- 1Graduate School of Life Sciences, Toyo University, Gunma, Japan
- 2Bio-Nano Electronics Research Center, Toyo University, Kawagoe, Japan
- 3Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, United States
Mrp (Multiple resistance and pH) antiporter was identified as a gene complementing an alkaline-sensitive mutant strain of alkaliphilic Bacillus halodurans C-125 in 1990. At that time, there was no example of a multi-subunit type Na+/H+ antiporter comprising six or seven hydrophobic proteins, and it was newly designated as the monovalent cation: proton antiporter-3 (CPA3) family in the classification of transporters. The Mrp antiporter is broadly distributed among bacteria and archaea, not only in alkaliphiles. Generally, all Mrp subunits, mrpA–G, are required for enzymatic activity. Two exceptions are Mrp from the archaea Methanosarcina acetivorans and the eubacteria Natranaerobius thermophilus, which are reported to sustain Na+/H+ antiport activity with the MrpA subunit alone. Two large subunits of the Mrp antiporter, MrpA and MrpD, are homologous to membrane-embedded subunits of the respiratory chain complex I, NuoL, NuoM, and NuoN, and the small subunit MrpC has homology with NuoK. The functions of the Mrp antiporter include sodium tolerance and pH homeostasis in an alkaline environment, nitrogen fixation in Schizolobium meliloti, bile salt tolerance in Bacillus subtilis and Vibrio cholerae, arsenic oxidation in Agrobacterium tumefaciens, pathogenesis in Pseudomonas aeruginosa and Staphylococcus aureus, and the conversion of energy involved in metabolism and hydrogen production in archaea. In addition, some Mrp antiporters transport K+ and Ca2+ instead of Na+, depending on the environmental conditions. Recently, the molecular structure of the respiratory chain complex I has been elucidated by others, and details of the mechanism by which it transports protons are being clarified. Based on this, several hypotheses concerning the substrate transport mechanism in the Mrp antiporter have been proposed. The MrpA and MrpD subunits, which are homologous to the proton transport subunit of complex I, are involved in the transport of protons and their coupling cations. Herein, we outline other recent findings on the Mrp antiporter.
Diversity of Na+/H+ Antiporters
The Na+/H+ antiporter is a secondary active transporter that utilizes the proton motive force to efflux intracellular sodium ions (Padan et al., 2005; Krulwich et al., 2011; Fuster and Alexander, 2014; Padan and Landau, 2016). It is a widely distributed membrane protein, and studies of it are being conducted in eukaryotic-derived NHE families as well as bacterial-derived NhaA families (Wakabayashi et al., 1997; Orlowski and Grinstein, 2004; Padan, 2014; Padan and Landau, 2016). The main physiological roles of the Na+/H+ antiporter are intracellular pH homeostasis and Na+ efflux. Na+ efflux by the Na+/H+ antiporter plays a critical role for sodium circulation inside and outside the cell because many bacteria, including marine bacteria, utilize both the proton motive force and sodium motive force (Zilberstein et al., 1982; Padan and Schuldiner, 1994; Vimont and Berche, 2000; Dibrov, 2005; Krulwich et al., 2011).
NHE, a mammalian Na+/H+ exchanger, is a group of 12-transmembrane membranes protein with multiple isoforms (Wakabayashi et al., 1997; Orlowski and Grinstein, 2011; Padan and Landau, 2016). The antiporters designated as NHE1–NHE5 are localized in the cell plasma membrane, while NHE6–NHE9 are present in the membranes of intracellular organelles (Ohgaki et al., 2011; Fuster and Alexander, 2014). Furthermore, NHE has a hydrophilic domain on the carboxyl-terminal side exposed to the cytoplasm. The interaction between this hydrophilic domain and calcineurin, which is a Ca2+-dependent serine/threonine protein phosphatase, is reportedly involved in intracellular pH homeostasis and is crucial for NHE ion transport activity (Wakabayashi et al., 1997; Pang et al., 2001; Hisamitsu et al., 2012). In NHE 1, it has been reported that enzymatic activity is activated in response to various stimuli including hormones, growth factors, and mechanical stress (Wakabayashi et al., 1997; Hisamitsu et al., 2012). Mammalian NHE has high homology with the bacterial NhaP family, while it has low homology with the bacterial NhaA family, a member of the bacterial Na+/H+ antiporter family (Waditee et al., 2001; Resch et al., 2011; Padan and Landau, 2016). In addition, the NhaP antiporter family has been shown to have a large hydrophilic domain at its carboxy-terminal side like NHE (Waditee et al., 2001; Mourin et al., 2017).
In general, bacteria have multiple Na+/H+ antiporters that are thought to exert appropriate responses to the ambient conditions of the growth environment and its associated stresses (Padan et al., 2005; Krulwich et al., 2011; Padan, 2014; Preiss et al., 2015). For example, Escherichia coli has three major Na+/H+ antiporters designated as NhaA, NhaB, and ChaA. It has been shown that NhaA is expressed as a response to the stress associated with alkaline pH and sodium ions in E. coli (Padan et al., 2001, 2005; Padan, 2014; Padan and Landau, 2016). Furthermore, NhaA is activated in alkaline pH while NhaB retains activity only in neutral pH; therefore, NhaA is thought to play a central role in the adaptation of E. coli to an alkaline environment. In addition to these findings, it has been shown that ChaA is a Ca2+/H+ antiporter and MdfA is a multidrug/proton antiporter that retains Na+/H+ antiport activity (Ivey et al., 1993; Shijuku et al., 2002; Lewinson et al., 2003).
Discovery of the Mrp Gene Cluster
Mrp was first discovered in work on an alkaline-sensitive strain of alkaliphilic Bacillus halodurans C-125 in Kudo et al. (1990). It was found that the gene cluster encoded a Na+/H+ antiporter (Hamamoto et al., 1994). The mrp gene cluster of B. halodurans C-125 comprises seven mrp genes (mrpABCDEFG), and the expressed proteins are predicted, from the amino acid sequence, to all be membrane proteins (Figure 1 and Table 1). The Mrp antiporter has been suggested to function as a complex of multiple membrane proteins (Kajiyama et al., 2007; Morino et al., 2008). Apart from the maintenance of cytoplasmic pH, the Mrp complex has various other physiological roles in different species, such as bile acid resistance in Bacillus subtilis and Vibrio cholera (Ito et al., 1999; Dzioba-Winogrodzki et al., 2009), Na+ homeostasis/tolerance in B. subtilis (Ito et al., 1999; Kosono et al., 1999; Ito et al., 2000), sporulation in B. subtilis (Kosono et al., 2000), plant infection in Sinorhizobium meliloti (Putnoky et al., 1998), pathogenesis in Pseudomonas aeruginosa (Kosono et al., 2005) and arsenic resistance in Agrobacterium tumefaciens (Kashyap et al., 2006).
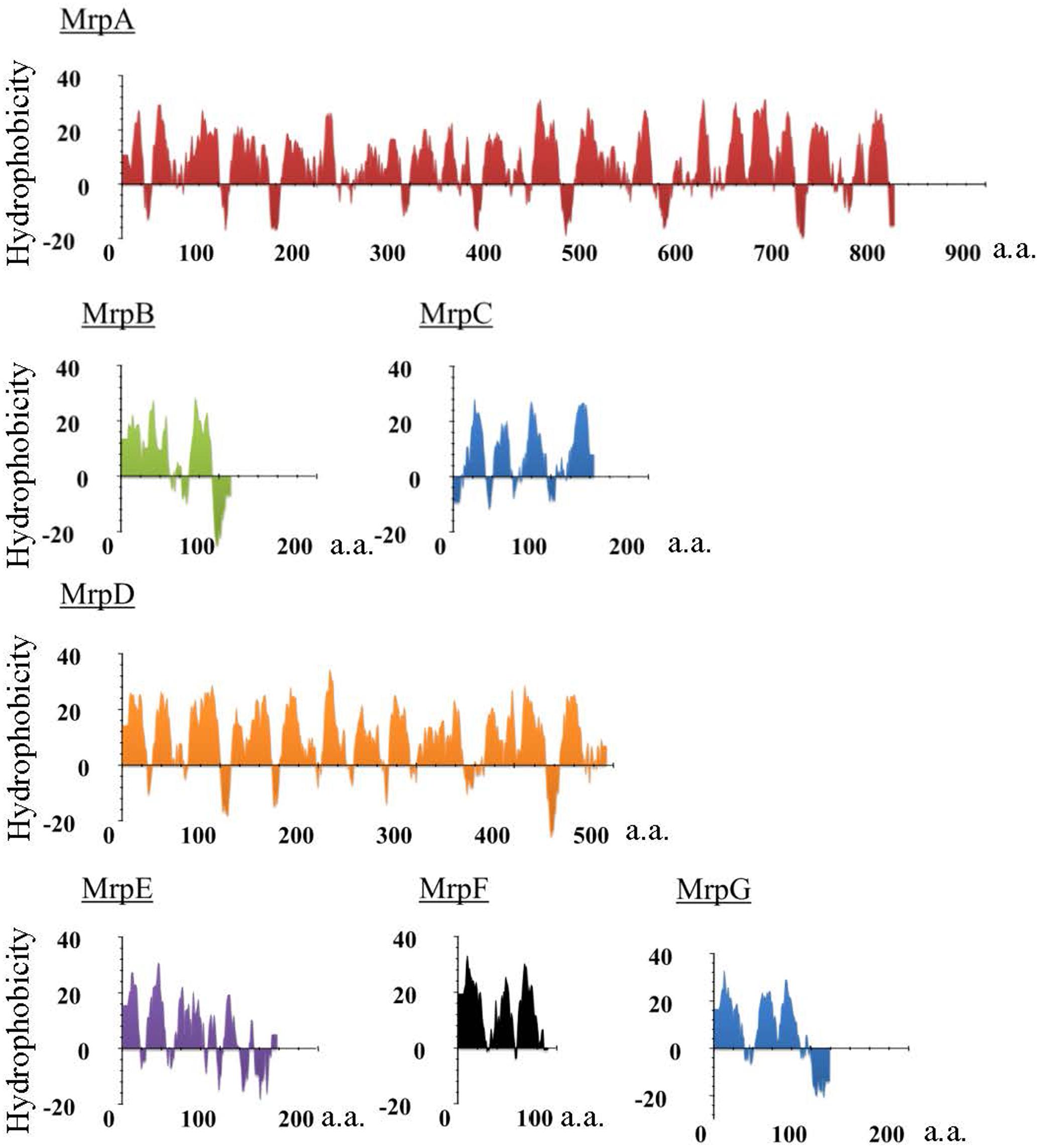
FIGURE 1. Hydropathy profile of the Mrp subunits derived from alkaliphilic B. pseudofirmus OF4. The hydropathy profile of the Mrp subunits derived from B. pseudofirmus OF4 was predicted using Kyte and Doolittle method. The vertical axis represents the degree of hydrophobicity, and the horizontal axis represents the number of amino acids (a.a.).
