- 1Departamento de Microbioloxía e Parasitoloxía, Instituto de Acuicultura, Universidade de Santiago de Compostela, Santiago de Compostela, Spain
- 2Department of Molecular Biology and Laboratory for Molecular Infection Medicine Sweden, Umeå Centre for Microbial Research, Umeå University, Umeå, Sweden
- 3Departamento de Bioloxía Funcional, Facultade de Bioloxía - CIBUS, Universidade de Santiago de Compostela, Santiago de Compostela, Spain
The marine pathogenic bacterium Photobacterium damselae subsp. damselae causes septicemia in marine animals and in humans. The pPHDD1 plasmid-encoded hemolysins damselysin (Dly) and phobalysin P (PhlyP), and the chromosome-encoded hemolysin phobalysin C (PhlyC) constitute its main virulence factors. However, the mechanisms by which expression of these three hemolysins is regulated remain unknown. Here we report the isolation of a mini-Tn10 transposon mutant which showed a strong impairment in its hemolytic activity. The transposon disrupted a putative sensor histidine kinase gene vda_000600 (rstB), which together with vda_000601 (rstA) is predicted to encode a putative two-component regulatory system. This system showed to be homologous to the Vibrio cholerae CarSR/VprAB and Escherichia coli RstAB systems. Reconstruction of the mutant by allelic exchange of rstB showed equal impairment in hemolysis, and complementation with a plasmid expressing rstAB restored hemolysis to wild-type levels. Remarkably, we demonstrated by promoter expression analyses that the reduced hemolysis in the rstB mutant was accompanied by a strong decrease in transcription activities of the three hemolysin genes dly (damselysin), hlyApl (phobalysin P) and hlyAch (phobalysin C). Thus, RstB, encoded in the small chromosome, regulates plasmid and chromosomal virulence genes. We also found that reduced expression of the three virulence genes correlated with a strong decrease in virulence in a sea bass model, demonstrating that RstB constitutes a master regulator of the three P. damselae subsp. damselae hemolysins and plays critical roles in the pathogenicity of this bacterium. This study represents the first evidence of a direct role of a RstAB-like system in the regulation of bacterial toxins.
Introduction
Photobacterium damselae subsp. damselae is a primary pathogen of a wide range of marine animals, including cetaceans, crustaceans, mollusks, reptiles and, most frequently, cultivated fish of economic importance in marine aquaculture (Fouz et al., 1992; Rivas et al., 2013b; Terceti et al., 2016). In addition, this pathogen can cause a severe necrotizing fasciitis in humans (Clarridge and Zighelboim-Daum, 1985; Yamane et al., 2004; Hundenborn et al., 2013). The route of entry of P. damselae subsp. damselae into animal hosts remains poorly investigated. There is sound evidence that water transmits the disease (Fouz et al., 2000), and skin injuries precede P. damselae subsp. damselae infections (Rivas et al., 2013b). Interestingly, it is known by experimental infection studies that only those strains virulent by the intraperitoneal infection route are also infective through water (Fouz et al., 2000). Previous studies demonstrated that the most virulent P. damselae subsp. damselae isolates were highly hemolytic and inflicted more severe hemorrhages in diseased fish (Fouz et al., 1992), thus setting a relation between hemolysis and virulence. Later, we discovered that highly hemolytic strains harbor the 153 kb virulence plasmid pPHDD1, which carries the hemolysin genes dly and hlyApl (Rivas et al., 2011). dly gene encodes damselysin, a phospholipase-D active against sphingomyelin (Kreger et al., 1987). hlyApl encodes the recently characterized pore-forming toxin phobalysin P (PhlyP) which stands for “photobacterial lysin encoded on a plasmid.” This toxin forms small membrane pores in the target cells causing efflux of K+ and entry of vital dyes (Rivas et al., 2015b). In addition, all the hemolytic strains contain hlyAch gene in chromosome I (Rivas et al., 2013a, 2014). hlyAch encodes phobalysin C (PhlyC), which shares 92% identity in its amino acid sequence with PhlyP. All three hemolysins are known to be secreted via type II secretion system (Rivas et al., 2015a). PhlyP and PhlyC exert an additive effect in hemolysis and virulence, whereas all three hemolysins act synergistically on erythrocytes, a phenomenon that accounts for maximum virulence in mice and fish (Rivas et al., 2013a). Recently, we have demonstrated that hemolysins increase bacterial adherence (Rivas et al., 2015b), which may be of paramount relevance in the early events of natural infection. In addition, tissue damage caused by hemolysins may provide a source of nutrients to further support bacterial infection.
Although the molecular basis of virulence in this pathogen has been extensively studied in recent years, little is known about how the expression of the three hemolysins is regulated. In order to produce virulence factors when required, pathogens must sense environmental changes such as osmolarity, temperature, nutrients and metal ions concentration among others, and thus regulate virulence genes accordingly. It is expected that P. damselae subsp. damselae, a bacterial pathogen that also possesses a free-living style, has such sensing systems to control hemolysin production.
Two-component regulatory systems (TCS) are particularly important in regulating gene expression in response to environmental signals. A typical TCS consists of a sensor histidine kinase (HK) and its cognate DNA-binding response regulator (RR) (Stock et al., 2000). Upon activation in response to a specific signal, the HK autophosphorylates a conserved histidine residue and this phosphoryl group will be transferred to the conserved aspartic acid of the cognate RR. Phosphorylation of the RR activates an output domain that can modulate gene expression (Gao et al., 2007). Most RRs are transcriptional factors, and once phosphorylated they bind to target promoters, activating or repressing transcription (Stock et al., 2000). Although several TCS are predicted to be encoded in the genome of P. damselae subsp. damselae, to date none of such systems has been characterized, and their possible role in the regulation of virulence factors is so far unknown.
In this study, we transposon-mutagenized P. damselae subsp. damselae strain RM-71 and screened for hemolytic defective clones. We found a mutant in the putative sensor histidine kinase gene rstB, one of the two genes of the rstAB operon that is predicted to encode a TCS, which was strongly impaired in hemolytic activity. Here, we demonstrate that rstB is involved in the transcriptional regulation of the plasmid genes dly and hlyApl and of the chromosomal gene hlyAch, and is essential for maximum hemolytic activity. The major role of rstB in the virulence of P. damselae subsp. damselae for fish is also demonstrated. This study represents the first report of a regulatory system of the P. damselae subsp. damselae virulence. In addition our results constitute the first evidence of the role of a RstAB-like system in regulation of bacterial toxins.
Materials and Methods
Bacterial Plasmids, Strains, and Culture Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. P. damselae subsp. damselae cells were grown at 25°C on tryptic soy agar (TSA) and broth (TSB) supplemented with NaCl up to 1% (TSA-1 and TSB-1, respectively) and supplemented with antibiotics when appropriate. For hemolysis assays on sheep blood agar plates (Oxoid), strains were cultured on TSA-1 plates and single colonies were inoculated on a blood agar plate, and hemolysis haloes were photographed after 15 h of growth at 25°C. E. coli was grown at 37°C in Luria-Bertani (LB) broth or LB agar. When necessary, antibiotics were used at the following final concentrations: kanamycin (Km) at 50 μg mL-1, gentamicin (Gm) at 15 μg mL-1, chloramphenicol (Cm) at 20 μg mL-1. For growth curves, at least three replicates per strain were grown in two independent experiments in 200 μl medium in a 96 well plate inoculated 1:100 from exponentially growing precultures (OD600∼0.02) and analyzed using a Biotek plate reader at 10 min intervals.
