- 1Laboratory of Marine Microbiology, College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Key Laboratory of Marine Chemistry Theory and Technology, Ministry of Education, Ocean University of China, Qingdao, China
The mud sediments of the eastern China marginal seas (ECMS) are deposited under different hydrodynamic conditions with different organic matter sources. These events have been demonstrated to exert significant influences on microbial communities and biogeochemical processes in surface sediments. However, the extent to which such effects occur in subsurface microbial communities remains unclear. In this study, both horizontal and vertical (five sites, each for eight layers) distributions of bacterial abundance and community composition in mud deposits of the South Yellow Sea (SYS) and East China Sea (ECS) were investigated by quantitative PCR and Illumina sequencing of the 16S rRNA gene. Both bacterial abundance and diversity were higher in the ECS than in the SYS, and tended to be higher in up than in deep layers. Proteobacteria (JTB255 marine benthic group), Acidobacteria and Bacteroidetes were dominant in the upper layers, whereas Lactococcus, Pseudomonas, and Dehalococcoidia were enriched in the deep layers. The bacterial communities in surface and subsurface sediments showed different inter-taxa relationships, indicating contrasting co-occurrence patterns. The bacterial communities in the upper layer samples clustered in accordance with mud zones, whereas those in the deep layer samples of all sites tended to cluster together. TOC δ13C and TON δ15N significantly affected the bacterial community composition, suggesting that the abundance and composition of organic matter played critical roles in shaping of sedimentary bacterial communities. This study provides novel insights into the distribution of subsurface bacterial communities in mud deposits of the ECMS, and provides clues for understanding the biogeochemical cycles in this area.
Introduction
Marginal seas are the transitional zones between the coastal and open oceans and occupy about 10% of the global ocean. These shallow, narrow and fast-deposition areas are reported to be major reservoirs of organic carbon burial in the marine system (Hedges and Keil, 1995), and have significant impacts on global biogeochemical cycles and even global climate changes to a considerable degree (Chen C.T.A. et al., 2004; Coelho et al., 2013). Correspondingly, sediments in these areas harbor a higher abundance of microbes than those in open oceans (Kallmeyer et al., 2012). These abundant microorganisms contribute significantly to the cycling of marine biogenic elements (Azam and Malfatti, 2007; Falkowski et al., 2008), especially carbon (Dyksma et al., 2016).
Bacterial communities were found to vary in different marginal sea sediments (Bowman and McCuaig, 2003; Bertics and Ziebis, 2009; Harrison et al., 2009; Zinger et al., 2011; Wang et al., 2012; Learman et al., 2016; Probandt et al., 2017). These can be explained by environmental heterogeneity (including sediment sources and hydrodynamic conditions) that can significantly influence the distribution of microbial communities and related biogeochemical processes. For example, organic matter has been demonstrated to be a driver of benthic microbial community structure across the Antarctic surface sediment (Learman et al., 2016). Meanwhile, shifts in bacterial community were observed in oil-contaminated and nitrogen-polluted sediments in the Antarctic near shore, Mediterranean Sea and East China Sea (ECS) (Powell et al., 2003; Polymenakou et al., 2006; Xiong et al., 2014), suggesting composition of organics and nutrients to be important factors as well. In addition, dissolved oxygen (DO) of the bottom water, temperature and sediment median grain size, have been detected to be vital factors shaping benthic bacterial communities in the Chinese marginal sea (Bohai Sea and Pearl Estuary), Arctic and North Sea (Wang et al., 2013; Liu et al., 2014; Zheng et al., 2014; Nguyen and Landfald, 2015; Probandt et al., 2017). Most studies mentioned above were based on surface sediments, but relatively few have focused on the vertical profile of bacterial communities in typical marginal sediments (Franco et al., 2007; Böer et al., 2009; Harrison et al., 2009; Liu et al., 2014). Depth related shifts in bacterial community in marginal sediments have been reported and were attributed to different factors such as contents of organic carbon, chlorophyll a and inorganic nutrients (Bowman and McCuaig, 2003; Böer et al., 2009; Harrison et al., 2009). None of these studies included a highly resolved vertical profile of sedimentary bacterial communities. Thus, the extent to which the benthic surface environmental heterogeneity affect subsurface microbial communities needs further investigation.
The eastern China marginal seas (ECMS) are typical eutrophic seas with different mud areas formed by sediments derived mainly from the Yellow River and Yangtze River. These mud areas are characterized by different sediment sources and hydrodynamic conditions resulted from complex water masses and ocean currents; therefore, they provide different environmental niches for microorganisms to survive. Accordingly, previous studies have shown distinct distribution patterns of functional microorganisms in different ECMS mud sediments (Yu et al., 2016; Gao et al., 2017). However, compositional distributions of total bacterial community in different mud sediments are currently unknown. We hypothesized that the total bacterial communities varied in surface sediments but converged in subsurface sediments in different mud areas of the ECMS. In this study, a high resolution vertical profile of bacterial abundance and community composition from five sites, each for eight layers, of the ECMS was provided. In addition, the bacterial co-occurrence patterns, which can help uncover potential inter-taxa relationships, in both surface and subsurface sediments were explored by using correlation based network analysis.
Materials and Methods
Study Site and Sampling
To compare sedimentary bacterial communities in different mud zones of the ECMS, five sites (SYS01, SYS02, ECS01, ECS02, and ECS03) distributed in four typical mud zones of the South Yellow Sea (SYS) and ECS were chosen. Locations of these samples have been reported by Yu et al. (2016). SYS01 and SYS02 are located in the SYS mud zone, where the deposits are mainly from sinking of the modern and old Yellow River-derived sedimentary organic matter (Hu et al., 2013). In addition, mud deposits in this area are considered as a result of the presence of cold water mass in summer, accompanied by seasonal weaken of the Yellow Sea Warm Current (Hu, 1984). Situated in the Yangtze River Estuary mud zone, ECS01 is mainly influenced by freshwater flowed out of the Yangtze River, which makes Yangtze River to be the dominant sediment source of ECS01 (Liu et al., 2007). ECS02 is located in the Zhe-Min mud zone. This area is influenced by a couple of alternatively predominant reversed currents (the Zhe-Min Coastal Current and Taiwan Warm Current); its sediments are mainly transported from the Yangtze River and the estuary mud zone along the Zhe-Min coast (Liu et al., 2007). ECS03 belongs to the distal Cheju Island mud zone, and its sediments are derived from both the Yangtze River and the old Yellow River, transported by the Yellow Sea Warm Current and river runoff from the Yangtze River (Liu et al., 2003).
The sediment samples were collected by a box corer during a cruise of R/V Dong Fang Hong 2 from 12 July to 2 August, 2013. Two PVC tubes were used to subsample the collected sediments at each site. One PVC core was immediately sliced at a 1-cm interval with a stainless-steel cutter and the sliced sediments were stored at -20°C (onboard) or -80°C (in laboratory) before organic matter measurement and DNA extraction. An aliquot of sediments at depths of 0–1, 12–13, and 32–33 cm from sites SYS01, SYS02, ECS02, and ECS03 was fixed with paraformaldehyde (2% final) in sterile plastic vessels and conserved in 1:1 PBS-ethanol at -20°C for 4′, 6-diamidino-2-phenylindole (DAPI) counting. The parallel core was prepared for pore water extraction. Pore water samples were collected by the Rhizon samplers at the cm-scale, poisoned by HgCl2 and stored at 4°C before dissolved inorganic nutrient measurement. For each core, eight sediment layers that were 0–1, 1–2, 2–3, 3–5, 7–8, 12–13, 22–23, and 32–33 cm (written as -0, -1, -2, -3, -5, -10, -20, and -30 cm, respectively) were chosen for microbiological analyses. Total organic carbon (TOC), total nitrogen (TN), stable carbon (TOC δ13C) and nitrogen isotopes (TON δ15N) in sediments, dissolved inorganic nutrients (NO3-, NO2-, NH4+, PO43-, SiO32- and SO42-) in pore water, salinity, DO and Chl a in bottom water were determined as previously described (Yu et al., 2016).
DNA Extraction
Genomic DNA was extracted from 0.25 g of sediment (wet weight) using the Power Soil DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, United States) and a FastPrep-24 cell disrupter (MP Biomedicals, Irvine, CA, United States) according to the manufacturer’s instructions. Quality and quantity of the extracted DNA were measured by a Nanodrop spectrophotometer ND-2000 (Thermo Fisher Scientific, United States). DNA was then subpackaged and stored at -80°C.
Quantification Analysis
Paraformaldehyde fixed sediment samples were diluted and homogenized with low-power ultrasonic wave at 20 W for 30 s. A volume of 50 μl sonicated sample was mixed with 10 mL PBS, collected on the 0.2-μm pore size filter (Isopore GTTP, Millipore), and stained with DAPI. For each sample, cell numbers were counted in ten random views under the fluorescence microscope. To exclude eukaryotic cells, only cells that are 0.5–5 μm in size were counted.
