- 1DAICENTRE (DBT-AIST International Centre for Translational and Environmental Research) and Bioinformatics Centre, Department of Microbiology, School of Life Sciences, Sikkim University, Gangtok, India
- 2National Centre for Microbial Resource, National Centre for Cell Science, Pune, India
Marcha, thiat, dawdim, hamei, humao, khekhrii, chowan, and phut are traditionally prepared dried starters used for production of various ethnic alcoholic beverages in North East states of India. The surveillance of mycobiome associated with these starters have been revealed by culture-dependent methods using phenotypic and molecular tools. We identified Wickerhamomyces anomalus, Pichia anomala, Saccharomycopsis fibuligera, Pichia terricola, Pichia kudriavzevii, and Candida glabrata by ITS-PCR. The diversity of yeasts and molds in all 40 samples was also investigated by culture-independent method using PCR-DGGE analysis. The average distributions of yeasts showed Saccharomyces cerevisiae (16.5%), Saccharomycopsis fibuligera (15.3%), Wickerhamomyces anomalus (11.3%), S. malanga (11.7%), Kluyveromyces marxianus (5.3%), Meyerozyma sp. (2.7%), Candida glabrata (2.7%), and many strains below 2%. About 12 strains of molds were also identified based on PCR-DGGE analysis which included Aspergillus penicillioides (5.0%), Rhizopus oryzae (3.3%), and sub-phylum: Mucoromycotina (2.1%). Different techniques used in this paper revealed the diversity and differences of mycobiome species in starter cultures of India which may be referred as baseline data for further research.
Introduction
Essence of alcoholic fermentation depends on different types of starters that copulate the uniqueness to organoleptic segmentations for ethnic values (Hesseltine, 1983; Steinkraus, 1996; Tamang et al., 2016b). Yeasts have several economic significances and have been used for centuries in the production of fermented foods and alcoholic beverages (Fleet, 2003; Tamang and Fleet, 2009; Jolly et al., 2017). In Asia, preparation of amylolytic (related to conversion of starch to sugar) and alcoholic (production of alcohol) starter is an innovative back sloping technique of cultivation of native microbiota in the form of dry, flattened, or round balls made up of rice/wheat for production of different traditional alcoholic beverages (Tamang, 2010), locally known as marcha in India, Nepal and Bhutan, benh men in Vietnam, bubod in the Philippines, chiu/chu/daque in China and Taiwan, loogpang in Thailand, ragi in Indonesia, and nuruk in Korea (Tamang, 2016). Traditional methods of preparation of Asian amylolytic dry starters are similar with slight variation in terms of wrapping materials, incubation period, size, and shapes of particular starters. Ethnic people practicing the age-old traditional preservation or sub-culturing amylolytic and alcohol-producing as well as flavor-enhancing fungi and bacteria have attracted many researchers to study the microbial diversity in such starters. In recent years, few researchers have reported the fungal and bacterial species using both culture-dependent and-independent techniques in some common starter cultures of Asia such as marcha of India (Tsuyoshi et al., 2005; Sha et al., 2017), daqua of China (Wang et al., 2008; Zheng et al., 2012; Lv et al., 2013; Chen et al., 2014; Xu et al., 2017), benh men of Vietnam (Dung et al., 2007; Thanh et al., 2008); nuruk of Korea (Jung et al., 2012), and dombea of Cambodia (Ly et al., 2018).
North East regions of India1 have several varieties of traditionally prepared and sun-dried starters prepared by different linguistic ethnic groups of people that include marcha of Sikkim, humao of Assam, hamei of Manipur, chowan of Tripura, thiat of Meghalaya, khekhrii of Nagaland, dowdim of Mizoram, and phut of Arunachal Pradesh (Figure 1). These starter cultures except khekhrii of Nagaland are traditionally prepared from soaked rice with some wild herbs, and then mixed with previously prepared starter powder (1–2%) as an inoculum (back-sloping). The mixtures are ground in a wooden mortal with addition of water to make a thick dough which are kneaded into round to flattened balls/cakes of different size and shape. Dough cakes are covered with fern fronds/paddy straws/jute sags, fermented at room temperature for 1–3 days; and fresh balls/cakes are sun dried for few days (Anupma et al., 2018). Khekhrii of Nagaland is prepared by naturally fermenting sprouted-rice grains and then sun-dried to use as dry starter culture to prepare zutho, local alcoholic beverage. Some species of yeasts Saccharomycopsis fibuligera, S. capsularis, Pichia anomala, P. burtonii, P. guilliermondii, P. fabianii, Trichosporon sp., Candida tropicalis, C. parapsilosis, C. montana, C. glabrata, Torulaspora delbrueckii, Saccharomyces cerevisiae, S. bayanus, and Wickerhamomyces anomalus were previously reported from some samples of marcha and hamei of India (Hesseltine and Kurtzman, 1990; Tamang and Sarkar, 1995; Tsuyoshi et al., 2005; Jeyaram et al., 2008, 2011, Sha et al., 2016, 2017).
One of the common methods for culture-dependent identification is by the analysis of the Internal Transcribed Spacer (ITS)1-5.8S-ITS2 region, which is widely applied in explorations of diversity of fungi associated with many traditional fermented foods (Caggia et al., 2001; Las Heras-Vazquez et al., 2003). ITS analysis may provide the fast and easy means for accurate identification at species level (Esteve-Zarzoso et al., 1999), due to greater sequence variation, the ITSl/ITS2 domains are more suited for species and strain identification than the 18s region (small subunit), the 5.8s region, and the 28s region (large subunit) (Iwen et al., 2002; Korabecna, 2007; Susan Slechta et al., 2012). However, the culture-dependent methods may not detect the whole microbial community in foods (Ercolini, 2004). The culture-independent methods such as PCR denaturing gradient gel electrophoresis (DGGE) analysis, are highly useful to detect the whole microbial communities in food samples (Chen et al., 2014; Puerari et al., 2015; Tamang et al., 2016a). PCR-DGGE analysis method has been designed to profile microbial communities directly from substrates including fermented foods, and is based on sequence-specific distinctions of 16SrRNA and 26SrRNAmplicons (Cocolin et al., 2000; Ercolini, 2004; Ercolini et al., 2004; Alegría et al., 2011).
No studies have been conducted on traditionally prepared starters of India except marcha (Tamang and Sarkar, 1995; Tsuyoshi et al., 2005; Sha et al., 2016, 2017), and hamei (Tamang et al., 2007; Jeyaram et al., 2008, 2011). Based on our preliminary analysis of microbial load in traditionally prepared starters of North East India, fungi mostly yeasts and filamentous molds (>106 cfu/g) predominate over bacteria. Hence, we aimed to study the mycobiome diversity in dried starters of India by culture-dependent and -independent methods to underline the continuous interest in the characterization of microbial consortia associate to poorly studied food fermentations to isolate new potential pro-technological and functional strains, to improve the conservation of microbial diversity, to characterize and limit spoilage microbes, microbial producers of toxic compounds, and pathogens (Capozzi and Spano, 2011; Russo et al., 2016; Tamang et al., 2016a,b; Gonelimali et al., 2018).
Materials and Methods
Sample Collection
Forty different samples of traditionally prepared starter (five samples of each starter) marcha of Sikkim, thiat of Meghalaya, hamei of Manipur, phut of Arunachal Pradesh, chowan of Tripura, dawdim of Mizoram, humao of Assam, and khekhrii of Nagaland were collected immediately after the preparation (fermentation and sun-dried drying) from local people of eight states of North East India, and were transferred to gamma irradiated sterile bottles, sealed, and stored in desiccator at room temperature for the further analysis.
Isolation of Microorganisms
Ten grams of sample was homogenized with 90 ml of 0.85% (w/v) sterile physiological saline in a stomacher lab-blender 400 (Seward, United Kingdom) for 1 min and serially diluted in the same diluents. Yeasts were isolated on yeast-malt extract agar (M424, HiMedia, India) and molds were isolated on potato dextrose agar (M096, HiMedia, India) supplemented with 10 IU ml-1 benzyl penicillin and 12 mg ml-1 streptomycin sulfate, and were incubated aerobically at 28°C for 3 days. Purity of the isolates was checked by streaking again on fresh agar plates of the same isolation medium, followed by microscopic examination. Isolation of yeast strains were typically based on morphotypes and criterion included size, color, shape, and appearance of fully grown culture on growth media. Colonies were counted as colony forming units (cfu)/g sample. Identified strains of yeasts were preserved in 20% glycerol at -20°C (Thapa and Tamang, 2004).
Culture-Dependent Approach for Diversity Analysis
Phenotypic and Biochemical Characterization
A total of 386 yeasts strains were isolated from 40 samples of eight different starters of North East India. Characterizations of yeasts were phenotypically tested on the basis of colony and cell morphology, sugar fermentation, and sugar assimilation tests. Cell morphology of actively growing yeast isolates was determined using a phase-contrast microscope (CH3-BH-PC; Olympus, Tokyo, Japan). Yeast cultures have been characterized on the basis of mycelium type, ascospore type, nitrate reduction, growth at 37 and 45°C, sugar fermentation, and sugar assimilation following the methods of Kurtzman et al. (2011).
Biolog System
Commercial Biolog Identification System (MicroLog TM System Release 4.2 User Guide 2001, Biolog, Inc.) based on the utilization of 95 substrates in 96-welled plate, were used for biochemical characterization of yeast isolates. Aliquots of the cultures were transferred to biolog plate wells and incubated at 37°C for 24–48 h, where positive results were recorded according to color changes. The results obtained were automatically read and analyzed using BiologMicrolog Reader and compared with the database of the Biolog Microlog database software (Biolog Inc.), which provided the most probable genera and species of the tested cultures.
