- 1Department of Pharmacology and Toxicology, College of Animal Husbandry and Veterinary Science, Henan Agricultural University, Zhengzhou, China
- 2Department of Animal Science, Henan Institute of Science and Technology, Xinxiang, China
Tet(M)-type proteins confer resistance to tetracycline and related antibiotics by interacting with the ribosome. Genes encoding Tet(M) have been found in a range of bacteria, including Escherichia coli. In the current study, conjugation experiments were performed between seven different tetracycline-resistant, azide-susceptible E. coli strains isolated from ducks and tetracycline-sensitive, azide-resistant E.coli J53. Transconjugants were obtained from two of the strains at a frequency of 1.2 × 10−8. PCR, southern blotting and sequencing demonstrated that tet(M) in the transconjugants was located on a ~50 kb IncHI2-type plasmid and was part of a composite transposon, designated Tn6539. This transposon is flanked by two IS26 elements in opposite orientation and contains the Tn3ΔtnpA+Δorf13-lp-tet(M)+gamma delta+tnpX+ΔtnpR sequences. The Δorf13-lp-tet(M) sequence was a highly conserved genetic fragment in E. coli harboring tet(M) and mainly located in the composite transposons flanked by IS6-family elements. In summary, Tn6539 is a new composite transposon capable of horizontal transfer of tet(M) among E. coli isolates.
Introduction
Resistance to tetracycline and related antibiotics can be conferred by efflux pumps, enzymatic inactivation and ribosomal protection proteins such as the ones encoded by tet(M). Tet(M) confers a wider spectrum of resistance to tetracyclines than efflux proteins except for Tet(B) and has the widest host range of any tetracycline resistance gene (Chopra and Roberts, 2001; Roberts and Schwarz, 2016). Currently, tet(M) has been found in more than 60 genera (http://faculty.washington.edu/marilynr/), likely because of the association of tet(M) with integrative and conjugative transposons, which facilitate horizontal transfer. For example, tet(M) was found to be associated with Tn916/Tn1545-like conjugative transposons in Enterococcus spp. (Cauwerts et al., 2007). Additionally, tet(M) was also found in Tn5801-like conjugative transposons in Staphylococcus aureus and Tn5397-like transposons in Clostridium difficile and Enterococcus faecium (Agersø et al., 2006; Vries et al., 2009). A tet(M) homolog was not found in clinical isolates of Escherichia coli until 2004 (Bryan et al., 2004), but has been described since then in isolates from different sources (Jones et al., 2006; Tuckman et al., 2007; Jurado-Rabadán et al., 2014; Gilrane et al., 2017). The E. coli isolates carrying tet(M) not only conferred resistance to oxytetracycline, tetracycline and doxycycline, but also to minocycline (Jones et al., 2006; Hu et al., 2013). At the same time, Tet(M) protein has the potential to acquire mutation leading to increased MICs of tigecycline (Linkevicius et al., 2015). Therefore, the spread of tet(M) has made tetracycline treatment for E. coli infections a clinical dilemma. However, which mobile genetic elements facilitate horizontal transfer of tet(M) among E. coli isolates is still poorly understood. To the best of our knowledge, only one report has described a mobile element containing tet(M) flanked by IS26 and ISVs1 in human (Jones et al., 2006).
Our previous study revealed that tet(M) plays a role in doxycycline resistance of E. coli isolated from ducks in China (Hu et al., 2013). In the current study, seven of the previously isolated doxycycline-resistant strains were used in conjugation experiments and in two cases, transfer of tet(M) was apparently mediated by a plasmid that harbored a new composite transposon, designated Tn6539. The structure of this transposon was elucidated by PCR, cloning, and sequencing experiments.
Materials and Methods
Bacterial Strains
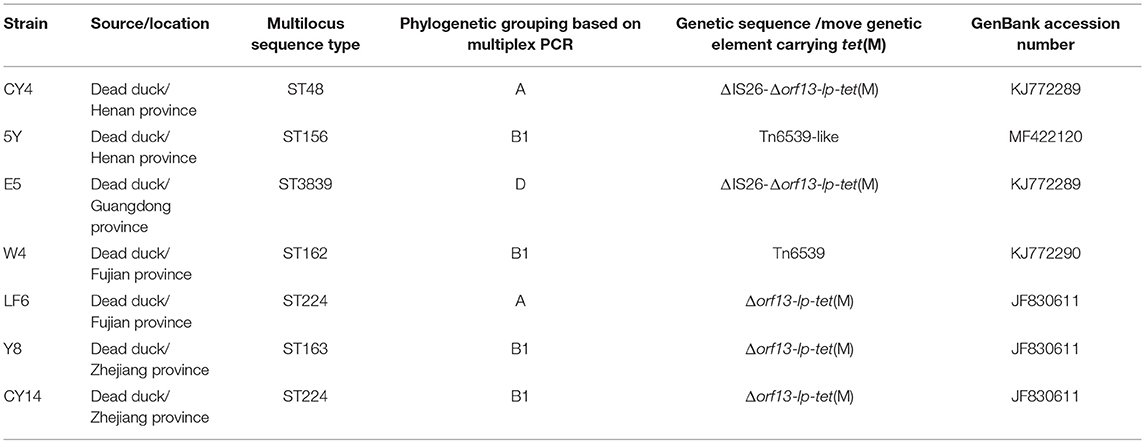
Seven full-length tet(M)-positive E. coli isolates CY4, E5, W4, LF6, Y8, CY14, and 5Y (Table 1) containing part of orf13, the promoter regions of tet(M), the Tet(M) leader peptide gene, and intact tet(M) were previously isolated from ducks (Hu et al., 2013). E. coli J53 and E. coli DH5a were used as the recipient strain in mating experiment and the host strain in cloning experiments, respectively.
