- 1Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
- 2Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals, College of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou, China
Assimilation of short-chain fatty acids (SCFAs) plays an important role in the survival and lipid biosynthesis of Mycobacteria. However, regulation of this process has not been thoroughly described. In the present work, we demonstrate that GlnR as a well-known nitrogen-sensing regulator transcriptionally modulates the AMP-forming propionyl-CoA synthetase (MsPrpE), and acetyl-CoA synthetases (MsAcs) is associated with SCFAs assimilation in Mycobacterium smegmatis, a model Mycobacterium. GlnR can directly activate the expression of MsprpE and Msacs by binding to their promoter regions based upon sensed nitrogen starvation in the host. Moreover, GlnR can activate the expression of lysine acetyltransferase encoding Mspat, which significantly decreases the activity of MsPrpE and MsAcs through increased acylation. Next, growth curves and resazurin assay show that GlnR can further regulate the growth of M. smegmatis on different SCFAs to control the viability. These results demonstrate that GlnR-mediated regulation of SCFA assimilation in response to the change of nitrogen signal serves to control the survival of M. smegmatis. These findings provide insights into the survival and nutrient utilization mechanisms of Mycobacteria in their host, which may enable new strategies in drug discovery for the control of tuberculosis.
Introduction
Intracellular nutrient utilization is a very important survival mechanism for pathogenic Mycobacteria such as Mycobacterium tuberculosis (Mtb) within the human host. In the host of macrophages, Mtb grows preferentially in phagosomes, a known nutritionally-constrained environment, where Mtb can exploit fatty acids and cholesterol as carbon and energy sources to survive (Pandey and Sassetti, 2008). In previous works, multiple lines of evidence support the idea that host fatty acids and cholesterol rather than carbohydrates serve as vital carbon sources for Mtb during infection and latency. Since the 1950s, Mtb was found to propagate in mammalian tissues preferentially by assimilating fatty acids as nutrients (Bloch and Segal, 1956). Thereafter, plenty of researches have revealed that the metabolism of fatty acid facilitate Mtb survival during infection (McKinney et al., 2000; Muñoz-Elías et al., 2006; Marrero et al., 2010). In addition, Mtb can also uptake cholesterol which is a crucial nutrient for Mtb survival within macrophages (Van der Geize et al., 2007; VanderVen et al., 2015) as well as various in vivo infection models (Chang et al., 2007; Pandey and Sassetti, 2008; Yam et al., 2009; Hu et al., 2010; Nesbitt et al., 2010).
In Mtb, fatty acids and cholesterol are degraded to short chain fatty acids (SCFAs, such as acetate and propionate) to be assimilated to generate energy or synthesize lipid (Yang et al., 2009; Wipperman et al., 2014). So far, assimilation of SCFAs and cholesterol is considered as a characteristic of Mtb (Cole et al., 1998). In brief, the majority of acetyl-CoA enters the TCA cycle to provide energy, and the remainder is used to synthesize mycolic acid through malonyl-CoA. Propionyl-CoA is assimilated into central carbon metabolism via the methylcitrate cycle and the methylmalonyl pathway (Savvi et al., 2008; Griffin et al., 2012). However, accumulation of propionyl-CoA is toxic and inhibitory to a few pivotal metabolic enzymes. Therefore, the metabolism of propionyl-CoA need to be tightly controlled (Carter and Alber, 2015). One known pathway for propionyl-CoA reduction is the synthesis of lipids such as phthiocerol dimycoserate (PDIM) (Wipperman et al., 2014), which contributes to sustaining infection and virulence. Consequently, the assimilation of SCFAs play a crucial role for the survival and virulence of Mtb.
In bacteria, SCFAs are assimilated via acyl-CoA intermediates which are generated by acyl-CoA synthetases catalysis, such as acetyl-CoA synthetase (Acs) and propionyl-CoA synthetase (PrpE). These enzymes were first characterized in Salmonella Typhimurium (Horswill and Escalante-Semerena, 1999). As a common route for propionyl-CoA synthesis in microorganisms, PrpE has also been investigated in other bacterial species (Rajashekhara and Watanabe, 2004; Kang et al., 2012; Yang et al., 2014). In addition to PrpE, Acs could also act as a secondary route for propionyl-CoA synthesis (Liu et al., 2014). Previous work has shown that the prpE deletion mutant could still use propionate as carbon and energy sources, since Acs could compensate for the lack of PrpE (Horswill and Escalante-Semerena, 1999).
During the assimilation of SCFAs, PrpE, or Acs are subject to regulation at multiple levels. On the transcriptional level, the regulatory mechanism of prpE is well understood in Gram-negative bacterium Escherichia coli, in which prpE is regulated by PrpR, a transcriptional activator for prpBCDE operon. Furthermore, similar phenomena were also found in Salmonella Typhimurium and Mtb (Palacios and Escalante-Semerena, 2000; Liu et al., 2014; Masiewicz et al., 2014). For acs, many direct and indirect regulators have been identified and studied in E. coli, these regulators can regulate the transcription of acs. For example, catabolite repression and the nucleoid proteins Fis and IHF act as direct factors, while several other indirect transcription factors were also identified (Kumari et al., 2000; Wolfe, 2005; Bernal et al., 2016). Meanwhile, in Gram-positive bacterium Bacillus subtilis, the acs gene is controlled by GTP-sensing transcription factor CodY and carbon flux regulator CcpA (Warner and Lolkema, 2003; Starai and Escalante-Semerena, 2004; Wolfe, 2005). For Mtb, LucA was identified recently as a lipid uptake coordinator to regulated the assimilation of both fatty acids and cholesterol when Mtb grow in macrophages (Nazarova et al., 2017).
Except for transcriptional modulation, post-translational modification (PTM) is an efficient mechanism for regulating gene expression and enzymatic activity in response to changing physiological conditions (Gardner et al., 2006). PrpE was found to undergo propionylation in vivo which inactivates its activity, while Acs is the first and best-known enzyme modulated by reversible lysine acetylation (RLA) in bacteria (Hayden et al., 2013). Recently, many scientists studied the mechanism of RLA regulating the activity of Acs in various bacteria, such as E. coli, Salmonella enterica, and Mtb (Starai et al., 2002; Castaño-Cerezo et al., 2011; Noy et al., 2014). Similar to Acs, PrpE of S. enterica is also regulated by reverse propionylation (Garrity et al., 2007). In Mtb, metabolism of fatty acid was found to be mediated by lysine acylation (Nambi et al., 2013). So far, much research progress has been made on the regulation of SCFAs assimilation, such as bacterial propionyl-CoA synthesis pathways. However, most of them concentrate on E. coli and Salmonella, whereas little is known in mycobacteria.
In this study, we scrutinized the regulatory mechanism of SCFAs assimilation in M. smegmatis at the transcriptional and post-translational levels. We found that important SCFA metabolic enzymes, i.e., propionyl-CoA synthetase (MsPrpE) and four AMP-forming Acs, are all activated at the transcriptional level by nitrogen-sensing regulator GlnR under nitrogen limited conditions. On the contrary, with nitrogen starvation, GlnR activates the expression of acetyltransferase coding gene (Mspat). Next, the highly expressed MsPat increase the acylation level of MsPrpE and MsAcs, resulting in significant decrease in enzymatic activity. Consequently, the results reveal that GlnR senses the change in environmental nitrogen signal to regulate the assimilation of SCFAs at the transcriptional and post-translational levels. These regulations directly impact the growth of M. smegmatis which utilizes different SCFAs and cholesterol as carbon sources. These findings provide insights into the mechanisms of SCFA assimilation, which affects the survival, detoxification, and infection of Mycobacteria.
Materials and Methods
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this work are listed in Table 1. Mycobacterium smegmatis mc2 155 wild type and its glnR deleted mutant (ΔglnR) were grown in LB broth (supplemented with 0.05% Tween 80) at 37°C, 200 rpm or on LB agar plates at 37°C. All E. coli strains were cultured in LB broth or on LB agar. When needed, M. smegmatis was cultured in Sauton’s medium [3.6 mM KH2PO4, 2 mM MgSO4, 10 mM citric acid, 0.1 mM ferric citrate, 6 μM ZnSO4, 0.2% glycerol, 0.015% Tyloxapol (pH7.0)] (Jenkins et al., 2013), supplemented with (NH4)2SO4 at 1 mM (nitrogen limiting) or 30 mM (nitrogen excess) after being washed twice in nitrogen-free medium. For growth curves, strains were grown in M9 media containing 12.6 mM Na2HPO4, 22 mM KH2PO4, 8 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, and 0.1% tyloxapol (Hayden et al., 2013), and the starting OD600 was 0.06. Provided carbon sources included 10 mM glucose or 10 mM sodium acetate, or 10 mM sodium propionate.
The glnR complemented strain was constructed referring to a previous work (Stover et al., 1991). In brief, glnR gene was amplified by PCR. The amplicon was inserted into shuttle vector pMV361, which was transformed into E. coli DH5α for recombinant vector screening and confirmation by PCR and sequencing. The verified plasmids were introduced into ΔglnR competent cell by electric-mediated protoplast transformation. Transformants were selected by its resistance to kanamycin and subsequently analyzed by qRT-PCR to verify integration of glnR gene into the M. smegmatis genome.
Cloning, Over-Expression, and Purification of Proteins
Msmeg_5784 (glnR), Msmeg_5458, Msmeg_6179 (MsacsA1), and Msmeg_5404 (MsprpE) genes were amplified from the genomic DNA of M. smegmatis, and Rv0818 was amplified from the genomic DNA of Mtb using a seamless cloning and assembly kit. The PCR products were homologous recombinated with pET-28a which has been digested with EcoRI and HindIII to generate a recombinant vector. The clone was confirmed by PCR and sequencing. The protein was expressed in E. coli BL21 (DE3) strain. A single colony was inoculated into 5 mL of LB medium containing 1% kanamycin and cultured at 37°C and 200 rpm overnight. The activated bacteria were then transferred to 50 mL of LB medium and induced with IPTG (0.7 mM) overnight until the OD600 reached 0.6.