Mini-Tn10 Mutagenesis and Identification of the Disrupted Gene
Mini-Tn10 mutagenesis was performed using the suicide conjugative plasmid pLOFKm (Herrero et al., 1990), with minor modifications as previously described (Rivas et al., 2015a). Genomic DNA from the clones with impaired hemolysis of strain RM-71 was purified with the genome DNA kit (Qbiogene), partially digested with BfuC1 and ligated to BamHI-digested plasmid pUC118. Ligation reactions were transformed into E. coli DH5α by electroporation (2.5 kV, 25 μF capacitance, and Pulse Controller Unit set to 200 Ω). Transformants were selected on LB agar plates supplemented with kanamycin and ampicillin. Inserts containing the kanamycin resistance gene of mini-Tn10 plus flanking chromosomal DNA, were amplified by PCR and sequenced. DNA sequences were obtained using a capillary DNA Sequencer ABI 3730xl (Applied Biosystems). The nucleotide sequence of the rstAB loci in RM-71 strain is available in the partially annotated whole genome shotgun sequence of this strain (GenBank Acc. No. LYBT01000056). Locus tags for rstA and rstB genes are, respectively, A0J47_03465 and A0J47_03460.
Mutant Construction and Gene Complementation
A non-polar deletion of rstB was constructed by allelic exchange using the Kmr suicide vector pNidKan as previously described (Rivas et al., 2015a), yielding the P. damselae subsp. damselae mutant strain MT151 (Table 1). For complementation of the rstB mutant, rstAB ORFs sequence together with the respective promoter sequence was amplified by PCR using Hi-Fidelity Kapa Taq, cloned into the CmR mobilizable vector pMRB24 and mobilized from E. coli S17-1-λpir into the MT151 mutant, to yield complemented strain MT157.
Hemolytic Assays With Bacterial ECPs
To obtain the extracellular products (ECPs), cultures of P. damselae subsp. damselae in TSB-1 were adjusted to an OD600 of 1, and 100 μl were spread with a sterile cotton swab over TSA-1 plates covered with cellophane, as previously described (Liu, 1957). Cells were incubated at 25°C for 48 h and washed off the cellophane using saline solution (0.85% [wt/vol] NaCl) and adjusted to an OD600 of 1. Cells were centrifuged at 15,000 ×g for 5 min and the supernatants were filtered through 0.22 μm-pore-size membranes and stored at -20°C until used. Quantitative hemolytic assays were carried out in triplicates using the method described by Bernheimer (1988), introducing minor modifications as previously described (Rivas et al., 2015a).
lacZ Transcriptional Fusions and β-Galactosidase Assays
DNA fragments corresponding to dly, hlyApl and hlyAch presumptive promoter regions extending from about 1 kb upstream of the ATG start codon to about 30 bp downstream of the start codon, were PCR-amplified and fused to a promoterless lacZ gene in the low-copy number reporter vector pHRP309. The transcriptional fusions dly::lacZ (pAJR51), hlyApl::lacZ (pAJR53) and hlyAch::lacZ (pAJR45), obtained in a previous study (Rivas et al., 2013a) were here mobilized by conjugation from E. coli S17-1-λpir into the parental RM-71 strain and into its rstB mutant derivative strain MT151. The P. damselae subsp. damselae strains carrying the promoter-lacZ fusion vectors were grown in TSB-1 and the β-galactosidase activities were measured by the method of Miller (1992). Three independent experiments of β-galactosidase activity measurement were carried out.
Optical Microscopy
Phase contrast microscopy was performed using stationary phase cultures. Bacteria were immobilized on LB pads containing 1% agarose. Image acquisition was performed using a Zeiss Axio Imager.Z2 microscope equipped with a Plan-Apochromat 63X phase contrast objective lens and an ORCA-Flash 4.0 LT digital CMOS camera (Hamamatsu), using the Zeiss Zen Blue software. For image processing and analysis the MicrobeJ plugin for Fiji was used (Ducret et al., 2016). Cell length and width from 570 to 700 cells per strain (three fields of 190–250 cells each) were measured and statistical significance was calculated using a t-test (unpaired).
Polymyxin B MIC Assay
To determine the polymyxin MIC of RM-71 and the rstB mutant MT151, strains were incubated 24 h at 25°C on TSA-1 plates in the presence of E-test gradient polymyxin B strips (bioMérieux).
Fish Virulence Assays
To test the influence of rstB gene deletion in the virulence of P. damselae subsp. damselae for fish, we conducted experimental infection challenges using sea bass (Dicentrarchus labrax) as a model, as previously described (Terceti et al., 2016). Fish were obtained from IGAFA (Illa de Arousa, Galicia, Spain). Groups of 10 fish (6 ± 1.2 g) per strain tested and per dose were acclimated in 100 l aquaria at 24°C for 1 week before the infections were performed. The virulence tests were conducted by intraperitoneal injection of bacterial suspensions. Fish were inoculated with 0.1 ml of bacterial suspensions of each strain in 0.85% NaCl solution at two different doses of 104 and 103 CFU/fish. As a control, a group of 10 fish was inoculated with 0.1 ml of sterile 0.85% NaCl solution. Fish mortality was recorded daily for 10 days post-challenge. Re-isolation and identification of the bacteria from the kidney of dead fish were performed. For this purpose, fish were aseptically dissected, kidney samples taken with sterile loops and seeded on TSA-1 and TCBS agar plates. Colonies were confirmed by the subsp. damselae-specific ureC gene PCR test as previously described (Osorio et al., 2000). The protocols of animal experimentation used in this study have been reviewed and approved by the Animal Ethic Committee of the Universidade de Santiago de Compostela.
