- 1School of Animal and Comparative Biomedical Sciences, The University of Arizona, Tucson, AZ, United States
- 2Department of Immunobiology, The University of Arizona, Tucson, AZ, United States
- 3Bio5 Institute for Collaborative Research, The University of Arizona, Tucson, AZ, United States
- 4Southern Arizona VA Health Care System, Tucson, AZ, United States
Morbidity and mortality attributed to Clostridium difficile infection (CDI) have increased over the past 20 years. Currently, antibiotics are the only US FDA-approved treatment for primary C. difficile infection, and these are, ironically, associated with disease relapse and the threat of burgeoning drug resistance. We previously showed that non-toxin virulence factors play key roles in CDI, and that colonization factors are critical for disease. Specifically, a C. difficile adhesin, Surface Layer Protein A (SlpA) is a major contributor to host cell attachment. In this work, we engineered Syn-LAB 2.0 and Syn-LAB 2.1, two synthetic biologic agents derived from lactic acid bacteria, to stably and constitutively express a host-cell binding fragment of the C. difficile adhesin SlpA on their cell-surface. Both agents harbor conditional suicide plasmids expressing a codon-optimized chimera of the lactic acid bacterium’s cell-wall anchoring surface-protein domain, fused to the conserved, highly adherent, host-cell-binding domain of C. difficile SlpA. Both agents also incorporate engineered biocontrol, obviating the need for any antibiotic selection. Syn-LAB 2.0 and Syn-LAB 2.1 possess positive biophysical and in vivo properties compared with their parental antecedents in that they robustly and constitutively display the SlpA chimera on their cell surface, potentiate human intestinal epithelial barrier function in vitro, are safe, tolerable and palatable to Golden Syrian hamsters and neonatal piglets at high daily doses, and are detectable in animal feces within 24 h of dosing, confirming robust colonization. In combination, the engineered strains also delay (in fixed doses) or prevent (when continuously administered) death of infected hamsters upon challenge with high doses of virulent C. difficile. Finally, fixed-dose Syn-LAB ameliorates diarrhea in a non-lethal model of neonatal piglet enteritis. Taken together, our findings suggest that the two synthetic biologics may be effectively employed as non-antibiotic interventions for CDI.
Introduction
The fastidiously anaerobic, Gram-positive bacillus Clostridium difficile is currently the largest contributor to healthcare-associated infections in US hospitals (McDonald et al., 2018). In the United States, an estimated 450,000 cases of C. difficile infections (CDI) occur annually (Lessa et al., 2015), costing the healthcare system ≥ $5.4 billion ( Desai et al., 2016). CDI symptoms range from mild/moderate diarrhea, which can progress to serious sequelae including pseudomembranous colitis. Antibiotic suppression of gut flora facilitates colonization by C. difficile spores that are commonly present in the environment, which germinate into vegetative cells that produce damaging toxins during late growth phase. Toxigenic (toxin-producing) C. difficile strains harbor a 19.6 kb genomic island called the Pathogenicity Locus (PaLoc), that encodes the glucosyltransferase toxins TcdA (309 kDa) and TcdB (267 kDa), which target host-cell G-proteins (Lyras et al., 2009). A third ADP ribosylase toxin CDT (Binary toxin) is present in some strains.
Clostridium difficile epidemiology has altered markedly in the past 20 years. Highly virulent strains, associated with severe disease, increased recurrence rate(s) and community onset, have become more prevalent (Ofori et al., 2018). Common human outbreak-associated strains are typed as North American Pulsed-Field type 1 (NAP1) and PCR ribotype 027 (NAP1/027) (Loo et al., 2005). Common veterinary strains (now also isolated from humans) belong to the NAP7/NAP8 clade (Ribotype 078) (Moono et al., 2016). There have been several worldwide outbreaks of NAP1/027/BI CDI since 2002 (Loo et al., 2005).
Bacterial adherence is an important C. difficile virulence attribute, with Surface-Layer proteins (SLPs; also known as cell-wall proteins, CWPs) playing key roles. C. difficile elaborates up to 29 different SLPs, which are displayed in para-crystalline architecture on the cell surface. C. difficile SLPs are also implicated in immune modulation; thus, they are critical non-toxin virulence factors (Bianco et al., 2011; Bruxelle et al., 2016). While SLPs have been proposed as anti-CDI vaccine candidates, many groups (including ours) have reported variability in SLP epitope antigenicity (Biazzo et al., 2013).
Surface-layer protein A (SlpA) and its orthologs are abundant members of the CWP complex in clostridia and lactobacilli. We previously published that pre-incubating human intestinal epithelial cells with C. difficile SlpA-enriched preparations, or purified SlpA, or bacteria with anti-SlpA antisera, reduced > 50% C. difficile adherence in a dose-dependent manner, implicating SlpA as a major adhesin (Merrigan et al., 2013). Notably, SlpA from a non-toxigenic C. difficile isolate blocked adhesion of the strain from which it was derived, as well as a phylogenetically unrelated, non-cognate strain. The degree of adherence inhibition was similar irrespective of the challenge isolate. SlpA is a heterodimer of high-and low-molecular weight (HMW and LMW) subunits. Both subunits bind independently to intestinal cells, with the LMW subunit displaying higher binding efficiency. This guided the engineering of our synthetic strains.
The typical requirement of antibiotics to precipitate CDI, as well as the remarkable efficacy of fecal microbiota transplants in treating refractory CDI, point to one unequivocal conclusion: colonization resistance is an effective and ‘natural’ method to combat CDI (Austin et al., 2014; Terveer et al., 2018). At a practical level, however, fecal transplantation may not be the ideal therapeutic option for all CDI patients (Wang et al., 2016; Gardiner et al., 2018). Alternate approaches that exploit colonization resistance for CDI prevention and/or cure are active areas of investigation (Allen-Vercoe and Petrof, 2013; Petrof and Khoruts, 2014). Probiotics, particularly lactic acid bacteria (LAB), have been considered as safe, palatable options to confer colonization resistance. LAB occupy the same gut niches as C. difficile in humans and rodents (Marco et al., 2007), and proliferate to the same or greater extent as C. difficile (Chiu et al., 2006). Although LAB can reduce symptoms in some patients, meta-analyses and large-cohort studies suggest variability in LAB protection against CDI (Mergenhagen et al., 2014; Goldenberg et al., 2017). The basis for this variability is unknown, and may reflect inconsistent gut colonization and persistence. Since LAB harbor SLP orthologs and can express heterologous SLP molecules on their surface (Raha et al., 2005; Zhu et al., 2010), we exploited these properties to engineer targeted synthetic biologics with enhanced colonization and immune elicitation properties.
Materials and Methods
Cell Lines
The human intestinal epithelial cell line C2BBe, a brush border-expressing Caco-2 sub-clone (Peterson and Mooseker, 1992), was used in this study and cultured as previously reported (Roxas et al., 2014).
Bacterial Strains and Plasmids
All strains and plasmid are described in Table 1. LAB were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). Specifically, Lactobacillus casei strain 334 (Orla-Jensen; Dellaglio et al., 2002), and Lactobacillus acidophilus strain 4356 (Roussel et al., 1993) were used for these studies. LAB were grown in De Man, Rogosa and Sharpe (MRS) broth (Duong et al., 2011) and incubated at 30°C in the presence of 5% CO2. Bacteria were cultured for 3–5 days to reach saturation [≥1.0 × 108 colony forming units (CFU) per mL].
Lactobacillus acidophilus and L. casei strains ferment the dextrose in MRS to distinct products, and the corresponding pH changes can be detected by including bromophenol blue into the media (MRS-BPB) (Lee and Lee, 2008). L. acidophilus, a homo-fermenter, metabolizes dextrose to lactic acid, and the plates remain violet/blue (pH > 4.6); L. casei, a hetero-fermenter converts dextrose to acetic acid, and the drop in pH (<3.0) results in a color change to yellow/white. Further, L. casei, unlike L. acidophilus, can ferment mannitol, and this can be verified by growth on Purple Broth Base (DifcoTM Becton, Dickinson and Company Sparks, Glencoe, MD, United States); the acidic change resulting from mannitol fermentation causes the pH indicator bromocresol purple to turn yellow.
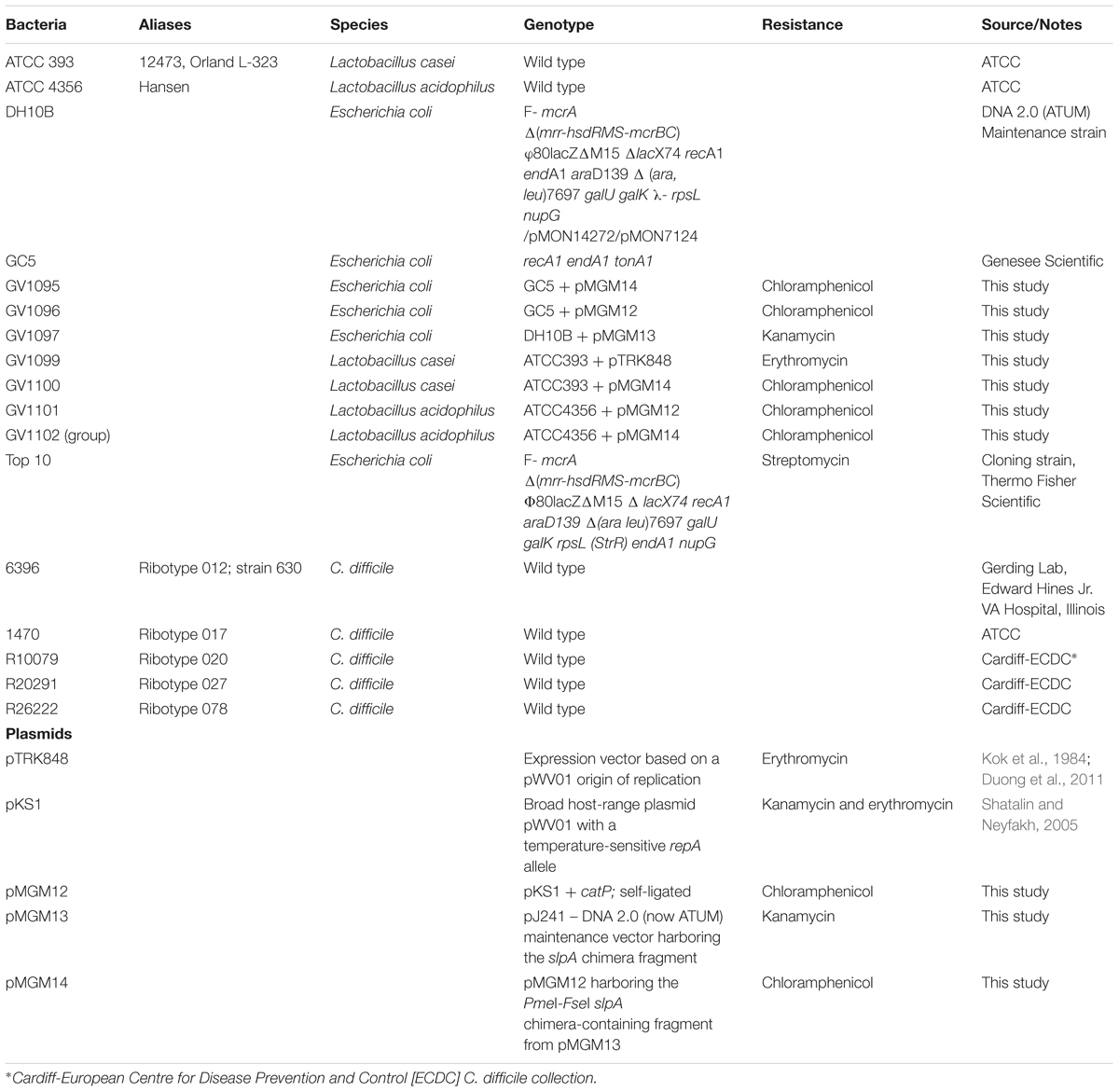
The slpA C. difficile/L. acidophilus “chimera” fragment (Figure 1A) was designed with a strong lactic-acid-bacterial (LAB) promoter [endogenous to the phosphoglycerate mutase (pgm) gene in plasmid pTRK848 (Duong et al., 2011)], a lactic-acid-bacterial Shine-Dalgarno (ribosome binding site) sequence (Duong et al., 2011), a signal sequence from a Lactobacillus acidophilus S-layer protein, a codon-optimized C. difficile strain 630 host-cell-binding fragment, and the L. acidophilus SlpA-ortholog cell-wall-binding domain(Michon et al., 2016). The entire fragment (F1) was chemically synthesized (DNA 2.0, now ATUM, Newark, CA, United States), and cloned into the DNA 2.0 maintenance vector pJ241 to yield pMGM13. A second DNA fragment (F2) comprising a broad host-range temperature-sensitive origin of replication (repA), and a chloramphenicol resistance gene (catP) was also chemically synthesized based on sequence information obtained from the broad host-range plasmid [pKS1, Steen et al., 2010]. This fragment was self-ligated to form pMGM11 (“empty vector”). The two synthesized fragments F1 and F2 harbored PmeI and FseI restriction sites at their termini; following digestion with those enzymes, linear F1 and F2 were ligated using T4 DNA ligase (Sigma, St. Louis, MO, United States) to generate pMGM14. Plasmid integrity of pMGM14 was confirmed by complete DNA sequencing. All plasmids were transformed into Escherichia coli strains DH10B, TOP10 or NCK1753 for maintenance or propagation purposes, or into E. coli GC5 prior to extraction for electroporation. All plasmids were PCR-verified prior to any use in LAB.
