were present in genomes of S. warneri, S. hominis, S. equorum, and S. cohnii were identified that are responsible for stress tolerance. Majority of these genes were responsible for protection against oxidative stress and rest were encoded to cope with heat and osmotic stresses. Moreover, all the endophytes genomes of Staphylococcus and Bacillus species contain genes responsible for auxin biosynthesis and siderophores production. Full details of these genes are available in Table S3. It was reported that members of genus Bacillus are most commonly found as endophytic bacteria in plants. Moreover, they play important role as a biocontrol agent against phytopathogens, stimulates plant growth, and also produce plant growth hormones such as auxin and gibberellin, as well as able to ameliorate drought stress (Forchetti et al., 2007). There are also several reports in literature where members of Staphylococcus were documented as endophytes such as in rice seed (Chaudhry and Patil, 2016), maize kernels (Liu et al., 2012), grapevine, and hybrid spruce (Collins et al., 2004).
In the present study, our goal was to generate genomic data resource of endophytic bacteria associated with healthy rice seeds (var. Pusa Basmati 1121) from India. As representatives, dominant members belonging to Firmicutes phylum were selected. Our genomic dataset which comprised of Staphylococcus and Bacillus genera and their phylogenomic analysis reported here will serve to catalyze future studies by providing a new lens to study their endophytic lifestyle and will help in deciphering novel biological insights of endophytic bacteria-host plant relationships.
Author Contributions
VC carried out isolation, characterization, and preservation of bacterial endophytes; VC, SS, and KB performed QC and DNA library preparation. VC and SS performed genome sequencing, analysis, and submission; PP and VC designed of the study and interpretation of data. PP coordinated the study and applied for funding. VC wrote the manuscript. All authors reviewed and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
VC acknowledge support from CSIR (CSIR-Nehru Science Postdoctoral Fellowship) and DST-SERB (SERB-National Post Doctoral Fellowship), New Delhi, India. We acknowledge the funding from “Expansion and modernization of Microbial Type Culture Collection and Gene Bank (MTCC)” project jointly supported by CSIR (Grant No. BSC0402) and DBT, Govt. of India (Grant No. BT/PR7368/INF/22/177/2012). We thank Director, CSIR-IMTECH, for encouragement and support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02115/full#supplementary-material
References
Auch, A. F., Von Jan, M., Klenk, H. P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2:117. doi: 10.4056/sigs.531120
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bacon, C. W., and Hinton, D. M. (2007). “Bacterial endophytes: the endophytic niche, its occupants, and its utility,” in Plant-Associated Bacteria, ed S. S. Gnanamanickam (Springer), 155–194.
Chaudhry, V., and Patil, P. B. (2016). Genomic investigation reveals evolution and lifestyle adaptation of endophytic Staphylococcus epidermidis. Sci. Rep. 6:19263. doi: 10.1038/srep19263
Collins, M. D., Lesley, H., Geoffrey, F., and Enevold, F. (2004). Corynebacterium caspium sp. nov., from a Caspian seal (Phoca caspica). Int. J. Syst. Evol. Microbiol. 54, 925–928. doi: 10.1099/ijs.0.02950-0
Farrar, K., Bryant, D., and Cope-Selby, N. (2014). Understanding and engineering beneficial plant-microbe interactions: plant growth promotion in energy crops. Plant Biotechnol. J. 12, 1193–1206. doi: 10.1111/pbi.12279
Forchetti, G., Masciarelli, O., Alemano, S., Alvarez, D., and Abdala, G. (2007). Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 76, 1145–1152. doi: 10.1007/s00253-007-1077-7
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Kaul, S., Sharma, T., and Dhar, M. K. (2016). “Omics” tools for better understanding the plant-endophyte interactions. Front. Plant Sci. 7:955. doi: 10.3389/fpls.2016.00955
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Lagesen, K., Hallin, P., Rødland, E. A., Stærfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Land, M., Hauser, L., Jun, S. R., Nookaew, I., Leuze, M. R., Ahn, T. H., et al. (2015). Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 15, 141–161. doi: 10.1007/s10142-015-0433-4
Liu, Y., Zuo, S., Xu, L., Zou, Y., and Song, W. (2012). Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch. Microbiol. 194, 1001–1012. doi: 10.1007/s00203-012-0836-8
Mizrahi-Man, O., Davenport, E. R., and Gilad, Y. (2013). Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS ONE 8:e53608. doi: 10.1371/journal.pone.0053608
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Sheibani-Tezerji, R., Naveed, M., Jehl, M. A., Sessitsch, A., Rattei, T., and Mitter, B. (2015). The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front. Microbiol. 6:440. doi: 10.3389/fmicb.2015.00440
Keywords: plant-microbe interactions, endophyte, rice seeds, NGS, genomics, firmicutes, Staphylococcus, Bacillus
Citation: Chaudhry V, Sharma S, Bansal K and Patil PB (2017) Glimpse into the Genomes of Rice Endophytic Bacteria: Diversity and Distribution of Firmicutes. Front. Microbiol. 7:2115. doi: 10.3389/fmicb.2016.02115
Received: 29 August 2016; Accepted: 15 December 2016;
Published: 05 January 2017.
Edited by:
John R. Battista, Louisiana State University, USAReviewed by:
Nikolai Ravin, Research Center for Biotechnology Russian Academy of Sciences, RussiaMichael Thomas Guarnieri, National Renewable Energy Laboratory, USA
Copyright © 2017 Chaudhry, Sharma, Bansal and Patil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prabhu B. Patil, cGJwYXRpbEBpbXRlY2gucmVzLmlu
 Vasvi Chaudhry
Vasvi Chaudhry Shikha Sharma
Shikha Sharma Kanika Bansal
Kanika Bansal Prabhu B. Patil
Prabhu B. Patil