Conjugation Experiments
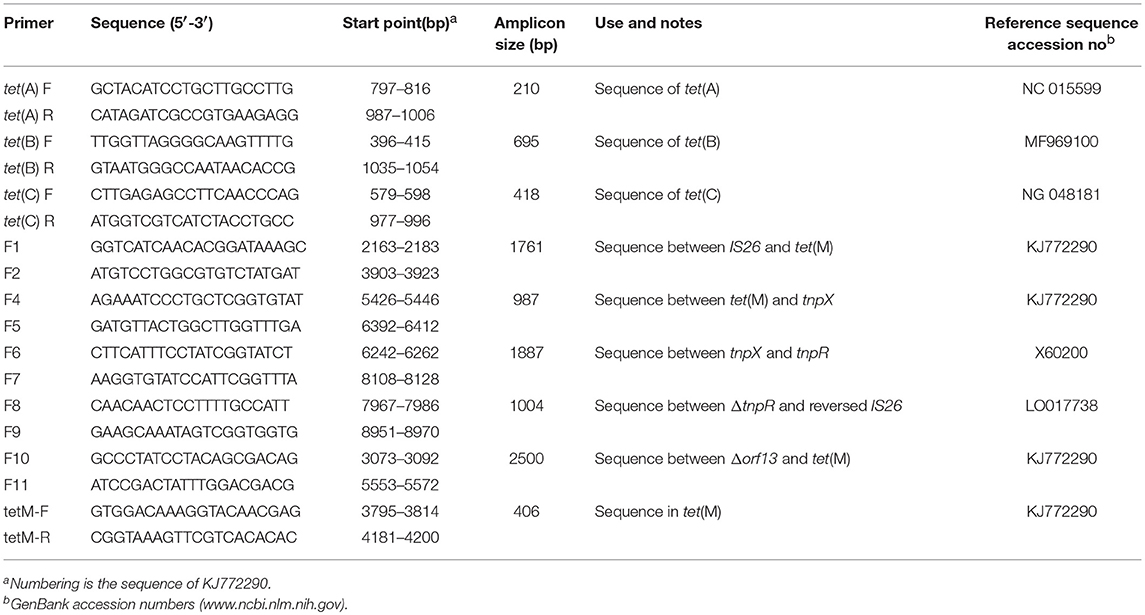
Seven E. coli isolates harboring full-length tet(M) were used for filter mating experiments with sodium azide-resistant E. coli J53 as a recipient as previously described (Devirgiliis et al., 2009). Briefly, overnight cultures of tetracycline-resistant, sodium azide-susceptible donor (E. coli isolate) and tetracycline-susceptible, sodium azide-resistant recipient (E. coli J53), grown in Luria-Bertani (LB) broth, were seeded at a 1:50 dilution in separate flasks of fresh LB broth. Following growth to log phase (OD600≈0.6) with shaking at the speed of 250 rpm at 37°C, 0.5 ml each of the donor and the recipient cultures were mixed and filtered through a 0.22-μm-pore-size membrane (Millipore Corp.) placed on prewarmed on MacConkey agar plates. After 24 h of incubation at 37°C, the cells were detached from filters in 5 ml of 1 × phosphate-buffered saline (pH7.4), and serial dilutions were plated on MacConkey agar plates containing 16 mg/L doxycycline and 100 mg/L sodium azide. Controls (unmixed donors and recipient cells) were treated in the same manner. Transconjugant colonies were recovered following incubation at 37°C for 24 to 72 h. Furthermore, amplification of the sequences of tet(A), tet(B), tet(C), and tet(M) in the transconjugants were performed using the primer sets in Table 2. Template DNA was prepared as follows. Amounts of 1.5 ml of transconjugant culture were pelleted, and cells were boiled in 200 μl of H2O for 15 min. After centrifugation, the supernatants were kept at −20°C. PCR was performed in a total volume of 50 μl, which contained 3 μl of supernatant, 50 pmol of each primer, 25 μl of PrimeSTAR® Max DNA Polymerase (TakaRa Biotechnology Co. Ltd, Japan). After an initial denaturation step of 3 min at 95°C, amplification was performed over 30 cycles, each one consisting of 30 s at 95°C, 30 s min at hybridization temperature [55°C for tet(A) and tet(C), 58°C for tet(B), and 52°C for tet(M)], and 1 min at 72°C, with a final extension step of 10 min at 72°C. The amplicons were sequenced using their corresponding primer set at commercial company (Shanghai Sangon Biological Engineering Technology and Services Co. Ltd, China). The sequences comparisons were performed using the BLASTtool available online at the National Center for Biotechnology Information of the National Library of Medicine (http://www.ncbi.nlm.nih.gov/blast/). Transconjugants carrying tet(M), but not tet(A), tet(B), or tet(C) were selected for further analysis.
