- 1State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 2National Engineering Research Center for Biotechnology, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
Saccharomyces cerevisiae immobilization is commonly used for efficient ethanol fuel production in industry due to the relatively higher ethanol stress resistance of S. cerevisiae in biofilms relative to planktonic cells. The mechanisms of biofilm formation and stress resistance, however, remain ambiguous. By analyzing biofilm and planktonic cell transcriptomes, this study observed that MIG1 (encoding a transcription factor) expression in cells increases during the biofilm formation process. To identify the role of MIG1 in yeast biofilm formation and the ethanol resistance of these cells, MIG1 was deleted and complemented in S. cerevisiae 1308. Results showed the MIG1 deletion mutant strain demonstrated weaker biofilm formation ability both on fibers and plastic than the wild-type and these could be restored by expressing MIG1 in deletion mutant. To verify the ability of MIG1 to regulate the expression of FLO genes, which encode adhesions responsible for yeast biofilm formation, FLO gene transcription levels were measured via qRT-PCR. Relative to wild-type S. cerevisiae, the adhesion genes FLO1, 5, and 9 which also demonstrate increased expression in the transcriptome of yeast cells during biofilm formation, but not FLO11, were down-regulated in the MIG1 mutant strain. Additionally, the MIG1 mutant lost a majority of its flocculation ability, which depended on cell-cell adhesions and its slightly invasive growth ability, dependent on cell-substrate adhesion. Deleting FLO1, 5, and 9 decreased biofilm formation on plastics, suggesting these FLO genes contribute to the biofilm formation process alongside FLO11. Moreover, the ethanol tolerance of yeast decreased in the MIG1 deletion mutant as well as the FLO11 deletion mutant, resulting in reduced biofilm formation during fermentation. It remains possible that in the later period of fermentation, when ethanol has accumulated, an over-expression of the FLO1, 5, and 9 genes regulated by MIG1 would enhanced cell-cell adhesions and thus protect cells in the outer layer of biofilms from ethanol, a function primarily dependent on cell-cell adhesions. This work offers a possible explanation for how biofilm formation is regulated during the immobilized fermentation process, and can enhance environmental tolerance in industrial production.
Introduction
Similar to human cities, biofilms are groups of microorganisms in which cells collaborate together and produce matrices of extracellular polymeric substance to survive (Kuhn et al., 2002). Biofilms protect cells from various external stimuli such as osmostress, heat shock, oxidative stress, and nutrient deficiencies (Blankenship and Mitchell, 2006). The difficulty of removing biofilms formed on medical devices and catheters in vivo by pathogenic microorganisms such as Pseudomonas aeruginosa, Candida albicans, and Staphylococcus aureus using antibiotic treatments presents a significant problem in medicine (Almirante et al., 2006). These characteristics, however, are beneficial in biofilm reactors. These reactors are formed by immobilized cells adhering to solid surfaces, and demonstrate excellent tolerance against substrates/products inhibition as well as higher reuse efficiency for immobilized strains in batch fermentation (Pal and Khanum, 2011). Repeated batch fermentations in biofilm reactors have been applied to efficiently produce ethanol fuel using S. cerevisiae with a high optimal conversion rate (Li et al., 2012). During the ethanol fermentation process, biofilms have returned higher ethanol yields and shorter fermentation times were observed relative to the planktonic cell fermentation process (Germec et al., 2015).
Biofilm formation occurs primarily through three phases: attachment, maturation, and dispersion. In response to certain environments, cells will attach to surfaces and begin producing exopolymeric substances (EPS, including polysaccharides, proteins, lipids and nucleic acids) to form a complex three-dimensional architecture (Suresh Kumar et al., 2007). Although the specific compositions of EPS remain unknown, EPS and especially their proteins have been found to support biofilm structures (Fong and Yildiz, 2015) and perform biochemical activities, such as the protease inhibitor found in Pseudomonas aeruginosa biofilm EPS that protects cells from proteolytic attacks (Tseng et al., 2018).
Adhesion genes, which contribute to the ability of cells to adhere to other cells or surfaces, are widely studied in biofilm formation (Stringer and Keely, 2001; Bojsen et al., 2012). S. cerevisiae carries a family of adhesive special surface glycoproteins encoded by FLO1, FLO5, FLO9, FLO10, and FLO11 (Teunissen and Steensma, 1995). They share similar but slightly different structures, and serve different functions (Halme et al., 2004). FLO1, FLO5, FLO9, and FLO10 confer cell-cell adherence and contribute to flocculation (Guo et al., 2000). FLO11 is responsible for cell-surface adhesions and required for agar invasive growth, pseudohyphae, and biofilm formation (Reynolds and Fink, 2001; Zara et al., 2005; Verstrepen and Klis, 2006); however, numerous types of these adhesions are present in different yeast species (Douglas et al., 2007). FLO11 is primarily recognized as the only FLO gene to confer surface adhesion and is thus required for several important developmental transitions including adherence to agar and plastic surfaces in many yeast strains, especially the widely studied S. cerevisiae Σ1278b (Guo et al., 2000; Reynolds and Fink, 2001). Differing from Σ1278b, the S288c strain requires FLO1 for biofilm formation as well as FLO11, and their regulations differ (Fichtner et al., 2007). In S. cerevisiae wine strains, FLO5 has been proven to drive adhesive properties which depend on surface adhesion ability (Di Gianvito et al., 2017). Several cell surface proteins besides FLO genes participate in yeast biofilm formation. Glycosylated cell surface proteins, encoded by CCW14 and YGP1, contribute to yeast biofilms. This may result from the hydrophobicity of the two proteins in haploid strains (Moreno-García et al., 2018).
Some biofilm findings have been based on cells exposed to stress conditions such as low pH and glucose (Reynolds and Fink, 2001; D’Urzo et al., 2014), heat shock (Grudniak et al., 2015), or oxidative stress and osmostress (Geier et al., 2008; Watanabe et al., 2013). By screening deletion mutants and overexpression strains (Andersen et al., 2014), a number of regulators have been shown to control yeast biofilm formation including MAPK (Madhani and Fink, 1997; Gagiano et al., 2003; Chavel et al., 2014), PKA (Villa et al., 2017), and main glucose repression (Lambrechts et al., 1996; Bester et al., 2012; Nakagawa et al., 2017) pathways. To understand the biofilm formation mechanisms observed in biofilm reactors during fermentation, several studies have employed biofilm and free cell comparisons (Li et al., 2015; Liu et al., 2016). These studies have uncovered the involvement of carbohydrates, amino acids, signal transduction, and oxidoreductase metabolism in biofilm formation. In the industry strain S. cerevisiae 1308 used in this laboratory, glycolysis and gluconeogenesis metabolism have been found to play key roles in the development of S. cerevisiae biofilms by comparing the transcriptomes of biofilms and free cells (Li et al., 2015).
MIG1, which encodes a C2H2 zinc finger protein, inhibits GAL gene expression in the presence of glucose and has been recognized as a main effector in the glucose repression pathway (Cao et al., 2011; Liu et al., 2011; Li et al., 2015). NRG1, which represses FLO11 by binding to its promoter, shares similar functions with MIG1 in the glucose repression pathway (Kuchin et al., 2002). When glucose concentrations drop, the activated SNF1 kinase complex can phosphorylate and inactivate repressors MIG1 and NRG1 (Bendrioua et al., 2014). FLO11 is supposed to be repressed by MIG1 as well as NRG1 in glucose repression (Gancedo, 1998; Winderickx et al., 2003). Conversely, an overexpression of MIG1 induces filamentous growth – a morphology confirmed to be primarily under the control of FLO genes (Karunanithi and Cullen, 2012). The effect of MIG1 on the expression of FLO11 and biofilm formation, however, has remained indeterminate in past research.
