- Guangdong Provincial Key Laboratory of Agro-Animal Genomics and Molecular Breeding, College of Animal Science, South China Agricultural University, Guangzhou, China
Endogenous retroviruses (ERVs) are retroviral sequences that remain fixed in the host genome, where they could play an important role. Some ERVs have been identified in insects and proven to have infectious properties. However, no information is available regarding Bombyx mori ERVs (BmERVs) to date. Here, we systematically identified 256 potential BmERVs in the silkworm genome via a whole-genome approach. BmERVs were relatively evenly distributed across each of the chromosomes and accounted for about 25% of the silkworm genome. All BmERVs were classified as young ERVs, with insertion times estimated to be less than 10 million years. Seven BmERVs possessing the env genes were identified. With the exception of the Orf133 Helicoverpa armigera nuclear polyhedrosis virus, the env sequences of BmERVs were distantly related to genes encoding F (Fa and Fb) and GP64 proteins from Group I and Group II NPVs. In addition, only the amino acid sequence of the BmERV-21 envelope protein shared a similar putative furin-like cleavage site and fusion peptide with Group II baculoviruses. All of the env genes in the seven BmERVs were verified to exist in the genome and be expressed in the midgut and fat bodies, which suggest that BmERVs might play an important role in the host biology.
Introduction
Endogenous retroviruses (ERVs) are remnants of ancient retroviral sequences that were once integrated into a host germ line and have remain fixed in the host genome for generations. Their genomic structure is composed of a central part with one to three major genes (gag, pol, and env) flanked by two long terminal repeats (LTRs) (Misseri et al., 2004). They could show infectious properties when possessing the env gene (Teysset et al., 1998). ERVs have been identified in the genomes of many vertebrate and invertebrate species (Terzian et al., 2001; Garcia-Etxebarria and Jugo, 2010, 2016; Bustamante Rivera et al., 2017). Due to structural similarity, ERVs can also be considered to be LTR retrotransposons (Havecker et al., 2004). However, the International Committee on Taxonomy of Viruses includes vertebrate ERVs in the family Retroviridae, while insect ERVs (IERVs) belong to the family Metaviridae (Fablet, 2014).
Classical vertebrate ERVs, such as ERV3 (human) (Bustamante Rivera et al., 2017), and ev21 and EAV-HP (chicken) (Wang et al., 2013; Feng et al., 2016), might play an important role in their hosts. Thus, there is continued interest in ERVs for the roles they could play in normal host biology and diseases, as well as the risks they pose in xenotransplantation (Chuong et al., 2016; Niu et al., 2017). Compared with vertebrate ERVs, IERVs have also been described for a long time but have received less attention. IERVs that have been identified include gypsy, 17-6, 297, ZAM, Idefix, nomad, and tirant in Drosophila melanogaster; Tv1 in Drosophila virilis; tom in Drosophila ananassae; TED in Trichoplusia ni; and yoyo in Ceratitis capitata (Terzian et al., 2001). Of these, both gypsy and ZAM have been found to have infectious properties (Kim et al., 1994; Leblanc et al., 2000). In addition, maternal transmission of gypsy may be influenced in the presence of the endosymbiotic bacterium Wolbachia (Touret et al., 2014). However, the role of IERVs in the host’s biological processes remains an interesting and open question.
Despite the prevalence of ERVs in insect genomes, to date, ERVs in silkworm have not been exploited. It has been found that lepidopteran insects diversified during the Early Cretaceous (Misof et al., 2014). The domesticated silkworm, Bombyx mori, is a lepidopteron model insect of economic importance. Initially, only several LTR retrotransposon elements were identified in B. mori (Abe et al., 2001), but it was later found that retrotransposable elements are the main structural component of the W chromosome (Abe et al., 2005). With the release of the silkworm genome sequence (Xia et al., 2004), LTR retrotransposons of domesticated silkworm were then systematically screened (Jin-Shan et al., 2005). However, thus far, B. mori ERVs (BmERVs) have not been thoroughly identified in the whole-genome sequence.
Previous research has reported that IERVs could become infectious agents via acquiring envelope-like gene from a class of insect baculoviruses (Malik et al., 2000). Nuclear polyhedron viruses (NPVs), as members of the Baculoviridae, have been mainly divided into Group I and Group II according to their different envelope gene (Zanotto et al., 1993). In the family Baculoviridae, two different envelope proteins named GP64 and F have been identified. NPVs in Group I use GP64 as their budded virus fusion protein, whereas NPVs in Group II lack GP64 and utilize F protein (Pearson and Rohrmann, 2002). The homologs of the F protein in Group I baculoviruses and F protein in Group II baculoviruses are, respectively, called Fb and Fa (Pearson and Rohrmann, 2002). However, the relationship of envelope proteins between BmERV and NPVs is still unclear.
In the present study, we took advantage of the newest silkworm genome sequence (from SilkDB V2.0) to perform a comprehensive mining and analysis of ERVs in B. mori. To our knowledge, this is the first systematic screening of ERVs in insect. The aim of this study was to carry out a detailed analysis of the BmERVs, extending our knowledge of these elements in invertebrates.
Materials and Methods
Identification and Annotation of BmERVs
The silkworm genome sequence was downloaded from SilkDB V2.01 and used as the input for BmERV identification with two pipelines (Figure 1). In the first pipeline, BmERVs were detected in the silkworm genome using the software LTRharvest (GenomeTools1.5.7) (Ellinghaus et al., 2008) and LTRdigest (Steinbiss et al., 2009). Pairs of putative LTRs that were separated by 1–15 kb and flanked by target site duplications were screened by LTRharvest. The threshold of LTR nucleotide similarity used in LTRharvest was greater than 80%; other parameters were set to their defaults. Internal sequence retroviral features of ERV candidates, including protein domains, polypurine tracts, and primer-binding sites, were predicted using LTRdigest with default parameters. A total of 314 retroviral protein domain profiles used for putative ERV domain annotation were downloaded from the Gypsy database.2 A second pipeline, MGEScan-LTR, was also employed to identify BmERVs using the default parameters. Each candidate containing at least three of the five canonical retroviral protein domains [Gag, PR, reverse transcriptase (RT), IN, and RH] was retained in the results from LTRharvest and MGEScan-LTR. Finally, the ERV candidates identified by the two pipelines were merged as the BmERV library.
