- 1Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 3Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 4Xisha/Nansha Ocean Observation and Research Station, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
Small non-coding RNAs (sRNAs) are important modulators of gene expression and are involved in the pathogenesis and survival of prokaryotes. However, few studies have been conducted with Vibrio alginolyticus, which limits our ability to probe the regulation of virulence and environmental adaptation by sRNAs in this opportunistic pathogen. In this study, the sRNA candidate srvg23535 was identified in V. alginolyticus ZJ-T. The precise transcript end, secondary structure, and sequence conservation were determined. A srvg23535 null mutant was constructed and characterized by using Phenotype MicroArray (PM) technology. In silico target prediction was conducted by IntaRNA and TargetRNA2. Subsequently, a 107 nt transcript was validated with a sigma70 promoter at the 5′ end and a Rho-independent terminator at the 3′ end. The sRNA srvg23535 had four stem-loop structures and was conserved among Vibrio harveyi, Vibrio parahaemolyticus, and Vibrio splendidus. Deletion of srvg23535 in V. alginolyticus ZJ-T led to a weaker utilization of D-mannose, D-melibiose, lactulose, and inosine as carbon sources but stronger utilization of L-cysteine as nitrogen source. Moreover, the srvg2353 mutant showed stronger resistance to osmotic stress but weaker resistance to pH stress. Additionally, a total of 22 common targets were identified and several were related to the observed phenotype of the mutant. This study indicated that the novel sRNA, srvg23535, is conserved and restricted to Vibrio spp., affecting the utilization of several carbon and nitrogen sources and the response to osmotic and pH stress. These results extend our understanding of sRNA regulation in V. alginolyticus and provide a significant resource for the further study of the precise target mRNAs of srvg23535, which may provide targets for antibacterial therapeutic or attenuated vaccines against Vibrio spp.
Introduction
Vibrio alginolyticus, a Gram-negative halophilic bacterium with worldwide distribution, is an opportunistic pathogen of marine animals and humans, which causes serious infections, such as otitis media, otitis externa, and gastroenteritis (Citil et al., 2015; Siddiqui et al., 2016). During invasion, pathogenic bacteria may be exposed to a variety of harsh environments, including high pH, high salinity, and starvation (Ulitzur, 1975; Roop et al., 2009). To adapt to rapid changes of environmental conditions, bacteria need to coordinate their gene expression quickly and precisely (Ruprecht et al., 2016). Many genes associated with infection and the ability of bacteria to survive in the host have been identified in V. alginolyticus (Ren et al., 2013; Xiong et al., 2014). However, empirical evidence of complex post-transcriptional regulation and coordinating gene expression that allow V. alginolyticus to adapt to the harsh environments is rare.
Small non-coding RNAs (sRNAs) are important post-transcriptional regulators in both eukaryotes and prokaryotes (Liu and Camilli, 2010). They usually function by base-pairing with mRNA to modulate stability and/or translation (Wagner and Romby, 2015). In bacteria, sRNAs have been reported to control a variety of biological processes, including metabolism, bacterial motility, biofilm formation, quorum sensing, iron homeostasis, and stress responses (Wagner and Romby, 2015; Liu W. et al., 2016), and, therefore, are associated with bacterial adaptation and virulence. Several reports have shown that Vibrio spp. encode sRNAs as well, which were mainly identified and studied in Vibrio cholerae and Vibrio harveyi (Nguyen and Jacq, 2014). These sRNAs include RyhB, which functions in iron homeostasis (Mey et al., 2005), MicX, which regulates the synthesis of outer membrane proteins (Davis and Waldor, 2007), and Qrrs, involved in quorum sensing (Tu and Bassler, 2007). Nevertheless, the function of sRNA in V. alginolyticus is poorly understood (Luo et al., 2016). The potential regulatory function of sRNAs in V. alginolyticus could be important in environmental adaptation, which probably affects the virulence of bacteria (Iustman et al., 2015).
The prediction and validation of bacterial sRNAs are two necessary steps before studies of function. With the development of high-throughput sequencing and bioinformatics, genome-wide searches based on the biocomputational prediction of non-coding RNA genes are widely used in the prediction of sRNAs both in eukaryotes and prokaryotes, and even archaea (Livny and Waldor, 2007). Predicted sRNAs are subsequently subjected to experimental physical validation, including Northern blot analysis, qPCR, and 5′ and 3′ rapid amplification of cDNA ends (RACE) (Sharma and Vogel, 2009). Given that most functional sRNAs are expressed independently, relying on a stable secondary structure, and are usually conserved among different species or even different genus, the study of promoter and terminator identification and conservation analysis are often conducted to promote function research (Argaman et al., 2001; Chen et al., 2002).
The Phenotype MicroArray (PM) platform for global analysis of cellular phenotypes was first developed by Bochner et al. (2001). The technology has been used to monitor most aspects of the cell function, including the utilization of nutrients, transportation, and catabolization (PM1-8), and sensitivity to environmental stresses, such as osmolytes, pH, and chemical agents (PM9-20) (Bochner, 2009). It has been developed to directly assay the effects of genetic changes in cells (Tohsato and Mori, 2008). For example, (p)ppGpp and c-di-GMP were found to be involved in antibiotic resistance in Mycobacterium smegmatis by analyzing the wild type and strains lacking the gene for either (p)ppGpp synthesis or c-di-GMP synthesis by PM analysis (Miller, 1998).
During our preliminary research, 194 sRNA candidates were predicted through RNA-seq by the presence or absence of the RNA molecular chaperone Hfq in V. alginolyticus ZJ-T (Yiqin et al., 2016). Consequently, several of them were identified by Northern blotting or qPCR. In this study, we reported the identification of srvg23535, which is novel and conserved among Vibrio spp. Additionally, we characterized its phenotype in the utilization of different carbon and nitrogen sources, and its response to osmotic and pH stress by using PM technology, and then predicted its genetic targets in silico. These data will enrich the study of sRNAs in V. alginolyticus, and help to discover more about the environmental adaptation of V. alginolyticus, which may provide new insights into its pathogenesis and assist in providing new antibacterial therapeutic targets or attenuated vaccines for Vibrio spp.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1.
V. alginolyticus was cultured in LBS [Luria–Bertani (LB) with 2% additional NaCl] medium at 30°C. Escherichia coli strains were cultured in LB medium (BD, United States) supplemented with appropriate antibiotics at 37°C. For the selection of transconjugants, TCBS medium (BD, United States) was used with 0.2% D-glucose and 5 μg ml-1 chloramphenicol (Cm). To select transconjugants that have undergone plasmid excision and allelic exchange, expression of the ccdB gene carried by the pSW7848 plasmid was induced by adding 0.2% L-arabinose to the medium. This gene encodes a topoisomerase which is lethal to Gram-negative bacteria (Loris et al., 1999).
Northern Blot Assay
The sRNA, srvg23535, was named according to the upstream gene locus. It was predicted by RNA-seq of V. alginolyticus ZJ-T and the hfq mutant strain Δhfq-T with the following four features: (i) the sRNA was expressed from an intergenic regions and the size was between 50 and 500 nt; (ii) the expression level was at least half of but not the same as that of the flanking genes; (iii) there was a sigma70 promoter at the 5′ end and a Rho-independent terminator at the 3′ end; and (iv) the secondary structure was stem loop structure. Subsequently, Northern blotting was conducted as described by Toffano-Nioche et al. (2012) with some modification to confirm the transcription of srvg23535. Briefly, V. alginolyticus ZJ-T was cultured in LBS and Zobell medium at the same time and bacterial cells were collected for RNA extraction from different time points along the growth curves. The RNA was separated with 8% PAGE and transferred to a Hybond N+ membrane. An oligonucleotide probe, NB-srvg23535 (Supplementary Table S1), was labeled at the 3′ end using terminal transferase (Fermentas, United States) and [α-32p] dCTP. RiboRuler Low Range RNA Ladder (Fermentas, United States) was run alongside the samples to allow an estimation of the transcript size. Briefly, within a certain size range, the nucleic acid migration distance (electrophoretic mobility) is inversely proportional to the logarithm of its size (Bishop et al., 1967; Gough and Gough, 1984). Thus, according to the size of different marker’s bands and their migration distance, a curve can be drawn with the migration distance as the horizontal axis and the logarithm of the size as the vertical axis. Then, according to the migration distance of the target sRNA, the size of it can be estimated. 5.0S RNA was used as a loading control with the NB-5.0S RNA probe (Supplementary Table S1).