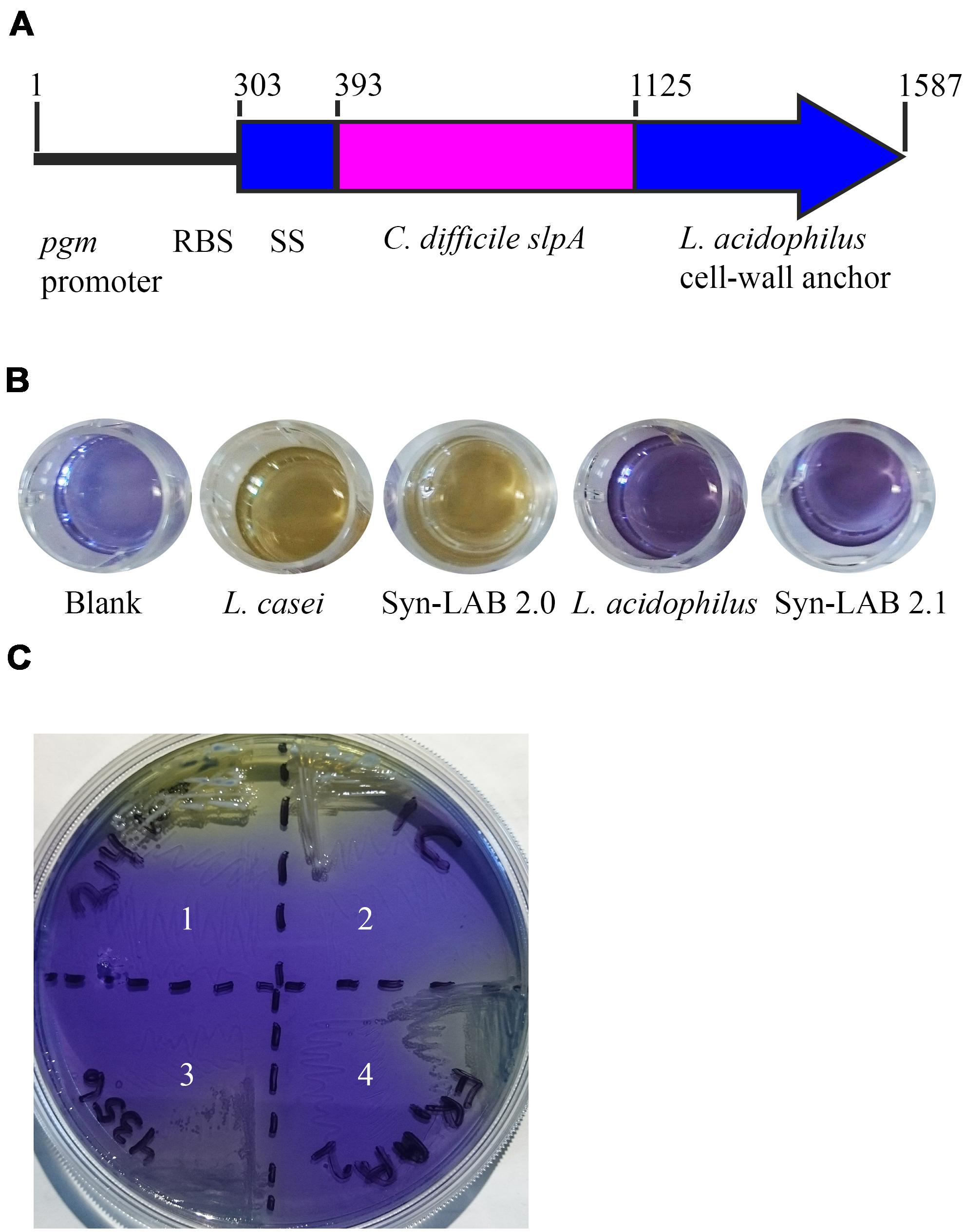
FIGURE 1. Design and confirmation of Syn-LAB strains. (A) Schematic of the slpA chimera. Expression is driven by the L. acidophilus (LA) pgm promoter and ribosome-binding site. The chimera encodes the LA signal sequence (SS), the codon-optimized C. difficile extracellular and antigenic SlpA fragment, and the LA Slp cell wall anchor. Syn-LAB strain confirmation via differential growth on mannitol-MRS broth (B) and mMRS-BPB agar (C). 1, L. casei parent strain; 2, Syn-LAB 2.0; 3, L. acidophilus parent strain; 4, Syn-LAB 2.1.
Lactic Acid Bacterial Transformation
The pMGM14 plasmid described above was first extracted from the E. coli storage strain, and 10 μg used for each LAB transformation. Lactobacillus acidophilus strain 4356 and Lactobacillus casei 343 were propagated in Mann-Rogosa-Sharpe (MRS) medium (Remel, Lenexa, KS, United States) per the method of Walker et al. (1996) and Kim et al. (2005). In brief, LAB strains were grown to saturation at 37°C in 5% CO2, sub-cultured at a 1:50 dilution, and re-grown using the same process two more times. To shear cell-wall proteins, and prepare the resulting protoplasts for electroporation, a fourth sub-culture was grown for 15 h in MRS supplemented with 1% glycine, and then sub-cultured in the same medium at a 1:50 dilution for an additional 6 h. Bacteria were harvested using centrifugation, and chilled pellets subjected to electroporation with 10 μg plasmid DNA, followed by recovery in MRS broth for 3 h, and growth on MRS-agar + 5 μg/mL chloramphenicol (Sigma, St. Louis, MO, United States) at 30°C in 5% CO2. Plasmid was extracted from one-half of a colony of multiple purified transformants, and verified via DNA sequence analysis. The remaining bacterial colonies were propagated in the presence of chloramphenicol at 42°C, the non-permissive temperature, at which repA is non-functional, thus selecting for integrants. For pMGM14-based transformants, putative integrants were purified, and assessed for chimeric slpA presence by PCR. All confirmations were performed at the non-permissive temperature. Three independently isolated and PCR-verified identical transformants were bio-banked for the L. casei strain (herein referred to as Syn-LAB 2.0 clones), and seventeen independently isolated and PCR-verified identical transformants for the L. acidophilus strain (herein referred to as Syn-LAB 2.1 clones). For in vitro studies only, all LAB strains were propagated at the non-permissive temperature with antibiotic selection as appropriate (5 μg/mL chloramphenicol). However, for all in vivo studies, while strains were propagated as above, no selection antibiotic was administered to the animals.
An identical procedure was used to generate empty-vector harboring LAB strains (Table 1); these strains were PCR-verified, and used only for in vivo studies. Plasmids were maintained episomally; the lack of homology with the LAB host limited the possibility of integration of vector sequences into the chromosome.
Transepithelial Electrical Resistance (TEER) Measurements
Polarized human intestinal epithelial cells (C2BBe; Peterson and Mooseker, 1992) were grown on 0.33-cm2 collagen-coated CorningTM Transwells (Thermo Fisher Scientific) for 14 days. Monolayers were treated apically with 1.42 × 107 CFU/well and 1.96 × 10 7 CFU/well of parental and Syn-LAB 2.0 respectively. Measurements were made every hour for 7 h, and at 24 h post-treatment using an epithelial volt-Ohm voltmeter (World Precision Instruments, Sarasota, FL, United States), and TEER calculated by applying Ohm’s Law. An identical setup was used when testing Syn-LAB 2.1 and its L. acidophilus parent strain.
Host Cell Survival Measurement
C2BBe monolayers were treated with media alone, the parent LAB strain, or the corresponding Slp chimera-expressing Syn-LAB derivatives, and host cell viability was assessed using the propidium iodide (PI) uptake assay as described previously (Roxas et al., 2012). Briefly, PI (2 μg/ml; Molecular Probes) was added to the treated cells, and fluorescence measured after 30 min using a microplate reader (Synergy HT; BioTek instruments, Winooski, VT, United States). To estimate maximal PI uptake, a set of wells were treated with 70% methanol prior to PI treatment.
Immunoblot Analyses
Surface layer proteins (Slp) were extracted from Lactobacillus parent and Syn-LAB strain saturated cultures (OD600nm= 1.5) using 0.2M glycine (pH2.2), as described by Calabi et al. (2001). For dot blot analyses, 31.25, 62.5, 125, 500 ng, 1 μg, and 2 μg of total protein were blotted on nitrocellulose membranes (Bio-Rad, Richmond, CA, United States). Slp extracts (5–10 μg) were also separated on 4–20% TGXTM pre-cast protein gels (Bio-Rad, Richmond, CA, United States). Separated proteins were transferred to 0.2-μm nitrocellulose membranes (Transblot Cell Apparatus, Bio-Rad). Blots were blocked with 5% non-fat milk in Tris-buffered saline containing Tween 20 (TBST) for 1 h, incubated with antiserum specific to C. difficile SlpA (raised against the C. difficile strain 630 SlpA (Merrigan et al., 2013) overnight at 4°C and in horseradish peroxidase-conjugated goat anti-rabbit antibody for 1 h at room temperature (Sigma–Aldrich, St. Louis, MO, United States). Membranes were washed five times for 5 min in blocking solution between each incubation step and developed with SuperSignal West Femto Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL, United States).
Immunofluorescence Microscopy
SlpA chimera expression in L. casei WT and Syn-LAB 2.0 and Syn-LAB 2.1 strains was evaluated via immunofluorescence staining using antiserum specific to C. difficile SlpA. Lactobacillus sp cultures were allowed to settle for 10 min in 12-well plates with poly-L-lysine-coated coverslips. Unattached bacteria were removed, and samples were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 20 min, permeabilized with 0.2% Triton X-100 in PBS for 15 min, quenched with 50 mM NH4Cl and 0.125M glycine in PBS for 15 min, and blocked with 5% IgG-free bovine serum albumin (BSA) in PBS for 1 h. Samples were incubated with antiserum specific to C. difficile SlpA overnight at 4°C, and then washed three times with 1% IgG-free BSA in PBS. Secondary antibodies (Alexa 488-conjugated goat anti-rabbit IgG antisera; Thermo Fisher Scientific, Waltham, MA, United States) were added at 8 μg/ml in 5% IgG-free BSA for 1 h. Samples were mounted in ProLong Diamond Antifade reagent (Thermo Fisher Scientific, Waltham, MA, United States). Intestinal tissue samples (ileum, cecum, and colon) from LVG Golden Syrian Hamsters (Charles River Laboratories, San Diego, CA, United States) treated with Syn-LAB 2.0 were frozen in OCT embedding medium (Tissue-Tek, Sakura Finetek, Torrance, CA, United States) and stored at -80°C. OCT-mounted tissue samples cut at 3 micron thickness were fixed in 4% paraformaldehyde in PBS (pH 7.4) for 20 min, and processed for SlpA immunofluorescence staining as described above. Samples were stained with 4,6-diamidino-2-phenylindole (DAPI) prior to mounting in ProLong Diamond Antifade reagent. Images were captured using EVOS® FL Imaging System (Thermo Fisher Scientific, Waltham, MA, United States) or DeltaVision Elite Deconvolution Microscope (GE Healthcare, Pittsburgh, PA, United States).
Flow Cytometry
Parent and transformed Lactobacillus sp strains were cultured in MRS broth as described above, and subjected to Gram’s staining to verify purity and morphology. Bacteria were pelleted by centrifugation at 4000 g for 2 min. Bacterial pellets were washed gently three times with blocking solution (2% IgG-free BSA in PBS) and then incubated with antiserum specific to C. difficile SlpA for 30 min. Secondary antibodies (Alexa Fluor 555-conjugated goat anti-rabbit IgG antisera; Thermo Fisher Scientific, Waltham, MA, United States) were added at 8 μg/ml in 2% IgG-free BSA for 30 min. Samples were washed three times with blocking solution after each antibody incubation step. Stained samples were re-suspended in blocking solution at 106cells/mL density and analyzed via flow cytometry using a BD FACSCANTO II machine (BD Biosciences, San Jose, CA, United States). List mode data files consisting of 10,000 events gated on FSC (forward scatter) vs. SSC (side scatter) were acquired and analyzed using FACSDiva 8.0.1 software (BD Biosciences, San Jose, CA, United States). Appropriate electronic compensation was adjusted by acquiring the cell populations stained with the fluorophore, as well as an unstained control.
Golden Syrian Hamster Studies
All hamster studies were approved by the Institutional Animal Care and Use Committee of the University of Arizona. The Golden Syrian hamster model was employed to study both colonization/shedding and protection conferred by Syn-LAB strains. For all studies, male hamsters (6–8 weeks; 90–110 g weight) were used.
Shedding studies: Prior to any treatment, hamster stool plated on MRS yielded no colonies, confirming that the animals were devoid of endogenous Lactobacillus bacteria. Animals received a daily dose of 108 Syn-LAB 2.0 or 108 Syn-LAB 2.1 respectively. Feeding and enumeration were continued for 6 days. For Syn-LAB 2.0-treated animals only, oral clindamycin (prescription solution; clindamycin sulfate; University of Arizona Pharmacy; 30 mg/kg) was administered on Day 4, and LAB detection monitored until Day 6. Fecal pellets were collected daily, and pellets were re-suspended in PBS, homogenized, serially diluted and plated on the appropriate Syn-LAB selective medium containing chloramphenicol. Colonies were detected only in stool samples from Syn-LAB-treated animals, and not from untreated controls. Shedding from all animals was statistically indistinguishable. For added confirmation, select colonies were 16S PCR-verified for L. casei as well as the presence of the chimeric slpA.
