- 1College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2Hubei Three Gorges Polytechnic, Yichang, China
- 3University College of Veterinary and Animal Sciences, The Islamia University of Bahawalpur Pakistan, Bahawalpur, Pakistan
- 4College of Animals Husbandry and Veterinary Medicine, Tibet Agriculture and Animal Husbandry University, Linzhi, China
The purpose of this study was to evaluate the antibacterial activity and safety of bacterias with probiotic potential isolated from free-ranging Tibetan yaks in high altitude regions of Tibet. For this purpose, one Leuconostoc pseudomesenteroides strain (named P1) and two Lactobacillus johnsonii and Lactobacillus mucosae strains (named LY1 and LY2), respectively, were isolated from fecal samples of Tibetan yaks. The antibacterial activity of the isolates was studied using Escherichia coli (E. coli ATCC 25922), Staphylococcus aureus (S. aureus ATCC 26112), and Salmonella enteritidis (S. enteritidis NCTC 13349) as indicator pathogens. The results showed that LY1 had high antibacterial efficacy against E. coli and S. enteritidis, while P1 had the most powerful bacteriostatic ability against S. aureus. PCR amplification showed that all the isolated strains were positive for Ent P2 (enterocin P-like bacteriocin) and exhibited a high tolerance to bile and low pH. Moreover, the safety of P1, LY1, and LY2 was determined through antibiotic resistance experiments, resistance gene testing, and hemolytic analysis while the antibacterial activity was assessed by in vitro and in vivo experiments. The LY2 strain was abandoned as a potential probiotic due to the detection of the vanA gene. The mice were fed from days 1 to 30 in six groups, the P1-1 (gavaged with P1 1 × 108 CFU/day), P1-2 (gavaged with P1 1 × 109 CFU/day), LY1-1 (gavaged with LY1 1 × 108 CFU/day), LY1-2 (gavaged with LY1 1 × 109 CFU/day), control (gavaged with an equal volume of vehicle), and blank control (gavaged with an equal volume of vehicle) groups. After 30 days, mice in the P1-1, P1-2, LY1-1, LY1-2, and control groups were intraperitoneal challenged with 1 × 108 CFU of E. coli (n = 10) in the abdomen. After 2 days of infection, the mice in the control group showed more severe damage in the liver, spleen and intestine than the mice in the P1-2 and LY1-2 groups. The mice in the P1-2 and LY1-2 groups had lower rates of diarrhea and mortality than other groups. In conclusion, bacteria with probiotic potential isolated from yaks may possibly be effective and safe antibacterial substances, providing a new treatment method to reduce the incidence of diarrhea associated with bacterial diseases in yaks.
Introduction
Infectious diseases are a threat for animal health and productivity in developing countries (Elhaig et al., 2016; Wen et al., 2016; Yilmaz et al., 2016; Mehmood et al., 2017). Diarrhea is a common problem in cattle that greatly affects the growth of animals and causes huge economic losses (Heuer et al., 2007). Diarrhea can be caused by many factors, such as bacteria and viruses. Many measures have been carried out to prevent diarrhea, including improvements in hygiene and feeding management systems, but it still occurs from time to time (Cho et al., 2013). Escherichia coli (E. coli), Salmonella enteritidis (S. enteritidis), and Staphylococcus aureus (S. aureus) are considered common pathogens that cause diarrhea (Dong et al., 2018). Antibiotics are widely used to treat diarrhea, but antibiotic use is considered a double-edged sword, as some problems have emerged with antibiotic usage, especially antibiotic resistance. Previous studies have shown that the misuse of antibiotics has become a serious problem, affecting microbial balance and increasing the incidence of antibiotic-associated diarrhea by 5 to 25% (Kaltenbach and Heitz, 2004). Probiotics are living microorganisms that not only improve the balance of intestinal microflora but also inhibit many pathogens (Food Agriculture Organization/World Health Organization [FAO/WHO], 2011). They are widely used in animal production because of their antibacterial activity and various biological characteristics. Some researchers have reported that Lactobacillus johnsonii BS15 (CCTCC M2013663) can be used to prevent non-alcoholic fatty liver disease in obese mice and improve growth performance along with meat quality (Wang H. et al., 2018). In addition, some studies have shown that L. johnsonii can improve lipid metabolism and intestinal microflora in chickens to prevent subclinical necrotic enteritis and reduce respiratory viral infections (Fonseca et al., 2017). Moreover, it has been reported that Leuconostoc mesenteroides can inhibit the growth of pathogens and can be used as a safe probiotic for further study (Makhloufi et al., 2013; Casado et al., 2014). However, some cases of bacteremia, liver damage and other clinical symptoms may be related to some Lactobacillus). Screening strains, as per previous reports (Rautio et al., 1999). The oral toxicity test in animals has been proposed as the basis for evaluating the safety of probiotics (Stine and Brown, 1996) for edible probiotics requires the following steps: (1) identifying the bacteria, (2) testing for probiotic potential through in vitro experiments, (3) proving that the probiotics are safe and effective, and (4) performing experiments in animals (Food Agriculture Organization/World Health Organization [FAO/WHO], 2006).
Yaks are long-haired bovines that live on high plateaus and in anoxic zones (Zhang et al., 2018; Wu et al., 2018a,b). Approximately 90% of the global yak population resides in the Qinghai, Tibet and Sichuan provinces of China (Li et al., 2015). Yaks are an important source of milk, meat and leather for local herders. However, it has been reported that diarrhea occurs in Tibetan yak calves. Therefore, the present study was designed to isolate bacteria with probiotic potential that can effectively inhibit diarrhea and reduce various losses associated with diarrhea in yaks.
In this study, the antibacterial activity of strains isolated from the feces of healthy Tibetan yaks was assessed with in vitro and in vivo experiments. The safety of the strains was evaluated with drug resistance experiments, drug resistance gene testing, hemolytic analysis and oral toxicity testing in mice. The purpose of this trial was to assess the safety of the isolated strains and to assess whether they can reduce the incidence of bacterial diseases in a mouse model. The results of this study may provide new ideas and agents for the treatment of diarrhea associated with bacterial diseases in yaks.
Materials and Methods
Sample Collection
A total of 30 fecal samples were randomly collected from different ages and genders of free-ranging yaks in the Nyingchi region of Tibet, China. The samples were kept at 4°C and transported to Huazhong Agricultural University, Wuhan, and they were then stored at -80°C for further experiments. In addition, E. coli, S. enteritidis, and S. aureus were provided by the Veterinary Science Center of Huazhong Agricultural University, China.
Isolation and Identification of Lactobacillus Strains
Three strains were isolated from the feces collected from healthy free-ranging Tibetan yaks. A portion (2 g) of each sample was blended in phosphate-buffered saline and shaken well for 30 min at 37°C. The supernatant was streaked on de Man, Rogosa and Sharpe (MRS, a selective agar for lactic acid bacteria) agar (Hangzhou Reagents, China) and then grown for 48 h at 37°C in an incubator under anaerobic conditions. Milky white and raised suspected lactic acid bacteria colonies were selected for purification and cultivation for three generations and then individual colonies were picked and inoculated in MRS broth for culture. For further validation, milky white raised colonies were tested by Gram staining and subjected to biochemical analysis. Finally, all isolated strains were confirmed and identified by genetic analysis using PCR and 16S rRNA sequencing. The genomic DNA of the Lactobacillus strains (2 mL Lactobacillus culture, 1 × 108 CFU/mL) was extracted using a Bacterial DNA Isolation Kit (Foregene, China). The universal PCR primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACTT-3′) were used to amplify the 16S rRNA gene (Wang L. et al., 2018). Furthermore, the PCR products were sequenced by the Qingke Biotech Company (Wuhan, China) and subjected to BLAST analysis on the NCBI website. The phylogenic tree was constructed using the neighbor-joining method with MEGA 6 software.