Results
Transposon Mutagenesis Identifies a rstAB-like Operon in P. damselae subsp. damselae With a Role in Hemolytic Activity
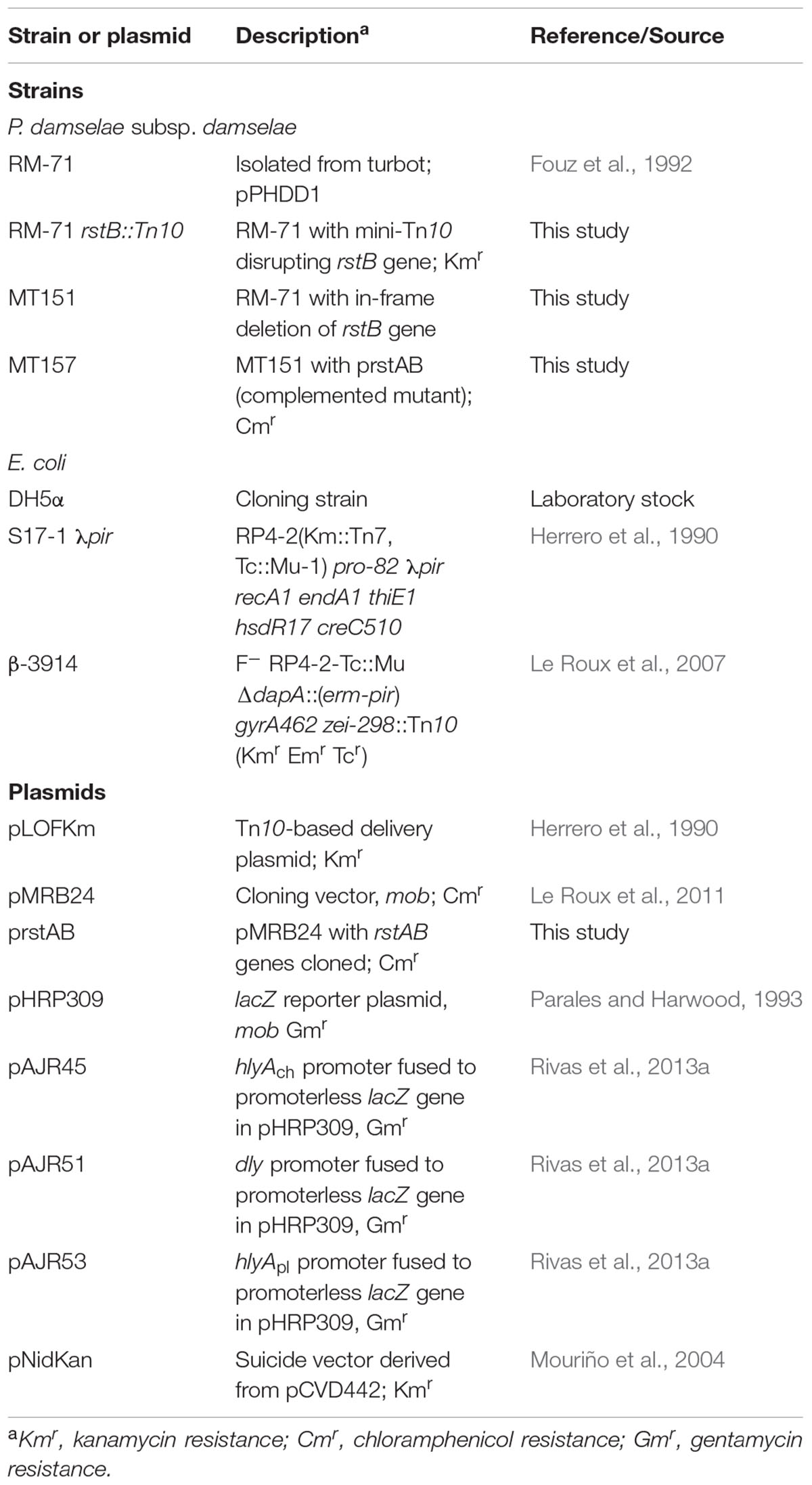
In order to gain an insight into how P. damselae subsp. damselae regulates its hemolytic activity, we screened a library of >2000 mini-Tn10 insertional mutants in the highly hemolytic and highly virulent pPHDD1-containing strain RM-71, and isolated a total of 12 mutants displaying severely affected hemolytic phenotypes. By genetic analyses we found that 11 out of the 12 mutants had the mini-Tn10 inserted either in one of the three hemolysin genes or in genes of the previously described type II secretion system that participates in hemolysin secretion (Rivas et al., 2015a), thus validating our screening method (data not shown). Interestingly, we found that the insertion in the remaining mutant clone (RM-71 rstB::Tn10) with a severely impaired hemolysis (Figure 1A) took place in a gene which was 100% identical to the vda_000600 locus of the P. damselae subsp. damselae type strain ATCC33539 (GenBank accession number ADBS00000000). This ORF, located in the small chromosome (ChrII) of P. damselae subsp. damselae, is predicted to constitute a putative two-gene operon together with the upstream gene vda_000601 (Figure 1B).
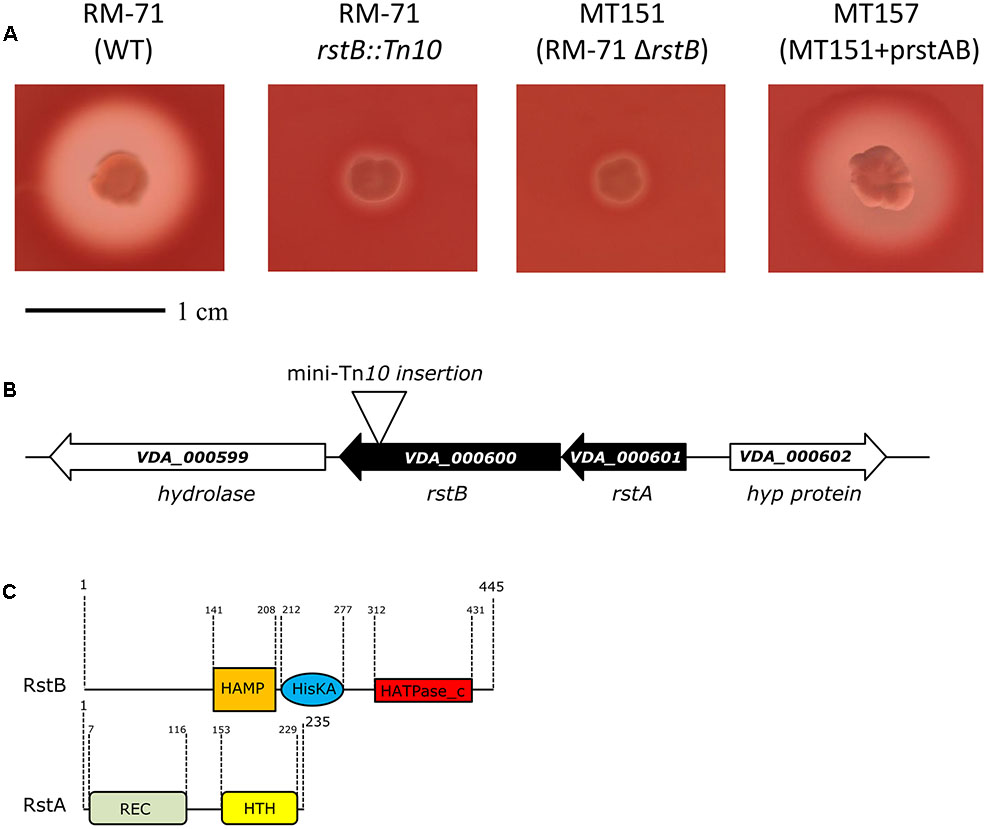
FIGURE 1. (A) Hemolytic phenotypes on sheep blood agar plates of P. damselae subsp. damselae parental strain RM-71 and derivatives. A mini-Tn10 transposon mutant in rstB gene (RM-71 rstB::Tn10) shows a reduction in hemolysis, a phenotype which is reproduced in the allelic-exchange ΔrstB mutant (MT151). Complementation of MT151 mutant with a plasmid expressing rstAB genes (strain MT157) restores the hemolytic phenotype to levels similar to the parental strain. Scale bar, 1 cm. (B) Physical map of the P. damselae subsp. damselae rstAB genes and flanking sequences, with the detailed point of insertion of the mini-Tn10 transposon in rstB gene. Gene numbers refer to gene annotation of the P. damselae subsp. damselae type strain ATCC 33539 complete genome (GenBank accession no. ADBS00000000). (C) Conserved domains of RstB and RstA proteins, showing the characteristic domains of bacterial two-component regulatory systems. RstB is the sensor histidine kinase, and RstA is the response regulator.
