- 1Department of Molecular Genetics, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, Netherlands
- 2Top Institute Food and Nutrition, Wageningen, Netherlands
Bacteria can deploy various mechanisms to combat environmental stresses. Many genes have previously been identified in Lactococcus lactis that are involved in sensing the stressors and those that are involved in regulating and mounting a defense against the stressful conditions. However, the expression of small regulatory RNAs (sRNAs) during industrially relevant stress conditions has not been assessed yet in L. lactis, while sRNAs have been shown to be involved in many stress responses in other bacteria. We have previously reported the presence of hundreds of putative regulatory RNAs in L. lactis, and have used high-throughput RNA sequencing (RNA-seq) in this study to assess their expression under six different stress conditions. The uniformly designed experimental set-up enabled a highly reliable comparison between the different stress responses and revealed that many sRNAs are differentially expressed under the conditions applied. The primary stress responses of L. lactis NCDO712 was benchmarked to earlier work and, for the first time, the differential expression was assessed of transfer RNAs (tRNAs) and the genes from the six recently sequenced plasmids of NCDO712. Although, we only applied stresses for 5 min, the majority of the well-known specific stress-induced genes are already differentially expressed. We find that most tRNAs decrease after all stresses applied, except for a small number, which are increased upon cold stress. Starvation was shown to induce the highest differential response, both in terms of number and expression level of genes. Our data pinpoints many novel stress-related uncharacterized genes and sRNAs, which calls for further assessment of their molecular and cellular function. These insights furthermore could impact the way parameters are designed for bacterial culture production and milk fermentation, as we find that very short stress conditions already greatly alter gene expression.
Introduction
Bacteria display many general as well as specific molecular responses to environmental changes. Sudden alterations in the environment can be of physical or chemical nature and can threaten the lifespan of a microbial cell, especially if the stress condition is too intense in time or intensity. The metabolic activity of bacteria used in industrial fermentations is altered upon stress in their for example acidification rates and flavor formation (Xie et al., 2004; Taïbi et al., 2011). Gaining insights in the effects of stress could improve the predictability, quality, and safety of fermentations.
The lactic acid bacterium (LAB) Lactococcus lactis is of eminent importance in the dairy industry, where it is used worldwide for the production of a large variety of cheeses and of buttermilk. The main function during milk fermentation of LAB such as L. lactis is to convert lactose into lactic acid. The consequent lowering of the pH leads to an increased shelf life of the fermented products as it prevents outgrowth of spoilage or pathogenic organisms. In addition, L. lactis provides texture, flavors, and aromas to the end products (Marilley and Casey, 2004; Smit et al., 2005). During their preparation as a starter culture, as well as in the actual fermentation process, fluctuations in temperature, osmolarity, pH, and nutrient availability cause significant stress to the L. lactis cells. They have evolved different response systems to sense and respond to potential lethal conditions and to defend themselves accordingly, in order to survive. Many of these mechanisms and the regulatory systems involved have been identified in L. lactis, on the basis of homology to proteins with known functions in other organisms and/or by experimental validation (Sanders et al., 1999; Smith et al., 2010). The potential role in stress of small non-coding regulatory RNAs (sRNAs) has not yet been assessed in L. lactis, while sRNAs have been shown to play an important function in a variety of stress conditions in other bacteria (Gottesman et al., 2006; Romby and Charpentier, 2010; Hoe et al., 2013).
Bacterial regulatory RNAs are generally non-coding transcripts that modulate gene expression post-transcriptionally (Waters and Storz, 2009). They are usually classified on whether or not genes are encoded on the strand opposite to the strand from which they derive. Non-coding RNAs that are located within intergenic regions (IGRs) are referred to as small RNAs (sRNAs) and roughly contain between 50 and 350 nucleotides. They are very heterogeneous in size and structure, and act in trans to target mRNAs (Gottesman and Storz, 2011; Storz et al., 2011). Transcripts that overlap in an antisense fashion with mRNAs from the opposite strand are called antisense RNAs (asRNAs) (Thomason and Storz, 2010; Georg and Hess, 2011). sRNAs and asRNAs with proven functions in regulating other RNAs and/or proteins can be considered regulatory RNAs. Base-pairing between a regulatory RNA and its target mRNA(s) can affect mRNA stability as well as translation, the latter by influencing the accessibility of the ribosomal binding site (RBS) on the target transcript (Morita and Aiba, 2011; Prevost et al., 2011; Bandyra et al., 2012; Papenfort and Vanderpool, 2015). Since the discovery of the first regulatory RNAs, starting with antisense RNAs involved in plasmid copy number control (Stougaard et al., 1981; Tomizawa et al., 1981) and the first genomically encoded sRNA MicF (Mizuno et al., 1984), many others have been described especially since recent advances have been in high-throughput RNA sequencing (RNA-seq) as well as in high-density tilling arrays (Nicolas et al., 2012). Various mechanisms of action have been elucidated since but determining the functions and mechanisms of action of novel sRNAs is still the major challenge. This may be illustrated by the abundant non-coding RNA 6S, which was discovered as early as 1967 (Hindley, 1967) but of which the function in modifying RNA polymerase activity was uncovered only in 2000 (Wassarman and Storz, 2000).
Several examples exist of sRNAs that perform a crucial role in the adaptation and survival of bacteria during stressful conditions. For instance, the sRNA RybB from E. coli and Salmonella is activated by the extracytoplasmic stress sigma factor, σE. Accumulation of misfolded outer membrane proteins (OMPs), for example in the stationary phase, can cause cell envelope stress. RybB downregulates many different omp mRNAs in order to prevent the synthesis of OMPs and to restore envelope homeostasis (Johansen et al., 2006; Papenfort et al., 2006; Papenfort and Vogel, 2009). Another striking example of how effective and diverse the scope of one sRNA can be is the bifunctional sRNA SgrS from E. coli and Salmonella. The transcription factor SgrR is triggered upon glucose-phosphate stress and activates SgrS transcription. The stress is immediately relieved by detoxification of the phosphosugar. This is mediated by SgrS base-pairing with yigL, blocking yigL degradation by RNase E and resulting in increased YigL expression (Papenfort et al., 2013). To decrease phosphosugar accumulation in the cell by PtsG and ManXYZ, SgrS base-pairs with and blocks translation of the mRNAs of manXYZ and ptsG, which are eventually degraded (Bobrovskyy and Vanderpool, 2014). The 5′ end of SgrS contains an ORF, sgrT. This SgrT peptide interacts with PtsG in such a way that it blocks the uptake of glucose (Wadler and Vanderpool, 2007).
Recently, we have re-annotated the genome of L. lactis MG1363 by extensive mining of differential RNA sequencing (dRNA-seq) data of samples taken at various points during growth as a batch culture (van der Meulen et al., 2016). This has led to the addition of 186 small non-coding RNAs and 60 antisense RNAs to the L. lactis genome annotation. Here we have treated the parent strain of L. lactis MG1363, L. lactis NCDO712, which carries a number of plasmids with industrial relevance (Tarazanova et al., 2016), to a number of industrial stresses. Specifically, the strain was exposed for a relatively short period of time of 5 min to cold, heat, acid, osmotic, oxidative or starvation stress to explore the organism's early transcriptome responses by strand-specific RNA-seq. The expression of recently annotated sRNAs and asRNAs was assessed and a significant number of them were observed to be differentially expressed under the various stress conditions. Moreover, extensive data mining allowed pinpointing many genes that are involved in the investigated, industry-related stress conditions.
Materials and Methods
Bacterial Strains and Media
Lactococcus lactis subsp. cremoris NCDO712 is an industrial strain containing six plasmids of which one, pLP712, carries lactose and casein utilization genes (Tarazanova et al., 2016). L. lactis MG1363 is its plasmid-free derivative (Gasson, 1983). L. lactis NCDO712 was grown as a standing culture at 30°C in M17 broth (Difco, Becton Dickinson, Le Pont de Claix, France) containing 0.5% (w/v) glucose (GM17), and on GM17 agar plates.
