- 1State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Culture Collection and Application, Guangdong Open Laboratory of Applied Microbiology, Guangzhou, China
Staphylococcus aureus is a pathogen associated with serious community and hospital-acquired diseases. The aim of this study was to investigate the prevalence of S. aureus from retail vegetables in China and then characterized S. aureus isolates by antibiotic resistance, staphylococcal enterotoxin genes, spa-typing and multi-locus sequence typing. Of 419 retail vegetable samples from 39 cities in China during 2011–2016, 24 (5.73%) samples were positive for S. aureus and the geometric mean was 3.85 MPN/g. The prevalence of S. aureus was highest in lettuce (13/84, 15.48%) followed by tomato (7/110, 6.36%), caraway (2/87, 2.30%), and cucumber (2/128, 1.56%), whereas other vegetables were free of S. aureus. A total of 30 isolates were analyzed. For antibiotics susceptibility test, most isolates (93.3%) were resistant to ampicillin and penicillin, whereas all isolates were susceptible to linezolid, trimethoprim/sulphamethoxazole 1:19, nitrofurantoin, rifampicin, and teicoplanin. All isolates (30/30, 100%) were resistant or intermediate resistant to more than three tested antibiotics, including 9 isolates (30%) were resisted more than 10 antibiotics. Five isolates were resistant to cefoxitin and carried mecA genes which confirmed as MRSA. Of the 18 investigated SE genes, the sem gene was the most frequently detected (86.7%) followed by the sec (83.3%), sep (70.0%), seg (56.7%), sel (53.3%), seh (50.0%), seq (50.0%), sej (46.7%), seb (36.7%), sen (36.7%), and ser (33.3%) genes were harbored by more than one third of the isolates, whereas the seo and seu were detected in only 6.75% of the isolates. MLST and spa typing observed high genetic diversity in S. aureus isolated from retail vegetable in China. ST59-t437 was the predominant types (3/5, 60%) of MRSA isolates, whereas ST188-t189 was the predominant types (7/25, 28%) of MSSA isolates. Our study reflects that the retail vegetable in China could be contaminated with S. aureus but the levels of S. aureus were not very excessive. In addition, these isolates had virulence potential, most of them were enterotoxigenic and multiple antimicrobial resistance, should be draw public attention. These data have signification implications for epidemiological and public health studies of this pathogen.
Introduction
Staphylococcus aureus is one of the most important pathogens and is responsible for various infections, such as wound infections and toxins-mediated syndromes as well as systemic and life-threatening diseases (Chambers and Deleo, 2009; Papadopoulos et al., 2018). Despite the ubiquitous distribution of S. aureus in nature, foods are still the important source of infection. Approximately 241,000 illnesses per year were causing foodborne disease in the United States by S. aureus (Scallan et al., 2011) and caused ~20–25% of foodborne bacterial outbreaks by S. aureus in China (Wang et al., 2014).
Differentiation between virulent and non-virulent strains is significant for evaluating the potential implications of the presence of this microorganism for food safety and public health. In general, S. aureus produced several virulence factors such as staphylococcal enterotoxins (SEs), leukocidin, exfoliatin, haemolysin, toxic shock syndrome toxin 1 (TSST-1) that can contribute in different ways to their pathogenicity. In which, SEs are heat stable proteins that are mainly associated with food poisoning outbreaks. It is shown that about 95% of staphylococcal food poisoning outbreaks were caused by the classical SE (SEA~SEE), and the remaining 55 of outbreaks were associated with other identified SEs (Altarazi et al., 2009). Furthermore, S. aureus also can be capable of acquiring antibiotic resistance determinants and exhibit resistance to multiple classes of antimicrobial agents. Methicillin-resistant S. aureus (MRSA) is practically resistant to all available β-lactam antimicrobial drugs. Nowadays, MRSA has been recognized as major cause of healthcare-associated infections worldwide and has been identified as an emerging pathogen outside the healthcare environment (Boucher, 2008).
Vegetables are essential to the human diet. Although the presence of S. aureus in retail food have been reported previously in China (Chao et al., 2015; Song et al., 2015; Yang et al., 2016a), very few studies focused on the retail vegetables, especial some types of vegetables, such as tomato, cucumber, lettuce, caraway which consumed raw popularly. Furthermore, the levels of contamination varied and influenced by food category, as well as geographical differences. It is necessary to investigate the qualitative and quantitative data of this bacterium in China to implement a system monitoring the prevalence patterns of S. aureus in food and environmental sources from different areas. Therefore, the aim of the present study was to investigate the prevalence and levels of S. aureus in retail vegetables from South China to North China and characterize these S. aureus isolates according to their antibiotic susceptibility profiles, enterotoxin genes, spa types and MLST types to determine their genetic background in China.
Materials and Methods
Sample Collection
Between July 2011 and June 2016, a total of 419 retail vegetable samples including 110 tomatoes, 128 cucumbers, 84 lettuces, 87 caraway and 10 other vegetables were collected from supermarkets, fairs and farmers' markets from 39 cities in 29 provinces and two directly controlled municipalities in China (Figure 1). The samples were placed in a cold box at a temperature approximately 4°C, tightly sealed with sterile plastic wrap, and transported to an accredited laboratory and subjected to microbiological analysis within 24 h.
Isolation and Identification of S. aureus
The isolation of S. aureus in the samples was determined using the most probable number (MPN) method according to GB 4789.10-2010 for food microbiological examination of S. aureus (National Food Safety Standards of China). Approximately 25 g of food sample was added to 225 mL of saline solution (Huankai, Guangzhou, China) for homogenization, then 1, 0.1, and 0.01 mL of each sample was inoculated in triplicate with trypticase soy broth (Huankai) supplemented with 10% NaCl and incubated at 37°C for 48 h, respectively. Loopfuls of the resulting cultures were streaked onto chromogenic S. aureus agar plates (Huankai), then incubated at 37°C for 24 h. Each positive sample selected 2–3 colonies with a pink color. After purification for 24 h at 37°C on NA plate (nutrient agar medium). Putative S. aureus isolates were tested for coagulase activity test by freeze-dried Rabbit Plasma (Huankai), and further confirmed by API STAPH identification test strips (bio Merieux, Marcy-1'Etoile, France) according to the manufacturer's instructions. The MPN value was determined on the basis of the number of positive tube(s) in each of the three sets using the MPN table.
