- 1Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Centre of Biotechnology and Microbiology, University of Peshawar, Peshawar, Pakistan
Wuyiencin is produced by Streptomyces albulus var. wuyiensis and used widely in agriculture to control a variety of fungal diseases, such as cucumber downy mildew, strawberry powdery mildew, and tomato gray mold. As an industrially-produced biopesticide, reducing production costs is very important for popularization of this approach. To obtain a rapidly growing strain that effectively shortens the fermentation time, we investigated the effects of knockout and overexpression of the wysPII gene, a member of the LuxR regulatory gene family, in S. albulus strain CK-15. The ΔwysPII mutant exhibited a reduced rate of growth and sporulation. The time taken to reach the greatest mycelial biomass was approximately 18 h shorter in the ooPII (wysPII overexpressing) strain compared with that of the wild-type (WT) strain. In addition, the time to reach the greatest wuyiencin production was 56 h in the ooPII strain compared with 62 h in the WT strain. Furthermore, wysPII was shown to act as an activator of morphological development without affecting wuyiencin production. Thus, the ooPII strain can be used to reduce costs and increase efficiency in industrial fermentation processes for wuyiencin production.
Introduction
Streptomyces are soil-dwelling, Gram-positive, filamentous bacteria, with a complex life cycle that involves formation of substrate mycelium, aerial mycelium, and spores (McCormick and Flardh, 2011). Many members of this bacterial lineage produce secondary metabolites including a variety of antibiotics (Bibb, 2005). Production of secondary metabolites is tightly regulated by complicated transcriptional processes involving multiple levels of regulation (Martín, 2004; Liu et al., 2013). Pathway-specific transcriptional regulation, which is the most fundamental level, is mediated by regulatory genes encoded within the respective biosynthetic gene clusters (Vicente et al., 2015). Some regulatory factors affect antibiotic production, while others affect morphological development and production of antibiotics via processes that are sometimes genetically coupled by shared transcriptional regulators (Hur et al., 2008; Ge et al., 2016). A few regulatory factors are involved only in morphological development, an example being MtrA, which is crucial for normal development of aerial hyphae of the model strain S. coelicolor (Cui et al., 2016; Zhang et al., 2017).
The LuxR family of regulators plays important roles in acyl-homoserine lactone-mediated quorum sensing in Gram-negative bacteria (Nasser and Sylvie, 2007), regulating many physiological processes, such as bioluminescence (Antunes et al., 2008), plasmid transfer (Danino et al., 2003), biofilm formation (Alonso-Hearn et al., 2010), as well as production of virulence factors (Passador et al., 1993), extracellular enzymes (Andersson et al., 2000), and secondary metabolites (Latifi et al., 1995). The LuxR superfamily comprises transcriptional regulators with a DNA-binding helix-turn-helix (HTH) motif in the C- and the N-terminal regions, and, frequently, an autoinducer-binding or response regulatory domain (De and Raaijmakers, 2009). HTH domains of approximately 65 amino acids are present in LuxR family transcriptional regulators. Most LuxR-type regulators activate transcription, but some function as repressors or have a dual role at different sites. These regulators control a large number of activities. LuxR-type HTH proteins can be activated by a two-component sensory transduction system in which the protein is activated by phosphorylation, normally on an aspartate residue (e.g., FixJ of Sinorhizobium meliloti) (Birck et al., 2002), or by binding to N-acyl-homoserine lactones (e.g., LuxR of Vibrio fischeri) (Fuqua et al., 1996). The LuxR/FixJ family also contains autonomous effector domain regulators, represented by GerE, which regulates spore formation in Bacillus subtilis (Ducros et al., 2001) and multiple ligand-binding regulators, exemplified by MalT in Escherichia coli, which functions as a maltose regulator (Schlegel et al., 2002). Most LuxR family members are identified as activators of antibiotic production and morphological development (Yu et al., 2012). LuxR family members include PimM, which positively regulates pimaricin biosynthesis in Streptomyces natalensis (Antón et al., 2007), NysRI, NysRII, and NysRIII, which regulate the S. noursei ATCC11455 nystatin biosynthetic pathway (Sekurova et al., 2004), TmcN, which is present in the tautomycetin biosynthetic pathway of Streptomyces sp. CK4412 (Hur et al., 2008), and GdmRI and GdmRII, which regulate the S. hygroscopicus 17997 geldanamycin biosynthetic pathway (He et al., 2008).
Wuyiencin is a nucleoside antibiotic produced by S. ahygroscopicus var. wuyiensis, first isolated from soil in the Wuyi Mountains, China (Wei et al., 1984). This strain was previously designated as S. ahygroscopicus, depending on presence or absence of hygroscopicity; however, based on further molecular identification and other characteristics, S. ahygroscopicus CK-15 was renamed S. albulus. As an efficient, broad-spectrum biological fungicide, wuyiencin is used in agriculture to control a variety of fungal diseases, such as cucumber downy mildew, strawberry powdery mildew, and tomato gray mold, with high efficiency and low toxicity compared with chemical pesticides (Zeng et al., 1996; Sun et al., 2003; Ge et al., 2015a). Wuyiencin meets national standards for organic food production (COFFC-R-0903-0070). Previously, we sequenced the genome of S. albulus var. wuyiensis strain CK-15 and analyzed the wuyiencin biosynthetic cluster (Ge et al., 2014, 2015b). WysR, a member of the PAS-LuxR family of regulators and present in this gene cluster, was identified as a transcriptional activator of wuyiencin biosynthesis. Overexpression of wysR increased the production of wuyiencin by almost threefold compared with the wild-type (WT) strain (Liu et al., 2014). WysR3, a novel member of the DeoR family of regulators, had no effect on antibiotic production, but acted as a repressor during morphological development. The ΔwysR3 strain grew faster than the WT, reaching the plateau stage of maximum biomass 12 h earlier in the flasks (Ge et al., 2016).
In this study, we characterized WysPII, a LuxR family protein encoded within the wuyiencin biosynthesis gene cluster. WysPII was shown to act as an activator during morphological development of S. albulus CK-15. Furthermore, a wysPII overexpression strain (ooPII) exhibited much more rapid wuyiencin production (shorter time to reach maximum) and increased growth compared with the WT strain. The ooPII strain is of significant industrial utility due to its potential to shorten the production process of wuyiencin by effectively reducing the time required for fermentation. These properties will promote the industrial application of wuyiencin.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Table 1 lists bacterial strains and plasmids used in this work. S. albulus var. wuyiensis strain CK-15 (China General Microbiological Culture Collection Center No. 0703; hereafter called S. wuyiensis CK-15) was cultured at 28°C on mannitol-soybean (MS) agar or in yeast extract malt extract (YEME) liquid medium (Hobbs et al., 1989). Fermentation medium contained (per 100 ml) 2 g soybean flour, 2 g glucose, 3 g corn starch, 300 mg CaCO3, and 400 mg (NH4)2SO4. E. coli DH5α was used for DNA manipulation and was grown in Luria–Bertani (LB) liquid broth or on LB-agar. The vector pEASY-T1-Simple was used for general cloning, pKC1139 was used as a disruption vector, and pSETC (Labes et al., 1997) for complementation and overexpression by standard procedures (Kieser, 2000; Liu et al., 2014). pSETC contains the constitutive and strong expression promoter PSF14 incorporated into the genome-integrating plasmid pSET152. Non-methylating E. coli strain ET12567/pUZ8002 was used for gene conjugation from E. coli to S. albulus CK-15. Rhodotorula rubra N-1 was used as the indicator strain in disk-agar wuyiencin diffusion assays. Apramycin (50 μg/ml) was used in LB medium as required.
DNA Manipulation and Sequence Analysis
Using a Streptomyces DNA Isolation Kit (GenMed Scientifics Inc., United States), total S. albulus CK-15 genomic DNA was isolated from mycelia after 72 h of cultivation on solid MS medium supplemented with 0.3% malt extract, 0.3% yeast extract, 0.5% tryptone, and 4% glucose, or from mycelia after 48 h of culture in YEME liquid medium. PCR amplification used 2×Taq PCR Mix (Bingda, China). The amplification conditions were: 95°C for 5 min; 32 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 2 min; and a final extension at 72°C for 10 min. Primers were designed using Primer Premier 5 and the DNA sequences were sequenced by BGI Tech (Beijing, China). DNAMAN software was used to compare nucleotide and protein sequences.
WysPII Deletion Mutant
A wysPII deletion mutant was generated by homologous recombination. Primers AF and AR were used to amplify the upstream region (1,504 bp) of wysPII (Table 2) and BF and BR to amplify the downstream region (1,357 bp). The resulting DNA fragments were ligated into pEASY-T1-Simple and digested with HindIII/XbaI or XbaI/EcoRI to yield plasmids pT1-A and pT1-B, respectively. The upstream and downstream region sequences were then ligated into pKC1139 to generate the disruption plasmid pKC1139-ΔwysPII, which was then transferred into S. albulus CK-15 by conjugation. Plates were incubated at 28°C for 16 h, then overlaid with sterile water (1 ml) containing 50 μg/ml apramycin and 25 μg/ml nalidixic acid. The plates were further incubated at 34°C until conjugants appeared. Double-crossover recombinants were selected on the basis of apramycin sensitivity followed by confirmation by PCR using primers TF and TR (Table 2).
Complementation of the wysPII Mutant
For complementation of wysPII disruptants, wysPII was amplified using the genomic DNA template with primers Com-F and Com-R. The fragment was digested with HindIII and EcoRI, and the product (2,856 bp) was ligated into the HindIII/EcoRI sites of pSETC to yield pSETC-wysPII. This plasmid was first transformed into E. coli ET12567 and then transferred into S. albulus CK-15 ΔwysPII by intergeneric conjugation to yield strain S. wuyiensis comPII. Apramycin-resistant transconjugants were confirmed by PCR. The phenotype of the resulting complementation strain was assessed by apramycin selection (50 μg/ml) on MYM medium.