Stress Treatments
All stress conditions were applied in two biological replicates by starting with two single colonies of L. lactis NCDO712 grown on GM17 agar plates. These were each used to inoculate 10 ml of fresh GM17 media and grown overnight. Each overnight culture was diluted 100-fold in a bottle with 500 ml GM17 and grown until an optical density at 600 nm (OD600) of 0.9 was reached. The content of each bottle was divided over seven 50-ml Greiner tubes and centrifuged at 4,000 g for 1.5 min. Subsequently, the cultures from each bottle were subjected to all conditions tested. The cell pellets were re-suspended in fresh GM17 containing 0.25% glucose (G*M17) and the following properties: control (G*M17 at 30°C), cold (G*M17 at 10°C), heat (G*M17 at 42°C), acid (G*M17 set at pH 4.5 with lactic acid), osmotic stress (G*M17 containing 2.5% NaCl), and oxidative stress (G*M17, shaking at 250 rpm). The cold stress was applied in an incubator while the heat stress was performed in a water bath to maintain the cold and heat conditions stable. For starvation stress, the cell pellets were re-suspended in filter-sterilized phosphate-buffered saline (PBS). The stress conditions were applied for 5 min, after which the cells were harvested by centrifugation at 10,000 rpm for 1 min, snap frozen in liquid nitrogen and stored at −80°C prior to RNA isolation.
RNA Isolation
RNA was isolated as described previously (van der Meulen et al., 2016). All procedures were executed at 4°C unless otherwise stated and all solutions were DEPC-treated and subsequently autoclaved. Frozen cell pellets were re-suspended in 400 μl TE-buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4) and added to 50 μl 10% sodium dodecyl sulfate (SDS), 500 μl phenol/chloroform (1:1 v/v), and 0.5 g glass beads (75–150 μm, Thermo Fischer Scientific, Rockford, IL, United States). The cells were disrupted by shaking 2 times for 45 s in a Biospec Mini-BeadBeater (Biospec Products, Bartlesville, OK, United States) with cooling on ice for 1 min between the shaking steps. Subsequently, the cell suspension was centrifuged at 14,000 rpm for 10 min. The upper phase containing the nucleic acids was treated with 500 μl chloroform and centrifuged as above. Nucleic acids in the water phase were precipitated by sodium acetate and ethanol. The nucleic acid pellet was re-suspended in 100 μl buffer consisting of 82 μl MQ, 10 μl 10x DNase I buffer, 5 μl RNase-free DNase I (Roche Diagnostics GmbH, Mannheim, Germany), and 3 μl RiboLock RNase inhibitor (Fermentas/Thermo Scientific, Vilnius, Lithuania), and treated for 30 min at 37°C. The RNA was then purified using standard phenol/chloroform extraction and sodium acetate/ethanol precipitation. RNA pellets were re-suspended in 50 μl elution buffer from the High Pure RNA Isolation Kit (Roche Diagnostics, Almere, the Netherlands) and stored at −80°C.
RNA Treatment, Library Preparation, and RNA Deep Sequencing
RNA concentration was measured with a Nanodrop ND-1000 (Thermo Fischer Scientific). As a measure of RNA quality, the integrity of the 16S/23S rRNA and the presence of any DNA contamination were assessed by using an Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany). cDNA library was prepared by employing a ScriptSeqTM Complete Kit for Bacteria (Epicentre, Madison, WI, United States) including Ribo-Zero™ for rRNA removal. The cDNA libraries were sequenced at Otogenetics Corporation (Norcross, GA, United States) on an Illumina HiSeq2000.
Data Analysis
Raw sequence reads were analyzed for quality and trimmed with a PHRED score >28. Read alignment was performed on the genomic DNA of L. lactic NCDO712 (nucleotide sequences of the chromosome and all six plasmids of NCDO712; Tarazanova et al., 2016) using Bowtie 2 (Langmead and Salzberg, 2012). RKPM values were used as an input for the T-REx analysis pipeline (de Jong et al., 2015) together with a text file describing the factors, contrasts, and classes. In the class file, genes from the NCDO712 plasmids were colored green while sRNAs were colored red (see Supplementary Information S1). T-REx was used to perform all statistical analyses (de Jong et al., 2015).
Data Access
The RNA-seq data is publically available in GEO under accession number GSE98499.
Results
RNA-Seq Reveals That Starvation Has a Large Impact on the Transcriptome of L. lactis
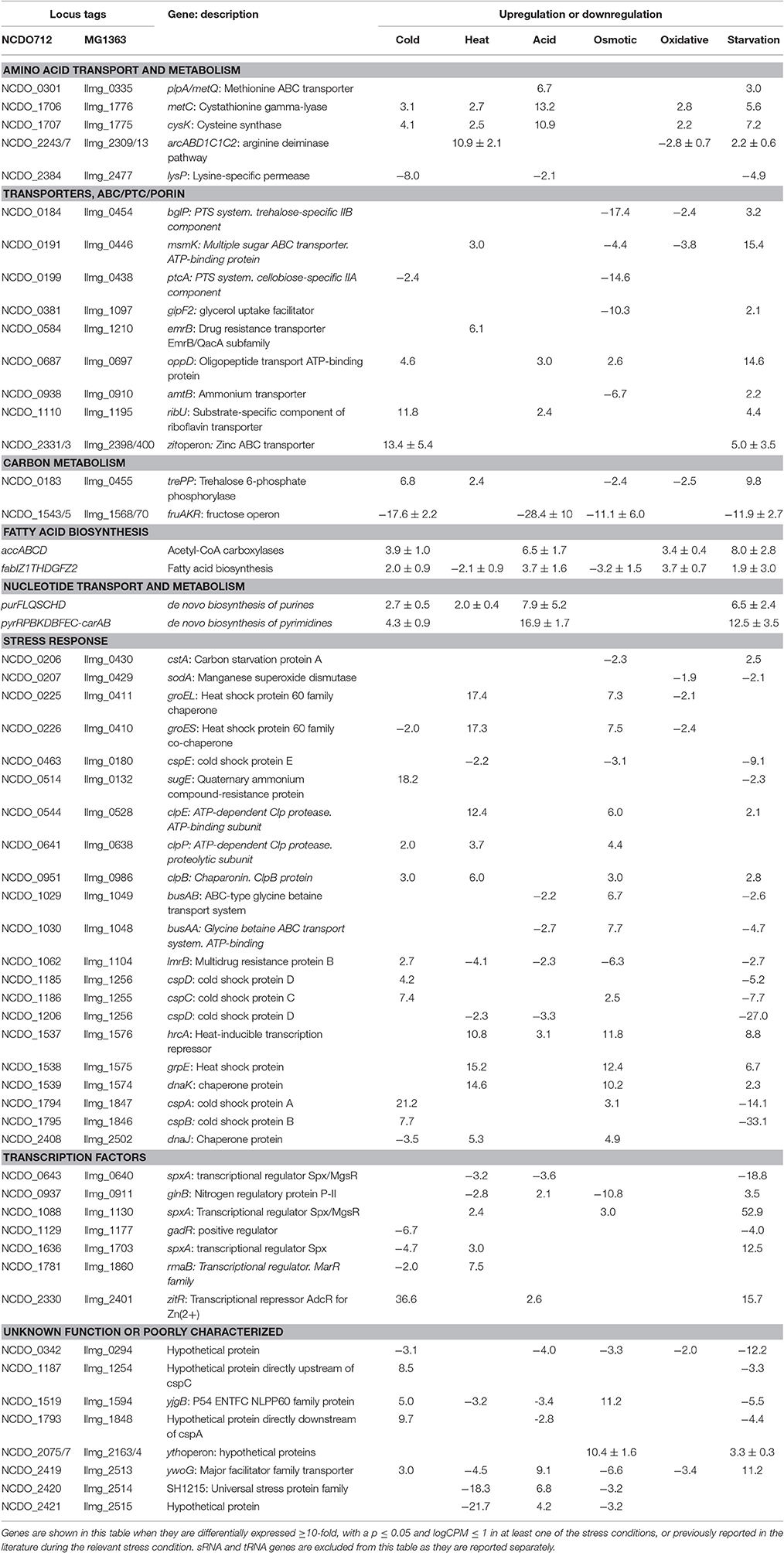
The transcriptomic response of L. lactis subsp. cremoris NCDO712 (hereafter named L. lactis NCDO712) after 5 min of cold, heat, acid, osmotic, oxidative, or starvation stress was determined by high-throughput RNA sequencing. This resulted in a total of 246M of reads of which 209M reads (85%) were successfully mapped on the genome and plasmids of L. lactis NCDO712. The libraries varied between 11M and 19M reads per individual sample (Figure 1A). The data was normalized using the T-REx software (de Jong et al., 2015) and plotted in a box plot of normalized signals for all samples (Figure 1B). A Principle Component Analysis (PCA) shows that the stress conditions are statistically well-distributed from each other (Figure 1C). The transcriptome of cells exposed to osmotic stress or to starvation were most different from that of the control. The absolute numbers of differentially expressed genes underpin these observations; the largest transcriptome changes were observed after starvation (756 genes involved), while oxidative stress had the least impact (91 affected genes). Table 1 gives an overview of the counts of the differentially expressed genes and Figure 2 shows the distribution of affected genes for each stress condition. To gain insight in the distribution of the stress-responsive genes, those of which the expression changed highly (fold change ≥ 5 and p ≤ 0.01) under all conditions were visualized in a heatmap. T-REx was used to pinpoint nine clusters that vary strongly in size and in the functions of the constituting genes. One cluster is rich in stress-related genes, while another one contains predominantly sRNAs. See Figure 3 for the heatmap and the complete list of cluster descriptions. Genes that were affected by a fold change ≥ 10-fold and p ≤ 0.01) are listed in Table 2 and are discussed in more detail below.