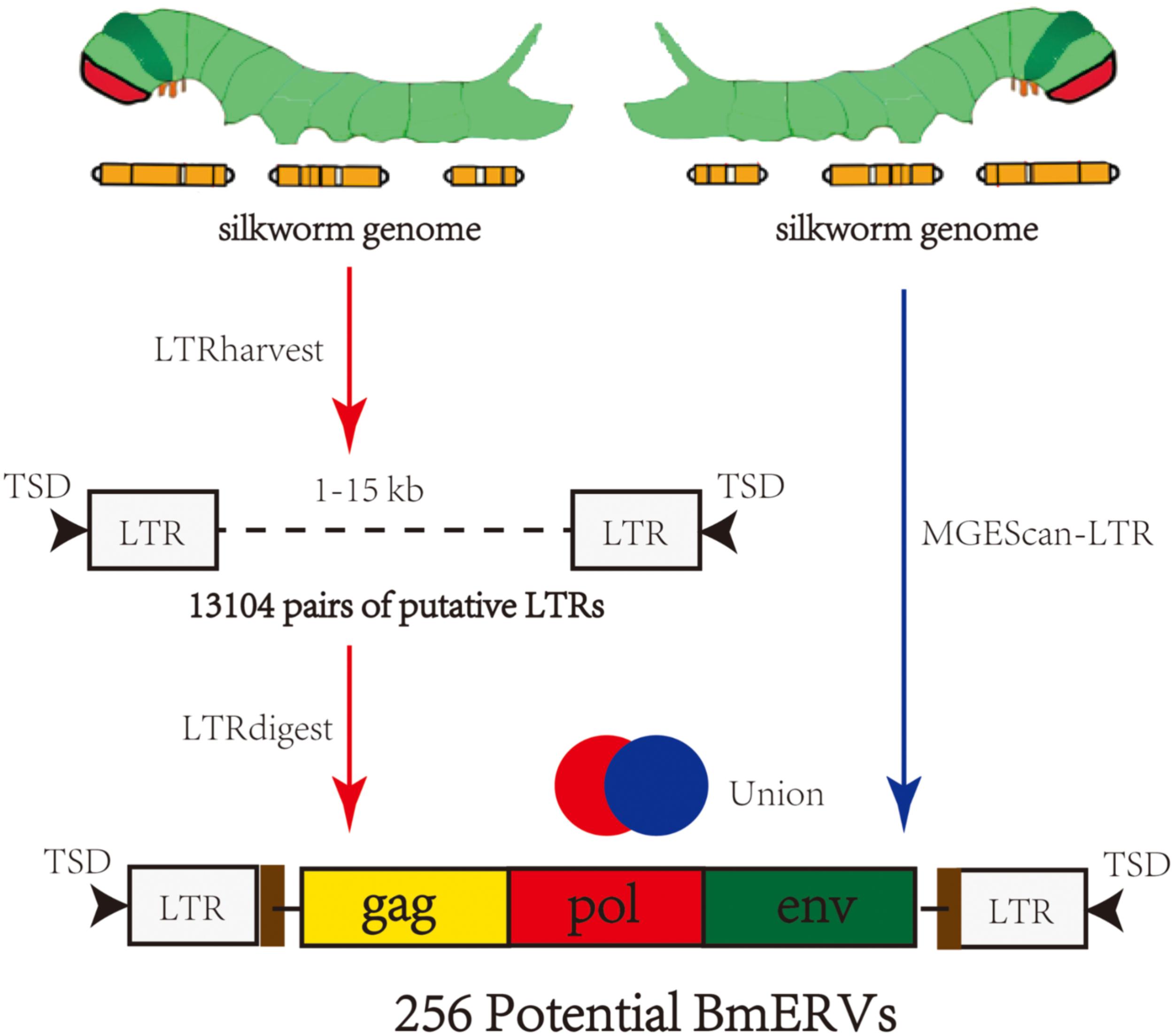
FIGURE 1. Summary of the strategy used to identify BmERVs. A total of 13,104 pairs of putative LTRs were identified by LTRharvest, and all of them were annotated using LTRdigest. A second pipeline, MGEScan-LTR, was employed to identify BmERVs with the default parameters. Each candidate containing at least three of the five canonical retroviral protein domains (Gag, PR, RT, IN, and RH) was extracted from the results of LTRharvest and MGEScan-LTR as potential BmERVs. Finally, we identified 256 potential BmERVs by merging the ERV candidates identified by the independent pipelines.
BmERV Classification and Phylogenetic Analysis
Sequences of the RT domain from BmERVs and known exogenous retroviruses and ERVs (Supplementary Table S1) were used for multiple alignment using MUSCLE (v3.8.31) (Edgar, 2004). A neighbor-joining phylogeny was built from the RT domain alignment using MEGA6 with 1,000 bootstrap replicates. The putative families of BmERVs were defined based on their support in phylogenetic trees.
Insertion Time of BmERVs
When the host neutral substitution rate is known, the integration age of an individual ERV can be estimated by measuring the pairwise distance between LTR sequences. To estimate the insertion time of BmERVs, we used the LTR divergence calculated as K/2r (Zhuo et al., 2013). K is the pairwise evolutionary distance of LTR pairs and r is a substitution rate using the Drosophila neutral rate of 15.6 × 10-9 substitutions per year (Xia et al., 2004).
RepeatMasker Analysis
Repetitive elements nested within the BmERV library were removed using Repbase (version 17.11) (Bao et al., 2015). The new BmERV library was subsequently used to run RepeatMasker3 on the silkworm genome using the sensitive Crossmatch alignment program with the default parameters.
Analysis of BmERV env Genes
BmERVs possessing an open reading frame (ORF) coding for an env-like genes, including BmERV-21, BmERV-83, BmERV-94, BmERV-137, BmERV-143, BmERV-145, and BmERV-228, were selected from the BmERV library. The fusogenic region of the BmERV envelope glycoprotein was analyzed using MegAlign (DNASTAR Lasergene.v7.1) and Weblogo.4 Several known IERVs including D. melanogaster ZAM virus (ZAM), D. melanogaster Idefix virus (Idefix), and D. melanogaster Gypsy virus (DmeGypV) were used as references. In addition, sequence alignments were compared between BmERV env genes and genes that code for envelope proteins (F and GP64) in Group I and Group II nuclear polyhedron viruses (NPVs) using MegAlign and MEGA6. The GenBank accession numbers of NPV strains are summarized as follows. Group I NPVs: Autographa californica nuclear polyhedrosis virus (AcMNPV) cloneC6 (L22858.1), AcMNPV E2 (KM667940.1), Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus (U75930.2), and Bombyx mori nuclear polyhedrosis virus (BmNPV) (strains Brazilian: KJ186100.1, Guangxi: JQ991011.1, India: JQ991010.1, JapanH4: LC150780.1, Korea C1:KF306215.1, Suzhou Cubic: JQ991009.1, United States T3: KF306215.1, Zhengjiang: JQ991008.1); Group II NPVs: Lymantria dispar multiple nucleopolyhedrovirus (LdMNPV)-RR01(KX618634.1), LdMNPV-Ab-a624 (KT626572.1), Spodoptera exigua multiple nucleopolyhedrovirus (SeMNPV) HT-SeG25(HG425347.2), SeMNPV VT-SEOX4 (HG425345.1), and Helicoverpa armigera nuclear polyhedrosis virus (HaSNPV) (NC_003094.2).
PCR and RT-PCR Amplification of env Genes in Bombyx mori
To verify the BmERVs identified by bioinformatics, the env genes of BmERVs were amplified with specific primers (Supplementary Table S2) using DNA template. RT-PCR was employed to detect the transcription of BmERV env genes by using the same primers with cDNA template. DNA and RNA samples were extracted from the fat bodies and midgut of the silkworm at the 5th-instar stage. Samples from five individuals were pooled in equal amounts. cDNA synthesis was performed with a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan) according to the manufacturer’s protocol. In this kit, the first step is to eliminate the genomic DNA using gDNA Eraser.