Challenge studies: These were performed in two modalities, a “fixed-dose” and a “continuous dose” format. All animals received clindamycin (prescription solution; clindamycin sulfate; University of Arizona Pharmacy; 30 mg/kg). 1000 spores of C. difficile strain CD630 was used in the challenge studies where indicated, and 108 CFU LAB was used wherever indicated. Group 1 animals received clindamycin on day -3 but no other intervention (black line in Figure 8). Group 2 hamsters received clindamycin (day -3) and C. difficile challenge (day 0), but no LAB treatment (blue line in Figure 8). Group 3 hamsters received L. casei parent strain/empty vector on days -6, -5, -4, -2, -1, and 0, and clindamycin on day -3, followed by C. difficile challenge on day 0 (magenta line in Figure 8). Group 4 hamsters received Syn-LAB 2.0 on days -6, -5, -4, -2, -1, and 0, clindamycin on day -3, and C. difficile challenge on day 0 (green line in Figure 8).
For continuous-dose studies, the clindamycin dose and timing was similar to that above, and only the “C. difficile,” “Empty Vector” and “LAB” groups as above were evaluated. Both “Empty Vector” and Syn-LAB 2.0 (“LAB”) were continuously dosed at 108 CFU per animal per day starting at Day -6 before infection, until death/euthanasia.
Where appropriate, infections commenced 72 h post-antibiotic administration, and the challenge strain used was C. difficile strain 630 (1000 spores; orally administered in PBS). Animals were monitored for disease symptoms (wet-tail, ruffled coat, lethargy) through the course of the studies. Moribund hamsters or those meeting the criteria for euthanasia were administered 270 mg/kg commercial euthanizing solution (Euthanasia III, MedPharma Inc, Pomona, CA, United States). Euthanized hamsters were dissected for visualization of gross pathology, and cecal contents harvested and plated on selective medium for recovery and molecular typing of C. difficile (using 16s-23s rDNA intergenic fragment profiling and comparison with the organisms used for infection). In all studies, and all groups, fecal pellets were also collected daily, re-suspended in PBS, homogenized, serially diluted and plated on C. difficile or L. casei selective medium as appropriate.
Immune response studies: for these experiments, age- and weight-matched Golden Syrian hamsters (at least 3 per group) were administered 108 CFU Syn-LAB 2.0 daily, or left untreated, for 21 days. Animals were then euthanized, whole blood harvested via cardiac puncture, and serum immediately retrieved after blood was centrifuged at 1000 g for 10 min. This material was aliquoted and stored at -80°C until further use. For immune response assessments, the same methodology as immunoblotting above was used, but serum from Syn-LAB or mock-treated animals was used as the source of primary antibody.
Neonatal Piglet Studies
All piglet studies were approved by the Institutional Animal Care and Use Committee of the University of Arizona; we assessed Syn-LAB 2.1 safety, tolerability and efficacy in protecting against C. difficile challenge. Newborn male and female piglets were obtained via assisted delivery from a local antibiotic-free, small-volume farm, and transferred to the University of Arizona Central Animal Facility within 2 h of birth. On Day 2 post-birth, piglets were treated with oral vancomycin (50 mg/kg; prescription solution, University of Arizona Pharmacy) to ablate any pre-existing C. difficile colonization. On day 6 post-birth, piglets were administered 1010 Syn-LAB 2.1 in milk replacer every 8 h. On day 7, a subset of animals was administered a non-lethal dose of 1000 C. difficile spores of strain 630. Monitoring included checks every 8 h thereafter, with weight, stimulus response and dehydrations scores recorded. Upon completion of the study, piglets were anesthetized with Ketamine/Xylazine, and then humanely euthanized with commercial euthanizing solution (Euthanasia III, MedPharma Inc, Pomona, CA, United States) followed by cardiac puncture. Histologic analyses included standard hematoxylin-eosin staining of colonic tissues following standard methodologies (Kiernan, 2008), and immunofluorescence staining of tissues with anti-C. difficile SlpA serum as described in detail above for visualization of Syn-LAB 2.0.
Statistical Analysis
Multiple statistical tests were employed and utilized the Excel-Stat application to determine significance for experiments involving quantitation. For growth and bacterial burden, Student’s t-tests were performed to compute differences between parental and Syn-LAB strains, and errors bars calculated from standard deviation(s). For in vivo studies, Kaplan–Meier survival curves were computed followed by Log-Rank tests for post hoc analyses.
Results
Construction of C. difficile/Lactic Acid Bacterium (LAB) SlpA Chimera-Encoding Plasmids and Generation of Syn-LAB Strains
A 7740-base pair (bp) shuttle vector was assembled from chemically synthesized fragments. The vector has the following features: a temperature-sensitive repA allele and a chimeric slpA (Figure 1A) that includes; (1) a strong L. acidophilus (LA) promoter [upstream sequences of the phosphoglycerate mutase (pgm) gene (base pairs 178339-178600 of the L. acidophilus NCFM genome, Genbank accession number CP000033.3 (Altermann et al., 2005; Duong et al., 2011) (2) an LA Shine-Dalgarno (ribosome binding site) sequence (CCTGCA); and (3) sequences encoding a chimeric SlpA that includes a LA signal sequence (amino acids 1–30 of LA SlpA) (Altermann et al., 2005), codon-optimized C. difficile strain 630 LMW SlpA host-cell-binding region (amino acids 1–243) (Karjalainen et al., 2002), and the LA cell-wall-binding domain (amino acids 291–444) (Altermann et al., 2005). Precise engineering of the entire 7740 base pair vector was confirmed by complete DNA sequencing.
Lactic acid bacterial strains were individually transformed with this shuttle vector. All transformants were recovered at the permissive temperature (30°C) where RepA is functional, confirmed by biochemical tests and selective plating (Figures 1B,C), and further confirmed by multiple PCR tests (not shown). Finally, transformants were propagated with selection at the non-permissive temperature (37–42°C) that allows for recovery of integrants. A total of 3 independently isolated Lactobacillus casei transformants were obtained (herein collectively referred to as Syn-LAB 2.0 clones). Seventeen independently isolated Lactobacillus acidophilus transformants were also obtained (herein referred to as Syn-LAB 2.1 clones). Syn-LAB clones of each LAB species were confirmed via PCR, phenotypic (growth; not shown) and biochemical tests (Figures 1B,C), and bio-banked. The unique carbohydrate fermentation profiles were exploited to readily distinguish between the two species: L. casei can ferment mannitol, and converts dextrose to acetic acid, and the corresponding media acidification manifested as a color change to yellow/white in the presence of appropriate pH indicators. L. acidophilus, on the other hand, does not ferment mannitol, and converts dextrose to lactic acid; the media remained purple/blue in the presence of the corresponding carbohydrates. Gram’s staining revealed that in contrast to the rod-shaped morphology of the parent L. casei strain, Syn-LAB 2.0 cells were shorter and curved (Figures 2C,E). Syn-LAB 2.1 bacteria were indistinguishable from the parent L. acidophilus strain (Figures 2D,F).
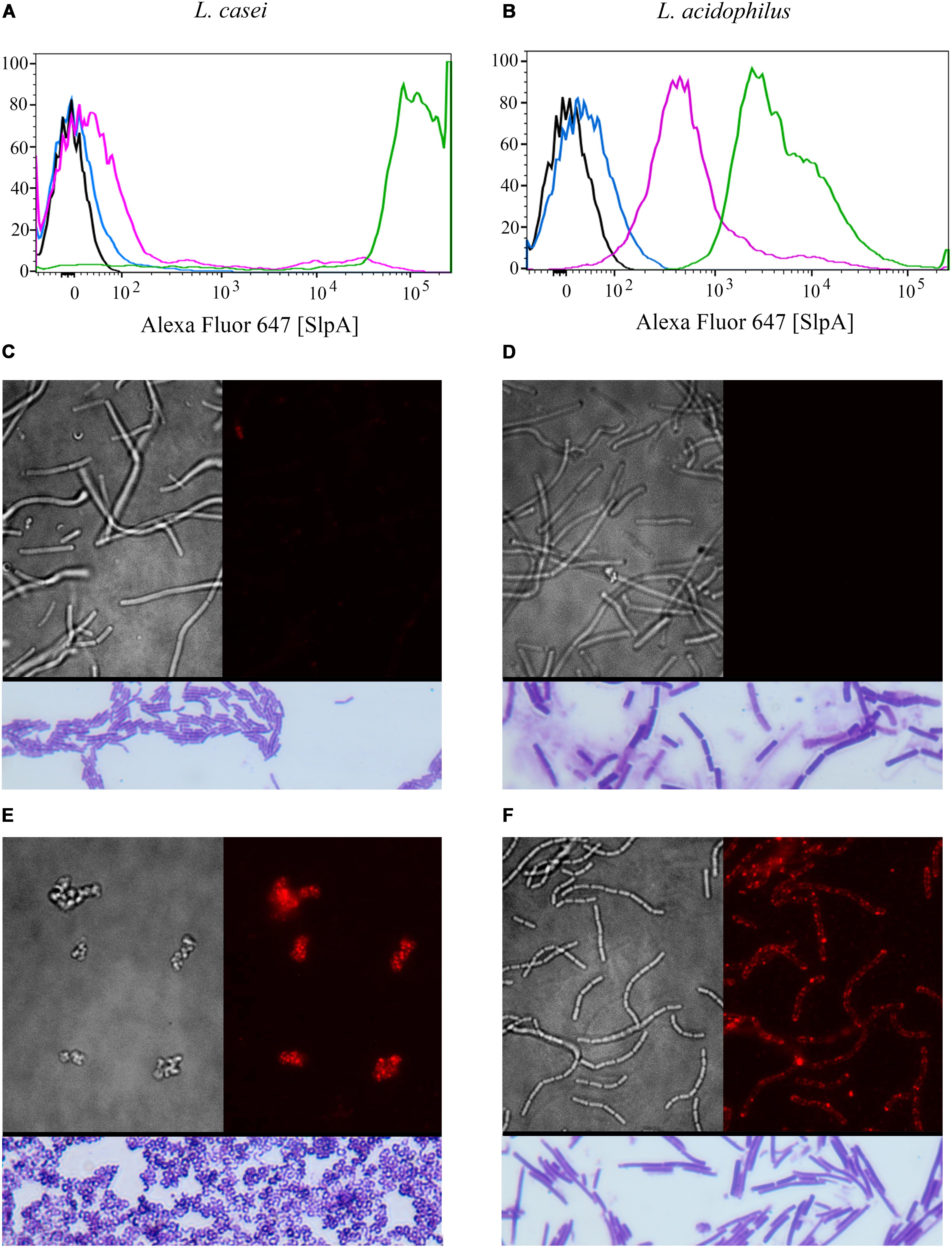
FIGURE 2. Syn-LAB 2.0 and Syn-LAB 2.1 surface-display chimeric SlpA. (A,B) Fluorescence-activating cell sorting analyses. (A) Black, unstained L. casei parent strain; Blue, unstained Syn-LAB 2.0; Magenta, SlpA-stained L. casei parent strain; Green, SlpA-stained Syn-LAB 2.0 (median fluorescence > 100,000). L. casei does not have a classic S-layer, therefore, Syn-LAB shift is unique. (B) Black = unstained L. acidophilus parent strain; Blue, unstained Syn-LAB 2.1; Magenta, SlpA-stained L. acidophilus parent strain; Green, SlpA-stained Syn-LAB 2.1 (median fluorescence > 4,000). L. acidophilus has a native S-layer, therefore, chimeric SlpA is detected as a discrete, strong fluorescence shift. (C–F) microscopy; (C) brightfield image, L. casei parent strain with minimal detectable SlpA fluorescence; (D) brightfield image, L. acidophilus parent strain with undetectable SlpA fluorescence; (E) immunofluorescence, Syn-LAB 2.0 with intense, punctate, SlpA staining, and (F) immunofluorescence, Syn-LAB 2.1 with intense SlpA staining. All strains were probed with a C. difficile -specific anti-SlpA serum. Images are representative of at least 20 fields and > 1000 bacteria visualized. All images were visualized with a high-resolution DeltaVision deconvolution microscope. Gram’s stained bacteria are shown in rectangles below each of (C–F).
One isolate each of Syn-LAB 2.0 and Syn-LAB 2.1 as well as the respective parent Lactobacillus sp strains were used with appropriate antibiotic selection for the in vitro studies presented below. For in vivo studies, Syn-LAB 2.0, Syn-LAB 2.1, a combination thereof, or “empty vector” harboring Lactobacillus sp. strains were tested; no selection antibiotics were used in animals.
Syn-LAB Strains Display Chimeric C. difficile SlpA
SlpA chimera expression was confirmed via multiple methodologies for both engineered biologics (Syn-LAB 2.0 and Syn-LAB 2.1). First, flow cytometry was used to determine the degree of heterologous (chimeric) SlpA surface display. In actively growing cultures, almost 100% of Syn-LAB 2.0 bacteria displayed the C. difficile SlpA chimera (confirmed by fluorescence shifts in the engineered isolate compared to the parent strains (Figure 2A). Similar results were obtained for Syn-LAB 2.1 (Figure 2B), confirming robust SlpA display in that biologic as well.
Second, chimeric SlpA expression and display was visualized via immunofluorescence using anti-C. difficile SlpA antiserum (Merrigan et al., 2013). In contrast to the parent L. casei strain (Figure 2C), the engineered Syn-LAB 2.0 derivative revealed dense SlpA staining (Figure 2E). Similarly, Syn-LAB 2.1 (Figure 2F), but not the parent L. acidophilus strain (Figure 2D), exhibited intense and punctate surface SlpA staining. Staining was specific since no signal was detected on either Syn-LAB strain with pre-immune serum (not shown).