RACE
The corresponding 5′ and 3′ end of srvg23535 were identified by 5′ and 3′ RACE, respectively. Briefly, V. alginolyticus ZJ-T was cultured overnight in LBS medium at 30°C with vigorous shaking. The overnight culture was diluted to OD600 0.01 with fresh LBS medium and grown until OD600 0.8. Total RNA was isolated by using RNAprep Pure Cell/Bacteria Kit (TIANGEN, China). 5′ and 3′ RACE was conducted by using a SMARTer® RACE 5′/3′ Kit (Clontech, United States) (Supplementary Table S1). Finally, at least 12 clones of 5′ or 3′ RACE action were sequenced to determine the ends of srvg23535.
Promoter, Terminator Characterization, and Novelty Assessment
According to the transcript sequence, the sigma70 promoter was predicated by BPROM (Solovyev and Salamov, 2011) searching the region 200 nt upstream of the 5′ end, while the Rho-independent terminator was predicated by ARNold (Lesnik et al., 2001) searching the region 200 nt downstream of the 3′ end. The transcript sequence was blasted into the Rfam database to assess the novelty (Griffithsjones et al., 2003).
Secondary Structure Characterization and Phylogenetic Analysis
The MFOLD (Zuker et al., 1999) program was used to predicate the secondary structure of srvg23535 based on the lowest folding energy. The sequence including the transcript and the terminator were blasted against the representative genomes of microbes. Then multiple sequences alignment was conducted using ClustalW. The phylogenetic tree was constructed from the aligned sequences using Kimura 2-parameter model with the neighbor-joining method, bootstrapped 1,000 times via MEGA6.0 software.
In silico Analysis of Target Genes
The target genes of srvg23535 were predicted by using Web-based programs: IntaRNA (Siqueira et al., 2016) and TargetRNA2 (Kery et al., 2014). The hybridization between the sRNA transcript sequence and the sequence comprising 150 nt upstream until 150 nt downstream of the start codon of each annotated gene was screened in the genome of V. alginolyticus ZJ-T. To consider an interaction as positive, we used the corresponding p < 0.05 of both programs, and energy < -13 kcal/mol with IntaRNA and synonym energy < -8 kcal/mol with TargetRNA2 were taken as the threshold. Finally, the common predictions of both programs were confirmed as the target candidates.
Gene Disruption
To generate the sRNA disruptant, the sequence from 46 bp before the 5′ end to 2 bp after the 3′ end was deleted from the chromosome of V. alginolyticus ZJ-T. The deletion was constructed by homologous recombination as described before with some modification (Yiqin et al., 2016). Briefly, two flanking fragments of srvg23535 (Figure 1A) were amplified with two pairs of primers, srvg23535-UP-F and -R and srvg23535-DOWN-F and -R, respectively, and the linearized pSW7848 was amplified with pSW7848-F and -R (Supplementary Table S1). srvg23535-UP-F and srvg23535-DOWN-R contained overlapping extensions with pSW7848-R and -F, respectively, and srvg23535-UP-R contained overlapping extensions with srvg23535-DOWN-F. The two flanking fragments were further assembled into the linearized pSW7848 by using a ClonExpress Multis One Step Cloning Kit (Vozyme, China), generating the recombinant plasmid pSW7848-Δsrvg23535 comprising the 1,084 bp upstream and 1,105 bp downstream regions of srvg23535 (Table 1), using E. coli П3813 as an intermediate host. The recombinant plasmid was transferred by conjugation from strain GEB883 (Table 1) to V. alginolyticus ZJ-T before allelic exchange as described above. The sRNA disruptant was then confirmed by sequencing and the strain was named ZJ-T-Δsrvg23535 (Figure 1 and Table 1).
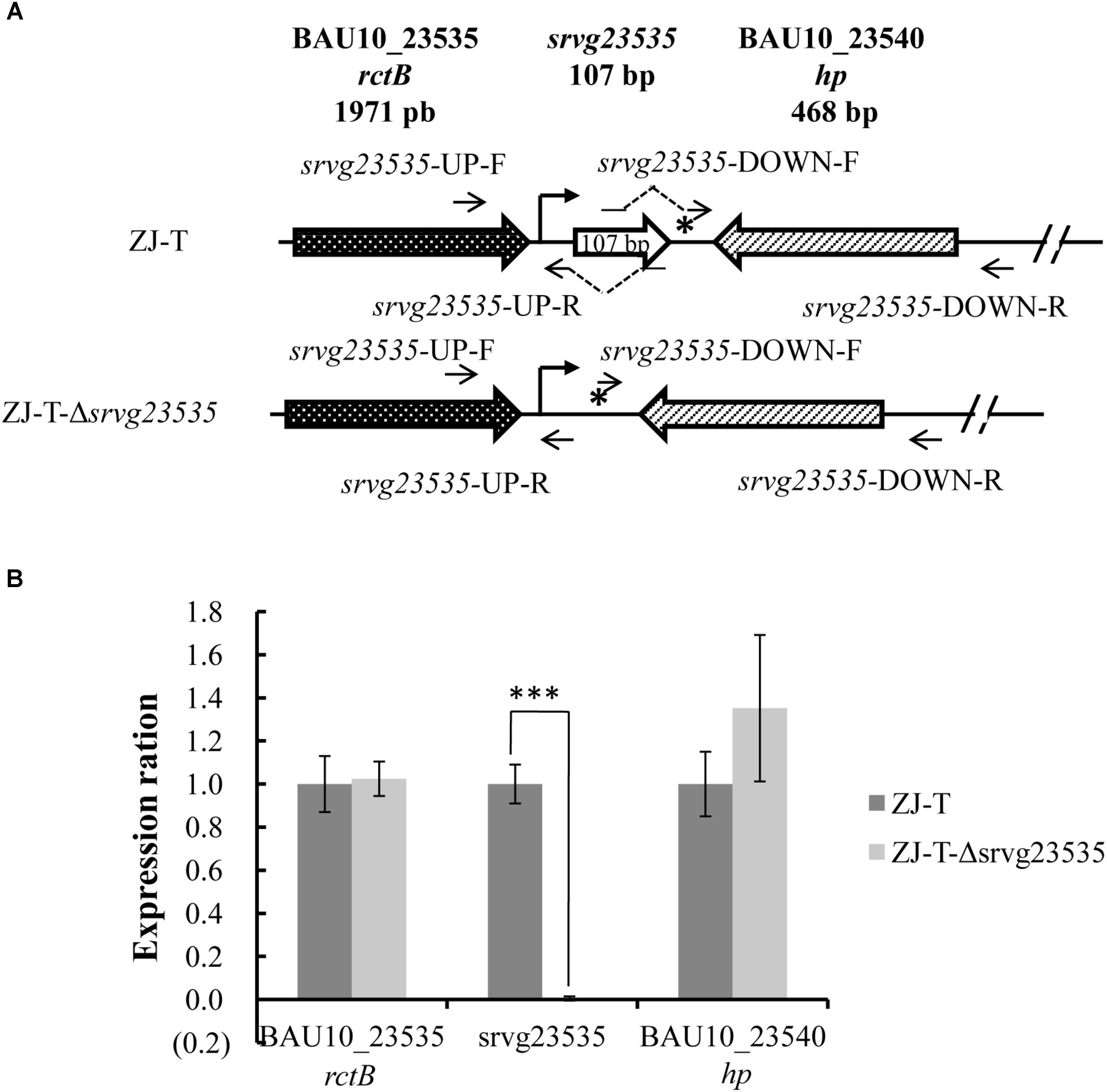
FIGURE 1. Structure of the srvg23535 locus and neighboring gene expression in V. alginolyticus ZJ-T and the mutant strain. (A) The sRNA srvg23535 is downstream of the rctB gene (locus ID = BAU10_23535) and upstream of a hypothetical protein gene (locus ID = BAU10_23540). A 107 nt transcript was determined by RACE. A putative Sigma70 promoter identified upstream of srvg23535 is indicated by an arrow and a Rho-independent terminator identified downstream of srvg23535 is indicated by a star. A total of 155 nt, from the 46 bp before the 5’ end to the 2 bp after the 3’ end was deleted, giving rise to ZJ-T-Δsrvg23535. (B) Relative expression of srvg23535, the upstream gene (rctB) and the downstream gene (hp) in the mutant strain compared to the wild type. Relative expression (normalized to the wild-type level for each gene) was determined by qPCR. Error bars correspond to the standard errors from three biological replicates. Statistically significant differences are indicated (∗∗∗p < 0.001).
