- 1State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2RT-Europe Research Center, Mosonmagyaróvár, Hungary
The number of multidrug-resistant strains of Riemerella anatipestifer continues to increase, and new strategies for the treatment of associated infections are necessary. Recently, numerous studies have shown that efflux pumps (EPs) play key roles in universal bacterial mechanisms that contribute to antibiotic resistance. In addition, studies have shown that the effects of antibiotics that are subjected to efflux can be reinforced by their combined use with efflux pump inhibitors (EPIs). Unfortunately, the role of the efflux system in R. anatipestifer remains barely understood. In this study, we evaluated the role of EPs and resistance genes in the resistance generated by clinical strains of R. anatipestifer to antibiotics. A set of 10 R. anatipestifer strains were characterized by drug resistance, associated resistance genes, and antibiotic profiles in the presence and absence of EPIs. Efflux activity was studied on a real time basis through a fluorometric method. Quantification of the levels of mRNA transcription of efflux pump genes (EPGs) was determined by RT-qPCR. Several approaches (detection of resistance genes, drug susceptibility testing, and growth kinetics analysis) were used to assess the correlation between the effect of the EPIs and the resistance levels. Analysis of the R. anatipestifer growth inhibition tests showed that the antibiotic activity was enhanced by the synergy of EPIs. Among the various resistance genes that confer antibiotic resistance, different minimum inhibitory concentrations (MICs) were observed. The different levels of resistance were reduced by EPIs. Real time fluorometry showed that all the R. anatipestifer strains presented inherent efflux activity, conferring varying levels of inhibition in the presence of EPIs. Moreover, 15 EPGs were overexpressed in the presence of antibiotics. The addition of EPIs to antibiotics led to downregulation in the expression of some EPGs and a simultaneous increase in drug resistance and sensitivity. These results demonstrated the contribution of these EPs in the resistant phenotype of the clinical strains of R. anatipestifer that are under investigation, independently of the resistant genotype of the respective strains. Intrinsic efflux activity was possibly linked to the evolution of resistance in multidrug-resistant isolates of R. anatipestifer. Furthermore, the inhibition of EPs by EPIs could enhance the clinical effects of antibiotics.
Introduction
Duck serositis, caused by Riemerella anatipestifer, is an important communicable disease (Zheng et al., 2012; Guo et al., 2017). R. anatipestifer is a gram-negative, rod-shaped, non-spore-forming, non-motile bacterium that can primarily infect ducks that are l−5 weeks of age, via the respiratory or digestive tracts (Chikuba et al., 2016). Infection with this pathogen leads to great economic losses owing to the significant weight loss observed in ducklings and high mortality rates of up to 75% (Wang Q. et al., 2017). Thus far, at least 21 different serotypes of R. anatipestifer have been identified, and cross-immunoprotection among different serotypes has been barely reported (Zhai et al., 2013; Gyuris et al., 2017). All these factors complicate the prevention of this disease.
The molecular mechanisms associated with this bacterial infection remain largely unknown, and its treatment depends mainly on chemotherapy. However, the chronic use of antimicrobials has increasingly aggravated the resistance in R. anatipestifer isolates (Chang et al., 2003; Zhong et al., 2009). In previous studies (Chang et al., 2003; Chen et al., 2010, 2012; Xing et al., 2015), various antibiotic susceptibility tests of the field isolates of R. anatipestifer are reported. Nevertheless, studies on the molecular mechanisms of R. anatipestifer isolates that are resistant to antibiotics are limited. The resistance mechanisms associated with drug efflux pumps (EPs) have not been reported (Li, X. Z. et al., 2016).
Since the early 1990s, when the bacterial EPs were investigated for their significant contribution to multidrug resistance (MDR), a large number of studies have focused on efflux-mediated antimicrobial resistance and efflux determinants (Li et al., 2015). In bacteria, resistance can take many forms. Bacterial drug EPs constitute a major mechanism of both natural and acquired resistance to a diverse range of clinically used antibiotics and biocides. Based on the energy source, number, size, and substrates of the transporters, the EPs can be divided into five families (Li, X. Z. et al., 2016, chapter 6) that have been associated with drug resistance: the multidrug and toxic-compound extrusion (MATE) family and the small multidrug resistance (SMR) family, which are singlet transporters and occur in all species; and three superfamilies, which include the resistance nodulation and cell division (RND) super family, the ATP-binding cassette (ABC) superfamily, and the major facilitator superfamily (MFS), which are multicomponent transporters that operate through a complex efflux machinery that often include the pump, accessory adaptor proteins, and outer membrane proteins.
Bacterial EPs play a major role in the development of bacterial MDR. Efflux pump inhibitors (EPIs) maybe the best solution to this problem, as they can reverse resistance in bacteria (Li, X. Z. et al., 2016, chapter 29). EPIs act against MDR EPs with a wide substrate spectrum, and they are expected to not only reverse resistance to a single drug but also to multiple clinically beneficial antimicrobials at the same time. However, many discovered EPIs have either failed the preclinical trials, have failed to reach clinical concentrations for their activity in vivo, or have toxicity problems (Li, X. Z. et al., 2016, chapter 28). The effects of some EPIs on antimicrobial susceptibilities have been studied in several microorganisms, including Escherichia coli (Opperman et al., 2014), Salmonella (Sutkuviene et al., 2013), Klebsiella pneumoniae (Chevalier et al., 2004), Mycobacterium tuberculosis (Coelho et al., 2015; Machado et al., 2017), Acinetobacter baumannii (Richmond et al., 2013), and Campylobacter (Kurincic et al., 2012). However, in regard to this, no systematic, comprehensive research has been conducted in R. anatipestifer.
In this study, we used the standard broth microdilution method to determine the antibiotic susceptibility of a wide range of antibiotics against R. anatipestifer isolates, collected from different regions of China between 2010 and 2017. The presence of resistance genes and integrons in R. anatipestifer were detected by PCR. Through comparative analysis of a resistant phenotype and genotype of R. anatipestifer strains, we determined the extent of resistance in some R. anatipestifer isolates toward antimicrobial agents. Characterization of the arrays of integrons is a useful epidemiological tool in the study of the evolution of MDR. Various functions, especially drug resistance, were mediated into hosts via mobile gene cassettes through specific excision and integration in integrons (van Essen-Zandbergen et al., 2007; Li Y. et al., 2018). For R. anatipestifer, five resistance gene cassettes (dhfrI–aadA2, aadA11, aadA5, aa6II–catB3–aadA1, and aadA1–dhfrA1) of class 1 integrons have been reported (Zheng et al., 2012). The resistance observed could not be explained solely by the presence of the drug-resistant genes or mutations (Jaillard et al., 2017; Uddin and Ahn, 2017). Some R. anatipestifer isolates, in which drug-resistant genes were not detected, still showed resistance and some even showed MDR. These findings led us to explore the activity of EPs and its association with drug resistance.
We chose two broad-spectrum EPIs: carbonyl cyanide m-chlorophenylhydrazone (CCCP; Li Y. F. et al., 2016) that acts as an energy blocker and can inhibit EPs that are driven by hydrogen ion gradients; and Phe-Arg-β-naphthylamide (PAβN; Lamers et al., 2013), a protonophore inhibitor that can affect membrane integrity. These EPIs perturb the bacterial efflux systems, such as MFS and RND. In this study, we presume that MFS or RND EPs may play a major role in resistance associated with antibiotics in R. anatipestifer isolates.
The main purpose of this study was to assess the role played by the EP systems in the resistant phenotype and genotype of R. anatipestifer strains that carry different drug-resistant genes. In order to achieve this, we explored the contribution of the efflux mechanisms in the overall resistance to seven different types of antibiotics, in nine resistant clinical isolates of R. anatipestifer from four distinct geographical areas of China. The following processes were performed: (1) analysis of the synergistic effect of the efflux inhibitors, CCCP and PAβN, on the minimum inhibitory concentrations (MICs) of the antibiotics; (2) analysis of real time efflux activity to confirm the existence of active EP systems in these strains, using ethidium bromide (EB) as an efflux substrate, and subsequent calculation of final fluorescence both in the presence and absence of each efflux inhibitor; and (3) analysis of the levels of mRNA transcription (n-fold) of selected efflux pump genes (EPGs) in the strains under investigation by RT-qPCR. The results revealed that the addition of efflux inhibitors enhanced the effects of antibacterial drugs, independently of the genotypic resistance of the R. anatipestifer strains. The present study indicated the validity of a synergistic combination of drugs with traditional therapy, despite the presence of drug-resistant genes. This strategy proved to be beneficial, as it reduced the level of resistance among the strains. Moreover, the findings of the present study strongly supported the potential of these efflux inhibitors as adjuvants in the chemotherapy of R. anatipestifer infections, and these findings helped to increase the understanding of the MDR mechanisms of R. anatipestifer.
Materials and Methods
Strains
The strains selected for this study are described in Table 1. All strains were preserved in the laboratory. The field isolate strains of R. anatipestifer (RA) were obtained from duck brain homogenates between March 2010 and December 2017, as previously described (Zheng et al., 2012). The samples were collected from diseased ducks in Shandong, Henan, Gongdong, and Anhui Provinces of China. The strains were categorized as multidrug resistant (resistant to at least three classes of antibiotics) and extensively drug resistant (MDR plus resistance to any fluoroquinolone and ampicillin, cefoxitin, or florfenicol) based on Table 2 data. The specific details of antimicrobial resistance phenotypes for each isolate are shown in Table 1. The RA01, ATCC11845, and E. coli ATCC25922 reference strains were obtained from the American Type Culture Collection (Virginia, USA) and were used as controls.
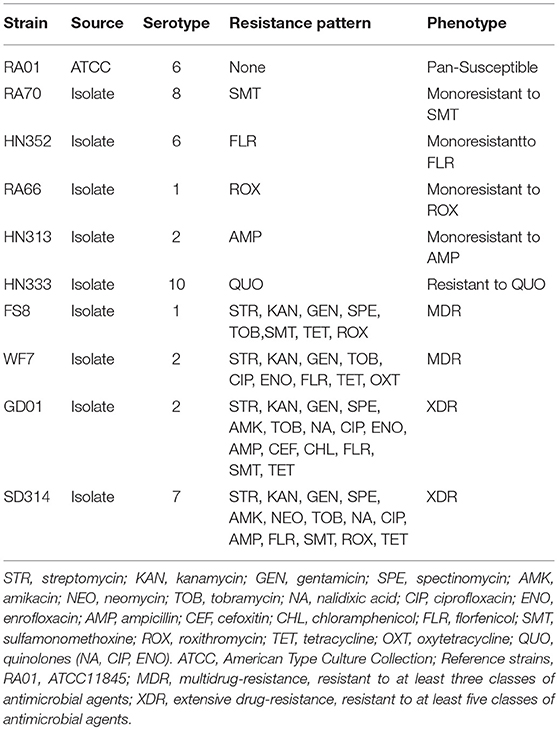
Table 1. Categorization of the R. anatipestifer strains that were studied according to their serotype, resistance pattern, and phenotype.