To explore the effect of transcription regulator MIG1 in yeast biofilm formation, the present study utilized transcription to analyze MIG1 and FLO genes expression changes in industrial yeast during biofilm formation, verifying the effects of these genes on biofilm formation. It was found that in addition to FLO11, three other FLO genes (FLO1, FLO5, and FLO9) are essential for S. cerevisiae biofilm formation during immobilization. Furthermore, MIG1 may function as a regulator of FLO1, 5 and 9 genes, as they responded to MIG1 expression changes during biofilm formation.
Materials and Methods
Yeast Strains and Growth Conditions
Saccharomyces cerevisiae 1308 (Chen et al., 2013) is a diploid industrial strain isolated from fermentative habitats and commonly grown in solid yeast extract peptone dextrose medium (1% yeast extract, 2% peptone, 2% glucose and 2% agar) at 30°C. In this study, yeast strain cultures were grown in liquid yeast extract peptone dextrose medium (1% yeast extract, 2% peptone, and 2% glucose). Fermentation experiments were performed in a fermentation medium containing 20% glucose, 0.4% peptone, 0.4% (NH4)2SO4, 0.3% yeast extract, 0.3% KH2PO4, 0.05% MgSO4, 0.005% ZnSO4⋅7H2O, and 0.005% FeSO4⋅7H2O. To select yeast transformants, G418 Sulfate (345180, Merck, Japan) was added at final concentrations of 400 and 800 μg/mL to solid yeast extract peptone dextrose medium (YPD).
Ethanol fermentations were performed by adding 1 mL overnight cultures to 250 mL flasks already containing 100 mL fermentation medium in the presence or absence of 4 g dry cotton fiber. Flasks were placed on a shaker activated at 250 rpm/min and maintained at 35°C. Continuous batch fermentation was conducted for the immobilized culture; in these, “waste broth” was removed and fresh broth was added as residual glucose was depleted (<1 g/L).
RNA Preparation, cDNA Library Construction and Transcription Profiling Data Analysis
Biofilm cells were isolated from cotton fibers via ultrasonication at three different stages during biofilm development. Planktonic and biofilm cells were collected and washed twice in PBS. Cell pellets were immediately frozen in liquid nitrogen and stored at -80°C. Three biological replicates were prepared from the samples taken under each condition. RNA was isolated from both free and biofilm S. cerevisiae cells using the methods previously described (Cao et al., 2006). A cDNA library was constructed using published methods (Li et al., 2015). The reads per kilobase transcriptome per million mapped reads method (RPKM; Mortazavi et al., 2008) was applied to calculate the expression levels of selected genes. Furthermore, the works of Audic and Claverie (1997) were used to determine the significance of the digital gene expression profiles. This study selected a level of FDR ≤0.001 and absolute value of Log2Ratio ≥1 as criteria for assessing the significance of differential gene expression.
Construction of Deletion and Complemented Mutants
Saccharomyces cerevisiae strain mutants were constructed by deleting corresponding genes in S. cerevisiae 1308, the selected industrial yeast strain, using the homologous recombination system (LFH-PCR: PCR-Synthesis of disruption cassettes with long flanking homology) according to published methodology (Nikawa and Kawabata, 1998). Knock-in component construction is described below. The PCR primers used in this study are listed in Table 1. PCR-generated DNA molecules (knock-in components) consisted of a KanMX marker cassette, as KanMX sequences show G418 resistance in S. cerevisiae and Kanamycin resistance in Escherichia coli. KanMX marker cassettes with long homologous regions (450–500 bp) flanking the target locus was then used for directed gene alterations in S. cerevisiae. Knock-in components were then transformed into competent S. cerevisiae 1308 cells using a Bio-Rad electroporation systems set at 1.5 kV, 25 mF with a 200 Ohm pulse controller. The sorbitol transformation method was used.
Complemented strain MIG1Δ + pMIG1 and FLO11Δ + pFLO11 were constructed by expressing MIG1 and FLO11 in strain MIG1Δ and FLO11Δ by plasmid pYX212-AurR respectively. Plasmids were constructed using ClonExpress® One Step Cloning Kit. MIG1 and FLO11 were amplified with primers pAurR-MIG1-F(R) and pAurR-FLO11-F(R) from S. cerevisiae genome, then it were gel purified and ligated to lineated linearized plasmid pYX212-AurR (using restriction enzyme XbaI and SalI) Plasmid were first constructed in E. coli DH-5α, and then transformed into S. cerevisiae 1308 mutants respectively. All the primers and sequence information can be found as Table 1.
qRT-PCR Analysis
RNA extractions and quality control experiments were performed as described in the previous section. Reverse transcription was performed using an AMV First Strand cDNA Synthesis Kit (Sangon Biotech) according to standard protocols. Primer 5 software was used to select the primers. The analyzed genes and primers used for analysis are listed in Table 2. Quantitative real-time PCR (qRT-PCR) assays were performed using SYBR Green PCR Master Mix (Applied Biosystems) in a StepOnePlus Real-Time PCR System. Reactions were performed according to manufacturer instructions, and three technical replicates with one negative control were performed for each sample. Gene transcription levels were determined according to the 2−ΔΔCT method, using 18s rRNA and FBA1 as reference genes for normalizing gene expression levels (Tofalo et al., 2014; Li et al., 2015; Nadai et al., 2015).
Biofilm Formation on Plastics
Yeast strains were grown in YPD overnight at 30°C. After collection and washing, cells were resuspended in YPD at an OD600 of 1 and transferred to the wells of a microtiter plate where they were incubated for 24 h at 30°C. Four replicate wells were used for each treatment. Biofilm-containing wells were washed twice in 200 μL PBS to remove free cells. Biofilms were then stained with 1% crystal violet, after which wells were washed repeatedly with water and photographed. For quantification, crystal violet was solubilized by adding 100 ml of acetic acid, plates were incubated for 15 min, and the absorbance at 570 nm was measured using a microplate reader.
Confocal Laser Scanning Microscopy (CLSM) Analysis
Biofilm cells growing on cotton fiber media were harvested and immediately stained with FUN-1 and Alexa Fluor 488-conjugated ConA (both from Molecular Probes, Inc., Eugene, OR). FUN-1 (excitation wavelength: 543 nm; emission wavelength: 560 nm) is converted to an orange-red molecule in metabolically active cells, while Alexa-Fluor 488-conjugated ConA (excitation wavelength: 488 nm; emission wavelength: 505 nm) binds to the glucose and mannose residues of cell-wall polysaccharides, displaying green fluorescence. Confocal images were captured using a Leica TCS SP5 II.
Flocculation Assay
Strains were grown in YPD medium for 12 h at 30°C. After being diluted to equivalent OD600 levels, each strain was placed in a separate test tube. The tubes were then vibrated thoroughly to suspend all contained cells and subsequently left to stand until all cells underwent sedimentation. Images were recorded at 5 min intervals, and flocculation ability was measured as the time required for sedimentation to complete.
Standard Plate-Wash Assay
Cells were grown on standard YPD agar plates for 3 days. Observations indicated all strains grew equally well in this environment. Next, each plate was added to 1 ml water and shaken at 50 rpm for 2 min. The water was then discarded and images of the colonies were recorded.