Antibiotic Susceptibility Testing
All confirmed S. aureus were tested for antibiotic susceptibility. It was using the Kirby–Bauer method (Bauer et al., 1966) which performed by standard disk diffusion on Mueller–Hinton agar incubated at 37°C for 24 h, following the guidelines of the Clinical and Laboratory Standards Institute (The Clinical and Laboratory Standards Institute, 2015). A total of 24 antibiotics (Oxoid, Basingstoke, UK) were classified into 14 different groups according to the WHO (Organization, 2007): amoxycillin/clavulanic acid (AMC, 30 μg), ampicillin (AMP, 10 μg), cefepime (FEP, 10 μg), Cefoxitin (FOX, 30 μg), penicillin G (P, 10U), ceftazidime (CAZ, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), kanamycin (K, 30 μg), streptomycin (S, 25 μg), chloramphenicol (C, 30 μg), clindamycin (DA, 2 μg), erythromycin (E, 15 μg), telithromycin (TEL, 15 μg), ciprofloxacin (CIP, 5 μg), Norfloxacin (NOR, 10 μg), tetracycline (TE, 30 μg), Linezolid (LZD, 30 μg), rifampicin (RD, 5 μg), Trimethoprim/sulphamethoxazole 1:19 (SXT, 25 μg), Quinupristin/dalfopristin (QD, 15 μg), Teicoplanin(TEC, 30 μg), Nitrofurantoin (F, 300 μg) and Fusidic acid (FD, 10 μg). Staphylococcus aureus ATCC25923 and Escherichia coli ATCC25922 were included for quality control (The Clinical and Laboratory Standards Institute, 2015). The presence of the mecA/mecC gene was studied by PCR in all cefoxitin resistant isolates (Pérez-Roth et al., 2001; Stegger et al., 2012).
Detection of Staphylococcal Enterotoxin Genes
All isolates were tested by PCR for the presence of 18 genes coding for staphylococcal enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, sej, sek, sel, sem, sen, seo, sep, seq, ser, and seu) (Varshney et al., 2009). The amplicons were subjected to electrophoresed on 1.5% agarose containing Goldview for 0.5 h at 120 V and visualized under a UV transilluminator gel imaging system (GE Healthcare, WI, USA). The images were saved as TIFF files for analysis.
spa-Typing
Sequence typing of the S. aureus protein A (spa) repeat region was amplified according to a published protocol (Shopsin et al., 1999). All isolates were analyzed using the primers spa-1113f (5′-TAAAGACGATCCTTCGGTGAGC-3′) and spa-1514r (5′-CAGCAGTAGTGCCGTTTGCTT-3′). The PCR amplification conditions were as follow: an initial cycle of 80°C for 5 min; 35 cycles of 94°C for 45 s, 60°C for 45 s, 72°C for 2 min and a final extension at 72°C for 10 min. The spa types were randomly assigned using the Spa Server website (http://spaserver2.ridom.de).
Multi Locus Sequencing Typing
The MLST scheme used to characterize S. aureus isolates is based on the sequence analysis of the following seven housekeeping genes: arcC (Carbamate kinase), aroE (Shikimate dehydrogenase), glpF (Glycerol kinase), gmk (Guanylate kinase), pta (Phosphate acetyltransferase), tpi (Triosephosphate isomerase), and yqil (Acetyle coenzyme A acetyltransferase) (Enright et al., 2013). The PCR amplification conditions were as follow: an initial cycle of 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 2 min and a final extension at 72°C for 10 min. The DNA fragments were purified by using a PCR purification kit (Qiagen, Genmany) and sequenced in each direction with Big Dye fluorescent terminators on an ABI 3730XL sequencer (Applied BioSystems). For each MLST locus, an allele number was given to each distinct sequence variant, and a distinct sequence type (ST) number was attributed to each distinct combination of alleles at the seven genes. Sequence types (STs) were determined by using the Staphylococcus aureus MLST database (https://pubmlst.org/saureus/). Sequence Type Analysis and Recombinational Tests software (S.T.A.R.T. ver.2; http://pubmlst.org/software/analysis/start2) was used to analyze the data of MLST.
Statistical Analysis
The bacterial numbers were converted to base-10 logarithms for statistical analysis. MPN values < 0.3 MPN/g were set to 0.15, and MPN values > 110 MPN/g were assigned the maximum value for this test (Motes et al., 1998). The chi-square test was used to determine differences in the prevalence and levels of S. aureus-positive samples between qualitative variables. All statistical analyses were performed using the SPSS v21.0 software package.