Overexpression of wysPII
pSTEC-wysPII was introduced into WT S. albulus CK-15 by conjugation to yield the S. albulus ooPII strain. The empty vector pSETC was also transferred into WT S. albulus CK-15 as a control. Apramycin-resistant transconjugants were confirmed by PCR using primers Am-F and Am-R, and the phenotype of the resulting overexpression strain was determined as above.
Wuyiencin Production Assay
Fresh mature seed solution was inoculated with spores (1.0 × 106) in 50 ml YEME medium and incubated at 28°C for 20 h. Seed cultures were transferred into 100 ml 10% fermentation medium and incubated at 28°C with agitation (220 rpm) for 64 h. The fermentation broth was filtered using Millipore express membrane filters (pore diameter 0.22 μm; Merck kGaA Darmstadt, Germany). The antibacterial effects of the broth against R. rubra were tested by measuring the diameter of bacteriostasis circles.
For the actual wuyiencin production time-course analysis, fermentation broth was collected by filtration using Millipore express membrane filters (pore diameter 0.22 μm; Merck) at 2-h intervals for 72 h. Wuyiencin concentrations in the broth were also determined using an Agilent 1100 high-performance liquid chromatography (HPLC) system with an SB-AQ column (4.6 mm × 250 mm i.d., 5 μm) at 25°C. The mobile phase was 1.4 g/L C2HCl3O2 (flow rate 1.5 ml/min) with detection at 254 nm. The wuyiencin standard was prepared using the following series of purification steps: oxalic acid precipitation, macroporous resin HP-20 adsorption, HW-40C and HW-40F resin adsorption and lyophilization. Wuyiencin had a retention time of 15 min in these conditions.
Measurement of Mycelial Biomass
Fresh mature seed solution was inoculated into 50 ml M3G medium [10 g (NH4)2SO4, 50 g glucose, 5.0 g yeast powder, 1.36 g KH2PO4, 0.8 g K2PO4⋅3H2O, 0.5 g MgSO4⋅7H2O, 0.04 g ZnSO4⋅7H2O, and 0.03 g FeSO4⋅7H2O, per liter, pH 7.0] and incubated at 28°C with agitation at 220 rpm for 48 h to produce a seed culture. The seed culture for each strain was then inoculated at 2% into 100 ml M3G medium and incubated at 28°C (220 rpm). Samples were collected every 6 h and dried to constant weight at 90°C on filter papers. The difference in weight of the paper before and after filtration represented the dry weight of mycelium in the fermented liquid sample. The strains (WT, ΔwysPII, comPII and ooPII) were also cultured at 28°C on MS, ISP 2 (4 g glucose, 10 g malt infusion, 4 g yeast extract, 17 g agar, per liter, pH 7.0–7.4) and ISP 3(20 g oats, 0.5 g K2HPO4, 0.2 g KNO3, 0.2 g MgSO4, 17 g agar, per liter) media to evaluate differences in the mycelial biomass of these strains at specific time-points.
Isolation of Total RNA and Reverse Transcription-Quantitative PCR
The WT strain and ooPII strains were grown at 28°C on MS medium plate for 72 h. The sample was first ground in a pre-cooled mortar, frozen in liquid nitrogen and powdered before Trizol Reagent (CWBIO) was added. The Ultrapure RNA Kit (CWBIO) was used to isolate total RNA according to the manufacturer’s instructions. The crude RNA samples were treated with the DNase I (TIANGEN) to degrade the chromosomal DNA.
cDNA reverse transcription was performed using the PrimeScript™ RT reagent Kit(TaKaRa) with gDNA Eraser. The PCR amplification was performed with cDNA as template and primers (Table 2) using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus), under the following conditions: 95°C for 30 s followed 40 cycles at 95°C for 15 s, 60°C for 40 s, and 72°C for 30 s. hrdB was amplified as the internal control. All cultures were analyzed by real-time PCR, with each specimen was copied in triplicate and RNA isolated from three independent cultures.
Deposition of Nucleotide Sequence
The sequences reported in this study have been deposited in GenBank with accession no. MG871748.
Results
Sequence Analysis of the wysPII Gene
Streptomyces albulus CK-15 genome sequencing revealed that the wysPII gene product (WysPII; 952 amino acids, calculated molecular mass 101.82 kDa) showed 80.10% sequence identity with the S. noursei ATCC11455 protein NysRII (AF263912.1), which is a putative 953-amino acid regulatory protein encoded in the nystatin biosynthesis gene cluster. The putative protein sequence translated from the nucleotide was further analyzed using NCBI BLASTp showed 44.23% sequence identity of WysPII with another putative transcriptional regulator which has been reported in the tetramycin ttmRII biosynthesis gene cluster. WysPII has a HTH DNA-binding domain at its N-terminus, and its C-terminus is homologous to COG3899, which is predicted to be an ATPase. It also contains a MalT family domain, which regulates the mal regulon (Schlegel et al., 2002).
Influence of wysPII Inactivation on Morphological Development and Antibiotic Production
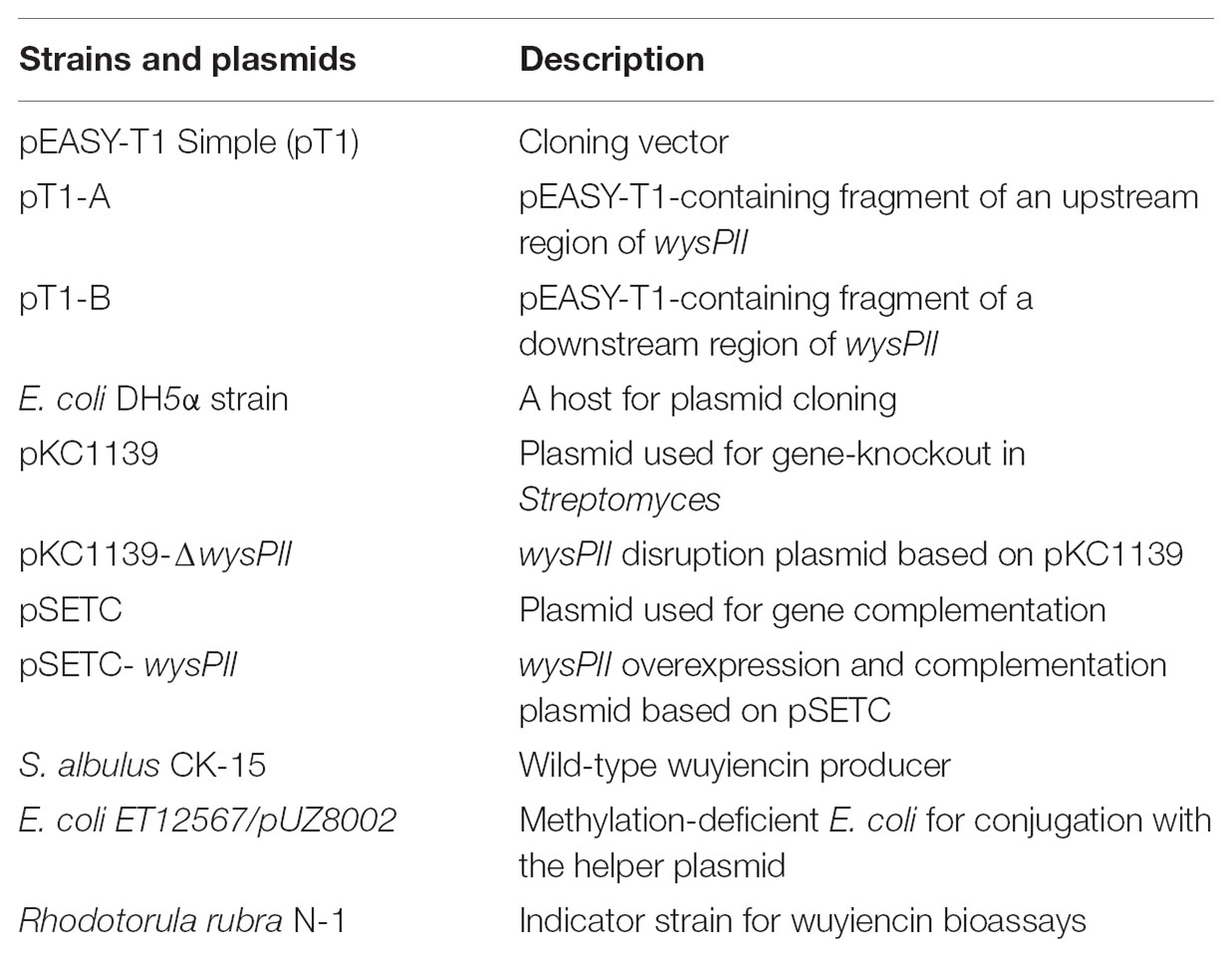
The roles of wysPII in wuyiencin production and morphological development were investigated in gene inactivation studies. Disruption vector pKC1139-ΔwysPII was transferred into S. albulus CK-15 by intergeneric conjugation and the wysPII gene was disrupted by homologous recombination (Figure 1A). pKC1139 is sensitive to temperature and replication is inhibited at temperatures >34°C; therefore, replication of the recombinant plasmid pKC1139-ΔwysPII in strain CK-15 was prevented in these conditions. Finally, a wysPII deletion mutant (S. albulus ΔwysPII) was selected by apramycin-resistance (Figure 1B).