Drug Susceptibility Testing and MIC Determination
The selection of antibiotics in the present study was made based on those most widely used in the duck industry (Zhong et al., 2009; Li Y. et al., 2016). These antibiotics were purchased from Solarbio (Beijing, China). The strains were tested to determine the MICs of 18 antibiotics (Table 2 and Table S1), according to the Clinical and Laboratory Standards Institute's (CLSI) 2-fold serial broth microdilution method (CLSI, 2016). Briefly, each R. anatipestifer isolate was seeded in trypticase soy broth (TSB) with 5% calf serum, shaken at 200 rpm (revolutions per minute) for 6–12 h, before being incubated at 37°C for 24 h with 5% CO2. A volume of 200 μL of the microbial suspension of 106 CFU/mL was added to each well, each antimicrobial agent was added in successive rows, to attain concentrations between 64 and 0.125 μg/mL. All studies were carried out in triplicates. The MIC was recognized as the lowest concentration of the antimicrobial agent that can inhibit visible growth of bacteria (CLSI, 2016). The RA01, ATCC11845, and E. coli ATCC25922 strains were used as controls. The 18 antibiotics tested included β-lactams (ampicillin and cefoxitin); aminoglycosides (kanamycin, streptomycin, gentamicin, spectinomycin, amikacin, neomycin, and tobramycin); quinolones (nalidixic acid, ciprofloxacin, and enrofloxacin); amphenicols (chloramphenicol and florfenicol); sulfonamides (sulfamonomethoxine); macrolides (roxithromycin); and tetracyclines (tetracycline and oxytetracycline).
Detection of Resistance Genes, Mutations Associated With Resistance, and Integrons
To prepare genomic DNA, the bacterial suspension was collected and heated at 100°C for 10 min and then incubated on ice for 10 min. The samples were then centrifuged (Thermo Pico17) at 13,800 × g for 5 min. The supernatants were collected and stored at 4°C until further use. The PCR amplification of quinolone resistance determining regions (QRDR); plasmid-mediated quinolone resistance (PMQR) genes; aminoglycoside resistance genes; beta-lactamase resistance genes; tetracycline resistance genes; amphenicol (chloramphenicol and florfenicol) resistance genes; sulfonamide resistance genes; blaNDM−1 and mcr-1 resistance genes; class 1, 2, and 3 integrase genes; and a variable region of class 1, 2, and 3 integrons was carried out using the primers presented in the Supplementary Material (Table S2). The PCR amplification was carried out using a Takara Taq Master Mix Kit (TakaRa, Dalian, China) for standard PCR. The annealing temperature for each pair of primers is shown in Table S2. The positive PCR products were sequenced by the Nanjing Genscript Biology Company. Sequence data were then analyzed by DNASTAR and the sequences were compared with reference sequences from NCBI GenBank.
Drug Susceptibility Testing of Antibiotics in the Presence and Absence of Efflux Inhibitors
To determine the impact of active EP systems on resistance of R. anatipestifer isolates, a panel of nine R. anatipestifer drug-resistant strains was characterized for drug resistance genes and antibiotic profiles, in the presence and absence of PAβN and CCCP. The final use concentrations of the EPIs used were PAβN at 40 μg/mL and CCCP at 5 μg/mL. The correlation between the effect of the EPIs on the MICs of the antibiotics was assessed by synergism assays (refer to the previously mentioned broth microdilution method). The determination of MICs and the interpretation of results were performed as previously described (Sun et al., 2012). A reduction in MIC of at least 4-fold was considered as indicative of efflux (Li, X. Z. et al., 2016).
To quantify the role of the EPs in the strains under investigation and to establish a clinical association, we investigated the time taken for the detection of growth rate of each strain subjected to the antibiotics, in the presence and in the absence of the EPIs. The assays were performed as follows. Phe-Arg-β-naphthylamide (40 μg/mL) and CCCP (5 μg/mL) were added to the culture medium. These concentrations failed to affect the growth of the bacteria. Each antibiotic was tested at half MIC (determined for each strain), a concentration that does not affect the growth of R. anatipestifer isolates. All experiments were performed at least thrice for verification. Each time, bacterial strains were grown with TSB at 37°C until the cells reached an optical density (OD) at 600 nm of 1.0 (1011–1012 cfu/mL; Ji et al., 2017; Li S. et al., 2018). A 2% inoculum (100 μL in 5 mL) of bacterial culture was added to fresh TSB. This suspension was incubated under shaking conditions (200 rpm) at 37°C. The OD at a wavelength of 600 nm was measured over a period of 12 h using a UV spectrophotometer (Beckman Coulter, DU730, USA). The evaluation of bacterial growth vs. growth in the presence of antibiotics vs. growth in a mixture of antibiotics and efflux inhibitor was performed, by comparing the ODs of each strain, starting at the first hour and at 1 h intervals thereafter. Thus, the degree to which PAβN and CCCP enhanced the potentiation of antibiotic activity was determined.
Ethidium Bromide Accumulation and Efflux Assay
For the standard EB accumulation assays, cells were grown to an OD600 of 1.0 (1011–1012 cfu/mL) and 200 μL of cells were loaded into 96-well black plates and mixed with EB for 60 min to determine EB accumulation. To evaluate the effects of the efflux inhibitors' impact on efflux activity based on the accumulation of EB, the EB-loaded cells were washed and resuspended in phosphate-buffered saline (PBS), either with or without the EPIs, PAβN (40 μg/mL) and CCCP (5 μg/mL). The plates were kept at room temperature for 15 min and then fluorescence detection was performed. Each assay lasted for 40 min, and the signal was acquired every 5 min. Specific EB concentrations are indicated in the legends of Figures S1, S2. Fluorescence was determined by a microplate reader (Varioskan LUX, Thermo Fisher, USA) at emission and excitation wavelengths of 605 and 525 nm, respectively (Liu et al., 2017). The raw data obtained was normalized by comparing the relative fluorescence units obtained under the conditions that promote efflux (in the presence of 5% calf serum and in the absence of efflux inhibitors) with the relative fluorescence units from the control, in which no efflux was evident (in the presence of efflux inhibitors with 5% calf serum). The raw data was also normalized to the EB-loaded cells to which 5% calf serum was added, as the relative fluorescence of these cells was considered to be equivalent to 1 (Machado et al., 2017).
RT-qPCR Analysis of Putative Efflux Pump Genes
Strains were grown to an OD600 of 1.0 (1011–1012 cfu/mL) in shake flasks containing TSB, 5% calf serum, and half the MIC of each antibiotic as follows: RA70 was exposed to sulfamethazine (SMT); HN352 was exposed to amikacin (AMK); RA66 was exposed to roxithromycin (ROX); HN313 was exposed to ampicillin (AMP); HN333 was exposed to QUO (ciprofloxacin or enrofloxacin); FS8 was exposed separately to SMT, ROX, and ciprofloxacin (CIP); WF7 was exposed separately to florfenicol (FLR), CIP, and oxytetracycline (OXT); GD01 was exposed separately to FLR, AMP, CIP, AMK, and chloramphenicol (CHL); SD314 was exposed separately to FLR, ROX, AMP, CIP, neomycin (NEO), and AMK. The antibiotics were chosen for testing based on the MIC changes for each strain, which was affected by the EPIs. Although there are many factors that promote drug resistance, such as drug inactivating enzymes, alteration of drug targets, mobile resistant genetic elements; in this study, since the MICs of the antibiotics were reduced in the presence of EPIs by at least 4-fold, this was considered indicative of efflux activity (see Tables 2, 4, Table S1).
Moreover, to evaluate the effects of the antibiotics alone and in combination with the efflux inhibitors on EP activity, we analyzed the expression of the EPGs exposed to the antibiotic alone or combined with the inhibitors PAβN (40 μg/mL) and CCCP (5 μg/mL). The relative expression of the EPGs was assessed by comparing the relative quantity of the respective mRNA in the presence of the antibiotic, and the antibiotic + sub-MIC concentration of the inhibitor, with those of the non-exposed strain. Total RNA was extracted using Invitrogen™ Trizol™ Max™ Bacterial RNA Isolation Kit (Thermo Fisher Scientific, USA). The relative expression of the EPGs RIA-1800, RIA-1853, RIA-0245, RIA-0257, RIA-0437, RIA-0577, RIA-0746, RIA-1554, RIA-1117, RIA-1118, RIA-1215, RIA-1993, RIA-0286, RIA-1069, and RIA-1614 was analyzed by RT-qPCR. The primers used for RT-qPCR are listed in the Supplementary Material (Table S3). Reverse transcription of complementary DNAs and quantitative reverse transcription-PCR were performed simultaneously, using an AceQ qPCR SYBR Green Master Mix Kit (Vazyme, Nanjing, China), a HiScript Q RT SuperMix for qPCR Kit (Vazyme, Nanjing, China), and a MX3000P instrument (Agilent Technologies, Santa Clara, USA).
The protocol that was used comprised the following amplification program: reverse transcription for 15 min at 50°C, followed by 2 min at 85°C, an initial denaturation step for 5 min at 95°C, and 40 cycles (95°C for 10 s and 60°C for 30 s). The specificity of all the primers was analyzed by using melting curves. The melting curves were recorded after the 40th cycle by increasing the temperature stepwise by 0.5°C every 5 s from 65 to 95°C. The comparative quantification cycle method was used to determine the relative mRNA expression of the efflux genes (Livak and Schmittgen, 2001). The relative expression of each target gene was determined by comparing the relative quantity of the respective mRNA in the presence of the antibiotic and in the presence of the antibiotic and efflux inhibitor to that of the non-exposed mRNA. Each strain was assayed in triplicates, using the RNA obtained from three independent cultures. A relative expression level equal to 1 indicated that the expression level was identical to the unexposed strain. Each sample cycle threshold (CT) mean was calculated and standard deviations were calculated for each mean CT value. Normalization of the mRNA levels was done using the R. anatipestifer recA housekeeping gene (Huang et al., 2017) as internal control for each experiment and presented as the mean-fold change (±SD) compared with the control. Furthermore, the 2−ΔΔCt formula was used to calculate the relative expression levels of the efflux genes. The real time PCR data were detected and analyzed by the MxPro Software (Mx3000p Quantitative PCR) according to default parameters, which generated the CT values for each reaction.
Ethics Statement
All animal were handled by following strict procedures according to the (indicate as appropriate). The study was approved by the Institutional Animal Care and Use Committee of the Lanzhou Veterinary Research Institute.
Statistical Analysis
The data were analyzed using two-way ANOVA multiple comparisons using the GraphPad Prism 6.0 Windows software. Levels of significance were set as follows: *P < 0.05 was considered statistically significant, and **P < 0.01, ***P < 0.001, and ****P < 0.0001 were all considered highly significant.
Results
Correlating Phenotype and Genotype Associated With Antibiotic Resistance
To establish associations among phenotypic drug resistance and resistance determinants, we searched for the presence of resistance genes, mutations in resistance genes, and integrons associated specifically with resistance to the antibiotics under investigation.