Results
Prevalence of S. aureus in Retail Vegetables in China
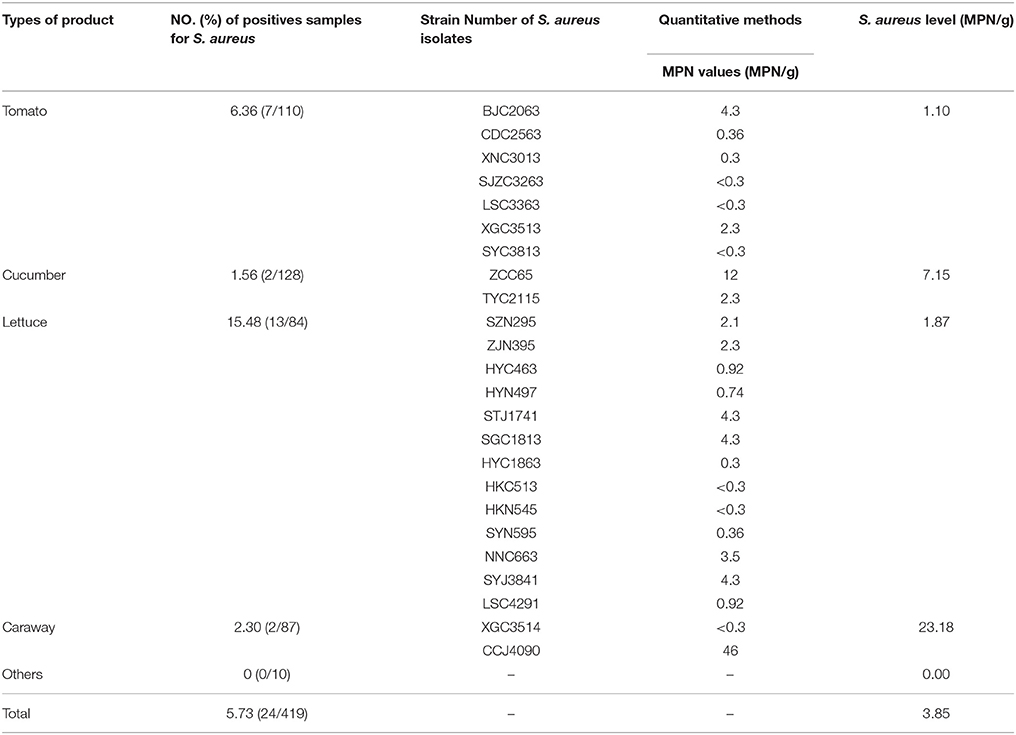
The results of the prevalence testing are summarized in Table 1. Overall, among 419 retail vegetables from 39 cities were examined, S. aureus were detected in 24 (5.73%) of samples from 20 cities and the geometric mean was 3.85 MPN/g. The prevalence of S. aureus was most common in lettuce (13/84, 15.48%) followed by tomato (7/110, 6.36%), caraway (2/87, 2.30%), and cucumber (2/128, 1.56%), whereas other vegetables (0/10, 0%) were free of S. aureus. Most of positive samples (91.67%) were less than 10 MPN/g by quantitative method, whereas none sample were reached 100 MPN/g.
Antibiotic Susceptibility Testing
The antibiotic susceptibility results of 30 S. aureus isolates are showed in Table 2. Overall, the isolates were susceptible to LZD, SXT, F, RD and TEC, except one isolate having intermediate resistance to RD and 7 isolates having intermediate resistance to TEC. For 24 antibiotics, most isolates (93.3%) were resistant to AMP and P, followed by TE (43.3%), E (40.0%), K (33.3%), AMC (26.7%), S (23.3%), C (23.3%), DA (23.3%), CIP (23.3%), TEL (20%), and others (< 20%). However, all isolates were resistant or intermediate resistant to more than three tested antibiotics, of which 9 isolates (30%) were resistant to more than 10 antibiotics. Five isolates which were resistant to FOX and carried mecA genes, including 3 isolates collected from lettuce and 2 isolates obtained from caraway, confirmed as MRSA. These MRSA isolates showed resistant to all selected β-Lactams. Except that, there is no significantly difference between MRSA and MSSA for most antimicrobial tested.
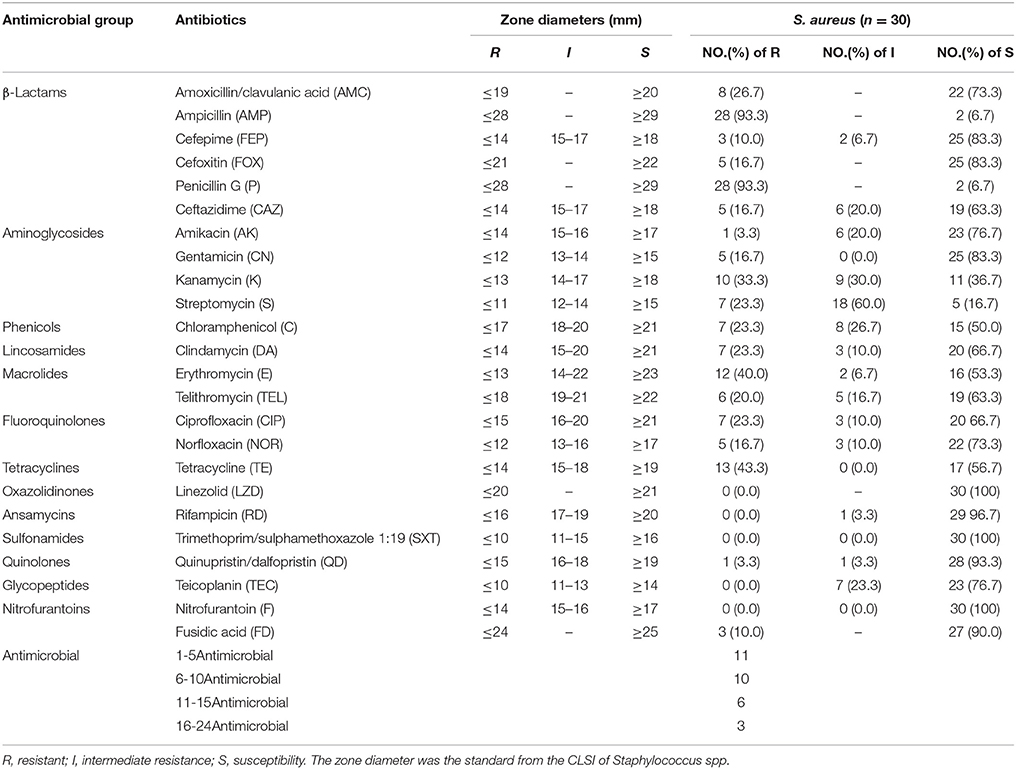
Table 2. Results of antimicrobial susceptibility tests of Staphylococcus aureus isolates obtained from retail vegetables in China.