Molecular Identification of Yeast Isolates
Identification of yeast isolates were done by ITS region sequencing wherein DNA extraction, PCR for ITS region, sequencing, and phylogenic affiliations were performed subsequently. Briefly, yeast DNA was extracted using ProMega DNA kit (ProMega). One gram of yeast cell pellet was suspended in lysis solution and incubated at 65°C for 15 min. Subsequently, the RNA was eliminated from the cellular lysate by administering the RNase solution following incubation at 35°C for 15 min. The residual proteins were removed by adding protein precipitation solution and centrifugation at maximum speed. Finally, the DNA was precipitated by adding isopropanol, which was purified with two washes of 70% ethanol. The quality of DNA was checked on 0.8% agarose gel and concentration was measured using Nano-Drop ND-1000 spectrophotometer (Nano Drop Technologies, Wilmington, DE, United States) as described by Banskar et al. (2016). The DNA was stored at -20°C until further processing. For amplification of the ITS region, the forward primer ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and reverse primer ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990) were used and PCR mixture and the thermal cycling protocol conditions were applied as described by (Esteve-Zarzoso et al., 1999). Products were analyzed on 1.5% agarose gel containing 0.7 mg/ml of ethidium bromide and visualized under UV light (UV source Gel-Doc 1000, Bio-Rad). Approximate size of amplicons was determined using standard molecular weight markers (Himedia-100-bp DNA Ladder) (Lv et al., 2013). All PCR-amplified products were purified and sequenced using ABI-DNA-Sequencer (ABI Genetic Analyser 3500, HITACHI, Japan). The sequences were compared with the GenBank database using the BLAST program (Altschul et al., 1990; Zhao and Chu, 2014). Sequences were visualized and edited using Chromas Version 1.452 (Pryce et al., 2003).
Culture-Independent Approach for Diversity Analysis
DNA Extraction, PCR Amplification From Starter Cultures
About 10 g of starters was homogenized in 90 ml of 0.85% w/v sterile physiological saline, and subsequently filtered through four layers of sterile cheese-cloth. The resulting filtered solutions were centrifuged at 14,000 g for 10 min at 4°C (Lv et al., 2013). Then, the pellets were subjected to DNA extraction using the ProMega DNA extraction kit (ProMega, United States) according to the manufacturer’s instructions. Quality of resultant DNA was checked on 0.8% agarose gel and concentration was measured using Nano-Drop ND-1000 spectrophotometer (Nano Drop Technologies, Wilmington, DE, United States) as previously described (Banskar et al., 2016). The 250 nucleotides of the 5′-end D1/D2 region of the 26SrRNA gene was amplified by PCR using the primer NL1 (5′-CGC CCG CGC GCG GGC GGG GCG GGG GCC ATA TCA ATA AGC GGA AAA G-3′) (the GC clamp sequence used is underlined) and a reverse primer LS2 (5′-ATT CCC AAA CAA CTC GAC TC-3′) (Cocolin et al., 2000; El Sheikha et al., 2009). PCR was performed in a final volume of 50 μl containing 10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dATP, dCTP, dGTP, and dTTP, 0.2 mM of the primers, and 1.25 IU Taq-DNA polymerase (Promega, United States) and 2 μl of the extracted DNA (approximately 50 ng) using Thermal Cyclers (Applied Biosystems, United States). The reactions were run for 30 cycles at 95°C for 60 s for denaturation, at 52°C for 45 s for annealing, and at 72°C for 60 s for extension and finally for 7 min at 72°C (Cocolin et al., 2002). The PCR products were analyzed on 2.0% agarose gel containing 0.5 μg/ml ethidium bromide and were visualized in UV source GelDoc (Bio-Rad) (Cocolin et al., 2000). The concentration was again measured using Nano-DropND-1000 spectrophotometer.
PCR-DGGE Fingerprinting and Sequencing of DGGE Eluted Bands
The PCR products were analyzed by DGGE using DCodeTM Universal Mutation Detection System (DGGEK-1001, CBS Scientific, San Diego, CA, United States) following the procedure of El Sheikha et al. (2009). Samples containing approximately equal amounts of PCR products were loaded into 8% w/v polyacrylamide gels (acrylamide:N,N′-methylene bisacrylamide, 37.5:1; Promega) in 1 × TAE buffer (40 mM Tris–HCl, pH 7.4, 20 mM sodium acetate, and 1.0 mM Na2-EDTA). All electrophoresis experiments were performed at 60°C using a denaturing gradient in the range of 30–50% (100% corresponded to 7 M urea and 40% v/v formamide; Promega) (Cocolin et al., 2002). The gels were electrophoresed at 20 V for 10 min and then at 80 V for 12 h (El Sheikha et al., 2009). The gels were stained with SYBR Gold for 30 min (reconstituted according to the manufacturer’s directions; Molecular Probes, Invitrogen, United States) and photographed in UV source GelDoc (Bio-Rad, United States) as described by Grizard et al. (2014). The DGGE bands were excised using sterile micro pipette tips. DNA of each band was eluted in 50 μl sterile water overnight at 4°C and 2 μl of the eluted DNA was reamplified as following the method of Cocolin et al. (2000). The PCR products which yielded only one band in DGGE electrophoresis were amplified with the primers without GC-clamp, purified and finally sequenced with the help of ABI-DNA-Sequencer (ABI Genetic Analyser 3500, HITACHI, Japan). The sequences were compared with the GenBank database using the BLAST program (Altschul et al., 1990; Zhao and Chu, 2014). The DNA sequences obtained from sequencing of total 202 bands was submitted to GeneBank.
Bioinformatics and Statistical Analysis
Quality of raw ITS region from yeast isolates and PCR-DGGE band sequencing data was checked with the help of Sequence Scanner software (Applied Bio systems, United States) and the data alignment and analysis were done with the help of SEQMANN software (DNASTAR, United States). After the data alignment, BLAST program was used for comparing DNA databases for sequence similarities available on the server3 (Altschul et al., 1990; Zhao and Chu, 2014). Construction of a phylogenetic tree by the neighbor-joining method (Saitou and Nei, 1987) was performed using the CLUSTAL W program (Thompson et al., 1994). Shannon index of general diversity (H) and the richness of the microbial community as microbial diversity indices were determined by following the method of Oguntoyinbo et al. (2011). Other graphical emphasis was done on igraph package in R Software (Csardi and Nepusz, 2006).
Nucleotide Accessions
The sequences obtained from ITS region sequencing of isolated 46 yeast strains have been deposited in the GenBank under accessions: KY587119–KY626335 and 26S rRNA gene of 202 bands excised from PCR-DGGE under accessions: KY594045–KY594246.
Results
Culture-Dependent Approach
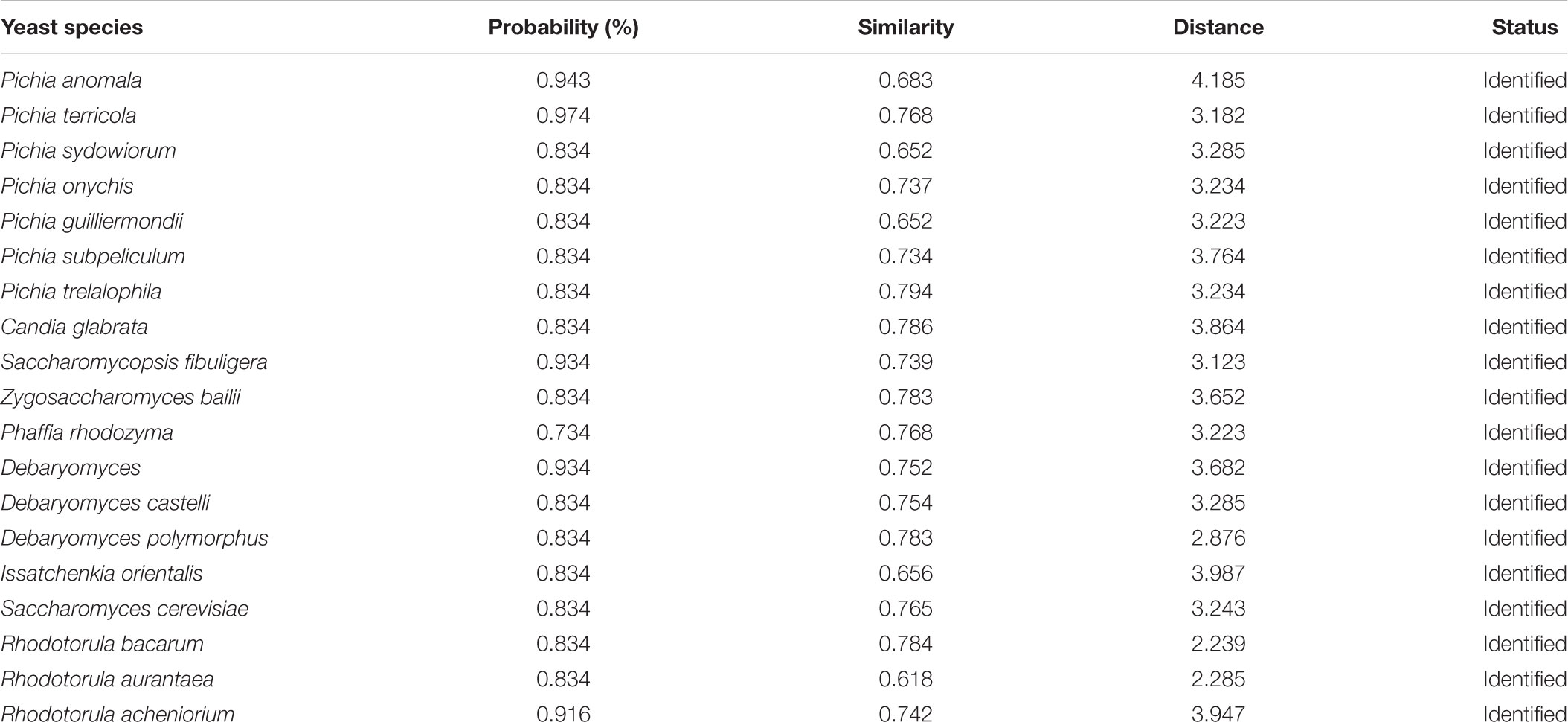
The average populations of yeast in all eight starters was 7.2 × 106 cfu/g (Table 1). Ascertaining the cultured diversity, a total of 386 yeasts strains were isolated from 40 samples and characterized by phenotypic assessment on the basis of colony morphology, cell morphology, sugar fermentation, and sugar assimilation tests (Table 2). Tentatively the following yeast genera were phenotypically identified using the taxonomical keys of Kurtzman et al. (2011) as Saccharomyces, Pichia, Candida, Issatchenkia, Kluyveromyces, Schizosaccharomyces, Saccharomycopsis, and Torulopsis (Table 2). Their metabolic capacities were also assessed by using the Biolog system. By comparing with the yeast database (MicroLog TM System Release 4.2 User Guide 2001, Biolog), the result revealed that maximum identified yeast species were associated with starter having ≥0.75% probability and ≥0.7 similarities index value (Table 3). The yeasts strain Pichia terricola showed highest ≥0.974% probability with ≥0.77 similarities index value. It was observed that the results from Biolog were revealing more diversity of yeasts than phenotypic characterization and it presented in Supplementary Data Sheet S1.