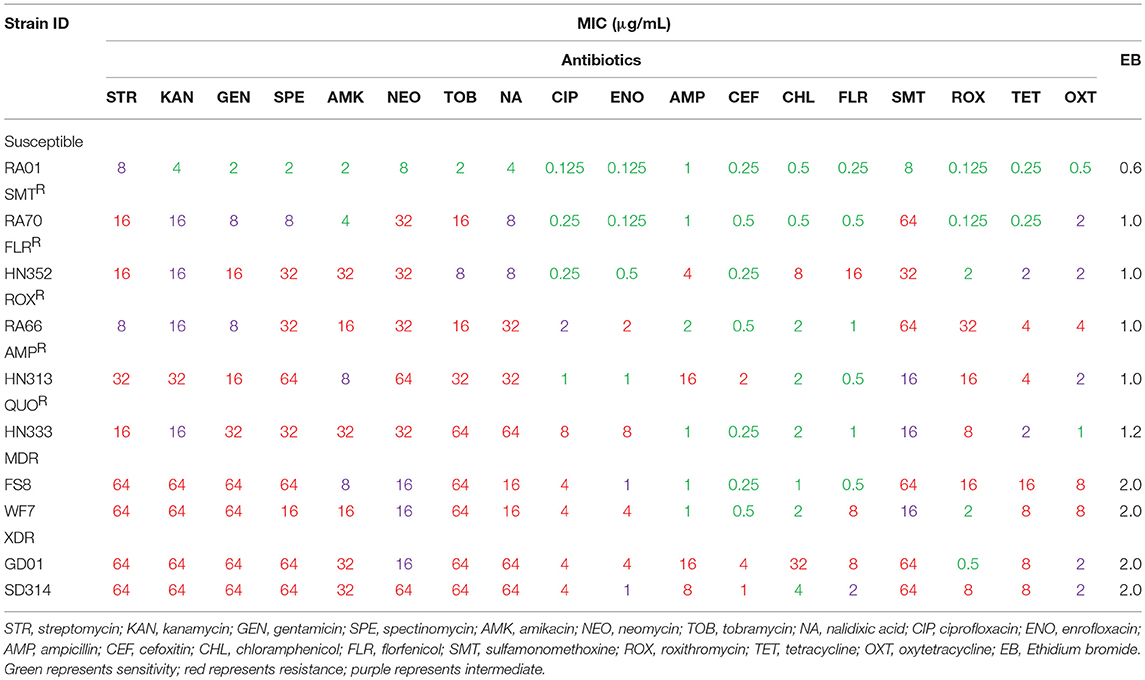
First, according to the MICs of each antibiotic (Table 2), 10 R. anatipestifer isolates from six different serotypes were selected, based on their drug resistance patterns (Table 1). The distribution of MIC values for each antibiotic is shown in Table 2. Multidrug-resistant isolates were resistant to at least three antimicrobial agents, and extensively drug-resistant isolates were multidrug-resistant strains that were additionally resistant to any fluoroquinolone and ampicillin, cefoxitin, or florfenicol. The R. anatipestifer isolates exhibited resistance to many antimicrobial agents, particularly aminoglycosides, fluoroquinolones, and sulfonamides, all of which are commonly used in ducks.
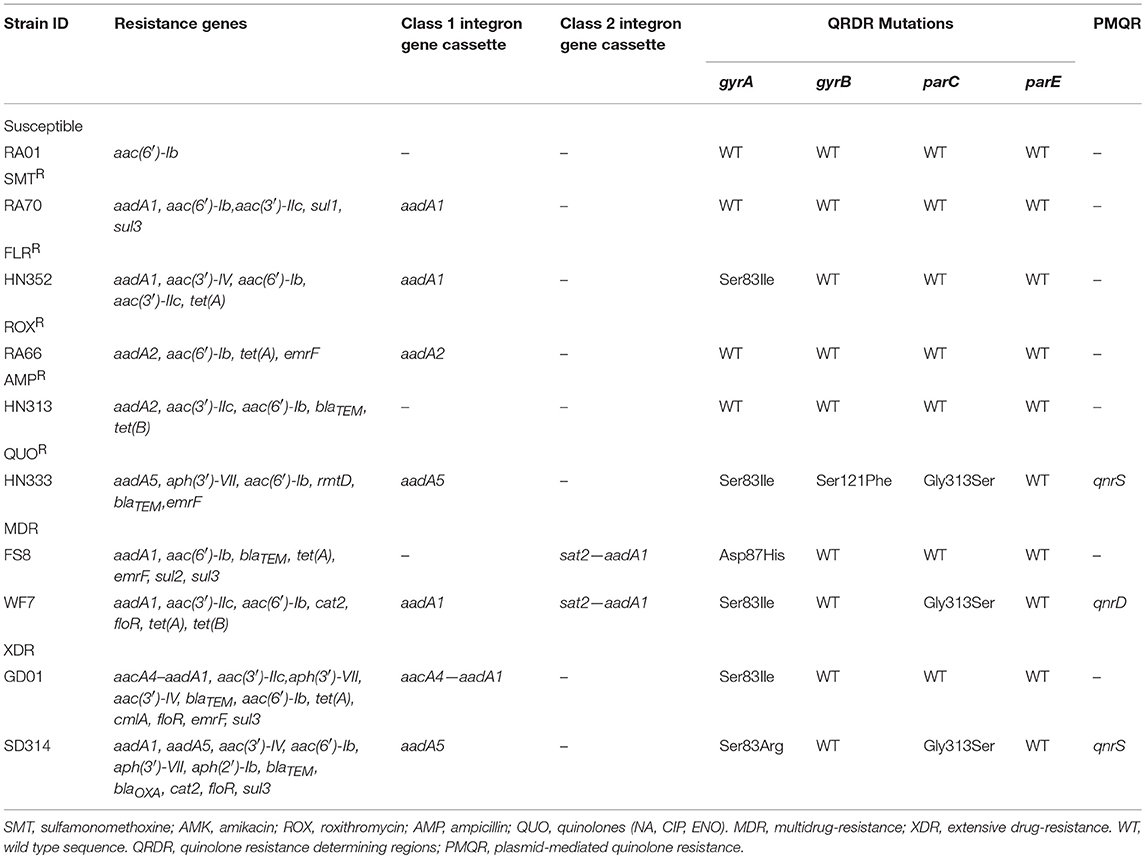
The distribution of resistance genes, integrons, and QRDR mutations is shown in Table 3. Among the aminoglycoside resistance genes, eight aminoglycoside modifying enzyme genes were identified. The aac(6′)-Ib gene that causes resistance to kanamycin, tobramycin, and amikacin was detected in all the isolates. This is the most common resistance gene found in clinical isolates that were resistant to aminoglycosides. The aadA1, aadA2, and aadA5 genes that confer resistance to spectinomycin and streptomycin, the aadA1 and aadA2 were detected in almost all of the isolates, but the aadA5 gene was detected in the HN333 and SD314 isolates only. The aac(3′)-IIc gene that causes resistance to gentamicin and tobramycin was detected in seven isolates. The aac(3′)-IV gene that mediates resistance to tobramycin, gentamicin, and neomycin was detected in the HN352, GD01, and SD314 isolates. The aph(3′)-VII and aph(2′)-Ib genes that cause resistance to amikacin, neomycin, and kanamycin were detected in the HN352, HN333, GD01, and SD314 isolates. Among the macrolide-resistance genes, only the emrF gene was detected in five isolates. Among the chloramphenicol- and florfenicol-resistance genes, cat2, cmlA, and floR were detected in the WF7, GD01, and SD314 R. anatipestifer isolates, respectively. Among the tetracycline and sulfonamide resistance genes, tet(A), tet(B), sul1, sul2, and sul3 were present in the RA70, RA66, HN313, FS8, WF7, GD01, and SD314 isolates. Among the beta-lactamase resistance genes, the blaTEM gene was detected in the HN313, HN333, FS8, and GD01 isolates. Only the blaOXAgene was detected in the SD314 isolate. One 16S rRNA methylase gene (rmtD) was detected in the HN333 isolate. Notably, two resistance genes (blaNDM−1and mcr-1) were not detected. Eight R. anatipestifer isolates harbored one or two class 1 or class 2 integrons. Four different cassette arrays (aadA1, aadA2, aadA5, and aacA4-aadA1) of the class 1 integron and the sat2-aadA1 gene cassette of the class 2 integron were discovered. The class 3 integron was not detected in the strains under investigation. The aadA1 and aac(6′)-Ib-floR gene cassettes that cause resistance to aminoglycoside antibiotics and chloramphenicol were detected in the WF7, GD01, and SD314 isolates.
Finally, in light of the MICs of quinolones and fluoroquinolones, ten R. anatipestifer isolates were selected for the detection of QRDR mutations (gyrA, gyrB, parC, and parE) and PMQR. The distribution of the QRDR gene mutations and PMQR genes are listed in Table 3. Four isolates exhibited the wild type genes gyrA, gyrB, parC, and parE. Three isolates had a single mutation in gyrA at codon 83 (Ser to Ile) or codon 87 (Asp to His). Two isolates had one parC mutation (Gly313 to Ser) and one gyrA hotspot mutation. One isolate had one gyrB mutation (Ser121 to Phe), one parC mutation (Gly313 to Ser), and one gyrA hotspot mutation. No mutations were detected in the parE genes. In regard to the detection of PMQR genes, three isolates were observed to carry two qnrS genes and one qnrD gene, respectively. The R. anatipestifer isolates, particularly HN333, GD01, and SD314, were resistant to quinolones in the broth microdilution assay at the highest concentrations (Table 2).
Briefly, 18 antibiotics out of the seven classes of antibiotics were evaluated, and we selected one representative antibiotic family for a subsequent study. For example, the resistance phenotype detected was mostly consistent with the detection of resistance genes among the majority of isolates, such as the aac(6′)-Ib or aph(3′)-VII gene. These genes were associated with amikacin and kanamycin resistance. The QRDR mutation and PMQR, which are quinolone resistance genes, were tested in all R. anatipestifer strains; the HN352 isolate carrying gyrA (Ser83 to Ile) was susceptible to quinolone, and FS8 isolate that was negative for PMQR was resistant to quinolones. The blaTEM gene confers ampicillin resistance and aac(6′)-Ib is an amikacin resistant gene, whereas the WF7 isolate that carries the aac(6′)-Ib gene was susceptible to amikacin, and SD314 isolates negative for blaTEM were resistant to ampicillin. Therefore, other resistance mechanisms may exist in R. anatipestifer.