Chimeric C. difficile SlpA Is Incorporated Into the Lactic Acid Bacterial Cell Surface
We also confirmed that the C. difficile-specific SlpA visualized in Figure 2 above was due to incorporation of the chimeric protein into the LAB cell wall. Sheared total Surface-Layer (S-layer) proteins from Syn-LAB 2.0 and Syn-LAB 2.1, and the isogenic parent strains (Merrigan et al., 2013), were separated via agarose gel electrophoresis (Figure 3A), and immunoblotted using the anti-SlpA antiserum. Both synthetic biologics, but not the isogenic parent strains, displayed altered total S-layer profiles, and a discrete unique band, appropriate to the expected size of the chimera, (Figure 3B). Mass spectrometry confirmed the presence of chimeric SlpA sequences (not shown). Due to the polyclonal nature of the antiserum other non-specifically reacting bands were also observed in the parent strains.
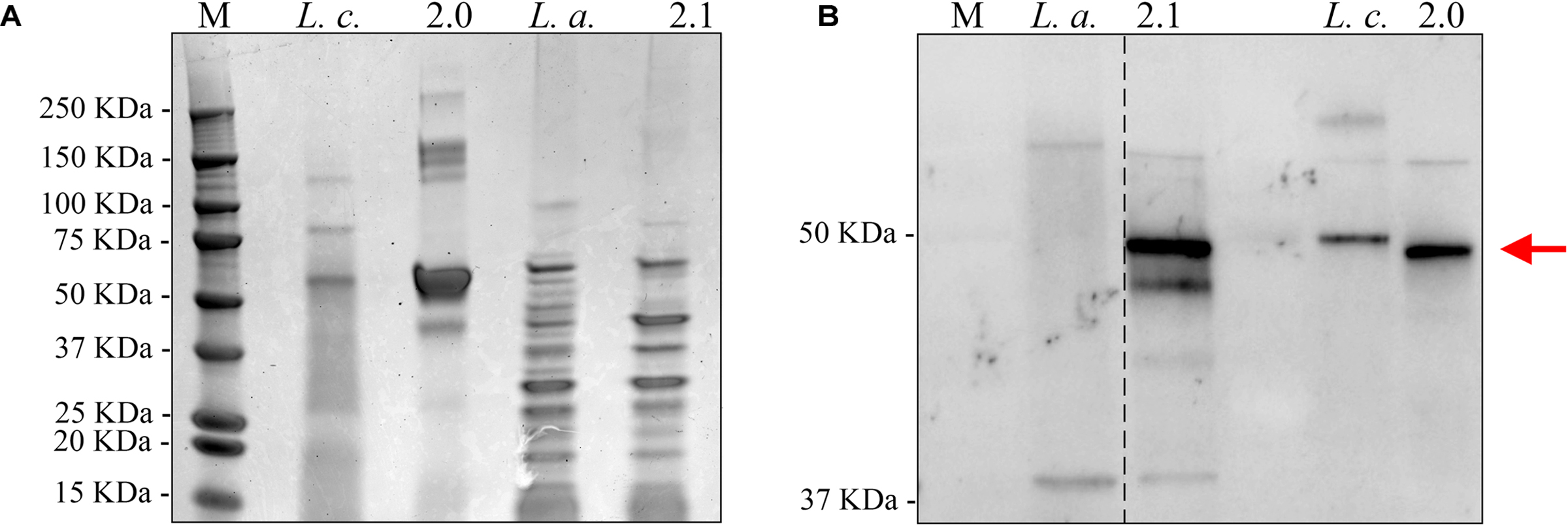
FIGURE 3. Chimeric C. difficile SlpA is incorporated into the lactic acid bacterial (LAB) cell wall. (A) Sheared total S-Layer Proteins from the L. casei parent strain (Lc) or Syn-LAB 2.0; and the L. acidophilus parent strain (La), or Syn-LAB 2.1. (B) Immunoblot probing the total Surface Layer Proteins of parent and Syn-LAB strains with antiserum specific to C. difficile SlpA. The red arrow points to unique, expected-size bands present only in Syn-LAB 2.0 and Syn-LAB 2.1 respectively. The dotted line indicates that the original immunoblot was edited to remove irrelevant lanes.
Chimeric SlpA Expression Preserves Epithelial Barrier Function
We performed a series of studies to rule out potential adverse effects of the engineered strains on intestinal epithelial cell health and function. First, potential impact of Syn-LAB strains on intestinal epithelial barrier function was assessed via trans-epithelial electrical resistance (TEER) measurements. Unexpectedly, the parent L. casei strain decreased TEER over a 7-h period, with changes becoming consistently apparent as early as 3 h post-application (Figure 4A). In contrast, addition of Syn-LAB 2.0 did not significantly alter TEER relative to mock-treated cells. Similarly, Syn-LAB 2.1, or the parent L. acidophilus strain, had no impact on the TEER of C2BBe cells (Figure 4B). This suggests that Syn-LAB 2.0 and Syn-LAB2.1 do not disrupt host epithelial barrier function.
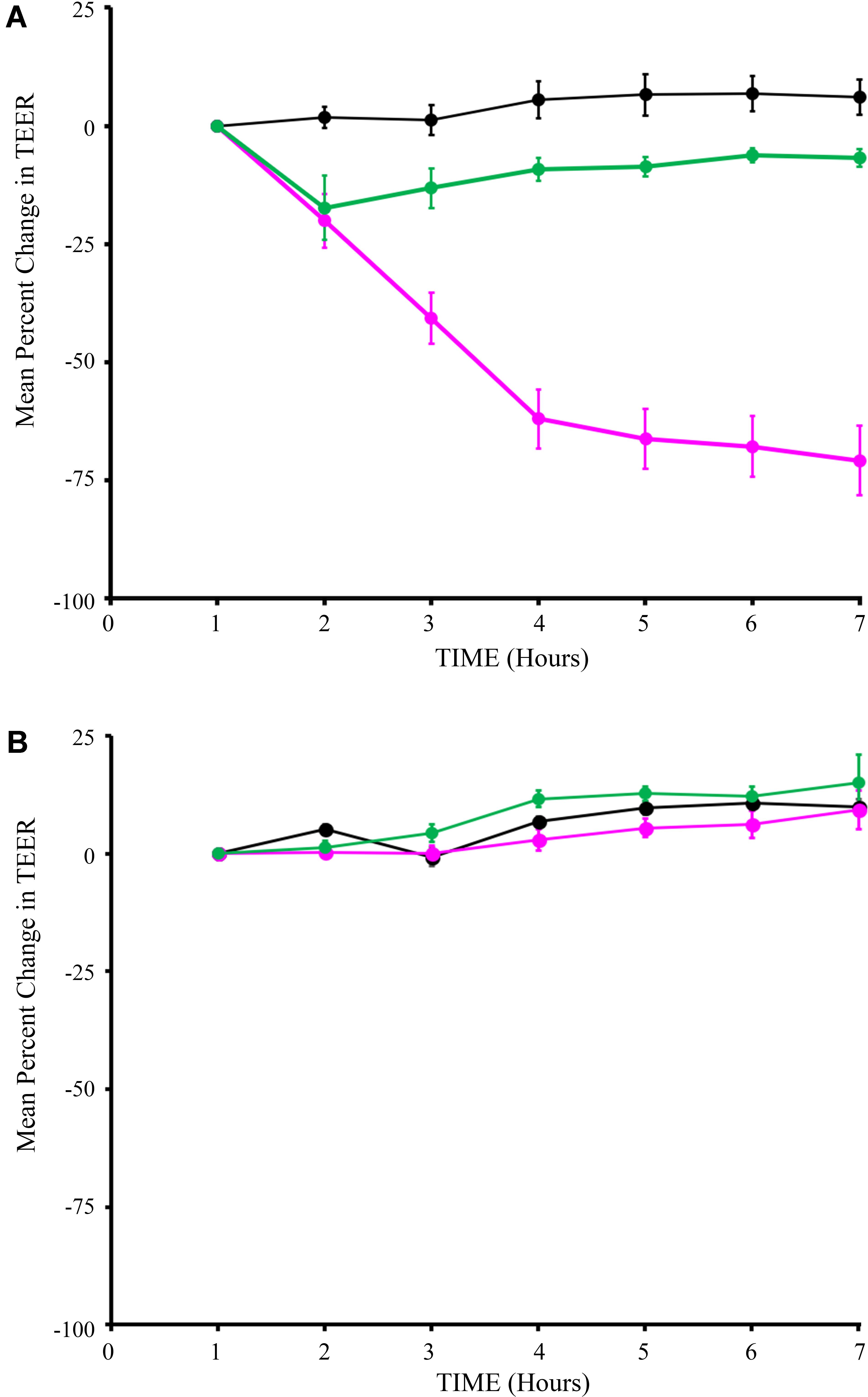
FIGURE 4. Syn-LAB 2.0 preserves epithelial barrier function. Transepithelial electrical resistance (TEER) measurements of C2BBe monolayers that were mock-treated (black line), or exposed to the parent L. casei strain (green line), or Syn-LAB 2.0 (magenta line; A), or parent L. acidophilus strain (green line), or Syn-LAB 2.1 (magenta line; B), for 8 h. Experiments were performed in at least three biological replicates. Host cells were treated with 100 bacteria per cell (MOI = 100).
Chimeric SlpA Display Does Not Compromise Host Cell Viability
Next, we assessed the impact of Syn-LAB on the survival of host intestinal epithelial cells. C2BBe cells were grown to confluence, and either mock-treated, or treated with L. casei, L. acidophilus, or the respective Syn-LAB derivatives for up to 8 h. Cell death was continuously monitored via propidium iodide (PI) uptake, a DNA-intercalating dye that only enters dying cells with compromised cell membranes. L. casei, L. acidophilus, and the respective chimera-expressing Syn-LAB derivatives did not significantly impact epithelial cell viability relative to mock-treated cells (Figures 5A,B).
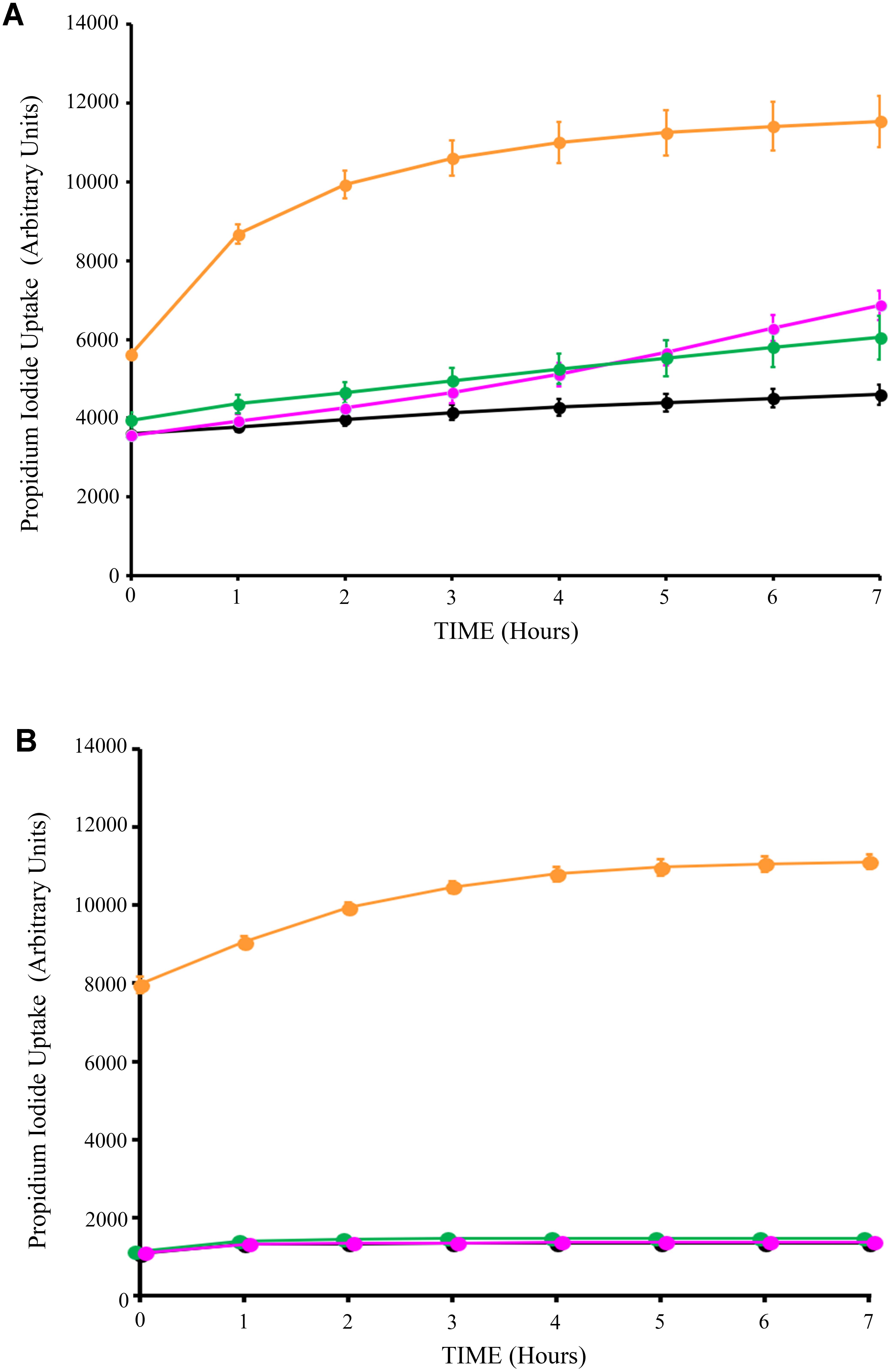
FIGURE 5. Lactic acid bacteria (LAB) treatment does not impact host-cell viability. Propidium iodide uptake assays of C2BBe monolayers mock-treated (black line), or exposed to the parent L. casei strain (green line), or Syn-LAB 2.0 (magenta line; A), or parent L. acidophilus strain (green line), or Syn-LAB 2.1 (magenta line; B), for 8 h. Orange line shows methanol-treated monolayers representative of maximal cell death. Experiments were performed in at least three biological replicates. Host cells were treated with 100 bacteria per cell (MOI = 100).