For protein expression, cells were harvested by centrifugation (8000 rpm, 10 min) and washed twice with PBS (pH8.0). The cells were suspended by 25 mL of PBS and sonicated. The debris was separated from the soluble fraction by centrifugation at 8,000 rpm and 4°C for 30 min. Supernatant was purified using Ni-NTA agarose column (Merck, Darmstadt, Germany). The column was washed with 10 mL of 10 mM imidazole in 50 mM NaH2PO4 and 300 mM NaCl, pH 8.0. The desired protein was eluted with 20–250 mM imidazole in 50 mM NaH2PO4 and 300 mM NaCl (pH 8.0). We analyzed the fractions by SDS-PAGE and polled the ones that contained the desired protein. The polled fractions were dialyzed against PBS containing 10% glycerol. The his tag of MsAcs and MsPrpE was removed by thrombin treatment at 4°C overnight. The protein was concentrated using centrifugation with a 30 K cutoff (Millipore, Billerica, MA, United States). All protein concentrations were measured using the BCA quantitative kit.
Over-Expression of His-Acs, His-GlnR, and His-PrpE From M. smegmatis
Msmeg_5784 (glnR), Msmeg_6179 (MsacsA1), and Msmeg_5404 (MsprpE) genes were amplified from the genomic DNA of M. smegmatis by PCR and then cloned into the shuttle vector pMV-261 which has the resistance of kanamycin yielding -5784, pMV-261-6179 and pmv-261-5404 (Stover et al., 1991). The overexpression plasmids of pMV-261-5784, pMV-261-6179, and pMV-261-5404 were confirmed by PCR and sequencing were introduced into M. smegmatis by electric-mediated protoplast transformation. The complementary strain of His-GlnR was obtained by introducing pMV-361-5784 recombined vector to glnR knockout (ΔglnR) strain. The overexpression M. smegmatis strains were obtained by kanamycin resistance screening and confirmed by protein SDS-PAGE.
The overexpression strains were harvested by centrifugation (8000 rpm, 10 min) and washed twice with PBS (pH8.0). The cells were resuspended in PBS and sonicated, and the debris were removed by centrifugation at 8,000 rpm 4°C for 30 min. The desired protein was purified using Ni-NTA agarose column as mentioned above. Samples were analyzed using SDS-PAGE and then subjected to Western blot analysis. We used two antibodies: HRP conjugated acetyl-lysine antibody (Cat# ICP0380) (Immunechem Pharmaceuticals, Burnaby, BC, Canada), and HRP conjugated His tag antibody (Abmart). The bound antibody was visualized using enhanced chemiluminescence method.
Electrophoretic Mobility Shift Assay (EMSA)
The putative promoter regions (upstream region about -300–60) containing GlnR-binding sites were amplified by PCR using the primers listed in Table 2, then biotin-labeled by PCR with biotin-modified universal primer (5′ biotin-AGCCAGTGGCGATAAG-3′) (Liao et al., 2014). The PCR products were identified by agarose gel electrophoresis and purified using a PCR purification kit (Shanghai Generay Biotech), and were then used as EMSA probes. The concentrations of probes were quantified with a microplate reader (Biotek, United States) and diluted to 5 ng/μL. EMSAs assay was carried out using the Chemiluminescent EMSA Kit (Beyotime Biotechnology, China). The binding solution (total volume was 10 μL) containing 1 μL probes, varying amounts of purified GlnR and 1 μL Gel-Shift binding buffer (50 μg/mL poly (dI-dC), 1 mM DTT, 10% glycerol, 1 mM EDTA, 50 mM NaCl, 25 mM MgCl2, 10 mM Tris–HCl, pH 8.0, 0.01% Nonidet P40) was incubated at room temperature for 20 min. After binding, the samples were separated on a non-denaturating PAGE gel in ice-bathed 0.5 × Tris-borate-EDTA (Shanghai Bioscience) at 170 V. Then the probe, protein and their compounds were transferred to a nylon membrane at 160 V for 30 min. Finally, the bands were detected by BeyoECL Plus (You et al., 2017).
Quantitative Real-Time PCR
Cells grown to exponential stage in nitrogen-limited medium were harvested by centrifugation at 4°C. Total RNA was prepared using RNeasy Mini Kit (Qiagen, Valencia, CA, United States). The RNA quality was analyzed by 1% agarose gel electrophoresis, then the RNA was quantified by microplate reader (BioTek, United States). The residual DNA was digested and RNA was reverse transcribed using a PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Shiga, Japan) for qRT-PCR. PCR reactions were performed using 2 × RealStar Green Fast Mixture (GenStar) with primers listed in Table 3. Each PCR reaction with 20 μL final volume contained 50 ng cDNA, and was performed on a CFX96 Real-Time System (Bio-Rad). The qPCR conditions were: 95°C for 5 min followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 30 s, and final extension at 72°C for 10 min (You et al., 2016).
In Vitro Protein Propionylation and Acetylation Assays
Propionylation assays were carried out in a 100 μL mixture that included buffer B (50 mM HEPES, pH 7.5), 2 μM MsAcs or MsPrpE, 20 μM Propionyl-CoA, and 2.7 μM MsPat with or without 1 mM cAMP. Acetylation assays were carried out in a 100 μL reaction mixture that contained buffer A (50 mM HEPES and 150 mM NaCl, pH 7.5), 2 μM MsAcs or MsPrpE, 100 μM Acetyl-CoA, and 2.7 μM MsPat with or without 1 mM cAMP. Reactions were incubated at 37°C for 2 h (Xu et al., 2011; Liu et al., 2016). After propionylation or acetylation, the MsPrpE or MsAcs was extracted from the reaction products using gel filtration and ultrafiltration (Xu et al., 2014). The propionylated or acetylated proteins were divided into two portions: one was used in SDS-PAGE Western blot to monitor the propionylation or acetylation, and the other was used to measure the change of enzymatic activity of MsPrpE or MsAcs (You et al., 2017).
Western Blot Assays
The concentration of the protein was measured using BCA Protein Assay Kit. Samples were isolated by SDS-PAGE (10% acrylamide) and then transferred to a PVDF membrane at 380 mA for 1 h. The PVDF membrane was blocked in BSA blocking buffer for 2 h and then incubated with acetyl-lysine antibody (dilution of 1:15,000) at 4°C overnight. The blot was washed three times (once 10 min) with TBST buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20), and the bound antibody was detected by ECL system (CTB, United States).
In Vitro MsPrpE and MsAcs Assays
MsPrpE catalyzes the first step of propionate metabolism by converting propionate to propionyl-CoA. The formated propionyl-CoA was measured using a biosynthetic assay (Liu et al., 2014). The reaction mixtures contain 50 mM Tris–HCl (pH 8.0), 100 mM sodium propionate (sodium acetate), 5 mM ATP, 3 mM MgCl2, 0.5 mM DTT, 300 mM hydroxylamine, and 2.5 mM CoA in a total volume of 200 μL. MsPrpE or MsAcs was added into the mixture and incubated at 37°C. The color agent containing 0.2 M TCA, 0.4 M FeCl36H2O, 5% (v/v) concentrated hydrochloric acid was added into the mixture, then incubating for 30 min to stop the reaction. The color generated was measured at 540 nm by microplate reader. The activity of MsPrpE was represented by the formation of propionylhydroxamate per protein amount and time (You et al., 2016).
Resazurin Assays
Resazurin was used to monitor the growth and viability of M. smegmatis in different carbon resources. Strains were washed twice in media without carbon, and then growing in M9 media with 10 mM glucose, 10 mM acetate, 10 mM propionate, 10 mM cholesterol, or no carbon. The initial OD of strains is 0.05, after cultured at 37°C and 100 rpm for 72 h. The culture was transferred into a 96-well plate. Resazurin solution (12.5 mg/ml final concentration; Sigma) was then added, and incubated for 10 h. Emitted fluorescence signal at 590 nm was recorded. After 10 h of resazurin addition, the values were normalized to the wild-type strain under the same growth condition. Meanwhile, samples without cells and with cells but without carbon source were used as negative controls (Hayden et al., 2013).
Mass Spectrometry Analysis of the Acylation Site
The acetylated and propionylated sites of MsAcsA1 and MsPrpE were analyzed by mass spectrometry (MS) according to our previous work (Liu et al., 2016). In brief, acetylated or propionylated MsAcsA1 or MsPrpE was analyzed by SDS-PAGE. The single protein band corresponding to MsAcsA1 or MsPrpE was cut from the gel and washed twice with 50% ethanol to remove the stain. Gel pieces were rehydrated using 10 ng/μL trypsin in 50 mM ammonium bicarbonate (Promega, Madison, WI, United States), and digestion was continued at 37°C for 12 h. Then, the digests were analyzed by LC-MS/MS. Full MS spectra (m/z range: 350–1700) were obtained in the resolution of 240,000. The five most intense ions were extracted in turn from each full MS spectrum for MS/MS fragmentation. Automatic gain control (AGC) setting for ion trap and orbitrap was 3E4 and 1E6. A Mascot search engine (v2.3.01, Matrix Science, United Kingdom) was used to check against the MsAcsA1 or MsPrpE sequence from “.mgf” files generated by Thermo Proteome Discoverer (Thermo Fisher Scientific). Within the searching process, these parameters were used with 10 ppm peptide mass tolerance, 0.5 Da fragment mass tolerance, and selected charge states (+2, +3, and +4). The program-identified peptides were verified manually.
Results
GlnR Regulates Gene Expression in SCFA Assimilation in Response to Nitrogen Condition
Through in silico analysis of the SCFAs metabolic pathway in M. smegmatis, we found one AMP-forming propionyl-CoA synthetase (MsPrpE, MSMEG_5404, EC:6.2.1.17) and four AMP-forming Acs (EC:6.2.1.1, hereinafter referred to MsAcsA1, MSMEG_6179; MsAcsA2, MSMEG_0718; MsAcsA3, MSMEG_3986; MsAcsA4, MSMEG_5650). For in vitro MsAcs assays, we found that all MsAcs had Acs activity (Figure 1A), and the activity of MsAcsA1 was highest. Meanwhile, MsPrpE, MsAcsA1, MsAcsA3, and MsAcsA4 also showed high propionyl-CoA synthetase activity (Figure 1B). In order to analyze the expression of Msacs and MsprpE genes in response to different fatty acids in M. smegmatis, we investigated the transcription activity of four Msacs and one MsprpE genes in cells grown in acetate and propionate using glucose as control. As seen in Figure 1C, the expression of MsacsA1 was the highest when acetate was the sole carbon source, which was consistent with the ACS activity in Figure 1A. Therefore, this result indicates that MsacsA1 could play a major role in acetate assimilation. In the uptake of propionate (Figure 1D), the MsprpE gene was highly expressed. Combining the enzymatic activity data in Figure 1B, it is suggested that MsprpE was responsible for propionate uptake. At the same time, MsacsA1 and MsacsA4 also played an important role in propionate metabolism. As AMP-forming Acs, MsAcsA1 is essential for M. smegmatis (Xu et al., 2011; Hayden et al., 2013; Chopra et al., 2014). Based upon existing data on acetate metabolism, we selected MsAcsA1 as a representative Msacs gene combining MsprpE to explore the mechanism of SCFA assimilation.