Prevalence and Distribution of Enterotoxin Genes
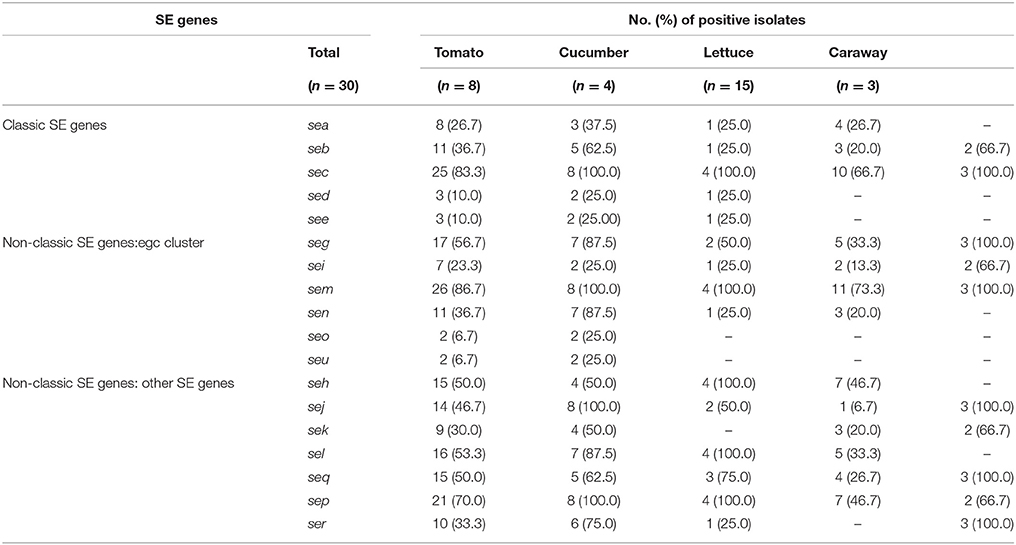
Among the 30 isolates analyzed, the isolates harbored at least one of the SE gene, including nine isolates carried more than 10 SE genes. Of the 18 investigated SE genes, the sem gene was the most frequently detected (86.7%) followed by the sec (83.3%), sep (70.0%), seg (56.7%), sel (53.3%), seh (50.0%), seq (50.0%), sej (46.7%), seb (36.7%), sen (36.7%), ser (33.3%), sek (30.0%), sea (26.7%), sei (23.3%), sed (10.0%), see (10.0%), seo (6.7%), and seu (6.7%) (Table 3). The classic SE genes (sea, seb, sec, sed, and see) showed 23.26% (50/215) of the detected genes, whereas the egc cluster (seg, sei, sem, sen, seo, and seu) accounted for 30.23% (65/215).
Molecular Characterization of S. aureus
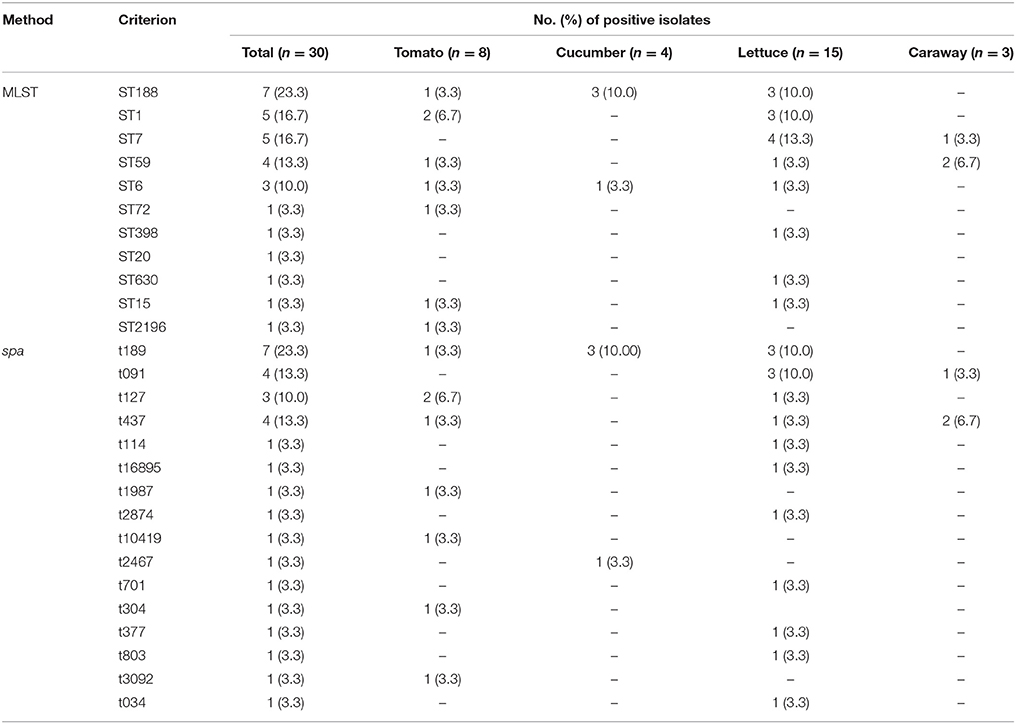
Spa-typing detected a total of 16 different types from 30 S. aureus, including one new type (t16985). The most common spa type was t189 (23.33%) followed by t091 (13.33%) and t437 (13.33%). Other spa types, such as t114, t1987, t2874, t10419, t2467, t701, t304, t377, t803, t3092, and t034, were both singletons independently of the isolates source (Table 4).