Quantitative RT-PCR (qPCR)
qPCR was conducted to test the potential effect of the srvg23535 deletion on neighboring gene expression (potential polar effects) as described below (Supplementary Table S1). Three single colonies of strains ZJ-T and ZJ-T-Δsrvg23535 were inoculated overnight in three independent cultures with LBS medium. Overnight cultures were then diluted to OD600 0.01 in LBS fresh medium and bacterial cells were collected by centrifugation when grown to early exponential phase (OD600 0.5). All reagents were from Takara Bio Inc. (Japan). Total RNA was isolated by using RNAiso Plus, and 1 μg of RNA was DNase I-treated and reverse transcribed with PrimeScriptTM RT reagent Kit with gDNA Eraser. PCR amplification was then carried out on a LightCycler480 (Roche, Switzerland) by using SYBR Premix Ex TaqTM II with primers specific to the genes to be assayed. Three housekeeping genes (recA, uvrA, and gyrA) were used as internal controls (Supplementary Table S1). Three technological assessments were conducted on each biological replicate. Relative gene expression was calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001) and normalized to the wild-type ZJ-T value. Statistical significance was determined by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
PM Assay
Phenotype MicroArray technology was used to identify the function of srvg23535 in the utilization of different carbon sources, nitrogen sources, response to osmotic stress, and response to pH stress with the plates PM01, PM03, PM09, and PM10, respectively (Bochner et al., 2001). The test components are shown in Supplementary Table S2. All the substrates tested were pre-dispensed and dried in 96-well plates, requiring only inoculation with bacteria suspended in a buffer containing a dye, such as tetrazolium violet. During growth, bacterial metabolism leads to the irreversible reduction of the dye with the production of a purple color that can be read as the change in absorbance over time (Khatri et al., 2013). The measurements of growth were conducted at 590 nm (Blumenstein et al., 2015).
The wild type and the srvg23535 disruption cells were streaked on LBS agar plates and grown overnight at 30°C. All PM procedures were performed according to the protocol “PM Procedures for E. coli and other GN Bacteria” (Biolog, Inc., 26 August 2005; Supplementary File S1) with some modification because V. alginolyticus achieved better growth with 3% NaCl. For each strain and each biological replicate, 260 μl Dye Mix A and 4,075 μl water containing 14% NaCl were added to the inoculating fluid of 1.2 × IF-0, resulting in a final volume of 26 ml 1.0 × IF-0. In addition, 240 μl Dye Mix A and 3,760 μl water containing 14% NaCl were added to the inoculating fluid of 1.2 × IF-10 and resulting in a final volume of 24 ml 1.0 × IF-10. Cells were transferred and suspended into 26 ml of 1.0 × IF-0 to achieve 85% T (transmittance) in the BIOLOG Turbidimeter. Each well of the PM01 plate (carbon sources) was inoculated with 100 μl of the 85% T cell suspension. As the PM03 experiment required an appropriate carbon source, a stock solution of 2 M sodium succinate was used as an additive as recommended in the PM procedures for Gram-negative bacteria. Finally, 120 μl of the 2 M sodium succinate (100×) was added to 12 ml of the 85% T cell suspension prepared for PM01 and mixed completely but gently. Then, each well of the PM03 plate (nitrogen sources) was inoculated with 100 μl of the prepared cell suspension. For PM09 and PM10 experiments, 120 μl of the 85% T cell suspension prepared for PM01 was transferred into the prepared 24 ml 1.0 × IF-10 + dye (1:200 dilution) and mixed completely but gently. Then, each well of the PM09 and PM10 plates (the response to osmotic and pH stress, respectively) were inoculated with 100 μl of the prepared cell suspension. All PM plates were sealed with parafilm and inoculated in the incubator at 30°C. OD590 was measured at regular time intervals using a Multiskan Ascent plate reader (Thermo Fisher Scientific, United States) to indicate the conversion of the tetrazolium dye for 5 days. Finally, three biological replicates were conducted and the read-outs were analyzed with graphs. A one-way analysis of covariance (ANCOVA) was used to analyze the effect of treatment (srvg23535 deletion) on the OD590 with time as a covariate.
Results
Computational Prediction and Experimental Identification of srvg23535 in V. alginolyticus
The sRNA candidate srvg23535 is shown in an Artemis window (Figure 2A). It is flanked by a forward expressed translation elongation factor gene rctB (BAU10_23535) and a reverse expressed hypothetical protein gene (BAU10_23540) with a size of 93 nt. Only a single band was detected and its apparent size of around 80 nt in the Northern blot is smaller than the size of 93 nt deduced from the RNA-seq experiment (Figures 2B,C). As the bacteria grew, srvg23535 was expressed consistently both in LBS and Zobell medium (Figures 2B,C).
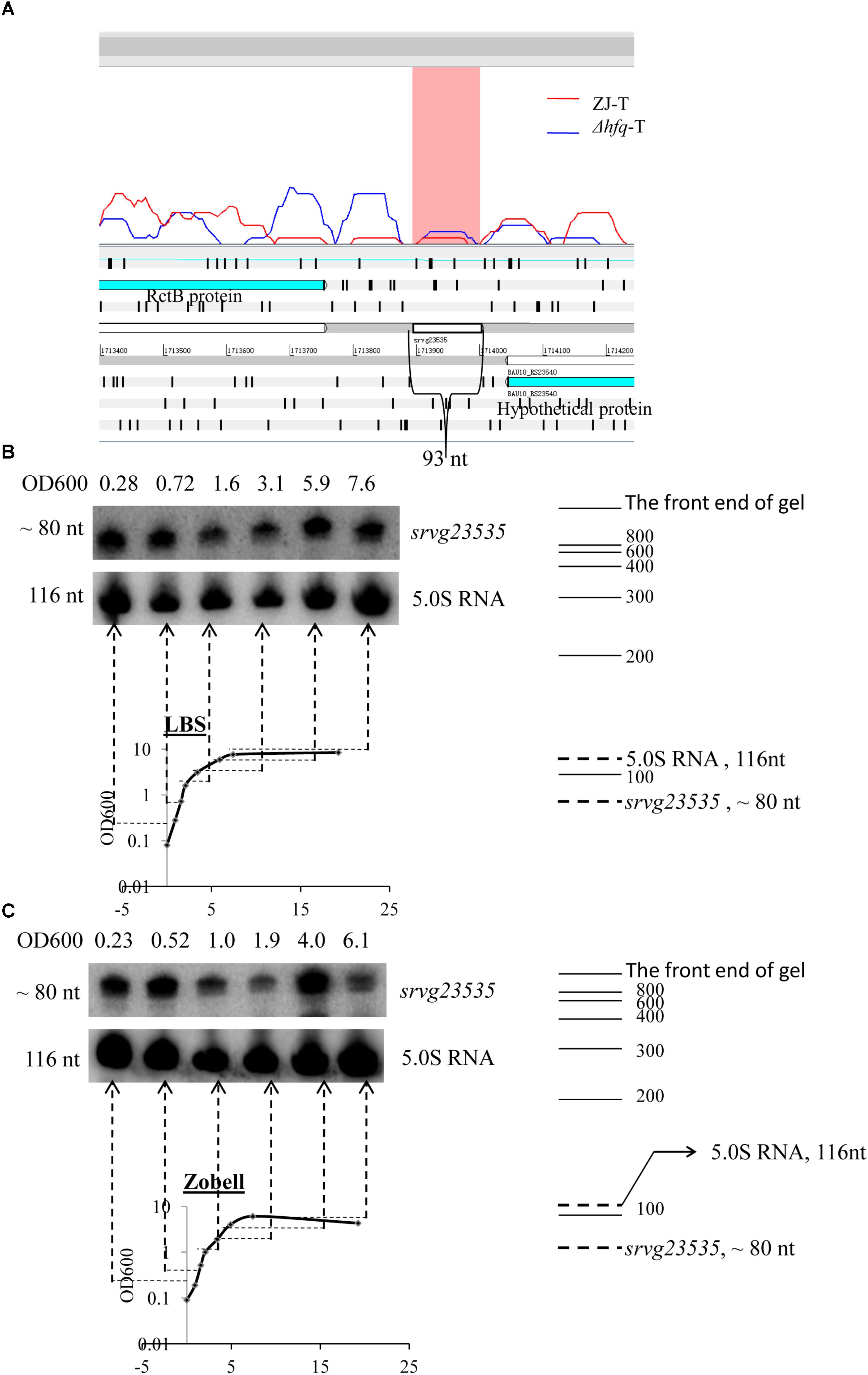
FIGURE 2. Locus feature and the expression of srvg23535. (A) An Artemis window shows the expression of srvg23535 flanking by rctB and an hp gene. The data was adapted from the RNA-seq results (Yiqin et al., 2016). (B) Northern blot of srvg23535 in LBS medium. The culture OD600 for each sample is indicated above each lane and the growth curve is presented underneath. The name of the sRNA is indicated on the right and the approximate size of the bands (determined by comparison with the position of RNA markers of known size) is indicated on the left. 5.0S RNA was used as a loading control. (C) Northern blot of srvg23535 in Zobell medium. The legend is the same as (B).