Fermentation Ethanol Resistance Test
First, yeast cells underwent a 3-day immobilization process on cotton fiber under the previously described immobilization conditions (see section ‘Planktonic cultivations and biofilm fermentation’). When the ethanol resistance test was started, the residual medium was discarded into flasks and fresh medium containing selected glucose concentrations was added (other ingredients were the same as above). Different ethanol volumes, ranging from 5% (v/v) to 15% (v/v), were added and 1 ml of samples were drawn from each flask at 4 h intervals. Samples underwent the following procedures: first, samples were centrifuged at 10,000 rpm for 1 min to sediment cells; next, the supernatant was transferred to another 1.5 ml centrifuge tube; and finally, the glucose concentration of the supernatant was tested using the DNS method (3, 5-Dinitrosalicylic acid).
Statistical Analysis
All experiments were done in at least triplicate. The data presented are the means of three or more experiments. Significant differences were determined by Student’s t-test (p < 0.05).
Results and Discussion
Transcriptional Changes in FLO and MIG1 Genes
Whole-genome expression profiling was performed to explore the gene expression differences between biofilm-forming cells and planktonic cells, comparing gene expression during the biofilm attachment (3 h), sessile growth (14 h), and biofilm maturation (30 h) periods (based on mRNA RPKM values). Genes whose expression change by over 2.0 fold were recognized as significantly regulated. Such RNA-Seq results have been proven accurate by Li et al. (2015). Comparing biofilm and planktonic cells, the FLO11 gene was up-regulated by 6.8-fold, 5.0-fold, and 18.4-fold in the three selected periods. FLO1, 5, and 9 were down-regulated at the attachment period, but up-regulated during the maturation period (biofilm vs. planktonic). The expression levels of these FLO genes increased during the biofilm formation process, especially in the maturation period. A different flocculation gene, FLO10, was down-regulated in all three periods (biofilm vs. planktonic), with its expression level in biofilm cells decreasing during biofilm formation (Figure 1A).
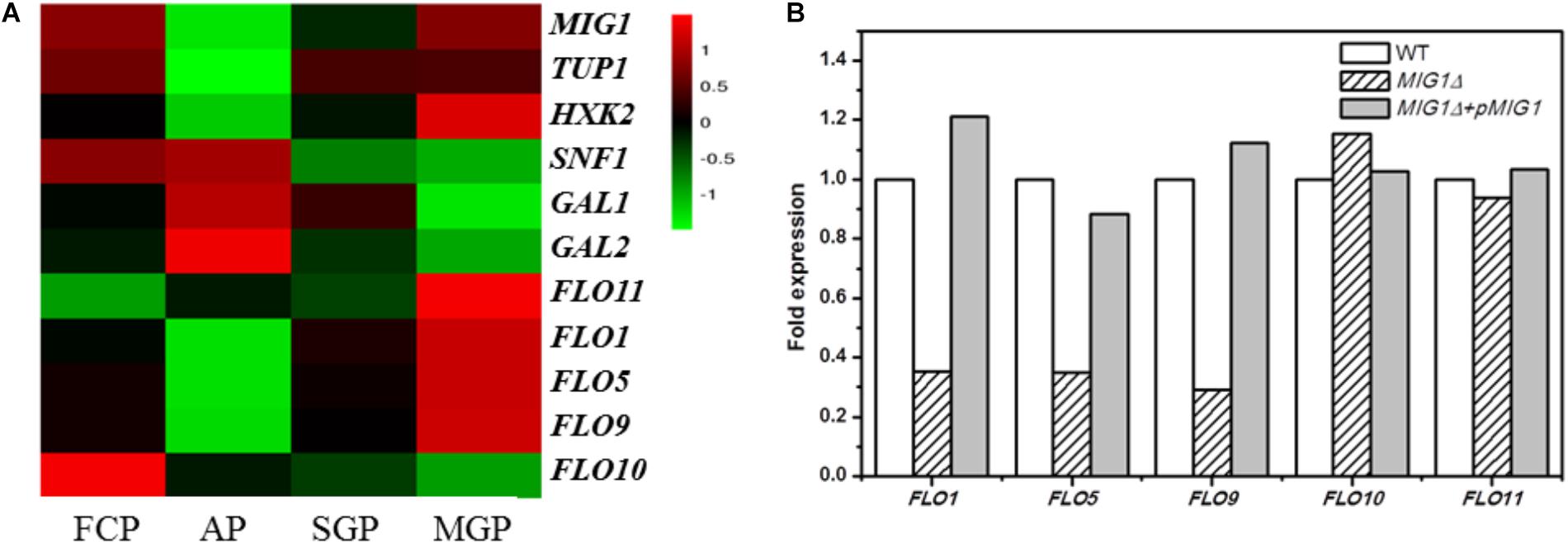
FIGURE 1. Transcriptional proofing and qRT-PCR results. (A) Transcriptional levels of MIG1, its regulated genes and FLO genes in FCP (free cells period), biofilm cells at AP (attachment period), SGP (sessile growth period), and BMP (biofilm maturation period). (B) qRT-PCR results. Relative expression of FLO genes in MIG1Δ and MIG1Δ + pMIG1 compared with wild-type respectively.
Among the investigated genes, the transcription factor gene MIG1 attracted attention with a significant, 27-fold down-regulation during the attachment period, whereas the FLO11 gene, hypothetically located downstream, was up-regulated throughout biofilm formation (biofilm vs. planktonic, as noted above). When biofilm cells were compared across the three periods, however, the expression levels of MIG1 increased alongside FLO11 (Figure 1A). When compared with biofilm cells at attachment, MIG1 was up-regulated 14- and 26-fold during the sessile growth and maturation periods, respectively. SNF1, the encoded primary subunit of the SNF1 kinase complex which can inactivate and repress MIG1 (Bendrioua et al., 2014), was down-regulated. This was especially true during the maturation period when MIG1 expression was maximized in biofilm cells, supporting the theory that MIG1 repression became stronger in later periods. GAL1 and GAL2, which are repressed by MIG1 (Cao et al., 2011), were not significantly down-regulated in biofilm cells during the maturation period relative to the attachment period (Figure 1A). Furthermore, the expression of HXK2, also repressed by MIG1 (Fichtner et al., 2007; Peláez et al., 2012), increased alongside MIG1 during biofilm formation (Figure 1A).
The down-regulated expression of FLO10 was consistent with the results of Verstrepen and Klis (2006), who found that FLO10 expression conferred weak flocculation. The up-regulation of FLO1, 5, and 9 during later periods suggests that cell-cell adhesions play a role in biofilm fermentation. In biofilm cells, the expression levels of FLO1, 5, 9, and 11 increased alongside MIG1 in all three periods of biofilm formation. The varying trends in FLO11 transcription levels resembled those of MIG1, contrary to previous reports that MIG1 represses FLO11 (Verstrepen and Klis, 2006). MIG1 can function as a transcriptional activator in some contexts, particularly in cells lacking the chromatin-remodeling protein encoded by TUP1 (Treitel and Carlson, 1995) which was down-regulated in the biofilm cells investigated here (Figure 1A). Additionally, the GAL1, GAL2, and HXK2 genes repressed by MIG1 were not down-regulated during MIG1 overexpression in the later periods of biofilm formation. These observations failed to illustrate the transcription repressor actions of MIG1 during biofilm formation. In addition, overexpressed MIG1 induced filamentous growth which morphology mainly controlled by FLO genes (Karunanithi and Cullen, 2012). Further studies will thus be required to uncover the role of MIG1 in FLO11 expression and yeast biofilm formation.