Eleven different STs were identified for all isolates from retail vegetables (Table 4). ST188 (7/30, 23.3%), ST1 (5/30, 16.7%), ST7 (5/30, 16.7%) and ST6 (3/30, 10%) were the frequent STs in this research. A phylogenetic tree based on the 7 concatenated MLST sequences (Figure 2) shows the relatedness between the vegetable isolates. Combining the STs and spa types, ST59-t437 was the predominant types (3/5, 60%) of MRSA isolates, whereas ST188-t189 was the predominant types (7/25, 28%) of MSSA isolates. Most isolates demonstrated high concordance between STs and spa types. Three ST6 isolates showed different spa types (ST6-t2467, ST6-t701, and ST6-t304) isolated from different places.
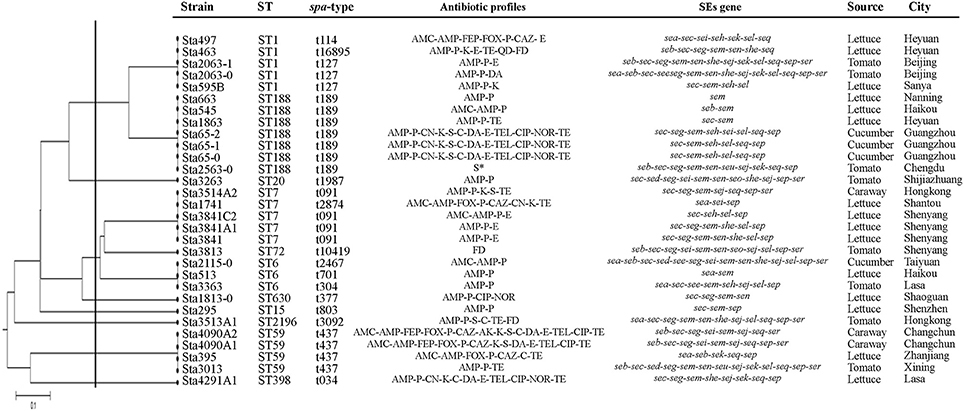
Figure 2. The UPGMA (unweighted pair group method with arithmetic mean) tree of the 7 multi-locus sequence typing loci of vegetable S. aureus isolates. S*, susceptible. This tree was generated using the S.T.A.R.T (version 2).
Discussion
The presence of S. aureus in vegetable have been reported are very fewer than other types of food. Furthermore, full-scale geographic investigation is necessary. In this study, we investigate retail vegetables from 39 cities in China, covering most provincial capitals of China, which showed that 24 positive samples were detected S. aureus including 4 MRSA-positive samples from 419 retail vegetables in 20 cities. It indicated that the retail vegetable in China could be contaminated with S. aureus. In China, there is no standard limit of S. aureus in vegetable. In the present study, most of positive samples (91.67%) were less than 10 MPN/g and only one sample reached 46 MPN/g from caraway. Thus, refer to the limit of S. aureus in other types of food (GB 29921-2013), on the whole, levels of S. aureus in retail vegetable in China were not very excessive. Compare with other surveys, our result was lower than those from retail raw meat, infant formula milk, lettuces, or aquatic products in China, which reported that the contamination of S. aureus was 24.2, 11.2, 10.0, and 37.2%, respectively (Seo et al., 2010; Wang et al., 2012, 2013; Rong et al., 2017). However, vegetables are essential to the human diet in China, especially with the rationalization of diet structure recent years. Furthermore, the contamination focus on lettuce, tomato, caraway, and cucumber in this study. These types of samples always consumed raw, which indicated consumers may be exposed to infection, and should be draw public attention.
In recent years, the spread of antibiotic resistance among strains of S. aureus is of great public and clinical concern in the treatment of staphylococcal infections (Shankar, 2012). Many researchers have reported resistance strains of S. aureus isolates from various food samples in different countries (Argudín et al., 2012; Yang et al., 2016b; Wang et al., 2017). In the current study, 76.7% of vegetable S. aureus isolates (23/30) were resistant or intermediate resistant to more than three tested antibiotics classes, of which 9 isolates were resistant to more than 10 antibiotics. At present, our data is higher than previous reports among various food resources in other countries (Kozytska et al., 2010; Shahraz et al., 2012; Spanu et al., 2012; Ge et al., 2017; Papadopoulos et al., 2018). In this study, 93.3% of isolates were resistant to ampicillin and penicillin, showing lower than the research by Hong et al. (2015) from leaf vegetables (96.3%) in Korea. Resistance to other antimicrobials, e.g., tetracycline, erythromycin, gentamicin, and ciprofloxacin, is similar to the reports of Chao et al. (2007), who observed that 49.4% of S. aureus isolates from food products were resistant to tetracycline, 24.1% to erythromycin, and 13.8% to gentamicin. Considering these antibiotics have been increasingly used in animal breeding or human treatment and exchange of antibiotic-resistant genes by the mobile genetic elements (MGEs) (Lindsay, 2014), it is not surprising that resistant strains become more common in the present. As we know, methicillin-resistant S. aureus (MRSA) represents a serious public health issue due to its ability to colonize and infect humans and animals, there were 5 MRSA strains isolated from 4 positive samples in this study. Cross contamination from environments may be the major reasons because animal-derived food products are widely known to be an important reservoir for MRSA (Petinaki and Spiliopoulou, 2012). However, the high antimicrobial resistance of S. aureus observed in this study should receive much attention. Moreover, controlled use of antimicrobials would limit the emergence of drug-resistant bacteria.