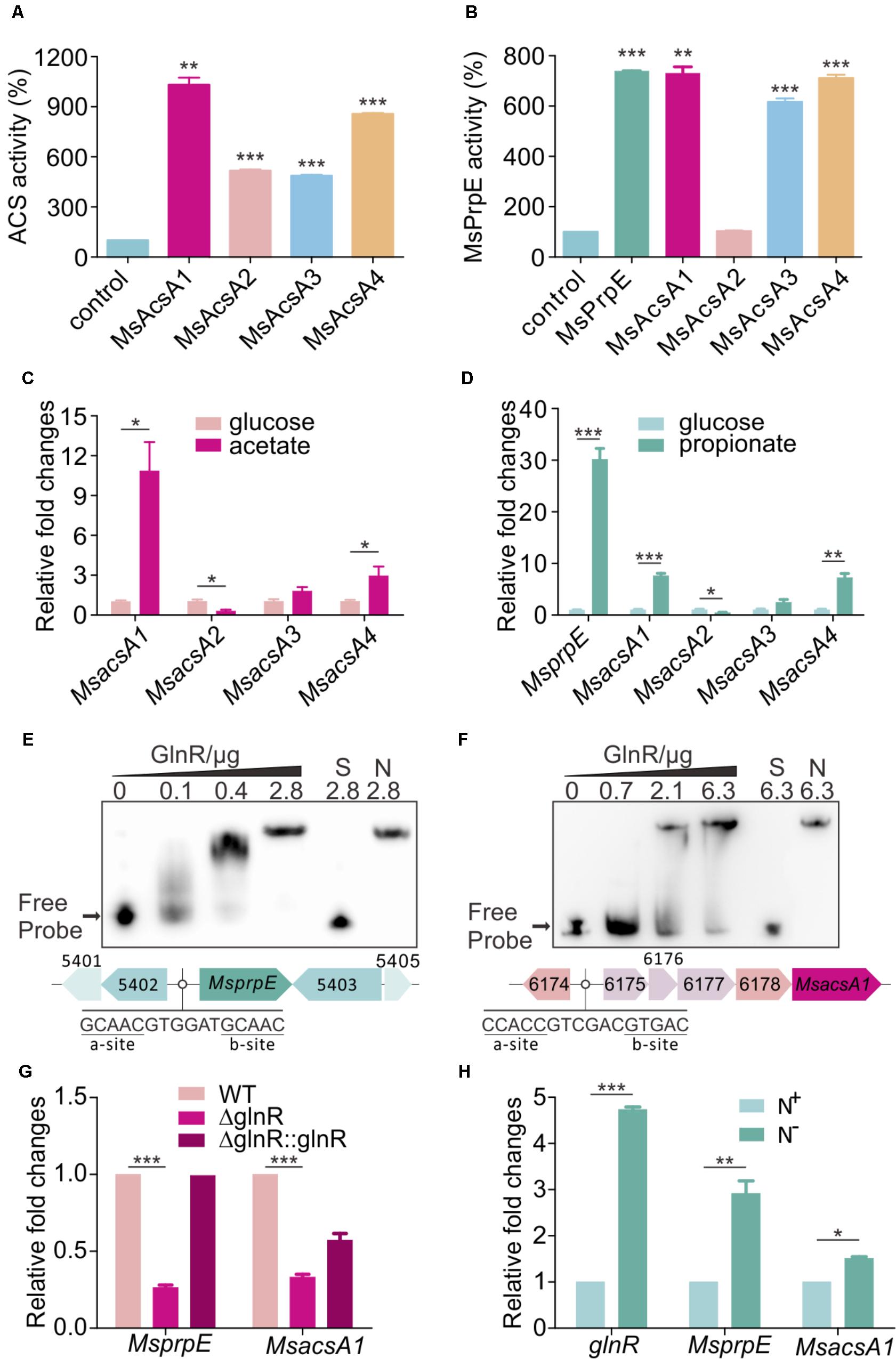
FIGURE 1. GlnR regulates the expression of Msacs and MsprpE in Mycobacterium smegmatis. (A) Acetyl-CoA synthetase activity of MsAcs. (B) Propionyl-CoA synthetase activity of MsPrpE and MsAcs. (C) Transcriptional levels of the Msacs genes in wild type (WT) strain grown in glucose or acetate as carbon sources. (D) Transcription levels of MsprpE and Msacs in WT grown in glucose or propionate as carbon sources. (E,F) Electrophoretic mobility shift assay (EMSA) identification of GlnR binding with the promoter region of MsprpE and MsacsA1. GlnR protein labeled with His tag incubate with probe (5 ng, MsprpE or MsacsA1). Unlabeled specific probe (S) (200-fold excess) or non-specific competitor DNA (sperm DNA) (N) were used as controls. (G) Differential expression analysis of glnR regulating MsprpE and MsacsA1. WT, ΔglnR (glnR knocked out mutant) and ΔglnR::glnR (glnR complementary) were cultured at 30°C to the middle exponential phase. (H) WT was cultured in Sauton medium supplemented with N+ and N-, following comparative analysis of glnR, MsprpE, or MsacsA1 gene in the excess (N+) or limited (N-) nitrogen condition. Error bars represent the standard error, n = 3. P-values were determined by two-tailed unpaired t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Using MEME/MAST and PREDetector to analyze the motif of MsprpE and Msacs genes (Liao et al., 2014), we found a classic GlnR binding box in the promoter region of these genes which consists of one a site and one b site separated by a six-nucleotide motif (a-n6-b) (Amon et al., 2009; Liao et al., 2014). In order to validate the binding of GlnR to this box, purified GlnR protein was diluted to different concentrations for the EMSA assay using a DNA probe containing those sites. The unlabeled specific probe (S) (200-fold excess) or non-specific competitor DNA (sperm DNA) (N) were used as controls. Results indicated that GlnR could bind with the promotor region of the MsprpE and four Msacs genes in M. smegmatis (Figures 1E,F and Supplementary Figure S1A). To investigate the regulation of GlnR to MsprpE and Msacs genes, we constructed a glnR deletion strain of M. smegmatis (ΔglnR) and glnR complementary strain (ΔglnR::glnR) for qPCR assay and analyzed the differential expression of those genes in the mutant strains. As shown in Figure 1G and Supplementary Figure S1B, the transcriptional level of prpE and all four Msacs genes sharply decreased compared to those in the wild type (WT). Meanwhile, the expressions of these genes were recovered in ΔglnR::glnR. The qPCR and EMSA results indicated that the MsprpE gene and the four Msacs genes were all directly subject to the regulation of GlnR in M. smegmatis. GlnR is known as a globe nitrogen regulator, whose expression is activated in nitrogen limited environments (Amon et al., 2008). To test whether nitrogen availability interferes with the regulation of GlnR on MsprpE and Msacs, we established a comparative model with Sauton medium supplementing excess (N+) or limited (N-) nitrogen. Results suggested that the expression of glnR was activated in response to nitrogen starvation, and the MsprpE and four Msacs genes were further up-regulated at the transcriptional level (Figure 1H and Supplementary Figure S1C). Since propionyl-CoA and acetyl-CoA are both vital intermediates for SCFA assimilation, SCFA assimilation in M. smegmatis may be linked to nitrogen metabolism via the regulation by GlnR.
Activity of MsPrpE and MsAcs Is Inhibited by MsPat-Mediated Acetylation/Propionylation
In a previous study, we found that Acs could be regulated by acetylation in S. erythraea (You et al., 2017). In addition, Acs in Mtb can also be regulated by reversible acetylation (Xu et al., 2011). We hypothesized that MsPrpE and MsAcs could be regulated by acylation (e.g., propionylation and acetylation) at the post-translational level. To directly validate this idea, we selected MsPrpE and MsAcsA1 for the propionylation and acetylation assays. Western blot analyses (Figures 2A,D) showed that MsAcsA1 and MsPrpE were propionylated by MsPat with propionyl-CoA (PrCoA) in the reaction system regardless of the presence of cAMP.