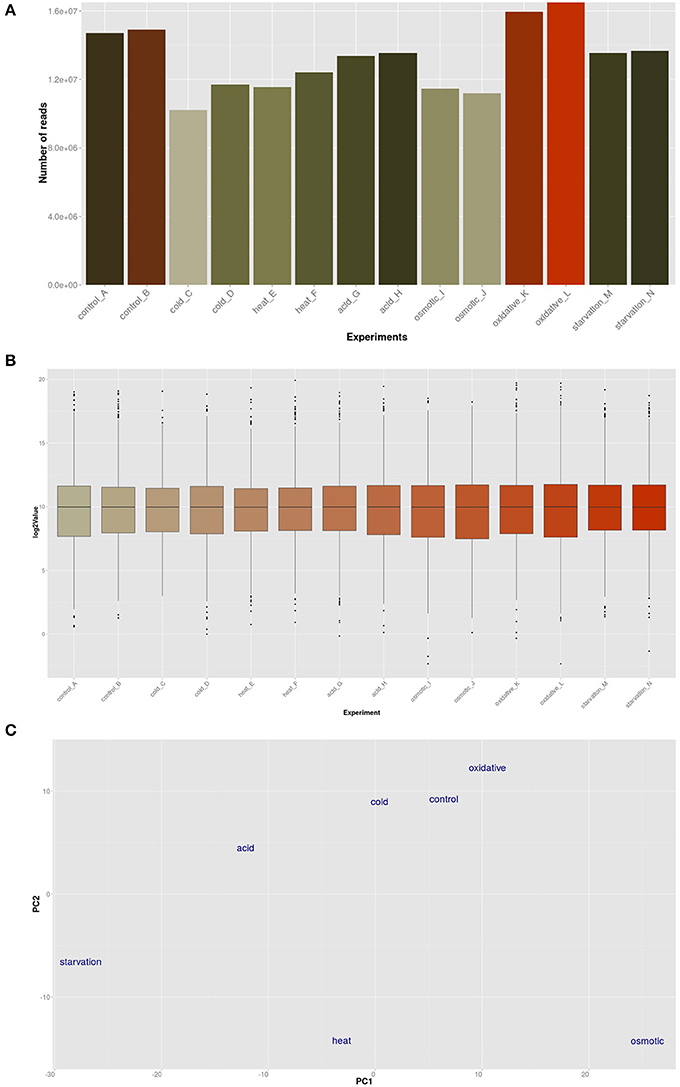
Figure 1. Global analysis by T-Rex (de Jong et al., 2015) of the RNA-seq data. (A) Total number of RNA-seq reads per sample; (B) Box-plot expression values of all experiments; (C) Principle Component Analysis (PCA) plot of the conditions employed in this study.
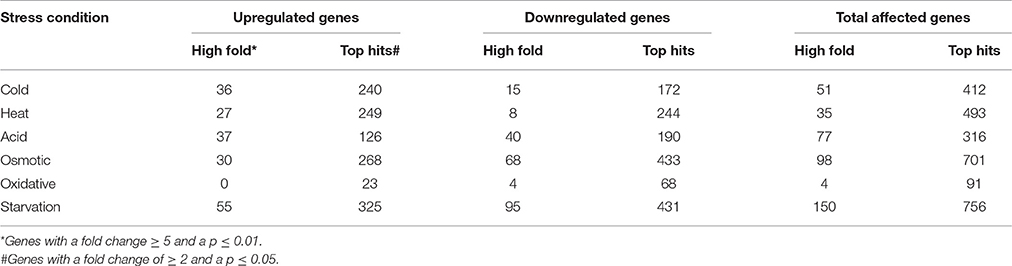
Table 1. Absolute numbers of differentially expressed genes after 5 min of exposure to the indicated stress.
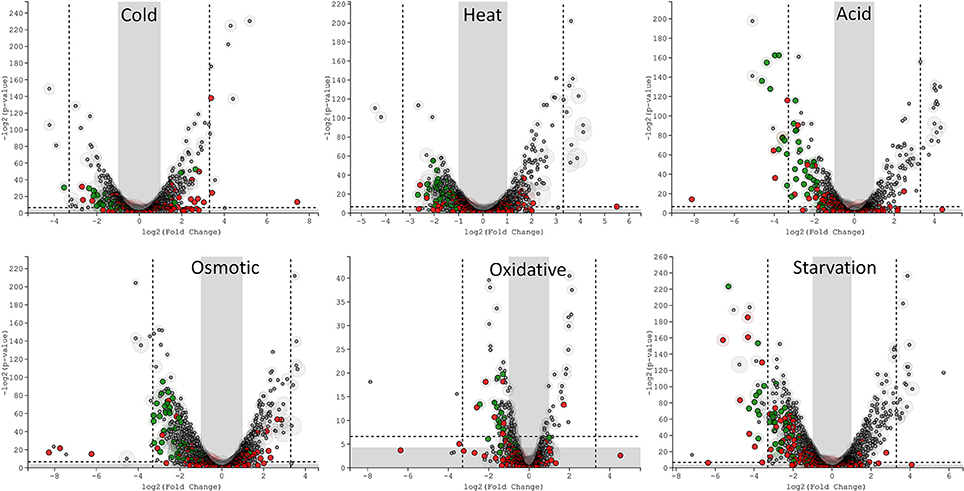
Figure 2. T-REx-generated Volcano plots of the six different stress conditions displaying significance vs. gene expression on the y and x axes respectively. Outside the gray areas: genes with fold change ≥ 2 and a p ≤ 0.05, outside the dashed lines: genes with fold change ≥ 5 and a p ≤ 0.01. Green circles: tRNA genes, red circles: sRNA genes, gray circles: all other genes.
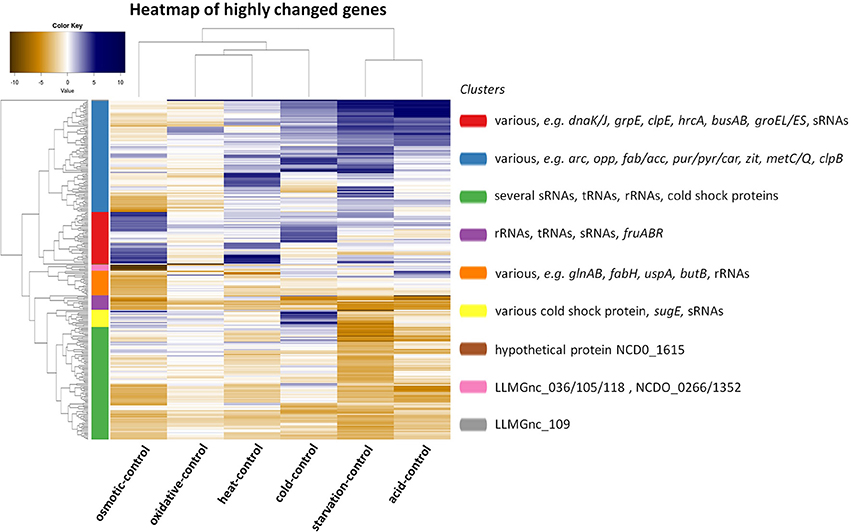
Figure 3. Heat map showing the correlation of a comparison between the indicated stress condition vs. its control and clusters of genes of which the expression has changed with a high fold (fold change ≥ 5 and p ≤ 0.01). Nine clusters were calculated by T-REx (de Jong et al., 2015) and dominant and stress-related genes are listed in clusters.