Antimicrobial Susceptibility Testing
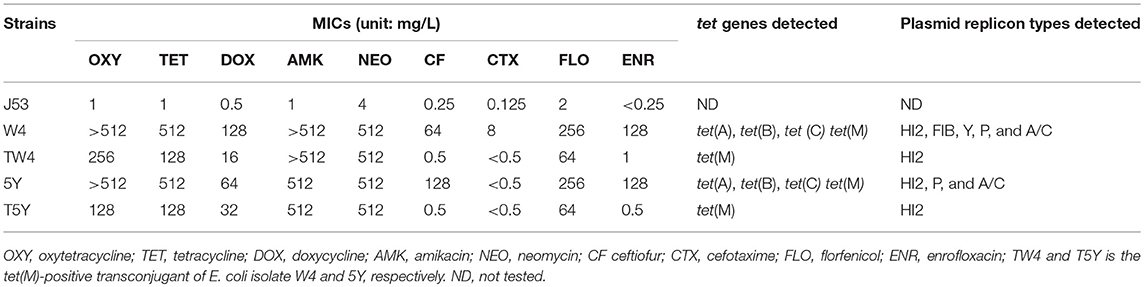
The susceptibility of tet(M)-positive transconjugants, their corresponding donors, and E. coli J53 to different antibiotics (Table 3) was determined by broth microdilution assays (Clinical Laboratory Standards Institute, 2014). Minimum inhibitory concentrations were determined on three independent occasions. E. coli ATCC 25922 was used for quality control in all susceptibility tests.
Plasmid Analysis
The incompatibility (Inc) groups of plasmids from transconjugants and their corresponding donors were determined by PCR-based replicon typing (Carattoli et al., 2005). Plasmids from the transconjugants and their corresponding donors (W4 and 5Y) were extracted using the QIAGEN Plasmid Midi Kit (Hilden, Germany) and subjected to electrophoresis on a 0.7% agarose gel. Plasmid size was estimated by comparison with reference plasmid DNAs of E. coli V517. The purified plasmids from W4 and 5Y were transferred to a nylon membrane (Roche Molecular Systems, Basel, Switzerland) and Southern blotting was carried out using a digoxigenin-labeled tet(M) PCR fragment [406 base pairs (bp), Table 2] as a probe.
Sequence Determinations
The tet(M)-positive conjugative plasmid pTW4 was digested with EcoRI and the fragments were ligated into the EcoRI-digested pBluescriptIISK(+). The recombined plasmids were electroporated into E. coli DH5α and white colonies were selected on LB/X-gal/IPTG agar supplemented with ampicillin (100 mg/L) and oxytetracycline (128 mg/L). The presence of tet(M) in transformants was confirmed by PCR using the primer set tetM-F and tetM-R (Table 2). The plasmid inserts were sequenced using M13 universal sequencing primers. The nucleotide and deduced protein sequences were analyzed with EditSeq and Megalign software (DNAstar, Madison, WI, USA). Sequence similarity and conserved domain searches were carried out using the BLASTtool available online at the National Center for Biotechnology Information of the National Library of Medicine (http://www.ncbi.nlm.nih.gov/blast/).
PCR Mapping Analysis
Based on the sequence of the obtained inserted fragment between hp and partial tnpX (6,544 bp, the left sequence of KJ772290 in Figure 2) and the complete sequence of Tn1000 (accession number X60200), the primer set F6 and F7 (Table 2) was designed to amplify the sequence between tnpX and the truncated tnpR. The amplicon was ligated to the 3′-terminus of the inserted fragment. The sequence between partial tnpR sequence (ΔtnpR) and right-terminal IS26 element was amplified using the primer set F8 and F9 designed according to the corresponding sequence of plasmid PRC557 (accession no. LO017738).
To monitor whether similar genetic organization exists in the six remaining E. coli isolates, PCR mapping was performed using the primer set F6 and F7, F8 and F9, and the primer sets designed using the software Primer Primer 5.0 (PREMIER Biosoft, American) on the obtained sequence in the plasmid pTW4 (accession no. KJ772290). The primer sets are shown in Table 2, Figure 2. All PCR amplicons were sequenced using their corresponding primer set at commercial company (Shanghai Sangon Biological Engineering Technology & Services Co. Ltd, China), and the results were compared to sequences in the GenBank database using the BLAST tool.
Molecular Typing and Phylogenetic Analysis
The seven E. coli isolates were typed by multilocus sequence typing (MLST) (http://mlst.warwick.ac.uk/mlst/) as previously described (http://www.shigatox.net/mlst) (Clermont et al., 2015). Briefly, seven housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) were amplified and sequenced. The corresponding sequence types (STs) were matched using the electronic database on the E. coli MLST website. Additionally, the phylogenetic groups of the seven isolates were determined based on PCR detection of the chuA and yiaA genes and DNA fragment TSPE4.C2 (Clermont et al., 2000).
Nucleotide Sequencing and Submission of tet(M) Sequences
The nucleotide sequences containing full-length tet(M) were deposited in GenBank (accession numbers KJ772290, KJ772289, and MF422120).
Results
Transfer of tet(M) and Multidrug Resistance
Two tet(M)-positive transconjugants (TW4 and T5Y) were obtained from donors W4 and 5Y at a frequency of 1.2 × 10−8. As shown in Table 3, W4, 5Y, TW4, and T5Y exhibited resistance to tetracycline, oxytetracycline, and doxycycline. The minimum inhibitory concentrations of TW4 and T5Y to the above-mentioned drugs were 8- to 256-fold higher than those of the recipient strain E. coli J53. TW4 and T5Y also exhibited resistance to amikacin, neomycin, and florfenicol, suggesting that the corresponding resistance markers were co-transferred into the recipient strain. In addition, the transconjugants carrying tet(A) and tet(C) were also found on the selection plates.