Transcriptional Start and End Points of srvg23535
Two bands were detected for 5′ RACE and one band was found for 3′ RACE (Figure 3A and Supplementary Figure S1). However, the sequence results showed that the larger band of 5′ RACE was by-product and non-specific product, and only one transcript was defined by RACE (Figure 3B), which was consistent with the results obtained from Northern blotting. The most abundant srvg23535 was 107 nt in length (Figure 3B), which is bigger than that deduced from RNA-seq (93 nt) and estimated by Northern blotting (around 80 nt).
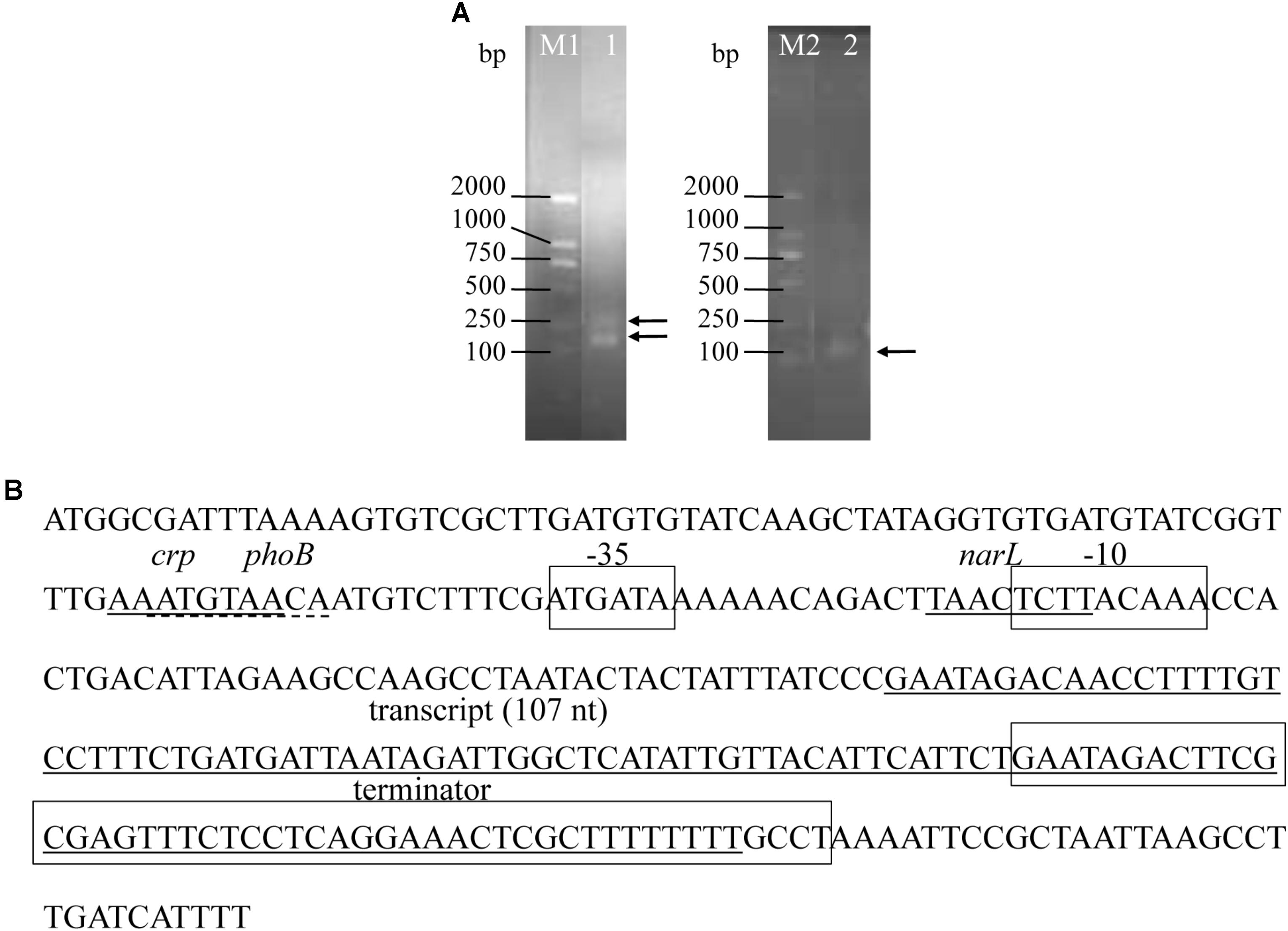
FIGURE 3. Precise transcriptional start and end points of srvg23535 were determined by the RACE experiments, and the promoter and terminator were predicated. (A) PCR results of 5′- and 3′-RACE. The bands were indicated by arrows. Lane M1: DNA Marker DL2000, it was cropped from Supplementary Figure S1A lane M; Lane 1: 5′-RACE of srvg23535, it was cropped from Supplementary Figure S1A lane 3; Lane M2: DNA Marker DL2000, it was cropped from Supplementary Figure S1B lane M; Lane 2: 3′-RACE of srvg23535, it was cropped from Supplementary Figure S1B lane 6. (B) Transcript size of srvg23535 was 107 nt. The first solid line covers the sequence of “AAATGTAA,” and indicates the crp transcriptional regulate factor; the first dashed line covers the sequence of “ATGTAACA,” and indicates the phoB transcriptional regulate factor; the second solid line covers the sequence of “TAACTCTT,” and indicates the narL transcriptional regulate factor; the third solid line covers the sequence form “GAATAGACAA” to “GCTTTTTTTT,” and indicates the total transcript of srvg23535; the first frame covers the sequence of “ATGATA,” and indicates the –35 box; the second frame covers the sequence of “TCTTACAAA,” and indicates the –10 box; the third frame covers the sequence from “GAATAGACTT” to “TTTTTTGCCT,” and indicates terminator of srvg23535.
Promoter, Terminator, and Novelty Analysis
One sigma70 promoter was predicated by BPROM and one Rho-independent terminator was predicated by ARNold (Figure 3B). Apart from the -10 and -35 boxes, three (a crp, a phoB, and a narL) transcriptional regulatory factors were predicted in the promoter (Figure 3B). Novelty assessment by blasting into the Rfam database showed that the sRNA was found first.
The Secondary Structure and the Conservation of srvg23535
A total of four stem-loop structures with a dG of -31.85 kJ may facilitate the high stability of srvg23535 (Figure 4A). Stem-loop 4, which probably acts as a putative transcriptional terminator, was predicted near the 3′ end region of srvg23535. The multi-sequence alignment of srvg23535 against representative genomes of microbes indicated that the sequences of stem-loop 1 and 3 had high similarity to other homologs. However, the transcript sequences were divided into four clusters by the sequence of stem-loop 2 and 4, which were consistent with the phylogenetic tree (Figure 4B). Furthermore, the phylogenetic tree showed that the srvg23535 was conserved among V. harveyi, Vibrio parahaemolyticus, and Vibrio splendidus, but not present in Vibrio tapetis, Vibrio aesturianus, V. cholerae or in species outside the Vibrio (Figure 4C).