MIG1 and FLO Genes Function in Biofilm Formation
To investigate the role of MIG1 in yeast biofilm formation, MIG1 deletion and complemented strains were generated from the wild-type strain. To verify the effects of flocculation genes on the ability of the selected industry strain to form biofilms, mutant strains FLO1Δ, FLO5Δ, FLO9Δ, and FLO11Δ were constructed. Confocal laser scanning microscopy (CLSM) was utilized to observe the biofilms formed on fibers, which live in self-produced matrices of hydrated EPS, primarily polysaccharides (Flemming and Wingender, 2010). FUN-1 and Alexa Fluor 488-conjugated ConA were used to dye live cells red-orange and polysaccharides green respectively. Biofilms were formed after 72 h on cotton fiber substrates. Attachments to the immobilization carrier were less obvious in MIG1Δ and FLO11Δ strains relative to the wild-type. It also can be observed that biofilms formed by MIG1Δ + pMIG1 and FLO11Δ + pFLO11 were restored (Figure 2A). As the attachments of FLO1Δ, FLO5Δ, and FLO9Δ on the immobilization carrier appeared identical to those of the wild-type, their data are not shown.
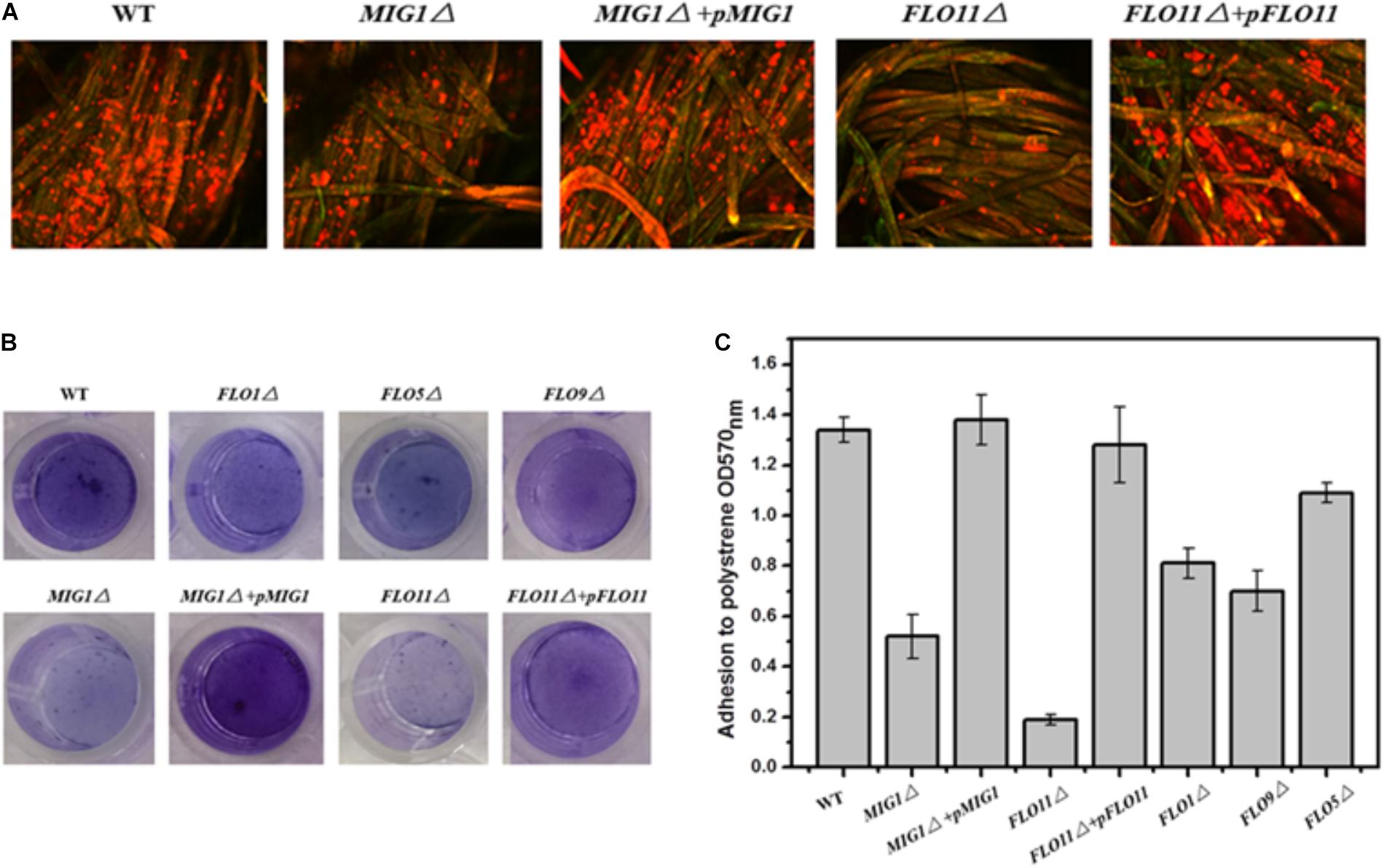
FIGURE 2. Biofilm formation. (A) CLSM images of wide-type, MIG1Δ, MIG1Δ + pMIG1, FLO11Δ and FLO11Δ + pFLO11 biofilms formed on fibers after 24 h fermentation. Strains were dyed orange-red by FUN-1 and polysaccharides were dyed green by Alexa Fluor 488-conjugated ConA. (B) Adherence abilities of wide-type, FLO1Δ, FLO5Δ, FLO9Δ, MIG1Δ, MIG1Δ + pMIG1, FLO11Δ, and FLO11Δ + pFLO11 strains were assayed after incubation in polystyrene 24 h. Wells were washed twice with PBS (in 200 μL) to move away free cells and stained with 1% crystal violet. Wells were washed repeatedly with water and photographed. (C) Adhesion was expressed as OD570 and was measured by solubilizing crystal violet in acetic acid.
The photographs recorded via CLSM were used to roughly evaluate the biofilms formed by these strains. To estimate and distinguish the biofilms of each S. cerevisiae strain, biofilm formation during growth in 96-well plates was quantified using the crystal violet staining method. All deletion mutant strains demonstrated decreases in biofilm formation compared to the wild-type (Figure 2B). FLO11Δ formed only 1/7 of the biofilm produced by the wild-type, the least among these mutant strains. The mutant strains in order decreasing biofilm formation were FLO9Δ > FLO1Δ > FLO5Δ > MIG1Δ > FLO11Δ. Two complemented strains MIG1Δ + pMIG1 and FLO11Δ + pFLO11 formed biofilms as much as wild-type (Figure 2C).
FLO11 has been widely recognized as important in cell-substrate interactions and biofilm formation in yeast. Other FLO genes influence cell–cell interactions and exert an invisible effect on biofilm formation (Guo et al., 2000; Halme et al., 2004). The reduction in biofilm formation in both FLO11Δ in these tests and Σ1278b confirms that FLO11 plays an important role in yeast biofilm formation. However, FLO1 was also required for biofilms in the S288c strain, and its regulation differed from FLO11 (Fichtner et al., 2007). In the 1308 strain used in this study, FLO1, 5, and 9 were all required for biofilm formation. This differed from Σ1278b and S288c (Douglas et al., 2007; Van Mulders et al., 2009). These observations indicated that disabling FLO1, 5, 9, and 11 genes, as well as MIG1, vitiates biofilm formation ability. Cell-cell and cell-surface adhesion abilities were both required for yeast biofilm formation.