Molecular Characteristics of Conjugative Plasmids
W4 and 5Y carried more than three plasmids (Figure 1). The sizes of the plasmids ranged from approximately 3.2 to more than 54.2 kb. Notably, TW4 and T5Y each carried only one plasmid (designated pTW4 and p5Y, respectively) of approximately 50 kb. Southern hybridization with the digoxigenin-labeled tet(M) PCR fragment indicated that isolates W4 and 5Y yielded one distinct signal located on this plasmid. PCR-based inc replicon typing showed that pTW4 and p5Y belonged to the HI2 type. The other plasmids in W4 and 5Y were positive for the HI2, FIB, Y, P, and A/C and the P, and A/C type, respectively.
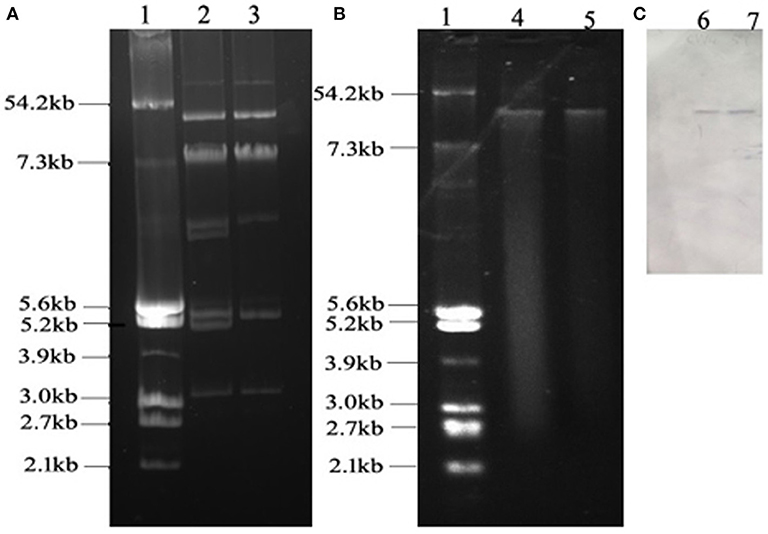
Figure 1. (A) Agarose electrophoresis of plasmids from E. coli strains W4 and 5Y. (B) Agarose electrophoresis of plasmids from transconjugant E. coli TW4 and T5Y. (C) Southern blot of plasmids from W4 and 5Y and labeled tet(M) fragment. Lane 1, reference plasmid DNAs of E. coli V517; lane 2, plasmids from W4; lane 3, plasmids from 5Y; lane 4, plasmid from TW4; lane 5, plasmid from T5Y; lane 6, southern blot, plasmids from W4; lane 7, southern blot, plasmids from 5Y.
Genetic Organization of tet(M) in Conjugative Plasmid and Sequence Analysis
The analysis of recombinant plasmids carrying EcoRI fragments from the conjugative plasmid pTW4 demonstrated that tet(M) was located on a fragment of approximately 6.5 kbp between hp and truncated tnpX (pTW4 plasmid in Figure 2). Sequence analysis indicated that a truncated orf13, the tet(M) leader peptide gene and tet(M) are located between bp 2989 and 5609 (numbering nucleotide position starts from the first base, G, in the sequence of KJ772290.). Sequence comparisons showed that the Δorf13-lp-tet(M) exhibited ≥99% sequence identity to the corresponding sequences present in the GenBank database. Among them, the Δorf13-lp-tet(M) exhibited 100% sequence identity to the corresponding sequence in the chromosome of Straphylococcus rostri (accession no. FN550102), Streptococcus spp. (CP008813 and CP003859), E. faecalis DENG (CP004081), and E. coli (CP021844).
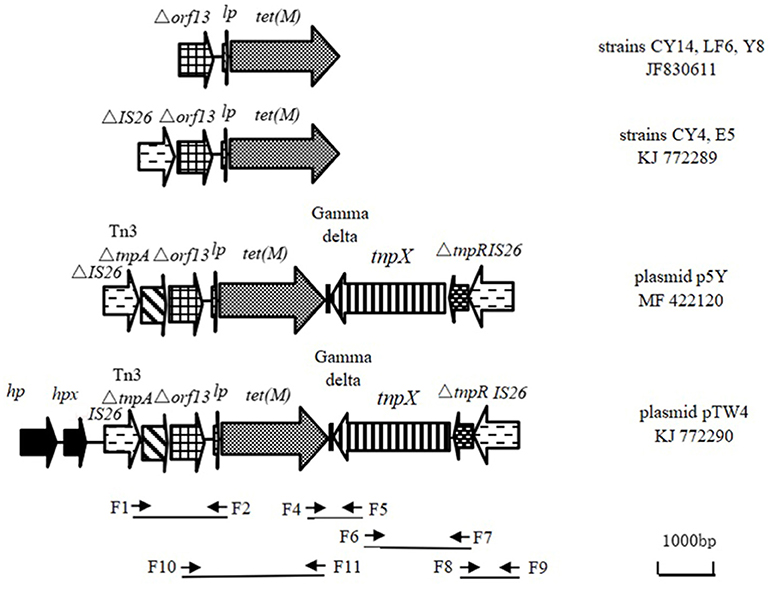
Figure 2. Location of tet(M) in clinical isolates of E. coli and graphical representation of primer pairs along the tested genetic structure KJ772290 (hp), gene encoding hypothetical protein; hpx, gene encoding putative protein X; lp, gene encoding Tet(M) leader peptide; fragment between A and B position in the sequence KJ772290 was obtained by cloning experiments. Arrows represent the orientation of each primer and relative positions of the primers along the tested linear sequence.