Generally speaking, staphylococcal enterotoxins (SEs) encoded by SE genes, which play an important role in the pathogenicity of the bacteria. In our study, 18 SE genes were tested and all isolates harbored at least one of the SE genes. Three genes, sem (86.7%), sec (83.3%), and seq (70.0%) were more frequently detected, whereas sed (10.0%), see (10.0%), seo (6.7%), and seu (6.7%) were detected at lower frequencies. From these types of SEs, it was suggested that about 95% of staphylococcal food-poisoning outbreaks were caused by strains carrying the classical SE (SEA-SEE) (Mashouf et al., 2015). Chao et al. (2015) have analyzed different sources of SE gene distributions, which found that the classic genes in both foodborne isolates and human origin isolates were significant higher than that in animal origin. In this study, 23.3% of the classic SE genes were detected, which means potential virulence as these isolates and potentially capable of causing an epidemic. Furthermore, the egc cluster (seg, sei, sem, sen, seo, seu) was widely distribute in clinic isolates and as a putative nursery of enterotoxin genes (Jarraud et al., 2001). Previous studies have showed that strains carried novel SE or SE-like genes could cause SFPs (McLauchlin et al., 2000; Argudín et al., 2010; Johler et al., 2015), it found 30.2% of detected genes of the egc cluster, higher than the classical SE genes, which is consist with previous reports (Smyth et al., 2005). However, most of SEs genes are carried and disseminated through mobile genetic elements that their spread among S. aureus isolates can modify their ability to cause disease and contribute to the evolution of this important pathogen. So the isolates harboring these genes were potential hazards to food safety.
MLST and spa typing observed high genetic diversity in S. aureus isolated from retail vegetable in China. In this study, it detected 16 different spa types and 11 different STs by two typing methods from 30 S. aureus isolates. The major STs, such as ST1, ST188, ST7, and ST6 in this study, was consist with (Song et al., 2015)'s result but was distinguish with previous studies, e.g., ST8 in US retail meat, ST1 in Chinese RTE food and ST152 in South Italy dairy product (Basanisi et al., 2017; Ge et al., 2017; Yang et al., 2018). It is indicated the genetically diversity with geographic origins and types. However, virulence genes and antibiotic profiles remarkably variation in same types. For instance, sta663 and sta65-0 belonged to ST188 from different cities, of which sta663 harbored sem gene and resistant to 2 antibiotics, whereas sta65-2 harbored sec-seg-sem-seh-sei-sel-seq-sep and resist for 12 antibiotics.
As shown in Figure 2, we observed a good agreement between MLST and spa typing result. Spa typing is a useful genotyping method for S. aureus. Compared with MLST typing, spa typing method was more discrimination. Three ST6 isolates showed different spa types (ST6-t2467, ST6-t701, and ST6-t304) isolated from different places. It also could distinguish antibiotic resistance in this study, which showed difference between the MRSA isolates (ST1-t114 and ST7-t2874) and MSSA isolates (ST1-t127 and ST7-t091). Furthermore, ST59-t437, which showed the predominant types of MRSA isolates in this study, was also occurs in Hong Kong, China, Vietnam, Japan, and Australia for community-associated MRSA (CA-MRSA) (Coombs et al., 2006; Tang et al., 2007; Ho et al., 2008; Higuchi et al., 2010; Wu et al., 2010). In China, Wang et al. (2004) was first reported this type of CA-MRSA. They found that 16 of 17 CA-MRSA isolates from children admitted to hospital with skin and soft tissue infections (SSTIs) from 1997 to 2002 belonged to ST59, and carried PVL genes. After that, more and more researches reports ST59 CA-MRSA. According to Chuang and Huang (2013) ‘s review, ST59 (and its single locus variant ST338) was the major lineage accounting for up to two-thirds of isolates, followed by ST910-IVa-t318 and ST1 in CA-MRSA. Therefore, it should pay great attention for this situation which found ST59-t437 was the predominant types of MRSA isolates in retail vegetable in our study.
In summary, our study is the first systematical investigation of prevalence and contamination level for S. aureus isolated from retail vegetable in China. Our reports showed that the retail vegetable in China could be contaminated with S. aureus but the levels of S. aureus were not excessive. Most vegetable isolates exhibited resistance to different antimicrobials. In addition, different toxins genes existed in many S. aureus isolates. Thus, improved effective measures should be implemented in the food processing to control contamination and exposure to S. aureus. Besides, some MRSA strains, relevant to major CA-MRSA clone in Asia, were found in this study. Thus, it should be draw public attention and further studies are required to ascertain major clone of MRSA from retail food in China.
Author Contributions
SW, JH, and TL conceived and designed the experiments. JH and FZ performed the experiments. SW and JH analyzed the data. LX, MC, and YD contributed reagents, materials, and analysis tools. SW, QW, and JZ contributed to the writing of the manuscript.