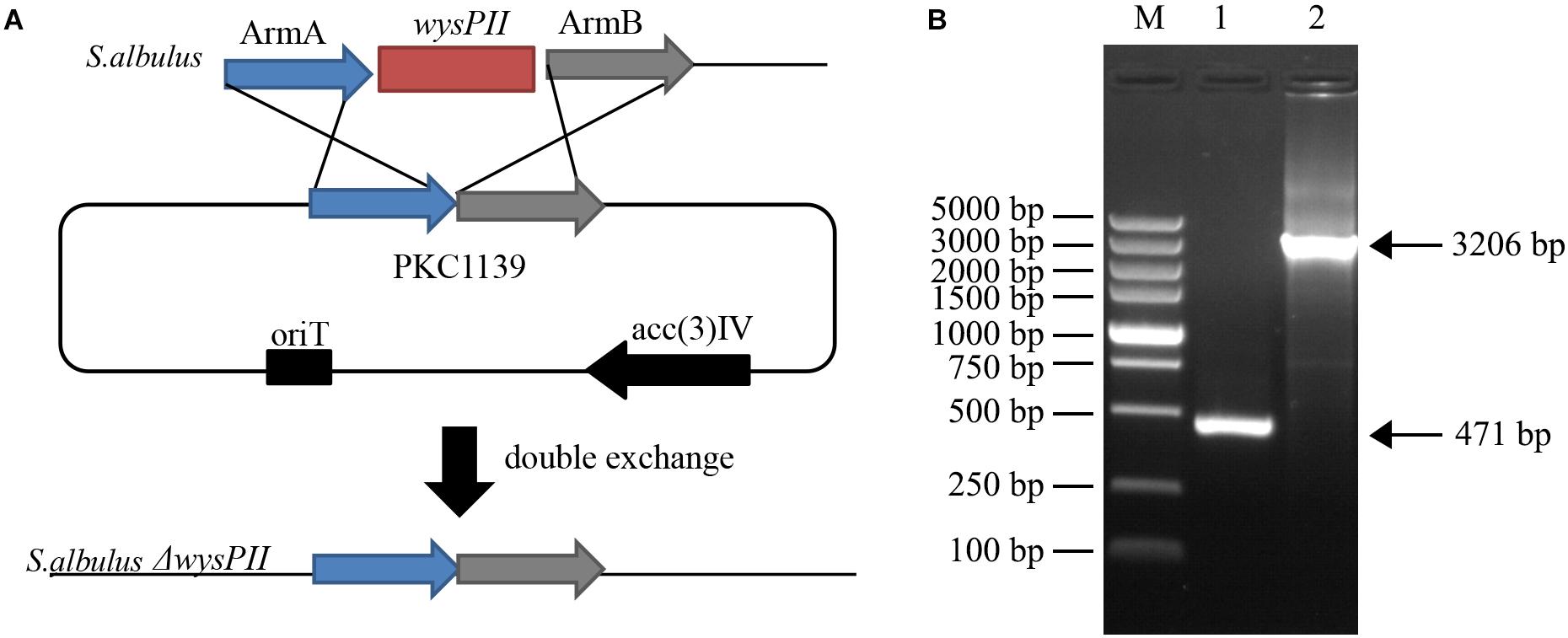
FIGURE 1. Identification of wysPII deletion (A) Gene replacement of wysPII in Streptomyces albulus CK-15. (B) Confirmation of the constructed ΔwysPII mutant by PCR. Lanes: M, BM 5,000-bp DNA ladder; 1, PCR verification with primers TF and TR using S. albulus ΔwysPII genomic DNA as the template; 2, PCR verification with primers TF and TR using S. albulus CK-15 genomic DNA as the template.
When grown on solid MS medium, the ΔwysPII mutant showed morphological changes that were apparent by both visual inspection and electron microscopy. Furthermore, the growth rate of the ΔwysPII was lower than that of the WT (Figures 2A,C). In addition, sporulation of ΔwysPII was reduced, and the ΔwysPII spores were much lighter in color than those of the WT strain. We also assessed production of wuyiencin in filtered fermentation broths of the WT and ΔwysPII strains. HPLC analysis revealed there were no differences in the wuyiencin production by the ΔwysPII and WT strains (1268.41 and 1280.93 mg/l, respectively; Figures 2D–F). Furthermore, crude extracts of the ΔwysPII broth were shown to inhibit R. rubra growth (Figure 2B). Thus, wysPII exerts a positive regulatory effect on the morphological development of S. albulus CK-15, but is not essential for wuyiencin biosynthesis.
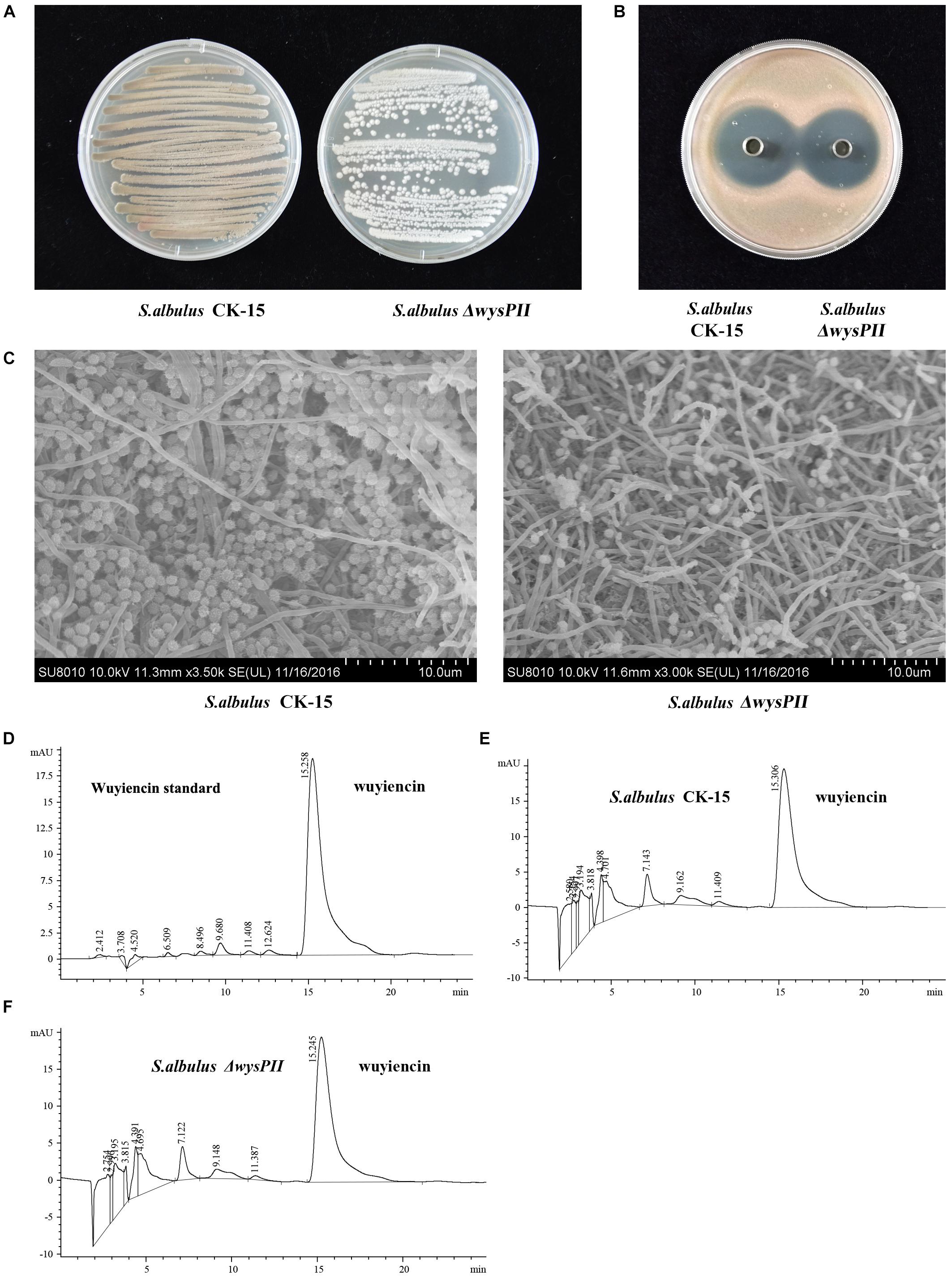
FIGURE 2. Effect of wysPII deletion on morphological development and wuyiencin production. (A) Phenotypes of 5-day-old S. albulus CK-15 and S. albulus ΔwysPII. (B) Antibacterial effect of S. albulus CK-15 and S. albulusΔwysPII fermentation culture supernatant. (C) Electron microscopy of S. albulus CK-15 and S. albulus ΔwysPII. (D) HPLC analysis of standard wuyiencin. (E) HPLC analysis of wuyiencin production levels in S. albulus CK-15. (F) HPLC analysis of wuyiencin production levels in S. albulusΔwysPII.
Trans-Complementation of the wysPII Mutant
We also investigated wysPII in complementation studies of the deletion mutant. A DNA fragment containing wysPII and the putative promoter was inserted into integrative vector pSETC to generate pSETC-wysPII, which was then transferred from E. coli ET12567 (pUZ8002) to ΔwysPII. Apramycin-resistant transconjugants were confirmed by PCR amplification of a 777-bp fragment. The expected PCR product was observed only in the complementation strain S. albulus comPII (Figure 3A). Production of wuyiencin by the complementation strain was 1276.30 mg/L equal to that by the WT (Figures 3B,D), and no notable visual phenotypic differences were observed between the strains (Figures 3C,E). Thus, complementation of ΔwysPII with wysPII restored the WT phenotype in terms of morphological development.