At the left end of the tet(M)-containing EcoRI fragment, codons 1–1,067 were indistinguishable from those found in plasmid pM160133 (CP022165) and plasmid pSCE516-1 (KX023262) from E. coli isolates from a New York patient and chickens in China. These isolates contained two ORFs in the same orientation as tet(M) encoding a hypothetical protein and putative protein X. Additionally, codons 2522–5609 showed 100% sequence identity to the corresponding sequences in the chromosome of E. coli (CP021840 and CP021844), which contain an ORF in the transcription orientation of tet(M) in addition to the Δorf13-lp-tet(M). This ORF located in the left of Δorf13-lp-tet(M) in the plasmid pTW4 was indistinguishable from the corresponding sequence of the 3′-terminal of the gene encoding Tn3-family transposase (TnpA) found in plasmid pM160133 (CP022165). Notably, an intact IS26 element including 14-bp perfect terminal inverted repeats and IS26 transposase was found between the gene encoding putative protein X and truncated Tn3 tnpA.
At the right end of the tet(M)-containing EcoRI fragment (basepairs 2522 to 65544), Δorf13-lp-tet(M)+gamma delta +incomplete tnpX were present in pTW4 and these sequences were 99% identical to the corresponding sequence in plasmids p41-3 (LC318054) and p15 (LC317981) from E. coli isolates from beef cattle in Japan. Obviously, the downstream of Δorf13-lp-tet(M) is a Tn1000 element. To determine whether a complete Tn1000 was present, we designed a forward primer for the obtained tnpX sequence and reverse primers for different positions along the sequence of Tn1000 (X60200). A fragment, 1,887 bp in size, between tnpX and ΔtnpR was obtained using primer set F6 and F7 (Table 2). Alignment of Δorf13-lp-tet(M) to part of the sequence of Tn1000 suggests that an unknown mobile genetic element is present upstream of ΔtnpR. IS26 elements typically exist in genomic DNA by forming regions containing antibiotic resistance genes flanked by two IS26 elements in the same or opposite orientations (Naas et al., 2001; Harmer and Hall, 2016). Since an intact IS26 element was detected in the tet(M)-containing EcoRI fragment, we further extended the sequence at its 3′-terminal end and obtained a fragment of 1,004 bp using primer set F8 and F9 (Table 2). Therefore, pTW4 contains IS26+Tn3ΔtnpA+Δorf13-lp-tet(M)+gamma delta+tnpX+ΔtnpR+ reverse IS26 (Figure 2). The presence of two intact IS26 elements suggests that the genetic organization carrying tet(M) is a previously unknown composite transposon which has now been registered as Tn6539 in the Transposon Nomenclature Database from UCL Eastman (http://transposon.lstmed.ac.uk/tn-registry).
Genetic Sequences Carrying tet(M) in Six Remaining Strains of E. coli and Homology Analysis
The genetic sequences carrying tet(M) in the tested strains are shown in Figure 2. Four sequences from seven E. coli isolates carry the genetic fragment of Δorf13-lp-tet(M). Among them, genomic DNAs from CY14, LF6, and Y8 only carry Δorf13-lp-tet(M) which showed 99.1% nucleotide sequence identity to our previously obtained the sequence carrying tet(M) [JF83061]. A truncated IS26 element was located upstream of Δorf13 in the genomic DNA from CY4 and E5 (KJ772289). Additionally, the conjugative plasmid p5Y harbors a highly similar composite transposon to that in pTW4.
The similarities of Tn6539 in plasmid pTW4 with the sequences from E. coli deposited in GenBank are shown in Figure 3. The Δorf13-lp-tet(M) fragment is located upstream or at the 3′-terminal end of part or the complete gene encoding Tn3-family transposase in the genomic DNA from the six E. coli isolates. The genomic DNAs from five of the six isolates carry intact IS6–family element(s). Among them, the chromosomal DNA from E. coli EC974 (CP021844) harbors a composite transposon consisting of orf13-lp-tet(M) and two IS15DIV elements belonging to the IS6-family in opposite orientations at its boundaries. Additionally, an Inc HI2–type plasmid pM160133 (CP022165) also carries a composite transposon with a similar genetic origination to that in the chromosome of E. coli EC974. The length of the genetic fragment flanked by two IS15DIVs elements was 22,182 bp and carried the resistance genes aadA2, dfrA12, and floR in addition to tet(M).