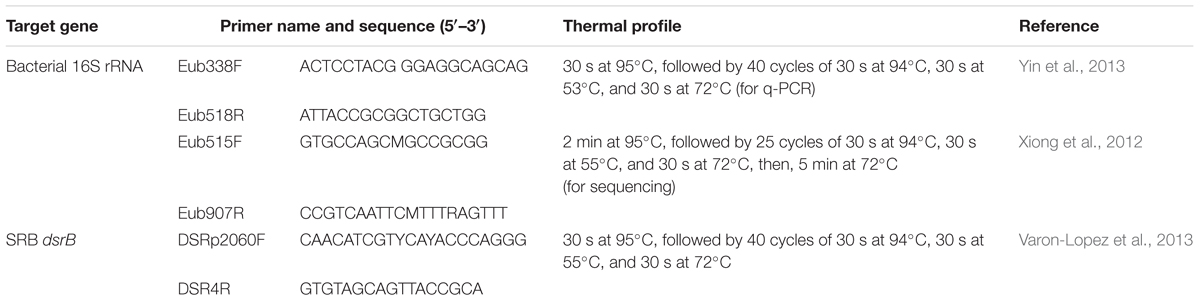
Quantitative PCR was performed to quantify the abundance of total bacteria and sulfate-reducing bacteria (SRB) in the samples using primers of the 16S rRNA gene and dissimilatory sulfite reductase β-subunit (dsrB) gene, respectively (Varon-Lopez et al., 2013; Yin et al., 2013). A 20 μl mixture contained 10 μl of SYBR Premix ExTaq II (2×), 0.4 μl of ROX Reference Dye II (50×) (TaKaRa, Tokyo, Japan), 0.2 μl of primers for each gene (10 μM), and 2 μl of template. Primers and thermal cycling steps are shown in Table 1. All assays were conducted in triplicate with negative controls using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States).
Standard curves were constructed by PCR amplifying a 10-fold serial dilution of plasmids containing target gene fragments. The amplification curves showed well linear relationships (R2> 0.999) and the amplification efficiencies were 91.65 and 95.28% for the bacterial 16S rRNA gene and dsrB gene, respectively. The single-peak melting curves and the only bond in gel electrophoresis guaranteed specificity of the qPCR analysis.
High Throughput Sequencing and Reads Processing
The PCR and sequencing were performed as previously described (Liang et al., 2015) with minor modifications. Primers Eub515F/Eub907R (Table 1) were used for bacterial 16S rRNA gene amplification. The forward and reverse primers were tagged with adapter, pad and linker sequences, and the reverse primer was linked with barcode sequences for pooling of multiple samples in one run of MiSeq sequencing. PCR was run on an ABI GeneAmp® 9700 cycler and thermal cycling steps are shown in Table 1. The PCR products of each sample were pooled, purified by an AxyPrepTM DNA Gel Extraction Kit (Axygen, Hangzhou, China) and quantified using a QuantiFluorTM-ST Solid Standard (Promega, Madison, WI, United States). Sequencing was conducted on a MiSeq Desktop Sequencer at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China.
The raw data were filtered according to the pipeline of Quantitative Insights into Microbial Ecology (QIIME1, Caporaso et al., 2010). Reads were assigned to samples according to their barcodes with no mismatch. The raw reads that had a quality score higher than 20 over a 5 bp window size and a minimum length of 100 bp (Kong, 2011) were retained. The pair-end reads were joined with at least a 50 bp overlap and less than 5% mismatches using FLASH (Magoc and Salzberg, 2011). A perl script daisychopper.pl (Gilbert et al., 2009) was used to randomly subsample sequences from each sample according to the least read numbers for equalizing sampling efforts. Operational taxonomic units (OTUs) clustering and taxonomy assignment were also performed in QIIME. Specifically, OTUs were defined at a 97% sequence similarity level, and then chimera sequences were detected and removed with UCHIME (Edgar et al., 2011) as recommended by QIIME tutorials. Taxonomy was assigned using the RDP Classifier v2.2 (Wang et al., 2007) against the SILVA v115 16S rRNA gene reference database2 with a minimum support threshold of 70%.
The Illumina sequences were deposited in the National Center for Biotechnology Information Short Read Archive database under SRP076973.
Statistical Analysis
The diversity indices, including Good’s coverage, Chao1 and Shannon index, were calculated for alpha diversity analysis. Molecular ecological network analyses were conducted by the package ‘Hmisc,’ ‘igraph,’ and ‘qvalue’ in R software (RDC TEAM, 2008) using bacterial groups at the family level with read numbers > 50 across all samples to simplify the network. Co-occurrence pairs with a Spearman’s correlation coefficient > 0.7 or < -0.7 and a P-value < 0.01 (Benjamini and Hochberg adjusted) were considered as a valid co-occurrence event. The R script was provided in Supplementary Material. Gephi (version 0.8.2 beta, Bastian et al., 2009) was used for network visualization. Linear discriminate analysis (LDA) effect size (LEfSe) (Segata et al., 2011) was used to identify taxa with significant differences between mud zones and depths at various taxonomic levels. For beta diversity, classification of bacterial communities was performed by principal coordinate analysis (PCoA) using Fast UniFrac (Lozupone et al., 2011). Pairwise analyses of similarities (ANOSIM) was performed in PRIMER 5 (Plymouth Marine Laboratory, West Hoe, Plymouth, United Kingdom). The relationships between phylotypes and environmental factors were evaluated by redundancy analysis (RDA) in CANOCO (Version 5.0, Microcomputer Power) with 9999 Monte Carlo permutation tests using square root-transformed data. Pearson correlation test was also used to evaluate correlations between percentage composition of taxa and environmental factors. In order to accurately estimate the correlations, only the top 20 phyla, top 30 classes, top 50 orders, top 50 families, top 50 genera were tested.
Results
Environmental Characterization
Detailed sediment and pore water environmental parameters have been described by Yu et al. (2016). In brief, at all sites, the NO3- concentration in pore water, TOC and TN content in sediment had a tendency to decrease with depth. Higher sedimentary C/N ratio and NO3- concentration were detected in SYS than in ECS samples. The pore water concentration of NH4+ at ECS01 was higher than that at other sites. Much depleted values of TON δ15N and TOC δ13C were observed in sediment samples of ECS01.
Direct Cell Counting and Quantitative PCR
The result of direct cell counting showed that microbial cell numbers in the ECMS varied from 3.17 × 108 to 4.19 × 109 cells g-1 (Figure 1A). The microbial cell numbers decreased with depth, and significant difference was observed between 0–1 cm samples and 32–33 cm samples (P < 0.05). Meanwhile, sediments from the ECS contained more microbial cells than those from the SYS (P < 0.01).
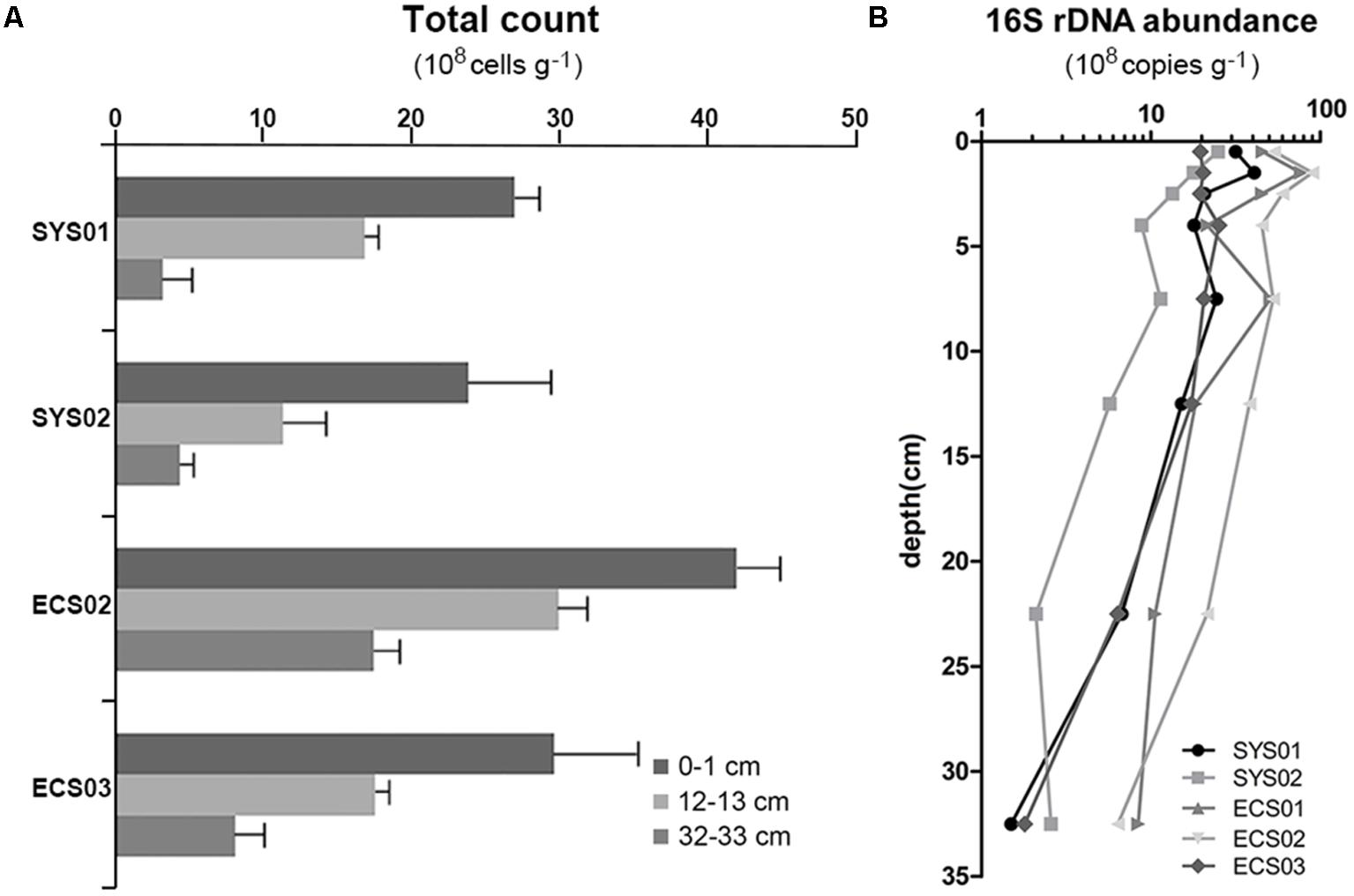
FIGURE 1. Total cell counts (A) and 16S rRNA gene abundance (B) of sediment cores from the Eastern China Marginal Seas.