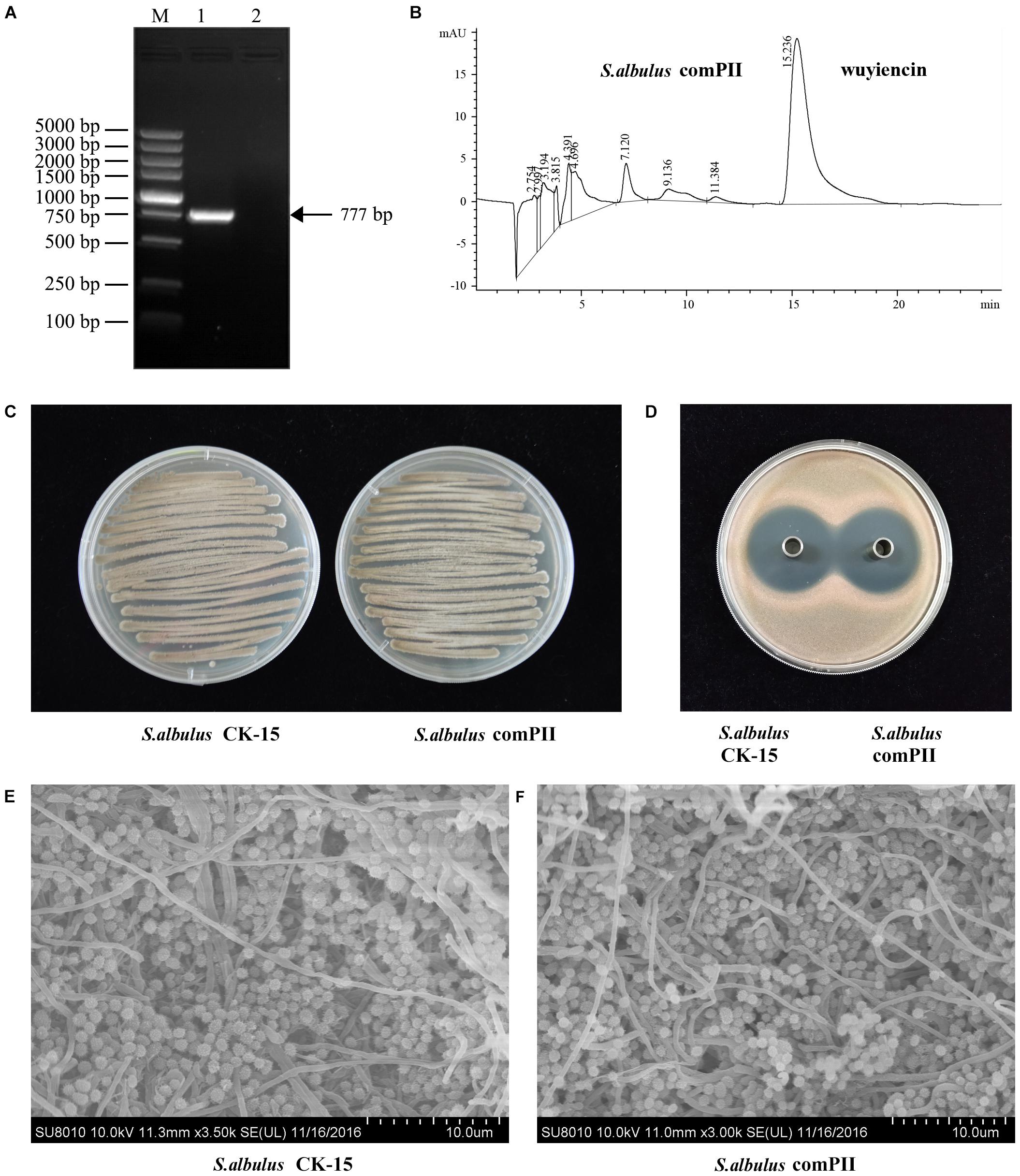
FIGURE 3. Effect of wysPII complementation on morphological development and wuyiencin production. (A) Confirmation of the complementation strain S. albulus comPII by PCR. Lanes: M, BM 5000-bp DNA ladder; 1, PCR verification with primers Am-F and Am-R using S. albulus comPII genomic DNA as the template; 2, PCR verification with primers Am-F and Am-R using S. albulus CK-15 genomic DNA as the template. (B) HPLC analysis of wuyiencin production levels in S. albulus comPII. (C) Phenotypes of 5-day-old S. albulus CK-15 and S. albulus comPII cultured on an MS plate. (D) Antibacterial effect of S. albulus CK-15 and S. albulus comPII fermentation culture supernatant. (E) Electron microscopy of S. albulus CK-15 and S. albulus comPII.
Effects of wysPII Overexpression on Morphological Development
pSTEC-wysPII, with wysPII adjacent to promoter PSF14, was then transferred from E. coli ET12567 (pUZ8002) into S. albulus CK-15. The overexpression strain was designated S. albulus ooPII. As a positive control, the empty vector pSETC was also introduced into S. albulus CK-15 (S. albulus pSETC). Apramycin-resistant transconjugants were confirmed by PCR. There were marked differences in the levels of sporulation of the ooPII transconjugants and the WT strain. Accelerated growth was observed for the ooPII strain compared with that of the WT (Figures 4A,C). However, there were no differences in the inhibitory effects of S. albulus pSETC extracts on R. rubra growth compared with those mediated by the ooPII strain (Figure 4B), and wuyiencin production of S. albulus pSETC was 1278.90 mg/L unchanged compared with the overexpression strain (Figures 4D,E).
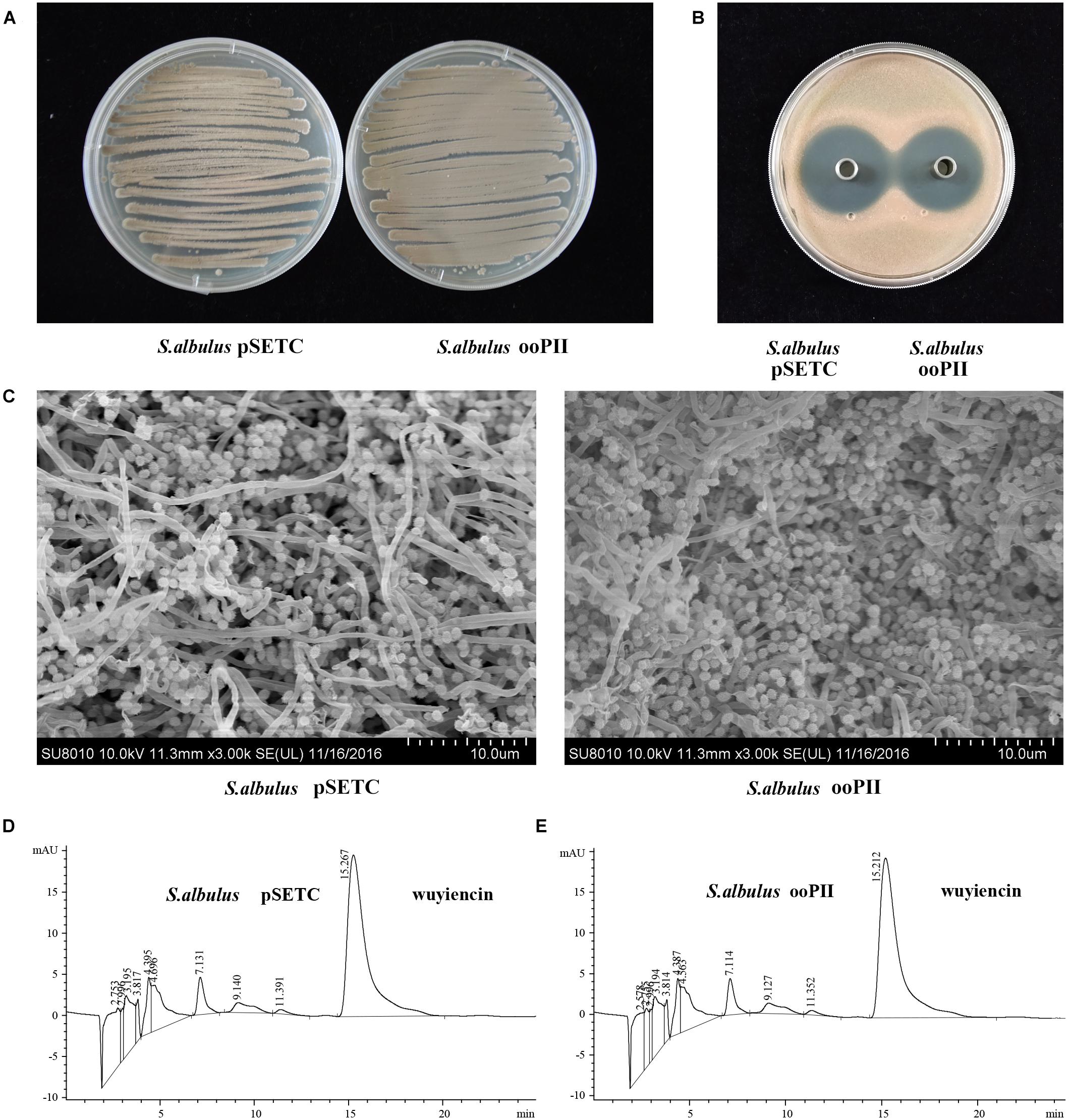
FIGURE 4. Effect of wysPII overexpression on morphological development and wuyiencin production. (A) Phenotypes of 5-day-old S. albulus pSETC and S. albulus ooPII. (B) Antibacterial effect of S. albulus pSETC and S. albulus ooPII fermentation culture supernatant. (C) Electron microscopy of S. albulus pSETC and S. albulus ooPII. (D) HPLC analysis of wuyiencin production levels in S. albulus pSETC. (E) HPLC analysis of wuyiencin production levels in S. albulus ooPII.