TABLE 2. Grouping of total isolates of yeasts from starters of North East India on the basis of fermentation, and assimilation of sugars and other phenotypic tests.
Out of 386 isolates, 46 representative isolates were grouped based on colony appearance, cell shape, type of mycelia and ascospores, pellicle formation, nitrate reduction, and growth at 37 and 45°C. Precisely, species level identification was done with molecular methods by ITS-region gene sequence analysis. We found that all cultures were identified in six species only as: Wickerhamomyces anomalus, Pichia anomala, Saccharomycopsis fibuligera, Pichia terricola, Pichia kudriavzevii, and Candida glabrata which was reported in Supplementary Data Sheet S2. The average distributions in all starters and molecular phylogenetic relationship with neighbor-joining method were shown in Figure 2. From the sequencing results of ITS region gene; it was observed that species richness (R) was higher in dawdim, hamei, thiat than marcha khekhrii, chowan, and phut (Table 4). Wickerhamomyces anomalus was dominant in all starters. The Shannon index (H) of yeasts isolates was higher in dawdim than other starters (Table 4).
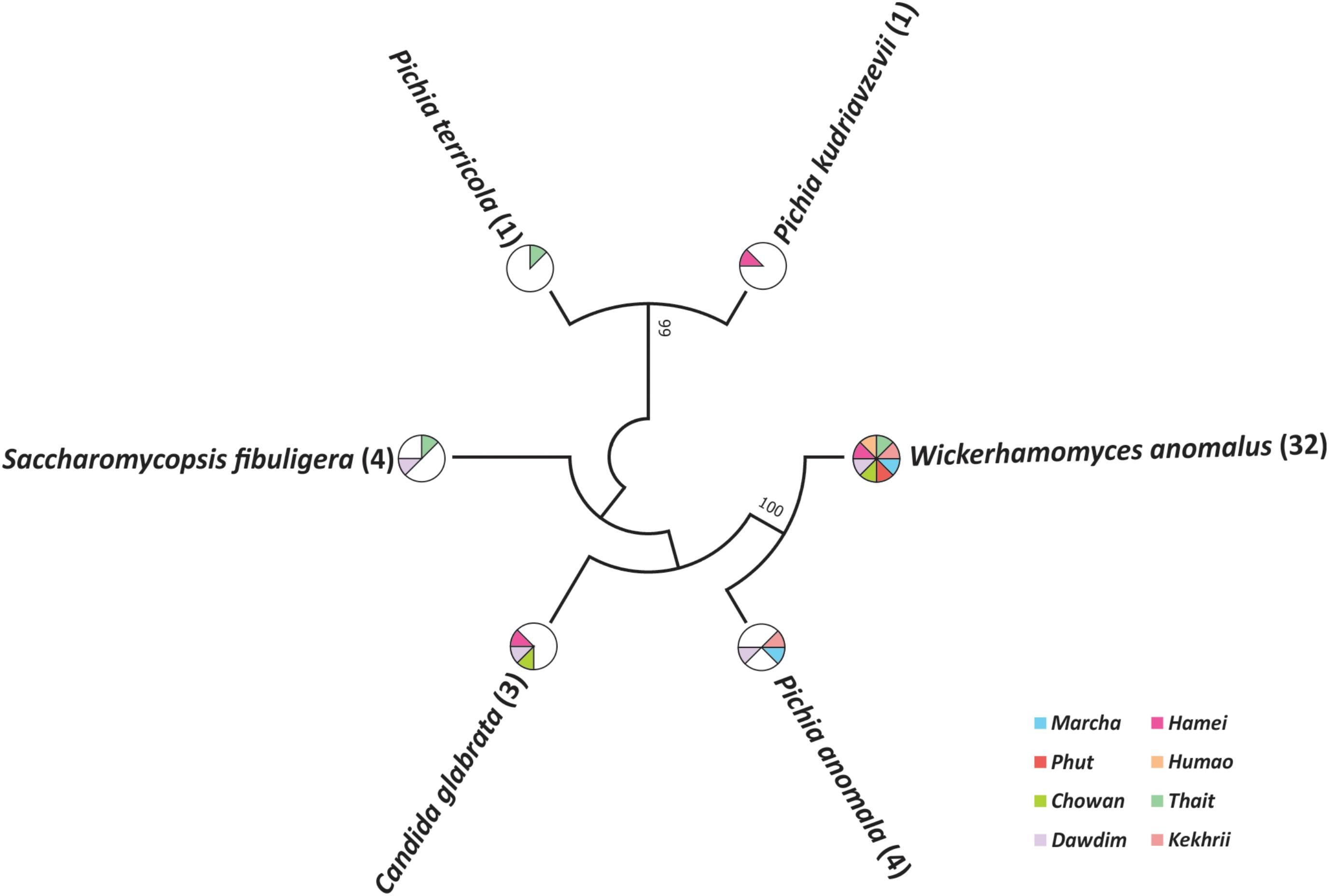
FIGURE 2. Molecular phylogenetic analysis of 46 yeast isolates recovered from a starters based on ITS region sequencing. The bootstrap consensus tree derived with 1000 replicates to neighbor-joining method and Kimura two-parameter model. Numbers on branches depict the percent occurrence of a given branch during 1000 replicates. The origin distribution patterns of these isolates were depicted in subsequent pi-charts.
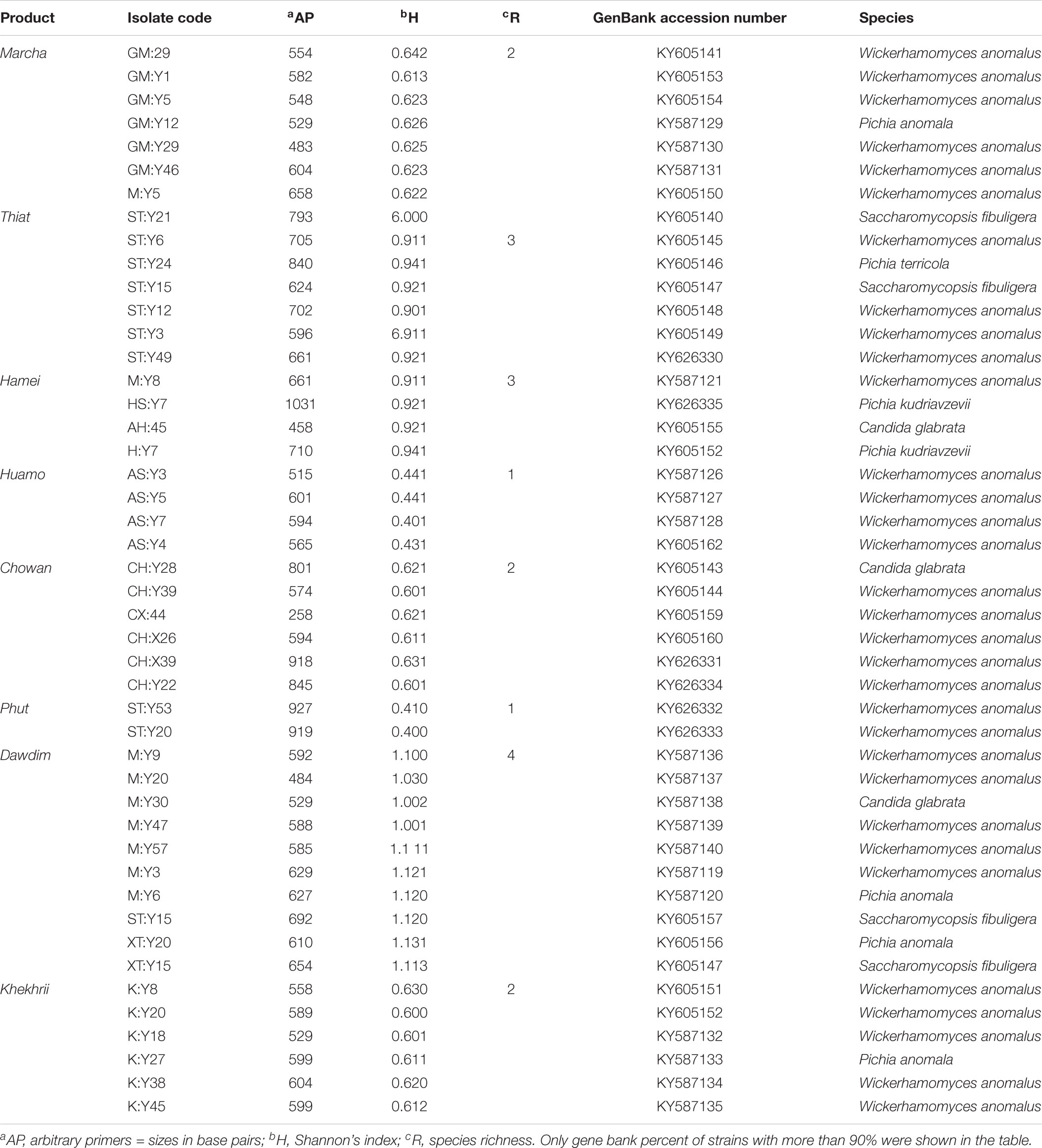
TABLE 4. Molecular characterization and identification results of 46 yeast strains from starters of North East India by PCR ITS1-5.8S ITS2.