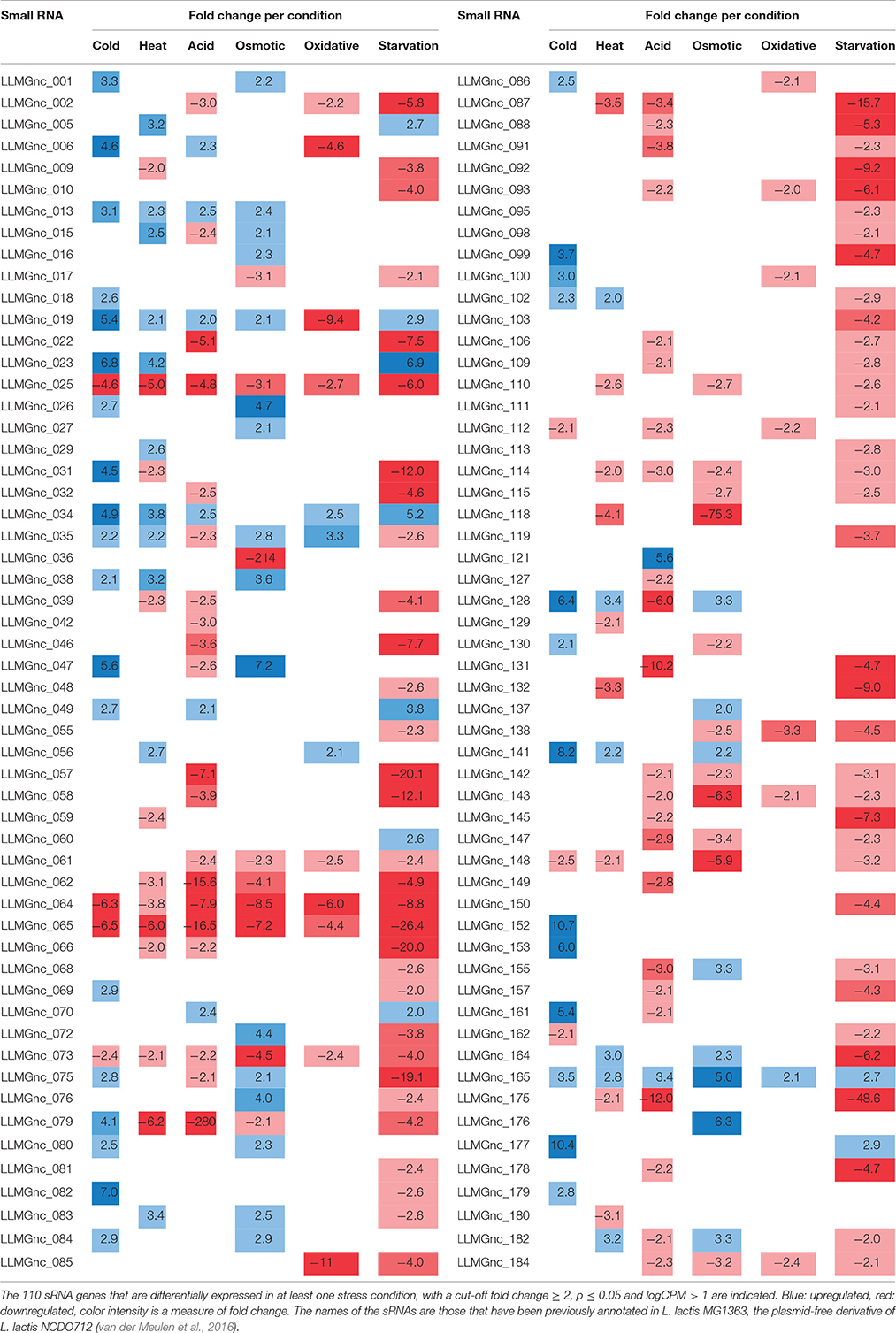
sRNAs Are Highly Affected After 5 Min of Stress Induction
We assessed the expression of the 186 sRNAs that have recently been identified in the genome of L. lactis MG1363 (van der Meulen et al., 2016). The majority of the sRNAs that were significantly changed after applying acid, oxidative, or starvation stress showed a decrease in expression. In contrast, after cold stress more sRNAs were upregulated instead of downregulated (see Figure 4). Of the 186 sRNAs, the expression of 110 was significantly changed after applying at least one stressor, while 42 sRNAs were differentially expressed under at least three stress conditions (Table 3). This list of sRNAs was restricted to those with a logCPM value >1. The expression of some of these 110 sRNA genes was highly affected under only one specific stress condition. For example, LLMGnc_152 (10.7-fold) and LLMGnc_153 (6.0-fold) were increased specifically after cold stress, while LLMGnc_176 (6.3-fold) showed a higher expression after salt stress. The sRNA LLMGnc_092 (−9.2-fold) was only decreased after starvation, while LLMGnc_025/064/065/073 were downregulated in all conditions.
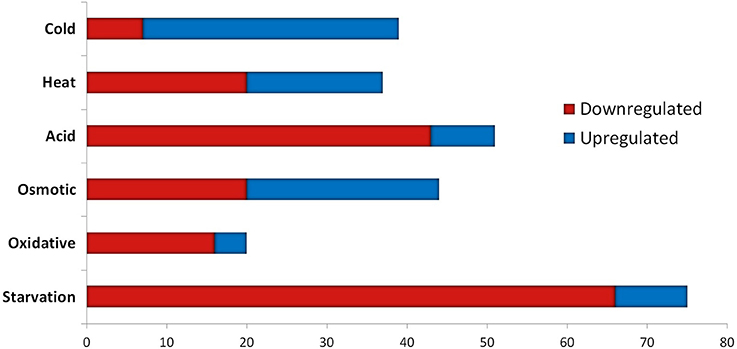
Figure 4. Overview of significantly differentially expressed sRNA genes under various stress conditions (cut-off fold change ≥ 2, p ≤ 0.05 and logCPM > 1).
Most Transfer RNAs Decrease Rapidly after a Short Pulse of Stress
Transfer RNAs (tRNAs) play a crucial role in the translation of mRNAs and it is important for cells to balance their tRNA levels, as well as to ensure optimal utilization of amino acids. Most of the L. lactis tRNAs are downregulated under all of the stresses applied. A number of tRNAs, such as NCDO_2402 (Val-CAG) and NCDO_2022 (Lys-TTT), are upregulated under some of the conditions. A distinct tRNA response is observed upon cold stress; seven tRNAs are upregulated by at least 2-fold. Exposure to acid or starvation stress induced the most severe changes in tRNA expression. See Figure 4 for a complete overview of tRNA expression under the various stress conditions.
Cold Stress Induces a Zinc Uptake System
During a 5-min cold stress at 10°C, 412 genes were differentially expressed (Table 1). The most highly upregulated transcripts include those from the zit operon, a gene cluster involved in Zn2+ uptake and regulation (Llull and Poquet, 2004). Also, expression of the gene sugE, encoding a presumed multidrug resistance protein, was increased (~18-fold), as well as genes involved in nucleotide synthesis (pur and pyr operons) and the fab and acc operons for saturated fatty acid biosynthesis. Transcripts encoding Cold Shock Proteins A, B, C, and D were upregulated at least 4-fold, as would be expected in cells under cold stress (Wouters et al., 1998). The gene of an uncharacterized protein (Llmg_1848) with high sequence similarity to a bacteriocin in other L. lactis species, is located downstream of cspA and was also upregulated by a factor of ~10. Downregulation was observed e.g., for the fructose utilization fru operon (~18-fold), the lysine-specific permease lysP gene (~8-fold) and the ribosomal RNA 5S (~9-fold). Thirty-nine sRNAs from intergenic regions were significantly affected, of which 12 were changed at least 5-fold (see Figure 4 and Table 3).