The Wuyiencin Production Time-Course
To compare the wuyiencin production time-course of the wysPII overexpression and WT strains, fermentation broth were collected at 2-h intervals by filtration using Millipore express membrane filters (pore diameter 0.22 μm; Merck) and analyzed by HPLC (Figure 5). The highest wuyiencin production by ooPII was detected after 56 h and 45.91% higher than that produced by the WT strain (1318.79 mg/L vs. 903.80 mg/L). The highest wuyiencin production by the WT strain was detected at 62 h (1307.53 mg/L). There was no significant difference in the greatest wuyiencin production between these two strains. Overexpression of wysR, which is a transcriptional activator of wuyiencin production, was also tested in the production time-course. The greatest wuyiencin production by ooR was observed at 60 h, which was almost threefold fasted than the time required by the WT strain.
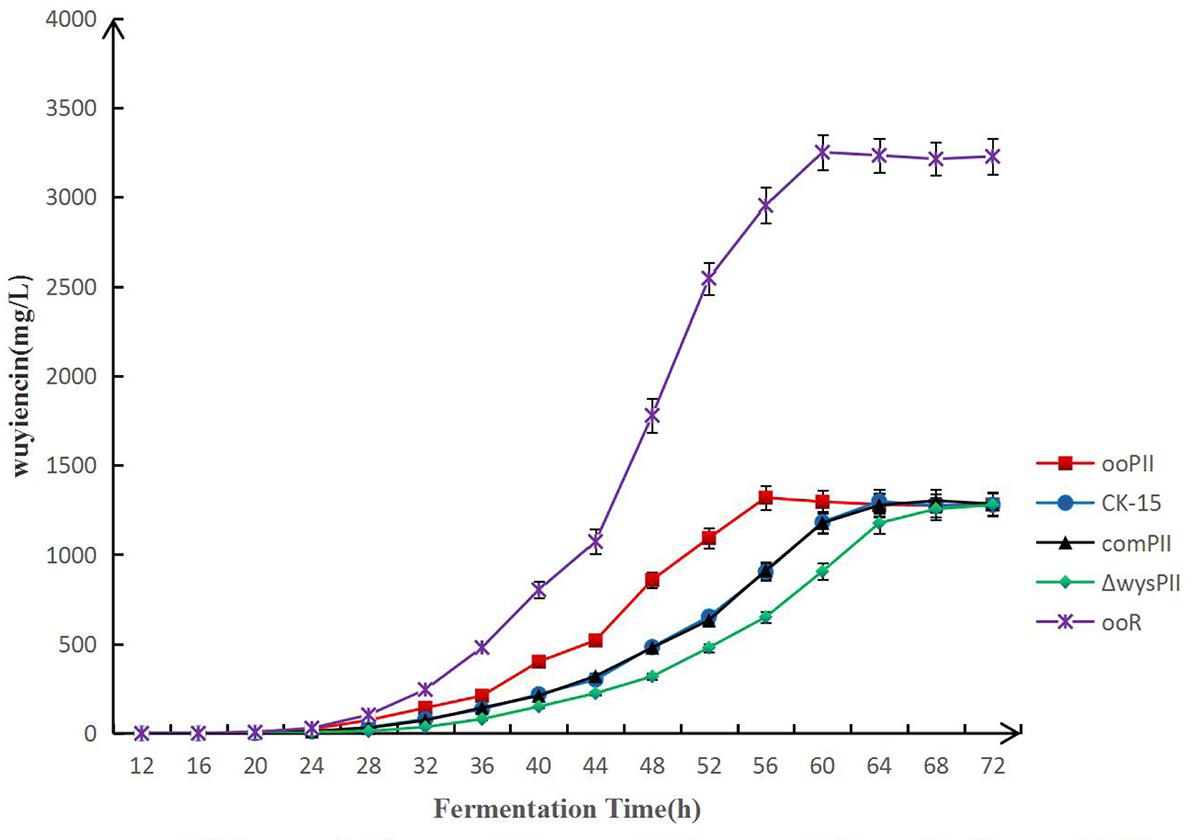
FIGURE 5. Time-course of wuyiencin production. Wuyiencin production by S. albulus CK-15, S. albulus ooPII, S. albulus ΔwysPII, S. albulus comPII and S. albulus ooR during fermentation for 72 h.
Effect of wysPII on Mycelial Biomass
To identify any effects of wysPII on mycelial biomass, we studied the morphological development from 48 to 120 h of the WT. The ΔwysPII, comPII, and ooPII strains were cultured on MS, ISP2, and ISP3 solid medium, respectively (Figures 6–8). Spores were produced by ooPII at 48 h, whereas the WT and comPII strains had only reached the mycelial period of development at this timepoint, and the ΔwysPII strain had few mycelia. Gray spores were observed in the ΔwysPII strain by 72 h, while the spores of other three strains remained white. To monitor growth, samples were collected every 6 h and the mycelial biomass was determined (Figure 9). For the ooPII strain, the latent phase was from 0 to18 h, and the logarithmic phase was from 18 to 60 h. The greatest mycelial biomass was observed at 72 h for the WT, but ooPII showed more rapid growth, reaching the maximum biomass at 54 h. In contrast, ΔwysPII exhibited delayed growth, reaching its maximum at 78 h.
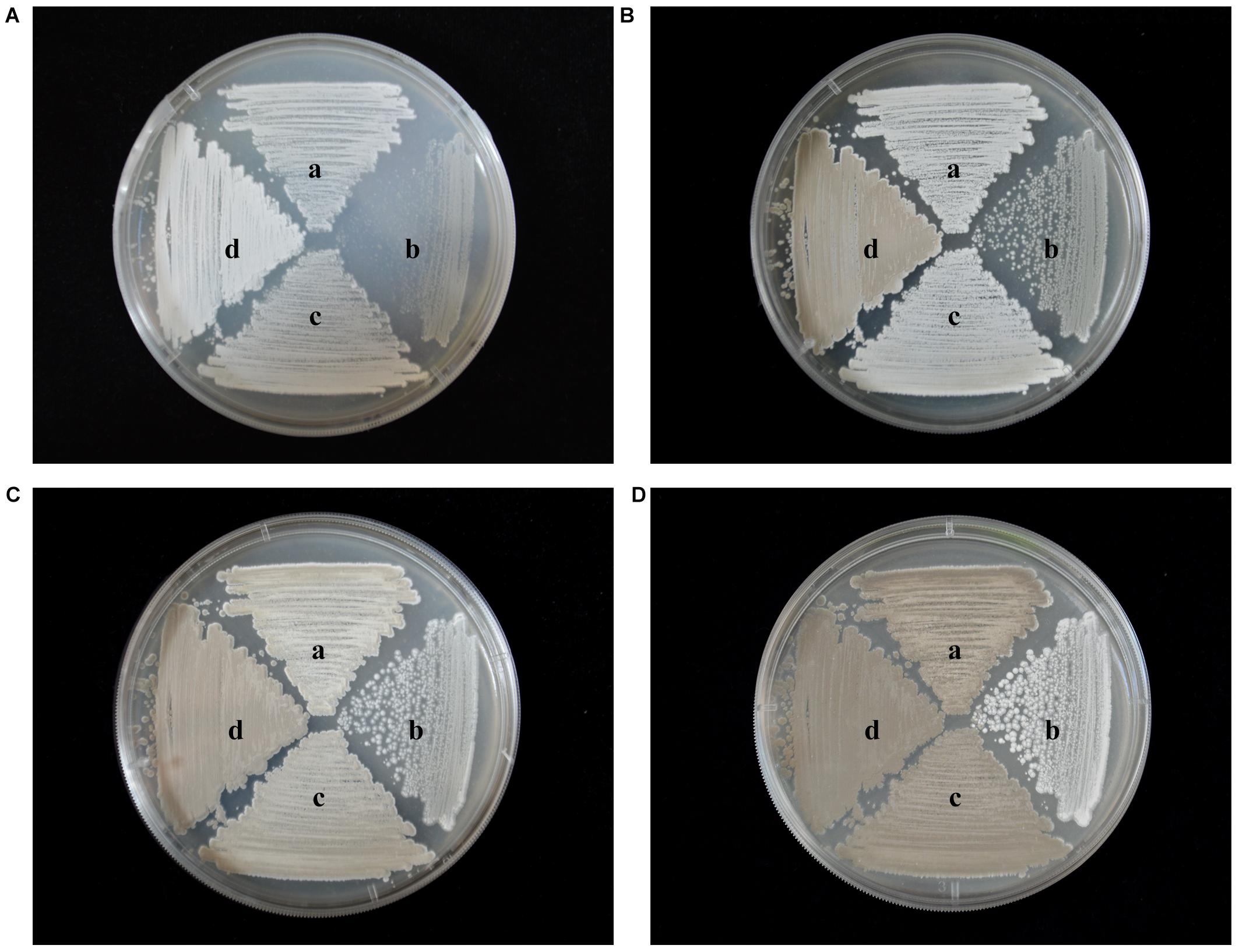
FIGURE 6. Morphological development of each strain on MS medium from 48 to 120 h. (A) The growth condition of different strains at 48 h. (B) Cultures of different strains after 72 h. (C) Cultures of different strains after 96 h. (D) Cultures of different strains after 120 h. (a) S. albulus CK-15, (b) S. albulus ΔwysPII, (c) S. albulus comPII, (d) S. albulus ooPII.