Quantification of Antibiotic Resistance in the Presence of Efflux Pump Inhibitors
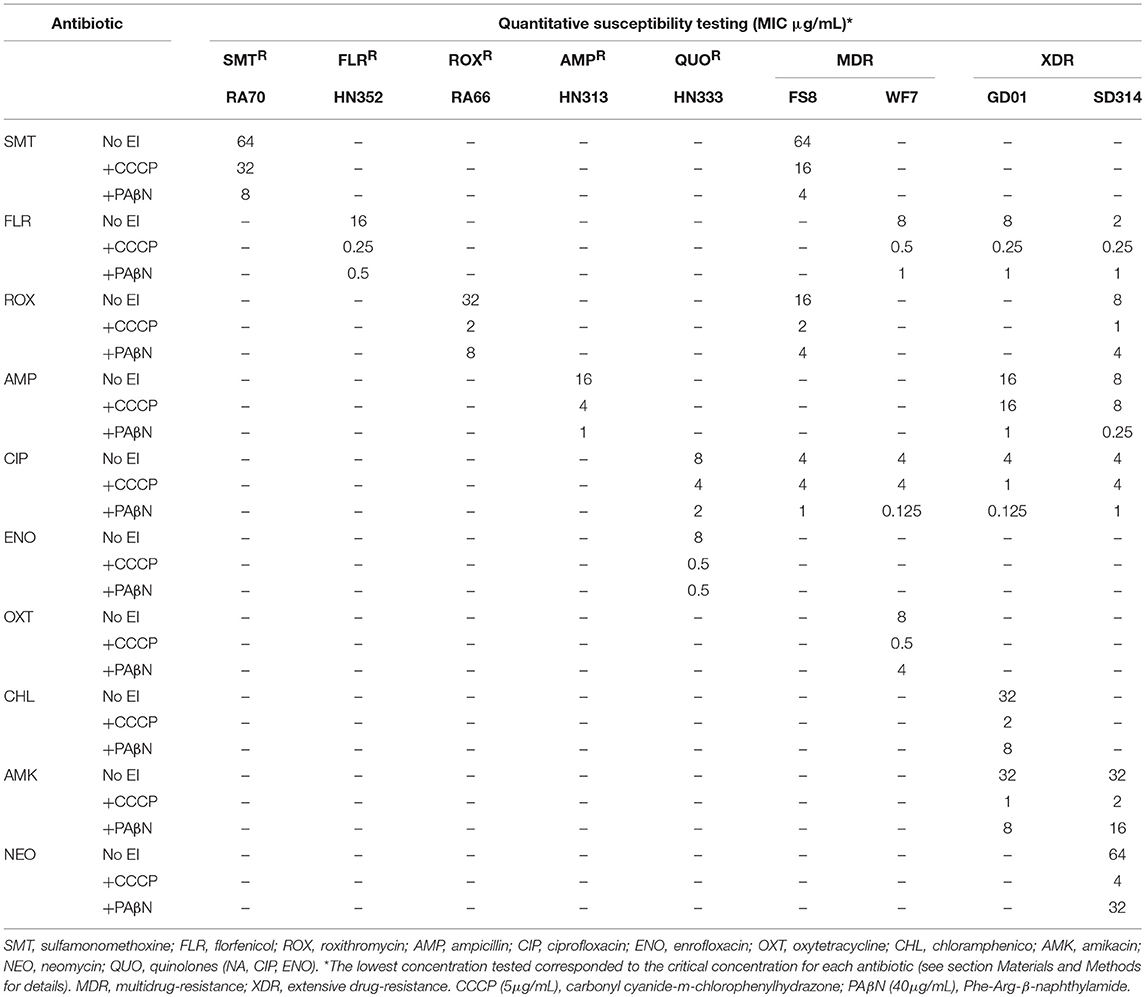
We quantified the MICs of antibiotics for specific strains in the presence or absence of EPIs. To achieve this, we selected representative clinical strains of R. anatipestifer from four distinct geographical areas. The MICs of the antimicrobial agents, both in the presence and absence of EPIs, were determined for each strain and the results are presented in Table 4. The MICs observed in the presence or absence of EPIs showed considerable differences among the seven groups of strains. Similarly, the MICs of the specific antimicrobial agents showed different MIC values. Therefore, we assumed that the different resistance levels observed for the same genotype or phenotype were due to the different levels of active efflux of the antibiotics. The existence of active efflux was evaluated through the reduction of the MICs for the antibiotics in the presence of the EPIs CCCP and PAβN, growth rate analyses, and EP gene expression.
Table 4 illustrates the strains that showed MIC values of the antibiotics with a reduction of 4-fold or above in the combination of CCCP and PAβN. The RA70 and FS8 strains that were resistant to SMT showed high level resistance (R = 64 μg/mL). The addition of PAβN reduced the MIC of SMT (8- to 16-fold) and the addition of CCCP also reduced the MIC of SMT (2- to 4-fold).
For HN352, WF7, GD01, and SD314, the MIC for FLR (R ≥ 2–16 μg/mL) was reduced more than 8-fold by the pump inhibitors. Regarding ROX resistance (R ≥ 8–32 μg/mL), the MIC values were reduced in the presence of CCCP in all ROX-resistant strains by 16-fold (RA66), 8-fold (FS8), and 8-fold (SD314); and in the presence of PAβN in all ROX-resistant strains by 4-fold (RA66), 4-fold (FS8), and 2-fold (SD314). The addition of PAβN was able to reduce the MIC of AMP (R ≥ 8–16 μg/mL) in HN313 (16-fold), GD01 (16-fold), and SD314 (32-fold). In contrast, the addition of CCCP failed to modify the MIC of AMP for GD01 and SD314; the same trend was not observed in HN313. Of the five strains (Table 4) resistant to CIP (R ≥ 4–8 μg/mL), with the exception of GD01 (with a 4-fold reduction), none were reduced in the presence of the CCCP inhibitor. However, in the CIP-resistant strains, the MIC values were reduced from 4- to 32-fold, depending on the strains. Resistance to ENO (R = 8 μg/mL) was reduced in strain HN333 alone, by 16-fold in the presence of CCCP and PAβN. The MIC for OXT (R = 8 μg/mL) in the WF7 strain was reduced by 16-fold in the presence of CCCP alone. Similar to GD01, the MIC for CHL (R ≥ 32 μg/mL) was reduced by 16-fold in the presence of CCCP. The effect of the PAβN inhibitor on the WF7 and GD01 strains was not obvious.
With regard to resistance to AMK (R = 32 μg/mL), the results showed that the addition of CCCP reduced the MIC of GD01 (32-fold) and SD314 (32-fold), and, to a lesser extent, PAβN also reduced the MIC of GD01 (4-fold) and SD314 (2-fold). Similarly, the MIC of NEO (R = 64 μg/mL) in the SD314 strain was reduced by 16-fold in the presence of CCCP and by 2-fold in the presence of PAβN. These data indicated the presence of efflux systems that are associated with drug resistance in R. anatipestifer.
Determination of the Intrinsic Efflux Capacity of R. anatipestifer Strains
Resistance, particularly MDR, takes many forms and is often a complex process. However, the primary response of most pathogens to antibiotic exposure requires a drug EP. A better understanding of the processes that are involved in studies that are more extensive would lead to improved treatment applications. To confirm the existence of active efflux systems in the strains under investigation, we assessed their ability to efflux EB by real time fluorometry (Viveiros et al., 2010; Amaral et al., 2011). To perform these assays, one representative from each drug-resistant group was selected as follows: RA70, mono-resistant to SMT; HN352, mono-resistant to FLR; RA66, mono-resistant to ROX; HN313, mono-resistant to AMP; HN333, resistant to QUO; FS8 and WF7, multidrug-resistant; and GD01 and SD314, extensively drug-resistant (Figures S1, S2). The R. anatipestifer strain RA01 was considered representative of drug-susceptible strains.
First, the lowest concentration that caused minimal accumulation of EB was determined. The lowest concentration that resulted in equilibrium between the influx and efflux of EB was 0.3 μg/mL for the reference strain RA01; 0.5 μg/mL for RA70, HN352, RA66, and HN313 mono-resistant strains; 0.6 μg/mL for the HN333 strain; and 1.0 μg/mL for the multidrug-resistant and extensively drug-resistant strains. These results indicated that the clinical antibiotic resistant R. anatipestifer strains could handle higher concentrations of EB than the reference strain and suggested the presence of more active EP systems in resistant strains. Figures S1, S2 illustrate the effects of the efflux of EB with different efflux activity efficiencies. The efflux of EB was also affected by the presence of 5% calf serum (compare the black dotted line with the colored curve on each graph), demonstrating that sufficient external nutrition of the cells was necessary to guarantee optimal efflux activity in R. anatipestifer. The assays showed that all strains under investigation presented efflux activity. This is an intrinsic characteristic of susceptible and drug-resistant strains of R. anatipestifer.
The varied abilities for the efflux of EB demonstrated by the strains were observed as follows: RA01 presented basal efflux activity; in the clinical GD01 and WF7 strains (Figure S2), efflux activity was more pronounced; the EB efflux activity of HN352, RA66, HN313, FS8, and SD314 strains was significant and has similar levels, which was less than that of the GD01 and WF7 strains; and RA70 (Figure S1) and HN333 (Figure S2) presented diverse efflux activity. In most of the strains, the efflux inhibitor that yielded the highest inhibitory effect was CCCP, followed by PAβN; however, in HN333, the highest EB accumulation rate was achieved following the addition of PAβN, as assessed by the fluorometric method. These results might be associated with the different environmental conditions and drugs to which these strains were subjected to in the clinical setting. The results also revealed that the two inhibitors were able to reduce real time efflux activity in the majority of the strains and provided further evidence that active efflux was inhibited by these compounds.
Effects of Antibiotics and Efflux Inhibitors on Efflux Pump Gene Expression
Putative EPGs were selected by searching for genes in the National Center for Biotechnology Information (NCBI) database (Wang et al., 2012, 2015; Song et al., 2016), using combinations of the following keywords: “efflux pump,” “R. anatipestifer,” “MFS,” “RND,” “SMR,” “MATE,” and “ABC.” In addition, a review of previous articles led to the selection of 15 putative genes for further analysis in the present study (Table S4). To validate our previous findings, we analyzed the contribution of EPGs to antibiotic resistance in the selected strains. The quantification of the EP mRNA levels of the R. anatipestifer clinical isolates is shown in Figures 1–4.
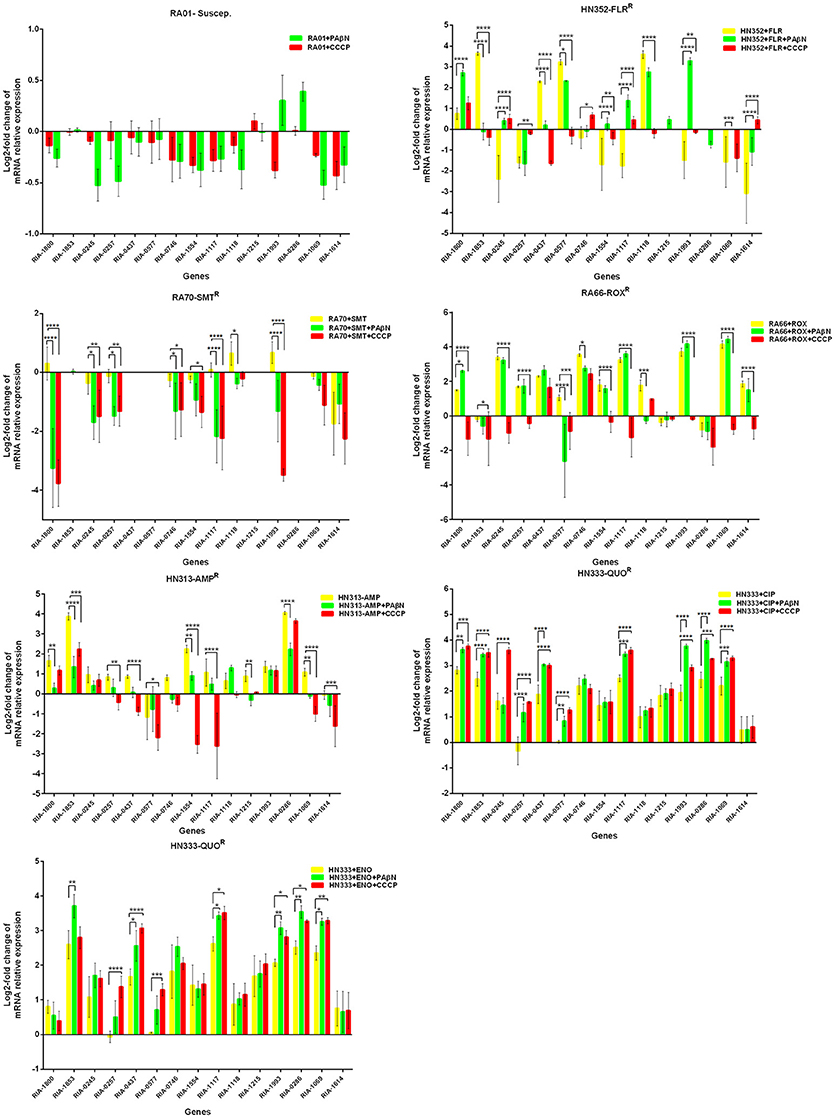
Figure 1. Quantification of the relative mRNA expression levels of a panel of EPGs in mono-resistant strains. Strains were grown in the presence of half MIC of each antibiotic; CCCP (5 μg/mL) and PAβN (40 μg/mL), showed no effects on the growth of R. anatipestifer isolates. The MICs of the antibiotics and EPIs were determined for each strain and are presented in Table 4. Change in the level of transcription of EPGs was measured as fold change normalized to recA gene expression and was subsequently calculated as log2-fold change that is relative to the untreated cell culture. Levels of significance were set as follows: *P < 0.05 was considered statistically significant and **P < 0.01, ***P < 0.001, and ****P < 0.0001 were all considered highly significant.