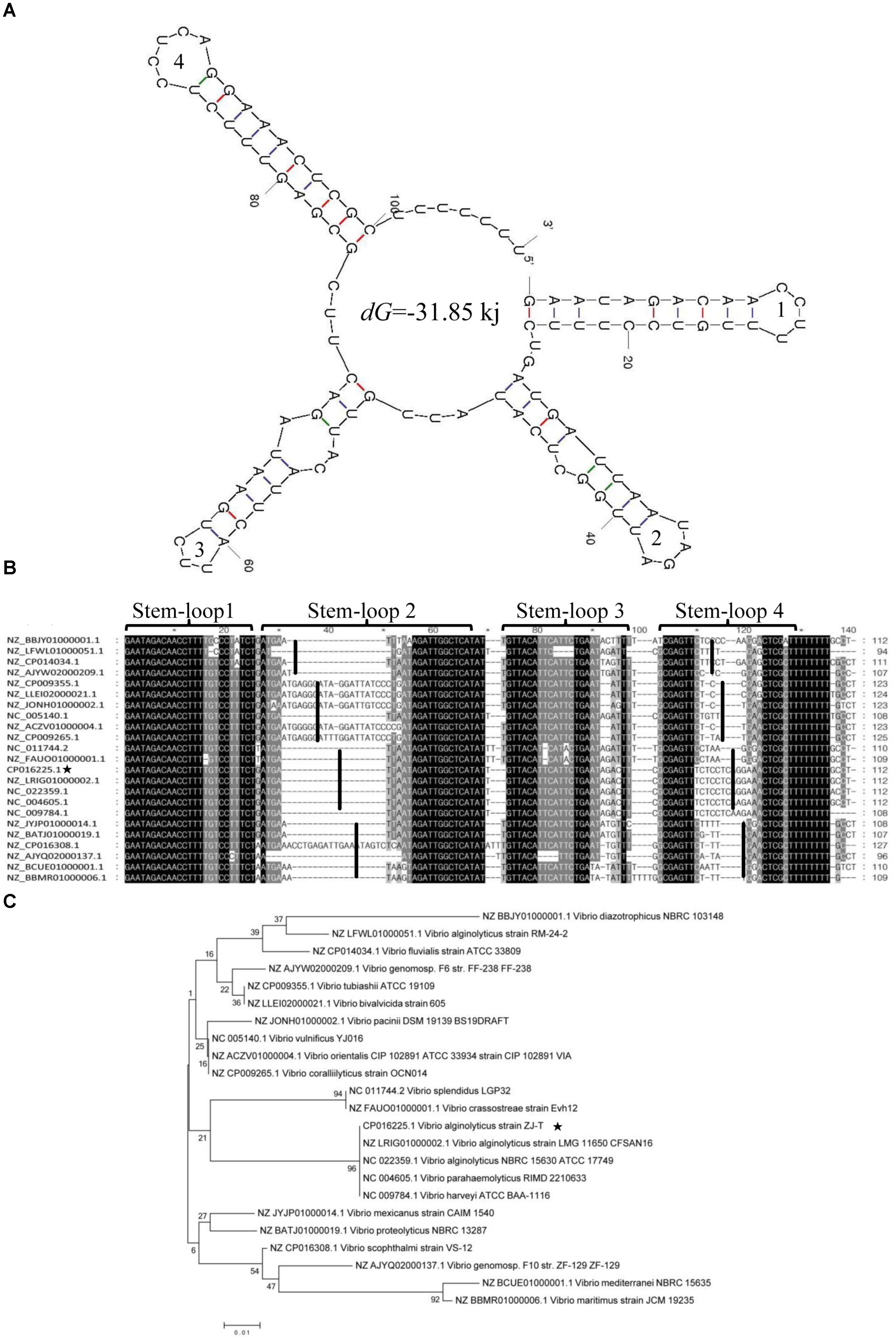
FIGURE 4. Secondary structure, multi-sequence alignment and phylogenetic analysis of srvg23535. (A) The srvg23535 was consisted of four stem-loop structures with a dG of –31.85 kJ, and the stem-loops were numbered. (B) The multi-sequence alignment result. The sequences were divided into four clusters by stem-loop 2 and 4. The sequences of stem-loop 1 and 3 were highly similar to other homologs. (C) The phylogenetic tree of srvg23535 with representative microbes aligned by using ClustalW. The numbers on the branch points mean the relative bootstrap (%, Bootstrap values ÷ 1000 × 100). The bar indicates 0.01 substitutions per sequence position.
sRNA Target Prediction
A total of 22 common targets were identified under the parameters set (Table 2 and Supplementary Table S3). Then we parsed the predicted mRNA targets based on their GO annotation and KEGG pathway. The results showed that the 22 targets were involved in a series of cellular processes, including metabolism, virulence, transport, and bacterial chemotaxis (Supplementary Table S3). However, six of the target genes were categorized as “hypothetical proteins” (Table 2 and Supplementary Table S3).
Construction of srvg23535 Null Mutant
A gene disruptant of srvg23535, from 46 bp before the 5′ end to 2 bp after the 3′ end, was constructed (Figure 1A). It was important to verify whether the deletion had a polar effect on the neighboring genes, thus we checked the expression by qPCR of all three genes (upstream, srvg23535, and the downstream gene) in wild type and ZJ-T-Δsrvg23535. As shown in Figure 1B, a significant difference of expression was observed only in the case of srvg23535: expectedly expression could not be detected in the mutant.
Functional Analysis of srvg23535
In total, 95 different carbon sources (PM01), 95 nitrogen sources (PM03), and tolerances to different osmolytes (PM09) and pH (PM10) were detected to identify the comprehensive role of srvg23535 by using PM technology. General results are shown in Supplementary Figures S2–S5, respectively. Significantly different phenotypes between the wild type and the mutant strain are described in the form of a heat map shown in Figure 5. The heat map was created with OD590 of the maximum of different time points. The details of phenotype differences are shown in Supplementary Table S4 and their original data is shown in Supplementary File S2.
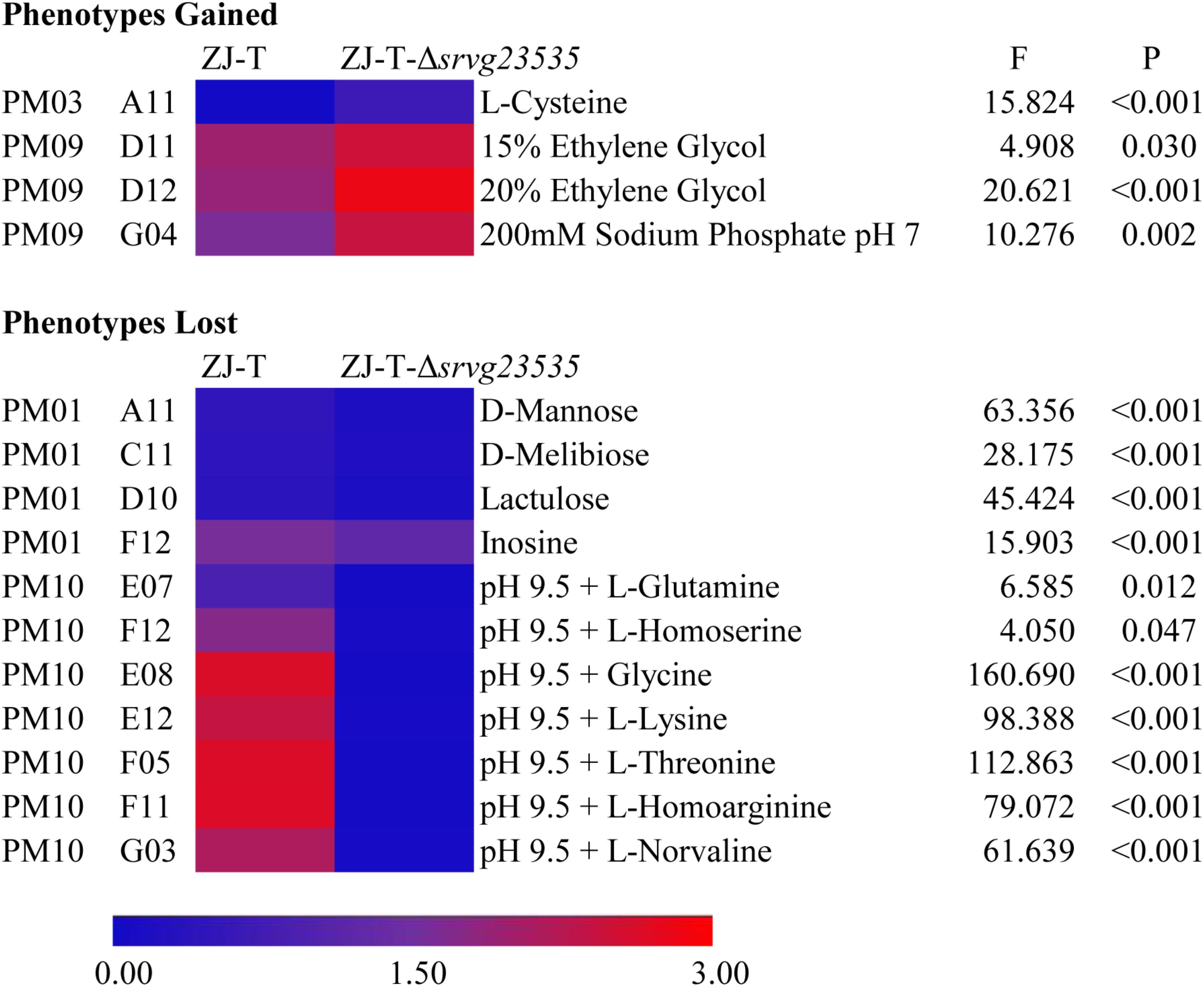
FIGURE 5. Gained and lost phenotypes in the absence of srvg23535 are described with a heat map (F- and P-values correspond to the results of one-way ANCOVA).
Carbon Utilization
Both the wild type ZJ-T and the srvg23535 mutant strain ZJ-T-Δsrvg23535 were able to utilize 60.0% (57/95) of the tested carbon sources. Compared to the wild type, the mutant has a longer lag phase or lost viability during stationary phase when bacteria were cultured with D-mannose, D-melibiose, lactulose or inosine (PM01 wells A11, C11, D10, and F12, respectively; one-way ANCOVA, F = 63.356, 28.175, 45.424, 15.903, respectively, and all P < 0.001) as carbon sources (Figure 5, Supplementary Figure S2, and Supplementary Table S4).