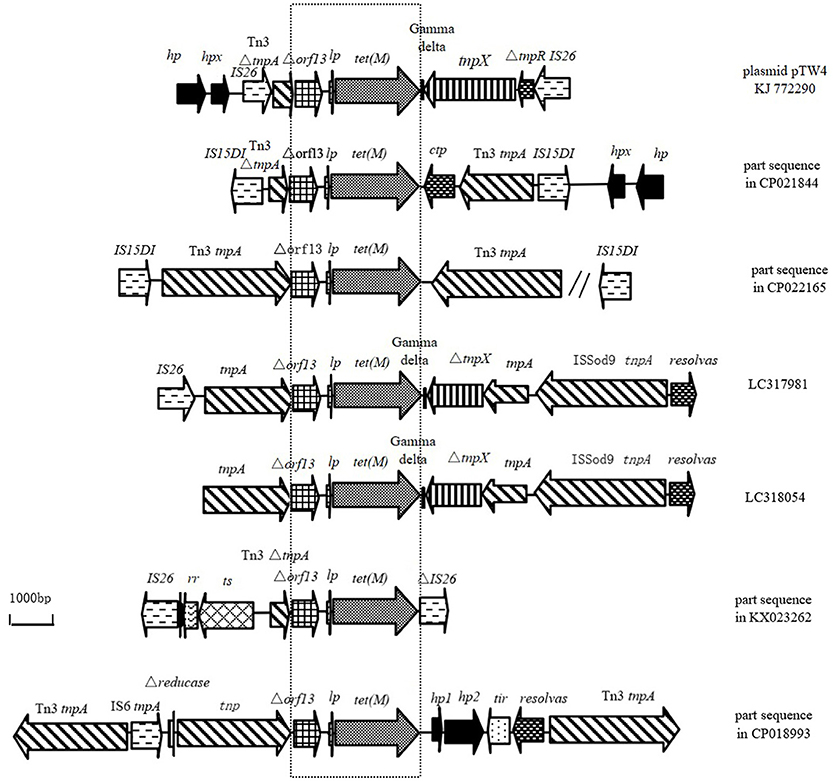
Figure 3. Structural features surrounding the sequence of Δorf13 +lp+tet(M) in KJ772290 compared to other sequences deposited in GenBank. (Similar regions are indicated by dotted lines; hp, gene encoding hypothetical protein; hpx, gene encoding putative protein X; lp, gene encoding TetM leader peptide; ctp, gene encoding conjugative transfer protein; symbol “//” the sequence of 13,102 bp between Tn3 tnpA and IS15D1; rr, gene encoding RadR regulator; ts, gene encoding chromate transporter).
Genetic Relatedness and Phylogenetic Analysis of Seven E. coli Isolates
MLST analysis showed that the seven tested isolates had six different sequence types. W4, 5Y, CY4, E5, and Y8 belong to ST162, ST156, ST48, ST3839, and ST163, respectively. CY14 and LF6 belong to ST224. Notably, E5 belongs to a new MLST profile (ST3839). Phylogenetic analysis showed that the seven strains belonged to group A (CY4 and LF6), group B1 (W4, 5Y, Y8, and CY14), and group D (E5) which is commonly considered as including pathogenic bacteria (Clermont et al., 2000).
Discussion
In this study, tet(M) was found in IncHI2-type conjugative plasmids from E. coli isolates W4 and 5Y. Although tet(M)-like genes are most commonly found in bacterial chromosome (Agersø et al., 2006; Vries et al., 2009), they also exist in conjugative plasmids from Campylobacter jejuni (Taylor et al., 1987), Neisseria meningitidis, Kingella denitrificans, Eikenella corrodens (Knapp et al., 1988), Clostridium perfringens (Yras and Rood, 1996), and E. coli (CP022165). Therefore, conjugative plasmids are an important vehicle facilitating the horizontal transfer of tet(M)-like genes between and across genera. IncHI2 plasmids play an important role in the acquisition and dissemination of antibiotic resistance genes among Gram-negative bacteria (Cain and Hall, 2012) and have been found to carry numerous classes of resistance genes including those conferring resistance to β-lactams (blaSHV, blaCTX−M, blaCMY, blaOXA, blaTEM, and NDM-1), quinolones [oqxAB, qnrA, qnrS, qnrB, and aac(6′)-Ib-cr], aminoglycosides (armA, aadA, aacA4, strA, and strB), amphenicols (catA1 and floR), trimethoprin (dhfr1), sulfonamides (sul1 and sul2), fosfomycin (fosA3), and colistin (mcr-1) (Chen et al., 2016; Fang et al., 2016; Hadjadj et al., 2017). Additionally, these multidrug-resistant IncHI2-type plasmids are broadly distributed among different bacteria including E. coli, Salmonella spp., Shigella flexnei, Klebesiella spp., and Enterobacter cloacae (Liu et al., 2015; Kieffer et al., 2017). Thus, the IncHI2-type conjugative plasmids carrying tet(M) found in this study may spread between and across genera in the future, threatening tetracycline treatment in clinical practice.