Antibiotic Susceptibility Assay
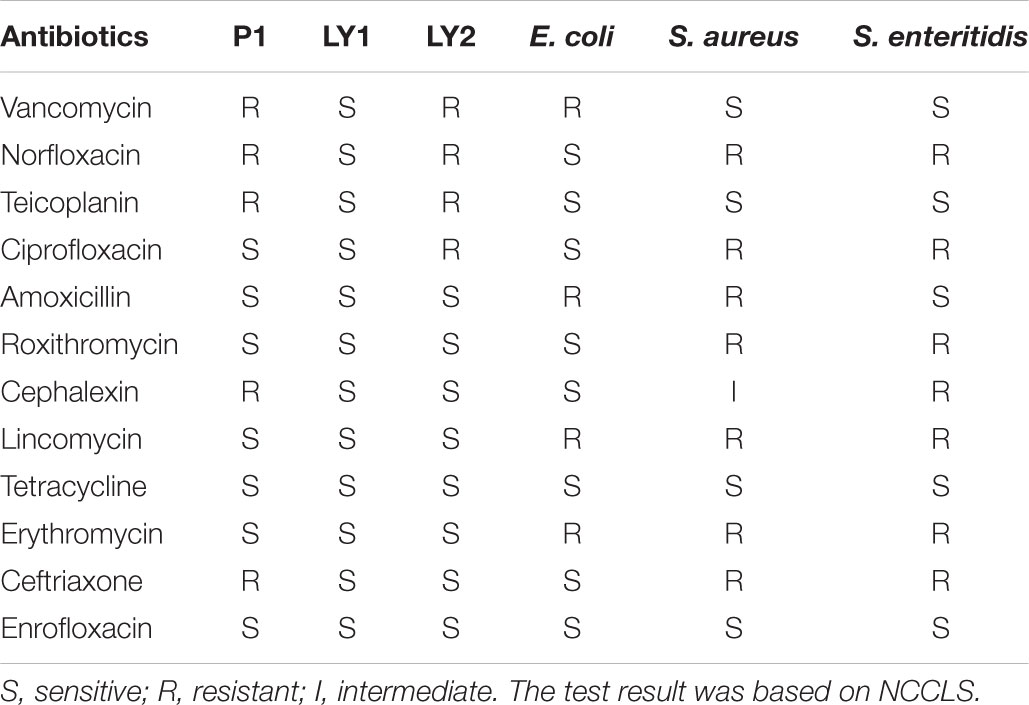
The drug sensitivity of isolated strains was tested with the disk diffusion method (Ghosh et al., 2015). The isolated strains and indicator strains (1 × 108 CFU/mL) were aspirated onto MRS agar plates and Luria-Bertani (LB) agar plates. The inhibition zone diameter was measured after 24 h of incubation at 37°C by using 12 drugs-sensitive tablets (vancomycin 30 μg, norfloxacin 10 μg, teicoplanin 30 μg, ciprofloxacin 5 μg, amoxicillin 25 μg, roxithromycin 15 μg, cephalexin 30 μg, lincomycin 2 μg, tetracycline 30 μg, erythromycin 15 μg, ceftriaxone 30 μg, and enrofloxacin 10 μg) from Hangwei Biotechnology Company, China (Mandal et al., 2016). Each antibiotic was graded as resistant (R), intermediate (I) or sensible (S) according to National Committee for Clinical Laboratory Standards (NCCLS) (National Committee for Clinical Laboratory Standards [NCCLS], 2003).
Acid and Bile Tolerance Properties
A good potential probiotic needs to be stable at a low pH and in the presence of high bile salts in the stomach and digestive tract (Khalil et al., 2018). Therefore, we carried out acid and bile resistance experiments using the Kobierecka’s method, with a few improvements (Kobierecka et al., 2017). Bacterial solutions (1 mL, 1 × 108 CFU/mL) were inoculated into MRS broth (7 mL) and incubated at 37°C. The survival rate was calculated with the viable plate counting method after 3 h. The MRS broth was adjusted to pH 2, pH 3, pH 4, and pH 5 with 1 M HCl, and the control broth was broth without HCl. To determine the survival rate of the strains under different bile salt concentrations, the strains were cultured for 24 h and spread evenly on MRS agar plates with bile salt concentrations of 0.1, 0.2, 0.3, 0.4, and 0.5% ox gall. Non-salted MRS agar plates served as controls. After 48 h, the viable colonies were counted.
In vitro Antibacterial Tests
The inhibition of the growth of pathogens is an important feature of probiotics (Khalil et al., 2018). The ability of the isolated strains to resist pathogenic bacteria was determined with an agar diffusion test (Zhang et al., 2016). E. coli (1 × 108 CFU/mL), S. enteritidis (1 × 108 CFU/mL) and S. aureus (1 × 108 CFU/mL) were used as the indicator pathogens in this experiment. The isolated strains were incubated at 37°C for 24 h in MRS broth and then 100 μL (1 × 108 CFU/mL) of bacterial solution was injected into LB agar plates punched with a 5 mm punch. The inhibition zone diameter was measured after 24 h.
Hemolytic Activity
All strains were cultured in MRS broth for 24 h, and then the bacterial solutions were streaked onto blood agar plates containing yak blood. The hemolytic activity of the strains was evaluated after 24 h at 37°C, and S. aureus was used as a positive control.
Determination of Growth Performance of Isolated Strains
All isolated strains were cultured in MRS broth for 24 h, and then the bacterial solutions (1 mL, 1 × 108 CFU/mL) were inoculated into MRS broth (7 mL). The OD600, CFU and pH were measured every 2 h up to 36 h (n = 3).
PCR Amplification and Detection of the Bacteriocin Gene
The DNA sequences of isolated strains were used as templates, and Ent P2 gene primers were used to detect the bacteriocin gene by PCR amplification as described by Kang and Lee (2005). The primer sequences for the Ent P2 gene were F (5′-GCTACGCGTTCATATGGTAATGGTG-3′) and R (5′-ATGTCCCATACCTGCCAAACCAGAAGC-3′). The primers were synthesized by Qingke Biotechnology Company (Wuhan, China). The PCR conditions were similar to Kang and Lee (2005), except that the annealing temperature was changed to 60°C.
PCR Detection of Antibiotic Resistance Genes
To identify the antibiotic resistance determinants, all the isolated strains were examined for the presence of tetracycline resistance genes [tet (L), tet (M), tet (O), and tet (S)] (Aarestrup et al., 2000) and glycopeptide-resistant genes [vanA (Dutkamalen et al., 1995) and vanB (Ramos-Trujillo et al., 2003)]. For detection of the possible presence of integron, a class 1 integrase-specific fragment of the Int1 gene was used (Sáenz et al., 2004). All the primer sequences used during the study are given in Supplementary Table S1.
Animal Toxicity Study
Mice were selected as experimental animals to evaluate the safety of the isolated strains. Briefly, a total of 20 male and 20 female KM mice (15–18 g) were housed under standard hygienic conditions at a temperature of 22 ± 2°C and 55 ± 2% humidity with a normal light-dark cycle. The animals were fed a basal diet and had free access to water during the entire experimental period. After 3 days of acclimation, the mice were randomly divided into 4 equal groups (n = 10). Mice in three of the experimental groups received P1, LY1, or LY2 at 1 × 109 CFU/day for 18 consecutive days. Mice in the control group were forcibly gavaged with an equal volume of vehicle. All animal protocols were carried out according to standard guidelines from the Good Laboratory Practice Guidelines (Hawkins, 1993). All the experiments were approved and reviewed by Institutional Animal Welfare and Research, Ethics Committee guidelines of Huazhong Agricultural University, Wuhan, China. The animals’ activity, behavior, hair luster, food intake, body weight and general health status were observed on a daily basis throughout the experiment. After 18 days of probiotic gavage, all the animals were sacrificed by cervical dislocation, and blood, liver and kidney samples were collected under sterile conditions. Statistical analysis of the data for multiple comparisons was performed by one-way analysis of variance followed by Duncan’s test. A level of P < 0.05 was considered statistically significant.
Bacterial Translocation
Bacterial translocation was analyzed in the blood, liver and kidneys (Zhou et al., 2000). Approximately 50 μL of blood was inoculated onto MRS agar plates and brain-heart infusion (BHI) agar plates for 48 h at 37°C. Tissue samples were homogenized in buffered peptone water (1 g/mL), and 100 mL of homogenates was cultured on MRS and BHI agar plates under the same conditions as those for the blood. The translocation rate was calculated as follows: translocation rate of bacteria = number of mice in which translocation was detected/total number of mice. Positive growth on agar plates was defined by a colony count of at least one. A chi-square test was used to analyze differences between rates (bacterial translocation incidence).