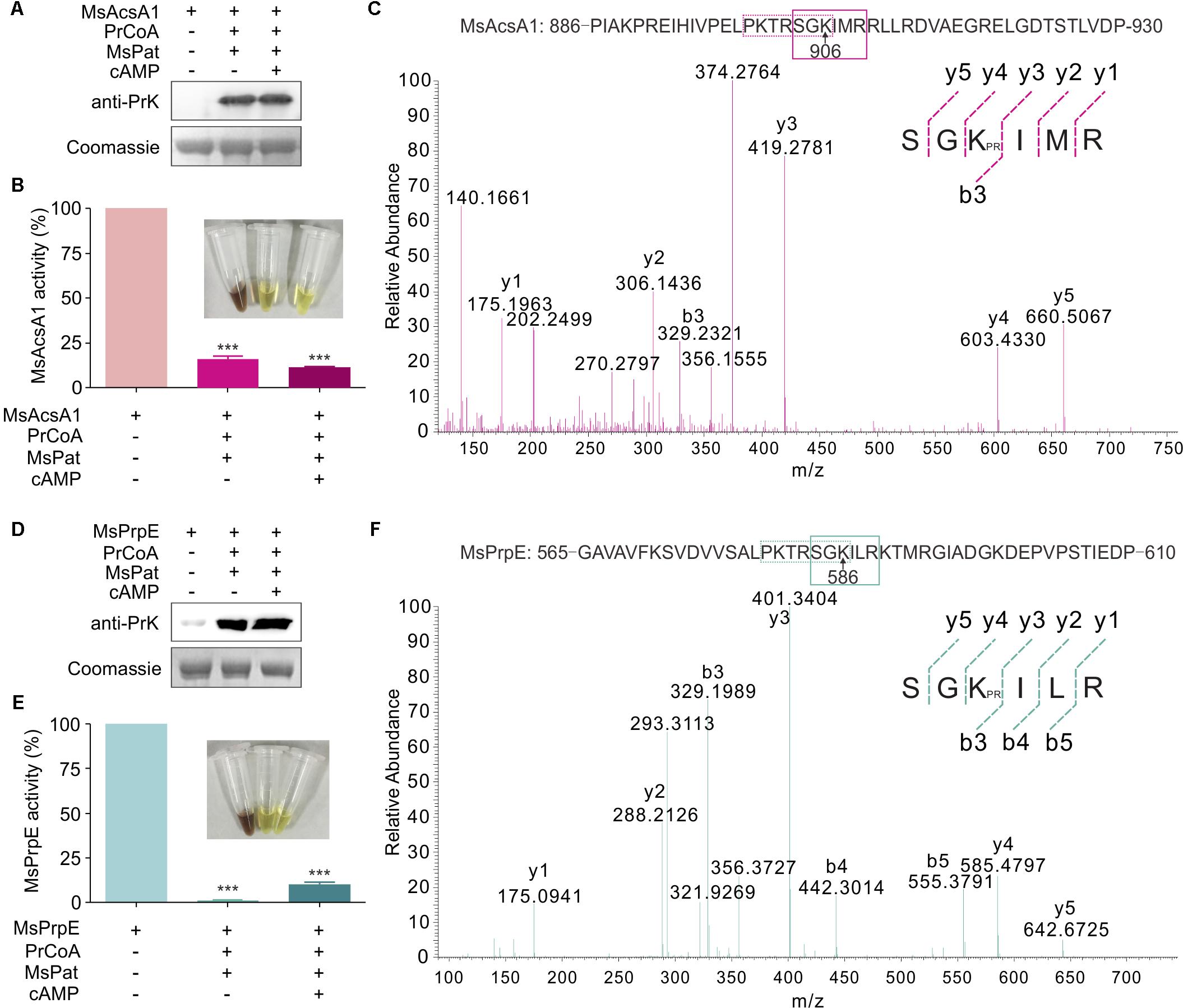
FIGURE 2. The activity of MsPrpE and MsAcsA1 are regulated by propionylation. (A) MsAcsA1 (2 μM) was incubated independently or with MsPat (2.7 μM), Propionyl-CoA (20 μM), and cAMP (1 mM) within a total volume of 100 μL at 37°C for 2 h, followed by SDS-PAGE analysis. The propionylation extent was verified using Western blot assay (Upper). Meanwhile, another PAGE gel was stained with Coomassie brilliant blue as control (Lower). (B) Enzymatic activity of MsAcsA1 using same condition with (A). (C) LC-MS/MS spectrum of a tryptic peptide (m/z = 419.3) obtained from propionylated MsAcsA1. This spectrum matched the peptide sequence (boxed) in MsAcsA1, where a mass shift occurred between b3 and y4 ions consistent with propionylation at the lysine residue (amino acid position 906). (D) MsPrpE was incubated alone or in the presence of MsPat with the same condition of (A), followed by SDS-PAGE analysis. The propionylation extent was verified by Western blot (Upper). (E) Enzymatic activity of MsPrpE and MsAcsA1 using same condition with (D). (F) LC-MS/MS spectrum of a tryptic peptide (m/z = 401.3) obtained from propionylated MsPrpE. This spectrum matched the peptide sequence (boxed) in MsPrpE, where a mass shift occurred between b3 and y4 ions consistent with propionylation at the lysine residue (amino acid position 586). Error bar represent the standard error, and all experiments replicate three times. ∗∗∗P < 0.001.
Many studies show PTM, such as propionylation and acetylation, play vital roles in the regulation of enzymatic activity (Xiong and Guan, 2012; Okanishi et al., 2014; Verdin and Ott, 2015). In this work, we found that the activity of both MsPrpE and MsAcsA1 were negatively modulated after propionylation, as MsPrpE was almost deactivated, whereas MsAcsA1 activity also decreased by more than 75%. Furthermore, the effect of propionylation on enzymatic activity was also evidenced by the change of color in (Figures 2B,E).
To identify the propionylation site(s) in MsAcs and MsPrpE, in vitro propionylated enzymes were separated by electrophoresis, and the bands corresponding to MsAcsA1 and MsPrpE were analyzed by LC-MS/MS. The results confirmed that a peptide with a sequence of SGKIMR (m/z = 419.3) contained a major propionylated lysine residue (Lys906) in MsAcsA1 (Figure 2C), and SGKILR (m/z = 401.3) with a propionylated lysine residue (Lys586) in MsPrpE (Figure 2F). Both lysine residues belong to the typical motif of PXXXXGK, which is recognized by bacterial acetyltransferases to acetylate the lysine residue in AMP-forming Acs (Starai et al., 2002). In the present study, we found that this motif could be recognized by acetyltransferases and subsequently acetylated and propionylated at its last lysine residue.
Western Blot also confirmed that in the presence of acetyl-CoA, MsPrpE and MsAcsA1 could be acetylated (Figures 3A,D). After acetylation, enzymatic activity sharply decreased as evidenced by disappearing colors, similar to the phenomenon observed in the propionylation assay (Figures 3B,E). The acetylation site in MsAcsA1 had been previously identified as SGKIMR (m/z = 419.3) using mass spectrometry (Xu et al., 2011), which was confirmed to be the same site (Lys906) in our propionylation analysis. In addition, LC-MS analysis suggested that the acetylation site (Lys586, SGKILR, m/z = 401.3) of MsPrpE coincided with the propionylation site (Figure 3C). Therefore, propionylation and acetylation could compete for substrates, thus affecting the relative concentration between propionyl-CoA and acetyl-CoA.
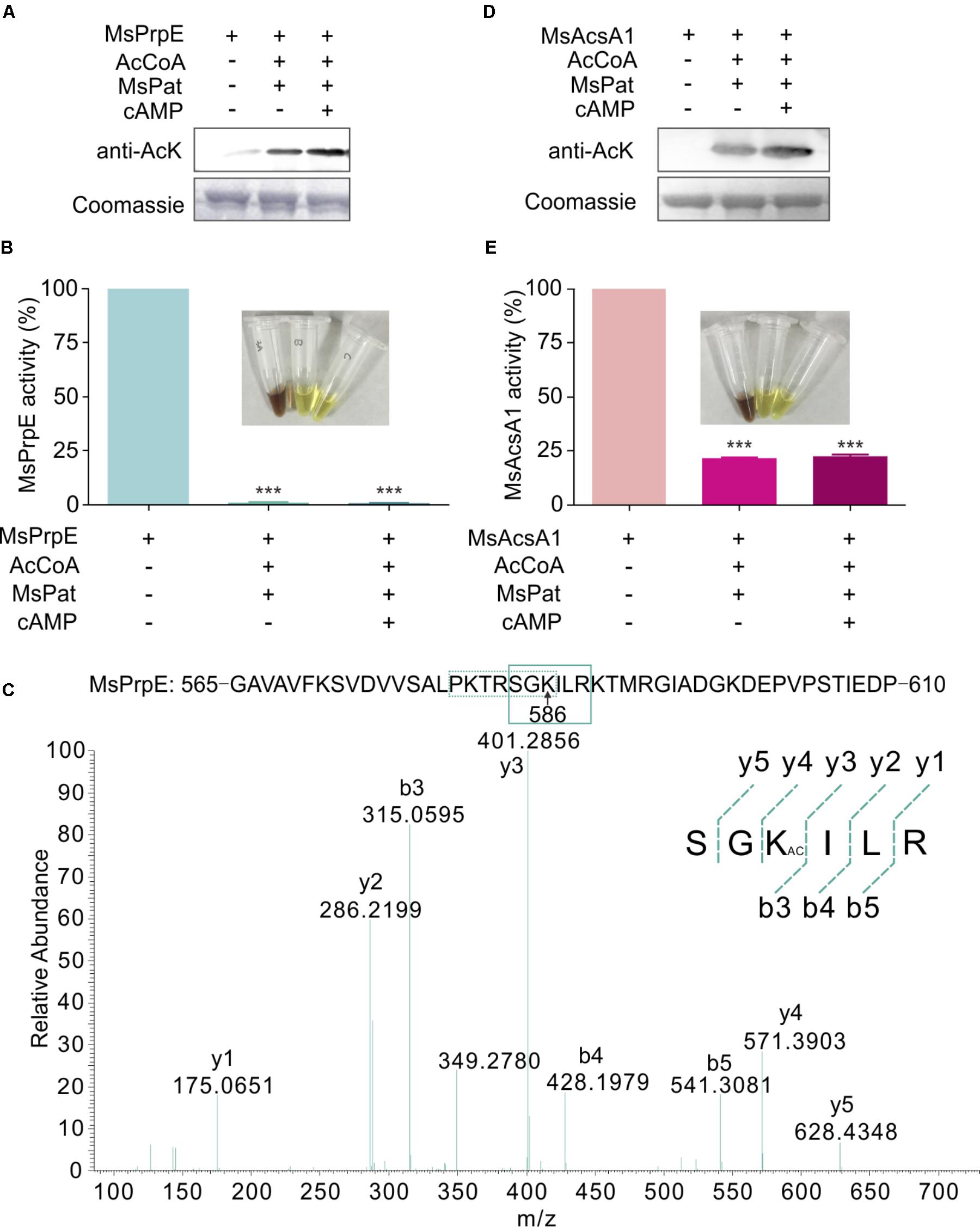
FIGURE 3. The activity of MsPrpE and MsAcsA1 are regulated by acetylation. (A) MsPrpE (2 μM) was incubated independently or with MsPat (2.7 μM), AcCoA (100 μM), and cAMP (1 mM) within a total volume of 100 μL at 37°C for 2 h, followed by SDS-PAGE analysis. The acetylation extent was verified using Western blot assay (Upper). Meanwhile, another PAGE gel was stained with Coomassie brilliant blue as control (Lower). (B) Enzymatic activity of MsPrpE using same condition with (A). (C) LC-MS/MS spectrum of a tryptic peptide (m/z = 401.3) obtained from acetylated MsPrpE. This spectrum matched the peptide sequence (boxed) in MsPrpE, where a mass shift occurred between b3 and y4 ions consistent with acetylation at the lysine residue (amino acid position 586). (D) MsAcsA1 was incubated alone or in the presence of MsPat with the same condition of (A), followed by SDS-PAGE analysis. The acetylation extent was verified using Western blot (Upper). (E) Enzymatic activity of MsAcsA1 using same condition with (A). Error bar represent the standard error, and all experiments replicate three times. ∗∗∗P < 0.001.