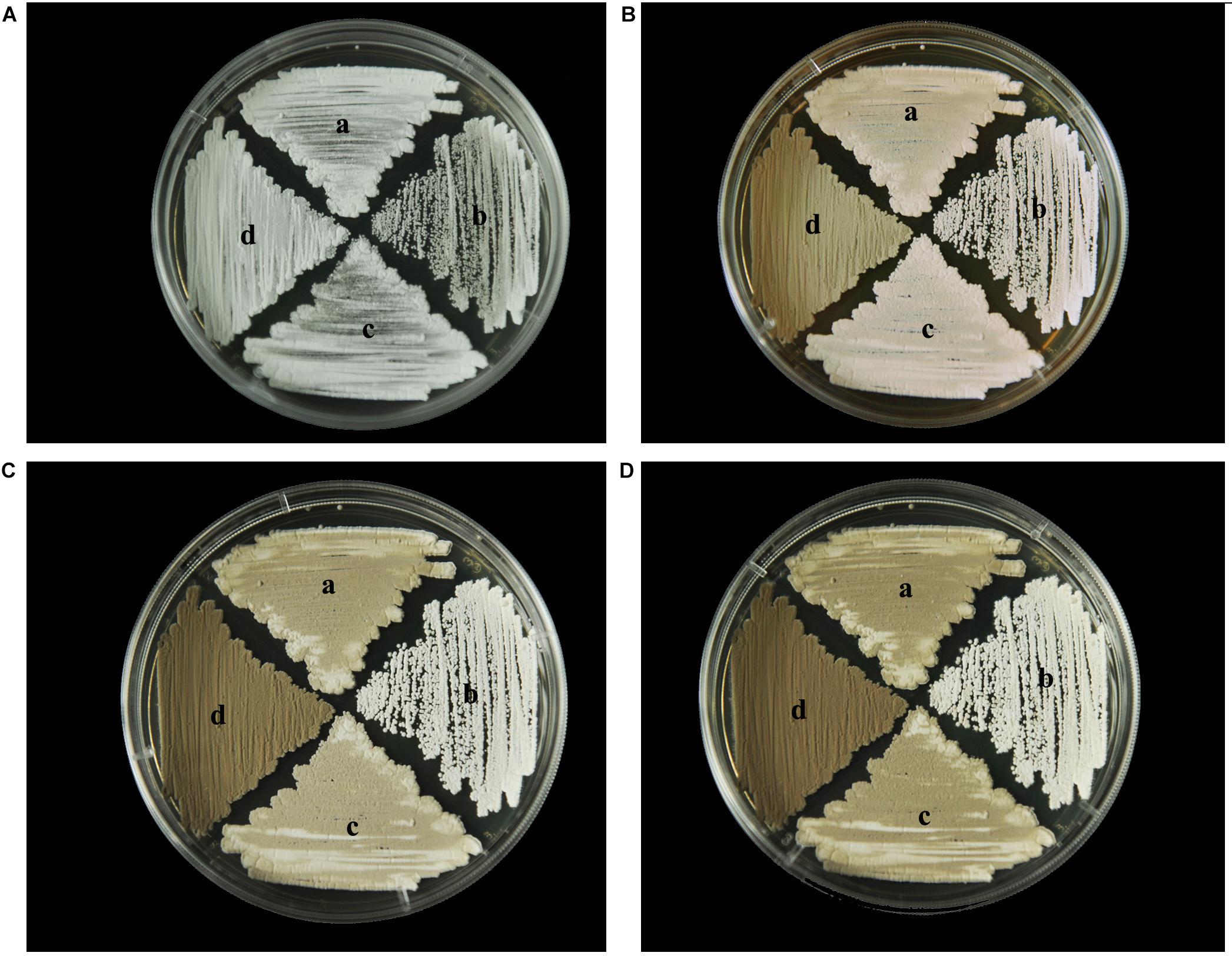
FIGURE 7. Morphological development of each strain on ISP2 medium from 48 to 120 h. (A) The growth of different strains at 48 h. (B) Cultures of different strains after 72 h. (C) Cultures of different strains after 96 h. (D) Cultures of different strains after 120 h. (a) S. albulus CK-15, (b) S. albulus ΔwysPII, (c) S. albulus comPII, (d) S. albulus ooPII.
FIGURE 8. Morphological development of each strain on ISP3 medium from 48 to 120 h. (A) The growth of different strains at 48 h. (B) Cultures of different strains after 72 h. (C) Cultures of different strains after 96 h. (D) Cultures of different strains after 120 h. (a) S. albulus CK-15, (b) S. albulus ΔwysPII, (c) S. albulus comPII, (d) S. albulus ooPII.
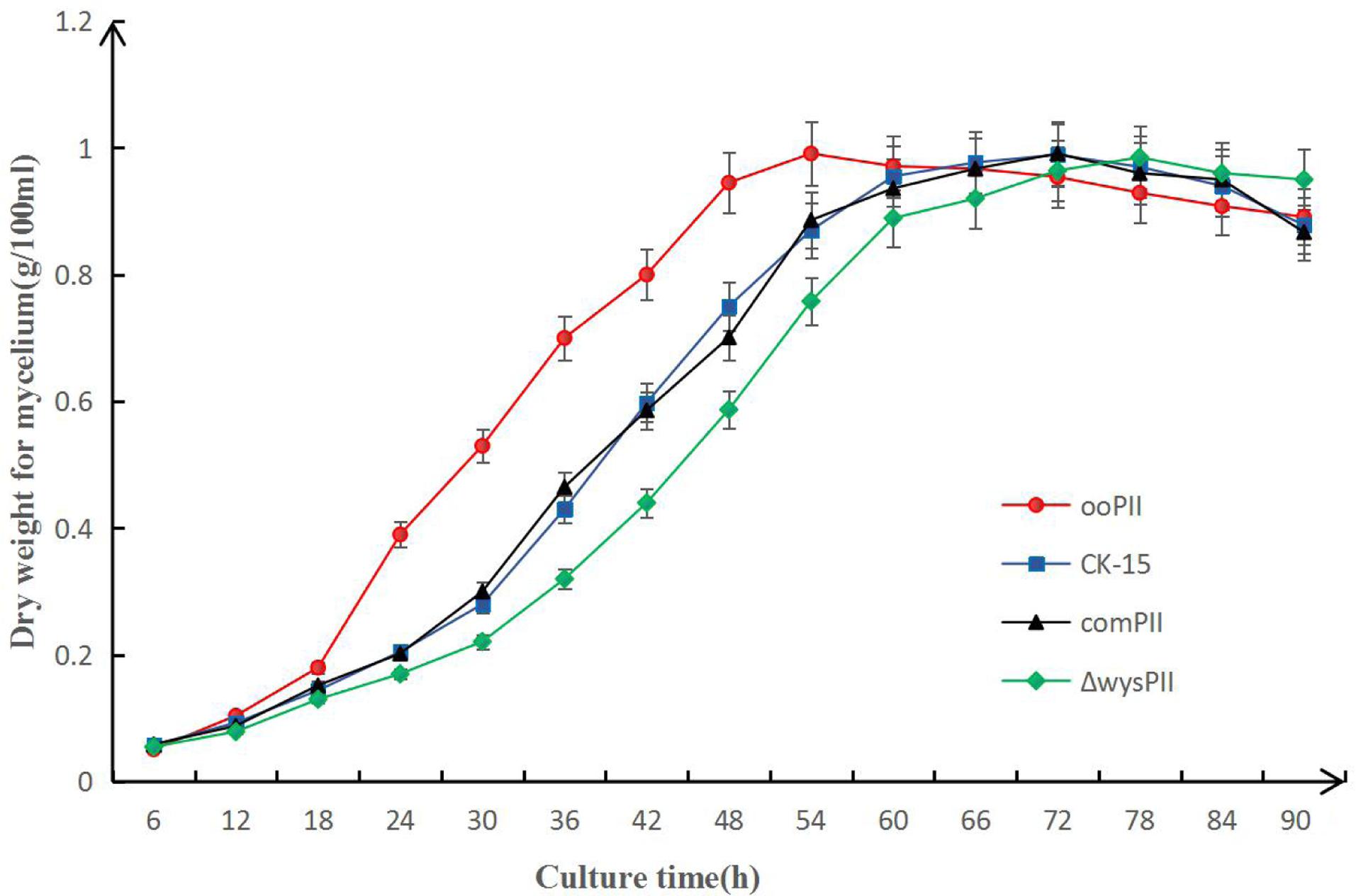
FIGURE 9. Result of mycelial biomass measurement. Biomass of S. albulus CK-15, S. albulus ooPII, S. albulus ΔwysPII, S. albulus comPII during culture for 90 h.
Developmental Gene Expression Levels in wysPII Overexpression Strain
To investigate the mechanism of the rapid growth and increased sporulation of the ooPII strain, we analyzed wysPII expression in the ooPII and WT strains by RT-qPCR. We also analyzed the expression of several genes of the bld, whi, and ram families, which are crucial for the morphological development of Streptomyces, especially for mycelial growth and sporulation (Keijser et al., 2000; Chater, 2001; Kim et al., 2014). All of the selected genes were highly overexpressed in the ooPII strain (Figure 10). The expression levels of seven of the genes (whiA, whiH, whiI, ramA, ramB, ramC, and ramR) were more than 10 times higher than those in the WT strain. Furthermore, it is worth noting that the expression levels of whiH and whiI in the ooPII strain were more than 20 times higher than those in the WT strain. These result suggested that wysPII activates these genes, which are crucial for the morphological development of Streptomyces.