Preventive Effects of the Strains Against Bacterial Diseases in Mice
To evaluate the effects of the strains on preventing bacterial diseases, a total of 30 male and 30 female KM mice (15–18 g) were housed under the same conditions as those described previously. The mice were assigned to 6 treatment groups, including the P1-1 (gavaged with P1 1 × 108 CFU/day), P1-2 (gavaged with P1 1 × 109 CFU/day), LY1-1 (gavaged with LY1 1 × 108 CFU/day), LY1-2 (gavaged with LY1 1 × 109 CFU/day), control (gavaged with an equal volume of vehicle), and blank control (gavaged with an equal volume of vehicle) groups. After 30 days, mice in the P1-1, P1-2, LY1-1, LY1-2, and control groups were intraperitoneal challenged with 1 × 108 CFU of E. coli in the abdominal region, while the mice in blank control group were intra-peritoneal challenged with an equal volume of vehicle. After 2 days of infection, the surviving animals were sacrificed via cervical dislocation, and the liver, duodenum and spleen were harvested. Small specimens of the liver, spleen and duodenum were placed in 10% neutral formalin and routinely processed by embedding in paraffin. Tissue sections (4–5 μm) were stained with hematoxylin and eosin and examined under a light microscope. The animals’ activity, behavior, death, diarrhea, and general health status were closely observed throughout the experimental period. The presence of diarrhea was observed as mucoid, watery soft stools, often with a wet area around the anus according to Bolick et al. (2014).
Results
Isolation and Identification of Potential Probiotics
We observed milky white and raised colonies on MRS agar and three strains were selected: P1, LY1, and LY2. All strains were Gram-positive and catalase-negative. In addition, 16S rRNA sequence analysis showed that P1 was 99% homologous to Leuconostoc pseudomesenteroides, LY1 was 99% homologous to L. johnsonii and LY2 was 99% homologous to Lactobacillus mucosae. The phylogenic tree was constructed using the neighbor-joining method with MEGA 6 software (Figure 1).
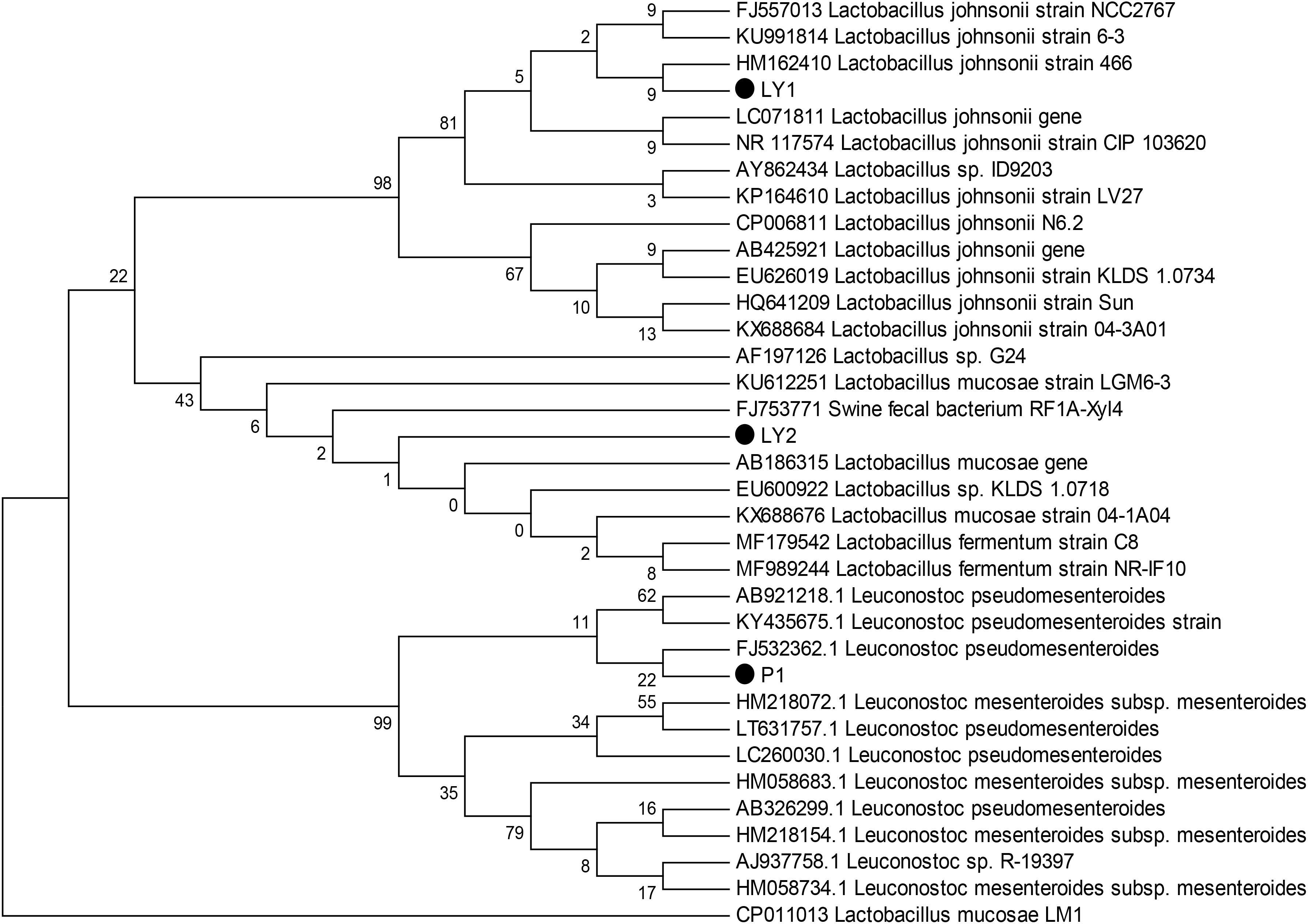
FIGURE 1. Phylogenetic tree based on the 16S rRNA gene sequences using the neighbor-joining method. The three isolated strains were P1, LY1, and LY2. The phylogenetic tree was statistically evaluated using 1000 bootstrap replicates.
Antibiotic Susceptibility Assay
P1, LY1, and LY2 showed a low degree of drug resistance (Table 1). LY1 was sensitive to all the antibiotics used in this study. Resistance rates of P1 and LY2 were lower than S. aureus and S. enteritidis as expected.
Acid and Bile Salts Tolerance
The isolates were incubated at pH 2, 3, 4, and 5 and at bile salt concentrations of 0.1, 0.2, 0.3, 0.4, and 0.5% at 37°C for 3 h. Strain P1 appeared to be tolerant to a pH of as low as 4 but then began to lose survival while strains LY1 and LY2 began to lose viability at pH < 5. On the other hand, the three strains’ survival rates ranged from 60.43 to 97.40% when less than 0.3% bile salt concentration (Figure 2). The survival rates were calculated as follows: survival rate = [c/co] ∗ 100%, where c and co represent the number of colonies in the experimental group and the control group, respectively.
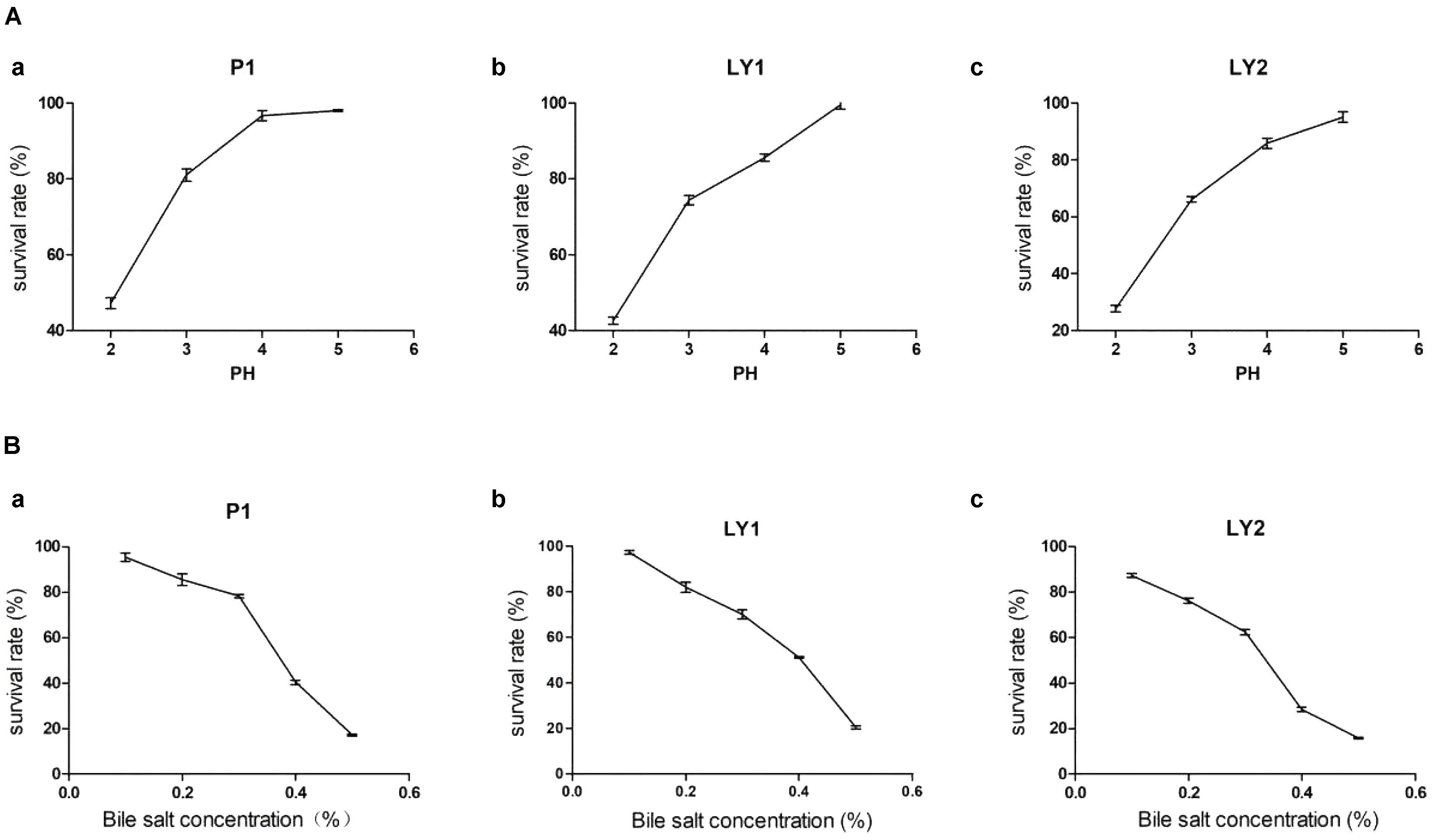
FIGURE 2. Acid and bile salts tolerance. (A) The tolerance of the isolated strains to acid. (a) P1. (b) LY1. (c) LY2. (B) The tolerance of the isolated strains to bile salts. (a) P1. (b) LY1. (c) LY2.