GlnR Controls MsPrpE and MsAcs Acylation by Regulating Mspat Transcription
Our previous works have shown that GlnR regulates acetate metabolism and nitrogen metabolism in S. erythraea at transcriptional and post-translational levels (Liao et al., 2015; You et al., 2016, 2017). In this study, a classic motif (a-n6-b) was also identified upstream of Mspat promoter (Figure 4A). Therefore, Mspat could be mediated by GlnR in M. smegmatis. To verify this hypothesis, an EMSA assay was conducted with a gradient of GlnR concentrations. Figure 4A suggests that GlnR strongly and specifically binds to the regulation region of Mspat. As shown previously, the regulation of MsprpE or Msacs by GlnR is activated by nitrogen limitation. Here, we further demonstrate that the expression of glnR, which is closely associated with Mspat regulation, is also influenced by nitrogen availability.
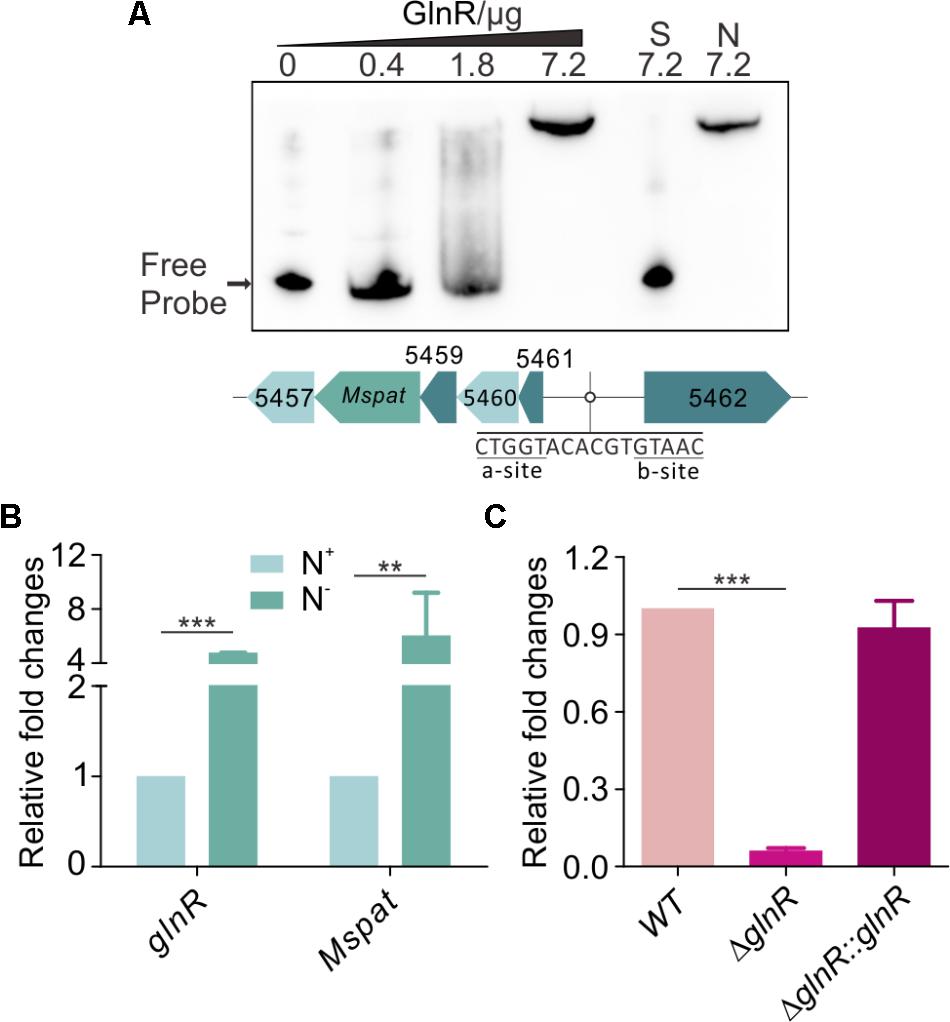
FIGURE 4. GlnR regulate the expression of Mspat in M. smegmatis. (A) EMSA identification of GlnR binding with the promoter region of Mspat. GlnR protein labeled with His tag with different gradient was incubated with probe (5 ng, Mspat). A 200-fold excess of unlabeled specific probe (S) or non-specific competitor DNA (sperm DNA) (N) were conducted as controls. (B) Comparative analysis of Mspat gene in the excess (N+) or limited (N-) nitrogen condition. (C) Differential expression analysis of glnR regulating Mspat. WT (wild type), ΔglnR (glnR knocked out mutant) and ΔglnR::glnR (glnR complementary) were cultured at 30°C to the middle exponential phase. Error bar represent the standard error, and all experiments replicate three times. ∗∗P < 0.01; ∗∗∗P < 0.001.
As shown in Figure 4B, under nitrogen deprived conditions, expression of both Mspat and glnR was up-regulated, which indicates that Mspat could be modulated by GlnR under limited nitrogen conditions. To verify this hypothesis, we conducted a comparative analysis of Mspat in WT, ΔglnR, and ΔglnR::glnR strains (Figure 4C). Expression of Mspat decreased dramatically when glnR was knocked out, and was restored with glnR complementary. Therefore, the Mspat gene was regulated by GlnR in limited nitrogen environments. Considering that MsPat could propionate and acetylate MsPrpE and MsAcs (Figures 2A–F, 3A–E), we speculated that GlnR regulated the acylation of MsPrpE and MsAcs in response to nitrogen availability. The influence of GlnR on acylation level of MsPrpE and MsAcs was estimated by Western blotting using anti-propionated-lysine (anti-PrK) and anti-acetylated-lysine (anti-AcK) antibodies. As shown in Figure 5A, a sharper decrease in the propionylation and acetylation level of MsPrpE and MsAcsA1 was observed in ΔglnR compared to WT strains. The results demonstrated that the acylation of MsPrpE and MsAcsA1 were under the regulation of GlnR. As discussed, the expression of glnR is subjected to nitrogen availability. Western blot results in Figure 5B indicate that MsPrpE and MsAcsA1 could be acylated only under nitrogen deprived conditions, which suggest that the acylation of MsPrpE and MsAcs are under the control of GlnR in response to nitrogen signal in the environment.
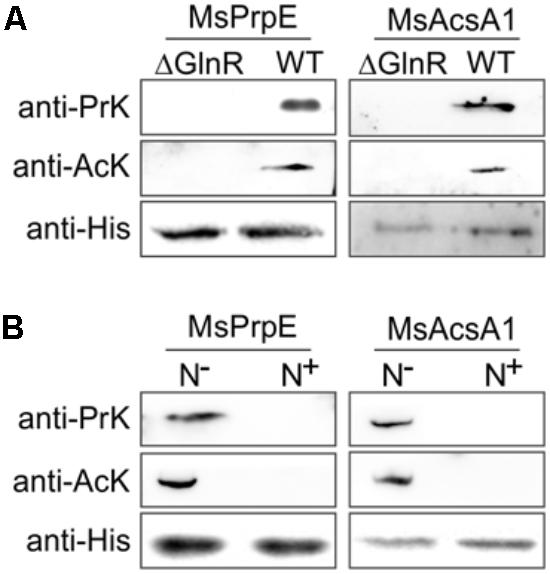
FIGURE 5. GlnR regulates acylation of the enzymes associated with short-chain fatty acid (SCFA) assimilation in M. smegmatis. (A) Equal His-PrpE or His-Acs protein (2 mg) purified from WT and ΔglnR strains of M. smegmatis cultured in LB medium were used to determine the propionylation level of MsPrpE (Upper) or acetylation level of MsAcs enzymes (Middle). Equal non-acylation proteins were used as controls (Lower). (B) Equal His-PrpE or His-Acs protein (1 mg) purified from M. smegmatis WT strain cultured in minimal Sauton medium with excess/limited nitrogen (N+/N-) were used to determine the Propionylation level of MsPrpE (Upper) or acetylation level of MsAcsA1 enzymes (Middle). Equal non-acylation proteins were used as controls (Lower).
Effect of GlnR on the Growth and Viability of M. smegmatis in Macrophages
To investigate whether the regulation from GlnR on MsPrpE and MsAcs influenced nutrient utilization in M. smegmatis, ΔglnR and ΔglnR::glnR strains were further cultured on 10 mM propionate, acetate, cholesterol, and glucose, respectively. When glnR gene was deleted, growth on 10 mM propionate or acetate was higher compared to WT strains (Figures 6A,B), indicating an important role of GlnR in the assimilation of SCFAs in M. smegmatis. On the one hand, GlnR enhanced the transcription of MsprpE and Msacs (Figures 1G,H and Supplementary Figure S1B), as glnR deletion led to decreased enzyme activity of MsPrpE and MsAcs. Moreover GlnR up-regulated Mspat, resulting in increased acylation of MsPrpE and MsAcs. Acylation of these two enzymes decreased their activity, which further inhibited the utilization of nutrients including propionate, acetate in M. smegmatis. This regulatory network impacts the production of pigments (Figures 2B,E, 3B,E). Besides SCFAs, GlnR could also regulate the growth of M. smegmatis in cholesterol (Figure 6C), which is known as a vital carbon source for the survival of M. smegmatis in its host. Nevertheless, utilization of glucose in M. smegmatis was not affected by GlnR (Figure 6D).