An in silico search for VDA_000600 and VDA_000601 orthologs in other Vibrionaceae species showed the highest identity with two ORFs encoded in chromosome I of Vibrio cholerae N16961, annotated as VC1319 (50% identity with VDA_000600) and VC1320 (65% identity with VDA_000601), a sensor histidine kinase and a DNA-binding RR, respectively. These two V. cholerae ORFs were coined with different names in two independent studies, CarSR (Bilecen and Yildiz, 2009) and VprAB (Herrera et al., 2014), respectively.
Among the Enterobacteriaceae, VDA_000600 showed its best match (32% identity) to E. coli RstB, and VDA_000601 best match (41% identical) was to E. coli DNA-binding RR RstA. The RstAB two-component system is part of the Mg+2-sensing PhoPQ regulon of E. coli (Ogasawara et al., 2007) and Salmonella enterica (Perez et al., 2009). Due to the duality of gene nomenclature in V. cholerae, and since most studies on this TCS were conducted with E. coli and S. enterica, we dubbed the P. damselae subsp. damselae genes following the original E. coli nomenclature. Therefore, VDA_000600 encodes RstB and VDA_000601 encodes RstA in P. damselae subsp. damselae.
We found that the P. damselae subsp. damselae rstA stop codon and the rstB translational initiation codon are separated by 14 bp, suggesting that they are organized in an operon where rstA is the first gene. rstB is predicted to encode a 445-amino-acid protein and contains three domains (Figure 1C): a histidine kinase-adenylyl cyclase-methyl-accepting protein and phosphatase domain (HAMP), a phosphoacceptor histidine kinase domain (HisKA), and an ATPase domain (HATPase_c). rstA is predicted to encode a 235-amino-acid protein and contains two domains: a signal receiver domain (REC) that includes the phosphoacceptor site, and a helix-turn-helix domain (HTH) for DNA-binding. We found that the P. damselae subsp. damselae RstB and RstA proteins contain the conserved Histidine 222 (H222) and Aspartate 55 (D55) residues, respectively, which correspond to the sites for phosphorylation in the V. cholerae homologs (Herrera et al., 2014) (Supplementary Figure S1). Altogether, the comparative analysis clearly suggests that P. damselae subsp. damselae vda_000600 and vda_000601 genes encode a RstAB-like TCS.
rstB Deletion Mutants Are Impaired in Hemolytic Activity
To confirm the association between the transposon mutation of rstB and the impaired hemolysis, a non-polar deletion of rstB was generated, yielding strain MT151. As a result, deletion of rstB caused the same hemolytic phenotype observed in the transposon mutant (Figure 1A). In order to obtain a quantitative measure of the role of rstB in the hemolytic activity, assays with sheep erythrocyte suspensions and bacterial ECPs were conducted. As a result, we found that deletion of rstB caused a decrease of the hemolytic activity from 325 hemolytic units (HU) produced by the parental (RM-71) to 2 HU of the ΔrstB mutant (MT151) (Figure 2). In order to complement the rstB mutant, we cloned the complete rstAB operon with its putative promoter sequence upstream rstA into the mobilizable, CmR plasmid pMRB24, yielding plasmid prstAB. We found that complementation of MT151 with this plasmid (yielding strain MT157) fully restored the hemolytic halo (Figure 1A) and HU of the parental strain (Figure 2). These results altogether clearly indicate that rstB plays a major role in the hemolytic activity in P. damselae subsp. damselae.
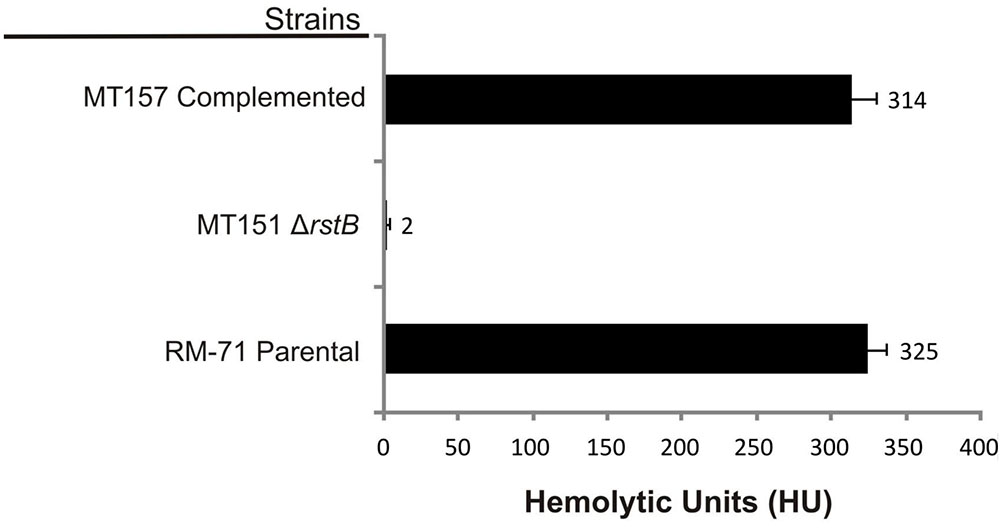
FIGURE 2. Hemolytic activities of parental strain RM-71, rstB deletion mutant MT151, and complemented strain MT157 (prstAB), determined with bacterial extracellular products (ECPs) and with suspensions of sheep erythrocytes. The rstB mutant is severely impaired in hemolysis, with a reduction of hemolytic units (HU) from 325 to 2 HU. The release of hemoglobin was determined at A540. One hemolytic unit is defined as the amount of hemolysin which lyses 50% of sheep erythrocytes. All assays were carried out in triplicate and mean values with standard deviation are shown.
Expression of dly, hlyApl and hlyAch Genes Is Controlled by RstB
Based on the above observations that P. damselae subsp. damselae rstB mutation strongly impairs hemolytic activity, we next examined whether RstB regulates the expression of the three hemolysin genes at the transcriptional level. For this, we evaluated changes in the expression of dly, hlyApl and hlyAch promoters cloned in vector pHRP309 upstream of a promoterless lacZ gene. Transcription was measured by determining β-galactosidase activities in parental and mutant strains. We observed that under standard TSB-1 culture conditions, deletion of rstB in the pPHDD1-harboring strain RM-71 caused a 15-fold, 10-fold, and 3-fold decrease in β-galactosidase levels of the dly::lacZ, hlyApl::lacZ, and hlyAch::lacZ fusions, respectively (Figure 3). These results clearly suggest that the hemolytic defect of the rstB mutant is due to an effect on hemolysin gene transcription. It is thus noteworthy that the rstAB system, encoded in the chromosome II of P. damselae subsp. damselae, regulates two plasmid-encoded hemolysins and a chromosome-I encoded hemolysin (Figure 4).