Expression Levels of FLO Genes in MIG1 Deletion and Complemented Strain Biofilm Cells
As the transcription repression activities of MIG1 were significantly down-regulated during the biofilm attachment period (biofilm vs. planktonic), the decrease in biofilm formation observed in MIG1Δ was unexpected. qRT-PCR tests were performed on both the wild-type, MIG1Δ and MIG1Δ + pMIG1 cells to examine the expression of FLO genes in biofilm-forming cells (Figure 1B). Expressions in the wild-type condition were defined as 100%, and expressions in mutant strains were calculated as relative numbers to evaluate the effects MIG1 during biofilm formation. The FLO1, 5, and 9 genes were down-regulated in MIG1Δ; however, no pronounced expression difference were observed in the FLO10 and FLO11 genes when 18s rRNA was used as the reference gene. This result was supported when FBA1 was used as an alternative reference gene (Supplementary Figure S1). Expression levels of these genes in MIG1 complemented strain were increased compared with genes in deletion mutant.
According to the qRT-PCR results, FLO1, 5, and 9 were down-regulated in MIG1Δ as compared to the wild-type. Expression levels of these genes were restored to original level in MIG1 complemented strain. This same expression pattern between MIG1 and the FLO1, 5, and 9 genes was observed in biofilm formation, suggesting these FLO genes were regulated by MIG1. Mature biofilms were divided into three parts: the outer, intermediate, and inner surface layers (Jiang et al., 2015). Although cells-surface adhesion is a primary step in biofilm formation and critical for forming the inner biofilm layer (Periasamy et al., 2012), cell–cell adhesions appeared more requisite in the intermediate and outer layers. The expressions of MIG1 and FLO1, 5, and 9 genes decreased during the attachment period but became over-expressed in the maturation period, indicating adhesions are required for biofilm formation. FLO11 was neither up- nor down-regulated in MIG1Δ or MIG1Δ + pMIG1 relative to the wild-type, indicating FLO11 was not regulated by transcription factor MIG1 in the yeast biofilm formation process.
MIG1 Regulated Yeast Flocculation Ability and Invasive Growth
The regulation MIG1 exerted on FLO1, 5, and 9 gene transcriptions indicated it may affect the yeast flocculation ability and adhesion-dependent invasive growth. Phenotype changes were observed between the mutant strains.
To illustrate the effect of MIG1 on flocculation ability, the MIG1and FLO11 mutant and complemented strains were compared (Figure 3A). The MIG1Δ strain lost a majority of its flocculation ability and MIG1Δ + pMIG1 restored the ability; conversely, FLO11 deletion and complemented had no influence on flocculation. A plate-wash test was performed to evaluate the impact of MIG1 on invasive cell growth (Figure 3B). In this test, FLO11Δ was observed to have the fewest colonies remaining on the agar plate, suggesting a weakened invasive ability. MIG1Δ also demonstrated slightly decreased invasive growth. The invasive growth was restored by overexpressing the two genes in its corresponding mutant strains respectively.
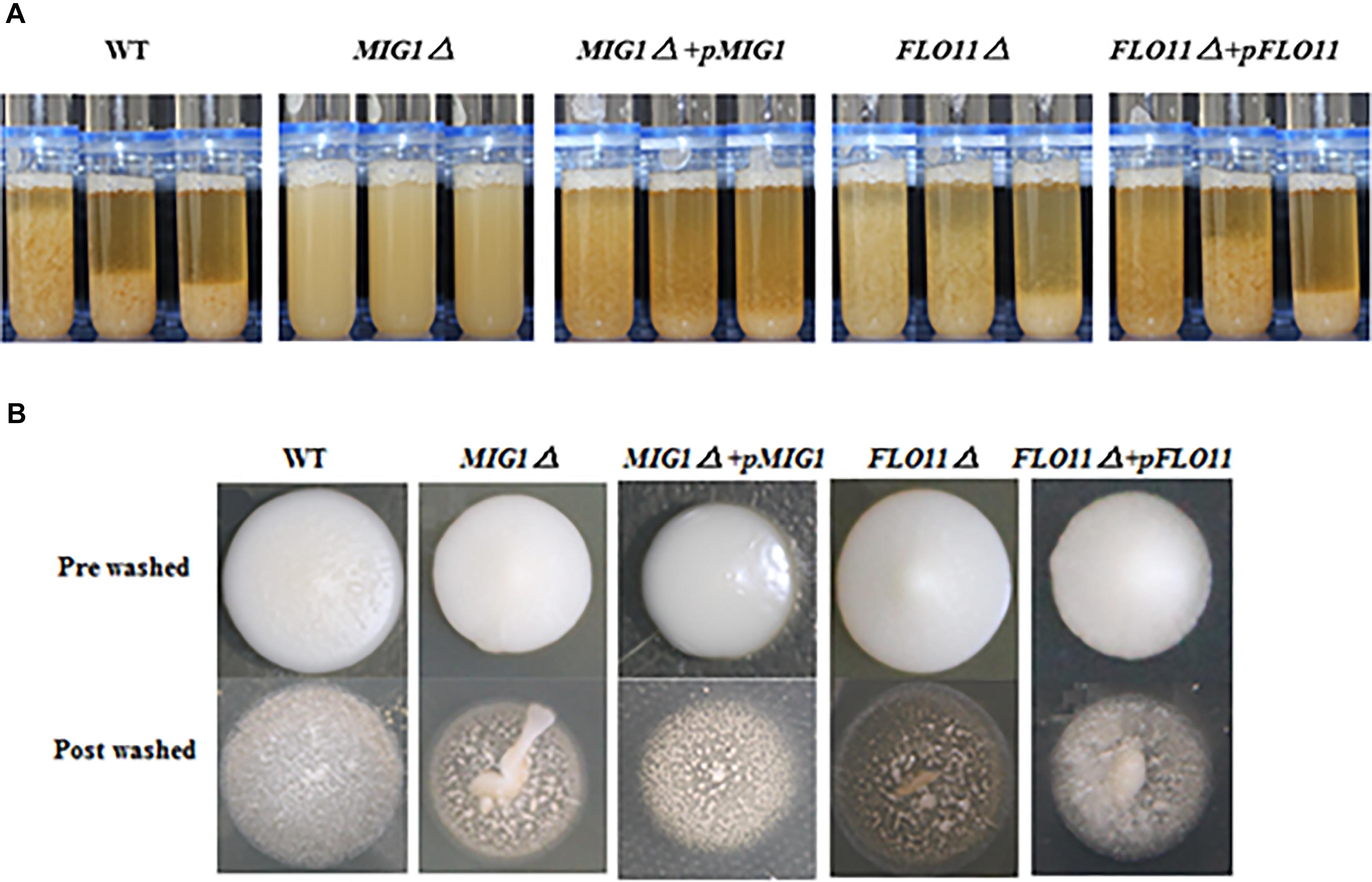
FIGURE 3. Invasive growth and flocculation ability. (A) Wild-type, MIG1Δ, MIG1Δ + pMIG1, FLO11Δ, and FLO11Δ + pFLO11 strains were photographed every 5 min. (B) Plate wash tests of wide-type, MIG1Δ, MIG1Δ + pMIG1, FLO11Δ, and FLO11Δ + pFLO11. Strains were grown on YPD agars for 3 days, same amount of water were added and plates were placed on the shaker for 10 min. Photos of pre-washed strains and post-washed strains were taken.