Heat Stress Induces, Next to Protein Chaperone Genes, the Arc Operon
At high temperatures, proteins may be at risk of denaturation and cells may encounter difficulties in the synthesis of new proteins (Nguyen et al., 1989; Parsell and Lindquist, 1993). The genes of several protein chaperones, such as GroEL, GroES, DnaJ, DnaK, and GrpE are usually upregulated after heat stress. The chaperones aid in protein folding and maturation. Their genes were upregulated ~14–18 times (except dnaJ, which was increased ~5-fold) after L. lactis was placed for 5 min at 42°C (see Table 2). The Clp protease genes clpE, clpP, and clpB were also upregulated, as was the gene with an unknown function upstream of clpE, llmg_0527. Surprisingly, expression of genes belonging to the arginine deaminase pathway (arc operon) were increased ~11-fold, while this operon was downregulated after 30 min of incubation of L. lactis IL1403 at 42°C in previous work (Xie et al., 2004). An operon predicted to be involved in the utilization of maltose (llmg_0485-llmg_0490), the ribose operon (rbsABCDK) and llmg_1210, predicted to encode the drug resistance transporter EmrB, were all upregulated. We also observed that the lacR-lacABCDEF gene cluster, located on the largest plasmid pLP712 of L. lactis NCDO712 (Wegmann et al., 2012) and involved in lactose utilization, was upregulated ~3-fold. The heat treatment caused a decrease by a factor of 2–3 of several rRNA species, as well as of most tRNA-transcripts (Figure 5). An operon (llmg_2513-llmg_2515) containing a gene for the universal stress protein A (UspA) and two uncharacterized genes was decreased most severely after heat shock, followed in severity of fold change by the multidrug resistance protein B gene (lmrB, llmg_1104). In total, 37 sRNAs were differentially expressed (Table 3). Expression of two of them was decreased over 5-fold: LLMGnc_065 (−6.0) and LLMGnc_079 (−6.2).
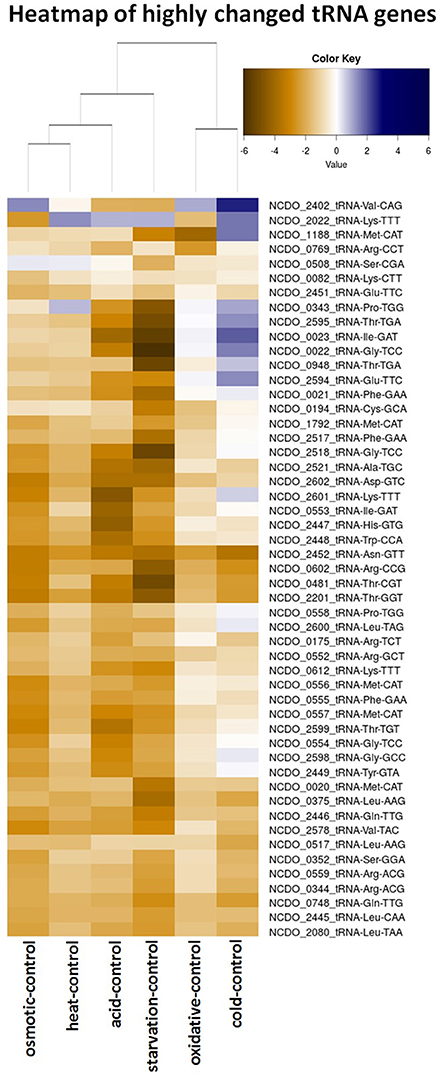
Figure 5. Heatmap of the expression of all tRNA genes from L. lactis NCDO712 under the various stress conditions. Expression values are presented in log2-fold, as depicted in the color key.
Acid Stress Induces Nucleotide Biosynthesis and Cysteine/Methionine Metabolism
Genes for de novo synthesis of pyrimidines (pyr) and purines (pur) were highly upregulated after 5 min of exposure to pH 4.5. Expression of these operons was also seen during starvation and, to a lesser extent, upon cold shock. Among the top upregulated genes were metCcysK encoding a cystathionine gamma-lyase and cysteine synthase, respectively, involved in cysteine and methionine metabolism. The genes llmg_0333-0340 were all affected by the acid stress applied, although they are not transcribed from the same operon. Among these genes are those of a putative methionine ABC transporter system (llmg_0335-0340 upregulated), and thiT (llmg_0334) encoding the thiamine transporter (Erkens and Slotboom, 2010) and a putative transcriptional regulator gene (llmg_0333) (both downregulated). The fab operon, responsible for the biosynthesis of saturated fatty acids (SFAs) for membrane phospholipids, was upregulated. Expression of the operon llmg_2513-2515 was increased, while these genes were strongly downregulated after heat (see above) and salt stress (see further). The putative Mn2+/Fe2+ transporter gene mntH, which is located on plasmid pNZ712, was upregulated 2.5-fold under pH 4.5 stress. Interestingly, the chromosomal gene mgtA, putatively specifying Mg2+ transport, was downregulated 3.7-fold. Upregulation was furthermore observed for llmg_1915-1917 (ykgEFG) and for the following genes with predicted functions; llmg_1066 (unknown), llmg_1133 (exonuclease), and llmg_1702 (glutathione reductase). The fruAKR operon for fructose transport, conversion of fructose 1-phosphate to fructose 1,6-bisphosphate and their regulation by the FruR repressor (Barriere et al., 2005), was downregulated on average ~28-fold. The fhuABCD operon for putative ferric siderophore transport was downregulated by a factor 3–7. Of the 51 sRNA genes of which the expression was significantly changed, only 8 were upregulated; among these was LLMGnc_121, which was exclusively upregulated after acid stress (Table 3).
Osmotic Stress Induces Chaperones and a Putative Stress-Responsive Regulator
Osmotic stress was induced by adding 2.5% NaCl to the cell culture for 5 min. Among the highest upregulated genes are those of an operon (llmg_2163 - llmg_2164) specifying a putative stress-responsive transcriptional regulator with a PspC domain (Llmg_2163). Both genes are ~10-fold upregulated; it was also induced upon overproduction in L. lactis of the membrane protein BcaP (Pinto et al., 2011) and after exposure to the bacteriocin Lcn972 (Martínez et al., 2007). A deletion mutant of llmg_2164 was shown to be very sensitive to NaCl (Roces et al., 2009). In E. coli, the psp operon is induced after application of various types of stresses including salt stress (Brissette et al., 1991). As expected, induction was seen of the genes specifying the glycine betaine ABC transport system BusAA-BusAB (~7- to 8-fold). Some of the responses observed during the exposure to the high concentration of salt were similar to those seen after heat stress. In particular, transcripts encoding the chaperones GroEL, GroES, DnaJ, DnaK, and GrpE were upregulated in the same fold change range. Induction of these proteins has been reported previously for both heat and salt stress (Kilstrup et al., 1997). Both operons for oligopeptide transport were also upregulated under the salt stress applied here.
Genes encoding transporters for various substrates were strongly downregulated, among which the PTS IIA component genes fruA and ptcA, that of the IIB PTS component, bglP, lmrB specifying multidrug resistance protein B (both llmg_0967 and llmg_1104), msmK, encoding a multiple sugar ABC transporter and amtB involved in ammonium transport. Also, the gene encoding the glycerol uptake facilitator protein GlpF2 was downregulated by ~10-fold, as was the lac operon on pLP712. As shown before (Xie et al., 2004), the potABCD operon involved in spermidine/putrescine transport and the fatty acid biosynthesis operons fab and acc were downregulated under osmotic stress. As mentioned above, the llmg_2513-2515 operon was downregulated. From the 44 significantly affected sRNA genes, the expression of LLMGnc_036 (214-fold down), LLMGnc_118 (75-fold down), and LLMGnc_176 (6.3-fold up) was specifically only changed after salt stress using a threshold of 5-fold change.
Shaking of a Culture of L. lactis Triggers Saturated Fatty Acid Biosynthesis Genes
From all stress conditions tested, oxidative stress applied by shaking of the culture resulted in the least number of differentially expressed genes (Table 1). The ones that did change did so with relatively minor fold changes. Among the few upregulated genes were those of the pathway for saturated fatty acid biosynthesis, including fabT, the transcriptional repressor of this route (Eckhardt et al., 2013). Downregulation was seen of arcABD1C1C2, llmg_1915-1917 (ykgEFG), and of genes involved in the uptake and/or utilization of maltose, trehalose, and lactose. The heat shock chaperone genes groEL and groES were downregulated ~2-fold. Unexpectedly, the gene for the manganese superoxide dismutase SodA, an enzyme well-known for its role in oxidative stress, was decreased 2-fold relative to the unstressed control. Twenty sRNAs were affected by at least 2-fold. LLMGnc_019 was downregulated 9.4-fold, while it was upregulated in all the other stress conditions (see Table 3).