With regard to the mono-resistant strains, when RA70 was exposed to SMT, a relatively small increase in the mRNA levels was observed for all the genes under investigation. When PAβN or CCCP was added, a significant reduction in the expression of four genes was evident, namely RIA-1800, RIA-1117, and RIA-1993. When the florfenicol-resistant strain, HN352 was exposed to FLR, among all the genes under investigation, four were observed to be overexpressed, namely RIA-1853, RIA-0437, RIA-0577, and RIA-1118. The addition of efflux inhibitors highlighted the contribution of different classes of EPs to different efflux levels in the various isolates used in the study. The presence of CCCP led to significantly reduced efflux in HN352 (with the exception of RIA-0245, RIA-1117, and RIA-1614). However, the addition of PAβN caused the greatest increase in the expression of RIA-1800, RIA-1117, and RIA-1993. Generally, the increase observed was significantly greater in the presence of CCCP (Figure 1).
In the case of RA66 exposed to ROX, the addition of PAβN and ROX highlighted the contribution of different classes of EPs to almost similar efflux levels that were observed in the HN352 isolates. In comparison to PAβN, CCCP caused a significant reduction in the efflux of RA66, in the following genes: RIA-1800, RIA-0245, RIA-0257, RIA-0577, RIA-1554, RIA-1117, RIA-1993, RIA-1069, and RIA-1614. In the HN313 strain exposed to AMP, five genes were found to be overexpressed, namely RIA-1800, RIA-1853, RIA-1554, RIA-1117, RIA-0286, and RIA-1069. The addition of CCCP caused a significant reduction in the expression of RIA-1554, RIA-1117, and RIA-1069. However, the addition of PAβN also caused a significant reduction in the expression of RIA-1800, RIA-1853, and RIA-0286. The QUO-resistant strain HN333, when exposed to CIP, showed significant overexpression of all the genes under investigation, with the exception of RIA-0257, RIA-0577, and RIA-1614. When exposed to enrofloxacin, HN333 showed significant overexpression of all the genes tested, with the exception of RIA-1800, RIA-0257, RIA-0577, and RIA-1614. In contrast, the addition of PAβN and CCCP caused a significant increase in the expression of some EPGs, such as RIA-1853, RIA-0437, RIA-1117, RIA-1993, RIA-0286, and RIA-1069 (Figure 1). The multidrug-resistant FS8 strains were exposed to SMT, ROX, and CIP, respectively, and similar patterns were observed among the mono-resistant strains. Similarly, the multidrug-resistant WF7 strains were exposed to FLR or CIP, and similar patterns were observed among the mono-resistant strains. Three out of 15 genes were overexpressed upon exposure to OXT; RIA-1069, particularly, showed the highest level of expression in WF7 strains. The addition of PAβN and CCCP to the multidrug-resistant FS8 and WF7 strains showed similar effects (Figure 2). The extensively drug-resistant GD01 (Figure 3) and SD314 (Figure 4) strains that were exposed to various antibiotics showed different efflux levels. Overall, RIA-1800, RIA-1118, and RIA-1993 were overexpressed in RA70 and FS8, independently of the SMT to which they were exposed. The RIA-0437 gene was overexpressed in HN352, WF7, GD01, and SD314 strains, independently of the amphenicols to which it was exposed. The RIA-0245, RIA-0746, RIA-1117, RIA-1993, and RIA-1069 genes were overexpressed in the RA66, FS8, and SD314 strains, independently of the ROX to which they were exposed. In addition, the RIA-1853, RIA-1554, and RIA-0286 genes were overexpressed in the HN313, GD01, and SD314 strains, independently of the AMP to which they were exposed. In contrast, RIA-0257, RIA-0577, and RIA-1614 showed the lowest levels of expression in ciprofloxacin-resistant strains and in response to ciprofloxacin antibiotics. When the extensively drug-resistant strains GD01 and SD314 were exposed to AMK, nine EPGs were found to be overexpressed. This suggested that the EPs of clinical isolates might be expressed at different levels in the presence of different antibiotics. To summarize, the addition of PAβN with various antibiotics affected the expression levels of EPs in a way similar to that observed with CCCP. Nevertheless, with the exception of QUO and AMP antibiotics, the reduction observed in expression following the addition of PAβN was not as high as that observed with CCCP.
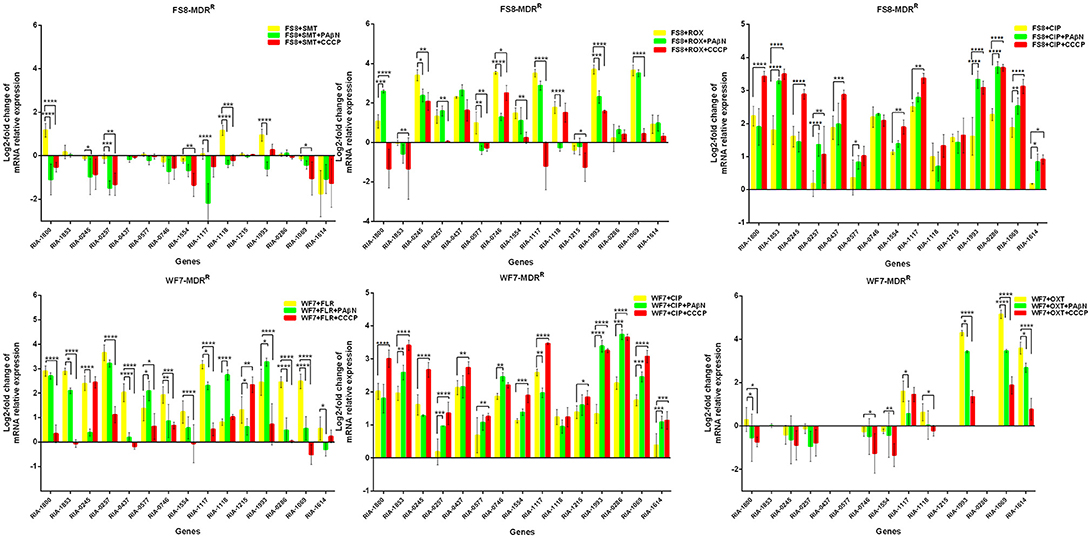
Figure 2. Quantification of the relative mRNA expression levels of a panel of EPGs in multidrug-resistant strains. Strains were grown in the presence of half MIC of each antibiotic; CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. The MICs of the antibiotics and EPIs were determined for each strain and are presented in Table 4. Change in the level of transcription of EPGs was measured as fold change normalized to recA gene expression and was subsequently calculated as log2-fold change that is relative to the untreated cell culture. Levels of significance were set as follows: *P < 0.05 was considered statistically significant, and **P < 0.01, ***P < 0.001, and ****P < 0.0001 were all considered highly significant.
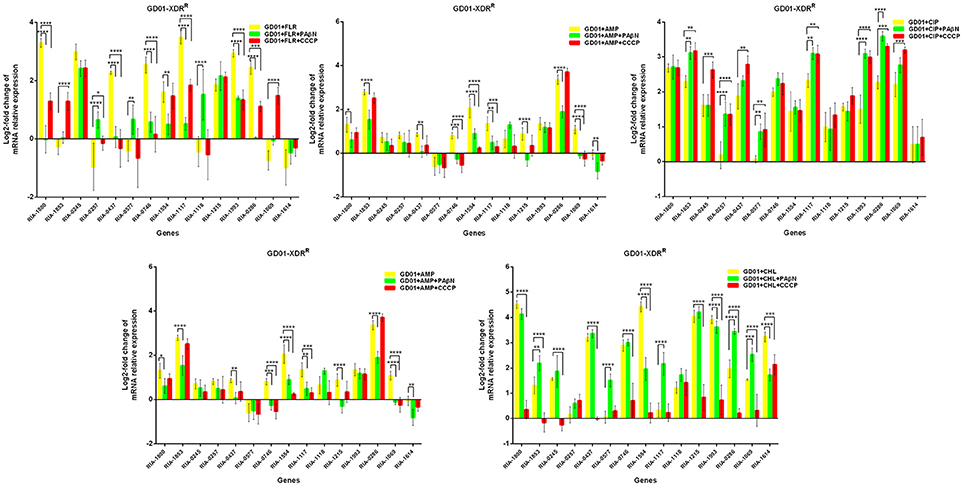
Figure 3. Quantification of the relative mRNA expression levels of a panel of EPGs in GD01 strains. Strains were grown in the presence of half MIC of each antibiotic; CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. The MICs of the antibiotics and EPIs were determined for each strain and are presented in Table 4. Change in the level of transcription of EPGs was measured as fold change normalized to recA gene expression and was subsequently calculated as log2-fold change that is relative to the untreated cell culture. Levels of significance were set as follows: *P < 0.05 was considered statistically significant, and **P < 0.01, ***P < 0.001, and ****P < 0.0001, were all considered highly significant.
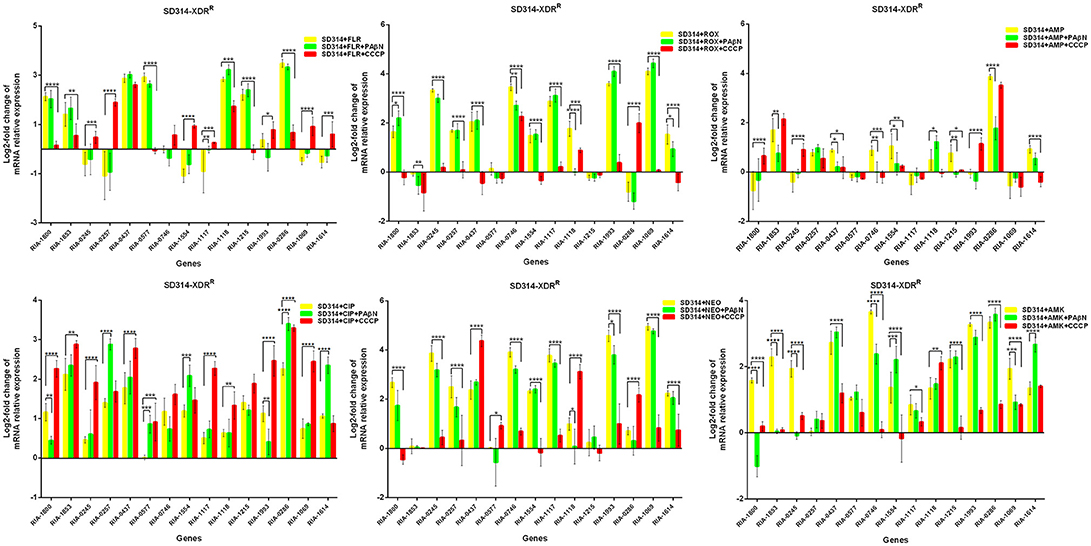
Figure 4. Quantification of the relative mRNA expression levels of a panel of EPGs in SD314 strains. Strains were grown in the presence of half MIC of each antibiotic; CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. The MICs of the antibiotics and EPIs were determined for each strain and are presented in Table 4. Change in the level of transcription of EPGs was measured as fold change normalized to recA gene expression and was subsequently calculated as log2-fold change that is relative to the untreated cell culture. Levels of significance were set as follows: *P < 0.05 was considered statistically significant, and **P < 0.01, ***P < 0.001, and ****P < 0.0001 were all considered highly significant.