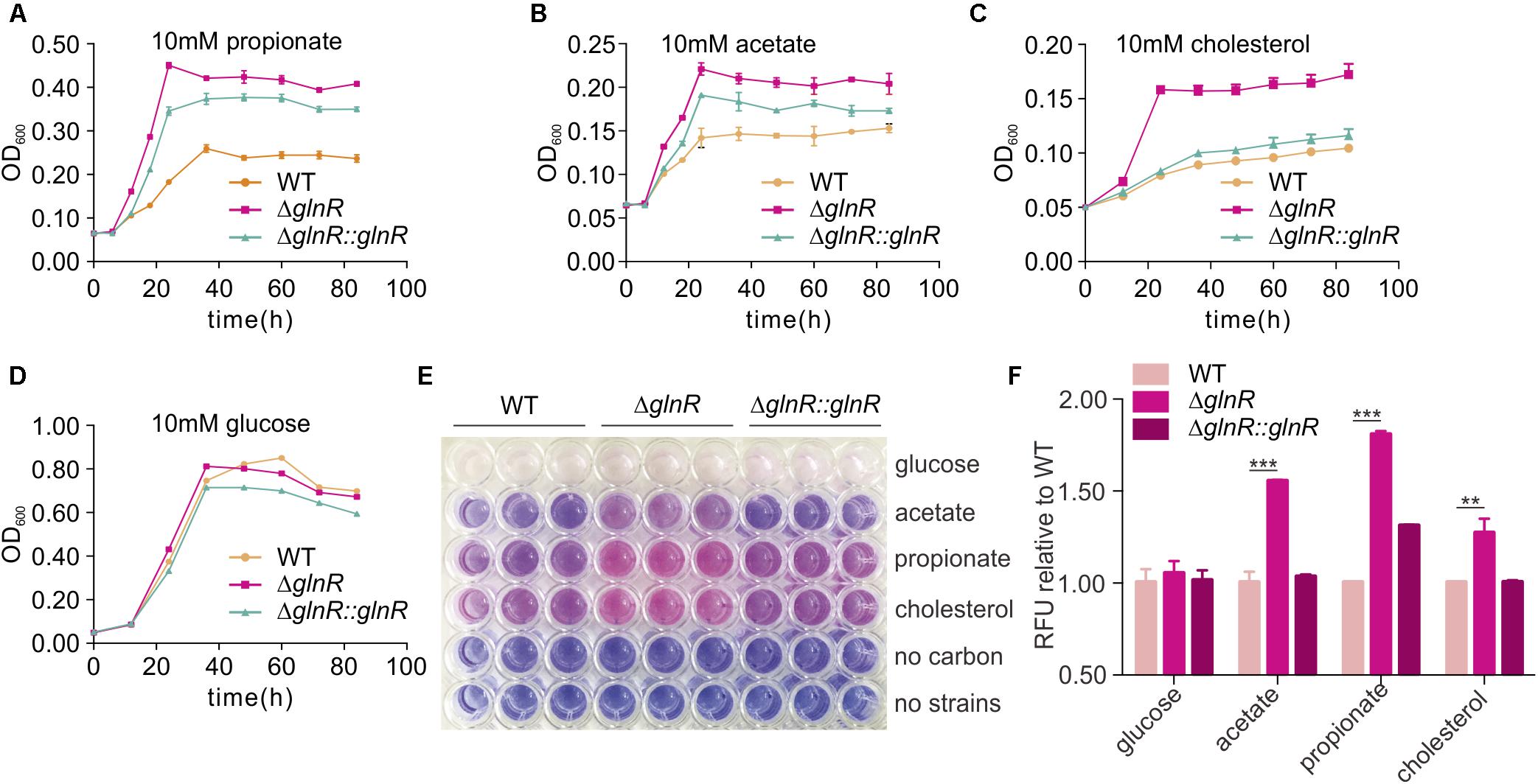
FIGURE 6. glnR gene mutation increase the growth and viability of M. smegmatis. (A–D) growth curves of the M. smegmatis WT, ΔglnR and ΔglnR::glnR strains growing in minimal medium with 10 mM propionate, acetate, cholesterol, or glucose, respectively. (E) the 96-well plate of 10 h incubation after resazurin addition. the metabolically active cells were indicated with pink color, whereas, the blue color represented resazurin reducing less and indicated low metabolic activity. (F) Quantification of resazurin fluorescence for cells growth. The fluorescence value at 10 h post-resazurin addition was normalized to the control of wild-type in different carbon condition. For (A–D) and (F), error bars denote the SD of three replicates. ∗∗P < 0.01; ∗∗∗P < 0.001.
Resazurin reduction assay was conducted to directly display the metabolic defects of M. smegmatis cultured with different SCFAs and cholesterol (Hayden et al., 2013). Resazurin as a blue compound which can be reduced by metabolically active cells. With resazurin reduced, it will be converted to a pink fluorescent substance which can be quantified to indicate the metabolic activity and cell number. For the resazurin test, target strains were cultured in a 96-well plate in propionate, acetate, cholesterol, or glucose-containing media. After 72 h of growth, resazurin was injected into each well and incubated for 10 h before visual examination of color. A significant decrease in the resazurin level was observed in WT, ΔglnR mutant, and complemented ΔglnR::glnR strains, as indicated by the pink color in Figure 6E and quantitated growth in Figures 6A–D. In propionate, acetate, and cholesterol media, the ΔglnR mutant strain reduced resazurin to greater extents compared to WT and ΔglnR::glnR. At the same time, the fluorescence of each strain was reported against that of WT in the same carbon source at 10 h after post-resazurin addition (Figure 6F). M. smegmatis metabolism was quantitated in different SCFAs and cholesterol as regulated by GlnR. In addition, in growth curve and resazurin analysis (Figures 6A,C,F), we found that the growth of ΔglnR::glnR strain is similar to WT in cholesterol as carbon, while it is different for ΔglnR::glnR strain growing in propionate. These results indicate that the amount of GlnR produced in complemented strain could be affected by different carbon. Phenotypic data demonstrate that GlnR responded to nitrogen level in the environment to regulate nutrient assimilation and utilization, which affected the viability and invasion of M. smegmatis in macrophage.
Discussion
Carbon and nitrogen are crucial nutrients for Mtb survival. Mtb harnesses energy from the oxidation of various carbon sources (e.g., glucose, glycerol) to pyruvate and ultimately CO2 in central metabolic pathways. In macrophages, alternative carbon sources from the host, e.g., fatty acids and cholesterol, are utilized during Mtb infection (Schnappinger et al., 2003; Muñoz-Elías et al., 2005, 2006; Munoz-Elias and McKinney, 2005, 2006; Pandey and Sassetti, 2008). In this process, SCFAs such as propionate and acetate are critical intermediate metabolites (Starai et al., 2003). All catabolic pathways for propionate and acetate require activation of SCFAs into corresponding SCFAcyl-CoA forms before they can be converted and enter central metabolism to synthesize lipids that are essential for the survival and virulence of Mycobacteria, e.g., PDIM and mycolic acid (Figure 7). Nitrogen is another vital energy source, which exists as proteins, nucleic acids, and cell wall components. In bacteria, carbon and nitrogen are controlled by α-ketoglutarate (αKG), which represses enzyme I of the phosphotransferase system and in turn regulates the rate of glucose uptake in accordance with nitrogen level (Shimizu, 2013). In this study, it is found that the central nitrogen regulator, GlnR, can sense nitrogen shortages and up-regulate MsprpE and Msacs accordingly to increase the assimilation of SCFAs. GlnR can also up-regulate the expression of Mspat in nitrogen deprived environments and increase the acylation of MsPrpE and MsAcs to decrease the assimilation of SCFAs. On the one hand, under nitrogen limited conditions, GlnR activates MsAcs/MsPrpE to facilitate the assimilation of SCFAs to generate acetyl-CoA/propionyl-CoA and replenish the pool of acyl-CoA. The GlnR-mediated activation of acetyl-CoA synthesis under nitrogen-limited conditions leads to an increase of the intracellular concentration of nitrogen acceptor molecules (αKG and oxaloacetate), thus allowing for efficient assimilation of ammonium (You et al., 2017). Note this process is only in response to nitrogen starvation. On the other hand, the activation of MsPat leads to acylation of MsAcs/MsPrpE with acetyl-CoA/propionyl-CoA as a donor at the PTM level. This regulation is not only related to nitrogen signal, but also the level of intracellular acetyl-CoA/propionyl-CoA. If the concentration of intracellular acyl-CoA is high, PTM will be activated to inhibit the activity of MsAcs/MsPrpE and prevent further accumulation of acetyl-CoA/propionyl-CoA. In contrast, low concentration of acyl-CoA inhibits the acylation of MsAcs/MsPrpE, which remains active in SCFAs assimilation. Consequently, GlnR responds to nitrogen limitation to regulate SCFAs assimilation, whereas abundant nitrogen inactivates GlnR and inhibits SCFA uptake through TPM and reduced consumption of acyl-CoA through the acylation of MsAcs/MsPrpE (Figure 7).
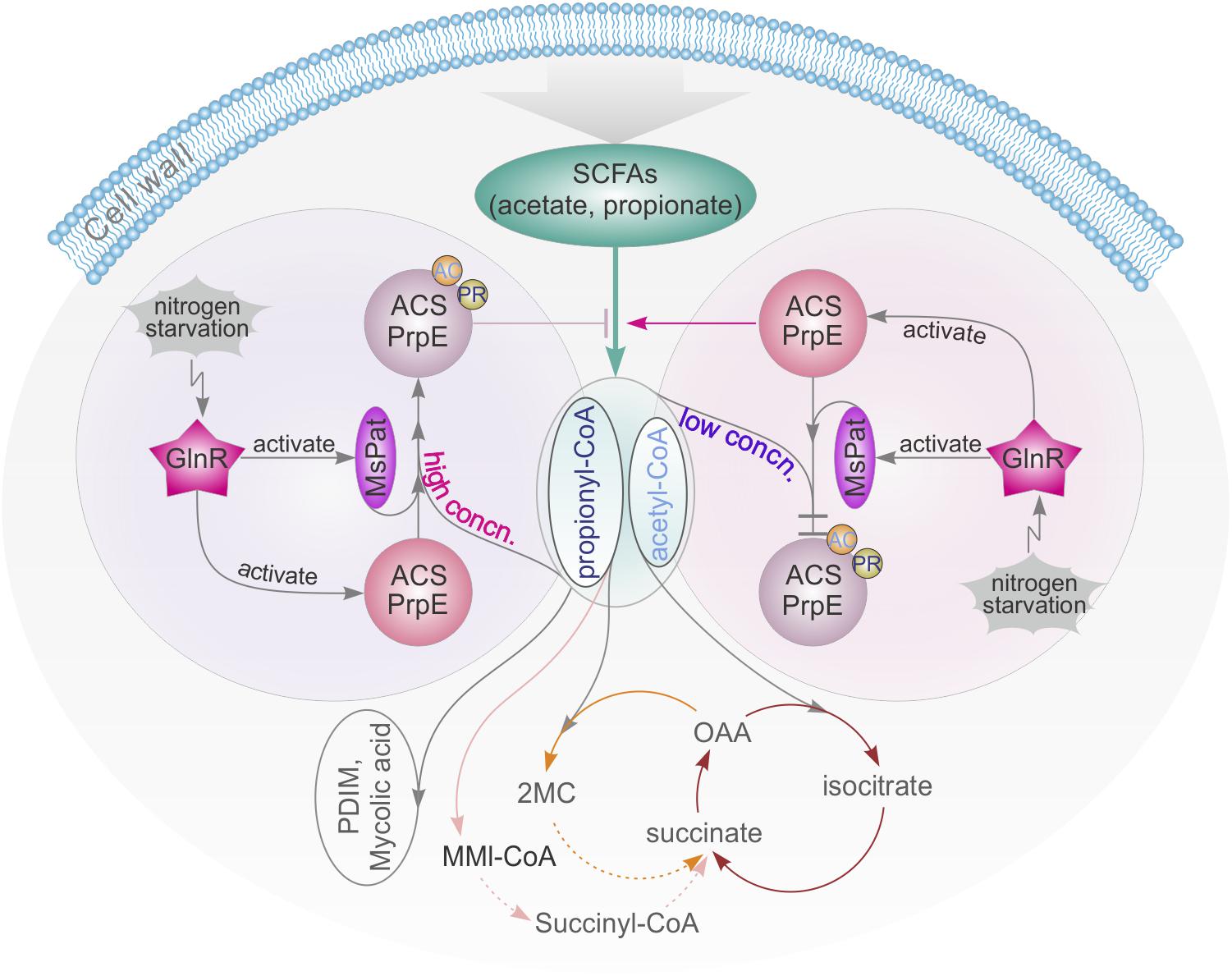
FIGURE 7. GlnR-mediated regulation of SCFAs assimilation revealed a tight connection of carbon and nitrogen metabolism in M. smegmatis. high concn., high concentration; low concn., low concentration; cycle with brown line: TCA cycle; cycle with yellow line: Methylcitrate cycle; cycle with pink line: Methylmalonyl CoA pathway; OAA, oxaloacetate; 2MC, 2 methylcitrate; MMI-CoA, Methylmalonyl-CoA; PDIM, phthiocerol dimycocerosate.