Starvation in PBS Resulted in the Most Dramatic Transcriptome Changes
Incubation of the cells for 5 min in PBS greatly affected the transcriptome of L. lactis, as witnessed by the large number of 150 genes of which the expression had changed significantly, and at least by 5-fold. Different biological functions were switched on or off in response to the sudden absence of basically all nutrients. For example, the operons for the de novo biosynthesis of nucleotides, pyr, pur, and car, were all highly upregulated. Moreover, the genes of several transport systems were upregulated in an apparent attempt to import a (any) carbon source (lactose: lac operon, multiple sugar ABC transporter: msmK, ribose: rbs operon, cellobiose/glucose: ptcC), ions (manganese: mtsA, zinc: zit operon), amino acids (arginine: arc, methionine: metQ, branched-chain amino acid transporter: ctrA/bcaP), and vitamins (riboflavin: rib operon). On the other hand, some uptake system genes were downregulated, such as thiT, specifying the thiamin transporter, and the fru operon for fructose uptake and utilization. Strong downregulation was also observed for the genes of all six cold shock proteins. Dozens of (predicted) transcriptional regulators were affected upon the starvation stress applied here, among which all of the spxA genes. Interestingly, the expression profiles were very different. For instance, spxA with locus tag llmg_0640 was decreased ~19-fold, while expression of llmg_1703 and llmg_1130 was increased 12.5- and 53-fold, respectively. Notably, the gene for the putative transcriptional repressor CadC, which is located on plasmid pSH73 (Tarazanova et al., 2016), was 4.9-fold upregulated. Besides a strong decrease in the expression of different tRNA genes, also transcripts for ribosomal proteins were downregulated. Starvation changed the expression of 75 sRNAs, of which the majority was downregulated. The nine sRNA genes that were upregulated after starvation were also increased under at least one of the other conditions tested here, with the exception of LLMGnc_060, which was only affected after starvation.
Discussion
High-throughput RNA sequencing was used in this study to examine the transcriptome changes caused by various industrially relevant stress conditions applied to L. lactis NCDO712. Previous studies using DNA microarray- and proteomics technologies have identified genes and proteins involved in the various environmental stress responses in L. lactis. However, little to no insight has been obtained so far as to which small regulatory RNAs (sRNAs) and antisense transcripts (asRNAs) are affected by stress, and to what extent. The strand specificity of DNA microarray probes does not allow detection using this technology of antisense transcripts. Also, conventional DNA microarrays usually do not carry tRNA probes and, of course, no probes for as yet undefined transcripts; high-density tilling arrays can detect unknown transcripts. RNA sequencing can be used to uncover all transcripts in an organism at a specific moment in time. It also provides a higher dynamic range for quantitative gene expression analysis than DNA microarrays, provides single-base resolution and suffers less from background noise signals (Wang et al., 2009). In the present study, we have applied a size cut-off of 50 nt to detect sRNAs.
Bacterial cells employed during starter culture or cheese production encounter stress conditions that are similar to the ones applied here, albeit not always as instant and short-lived (5 min induction in our experiments). We chose such a very brief duration of the stressors because we were interested in the ensuing very first transcriptional responses, while in previous studies stress conditions were applied for 10 min to up to 4 h (Sanders et al., 1999). The longer the exposure time, the more secondary effects are activated that obscure the actual first response. The expression of stress-induced genes can quickly build up to a certain level, after which it decreases again. This is, for instance, observed for the L. lactis hrcA, groESL, dnaJ, and dnaK genes, which all reach a maximum expression level after 15 min of heat shock (Arnau et al., 1996). A study in Salmonella typhimurium shows a detailed overview of sRNA expression over time, by sequencing of Hfq-bound transcripts. While some sRNAs were expressed throughout growth, others were only dominant at one specific growth phase (Chao et al., 2012). Growth-phase and stress-dependent expression of sRNAs in L. lactis were only conducted for a selection of sRNAs in our previous work (van der Meulen et al., 2016). Here, we focused our analyses on the changes in expression levels of sRNA genes and of tRNA genes. Albeit that the current study was of a fundamental nature, some of the results presented here might ultimately be used in starter culture production or milk fermentation by applying short pulses of stress to the bacteria.
We observed that a number of operons and individual genes were highly affected by three or more stress conditions. For example, the pur and pyr operons for the de novo synthesis of purines and pyrimidines were upregulated after cold, acid and starvation stress. The fruAKR was downregulated upon cold, acid, osmotic, and starvation stress. The fru operon was previously reported to be upregulated in response to cell envelope stress by the bacteriocin Lcn972 in L. lactis (Martínez et al., 2007), and upregulated in Lactococcus garvieae after cold stress (Aguado-Urda et al., 2013). Therefore, we could argue that fru is highly reactive to different stressors. The metC-cysK operon was upregulated under all stress conditions applied here except salt addition. The highest effect was observed in acid stress. The Lactobacillus plantarum metC-cysK genes were upregulated after exposure to 0.1% porcine bile (Bron et al., 2006), but downregulated by p-coumaric acid (Reverón et al., 2012). Genes from the saturated fatty acid biosynthesis pathway (fab and acc) were upregulated after oxidative, cold and acid stress, while they were downregulated after starvation, heat and salt stress. For cold stress, however, one would have expected to see a decrease in SFAs, to maintain membrane fluidity at lower temperatures (Tsakalidou and Papadimitriou, 2011). The differences observed during oxidative stress might not be caused by higher levels of oxygen, but rather by the shaking that was used to induce oxidative stress. Previously, it was reported that cell envelope stress caused by membrane protein overproduction also affects fab. The exact response in this study depended on the identity of the overproduced membrane protein, leading to either an increase or a decrease in fab expression (Marreddy et al., 2011). We therefore propose that fatty acid biosynthesis in L. lactis is highly adaptive to various stressors and might rapidly fluctuate in time after the stress has been applied.
Major groups of defense mechanisms were significantly induced, despite the short exposure times employed. These include transcripts encoding protein chaperones such as GroEL, GroES, DnaK, DnaJ, and GrpE, which were induced during heat and salt stress. These conditions also induced protease genes such as clpE, clpP, and clpB. Cold shock induces cspA, cspB, cspC, and cspD specifying the major cold shock proteins binding to DNA or RNA (Ermolenko and Makhatadze, 2002). During cold stress, the zit operon for the uptake of Zn2+ was highly upregulated, suggesting that zinc ions play an important role during cold stress, possibly as a cofactor for cold stress-related proteins, and/or have an effect on membrane fluidity. Osmotic stress expectedly induced the expression of the genes of the transport proteins BusAA-BusAB. However, no significant induction of gadCB was observed after osmotic and heat stress. Strong downregulation of glpF2 was observed specifically after osmotic stress, while it was slightly increased after starvation. The glycerol uptake facilitator protein GlpF2 from in L. plantarum was shown to facilitate the diffusion of water, dihydroxyacetone and glycerol (Bienert et al., 2013), and could be an important factor for osmotic homeostasis in the cell. The upregulation of the arc operon after incubation for 5 min at 42°C was unexpected, since it has been reported before that arc decreases upon 30 min of heat stress at 42°C (Xie et al., 2004). This might be explained by differences in the heat exposure times and/or the specific strains used. Also, the arc operon is under complex regulation, with several protein regulators being involved [CcpA, ArgR/AhrC and, possibly, CodY (den Hengst et al., 2005; Zomer et al., 2007; Larsen et al., 2008)], which apparently leads to a rather dynamic expression profile (J.P. Pinto, PhD thesis, Groningen, 2015). Another unexpected result was the decrease of sodA expression during oxidative stress, while this gene is usually reported to be strongly induced under these circumstances (Sanders et al., 1995; Miyoshi et al., 2003). The short time of shaking (5 min) did perhaps not allow building up stressful oxygen levels. A more pronounced effect could have been generated with baffled shake flasks or by active addition of oxygen to the flasks, although these conditions would be far from industrial reality. Stress-related operons and genes reported before and observed here are summarized in Figure 6.