Evaluation of Growth Kinetics of the Synergistic Effect Between Efflux Inhibitors and Antibiotics
According to previous reports, the combination of an antibiotic and an efflux inhibitor at subminimal inhibitory concentrations delays the growth of M. tuberculosis drug-resistant strains (Coelho et al., 2015; Machado et al., 2017). Therefore, in R. anatipestifer drug-resistant strains, the EP is also believed to contribute to the overall level of antibiotic resistance described above. Furthermore, the delay in the growth of each drug-resistant strain, due to the stress imposed by the combination of an antibiotic and an efflux inhibitor at subminimal inhibitory concentrations, will render them more susceptible to the effects of the antibiotic. To test this hypothesis, we conducted drug susceptibility tests for the selected R. anatipestifer strains.
First, the optimal concentrations of PAβN and CCCP at which the growth of bacteria was not influenced were selected. The results showed that the R. anatipestifer in the tube containing the antibiotic and EPI grew more slowly than only in the presence of the antibiotic during the evaluation period of 12 h. As evident from the growth curves (Figures 5, 6), with the exception of amphenicols, tetracyclines, and aminoglycosides, all the antimicrobials showed no significant differences in the delay of growth curves between CCCP and PAβN (P > 0.05). In comparison with SMT only, the combination of sulfonamides with efflux inhibitors yielded a delay in the growth rates of RA70 and FS8 strains. Similarly, the delay in growth rates in the presence of ciprofloxacin or enrofloxacin indicated a potentiating effect of efflux inhibitors on the activity of the antibiotics. Notably, CCCP and PAβN showed similar effects on the quinolone-resistant isolates. The combination of ROX and an efflux inhibitor induced a delay in the growth of the strains. The effects of CCCP were relatively superior to those of PAβN. The combination of ampicillin and an efflux inhibitor also caused a delay in the growth of the strains; however, PAβN showed superior effects when compared with CCCP.
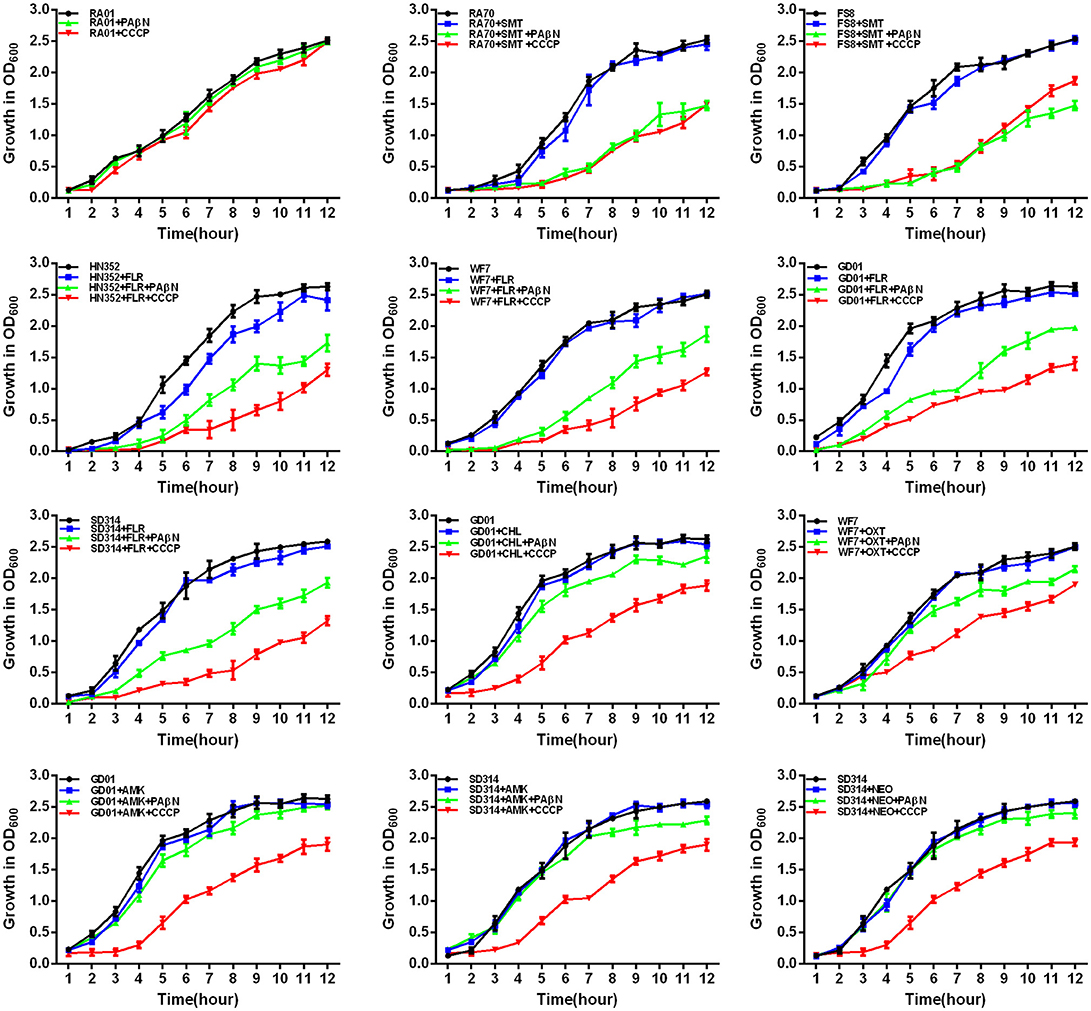
Figure 5. Quantitative drug susceptibility testing of the antibiotics for the R. anatipestifer strains, in the presence or absence of each EPI. CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. Strains were grown in the presence of half MIC of each antibiotic as follows: RA70 (32 μg/mL) and FS8 (32 μg/mL) were exposed to SMT, HN352 (8 μg/mL), WF7 (4 μg/mL), GD01 (4 μg/mL), and SD314 (1 μg/mL) were exposed to FLR; GD01 (16 μg/mL) was exposed to CHL; WF7 (4 μg/mL) was exposed to OXT; GD01(16 μg/mL) and SD314 (16 μg/mL) were exposed to AMK; SD314 (32 μg/mL) was exposed to NEO. The figure showed that the R. anatipestifer strains grew slowly (between 1 and 12 h) because of the synergistic effect of the antibiotics and EPIs.
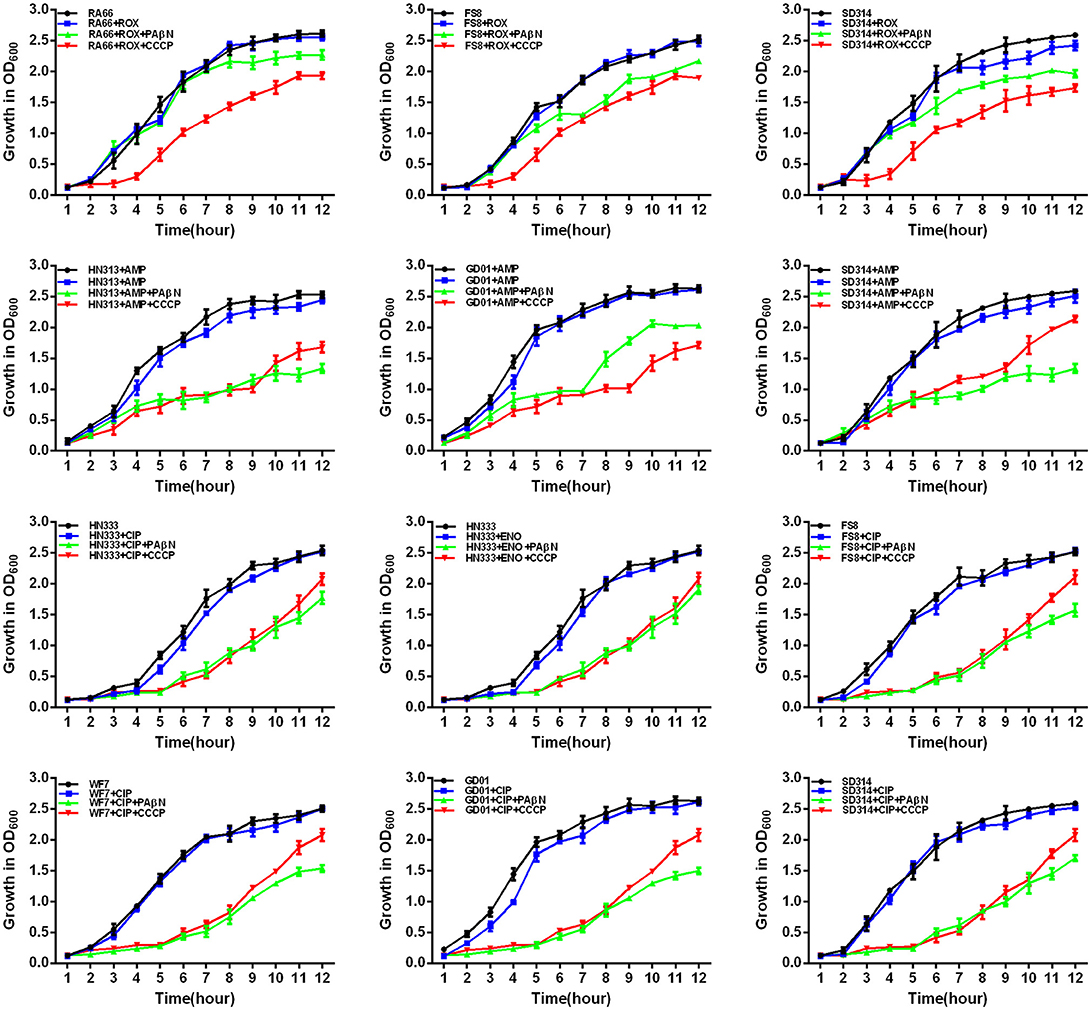
Figure 6. Quantitative drug susceptibility testing of antibiotics for the R. anatipestifer strains, in the presence or absence of each EPI. CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. Strains were grown in the presence of half MIC of each antibiotic as follows: RA66 (16 μg/mL), FS8 (8 μg/mL), and SD314 (4 μg/mL) were exposed to ROX; HN313 (8 μg/mL), GD01 (8 μg/mL), and SD314 (4 μg/mL) were exposed to AMP; HN333 (4 μg/mL) was exposed to ENO; HN333 (4 μg/mL), FS8 (2 μg/mL), WF7 (2 μg/mL), GD01 (2 μg/mL), and SD314 (2 μg/mL) were exposed to CIP. The figure showed that the R. anatipestifer strains grew slowly (between 1 and 12 h) because of the synergistic effect of the antibiotics and EPIs.
Notably, aminoglycosides plus PAβN showed no effect on the growth of the strains, whereas when in combination with CCCP, a significant reduction in the growth of the strains was evident (P < 0.05). These results showed that the antibiotic activity was definitely enhanced in the presence of an efflux inhibitor, as demonstrated by the delay in the growth of the strains.