Antibacterial Tests in vitro
All the isolated strains showed inhibition activity against E. coli, S. aureus, and S. enteritidis in the well diffusion assay (Figure 3). The inhibition zone diameter ranged from 17.17 to 22.60 mm. LY1 exhibited greater inhibition of E. coli, with a diameter of 21.97 mm (Figure 4A). P1 showed the strongest inhibitory effect against S. aureus, with a diameter of 17.17 mm (Figure 4B). LY1 had the strongest inhibitory effect on S. enteritidis, with a zone of inhibition of 22.60 mm (Figure 4C).
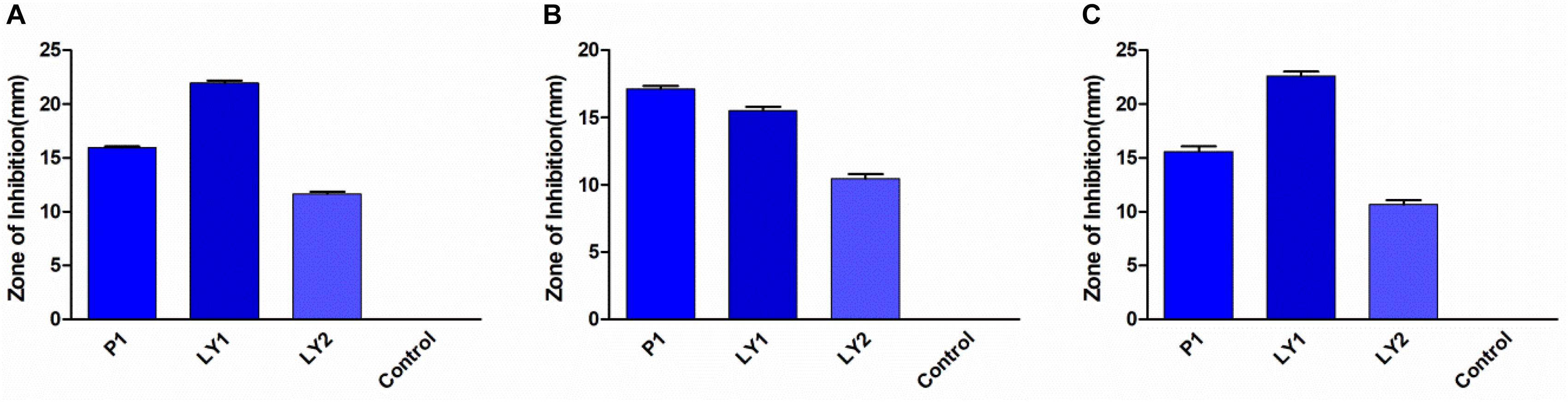
FIGURE 3. The results of the antibacterial experiment. (A) The inhibition zone diameters of P1, LY1, and LY2 against E. coli. (B) The inhibition zone diameters of P1, LY1, and LY2 against S. aureus. (C) The inhibition zone diameters of P1, LY1, and LY2 against S. enteritidis.
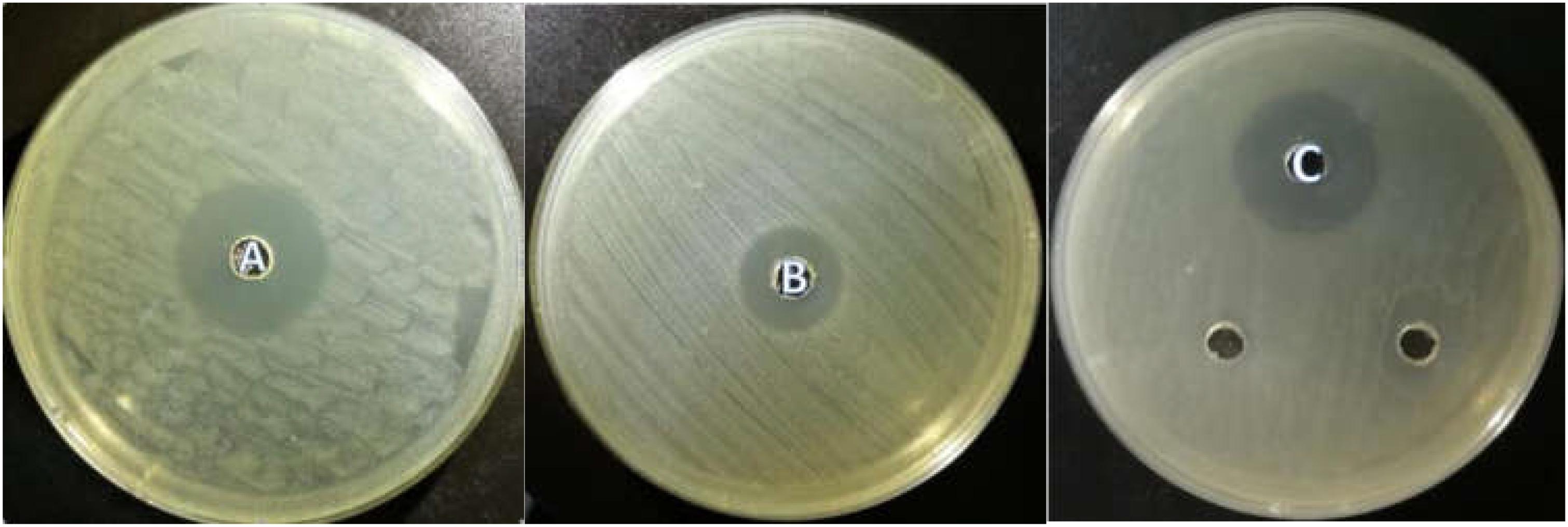
FIGURE 4. (A) The inhibition zone diameter of LY1 against E. coli. (B) The inhibition zone diameter of P1 against S. aureus. (C) The inhibition zone diameter of LY1 against S. enteritidis.
Isolated Strains Growth Time Curve
The results about isolated strains growth time curve are shown in Figures 5A–C. The growth of microorganisms in liquid cultures assayed by OD600 clearly showed that the logarithmic growth phases of P1 and LY1 were 2–12 and 4–14 h, respectively. At the same time, the pH values for P1 and LY1 clearly dropped. The CFU of P1 began to decrease after 24 h, and the OD600 value did not decrease. In the logarithmic phase of LY2, the pH value rapidly decreased at approximately 4–12 h, entered the plateau phase, and ultimately declined after 24 h, but LY1 entered the decline stage approximately 22 h later.