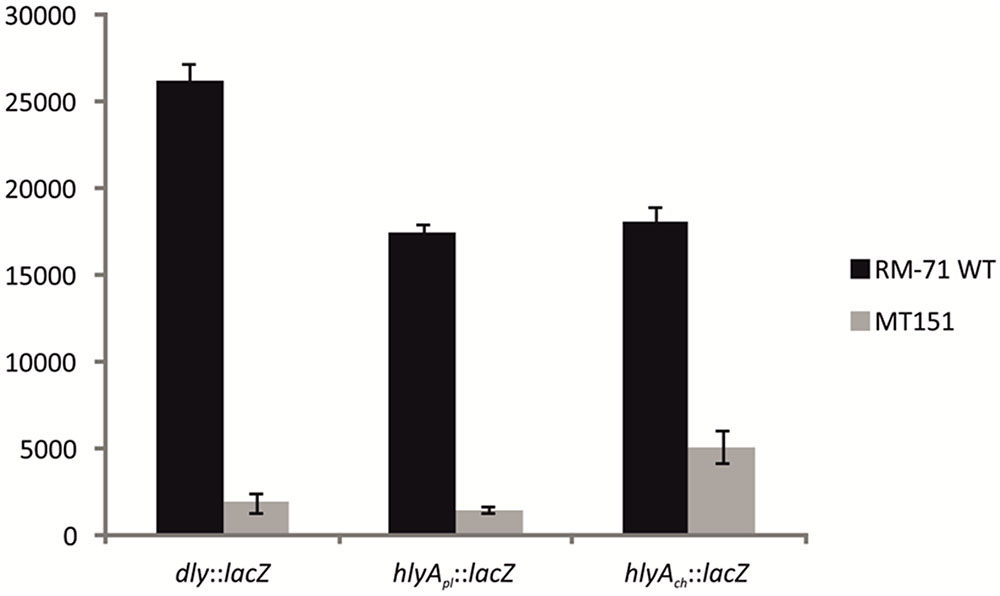
FIGURE 3. Transcriptional activity (β-galactosidase units) of the lacZ fusions to the dly, hlyApl, and hlyAch promoters, under parental (RM-71) and ΔrstB mutant (MT151) backgrounds. All experiments were carried out in triplicate. Mean values with standard deviations denoted by error bars are shown.
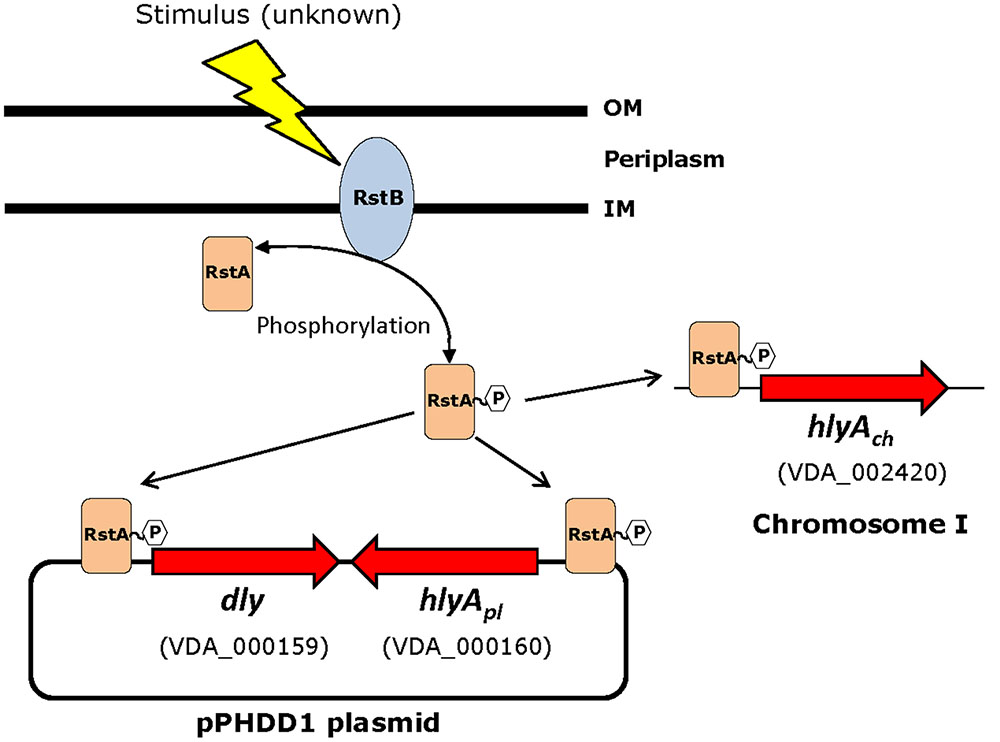
FIGURE 4. Schematic depicting the three hemolysin genes reported in this study to be part of the RstAB regulon of P. damselae subsp. damselae. Note that two hemolysin genes (dly and hlyApl) are encoded in pPHDD1 plasmid whereas hlyAch is encoded in chromosome I. Gene numbers refer to the annotated genome of the type strain ATCC33539.
A previous study reported that RstA-regulated genes in E. coli share a conserved consensus sequence in their promoters, the so-called RstA box (TACATNTNGTTACA) (Ogasawara et al., 2007). We in silico analyzed the nucleotide sequences of dly, hlyApl and hlyAch gene promoters for presence of conserved motifs that might act as RstA-binding sequences (Supplementary Figure S2). The three promoters diverged notably in their nucleotide sequences, and even though PhlyP and PhlyC amino acid sequences are 92% identical their respective promoters bear little similarity (Rivas et al., 2013a). Interestingly, we detected a short conserved sequence among the three promoters that shared some residues in common with the E. coli consensus RstA box (Supplementary Figure S2).
The rstB Mutant Exhibited Normal Growth and Cell Morphology, and Showed No Detectable Changes in Polymyxin Sensitivity
To determine the importance of rstB in P. damselae subsp. damselae physiology beyond its role in hemolysin gene regulation, we analyzed the effect of rstB gene deletion on growth. We observed that absence of rstB had no apparent effect on growth in TSB-1 medium, as both mutant and complemented cultures reached identical optical densities as the wild-type (Figure 5A). Microscopy analysis revealed that there were also no morphological differences -i.e., cell width and length- between these strains (Figures 5B–D).
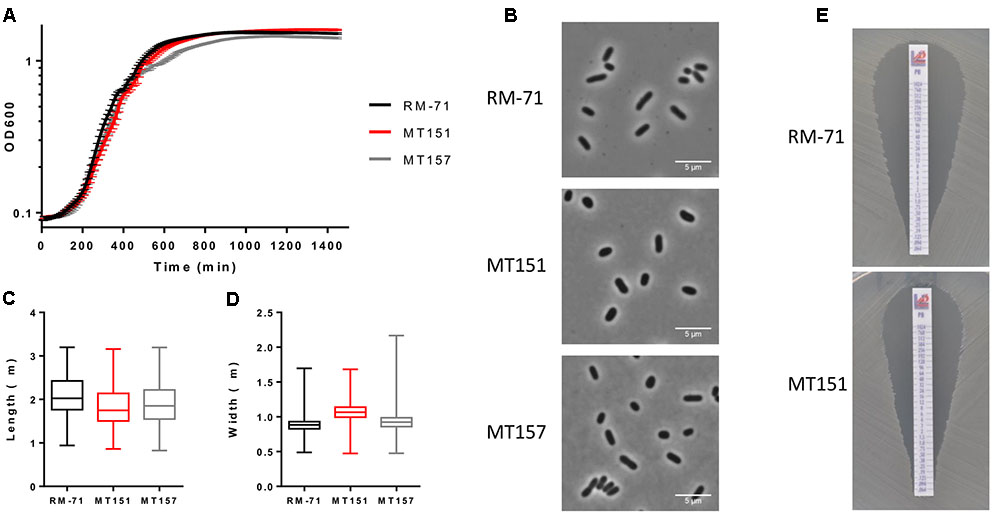
FIGURE 5. (A) Growth curves of RM-71, rstB mutant (MT151) and complemented strain (MT157) in TSB-1 medium. (B) Phase contrast images of cells grown to stationary phase in TSB-1. Scale bars 5 μm. (C) Box plot graphs showing the comparison of cell length in stationary phase cultures. Whiskers indicate min and max values. (D) Box plot graphs showing the comparison of cell width in stationary phase cultures. Whiskers indicate min and max values. (E) E-test for polymyxin sensitivity showed no differences between RM-71 and the rstB mutant MT151.