Nitrogen Utilization
Generally, ZJ-T and ZJ-T-Δsrvg23535 almost showed the same availability and utilization of the tested nitrogen sources in PM03, except that ZJ-T-Δsrvg23535 gained sharp growth after 3 days with L-cysteine as a nitrogen source (PM03 well A11; one-way ANCOVA, F = 15.824 and P < 0.001), whereas ZJ-T could not grow (Figure 5, Supplementary Figure S3, and Supplementary Table S4).
Osmolytes Tolerance
The PM09 data showed that as the concentration of ethylene glycol increased (PM09 wells D09 to D12), ZJ-T-Δsrvg23535 obtained a higher viability at stationary phase, showing a stronger tolerance to ethylene glycol, especially to 15% and 20% ethylene glycol (PM09 well D11 and D12; one-way ANCOVA, F = 4.908 and 20.621, respectively, and P = 0.030 and <0.001, respectively) (Figure 5 and Supplementary Table S4). The mutant obtained a higher viability at stationary phase when exposed to 200 mM sodium phosphate at a pH = 7 (PM09 well G04; one-way ANCOVA, F = 10.276, and P = 0.002) (Figure 5 and Supplementary Table S4).
Survival in Different pH Environments
PM10 data indicated that, compared to the wild type, the mutant had a longer lag phase when grown at pH 9.5 with L-glutamine or L-homoserine (PM10 wells E07 and F12, respectively; one-way ANCOVA, F = 6.585 and 4.050, respectively, and P = 0.012 and 0.047, respectively) (Figure 5 and Supplementary Table S4), and totally lost growth at pH 9.5 with glycine, L-lysine, L-threonine, L-homoarginine or L-norvaline (PM10 wells E08, E12, F05, F11, G03, respectively; one-way ANCOVA, F = 160.690, 98.388, 112.863, 79.072, and 61.639, respectively, and all P < 0.001) (Supplementary Figure S5 and Supplementary Table S4).
Discussion
We used an RNA-seq strategy to identify candidate sRNAs of V. aglinolyticus ZJ-T and successfully identified srvg23535. Northern blotting showed that srvg23535 was around 80 nt and displayed growth-phase-independent expression both in LBS and Zobell medium, indicating that srvg23535 would not be functional phase-dependent probably without involving quorum sensing and biofilm formation (Nguyen and Jacq, 2014). Subsequently, the precise boundary of srvg23535 was determined by using 5′ and 3′ RACE which identified a 107 nt transcript. The smaller transcript size estimated from Northern blot is probably the mature form of srvg23535 since RNA processing plays a critical role in gene regulation (Duan et al., 2007). Or, it may probably be caused by the size calculation method of Northern blot which has limitation in measure of low or high molecular weight of fragments (Bishop et al., 1967; Gough and Gough, 1984). The Rfam database is a collection of RNA families (Kalvari et al., 2017), which can be used to assess the novelty of sRNAs. By this approach, srvg23535 was showed to be first found, leading it more valuable to be characterized.
It has become clear that sRNA can have a conserved sequence and structure among homologs, resulting in a comprehensive identification of sRNAs by using comparative genomics (Peer and Margalit, 2011). According to the transcript determined by RACE, the sRNA conservation was analyzed with a phylogenetic tree. The result showed that srvg23535 is conserved among several Vibrio species, such as V. harveyi, V. parahaemolyticus, and V. splendidus, but not found in V. tapetis, V. aesturianus, and V. cholerae or outside the Vibrio spp. Similarly, Tsai et al. (2014) have identified seven novel sRNAs in Deinococcus radiodurans and conservation analysis confirmed that the sRNAs were restricted to the Deinococcaceae family. However, although sRNAs have a similar sequence, they may perform different roles in different species with low similarity in the target-binding regions (Peer and Margalit, 2011). Thereby, srvg23535 is identified among several Vibrio spp., the sequence and the stem-loop structures are divided into four clusters probably relating to different functions in different species.
Based on the transcript sequence, the promoter was characterized. Three transcriptional regulatory factors (a crp, a phoB, and a narL) were predicted in the sigma70 promoter, indicating that srvg23535 could be activated by cAMP, phosphorylation, and nitrite and nitrate ions. It has been shown that the cAMP receptor protein (Crp) binds to cAMP, which causes a conformational change that allows Crp to bind tightly to the crp site in the promoter of the genes it controls. These genes are primarily involved in the transport and catabolism of sugars and amino acids (De and Gottesman, 2009). De and Gottesman (2009) showed that CyaR sRNA repressed the expression of ompX, yqaE, nadE, and luxS via Crp activation, providing a link between catabolite repression, quorum sensing, and nitrogen assimilation in E. coli. PhoB is a response regulator of the two-component signal transduction system. When PhoB is activated by phosphorylation, it binds to a pho box, such as phoB, in the promoter thereby promoting the transcription of genes whose products are involved in phosphorus uptake and metabolism (Neidhardt, 1996). NarL is triggered by nitrite and nitrate ions. Phosphorylated NarL binds to specific sequences within target promoters to regulate transcription initiation (Browning et al., 2004). These results suggest that srvg23535 is probably involved in the transport and metabolism of carbon, nitrogen, and phosphorus sources.
Phenotypic regulation of srvg23535 was systematically analyzed using PM technology between the wild type and the deletion mutant strain. In addition, target prediction was conducted to link phenotypic regulation and genetic regulation. sRNAs have been extensively reported to be involved in a variety of cellular processes (Gripenland et al., 2010; Papenfort and Vogel, 2014; Wagner and Romby, 2015). In this study, we found that the srvg23535 mutant displayed growth defects when bacteria were cultured with D-mannose, D-melibiose, lactulose or inosine as the carbon sources. The absence of srvg23535 may affect the transport or/and the degradation of these carbon sources. However, only a maltose acetyltransferase coding gene maa (BAU10_18400) was predicated as the common target involved in carbon metabolism. The mutant gained growth sharply after 3 days when the bacteria were cultured with L-cysteine as a nitrogen source, whereas the wild type could not grow in this condition. Consistent with such a phenotype, the target gene was predicted to be araC (BAU10_22390), which encodes an aminotransferase and is involved in L-cysteine lyase. In addition, the regulation of the utilization of carbon and nitrogen sources may also relate to the promoter feature as described before. During invasion and survival in the host, bacteria are exposed to different conditions (Ulitzur, 1975). A total of 44 sRNAs in Geobacillus thermoleovorans were found to be involved in high-temperature adaptation by Tan and Alam (2010). Here, we found that the srvg23535 mutant showed a stronger tolerance to 15% ethylene glycol, 20% ethylene glycol, and 200 mM sodium phosphate at pH 7. However, compared to the wild type, the mutant had a weaker survival or even totally lost growth at pH 9.5 with L-glutamine, L-homoserine, glycine, L-lysine, L-threonine, L-homoarginine or L-norvaline. The two-component system and ABC transporter system are widely reported to regulate osmotic stress (Mizuno and Mizushima, 1990; Yan et al., 2013). Additionally, the two-component system is also reported to be involved in pH stress (Nakayama and Watanabe, 1995). Three genes, including an ABC transporter permease encoding gene (BAU10_15400, ABC transporter system), a chemotaxis protein-encoding gene (BAU10_22080, two-component system), and a histidine kinase encoding gene (BAU10_22015, two-component system) were predicted as being regulated by srvg23535 and probably contributed to a stronger resistance to ethylene glycol osmotic stress and phosphate toxicity, and a weaker survival in different pH environments. Furthermore, exsE, a type III secretion regulator, was predicated as the target of srvg23535 which indicates that srvg23535 is probably involved in the pathogenicity of V. alginolyticus. ExsE has been reported as a regulator of the expression of type III secretion system, which generally contributes to the adhesion in pathogens (Rietsch et al., 2005; Erwin et al., 2012; Liu J. et al., 2016). In Vibrio species, including V. alginolyticus, exsE has been suggested to be a negative regulator of T3SS expression (Erwin et al., 2012; Liu J. et al., 2016).