Results
De Novo Detection of BmERVs in the Silkworm Genome
Two different pipelines were employed for mining BmERVs in the silkworm genome (Figure 1). First, BmERVs were detected by LTRharvest and annotated with LTRdigest. A total of 13,104 ERV candidates with predicted LTR pairs in the silkworm genome were identified with LTRharvest. These candidates were filtered down to 175 ERV candidates by LTRdigest. Additionally, MGEScan revealed 230 ERV candidates. After merging the ERV candidates identified by the two pipelines, we identified a total of 256 BmERVs in the silkworm genome (Figure 1). The full -lengths of these 256 BmERVs varied from 1,912 to 20,275 bp, and their LTR lengths ranged from 101 to 687 bp (details of the 256 BmERVs are shown in Supplementary Data Sheet S1). Among the detected BmERVs, the most common structures were LTR-gag-pro-pol-LTR and LTR-pol-LTR (Table 1). However, only seven BmERVs contained the env gene and three BmERVs contained the complete structure of an insect retrovirus, but none of these had an entire intact sequence (Table 1 and Supplementary Figure S1). This result suggested that entire functional BmERVs might not exist in B. mori.
Phylogenetic Analysis and Classification of BmERVs
To classify the detected BmERVs, the RT domain sequences of BmERVs, vertebrate retroviruses, and IERVs were used to build a multiple alignment and compute phylogenetic trees using the neighbor-joining method implemented in MEGA 6. Of the 256 BmERVs identified in this sutdy, 68 BmERVs had a RT domain conserved enough to be aligned confidently for phylogenetic analysis. According to the phylogenetic tree analysis, BmERVs were classed into three groups and nine families (Figure 2 and Supplementary Data Sheet S2). Most of the BmERVs included in the phylogenetic analysis belonged to the class I group and were classified into four families (Supplementary Data Sheet S2). Additionally, 176 BmERVs were marked as unclassified members (Supplementary Data Sheet S2). On the whole, class I and class II BmERVs were related to IERVs from errantivirus and metavirus (Figure 2). There was a close relationship between BmERV-47/187 and the insect semotivirus roo (Figure 2). Surprisingly, class III BmERVs were related to vertebrate retroviruses (Figure 2).
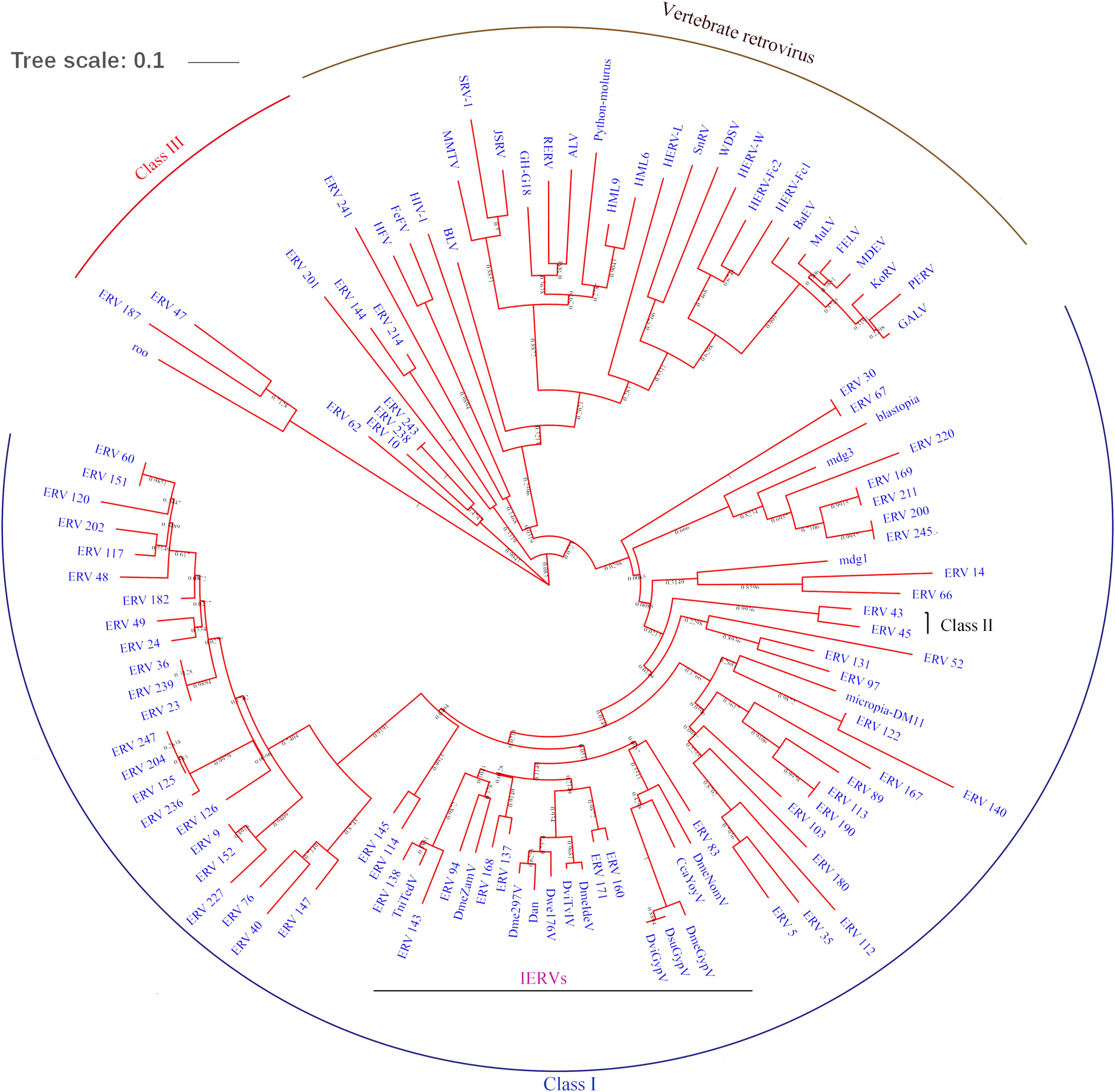
FIGURE 2. RT region-based phylogenetic tree of BmERVs. A total of 68 BmERVs possessing a RT domain were included. Other retrovirus used in this analysis included vertebrate retrovirus, insect LTR-retrotransposons, and IERVs (Supplementary Table S1). Topology was based on the neighbor-joining method with 1,000 bootstraps.
Census of the BmERV Population in the Silkworm Genome
To fully assess the abundance of BmERV-derived sequences, RepeatMasker was employed to annotate the silkworm genome using all of the BmERV and non-redundant ERV sequences deposited in Repbase. The BmERV-related sequences annotated by RepeatMasker occupied 25.56% of the silkworm genome (Supplementary Data Sheet S3). BmERVs were relatively evenly distributed across the individual chromosomes (Figure 3). Chromosome 2 possesses the most BmERVs (Figure 3).