The loss of flocculation ability and decreased expression of the FLO1, 5, and 9 genes in MIG1Δ confirmed that MIG1 play an important role in cell–cell adhesion by regulating the expression levels of those genes. FLO11 was not required for cell–cell adhesion, as reported by Fichtner et al. (2007). Less obviously, the decrease in invasive growth shown by MIG1Δ may have been caused by a reduction in the invasive growth glycoproteins encoded by FLO1, 5, and 9 which regulated biofilm formation with two possible aspects: first, FLO1, 5, and 9-dependent cell–cell adhesions were required for the intermediate and outer layers of mature biofilm, and these genes were overexpressed during appropriate formation periods. Second, the expression levels of FLO1, 5, and 9 influenced cell-surface adhesion, suggesting these FLO genes influence the maintenance of stability in inner layers.
MIG1 Function on Ethanol Resistance
In fermentation experiments, conducted to investigate the abilities of mutant strain biofilms to survive during the accumulation of high ethanol concentrations, strain growth was inhibited and glucose consumption rates decreased when ethanol concentrations were high. The residual glucose concentrations were measured to demonstrate the environmental resistance ability of each strain.
In the 0 and 5% ethanol groups, glucose was depleted within 8 h. Although the glucose consumption rates of these strains in 10% were slower than in 5% group, there was no obvious difference among the three strains in a same group (Figures 4A,B); however, the consumption rates of all strains were slower in 10% ethanol relative to 5%. In the 10% ethanol group, both wild-type and MIG1Δ strains depleted their glucose supply at approximately 12 h, whereas FLO11Δ took 16 h. The glucose consumption rates of MIG1Δ and FLO11Δ were both slower than that of the wild-type (Figure 4C). When ethanol concentrations reached 15%, the wild-type strain had depleted all available glucose after 16 h. At 24 h, however, there were 2 g/L and 12 g/L glucose remaining in the MIG1Δ and FLO11Δ samples, respectively (Figure 4D). MIG1 and FLO11 complemented strains depleted glucose faster than its deletion strains in 10 and 15% ethanol groups.
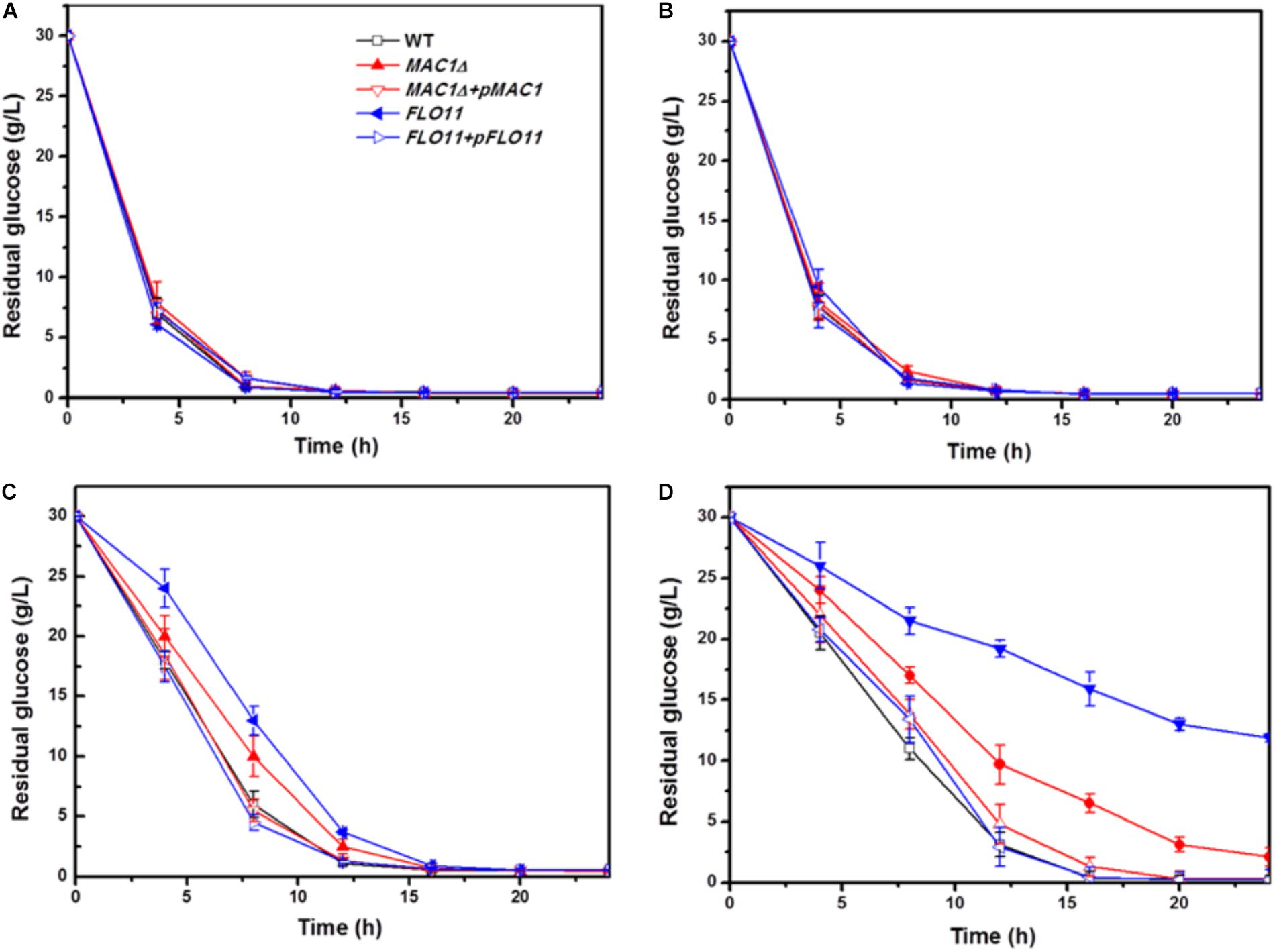
FIGURE 4. Glucose concentration change during fermentation in different ethanol concentration. (A–D) Residual concentrations of glucose were measured under 0, 5, 10, and 15% ethanol concentrations, in biofilm state fermentation.
Previous studies have shown that FLO11 contributes to biofilm formation, and is therefore required for environmental resistance (Váchová et al., 2011). The present results confirmed that FLO11 deficiency abated the ethanol tolerance of cell strains in biofilm fermentation. Flocculation, dependent on cell–cell adhesion, plays an important role in the ethanol resistance of cells as well as biofilm formation in many yeast strains. Flocculation has been restored by an overexpression of FLO11 in laboratory strain S288C, protecting inner cells from multiple stresses including high ethanol concentrations (Smukalla et al., 2008). Furthermore, flocculation strains showed high levels of FLO5 expression and significant resistance to ethanol stress (Tofalo et al., 2014). Cell–cell adhesions, which occur during the later biofilm formation stages, play important roles in ethanol resistance and cell-surface adhesion. The decreased ethanol resistance of MIG1Δ may have resulted from decreased cell–cell and cell–surface adhesions. FLO11Δ showed weaker biofilm formation and ethanol resistance than MIG1Δ, suggesting cell–surface adhesions are a primary factor in ethanol resistance.
Conclusion
This study focused on elucidating the roles played by the MIG1 (transcription factor) and FLO (adhesion) genes in yeast biofilm formation and ethanol resistance. Results showed FLO11, which confers cell-surface adhesion abilities, played an important role in yeast biofilm formation but was not repressed by transcription factor MIG1. MIG1 regulating the expression levels of FLO1, 5 and 9 which affected yeast biofilm formation respectively. According to these findings, this work presents the possibility that FLO1, 5, and 9 gene expressions were increased at later stages and high ethanol concentrations, regulated by MIG1, in order to protect cells by keeping or increasing the outer layer of biofilms via enhanced cell-cell adhesions. The present work serves as a basis for future studies to examine the complex network systems that regulate S. cerevisiae biofilm formation and maintenance, as more work will be necessary to elucidate the regulation pathways by which MIG1 influences these FLO genes in S. cerevisiae biofilm development.