Pathogens must adapt to various conditions to invade and survive in the host with a series of strategies by precise gene expression (Wagner and Romby, 2015). The regulation of metabolism and stress response are reported to be two main strategies of bacterial environmental adaptation (Yiqin et al., 2016). For example, prfA was reported to regulate glucose-1-phosphate utilization which affected the virulence of Listeria monocytogenes (Makarova et al., 2011). Salmonella spp. have evolved a variety of acid resistance mechanisms, including acid tolerance response (Van, 2010), acid resistance (Handa et al., 2000), and changes in cell membrane composition (Scherzinger et al., 1991) to survive in acid conditions, which affect their virulence. Therefore, these results supply new evidence in the study of the relationship between environmental adaptation and virulence mechanisms by the regulation of sRNA in V. alginolyticus. However, more work needs to be done to find the precise target genes of srvg23535 and to fully understand the regulatory mechanism, thereby providing new antibacterial therapeutic targets or attenuated vaccine targets against Vibrio spp.
Conclusion
This study comprehensively identified and characterized a novel sRNA srvg23535 in V. alginolyticus. It is conserved among several Vibrio spp. with four stem-loop structures. Furthermore, according to the PM analysis, srvg23535 affects the utilization of several carbon and nitrogen sources and even the response to some stress conditions. Although target prediction was conducted, not all the predicted targets are consistent with the phenotypes. As a matter of future study, we recommend that the interaction of srvg23535 and the predicted target genes are verified, and their expression is compared by RNA-seq to assess the genotypic regulation.
Author Contributions
YD conceived the study, analyzed the data, and wrote the manuscript. CHC, HM, and JW performed the experiments. YS, SL, and ZG critically revised the manuscript. JF and CC contributed the reagents. All authors read and approved the manuscript for publication.
Funding
This work was supported by grants from Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2017YB27 and 2017YB01), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2017HY-ZD1007), the Special Fund for Strategic Pilot Technology of Chinese Academy of Sciences (XDA13020302), the China Agriculture Research System (CARS-48), and National Natural Science Foundation of China (31502210).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Annick Jacq for kindly helping in the experimental operation, International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript and the Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences for partial funding.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02394/full#supplementary-material
FIGURE S1 | The original gels of RACE results. (A) PCR results of 5′-RACE. Lane M: DNA Marker DL2000; Lane 3: 5′-RACE of srvg23535; Remaining lanes are 5′-RACE results of other sRNAs those have been done at the same time. (B) PCR results of 3′-RACE. Lane M: DNA Marker DL2000; Lane 6: 3′-RACE of srvg23535; Remaining lanes are 3′-RACE results of other sRNAs those have been done at the same time.
FIGURE S2 |(A–H) Effect of srvg23535 deletion on the utilization of 95 various carbon sources which were test by the PM01 plate (the results are represented as the average values, n = 3).
FIGURE S3 |(A–H) Effect of srvg23535 deletion on the utilization of 95 various nitrogen sources which were test by the PM03 plate (the results are represented as the average values, n = 3).
FIGURE S4 |(A–H) Effect of srvg23535 deletion on the growth responses to osmolytes stress which were test by the PM09 plate (the results are represented as the average values, n = 3).
FIGURE S5 |(A–H) Effect of srvg23535 deletion on the growth responses to different pH stress which were test by the PM10 plate (the results are represented as the average values, n = 3).
TABLE S1 | Nucleotide sequences of primers or probes used in this study.
TABLE S2 | Tested components of the PM01, PM03, PM09, and PM10 plates.
TABLE S3 | Details of common targets predicted by IntaRNA and target RNA2.
TABLE S4 | Details of phenotype differences generated by PM technology (the results of the graphs are represented as the average ± SE, n = 3).
FILE S1 | PM Procedures for E. coli and other GN Bacteria.
FILE S2 | The original data of phenotype differences.
References
Argaman, L., Hershberg, R., Vogel, J., Bejerano, G., Wagner, E. G., Margalit, H., et al. (2001). Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11, 941–950. doi: 10.1016/S0960-9822(01)00270-6
Bishop, D. H., Claybrook, J. R., and Spiegelman, S. (1967). Electrophoretic separation of viral nucleic acids on polyacrylamide gels. J. Mol. Biol. 26, 373–387. doi: 10.1016/0022-2836(67)90310-5
Blumenstein, K., Macayasanz, D., Martín, J. A., Albrectsen, B. R., and Witzell, J. (2015). Phenotype microarrays as a complementary tool to next generation sequencing for characterization of tree endophytes. Front. Microbiol. 6:1033. doi: 10.3389/fmicb.2015.01033
Bochner, B. (2009). Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33, 191–205. doi: 10.1016/j.apmr.2008.10.013
Bochner, B. R., Gadzinski, P., and Panomitros, E. (2001). Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11, 1246–1255. doi: 10.1101/gr.186501
Browning, D. F., Cole, J. A., and Busby, S. J. (2004). Transcription activation by remodelling of a nucleoprotein assembly: the role of NarL at the FNR-dependent Escherichia coli nir promoter. Mol. Microbiol. 53, 203–215. doi: 10.1006/jmaa.1999.6641
Chang, C., Jin, X., and Chaoqun, H. (2009). Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling 25, 525–531. doi: 10.1080/08927010902964578
Chen, S., Lesnik, E. A., Hall, T. A., Sampath, R., Griffey, R. H., Ecker, D. J., et al. (2002). A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65, 157–177. doi: 10.1016/S0303-2647(02)00013-8
Citil, B. E., Derin, S., Sankur, F., Sahan, M., and Citil, M. U. (2015). Vibrio alginolyticus associated chronic myringitis acquired in mediterranean waters of turkey. Case Rep. Infect. Dis. 2015:187212–187215. doi: 10.1155/2015/187212
Davis, B. M., and Waldor, M. K. (2007). RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 65, 373–385. doi: 10.1111/j.1365-2958.2007.05796.x
De, L. N., and Gottesman, S. (2009). The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191, 461–476. doi: 10.1128/JB.01157-08
Duan, R., Pak, C., and Jin, P. (2007). Single nucleotide polymorphism associated with mature mir-125a alters the processing of pri-mirna. Hum. Mol. Genet. 16, 1124–1131. doi: 10.1093/hmg/ddm062
Erwin, D. P., Nydam, S. D., and Call, D. R. (2012). Vibrio parahaemolyticus ExsE is requisite for initial adhesion and subsequent type III secretion system 1-dependent autophagy in Hela cells. Microbiology 158, 2303–2314. doi: 10.1099/mic.0.059931-0
Gough, E. J., and Gough, N. M. (1984). Diect calaon of the szes of DNA fragments separated by gel electrophoresis using programmes witten for a pocket caculator. Nucleic Acids Res. 12, 845–853. doi: 10.1093/nar/12.1Part2.845
Griffithsjones, S., Bateman, A., Marshall, M., Khanna, A., and Eddy, S. R. (2003). Rfam: an RNA family database. Nucleic Acids Res. 31, 439–441. doi: 10.1093/nar/gkg006
Gripenland, J., Netterling, S., Loh, E., Tiensuu, T., Toledoarana, A., and Johansson, J. (2010). RNAs: regulators of bacterial virulence. Nat. Rev. Microbiol. 8, 857–866. doi: 10.1038/nrmicro2457
Handa, N., Ichige, A., Kusano, K., and Kobayashi, I. (2000). Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182, 2218–2229. doi: 10.1128/JB.182.8.2218-2229.2000
Iustman, L. J. R., Tribelli, P. M., Ibarra, J. G., Catone, M. V., Venero, E. C. S., and López, N. I. (2015). Genome sequence analysis of Pseudomonas extremaustralis provides new insights into environmental adaptability and extreme conditions resistance. Extremophiles 19, 207–220. doi: 10.1007/s00792-014-0700-7
Kalvari, I., Argasinska, J., Quinonesolvera, N., Nawrocki, E. P., Rivas, E., Eddy, S. R., et al. (2017). Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342. doi: 10.1093/nar/gkx1038
Kery, M. B., Feldman, M., Livny, J., and Tjaden, B. (2014). TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 42, W124–W129. doi: 10.1093/nar/gku317
Khatri, B., Fielder, M., Jones, G., Newell, W., Abuoun, M., and Wheeler, P. R. (2013). High throughput phenotypic analysis of Mycobacterium tuberculosis and Mycobacterium bovis strains’ metabolism using biolog phenotype microarrays. PLoS One 8:e52673. doi: 10.1371/journal.pone.0052673
Le Roux, F., Binesse, J., Saulnier, D., and Mazel, D. (2007). Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73, 777–784. doi: 10.1128/AEM.02147-06
Lesnik, E. A., Sampath, R., Levene, H. B., Henderson, T. J., Mcneil, J. A., and Ecker, D. J. (2001). Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 29, 3583–3594. doi: 10.1093/nar/29.17.3583
Liu, J., Lu, S. Y., Orfe, L. H., Ren, C. H., Hu, C. Q., Call, D. R., et al. (2016). Exse is a negative regulator for t3ss gene expression in vibrio alginolyticus. Front. Cell. Infect. Microbiol. 6:177. doi: 10.3389/fcimb.2016.00177
Liu, W., Bertrand, M., Chaneac, C., and Achouak, W. (2016). TiO2 nanoparticles alter iron homeostasis in Pseudomonas brassicacearum as revealed by PrrF sRNA modulation. Environ. Sci. Nano 3, 1473–1482. doi: 10.1039/C6EN00316H
Liu, J. M., and Camilli, A. (2010). A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13, 18–23. doi: 10.1016/j.mib.2009.11.004
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livny, J., and Waldor, M. K. (2007). Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 10, 96–101. doi: 10.1016/j.mib.2007.03.005
Loris, R., Dao-Thi, M. H., Bahassi, E. M., Melderen, L. V., Poortmans, F., Liddington, R., et al. (1999). Crystal structure of CcdB, a topoisomerase poison from E. coli 1. J. Mol. Biol. 285, 1667–1677. doi: 10.1006/jmbi.1998.2395
Luo, G., Huang, L., Su, Y., Qin, Y., Xu, X., Zhao, L., et al. (2016). FlrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 5:e85. doi: 10.1038/emi.2016.82
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Mey, A. R., Craig, S. A., and Payne, S. M. (2005). Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73, 5706–5719. doi: 10.1128/IAI.73.9.5709-5719.2005
Miller, R. V. (1998). Bacterial gene swapping in nature. Sci. Am. 278, 66–71. doi: 10.1038/scientificamerican0198-66
Mizuno, T., and Mizushima, S. (1990). Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mole Microbiol. 4, 1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x
Nakayama, S., and Watanabe, H. (1995). Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177, 5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995
Neidhardt, F. C. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM Press.