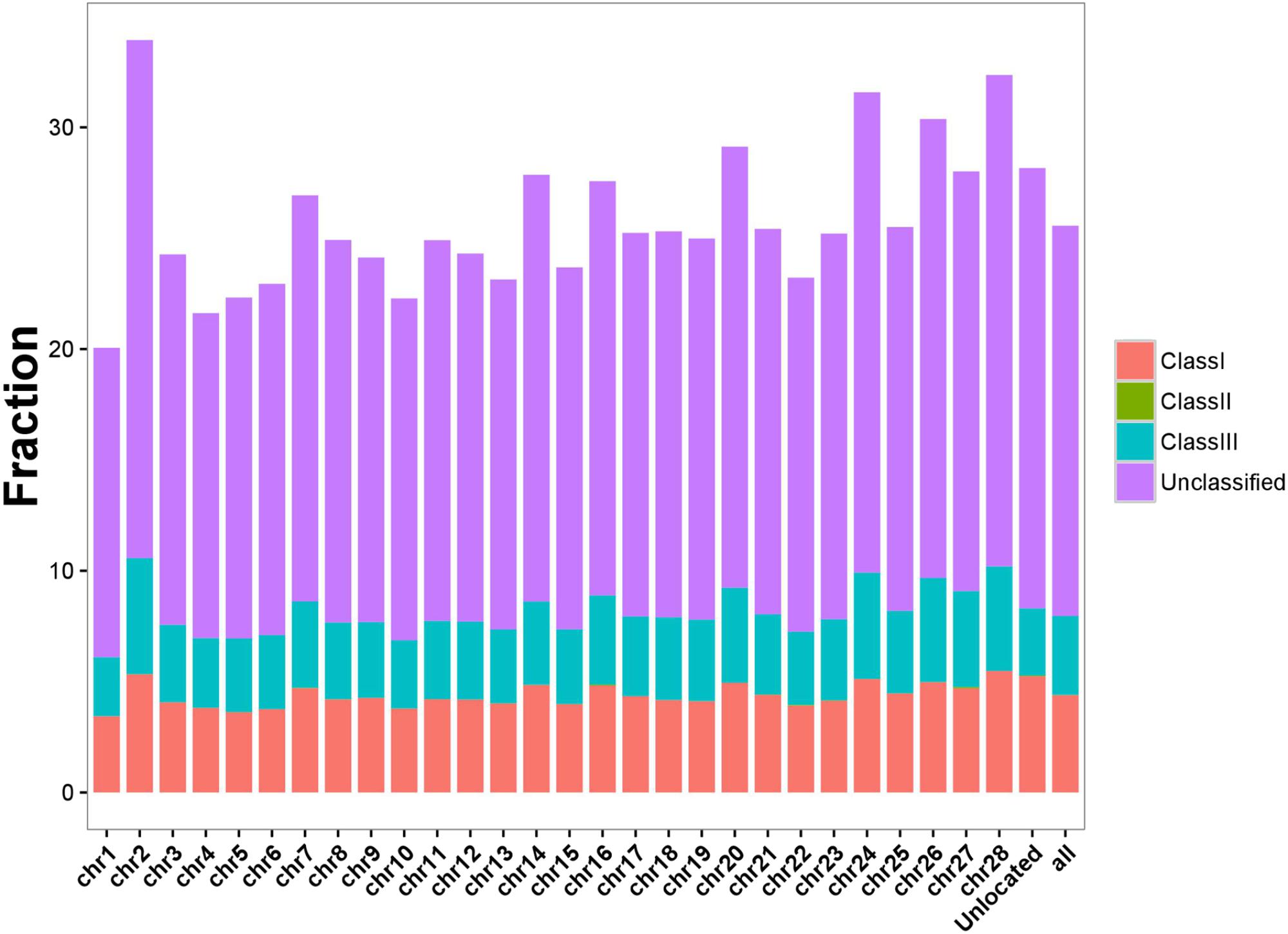
FIGURE 3. BmERV abundance in different chromosomes. Different BmERV classes are labeled with different colors. The abundance is illustrated using the percentage of the chromosome sequences. “Unlocated” represents BmERV sequences that are not located on a particular chromosome. “All” indicates the percentage of the whole silkworm genome occupied by BmERVs.
Insertion Time of BmERVs in the Silkworm Genome
As the neutral substitution rate of B. mori is still unknown, we applied a rate of neutral substitution in fruitfly that has been previously used for estimating the origination time of silkworm transposable elements (Xia et al., 2004) to date the integration of the BmERVs. Our results showed that BmERVs integrated relatively recently, estimated at less than 10 million years ago (MYA) (Figure 4), according to published standards (Garcia-Etxebarria and Jugo, 2016): young ERVs (up to 10–22 MYA), middle-aged ERVs (between 10–22 MYA and 20–43 MYA), and ancient ERVs (more than 43 MYA). The insertion time of most BmERVs was less than 4 MYA (Figure 4). Indeed, BmERV-255 was the oldest among the BmERVs, but only inserted about 8.23 MYA (Supplementary Data Sheet S4). In addition, 56 of the BmERVs had strictly identical LTR pairs, indicating that these BmERVs inserted very recently (Supplementary Data Sheet S4). These data suggest that ERV integration events in B. mori occurred recently.
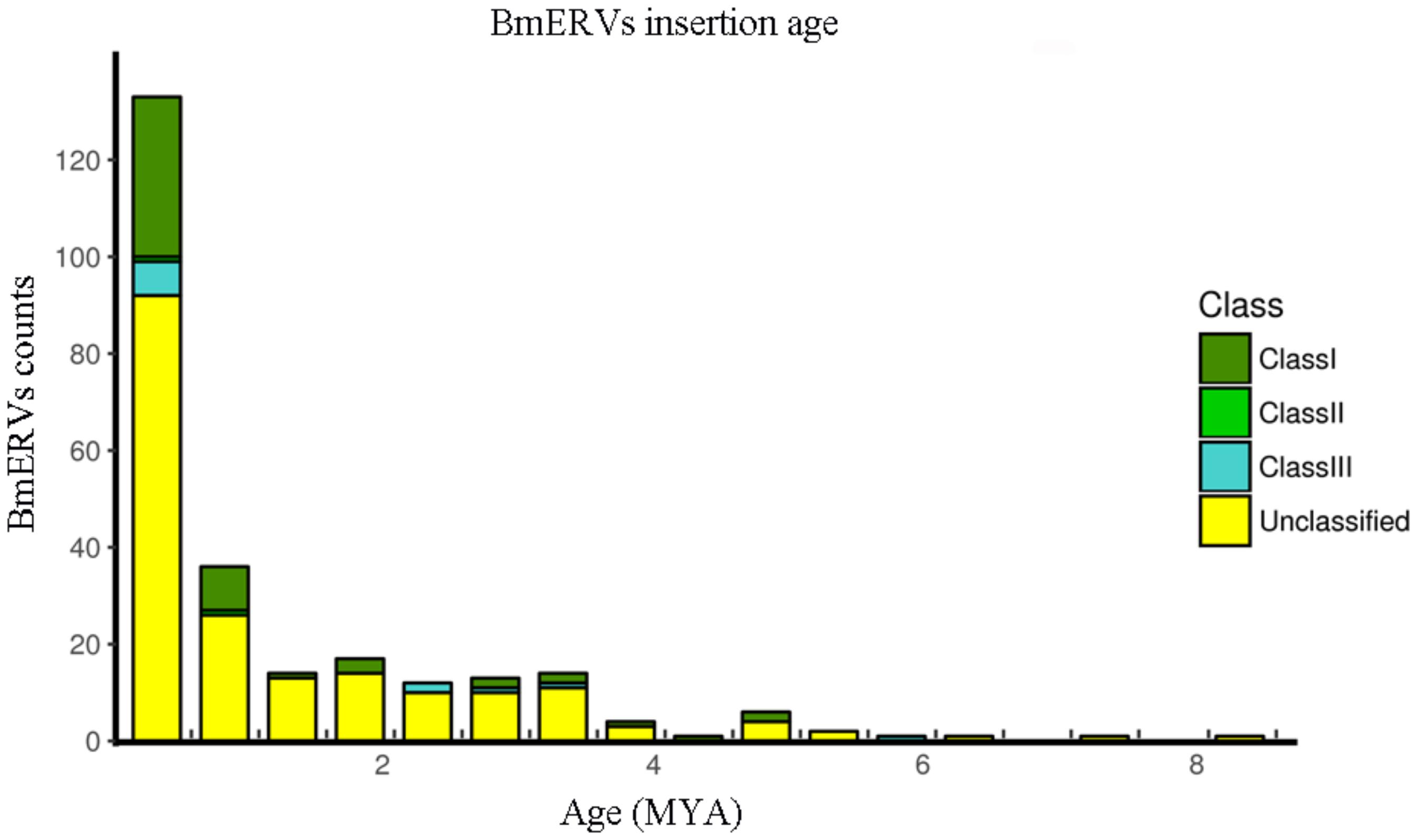
FIGURE 4. Insertion time of BmERVs. The numbers of different BmERV classes are shown in different colors. Insertion time was estimated by LTR pair divergence. Abbreviation: MYA, million years ago.