Availability of Data and Material
The datasets supporting the conclusions of this article are included within the article and its additional files.
Author Contributions
LY participated in the design of the study, constructed the strains, participated in the fermentation experiments, drafted the manuscript, and revised the manuscript. CZ participated in the design of the study, the fermentation experiments, and drafted the manuscript. YC participated in the design of the study. HY conceived the study and participated in its design. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Basic Research Program of China (973) (2013CB733602), the Major Research Plan of the National Natural Science Foundation of China (21390204), the Key Program of the National Natural Science Foundation of China (21636003), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R28), Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM), the Technology Support Program of Jiangsu (Grant No. BE2014715), the Outstanding Youth Foundation of Jiangsu (Grant No. SBK2017010373), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01860/full#supplementary-material
FIGURE S1 | Relative expression of FLO genes in MIG1Δ compared with wild-type respectively using FBA1 as reference gene.
References
Almirante, B., Rodríguez, D., Cuencaestrella, M., Almela, M., Sanchez, F., Ayats, J., et al. (2006). Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 44, 1681–1685. doi: 10.1128/JCM.44.5.1681-1685.2006
Andersen, K. S., Bojsen, R., Sørensen, L. G. R., Nielsen, M. W., Lisby, M., Folkesson, A., et al. (2014). Genetic basis for Saccharomyces cerevisiae biofilm in liquid medium. G3 4, 1671–1680. doi: 10.1534/g3.114.010892
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Bendrioua, L., Smedh, M., Almquist, J., Cvijovic, M., Jirstrand, M., Goksör, M., et al. (2014). Yeast AMP-activated protein kinase monitors glucose concentration changes and absolute glucose levels. J. Biol. Chem. 289, 12863–12875. doi: 10.1074/jbc.M114.547976
Bester, M. C., Dan, J., and Bauer, F. F. (2012). Many Saccharomyces cerevisiae cell wall protein encoding genes are coregulated by Mss11, but cellular adhesion phenotypes appear only Flo protein dependent. G3 2, 131–141. doi: 10.1534/g3.111.001644
Blankenship, J. R., and Mitchell, A. P. (2006). How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9, 588–594. doi: 10.1016/j.mib.2006.10.003
Bojsen, R. K., Andersen, K. S., and Regenberg, B. (2012). Saccharomyces cerevisiae — a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 65, 169–182. doi: 10.1111/j.1574-695X.2012.00943.x
Cao, H., Yue, M., Li, S., Bai, X., Zhao, X., and Du, Y. (2011). The impact of MIG1 and/or MIG2 disruption on aerobic metabolism of succinate dehydrogenase negative Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 89, 733–738. doi: 10.1007/s00253-010-2894-7
Cao, Y., Cao, Y., Xu, Z., Ying, K., Li, Y., Xie, Y., et al. (2006). cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob. Agents Chemother. 26, 404–404. doi: 10.1128/AAC.49.2.584-589.2005
Chavel, C. A., Caccamise, L. M., Li, B., and Cullen, P. J. (2014). Global regulation of a differentiation MAPK pathway in yeast. Genetics 198, 1309–1328. doi: 10.1534/genetics.114.168252
Chen, Y., Liu, Q., Zhou, T., Li, B., Yao, S., Li, A., et al. (2013). Ethanol production by repeated batch and continuous fermentations by Saccharomyces cerevisiae immobilized in a fibrous bed bioreactor. J. Microbiol. Biotechnol. 23, 511–517. doi: 10.4014/jmb.1209.09066
Di Gianvito, P., Tesnière, C., Suzzi, G., Blondin, B., and Tofalo, R. (2017). FLO 5 gene controls flocculation phenotype and adhesive properties in a Saccharomyces cerevisiae sparkling wine strain. Sci. Rep. 7:10786. doi: 10.1038/s41598-017-09990-9
Douglas, L. M., Li, L., Yang, Y., and Dranginis, A. M. (2007). Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 6, 2214–2221. doi: 10.1128/EC.00284-06
D’Urzo, N., Martinelli, M., Pezzicoli, A., De Cesare, V., Pinto, V., Margarit, I., et al. (2014). Acidic pH strongly enhances in vitro biofilm formation by a subset of hypervirulent ST-17 Streptococcus agalactiae strains. Appl. Environ. Microbiol. 80, 2176–2185. doi: 10.1128/AEM.03627-13
Fichtner, L., Schulze, F., and Braus, G. H. (2007). Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Mol. Microbiol. 66, 1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Fong, J. N., and Yildiz, F. H. (2015). Biofilm matrix proteins. Microbiol. Spectr. 3:MB-0004-2014. doi: 10.1128/microbiolspec.MB-0004-2014
Gagiano, M., Bester, M., Dyk, D. V., Franken, J., Bauer, F. F., and Pretorius, I. S. (2003). Mss11p is a transcription factor regulating pseudohyphal differentiation, invasive growth and starch metabolism in Saccharomyces cerevisiae in response to nutrient availability. Mol. Microbiol. 47, 119–134. doi: 10.1046/j.1365-2958.2003.03247.x
Geier, H., Mostowy, S., Cangelosi, G. A., Behr, M. A., and Ford, T. E. (2008). Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 74, 1798–1804. doi: 10.1128/AEM.02066-07
Germec, M., Turhan, I., Karhan, M., and Demirci, A. (2015). Ethanol production via repeated-batch fermentation from carob pod extract by using Saccharomyces cerevisiae in biofilm reactor. Fuel 161, 304–311. doi: 10.1016/j.fuel.2015.08.060
Grudniak, A. M., Wlodkowska, J., and Wolska, K. I. (2015). Chaperone DnaJ influences the formation of biofilm by Escherichia coli. Pol. J. Microbiol. 64, 279–283. doi: 10.5604/01.3001.0009.2123
Guo, B., Styles, C. A., Feng, Q., and Fink, G. R. (2000). A Saccharomyces gene family involved in invasive growth, cell–cell adhesion, and mating. Proc. Natl. Acad. Sci. U.S.A. 97, 12158–12163. doi: 10.1073/pnas.220420397
Halme, A., Bumgarner, S., Styles, C., and Fink, G. R. (2004). Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116, 405–415. doi: 10.1016/S0092-8674(04)00118-7
Jiang, S., Huang, X., Zhang, C., Cai, Z., and Zou, T. (2015). Morphological and proteomic analyses of the biofilms generated by Streptococcus mutans, isolated from caries-active and caries-free adults. J. Dent. Sci. 10, 206–215. doi: 10.1016/j.jds.2014.09.001
Karunanithi, S., and Cullen, P. J. (2012). The filamentous growth MAPK pathway responds to glucose starvation through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics 192, 869–887. doi: 10.1534/genetics.112.142661
Kuchin, S., Vyas, V. K., and Carlson, M. (2002). Snf1 protein kinase and the repressors NRG1 and NRG2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22, 3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002
Kuhn, D. M., George, T., Chandra, J., Mukherjee, P. K., and Ghannoum, M. A. (2002). Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46, 1773–1780. doi: 10.1128/AAC.46.6.1773-1780.2002