Syn-LABs Are Safe and Tolerable in Multiple Animal Models, and Robustly Colonize the Mammalian Gastrointestinal Tract (GIT)
PCR- and microbiologically verified pure cultures of Syn-LAB strain 2.0 was used for these studies, and prepared for in vivo dosing at ∼108 bacteria per 200 μL volume of PBS. To conserve animals, the shedding/colonization assessments were performed as pilot studies. For broad assessment of Syn-LAB shedding, two animal models (Golden Syrian hamsters and neonatal piglets) were utilized, with each model testing one of the two Syn-LAB strains respectively.
Golden Syrian hamsters received either no treatment (control), or a single dose of the broad-spectrum antibiotic clindamycin, followed by 6 daily doses of the L. casei-based Syn-LAB 2.0 (antibiotic “pre-treatment” group), or 3 days of Syn-LAB 2.0, followed by a single dose of oral clindamycin (as above), followed by another 3 days of Syn-LAB 2.0 (antibiotic “mid-cycle treatment” group). Syn-LAB treated hamsters started shedding the biologic on Day 1 post-administration; this continued until the end of the study (similar studies were performed for Syn-LAB 2.1, with identical observations; not shown). Hamsters that received clindamycin on Day 4 post-Syn-LAB showed no evidence of the biologic on Day 5, confirming the in vivo susceptibility of Syn-LAB 2.0 to standard antimicrobial therapy (Figure 6A). However, when Syn-LAB administration was restarted, shedding resumed at a magnitude similar to that observed prior to antimicrobial therapy (Figure 6A). Importantly, all hamsters were similarly colonized, with Syn-LAB fecal titers reaching, or exceeding, 105 colony forming units/gram stool on Day 3 post-treatment (Figure 6B). Immunofluorescence studies of colonic tissues harvested post-necropsy revealed dense luminal staining only from Syn-LAB 2.0 treated hamsters (Figure 6C, right panel) as compared with mock-treated animals (Figure 6C, left panel), confirming C. difficile SlpA expression in the hamster gastrointestinal tract. Finally, Syn-LAB 2.0 was avidly consumed by all hamsters, with no requirement for a pre- or post-dosing sweetened electrolyte “chaser.” Safety and tolerability were also confirmed via lack of any adverse effects in any Syn-LAB-treated hamsters, as well as appropriate activity and alertness throughout the study.
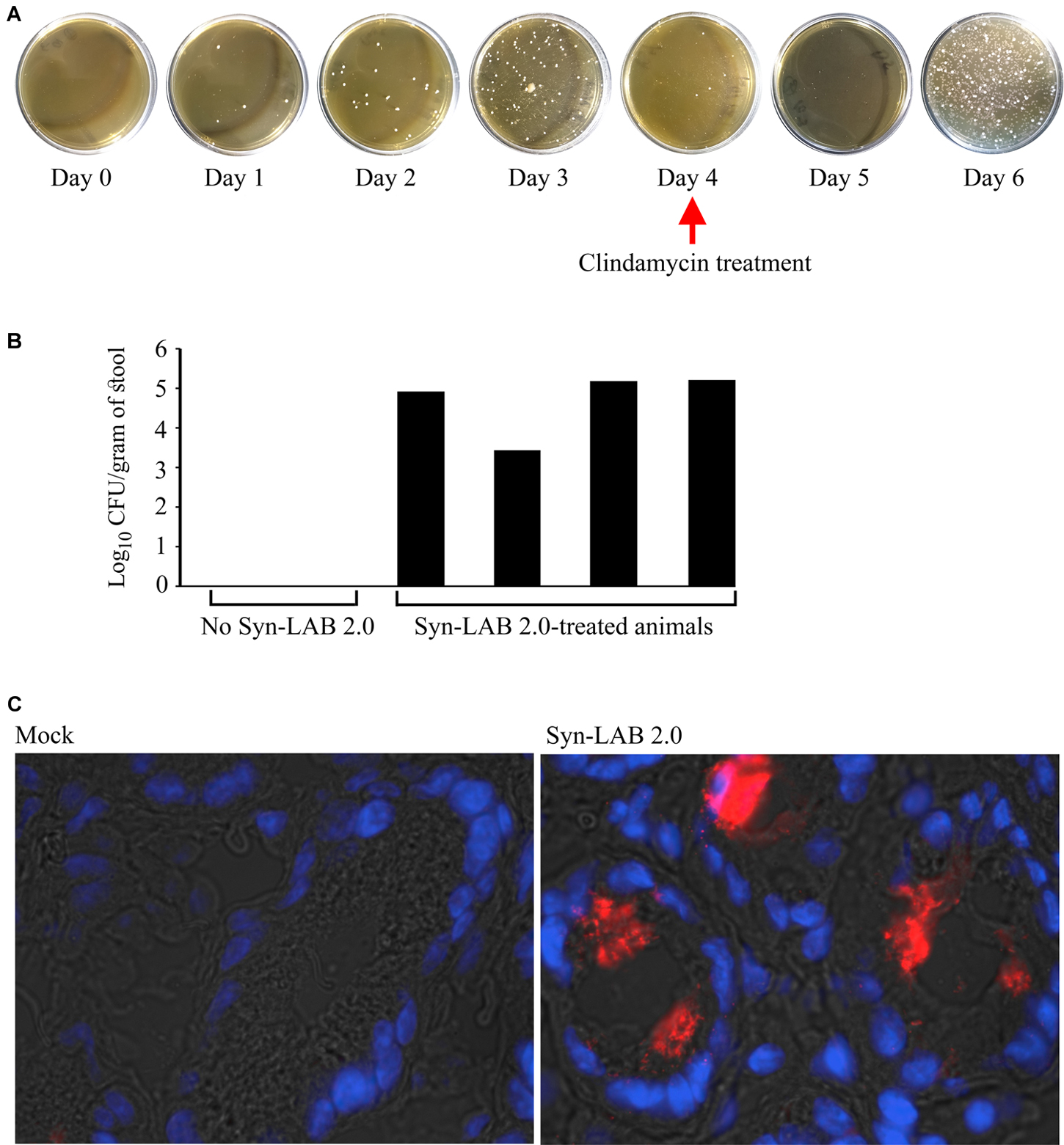
FIGURE 6. Syn-LAB colonization of Golden Syrian Hamsters. (A) Syn-LAB 2.0 colonizes the hamster gastrointestinal tract, and is shed as early as 1 day post-administration. Hamster stool pellets from animals continuously administered Syn-LAB 2.0 at 108CFU/day cultured on MRS-chloramphenicol. Antibiotic treatment on Day 4 ablates Syn-LAB 2.0 shedding only for 1 day, followed by continued detection from Day 5 onward. Data are representative of 8 infected animals. (B) Syn-LAB 2.0 shedding is comparable in all treated animals (black bars), but not detectable in control, untreated, animals. (C) immunofluorescence analysis using anti-C. difficile SlpA serum reveals intense staining in the colonic lumen of Syn-LAB 2.0-fed animals (right), but not mock-treated animals (left). Images are representative at least 10 fields visualized per section.
Continuous Syn-LAB Administration Induces an Anti-C. difficile SlpA Immune Response
While the primary goal was to design biologic agents that could competitively occupy C. difficile attachment sites in the gut, we also explored the possibility of an anti-SlpA immune response following long-term Syn-LAB administration. Golden Syrian hamsters were continuously administered Syn-LAB 2.0 as a once-daily 108 CFU dose for 55 days. Hamsters shed the biologic consistently throughout the process confirming that they were appropriately colonized. Age- and weight-matched control hamsters received no treatment. At the end of the study, hamsters were humanely euthanized, whole blood collected, and immunoblot-based analyses performed to assess anti-Syn-LAB immune response. Serum from Syn-LAB 2.0-treated hamsters (Figure 7A, top right panel), but not from mock-treated animals (Figure 7A, top left panel), detected C. difficile strain 630 SlpA in a dose-dependent manner. Presence of Slp proteins in the corresponding membranes was verified by re-probing the blots with a SlpA-specific antiserum previously generated in our laboratory (Figure 7A, lower panels). Finally, the same experiments were performed using Slp preparations from clinically-relevant isolates of diverse C. difficile ribotypes (012, 017, 020, 027, 078). Reactivity was observed only when serum from Syn-LAB-treated hamsters was used (Figure 7B). This suggested that the Syn-LAB SlpA moiety elicited a cross-reactive immune response (recognition of non-cognate C. difficile SlpA).
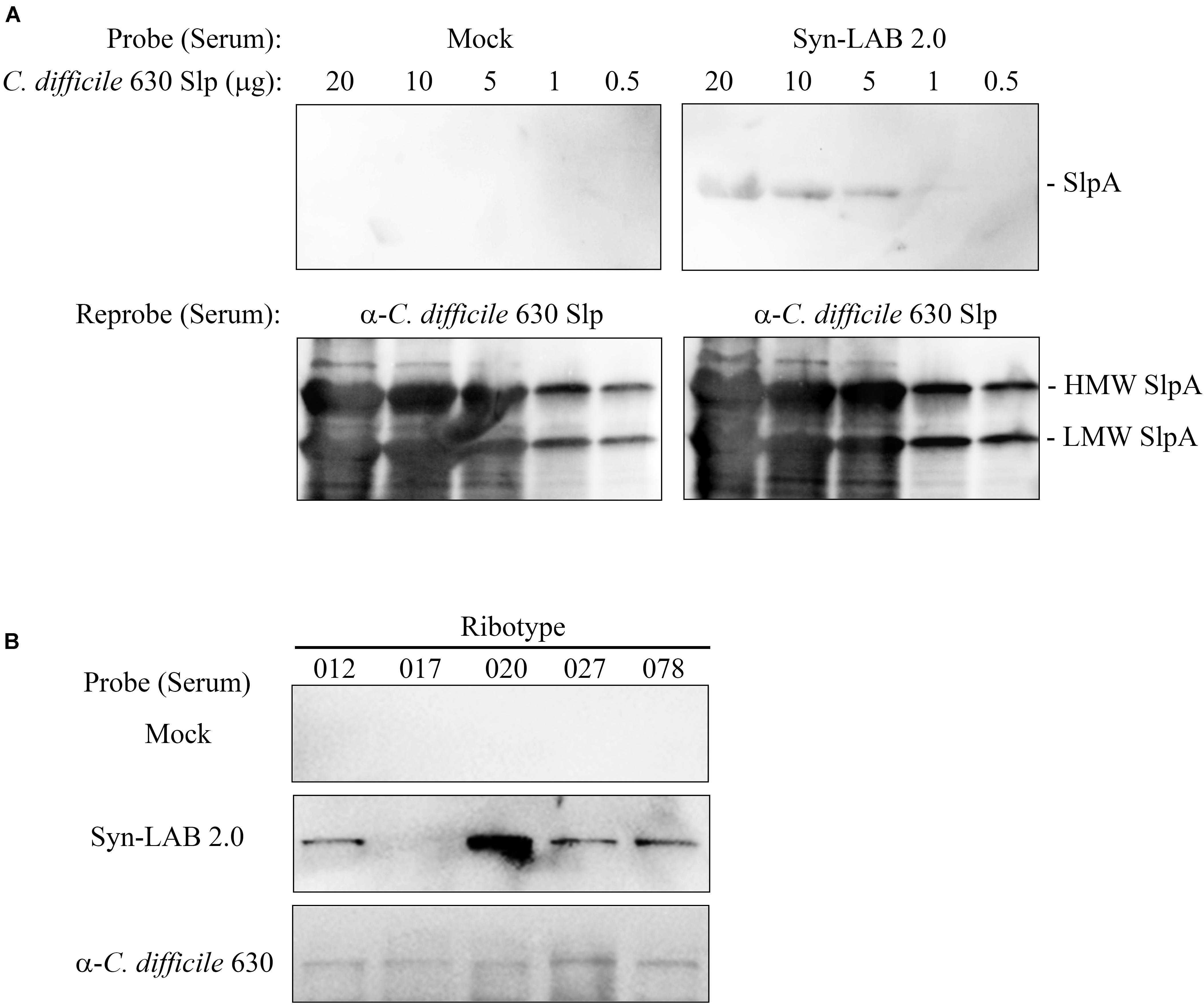
FIGURE 7. Syn-LAB 2.0 elicits an anti-C. difficile SlpA immune response. (A) Top panels, dose-response immunoblots of S-layer proteins (20 to 0.5 μg) from C. difficile strain 630 probed with serum from a Syn-LAB 2.0-treated animal (right), or serum from an untreated, age- and weight-matched hamster (left). Bottom panels, to verify efficient separation and transfer of the S-layer proteins, the membranes were stripped and re-probed with a polyclonal C. difficile anti-SlpA antiserum. Both C. difficile SlpA subunits were detected (arrows). (B) S-layer proteins from C. difficile clinical isolates of diverse ribotypes probed with serum from an untreated animal (upper row), a Syn-LAB 2.0-treated animal (middle row), or with anti-C. difficile strain 630 SlpA serum (bottom row).