Discussion
Antimicrobials target several essential cellular functions in bacteria, including the biosynthetic pathways of the cell wall, proteins, and nucleic acids, thereby, producing inhibitory and even lethal effects on bacterial survival (Li, X. Z. et al., 2016). In contrast, microorganisms possess remarkable capacities to counteract the action of antimicrobial agents, thereby, conferring resistance. The mechanisms of resistance mainly entail the production of drug inactivating enzymes, alteration of drug targets, mobile resistant genetic elements, and prevention of drug access; this last mechanism refers to the functions of drug efflux and influx (Du Toit, 2017). Efflux is a key mechanism of cellular responses to varied circumstances (Chitsaz and Brown, 2017) and plays a main role in a range of bacterial behaviors, including biofilm formation, quorum sensing, pathogenicity, and virulence. This mechanism allows bacterial cells to regulate their inner environments through the efflux of toxic substances, such as antimicrobial agents and metabolic products.
In China, unfortunately, many domestic ducks frequently share an environment with wild waterfowl, livestock, and humans of the same district. Furthermore, the widespread use of antibiotics to treat related infections has resulted in MDR in R. anatipestifer. According to the above theory, it is useful to understand the mechanism by which R. anatipestifer develops resistance and how this can improve treatment (Wang Y. et al., 2017).
The development of drug resistance in R. anatipestifer has long been correlated with the evolution of resistance genes that code for the drug targets. Several studies have been based on the simple assessment and evaluation of the levels of expression of R. anatipestifer EPGs (Li Y. F. et al., 2016; Zhang et al., 2017) and few have determined the effects of efflux inhibitors on the MICs of the antibiotics that are generally used to treat R. anatipestifer. Therefore, in the present work, our first aim was to determine whether the combination of antibiotics and an EPI acts synergistically against a set of drug-resistant R. anatipestifer strains. The second aim was to reveal the contribution of EP systems to overall drug resistance in these strains.
Based on our clinical investigations, 44 R. anatipestifer strains were found to be resistant (data not shown). We selected a set of R. anatipestifer strains with different phenotypes and genotypes to study the relationship between the effects of antibiotics and putative EPs that act synergistically. When compared with a few previous reports about the resistance mechanisms of R. anatipestifer, the present study focused on the distribution of resistance genes, integrons, and QRDR mutations in R. anatipestifer isolates, as shown in Table 3.
Notably, the present study reported that PMQR genes were detected in HN333 (qnrS), WF7 (qnrD), and SD314 (qnrS) isolates. To the best of our knowledge, this is the first report of PMQR resistance genes in R. anatipestifer. The chloramphenicol and florfenicol resistance genes cat and floR detected in this study were also reported in R. anatipestifer isolates from Taiwan (Chen et al., 2010, 2012). The aminoglycoside resistance genes, blaTEM−1 (the beta-lactamase resistance gene), cmlA (chloramphenicol- and florfenicol-resistance gene), tetracycline and sulfonamide resistance genes were also detected in the R. anatipestifer isolates from South China by Sun et al. (2012). Zhu et al. (2018) detected the tet(A), tet(M), tet(Q), tet(O), tet(B), and tet(O/W/32/O) genes in the R. anatipestifer isolates from different regions of China (Zhu et al., 2018). In our study, only tet(A) and tet(B) genes were detected. These results show that R. anatipestifer has a high diversity of drug resistance genes.
At present, we do not know the exact mechanism by which these compounds inhibit EP activity in R. anatipestifer. In order to correlate the data obtained from the MIC determination and efflux activity, we applied a broth microdilution method and growth kinetics techniques to evaluate the synergistic effect between the efflux inhibitors and antibiotics. R. anatipestifer has been shown to possess various antibiotic resistance mechanisms. It was hypothesized that the strains that yield higher MICs of the antibiotics would display increased levels of efflux, particularly for their MDR phenotype. However, the clinical isolates also possessed multiple antibiotic resistance genes (see Table 3) that presumably dissimulate the effects of efflux on the phenotype, when was gauged by the MIC. The addition of EPIs highlighted the contribution of EPs to efflux levels in the isolates. In the GD01, SD314, and RA66 isolates, the addition of CCCP, an inhibitor that dissipates the proton motive force required by several EPs, caused a remarkable increase in the accumulation of various antibiotics, such as AMK, NEO, and ROX. This effect led to the slow growth of bacteria and reduced MICs (see Table 1, Figures 5, 6). These findings suggested that reduced efflux and increased antibiotic activity could be attributed to the inhibition of active efflux. The addition of PAβN, an inhibitor of efflux transporters mainly of RND transporters, such as E. coli AcrAB-TolC and P. aeruginosa MexAB-OprM (Li, X. Z. et al., 2016, chapter 1), also promoted the reduction of the levels of efflux. This was mainly manifested in the synergistic effect of sulfonamide and quinolone antibiotics in the RA70, multidrug-resistant, and extensively drug-resistant strains that caused the slow growth of bacteria and the reduction in MICs (see Table 4, Figures 5, 6), since PAβN is not a specific inhibitor of RND EPs. However, this reduction was not as great as that seen with CCCP, suggesting that either the RND pumps associated with efflux in R. anatipestifer were limited or CCCP mediated other cellular activities in R. anatipestifer. Furthermore, the efflux inhibitors possibly acted as helper compounds of antibiotic activity in R. anatipestifer treatment. Thus, the antibiotics could concentrate and penetrate the bacteria, thereby, increasing their effective concentrations. The combined use of antibiotics and EPIs in the control and prevention of R. anatipestifer therapy would be beneficial for the development of new therapeutic strategies for R. anatipestifer infection.
For the analysis of putative EP gene expression, we first compared the mRNA levels of the R. anatipestifer clinical strains with the antibiotic susceptible reference strain (RA01) following exposure to the antibiotics and EPIs. All strains presented an intricate resistance pattern to the antibiotics tested (Tables 1, 2), due to the presence of the resistance gene (Table 3), coupled with a component of efflux, as demonstrated by the reduction in resistance levels with the EPIs (Table 4). No EP gene was overexpressed in the RA01 strain (used as a reference; Figure 1). The EPGs, RIA-1853 and RIA-0286, were overexpressed in the presence of AMP and AMK, demonstrating the contribution of these EPs to the resistance phenotype of the HN313, GD01, and SD314 strains (Figures 1, 3, 4). The addition of CCCP to AMK caused a significant reduction in the expression of RIA-1853 and RIA-0286. Furthermore, the addition of PAβN to AMP also significantly reduced the expression of RIA-1853 and RIA-0286.
The EPGs, RIA-1993, RIA-1069, and RIA-1614, were overexpressed in the presence of OXT in the WF7 strain. Following the addition of CCCP or PAβN to OXT, a significant reduction was observed in the expression of those genes. These EPGs might be associated with aminoglycoside, β-lactam, and tetracycline resistance in particular strains. We noticed a general overexpression of almost all the EPGs in the resistant strains upon exposure to quinolones. These results indicated that the activity of R. anatipestifer EPs was multifunctional and was associated with the outflow of drugs in response to a particular gene.
The RT-qPCR data combined with the real time growth kinetics and the results of MICs showed that the response of the resistant clinical strains were efflux-mediated. Furthermore, we found a linear correlation between EP gene expression and the reduction in antibiotic resistance induced by the EPIs, such as that observed on exposure to aminoglycosides. In addition, we noticed that some genes were upregulated upon exposure to the combination of antibiotics and EPIs (see Figure 1, for example HN333-QUO), this might reflect another mechanism of action of the EPIs in R. anatipestifer. These findings also suggested that the EPs of clinical strains of R. anatipestifer had diverse substrate specificities. The results suggested that the RND and MFS EPs might play a major role in R. anatipestifer isolates. To explore the details of the mechanisms of EP resistance and to test these hypotheses, further experiments would be necessary.
In summary, to our knowledge, this is the first report to investigate the expression levels of EPGs. The rationale and procedures employed have proved to be useful in evaluating the widespread existence of active efflux systems in drug-resistant R. anatipestifer strains. This study supported the concept that intrinsic efflux activity also contributes to overall resistance in drug-resistant clinical strains of R. anatipestifer. Our results also showed that EPIs, such as PAβN and CCCP, inhibited efflux activity to enhance the clinical effects of antibiotics. It is noteworthy that during the process of clinical treatment, antibiotics were mixed with various antibacterial effects, or the effects of antibiotics and EPIs were superimposed on each other. This study has demonstrated the utility of EPIs in the screening for novel therapeutic options to combat drug resistance in R. anatipestifer and has shown that the combination of antibiotics and EPIs should be further investigated for effective clinical applications.
Author Contributions
QC and FZ conceived and designed the study. QC, GJ, SL, and XG performed the experiments. QC, XG, FZ, LS, SS, and YL analyzed the date. QC wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31602087, 31572532, and 31602051), Youth Science and Technology Fund of Gansu Province (1606RJYA281), Science and Technology Plan Project of Gansu Province (17YF1WA170) and Bacterial Disease in Grazing animal Team.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02136/full#supplementary-material
Figure S1. Assessment of the effect of EPIs on the accumulation of EB on the Riemerella anatipestifer strains used in the study. Fluorometric EB efflux assay. Ethidium bromide was used at the equilibrium concentration for each strain as follows: 0.3 μg/mL for RA01; 0.5 μg/mL for RA70, HN352, RA66, and HN313 strains. Each EPI was tested at half MIC; CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. Dotted line means strains assayed with 5% calf serum + PBS; red line means strains assayed with PBS; green line means strains assayed with 5% calf serum + PBS + PAβN; yellow line means strains assayed with 5% calf serum + PBS + CCCP.
Figure S2. Assessment of the effect of EPIs on the accumulation of EB on the Riemerella anatipestifer strains used in the study. Fluorometric EB efflux assay. Ethidium bromide was used at the equilibrium concentration for each strain as follows: 0.6 μg/mL for the HN333 strain; and 1.0 μg/mL for FS8, WF7, GD01, and SD314 strains. Each EPI was tested at half MIC; CCCP (5 μg/mL) and PAβN (40 μg/mL) showed no effects on the growth of R. anatipestifer isolates. Dotted line means strains assayed with 5% calf serum + PBS; red line means strains assayed with PBS; green line means strains assayed with 5% calf serum + PBS + PAβN; yellow line means strains assayed with 5% calf serum + PBS + CCCP.
Table S1. Minimal iinhibitory concentration interpretive standards for R. anatipestifer strains studied.
Table S2. PCR primers used in this study.
Table S3. Table S3 RT-qPCR primers used in this study.
Table S4. Putative efflux transporters described in Riemerella anatipestifer and their substrates.
References
Amaral, L., Cerca, P., Spengler, G., Machado, L., Martins, A., Couto, I., et al. (2011). Ethidium bromide efflux by Salmonella: modulation by metabolic energy, pH, ions and phenothiazines. Int. J. Antimicrob. Agents 38, 140–145. doi: 10.1016/j.ijantimicag.2011.03.014
Chang, C. F., Lin, W. H., Yeh, T. M., Chiang, T. S., and Chang, Y. F. (2003). Antimicrobial susceptibility of Riemerella anatipestifer isolated from ducks and the efficacy of ceftiofur treatment. J. Vet. Diagn. Invest. 15, 26–29. doi: 10.1177/104063870301500106
Chen, Y. P., Lee, S. H., Chou, C. H., and Tsai, H. J. (2012). Detection of florfenicol resistance genes in Riemerella anatipestifer isolated from ducks and geese. Vet. Microbiol. 154, 325–331. doi: 10.1016/j.vetmic.2011.07.012
Chen, Y. P., Tsao, M. Y., Lee, S. H., Chou, C. H., and Tsai, H. J. (2010). Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol. 39, 333–338. doi: 10.1080/03079457.2010.507761
Chevalier, J., Bredin, J., Mahamoud, A., Malléa, M., Barbe, J., and Pagès, J. M. (2004). Inhibitors of antibiotic efflux in resistant Enterobacter aerogenes and Klebsiella pneumoniae strains. Antimicrob Agents Chemother. 48, 1043–1046. doi: 10.1128/AAC.48.3.1043-1046.2004
Chikuba, T., Uehara, H., Fumikura, S., Takahashi, K., Suzuki, Y., Hoshinoo, K., et al. (2016). Riemerella anatipestifer infection in domestic ducks in Japan 2014. J. Vet. Med. Sci. 78, 1635–1638. doi: 10.1292/jvms.16-0278
Chitsaz, M., and Brown, M. H. (2017). The role played by drug efflux pumps in bacterial multidrug resistance. Essays Biochem. 61, 127–139. doi: 10.1042/EBC20160064
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement. CLSI document M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute.