Quantitative PCR was also used to detect the abundance of the bacterial 16S rRNA gene, which showed a range of abundance between 1.5 × 108 (SYS01-30) to 9.0 × 109 copies g-1 (ECS02-1) (Figure 1B). Similar to the result of direct cell counting, samples from sites located on the south tended to have higher copy numbers of the bacterial 16S rRNA gene. Vertically, the abundance fluctuated at a high level within the top 5 cm, and then declined gradually with depth. The 16S rRNA gene abundance was significantly correlated with PO43- (r = -0.466, P = 0.007), TN (r = 0.463, P = 0.003), TON δ15N (r = -0.439, P = 0.005), and C/N (r = -0.321, P = 0.043). Copy number of dsrB gene varied in the range of 2.7 × 106 to 9.6 × 107 copies g-1, and no significant difference was observed among studied sites or depths.
Bacterial Diversity and Richness
A total of 1,472,593 overlapped reads across the 40 samples were generated through Illumina sequencing, and 1,190,856 reads were left after quality control (Supplementary Table S1). Read numbers in each sample were limited to 24,048 after rarefaction for further analyses. All sequences yielded 10,746 OTUs at a 97% sequence similarity level (Supplementary Data Sheet S2). The Good’s coverage values ranged from 91.99 to 98.17% across samples, indicating that sequences generated from these samples could represent most of the bacterial community in the studied sites. Sites located in the ECS had higher Chao1 and Shannon indices than those in the SYS (P < 0.05). Site ECS03 located in the Distal Cheju Island mud area had the highest bacterial diversity in terms of Shannon index (P < 0.05). As for depth, Shannon diversity of the surface samples (0–2 cm) was higher than that of the 30 cm samples (P < 0.05). A negative correlation was found between Shannon diversity and C/N (r = -0.544, P = 0.0003).
Taxonomic Description
In total, 47 phyla were observed in the 40 ECMS sediment samples. The most dominant phylum was Proteobacteria occupying 45.62% of all sequences. This was followed by Planctomycetes, Chloroflexi, Acidobacteria, Bacteroidetes, Firmicutes, Nitrospirae, candidate division WS3, Gemmatimonadetes and Actinobacteria, which jointly accounted for 44.43% of all sequences. Within Proteobacteria, Deltaproteobacteria (21.93%) and Gammaproteobacteria (18.79%) were the most abundant classes. In addition, eight minor phyla (SM2F11, WCHB1-60, OC31, CKC4, candidate division KB1, GOUTA4, Thermotogae, and GAL08) were represented each by less than 50 sequences.
The bacterial community compositions varied among depths and sites (Figure 2). The proportions of Proteobacteria, Acidobacteria, and Bacteroidetes were high in surface sediments, whereas Chloroflexi and Firmicutes tended to be enriched in deep layers. The sample similarity analysis based on the Bray–Curtis dissimilarity at the genus level showed that samples were clustered into two groups (Supplementary Figure S1). The boundary between these two groups was plotted as a dotted line in Figure 2, illustrating the separation of surface and deep bacterial communities. To discover bacterial groups with significant differences between these two sediment types, LEfSe was conducted from the phylum to genus levels with a LDA threshold of 3.5. The results of LEfSe confirmed the tendency shown at the phylum and genus levels and revealed that Lactococcus (genus of Firmicutes, dominated by Lactococcus piscium), Pseudomonas (genus of Pseudomonadales, dominated by Pseudomonas azotoformans and P. fragi) and Dehalococcoidia (class of Chloroflexi) were significantly abundant in deep layers, whereas Acidobacteria (class) and JTB255 marine benthic group (JTB255-MBG, a family of Xanthomonadales) preferred surface layers (Figure 3). In addition, LEfSe with a LDA value of 3.5 was also used to predict effect differences in bacterial groups among different sites (Supplementary Figure S2). A total of 28 bacterial groups, including five phyla, five classes, six orders, seven families, and five genera, were highlighted to be the specialized taxa for each site. Syntrophobacterales from the family to genus levels and Desulfobacteraceae at the family level were enriched at SYS01. The abundance of Firmicutes, Chloroflexi, and Pseudomonadales were significantly higher at SYS02. JTB255-MBG was enriched at ECS01, making the proportion of Proteobacteria reached to its top at this site. Bacteroidetes (from phylum to class levels) was enriched at ECS02. ECS03 had a higher abundance of Planctomycetes and Deltaproteobacteria than other sites.
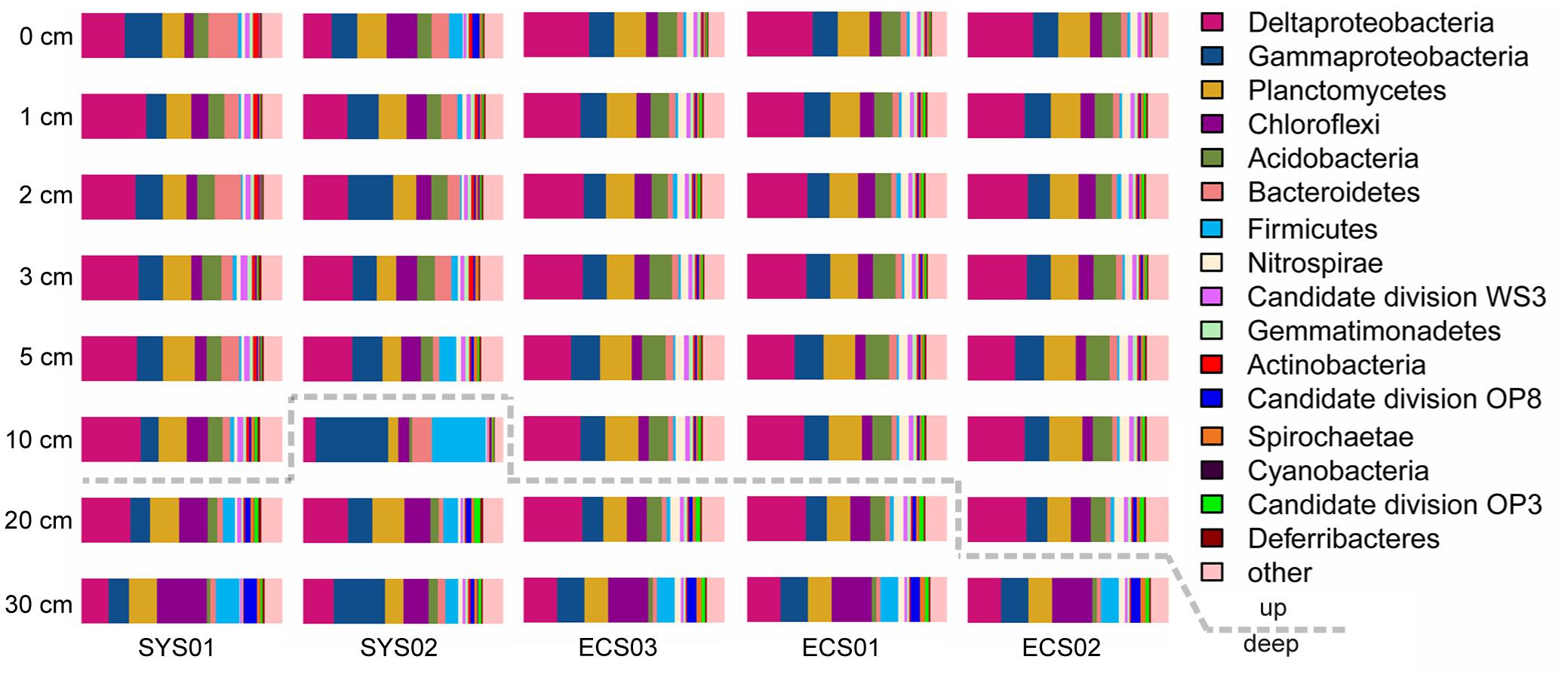
FIGURE 2. Relative abundance of dominant phyla (Deltaproteobacteria and Gammaproteobacteria) in different sediment samples. Dotted line divided the two groups defined by the Bray–Curtis similarity analysis at the genus level shown in Supplementary Figure S1.