IS26, a member of the IS6 insertion sequence family, contributes to the dissemination of antibiotic resistance genes in Gram-negative bacteria mainly by forming composite transposons. Most composite transposons contain a center region harboring resistance genes and two IS26 elements in the same or opposite orientations at its bounders (Harmer and Hall, 2016). In Gram-negative bacteria, composition transposons are usually found with IS26 elements at their borders, but not flanked by 8-bp target site duplication (TSD), a hall marker of IS26 integration (Curiao et al., 2011; Zienkiewicz et al., 2013). Therefore, researchers considered that the composite transposons were formed by homologous recombination rather than a transposition. In 2014, a new model for IS26-mediated mobilization of resistance genes has been proposed by Harmer et al. (2014), whereby an IS26 element together with resistance gene(s), called a translocatable units (TU), is considered as a new family mobile genetic element. TU released from the genetic sequences can recognized another IS26 as target and is incorporated immediately adjacent to it in the same orientation. This results in IS26 element arraying in tandem and the formation of composite transposons. The frequency of cointegrate formation mediated by TU was 60-fold higher than that mediated by a single IS26, which indicated that an intact IS26 element in genetic sequences owns a stronger ability to recruit another IS26-mediated resistance genes. Furthermore, it was confirmed that TU integration could occur by Tnp (transposase in IS26 element)-catalyzed reaction or RecA-dependent homologous recombination. However, Tnp-catalyzed reaction was 100-fold more efficient than RecA-dependent homologous recombination (Harmer and Hall, 2015). In addition, He et al. (2015) analyzed the distinct patterns of sequences flanking 70 IS26 copies in eight genomes from Carbapenemase-producing Enterobacteriaceae and found that IS26 promoted rearrangement of resistance plasmids by both inter- and intramolecular replicative transposition (He et al., 2015). In the current study, Tn6539 was found to be a composite transposon containing Δorf13-lp-tet(M) and two IS26 elements in opposite orientations at its borders. This type of composite transposon was first found in a multidrug-resistant plasmid from an E. coli MG-1 and named Tn2000. The center region flanked by two IS26 elements in Tn2000 is a class 1 integrin, In53, carrying nine functional resistance gene cassettes (Naas et al., 2001). Structures of composite transposons similar to that of Tn6539 were found in plasmids from E. coli isolates from different sources (Literacka et al., 2009; Smet et al., 2010; Yang et al., 2014), but the molecular mechanism of their movement among Gram-negative bacteria remains unknown and requires further analysis.
Conclusion
We described the organization of a novel composite transposon Tn6539 in an IncHI2-type conjugative plasmid from two E. coli isolates with different genetic backgrounds. The transposon carries tet(M) and two intact IS26 elements in opposite orientations at its bounders.
Author Contributions
GH and LY conceived and designed the experiments. YS, YL, HW, and LW performed the experiments. JL, YP, and YS analyzed the data. DH and HW contributed reagents materials analysis tools. YS and GH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by the National Key Research and Development Program of China (2016YFD05101304) and National Natural Science Foundation of China-Henan Talent Training Joint fund (U1504327).
References
Agersø, Y., Pedersen, A. G., and Aarestrup, F. M. (2006). Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J. Antimicrob. Chemother. 57, 832–839. doi: 10.1093/jac/dkl069
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004
Cain, A. K., and Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Cauwerts, K., Decostere, K., Graef, E. M. D., Haesebrouck, F., and Pasmans, F. (2007). High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Dis. 36, 395–399. doi: 10.1080/03079450701589167
Chen, W. Y., Fang, T. Z., Zhou, X. J., Zhang, D. F., Shi, X. M., and Shi, C. L. (2016). IncHI2 Plasmids are predominant in antibiotic-resistant Salmonella isolates. Front. Microbiol. 7:1566. doi: 10.3389/fmicb.2016.01566
Chopra, I., and Roberts, M. Y. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Bio Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Clermont, O., Bonacorsi, S., and Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clermont, O., Gordon, D., and Denamur, E. (2015). Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiolgy 161, 980–988. doi: 10.1099/mic.0.000063
Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement M100-S24. Wayne, PA: CLSI.
Curiao, T., Cantón, R., Garcillán-Barcia, M. P., Cruz, F., Baquero, F., and Coque, T. M. (2011). Association of composite IS26-sul3 elements with highly transmissible inci1 plasmids in extended-spectrum-β-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother. 55, 2451–2457. doi: 10.1128/AAC.01448-10
Devirgiliis, C., Coppola, D., Barile, S., Colonna, B., and Perozzi, G. (2009). Characterization of the Tn916 conjugative transposon in a food-borne strain of Lactobacillus paracase. Appl. Environ. Microbiol. 75, 3866–3871. doi: 10.1128/AEM.00589-09
Fang, L. X., Li, X. P., Li, L., Li, S. M., Liao, X. P., Sun, J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312. doi: 10.1038/srep25312