Comparison of BmERV env Genes With F and GP64 Genes From NPVs
To explore the relationship between env gene of BmERVs and genes encoding the envelope protein (Env) in NPVs, we analyzed the evolutionary relationships and homology among these genes. Fa and Fb represent the F protein of Group II NPVs and the homologs of the F protein from Group I NPVs, respectively. The results showed that the env sequences of BmERVs were distantly related to genes encoding F and GP64 proteins from Group I and Group II NPVs (Figures 5A,B). Surprisingly, one of the phylogenetic lineages contained BmERVs env genes and HaSNPVgOrf133 (Figure 5A). Indeed, there were a few regions of the alignment where BmERVs env genes were not similar to each other. The BmERV env genes shared low homology with the F and GP64 genes from Group I and Group II NPVs (Supplementary Figure S2).

FIGURE 5. Phylogenetic analysis based on the env genes of BmERVs and the F and GP64 genes from NPVs. Group I NPVs including AcMNPV, OpMNPV, and BmNPVs (strains Brazilian, Guangxi, etc.) possess GP64 and the homologs of F (Fb). Group II NPVs have F (Fa) but lack GP64. (A) Phylogenetic analysis of the env gene of BmERVs and F genes from Group I NPVs (Fb) and Group II NPVs (Fa). (B) Phylogenetic analysis of the env gene of BmERVs and GP64 of Group I NPVs. About 500 bp of the env-fragments were aligned to generate the phylogenetic tree. AcMNPV, Autographa californica nuclear polyhedrosis virus; OpMNPV, Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus; BmNPV, Bombyx mori nuclear polyhedrosis virus; HaSNPV, Helicoverpa armigera nuclear polyhedrosis virus; LdMNPV, Lymantria dispar multiple nucleopolyhedrovirus; SeMNPV, Spodoptera exigua multiple nucleopolyhedrovirus.
Interestingly, we found that the amino acid sequences of BmERV-21 shared a region of similarity with the fusion protein of Group II NPVs and known insect ERVs, which included the furin cleavage signal and downstream predicted fusion peptide (Figure 6A). Logo representation of the furin-like consensus motif (RxxR) and peptide fusion consensus sequences (GxxxxxGxxxKxxxGxxDxxD) between BmERV-21 and Idefix, ZAM, LdMNPV, and SeMNPV are shown in Figure 6B, suggesting that the env gene of BmERV-21 might encode a fusion protein. However, similar fusion peptide sequences were not found in BmERV-83, BmERV-94, BmERV-137, BmERV-143, BmERV-145, BmERV-228, or the fusion proteins (GP64 and Fb) of BmNPVs.
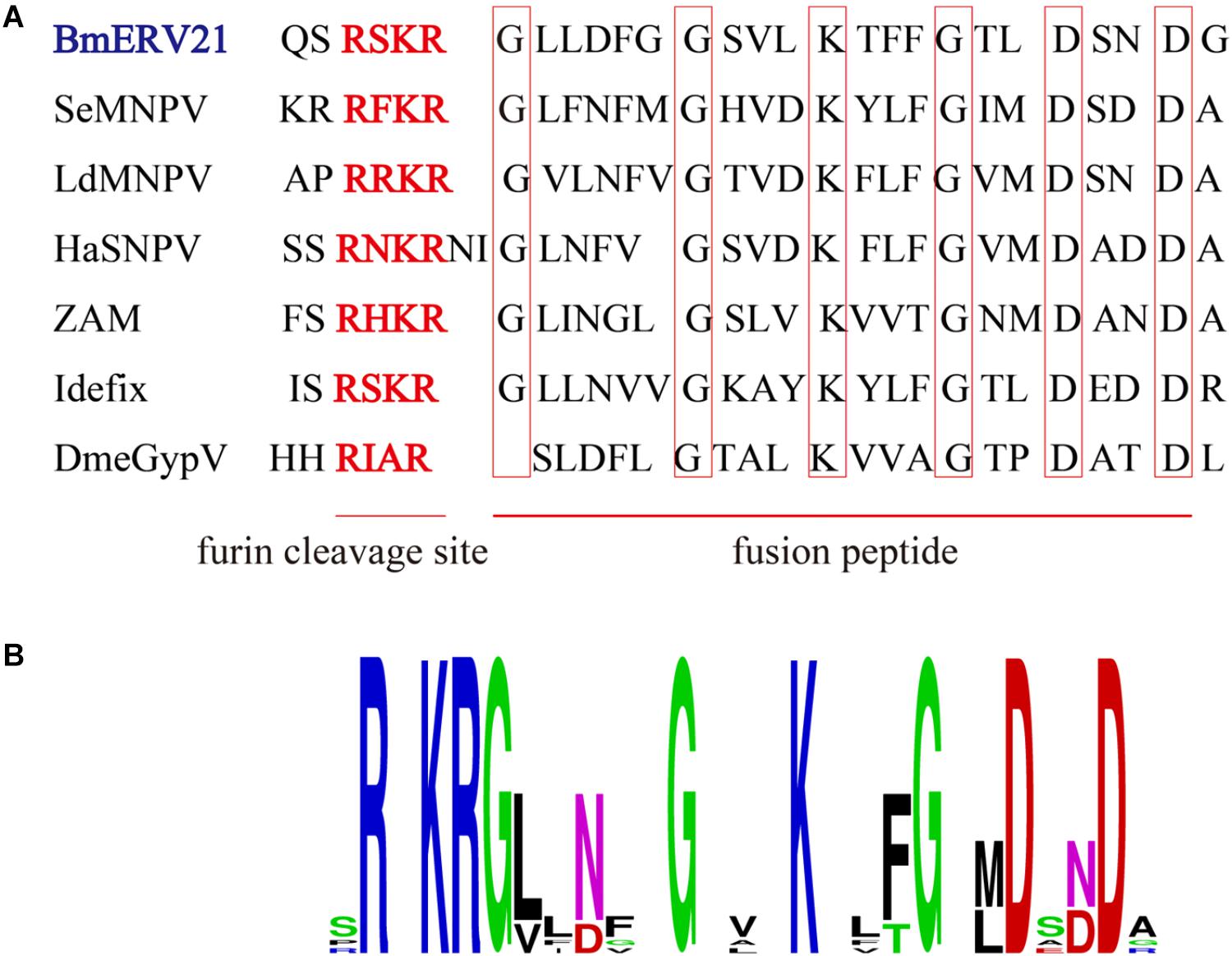
FIGURE 6. Conserved amino acid sequence block in the Env protein of BmERVs and IERVs and Group II NPV F proteins. (A) Multiple alignment of the conserved amino acid sequence block showed a consensus furin cleavage site RxxR and the peptide fusion consensus sequence in the Env protein of BmERV-21, ZAM, Idefix, DmeGypV, and the F protein of SeMNPV, LdMNPV, and HaSNPV. (B) Logo visualization of the furin cleavage site and peptide fusion consensus sequence performed using WEBLOGO (weblogo.berkeley.edu/logo.cgi).