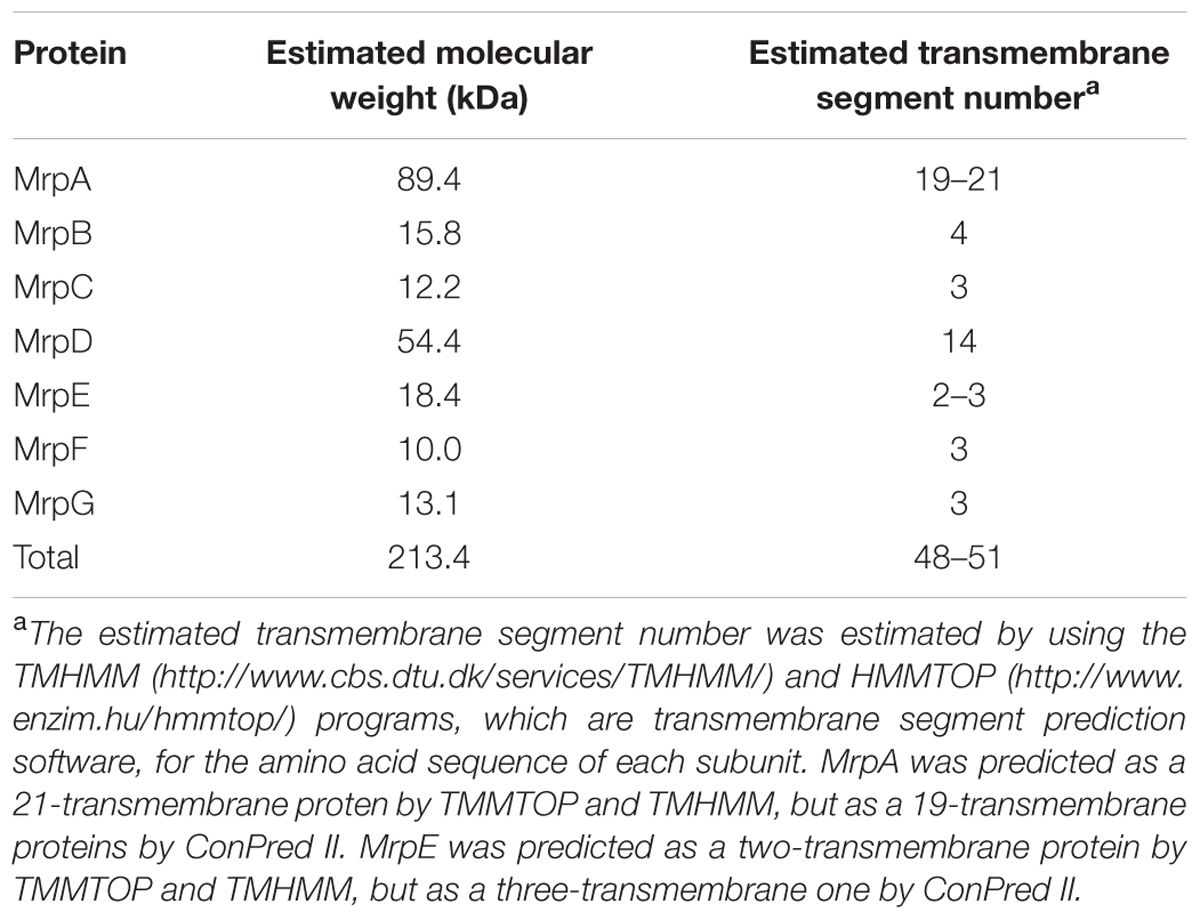
TABLE 1. Molecular weight of each Mrp subunit derived from the B. pseudofirmus OF4 strain and the estimated number of transmembrane regions.
Phylogenetic Analysis of the Mrp Gene Cluster
The Mrp antiporter has been found in alkaliphilic bacteria as well as in many other bacteria and archaea. Genome analyses in a wide range of microorganisms clarified that the structure of the mrp gene cluster is diverse (Figure 2) (Swartz et al., 2005; Krulwich et al., 2009). Because of its distinctive properties, Mrp antiporter systems have been classified in their own category, cation: proton antiporter-3 (CPA3), in the transporter classification system (Saier et al., 2009, 2016). So far, the mrp gene cluster has been classified into three groups. Group 1 antiporters are composed of seven mrp genes, and it is found in many Bacillus spp. and in Staphylococcus aureus. Group 2 has a mrp gene cluster (mrpA’CDEFG) of six genes. This group belongs to bacteria such as Pseudomonas aeruginosa and Vibrio cholerae, in which it appears that the mrpA gene is fused with the mrpB gene encoding a fusion protein (Kosono et al., 2005; Swartz et al., 2005; Dzioba-Winogrodzki et al., 2009). Sinorhizobium meliloti has two sets of mrp (alias pha) gene clusters, one belongs to Group 1 (Pha2) and the other belongs to Group 2 (Pha1) (Putnoky et al., 1998; Yamaguchi et al., 2009). The mrp gene cluster belonging to Group 3 has each subunit, but the gene order is irregular. For example, the mrp of cyanobacteria has two mrpB genes, and the gene sequence in the gene cluster is as follows: mrpCDCDEFGBB (Waditee et al., 2001).
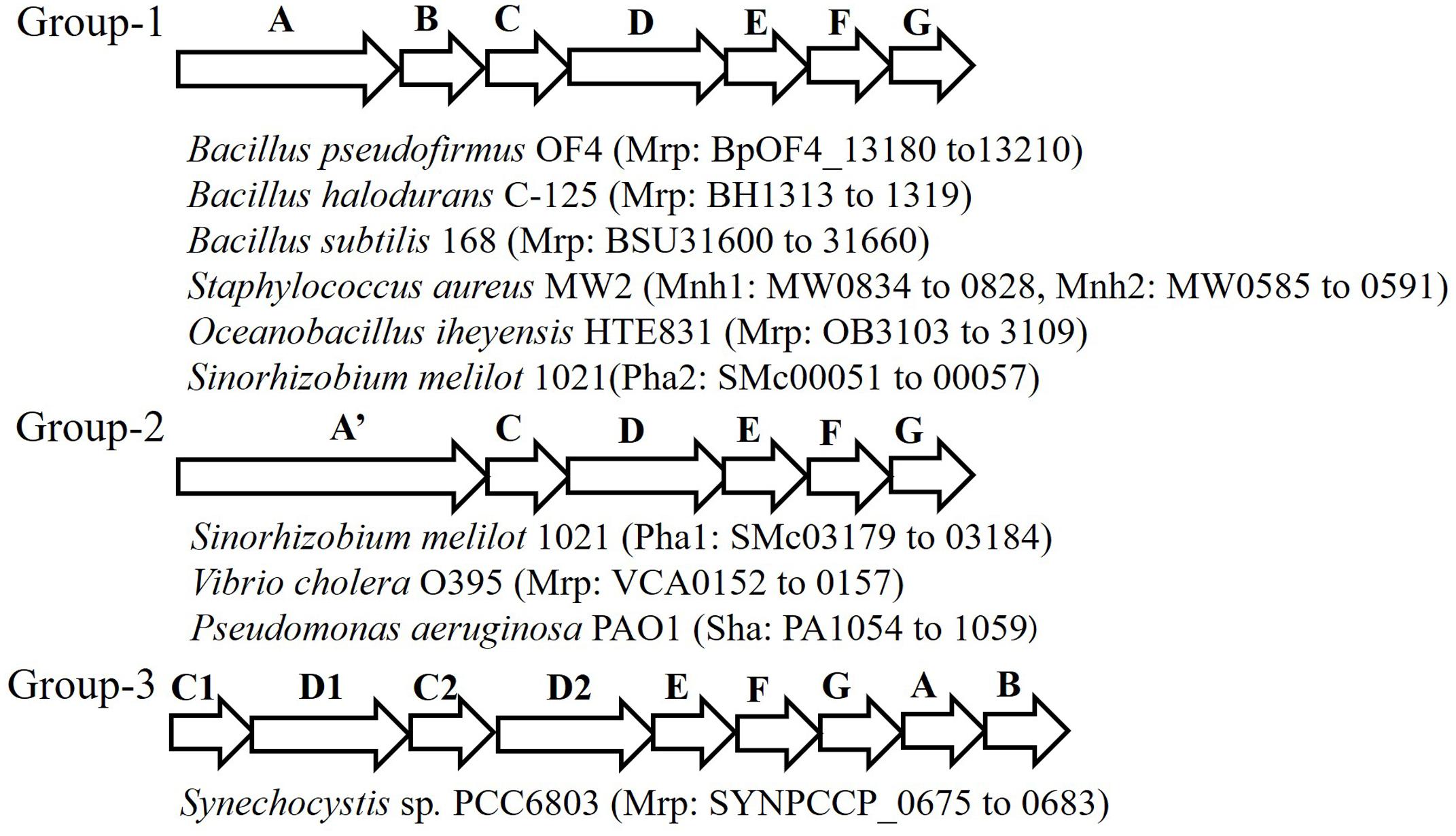
FIGURE 2. Schematic of the diversity of the mrp gene cluster in several bacteria. This figure depicts the genetic structure of the mrp gene cluster of each bacterium. The locus tag numbers of each Mrp subunit gene are listed after the bacterial name. mrpA’ is a gene involving the fusion of mrpA and mrpB.