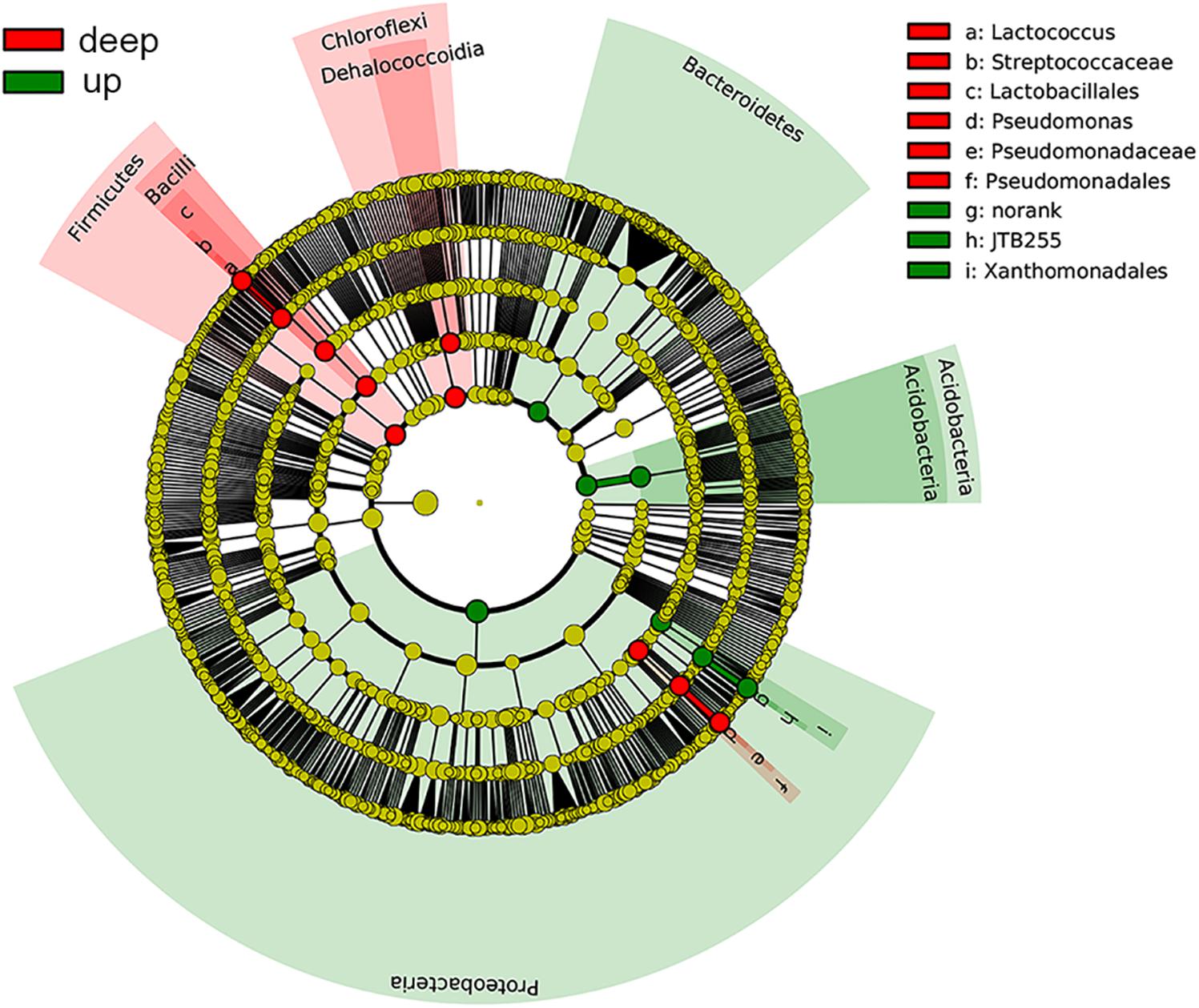
FIGURE 3. Cladograms showing the differences in relative abundance of bacterial groups between up and deep layers according to the LEfSe analysis with a LDA threshold of 3.5. Taxa with significant differences were highlighted by colored circles and shadings.
Inter-taxa relationship network was structured based on bacterial families whose read numbers were more than 50 across all samples. A total of 242 nodes and 2289 edges were presented in the network with a threshold of ±0.7 for Spearman’s coefficient and 0.01 for P-value (Figure 4). The bacterial families from Proteobacteria, Acidobacteria, and Planctomycetes displayed wide correlations with others. Proteobacteria made up more than 1/3 of the nodes in the network. The families tended to networking into two modules, and hubs of the two modules belonged to Proteobacteria, Bacteroidetes, Lentisphaerae, and Chloroflexi, and Proteobacteria, Firmicutes, and Planctomycetes, respectively. Twenty-six negative correlations were observed and distributed mainly between Proteobacteria and Chloroflexi.
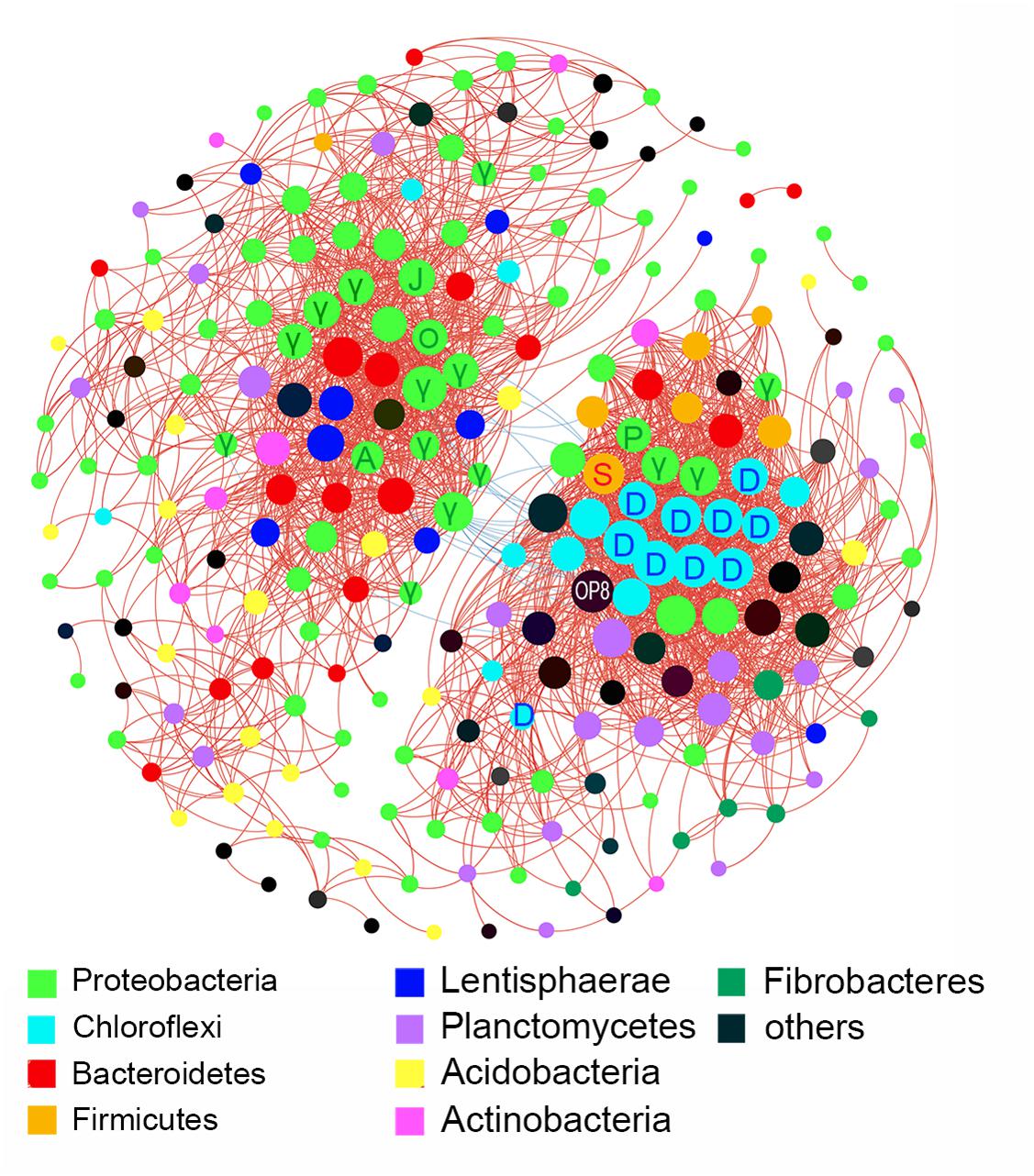
FIGURE 4. Inter-taxa relation network with thresholds of ±0.7 for Spearman’s coefficient and 0.01 for P-value. Correlations between families were represented by colored lines between nodes (red for positive, blue for negative). Size of node depended on the number of connections. A, Alteromonadaceae; D, Dehalococcoidia; γ, Gammaproteobacteria; J, JTB255; O, Oceanospirillaceae; P, Pseudomonadaceae; S, Streptococcaceae.