Integration and Effective Transcription of the BmERV env Genes
To further verify whether BmERVs env genes exist in the silkworm genome and are expressed in silkworm tissues, RCR and RT-PCR were employed to detect BmERV env genes using DNA and cDNA templates extracted from the midguts and fat bodies of B. mori, respectively. DNA samples from both of midguts and fat bodies produced a band of BmERVs provirus env; however, BmERV-145 showed a weaker band than other strains (Figure 7A). PCR products were used for direct sequencing analysis. The sequencing results also identified the BmERV env sequences, in accordance with the sequences identified by bioinformatics (data not shown). The results demonstrated that BmERV env genes indeed integrated into the silkworm genome. Furthermore, RT-PCR results showed that the env genes of BmERV-83, BmERV-94, and BmERV-143 were expressed abundantly in the midguts and fat bodies (Figure 7B). The expression of the env genes of BmERV-21 and BmERV-137 was also detected in midguts and fat bodies (Figure 7B). However, BmERV-145 and BmERV-228 env expression was relatively weaker in midguts and fat bodies (Figure 7B). The results suggest that BmERV elements can be effectively transcribed in silkworm tissues.
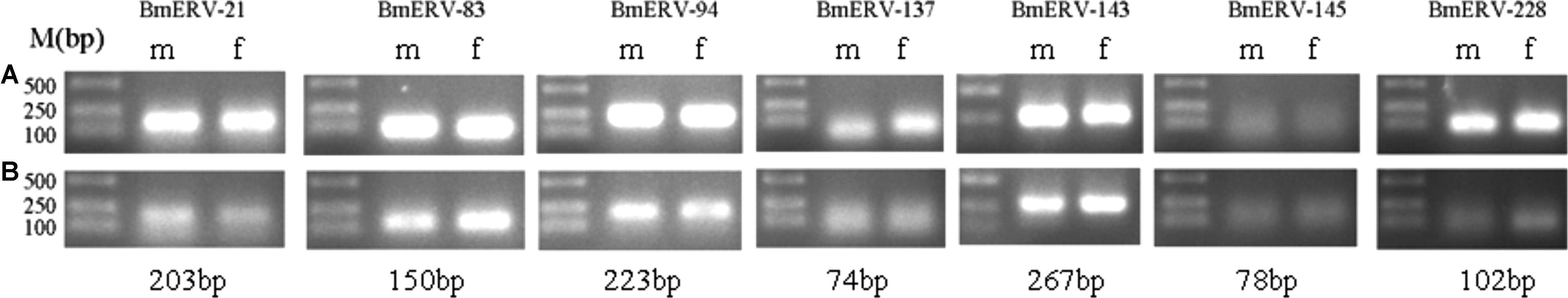
FIGURE 7. Integration and effective transcription of the BmERV env genes in the tissues of Bombyx mori. (A) PCR with DNA templates was used to verify the presence of env genes in the silkworm genome. Tissues samples of DaZao were extracted from midguts (m) and fat bodies (f). (B) RT-PCR was employed to detect the expression of the env genes of BmERV-21, BmERV-83, BmERV-94, BmERV-137, BmERV-143, BmERV-145, and BmERV-228 using cDNA templates.
Discussion
This study attempted to systematically identify and characterize ERVs in the silkworm genome. By combining two different de novo mining strategies, we identified 256 potential BmERVs in the silkworm genome. The silkworm genome is composed of 28 chromosomes with female heterogametic constitution: ZZ for male and ZW for female. The existing silkworm genome data does not include W chromosome sequences (Xia et al., 2014); however, it have been found that many retrotransposon elements have accumulated on the W chromosome of B. mori (Abe et al., 2000, 2001, 2005). Thus, more BmERVs will likely be identified when B. mori W chromosome sequence data are published.
Phylogenetic analysis based on the RT region was used to cluster BmERVs into nine putative families and three groups. As expected, we found most of the BmERVs were related to IERVs such as DmeZamV and DmeGypV, according to the phylogenetic analysis. However, BmERVs of class III were related to vertebrate retroviruses. In D. melanogaster, phylogenetic analysis shows that three kinds of retrotransposons are similar to class I, II, and III retroviruses, respectively (Llorens C. et al., 2008). LTR retrotransposons or retroviruses in insects and vertebrate retroviruses have a common evolutionary history and should be considered in parallel (Nefedova and Kim, 2017).
Due to using a substitution rate of Drosophila, and the homogenization of the LTR pairs caused by the effect of recombination or gene conversion events, the insertion time proposed for BmERVs is only an estimate. Similarly, the rate of neutral substitution in fruitfly has been previously used for estimating the origination time of silkworm transposable elements (Xia et al., 2004). The oldest bovine ERV was inserted between 126 and 58 MYA (Garcia-Etxebarria and Jugo, 2010). The insertion time of ERV elements in humans and in chimpanzees is 19.5–8.9 MYA and 33–15.8 MYA, respectively (Garcia-Etxebarria and Jugo, 2010). However, our data show that all of the BmERVs are young members, with the oldest insertion dated to approximately 8 MYA. Thus, BmERVs inserted into the silkworm genome recently.