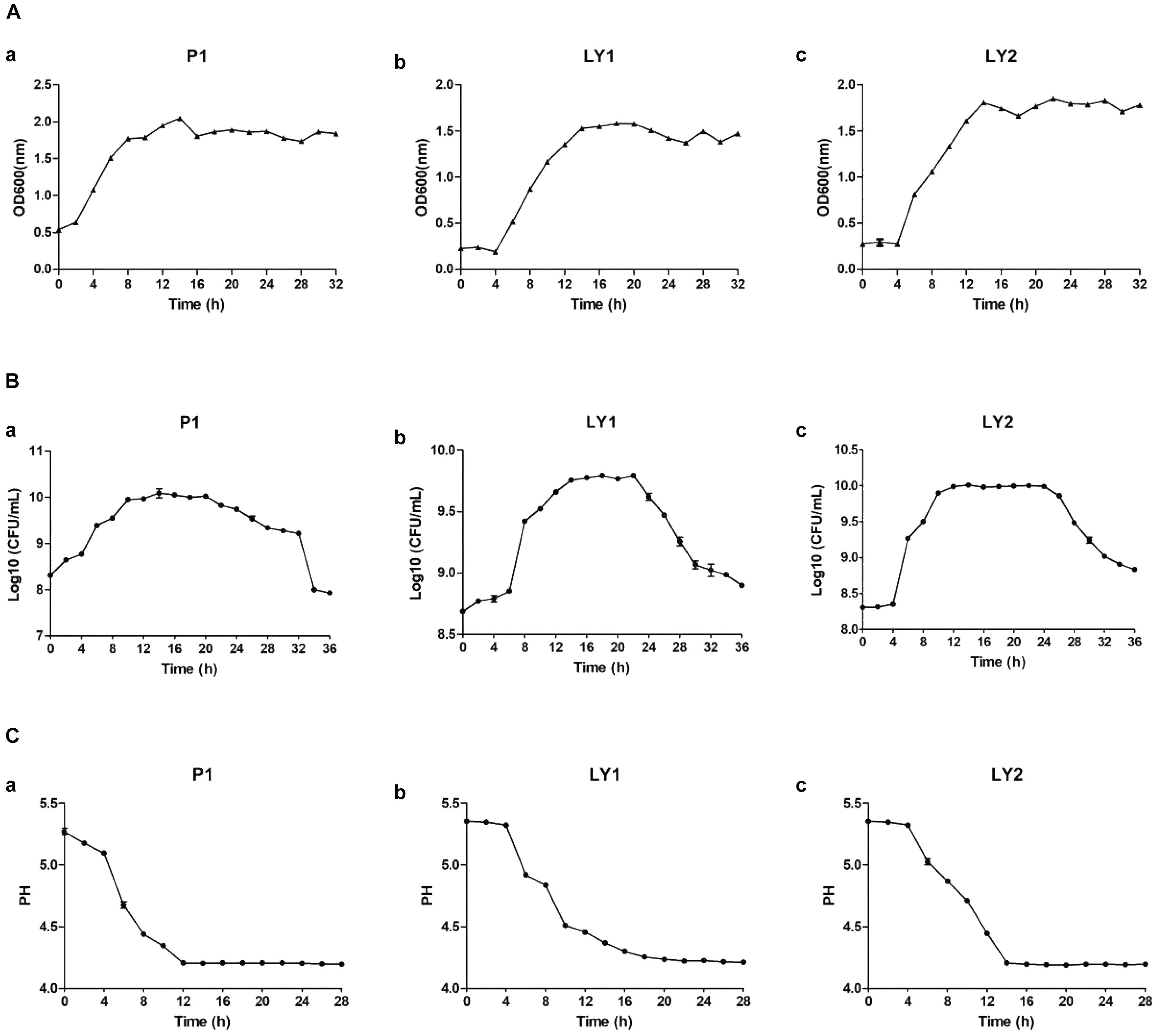
FIGURE 5. Measurement of OD600 values, colony-forming units and pH. (A) OD600 values measured every 2 h for a total of 32 h. (a) P1. (b) LY1. (c) LY2. (B) The colony-forming units were analyzed every 2 h for a total of 32 h, and the number of colonies was calculated as the log10 value of CFU per milliliter. (a) P1. (b) LY1. (c) LY2. (C) pH measurements taken every 2 h for a total of 26 h. (a) P1. (b) LY1. (c) LY2.
Hemolytic Activity
After 24 h, the hemolytic activity of the strains was judged by observing the hemolytic rings on blood agar plates. Complete lysis (β-hemolysis) was caused by the positive control strain, whereas P1, LY1, and LY2 were not involved in the lysis of erythrocytes (γ-hemolysis).
PCR Amplification of the Bacteriocin Gene
All isolated strains were positive for Ent P2, and the template DNA and Ent P2 primers produced amplicons of approximately 130 bp (Figure 6).
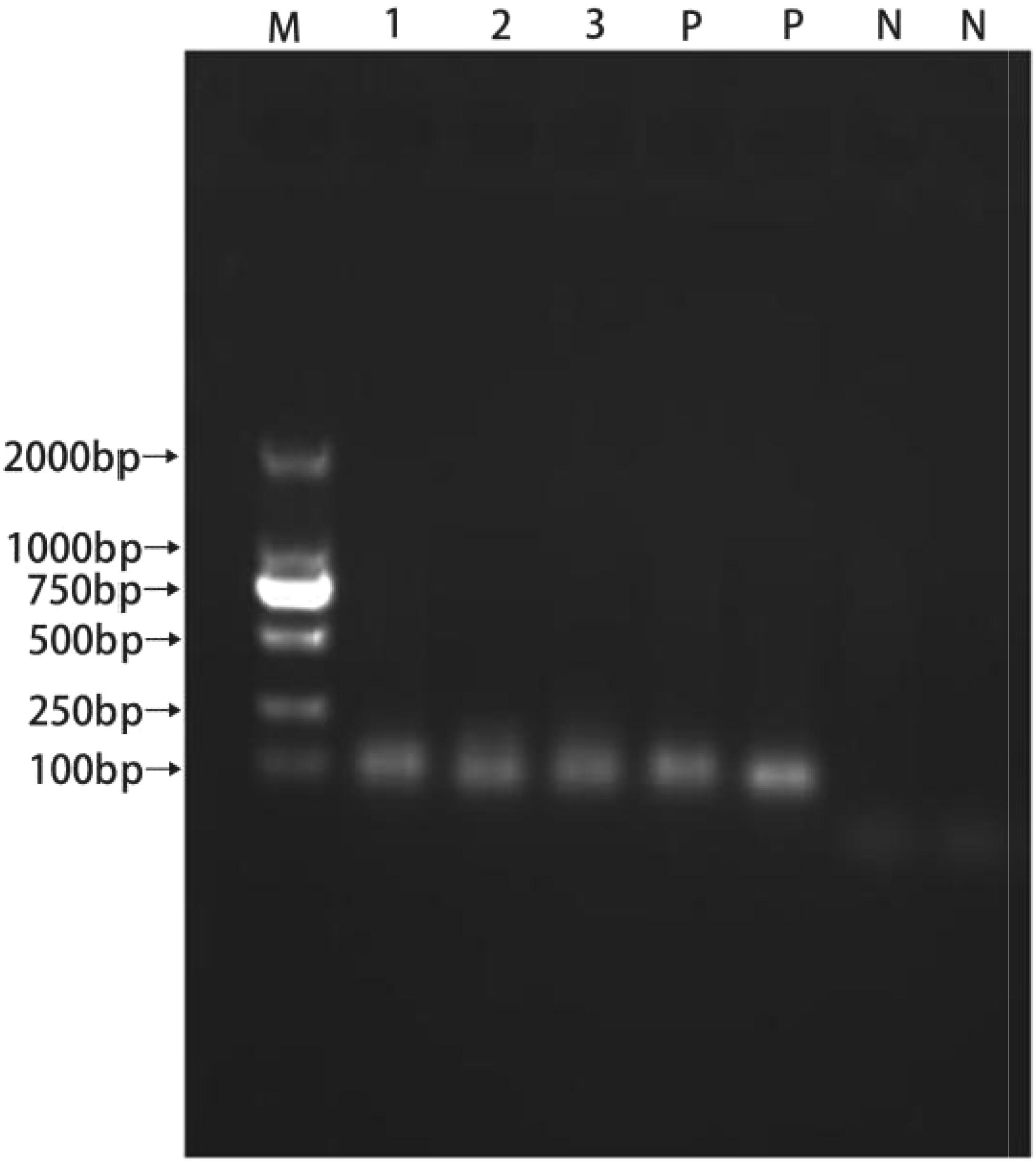
FIGURE 6. PCR products resulting from amplification with the enterocin P2 primers. (1) P1; (2) LY; (3) LY2; (P) positive control; (N) negative control.