A total of 36 pairs of bacterial groups and environmental factors were observed to have significant correlations (P < 0.01 and |r| > 0.6) (Supplementary Table S2). Sixteen taxa belonging to Chloroflexi, Planctomycetes, Spirochaetae, candidate division OP8 and Deltaproteobacteria were significantly correlated with sediment depth. Nine taxa belonging to Deltaproteobacteria, Gammaproteobacteria and BD2-11 terrestrial group (order of Gemmatimonadetes) and seven taxa belonging to Bacteroidetes, Nitrospirae, and Planctomyces showed significant correlations with TON δ15N and TOC, respectively.
Community Comparison at the OTU Level and Environmental Factors Explaining Community Variations
The samples clustered basically according to different mud zones as shown in the PCoA (Figure 5A), which considered both the topology of evolutionary trees and abundance of OTUs. The upper layer samples clustered in accordance with mud zones, whereas the deep layer samples of all sites tended to cluster together. To be specific, the upper layer samples (0, 1, 2, 3, and 5 cm) of each site clustered tightly with the exception that those at SYS02 were slightly scattered. Contrastingly, the deep layer samples (20 and 30 cm) of each site were more similar with each other. Interestingly, sediments at the 10-cm layer displayed different clustering relationships in different sites. At ECS02 and ECS03, the 10-cm layer resembled more closely the upper layer samples of the same site, whereas at SYS01, SYS02, and ECS01, they tended to group with the deep layer samples. The two-way ANOSIM revealed that sediment depth (global R = 0.835, p < 0.001) could explain more variances than mud zones (global R = 0.690, p < 0.001). RDA analysis was performed and uncovered that seven environmental factors had significantly influences, which jointly accounted for 70.2% of the total variation. TOC δ13C (F = 5.4, P = 0.001) contributed the most with 29.0%, followed by TON δ15N (F = 5.2, P = 0.001), TOC (F = 4.3, P = 0.001), PO43- (F = 3.1, P = 0.002), NH4+ (F = 2.3, P = 0.01), TN (F = 2.3, P = 0.017), and C/N (F = 2.3, P = 0.023). No significant correlation was observed between NO3-, NO2-, or SiO32- and the communities. Influences of the top seven environmental factors on bacterial communities were shown in Figure 5B. NH4+ seemed to exert significant impacts on structuring bacterial communities of ECS01. The up sediments of SYS01 appeared to be separated from other samples by TOC.
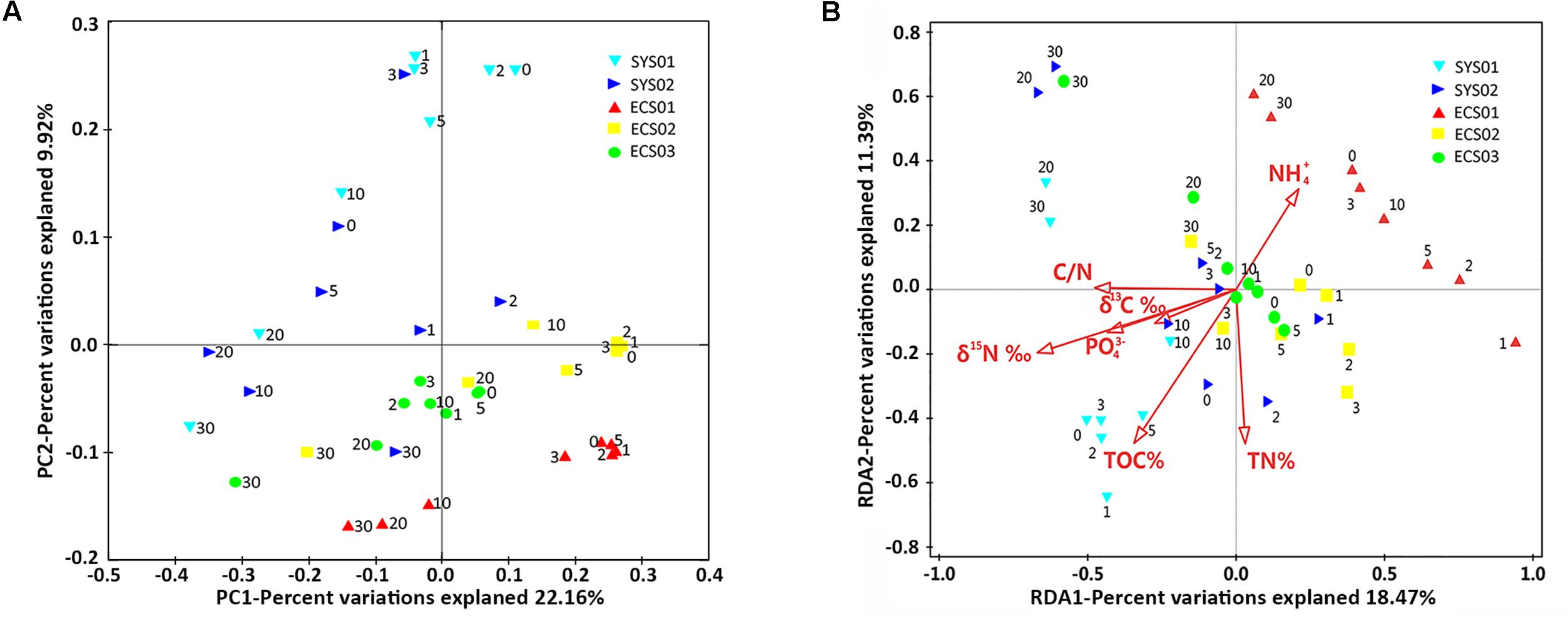
FIGURE 5. Community analysis at the OTU level. (A) Unweighted PCoA plot with PC1 and PC2. (B) RDA analysis illustrating the relationship between bacterial community at the OTU level and top environmental variables.
Discussion
Bacterial Abundance in Sediments of the ECMS and Potential Environmental Drivers
In this study, direct cell counting was implemented to evaluate the microbial abundance in sediments of the ECMS. According to Liu et al. (2015), the bacterial abundance was two to three orders of magnitude higher than the archaeal abundance in sediment of the ECMS. Thus, the counted cell numbers are approximately equal to the bacterial cell numbers. The cell counts were consistent with the result of 16S rRNA gene quantification. Sedimentary bacterial abundance in the ECMS was in the same range with that reported in the SYS (Liu et al., 2015), and was slightly lower than that in bioturbated coastal sediments from the Catalina Island (Bertics and Ziebis, 2009; Plotieau et al., 2013). By contrast, this abundance was higher than that in the Eastern Mediterranean Sea (Polymenakou et al., 2006) and in ODP sites from the Okinawa Trough and Peru margin (Mauclaire et al., 2004; Jean et al., 2005). Different environmental features may explain some of this variance in bacterial abundance across different areas. The bacterial abundance in sediments from the southern mud areas especially at site ECS01 and ECS02 was significantly higher (Figure 1B). These two sites are located just outside of the Yangtze River Estuary and Zhe-Min coasts and are readily influenced by terrigenous nutrient input from land, which could lead to the observed high bacterial abundance.
The abundance of bacteria steadily decreased with sediment depth, in well agreement with the global distribution pattern of benthic microbial abundance (Kallmeyer et al., 2012). Generally, aerobic bacterial respiration consumed DO rapidly in upper layers of the eutrophic sediment, which would result in reduced rate of microbial carbon oxidation in deep sediments and subsequent decrease in bacterial abundance (Røy et al., 2012). Availability of organic matter, as the main electron donor in marine sediments, may also affect bacterial abundance. In this study, the 16S rRNA gene abundance was positively correlated with TN (P < 0.01), negatively correlated with C/N (P < 0.01) and TON δ15N (P < 0.05). These results could partly contribute to the negative correlation between Shannon diversity and C/N, and revealed that the content, source and composition of organic matter are important in determining the abundance of benthic bacteria in the ECMS, with fresh and marine organic matter (higher TN and lower C/N ratio) supporting higher bacterial abundance.
The abundance of dsrB examined in this study was similar to that in the Pearl River estuary (Jiang et al., 2009), Bohai Sea and Yellow Sea (Liu et al., 2015), but lower than that in the Baltic Sea (Leloup et al., 2009) and Blake Sea (Leloup et al., 2007). As sulfate-reducing prokaryotes are anaerobic, the high level of DO in overlaying water of the ECMS sediments, compared with that in the Baltic Sea and Black Sea, may contribute to these variations. No significant differences in copy numbers of dsrB gene were observed among studied sites or depths. This was consistent with the invariable SO42- contents observed in the pore waters (Yu et al., 2016).
Distribution Patterns of Bacterial Community in Sediments of the ECMS and Potential Environmental Drivers
Limited studies of benthic microbial community in the ECMS were focused only on surface sediments. They showed that sediment sources, hydrodynamic conditions and concentration of nutrients might be the crucial factors in shaping bacterial communities (Liu et al., 2014; Xiong et al., 2014). To uncover the extent of influence of such effects on subsurface bacterial communities, the vertical profile of bacterial communities in mud sediments of the ECMS was sampled in this study. We found that bacterial communities of the up and deep sediment layers in the ECMS exhibited contrasting distribution patterns across sites.