Syn-LABs Protect Syrian Golden Hamsters From C. difficile-Induced Death
Since single-species probiotics are thought to have limited ability to protect against CDI (Wullt et al., 2003; Vernaya et al., 2017), we used a combination of Syn-LAB 2.0 and Syn-LAB 2.1 in hamster protection studies. A mixed culture of the biologics (108 CFU total) was administered to antibiotic-sensitized Golden Syrian hamsters either as a fixed dose (FD) formulation (6 doses) or as a continuous dose (CD) formulation (3 days prior to clindamycin until the end of the study). Syn-LAB-treated hamsters were compared to those receiving C. difficile alone, or those administered LAB containing the empty vector. Challenge studies used a high inoculum (∼1000 spores) of C. difficile strain 630 [a virulent, outbreak-associated isolate (Merrigan et al., 2013)].
Fixed dosing of the Syn-LAB combination significantly delayed death of hamsters compared to mock-treated animals, as well as those administered the empty-vector-harboring strains (Figure 8A). Continuous administration of Syn-LABs afforded statistically significant protection against CDI throughout the course of 12 days of infection (Figure 8B). Specifically, and as compared with untreated hamsters, protection was highly significant at multiple time points during the infection course (p = 0.0091, p = 0.0014, p = 0.0005, p = 0.0005 at 6, 8, 11 and 12 days post-infection respectively). This was in contrast to the protection afforded when hamsters were administered the parent LAB strain harboring the empty vector (p = 0.1147, p = 0.0147, p = 0.0147, p = 0.0147 on Days 6, 8, 11, and 12 post-infection respectively; 99% confidence interval for significance). Additionally, parent strain-treated hamsters succumbed to disease earlier in the infectious course, and were more often found moribund, with symptoms consistent with fulminant CDI (profound wet-tail, lethargy, sternal recumbency and cecal hemorrhage).
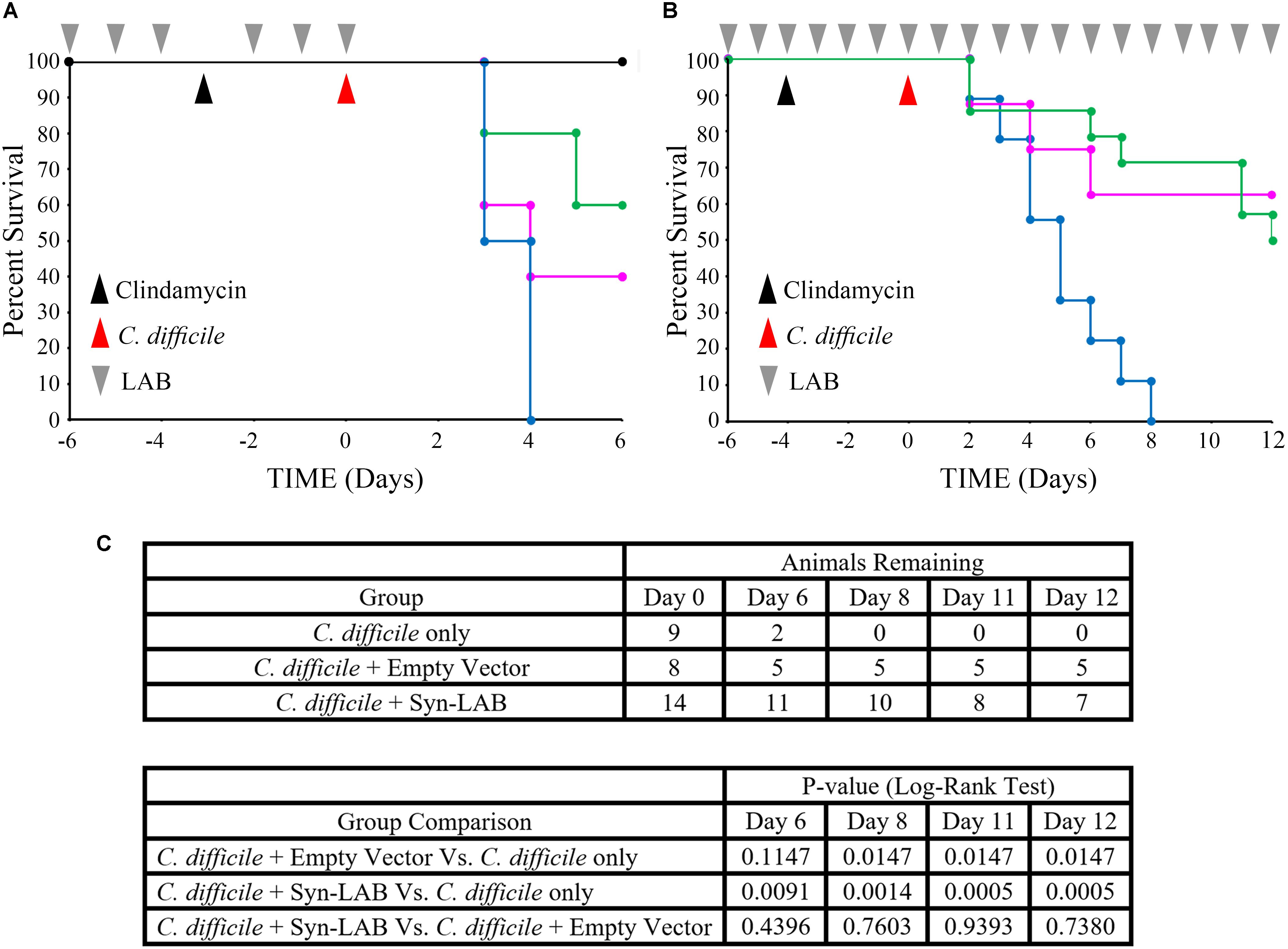
FIGURE 8. Syn-LAB biologics protect hamsters from lethal CDI. (A,B) Co-administration of Syn-LAB 2.0 and 2.1 protects Golden Syrian hamsters from CDI. (A), pilot study; Kaplan–Meier survival plot. Pre-treatment with Syn-LABs (6 once-daily doses of 1 × 1010 CFU each) delays death of hamsters infected with 1000 C. difficile spores. Black line, untreated, uninfected animals; blue line, C. difficile-infected animals; magenta line, animals treated with parent LAB strains harboring empty vector, and then infected; green line, animals treated with Syn-Lab 2.0+2.1 and then infected. (B), powered study, Kaplan–Meier survival plot. Continuous Syn-LAB 2.0+2.1 dosing (pre- and post-infection) prevents death of C. difficile-infected hamsters. blue line, C. difficile-infected animals; magenta line = animals treated with parent LAB strains harboring empty vector, and then infected; green line = animals treated with Syn-Lab 2.0+2.1 and then infected. (C) Group comparisons and statistical test results. Number of surviving animals in powered study (top), and Log-Rank tests (bottom). p ≤ 0.01 = significant.
Re-administration of clindamycin to Syn-LAB-treated, C. difficile-challenged hamsters 14 days post infection did not result in disease or mortality (not shown). This suggested that Syn-LAB-mediated colonization resistance also ablated C. difficile persistence. Taken together, Syn-LAB administration was highly protective in the hamster model described above.
Fixed-Dose Syn-LAB Administration Protects Neonatal Piglets From C. difficile-Induced Diarrhea
Preliminary neonatal piglet studies used healthy, newborn animals treated with vancomycin on Day 4 post-farrowing to ensure elimination of any carryover C. difficile bacteria from the farm. Control piglets were given PBS followed by C. difficile challenge, whereas treated piglets were administered three 108 CFU doses of the fast-growing Syn-LAB 2.1 strain over 24 h. Syn-LAB 2.1 was detected as early as 24 h after the first dose, and shedding continued until the end of the study (not shown).
In this model, CDI [1000 spores for piglets (Steele et al., 2010)], resulted in profuse diarrhea (Figure 9A, left panel, stool score of 1). Diarrheic symptoms in these piglets continued unabated for at least 3 days, at which time the accumulated dehydration and inappetance criteria necessitated euthanasia. Microscopic examination of colonic tissues from infected piglets revealed gross hemorrhage with an abundance of inflammatory infiltrates (Figure 9B, middle panel). However, piglets that received a 1-day administration of Syn-LAB 2.1 had well-formed stool (Stool score of 3–4; Figure 9A, right panel), as well as normal activity and appetite. Colonic tissue from these animals showed markedly less hemorrhage and inflammatory damage compared to those from piglets with CDI alone (Figure 9B, right panel).
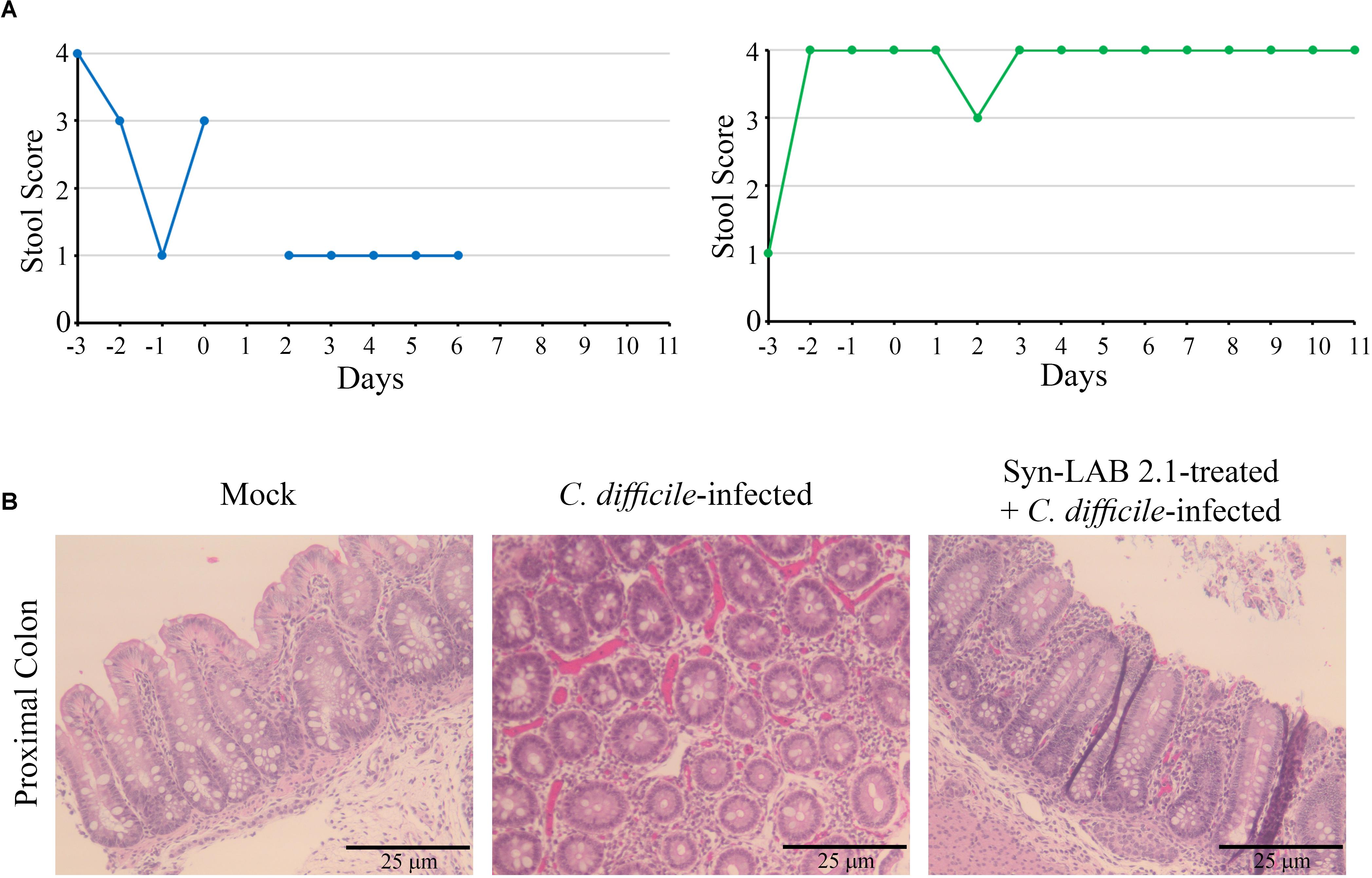
FIGURE 9. Syn-LAB 2.1 protects piglets from CDI diarrhea. Pilot, non-lethal CDI model. (A) Stool consistency scoring; 1 = diarrheic, 4 = fully formed. Left, C. difficile-infected animal, with consistently low stool scores indicating unremitting diarrhea. Right, Syn-LAB 2.1 treated + C. difficile-infected animal, with consistently high stool score, indicating no diarrhea. The example shown is representative of 3 animals studied. The animal in the left panel was euthanized after the last time-point shown due to unremitting diarrhea, increasing dehydration and inappetence. Similar results for the other two animals. All infected animals in the group received 1000 spores of C. difficile. (B) microscopic examination of proximal colon tissues from piglets. Left, hematoxylin-eosin staining of tissue from uninfected animal showing normal epithelium, little/no inflammatory infiltrate and no overt damage or necrosis. Middle, C. difficile-infected animal, revealing gross hemorrhage with an abundance of inflammatory infiltrates. Right, Syn-LAB 2.1-treated and C. difficile-infected animal tissue showing marked reduction in both overt hemorrhage and inflammation.