Presence of Resistance Genes Analyzed by PCR
The tetracycline resistance-determining genes tet (L), tet (S), tet (O), and tet (M) were not found in any of the isolated strains, and the strains did not contain the integron Int1 gene. It was found that LY2 contained the vanA gene, but none of the strains had the vanB gene.
Animal Toxicity Study
During the experiment, no animals were recorded to have symptoms such as disease, death or diarrhea. Animals in P1, LY1 and control groups did not show significant differences in behavior, hair, mental status or feed intake (data not shown). However, the data in LY2 group was abandoned because mice exhibited lethargy and unresponsiveness. The appearance of the internal organs and intestinal segments was normal, and no ulcers or adhesions were observed by macroscopic evaluation. As shown in Figure 7, there was no significant difference in the body weight changes between the control, P1 and LY1 groups.
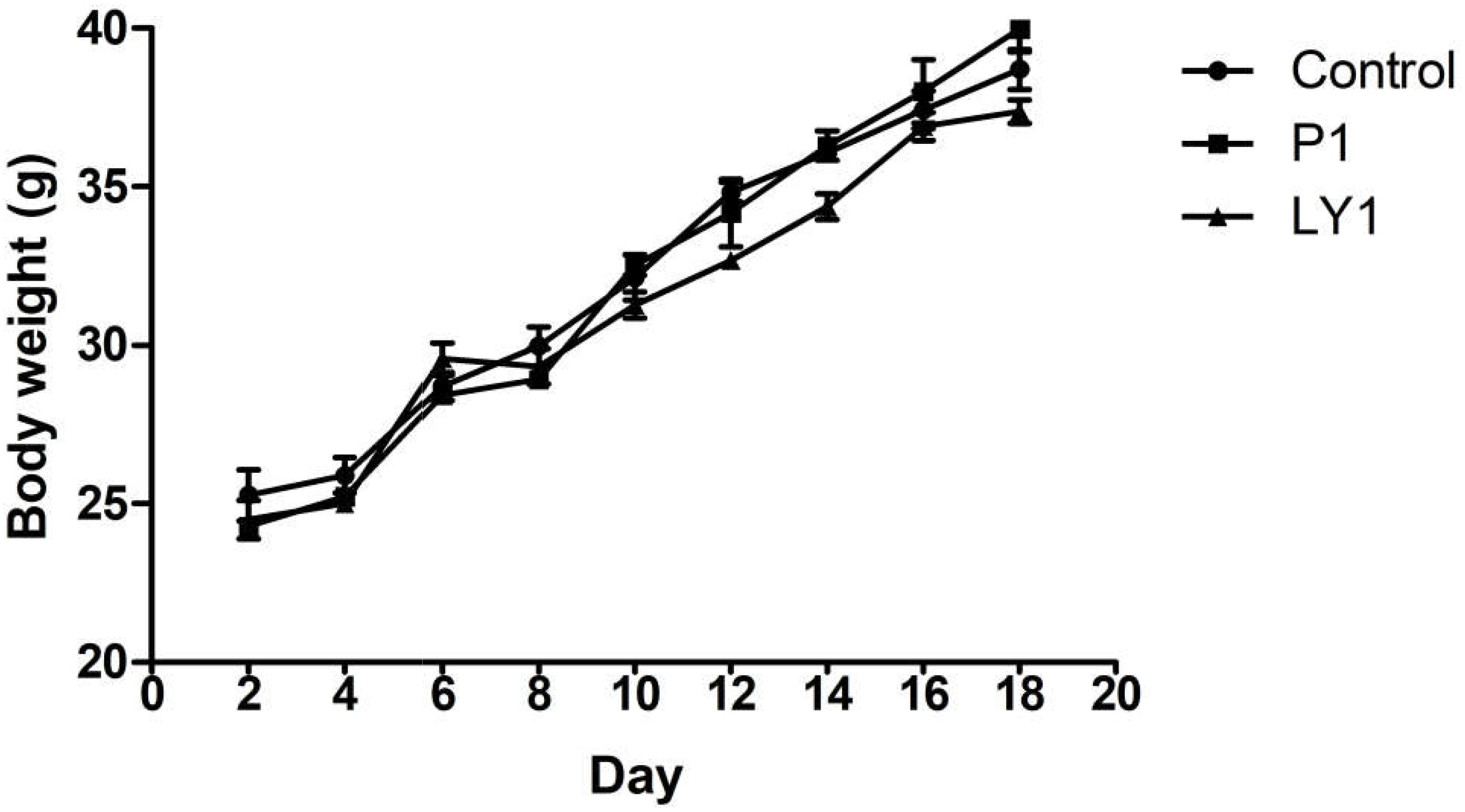
FIGURE 7. Body weight changes in the mice of the control and probiotic-treated groups. ● Represent the changes of body weight in the mice of the control group; ■ represent the changes of body weight in the mice of the P1 group; ▲ represent the changes of body weight in the mice of the LY1 group. The values are presented as the mean ± SD (n = 10). A level of P < 0.05 was considered statistically significant.
Translocation Rates of Bacteria
There was no statistically difference in the incidence of translocation of bacteria to the liver or kidney between the control group and any of the experimental, and no bacteremia was observed in any of the groups.
Strains Preventing Bacterial Diseases
The mice in the control group showed the most serious disease symptoms: matted hair, poor mental state, high rate of diarrhea and death. Compared with mice in the control, P1-1 and LY1-1 groups, mice in the P1-2 and LY1-2 groups showed the lowest rates of diarrhea and mortality (Table 2). In this experiment, clear degeneration and congestion were detected within the livers of mice in the control group but not within those of mice in the blank control group. Mice in the P1-2 and LY1-2 groups exhibited reduced degeneration and congestion (Supplementary Figure S1). In terms of the spleen, mice in the control group showed severe lymphopenia, a large increase in red blood cells and a loss of normal structure compared with mice in the blank control group. The mice in the P1-2 and LY1-2 groups showed significant recovery compared to the mice in the control group (Supplementary Figure S2). In addition, degeneration was observed in the intestinal villi of the duodenum in the control group mice, whereas there was no degeneration in the P1-2 and LY1-2 group mice (Supplementary Figure S3).
Discussion
At present, antibiotics are widely used in the animal husbandry, humans, agriculture and aquaculture, but some problems have also emerged, especially antibiotic resistance (Tenover and Hughes, 1996). Diarrhea is a complex disease that can easily be caused by changes in the environment or other unfavorable conditions, and it is one of the major causes of economic losses in the cattle industry. E. coli, S. enteritidis, and S. aureus are common pathogens causing diarrhea (Yongil and Kyoungjin, 2014). Probiotics can maintain the balance of the intestinal flora, inhibit the colonization and survival of pathogens in the intestine and prevent inflammation (Archambaud et al., 2012; Yu et al., 2015). The purpose of our study was to find a safe and effective way to reduce the incidence of bacterial diseases in yaks. Recently, a large number of yaks have died due to bacterial infections. Yak meat and milk are necessities of daily life for Tibetans, so the use of antibiotics may pose a health risk to local herdsmen. Hence, the present study could be used as a reference for the prevention and control of bacterial infection in the Tibetan Plateau of China (Wu et al., 2017). The results show that LY1 has a strong antagonistic effect against E. coli and S. enteritidis. However, P1 had a strong antagonistic effect against S. aureus.
Bactericidal peptides can be produced by enterocins, including enterocins A, B, P, and L50, which are class II bacteriocins (Aymerich et al., 1996). Enterocin P is unique with a wide inhibitory spectrum (Herranz et al., 2001). In this study, all the isolated strains were positive for the Ent P2 gene and showed positive antagonism against pathogenic bacteria. The key for probiotics to play a role in promoting the physiological health of the host is the ability to survive the intestine (Kaushik et al., 2009). Therefore, the isolated strain must be able to survive under low pH and high bile salt concentrations (Mandal et al., 2016). Isolated strains were tested for acidity and bile salt tolerance, and the results showed that they were highly tolerant, especially P1 and LY1.