Bacterial communities of the upper layers in each site were clearly separated (Figure 5A) and were found to be influenced by different environmental factors, such as TOC, NH4+ and PO43- (Figure 5B). TOC separated the upper layer sediment of SYS01 from others, confirming the role of TOC as an important factor in shaping relative abundance of benthic bacterial groups (Jorgensen et al., 2012; Liu et al., 2014). NH4+ distinguished the bacterial communities in ECS01 sediments especially the upper layer samples from others, confirming that sedimentary bacterial community structures can be effected by nitrogen pollution (Xiong et al., 2014).
These distribution patterns are reflected by differential correlations between taxonomic groups and environment factors, in particular organic matter and nutrients (Supplementary Table S2). For example, Flavobacteriaceae, a major group of Bacteroidetes, showed a significantly positive relationship with TOC (r > 0.6 and P < 0.01), and this agreed with their chemoorganotrophic lifestyle functioning especially in degrading high molecular weight dissolved organic matter, such as polysaccharides (Bennke et al., 2016; Teeling et al., 2016). Planctomyces also preferred high concentration of nitrogen and organic carbon substrates. However, Nitrospirae preferred relatively oligotrophic environments as evidenced by the significantly negative relationship with TOC. Nitrospirae are nitrite oxidizing bacteria functioning in aerobic nitrite oxidization. However, they have been found to widely distribute in anaerobic marine sediments (Liu et al., 2014; Nunoura et al., 2016; Chen et al., 2017). Whether sedimentary Nitrospirae are inactive or have other uncharacterized physiologies needs further investigation. JTB255-MBG, Acidiferrobacter and BD2-11 terrestrial group (belonging to Gemmatimonadetes) preferred substrates with a low content of TON δ15N, while Syntrophobacterales was opposite. These results confirmed that varied sediment sources could provide different environmental niches for the growth of different bacterial communities.
Different from the scattered distribution of upper layer samples, the deep layer (20 and 30-cm layer) samples showed a closer clustering relationship regardless of studied sites (Figure 5A). TN and TOC seemed to play roles in converging the deep layer samples of different mud zones (Figure 5B). However, it was noteworthy that these environmental factors fluctuated more widely among sites than among depths of one site. Thus, they might be not the direct driving force that clustered the deep layer samples of studied sites. As mentioned above, these factors mainly influenced the relative abundance of up sediment-dominant Flavobacteriaceae, JTB255-MBG and Acidiferrobacter. These impacts would disappear in deep sediment layers with a lower abundance of up sediment-dominant taxa. Subsequently, the deep layer samples clustered closely. Indeed, DO under the top 1 μm sediments decreased sharply from ∼120–250 μM to an undetectable level in the study area (Yu et al., 2016). Thus, it might be inferred that DO, or redox state, was the crucial factor contributing to the differences between up and deep layer communities.
It was interesting to note that the 10 and 20-cm layer samples displayed different clustering patterns in each site. We speculated that this discrepancy might be attributed to site-specific hydrodynamic conditions, although no environment factor detected here could explain this difference. For example, ECS02 was influenced by the Taiwan Warm Current, resulting in a higher summer flow velocity in this site (Lim et al., 2007; Liu et al., 2007), while SYS02 was located in the bottom of the Yellow Sea Trough covered by the Yellow Sea Cold Water Mass, causing a lower flow velocity and deposition rate than other sites detected (Huh and Su, 1999; Yang et al., 2003; Chen Z. et al., 2004). These differences in fluid dynamics lead to varied particle size and redox profiles across different sites, thus influencing the cluster of 10 and 20-cm layer samples.
Bacterial Community Compositions in Sediments of the ECMS
The sedimentary bacterial community composition in the ECMS was in high accordance with previous studies of the same area (Xiong et al., 2014; Liu et al., 2015) and other marginal seas (Zinger et al., 2011; Wang et al., 2012; Sun et al., 2013). In comparison with those from the deep sea or coastal areas adjacent to open oceans (Schauer et al., 2010; Dyksma et al., 2016; Walsh et al., 2016), a higher ratio of Deltaproteobacteria to Gammaproteobacteria was observed in all sediment depths of this study. Deltaproteobacteria and Gammaproteobacteria (average 48.1 and 41.2%, respectively) were the major predominant classes of Proteobacteria, and they have been demonstrated to play important roles in organic matter mineralization and dark carbon fixation, respectively, in coastal sediments (Thamdrup and Canfield, 1996; Dyksma et al., 2016). The high abundance of Deltaproteobacteria relative to Gammaproteobacteria may reflect specific response of marginal bacterial communities to terrigenous organic inputs, and indicate a higher potential of organic matter mineralization than carbon fixation in the ECMS sediments.
There were significant differences in diversity and composition of bacterial communities between up and deep sediment samples (Supplementary Figure S1 and Figure 5A). The upper layers owned a more diverse community than the deep layers. Species capable of thriving under aerobic and anaerobic environments can coexist at the shallow sediment, thus resulting in the high diversity. LEfSe analysis showed that Lactococcus and Pseudomonas of Gammaproteobacteria, and members of Dehalococcoidia (class of Chloroflexi) were enriched in the deep sediment samples (Figure 3). The former two genera were usually found in non-marine environments and played significant roles in spoilage of meat, dairy and fish (Williams et al., 1990; Champagne et al., 1994; Sakala et al., 2002). The proportions of these two genera were higher in several deep layer samples, which might be related to the general decrease in bacterial abundance with depth combined with their presence as contaminants in the extraction kit (Salter et al., 2014). However, sampling and DNA extraction methods in the present study were in accordance with methods used in previous studies on different samples (Nguyen and Landfald, 2015; Chen et al., 2017), in which case these potential contaminant genera were not detected. Thus, it is also possible that they might exist in deep layer sediments and feed on organics, such as remnants of marine animals. Dehalococcoidia were widely distributed in deep marine sediments (Durbin and Teske, 2011; Jorgensen et al., 2012). They exhibited significant correlations among each other and formed the deep-abundant module in the network analysis illustrating inter-taxa relationships (Figure 4). Different subgroups of Dehalococcoidia can inhabit different ecological niches (Bowman and McCuaig, 2003; Tas et al., 2010; Durbin and Teske, 2011; Wasmund et al., 2015), and they potentially own versatile ecological functions such as CO2 fixation, dimethyl sulfoxide utilization, aromatics and fatty acids oxidization, and acetate production (Hug et al., 2013; Wasmund et al., 2014). The observed positive co-occurrence patterns within members of this class may suggest that these variable physiological features can be highly dependent and integrated, or help relieve interspecific competitions. The deep-abundant module was also involved in correlations among Dehalococcoidia and several other bacterial clades, including strictly anaerobic Spirochaetaceae, Sva0485 and low-oxygen-adapted candidate phylum OP8 (Bowman and McCuaig, 2003; Farag et al., 2014), Pseudomonadaceae and Streptococcaceae, implicating biogeochemical complexity in the deep marine sediments.
Comparatively, correlations among Acidobacteria, Bacteroidetes, Proteobacteria, and Lentisphaerae constituted the up-abundant module in the network, and the former three taxa were shown to have significantly higher proportions in the upper layers by LEfSe. JTB255-MBG, a member of order Xanthomonadales belonging to Gammaproteobacteria, was the most abundant clades (average 35%) in the upper layer sediments examined in this study. In the network, JTB255-MBG showed a high degree of connectivity with other surface-abundant taxa, indicating that growth of JTB255-MBG may be highly dependent on other bacterial clades, which may provide potential insights in developing new cultivating strategies for obtaining a pure isolate of this clade. Strains of Lentisphaerae were detected to produce transparent exopolymers (TEP) (Cho et al., 2004), a key factor of biofilm initiation and outgrowth (Berman and Passow, 2007). The involvement of Lentisphaerae and other marine biofilm residents, including Rhodopirellula, Oceanospirillaceae, Alteromonadaceae, Acidobacteria, Planctomyces, OM190, and Bacteroidetes (Bengtsson and Øvreås, 2010; Eichorst et al., 2011; Ruvindy et al., 2015; Lawes et al., 2016) in the up-abundant module indicated that biofilm may regulate bacterial interactions in the upper layer sediment.
Conclusion
This study presented a detailed description of spatial and depth-related distribution patterns of bacterial communities in sediments of the ECMS. Abundance, diversity, and community structure varied significantly with sediment depth. The up and deep bacterial communities displayed different distribution patterns. The upper layer samples clustered in accordance with mud zones, whereas the deep layer samples of all sites tended to cluster together. TOC δ13C and TON δ15N significantly affected the bacterial community composition, suggesting that abundance and composition of organic matter played critical roles in shaping bacterial communities. Moreover, bacterial communities in the shallow and deep sediments showed different inter-taxa relationships, indicating different co-occurrence patterns in surface and subsurface sediments. This study provided a detailed outline of subsurface bacterial communities in mud deposits of the ECMS for the first time, and provided clues for uncovering biogeochemical cycles in this area.