In addition, the proportion of BmERVs in the genome was also different from that in vertebrates. ERVs often occupy a substantial fraction of genomes, accounting for about 5% of bat, 10% of mouse, and 8% of human genome sequences (Zhuo et al., 2013). In this work, using a rigorous set of criteria, we estimated that BmERVs occupied ∼25% of the silkworm genome, which is significantly higher than the proportion identified in vertebrates. IERVs have been mostly reported in Drosophila, but to our knowledge their quantity in genome-wide is still unknown. The repeat elements in the silkworm genome represent approximately 43.6%, which is significantly higher than that detected in other insect species including 12 Drosophila species (Osanai-Futahashi et al., 2008; Xia et al., 2014). We speculated that ERV elements in B. mori would also represent a larger proportion of the genome than those in Drosophila. Nevertheless, in this study, no full-length BmERVs were identified in the silkworm genome. On the contrary, in other insects, several ERVs have been identified, and most of them possess a complete retroviral structure (Pelisson et al., 2002; Akkouche et al., 2012). Thus far, only the well-known gypsy and ZAM elements of D. melanogaster have been shown to possess infectious properties (Leblanc et al., 2000; Llorens J.V. et al., 2008). In general, the infectious ability of IERVs is thought to be associated with the expression of Env encoded by the env gene (Teysset et al., 1998).
However, only seven BmERVs possessing env genes were identified in the silkworm genome. Furthermore, according to the sequences identified by bioinformatics, we found that the BmERV env genes did not contain a full-length ORF. It is believed that some LTR retrotransposons which lacked the env gene integrated into the dsDNA genome of a baculovirus and “captured” env gene from baculoviruses (Malik et al., 2000). Two different envelope proteins, named GP64 and F, have been identified in the family Baculoviridae (Pearson and Rohrmann, 2002). In general, Group I baculoviruses possess GP64 and the homologs of the F protein (Fb), whereas Group II baculoviruses have the F protein (Fa) but lack GP64 (Pearson and Rohrmann, 2002). Indeed, IERV Env proteins showed significant homology to the fusion protein FP of Group II baculoviruses and also have fusogenic properties (Rohrmann and Karplus, 2001; Misseri et al., 2003, 2004). However, only the BmERV-21 amino acid sequence shared a similar putative furin-like cleavage site and fusion peptide with Group II baculoviruses. Further experimental detection is needed to confirm whether the sequences of BmERV-21 and its env gene are complete. To date, no complete BmERV sequence has been verified in living tissues or cell lines, not to mention the functional research. In addition, except for HaSNPVgOrf133, we found that the env sequences of BmERVs were distantly related to genes encoding F (Fa and Fb) and GP64 protein from Group I and Group II NPVs. Thus, BmERVs might have “captured” an env gene from HaSNPV, and this event would have happened a long time ago. We speculated that B. mori might have been infected by various baculoviruses before domestication. But for the moment, B. mori does not seem to be infected with HaSNPV. Compared to wild insects, B. mori is a completely domesticated insect with a good living environment and, as such, could be largely exempt from long-term baculovirus infection. Only healthy individuals are used to produce and reproduce. Moreover, we estimated that ERV integration events in B. mori occurred recently. Therefore, we speculate that the domestication and recent integration events might account for only seven env genes being detected in the silkworm genome. If the ERVs of wild silkworm were identified and compared with those of BmERVs, a greater understanding of the evolution of BmERVs would be achieved.
ERVs occupy a significant portion of the host genome. The functions of ERVs are of particular interest, serving as a regulatory factor to influence host gene expression (Suntsova et al., 2015). Additionally, functional ERV genes such as env encode active proteins, which may exert important physiological functions in the host (Suntsova et al., 2015). Our results confirmed that the BmERVs env genes existed in the silkworm genome and exhibited transcriptional activity in the midguts and fat bodies. Thus, BmERV env genes might also play an important role in host biology, especially in antiviral defense. In fact, there have been calls for attention to the role of IERVs in the immune response (Fablet, 2014).
Taken together, this is indeed the first genome-wide approach for the screening and characterization of BmERVs. Further studies are thus needed to verify whether the BmERVs possess complete structures and whether infectious properties exist in vivo. Such studies would elucidate the roles of BmERVs in B. mori.
Author Contributions
MF participated in the design of the study and drafted the manuscript. MF and XW collected and analyzed the data. FR helped in the sample collection. NZ and YZ performed PCR and RT-PCR. JS participated in the design and coordination of the study, and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by China Postdoctoral Science Foundation (Grant Number 2017M622714), the Natural Science Foundation of Guangdong Province (Grant Number 2018A030310210), and the National Natural Science Foundation of China (Grant Number 31372373).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Gene Denovo Corp. for its help in bioinformatics analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01732/full#supplementary-material
FIGURE S1 | Structure of 256 BmERVs.
FIGURE S2 | Homology comparison between env of BmERVs and the F and GP64 genes of NPVs. (A) Homology of env of BmERVs and F genes from Group I NPVs (Fb) and Group II NPVs (Fa). (B) Homology of env of BmERVs and GP64 of Group I NPVs.
TABLE S1 | Reverse transcriptase (RT) domain from vertebrate retroviruses and insect LTR- retrotransposons used in this study.
TABLE S2 | Primers used for amplifying the env genes of BmERV.
DATA SHEET S1 | Details of all of BmERVs.
DATA SHEET S2 | Classification of BmERVs.
DATA SHEET S3 | Census of BmERVs in the silkworm chromosome.
DATA SHEET S4 | Insertion time of BmERVs in the silkworm genome.