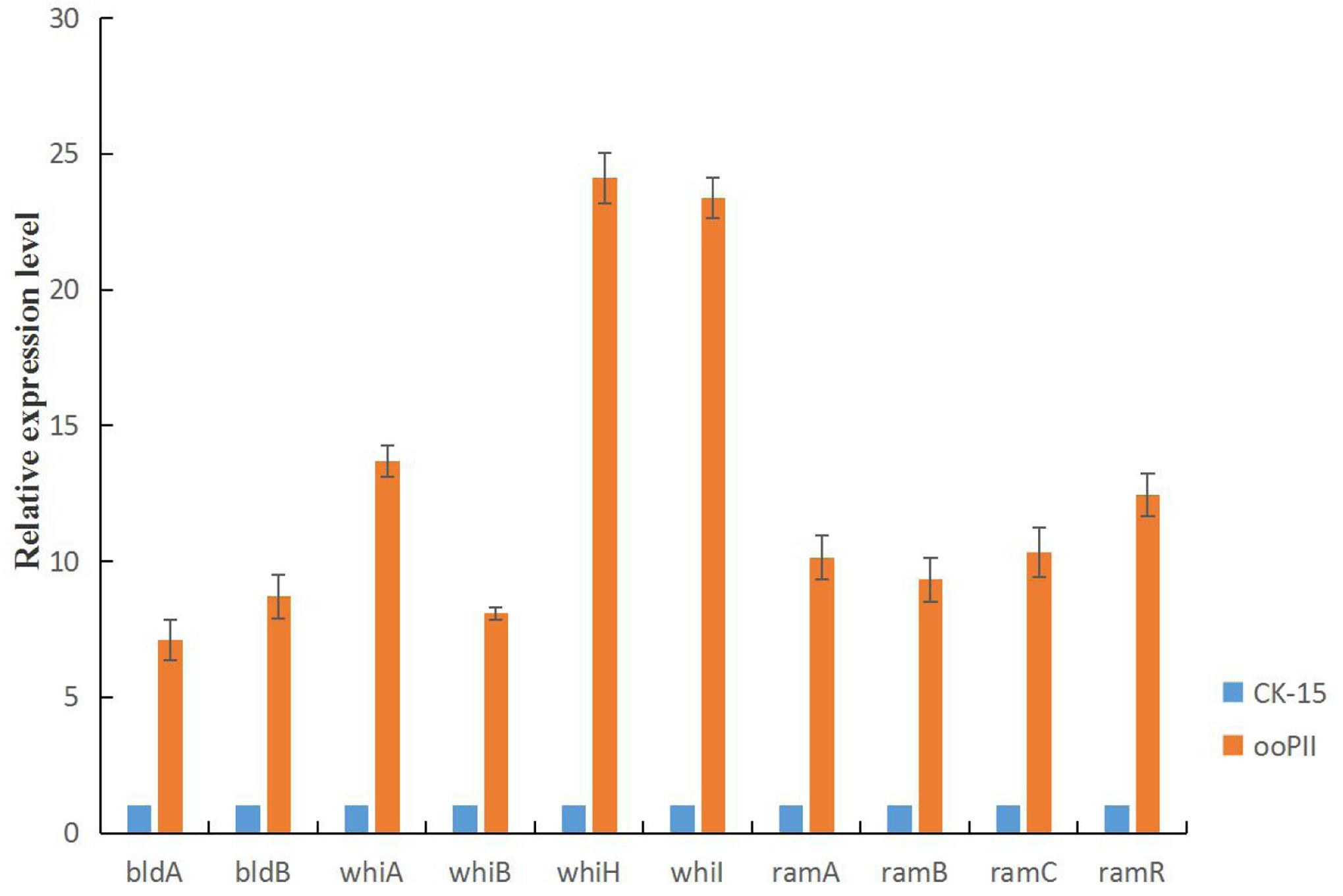
FIGURE 10. Developmental gene expression in S. albulus CK-15 and S. albulus ooPII by RT-qPCR. The RNA samples were isolated from 72-h cultures on MS medium. The relative values of S. albulus CK-15 are designated as 1. Data represent the mean ± standard deviation of three independent experiments.
Discussion
Morphological development of Streptomyces has been studied mainly in S. coelicolor (Zhang et al., 2017). Although developmental regulation in Streptomyces is not completely understood, bld and whi genes have been shown to be involved (Flardh and Buttner, 2009). Mutations in bld block formation of aerial mycelia, resulting in a ‘bald’ appearance, whereas mutations in whi block steps in the conversion of aerial mycelia to mature, gray spores, and whi mutants appear white (Chater, 2011) because the mutants fail to produce the gray polyketide pigment that is visible in the mature WT spores (Den Hengst et al., 2010). In S. coelicolor, SapB, a small, hydrophobic, morphogenetic peptide, is crucial for differentiation into aerial hyphae and the ram gene cluster is indispensable for SapB production (Kodani et al., 2004). In this study, the wysPII mutant exhibited slower growth and delayed sporulation compared with WT S. wuyiensis CK-15. In addition, the ΔwysPII strain produced white spores, while those produced by the WT strain were gray. The ΔwysPII strain showed the same phenotypic features as mutants of the whi family, which is inconsistent with the effects of mutations reported in other LuxR family regulators. All the selected genes (bldA, bldB, whiA, whiB, whiH, whiI, ramA, ramB, ramC, and ramR) were highly overexpressed in the ooPII strain (Figure 10), especially whiH and whiI. This result suggests that wysPII activates these genes, which are crucial for the morphological development of Streptomyces. Furthermore, the ooPII strain grew much faster than the WT strain and exhibited increased sporulation. Most LuxR family regulators have been reported as activators of antibiotic production in Streptomyces (Yu et al., 2012). S. natalensis strains expressing a mutant form of PimR, which is a putative regulator of pimaricin production, exhibited the same growth and morphological characteristics as the WT strain (Antón et al., 2004). WysR is a luxR family transcriptional activator of wuyiencin biosynthesis and morphological development (Liu et al., 2014). Very few LuxR family members have been reported to regulate the morphological development of bacteria (Vicente et al., 2014). In a study of positive regulators of tetramycin biosynthesis in S. ahygroscopicus, colonies of ttmRII mutants showed less gray pigmentation and no regulatory effect on antibiotic production was observed (Cui et al., 2016).
WysPII is a multidomain protein, which was found to contain a MalT family regulator domain, with the C-terminal HTH motif that is a positive activator of transcription of the mal regulon in E. coli K-12 (Panagiotidis et al., 1998). Mutants without malT are unable to grow on maltose, and strains with mutations in this domain show constitutive expression of the mal gene (Schlegel et al., 2002). However, the WT, ooPII and ΔwysPII strains exhibited growth retardation and reduced sporulation when cultured with maltose as the only carbon source. Thus, the malT may have no obvious effect on the morphological development of S. albulus CK-15.
The results of the present study indicate that wysPII functions as an activator of morphological development, with wysPII overexpression leading to more rapid growth than that of the WT and wysPII deletion leading to retardation of morphological development. In the wysPII overexpressing strain, large quantities of spores were produced by 72 h when cultured on solid MS medium, while spore production by the WT strain was only observed after 96 h (Figure 6). In liquid culture, the wysPII overexpressing strain grew faster and reached maximum biomass at 54 h, approximately 18 h faster than the WT (Figure 9). In terms of wuyiencin production, the maximum levels were generated by the ooPII strain at 56 h, while this occurred at 62 h in the WT strain (Figure 5). Improving the efficiency and reducing the cost of wuyiencin production is essential to promote of its use as an agricultural biopesticide to control plant fungal diseases; the use of overexpressing strains may be advantageous in industrial production processes.
Author Contributions
KZ conceived and supervised the studies. BL and BG performed all experiments and wrote the paper. JM, QW, AK, and LS analyzed the data.
Funding
This work was supported by grants from the Special Fund for Basic Scientific Research of the Chinese Academy of Agricultural Sciences in China (Y2017JC12 and S2018XM12), the National Natural Science Foundation of China (31772215), and the National Key Research & Development (R&D) Plan in China (2017YFD0201100).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alonso-Hearn, M., Eckstein, T. M., and Sommer, S. (2010). A Mycobacterium avium subsp. paratuberculosis LuxR regulates cell envelope and virulence. Innate Immun. 16, 235–247. doi: 10.1177/1753425909339811
Andersson, R. A., Eriksson, A. R. B., Heikinheimo, R., Mäe, A., Pirhonen, M., Kõiv, V., et al. (2000). Quorum sensing in the plant pathogen Erwinia carotovora subsp. carotovora: the role of expREcc. Mol. Plant Microbe Interact. 13, 384–393. doi: 10.1094/MPMI.2000.13.4.384
Antón, N., Mendes, M. V., Martín, J. f., and aparicio, j. f. (2004). Identification of pimr as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J. Bacteriol. 186, 2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004
Antón, N., Santos-Aberturas, J., Mendes, M. V., Guerra, S. M., Martín, J. F., and Aparicio, J. F. (2007). PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 153, 3174–3183. doi: 10.1099/mic.0.2007/009126-0
Antunes, L. C. M., Ferreira, R. B. R., Lostroh, C. P., and Greenberg, E. P. (2008). A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. 190, 4392–4397. doi: 10.1128/JB.01443-07
Bibb, M. J. (2005). Regulation of secondary metabolism in Streptomycetes. Curr. Opin. Microbiol. 8, 208–215. doi: 10.1016/j.mib.2005.02.016
Birck, C., Malfois, M., Svergun, D., and Samama, J. P. (2002). Insights into signal transduction revealed by the low resolution structure of the FixJ response regulator. J. Mol. Biol. 321, 447–457. doi: 10.1016/S0022-2836(02)00651-4
Chater, K. (2011). “Differentiation in Streptomyces: the properties and programming of diverse cell-types,” in Streptomyces: Molecular Biology and Biotechnology, ed. P. Dyson (Poole: Caister Academic Press), 43–86.