Funding
We would like to acknowledge the financing support of the National Key R&D Program of China (2017YFC1600101), the Science and Technology Projects of Guangzhou (201504010036), China's Post-doctoral Science Fund (2017M612623), and GDAS' Special Project of Science and Technology Development (2017GDASCX-0817).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Altarazi, Y. H., Albetar, M. A., and Alaboudi, A. R. (2009). Biotyping and enterotoxigenicity of Staphylococci isolated from fresh and frozen meat marketed in Jordan. Food Res. Int. 42, 374–379. doi: 10.1016/j.foodres.2009.01.005
Argudín, M. A., Mendoza, M. C., González-Hevia, M. A., Bances, M., Guerra, B., and Rodicio, M. R. (2012). Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Env. Microbiol. 78, 2930–2935. doi: 10.1128/AEM.07487-11
Argudín, M. Á., Mendoza, M. C., and Rodicio, M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel). 2, 1751–1773. doi: 10.3390/toxins2071751
Basanisi, M. G., La, B. G., Nobili, G., Franconieri, I., and La, S. G. (2017). Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 6, 141–146. doi: 10.1016/j.fm.2016.10.020
Bauer, A. W., Kirby, W., Sherris, J. C., and turck, Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Boucher, H. W. (2008). Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl 5.), S344. doi: 10.1086/533590
Chambers, H. F., and Deleo, F. R. (2009). Waves of Resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol 7, 629–641. doi: 10.1038/nrmicro2200
Chao, G., Bao, G., Cao, Y., Yan, W., Yan, W., Zhang, X., et al. (2015). Prevalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food Microbiol. 211, 142–147. doi: 10.1016/j.ijfoodmicro.2015.07.018
Chao, G., Zhou, X., Jiao, X., Qian, X., and Xu, L. (2007). Prevalence and antimicrobial resistance of foodborne pathogens isolated from food products in China. Foodborne Pathog. Dis. 4, 277–284. doi: 10.1089/fpd.2007.0088
Chuang, Y. Y., and Huang, Y. C. (2013). Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect. Dis. 13, 698–708. doi: 10.1016/S1473-3099(13)70136-1
Enright, M. C., Day, N. P. J., Davies, C. E., Peacock, S. J., and Spratt, B. G. (2013). Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 51, 306–310. doi: 10.1128/JCM.02421-12
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Higuchi, W., Hung, W. C., Takano, T., Iwao, Y., Ozaki, K., Isobe, H., et al. (2010). Molecular characteristics of the taiwanese multiple drug-resistant st59 clone of panton-valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus from pediatric cellulitis. J. Infect. Chemother. 16, 144–149. doi: 10.1007/s10156-010-0029-9
Ho, P. L., Chuang, S. K., Choi, Y. F., Lee, R. A., Lit, A. C., Ng, T. K., et al. (2008). Community-associated methicillin-resistant and methicillin-sensitive Staphylococcus aureus: skin and soft tissue infections in Hong Kong. Diagn. Microbiol. Infect. Dis. 61, 245–250. doi: 10.1016/j.diagmicrobio.2007.12.015
Hong, J., Kim, Y., Kim, J., Heu, S., Kim, S. R., Kim, K. P., et al. (2015). Genetic diversity and antibiotic resistance patterns of Staphylococcus aureus isolated from leaf vegetables in korea. J. Food Sci. 80, M1526–M1531. doi: 10.1111/1750-3841.12909
Jarraud, S., Peyrat, M. A., Lim, A., Tristan, A., Bes, M., Mougel, C., et al. (2001). egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166, 669–677. doi: 10.4049/jimmunol.166.1.669
Johler, S., Giannini, P., Jermini, M., Hummerjohann, J., Baumgartner, A., and Stephan, R. (2015). Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins (Basel). 7, 997–1004. doi: 10.3390/toxins7030997
Kozytska, S., Stauss, D., Pawlik, M. C., Hensen, S., Eckart, M., Ziebuhr, W., et al. (2010). Identification of specific genes in Staphylococcus aureus strains associated with bovine mastitis. Vet. Microbiol. 145, 360-365. doi: 10.1016/j.vetmic.2010.03.020
Lindsay, J. A. (2014). Evolution of Staphylococcus aureus and MRSA during outbreaks. Infect. Genet. Evol. 21, 548–553. doi: 10.1016/j.meegid.2013.04.017
Mashouf, R. Y., Hosseini, S. M., Mousavi, S. M., and Arabestani, M. R. (2015). Prevalence of Enterotoxin Genes and Antibacterial Susceptibility Pattern of Staphylococcus aureus strains isolated from animal originated foods in west of Iran. Oman. Med. J. 30, 283–290. doi: 10.5001/omj.2015.56
McLauchlin, J., Narayanan, G. L., Mithani, V., and O'Neill, G. (2000). The detection of enterotoxins and toxic shock syndrome toxin genes in Staphylococcus aureus by polymerase chain reaction. J. Food Prot. 63, 479–488. doi: 10.4315/0362-028X-63.4.479
Motes, M. L., DePaola, A., Cook, D., Veazey, J., Hunsucker, J., Garthright, W., et al. (1998). Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and atlantic coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64, 1459–1465.
Organization, W. H. (2007). Critically Important Antimicrobials for Human Medicine: Categorization for the Development of Risk Management Strategies to Contain Antimicrobial Resistance Due to Non-human Antimicrobial Use: Report of the Second WHO Expert Meeting, Copenhagen.
Papadopoulos, P., Papadopoulos, T., Angelidis, A. S., Boukouvala, E., Zdragas, A., Papa, A., et al. (2018). Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus (MRSA) along the production chain of dairy products in north-western Greece. Food Microbiol. 69, 43–50. doi: 10.1016/j.fm.2017.07.016
Pérez-Roth, E., Claverie-Martín, F., Villar, J., and Méndez-Alvarez, S. (2001). Multiplex PCR for Simultaneous Identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39, 4037–4041. doi: 10.1128/JCM.39.11.4037-4041.2001
Petinaki, E., and Spiliopoulou, I. (2012). Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin. Microbiol. Infect. 18, 626–637. doi: 10.1111/j.1469-0691.2012.03881.x
Rong, D., Wu, Q., Xu, M., Zhang, J., and Yu, S. (2017). Prevalence, virulence genes, antimicrobial susceptibility, and genetic diversity of Staphylococcus aureus from retail aquatic products in China. Front. Microbiol. 8:714. doi: 10.3389/fmicb.2017.00714
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M.-A., Roy, S. L., et al. (2011). Foodborne illness acquired in the united states—major pathogens. Emerging Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Seo, Y. H., Jang, J. H., and Moon, K. D. (2010). Occurrence and characterization of enterotoxigenic Staphylococcus aureus isolated from minimally processed vegetables and sprouts in Korea. Food Sci. Biotechnol. 19, 313–319. doi: 10.1007/s10068-010-0045-7
Shahraz, F., Dadkhah, H., Khaksar, R., Mahmoudzadeh, M., Hosseini, H., Kamran, M., et al. (2012). Analysis of antibiotic resistance patterns and detection of mecA gene in Staphylococcus aureus isolated from packaged hamburger. Meat Sci. 90, 759–763. doi: 10.1016/j.meatsci.2011.11.009
Shankar, P. R. (2012). The evolving threat of antimicrobial resistance: options for action. Aust Med. J. 5, 592–595.
Shopsin, B., Gomez, M., Montgomery, S. O., Smith, D. H., Waddington, M., Dodge, D. E., et al. (1999). Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37, 3556–3563.