Footnotes
- ^ http://silkworm.genomics.org.cn/
- ^ http://www.gydb.org/index.php/Main_Page
- ^ http://www.repeatmasker.org/
- ^ http://weblogo.berkeley.edu/logo.cgi
References
Abe, H., Mita, K., Yasukochi, Y., Oshiki, T., and Shimada, T. (2005). Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet. Genome Res. 110, 144–151. doi: 10.1159/000084946
Abe, H., Ohbayashi, F., Shimada, T., Sugasaki, T., Kawai, S., Mita, K., et al. (2000). Molecular structure of a novel gypsy-Ty3-like retrotransposon (Kabuki) and nested retrotransposable elements on the W chromosome of the silkworm Bombyx mori. Mol. Gen. Genet. 263, 916–924. doi: 10.1007/s004380000270
Abe, H., Ohbayashi, F., Sugasaki, T., Kanehara, M., Terada, T., Shimada, T., et al. (2001). Two novel Pao-like retrotransposons (Kamikaze and Yamato) from the silkworm species Bombyx mori and B. mandarina: common structural features of Pao-like elements. Mol. Genet. Genomics 265, 375–385. doi: 10.1007/s004380000428
Akkouche, A., Rebollo, R., Burlet, N., Esnault, C., Martinez, S., Viginier, B., et al. (2012). tirant, a newly discovered active endogenous retrovirus in Drosophila simulans. J. Virol. 86, 3675–3681. doi: 10.1128/JVI.07146-11
Bao, W., Kojima, K. K., and Kohany, O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6:11. doi: 10.1186/s13100-015-0041-9
Bustamante Rivera, Y. Y., Brutting, C., Schmidt, C., Volkmer, I., and Staege, M. S. (2017). Endogenous retrovirus 3 - history, physiology, and pathology. Front. Microbiol. 8:2691. doi: 10.3389/fmicb.2017.02691
Chuong, E. B., Elde, N. C., and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087. doi: 10.1126/science.aad5497
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ellinghaus, D., Kurtz, S., and Willhoeft, U. (2008). LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9:18. doi: 10.1186/1471-2105-9-18
Fablet, M. (2014). Host control of insect endogenous retroviruses: small RNA silencing and immune response. Viruses 6, 4447–4464. doi: 10.3390/v6114447
Feng, M., Tan, Y., Dai, M., Li, Y., Xie, T., Li, H., et al. (2016). Endogenous retrovirus ev21 dose not recombine with ALV-J and induces the expression of ISGs in the HOST. Front. Cell. Infect. Microbiol. 6:140. doi: 10.3389/fcimb.2016.00140
Garcia-Etxebarria, K., and Jugo, B. M. (2010). Genome-wide detection and characterization of endogenous retroviruses in Bos taurus. J. Virol. 84, 10852–10862. doi: 10.1128/JVI.00106-10
Garcia-Etxebarria, K., and Jugo, B. M. (2016). Genome-wide reexamination of endogenous retroviruses in Rattus norvegicus. Virology 494, 119–128. doi: 10.1016/j.virol.2016.04.006
Havecker, E. R., Gao, X., and Voytas, D. F. (2004). The diversity of LTR retrotransposons. Genome Biol. 5:225. doi: 10.1186/gb-2004-5-6-225
Jin-Shan, X., Qing-You, X., Jun, L., Guo-Qing, P., and Ze-Yang, Z. (2005). Survey of long terminal repeat retrotransposons of domesticated silkworm (Bombyx mori). Insect Biochem. Mol. Biol. 35, 921–929. doi: 10.1016/j.ibmb.2005.03.014
Kim, A., Terzian, C., Santamaria, P., Pelisson, A., Purd’homme, N., and Bucheton, A. (1994). Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 91, 1285–1289. doi: 10.1073/pnas.91.4.1285
Leblanc, P., Desset, S., Giorgi, F., Taddei, A. R., Fausto, A. M., Mazzini, M., et al. (2000). Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 74, 10658–10669. doi: 10.1128/JVI.74.22.10658-10669.2000
Llorens, C., Fares, M. A., and Moya, A. (2008). Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 8:276. doi: 10.1186/1471-2148-8-276
Llorens, J. V., Clark, J. B., Martinez-Garay, I., Soriano, S., De Frutos, R., and Martinez-Sebastian, M. J. (2008). Gypsy endogenous retrovirus maintains potential infectivity in several species of Drosophilids. BMC Evol. Biol. 8:302. doi: 10.1186/1471-2148-8-302
Malik, H. S., Henikoff, S., and Eickbush, T. H. (2000). Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 10, 1307–1318. doi: 10.1101/gr.145000
Misof, B., Liu, S., Meusemann, K., Peters, R. S., Donath, A., Mayer, C., et al. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. doi: 10.1126/science.1257570
Misseri, Y., Cerutti, M., Devauchelle, G., Bucheton, A., and Terzian, C. (2004). Analysis of the Drosophila gypsy endogenous retrovirus envelope glycoprotein. J. Gen. Virol. 85, 3325–3331. doi: 10.1099/vir.0.79911-0
Misseri, Y., Labesse, G., Bucheton, A., and Terzian, C. (2003). Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol. 11, 253–256. doi: 10.1016/S0966-842X(03)00119-7
Nefedova, L., and Kim, A. (2017). Mechanisms of LTR-retroelement transposition: lessons from Drosophila melanogaster. Viruses 9:E81. doi: 10.3390/v9040081
Niu, D., Wei, H. J., Lin, L., George, H., Wang, T., Lee, I. H., et al. (2017). Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 357, 1303–1307. doi: 10.1126/science.aan4187
Osanai-Futahashi, M., Suetsugu, Y., Mita, K., and Fujiwara, H. (2008). Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 38, 1046–1057. doi: 10.1016/j.ibmb.2008.05.012
Pearson, M. N., and Rohrmann, G. F. (2002). Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 76, 5301–5304. doi: 10.1128/JVI.76.11.5301-5304.2002
Pelisson, A., Mejlumian, L., Robert, V., Terzian, C., and Bucheton, A. (2002). Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem. Mol. Biol. 32, 1249–1256. doi: 10.1016/S0965-1748(02)00088-7
Rohrmann, G. F., and Karplus, P. A. (2001). Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol. Biol. 1:1. doi: 10.1186/1471-2148-1-1
Steinbiss, S., Willhoeft, U., Gremme, G., and Kurtz, S. (2009). Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013. doi: 10.1093/nar/gkp759
Suntsova, M., Garazha, A., Ivanova, A., Kaminsky, D., Zhavoronkov, A., and Buzdin, A. (2015). Molecular functions of human endogenous retroviruses in health and disease. Cell. Mol. Life Sci. 72, 3653–3675. doi: 10.1007/s00018-015-1947-6
Terzian, C., Pelisson, A., and Bucheton, A. (2001). Evolution and phylogeny of insect endogenous retroviruses. BMC Evol. Biol. 1:3. doi: 10.1186/1471-2148-1-3
Teysset, L., Burns, J. C., Shike, H., Sullivan, B. L., Bucheton, A., and Terzian, C. (1998). A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J. Virol. 72, 853–856.
Touret, F., Guiguen, F., and Terzian, C. (2014). Wolbachia influences the maternal transmission of the gypsy endogenous retrovirus in Drosophila melanogaster. mBio 5:e01529-14. doi: 10.1128/mBio.01529-14
Wang, Z., Qu, L., Yao, J., Yang, X., Li, G., Zhang, Y., et al. (2013). An EAV-HP insertion in 5′ Flanking region of SLCO1B3 causes blue eggshell in the chicken. PLoS Genet. 9:e1003183. doi: 10.1371/journal.pgen.1003183
Xia, Q., Li, S., and Feng, Q. (2014). Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 59, 513–536. doi: 10.1146/annurev-ento-011613-161940
Xia, Q., Zhou, Z., Lu, C., Cheng, D., Dai, F., Li, B., et al. (2004). A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306, 1937–1940. doi: 10.1126/science.1102210
Zanotto, P. M., Kessing, B. D., and Maruniak, J. E. (1993). Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 62, 147–164. doi: 10.1006/jipa.1993.1090
Keywords: endogenous retroviruses, Bombyx mori, NPVs, env, genome
Citation: Feng M, Wang X, Ren F, Zhang N, Zhou Y and Sun J (2018) Genome-Wide Characterization of Endogenous Retroviruses in Bombyx mori Reveals the Relatives and Activity of env Genes. Front. Microbiol. 9:1732. doi: 10.3389/fmicb.2018.01732
Received: 23 April 2018; Accepted: 11 July 2018;
Published: 03 August 2018.
Edited by:
Ping An, Frederick National Laboratory for Cancer Research (NIH), United StatesReviewed by:
Antoinette Van Der Kuyl, University of Amsterdam, NetherlandsTara Patricia Hurst, Abcam, United Kingdom
Copyright © 2018 Feng, Wang, Ren, Zhang, Zhou and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingchen Sun, Y3lmekBzY2F1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Min Feng
Min Feng Xiong Wang†
Xiong Wang† Jingchen Sun
Jingchen Sun