Author Contributions
YQ carried out sample collecting, laboratory work, data analysis, and drafted the manuscript. JL conceived the study, revised and finalized the manuscript. MZ and X-HZ participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2016YFA0601303) and the National Natural Science Foundation of China through grants 41730530, 41521064, 41476112, and 41506154.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all of the scientists and crew members on the R/V Dong Fang Hong 2 during the expedition for their great efforts and help in sample collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01128/full#supplementary-material
Footnotes
References
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: an open source software for exploring and manipulating networks. ICWSM 8, 361–362. doi: 10.13140/2.1.1341.1520
Bengtsson, M. M., and Øvreås, L. (2010). Planctomycetes dominate biofilms on surfaces of the kelp Laminaria hyperborea. BMC Microbiol. 10:261. doi: 10.1186/1471-2180-10-261
Bennke, C. M., Krüger, K., Kappelmann, L., Huang, S., Gobet, A., Schüler, M., et al. (2016). Polysaccharide utilisation loci of Bacteroidetes from two contrasting open ocean sites in the North Atlantic. Environ. Microbiol. 18, 4456–4470. doi: 10.1111/1462-2920.13429
Berman, T., and Passow, U. (2007). Transparent exopolymer particles (TEP): an overlooked factor in the process of biofilm formation in aquatic environments. Nat. Proc. doi: 10.1038/npre.2007.1182.1
Bertics, V. J., and Ziebis, W. (2009). Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J. 3, 1269–1285. doi: 10.1038/ismej.2009.62
Böer, S. I., Hedtkamp, S. I., van Beusekom, J. E., Fuhrman, J. A., Boetius, A., and Ramette, A. (2009). Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 3, 780–791. doi: 10.1038/ismej.2009.29
Bowman, J. P., and McCuaig, R. D. (2003). Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69, 2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Champagne, C. P., Laing, R. R., Roy, D., Mafu, A. A., Griffiths, M. W., and White, C. (1994). Psychrotrophs in dairy products: their effects and their control. Crit. Rev. Food Sci. 34, 1–30. doi: 10.1080/10408399409527648
Chen, C. T. A., Andreev, A., Kim, K. R., and Yamamoto, M. (2004). Roles of continental shelves and marginal seas in the biogeochemical cycles of the North Pacific Ocean. J. Oceanogr. 60, 17–44. doi: 10.1023/B:JOCE.0000038316.56018.d4
Chen, Z., Saito, Y., Kanai, Y., Wei, T., Li, L., Yao, H., et al. (2004). Low concentration of heavy metals in the Yangtze estuarine sediments, China: a diluting setting. Estuar. Coast. Shelf Sci. 60, 91–100. doi: 10.1016/j.ecss.2003.11.021
Chen, Y., Liu, Y., and Wang, X. (2017). Spatiotemporal variation of bacterial and archaeal communities in sediments of a drinking reservoir, Beijing, China. Appl. Microbiol. Biotechnol. 101, 3379–3391. doi: 10.1007/s00253-016-8019-1
Cho, J. C., Vergin, K. L., Morris, R. M., and Giovannoni, S. J. (2004). Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum. Lentisphaerae. Environ. Microbiol. 6, 611–621. doi: 10.1111/j.1462-2920.2004.00614.x
Coelho, F. J., Santos, A. L., Coimbra, J., Almeida, A., Cunha,Â., Cleary, D. F., et al. (2013). Interactive effects of global climate change and pollution on marine microbes: the way ahead. Ecol. Evol. 3, 1808–1818. doi: 10.1002/ece3.565
Durbin, A. M., and Teske, A. (2011). Microbial diversity and stratification of South Pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., Meier, D., Meyerdierks, A., et al. (2016). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Eichorst, S. A., Kuske, C. R., and Schmidt, T. M. (2011). Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77, 586–596. doi: 10.1128/AEM.01080-10
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Farag, I. F., Davis, J. P., Youssef, N. H., and Elshahed, M. S. (2014). Global patterns of abundance, diversity and community structure of the Aminicenantes (candidate phylum OP8). PLoS One 9:3. doi: 10.1371/journal.pone.0092139
Franco, M. A., de Mesel, I., Demba, D. M., van der Gucht, K., van Gansbeke, D., van Rijswijk, P., et al. (2007). Effect of phytoplankton bloom deposits on benthic bacterial communities in two contrasting sediments in the southern North Sea. Aquat. Microb. Ecol. 48, 241–254. doi: 10.3354/ame048241
Gao, M., Liu, J., Qiao, Y., Zhao, M., and Zhang, X. H. (2017). Diversity and abundance of the denitrifying microbiota in the sediment of eastern china marginal seas and the impact of environmental factors. Microb. Ecol. 73, 601–615. doi: 10.1007/s00248-016-0906-6
Gilbert, J. A., Field, D., Swift, P., Newbold, L., Oliver, A., Smyth, T., et al. (2009). The seasonal structure of microbial communities in the western English Channel. Environ. Microbiol. 11, 3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x
Harrison, B. K., Zhang, H., Berelson, W., and Orphan, V. J. (2009). Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California). Appl. Environ. Microbiol. 75, 1487–1499. doi: 10.1128/AEM.01812-08
Hedges, J. I., and Keil, R. G. (1995). Sedimentary organic matter preservation: an assessment and speculative synthesis. Mar. Chem. 49, 81–115. doi: 10.1016/0304-4203(95)00008-F
Hu, D. X. (1984). Upwelling and sedimentation dynamics. Chin. J. Oceanol. Limn. 2, 12–19. doi: 10.1007/BF02888388
Hu, L., Shi, X., Guo, Z., Wang, H., and Yang, Z. (2013). Sources, dispersal and preservation of sedimentary organic matter in the Yellow Sea: the importance of depositional hydrodynamic forcing. Mar. Geol. 335, 52–63. doi: 10.1016/j.margeo.2012.10.008
Hug, L., Castelle, C., Wrighton, K., Thomas, B., Sharon, I., Frischkorn, K., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. doi: 10.1186/2049-2618-1-22
Huh, C. A., and Su, C. C. (1999). Sedimentation dynamics in the East China Sea elucidated from Pb-210, Cs-137 and Pu-239, Pu-240. Mar. Geol. 160, 183–196. doi: 10.1016/S0025-3227(99)00020-1
Jean, J. S., Chiang, T. Y., Wei, K. Y., Jiang, W. T., Liu, C. C., and Tsai, Y. P. (2005). Bacterial activity and their physiological characteristics in the sediments of ODP Holes 1202A and 1202D, Okinawa Trough, Western Pacific. Terr. Atmos Oceanic Sci. 16, 113–136. doi: 10.3319/TAO.2005.16.1.113(OT)
Jiang, L., Zheng, Y., Peng, X., Zhou, H., Zhang, C., Xiao, X., et al. (2009). Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 70, 249–262. doi: 10.1111/j.1574-6941.2009.00758.x
Jorgensen, S. L., Hannisdal, B., Lanzén, A., Baumberger, T., Flesland, K., Fonseca, R., et al. (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc. Natl. Acad. Sci. U.S.A. 109, E2846–E2855. doi: 10.1594/PANGAEA.786687
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kong, Y. (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153. doi: 10.1016/j.ygeno.2011.05.009
Lawes, J. C., Neilan, B. A., Brown, M. V., Clark, G. F., and Johnston, E. L. (2016). Elevated nutrients change bacterial community composition and connectivity: high throughput sequencing of young marine biofilms. Biofouling 32, 57–69. doi: 10.1080/08927014.2015.1126581
Learman, D. R., Henson, M. W., Thrash, J. C., Temperton, B., Brannock, P. M., Santos, S. R., et al. (2016). Biogeochemical and microbial variation across 5500 km of Antarctic surface sediment implicates organic matter as a driver of benthic community structure. Front. Microbiol. 7:284. doi: 10.3389/fmicb.2016.00284
Leloup, J., Fossing, H., Kohls, K., Holmkvist, L., Borowski, C., and Jørgensen, B. B. (2009). Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ. Microbiol. 11, 1278–1291. doi: 10.1111/j.1462-2920.2008.01855.x
Leloup, J., Loy, A., Knab, N. J., Borowski, C., Wagner, M., and Jørgensen, B. B. (2007). Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 9, 131–142. doi: 10.1111/j.1462-2920.2006.01122.x
Liang, Y., Jiang, Y., Wang, F., Wen, C., Deng, Y., Xue, K., et al. (2015). Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. ISME J. 9, 2561–2572. doi: 10.1038/ismej.2015.78
Lim, D. I., Choi, J. Y., Jung, H. S., Rho, K. C., and Ahn, K. S. (2007). Recent sediment accumulation and origin of shelf mud deposits in the Yellow and East China Seas. Prog. Oceanogr. 73, 145–159. doi: 10.1016/j.pocean.2007.02.004