Previous studies reported that V. cholerae vprAB mutants (homologs of P. damselae subsp. damselae RstAB) exhibited sensitivity to the cationic antimicrobial peptide polymyxin B (Herrera et al., 2014; Bilecen et al., 2015). To date, the influence of polymyxin B in P. damselae subsp. damselae viability had not been explored. In order to test the response of P. damselae subsp. damselae parental strain and rstB mutant to polymyxin B, we determined the MIC for this compound using gradient polymyxin B E-strips in TSA-1 agar medium. We found that both the parental strain RM-71 and the rstB mutant MT151 were highly sensitive to polymyxin B, displaying a MIC of 0.125 μg/mL (Figure 5E). Contrary to what has been reported in V. cholerae, deletion of rstB does not influence polymyxin sensitivity in this pathogen.
RstB Plays a Major Role in Virulence for Fish
In a previous study we demonstrated that dly, hlyApl and hlyAch genes play a key role in the virulence of P. damselae subsp. damselae in fish (Rivas et al., 2013a). Since we have here demonstrated that rstB is involved in the regulation of these three toxin genes, we wanted to evaluate its contribution to P. damselae subsp. damselae virulence. For this purpose, virulence tests were conducted in sea bass (Dicentrarchus labrax) as a model fish, inoculating the parental strain RM-71 as well as its rstB derivative mutant MT151. The challenges were conducted in fish kept at a water temperature of 24°C and using two different doses of 104 and 103 CFU/fish, and 10 fish for each dose and strain. The parental strain RM-71 was virulent for sea bass, with mortality rates of 80 and 60% at each dose, respectively (Figure 6). Noteworthy, this strain caused the death of 80% of the animals in less than 48 h at the higher dose (104 CFU/fish). On the contrary, the rstB mutant strain MT151 did not cause any mortality at the two assayed doses (104 and 103 CFU/fish). P. damselae subsp. damselae RM-71 could be re-isolated from the kidney of all dead fish post-challenge, and colonies were confirmed by phenotypical tests and by a colony-PCR targeted to the P. damselae subsp. damselae-specific ureC gene (data not shown). These results thus demonstrate that mutation of rstB strongly impairs virulence of this pathogen for fish.
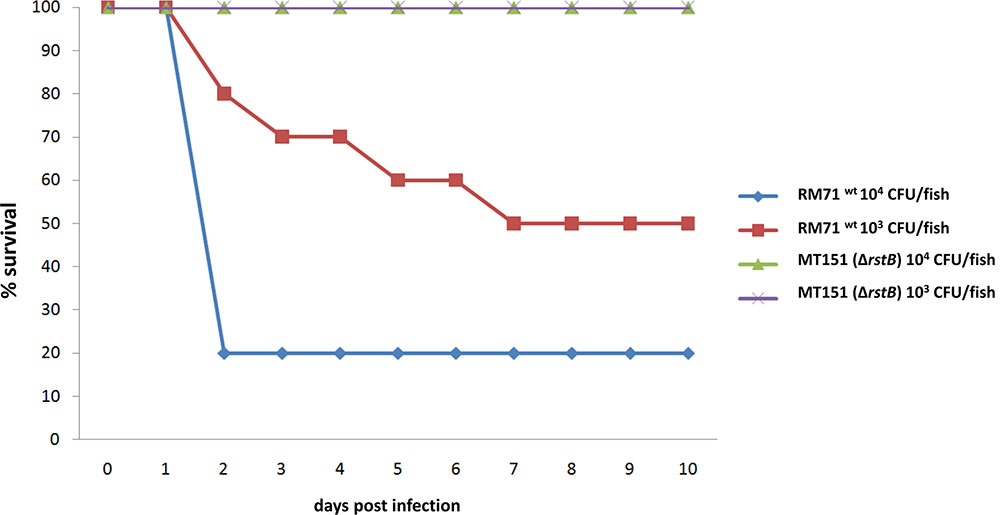
FIGURE 6. Survival (%) of sea bass intraperitoneally challenged with two different doses (104 CFU/fish and 103 CFU/fish) of the P. damselae subsp. damselae parental strain (RM-71) and the ΔrstB mutant MT151. For each dose assayed, a total of 10 fish were inoculated.
Discussion
Several TCS have been reported to regulate toxin expression in species of the Vibrionaceae. This is the case of the V. cholerae ToxRS and VarS/VarA systems that enhance the expression of ToxT, a transcriptional factor that positively regulates cholera toxin production (DiRita et al., 1991; Jang et al., 2011). Also, the V. parahaemolyticus ToxRS system regulates effector proteins of the type III secretion system (Whitaker et al., 2012).
In this study we identified rstB, the predicted histidine-kinase gene of the RstAB-like two-component system that positively regulates the three hemolysin genes of P. damselae subsp. damselae. To the best of our knowledge this represents the first evidence of a RstAB homologous system that regulates toxin genes in a pathogenic bacterium. The singularity of our findings is reinforced by the fact that one of the regulated genes is in chromosome I (hlyAch), whereas the two other regulated genes (dly and hlyApl) are encoded within a horizontally acquired large virulence plasmid (pPHDD1). It is thus interesting to note that rstB, encoded in the small chromosome of P. damselae subsp. damselae, can both regulate plasmid and chromosomal loci. Homologues of RstAB have received little attention to date, but some knowledge has been gained in the discovery of the RstAB regulon, and specific genes whose expression is regulated directly by the RR RstA have been identified. The RstAB regulon seems to be little predictive: whereas E. coli RstAB positively regulates the three genes yfiA, entE and cspC (Oshima et al., 2002), expression of yfiA and cspC in S. enterica remained unaffected both under over-expression of rstA and under deletion of rstA (Cabeza et al., 2007). A recent study evidenced that a S. enterica rstB mutant had reduced expression levels of genes related to pyrimidine metabolism (udp and cdd genes), enterobactin biosynthesis (entA, entB, entE, and entF) and ferrous iron transport (feoA, feoB) (Tran et al., 2016). These results thus suggest that the RstAB TCS of different species, although phylogenetically closely-related, govern the expression of distinct groups of genes in different bacteria.
Within Vibrionaceae, the first evidence for the role of RstAB homologues came after the identification of V. cholerae genes VC1319 and VC1320. These genes were dubbed carS and carR respectively in a study aimed at identifying genes regulated by calcium, and it was determined that CarSR negatively regulated biofilm formation and expression of the vps polysaccharide genes (Bilecen and Yildiz, 2009). Later, it was found that carSR transcription was negatively regulated in response to an external increase of Ca2+, and CarSR negatively regulated vps gene expression and biofilm formation (Bilecen et al., 2015). In another study, the same V. cholerae genes alternatively dubbed VprB/VprA, were found to regulate expression of the almEFG operon, which encodes proteins necessary for glycine modification of lipid A and required for colonization of a mammalian host (Herrera et al., 2014). This addition of glycine residues to the lipid A domain of lipopolysaccharide conferred polymyxin resistance. We have demonstrated here that the P. damselae subsp. damselae rstB mutant exhibits the same sensitivity to the cationic antimicrobial peptide polymyxin as the parental strain. Since no homologs of almEFG genes are found in the complete genome of the P. damselae subsp. damselae reference strain ATCC33539, it is likely that P. damselae subsp. damselae does not share the lipid A modification function with V. cholerae.