Staphylococcus aureus has been shown to have two sets of group 1 type mrp (alias mnh) gene clusters, mnh1 and mnh2. mnh1 has been found to encode the Na+/H+ antiporter; however, the function of the product encoded by mnh2 remains unknown (Swartz et al., 2007). Similarly, genomic analyses have revealed that alkaliphilic Bacillus clausii and the marine bacterium Oceanobacillus iheyensis have two sets of mrp gene clusters (Krulwich and Ito, 2013). However, there are no reported examples of the physiological and functional differences between them. In addition, analysis of many microbial genomes has revealed three mrp gene clusters. For example, Microbacterium sp. TS-1 has three sets of Mrp gene clusters, two of them (locus tags, MTS1_01879-01874 and MTS1_02182-02187) belong to Group 2 and the third one (locus tags, MTS1_02374-02382; mrpFGBCDDAE) belongs to Group 3 (Fujinami et al., 2013) and hyperthermophilic archaeon, Thermococcus onnurineus NA1 has three sets of Mrp gene clusters, all of which (locus tags; TON_0272-0266, TON_1574-1580, TON_1025-1031) belong to Group 1 (Lim et al., 2010).
The mrp gene cluster of anaerobic bacteria has a gene structure that is markedly different from that of aerobic bacterial-derived mrp gene clusters. For example, the mrp gene cluster of Natranaerobius thermophilus retains three overlapping mrpB genes (Mesbah et al., 2009). In addition, in the mrp gene cluster derived from Synechocystis sp. PCC 6803, duplication of the mrpD and mrpC genes as well as the mrpB gene is observed (Krulwich et al., 2009). Similar gene arrangements have been reported in other cyanobacteria (Fukaya et al., 2009).
Relationship Between Mrp Antiporter and Respiratory Chain Complex I
MrpA and MrpD subunits have homology with the respiratory chain complex I subunit (Figures 3, 4) (Mathiesen and Hägerhäll, 2003; Moparthi and Hägerhäll, 2011; Moparthi et al., 2014). The respiratory chain complex I is a protein complex belonging to the electron transport system, which oxidizes NADH supplied from the TCA cycle, among other sources. It reduces quinone and effluxes protons from the cell. The NuoL, NuoM, and NuoN subunits, which are subunits of the respiratory chain complex I in E. coli, have been analyzed because of homology with the Mrp antiporter subunit (Nakamaru-Ogiso et al., 2003a,b, 2010; Torres-Bacete et al., 2007; Ohnishi et al., 2010; Torres-Bacete et al., 2011; Sperling et al., 2016; Morino et al., 2017). These three Nuo subunits have highly conserved glutamic acid residues and lysine residues (Figure 4), which have been suggested to be the core of the proton transport pathway, based on the crystal structure of E. coli (Baranova et al., 2007; Efremov and Sazanov, 2011; Sazanov, 2014). In MrpA and MrpD subunits, these charged residues are highly conserved, and it has been reported that glutamate residues are also conserved at the same position in B. subtilis and B. pseudofirmus OF4. Mrp antiporters have been shown to be essential for antiport activity in various settings (Kosono et al., 2005; Kajiyama et al., 2009; Morino et al., 2010).
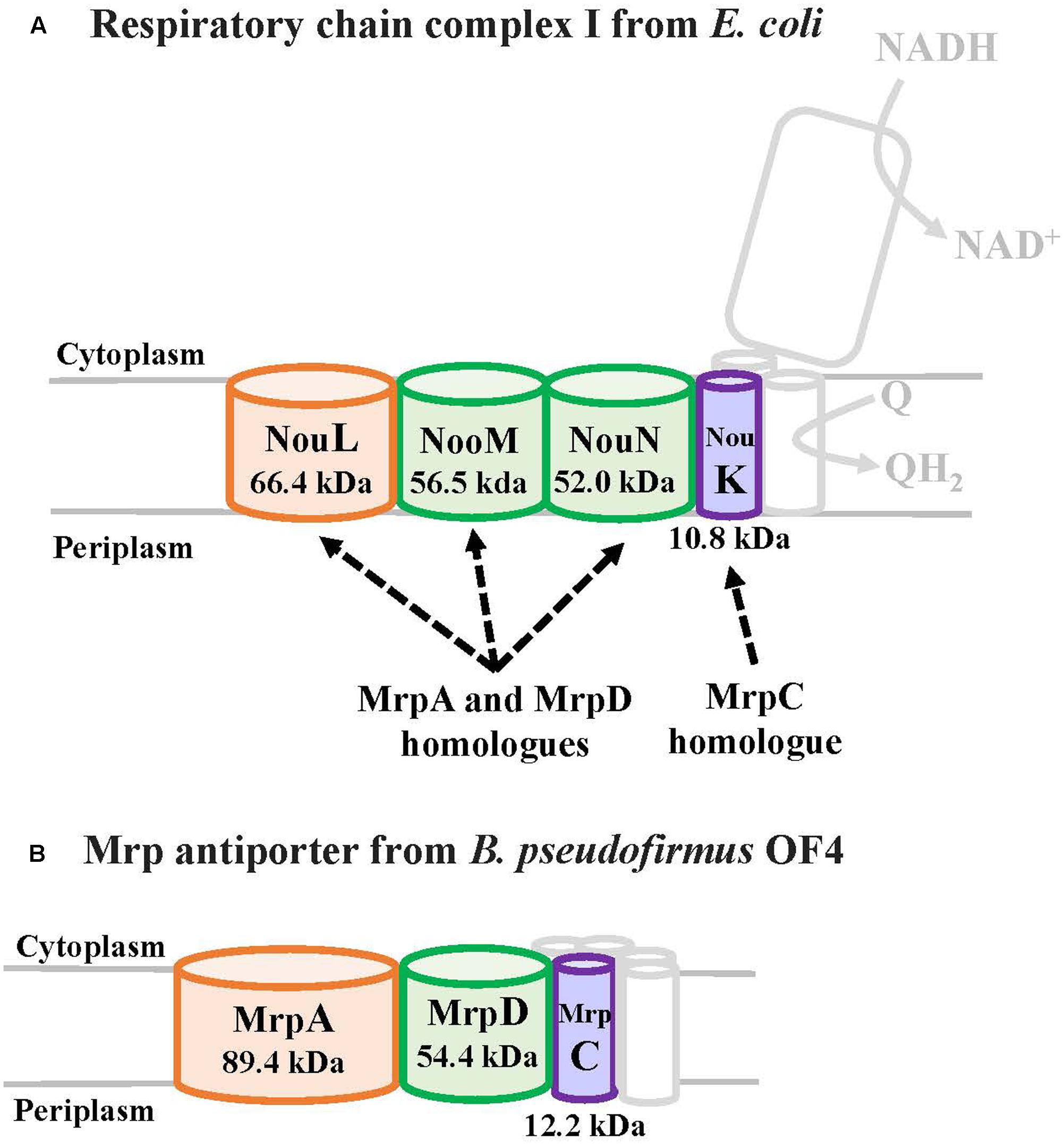
FIGURE 3. The E. coli respiratory chain complex I subunit with homology to the Mrp subunit. (A) Schematic diagram of respiratory chain complex I. The NuoL, NuoM, and NuoN subunits of the E. coli respiratory chain complex I have homology with the MrpA and MrpD subunits of the Mrp antiporter, and the NuoK subunits also have partial homology with the MrpC subunit (Mathiesen and Hägerhäll, 2003). (B) Schematic diagram of Mrp antiporter.

FIGURE 4. Common regions preserved between the B. pseudofirmus OF4 Mrp antiporter and the E. coli complex I subunit. MrpA, MrpD, NuoL, NuoM, and NuoN proteins having homology to each other are described. The NDH-1 domains conserved among them are represented in gray. The glutamic acid residue (–) and the lysine residue (+) conserved in the NDH-1 region are shown in the schematic. The C-terminal region of MrpA, in black, has been found to be systematically related to the MrpB protein by PSI-Blast analysis (Mathiesen and Hägerhäll, 2003).
The crystal structure of the E. coli respiratory chain complex I revealed that the NuoL subunit had a long helical chain at its carboxy terminus (Efremov and Sazanov, 2011; Sazanov, 2014). Analysis of a long-chain, helix-deficient strain of the NuoL subunit in E. coli respiratory chain complex I indicated that this helix is indispensable for proton transport, complex formation, and NADH oxidation (Ohnishi et al., 2010; Efremov and Sazanov, 2011; Torres-Bacete et al., 2011; Sazanov, 2014). This suggested that it functions as a “piston” that couples oxidation and quinone reduction to proton transport. The MrpA subunit has an additional transmembrane region at the carboxy terminus similar to the NuoL subunit. In addition, part of the MrpA carboxy terminus has high sequence homology with MrpB, as shown by PSI-Blast analysis; it is speculated that it is a characteristic region only of the Mrp antiporter (Krulwich et al., 2009). Recently, it was reported that the MrpA carboxy-terminal region of B. pseudofirmus OF4 has indispensable roles in antiport function (Morino et al., 2017).
Features of the Mrp Antiporter from Alkaliphilic Bacillus pseudofirmus OF4
Within the Mrp antiporter family, the B. pseudofirmus OF4-derived Mrp antiporter (Bp–Mrp) has undergone advanced functional and structural analyses that has revealed: (1) formation of the complex and role of each subunit; (2) identification of amino acid residues with important structural and functional roles, as determined by site-specific functional analysis; (3) analysis of the specific C-terminal region of MrpA; and (4) purification and reconstitution of the Bp–Mrp antiporter.