Liu, J., Liu, X., Wang, M., Qiao, Y., Zheng, Y., and Zhang, X.-H. (2015). Bacterial and archaeal communities in sediments of the North Chinese Marginal Seas. Microb. Ecol. 70, 105–117. doi: 10.1007/s00248-014-0553-8
Liu, J., Yang, H., Zhao, M., and Zhang, X.-H. (2014). Spatial distribution patterns of benthic microbial communities along the Pearl Estuary, China. Syst. Appl. Microbiol. 37, 578–589. doi: 10.1016/j.syapm.2014.10.005
Liu, J., Zhu, R., and Li, G. (2003). Rock magnetic properties of the fine-grained sediment on the outer shelf of the East China Sea: implication for provenance. Mar. Geol. 193, 195–206. doi: 10.1016/S0025-3227(02)00660-6
Liu, J. P., Xu, K. H., Li, A. C., Milliman, J. D., Velozzi, D. M., Xiao, S. B., et al. (2007). Flux and fate of Yangtze River sediment delivered to the East China Sea. Geomorphology 85, 208–224. doi: 10.1016/j.geomorph.2006.03.023
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mauclaire, L., Zepp, K., Meister, P., and McKenzie, J. (2004). Direct in situ detection of cells in deep-sea sediment cores from the Peru Margin (ODP Leg 201, Site 1229). Geobiology 2, 217–223. doi: 10.1111/j.1472-4677.2004.00035.x
Nguyen, T. T., and Landfald, B. (2015). Polar front associated variation in Prokaryotic community structure in Arctic shelf seafloor. Front. Microbiol. 6:17. doi: 10.3389/fmicb.2015.00017
Nunoura, T., Takaki, Y., Shimamura, S., Kakuta, J., Kazama, H., Hirai, M., et al. (2016). Variance and potential niche separation of microbial communities in subseafloor sediments off Shimokita Peninsula, Japan. Environ. Microbiol. 18, 1889–1906. doi: 10.1111/1462-2920.13096
Plotieau, T., Lavitra, T., Gillan, D. C., and Eeckhaut, I. (2013). Bacterial diversity of the sediments transiting through the gut of Holothuria scabra (Holothuroidea; Echinodermata). Mar. Biol. 160, 3087–3101. doi: 10.1007/s00227-013-2297-2
Polymenakou, P. N., Tselepides, A., Stephanou, E. G., and Bertilsson, S. (2006). Carbon speciation and composition of natural microbial communities in polluted and pristine sediments of the Eastern Mediterranean Sea. Mar. Pollut. Bull. 52, 1396–1405. doi: 10.1016/j.marpolbul.2006.03.021
Powell, S. M., Bowman, J. P., Snape, I., and Stark, J. S. (2003). Microbial Community variation in pristine and polluted near shore Antarctic sediments. FEMS Microbiol. Ecol. 45, 135–145. doi: 10.1016/S0168-6496(03)00135-1
Probandt, D., Knittel, K., Tegetmeyer, H. E., Ahmerkamp, S., Holtappels, M., and Amann, R. (2017). Permeability shapes bacterial communities in sublittoral surface sediments. Environ. Microbiol. 19, 1584–1599. doi: 10.1111/1462-2920.13676
RDC TEAM (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Røy, H., Kallmeyer, J., Adhikari, R., Pockalny, R., Jørgensen, B., and D’Hondt, S. (2012). Aerobic microbial respiration in 86-million-year-old deep-sea red clay. Science 336, 922–925. doi: 10.1126/science.1219424
Ruvindy, R., White, R. A. I. I. I., Neilan, B. A., and Burns, B. P. (2015). Unravelling core microbial metabolisms in the hypersaline microbial mats of shark bay using high-throughput metagenomics. ISME J. 10, 183–196. doi: 10.1038/ismej.2015.87
Sakala, R. M., Hayashidani, H., Kato, Y., Kaneuchi, C., and Ogawa, M. (2002). Isolation and characterization of Lactococcus piscium strains from vacuum-packaged refrigerated beef. J. Appl. Microbiol. 92, 173–179. doi: 10.1046/j.1365-2672.2002.01513.x
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., and Cookson, W. O. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. doi: 10.1186/s12915-014-0087-z
Schauer, R., Bienhold, C., Ramette, A., and Harder, J. (2010). Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 4, 159–170. doi: 10.1038/ismej.2009.106
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Sun, M. Y., Dafforn, K. A., Johnston, E. L., and Brown, M. V. (2013). Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ. Microbiol. 15, 2517–2531. doi: 10.1111/1462-2920.12133
Tas, N., van Eekert, M. H., de Vos, W. M., and Smidt, H. (2010). The little bacteria that can – diversity, genomics and ecophysiology of ‘Dehalococcoides’ spp. in contaminated environments. Microb. Biotechnol. 3, 389–402. doi: 10.1111/j.1751-7915.2009.00147.x
Teeling, H., Fuchs, B. M., Bennke, C. M., Krüger, K., and Chafee, M. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. elife 5:e11888. doi: 10.7554/eLife.11888.001
Thamdrup, B., and Canfield, D. E. (1996). Pathways of carbon oxidation in continental margin sediments off central Chile. Limnol. Oceanogr. 41, 1629–1650. doi: 10.4319/lo.1996.41.8.1629
Varon-Lopez, M., Dias, A. C., Fasanella, C. C., Durrer, A., Melo, I. S., Kuramae, E. E., et al. (2013). Sulphur-oxidizing and sulphatereducing communities in Brazilian mangrove sediments. Environ. Microbiol. 16, 845–855. doi: 10.1111/1462-2920.12237
Walsh, E. A., Kirkpatrick, J. B., Rutherford, S. D., Smith, D. C., Sogin, M., and D’Hondt, S. (2016). Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989. doi: 10.1038/ismej.2015.175
Wang, L., Liu, L., Zheng, B., Zhu, Y., and Wang, X. (2013). Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar. Pollut. Bull. 72, 181–187. doi: 10.1016/j.marpolbul.2013.04.005
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, Y., Sheng, H. F., He, Y., Wu, J. Y., Jiang, Y. X., Tam, N. F., et al. (2012). Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 78, 8264–8271. doi: 10.1128/AEM.01821-12
Wasmund, K., Algora, C., Müller, J., Krüger, M., Lloyd, K. G., Reinhardt, R., et al. (2015). Development and application of primers for the class Dehalococcoidia (phylum Chloroflexi) enables deep insights into diversity and stratification of subgroups in the marine subsurface. Environ. Microbiol. 17, 3540–3556. doi: 10.1111/1462-2920.12510
Wasmund, K., Schreiber, L., Lloyd, K. G., Petersen, D. G., Schramm, A., Stepanauskas, R., et al. (2014). Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J. 8, 383–397. doi: 10.1038/ismej.2013.143
Williams, A. M., Fryer, J. L., and Collins, M. D. (1990). Lactococcus piscium sp. nov. a new Lactococcus species from salmonid fish. FEMS Microbiol. Lett. 68, 109–113. doi: 10.1111/j.1574-6968.1990.tb04132.x
Xiong, J., Liu, Y., Lin, X., Zhang, H., Zeng, J., Hou, J., et al. (2012). Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ. Microbiol. 14, 2457–2466. doi: 10.1111/j.1462-2920.2012.02799.x
Xiong, J., Ye, X., Wang, K., Chen, H., Hu, C., Zhu, J., et al. (2014). Biogeography of the sediment bacterial community responds to a nitrogen pollution gradient in the East China Sea. Appl. Environ. Microbiol. 80, 1919–1925. doi: 10.1128/AEM.03731-13
Yang, S. Y., Jung, H. S., Lim, D. I., and Li, C. X. (2003). A review on the provenance discrimination of sediments in the Yellow Sea. Earth Sci. Rev. 63, 93–120. doi: 10.1016/S0012-8252(03)00033-3
Yin, Q., Fu, B., Li, B., Shi, X., Inagaki, F., and Zhang, X.-H. (2013). Spatial variations in microbial community composition in surface seawater from the ultra-oligotrophic center to rim of the South pacific gyre. PLoS One 8:e55148. doi: 10.1371/journal.pone.0055148
Yu, S., Yao, P., Liu, J., Zhao, B., and Zhang, G. (2016). Diversity, abundance, and niche differentiation of ammonia-oxidizing prokaryotes in mud deposits of the eastern china marginal seas. Front. Microbiol. 7:37. doi: 10.3389/fmicb.2016.00137
Zheng, B., Wang, L., and Liu, L. (2014). Bacterial community structure and its regulating factors in the intertidal sediment along the Liaodong Bay of Bohai Sea, China. Microbiol. Res. 169, 585–592. doi: 10.1016/j.micres.2013.09.019
Keywords: bacterial communities, eastern China marginal seas, spatial distribution, diversity, mud deposits
Citation: Qiao Y, Liu J, Zhao M and Zhang X-H (2018) Sediment Depth-Dependent Spatial Variations of Bacterial Communities in Mud Deposits of the Eastern China Marginal Seas. Front. Microbiol. 9:1128. doi: 10.3389/fmicb.2018.01128
Received: 08 February 2017; Accepted: 14 May 2018;
Published: 31 May 2018.
Edited by:
Stefan M. Sievert, Woods Hole Oceanographic Institution, United StatesReviewed by:
Maria Pachiadaki, Bigelow Laboratory for Ocean Sciences, United StatesMeng Li, Shenzhen University, China
Copyright © 2018 Qiao, Liu, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiwen Liu, bGl1aml3ZW5Ab3VjLmVkdS5jbg==
 Yanlu Qiao1,2
Yanlu Qiao1,2 Jiwen Liu
Jiwen Liu Meixun Zhao
Meixun Zhao Xiao-Hua Zhang
Xiao-Hua Zhang