Gilrane, V. L., Lobo, S., Huang, W. H., Zhuge, J., Yin, C. H., Chen, D., et al. (2017). Complete genome sequence of a colistin-resistant Escherichia coli strain harboring mcr-1 on an IncHI2 plasmid in the United States. Genome Announcements 5, e01095–e01017. doi: 10.1128/genomeA.01095-17
Hadjadj, L., Riziki, T., Zhu, Y., Li, J., Diene, S. M., and Rolain, J. M. (2017). Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Gene 8:394. doi: 10.3390/genes8120394
Harmer, C. J., and Hall, R. M. (2015). IS26-mediated precise excision of the IS26-aphA1a translocatable unit. MBIO 6, e01866–e01815. doi: 10.1128/mBio.01866-15
Harmer, C. J., and Hall, R. M. (2016). IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1, e00038–e00016. doi: 10.1128/mSphere.00038-16
Harmer, C. J., Moran, R. A., and Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBIO 5, e0180–e0114. doi: 10.1128/mBio.01801-14
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBIO 6:e00762. doi: 10.1128/mBio.00762-15
Hu, G. Z., Pan, Y. S., Wu, H., Hu, H., Xu, R., Yuan, L., et al. (2013). Prevalence of tetracycline resistance genes and identification of tet(M) in clinical isolates of Escherichia coli from sick ducks in China. J. Med. Microbiol. 62, 851–858. doi: 10.1099/jmm.0.051896-0
Jones, C. H., Tuckman, M., Murphy, E., and Bradford, P. A. (2006). Identification and sequence of a tet(M) tetracycline resistance determinant homologue in clinical isolates of Escherichia coli. J. Bacteriol. 188, 7151–7164. doi: 10.1128/JB.00705-06
Jurado-Rabadán, S., Fuente, R., Ruiz-Santa-Quiteria, J. A., Orden, J. A., de Vries, L. E., and Agersø, Y. (2014). Detection and linkage to mobile genetic elements of tetracycline resistance gene tet(M) in Escherichia coli isolates from pigs. BMC Vet. Res. 10:155. doi: 10.1186/1746-6148-10-155
Kieffer, N., Aires-de-Sousa, M., Nordmann, P., and Poirel, L. (2017). High rate of MCR-1–producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg Infect. Dis. 23, 2023–2029. doi: 10.3201/eid2312.170883
Knapp, J. S., Johnson, S. R., Zemilman, J. M., Roberts, M. C., and Morse, S. A. (1988). High-level tetracycline resistance resulting from TetM in strains of Neisseria spp., Kingella denitrificans, and Eikenella corrodens. Antimicrob. Agents Chemother. 32, 765–767. doi: 10.1128/AAC.32.5.765
Linkevicius, M., Sandegren, L., and Andersson, D. I. (2015). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60, 789–796. doi: 10.1128/AAC.02465-15
Literacka, E., Bedenic, B., Baraniak, A., Fiett, J., Tonkic, M., Jajic-Bencic, I., et al. (2009). blaCTX−M genes in Escherichia coli strains from Croatian Hospitals are located in new (blaCTX−M−3a) and widely spread (blaCTX−M−3a and blaCTX−M−15) genetic structures. Antimicrob Agents Chemother. 53, 1630–1635. doi: 10.1128/AAC.01431-08
Liu, G. L., Qin, S. S., Xu, H., Xu, L. J., Zhao, D., Liu, X. C., et al. (2015). New Delhi metallo-β-lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan province, China. PLoS ONE 10:e0135044. doi: 10.1371/journal.pone.0135044
Naas, T., Mikami, Y., Imai, T., Poirel, L., and Nordmann, P. (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 1, 235–249. doi: 10.1128/JB.183.1.235-249.2001
Roberts, M. C., and Schwarz, S. (2016). Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J. Environ. Qual. 45, 576–592. doi: 10.2134/jeq2015.04.0207
Smet, A., Nieuwerburgh, F. V., Vandekerckhove, T. T. M., Martel, A., Deforce, D., Butaye, P., et al. (2010). Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLOS ONE 5:e11202. doi: 10.1371/journal.pone.0011202
Taylor, D. E., Hiratsuka, K., Ray, H., and Manavathu, E. K. (1987). Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J. Bacteriol. 169, 2984–2989. doi: 10.1128/jb.169.7.2984-2989.1987
Tuckman, M., Petersen, P. J., Howe, A. Y. M., Orlowski, M., Mullen, S., Chan, K., et al. (2007). Occurrence of tetracycline resistance genes among Escherichia coli isolates from the phase 3 clinical trials for tigecycline. Antimicrob Agents Chemother. 51, 3205–3211. doi: 10.1128/AAC.00625-07
Vries, L. E., Christensen, H., Skov, R. L., Aarestrup, F. M., and Agersø, Y. (2009). Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 64, 490–500. doi: 10.1093/jac/dkp214
Yang, X. Y., Liu, W. L., Liu, Y. Y., Wang, J., Lv, L. C., Chen, X. J., et al. (2014). F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX−−M−55/−14/−65 in Escherichia coli from chickens in China. Front. Microbiol. 5:688. doi: 10.3389/fmicb.2014.00688
Yras, D. L., and Rood, J. I. (1996). Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob. Agents Chemother. 40, 2500–2504. doi: 10.1128/AAC.40.11.2500
Keywords: composite transposonTn6539, tet(M) gene, IS26 element, IncHI2-type plasmid, Escherichia coli isolates
Citation: Sun Y, Liu Y, Wu H, Wang L, Liu J, Yuan L, Pan Y, He D and Hu G (2019) IS26-Flanked Composite Transposon Tn6539 Carrying the tet(M) Gene in IncHI2-Type Conjugative Plasmids From Escherichia coli Isolated From Ducks in China. Front. Microbiol. 9:3168. doi: 10.3389/fmicb.2018.03168
Received: 25 June 2018; Accepted: 07 December 2018;
Published: 15 January 2019.
Edited by:
Charles W. Knapp, University of Strathclyde, United KingdomReviewed by:
Michael P. Ryan, University of Limerick, IrelandRolf Dieter Joerger, University of Delaware, United States
Copyright © 2019 Sun, Liu, Wu, Wang, Liu, Yuan, Pan, He and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gong-zheng Hu, eWFvbGlsYWJAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ya-wei Sun
Ya-wei Sun Ying-ying Liu
Ying-ying Liu Hua Wu1†
Hua Wu1† Li Yuan
Li Yuan Gong-zheng Hu
Gong-zheng Hu