Formation of Bp–Mrp Complex and the Role of Each Mrp Subunit
Bp–Mrp was estimated to form a membrane protein complex expressed from seven mrp genes. Bp–Mrp expressed in E. coli was separated by Blue native PAGE (BN-PAGE); subsequently, each Mrp subunit was detected by Western blotting to investigate whether the Mrp antiporter successfully formed a complex (Morino et al., 2008). The results confirmed formation of a Mrp complex (220 kDa), estimated to be a monomer consisting of all subunits, as well as a MrpABCDEFG complex (400 kDa), estimated to be a dimer. A MrpABCD subcomplex comprising MrpA, B, C, and D subunits was also detected; this subcomplex was shown not to be catalytically active (Morino et al., 2008).
Mutants were also constructed, each with the deletion of a single mrp, to enable investigation of the role of each Mrp subunit in complex formation (Morino et al., 2008). The results showed that, in the membrane fraction of the mrpD deletion mutant, no other Mrp subunits were detected. On the other hand, Mrp subunits other than MrpE could be detected in the membrane of the mrpE-deficient mutant. From BN-PAGE analysis, it was confirmed that the Mrp subunits other than MrpE form a complex in the mrpE deletion mutant. These results suggested that the MrpD subunit is important in the formation of the Bp–Mrp complex. It may have a role as a scaffold when other Mrp subunits are expressed in the cell membrane. By contrast, the MrpE subunit appears to be incorporated in the final step of complex formation and possibly plays an important role in ensuring that the Mrp complex can exert its full activity. However, in B. subtilis, it was reported that MrpE is dispensable for ion transport activity (Yoshinaka et al., 2003; Morino et al., 2008).
Site-Directed Amino Acid Substitution Mutagenesis and Identification of Residues in the Bp–Mrp Antiporter Important for Ion Transport
The Bp–Mrp antiporter was studied to identify amino acid residues within it that are important for ion transport and Mrp complex formation. Site-specific mutations were introduced at amino acid residues conserved between Mrp homologs. In MrpA and MrpD subunits, mutations were also introduced at amino acid residues conserved among the NuoL, NuoM, and NuoN subunits of the homologous E. coli respiratory chain complex I (Morino et al., 2010). The mutants were expressed in the E. coli KNabc strain, in which three major Na+/H+ antiporter genes (nhaA, nhaB, and chaA) are deleted; subsequently, the mutants were tested for sodium sensitivity, antiport activity, and their complex formation ability. Each amino acid substitution mutant could be classified into one of eight categories from each phenotype. Figure 5 shows a summary of the phenotype at each mutation site (Morino et al., 2010, 2017). Mutants classified into categories 1 and 2 have been shown to affect Mrp complex formation. Mutants classified into categories 3–7 were confirmed to undergo complex formation but resulted in a decrease in Na+/H+ antiport activity and a decrease in the sodium-sensitive complementary activity of E. coli KNabc.
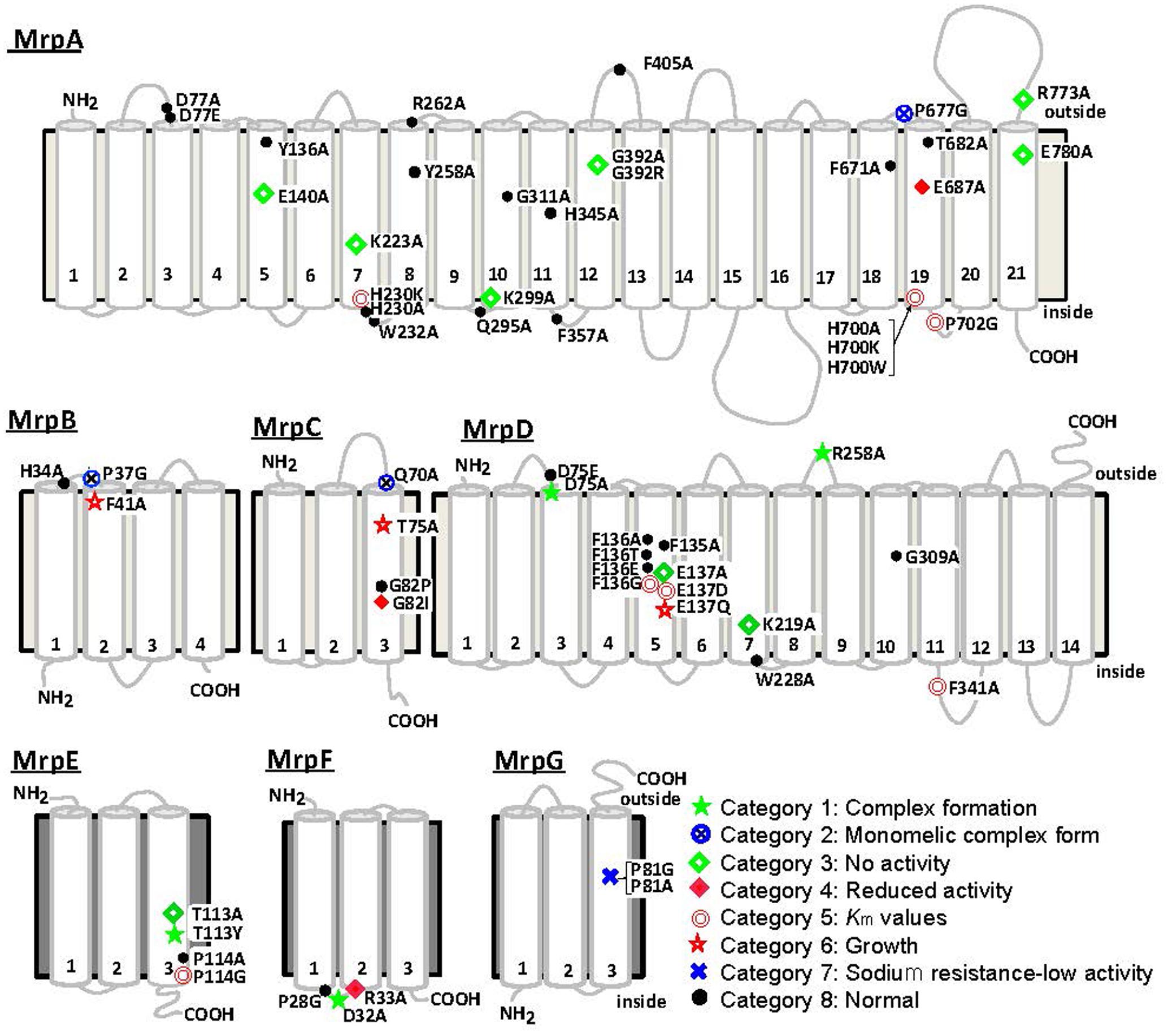
FIGURE 5. Transmembrane topology of Bp–Mrp proteins and positions of mutations. Transmembrane segments predicted by ConPred II, HMMTOP, and TMHMM (available online) were used in the analyses of the secondary structure predictions for each Mrp subunit.  , positions at which the mutations complemented an antiporter-deficient E. coli KNabc transformant and exhibited normal Na+/H+ antiport activity;
, positions at which the mutations complemented an antiporter-deficient E. coli KNabc transformant and exhibited normal Na+/H+ antiport activity;  , mutations that affected the level of Mrp proteins in the membrane;
, mutations that affected the level of Mrp proteins in the membrane;  , mutations that led to loss of the Na+/H+ antiport activity and loss of Na+ tolerance;
, mutations that led to loss of the Na+/H+ antiport activity and loss of Na+ tolerance;  , mutations that decreased the Na+/H+ antiport activity, without an effect on growth of the E. coli transformant;
, mutations that decreased the Na+/H+ antiport activity, without an effect on growth of the E. coli transformant;  , mutations that affected the Km values of Mrp-dependent antiport activity;
, mutations that affected the Km values of Mrp-dependent antiport activity;  , mutations that affected transformant cell growth;
, mutations that affected transformant cell growth;  , mutations that affected Mrp complex formation;
, mutations that affected Mrp complex formation;  , the two mutants in MrpG-P81 that had a unique phenotype.
, the two mutants in MrpG-P81 that had a unique phenotype.
In category 1, MrpD-D75A, MrpD-R258A, MrpE-T113Y, and MrpF-D32A were studied, and their Na+/H+ antiport activity was found to be completely lost, with no Mrp complex detected.
In category 2, MrpA-P677G, MrpB-P37G, and MrpC-Q70A mutations were associated with the retention of Na+/H+ antiport activity but failure to show formation of the Mrp complex monomer in BN-PAGE analysis. These mutations were assumed to destabilize the interaction between the MrpABCD subcomplex and each of the MrpE, MrpF, and MrpG subunits.
MrpA-E140A, MrpA-K223A, MrpA-K299A, MrpA-G392R, MrpA-R773A, MrpA-E780A, MrpD-E137A, MrpD-K219A, and MrpE-T113A, which are classified into category 3, retained the Mrp complex but Na+/H+ antiport activity was completely lost.
MrpC-G82I and MrpF-R33A, classified into category 4, exhibited Na+/H+ antiport activity that was decreased by approximately 70% compared with wild-type activity.
In category 5, the apparent Km for Na+ of Na+/H+ antiport activity increased in MrpA-H230K, MrpA-H700A, MrpA-H700K, MrpA-H700W, MrpA-P702G, MrpD-F136G, MrpD-E137D, MrpD-F341A, and MrpE-P114G. Because MrpA-H230, MrpA-H700, MrpA-P702, and MrpD-F136 are adjacent to charged residues essential for activity (MrpA-K223, MrpA-E687, MrpD-E137), along with these charged residues, it is assumed that they are involved in ion transport along with these chargeable residues. Although the functional roles of MrpD-F341 and MrpE-P114 are unknown, it is inferred that the low-molecular-weight subunit MrpE may also be involved in ion transport.