Smyth, D. S., Hartigan, P. J., Meaney, W. J., Fitzgerald, J. R., Deobald, C. F., Bohach, G. A., et al. (2005). Superantigen genes encoded by the egc cluster and SaPIbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J. Med. Microbiol. 54, 401–411. doi: 10.1099/jmm.0.45863-0
Song, M., Bai, Y., Xu, J., Carter, M. Q., Shi, C., and Shi, X. (2015). Genetic diversity and virulence potential of Staphylococcus aureus isolates from raw and processed food commodities in Shanghai. Int. J. Food Microbiol. 195, 1–8. doi: 10.1016/j.ijfoodmicro.2014.11.020
Spanu, V., Spanu, C., Virdis, S., Cossu, F., Scarano, C., and De Santis, E. P. (2012). Virulence factors and genetic variability of Staphylococcus aureus strains isolated from raw sheep's milk cheese. Int. J. Food Microbiol. 153, 53–57. doi: 10.1016/j.ijfoodmicro.2011.10.015
Stegger, M., Andersen, P. S., Kearns, A., Pichon, B., Holmes, M. A., Edwards, G., et al. (2012). Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA LGA251. Clini. Microbiol. Infect. 18, 395–400. doi: 10.1111/j.1469-0691.2011.03715.x
Tang, C. T., Tho, N. D., Hoa, N. T., Phuong, N. T. M., Le, V. T., Diep, T. S., et al. (2007). An outbreak of severe infections with community-acquired mrsa carrying the panton-valentine leukocidin following vaccination. PLoS ONE 2:e822. doi: 10.1371/journal.pone.0000822
The Clinical and Laboratory Standards Institute (CLSI) (2015). “Performance standards for antimicrobial susceptibility testing,” in Twenty-Fifth Informational Supplement. Approved Standard-M02-A12. Wayne, PA: The Clinical and Laboratory Standards Institute.
Varshney, A. K., Mediavilla, J. R., Robiou, N., Guh, A., Wang, X., Gialanella, P., et al. (2009). Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl. Env. Microbiol. 75:6839. doi: 10.1128/A.E.M.00272-09
Wang, C. C., Lo, W. T., Chu, M. L., and Siu, L. K. (2004). Epidemiological typing of community-acquired methicillin-resistant Staphylococcus aureus isolates from children in Taiwan. Clini. Infect. Dis. 39, 481–487. doi: 10.1086/422642
Wang, W., Baloch, Z., Jiang, T., Zhang, C., Peng, Z., Li, F., et al. (2017). Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 8:2256. doi: 10.3389/fmicb.2017.02256
Wang, X., Li, G., Xia, X., Yang, B., Xi, M., and Meng, J. (2014). Antimicrobial susceptibility and molecular typing of methicillin-resistant staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog. Dis. 11:281. doi: 10.1089/fpd.2013.1643
Wang, X., Meng, J., Zhang, J., Zhou, T., Zhang, Y., Yang, B., et al. (2012). Characterization of Staphylococcus aureus isolated from powdered infant formula milk and infant rice cereal in China. Int. J. Food Microbiol. 153, 142-147. doi: 10.1016/j.ijfoodmicro.2011.10.030
Wang, X., Tao, X., Xia, X., Yang, B., Xi, M., Meng, J., et al. (2013). Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail raw chicken in China. Food Control 29, 103–106. doi: 10.1016/j.foodcont.2012.06.002
Coombs, G. W., Pearson, J. C., O'Brien, F. G., Murray, R. J., Grubb, W. B., and Christiansen, K. G. (2006). Methicillin-resistant Staphylococcus aureus Clones, Western Australia. Emerg. Infect. Dis. 12, 241–247. doi: 10.3201/eid1202.050454
Wu, D. J., Geng, W. J., Yang, Y. H., and Shen, X. Z. (2010). OL-049 Epidemiology and molecular characteristics of community-associated methicillin-resistant and methicillin-susceptible Staphylococcus aureus from skin/soft tissue infections in Beijing Children's Hospital, China. Int. J. Infect. Dis. 14, S22–S22. doi: 10.1016/S1201-9712(10)60061-6
Yang, S., Pei, X., Wang, G., Yan, L., Hu, J., Li, Y., et al. (2016a). Prevalence of food-borne pathogens in ready-to-eat meat products in seven different Chinese regions. Food Control 65, 92–98. doi: 10.1016/j.foodcont.2016.01.009
Yang, X., Yu, S., Wu, Q., Zhang, J., Wu, S., and Rong, D. (2018). Multilocus sequence typing and virulence-associated gene profile analysis of Staphylococcus aureus isolates from retail ready-to-eat food in China. Front. Microbiol. 9:197. doi: 10.3389/fmicb.2018.00197
Keywords: S. aureus, vegetable, antimicrobial resistance, enterotoxin, MLST, spa typing
Citation: Wu S, Huang J, Wu Q, Zhang F, Zhang J, Lei T, Chen M, Ding Y and Xue L (2018) Prevalence and Characterization of Staphylococcus aureus Isolated From Retail Vegetables in China. Front. Microbiol. 9:1263. doi: 10.3389/fmicb.2018.01263
Received: 14 February 2018; Accepted: 24 May 2018;
Published: 14 June 2018.
Edited by:
Chunbao Li, Nanjing Agricultural University, ChinaReviewed by:
Beatrix Stessl, Veterinärmedizinische Universität Wien, AustriaAnca Ioana Nicolau, Dunarea de Jos University, Romania
Copyright © 2018 Wu, Huang, Wu, Zhang, Zhang, Lei, Chen, Ding and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t
†These authors have equally contributed to this work.
 Shi Wu
Shi Wu Jiahui Huang1†
Jiahui Huang1† Jumei Zhang
Jumei Zhang Moutong Chen
Moutong Chen Yu Ding
Yu Ding