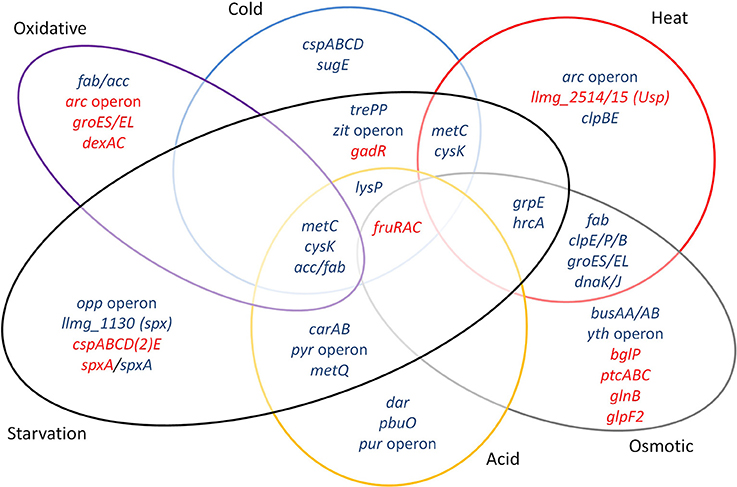
Figure 6. Venn diagram assembled from highly differentially expressed genes and stress-related operons and genes reported previously and observed in this study. Blue font: upregulated genes, red font: downregulated genes.
The plasmids of L. lactis NCDO712 have recently been sequenced and annotated, allowing examining the responses of their genes to the various stresses applied here. Indeed, various plasmid genes were affected under stress, such as the lactose utilization lac operon on pLP712, the putative Mn2+/Fe2+ transporter specified by mntH (pNZ712; acid stress) and the gene for a possible transcriptional repressor, cadC (pSH73; starvation). Since we applied a very short time of 5 min of stress exposure it is unlikely that these differences were the result of a change in plasmid copy numbers.
Transfer RNAs (tRNAs) are crucial components in translation. The rate of translation of a certain codon is directly coupled to the amount of cognate tRNA in the cell (Varenne et al., 1984), which is a measure of gene expression potential of the bacterial cell. Recently, the tRNAome of L. lactis was determined including the positions of 16 post-transcriptional modifications (Puri et al., 2014). Protein-overexpression stress employed in that study led to changes in tRNA expression required for up-regulation of housekeeping genes. An increase in tRNAs that would reflect the codon usage of the gene of the overexpressed protein was not seen. In the study presented here we generally observe a decrease of most of the tRNAs when L. lactis is placed under stress. Heat, cold, and oxidative stress seems to affect cellular tRNA transcript levels the least; we noted that cold stress induces the expression of seven tRNA genes. The RNA-seq data did not allow uncovering the actual charging or state of modification of the tRNAs.
Of the 186 sRNA genes currently annotated in L. lactis more than half were shown to be differentially expressed in response to one or more of the six different stress conditions that we employed. Since functional characterization has only been performed for a small number of these sRNAs (van der Meulen et al., 2016), only a few conclusions can be drawn as of yet. Some of the downregulated sRNAs might perform housekeeping functions and would not be required under conditions in which cells do no longer grow. On the other hand, sRNAs that are induced might respond to the stressor either to bring the cells into a protective state of slower or no growth, or they might act more specifically in order to overcome the harmful environmental change. Research on the latter group of sRNAs could increase our insights in their functioning during industrially relevant stress conditions and is currently ongoing.
Author Contributions
SM and JK designed the experiments; SM performed the experiments; SM and AJ analyzed the data; SM and JK wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We kindly thank the members of the functional fermentation team within the Top Institute Food and Nutrition (TIFN). In particular, we would like to thank Herwig Bachmann, Eddy Smid, Arjen Nauta, Claire Price, and Hans Brandsma for helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01704/full#supplementary-material
Supplementary Information S1. Excel table containing the input files for T-Rex.
References
Aguado-Urda, M., Gibello, A., del Mar Blanco, M., Fernández-Garayzábal, J. F., López-Alonso, V., and López-Campos, G. H. (2013). Global transcriptome analysis of Lactococcus garvieae strains in response to temperature. PLoS ONE 8:e79692. doi: 10.1371/journal.pone.0079692
Arnau, J., Sorensen, K. I., Appel, K. F., Vogensen, F. K., and Hammer, K. (1996). Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology 142(Pt 7), 1685–1691. doi: 10.1099/13500872-142-7-1685
Bandyra, K. J., Said, N., Pfeiffer, V., Górna, M. W., Vogel, J., and Luisi, B. F. (2012). The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase, E. Mol. Cell 47, 943–953. doi: 10.1016/j.molcel.2012.07.015
Barriere, C., Veiga-da-Cunha, M., Pons, N., Guedon, E., van Hijum, S. A., Kok, J., et al. (2005). Fructose utilization in Lactococcus lactis as a model for low-GC gram-positive bacteria: its regulator, signal, and DNA-binding site. J. Bacteriol. 187, 3752–3761. doi: 10.1128/JB.187.11.3752-3761.2005
Bienert, G. P., Desguin, B., Chaumont, F., and Hols, P. (2013). Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem. J. 454, 559–570. doi: 10.1042/BJ20130388
Bobrovskyy, M., and Vanderpool, C. K. (2014). The small RNA SgrS: roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 4:61. doi: 10.3389/fcimb.2014.00061
Brissette, J. L., Weiner, L., Ripmaster, T. L., and Model, P. (1991). Characterization and sequence of the Escherichia coli stress-induced psp operon. J. Mol. Biol. 220, 35–48. doi: 10.1016/0022-2836(91)90379-K
Bron, P. A., Molenaar, D., de Vos, W. M., and Kleerebezem, M. (2006). DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100, 728–738. doi: 10.1111/j.1365-2672.2006.02891.x
Chao, Y., Papenfort, K., Reinhardt, R., Sharma, C. M., and Vogel, J. (2012). An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019. doi: 10.1038/emboj.2012.229
de Jong, A., van der Meulen, S., Kuipers, O. P., and Kok, J. (2015). T-REx: transcriptome analysis webserver for RNA-seq Expression data. BMC Genomics 16:663. doi: 10.1186/s12864-015-1834-4
den Hengst, C. D., van Hijum, S. A., Geurts, J. M., Nauta, A., Kok, J., and Kuipers, O. P. (2005). The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280, 34332–34342. doi: 10.1074/jbc.M502349200
Eckhardt, T. H., Skotnicka, D., Kok, J., and Kuipers, O. P. (2013). Transcriptional regulation of fatty acid biosynthesis in Lactococcus lactis. J. Bacteriol. 195, 1081–1089. doi: 10.1128/JB.02043-12
Erkens, G. B., and Slotboom, D. J. (2010). Biochemical characterization of ThiT from Lactococcus lactis: a thiamin transporter with picomolar substrate binding affinity. Biochemistry 49, 3203–3212. doi: 10.1021/bi100154r
Ermolenko, D., and Makhatadze, G. (2002). Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59, 1902–1913. doi: 10.1007/PL00012513
Gasson, M. J. (1983). Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9.
Georg, J., and Hess, W. R. (2011). cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75, 286–300. doi: 10.1128/MMBR.00032-10
Gottesman, S., McCullen, C. A., Guillier, M., Vanderpool, C. K., Majdalani, N., Benhammou, J., et al. (2006). Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 71, 1–11. doi: 10.1101/sqb.2006.71.016
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Hindley, J. (1967). Fractionation of 32 P-labelled ribonucleic acids on polyacrylamide gels and their characterization by fingerprinting. J. Mol. Biol. 30, 125–136. doi: 10.1016/0022-2836(67)90248-3
Hoe, C., Raabe, C. A., Rozhdestvensky, T. S., and Tang, T. (2013). Bacterial sRNAs: regulation in stress. Int. J. Med. Microbiol. 303, 217–229. doi: 10.1016/j.ijmm.2013.04.002
Johansen, J., Rasmussen, A. A., Overgaard, M., and Valentin-Hansen, P. (2006). Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364, 1–8. doi: 10.1016/j.jmb.2006.09.004
Kilstrup, M., Jacobsen, S., Hammer, K., and Vogensen, F. K. (1997). Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus lactis. Appl. Environ. Microbiol. 63, 1826–1837.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Larsen, R., van Hijum, S. A., Martinussen, J., Kuipers, O. P., and Kok, J. (2008). Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Appl. Environ. Microbiol. 74, 4768–4771. doi: 10.1128/AEM.00117-08
Llull, D., and Poquet, I. (2004). New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl. Environ. Microbiol. 70, 5398–5406. doi: 10.1128/AEM.70.9.5398-5406.2004
Marilley, L., and Casey, M. (2004). Flavours of cheese products: metabolic pathways, analytical tools and identification of producing strains. Int. J. Food Microbiol. 90, 139–159. doi: 10.1016/S0168-1605(03)00304-0
Marreddy, R. K., Pinto, J. P., Wolters, J. C., Geertsma, E. R., Fusetti, F., Permentier, H. P., et al. (2011). The response of Lactococcus lactis to membrane protein production. PLoS ONE 6:e24060. doi: 10.1371/journal.pone.0024060
Martínez, B., Zomer, A. L., Rodríguez, A., Kok, J., and Kuipers, O. P. (2007). Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64, 473–486. doi: 10.1111/j.1365-2958.2007.05668.x
Miyoshi, A., Rochat, T., Gratadoux, J., Le Loir, Y., Oliveira, S. C., Langella, P., et al. (2003). Oxidative stress in Lactococcus lactis. Genet. Mol. Res. 2, 348–359.