MrpB-F41A and MrpC-T75A, classified into category 6, retained normal Na+/H+ antiport activity but could not completely complement the sodium sensitivity in E. coli KNabc.
Na+/H+ antiport activity was completely inactivated in MrpG-P81A, classified into category 7. Surprisingly, however, the sodium sensitivity of E. coli KNabc could be complemented similarly to that of the wild type (see below).
The amino acid substitution mutants that showed the same phenotype as the wild type were designated into category 8.
In MrpA-E140, MrpA-K223, MrpD-E137A, and MrpD-K219A, there was conservation of not only the MrpA and MrpD subunits but also the respiratory chain complex I subunit. They are extremely important for Na+/H+ antiport activity, and it was speculated from the complex I crystal structure that the respiratory chain complex I also participates in ion transport. In addition, MrpG-P81A in category 7 did not retain Na+/H+ antiport activity, but it was able to complement the sodium sensitivity of the E. coli KNabc. This suggested that the Mrp antiporter of MrpG-P81A has Na+ efflux capacity coupled with the transport of ions other than protons. For example, membrane potential-driven sodium ion excretion may occur concomitantly with the transport of anions. However, the phenotype of MrpG-P81A, including the possibility of having other transporting substrates, is only a hypothesis at this point; therefore, more detailed analyses are needed.
Functional Analysis of the Carboxyl-Terminal Region of the MrpA Subunit of the Bp–Mrp Antiporter
The C-terminal region of MrpA, which has similarity to the MrpB subunit, is conserved. This region of MrpA is not preserved in the respiratory chain complex I subunit; therefore, it is predicted to have unique functions and roles in the Mrp antiporter. Site-specific mutations involving substitutions at highly conserved amino acid residues located in the C-terminal region of Bp–MrpA were introduced (Morino et al., 2017). Two glutamic acid residues are conserved in the C-terminal region of MrpA, as has been reported by Kosono et al. (2006) using B. subtilis Mrp. In the Bp–Mrp antiporter, these acidic residues (MrpA-E687 and MrpA-E778) are also essential for Na+/H+ antiport activity. In addition, MrpA-P683G retained normal Na+/H+ antiport activity; however, the monomeric MrpABCDEFG complex could not be detected by BN-PAGE analysis. The fact that the same phenotype is also found in MrpB-P37G and MrpC-Q70A suggested that the C-terminal region of MrpA is a region through which interactions with low-molecular-weight Mrp subunits, MrpB and MrpC, occur. In addition, it was observed that Na+/H+ antiport activity decreased in MrpA-P702G and MrpA-R773A mutants, suggesting that the C-terminal region of MrpA has an important function in ion transport.
The above results also showed that the C-terminal region of MrpA has important functions not only in ion transport but also in interactions between subunits. Furthermore, the C-terminal region of MrpA is a region unique to the Mrp antiporter and is suggested to be involved in Na+/H+ antiport activity.
Purification and Reconstitution of the Bp–Mrp Antiporter
Reports have been published on structural analyses of various protein complexes by techniques such as single-particle analysis by the observation of high-purity samples under an electron microscope. For example, in the respiratory chain complex I, an L-shaped structure has been observed under an electron microscope (Holt et al., 2003). The structure of very large macromolecules, such as the H-ring, which is a component of the basal body of bacterial flagella, has also been clarified by microscopic observation (Terashima et al., 2010). High-purity samples of target proteins and complexes thereof are indispensable for such advanced structural analysis. As such, purification of the Mrp antiporter derived from B. pseudofirmus OF4 was investigated. The Mrp antiporter expressed in E. coli was purified by TALON resin and reconstituted into an artificial lipid membrane for further confirmation of its Na+/H+ antiport activity. In the reconstituted membrane, the proton motive force required for Mrp antiporter activation was generated by FoF1-ATPase derived from Bacillus sp. PS3, which was simultaneously reconstituted. This report is the first to describe the successful reconstitution of purified bacterial-derived Mrp antiporter into proteoliposomes retaining Na+/H+ antiport activity (Morino et al., 2014).
Mrp Antiporters from Gram-Positive Bacteria Other than Alkaliphilic Bacillus spp.
It was shown that the mrp (alias sha) gene cluster of B. subtilis encodes a Na+/H+ antiporter and plays a major role in the mechanism of sodium tolerance of B. subtilis (Ito et al., 1999; Kosono et al., 1999). Various mrp-deficient strains have been produced in B. subtilis, and it has been reported from their analysis that the mrpF gene contributes to bile acid tolerance (Ito et al., 1999). Furthermore, it has been reported that sodium efflux capacity is retained in a mrpE gene-deficient strain (Yoshinaka et al., 2003; Morino et al., 2008). S. aureus Mrp is expected to be a target protein of a novel antibiotic because since growth inhibition of S. aureus is suppressed by inhibiting translation of the mrpD gene using antisense RNA (Ji et al., 2001).
Polyextremophiles such as Natranaerobius thermophilus are halophilic, alkaliphilic, and thermophilic bacteria that grow optimally at 3.5 M Na+, pH 9.5, and 53°C–55°C (Mesbah et al., 2009). This bacterium has at least eight electrogenic Na+(K+)/H+ antiporters. One of them, Nt-Nha, has homology with MrpA and MrpD, the two large subunits of group 1. In previous studies, none of the Mrp antiporters exhibited antiport activity with MrpA or MrpD alone. However, this Nt-Nha alone showed Na+ (K+)/H+ antiport activity. This supports the suggestion that MrpA and MrpD are critical for the ion transport pathway for antiporters in the CPA 3 family (Krulwich et al., 2009). Recently, study of the Mrp complex of Methanosarcina acetivorans from the archaeal domain suggested that MrpA is essential for antiport activity and that the MrpA/MrpD subcomplex is critical for catalyzing Na+/H+ antiport activity (Jasso-Chavez et al., 2017). This is the second example showing that the Mrp complex exhibits antiport activity even without all its subunits. The consequence of this observation is discussed in Section “Mrp Antiporters from Archaea”.
Mrp Antiporters from Gram-Negative Bacteria
Sinorhizobium meliloti has two sets of mrp (pha) gene clusters, one belongs to Group 1 (Pha2) and the other belongs to Group 2 (Pha1). The pha1 gene cluster (SMc03179 to 03184) was identified as a mutation insertion site in a potassium-sensitive strain of the root nodule bacterium Sinorhizobium meliloti (Putnoky et al., 1998). Sinorhizobium, a symbiotic bacterium, retains potassium-dependent alkaline pH homeostasis ability; however, pha1 deficiency reportedly causes a loss of alkaline environmental adaptability (Putnoky et al., 1998). Detailed analysis revealed that the pha1 gene cluster derived from Sinorhizobium encodes a K+ (Na+)/H+ antiporter (Putnoky et al., 1998; Yamaguchi et al., 2009).
The mrp (sha) gene cluster has also been found in Pseudomonas aeruginosa, and it reportedly encodes a Na+/H+ antiporter. Furthermore, inactivation of the mrp gene cluster in P. aeruginosa PAO1 has been reported to cause reduced pathogenicity (Kosono et al., 2005).
In a study of the group 2 Mrp antiporter of Vibrio cholerae, expressed in a major Na+/H+ antiporter-deficient E. coli strain, EP432, this antiporter had Na+ (Li+, K+)/H+ antiport activity with optimal pH at pH 9–9.5 and also showed bile acid resistance in E. coli (Dzioba-Winogrodzki et al., 2009). A deletion mutant of the group 2 mrp gene cluster from V. cholerae revealed mutant physiological defects in nitrogen metabolism, cell motility, and biofilm formation (Aagesen et al., 2016).
In a study of the group 1 Mrp antiporter of Thermomicrobium roseum expressed in a Na+/H+ antiporter-deficient E. coli strain, KNabc, it was surprisingly found that this antiporter does not catalyze monovalent cation/proton antiport similar to the Mrp antiporters studied to date but catalyzes Ca2+/H+ antiport in E. coli membrane vesicles (Morino and Ito, 2012). This bacterium was isolated from an alkaline hot spring in Yellowstone National Park (Jackson et al., 1973).
The gene cluster of a halotolerant cyanobacterium, Aphanothece halophytica mrp (Ah-mrp), which belongs to group 3, has a characteristic genetic structure that retains two mrpD genes in an unusual gene order (mrpCD1D2EFGAB). Study of a sodium-sensitive mutant E. coli expressing Ah-mrp showed that the cyanobacterial Mrp antiporter functions as a Na+/H+ antiporter and also contributes to sodium tolerance (Fukaya et al., 2009). Another cyanobacterium, Anabaena sp. strain PCC 7120, has a group 1 Mrp antiporter. Growth and photosynthesis were inhibited in a mrpA mutant cyanobacterial strain (Blanco-Rivero et al., 2009).
It has been reported that the group 1 Mrp antiporters of the halotolerant alkaliphile Halomonas sp. Y2 and the halophilic and alkaliphilic Halomonas zhadongensis had Na+ (Li+, K+)/H+ antiporter functions under alkaline conditions (Meng et al., 2014; Cheng et al., 2016).
Mrp Antiporters from Archaea
Many Mrp complexes are annotated not only from bacterial genomes but also from archaea (Swartz et al., 2005). The Mrp antiporter from the methanogen Methanosarcina acetivorans C2A is composed of a group 1 type of gene cluster comprising seven genes (mrpABCDEFG). This Mrp complex plays an essential role in efficient ATP synthesis and optimal growth under conditions with low concentrations of acetic acid in the environment (Jasso-Chavez et al., 2013). Deficiency of a major Na+/H+ antiporter in E. coli cells expressing only MrpA from M. acetivorans was still associated with Na+/H+ antiport activity, although the Km value was as low as ca. 50 mM (Jasso-Chavez et al., 2017). The details of these transport mechanisms have not yet been reported.