Chater, K. F. (2001). Regulation of sporulation in Streptomyces coelicolor A3 (2): a checkpoint multiplex? Curr. Opin. Microbiol. 4, 667–673. doi: 10.1016/S1369-5274(01)00267-3
Cui, H., Ni, X. P., Liu, S. J., Sun, Z. P., Ren, J., Su, J. Q., et al. (2016). Characterization of three positive regulators for tetramycin biosynthesis in Streptomyces ahygroscopicus. FEMS Microbiol. Lett. 363:fnw109. doi: 10.1093/femsle/fnw109
Danino, V. E., Wilkinson, A., Edwards, A., and Downie, J. A. (2003). Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 50, 511–525. doi: 10.1046/j.1365-2958.2003.03699.x
De, B. I., and Raaijmakers, J. M. (2009). Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl. Environ. Microb. 75, 4753–4761. doi: 10.1128/AEM.00575-09
Den Hengst, C. D., Tran, N. T., Bibb, M. J., Chandra, G., Leskiw, B. K., and Buttner, M. J. (2010). Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78, 361–379. doi: 10.1111/j.1365-2958.2010.07338.x
Ducros, V. M., Lewis, R. J., Verma, C. S., Dodson, E. J., Leonard, G., Turkenburg, J. P., et al. (2001). Crystal structure of GerE, the ultimate transcriptional regulator of spore formation in Bacillus subtilis. J. Mol. Biol. 306, 759–771. doi: 10.1006/jmbi.2001.4443
Flardh, K., and Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. doi: 10.1038/nrmicro1968
Fuqua, C., Winans, S. C., and Greenberg, E. P. (1996). Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751. doi: 10.1146/annurev.micro.50.1.727
Ge, B. B., Cheng, Y., Liu, Y., Liu, B. H., and Zhang, K. C. (2015a). Biological control of Botrytis cinerea on tomato plants using Streptomyces ahygroscopicus strain CK-15. Lett. Appl. Microbiol. 61, 596–602. doi: 10.1111/lam.12500
Ge, B. B., Liu, Y., Liu, B., and Zhang, K. C. (2015b). Draft genome sequence of Streptomyces ahygroscopicus subsp. wuyiensis CK-15, isolated from soil in Fujian Province, China. Genome Announc. 3:e01125-15. doi: 10.1128/genomeA.01125-15
Ge, B. B., Liu, Y., Liu, B., Zhao, W. J., and Zhang, K. C. (2016). Characterization of novel DeoR-family member from the Streptomyces ahygroscopicus strain CK-15 that acts as a repressor of morphological development. Appl. Microbiol. Biotechnol. 100, 8819–8828. doi: 10.1007/s00253-016-7661-y
Ge, B. B., Yang, Z. J., Tan, B. B., Liu, Y. Y., Liu, Y., Sun, L., et al. (2014). Cloning and analysis of wuyiencin partial biosynthetic gene cluster of Streptomyces ahygroscopicus var. wuyiensis CK-15. Chin. J. Biol. Control. 30, 678–684.
He, W. Q., Lei, J., Liu, Y. Y., and Wang, Y. G. (2008). The LuxR family members GdmRI and GdmRII are positive regulators of geldanamycin biosynthesis in Streptomyces hygroscopicus 17997. Arch. Microbiol. 189, 501–510. doi: 10.1007/s00203-007-0346-2
Hobbs, G., Frazer, C. M., Gardner, D. C., Cullum, J. A., and Oliver, S. G. (1989). Dispersedgrowth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 31, 272–277. doi: 10.1007/BF00258408
Hur, Y. A., Choi, S. S., Sherman, D. H., and Kim, E. S. (2008). Identification of TmcN as a pathway-specific positive regulator of tautomycetin biosynthesis in Streptomyces sp. CK4412. Microbiology 154, 2912–2919. doi: 10.1099/mic.0.2008/018903-0
Keijser, B. J., van Wezel, G. P., Canters, G. W., Kieser, T., and Vijgenboom, E. (2000). The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2, 565–574.
Kim, J. M., Won, H. S., and Kang, S. O. (2014). The C-terminal domain of the transcriptional regulator BldD from Streptomyces coelicolor A3 (2) constitutes a novel fold of winged-helix domains. Proteins 82, 1093–1098. doi: 10.1002/prot.24481
Kodani, S., Hudson, M. E., Durrant, M. C., Buttner, M. J., Nodwell, J. R., and Willey, J. M. (2004). The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 101, 11448–11453. doi: 10.1073/pnas.0404220101
Labes, G., Bibb, M., and Wohlleben, W. (1997). Isolation and characterization of a strong promoter element from the Streptomyces ghanaensis phage 119 using the gentamicin resistance gene (aacC1) of Tn1696 as reporter. Microbiology 143, 1503–1512. doi: 10.1099/00221287-143-5-1503
Latifi, A., Winson, M. K., Foglino, M., Bycroft, B. W., Stewart, G. S., Lazdunski, A., et al. (1995). Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17, 333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x
Liu, G., Chater, K. F., Chandra, G., Niu, G. Q., and Tan, H. R. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
Liu, Y. Y., Ryu, H., Ge, B. B., Pan, G. H., Sun, L., Park, K., et al. (2014). Improvement of wuyiencin biosynthesis in Streptomyces wuyiensis CK-15 by identification of a key regulator. WysR. J. Microbiol. Biotechnol. 24, 1644–1653. doi: 10.4014/jmb.1405.05017
Martín, J. F. (2004). Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J. Bacteriol. 186, 5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004
McCormick, J. R., and Flardh, K. (2011). Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36, 206–231. doi: 10.1111/j.1574-6976.2011.00317.x
Nasser, W., and Sylvie, R. (2007). New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem. 387, 381–390. doi: 10.1007/s00216-006-0702-0
Panagiotidis, C. H., Boos, W., and Shuman, H. A. (1998). The ATP-binding cassette subunit of the maltose transporter MalK antagonizes MalT, the activator of the Escherichia coli mal regulon. Mol. Microbiol. 30, 535–546. doi: 10.1046/j.1365-2958.1998.01084.x
Passador, L., Cook, J. M., Gambello, M. J., Rust, L., and Iglewski, B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260, 1127–1131. doi: 10.1126/science.8493556
Schlegel, A., Bohm, A., Lee, S. J., Peist, R., Decker, K., and Boos, W. (2002). Network regulation of the Escherichia coli maltose system. J. Mol. Microbiol. Biotechnol. 4, 301–307.
Sekurova, O. N., Brautaset, T., Sletta, H., Borgos, S. E., Jakobsen, M. ØM., Ellingsen, T. E., et al. (2004). In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186, 1345–1354. doi: 10.1128/JB.186.5.1345-1354.2004
Sun, Y. Z., Zeng, H. M., Shi, Y. P., and Li, G. (2003). Mode of action of Wuyiencin on Botrytis cinerea. Acta Phytopathol. Sin. 33, 434–438. doi: 10.1111/lam.12500
Vicente, C. M., Payero, T. D., Santos-Aberturas, J., Barreales, E. G., Pedro, A., and Aparicio, G. F. (2015). Pathway-specific regulation revisited: cross-regulation of multiple disparate gene clusters by PAS-LuxR transcriptional regulators. Appl. Microbiol. Biotechnol. 99, 5123–5135. doi: 10.1007/s00253-015-6472-x
Vicente, C. M., Santos- Aberturas, J., Pavero, T. D., Barreales, E. G., de Pedro, A., and Aparicio, J. F. (2014). PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 98, 9311–9324. doi: 10.1007/s00253-014-5998-7
Wei, R. Q., Lin, D. C., and Chen, Z. H. (1984). Identification of the producing strain of agricultural antibiotic Bo-10. Acta Microbiol. Sin. 24, 401–402.
Yu, Q., Bai, L. Q., Zhou, X. F., and Deng, Z. X. (2012). Inactivation of the positive LuxR-type oligomycin biosynthesis regulators OlmRI and OlmRII increases avermectin production in Streptomyces avermitilis. Chin. Sci. Bull. 57, 869–876. doi: 10.1007/s11434-011-4865-5
Zeng, H. M., Lin, D. X., and Shi, Y. P. (1996). Mutation of the protoplast of Wuyimycin producing strain. Chin. J. Biol. Control. 12, 48–48.
Keywords: Streptomyces albulus, wysPII, regulatory genes, LuxR family, morphological development
Citation: Liu B, Ge B, Ma J, Wei Q, Khan AA, Shi L and Zhang K (2018) Identification of wysPII as an Activator of Morphological Development in Streptomyces albulus CK-15. Front. Microbiol. 9:2550. doi: 10.3389/fmicb.2018.02550
Received: 20 July 2018; Accepted: 05 October 2018;
Published: 25 October 2018.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Kozo Ochi, Hiroshima Institute of Technology, JapanTomislav Cernava, Graz University of Technology, Austria
Copyright © 2018 Liu, Ge, Ma, Wei, Khan, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kecheng Zhang, emhhbmdrZWNoZW5nQHNpbmEuY29t
†These authors have contributed equally to this work
 Binghua Liu
Binghua Liu Beibei Ge
Beibei Ge Jinjin Ma1
Jinjin Ma1 Abid Ali Khan
Abid Ali Khan Kecheng Zhang
Kecheng Zhang