Coelho, T., Machado, D., Couto, I., Maschmann, R., Ramos, D., Von Groll, A., et al. (2015). Enhancement of antibiotic activity by efflux inhibitors against multidrug resistant Mycobacterium tuberculosis clinical isolates from Brazil. Front. Microbiol. 6:330. doi: 10.3389/fmicb.2015.00330
Du Toit, A. (2017). Bacterial physiology: efflux pumps, fitness and virulence. Nat. Rev. Microbiol. 15, 512–513. doi: 10.1038/nrmicro.2017.97
Guo, Y., Hu, D., Guo, J., Wang, T., Xiao, Y., Wang, X., et al. (2017). Riemerella anatipestifer type IX secretion system is required for virulence and gelatinase secretion. Front. Microbiol. 8:2553. doi: 10.3389/fmicb.2017.02553
Gyuris, É., Wehmann, E., Czeibert, K., and Magyar, T. (2017). Antimicrobial susceptibility of Riemerella anatipestifer strains isolated from geese and ducks in Hungary. Acta Vet. Hung. 65, 153–165. doi: 10.1556/004.2017.016
Huang, L., Yuan, H., Liu, M. F., Zhao, X. X., Wang, M. S., Jia, R. Y., et al. (2017). Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8:297. doi: 10.3389/fmicb.2017.00297
Jaillard, M., Van Belkum, A., Cady, K. C., Creely, D., Shortridge, D., Blanc, B., et al. (2017). Correlation between phenotypic antibiotic susceptibility and the resistome in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 50, 210–218. doi: 10.1016/j.ijantimicag.2017.02.026
Ji, G., Chen, Q. W., Gong, X. W., Liu, Y. S., and Zheng, F. Y. (2017). Determination of the optimal testing conditions for the MICs of common antibiotics against Riemerella anatipestifer. Heilongjiang Anim. Sci. Vet. Med. 5, 32–36, 40 (in Chinese). doi: 10.13881/j.cnki.hljxmsy.2017.0388
Kurincic, M., Klancnik, A., and Smole Mozina, S. (2012). Effects of efflux pump inhibitors on erythromycin, ciprofloxacin, and tetracycline resistance in Campylobacter spp. isolates. Microb. Drug Resist. 18, 492–501. doi: 10.1089/mdr.2012.0017
Lamers, R. P., Cavallari, J. F., and Burrows, L. L. (2013). The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAbetaN) permeabilizes the outer membrane of gram-negative bacteria. PLoS ONE 8:e60666. doi: 10.1371/journal.pone.0060666
Li, S., Gong, X., Chen, Q., Zheng, F., Ji, G., and Liu, Y. (2018). Threshold level of Riemerella anatipestifer crossing blood-brain barrier and expression profiles of immune-related proteins in blood and brain tissue from infected ducks. Vet. Immunol. Immunopathol. 200, 26–31. doi: 10.1016/j.vetimm.2018.04.005
Li, X. Z., Elkins, C. A., and Zgurskaya, H. I. (2016). Efflux-Mediated Antimicrobial Resistance in Bacteria. Springer International Publishing. doi: 10.1007/978-3-319-39658-3
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Li, Y., Yang, L., Fu, J., Yan, M., Chen, D., and Zhang, L. (2018). Genotyping and high flux sequencing of the bacterial pathogenic elements - integrons. Microb. Pathog. 116, 22–25. doi: 10.1016/j.micpath.2017.12.073
Li, Y., Zhang, Y., Ding, H., Mei, X., Liu, W., Zeng, J., et al. (2016). In vitro susceptibility of four antimicrobials against Riemerella anatipestifer isolates: a comparison of minimum inhibitory concentrations and mutant prevention concentrations for ceftiofur, cefquinome, florfenicol, and tilmicosin. BMC Vet. Res. 12:250. doi: 10.1186/s12917-016-0796-3
Li, Y. F., Jiang, H.-X., Xiang, R., Sun, N., Zhang, Y.-N., Zhao, L.-Q., et al. (2016). Effects of two efflux pump inhibitors on the drug susceptibility of Riemerella anatipestifer isolates from China. J. Integr. Agric. 15, 929–933. doi: 10.1016/S2095-3119(15)61031-0
Liu, J., Guo, L., Liu, J., Zhang, J., Zeng, H., Ning, Y., et al. (2017). Identification of an efflux transporter lmrb regulating stress response and extracellular polysaccharide synthesis in Streptococcus mutans. Front. Microbiol. 8:962. doi: 10.3389/fmicb.2017.00962
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Machado, D., Coelho, T. S., Perdigão, J., Pereira, C., Couto, I., Portugal, I., et al. (2017). Interplay between mutations and efflux in drug resistant clinical isolates of Mycobacterium tuberculosis. Front. Microbiol. 8:711. doi: 10.3389/fmicb.2017.00711
Opperman, T. J., Kwasny, S. M., Kim, H. S., Nguyen, S. T., Houseweart, C., D'souza, S., et al. (2014). Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 58, 722–733. doi: 10.1128/AAC.01866-13
Richmond, G. E., Chua, K. L., and Piddock, L. J. (2013). Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzamide). J. Antimicrob. Chemother. 68, 1594–1600. doi: 10.1093/jac/dkt052
Song, X. H., Zhou, W. S., Wang, J. B., Liu, M. F., Wang, M. S., Cheng, A. C., et al. (2016). Genome sequence of Riemerella anatipestifer strain RCAD0122, a multidrug-resistant isolate from ducks. Genome Announc. 4:e00332-16. doi: 10.1128/genomeA.00332-16
Sun, N., Liu, J. H., Yang, F., Lin, D. C., Li, G. H., Chen, Z. L., et al. (2012). Molecular characterization of the antimicrobial resistance of Riemerella anatipestifer isolated from ducks. Vet. Microbiol. 158, 376–383. doi: 10.1016/j.vetmic.2012.03.005
Sutkuviene, S., Mikalayeva, V., Pavan, S., Berti, F., and Daugelavicius, R. (2013). Evaluation of the efficiency of synthesized efflux pump inhibitors on Salmonella enterica ser. typhimurium cells. Chem. Biol. Drug Des. 82, 438–445. doi: 10.1111/cbdd.12173
Uddin, M. J., and Ahn, J. (2017). Associations between resistance phenotype and gene expression in response to serial exposure to oxacillin and ciprofloxacin in Staphylococcus aureus. Lett. Appl. Microbiol. 65, 462–468. doi: 10.1111/lam.12808
van Essen-Zandbergen, A., Smith, H., Veldman, K., and Mevius, D. (2007). Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J. Antimicrob. Chemother. 59, 746–750. doi: 10.1093/jac/dkl549
Viveiros, M., Rodrigues, L., Martins, M., Couto, I., Spengler, G., Martins, A., et al. (2010). Evaluation of efflux activity of bacteria by a semi-automated fluorometric system. Methods Mol. Biol. 642, 159–172. doi: 10.1007/978-1-60327-279-7_12
Wang, Q., Chen, M., and Zhang, W. (2017). A two-component signal transduction system contributes to the virulence of Riemerella anatipestifer. J. Vet. Sci. 19, 260–270. doi: 10.4142/jvs.2018.19.2.260
Wang, X., Ding, C., Wang, S., Han, X., and Yu, S. (2015). Whole-genome sequence analysis and genome-wide virulence gene identification of Riemerella anatipestifer strain Yb2. Appl. Environ. Microbiol. 81, 5093–5102. doi: 10.1128/AEM.00828-15
Wang, X., Zhu, D., Wang, M., Cheng, A., Jia, R., Zhou, Y., et al. (2012). Complete genome sequence of Riemerella anatipestifer reference strain. J. Bacteriol. 194, 3270–3271. doi: 10.1128/JB.00366-12
Wang, Y., Lu, T., Yin, X., Zhou, Z., Li, S., Liu, M., et al. (2017). A Novel RAYM_RS09735/RAYM_RS09740 two-component signaling system regulates gene expression and virulence in Riemerella anatipestifer. Front. Microbiol. 8:688. doi: 10.3389/fmicb.2017.00688
Xing, L., Yu, H., Qi, J., Jiang, P., Sun, B., Cui, J., et al. (2015). ErmF and ereD are responsible for erythromycin resistance in Riemerellaanatipestifer. PLoS ONE 10:e0131078. doi: 10.1371/journal.pone.0131078
Zhai, Z., Li, X., Xiao, X., Yu, J., Chen, M., Yu, Y., et al. (2013). Immunoproteomics selection of cross-protective vaccine candidates from Riemerella anatipestifer serotypes 1 and 2. Vet. Microbiol. 162, 850–857. doi: 10.1016/j.vetmic.2012.11.002
Zhang, X., Wang, M. S., Liu, M. F., Zhu, D. K., Biville, F., Jia, R. Y., et al. (2017). Contribution of RaeB, a putative RND-type transporter to aminoglycoside and detergent resistance in Riemerella anatipestifer. Front. Microbiol. 8:2435. doi: 10.3389/fmicb.2017.02435
Zheng, F., Lin, G., Zhou, J., Cao, X., Gong, X., Wang, G., et al. (2012). Discovery and characterization of gene cassettes-containing integrons in clinical strains of Riemerella anatipestifer. Vet. Microbiol. 156, 434–438. doi: 10.1016/j.vetmic.2011.11.002
Zhong, C. Y., Cheng, A. C., Wang, M. S., Zhu, D. K., Luo, Q. H., Zhong, C. D., et al. (2009). Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53, 601–607. doi: 10.1637/8552-120408-ResNote.1
Keywords: Riemerella anatipestifer, efflux pump, efflux pump inhibitors, synergism, drug resistance
Citation: Chen Q, Gong X, Zheng F, Ji G, Li S, Stipkovits L, Szathmary S and Liu Y (2018) Interplay Between the Phenotype and Genotype, and Efflux Pumps in Drug-Resistant Strains of Riemerella anatipestifer. Front. Microbiol. 9:2136. doi: 10.3389/fmicb.2018.02136
Received: 04 April 2018; Accepted: 21 August 2018;
Published: 01 October 2018.
Edited by:
Henrietta Venter, University of South Australia, AustraliaReviewed by:
Daniel Pletzer, University of British Columbia, CanadaMiguel Viveiros, Universidade NOVA de Lisboa, Portugal
Copyright © 2018 Chen, Gong, Zheng, Ji, Li, Stipkovits, Szathmary and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuying Zheng, emhlbmdmdXlpbmdAY2Fhcy5jbg==
Yongsheng Liu, bGl1eW9uZ3NoZW5nQGNhYXMuY24=
 Qiwei Chen
Qiwei Chen Xiaowei Gong1
Xiaowei Gong1 Susan Szathmary
Susan Szathmary Yongsheng Liu
Yongsheng Liu