Lambrechts, M. G., Bauer, F. F., Marmur, J., and Pretorius, I. S. (1996). Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 8419–8424. doi: 10.1073/pnas.93.16.8419
Li, B., Chen, Y., Chen, X., Liu, D., Niu, H., Xiong, J., et al. (2012). A novel immobilization method for nuclease P on macroporous absorbent resin with glutaraldehyde cross-linking and determination of its properties. Process Biochem. 47, 665–670. doi: 10.1016/j.procbio.2012.01.008
Li, Z. J., Chen, Y., Liu, D., Zhao, N., Cheng, H., Ren, H. F., et al. (2015). Involvement of glycolysis/gluconeogenesis and signaling regulatory pathways in Saccharomyces cerevisiae biofilms during fermentation. Front. Microbiol. 6:139. doi: 10.3389/fmicb.2015.00139
Liu, D., Xu, J., Wang, Y., Chen, Y., Shen, X., Niu, H., et al. (2016). Comparative transcriptomic analysis of Clostridium acetobutylicum, biofilm and planktonic cells. J. Biotechnol. 218, 1–12. doi: 10.1016/j.jbiotec.2015.11.017
Liu, G. L., Wang, D. S., Wang, L. F., Zhao, S. F., and Chi, Z. M. (2011). Mig1 is involved in mycelial formation and expression of the genes encoding extracellular enzymes in Saccharomycopsis fibuligera A11. Fungal Genet. Biol. 48, 904–913. doi: 10.1016/j.fgb.2011.04.008
Madhani, H. D., and Fink, G. R. (1997). Combinatorial control required for the specificity of yeast MAPK signaling. Science 275, 1314–1317. doi: 10.1126/science.275.5304.1314
Moreno-García, J., Coi, A. L., Zara, G., García-Martínez, T., Mauricio, J. C., and Budroni, M. (2018). Study of the role of the covalently linked cell wall protein (Ccw14p) and yeast glycoprotein (Ygp1p) within biofilm formation in a flor yeast strain. FEMS Yeast Res. 18:foy005. doi: 10.1093/femsyr/foy005
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nadai, C., Campanaro, S., Giacomini, A., and Corich, V. (2015). Selection and validation of reference genes for quantitative real-time PCR studies during Saccharomyces cerevisiae alcoholic fermentation in the presence of sulfite. Int. J. Food Microbiol. 215, 49–56. doi: 10.1016/j.ijfoodmicro.2015.08.012
Nakagawa, Y., Arai, Y., Toda, Y., Yamamura, H., Okuda, T., Hayakawa, M., et al. (2017). Glucose repression of FLO11 gene expression regulates pellicle formation by a wild pellicle-forming yeast strain isolated from contaminated wine. Biotechnol. Biotechnol. Equip. 31, 1–8. doi: 10.1080/13102818.2016.1246203
Nikawa, J., and Kawabata, M. (1998). PCR- and ligation-mediated synthesis of marker cassettes with long flanking homology regions for gene disruption in Saccharomyces cerevisiae. Nucleic Acids Res. 26, 860–861. doi: 10.1093/nar/26.3.860
Pal, A., and Khanum, F. (2011). Covalent immobilization of xylanase on glutaraldehyde activated alginate beads using response surface methodology: characterization of immobilized enzyme. Process Biochem. 46, 1315–1322. doi: 10.1016/j.procbio.2011.02.024
Peláez, R., Fernández-García, P., Herrero, P., and Moreno, F. (2012). Nuclear import of the yeast hexokinase 2 protein requires α/β-importin-dependent pathway. J. Biol. Chem. 287, 3518–3529. doi: 10.1074/jbc.M111.317230
Periasamy, S., Joo, H. S., Duong, A. C., Bach, T. H. L., Tan, V. Y., Chatterjee, S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286. doi: 10.1073/pnas.1115006109
Reynolds, T. B., and Fink, G. R. (2001). Bakers’ yeast, a model for fungal biofilm formation. Science 291, 878–881. doi: 10.1126/science.291.5505.878
Smukalla, S., Caldara, M., Pochet, N., Beauvais, A., Guadagnini, S., Yan, C., et al. (2008). FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 726–737. doi: 10.1016/j.cell.2008.09.037
Stringer, J. R., and Keely, S. P. (2001). Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69, 627–639. doi: 10.1128/IAI.69.2.627-639.2001
Kumar, A. S., Mody, K., and Jha, B. (2007). Bacterial exopolysaccharides–a perception. J. Basic Microbiol. 47, 103–117. doi: 10.1002/jobm.200610203
Teunissen, A. W., and Steensma, H. Y. (1995). Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11, 1001–1013. doi: 10.1002/yea.320111102
Tofalo, R., Perpetuini, G., Di Gianvito, P., Schirone, M., Corsetti, A., and Suzzi, G. (2014). Genetic diversity of FLO1 and FLO5 genes in wine flocculent Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 191, 45–52. doi: 10.1016/j.ijfoodmicro.2014.08.028
Treitel, M. A., and Carlson, M. (1995). Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. U.S.A. 92, 3132–3136. doi: 10.1073/pnas.92.8.3132
Tseng, B. S., Reichhardt, C., Merrihew, G. E., Araujo-Hernandez, S. A., Harrison, J. J., MacCoss, M. J., et al. (2018). A biofilm matrix-associated protease inhibitor protects Pseudomonas aeruginosa from proteolytic attack. mBio 9:e00543-18. doi: 10.1128/mBio.00543-18
Váchová, L., Stovícek, V., Hlavácek, O., Chernyavskiy, O., Stepánek, L., Kubínová, L., et al. (2011). Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. Commun. Integr. Biol. 194, 679-687. doi: 10.1083/jcb.201103129
Van Mulders, S. E., Christianen, E., Saerens, S. M., Daenen, L., Verbelen, P. J., Willaert, R., et al. (2009). Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9, 178–190. doi: 10.1111/j.1567-1364.2008.00462.x
Verstrepen, K. J., and Klis, F. M. (2006). Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60, 5–15. doi: 10.1111/j.1365-2958.2006.05072.x
Villa, F., Cappitelli, F., Cortesi, P., and Kunova, A. (2017). Fungal biofilms: targets for the development of novel strategies in plant disease management. Front. Microbiol. 8:654. doi: 10.3389/fmicb.2017.00654
Watanabe, J., Uehara, K., and Mogi, Y. (2013). Adaptation of the osmotolerant yeast Zygosaccharomyces rouxii to an osmotic environment through copy number amplification of FLO11D. Genetics 195, 393–405. doi: 10.1534/genetics.113.154690
Winderickx, J., Holsbeeks, I., Lagatie, O., Giots, F., Thevelein, J., and Han, D. W. (2003). From feast to famine; adaptation to nutrient availability in yeast. Top. Curr. Genet. 1, 305–386. doi: 10.1007/3-540-45611-2_7
Keywords: Saccharomyces cerevisiae, transcription factor MIG1, flocculation genes, biofilm, ethanol resistance
Citation: Yang L, Zheng C, Chen Y and Ying H (2018) FLO Genes Family and Transcription Factor MIG1 Regulate Saccharomyces cerevisiae Biofilm Formation During Immobilized Fermentation. Front. Microbiol. 9:1860. doi: 10.3389/fmicb.2018.01860
Received: 05 May 2018; Accepted: 24 July 2018;
Published: 23 August 2018.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyCarolina Henritta Pohl, University of the Free State, South Africa
Copyright © 2018 Yang, Zheng, Chen and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Chen, Y2hlbnlvbmcxOTgyQG5qdGVjaC5lZHUuY24= Hanjie Ying, eWluZ2hhbmppZUBuanRlY2guZWR1LmNu
†These authors have contributed equally to this work
 Leyun Yang
Leyun Yang Cheng Zheng1,2†
Cheng Zheng1,2† Yong Chen
Yong Chen Hanjie Ying
Hanjie Ying