The conservation of the candidate phosphorylation sites of RstBH222 and RstAD55, coincident with the same conserved residues in their RR and histidine kinase orthologs (Gao and Stock, 2009; Herrera et al., 2014) (Supplementary Figure S1) support the consideration of RstAB as the first putative TCS identified to date in P. damselae. We demonstrated here that RstB is necessary for optimal expression of the dly, hlyApl and hlyAch genes, and this regulation is likely exerted at the transcriptional level. Although the PhlyP and PhlyC amino acid sequences are 92% identical, the nucleotide sequences of their respective promoters bear little similarity (Rivas et al., 2013a). Still, we were able to detect a putative consensus sequence common to the three hemolysin gene promoters that might constitute a candidate for RstA binding (a RstA box) (Supplementary Figure S2), and shared similarity to the consensus RstA box of E. coli (Ogasawara et al., 2007). Evidences with other bacterial species revealed that the RstAB two-component system regulates gene functions by acting at the transcriptional level. This is the case of E. coli, where RstA is known to bind to a narrow region of 200 bp upstream the csgD promoter and regulate its transcription (Ogasawara et al., 2010). In addition, RstAB homologs can exert regulatory functions acting at other levels, as is the case of Salmonella enterica, where RstA promoted a reduction of RpoS cellular levels by inducing RpoS degradation (Cabeza et al., 2007).
The three hemolysin genes regulated by RstAB play a crucial role in virulence of P. damselae subsp. damselae for mammals and for fish and are responsible for hemolytic and cytotoxic activity (Kreger et al., 1987; Rivas et al., 2013a, 2015b). We have shown here that deletion of rstB strongly impairs virulence in the sea bass fish model. To date, only a few reports have demonstrated a direct role of RstAB in virulence. A Yersinia pseudotuberculosis rstA mutant was less virulent than the parental strain for mice (Flamez et al., 2008), but the potential virulence factors that are regulated by rstAB remain unknown. In V. cholerae, VprAB were found to be necessary for optimal intestinal colonization in a suckling mouse model, likely due to the role of VprAB as regulators of lipidA modification (Herrera et al., 2014). Lately, it was reported that rstAB mutations reduced virulence of avian pathogenic E. coli in a bird animal model (Gao et al., 2015), but these authors did not identify which virulence genes were regulated by RstA.
Expression of the rstAB genes is known to be under the control of the Mg2+-sensing PhoQP two-component system in E. coli (Ogasawara et al., 2007) and in Salmonella enterica (Perez et al., 2009). However, the specific stimuli that trigger the activation (autophosphorylation) of the sensor histidine kinase RstB in Enterobacteriaceae and of CarS/VpR in V. cholerae still remain undeciphered. Herrera et al. (2014) found that vprA promoter expression responded to biological signals as bile, acidic pH and sublethal concentrations of polymyxin B, but did not elucidate which signals trigger VprB phosphorylation. Similarly, the stimulus that triggers the RstAB-dependent hemolysin promoter activation in P. damselae subsp. damselae is so far unknown. P. damselae subsp. damselae has likely evolved adaptive responses to maximize hemolysin production into the host but not in its free-living planktonic stage. Ongoing studies aimed at elucidating the sensory signals that trigger the RstAB-dependent regulation of the expression of the three major virulence factors in P. damselae subsp. damselae are expected to reveal novel cues on this fascinating model of pathogenesis.
Author Contributions
MT, AR, LA, and MN performed the experiments. AR, LA, and FC helped in data analysis and manuscript editing. CO designed and directed the research and wrote the manuscript.
Funding
Research in CRO laboratory is supported by grants AGL2013-48353-R, and AGL2016-79738-R, both from the Ministry of Economy and Competitiveness (MINECO) of Spain (cofunded by the FEDER Programme from the European Union). MT is supported by a predoctoral grant of the Brazilian Ministry of Education and CAPES (Coordenaçao de Aperfeiçoamento de Pessoal de Nível Superior). AR is supported by a Postdoctoral Fellowship (Axudas á Etapa Inicial de Formación Posdoutoral do Plan Galego de Investigación, Innovación e Crecemento 2011–2015, Plan I2C) of Xunta de Galicia, Spain. Research in FC laboratory is supported by the Molecular Infection Medicine Sweden (MIMS), the Knut and Alice Wallenberg Foundation and the Swedish Research Council. LA is supported by Grant JCK-1326 from the Kempe Foundation (Kempestiftelserna).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Bernardo Fernández Souto and the Instituto Galego de Formación en Acuicultura (IGaFA) (Illa de Arousa, Galicia, Spain) for their valuable support in providing sea bass for the virulence challenges.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00582/full#supplementary-material
References
Bernheimer, A. W. (1988). Assay of hemolytic toxins. Methods Enzymol. 165, 213–217. doi: 10.1016/S0076-6879(88)65033-6
Bilecen, K., Fong, J. C. N., Cheng, A., Jones, C. J., Zamorano-Sánchez, D., and Yildiz, F. (2015). Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infect. Immun. 83, 1199–1209. doi: 10.1128/IAI.02700-14
Bilecen, K., and Yildiz, F. (2009). Identification of a calcium-controlled negative regulatory system affecting Vibrio cholerae biofilm formation. Environ. Microbiol. 11, 2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x
Cabeza, M. L., Aguirre, A., Soncini, F. C., and García Véscovi, E. (2007). Induction of RpoS degradation by the two-component system regulator RstA in Salmonella enterica. J. Bacteriol. 189, 7335–7342. doi: 10.1128/JB.00801-07
Clarridge, J. E., and Zighelboim-Daum, S. (1985). Isolation and characterization of two hemolytic phenotypes of Vibrio damsela associated with a fatal wound infection. J. Clin. Microbiol. 21, 302–306.