Culture-Independent Approach
In this study, we targeted D1 and D2 domain of 26S rRNA gene (large ribosomal subunit) of fungi from 40 samples of starters using PCR-DGGE fingerprint analysis. We used NL-1 forward primer and a new LS2 reverse primer to amplify the portion of 26S rRNA gene. These primers amplified a product of approximately 250 bp covering most of the D1 expansion loop. In PCR-DGGE fingerprint, diversity map distributions in the form of band patterns of yeasts and molds had been observed in different starters (Figure 3). Total 202 DGGE bands were selected on the basis of visualizing the prominent and differential band patterns inside the gels, after analysis of raw sequenced data with the help of BLAST comparison in GenBank as presented in Supplementary Data Sheet S2. More than 98% similar identity with the closest species of yeasts and molds has different phylum and genus level distribution pattern in different starters (Figure 4). Interestingly, we observed the distinct species were more than the shared species and phut was found to have highest diversity (Figure 5).
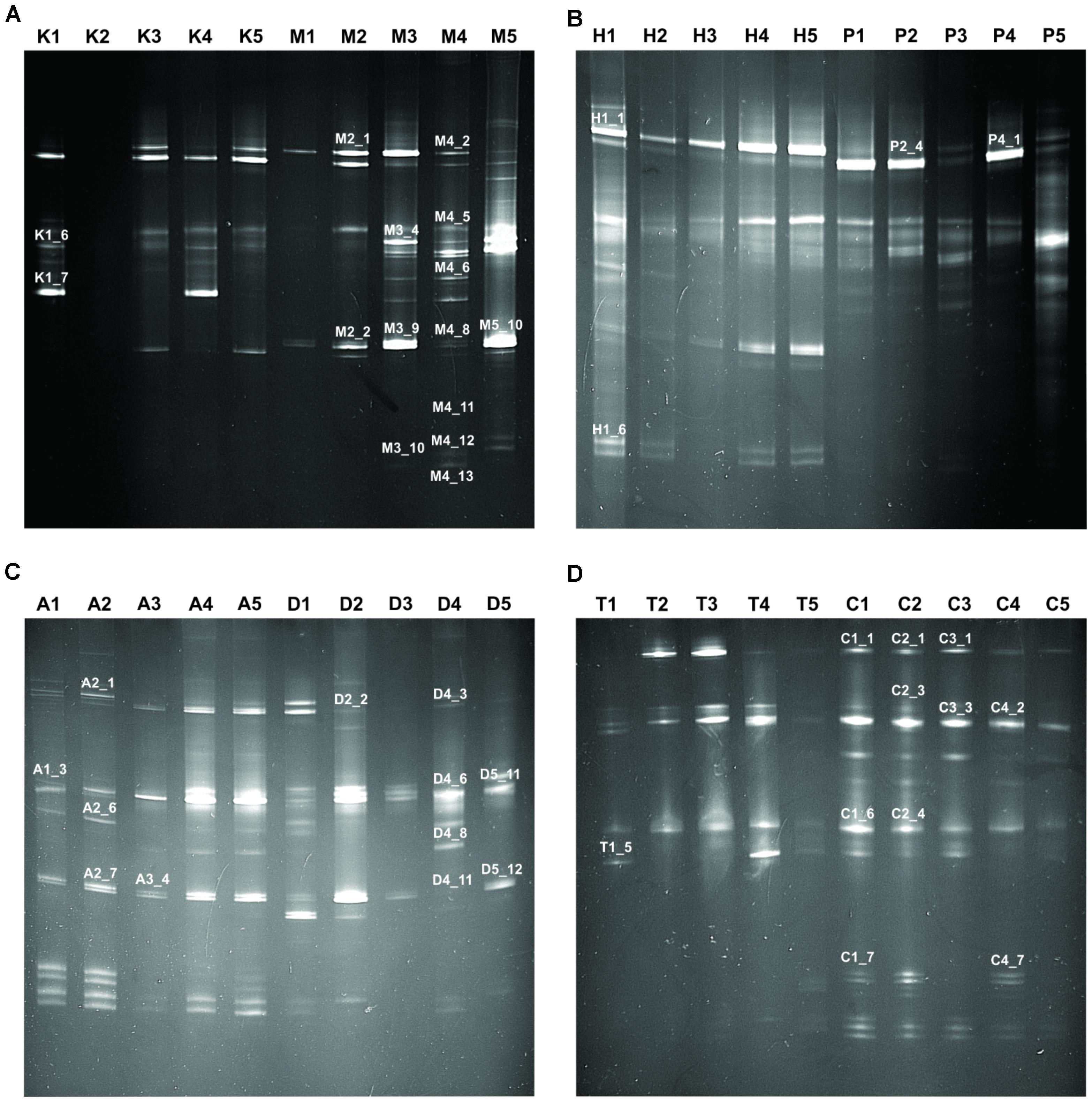
FIGURE 3. Fingerprint of PCR-DGGE analysis of different samples. Total 40 samples were taken for PCR-DGGE wherein five samples from each amylolytic starter used for fingerprinting. Samples and respective band patterns are demonstrated like: (A) Khekrii (K) and Marcha (M); (B) Hamai (H) and Humao (P); (C) Phut (A) and Dawdim (D); (D) Thiat (T) and Chowan (C). Representation of band numbers of respective bands on fingerprint were those which showed ≥98% sequence identity to GeneBank nucleotide database.
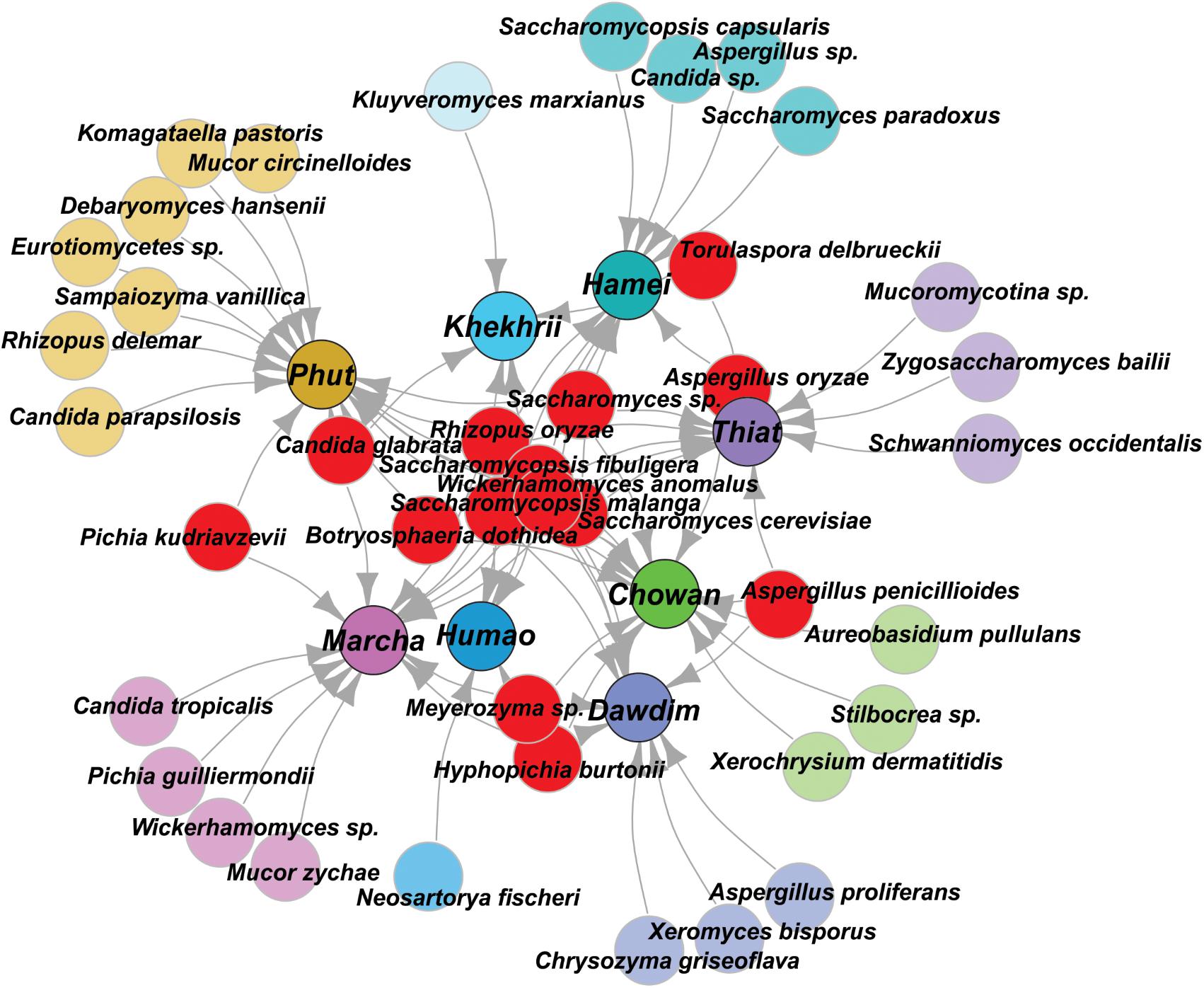
FIGURE 4. Graphical representation of all species identified in PCR-DGGE of 26SrRNA gene after sequencing. Shared species were represented in red color, and sample specific unique species were represented in respective colors to the starter samples and arrow indicated the origin distribution patterns of these isolates.