Previous research has revealed that the metabolisms of carbon and nitrogen are interconnected (Fisher and Sonenshein, 1991; Wacker et al., 2003; Choi and Saier, 2005). The regulatory network of GlnR in different bacteria from the actinomycete group has also been extensively studied (Burkovski, 2007; Tiffert et al., 2011; Liao et al., 2015; You et al., 2017). In these bacteria, GlnR is activated at the nitrogen starvation status to regulate its target genes. The same mechanism has also been found in M. smegmatis, in which GlnR up-regulated amtB, amt1, glnA, and nirBD in response to nitrogen limitation (Jenkins et al., 2013; Gouzy et al., 2014). In this study, we extended the regulatory network of GlnR to assimilation of SCFAs in Mycobacteria, and demonstrated that GlnR acts as a bridge in a complex cross-talk regulatory network to maintain the homeostasis of carbon and nitrogen metabolisms (Figure 7).
The inability of many bacteria, including Mycobacteria, to assimilate propionyl-CoA inhibits their growth even in the presence of other carbon sources. This could explain why accumulation of intermediates often blocks other key pathways in these bacteria (Griffin et al., 2012): the accumulation of propionate or propionyl-CoA is toxic, thus needs to be controlled at a suitable level for detoxification. Previous work has shown that detoxification of propionyl-CoA relies on the activity of the methylcitrate cycle or the methylmalonyl pathway, or incorporation of propionyl-CoA into methyl-branched lipids in the cell wall (Lee et al., 2013). In addition, propionyl-CoA can also be used as a donor of propionyl groups for protein modification, which also provides an effective “sink” for excess propionyl-CoA (Singhal et al., 2015). In this study, we found that GlnR can further regulate Mspat to influence propionylation to control the amount of propionyl-CoA. In addition, MsAcsA1 and MsPrpE share the same acylation sites, which affects the balance between propionyl-CoA and acetyl-CoA, thus impacts propionyl-CoA removal. This result provides a new perspective in understanding the detoxification process in Mtb, which may benefit control of this pathogen in humans in the long run.
Besides SCFAs, Mtb mainly utilizes cholesterol as another carbon source from the host for survival during infection (Pandey and Sassetti, 2008). After internalizing into macrophages, M. smegmatis must sustain attacks from phagosomes (Anes et al., 2006), which makes assimilation and utilization of nutrients in macrophages vital for its survival. Here, it is found that glnR deletion (ΔglnR) can significantly increase the viability of M. smegmatis using cholesterol as the sole carbon source, though the roles of GlnR in cholesterol regulation remain unclear.
So far, the activation mechanism of GlnR remains unknown (Jenkins et al., 2013). Here, we found that there are different growth status for ΔglnR::glnR strain growing in cholesterol or propionate. These results indicate that the activation and production of GlnR in complemented strain may be impacted by different carbon due to glnR inserted in M. smegmatis genome as a locus other than it in WT.
As a model system of Mycobacterium, M. smegmatis shares many characteristics with its pathogenic relatives, including basic cellular processes and carbon metabolism features, which are complemented with additional pathways adapted to pathogenicity (Rastogi et al., 2001; Vissa et al., 2009; Chopra et al., 2014). We have found GlnR could directly bind with the regulatory region of Mtbacs (Rv3667) and Mtbpat (Rv0998) in Mtb (Supplementary Figures S2A,B). Therefore, GlnR could regulate Mtbpat and Mtbacs to influence the pathways of SCFAs assimilation in Mtb, in a similar mechanism to that found in M. smegmatis.
Conclusion
We have established that the GlnR-mediated regulation of SCFA assimilation and cholesterol utilization in Mycobacteria is in response to nitrogen signal within the host. These findings could provide a deeper understanding of Mtb interaction with its host and its pathogenicity mechanisms for devising more effective control strategies of the pathogen.
Author Contributions
X-XL, W-BL, and B-CY designed the research. X-XL and M-JS performed the experiments. All authors contributed to new reagents and analytic tools, analyzed data, and wrote the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31730004, 21335003, and 21575089) and Natural Science Foundation of Shanghai (14ZR1409600).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01311/full#supplementary-material
FIGURE S1 | GlnR direct regulated Msacs genes. (A) EMSA identification of GlnR binding with the promoter region of MsacsA2, MsacsA3, and MsacsA4. (B) Differential expression analysis of glnR regulating MsacsA2, MsacsA3, and MsacsA4. (C) Comparative analysis of MsacsA2, MsacsA3, and MsacsA4 gene in the excess (N+) or limited (N-) nitrogen condition.
FIGURE S2 | GlnR (Rv0818) directly bind with the regulatory region of Mtbacs (Rv3667) (A) and Mtbpat (Rv0998) (B) in Mtb.
References
Amon, J., Bräu, T., Grimrath, A., Hänssler, E., Hasselt, K., Höller, M., et al. (2008). Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-type regulator GlnR. J. Bacteriol. 190, 7108–7116. doi: 10.1128/JB.00855-08
Amon, J., Titgemeyer, F., and Burkovski, A. (2009). A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 17, 20–29. doi: 10.1159/000159195
Anes, E., Peyron, P., Staali, L., Jordao, L., Gutierrez, M. G., Kress, H., et al. (2006). Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell. Microbiol. 8, 939–960. doi: 10.1111/j.1462-5822.2005.00675.x
Bernal, V., Castano-Cerezo, S., and Canovas, M. (2016). Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl. Microbiol. Biotechnol. 100, 8985–9001. doi: 10.1007/s00253-016-7832-x
Bloch, H., and Segal, W. (1956). Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72, 132–141.
Burkovski, A. (2007). Nitrogen control in Corynebacterium glutamicum: proteins, mechanisms, signals. J. Microbiol. Biotechnol. 17, 187–194.
Carter, M. S., and Alber, B. E. (2015). Transcriptional regulation by the short-chain fatty acyl coenzyme A regulator (ScfR) PccR controls propionyl coenzyme A assimilation by Rhodobacter sphaeroides. J. Bacteriol. 197, 3048–3056. doi: 10.1128/JB.00402-15
Castaño-Cerezo, S., Bernal, V., Blanco-Catalá, J., Iborra, J. L., and Cánovas, M. (2011). cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 82, 1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x
Chang, J. C., Harik, N. S., Liao, R. P., and Sherman, D. R. (2007). Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. J. Infect. Dis. 196, 788–795. doi: 10.1086/520089
Choi, S. K., and Saier, M. H. Jr. (2005). Regulation of sigL expression by the catabolite control protein CcpA involves a roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism. J. Bacteriol. 187, 6856–6861. doi: 10.1128/JB.187.19.6856-6861.2005
Chopra, T., Hamelin, R., Armand, F., Chiappe, D., Moniatte, M., and McKinney, J. D. (2014). Quantitative mass spectrometry reveals plasticity of metabolic networks in Mycobacterium smegmatis. Mol. Cell. Proteomics 13, 3014–3028. doi: 10.1074/mcp.M113.034082
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Fisher, S. H., and Sonenshein, A. L. (1991). Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu. Rev. Microbiol. 45, 107–135. doi: 10.1146/annurev.mi.45.100191.000543
Gardner, J. G., Grundy, F. J., Henkin, T. M., and Escalante-Semerena, J. C. (2006). Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis. J. Bacteriol. 188, 5460–5468. doi: 10.1128/JB.00215-06
Garrity, J., Gardner, J. G., Hawse, W., Wolberger, C., and Escalante-Semerena, J. C. (2007). N-lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282, 30239–30245. doi: 10.1074/jbc.M704409200
Gouzy, A., Poquet, Y., and Neyrolles, O. (2014). Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 12, 729–737. doi: 10.1038/nrmicro3349
Griffin, J. E., Pandey, A. K., Gilmore, S. A., Mizrahi, V., McKinney, J. D., Bertozzi, C. R., et al. (2012). Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19, 218–227. doi: 10.1016/j.chembiol.2011.12.016
Hayden, J. D., Brown, L. R., Gunawardena, H. P., Perkowski, E. F., Chen, X., and Braunstein, M. (2013). Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology 159(Pt 9), 1986–1999. doi: 10.1099/mic.0.068585-0
Horswill, A. R., and Escalante-Semerena, J. C. (1999). The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145(Pt 6), 1381–1388. doi: 10.1099/13500872-145-6-1381
Hu, Y., van der Geize, R., Besra, G. S., Gurcha, S. S., Liu, A., Rohde, M., et al. (2010). 3-Ketosteroid 9alpha-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 75, 107–121. doi: 10.1111/j.1365-2958.2009.06957.x
Jenkins, V. A., Barton, G. R., Robertson, B. D., and Williams, K. J. (2013). Genome wide analysis of the complete GlnR nitrogen-response regulon in Mycobacterium smegmatis. BMC Genomics 14:301. doi: 10.1186/1471-2164-14-301
Jenkins, V. A., Robertson, B. D., and Williams, K. J. (2012). Aspartate D48 is essential for the GlnR-mediated transcriptional response to nitrogen limitation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 330, 38–45. doi: 10.1111/j.1574-6968.2012.02530.x