According to the FAO/WHO report, although Lactobacillus is generally considered safe, they may cause some side effects (Food Agriculture Organization/World Health Organization [FAO/WHO], 2002). Therefore, it is recommended that antibiotic resistance and toxicity studies be used for safety testing. According to reports, some probiotics may cause hemolysis (Spinosa, 2009). The hemolytic properties of the isolated strains were tested, and no hemolytic activity was recorded for P1, LY1, or LY2. As a health threat, antibiotic resistance genes may be transmitted to bacteria in the gut by probiotics. Therefore, detection of the antibiotic resistance of probiotics is considered to be an important part of the safety testing of probiotics in vitro (European Food Safety Authority [EFSA], 2008). It has been reported that the tet gene present in some Lactobacillus strains may be transferred to other bacteria (Toomey et al., 2009). In the detection of resistance genes, the van gene deserves attention because vancomycin can be the last antibiotic used to cure infections caused by bacteria that are resistant to antibiotics (Woodford et al., 1995). In this study, the resistance test of the isolated strains and the indicator bacteria showed that the resistance of the indicator bacteria was higher than that of the isolated strains. The presence of the genes tet (L), tet (M), tet (O), tet (S), vanA, vanB, and Int1 was analyzed in the experiment, and neither P1 nor LY1 contained these genes. For LY2, only the vanA gene was detected.
Although these data illustrated the safety of the strains in vitro, we decided to also test the oral toxicity in animals to evaluate their safety in vivo. After being gavaged with the potential probiotic strains P1, LY1, and LY2 at high doses for 18 days, mice in P1 and LY1 presented no symptoms of illness, death or infection were recorded. However, LY2 was discarded due to vanA gene and poor mental state of the mice. Bacterial translocation is an important aspect of the detection of probiotic toxicity (Steffen and Berg, 1983). No bacteremia was recorded in any of the groups, and the detection rates of bacteria in the liver and kidney were similar between the control group and the experimental groups. Therefore, the occurrence of this translocation may not be related to the use of probiotics. In bacterial diseases prevention test in mice, P1 and LY1 were effective in reducing diarrhea associated with bacterial diseases. In conclusion, P1 and LY1 have strong antibacterial properties and probiotic potential and proved to be safe in the in vivo and in vitro experiments. Therefore, P1 and LY1 could be used to reduce diarrhea associated with bacterial diseases. Meanwhile, it probably has better clinical results if these two probiotics combined when administered because the two remaining strains have different antibacterial activities. Further experiments should be performed before these strains are used in clinical practice.
Author Contributions
JL, YW, AL, and HZ provided the research idea. KM, XJ, MI, AL, JJ, MW, LZ, and YW contributed reagents, materials, and analysis tools. YW and AL wrote the manuscript. JL, HZ, and KM revised the manuscript. All authors participated in writing and reviewing the manuscript.
Funding
This study was supported by the Tibet Autonomous Region Science Fund and the Chinese Agricultural Research Systems (CARS-37). Tibet Autonomous Region Science Fund (ZDZX2018000043).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02987/full#supplementary-material
References
Aarestrup, F. M., Agerso, Y., Gernersmidt, P., Madsen, M., and Jensen, L. B. (2000). Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans inthe community, chickens, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37, 127–137. doi: 10.1016/S0732-8893(00)00130-9
Archambaud, C., Nahori, M. A., Soubigou, G., Bécavin, C., Laval, L., and Lechat, P. (2012). Impact of lactobacilli on orally acquired listeriosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16684–16689. doi: 10.1073/pnas.1212809109
Aymerich, T., Holo, H., Havarstein, L. S., Hugas, M., Garriga, M., and Nes, I. F. (1996). Biochemical and genetic characterization of enterocin a from Enterococcus faecium, a new Antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62, 1676–1682.
Bolick, D. T., Kolling, G. L., Nd, M. J., de Oliveira, L. A., Tung, K., Philipson, C., et al. (2014). Zinc deficiency alters host response and pathogen virulence in a mouse model of entero aggregative Escherichia coli-induced diarrhea. Gut Microbes 5, 618–627. doi: 10.4161/19490976.2014.969642
Casado, M. M. C., Benomar, N., Lerma, L. L., Gálvez, A., and Abriouel, H. (2014). Antibiotic resistance of Lactobacillus pentosus and Leuconostoc pseudomesenteroides isolated from naturally-fermented Aloreña table olives throughout fermentation process. Int. J. Food Microbiol. 172, 110–118. doi: 10.1016/j.ijfoodmicro.2013.11.025
Cho, Y. I., Han, J. I., Wang, C., Cooper, V., Schwartz, K., and Engelken, T. (2013). Case-control study of microbiological etiology associated with calf diarrhea. Vet. Microbiol. 166, 375–385. doi: 10.1016/j.vetmic.2013.07.001
Dong, H. L., Zhang, H., Li, K., Khalid, M., Mujeeb, U. R., Fazul, N., et al. (2018). Prevalence and potential risk factors for Escherichia coli isolated from tibetan piglets with white score diarrhea. Pak. J. Zool. 50, 57–63. doi: 10.17582/journal.pjz/2018.50.1.57.63
Dutkamalen, S., Evers, S., and Courvalin, P. (1995). Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant Enterococci by PCR. J. Clin. Microbiol. 33, 24–27.
Elhaig, M. M., Selim, A., Mahmoud, M. M., and El-Gayar, E. K. (2016). Molecular confirmation of Trypanosoma evansi and Babesia bigemina in cattle from lower Egypt. Pak. Vet. J. 36, 409–414.
European Food Safety Authority [EFSA] (2008). Panel on Additives, and Products or Substances used in Animal Feed [FEEDAP]. Technical guidance – Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 732, 1–15. doi: 10.2903/j.efsa.2008.732
Fonseca, W., Lucey, K., Jang, S., Fujimura, K. E., Rasky, A., Ting, H.-A., et al. (2017). Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol. 10, 1569–1580. doi: 10.1038/mi.2017.13
Food Agriculture Organization/World Health Organization [FAO/WHO] (2002). Guidelines for the Evaluation of Probiotics in Food: Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Rome: FAO.
Food Agriculture Organization/World Health Organization [FAO/WHO] (2006). Guidelines for the Evaluation of Probiotics in Food.Report of a Joint FAO/WHO Working Group. Rome: FAO.
Food Agriculture Organization/World Health Organization [FAO/WHO] (2011). Expert Consultation on the Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Cordoba: FAO.
Ghosh, K., Ray, M., Adak, A., Halder, S. K., Das, A., and Jana, A. (2015). Role of probiotic Lactobacillus fermentum KKL1 in the preparation of a rice based fermented beverage. Bioresour. Technol. 188, 161–168. doi: 10.1016/j.biortech.2015.01.130
Hawkins, R. I. (1993). “Good laboratory practice,” in Experimental Toxicology, The Basis Issues, 2nd Edn, eds D. Anderson and D. M. Corning (Cambridge: Royal Society of Chemistry), 523–541.
Herranz, C., Cintas, L., Hernandez, P., Moll, G., and Driessen, A. (2001). Enterocin P causes potassium ion efflux from Enterococcus faecium T136 cells. Antimicrob. Agents Chemother. 45, 901–904. doi: 10.1128/AAC.45.3.901-904.2001
Heuer, C., Healy, A., and Zerbini, C. (2007). Economic effects of exposure to bovine viral diarrhea virus on dairy herds in New Zealand. J. Dairy Sci. 90, 5428–5438. doi: 10.3168/jds.2007-0258
Kaltenbach, G., and Heitz, D. (2004). Antibiotic-associated diarrhea in the elderly. Rev. Med. Intern. 25, 46–53. doi: 10.1016/j.revmed.2003.10.002
Kang, J. H., and Lee, M. S. (2005). Characterization of a bacteriocin produced by Enterococcus faecium GM-1 isolated from an infant. J. Appl. Microbiol. 98, 1169–1176. doi: 10.1111/j.1365-2672.2005.02556.x
Kaushik, J. K., Kumar, A., Duary, R. K., Mohanty, A. K., Grover, S., and Batish, V. K. (2009). Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One 4:e8099. doi: 10.1371/journal.pone.0008099
Khalil, E. S., AbdManap, M. Y., Mustafa, S., Alhelli, A. M., and Shokryazdan, P. (2018). Probiotic properties of exopolysaccharide-producing Lactobacillus strains isolated from Tempoyak. Molecules 23:398. doi: 10.3390/molecules23020398
Kobierecka, P. A., Wyszyńska, A. K., Aleksandrzak-Piekarczyk, T., Kuczkowski, M., Tuzimek, A., Piotrowska, W., et al. (2017). In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. Microbiol. Open 6:e00512. doi: 10.1002/mbo3.512
Li, J., Li, K., Shahzad, M., Han, Z., Nabi, F., and Gao, J. (2015). Seroprevalence of Bluetongue virus in domestic yaks (Bosgrunniens) in Tibetan regions of China based on circulating antibodies. Trop. Anim. Health Prod. 47, 1221–1223. doi: 10.1007/s11250-015-0853-0
Makhloufi, K. M., Carrémlouka, A., Peduzzi, J., Lombard, C., Reenen, C. A. V., and Dicks, L. M. T. (2013). Characterization of Leucocin B-KM432Bz from Leuconostoc pseudomesenteroides Isolated from Boza, and Comparison of its Efficiency to Pediocin PA-1. PLoS One 8:e70484. doi: 10.1371/journal.pone.0070484
Mandal, H., Jariwala, R., and Bagchi, T. (2016). Isolation and characterization of lactobacilli from human faeces and indigenous fermented foods for their potential application as probiotics. Can. J. Microbiol. 62, 349–359. doi: 10.1139/cjm-2015-0576
Mehmood, K., Hui, Z., Sabir, A. J., Abbas, R. Z., Ijaz, M., Durrani, A. Z., et al. (2017). A review on epidemiology, global prevalence and economical losses of fasciolosis in ruminants. Microb. Pathog. 109, 253–262. doi: 10.1016/j.micpath.2017.06.006
National Committee for Clinical Laboratory Standards [NCCLS] (2003). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 6th Edn. Wayne, PA: NCCLS.