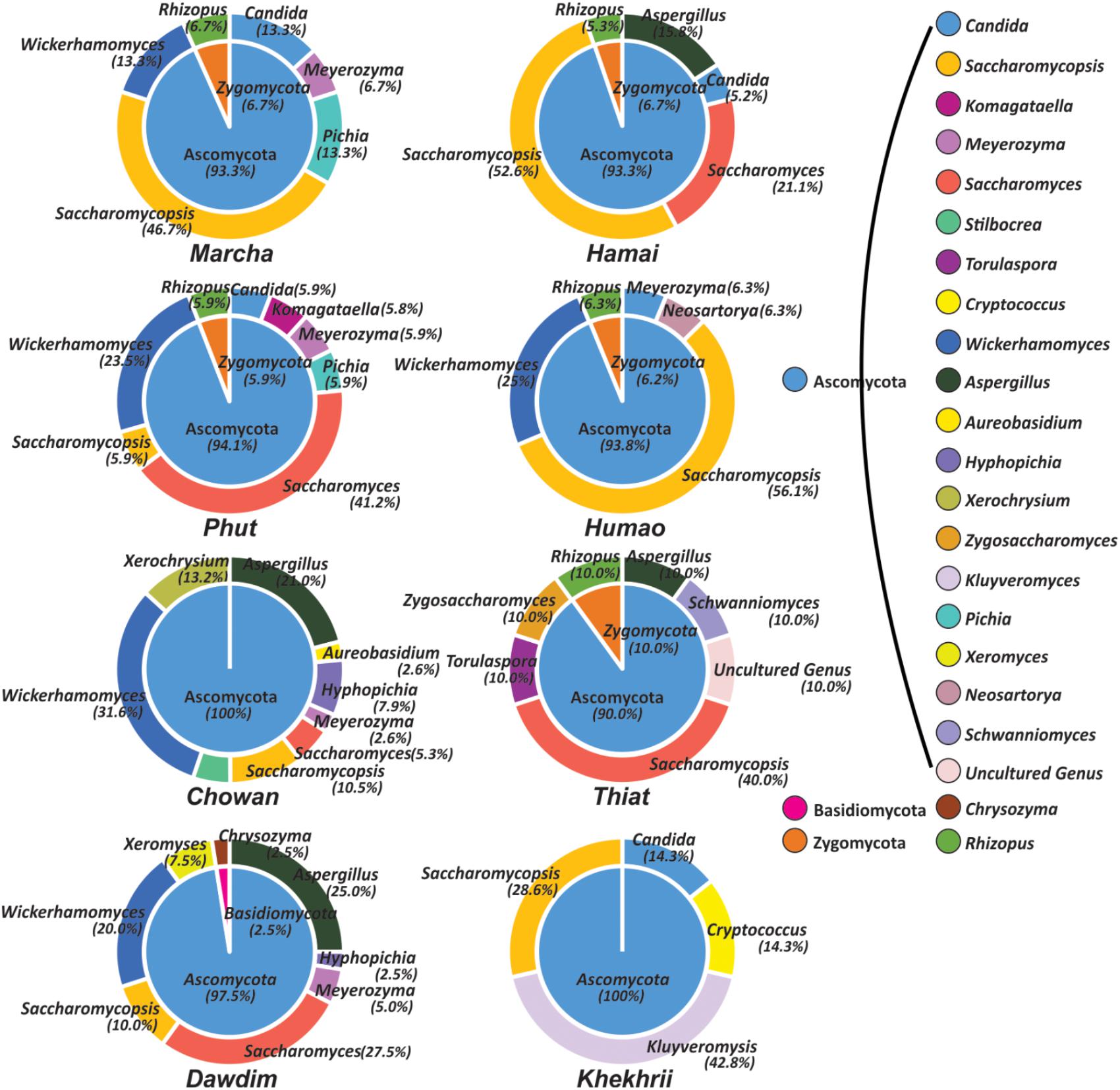
FIGURE 5. Genus and phylum level distribution of yeast and molds diversity in starters. Genus and respective phylum presented here was based on 98% identity cutoff value to the GeneBank database.
All these different techniques revealed the diversity and their differences of mycobiome species in different starters (Figure 6). Notably, the average distributions of yeasts in all samples were summarized as Saccharomyces cerevisiae (16.5%), Saccharomycopsis fibuligera (15.3%), Wickerhamomyces anomalus (11.3%), S. malanga (11.7%), Kluyveromyces marxianus (5.3%), Meyerozyma sp. (2.7%), Candida glabrata (2.7%), Saccharomyces sp. (1.3%), Hyphopichia burtonii (1.2%), Schwanniomyces occidentalis (1.1%), Pichia kudriavzevi (1.0%), Torulaspora delbrueckii (1.0%), Zygosaccharomyces bailii (1.0%), Pichia guilliermondii (1.0%), Candida parapsilosis (0.4%), Komagataella pastoris (0.3%), S. capsularis (0.6%), S. Paradoxus (0.6%), and C. tropicalis (0.1%). Similarly, the average distributions of molds in amylolytic starters were Aspergillus penicillioides (5.0%), Rhizopus oryzae (3.3%), sub-phylum: Mucoromycotina (2.1%), Cryptococcus amylolentus (1.7%), Xerochrysium dermatitidis (1.6%), Aspergillus oryzae (1.3%), Neosartorya fischeri (0.8%), A. proliferans (0.6%), Chrysozyma griseoflava (0.6%), Stilbocreasp. (0.6%), Mucor circinelloides (0.5%), Aureobasidium pullulans (0.4%), and Xeromyces bisporus (0.3%).
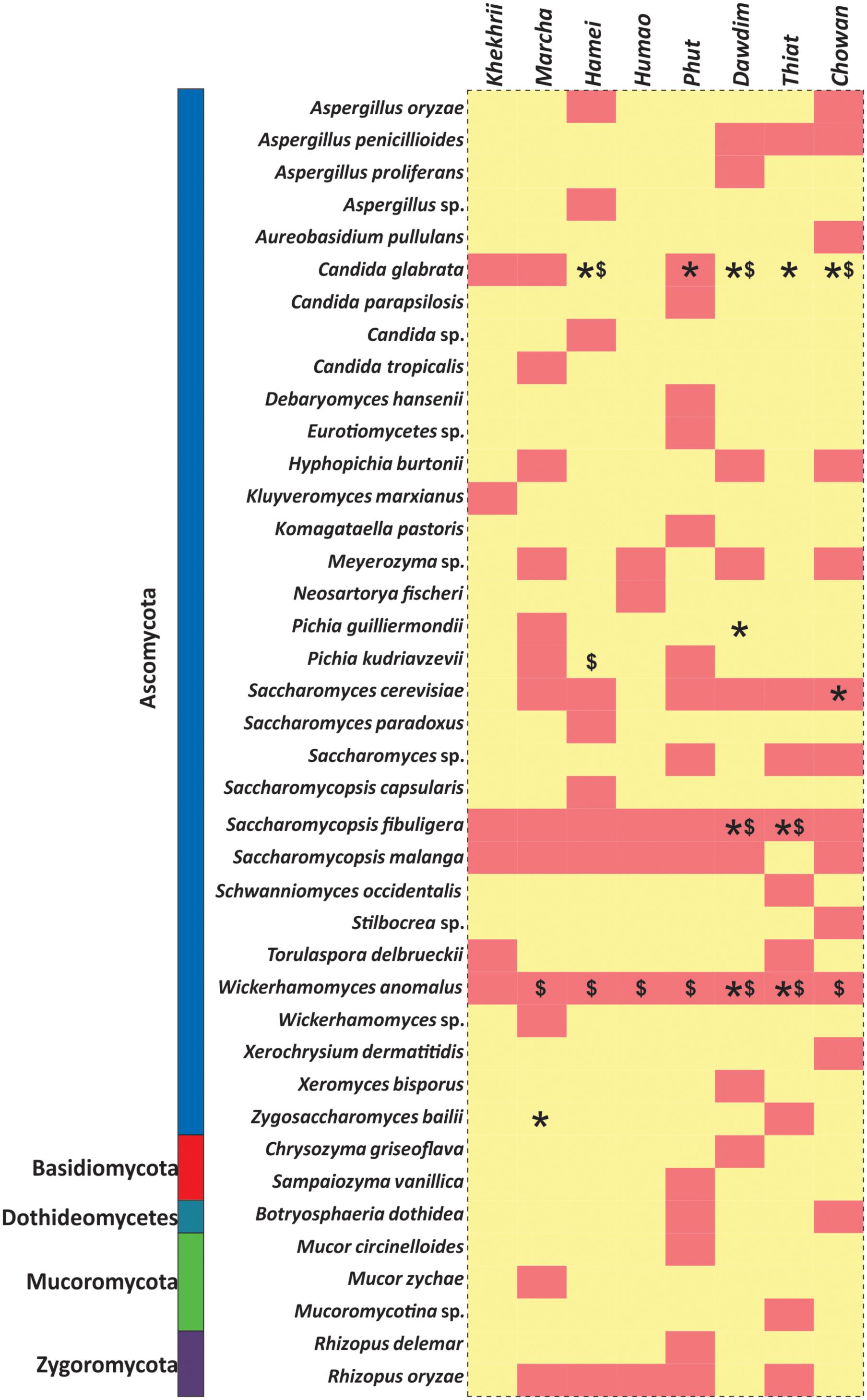
FIGURE 6. Heatmap showing the consensus species diversity observed during PCR-DGGE, Biolog identification hits and ITS region gene sequencing of yeast isolates. We used presence–absence value of PCR-DGGE species data to generate heatmap whereas red color indicates the presence and in other hand yellow color represents absence value. Other datasets were mapped over the heatmap like: Biolog identification (∗) and ITS-region gene sequencing of yeast isolates ($).
Discussion
Due to geographical locations, starters may have different and distinct mycobiome species diversity (Jeyaram et al., 2011). Going forward with this hypothesis, we examined and produced extensive surveillance report in different starters used in Indian alcoholic beverage production as an ethnic constituent. Results from Biolog system, where the profile of growth responses provides a metabolic fingerprint for each isolate (Praphailong et al., 1997), showed more diversity of yeasts in starters of North East India than phenotypic characterization based on probability and similarities index value. Even with high reliability rates, both phenotypic and Biolog tests did not coincide with the molecular reference tests for the majority of the isolates: when the identification results by Biolog were compared to 18S rRNA gene sequencing and species-specific PCR reactions (Nisiotou and Nychas, 2007). It has been previously reported that the ITS region gene analysis is a reliable routine technique for the differentiation of yeasts at species level (Clemente-Jimenez et al., 2004; Combina et al., 2005; Zott et al., 2008). Considering that species-specific PCR protocols target specific genes of genera and species, the reliability of ITS region gene sequences was considered to be 100% (Moraes et al., 2013). Another advantage of molecular culture-dependent method, which includes ITS, is that it allows a collection of pure cultures that may be used for further selection of suitable yeast strains to improve quality of alcoholic beverages (Lv et al., 2013).