Mizuno, T., Chou, M. Y., and Inouye, M. (1984). A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U.S.A. 81, 1966–1970. doi: 10.1073/pnas.81.7.1966
Morita, T., and Aiba, H. (2011). RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 25, 294–298 doi: 10.1101/gad.2030311
Nguyen, V. T., Morange, M., and Bensaude, O. (1989). Protein denaturation during heat shock and related stress. Escherichia coli beta-galactosidase and Photinus pyralis luciferase inactivation in mouse cells. J. Biol. Chem. 264, 10487–10492.
Nicolas, P., Mader, U., Dervyn, E., Rochat, T., Leduc, A., Pigeonneau, N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. doi: 10.1126/science.1206848
Papenfort, K., Pfeiffer, V., Mika, F., Lucchini, S., Hinton, J. C., and Vogel, J. (2006). σE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x
Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C. K., and Vogel, J. (2013). Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153, 426–437. doi: 10.1016/j.cell.2013.03.003
Papenfort, K., and Vanderpool, C. K. (2015). Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 39, 362–378. doi: 10.1093/femsre/fuv016
Papenfort, K., and Vogel, J. (2009). Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 160, 278–287. doi: 10.1016/j.resmic.2009.03.004
Parsell, D., and Lindquist, S. (1993). The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Pinto, J. P., Kuipers, O. P., Marreddy, R. K., Poolman, B., and Kok, J. (2011). Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS ONE 6:e21873. doi: 10.1371/journal.pone.0021873
Prevost, K., Desnoyers, G., Jacques, J. F., Lavoie, F., and Masse, E. (2011). Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 25, 385–396. doi: 10.1101/gad.2001711
Puri, P., Wetzel, C., Saffert, P., Gaston, K. W., Russell, S. P., Cordero Varela, J. A., et al. (2014). Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol. 93, 944–956. doi: 10.1111/mmi.12710
Reverón, I., Rivas, B., Muñoz, R., and Felipe, F. (2012). Genome-wide transcriptomic responses of a human isolate of Lactobacillus plantarum exposed to p-coumaric acid stress. Mol. Nutr. Food Res. 56, 1848–1859. doi: 10.1002/mnfr.201200384
Roces, C., Campelo, A. B., Veiga, P., Pinto, J. P., Rodríguez, A., and Martínez, B. (2009). Contribution of the CesR-regulated genes llmg0169 and llmg2164-2163 to Lactococcus lactis fitness. Int. J. Food Microbiol. 133, 279–285. doi: 10.1016/j.ijfoodmicro.2009.06.002
Romby, P., and Charpentier, E. (2010). An overview of RNAs with regulatory functions in gram-positive bacteria. Cell. Mol. Life Sci. 67, 217–237. doi: 10.1007/s00018-009-0162-8
Sanders, J. W., Leenhouts, K. J., Haandrikman, A. J., Venema, G., and Kok, J. (1995). Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J. Bacteriol. 177, 5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995
Sanders, J. W., Venema, G., and Kok, J. (1999). Environmental stress responses in Lactococcus lactis. FEMS Microbiol. Rev. 23, 483–501. doi: 10.1111/j.1574-6976.1999.tb00409.x
Smit, G., Smit, B. A., and Engels, W. J. (2005). Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29, 591–610. doi: 10.1016/j.fmrre.2005.04.002
Smith, W. M., Dykes, G. A., Soomro, A. H., and Turner, M. S. (2010). “Molecular mechanisms of stress resistance in Lactococcus lactis,” in Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, Vol. 2, ed A. Méndez-Vilas (Badajoz: Formatex Research Centre), 1106–1118.
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Stougaard, P., Molin, S., and Nordstrom, K. (1981). RNAs involved in copy-number control and incompatibility of plasmid R1. Proc. Natl. Acad. Sci. U.S.A. 78, 6008–6012. doi: 10.1073/pnas.78.10.6008
Taïbi, A., Dabour, N., Lamoureux, M., Roy, D., and LaPointe, G. (2011). Comparative transcriptome analysis of Lactococcus lactis subsp. cremoris strains under conditions simulating Cheddar cheese manufacture. Int. J. Food Microbiol. 146, 263–275. doi: 10.1016/j.ijfoodmicro.2011.02.034
Tarazanova, M., Beerthuyzen, M., Siezen, R., Fernandez-Gutierrez, M. M., de Jong, A., van der Meulen, S., et al. (2016). Plasmid complement of Lactococcus lactis NCDO712 reveals a novel pilus gene cluster. PLoS ONE 11:e0167970. doi: 10.1371/journal.pone.0167970
Thomason, M. K., and Storz, G. (2010). Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 44, 167–188. doi: 10.1146/annurev-genet-102209-163523
Tomizawa, J., Itoh, T., Selzer, G., and Som, T. (1981). Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. U.S.A. 78, 1421–1425. doi: 10.1073/pnas.78.3.1421
Tsakalidou, E., and Papadimitriou, K. (2011). Stress Responses of Lactic Acid Bacteria. New York, NY: Springer Science & Business Media.
van der Meulen, S. B., de Jong, A., and Kok, J. (2016). Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism. RNA Biol. 13, 353–366. doi: 10.1080/15476286.2016.1146855
Varenne, S., Buc, J., Lloubes, R., and Lazdunski, C. (1984). Translation is a non-uniform process: effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180, 549–576. doi: 10.1016/0022-2836(84)90027-5
Wadler, C. S., and Vanderpool, C. K. (2007). A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U.S.A. 104, 20454–20459. doi: 10.1073/pnas.0708102104
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wassarman, K. M., and Storz, G. (2000). 6S RNA regulates, E. coli RNA polymerase activity. Cell 101, 613–623. doi: 10.1016/S0092-8674(00)80873-9
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Wegmann, U., Overweg, K., Jeanson, S., Gasson, M., and Shearman, C. (2012). Molecular characterization and structural instability of the industrially important composite metabolic plasmid pLP712. Microbiology 158, 2936–2945. doi: 10.1099/mic.0.062554-0
Wouters, J. A., Sanders, J. W., Kok, J., de Vos, W. M., Kuipers, O. P., and Abee, T. (1998). Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology 144(Pt 10), 2885–2893. doi: 10.1099/00221287-144-10-2885
Xie, Y., Chou, L. S., Cutler, A., and Weimer, B. (2004). DNA Macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 70, 6738–6747. doi: 10.1128/AEM.70.11.6738-6747.2004
Keywords: RNA-Seq, sRNAs, transcriptomics, environmental stress, L. lactis
Citation: van der Meulen SB, de Jong A and Kok J (2017) Early Transcriptome Response of Lactococcus lactis to Environmental Stresses Reveals Differentially Expressed Small Regulatory RNAs and tRNAs. Front. Microbiol. 8:1704. doi: 10.3389/fmicb.2017.01704
Received: 23 May 2017; Accepted: 23 August 2017;
Published: 14 September 2017.
Edited by:
Rachel Susan Poretsky, University of Illinois at Chicago, United StatesReviewed by:
Sinead M. Waters, Teagasc, The Irish Agriculture and Food Development Authority, IrelandDave Siak-Wei Ow, Bioprocessing Technology Institute (A*STAR), Singapore
Copyright © 2017 van der Meulen, de Jong and Kok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Kok, amFuLmtva0BydWcubmw=
 Sjoerd B. van der Meulen
Sjoerd B. van der Meulen Anne de Jong1,2
Anne de Jong1,2 Jan Kok
Jan Kok