Nguyen, A. N., Disconzi, E., Charrière, G. M., Destoumieux-Garzón, D., Bouloc, P., Le Roux, F., et al. (2018). csrB gene duplication drives the evolution of redundant regulatory pathways controlling expression of the major toxic secreted metalloproteases in Vibrio tasmaniensis LGP32. mSphere 3:e00582-18. doi: 10.1128/mSphere.00582-18
Nguyen, A. N., and Jacq, A. (2014). Small RNAs in the Vibrionaceae: an ocean still to be explored. Wiley Interdiscip. Rev. RNA 5, 381–392. doi: 10.1002/wrna.1218
Papenfort, K., and Vogel, J. (2014). Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front. Cell. Infect. Microbiol. 4:91. doi: 10.3389/fcimb.2014.00091
Peer, A., and Margalit, H. (2011). Accessibility and evolutionary conservation mark bacterial small-rna target-binding regions. J. Bacteriol. 193, 1690–1701. doi: 10.1128/JB.01419-10
Ren, C., Hu, C., Jiang, X., Sun, H., Zhao, Z., Chen, C., et al. (2013). Distribution and pathogenic relationship of virulence associated genes among Vibrio alginolyticus from the mariculture systems. Mol. Cell. Probes 27, 164–168. doi: 10.1016/j.mcp.2013.01.004
Rietsch, A., Vallet-Gely, I., Dove, S. L., and Mekalanos, J. J. (2005). ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011. doi: 10.1073/pnas.0503005102
Roop, R. M., Gaines, J. M., Anderson, E. S., Caswell, C. C., and Martin, D. W. (2009). Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med. Microbiol. Immun. 198, 221–238. doi: 10.1007/s00430-009-0123-8
Ruprecht, V., Monzo, P., Ravasio, A., Yue, Z., Makhija, E., Strale, P. O., et al. (2016). How cells respond to environmental cues-insights from bio-functionalized substrates. J. Cell Sci. 130, 51–61. doi: 10.1242/jcs.196162
Scherzinger, E., Haring, V., Lurz, R., and Otto, S. (1991). Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA. RepB and RepC proteins. Nucleic Acids Res. 19, 1203–1211. doi: 10.1093/nar/19.6.1203
Sharma, C. M., and Vogel, J. (2009). Experimental approaches for the discovery and characterization of regulatory small RNA. Curr. Opin. Microbiol. 12, 536–546. doi: 10.1016/j.mib.2009.07.006
Siddiqui, R., Alam, M. M., Naser, M. N., Otomo, Y., Yasmin, M., Nessa, J., et al. (2016). Prevalence of Vibrio alginolyticus in sediment samples of river and coastal areas of Bangladesh. Bangladesh J. Microbiol. 29, 1–6. doi: 10.3329/bjm.v29i1.28421
Siqueira, F. M., Morais, G. L. D., Higashi, S., Beier, L. S., Breyer, G. M., Godinho, C. P. D. S., et al. (2016). Mycoplasma non-coding RNA: identification of small RNAs and targets. BMC Genomics 17:743. doi: 10.1186/s12864-016-3061-z
Solovyev, V., and Salamov, A. (2011). in Automatic Annotation of Microbial Genomes and Metagenomic Sequences, ed. W. L. Robert (NewYork, NY: Nova Science press), 1–18.
Tan, H. S., and Alam, M. (2010). sRNAs are Involved in High Temperature Adaptation of Geobacillus thermoleovorans. Heidelberg: EMBO Symposium Non-coding genome.
Toffano-Nioche, C., Nguyen, A. N., Kuchly, C., Ott, A., Gautheret, D., Bouloc, P., et al. (2012). Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus. RNA 18, 2201–2219. doi: 10.1261/rna.033324.112
Tohsato, Y., and Mori, H. (2008). Phenotype profiling of single gene deletion mutants of E. coli using biolog technology. Genome Inform. 21, 42–52.
Tsai, C., Liao, R., Chou, B., and Contreras, L. M. (2014). Transcriptional analysis of Deinococcus radiodurans reveal novel srnas that are differentially expressed under ionizing radiation. Appl. Environ. Microb. 81, 1754–1764. doi: 10.1128/AEM.03709-14
Tu, K. C., and Bassler, B. L. (2007). Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 21, 221–233. doi: 10.1101/gad.1502407
Ulitzur, S. (1975). Effect of temperature, salts, pH, and other factors on the development of peritrichous flagella in Vibrio alginolyticus. Arch. Microbiol. 104, 285–288. doi: 10.1007/BF00447338
Val, M.-E., Skovgaard, O., Ducos-Galand, M., Bland, M. J., and Mazel, D. (2012). Genome engineering in Vibrio cholerae: a feasible approach to address biological issues. PLoS Genet. 8:e1002472. doi: 10.1371/journal.pgen.1002472
Van, M. L. (2010). Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13, 781–785. doi: 10.1016/j.mib.2010.10.006
Wagner, E. G., and Romby, P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. doi: 10.1016/bs.adgen.2015.05.001
Xiaochun, H., Chang, C., Chunhua, R., Yingying, L., Yiqin, D., Yiying, Y., et al. (2017). Identification and characterization of a locus putatively involved in colanic acid biosynthesis in Vibrio alginolyticus ZJ-51. Biofouling 34, 1–14. doi: 10.1080/08927014.2017.1400020
Xiong, P., Peng, X., Wei, S., Chen, Y., Zhao, H., Tang, S., et al. (2014). Virulence-related genes of Vibrio alginolyticus and its virulence in mice. Wei Sheng Wu Xue Bao 54, 80–88.
Yan, Q., Power, K. A., Shane, C., Edward, F., Gopinath, G. R., Grim, C. J., et al. (2013). Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front. Microbiol. 4:256. doi: 10.3389/fmicb.2013.00256
Keywords: small non-coding RNAs, srvg23535, Vibrio alginolyticus, identification, Phenotype MicroArray technology
Citation: Deng Y, Su Y, Liu S, Guo Z, Cheng C, Ma H, Wu J, Feng J and Chen C (2018) Identification of a Novel Small RNA srvg23535 in Vibrio alginolyticus ZJ-T and Its Characterization With Phenotype MicroArray Technology. Front. Microbiol. 9:2394. doi: 10.3389/fmicb.2018.02394
Received: 07 July 2018; Accepted: 18 September 2018;
Published: 05 October 2018.
Edited by:
Eric Altermann, AgResearch, New ZealandReviewed by:
David R. Nelson, New York University, United StatesAkiko Soma, Chiba University, Japan
Copyright © 2018 Deng, Su, Liu, Guo, Cheng, Ma, Wu, Feng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Feng, anVhbmZlbmdAc2NzZnJpLmFjLmNu Chang Chen, Y2hlbi5jaGFuZ0BzY3Npby5hYy5jbg==
 Yiqin Deng
Yiqin Deng Youlu Su
Youlu Su Songlin Liu
Songlin Liu Zhixun Guo
Zhixun Guo Changhong Cheng1
Changhong Cheng1 Jinjun Wu
Jinjun Wu Juan Feng
Juan Feng