In this study, Wickerhamomyces anomalus, Pichia anomala, Saccharomycopsis fibuligera, and Candida glabrata were identified in starters using ITS analysis. The previous studies also reported Candida glabrata, Pichia anomala, and Saccharomycopsis fibuligera from marcha based on 18S rDNA sequences (Tsuyoshi et al., 2005). It has been reported that Candida glabrata, which is a moderate alcohol producer, has also been recovered in kodo ko jaanr, ethnic fermented finger millet beverage prepared by using marcha (Thapa and Tamang, 2004) and some traditional Vietnamese starters (Dung et al., 2007), indicating that it is involved in alcohol production. Non-Saccharomyces yeasts may contribute to flavor or aroma formation in the alcoholic beverage (Rojas et al., 2001; Fleet, 2003; Moreira et al., 2005; Dung et al., 2006; Jolly et al., 2017). Saccharomycopsis fibuligera, Saccharomyces cerevisiae, Wickerhamomyces anomala, Pichia sp., and Candida sp. are the most common yeasts present in rice-based starters of Asia (Lee and Fujio, 1999; Xie et al., 2007; Jeyaram et al., 2008). Interestingly, Wickerhamomyces anomalus, probably the most abundant yeast, was reported for the first time from all the eight amylolytic starters of North East India using ITS-PCR method. The multiple sequence alignment of the ITS region gene sequences of Wickerhamomyces anomalus may be used for many purposes including inferring the presence of ancestral relationships between the sequences (Rampersad, 2014). It may be noted that protein sequences that are structurally very similar can be evolutionarily distant which is referred to as distant homology (Li and Durbin, 2010).
Genomic DNA extracted directly from samples of dried starters of India using the PCR-DGGE tools showed diversity of yeasts Wickerhamomyces anomalus, Saccharomyces cerevisiae, S. malanga, S. paradoxus, Saccharomycopsis fibuligera, Sm. Capsularis, Candida glabrata, C. tropicalis, Meyerozyma sp., Pichia guilliermondii, and P. kudriavzevi. Some researchers have reported the microbial community in some traditionally prepared dried starters for production of alcoholic beverages using PCR-DGGE analysis such as principal amylase-producer yeast Sm. fibuligera and ethanol-producers S. cerevisiae in banh men of Vietnam (Thanh et al., 2008), nuruk of Korea (Jung et al., 2012), and yaa qu and hong qu of China (Lv et al., 2012, 2013; Chen et al., 2014), respectively. Sm. fibuligera secretes considerable amount of α-amylase, glucoamylase, acid proteases, and β-glucosidase, which are applied in the fermentation industry (Chi et al., 2009).
The dominance of S. cerevisiae in marcha, thiat, dawdim, and phut might be due to its competitive growth in the presence of fermentable sugars and its ethanol tolerance may be due to fast growth during various alcoholic fermentations (Dung et al., 2006, 2007; Jeyaram et al., 2008). S. cerevisiae has also found to be one of the dominant yeasts in all starters of North East India, because of its competitive growth under strict anaerobic conditions and its tolerance to ethanol (Romano et al., 2006). Wickerhamomyces anomalus, a regular component in several types of Asia-Pacific alcohol fermentation starters (Limtong et al., 2002; Thanh et al., 2008), was detected in all analyzed samples. P. guilliermondii which was observed in marcha was also reported from wheat-based qu for Chinese Shaoxing rice wine (Xie et al., 2007) and hamei of Manipur in India (Jeyaram et al., 2008), which can produce volatile phenols and esters in the initial stages of alcoholic fermentation (Moreira et al., 2005). Pichia kudriavzevii, Wickerhamomyces anomalus, S. malanga, Kluyveromyces marxianus, Torulaspora delbrueckii, Hyphopichia burtonii, S. capsularis, and Debaryomyces hansenii were also reported from other Asian starters for the production of flavor and ethanol (Dung et al., 2006; Xie et al., 2007; Thanh et al., 2008; Zhang et al., 2008; Jung et al., 2012; Lv et al., 2013; Chen et al., 2014). Zygosaccharomyces bailii is widely present in various food fermentations, such as wine, tea, and vinegar fermentations (Garavaglia et al., 2015), and also produced various flavor compounds including alcohol in Chinese Maotai liquor (Xu et al., 2017).
In chowan, few pathogenic fungi were also detected such as Xerochrysium dermatitidis, which is a pathogenic fungus causing skin diseases (Pitt et al., 2013); and Aureobasidium pullulans, a ubiquitous black, yeast-like human fungal pathogen found in soil, water, air, and limestone (Chan et al., 2011). These pathogenic fungi may be contaminated through various raw substrates including wild herbs, water, etc. during crude preparation of chowan by village people in Tripura. The presence of sub-phylum: Mucoromycotina, which is the earliest mutualistic symbiosis fungus with Haplomitriopsida liverworts (Field et al., 2015),probably passed through the plants used during preparation of thiat.
Besides yeast community, some molds Rhizopus spp. and Aspergillus spp. were also detected by PCR-DGGE analysis in starters except in khekhrii samples of Nagaland (prepared by naturally fermenting germinated sprouted rice grains). Species of Rhizopus spp. and Aspergillus were reported from many Asian amylolytic starters (Tamang et al., 1988; Oda et al., 2006; Yang et al., 2013; Zhu and Tramper, 2013). The distributions of yeasts communities in amylolytic starters of North East India were higher in comparison to molds, this may be due to low temperatures of that particular environment in North East India and also the substrates used for fermentation (Chi et al., 2009). These traditional starters are the result of long-term selection for preserving and cultivation the amylolytic and alcohol-producing native yeasts and fungi by ethnic people which has been practicing the traditional process for centuries (Tamang, 2010; Londoño-Hernández et al., 2017). The DGGE analysis has some disadvantages due to its inability to determine the relative abundance of dominant species, differentiate between viable and nonviable cells, and difficulties in interpretation of multi-bands (Nam et al., 2012; Dolci et al., 2015). Besides, DNA extraction efficiencies vary between microorganisms since DGGE band intensity is not always correlated with population density (Ercolini, 2004; Prakitchaiwattana et al., 2004; Lv et al., 2013). Sub-culturing or back sloping of desirable inocula from previous batch during the traditional preparation of starters under uncontrolled fermentation may pose health risks (Rossetti et al., 2009). However, combination of culture-dependent and -independent analysis may be used to assess the safety of the microbiota associated with spontaneous/natural fermentation that may help to predict the possible risks for human health (Capozzi et al., 2007; Van Hijum et al., 2013).
Conclusion
Starter making technology reflects the traditional method of “sub-culturing” of desirable inocula from previous batch to new culture using rice as base substrates by back-sloping, in North East India. Selection of ethnic starters from different geographical regions with diverse mycobiome is gaining the importance of species diversity as indigenous property. We performed one of the successful trials to find out the mycobiome associated with eight different dried starters of North East India analyzed by ITS-PCR and PCR-DGGE techniques. These results may enrich our knowledge of cultivable indigenous mycobiota present in the starters (amylolytic and alcoholic) of Asia that may be used to promote the production technology of unique ethnic alcoholic beverages high quality and typical attributes; moreover, data of starters of India can be used as reference data base for the further research.
Author Contributions
SS contributed to this present work as a part of his research work. MS and KJ helped and assisted in some molecular work Bioinformatics analysis. AS, YS, and JT framed and prepared this paper critically with final approval of JT.
Funding
This work was supported by Department of Biotechnology, Ministry of Science and Technology, New Delhi (BT/488NE/TBP/2013, dated: 11.08.2014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledged the financial support of Department of Biotechnology, Govt of India. SS is grateful to DBT for award of JRF/SRF in the project sanctioned to JT and NCMR, Pune, for some technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02237/full#supplementary-material
DATA SHEET S1 | GenBank accessions number of identified species of yeasts.
DATA SHEET S2 | Biolog identification of yeast strains isolated from different amylolytic starters of North East India.
Footnotes
- ^http://www.northeasttourism.gov.in
- ^http://www.technelysium.com.au/chromas.html
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Alegría,Á., González, R., Díaz, M., and Mayo, B. (2011). Assessment of microbial populations dynamics in a blue cheese by culturing and denaturing gradient gel electrophoresis. Curr. Microbiol. 62, 888–893. doi: 10.1007/s00284-010-9799-7
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anupma, A., Pradhan, P., Sha, S. P., and Tamang, J. P. (2018). Traditional skill of ethnic people of the Eastern Himalayas and North East India in preserving microbiota as dry amylolytic starters. Indian J. Tradit. Knowl. 17, 184–190.