In hyperthermophilic archaea, Mrp is reported to be involved in the metabolic system of hydrogen production (Kim et al., 2010; Lim et al., 2010, 2014; Schut et al., 2013; Boyd et al., 2014). It is known that a hydrogenase involved in hydrogen production of Pyrococcus furiosus and Thermococcus onnurineus NA1 is composed of a [NiFe] hydrogenase domain (Mbh) and Mrp type Na+/H+ antiporter domain. However, there have been no reports of measurement of Mrp antiport activity in these strains/species (Schut et al., 2013). Given the considerable interest in this issue, it is anticipated that the details of the Mrp antiporter that is involved in archaeal energy production will soon be clarified.
Prediction of the Ion Transport Route in the Mrp Antiporter
Figure 6 describes the prediction of the ion transport pathway of the Mrp antiporter (Moparthi and Hägerhäll, 2011; Moparthi et al., 2011; Sazanov, 2015). Owing to homology with the respiratory chain complex I subunit, it is expected that the Mrp antiporter is involved in an ion transport pathway via the MrpA and MrpD subunits. MrpA has the closest homology to the NuoL subunit of complex I and MrpD has the closest homology to complex I NuoM and NuoN subunits. Because the nuoL-deficient strain does not transport Na+, it was suggested that the NuoL subunit is involved in Na+ transport (Marreiros et al., 2014). Moreover, Na+ transport was previously demonstrated by the NuoL subunit (Steuber, 2003; Gemperli et al., 2007). Moparthi et al. (2011) reported that the phenotypes of an mrpA-deficient strain and mrpD-deficient strain of B. subtilis are complemented by expressing, respectively, NuoL and NuoN of the respiratory chain complex I of E. coli. These observations prompted them to propose that MrpA transports Na+, whereas MrpD transports H+ in the opposite direction, resulting in antiport activity (Moparthi et al., 2011) (Figure 6A).
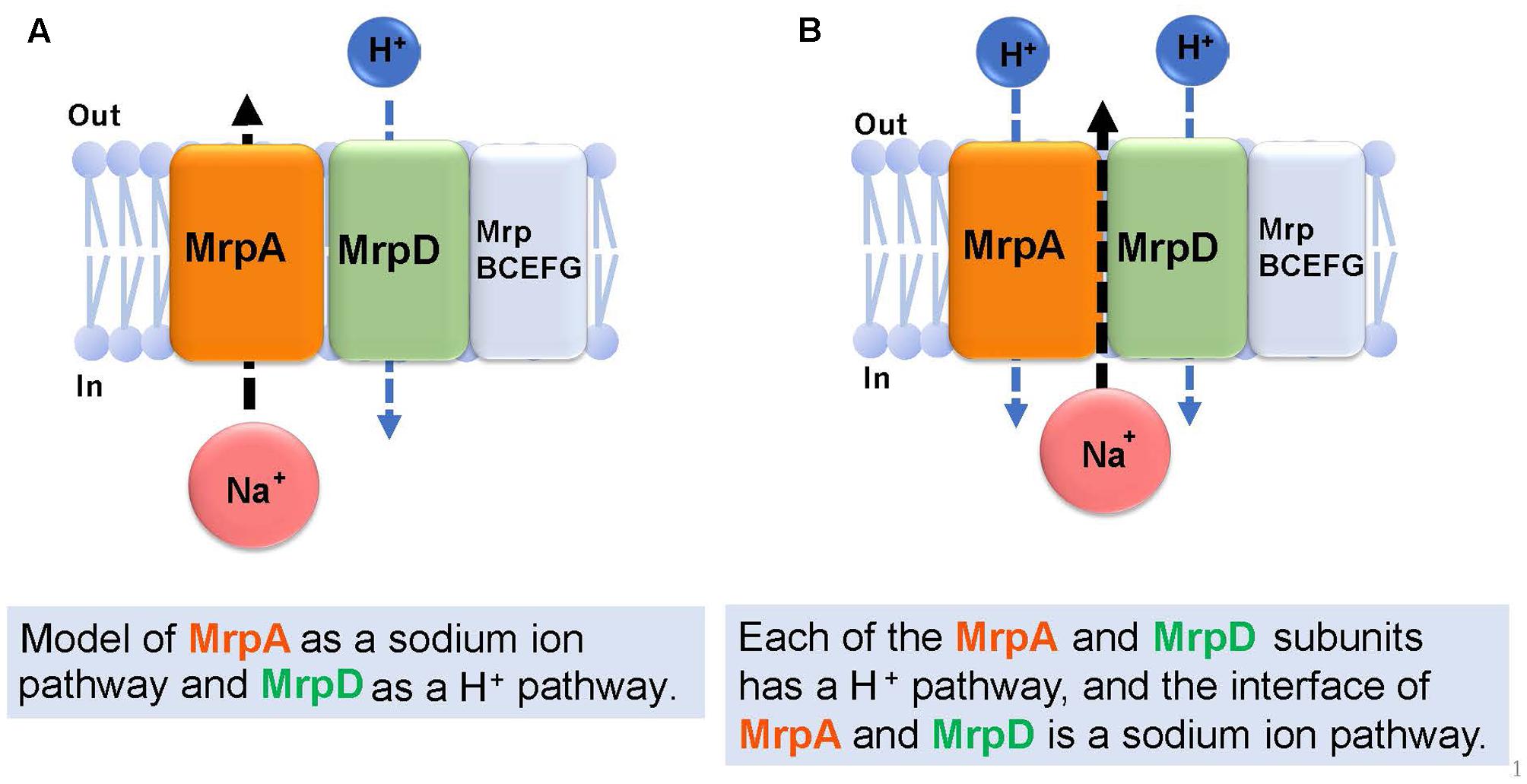
FIGURE 6. Ion transport pathway model of the Mrp antiporter. Based on previous studies, there are two kinds of ion transport models for the Mrp antiporter. The first model (A) proposes MrpA as a Na+ pathway and MrpD as a H+ pathway (Moparthi et al., 2011). The second model (B) proposes that each of the MrpA and MrpD subunits has a H+ pathway, and the interface of MrpA and MrpD is a Na+ pathway (Sazanov, 2014).
Sazanov reported that, at the interface between the transmembrane region (TM 5) of the MrpA subunit and the transmembrane region (TM 12) of the MrpD subunit, a Na+ transport pathway forms, which was confirmed from a homology model of the MrpA and MrpD subunits constructed from the results of crystal structure analyses of NuoL, NuoM, and NuoN (Sazanov, 2015). This model proposes that highly conserved glutamic acid residues in the NDH-1 motif that is common to the NuoL, NuoM, NuoN, MrpA, and MrpD subunits, function as cation binding sites (Figure 6B).
Prospects for the Future
It is expected that the details of Mrp antiporter complexes and their functional properties as revealed by recent studies will help to reveal the mechanisms of adaptation to environmental conditions not only in alkaliphilic bacteria but also in many other bacteria. The Mrp antiporter plays a major role in the environmental adaptation of a wide variety of bacteria, including pathogenic ones. Furthermore, because Mrp is only found in prokaryotes, studies may lead to the development of inhibitors of the roles of Mrp antiporters that are important in the host.
Author Contributions
The idea for this review paper was proposed by MI, MM, and TK. The paper was written by MI and TK.
Funding
This work was supported in part by research grant GM28454 from the National Institute of General Medical Sciences (to TK) as well as JSPS KAKENHI Grant Number 15K07012 (to MI).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aagesen, A. M., Schubiger, C. B., Hobson, E. C., Dibrov, P., and Hase, C. C. (2016). Effects of chromosomal deletion of the operon encoding the multiple resistance and pH-related antiporter in Vibrio cholerae. Microbiology 162, 2147–2158. doi: 10.1099/mic.0.000384
Baranova, E. A., Morgan, D. J., and Sazanov, L. A. (2007). Single particle analysis confirms distal location of subunits NuoL and NuoM in Escherichia coli complex I. J. Struct. Biol. 159, 238–242. doi: 10.1016/j.jsb.2007.01.009
Blanco-Rivero, A., Leganes, F., Fernandez-Valiente, E., and Fernandez-Pinas, F. (2009). mrpA (all1838), a gene involved in alkali and Na+ sensitivity, may also have a role in energy metabolism in the cyanobacterium Anabaena sp. strain PCC 7120. J. Plant Physiol. 166, 1488–1496. doi: 10.1016/j.jplph.2009.03.007
Boyd, E. S., Schut, G. J., Adams, M. W., and Perers, J. W. (2014). Hydrogen metabolism and the evolution of biological respiration. Microbe 9, 361–367. doi: 10.1128/microbe.9.361.1
Cheng, B., Meng, Y., Cui, Y., Li, C., Tao, F., Yin, H., et al. (2016). Alkaline response of a halotolerant alkaliphilic halomonas strain and functional diversity of its Na+ (K+)/H+ antiporters. J. Biol. Chem. 291, 26056–26065. doi: 10.1074/jbc.M116.751016
Dibrov, P. (2005). The sodium cycle in Vibrio cholerae: riddles in the dark. Biochemistry 70, 150–153. doi: 10.1007/s10541-005-0094-3
Dzioba-Winogrodzki, J., Winogrodzki, O., Krulwich, T. A., Boin, M. A., Hase, C. C., and Dibrov, P. (2009). The Vibrio cholerae Mrp system: cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J. Mol. Microbiol. Biotechnol. 16, 176–186. doi: 10.1159/000119547
Efremov, R. G., and Sazanov, L. A. (2011). Structure of the membrane domain of respiratory complex I. Nature 476, 414–420. doi: 10.1038/nature10330
Fujinami, S., Takeda, K., Onodera, T., Satoh, K., Sano, M., Narumi, I., et al. (2013). Draft genome sequence of sodium-independent alkaliphilic Microbacterium sp. strain TS-1. Genome Announc. 1:e01043-31. doi: 10.1128/genomeA.01043-13
Fukaya, F., Promden, W., Hibino, T., Tanaka, Y., Nakamura, T., and Takabe, T. (2009). An Mrp-like cluster in the halotolerant cyanobacterium Aphanothece halophytica functions as a Na+/H+ antiporter. Appl. Environ. Microbiol. 75, 6626–6629. doi: 10.1128/AEM.01387-09
Fuster, D. G., and Alexander, R. T. (2014). Traditional and emerging roles for the SLC9 Na+/H+ exchangers. Pflugers Arch. 466, 61–76. doi: 10.1007/s00424-013-1408-8
Gemperli, A. C., Schaffitzel, C., Jakob, C., and Steuber, J. (2007). Transport of Na+ and K+ by an antiporter-related subunit from the Escherichia coli NADH dehydrogenase I produced in Saccharomyces cerevisiae. Arch. Microbiol. 188, 509–521. doi: 10.1007/s00203-007-0272-3
Hamamoto, T., Hashimoto, M., Hino, M., Kitada, M., Seto, Y., Kudo, T., et al. (1994). Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14, 939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x
Hisamitsu, T., Nakamura, T. Y., and Wakabayashi, S. (2012). Na+/H+ exchanger 1 directly binds to calcineurin A and activates downstream NFAT signaling, leading to cardiomyocyte hypertrophy. Mol. Cell. Biol. 32, 3265–3280. doi: 10.1128/MCB.00145-12
Holt, P. J., Morgan, D. J., and Sazanov, L. A. (2003). The location of NuoL and NuoM subunits in the membrane domain of the Escherichia coli complex I: implications for the mechanism of proton pumping. J. Biol. Chem. 278, 43114–43120. doi: 10.1074/jbc.M308247200
Ito, M., Guffanti, A. A., Oudega, B., and Krulwich, T. A. (1999). mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181, 2394–2402.