Discussion
With the emergence of outbreak-associated strains in the past decade, CDI has become a problem of considerable magnitude in terms of human and economic costs (Viswanathan et al., 2010). The protection offered by a healthy microbiota, known as colonization resistance, is the most effective foil against CDI (Seekatz et al., 2018). Although antibiotic-mediated dysbiosis is the most typical precipitating factor for CDI, traditional therapeutic options rely on administration of more antibiotics (McDonald et al., 2018). Apart from concerns of increased antibiotic resistance in C. difficile, this approach aggravates intestinal dysbiosis and, in a subset of infected individuals, results in recurrent disease (Viswanathan et al., 2010). Therefore, strategies that exploit colonization resistance to prevent or treat CDI can not only be effective in mitigating disease, but also address the underlying dysbiosis.
Two broad therapeutic approaches that exploit colonization resistance are fecal microbiota transplantation (FMT) and probiotic administration. In randomized trials, FMT efficacy ranges from ∼50% to 90% based on delivery and number of infusions (van Nood et al., 2013; Youngster et al., 2014; Cammarota et al., 2015; Kelly et al., 2016; Lee et al., 2016), but this procedure is logistically challenging and could pose undefined risks to patients (Wang et al., 2016; Gardiner et al., 2018); as such, it is recommended only for patients that repeatedly fail antibiotic therapy (at least 3 CDI episodes; IDSA-SHEA guidelines (McDonald et al., 2018). While probiotics are more palatable and pose fewer risks, they show variable efficacy in treating CDI (Rezaie and Pimentel, 2014; McFarland, 2015; Barker et al., 2017; Goldenberg et al., 2017; Alberda et al., 2018). Some studies have shown probiotic efficacy when used in patients with no CDI history, but differences in formulation, dose, dosing duration and species composition preclude strong conclusions being drawn in favor of probiotics as CDI interventions. Indeed, the latest IDSA-SHEA recommendations for CDI intervention do not mention probiotics as a treatment option, and no recommendation is made for the agents in primary disease prevention (McDonald et al., 2018). Our goal was to develop a biologic agent for colonization resistance against CDI with consistent and robust efficacy against CDI, but with a safety profile comparable to extensively used probiotics. We, therefore, sought to engineer the “Generally Regarded as Safe” (GRAS) organisms Lactobacillus casei and Lactobacillus acidophilus to express C. difficile surface adhesins and, thereby, competitively exclude the pathogen from intestinal surfaces.
Lactobacilli can be extraordinarily recalcitrant to manipulation; this is both an advantage and a liability. The difficulties encountered in introducing or extracting DNA from the various species portends well for the use of the organisms in probiotic preparations due to reduced risk of horizontal acquisition of antibiotic resistance genes from endogenous microbiota. However, laboratory manipulation poses unique challenges. Indeed, only electroporation with high amounts (≥10 μg) of DNA, and a strain-specific optimized protocol, is the recommended method to transform Lactobacillus sp; despite this, reported efficiencies may be as low as 1 transformant/μg DNA (Vogel and Ehrmann, 1996). On balance, however, Lactobacillus casei and Lactobacillus acidophilus offer unique advantages that can be exploited for CDI treatment. L. casei can suppress the inflammatory cytokines produced in response to CDI (Boonma et al., 2014), upregulate mucin gene expression (Mattar et al., 2002), and also appears to confer human subjects some protection from CDI when administered as a fermented drink (Wong et al., 2014; Alberda et al., 2018). L. acidophilus has been shown to decrease C. difficile toxin gene expression and also protect animals in a murine CDI model (Yun et al., 2014). However, data regarding the strain-specific benefits of these organisms, or their consistently beneficial use in diverse patient cohorts, are scarce. For Syn-LAB engineering, we used pure, genome-sequenced, antibiotic sensitive strains of both genera. Lactobacilli are notoriously recalcitrant to taxonomic classification, and phenotypic or biochemical identification, and therefore need to be subjected to extensive molecular identification to confirm species purity (Pal et al., 2012). The Syn-LAB 2.0 antecedent strain is morphologically and biochemically distinguishable from the Syn-LAB 2.1 parent, allowing for clear discrimination of the strains.
The CD/LAB chimeric SlpA was robustly expressed in both Lactobacillus species, integrated into the cell wall, and displayed on the surface; thus, the surface display of the chimera was not hindered by presence of the native LAB S-layer. We assessed Syn-LAB dosing and maintenance, and its ability to protect against a high dose of C. difficile spore infection (> 1000 spores) in the lethal hamster model of infection [a spore dose of ∼100 causes 100% lethality (Sattar et al., 2015)]. Continuous Syn-LAB dosing was easily achieved in hamsters, and the bacteria were readily detected in high numbers in the stool as early as 1 day post-administration. With fixed dosing, however, the biologics gradually declined in numbers in the stool, and were below the level of detection within 5 days post-administration (not shown). Such tunable maintenance of the biologics in the gut would be sparing of the endogenous microbiota, whose reestablishment after antibiotic insult is critical to CDI relief.
Even with fixed, pre-challenge dosing (6 doses), the engineered Syn-LAB 2.0 strain afforded protection against CDI. With continuous dosing of the biologic agent, however, there was significant protection against disease as well as mortality, even in the absence of any plasmid maintenance antibiotics (none were used in our animal studies). Treatment with the parent LAB strains was partially protective with continuous dosing compared to no treatment at all; this was not statistically significant compared to untreated animals, and was more variable, correlating with observations in human clinical studies where probiotic use does not consistently protect against CDI (Mills et al., 2018).
Beyond colonization resistance, the elicitation of anti-C. difficile SlpA antibody response in Syn-LAB-treated hamsters is noteworthy. In previous studies, recombinant SlpA-vaccinated mice exhibited a modest decrease in subsequent fecal C. difficile shedding, and an anti-SlpA response afforded partial protection against CDI (Biazzo et al., 2013; Bruxelle et al., 2016). Thus, Syn-LAB strains could afford long-term protection from new- or re-infection with diverse C. difficile strains in the community or healthcare setting. The use of these targeted biologics, therefore, via a once-daily oral administration as we tested herein, is likely to be suitable for multiple hosts.
Unlike the lethal hamster model, piglets are natural hosts to CDI, and display symptoms similar to human infections. Recent studies have highlighted CDI burden in agriculture, and its impact on the swine industry (Grzeskowiak et al., 2016; Stein et al., 2017; Kim et al., 2018). Our preliminary studies in this model are promising: as with the hamster studies, Syn-LAB was delivered and maintained in the piglets with ease, with the biologic being shed in the stool consistently. Even with a single-day FD regimen, the piglets were protected against CDI-induced diarrhea, in contrast to animals given C. difficile alone.
Conclusion
Individuals (or animals) who are not appropriate candidates for anti-CDI immunization, those anticipating extended-duration antibiotic treatment, or those in long-term care facilities (LTCFs) where the risk of acquiring CDI is high may benefit from Syn-LAB-type agents. Further, asymptomatic carriage of C. difficile is considered to be a major factor in pre-disposing patients to active CDI (Blixt et al., 2017; Caroff et al., 2017), and Syn-LAB administration may substantially reduce this risk.
Limitations of this Study
The studies as presented support Syn-LAB biologics as powerful new tools to prevent CDI in multiple mammalian systems. However, we recognize limitations that will need to be addressed by fully powered studies prior to completion of Syn-LAB pre-clinical testing. First, Syn-LAB efficacy will need to be tested against an even wider panel of recent C. difficile clinical isolates than those shown in Figure 7. However, the strong serum reactivity from Syn-LAB-treated animals against diverse, clinically relevant C. difficile ribotypes is encouraging. Second, Syn-LAB 2.0 and 2.1 may differ in their individual propensities to protect against CDI, and this needs to be investigated further. Third, we need to evaluate if the anti-SlpA antibody response results in protective immunity against C. difficile challenge, and parse this from colonization resistance provided by the biologic itself. Finally, the ability of Syn-LABs as a therapeutic (and not just a prophylactic as presented herein) remains to be tested – and this will involve efforts to determine biologic efficacy at different time-points post-infection. These limitations notwithstanding, our studies show strong evidence that the engineered Syn-LAB strains have considerable potential as primary or adjunct therapeutic agents against CDIs in multiple mammalian systems.
Author Contributions
GV and VV conceptualized and funded these studies, wrote the manuscript, and provided full project oversight. JL, JK, MM, and JLR designed, performed, optimized, and interpreted the experiments. CA, FA, AC, RC-W, AM, RCM, RM, and SR performed, supported and fully participated in all animal studies as well as some in vitro confirmation studies (FA and RM). All the authors read the manuscript and provided feedback.
Funding
This research project was supported by the National Institutes of Health (GV; AI121590). Work in the Vedantam laboratory is also supported by the US Department of Veterans Affairs (1I01BX001183-01) and the USDA CSREES Hatch Program (ARZT-570410-A-02-139; GV). The pilot piglet studies reported herein were supported by an Asset Development Award from Tech Launch Arizona.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Dr. Todd Klaenhammer for the kind gift of the pTRK848 plasmid. Many thanks to all the current and former members of the Vedantam and Viswanathan laboratories, specifically Michele Chu and Andrea Treptow. The help and support of the University of Arizona Cancer Center ARL Cytometry Core Facility (Grant-CA 023074) is gratefully acknowledged. We also thank Dr. Tod McCauley for helpful discussions.
References
Alberda, C., Marcushamer, S., Hewer, T., Journault, N., and Kutsogiannis, D. (2018). Feasibility of a Lactobacillus casei drink in the intensive care unit for prevention of antibiotic associated diarrhea and Clostridium difficile. Nutrients 10:E539. doi: 10.3390/nu10050539
Allen-Vercoe, E., and Petrof, E. O. (2013). Artificial stool transplantation: progress towards a safer, more effective and acceptable alternative. Expert Rev. Gastroenterol. Hepatol. 7, 291–293. doi: 10.1586/egh.13.16
Altermann, E., Russell, W. M., Azcarate-Peril, M. A., Barrangou, R., Buck, B. L., McAuliffe, O., et al. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U.S.A. 102, 3906–3912. doi: 10.1073/pnas.0409188102
Austin, M., Mellow, M., and Tierney, W. M. (2014). Fecal microbiota transplantation in the treatment of Clostridium difficile infections. Am. J. Med. 127, 479–483. doi: 10.1016/j.amjmed.2014.02.017
Barker, A. K., Duster, M., Valentine, S., Hess, T., Archbald-Pannone, L., Guerrant, R., et al. (2017). A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J. Antimicrob. Chemother. 72, 3177–3180. doi: 10.1093/jac/dkx254
Bianco, M., Fedele, G., Quattrini, A., Spigaglia, P., Barbanti, F., Mastrantonio, P., et al. (2011). Immunomodulatory activities of surface-layer proteins obtained from epidemic and hypervirulent Clostridium difficile strains. J. Med. Microbiol. 60(Pt 8), 1162–1167. doi: 10.1099/jmm.0.029694-0
Biazzo, M., Cioncada, R., Fiaschi, L., Tedde, V., Spigaglia, P., Mastrantonio, P., et al. (2013). Diversity of cwp loci in clinical isolates of Clostridium difficile. J. Med. Microbiol. 62(Pt 9), 1444–1452. doi: 10.1099/jmm.0.058719-0
Blixt, T., Gradel, K. O., Homann, C., Seidelin, J. B., Schonning, K., Lester, A., et al. (2017). Asymptomatic carriers contribute to nosocomial Clostridium difficile infection: a cohort study of 4508 patients. Gastroenterology 152, 1031–1041.e2. doi: 10.1053/j.gastro.2016.12.035
Boonma, P., Spinler, J. K., Venable, S. F., Versalovic, J., and Tumwasorn, S. (2014). Lactobacillus rhamnosus L34 and Lactobacillus casei L39 suppress Clostridium difficile-induced IL-8 production by colonic epithelial cells. BMC Microbiol. 14:177. doi: 10.1186/1471-2180-14-177
Bruxelle, J. F., Mizrahi, A., Hoys, S., Collignon, A., Janoir, C., and Pechine, S. (2016). Immunogenic properties of the surface layer precursor of Clostridium difficile and vaccination assays in animal models. Anaerobe 37, 78–84. doi: 10.1016/j.anaerobe.2015.10.010
Calabi, E., Ward, S., Wren, B., Paxton, T., Panico, M., Morris, H., et al. (2001). Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40, 1187–1199. doi: 10.1046/j.1365-2958.2001.02461.x
Cammarota, G., Masucci, L., Ianiro, G., Bibbo, S., Dinoi, G., Costamagna, G., et al. (2015). Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 41, 835–843. doi: 10.1111/apt.13144
Caroff, D. A., Yokoe, D. S., and Klompas, M. (2017). Evolving insights into the epidemiology and control of Clostridium difficile in hospitals. Clin. Infect. Dis. 65, 1232–1238. doi: 10.1093/cid/cix456
Chiu, C. H., Lu, T. Y., Tseng, Y. Y., and Pan, T. M. (2006). The effects of Lactobacillus-fermented milk on lipid metabolism in hamsters fed on high-cholesterol diet. Appl. Microbiol. Biotechnol. 71, 238–245. doi: 10.1007/s00253-005-0145-0
Dellaglio, F., Felis, G. E., and Torriani, S. (2002). The status of the species Lactobacillus casei (Orla-Jensen 1916) Hansen and Lessel 1971 and Lactobacillus paracasei Collins et al. 1989. Request for an opinion. Int. J. Syst. Evol. Microbiol. 52(Pt 1), 285–287. doi: 10.1099/00207713-52-1-285
Desai, K., Gupta, S. B., Dubberke, E. R., Prabhu, V. S., Browne, C., and Mast, T. C. (2016). Epidemiological and economic burden of Clostridium difficile in the United States: estimates from a modeling approach. BMC Infect. Dis. 16:303. doi: 10.1186/s12879-016-1610-3
Duong, T., Miller, M. J., Barrangou, R., Azcarate-Peril, M. A., and Klaenhammer, T. R. (2011). Construction of vectors for inducible and constitutive gene expression in Lactobacillus. Microb. Biotechnol. 4, 357–367. doi: 10.1111/j.1751-7915.2010.00200.x
Gardiner, B. J., Thorpe, C. M., Pinkham, N. V., McDermott, L. A., Walk, S. T., and Snydman, D. R. (2018). A repeat offender: recurrent extraintestinal Clostridium difficile infection following fecal microbiota transplantation. Anaerobe 51, 68–72. doi: 10.1016/j.anaerobe.2018.04.007
Goldenberg, J. Z., Yap, C., Lytvyn, L., Lo, C. K., Beardsley, J., Mertz, D., et al. (2017). Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst. Rev. 12:CD006095. doi: 10.1002/14651858.CD006095.pub4
Grzeskowiak, L., Zentek, J., and Vahjen, W. (2016). Determination of the extent of Clostridium difficile colonisation and toxin accumulation in sows and neonatal piglets. Anaerobe 40, 5–9. doi: 10.1016/j.anaerobe.2016.04.012
Karjalainen, T., Saumier, N., Barc, M. C., Delmee, M., and Collignon, A. (2002). Clostridium difficile genotyping based on slpA variable region in S-layer gene sequence: an alternative to serotyping. J. Clin. Microbiol. 40, 2452–2458. doi: 10.1128/JCM.40.7.2452-2458.2002
Kelly, C. R., Khoruts, A., Staley, C., Sadowsky, M. J., Abd, M., Alani, M., et al. (2016). Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann. Intern. Med. 165, 609–616. doi: 10.7326/M16-0271
Kiernan, J. A. (2008). Histological and Histochemical Methods: theory and Practice, 4th Edn. Bloxham: Scion Publishing Ltd.