Banskar, S., Bhute, S. S., Suryavanshi, M. V., Punekar, S., and Shouche, Y. S. (2016). Microbiome analysis reveals the abundance of bacterial pathogens in Rousettus leschenaultii guano. Sci. Rep. 6:36948. doi: 10.1038/srep36948
Caggia, C., Restuccia, C., Pulvirenti, A., and Giudici, P. (2001). Identification of Pichia anomala isolated from yoghurt by RFLP of the ITS region. Int. J. Food Microbiol. 71, 71–73. doi: 10.1016/S0168-1605(01)00556-6
Capozzi, V., and Spano, G. (2011). Food microbial biodiversity and “microbes of protected origin”. Front. Microbiol. 2:237. doi: 10.3389/fmicb.2011.00237
Capozzi, V., Fragasso, M., Romaniello, R., Berbegal, C., Russo, P., and Spano, G. (2017). Spontaneous food fermentations and potential risks for human health. Fermentation 3:49. doi: 10.3390/fermentation3040049
Chan, G. F., Puad, M. S. A., Chin, C. F., and Rashid, N. A. A. (2011). Emergence of Aureobasidium pullulans as human fungal pathogen and molecular assay for future medical diagnosis. Folia Microbiol. 56, 459–467. doi: 10.1007/s12223-011-0070-9
Chen, B., Wu, Q., and Xu, Y. (2014). Filamentous fungal diversity and community structure associated with the solid state fermentation of Chinese Maotai-flavor liquor. Int. J. Food Microbiol. 179, 80–84. doi: 10.1016/j.ijfoodmicro.2014.03.011
Chi, Z., Chi, Z., Liu, G., Wang, F., Ju, L., and Zhang, T. (2009). Saccharomycopsis fibuligera and its applications in biotechnology. Biotechnol. Adv. 27, 423–431. doi: 10.1016/j.biotechadv.2009.03.003
Clemente-Jimenez, J. M., Mingorance-Cazorla, L., Martıìnez-Rodrıìguez, S., Las Heras-Vázquez, F. J., and Rodrıìguez-Vico, F. (2004). Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol. 21, 149–155. doi: 10.1016/S0740-0020(03)00063-7
Cocolin, L., Aggio, D., Manzano, M., Cantoni, C., and Comi, G. (2002). An application of PCR-DGGE analysis to profile the yeast populations in raw milk. Int. Dairy J. 12, 407–411. doi: 10.1016/S0958-6946(02)00023-7
Cocolin, L., Bisson, L. F., and Mills, D. A. (2000). Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol. Lett. 189, 81–87. doi: 10.1111/j.1574-6968.2000.tb09210.x
Combina, M., Mercado, L., Borgo, P., Elia, A., Jofre, V., Ganga, A., et al. (2005). Yeasts associated to Malbec grape berries from Mendoza, Argentina. J. Appl. Microbiol. 98, 1055–1061. doi: 10.1111/j.1365-2672.2005.02540.x
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Int. J. Complex Syst. 1695, 1–9.
Dolci, P., Alessandria, V., Rantsiou, K., and Cocolin, L. (2015). “Advanced methods for the identification, enumeration, and characterization of microorganisms in fermented foods,” in Advances in Fermented Foods and Beverages, ed. W. H. Holzapfel (London: Elsevier), 157–176. doi: 10.1016/b978-1-78242-015-7-4
Dung, N. T. P., Rombouts, F. M., and Nout, M. J. R. (2006). Functionality of selected strains of moulds and yeasts from vietnamese rice wine starters. Food Microbiol. 23, 331–340. doi: 10.1016/j.fm.2005.05.002
Dung, N. T. P., Rombouts, F. M., and Nout, M. J. R. (2007). Characteristics of some traditional vietnamese starch-based rice wine fermentation starters (men). LWT Food Sci. Technol. 40, 130–135. doi: 10.1016/J.LWT.2005.08.004
El Sheikha, A. F., Condur, A., Métayer, I., Le Nguyen, D. D., Loiseau, G., and Montet, D. (2009). Determination of fruit origin by using 26S rDNA fingerprinting of yeast communities by PCR-DGGE: preliminary application to Physalis fruits from Egypt. Yeast 26, 567–573. doi: 10.1002/yea.1707
Ercolini, D. (2004). PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56, 297–314. doi: 10.1016/J.MIMET.2003.11.006
Ercolini, D., Mauriello, G., Blaiotta, G., Moschetti, G., and Coppola, S. (2004). PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J. Appl. Microbiol. 96, 263–270. doi: 10.1046/j.1365-2672.2003.02146.x
Esteve-Zarzoso, B., Belloch, C., Uruburu, F., and Querol, A. (1999). Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49, 329–337. doi: 10.1099/00207713-49-1-329
Field, K. J., Rimington, W. R., Bidartondo, M. I., Allinson, K. E., Beerling, D. J., Cameron, D. D., et al. (2015). First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO2. New Phytol. 205, 743–756. doi: 10.1111/nph.13024
Fleet, G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. doi: 10.1016/S0168-1605(03)00245-9
Garavaglia, J., de Souza Schneider, R. D. C., Mendes, S. D. C., Welke, J. E., Zini, C. A., Caramão, E. B., et al. (2015). Evaluation of Zygosaccharomyces bailii BCV 08 as a co-starter in wine fermentation for the improvement of ethyl esters production. Microbiol. Res. 173, 59–65. doi: 10.1016/J.MICRES.2015.02.002
Gonelimali, F. D., Lin, J., Miao, W., Xuan, J., Charles, F., Chen, M., et al. (2018). Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front. Microbiol. 9:1639. doi: 10.3389/fmicb.2018.01639
Grizard, S., Dini-Andreote, F., Tieleman, B. I., and Salles, J. F. (2014). Dynamics of bacterial and fungal communities associated with eggshells during incubation. Ecol. Evol. 4, 1140–1157. doi: 10.1002/ece3.1011
Hesseltine, C. W. (1983). Microbiology of oriental fermented foods. Annu. Rev. Microbiol. 37, 575–601. doi: 10.1146/annurev.mi.37.100183.003043
Hesseltine, C. W., and Kurtzman, C. P. (1990). Yeasts in amylolytic food starters. Anal. Inst. Biol. Univ. Nac. Autón. México Ser. Bot. 60, 1–7. doi: 10.1016/B978-0-444-52149-1.00187-7
Iwen, P. C., Hinrichs, S. H., and Rupp, M. E. (2002). Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40, 87–109. doi: 10.1080/mmy.40.1.87.109
Jeyaram, K., Singh, W., Capece, A., and Romano, P. (2008). Molecular identification of yeast species associated with “Hamei” — a traditional starter used for rice wine production in Manipur. India. Int. J. Food Microbiol. 124, 115–125. doi: 10.1016/j.ijfoodmicro.2008.02.029
Jeyaram, K., Tamang, J. P., Capece, A., and Romano, P. (2011). Geographical markers for Saccharomyces cerevisiae strains with similar technological origins domesticated for rice-based ethnic fermented beverages production in North East India. Antonie Van Leeuwenhoek 100, 569–578. doi: 10.1007/s10482-011-9612-z
Jolly, N. P., Augustyn, O. P. H., and Pretorius, I. S. (2017). The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Vitic. 27, 15–39. doi: 10.21548/27-1-1475
Jung, M. J., Nam, Y. D., Roh, S. W., and Bae, J. W. (2012). Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 30, 112–123. doi: 10.1016/j.fm.2011.09.008
Korabecna, M. (2007). The variability in the fungal ribosomal DNA (ITS1, ITS2, and 5.8 S rRNA Gene): its biological meaning and application in medical mycology. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2,783–787.
Kurtzman, C. P., Fell, J. W., and Boekhout, T. (2011). The Yeasts: A Taxonomic Study, 5th Edn. Amsterdam: Elsevier Science Publisher.
Las Heras-Vazquez, F. J., Mingorance-Cazorla, L., Clemente-Jimenez, J. M., and Rodriguez-Vico, F. (2003). Identification of yeast species from orange fruit and juice by RFLP and sequence analysis of the 5.8S rRNA gene and the two internal transcribed spacers. FEMS Yeast Res. 3, 3–9. doi: 10.1111/j.1567-1364.2003.tb00132.x
Lee, A. C., and Fujio, Y. (1999). Microflora of banh men, a fermentation starter from Vietnam. World J. Microbiol. Biotechnol. 15, 51–55. doi: 10.1023/A:1008897909680
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Limtong, S., Sintara, S., and Suwannarit, P. (2002). Yeast diversity in Thai traditional alcoholic starter. Kasetsart J. Nat. Sci. 36, 149–158.
Londoño-Hernández, L., Ramírez-Toro, C., Ruiz, H. A., Ascacio-Valdés, J. A., Aguilar-Gonzalez, M. A., Rodríguez-Herrera, R., et al. (2017). Rhizopus oryzae–ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 257, 110–127. doi: 10.1016/j.ijfoodmicro.2017.06.012
Lv, X., Weng, X., and Huang, R. (2012). Research on biodiversity of yeasts associated with Hongqu glutinous rice wine starters and the traditional brewing process. J. Chin. Inst. Food Sci. Technol. 12, 182–190.