Ito, M., Guffanti, A. A., Wang, W., and Krulwich, T. A. (2000). Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J. Bacteriol. 182, 5663–5670. doi: 10.1128/JB.182.20.5663-5670.2000
Ivey, D. M., Guffanti, A. A., Zemsky, J., Pinner, E., Karpel, R., Padan, E., et al. (1993). Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J. Biol. Chem. 268, 11296–11303.
Jackson, T. J., Ramaley, R. F., and Meinsche, W. G. (1973) Thermomicrobium, a new genus of extremely thermophilic bacteria. Int. J. Syst. Bacteriol. 23, 28–36. doi: 10.1099/00207713-23-1-28
Jasso-Chavez, R., Apolinario, E. E., Sowers, K. R., and Ferry, J. G. (2013). MrpA functions in energy conversion during acetate-dependent growth of Methanosarcina acetivorans. J. Bacteriol. 195, 3987–3994. doi: 10.1128/JB.00581-13
Jasso-Chavez, R., Diaz-Perez, C., Rodriguez-Zavala, J. S., and Ferry, J. G. (2017). Functional role of MrpA in the MrpABCDEFG Na+/H+ antiporter complex from the Archaeon Methanosarcina acetivorans. J. Bacteriol. 199:e00662-16. doi: 10.1128/JB.00662-16
Ji, Y., Zhang, B., Van Horn, S. F., Warren, P., Woodnutt, G., Burnham, M. K., et al. (2001). Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293, 2266–2269. doi: 10.1126/science.1063566
Kajiyama, Y., Otagiri, M., Sekiguchi, J., Kosono, S., and Kudo, T. (2007). Complex formation by the mrpABCDEFG gene products, which constitute a principal Na+/H+ antiporter in Bacillus subtilis. J. Bacteriol. 189, 7511–7514. doi: 10.1128/JB.00968-07
Kajiyama, Y., Otagiri, M., Sekiguchi, J., Kudo, T., and Kosono, S. (2009). The MrpA, MrpB and MrpD subunits of the Mrp antiporter complex in Bacillus subtilis contain membrane-embedded and essential acidic residues. Microbiology 155, 2137–2147. doi: 10.1099/mic.0.025205-0
Kashyap, D. R., Botero, L. M., Lehr, C., Hassett, D. J., and Mcdermott, T. R. (2006). A Na+:H+ antiporter and a molybdate transporter are essential for arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188, 1577–1584. doi: 10.1128/JB.188.4.1577-1584.2006
Kim, Y. J., Lee, H. S., Kim, E. S., Bae, S. S., Lim, J. K., Matsumi, R., et al. (2010). Formate-driven growth coupled with H2 production. Nature 467, 352–355. doi: 10.1038/nature09375
Kosono, S., Haga, K., Tomizawa, R., Kajiyama, Y., Hatano, K., Takeda, S., et al. (2005). Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: its involvement in pathogenesis. J. Bacteriol. 187, 5242–5248. doi: 10.1128/JB.187.15.5242-5248.2005
Kosono, S., Kajiyama, Y., Kawasaki, S., Yoshinaka, T., Haga, K., and Kudo, T. (2006). Functional involvement of membrane-embedded and conserved acidic residues in the ShaA subunit of the multigene-encoded Na+/H+ antiporter in Bacillus subtilis. Biochim. Biophys. Acta 1758, 627–635. doi: 10.1016/j.bbamem.2006.04.012
Kosono, S., Morotomi, S., Kitada, M., and Kudo, T. (1999). Analyses of a Bacillus subtilis homologue of the Na+/H+ antiporter gene which is important for pH homeostasis of alkaliphilic Bacillus sp. C-125. Biochim. Biophys. Acta 1409, 171–175. doi: 10.1016/S0005-2728(98)00157-1
Kosono, S., Ohashi, Y., Kawamura, F., Kitada, M., and Kudo, T. (2000). Function of a principal Na+/H+ antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 182, 898–904. doi: 10.1128/JB.182.4.898-904.2000
Krulwich, T. A., Hicks, D. B., and Ito, M. (2009). Cation/proton antiporter complements of bacteria: Why so large and diverse? Mol. Microbiol. 74, k257–260. doi: 10.1111/j.1365-2958.2009.06842.x
Krulwich, T. A., and Ito, M. (2013). “Prokaryotic alkaliphiles,” in The Prokaryotes, 4th Edn, eds E. Rosenberg, E. F. Delong, F. Thompson, S. Lory, and E. Stackebrandt (New York, NY: Springer).
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343. doi: 10.1038/nrmicro2549
Kudo, T., Hino, M., Kitada, M., and Horikoshi, K. (1990). DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J. Bacteriol. 172, 7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990
Lewinson, O., Adler, J., Poelarends, G. J., Mazurkiewicz, P., Driessen, A. J., and Bibi, E. (2003). The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. U.S.A. 100, 1667–1672. doi: 10.1073/pnas.0435544100
Lim, J. K., Kang, S. G., Lebedinsky, A. V., Lee, J. H., and Lee, H. S. (2010). Identification of a novel class of membrane-bound [NiFe]-hydrogenases in Thermococcus onnurineus NA1 by in silico analysis. Appl. Environ. Microbiol. 76, 6286–6289. doi: 10.1128/AEM.00123-10
Lim, J. K., Mayer, F., Kang, S. G., and Muller, V. (2014). Energy conservation by oxidation of formate to carbon dioxide and hydrogen via a sodium ion current in a hyperthermophilic archaeon. Proc. Natl. Acad. Sci. U.S.A. 111, 11497–11502. doi: 10.1073/pnas.1407056111
Marreiros, B. C., Batista, A. P., and Pereira, M. M. (2014). Respiratory complex I from Escherichia coli does not transport Na+ in the absence of its NuoL subunit. FEBS Lett. 588, 4520–4525. doi: 10.1016/j.febslet.2014.10.030
Mathiesen, C., and Hägerhäll, C. (2003). The ‘antiporter module’ of respiratory chain complex I includes the MrpC/NuoK subunit – a revision of the modular evolution scheme. FEBS Lett. 549, 7–13. doi: 10.1016/S0014-5793(03)00767-1
Meng, L., Hong, S., Liu, H., Huang, H., Sun, H., Xu, T., et al. (2014). Cloning and identification of Group 1 mrp operon encoding a novel monovalent cation/proton antiporter system from the moderate halophile Halomonas zhaodongensis. Extremophiles 18, 963–972. doi: 10.1007/s00792-014-0666-5
Mesbah, N. M., Cook, G. M., and Wiegel, J. (2009). The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+ (K+)/H+ antiporters. Mol. Microbiol. 74, 270–281. doi: 10.1111/j.1365-2958.2009.06845.x
Moparthi, V. K., and Hägerhäll, C. (2011). “Recruitment of the antiporter module — a key event in complex I evolution,” in A Structural Perspective on Complex I, ed. L. A. Sazanov (Berlin: Springer), 123–143.
Moparthi, V. K., Kumar, B., Al-Eryani, Y., Sperling, E., Gorecki, K., Drakenberg, T., et al. (2014). Functional role of the MrpA- and MrpD-homologous protein subunits in enzyme complexes evolutionary related to respiratory chain complex I. Biochim. Biophys. Acta 1837, 178–185. doi: 10.1016/j.bbabio.2013.09.012
Moparthi, V. K., Kumar, B., Mathiesen, C., and Hägerhäll, C. (2011). Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis. Biochim. Biophys. Acta 1807, 427–436. doi: 10.1016/j.bbabio.2011.01.005
Morino, M., and Ito, M. (2012). Functional expression of the multi-subunit type calcium/proton antiporter from Thermomicrobium roseum. FEMS Microbiol. Lett. 335, 26–30. doi: 10.1111/j.1574-6968.2012.02634.x
Morino, M., Natsui, S., Ono, T., Swartz, T. H., Krulwich, T. A., and Ito, M. (2010). Single site mutations in the hetero-oligomeric Mrp antiporter from alkaliphilic Bacillus pseudofirmus OF4 that affect Na+/H+ antiport activity, sodium exclusion, individual Mrp protein levels, or Mrp complex formation. J. Biol. Chem. 285, 30942–30950. doi: 10.1074/jbc.M110.118661
Morino, M., Natsui, S., Swartz, T. H., Krulwich, T. A., and Ito, M. (2008). Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J. Bacteriol. 190, 4162–4172. doi: 10.1128/JB.00294-08
Morino, M., Ogoda, S., Krulwich, T. A., and Ito, M. (2017). Differences in the phenotypic effects of mutations in homologous MrpA and MrpD subunits of the multi-subunit Mrp-type Na+/H+ antiporter. Extremophiles 21, 51–64. doi: 10.1007/s00792-016-0877-z
Morino, M., Suzuki, T., Ito, M., and Krulwich, T. A. (2014). Purification and functional reconstitution of a seven-subunit mrp-type Na+/H+ antiporter. J. Bacteriol. 196, 28–35. doi: 10.1128/JB.01029-13
Mourin, M., Schubiger, C. B., Resch, C. T., Hase, C. C., and Dibrov, P. (2017). Physiology of the Vc-NhaP paralogous group of cation-proton antiporters in Vibrio cholerae. Mol. Cell. Biochem. 428, 87–99. doi: 10.1007/s11010-016-2919-3
Nakamaru-Ogiso, E., Kao, M. C., Chen, H., Sinha, S. C., Yagi, T., and Ohnishi, T. (2010). The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J. Biol. Chem. 285, k39070–39078. doi: 10.1074/jbc.M110.157826
Nakamaru-Ogiso, E., Sakamoto, K., Matsuno-Yagi, A., Miyoshi, H., and Yagi, T. (2003a). The ND5 subunit was labeled by a photoaffinity analogue of fenpyroximate in bovine mitochondrial complex I. Biochemistry 42, 746–754.