DiRita, V. J., Parsot, C., Jander, G., and Mekalanos, J. J. (1991). Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 88, 5403–5407. doi: 10.1073/pnas.88.12.5403
Ducret, A., Quardokus, E., and Brun, Y. V. (2016). MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1:16077. doi: 10.1038/nmicrobiol.2016.77
Flamez, C., Ricard, I., Arafah, S., Simonet, M., and Marceau, M. (2008). Phenotypic analysis of Yersinia pseudotuberculosis 32777 response regulator mutants: new insights into two-component system regulon plasticity in bacteria. Int. J. Med. Microbiol. 298, 193–207. doi: 10.1016/j.ijmm.2007.05.005
Fouz, B., Larsen, J. L., Nielsen, B., Barja, J. L., and Toranzo, A. E. (1992). Characterization of Vibrio damsela strains isolated from turbot Scophthalmus maximus in Spain. Dis. Aquat. Organ. 12, 155–166. doi: 10.3354/dao012155
Fouz, B., Toranzo, A. E., Milán, M., and Amaro, C. (2000). Evidence that water transmits the disease caused by the fish pathogen Photobacterium damselae subsp. damselae. J. Appl. Microbiol. 88, 531–535. doi: 10.1046/j.1365-2672.2000.00992.x
Gao, Q., Ye, Z., Wang, X., Mu, X., Gao, S., and Liu, X. (2015). RstA is required for the virulence of an avian pathogenic Escherichia coli O2 strain E058. Infect. Genet. Evol. 29, 180–188. doi: 10.1016/j.meegid.2014.11.022
Gao, R., Mack, T. R., and Stock, A. M. (2007). Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32, 225–234. doi: 10.1016/j.tibs.2007.03.002
Gao, R., and Stock, A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154. doi: 10.1146/annurev.micro.091208.073214
Herrera, C. M., Crofts, A. A., Henderson, J. C., Cassandra Pingali, S., Davies, B. W., and Trent, S. M. (2014). The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio 5:e02283-14. doi: 10.1128/mBio.02283-14
Herrero, M., de Lorenzo, V., and Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990
Hundenborn, J., Thurig, S., Kommerell, M., Haag, H., and Nolte, O. (2013). Severe wound infection with Photobacterium damselae ssp. damselae and Vibrio harveyi, following a laceration injury in marine environment: a case report and review of the literature. Case Rep. Med. 2013:610632. doi: 10.1155/2013/610632
Jang, J., Jung, K. T., Park, J., Yoo, C. K., and Rhie, G. E. (2011). The Vibrio cholerae VarS/VarA two-component system controls the expression of virulence proteins through ToxT regulation. Microbiology 157, 1466–1473. doi: 10.1099/mic.0.043737-0
Kreger, A. S., Bernheimer, A. W., Etkin, L. A., and Daniel, L. W. (1987). Phospholipase D activity of Vibrio damsela cytolysin and its interaction with sheep erythrocytes. Infect. Immun. 55, 3209–3212.
Le Roux, F., Binesse, J., Saulnier, D., and Mazel, D. (2007). Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784. doi: 10.1128/AEM.02147-06
Le Roux, F., Davis, B. M., and Waldor, M. K. (2011). Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 39, 1004–1013. doi: 10.1093/nar/gkq852
Liu, P. V. (1957). Survey of hemolysin production among species of pseudomonads. J. Bacteriol. 74, 718–727.
Miller, J. H. (1992). A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Mouriño, S., Osorio, C. R., and Lemos, M. L. (2004). Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186, 6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004
Ogasawara, H., Hasegawa, A., Kanda, E., Miki, T., Yamamoto, K., and Ishihama, A. (2007). Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189, 4791–4799. doi: 10.1128/JB.00319-07
Ogasawara, H., Yamada, K., Kori, A., Yamamoto, K., and Ishihama, A. (2010). Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156, 2470–2483. doi: 10.1099/mic.0.039131-0
Oshima, T., Aiba, H., Masuda, Y., Kanaya, S., Sugiura, M., Wanner, B. L., et al. (2002). Transcriptome analysis of all two-component regulatory systems mutants of Escherichia coli K-12. Mol. Microbiol. 46, 281–291. doi: 10.1046/j.1365-2958.2002.03170.x
Osorio, C. R., Toranzo, A. E., Romalde, J. L., and Barja, J. L. (2000). Multiplex PCR assay for ureC and 16S rRNA genes clearly discriminates between both subspecies of Photobacterium damselae. Dis. Aquat. Organ. 40, 177–183. doi: 10.3354/dao040177
Parales, R. E., and Harwood, C. S. (1993). Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram- bacteria. Gene 133, 23–30. doi: 10.1016/0378-1119(93)90220-W
Perez, J. C., Shin, D., Zwir, I., Latifi, T., Hadley, T. J., and Groisman, E. A. (2009). Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:e1000428. doi: 10.1371/journal.pgen.1000428
Rivas, A. J., Balado, M., Lemos, M. L., and Osorio, C. R. (2011). The Photobacterium damselae subsp. damselae hemolysins damselysin and HlyA are encoded within a new virulence plasmid. Infect. Immun. 79, 4617–4627. doi: 10.1128/IAI.05436-11
Rivas, A. J., Balado, M., Lemos, M. L., and Osorio, C. R. (2013a). Synergistic and additive effects of chromosomal and plasmid-encoded hemolysins contribute to hemolysis and virulence in Photobacterium damselae subsp. damselae. Infect. Immun. 81, 3287–3299. doi: 10.1128/IAI.00155-13
Rivas, A. J., Labella, A., Borrego, J. J., Lemos, M. L., and Osorio, C. R. (2014). Evidences for horizontal gene transfer, gene duplication and genetic variation as driving forces of the diversity of haemolytic phenotypes in Photobacterium damselae subsp. damselae. FEMS Microbiol. Lett. 355, 152–162. doi: 10.1111/1574-6968.12464
Rivas, A. J., Lemos, M. L., and Osorio, C. R. (2013b). Photobacterium damselae subsp. damselae, a bacterium pathogenic for marine animals and humans. Front. Microbiol. 4:283. doi: 10.3389/fmicb.2013.00283
Rivas, A. J., Vences, A., Husmann, M., Lemos, M. L., and Osorio, C. R. (2015a). Photobacterium damselae subsp. damselae major virulence factors Dly, plasmid-encoded HlyA, and chromosome-encoded HlyA are secreted via the type II secretion system. Infect. Immun. 83, 1246–1256. doi: 10.1128/IAI.02608-14
Rivas, A. J., Von Hoven, G., Neukirch, C., Meyenburg, M., Qin, Q., Füser, S., et al. (2015b). Phobalysin, a small ß-pore-forming toxin of Photobacterium damselae subsp. damselae. Infect. Immun. 83, 4335–4348. doi: 10.1128/IAI.00277-15
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Terceti, M. S., Ogut, H., and Osorio, C. R. (2016). Photobacterium damselae subsp. damselae, an emerging fish pathogen in the Black Sea: evidences of a multiclonal origin. Appl. Environ. Microbiol. 82, 3736–3745. doi: 10.1128/AEM.00781-16
Tran, T. K., Han, Q. Q., Shi, Y., and Guo, L. (2016). A comparative proteomic analysis of Salmonella typhimurium under the regulation of the RstA/RstB and PhoP/PhoQ systems. Biochim. Biophys. Acta 1864, 1686–1695. doi: 10.1016/j.bbapap.2016.09.003
Whitaker, W. B., Parent, M. A., Boyd, A., Richards, G. P., and Boyd, E. F. (2012). The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect. Immun. 80, 1834–1845. doi: 10.1128/IAI.06284-11
Keywords: RstAB, hemolysin, Photobacterium damselae, damselysin, phobalysin, vibriosis, CarSR
Citation: Terceti MS, Rivas AJ, Alvarez L, Noia M, Cava F and Osorio CR (2017) rstB Regulates Expression of the Photobacterium damselae subsp. damselae Major Virulence Factors Damselysin, Phobalysin P and Phobalysin C. Front. Microbiol. 8:582. doi: 10.3389/fmicb.2017.00582
Received: 26 December 2016; Accepted: 21 March 2017;
Published: 10 April 2017.
Edited by:
Alison Buchan, University of Tennessee, USAReviewed by:
Jia Wen, Duke University, USAAlexandre Soares Rosado, Federal University of Rio de Janeiro, Brazil
Copyright © 2017 Terceti, Rivas, Alvarez, Noia, Cava and Osorio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos R. Osorio, Y3Iub3NvcmlvQHVzYy5lcw==
†These authors have contributed equally to this study.
 Mateus S. Terceti
Mateus S. Terceti Amable J. Rivas
Amable J. Rivas Laura Alvarez
Laura Alvarez Manuel Noia3
Manuel Noia3 Carlos R. Osorio
Carlos R. Osorio