Lv, X. C., Huang, X. L., Zhang, W., Rao, P. F., and Ni, L. (2013). Yeast diversity of traditional alcohol fermentation starters for Hong Qu glutinous rice wine brewing, revealed by culture-dependent and culture-independent methods. Food Control 34, 183–190. doi: 10.1016/J.FOODCONT.2013.04.020
Ly, S., Mith, H., Tarayre, C., Taminiau, B., Daube, G., Fauconnier, M. L., et al. (2018). Impact of microbial composition of Cambodian traditional dried starters (Dombea) on flavor compounds of rice wine: combining amplicon sequencing with HP-SPME-GCMS. Front. Microbiol. 9:894. doi: 10.3389/fmicb.2018.00894
Moraes, P. M., Perin, L. M., Júnior, A. S., and Nero, L. A. (2013). Comparison of phenotypic and molecular tests to identify lactic acid bacteria. Braz. J. Microbiol. 44, 109–112. doi: 10.1590/S1517-83822013000100015
Moreira, N., Mendes, F., Hogg, T., and Vasconcelos, I. (2005). Alcohols, esters and heavy sulphur compounds production by pure and mixed cultures of apiculate wine yeasts. Int. J. Food Microbiol. 103, 285–294. doi: 10.1016/j.ijfoodmicro.2004.12.029
Nam, Y. D., Lee, S. Y., and Lim, S. I. (2012). Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 155, 36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013
Nisiotou, A. A., and Nychas, G. J. E. (2007). Yeast populations residing on healthy or botrytis-infected grapes from a vineyard in Attica. Greece Appl. Environ. Microbiol. 73, 2765–2768. doi: 10.1128/AEM.01864-06
Oda, K., Kakizono, D., Yamada, O., Iefuji, H., Akita, O., and Iwashita, K. (2006). Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 72, 3448–3457. doi: 10.1128/AEM.72.5.3448-3457.2006
Oguntoyinbo, F. A., Tourlomousis, P., Gasson, M. J., and Narbad, A. (2011). Analysis of bacterial communities of traditional fermented West African cereal foods using culture independent methods. Int. J. Food Microbiol. 145, 205–210. doi: 10.1016/j.ijfoodmicro.2010.12.025
Pitt, J. I., Lantz, H., Pettersson, O. V., and Leong, S. L. L. (2013). Xerochrysium gen. nov. and Bettsia, genera encompassing xerophilic species of Chrysosporium. IMA Fungus 4, 229–241. doi: 10.5598/imafungus.2013.04.02.08
Prakitchaiwattana, C., Fleet, G., and Heard, G. (2004). Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 4, 865–877. doi: 10.1016/j.femsyr.2004.05.004
Praphailong, W., Van Gestel, M., Fleet, G. H., and Heard, G. M. (1997). Evaluation of the Biolog system for the identification of food and beverage yeasts. Lett. Appl. Microbiol. 24, 455–459. doi: 10.1046/j.1472-765X.1997.00057.x
Pryce, T. M., Palladino, S., Kay, I. D., and Coombs, G. W. (2003). Rapid identification of fungi by sequencing the I T S l and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41, 369–381. doi: 10.1080/13693780310001600435
Puerari, C., Magalhães-Guedes, K. T., and Schwan, R. F. (2015). Physicochemical and microbiological characterization of chicha, a rice-based fermented beverage produced by Umutina Brazilian Amerindians. Food Microbiol. 46, 210–217. doi: 10.1016/j.fm.2014.08.009
Rampersad, S. N. (2014). ITS1, 5.8S and ITS2 secondary structure modelling for intra-specific differentiation among species of the Colletotrichum gloeosporioides sensu lato species complex. Springerplus 3:684. doi: 10.1186/2193-1801-3-684
Rojas, V., Gil, J. V., Piñaga, F., and Manzanares, P. (2001). Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 70, 283–289. doi: 10.1016/S0168-1605(01)00552-9
Romano, P., Capece, A., and Jespersen, L. (2006). “Taxonomic and ecological diversity of food and beverage yeasts,” in Yeasts in Food and Beverages, eds A. Querol and G. H. Fleet (Berlin: Springer), 13–53.
Rossetti, L., Carminati, D., Zago, M., and Giraffa, G. (2009). A qualified presumption of safety approach for the safety assessment of Grana Padano whey starters. Int. J. Food Microbiol. 130, 70–73. doi: 10.1016/j.ijfoodmicro.2009.01.003
Russo, P., Capozzi, V., Spano, G., Corbo, M. R., Sinigaglia, M., and Bevilacqua, A. (2016). Metabolites of microbial origin with an impact on health: ochratoxin a and biogenic amines. Front. Microbiol. 7:482. doi: 10.3389/fmicb.2016.00482
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sha, S. P., Anupama, A., Pradhan, P., Prasad, G. S., and Tamang, J. P. (2016). Identification of yeasts by polymerase-chain-reaction-mediated denaturing gradient gel electrophoresis in marcha, an ethnic amylolytic starter of India. J. Ethn. Foods 3, 292–296. doi: 10.1016/J.JEF.2016.11.009
Sha, S. P., Jani, K., Sharma, A., Anupma, A., Pradhan, P., Shouche, Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7:10967. doi: 10.1038/s41598-017-11609-y
Steinkraus, K. H. (1996). Handbook of Indigenous Fermented Food, 2nd Edn. New York, NY: Marcel Dekker, Inc.
Susan Slechta, E., Hohmann, S. L., Simmon, K., and Hanson, K. E. (2012). Internal transcribed spacer region sequence analysis using SmartGene IDNS software for the identification of unusual clinical yeast isolates. Medical. Mycol. 50, 458–466. doi: 10.3109/13693786.2011.630683
Tamang, J. P. (2010). Himalayan Fermented Foods: Microbiology, Nutrition, and Ethnic Values. New York, NY: CRC Press, Taylor & Francis Group.
Tamang, J. P. (2016). Ethnic Fermented Foods and Alcoholic Beverages of Asia. New Delhi: Springer India. doi: 10.1007/978-81-322-2800-4
Tamang, J. P., Dewan, S., Tamang, B., Rai, A., Schillinger, U., and Holzapfel, W. H. (2007). Lactic acid bacteria in Hamei and Marcha of North East India. Indian Microbiol. 47, 119–125. doi: 10.1007/s12088-007-0024-8
Tamang, J. P., and Fleet, G. H. (2009). “Yeasts diversity in fermented foods and beverages,” in Yeasts Biotechnology: Diversity and Applications, eds T. Satyanarayana and G. Kunze (New York, NY: Springer), 169–198. doi: 10.1007/978-1-4020-8292-4_9
Tamang, J. P., and Sarkar, P. K. (1995). Microflora of murcha: an amylolytic fermentation starter. Microbios 81, 115–122.
Tamang, J. P., Sarkar, P. K., and Hesseltine, C. W. (1988). Traditional fermented foods and beverages of Darjeeling and Sikkim–a review. J. Sci. Food Agric. 44, 375–385. doi: 10.1002/jsfa.2740440410
Tamang, J. P., Shin, D. H., Jung, S. J., and Chae, S. W. (2016a). Functional properties of microorganisms in fermented foods. Front. Microbiol. 7:578. doi: 10.3389/fmicb.2016.00578
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016b). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Thanh, V. N., Mai, L. T., and Tuan, D. A. (2008). Microbial diversity of traditional Vietnamese alcohol fermentation starters (banh men) as determined by PCR-mediated DGGE. Int. J. Food Microbiol. 128, 268–273. doi: 10.1016/j.ijfoodmicro.2008.08.020
Thapa, S., and Tamang, J. P. (2004). Product characterization of kodo ko jaanr: fermented finger millet beverage of the Himalayas. Food Microbiol. 21, 617–622. doi: 10.1016/J.FM.2004.01.004
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tsuyoshi, N., Fudou, R., Yamanaka, S., Kozaki, M., Tamang, N., Thapa, S., et al. (2005). Identification of yeast strains isolated from marcha in Sikkim, a microbial starter for amylolytic fermentation. Int. J. Food Microbiol. 99, 135–146. doi: 10.1016/j.ijfoodmicro.2004.08.011
Van Hijum, S. A., Vaughan, E. E., and Vogel, R. F. (2013). Application of state-of-art sequencing technologies to indigenous food fermentations. Curr. Opin. Biotechnol. 24, 178–186. doi: 10.1016/j.copbio.2012.08.004
Wang, C., Shi, D., and Gong, G. (2008). Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J. Microbiol. Biotechnol. 24, 2183–2190. doi: 10.1007/s11274-008-9728-0
White, T., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. Innis, D. Gelfand, J. Sninsky, and T. White (San Diego: Academic Press), 315–322.
Xie, G., Li, W., Lu, J., Cao, Y., Fang, H., Zou, H., et al. (2007). Isolation and Identification of representative fungi from Shaoxing rice wine wheat Qu using a polyphasic approach of culture-based and molecular-based methods. J. Inst. Brew. 113, 272–279. doi: 10.1002/j.2050-0416.2007.tb00287.x
Xu, Y., Zhi, Y., Wu, Q., Du, R., and Xu, Y. (2017). Zygosaccharomyces bailii is a potential producer of various flavor compounds in Chinese Maotai-flavor liquor fermentation. Front. Microbiol. 8:2609. doi: 10.3389/fmicb.2017.02609
Yang, S., Choi, S. J., Kwak, J., Kim, K., Seo, M., Moon, T. W., et al. (2013). Aspergillus oryzae strains isolated from traditional Korean nuruk: fermentation properties and influence on rice wine quality. Food Sci. Biotechnol. 22, 425–432. doi: 10.1007/s10068-013-0097-6
Zhang, Z., Chang, X., and Zhong, Q. (2008). Liquor Qu fungus system and enzymatic system character and microbial dynamic variety during vintage. Liquor Mak. 5, 24–29.
Zhao, K., and Chu, X. (2014). G-BLASTN: accelerating nucleotide alignment by graphics processors. Bioinformatics 30, 1384–1391. doi: 10.1093/bioinformatics/btu047
Zheng, X.-W., Yan, Z., Han, B. Z., Zwietering, M. H., Samson, R. A., Boekhout, T., et al. (2012). Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 31, 293–300. doi: 10.1016/j.fm.2012.03.008
Zhu, Y., and Tramper, J. (2013). Koji – where East meets West in fermentation. Biotechnol. Adv. 31, 1448–1457. doi: 10.1016/j.biotechadv.2013.07.001
Keywords: mycobiome, dried starters, PCR-DGGE analysis, yeasts, filamentous molds
Citation: Sha SP, Suryavanshi MV, Jani K, Sharma A, Shouche Y and Tamang JP (2018) Diversity of Yeasts and Molds by Culture-Dependent and Culture-Independent Methods for Mycobiome Surveillance of Traditionally Prepared Dried Starters for the Production of Indian Alcoholic Beverages. Front. Microbiol. 9:2237. doi: 10.3389/fmicb.2018.02237
Received: 12 April 2018; Accepted: 03 September 2018;
Published: 26 September 2018.
Edited by:
Vittorio Capozzi, University of Foggia, ItalyReviewed by:
Xiaolan Wang, Jiangxi Normal University, ChinaChibundu Ngozi Ezekiel, Babcock University, Nigeria
Copyright © 2018 Sha, Suryavanshi, Jani, Sharma, Shouche and Tamang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jyoti Prakash Tamang, anlvdGlfdGFtYW5nQGhvdG1haWwuY29t
 Shankar Prasad Sha
Shankar Prasad Sha Mangesh Vasant Suryavanshi
Mangesh Vasant Suryavanshi Kunal Jani
Kunal Jani Avinash Sharma
Avinash Sharma Yogesh Shouche
Yogesh Shouche Jyoti Prakash Tamang
Jyoti Prakash Tamang