Nakamaru-Ogiso, E., Seo, B. B., Yagi, T., and Matsuno-Yagi, A. (2003b). Amiloride inhibition of the proton-translocating NADH-quinone oxidoreductase of mammals and bacteria. FEBS Lett. 549, 43–46.
Ohgaki, R., Van, I. S. C., Matsushita, M., Hoekstra, D., and Kanazawa, H. (2011). Organellar Na+/H+ exchangers: novel players in organelle pH regulation and their emerging functions. Biochemistry 50, 443–450. doi: 10.1021/bi101082e
Ohnishi, T., Nakamaru-Ogiso, E., and Ohnishi, S. T. (2010). A new hypothesis on the simultaneous direct and indirect proton pump mechanisms in NADH-quinone oxidoreductase (complex I). FEBS Lett. 584, 4131–4137. doi: 10.1016/j.febslet.2010.08.039
Orlowski, J., and Grinstein, S. (2004). Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 447, 549–565. doi: 10.1007/s00424-003-1110-3
Orlowski, J., and Grinstein, S. (2011). Na+/H+ exchangers. Compr. Physiol. 1, 2083–2100. doi: 10.1002/cphy.c110020
Padan, E. (2014). Functional and structural dynamics of NhaA, a prototype for Na+ and H+ antiporters, which are responsible for Na+ and H+ homeostasis in cells. Biochim. Biophys. Acta 1837, 1047–1062. doi: 10.1016/j.bbabio.2013.12.007
Padan, E., Bibi, E., Ito, M., and Krulwich, T. A. (2005). Alkaline pH homeostasis in bacteria: new insights. Biochim. Biophys. Acta 1717, 67–88. doi: 10.1016/j.bbamem.2005.09.010
Padan, E., and Landau, M. (2016). Sodium-proton (Na+/H+) antiporters: properties and roles in health and disease. Met. Ions Life Sci. 16, 391–458. doi: 10.1007/978-3-319-21756-7_12
Padan, E., and Schuldiner, S. (1994). Molecular physiology of the Na+/H+ antiporter in Escherichia coli. J. Exp. Biol. 196, 443–456.
Padan, E., Venturi, M., Gerchman, Y., and Dover, N. (2001). Na+/H+ antiporters. Biochim. Biophys. Acta 1505, 144–157. doi: 10.1016/S0005-2728(00)00284-X
Pang, T., Su, X., Wakabayashi, S., and Shigekawa, M. (2001). Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J. Biol. Chem. 276, 17367–17372. doi: 10.1074/jbc.M100296200
Preiss, L., Hicks, D. B., Suzuki, S., Meier, T., and Krulwich, T. A. (2015). Alkaliphilic bacteria with impact on industrial applications, concepts of early life forms, and bioenergetics of ATP synthesis. Front. Bioeng. Biotechnol. 3:75. doi: 10.3389/fbioe.2015.00075
Putnoky, P., Kereszt, A., Nakamura, T., Endre, G., Grosskopf, E., Kiss, P., et al. (1998). The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28, 1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x
Resch, C. T., Winogrodzki, J. L., Hase, C. C., and Dibrov, P. (2011). Insights into the biochemistry of the ubiquitous NhaP family of cation/H+ antiporters. Biochem. Cell Biol. 89, 130–137. doi: 10.1139/o10-149
Saier, M. H. Jr., Reddy, V. S., Tsu, B. V., Ahmed, M. S., Li, C., and Moreno-Hagelsieb, G. (2016). The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379. doi: 10.1093/nar/gkv1103
Saier, M. H. Jr., Yen, M. R., Noto, K., Tamang, D. G., and Elkan, C. (2009). The transporter classification database: recent advances. Nucleic Acids Res. 37, D274–D278. doi: 10.1093/nar/gkn862
Sazanov, L. A. (2014). The mechanism of coupling between electron transfer and proton translocation in respiratory complex I. J. Bioenerg. Biomembr. 46, 247–253. doi: 10.1007/s10863-014-9554-z
Sazanov, L. A. (2015). A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388. doi: 10.1038/nrm3997
Schut, G. J., Boyd, E. S., Peters, J. W., and Adams, M. W. (2013). The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203. doi: 10.1111/j.1574-6976.2012.00346.x
Shijuku, T., Yamashino, T., Ohashi, H., Saito, H., Kakegawa, T., Ohta, M., et al. (2002). Expression of chaA, a sodium ion extrusion system of Escherichia coli, is regulated by osmolarity and pH. Biochim. Biophys. Acta 1556, 142–148. doi: 10.1016/S0005-2728(02)00345-6
Sperling, E., Gorecki, K., Drakenberg, T., and Hagerhall, C. (2016). Functional differentiation of antiporter-like polypeptides in complex I; a site-directed mutagenesis study of residues conserved in MrpA and NuoL but not in MrpD, NuoM, and NuoN. PLOS ONE 11:e0158972. doi: 10.1371/journal.pone.0158972
Steuber, J. (2003). The C-terminally truncated NuoL subunit (ND5 homologue) of the Na+-dependent complex I from Escherichia coli transports Na+. J. Biol. Chem. 278, 26817–26822. doi: 10.1074/jbc.M301682200
Swartz, T. H., Ikewada, S., Ishikawa, O., Ito, M., and Krulwich, T. A. (2005). The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9, 345–354.
Swartz T. H., Ito, M., Ohira, T., Natsui, S., Hicks, D. B., and Krulwich, T. A. (2007). Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 189, 3081–3090. doi: 10.1128/JB.00021-07
Terashima, H., Koike, M., Kojima, S., and Homma, M. (2010). The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J. Bacteriol. 192, 5609–5615. doi: 10.1128/JB.00720-10
Torres-Bacete, J., Nakamaru-Ogiso, E., Matsuno-Yagi, A., and Yagi, T. (2007). Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1: conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 282, 36914–36922. doi: 10.1074/jbc.M707855200
Torres-Bacete, J., Sinha, P. K., Matsuno-Yagi, A., and Yagi, T. (2011). Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J. Biol. Chem. 286, 34007–34014. doi: 10.1074/jbc.M111.260968
Vimont, S., and Berche, P. (2000). NhaA, an Na+/H+ antiporter involved in environmental survival of Vibrio cholerae. J. Bacteriol. 182, 2937–2944. doi: 10.1128/JB.182.10.2937-2944.2000
Waditee, R., Hibino, T., Tanaka, Y., Nakamura, T., Incharoensakdi, A., and Takabe, T. (2001). Halotolerant cyanobacterium Aphanothece halophytica contains an Na+/H+ antiporter, homologous to eukaryotic ones, with novel ion specificity affected by C-terminal tail. J. Biol. Chem. 276, 36931–36938. doi: 10.1074/jbc.M103650200
Wakabayashi, S., Shigekawa, M., and Pouyssegur, J. (1997). Molecular physiology of vertebrate Na+/H+ exchangers. Physiol. Rev. 77, 51–74.
Yamaguchi, T., Tsutsumi, F., Putnoky, P., Fukuhara, M., and Nakamura, T. (2009). pH-dependent regulation of the multi-subunit cation/proton antiporter Pha1 system from Sinorhizobium meliloti. Microbiology 155, 2750–2756. doi: 10.1099/mic.0.028563-0
Yoshinaka, T., Takasu, H., Tomizawa, R., Kosono, S., and Kudo, T. (2003). A shaE deletion mutant showed lower Na+ sensitivity compound to other deletion mutants in the Bacillus subtilis sodium/hydrogen antiporter (Sha) system. J. Biosci. Bioeng. 95, 306–309. doi: 10.1016/S1389-1723(03)80035-X
Keywords: alkaliphile, cation/proton antiporter, Mrp, complex I, multi-subunit antiporter, Bacillus, Thermomicrobium
Citation: Ito M, Morino M and Krulwich TA (2017) Mrp Antiporters Have Important Roles in Diverse Bacteria and Archaea. Front. Microbiol. 8:2325. doi: 10.3389/fmicb.2017.02325
Received: 27 August 2017; Accepted: 10 November 2017;
Published: 23 November 2017.
Edited by:
Baolei Jia, Chung-Ang University, South KoreaReviewed by:
Saori Kosono, The University of Tokyo, JapanSung Gyun Kang, Korea Institute of Ocean Science and Technology, South Korea
Copyright © 2017 Ito, Morino and Krulwich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahiro Ito, bWFzYWhpcm8uaXRvQHRveW8uanA=
 Masahiro Ito
Masahiro Ito Masato Morino1,3
Masato Morino1,3 Terry A. Krulwich
Terry A. Krulwich