Kang, Z., Wang, X., Li, Y., Wang, Q., and Qi, Q. (2012). Small RNA RyhB as a potential tool used for metabolic engineering in Escherichia coli. Biotechnol. Lett. 34, 527–531. doi: 10.1007/s10529-011-0794-2
Kumari, S., Beatty, C. M., Browning, D. F., Busby, S. J., Simel, E. J., Hovel-Miner, G., et al. (2000). Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182, 4173–4179. doi: 10.1128/JB.182.15.4173-4179.2000
Lee, W., VanderVen, B. C., Fahey, R. J., and Russell, D. G. (2013). Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 288, 6788–6800. doi: 10.1074/jbc.M112.445056
Liao, C. H., Yao, L., Xu, Y., Liu, W. B., Zhou, Y., and Ye, B. C. (2015). Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 112, 15630–15635. doi: 10.1073/pnas.1508465112
Liao, C. H., Yao, L. L., and Ye, B. C. (2014). Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators GlnR, DasR, and CRP. Mol. Microbiol. 94, 1065–1084. doi: 10.1111/mmi.12818
Liu, F., Gu, J., Wang, X., Zhang, X. E., and Deng, J. (2014). Acs is essential for propionate utilization in Escherichia coli. Biochem. Biophys. Res. Commun. 449, 272–277. doi: 10.1016/j.bbrc.2014.05.015
Liu, X. X., Liu, W. B., and Ye, B. C. (2016). Regulation of a protein acetyltransferase in Myxococcus xanthus by the coenzyme NADP. J. Bacteriol. 198, 623–632. doi: 10.1128/JB.00661-15
Marrero, J., Rhee, K. Y., Schnappinger, D., Pethe, K., and Ehrt, S. (2010). Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824. doi: 10.1073/pnas.1000715107
Masiewicz, P., Wolañski, M., Brzostek, A., Dziadek, J., and Zakrzewska-Czerwiñska, J. (2014). Propionate represses the dnaA gene via the methylcitrate pathway-regulating transcription factor, PrpR, in Mycobacterium tuberculosis. Antonie Van Leeuwenhoek 105, 951–959. doi: 10.1007/s10482-014-0153-0
McKinney, J. D., Höner zu Bentrup, K., Muñoz-Elías, E. J., Miczak, A., Chen, B., Chan, W. T., et al. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. doi: 10.1038/35021074
Munoz-Elias, E. J., and McKinney, J. D. (2005). Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644. doi: 10.1038/nm1252
Munoz-Elias, E. J., and McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22. doi: 10.1111/j.1462-5822.2005.00648.x
Muñoz-Elías, E. J., Timm, J., Botha, T., Chan, W. T., Gomez, J. E., and McKinney, J. D. (2005). Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect. Immun. 73, 546–551. doi: 10.1128/IAI.73.1.546-551.2005
Muñoz-Elías, E. J., Upton, A. M., Cherian, J., and McKinney, J. D. (2006). Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x
Nambi, S., Gupta, K., Bhattacharyya, M., Ramakrishnan, P., Ravikumar, V., Siddiqui, N., et al. (2013). Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J. Biol. Chem. 288, 14114–14124. doi: 10.1074/jbc.M113.463992
Nazarova, E. V., Montague, C. R., La, T., Wilburn, K. M., Sukumar, N., Lee, W., et al. (2017). Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. eLife 6:e26969. doi: 10.7554/eLife.26969
Nesbitt, N. M., Yang, X., Fontán, P., Kolesnikova, I., Smith, I., Sampson, N. S., et al. (2010). A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 78, 275–282. doi: 10.1128/IAI.00893-09
Noy, T., Xu, H., and Blanchard, J. S. (2014). Acetylation of acetyl-CoA synthetase from Mycobacterium tuberculosis leads to specific inactivation of the adenylation reaction. Arch. Biochem. Biophys. 55, 42–49. doi: 10.1016/j.abb.2014.04.004
Okanishi, H., Kim, K., Masui, R., and Kuramitsu, S. (2014). Lysine propionylation is a prevalent post-translational modification in Thermus thermophilus. Mol. Cell. Proteomics 13, 2382–2398. doi: 10.1074/mcp.M113.035659
Palacios, S., and Escalante-Semerena, J. C. (2000). prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182, 905–910. doi: 10.1128/JB.182.4.905-910.2000
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Rajashekhara, E., and Watanabe, K. (2004). Propionyl-coenzyme A synthetases of Ralstonia solanacearum and Salmonella choleraesuis display atypical kinetics. FEBS Lett. 556, 143–147. doi: 10.1016/S0014-5793(03)01394-2
Rastogi, N., Legrand, E., and Sola, C. (2001). The mycobacteria: an introduction to nomenclature and pathogenesis. Rev. Sci. Tech. 20, 21–54. doi: 10.20506/rst.20.1.1265
Savvi, S., Warner, D. F., Kana, B. D., McKinney, J. D., Mizrahi, V., and Dawes, S. S. (2008). Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 190, 3886–3895. doi: 10.1128/JB.01767-07
Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., et al. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704. doi: 10.1084/jem.20030846
Shimizu, K. (2013). Regulation systems of bacteria such as Escherichia coli in response to nutrient limitation and environmental stresses. Metabolites 4, 1–35. doi: 10.3390/metabo4010001
Singhal, A., Arora, G., Virmani, R., Kundu, P., Khanna, T., Sajid, A., et al. (2015). Systematic analysis of mycobacterial acylation reveals first example of acylation-mediated regulation of enzyme activity of a bacterial phosphatase. J. Biol. Chem. 290, 26218–26234. doi: 10.1074/jbc.M115.687269
Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., and Escalante-Semerena, J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392. doi: 10.1126/science.1077650
Starai, V. J., and Escalante-Semerena, J. C. (2004). Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61, 2020–2030. doi: 10.1007/s00018-004-3448-x
Starai, V. J., Takahashi, H., Boeke, J. D., and Escalante-Semerena, J. C. (2003). Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163, 545–555.
Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., et al. (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. doi: 10.1038/351456a0
Tiffert, Y., Franz-Wachtel, M., Fladerer, C., Nordheim, A., Reuther, J., Wohlleben, W., et al. (2011). Proteomic analysis of the GlnR-mediated response to nitrogen limitation in Streptomyces coelicolor M145. Appl. Microbiol. Biotechnol. 89, 1149–1159. doi: 10.1007/s00253-011-3086-9
Van der Geize, R., Yam, K., Heuser, T., Wilbrink, M. H., Hara, H., Anderton, M. C., et al. (2007). A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952. doi: 10.1073/pnas.0605728104
VanderVen, B. C., Fahey, R. J., Lee, W., Liu, Y., Abramovitch, R. B., Memmott, C., et al. (2015). Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog. 11:e1004679. doi: 10.1371/journal.ppat.1004679
Verdin, E., and Ott, M. (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264. doi: 10.1038/nrm3931
Vissa, V. D., Sakamuri, R. M., Li, W., and Brennan, P. J. (2009). Defining mycobacteria: shared and specific genome features for different lifestyles. Indian J. Microbiol. 49, 11–47. doi: 10.1007/s12088-009-0006-0
Wacker, I., Ludwig, H., Reif, I., Blencke, H. M., Detsch, C., and Stülke, J. (2003). The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149(Pt 10), 3001–3009. doi: 10.1099/mic.0.26479-0
Warner, J. B., and Lolkema, J. S. (2003). CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67, 475–490. doi: 10.1128/MMBR.67.4.475-490.2003
Wipperman, M. F., Sampson, N. S., and Thomas, S. T. (2014). Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. Biol. 49, 269–293. doi: 10.3109/10409238.2014.895700
Wolfe, A. J. (2005). The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50. doi: 10.1128/MMBR.69.1.12-50.2005
Xiong, Y., and Guan, K. L. (2012). Mechanistic insights into the regulation of metabolic enzymes by acetylation. J. Cell Biol. 198, 155–164. doi: 10.1083/jcb.201202056
Xu, H., Hegde, S. S., and Blanchard, J. S. (2011). Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 50, 5883–5892. doi: 10.1021/bi200156t
Xu, J. Y., You, D., Leng, P. Q., and Ye, B. C. (2014). Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J. Biol. Chem. 289, 27034–27045. doi: 10.1074/jbc.M114.579078
Yam, K. C., D’Angelo, I., Kalscheuer, R., Zhu, H., Wang, J. X., Snieckus, V., et al. (2009). Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 5:e1000344. doi: 10.1371/journal.ppat.1000344
Yang, J. E., Choi, Y. J., Lee, S. J., Kang, K. H., Lee, H., Oh, Y. H., et al. (2014). Metabolic engineering of Escherichia coli for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from glucose. Appl. Microbiol. Biotechnol. 98, 95–104. doi: 10.1007/s00253-013-5285-z
Yang, X., Nesbitt, N. M., Dubnau, E., Smith, I., and Sampson, N. S. (2009). Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry 48, 3819–3821. doi: 10.1021/bi9005418
You, D., Wang, M. M., and Ye, B. C. (2017). Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 103, 845–859. doi: 10.1111/mmi.13595
Keywords: propionyl-CoA/acetyl-CoA synthetase, GlnR, short-chain fatty acid, nitrogen metabolism, post-translational modification, acylation
Citation: Liu X-X, Shen M-J, Liu W-B and Ye B-C (2018) GlnR-Mediated Regulation of Short-Chain Fatty Acid Assimilation in Mycobacterium smegmatis. Front. Microbiol. 9:1311. doi: 10.3389/fmicb.2018.01311
Received: 28 March 2018; Accepted: 29 May 2018;
Published: 22 June 2018.
Edited by:
Jörg Stülke, Georg-August-Universität Göttingen, GermanyReviewed by:
Harold J. Schreier, University of Maryland, Baltimore, United StatesAirat R. Kayumov, Kazan Federal University, Russia
Copyright © 2018 Liu, Shen, Liu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Bing Liu, bHdiQGVjdXN0LmVkdS5jbg== Bang-Ce Ye, YmN5ZUBlY3VzdC5lZHUuY24=
†These authors have contributed equally to this work.
 Xin-Xin Liu
Xin-Xin Liu Meng-Jia Shen
Meng-Jia Shen Wei-Bing Liu
Wei-Bing Liu Bang-Ce Ye
Bang-Ce Ye