Kim, H. Y., Cho, A., Kim, J. W., Kim, H., and Kim, B. (2018). High prevalence of Clostridium difficile PCR ribotype 078 in pigs in Korea. Anaerobe 51, 42–46. doi: 10.1016/j.anaerobe.2018.03.012
Kim, Y. H., Han, K. S., Oh, S., You, S., and Kim, S. H. (2005). Optimization of technical conditions for the transformation of Lactobacillus acidophilus strains by electroporation. J. Appl. Microbiol. 99, 167–174. doi: 10.1111/j.1365-2672.2005.02563.x
Kok, J., van der Vossen, J. M., and Venema, G. (1984). Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48, 726–731.
Lee, C. H., Steiner, T., Petrof, E. O., Smieja, M., Roscoe, D., Nematallah, A., et al. (2016). Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA 315, 142–149. doi: 10.1001/jama.2015.18098
Lee, H. M., and Lee, Y. (2008). A differential medium for lactic acid-producing bacteria in a mixed culture. Lett. Appl. Microbiol. 46, 676–681. doi: 10.1111/j.1472-765X.2008.02371.x
Lessa, F. C., Mu, Y., Bamberg, W. M., Beldavs, Z. G., Dumyati, G. K., Dunn, J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834. doi: 10.1056/NEJMoa1408913
Loo, V. G., Poirier, L., Miller, M. A., Oughton, M., Libman, M. D., Michaud, S., et al. (2005). A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353, 2442–2449. doi: 10.1056/NEJMoa051639
Lyras, D., O’Connor, J. R., Howarth, P. M., Sambol, S. P., Carter, G. P., Phumoonna, T., et al. (2009). Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179. doi: 10.1038/nature07822
Marco, M. L., Bongers, R. S., de Vos, W. M., and Kleerebezem, M. (2007). Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73, 124–132. doi: 10.1128/AEM.01475-06
Mattar, A. F., Teitelbaum, D. H., Drongowski, R. A., Yongyi, F., Harmon, C. M., and Coran, A. G. (2002). Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr. Surg. Int. 18, 586–590. doi: 10.1007/s00383-002-0855-7
McDonald, L. C., Gerding, D. N., Johnson, S., Bakken, J. S., Carroll, K. C., Coffin, S. E., et al. (2018). Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the infectious diseases society of America (IDSA) and society for healthcare epidemiology of America (SHEA). Clin. Infect. Dis. 66, 987–994. doi: 10.1093/cid/ciy149
McFarland, L. V. (2015). Probiotics for the primary and secondary prevention of C. difficile infections: a meta-analysis and systematic review. Antibiotics 4, 160–178. doi: 10.3390/antibiotics4020160
Mergenhagen, K. A., Wojciechowski, A. L., and Paladino, J. A. (2014). A review of the economics of treating Clostridium difficile infection. Pharmacoeconomics 32, 639–650. doi: 10.1007/s40273-014-0161-y
Merrigan, M. M., Venugopal, A., Roxas, J. L., Anwar, F., Mallozzi, M. J., Roxas, B. A., et al. (2013). Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One 8:e78404. doi: 10.1371/journal.pone.0078404
Michon, C., Langella, P., Eijsink, V. G., Mathiesen, G., and Chatel, J. M. (2016). Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Fact. 15:70. doi: 10.1186/s12934-016-0468-9
Mills, J. P., Rao, K., and Young, V. B. (2018). Probiotics for prevention of Clostridium difficile infection. Curr. Opin. Gastroenterol. 34, 3–10. doi: 10.1097/MOG.0000000000000410
Moono, P., Foster, N. F., Hampson, D. J., Knight, D. R., Bloomfield, L. E., and Riley, T. V. (2016). Clostridium difficile Infection in production animals and avian species: a review. Foodborne Pathog. Dis. 13, 647–655. doi: 10.1089/fpd.2016.2181
Ofori, E., Ramai, D., Dhawan, M., Mustafa, F., Gasperino, J., and Reddy, M. (2018). Community-acquired Clostridium difficile: epidemiology, ribotype, risk factors, hospital and intensive care unit outcomes, and current and emerging therapies. J. Hosp. Infect. 99, 436–442. doi: 10.1016/j.jhin.2018.01.015
Pal, K., Szen, O., Kiss, A., and Naar, Z. (2012). Comparison and evaluation of molecular methods used for identification and discrimination of lactic acid bacteria. J. Sci. Food Agric. 92, 1931–1936. doi: 10.1002/jsfa.5564
Peterson, M. D., and Mooseker, M. S. (1992). Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J. Cell Sci. 102(Pt 3), 581–600.
Petrof, E. O., and Khoruts, A. (2014). From stool transplants to next-generation microbiota therapeutics. Gastroenterology 146, 1573–1582. doi: 10.1053/j.gastro.2014.01.004
Raha, A. R., Varma, N. R., Yusoff, K., Ross, E., and Foo, H. L. (2005). Cell surface display system for Lactococcus lactis: a novel development for oral vaccine. Appl. Microbiol. Biotechnol. 68, 75–81. doi: 10.1007/s00253-004-1851-8
Rezaie, A., and Pimentel, M. (2014). Probiotics for antibiotic-associated diarrhea: PLACIDE swings the pendulum. Gastroenterology 146, 1822–1823. doi: 10.1053/j.gastro.2014.04.011
Roussel, Y., Colmin, C., Simonet, J. M., and Decaris, B. (1993). Strain characterization, genome size and plasmid content in the Lactobacillus acidophilus group (Hansen and Mocquot). J. Appl. Bacteriol. 74, 549–556.
Roxas, J. L., Ryan, K., Vedantam, G., and Viswanathan, V. K. (2014). Enteropathogenic Escherichia coli dynamically regulates EGFR signaling in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G374–G380. doi: 10.1152/ajpgi.00312.2013
Roxas, J. L., Wilbur, J. S., Zhang, X., Martinez, G., Vedantam, G., and Viswanathan, V. K. (2012). The enteropathogenic Escherichia coli-secreted protein EspZ inhibits host cell apoptosis. Infect. Immun. 80, 3850–3857. doi: 10.1128/IAI.00335-12
Sattar, A., Thommes, P., Payne, L., Warn, P., and Vickers, R. J. (2015). SMT19969 for Clostridium difficile infection (CDI): in vivo efficacy compared with fidaxomicin and vancomycin in the hamster model of CDI. J. Antimicrob. Chemother. 70, 1757–1762. doi: 10.1093/jac/dkv005
Seekatz, A. M., Theriot, C. M., Rao, K., Chang, Y. M., Freeman, A. E., Kao, J. Y., et al. (2018). Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe doi: 10.1016/j.anaerobe.2018.04.001 [Epub ahead of print].
Shatalin, K. Y., and Neyfakh, A. A. (2005). Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol. Lett. 245, 315–319. doi: 10.1016/j.femsle.2005.03.029
Steele, J., Feng, H., Parry, N., and Tzipori, S. (2010). Piglet models of acute or chronic Clostridium difficile illness. J. Infect. Dis. 201, 428–434. doi: 10.1086/649799
Steen, E. J., Kang, Y., Bokinsky, G., Hu, Z., Schirmer, A., McClure, A., et al. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562. doi: 10.1038/nature08721
Stein, K., Egan, S., Lynch, H., Harmanus, C., Kyne, L., Herra, C., et al. (2017). PCR-ribotype distribution of Clostridium difficile in Irish pigs. Anaerobe 48, 237–241. doi: 10.1016/j.anaerobe.2017.10.004
Terveer, E. M., van Beurden, Y. H., Goorhuis, A., Mulder, C. J. J., Kuijper, E. J., Keller, J. J., et al. (2018). Faecal microbiota transplantation in clinical practice. Gut 67, 196–197. doi: 10.1136/gutjnl-2017-313909
van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
Vernaya, M., McAdam, J., and Hampton, M. D. (2017). Effectiveness of probiotics in reducing the incidence of Clostridium difficile-associated diarrhea in elderly patients: a systematic review. JBI Database System. Rev. Implement. Rep. 15, 140–164. doi: 10.11124/JBISRIR-2016-003234
Viswanathan, V. K., Mallozzi, M. J., and Vedantam, G. (2010). Clostridium difficile infection: an overview of the disease and its pathogenesis, epidemiology and interventions. Gut Microbes 1, 234–242. doi: 10.4161/gmic.1.4.12706
Vogel, R. F., and Ehrmann, M. (1996). Genetics of lactobacilli in food fermentations. Biotechnol. Annu. Rev. 2, 123–150. doi: 10.1016/S1387-2656(08)70008-5
Walker, D. C., Aoyama, K., and Klaenhammer, T. R. (1996). Electrotransformation of Lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138, 233–237. doi: 10.1111/j.1574-6968.1996.tb08163.x
Wang, S., Xu, M., Wang, W., Cao, X., Piao, M., Khan, S., et al. (2016). Systematic review: adverse events of fecal microbiota transplantation. PLoS One 11:e0161174. doi: 10.1371/journal.pone.0161174
Wong, S., Jamous, A., O’Driscoll, J., Sekhar, R., Weldon, M., Yau, C. Y., et al. (2014). A Lactobacillus casei Shirota probiotic drink reduces antibiotic-associated diarrhoea in patients with spinal cord injuries: a randomised controlled trial. Br. J. Nutr. 111, 672–678. doi: 10.1017/S0007114513002973
Wullt, M., Hagslatt, M. L., and Odenholt, I. (2003). Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: a double-blind, placebo-controlled trial. Scand. J. Infect. Dis. 35, 365–367. doi: 10.1080/00365540310010985
Youngster, I., Sauk, J., Pindar, C., Wilson, R. G., Kaplan, J. L., Smith, M. B., et al. (2014). Fecal microbiota transplant for relapsing Clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clin. Infect. Dis. 58, 1515–1522. doi: 10.1093/cid/ciu135
Yun, B., Oh, S., and Griffiths, M. W. (2014). Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 97, 4745–4758. doi: 10.3168/jds.2014-7921
Keywords: Clostridium difficile, infectious diarrhea, synthetic biology, surface layer protein, Lactobacillus
Citation: Vedantam G, Kochanowsky J, Lindsey J, Mallozzi M, Roxas JL, Adamson C, Anwar F, Clark A, Claus-Walker R, Mansoor A, McQuade R, Monasky R, Ramamurthy S, Roxas B and Viswanathan VK (2018) An Engineered Synthetic Biologic Protects Against Clostridium difficile Infection. Front. Microbiol. 9:2080. doi: 10.3389/fmicb.2018.02080
Received: 22 May 2018; Accepted: 14 August 2018;
Published: 05 September 2018.
Edited by:
Joseph Sorg, Texas A&M University, United StatesReviewed by:
Rita Tamayo, The University of North Carolina at Chapel Hill, United StatesSarah Anna Kuehne, University of Birmingham, United Kingdom
Copyright © 2018 Vedantam, Kochanowsky, Lindsey, Mallozzi, Roxas, Adamson, Anwar, Clark, Claus-Walker, Mansoor, McQuade, Monasky, Ramamurthy, Roxas and Viswanathan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gayatri Vedantam, Z2F5YXRyaUBlbWFpbC5hcml6b25hLmVkdQ== V. K. Viswanathan, dmt2QGVtYWlsLmFyaXpvbmEuZWR1
†These authors have contributed equally to this work
 Gayatri Vedantam
Gayatri Vedantam Joshua Kochanowsky
Joshua Kochanowsky Jason Lindsey
Jason Lindsey Michael Mallozzi
Michael Mallozzi Jennifer Lising Roxas
Jennifer Lising Roxas Chelsea Adamson1
Chelsea Adamson1 Andrew Clark
Andrew Clark Asad Mansoor
Asad Mansoor V. K. Viswanathan
V. K. Viswanathan