Ramos-Trujillo, E., Pérez-Roth, E., Méndez-Alvarez, S., and Claverie-Martín, F. (2003). Multiplex PCR for simultaneous detection of enterococcal genes vanA and vanB and staphylococcal genes mecA, ileS-2 and femB. Int. Microbiol. 6, 113–115. doi: 10.1007/s10123-003-0118-z
Rautio, M., Jousimiessomer, H., Kauma, H., Pietarinen, I., Saxelin, M., and Tynkkynen, S. (1999). Liver abscess due to a Lactobacillus rhamnosus Strain Indistinguishable from L. rhamnosus Strain GG. Clin. Infect. Dis. 28, 1159–1160. doi: 10.1086/514766
Sáenz, Y., Briñas, L., Domínguez, E., Ruiz, J., Zarazaga, M., and Vila, J. (2004). Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 48, 3996–4001. doi: 10.1128/AAC.48.10.3996-4001.2004
Spinosa, M. R. (2009). The trouble in tracing opportunistic pathogens: cholangitis due to Bacillus in a french hospital caused by a strain related to an italian probiotic? Microb. Ecol. Health Dis. 12, 99–101. doi: 10.1080/089106000435491
Steffen, E. K., and Berg, R. D. (1983). Relationship between cecal population levels of indigenous bacteria and translocation to the mesenteric lymph nodes. Infect. Immun. 39, 1252–1259.
Stine, K. E., and Brown, T. M. (1996). “Measuring toxicity and assessing risk,” in Principles of Toxicology, eds K. E. Stine and T. M. Brown (Whitefish, MT: CRC Lewis Publishers), 1–10.
Tenover, F. C., and Hughes, J. M. (1996). The challenges of emerging infectious diseases. development and spread of multiply-resistant bacterial pathogens. JAMA 275, 300–304. doi: 10.1001/jama.1996.03530280052036
Toomey, N., Monaghan,Á, Fanning, S., and Bolton, D. (2009). Transfer of antibiotic resistance marker genes between lactic acid bacteria in model rumen and plant environments. Appl. Environ. Microbiol. 75, 3146–3152. doi: 10.1128/AEM.02471-08
Wang, H., Ni, X., Qing, X., Liu, L., Xin, J., and Luo, M. (2018). Probiotic Lactobacillus johnsonii BS15 improves blood parameters related to immunity in broilers experimentally infected with subclinical necrotic enteritis. Front. Microbiol. 9:49. doi: 10.3389/fmicb.2018.00049
Wang, L., Zhang, H., Rehman, M. U., Mehmood, K., Jiang, X., and Iqbal, M. (2018). Antibacterial activity of Lactobacillus plantarum isolated from Tibetan yaks. Microb. Pathog. 115, 293–298. doi: 10.1016/j.micpath.2017.12.077
Wen, X. B., Jiang, H. T., Zhang, Y. L., Lang, X. Y., Liu, J., and Ni, H. B. (2016). Rapid and sensitive diagnosis of cattle anaplasmosis by loop-mediated isothermal amplification (LAMP). Pak. Vet. J. 36, 174–178.
Woodford, N., Johnson, A. P., Morrison, D., and Speller, D. C. (1995). Current perspectives on glycopeptide resistance. Clin. Microbiol. Rev. 8, 585–615. doi: 10.1128/CMR.8.4.585
Wu, Q., Dong, H., Zhang, H., Mehmood, K., Tian, F., Ijaz, M., et al. (2018a). Hypoxia promotes VEGF and HIF-1α expressions in endometrial epithelium cells of yaks (Bosgrunniens) in Qinghai-Tibet plateau. Int. J. Agric. Biol. 20, 911–915.
Wu, Q., Zhang, H., Mehmood, K., Li, K., Chang, Z., Jiang, X., et al. (2018b). Biological characteristics and phylogenetic analysis of lactic acid bacteria isolated from free-range yaks (Bosgrunniens) in Qinghai-Tibet plateau. Int. J. Agric. Biol. 20, 902–906.
Wu, Q. X., Zhang, H., Khalid, M., Li, K., Chang, Z. Y., Jiang, X., et al. (2017). Biological characteristics and phylogenetic analysis of lactic acid bacteria isolated from free-range yaks (Bosgrunniens)in Qinghai-tibet Plateau. Int. J. Agric. Biol. 20, 902–906.
Yilmaz, R., Cangul, I. T., Onat, K., Akkoc, A., Ozyigit, M. O., and Akdesir, E. (2016). Histopathological, immunohistochemical and bacteriological characterization of Mycoplasma bovis pneumonia in cattle. Pak. Vet. J. 36, 316–321.
Yongil, C., and Kyoungjin, Y. (2014). An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J. Vet. Sci. 15, 1–17. doi: 10.4142/jvs.2014.15.1.1
Yu, Q., Yuan, L., Deng, J., and Yang, Q. (2015). Lactobacillus protects the integrity of intestinal epithelial barrier damaged by pathogenic bacteria. Front. Cell. Infect. Microbiol. 5:26. doi: 10.3389/fcimb.2015.00026
Zhang, H., Dong, H. L., Khalid, M., Li, K., Fazul, N., Chang, Z. Y., et al. (2018). Physiological variations among blood parameters of domestic cats at high- and low-altitude regions of China. Arch. Physiol. Biochem. 124, 458–460. doi: 10.1080/13813455.2018.1423623
Zhang, H., Rehman, M. U., Li, K., Luo, H., Lan, Y., and Nabi, F. (2016). Antimicrobial resistance of Escherichia coli isolated from tibetan piglets suffering from white score diarrhea. Pak. Vet. J. 37, 43–46.
Zhou, J. S., Shu, Q., Rutherfurd, K. J., Prasad, J., Birtles, M. J., and Gopal, P. K. (2000). Safety assessment of potential probiotic lactic acid bacterial strains Lactobacillus rhamnosus HN001, Lb. acidophilus HN017, and Bifidobacterium lactis HN019 in BALB/c mice. Int. J. Food Microbiol. 56, 87–96. doi: 10.1016/S0168-1605(00)00219-1
Keywords: antibacterial activity, safety, Lactobacillus strains, Leuconostoc pseudomesenteroides, yaks
Citation: Wang Y, Li A, Jiang X, Zhang H, Mehmood K, Zhang L, Jiang J, Waqas M, Iqbal M and Li J (2018) Probiotic Potential of Leuconostoc pseudomesenteroides and Lactobacillus Strains Isolated From Yaks. Front. Microbiol. 9:2987. doi: 10.3389/fmicb.2018.02987
Received: 24 May 2018; Accepted: 19 November 2018;
Published: 04 December 2018.
Edited by:
Pilar García, Spanish National Research Council (CSIC), SpainReviewed by:
Valentina Bernini, Università degli Studi di Parma, ItalyJohn F. Kokai-Kun, Synthetic Biologics, Inc., United States
Copyright © 2018 Wang, Li, Jiang, Zhang, Mehmood, Zhang, Jiang, Waqas, Iqbal and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiakui Li, bGlqazIxMEBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Yaping Wang
Yaping Wang Aoyun Li1†
Aoyun Li1† Hui Zhang
Hui Zhang Lihong Zhang
Lihong Zhang Jiakui Li
Jiakui Li